
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 03 April 2024
Sec. Vaccines and Molecular Therapeutics
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1277447
This article is part of the Research Topic The next generation of poxvirus-based vaccines: Current developments and advances View all 3 articles
Modified vaccinia virus Ankara (MVA) has been widely tested in clinical trials as recombinant vector vaccine against infectious diseases and cancers in humans and animals. However, one biosafety concern about the use of MVA vectored vaccine is the potential for MVA to recombine with naturally occurring orthopoxviruses in cells and hosts in which it multiplies poorly and, therefore, producing viruses with mosaic genomes with altered genetic and phenotypic properties. We previously conducted co-infection and superinfection experiments with MVA vectored influenza vaccine (MVA-HANP) and a feline Cowpox virus (CPXV-No-F1) in Vero cells (that were semi-permissive to MVA infection) and showed that recombination occurred in both co-infected and superinfected cells. In this study, we selected the putative recombinant viruses and performed genomic characterization of these viruses. Some putative recombinant viruses displayed plaque morphology distinct of that of the parental viruses. Our analysis demonstrated that they had mosaic genomes of different lengths. The recombinant viruses, with a genome more similar to MVA-HANP (>50%), rescued deleted and/or fragmented genes in MVA and gained new host ranges genes. Our analysis also revealed that some MVA-HANP contained a partially deleted transgene expression cassette and one recombinant virus contained part of the transgene expression cassette similar to that incomplete MVA-HANP. The recombination in co-infected and superinfected Vero cells resulted in recombinant viruses with unpredictable biological and genetic properties as well as recovery of delete/fragmented genes in MVA and transfer of the transgene into replication competent CPXV. These results are relevant to hazard characterization and risk assessment of MVA vectored biologicals.
The Orthopoxvirus genus belongs to the Poxviridae family. The orthopoxviruses (OPXV) are viruses with large linear double stranded DNA genome (170 to 250kbp) (1). OPXV can infect vertebrates and insects (2). Among the OPXV that cause human diseases, Variola virus (VARV), vaccinia-like virus, Cowpox virus (CPXV) and Monkeypox virus (MPXV) are the most common (3–6). VARV is the causative agent of smallpox, a deadly viral disease, which was eradicated in 1980 as a result of a massive vaccination campaign (7). During the smallpox campaign, Vaccinia virus (VACV) was used as the smallpox vaccine and several VACV strains have been developed and used in different countries, such as New York City Board of Health (NYCBH) was used in the America, Tian tan in China, Ankara in Turkey, and Lister and modified vaccinia virus Ankara (MVA) in Europe (8–10).
MVA was administrated to over 120,000 people in Germany with no reported major side effects (10–13). MVA was derived from Chorioallantois vaccinia virus Ankara (CVA) by over 570 passages in primary chicken embryo fibroblasts (14). In this process, CVA genome suffered modifications including six large deletions and other mutations that lead to the reduction of the genome from 208kbp in CVA to 170kbp in MVA (15, 16). These mutations affected many genes involved in virus–host interaction and other genes responsible for evasion of the host immune response (16, 17).
As a result, MVA is unable to replicate productively in most mammalian cell lines (15, 17–21). Although some mammalian cell lines have been reported as permissive to MVA, such as Baby Hamster Kidney (BHK-21) cells (21, 22), and semi-permissive to MVA, such as Vero cells (African green monkey kidney epithelial cells) (15, 20). Recently, it was shown that a repair of the inactivated C16R/B22R in conjunction with the restoration of the deleted C12L gene restores production infection of many human cells with MVA (23). The host range restriction of MVA is considered the major biosafety advantage for its use as a vaccine vector along with its immunogenicity, its incapability to cause illness in vivo and its safety record (24–26).
Since the nineties, MVA has been widely tested in clinical trials as recombinant vector for vaccination against infectious diseases and cancers in both humans and animals (27, 28). Today, several MVA vaccines against HIV (29, 30), Ebola (31–34), respiratory syncytial virus (35), Middle East Respiratory Syndrome (36), cytomegalovirus (37), influenza (38, 39), tuberculosis (40) and malaria (41–43) are in different phases of clinical trials. MVA-BN (JYNNEUS or Imvanex) is licensed as a vaccine against Mpox and smallpox in both Europe and USA, and is currently being used for immunization against current global Mpox outbreak (44, 45). Even though MVA is already deployed as a vaccine and is a promising viral vector, there are still some biosafety aspects that should be considered during the biological hazard characterization of MVA and recombinant MVA (rMVA). One is the potential for MVA or rMVA to recombine with naturally occurring OPXV, which could lead to the rescue of interrupted or deleted genes in MVA or to transfer of the transgene to multiplication competent OPXV (46). Hence, the recombination could result in the emergence of novel mosaic viruses with atypical virulence and host range characteristics. Therefore, studies analyzing the potential of recombination between MVA and OPXVs are relevant to hazard characterization of MVA vectored vaccines. Co-infection and superinfection experiments constitute cell culture-based models to examine recombination of MVA with other OPXV during co-infection of semi-permissive cells as well as evaluate the possibility of superinfection exclusion in preventing recombination.
Recombination is not a rare event between OPXV. Natural occurring recombination events between OPXV have been reported (47–57). However, the possibility of recombination was considered negligible due to the low likelihood of co-localization of the viruses in the same cell, poor or no multiplication of MVA in many mammalian cells and superinfection exclusion (58, 59). The circulation of naturally zoonotic OPXV, such as CPXV and MPXV, has increased in recent years (6). Several cases of human cowpox infections caused by infected animals have been reported in Europe (49, 60–68) and the global outbreak of human Mpox has been reported in 110 countries (69) followed by increased vaccination with MVA. Therefore, the possibility of co-localization and recombination could have increased. We highlight that there is a need to better understand the mechanism of recombination and possible public health threat of recombination of two poxviruses after co- and superinfection.
In a previous study, we have demonstrated recombination in Vero cells co-infected and superinfected with recombinant MVA expressing the influenza virus haemagglutinin (HA) and nucleoprotein (NP) genes (MVA-HANP) and feline CPXV (27). In the present study, we sequenced the genome of putative recombinant viruses produced in semi-permissive Vero cells co-infected and superinfected with MVA-HANP and feline CPXV-No-F1, conducted genomic characterization of the parental and the putative recombinant viruses and mapped genome-wide recombination events in the recombinant viruses.
The co-infection and superinfection experiments with MVA-HANP and the naturally circulating Fennoscandian feline cowpox strain (CPXV-No-F1) were performed in Vero cells as previously described (27). The origin of the cowpox strain was described elsewhere (68). MVA-HANP was kindly provided by Dr. Bernard Moss, National Institutes of Health, USA. MVA-HANP contains the influenza virus HA (A/PR/8/34) and NP gene inserts (70). MVA-HANP was propagated in BHK-21 cells (ATCC CCL-10). CPXV-No-F1 and recombinant viruses were cultured on Vero cells (ATCC CCL-81). Vero cells and BHK-21 cells were propagated in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS). The cell cultures were maintained in a humidified 5% CO2 atmosphere at 37°C.
Vero cells are semi-permissive to MVA-HANP (15, 20) and permissive to CPXV (71). Vero cells were co-infected with MVA-HANP and CPXV-No-F1 at a multiplicity of infection (MOI) of 5 infection units per cell for each virus strain (Supplementary Figure 1). Superinfection with CPXV and MVA-HANP in Vero cells was performed in four experiments (Supplementary Figure 1). Vero cells were infected with CPXV-No-F1 at a MOI of 5. The infected Vero cells were superinfected with MVA-HANP at same MOI (of 5) after 4-hours (superinfection 1) or 6-hours post primary infection (ppi) (superinfection 3). The cells were incubated for 72 hours at 37°C. The same procedure was repeated for superinfection 2 and 4, but the primary infection was with MVA-HANP and the secondary infection with CPXV-No-F1 after 4-hours (superinfection 2) or 6-hours ppi (superinfection 4). After 72 hours ppi, the cells were harvested, freeze-thawn three times and sonicated. The co-infection and superinfection experiments were performed at the Poxvirus Laboratory (Biosafety Level-2), Department of Medical Biology, UiT - The Arctic University of Norway. The experiments were carried out at the appropriate containment levels.
The putative recombinant viruses were identified and selected in Vero Cells. The selection was based on (1) the expression of the Influenza virus HA protein and (2) plaque phenotype. The putative recombinant viruses that formed different plaque phenotype from the parental viruses were selected. The sonicated cells suspensions were inoculated on Vero cells and the viruses were plaque-purified several times before plaque amplification. The stock of the viruses was prepared from plaque purified viruses. The putative recombinant viruses carrying the influenza virus HA protein were detected by immunostaining, as described previously (22). Moreover, the plaque phenotype of the parental viruses was also examined in Vero cells.
Viral DNA of the plaque purified putative recombinant viruses and the parental virus MVA-HANP was isolated using QIAGEN Genomic-tip 100/G and QIAGEN Genomic DNA Buffer Set, following the manufacturer’s instructions (Qiagen, Hilden, Germany). The genomes were sequenced with Illumina MiSeq (Illumina Inc., San Diego, CA, United States) using reagent kit v3 with 2 × 300 bp paired-end reads and Oxford Nanopore Technology GridION (ONT; Oxford, United Kingdom), as previously described (49). Nanopore and Illumina library preparation have been described elsewhere (49). Nanopore sequencing was performed at the Genomics Support Centre Tromsø at UiT—The Arctic University of Norway and Illumina sequencing was conducted by the Norwegian Sequencing Centre, Oslo.
The genome assembly was performed using SPAdes v3.15.3 (72), as previously described (49). For the assembly of MVA-HANP, the parameter trusted-contigs and the reference genome MVA were used. The viral genomes were annotated using the Genome Annotation Transfer Utility (GATU) (73), as previously reported (49). Vaccinia virus Copenhagen (VACV-Cop), Choriollantois vaccinia virus Ankara (CVA) and MVA were used as reference genomes for the genome annotation of MVA-HANP. VACV strains were retrieved from the Viral Orthologous Clusters (VOCs) database (74). The parental viruses, CPXV-No-F1 (Genbank accession number OP125538) and MVA-HANP, were used as reference genomes for the genome annotation of the putative recombinant viruses. The complete genome of the parental CPXV-No-F1 has been published elsewhere (68).
The putative recombinant viral genomes were analyzed for possible recombination events using recombination detection program 4 (RPD4) (75) and SimPlot v3.5.1 (76) as described previously (49). The progeny virus genomes were aligned to the parental viruses and other CPXV strains (CPXV-Br and CPXV-Gri) with MAFFT v1.4.0 (77) implemented in Geneious Prime 2020.2.4. The CXPV strains were retrieved from the Viral Orthologous Clusters (VOCs) database (74). The gaps were not removed from the alignments. The recombination events identified by 5 of 7 methods (RDP (78), GENECONV (79), Bootscan (80), MaxChi (81), Chimaera (82), SiScan (83), and 3Seq (84)) with significant p-values (p ≤ 0.01) were considered potential recombinant events. Furthermore, the breakpoints in the recombinant genomes were checked manually in the alignments in case both programs could not detect the recombination event.
The plaque phenotype caused by the putative recombinant viruses and the parental viruses were examined in Vero cells. The parental CPXV-No-F1 forms medium, semilytic plaques. Whereas MVA-HANP do not produce plaques; however, some Vero cells have positive immunostaining for the HA transgene as a result of MVA-HANP limited infection and expression of its HA transgene (Figure 1).
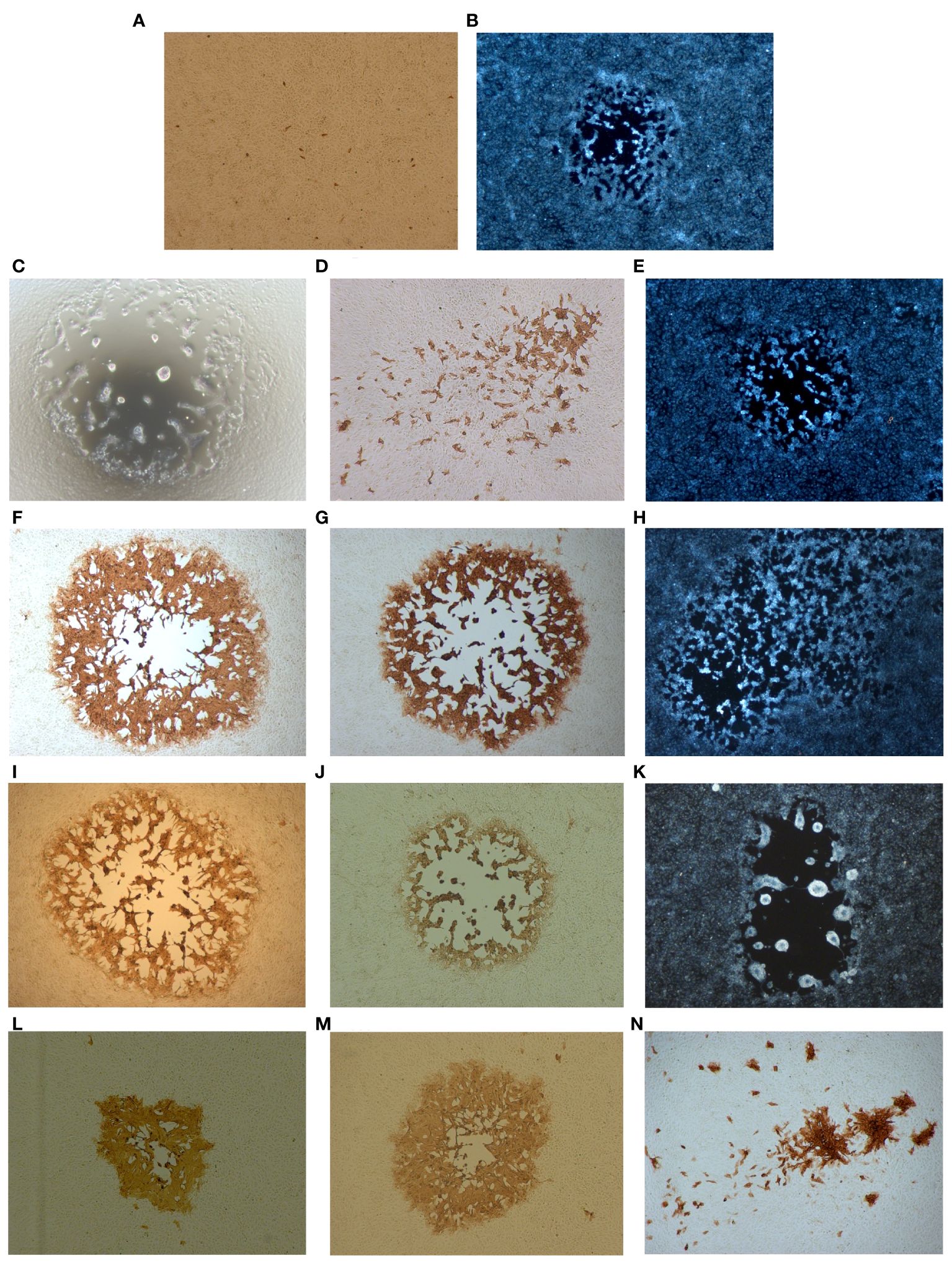
Figure 1 Plaque phenotypes of the parental viruses and the putative recombinant viruses. Confluent Vero cells were infected with the respective viruses and HA expression was monitored 48 h.p.i. by immunoperoxidase staining of fixed cells. (A) The parental virus MVA-HANP. (B) The parental virus CPXV-No-F1. (C) V-Rec1 from co-infection MVA-HANP and CPXV-No-F1. (D, E) V-Rec2 and V-Rec3 from superinfection 1 (primary infection with CPXV-No-F1 and secondary infection with MVA-HANP at 4h post primary infection, ppi). (F–H) V-Rec4, V-Rec5 and V-Rec6 from superinfection 2 (primary infection with MVA-HANP and secondary infection with CPXV-No-F1 at 4h ppi). (I–M) V-Rec7, V-Rec8, V-Rec9, V-Rec10 and V-Rec11 from superinfection 3 (primary infection with CPXV-No-F1 and secondary infection with MVA-HANP at 6h ppi). (N) V-Rec 12 from superinfection 4 (primary infection with MVA-HANP and secondary infection with CPXV-No-F1 at 6h ppi). All panels show representative fields at approximately 200× magnification.
Different plaques phenotypes and transgene-expressing plaques were observed in co-infected Vero cells. One putative recombinant virus was isolated from co-infected Vero cells, which was named V-Rec1 (Supplementary Table 1). The plaque of this recombinant virus is large, lytic, and characterized by syncytium formation and cell lysis (Figure 1C). In addition, this recombinant forms HA transgene negative plaques.
Similarly, different plaque phenotypes were observed in superinfected Vero cells regardless the virus strain used for the primary infection and the timing (4 and 6 hours ppi). Additionally, both the transgene-expressing and non-transgene-expressing viruses display different plaque phenotypes (Figures 1D–N). Two, three, five and one viruses were selected from superinfection 1, superinfection 2, superinfection 3 and superinfection 4, respectively (Supplementary Figure 1). We referred to these putative recombinant viruses as V-Rec2, V-Rec3, V-Rec4, V-Rec5, V-Rec6, V-Rec7, V-Rec8, V-Rec9, V-Rec10, V-Rec11 and V-Rec12 (Supplementary Table 1).
V-Rec2 from the superinfection 1 expresses the HA transgene and produced small and non-lytic plaques with secondary spread (comet formation) (Figure 1D). Whereas V-Rec3 from the same experiment is transgene negative and forms medium and lytic plaques (Figure 1E). The plaques of V-Rec4 and V-Rec5 from superinfection 2 are large, lytic and transgene positive. Unlike V-Rec4 and V-Rec5, V-Rec6 is transgene negative and forms medium and semilytic plaques with extensive secondary spread (Figures 1F–H).
V-Rec7, V-Rec8 and V-Rec9 from superinfection 3 form large and lytic plaques (Figures 1I–K). However, compared to V-Rec7 and V-Rec8, V-Rec9 do not express the transgene and forms syncytia. The other two putative recombinant viruses were chosen from superinfection 3, the progeny virus V-Rec10 and V-Rec11. Both viruses express the HA transgene and form semilytic plaques (Figures 1L, M). However, the size of their plaques are different. V-Rec11 produces large plaques, whereas V-Rec10 generates medium plaques. V-Rec12 from superinfection 4 expresses the HA transgene and displays small and non-lytic plaques with comet formation (Figure 1N).
The complete genomes of twelve putative recombinant viruses and the parental virus MVA-HANP were sequenced, assembled, and annotated (Figure 2). The whole genome sequences of MVA-HANP and V-Rec1 - V-Rec12 are available in GenBank, with Accession Number: OQ818667 and OQ822790 - OQ822801, respectively. The assembled genome of the parental MVA-HANP has a length of 181,712 bp, with inverted terminal repeats (ITR) of 9.8 kbp. Genome annotation predicted 199 coding sequences (CDS) in MVA-HANP genome (Supplementary Table 2). The sequencing analysis of the parental MVA-HANP showed that the double expression cassette containing the influenza HA and NP transgenes was inserted in A51R/A55R hybrid gene. However, the assembly of MVA-HANP showed that there are two populations of MVA-HANP, one with the complete double expression cassette (181,712 bp) and another with an incomplete double expression cassette (178,579 bp). A small part of NP transgene (~6%), whole HA transgene and downstream MVA-HANP flanking sequence (containing VACV-Cop A56R gene) are deleted in the latter. It only contains major part of the NP transgene (94%) and the upstream flanking MVA sequence.
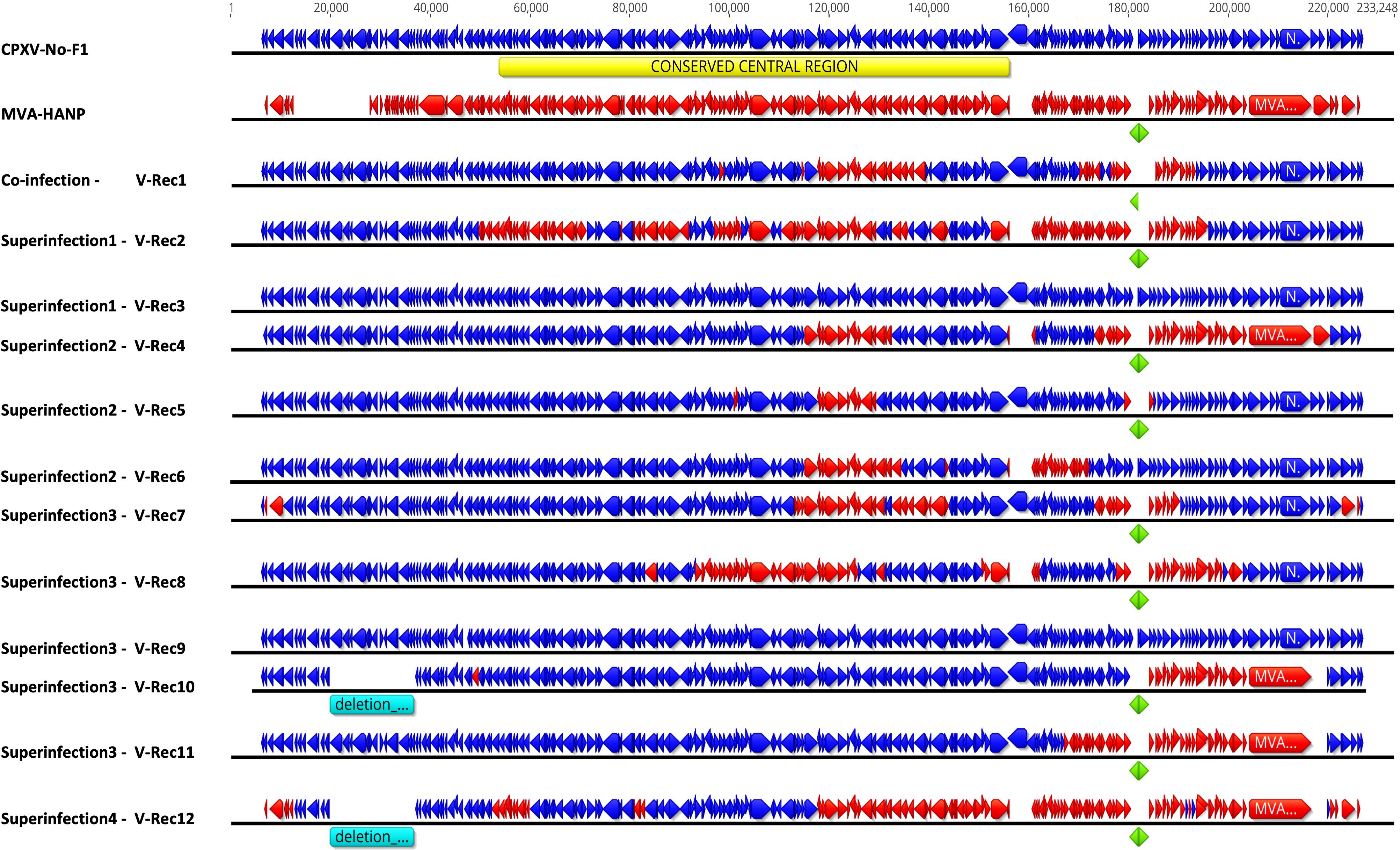
Figure 2 Genome map of the parental viruses (CPXV-No-F1 and MVA-HANP) and the putative recombinant viruses. The putative recombinant viruses were produced in Vero cells either co-infected or superinfected with CPXV-No-F1 and MVA-HANP. Superinfection 1, primary infection with CPXV-No-F1 and secondary infection with MVA-HANP at 4h post primary infection (ppi); Superinfection 2, primary infection with MVA-HANP and secondary infection with CPXV-No-F1 at 4h ppi; Superinfection 3, primary infection with CPXV-No-F1 and secondary infection with MVA-HANP at 6h ppi; Superinfection 4, primary infection with MVA-HANP and secondary infection with CPXV-No-F1 at 6h ppi. Blue blocks represent the coding sequences (CDS) from CPXV-No-F1. Red blocks represent CDS from MVA-HANP. Green blocks represent the influenza virus hemagglutinin (HA) and nucleoprotein (NP) transgenes. Turquoise blocks represent deleted regions in the recombinant viruses. Yellow block represents the conserved central region (VACV-Cop F4L – VACV-Cop A24R) in orthopoxvirus genomes.
The genome size and the number of predicted CDS of the parental and the progeny virus genomes are shown in the Table 1 and Supplementary Tables 2, 3. The length of progeny virus genomes was not uniform, ranging from 176.9 kbp to 221 kbp. Three putative recombinant viruses (V-Rec3, V-Rec5 and V-Rec9) have similar genome size to that of the parental CPXV-No-F1 and one recombinant virus (V-Rec12) possesses a smaller genome than that of the parental MVA-HANP (Table 1). The ITR of the progeny viruses ranged from 4.7 kbp to 8.3 kbp.
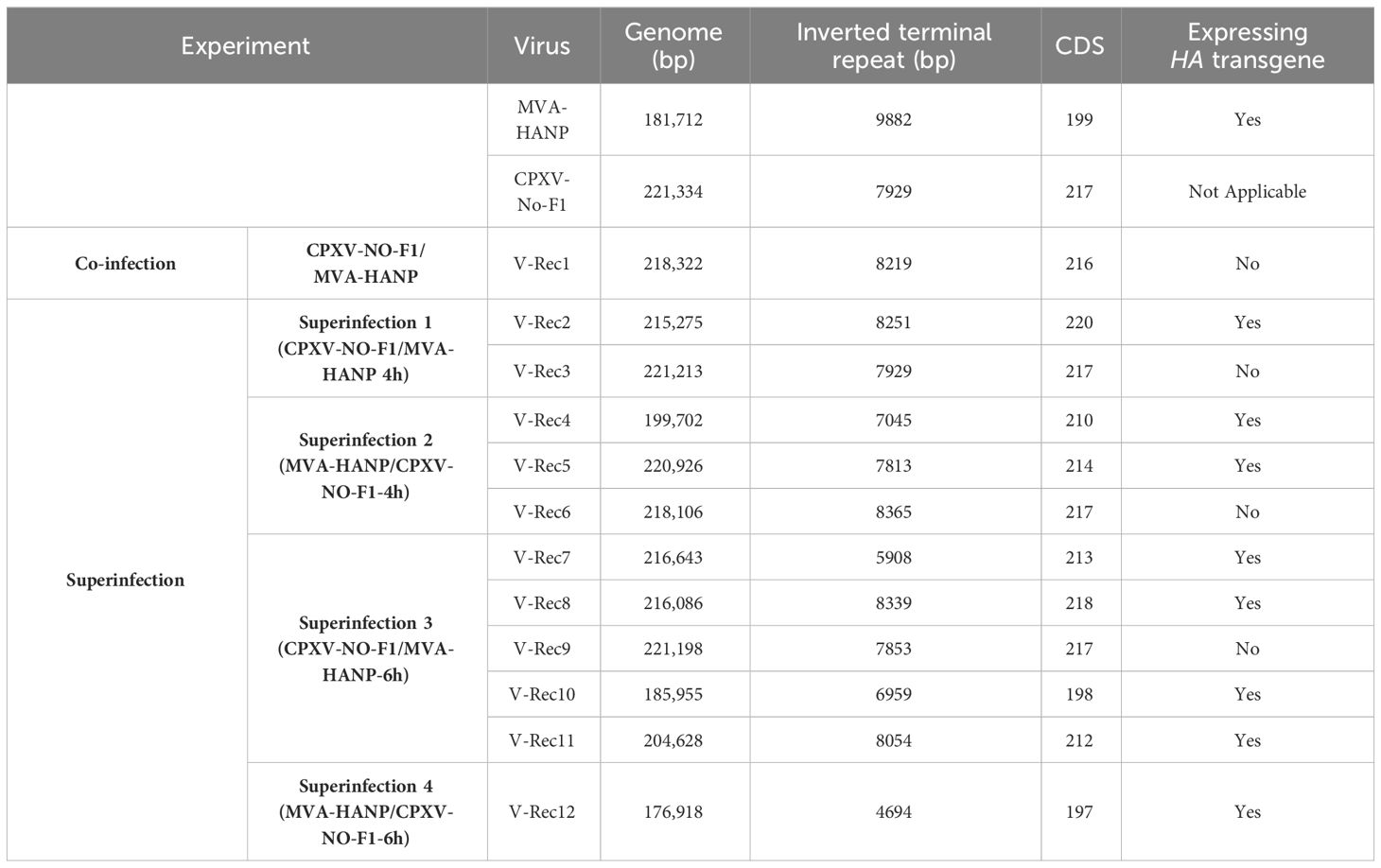
Table 1 Genome size and number of predicted CDS of CPXV-No-F1, MVA-HANP and the putative recombinant viruses.
The number of predicted CDS in the genomes of the putative recombinant viruses varied from 197 to 220 CDS (Table 1). The genomes of the putative recombinant viruses included more CDS than the parental MVA-HANP, with exception of V-Rec10 and V-Rec12. Furthermore, there were three viruses (V-Rec3, V-Rec6 and V-Rec9) that contained the same number of predicted CDS as the parental CPXV-No-F1.
In order to localize the possible recombination events in the putative recombinant viruses, the twelve putative recombinant viruses were examined for recombination. The analysis confirmed recombination across the genome of eleven recombinant viruses (Supplementary Table 4). Only one virus, V-Rec9 from superinfection 3, does not show any recombination event based on the recombination analysis. The location of recombinant events in the genome of the progeny viruses are random, there are no recombination distribution pattern along their genomes. The recombination events occur both in the conserved central region (VACV-Cop F4L to VACV-Cop A24L) and the variable regions, including ITR (Supplementary Table 5; Figure 2). The number of recombinant breakpoints distribute in the central region varied from 0 to 16, whereas the number of breakpoints in the variable regions ranges from 1 to 6. The genomes of the recombinant viruses are a mosaic of the two parental strains. The lengths of the DNA segments exchanged between the parental viruses range from approximately 200 bp to 36 kbp (Figure 2). The percentage of DNA derived from the parental strains in the recombinant viruses is variable (Figure 3). The majority of the recombinant viruses have more DNA from the parental CPXV-No-F1 (∼45,9%-99,9%) than that from the parental MVA-HANP (∼0,1%-54,1%); nevertheless, some of them contain the HA and NP transgenes. Only two recombinant viruses, V-Rec2 and V-Rec12, contain more DNA from the parental MVA-HANP (Figure 3; Supplementary Table 5).
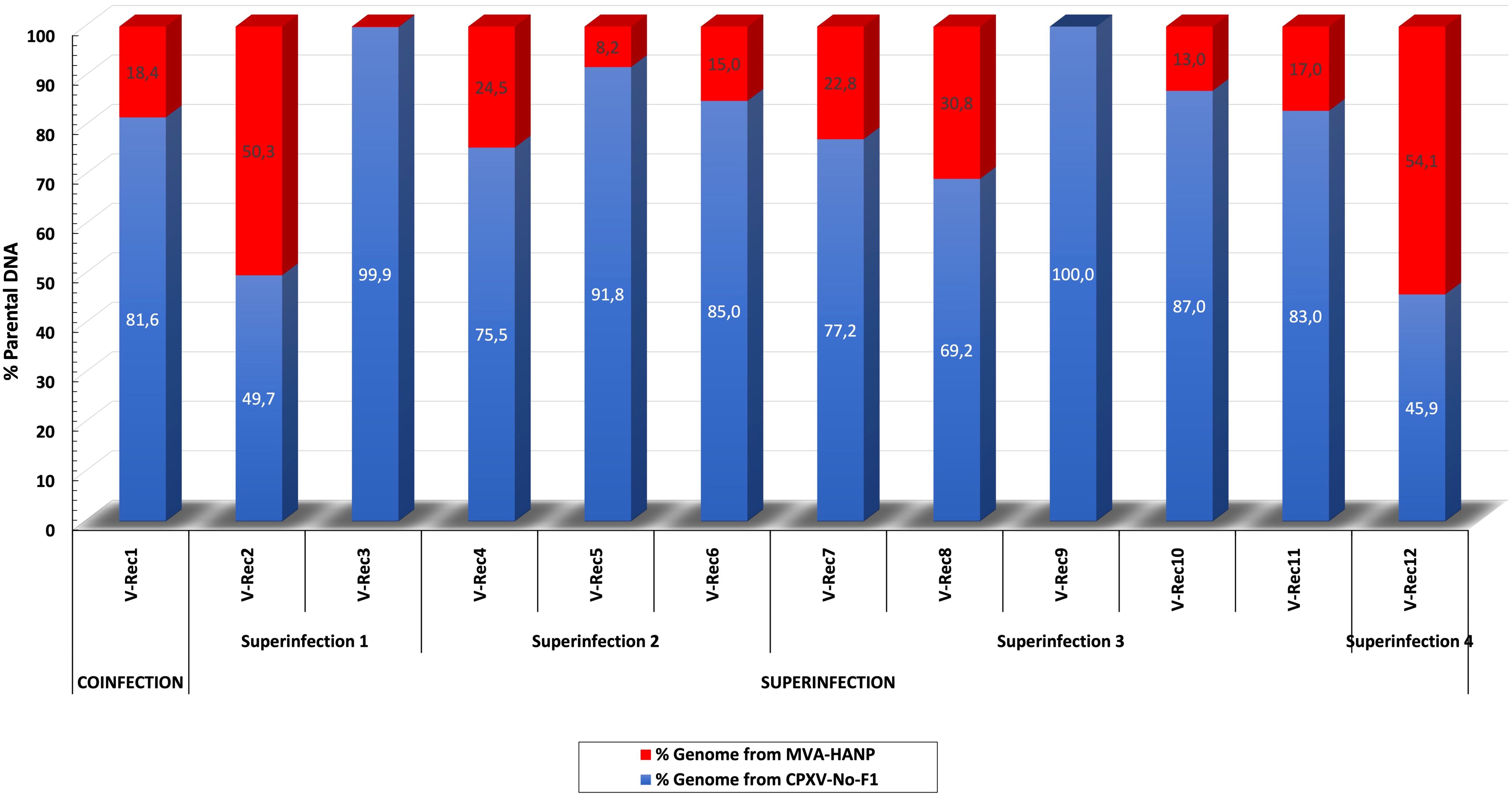
Figure 3 Percentage of DNA derived from the parental viruses (CPXV-No-F1 and MVA-HANP) in the putative recombinant viruses. The putative recombinant viruses were produced in Vero cells co-infected and superinfected with CPXV-No-F1 and MVA-HANP. Superinfection 1, primary infection with CPXV-No-F1 and secondary infection with MVA-HANP at 4h post primary infection (ppi); Superinfection 2, primary infection with MVA-HANP and secondary infection with CPXV-No-F1 at 4h ppi; Superinfection 3, primary infection with CPXV-No-F1 and secondary infection with MVA-HANP at 6h ppi; Superinfection 4, primary infection with MVA-HANP and secondary infection with CPXV-No-F1 at 6h ppi. Blue blocks represent the coding sequences (CDS) from CPXV-No-F1.
The parental MVA-HANP harbored a double expression cassette containing the influenza HA and NP transgenes. The sequencing analysis of the eight HA transgene expressing recombinant viruses (V-Rec2, V-Rec4, V-Rec5, V-Rec7, V-Rec8, V-Rec10, V-Rec11 and V-Rec12) confirmed that these viruses contain an intact double expression cassette and its flanking sequences. The sequencing analysis of non-HA-transgene expressing recombinant viruses (V-Rec1, V-Rec3, and V-Rec6) revealed that the HA-NP insert is absent in V-Rec3 and V-Rec6, but not in V-Rec1. Interestingly, the latter (V-Rec1) contains part of the double expression cassette. It comprises major part of the NP transgene (94%) and the upstream flanking MVA sequence (Figure 4A) similar to the incomplete MVA-HANP. The presence of the incomplete MVA-HANP population with a partial deleted double cassette expression was corroborated by the occurrence of recombinant virus V-Rec1.
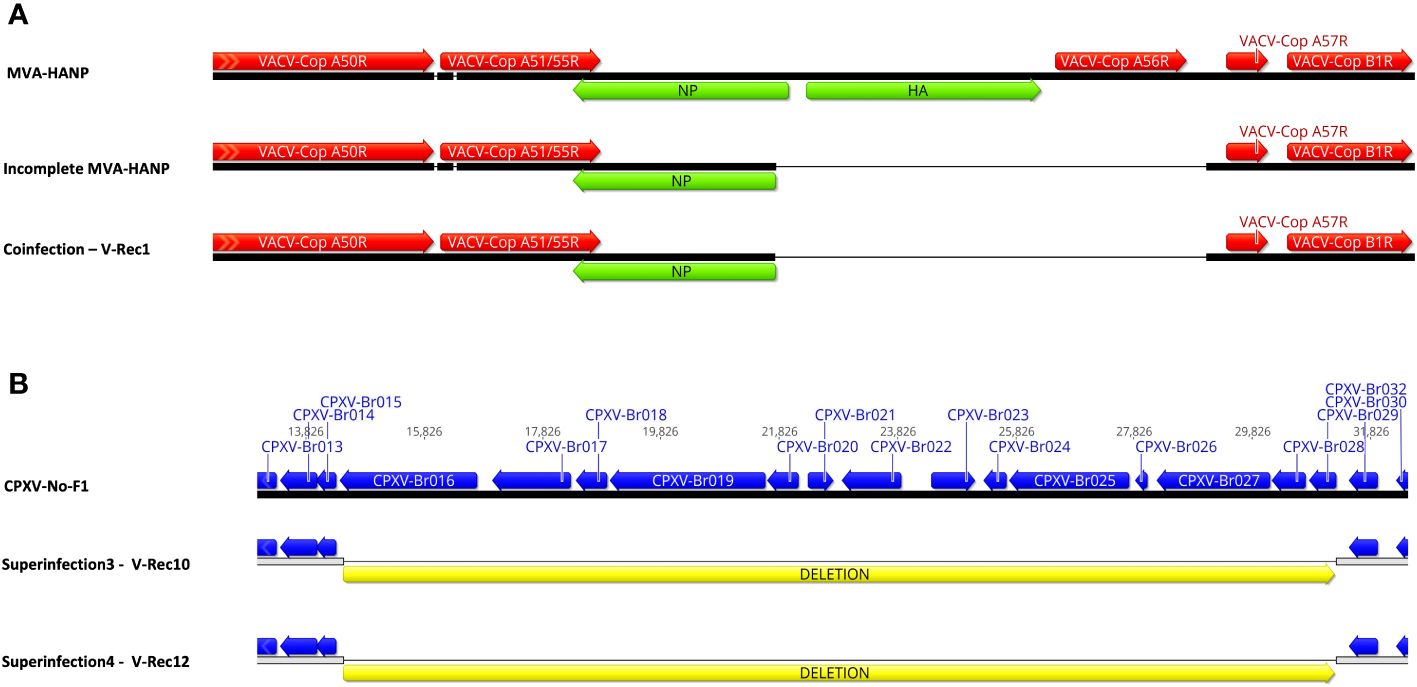
Figure 4 Comparison of the recombinant viruses with the parental virus. (A) Comparison of the recombinant region of V-Rec1 with MVA-HANP and incomplete MVA-HANP. V-Rec1 was produced in Vero cells co-infected with CPXV-No-F1 and MVA-HANP. Green blocks represent the influenza virus hemagglutinin (HA) and nucleoprotein (NP) transgenes. Red blocks represent the coding sequences (CDS) from MVA-HANP. (B) Deletion in V-Rec10 and V-Rec12. V-Rec10 was produced in Vero cells infected with CPXV-No-F1 and superinfected with MVA-HANP at 6h post primary infection (ppi) (superinfection 3). V-Rec12 was produced in Vero cells infected with MVA-HANP and superinfected with CPXV-No-F1 at 6h ppi (superinfection 4). Yellow blocks represent the deleted sequence in the progeny viruses.
In addition to the recombination events, our analyses revealed that the eleven viruses underwent other genetic variations, such as deletions. Although most of them are located outside of the CDS regions (Supplementary Table 6). A large deletion of 16,761 bp is found close to the left terminal genomic region of V-Rec10 and V-Rec12 (Figure 4B). The deleted sequence comprises from CXPV-BR016 gene to CPXV-BR029 gene. Most of these genes encode proteins involve in host range. Additional small deletions and insertions are also detected within these two recombinant genomes (Supplementary Table 6). The other recombinant viruses also contain deletions, insertions and/or non-synonymous single-nucleotide mutations (nsSNMs). Interestingly, we found two nsSNMs in VACV-Cop N2L and VACV-Cop K2L genes of non-recombinant virus V-Rec9 (Supplementary Table 6). The nsSNM in the VACV-Cop K2L gene resulted in the introduction of a premature stop codon and consequently the truncation of the protein.
As mentioned before there are two recombinant viruses with >50% of their genome derived from MVA-HANP. These two recombinant viruses rescued disrupted/deleted genes and, furthermore, gained genes that were absence in MVA-HANP. V-Rec12 rescued CPXV-Br009, CPXV-Br032, CPXV-Br033, CPXV-Br034, CPXV-Br035 and CPXV-Br036 genes (homologs to VACV-Cop C16L, C5L, C4L, C3L, C2L and C1L genes, respectively) that were lost in MVA during the attenuation process. Additionally, four fragmented/disrupted genes, CPXV-Br037, CPXV-Br039, CPXV-Br043 and CPXV-Br078 (homologs to VACV-Cop N1L, K1L, K5L and O1L genes, respectively), were rescued in the recombinant virus V-Rec12 by recombination. Although O1L gene is also fragmented in VACV-CVA. Moreover, the recombinant virus gained seven more genes that were absent in MVA and VACV-CVA strains: CPXV-Br010, CPXV-Br011, CPXV-Br012, CPXV-Br013, CPXV-Br014, CXPV-Br015 genes and one gene (No-F1-009 gene) that encodes BTB Kelch-domain containing protein. Compared to V-Rec12, V-Rec2 gained six additional genes (CPXV-Br002, CPXV-Br016, CPXV-Br017, CPXV-Br018, CPXV-Br19 and CPXV-Br020 genes) by recombination. Furthermore, the recombination events in V-Rec2 rescued deleted genes: CPXV-Br005, CPXV-Br009, CPXV-Br032, CPXV-Br033, CPXV-Br034, CPXV-Br035, CPXV-Br036, CPXV-Br039, CPXV-Br040, CPXV-Br213, CPXV-Br217 and CPXV-Br226 genes (homologs to VACV-Cop B28R, C16L, C5L, C4L, C3L, C2L, C1L, M1L, M2L, B20R, C12L and C22L genes). Additionally, twelve fragmented/disrupted genes in MVA were also rescued in this recombinant virus (CPXV-Br003, CPXV-Br006, CPXV-Br008, CPXV-Br023, CPXV-Br025, CPXV-Br027, CPXV-Br041, CPXV-Br078, CPXV-Br207, CPXV-Br212, CXPV-Br223 and CPXV-Br227 genes).
In our previous study, we performed co-infection and superinfection experiments with a naturally circulating Fennoscandian CPXV-No-F1 and MVA carrying an influenza HA and NP transgene in semi-permissive Vero cells. We have showed that recombination between these viruses occurred in both co-infected and superinfected Vero cells. Although the likelihood of recombination in co-infected and superinfected Vero cells was always considered low because (1) Vero cells are semi-permissive to MVA (15, 20) and (2) mechanisms as superinfection exclusion prevent the superinfection of the infected cell with a second virus (58, 59). A previous study has reported recombination between human CPXV (hCPXV) and MVA-HANP in co-infected BHK-21 cells (22). Compared to Vero cells, those cells are fully permissive to MVA-HANP and CPXV (20). Viral DNA replication in non-permissive cells to MVA infection is not blocked (85), therefore the likelihood of recombination increases since recombination and viral DNA replication are connected (86, 87). In our superinfection experiments, the time gap between the first and the second infection was 4h and 6h ppi to ensure the establishment of primary infection since it has been reported that VACV DNA synthesis in HeLa cells starts 2 hours postinfection (88). Although the viral DNA synthesis of the primary virus is not needed for superinfection exclusion (59, 89). A study demonstrated the time of superinfection of 4 and 6 hours after primary infection with VACV produced 90% and 99% exclusion of superinfecting virus, respectively (89).
The recombination between MVA-HANP and CPXV-No-F1 generated mosaic genomes containing genomic material from both parental viruses. Several progeny viruses displayed plaque morphologies different from that of the parental viruses. This was also observed in the progeny viruses arising from superinfected Vero cells with MVA-HANP and CPXV-No-F1, using 2h ppi (27). Similarly, the recombinant viruses from BHK-21 cells co-infected with MVA-HANP and hCPXV displayed non-parental and parental traits with respect to plaque phenotype, in vitro host range and cytopathogenicity (22, 71).
Our recombinant viruses showed different plaque phenotypes. It has been reported that the proteins encoded by VACV-Cop F5L, F11L, F12L, F13L, A33R, A34R, A36R, A56R, and B5R genes may be involved in determining the plaque morphology (90–95). Some of these genes were fragmented (i.e. VACV-Cop F5L and VACV-Cop F11L gene) or encompassed some deletions (i.e. VACV-Cop A36R) in MVA (16). Two of our recombinant viruses (V-Rec2 and V-Rec12) formed small and non-lytic plaques. These recombinant viruses contained the defective VACV-Cop F5L, F11L and VACV-Cop A36R genes from MVA-HANP. F5L and F11L proteins are required to form normal plaques. They increase the plaque size and only F5L protein promotes the formation of central plaque clearing (90–92). The deletion of the gene encoding A36R protein decreased the plaque size (96). Additionally, these two recombinant viruses displayed plaques with comet formation, similar to those of the recombinant virus V-Rec6. The genes associated to the formation of comet-shaped plaques are VACV-Cop A33R, A34R and B5R (93, 94, 97). Those recombinant viruses contained VACV-Cop A33R, A34R and B5R genes from MVA-HANP, except for recombinant virus V-Rec6 that contained VACV-Cop B5R gene derived from CPXV-No-F1. Other recombinant viruses that only have VACV-Cop B5R gene from MVA-HANP, as well as VACV-Cop A33R and VACV-Cop A34R genes from CPXV-No-F1, did not show plaques with comet formation.
Only two viruses produced syncytial plaques: recombinant virus V-Rec1 and non-recombinant V-Rec9. The latter produces a truncated K2L protein due to the introduction of an earlier stop codon in the VACV-Cop K2L gene as a result of a nsSNM. It has been reported that the lack of K2L protein causes the fusion of infected cells (98–100). This protein forms a complex with A56 protein, and the heterodimer prevents syncytia formation (101) (98–100, 102). The VACV-Cop A56R gene of V-Rec9 was intact; in contrast, this gene was deleted in the recombinant virus V-Rec1.
Several recombinant viruses were transgene positive. The proportion of HA-expressing recombinant viruses in the co-infected and superinfected Vero cells were reported elsewhere (27). The HA-expressing recombinant viruses had the complete double expression cassette, while the non-HA-expressing recombinant virus V-Rec1 contained an incomplete expression cassette without the HA transgene similar to the incomplete MVA-HANP. It seems that this progeny virus was the result of recombination of CPXV-No-F1 and the incomplete MVA-HANP. The instability of the transgene in MVA-HANP as well as in recombinant progeny viruses from BHK-21 cells co-infected with MVA-HANP and CPXV-No-H1 has been previously reported. MVA-HANP and the recombinant viruses were genetically unstable and lost the transgene during cell culture passages (22, 71). The instability of the transgene is one of the major concerns in the production of viral vector vaccines because any mutation in the expression cassette could lead to unpredicted characteristics. Furthermore, the transgene is used as a tag to monitor the expression cassette in the recombinant progeny viruses. Therefore, the loss or partial loss of the transgene hinders the tracking of released recombinant progeny viruses. Another concern about the use of MVA-HANP is to transfer the transgene into a multiplication competent OPXV (46). In this study, we have shown that the transgenes were transferred to the recombinant viruses with a genome mainly derived from CPXV-No-F1. Furthermore, the recombinant progeny viruses with the transgene displayed new and non-parental plaque phenotypes. A biological characterization of the recombinant viruses is required to investigate their host range, multiplication curves at low and high moi, cell tropism, transmissibility and virulence.
Compared to CVA, MVA had lost several genes and 25 genes were fragmented and/or suffered mutations during the attenuation process, such as the host range genes VACV-Cop K1L and C12L (16, 18). The recombination of MVA with a multiplication competent OPXV may lead to the restoration of disrupted/deleted genes in MVA. In this study, we observed that two of our recombinant viruses, that had >50% DNA from MVA-HANP, rescued deleted and fragment host range genes, such CXPV-Br009 (VACV-Cop C16L) and CPXV-Br041 (VACV-cop K1L) (23, 103). Even these recombinant viruses gained new host range genes, crmB/CPXV-Br226 (VACV-Cop B28R) and vCD30/CPXV015 (103, 104). The recombination of MVA with a wild OPXV was considered negligible since smallpox was eradicated. However, the circulation of the naturally OPXV (3, 49, 68, 105–107) and the emergence of new OPXV in the last few years (50, 108, 109) have increased the likelihood of recombination between MVA and a naturally replication competent OPXV during co-infection or superinfection of the same cell or host. For instance, the ongoing global outbreak of Mpox and prophylactic or post-exposure vaccination with MVA-BN provide a good scenario for co-infection/superinfection and subsequent recombination between MVA and MPXV. In addition, several orthopoxvirus outbreaks in humans have been reported worldwide (6, 110). Our study has shown that insertion of the transgene into the genome of a co-infecting or superinfecting OPXV was specific, but recombination in other parts of the genome were nonspecific and unpredictable. In addition, some of the recombinant viruses lost some or part of the transgenic cassette even when they were inserted into the intended genomic regions, and the loss of transgene may preclude tracking of recombinant viruses. To evaluate the potential for recombination and robust monitoring of potential recombinant viruses, hazard characterization and risk assessment of MVA vectored biologicals should include genome wide characterization.
Finally, it is important to highlight the limitations of this study. First, the high MOI used in our co-infection and superinfection experiments may not be achievable under natural co-infection/superinfection. Second, our experiments were done in cell cultures and an extrapolation cannot be made to animals with an intact immune system Vero cells in particular is a continuous cell line lacking type 1 interferon gene although it still has interferon receptors (111). Third, our use of selection markers like plaque phenotype, expression or non-expression of the transgene may have introduced a selection bias and thus underestimate the pattern and diversity of recombination between co-infecting or superinfecting viruses. Fourth, the likelihood of recombination in co-infected and superinfected cells is low; however, the possibility of co-localization has raised because the circulation of naturally zoonotic OPXV (e.g. CPXV and MPXV) has increased in recent years as well as the vaccination with MVA. Fifth, this study did not calculate recombination frequency (RF) with respect to the transgene cassette (112), and the number of recombinant viruses selected was low to statistically infer genome-wide patterns and diversity of recombination between CPXV and MVA-vectored vaccine during co-infecting and superinfecting scenarios. Future studies will address these limitations through plaque independent deep sequencing analysis of cell cultures and animals co-infected or superinfected with MVA and CPXV under conditions that better reflects natural infection.
In conclusion, superinfection exclusion and low permissivity of Vero cells to MVA did not prevent the recombination between MVA vectored vaccines and the naturally circulating CPXV during superinfection of cells. The recombination between MVA-HANP and the naturally circulating CPXV-No-F1 in co-infected and superinfected Vero cells lead to the generation of progeny viruses with non-parental biological and genetic characteristic as well as the regaining of delete/fragmented genes in MVA-HANP, transfer of the transgene into CPXV and introgression of other MVA genes to CPXV. This is a proof-of-concept study and future studies will examine the risk of recombination between MVA or rMVA and naturally circulating OPXV during co-infection and superinfection by determining the likelihood of recombination and its consequences.
The datasets presented in this study are deposited in the Genbank (https://www.ncbi.nlm.nih.gov/genbank/), accession numbers can be found below: OQ818667, OQ822790, OQ822791, OQ822792, OQ822793, OQ822794, OQ822795, OQ822796, OQ822797, OQ822798, OQ822799, OQ822800 and OQ822801.
DD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. UM: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. AB: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. AN: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. MO: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the University of Tromsø, the Arctic University of Norway (project A212100108) and the National Graduate School in Infection Biology and Antimicrobials (grant no. 249062). Article processing charge was paid UiT - The Arctic University of Norway.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1277447/full#supplementary-material
1. Hendrickson RC, Wang C, Hatcher EL, Lefkowitz EJ. Orthopoxvirus genome evolution: the role of gene loss. Viruses. (2010) 2:1933–67. doi: 10.3390/V2091933
2. MacLachlan NJ, Dubovi EJ eds. “Poxviridae”. In: Fenner’s Veterinary Virology. Academic Press, Boston. p. 157–74. doi: 10.1016/B978-0-12-800946-8.00007-6
3. Silva NIO, de Oliveira JS, Kroon EG, Trindade G de S, Drumond BP. Here, there, and everywhere: the wide host range and geographic distribution of zoonotic orthopoxviruses. Viruses. (2021) 13:1–20. doi: 10.3390/V13010043
4. Vora NM, Li Y, Geleishvili M, Emerson GL, Khmaladze E, Maghlakelidze G, et al. Human infection with a zoonotic orthopoxvirus in the country of Georgia. N Engl J Med. (2015) 372:1223. doi: 10.1056/NEJMOA1407647
5. Reynolds MG, Guagliardo SAJ, Nakazawa YJ, Doty JB, Mauldin MR. Understanding orthopoxvirus host range and evolution: from the enigmatic to the usual suspects. Curr Opin Virol. (2018) 28:108–15. doi: 10.1016/J.COVIRO.2017.11.012
6. Diaz JH. The disease ecology, epidemiology, clinical manifestations, management, prevention, and control of increasing human infections with animal orthopoxviruses. Wilderness Environ Med. (2021) 32:528–36. doi: 10.1016/J.WEM.2021.08.003
7. Strassburg MA. The global eradication of smallpox. Am J Infect Control. (1982) 10:53–9. doi: 10.1016/0196-6553(82)90003-7
8. Jacobs BL, Langland JO, Kibler KV, Denzler KL, White SD, Holechek SA, et al. Vaccinia virus vaccines: past, present and future. Antiviral Res. (2009) 84:1. doi: 10.1016/J.ANTIVIRAL.2009.06.006
9. Sánchez-Sampedro L, Perdiguero B, Mejías-Pérez E, García-Arriaza J, Di Pilato M, Esteban M. The evolution of poxvirus vaccines. Viruses. (2015) 7:1726–803. doi: 10.3390/v7041726
10. Mayr A. Smallpox vaccination and bioterrorism with pox viruses. Comp Immunol Microbiol Infect Dis. (2003) 26:423–30. doi: 10.1016/S0147-9571(03)00025-0
11. Pittman PR, Hahn M, Lee HS, Koca C, Samy N, Schmidt D, et al. Phase 3 efficacy trial of modified vaccinia ankara as a vaccine against smallpox. N Engl J Med. (2019) 381:1897–908. doi: 10.1056/nejmoa1817307
12. Mahnel H, Mayr A. Experiences with immunization against orthopox viruses of humans and animals using vaccine strain MVA. Berl Munch Tierarztl Wochenschr. (1994) 107:253–6.
13. Mayr A, Stickl H, Müller HK, Danner K, Singer H. [The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author’s transl)]. Zentralbl Bakteriol B. (1978) 167:375–90.
14. Mayr A, Hochstein-Mintzel V, Stickl H. Abstammung, Eigenschaften und Verwendung des attenuierten Vaccinia-Stammes MVA. Infection. (1975) 3:6–14. doi: 10.1007/BF01641272
15. Meyer H, Sutter G, Mayr A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J Gen Virol. (1991) 72:1031–8. doi: 10.1099/0022-1317-72-5-1031
16. Antoine G, Scheiflinger F, Dorner F, Falkner FG. The complete genomic sequence of the modified vaccinia Ankara strain: Comparison with other orthopoxviruses. Virology. (1998) 244:365–96. doi: 10.1006/viro.1998.9123
17. Blanchard TJ, Alcamí A, Andrea P, Smith GL. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: Implications for use as a human vaccine. J Gen Virol. (1998) 79:1159–67. doi: 10.1099/0022-1317-79-5-1159
18. Carroll MW, Moss B. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: Propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology. (1997) 238:198–211. doi: 10.1006/viro.1997.8845
19. Jordan I, Horn D, Oehmke S, Leendertz FH, Sandig V. Cell lines from the Egyptian fruit bat are permissive for modified vaccinia Ankara. Virus Res. (2009) 145:54–62. doi: 10.1016/j.virusres.2009.06.007
20. Okeke MI, Nilssen Ø, Traavik T. Modified vaccinia virus Ankara multiplies in the rat IEC-6 cells and limited production of mature virions occurs in other mammalian cell lines. J Gen Virol. (2006) 87:21–7. doi: 10.1099/vir.0.81479-0
21. Drexler I, Heller K, Wahren B, Erfle V, Sutter G. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J Gen Virol. (1998) 79:347–52. doi: 10.1099/0022-1317-79-2-347
22. Hansen H, Okeke MI, Nilssen Ø, Traavik T. Recombinant viruses obtained from co-infection in vitro with a live vaccinia-vectored influenza vaccine and a naturally occurring cowpox virus display different plaque phenotypes and loss of the transgene. Vaccine. (2004) 23:499–506. doi: 10.1016/J.VACCINE.2004.06.032
23. Peng C, Moss B. Repair of a previously uncharacterized second host-range gene contributes to full replication of modified vaccinia virus Ankara (MVA) in human cells. Proc Natl Acad Sci U.S.A. (2020) 117:3759–67. doi: 10.1073/pnas.1921098117
24. Suter M, Meisinger-Henschel C, Tzatzaris M, Hülsemann V, Lukassen S, Wulff NH, et al. Modified vaccinia Ankara strains with identical coding sequences actually represent complex mixtures of viruses that determine the biological properties of each strain. Vaccine. (2009) 27:7442–50. doi: 10.1016/j.vaccine.2009.05.095
25. Earl PL, Americo JL, Wyatt LS, Espenshade O, Bassler J, Gong K, et al. Rapid protection in a monkeypox model by a single injection of a replication-deficient vaccinia virus. Proc Natl Acad Sci U.S.A. (2008) 105:10889–94. doi: 10.1073/pnas.0804985105
26. Guerra S, González JM, Climent N, Reyburn H, López-Fernández LA, Nájera JL, et al. Selective induction of host genes by MVA-B, a candidate vaccine against HIV/AIDS. J Virol. (2010) 84:8141–52. doi: 10.1128/jvi.00749-10
27. Okeke MI, Okoli AS, Diaz D, Offor C, Oludotun TG, Tryland M, et al. Hazard characterization of modified vaccinia virus ankara vector: What are the knowledge gaps? Viruses. (2017) 9:1–29. doi: 10.3390/v9110318
28. Orlova OV, Glazkova DV, Bogoslovskaya EV, Shipulin GA, Yudin SM. Development of modified vaccinia virus ankara-based vaccines: advantages and applications. Vaccines. (2022) 10:1–16. doi: 10.3390/vaccines10091516
29. Joachim A, Nilsson C, Aboud S, Bakari M, Lyamuya EF, Robb ML, et al. Potent functional antibody responses elicited by HIV-I DNA priming and boosting with heterologous HIV-1 recombinant MVA in healthy Tanzanian adults. PloS One. (2015) 10:1–15. doi: 10.1371/journal.pone.0118486
30. Nilsson C, Godoy-Ramirez K, Hejdeman B, Bråve A, Gudmundsdotter L, Hallengärd D, et al. Broad and potent cellular and humoral immune responses after a second late HIV-modified vaccinia virus ankara vaccination in HIV-DNA-primed and HIV-modified vaccinia virus ankara-boosted swedish vaccinees. AIDS Res Hum Retroviruses. (2014) 30:299–311. doi: 10.1089/aid.2013.0149
31. Milligan ID, Gibani MM, Sewell R, Clutterbuck EA, Campbell D, Plested E, et al. Safety and immunogenicity of novel adenovirus type 26-and modified vaccinia Ankara-vectored Ebola vaccines: A randomized clinical trial. JAMA - J Am Med Assoc. (2016) 315:1610–23. doi: 10.1001/jama.2016.4218
32. Tapia MD, Sow SO, Lyke KE, Haidara FC, Diallo F, Doumbia M, et al. Use of ChAd3-EBO-Z Ebola virus vaccine in Malian and US adults, and boosting of Malian adults with MVA-BN-Filo: a phase 1, single-blind, randomised trial, a phase 1b, open-label and double-blind, dose-escalation trial, and a nested, randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. (2016) 16:31–42. doi: 10.1016/S1473-3099(15)00362-X
33. Callendret B, Vellinga J, Wunderlich K, Rodriguez A, Steigerwald R, Dirmeier U, et al. A prophylactic multivalent vaccine against different filovirus species is immunogenic and provides protection from lethal infections with Ebolavirus and Marburgvirus species in non-human primates. PloS One. (2018) 13:1–24. doi: 10.1371/journal.pone.0192312
34. Fuentes S, Ravichandran S, Coyle EM, Klenow L, Khurana S. Human antibody repertoire following ebola virus infection and vaccination. iScience. (2020) 23:1–12. doi: 10.1016/j.isci.2020.100920
35. Jordan E, Lawrence SJ, Meyer TPH, Schmidt D, Schultz S, Mueller J, et al. Broad antibody and cellular immune response from a phase 2 clinical trial with a novel multivalent poxvirus-based respiratory syncytial virus vaccine. J Infect Dis. (2021) 223:1062–72. doi: 10.1093/infdis/jiaa460
36. Koch T, Dahlke C, Fathi A, Kupke A, Krähling V, Okba NMA, et al. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect Dis. (2020) 20:827–38. doi: 10.1016/S1473-3099(20)30248-6
37. Aldoss I, la Rosa C, Baden LR, Longmate J, Ariza-Heredia EJ, Rida WN, et al. Poxvirus vectored cytomegalovirus vaccine to prevent cytomegalovirus viremia in transplant recipients: A phase 2, randomized clinical trial. Ann Intern Med. (2020) 172:306–16. doi: 10.7326/M19-2511
38. Kreijtz JHCM, Goeijenbier M, Moesker FM, van den Dries L, Goeijenbier S, De Gruyter HLM, et al. Safety and immunogenicity of a modified-vaccinia-virus-Ankara-based influenza A H5N1 vaccine: A randomised, double-blind phase 1/2a clinical trial. Lancet Infect Dis. (2014) 14:1196–207. doi: 10.1016/S1473-3099(14)70963-6
39. Puksuriwong S, Ahmed MS, Sharma R, Krishnan M, Leong S, Lambe T, et al. Modified vaccinia Ankara-vectored vaccine expressing nucleoprotein and matrix protein 1 (M1) activates mucosal M1-specific T-Cell immunity and tissue-resident memory T Cells in human nasopharynx-associated lymphoid tissue. J Infect Dis. (2020) 222:807–19. doi: 10.1093/infdis/jiz593
40. Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: A randomised, placebo-controlled phase 2b trial. Lancet. (2013) 381:1021–8. doi: 10.1016/S0140-6736(13)60177-4
41. Hodgson SH, Ewer KJ, Bliss CM, Edwards NJ, Rampling T, Anagnostou NA, et al. Evaluation of the efficacy of ChAd63-MVA vectored vaccines expressing circumsporozoite protein and ME-TRAP against controlled human malaria infection in malaria-naive individuals. J Infect Diseases. (2015) 211:1076–86. doi: 10.1093/infdis/jiu579
42. Biswas S, Choudhary P, Elias SC, Miura K, Milne KH, De Cassan SC, et al. Assessment of humoral immune responses to blood-stage malaria antigens following ChAd63-MVA immunization, controlled human malaria infection and natural exposure. PloS One. (2014) 9:1–17. doi: 10.1371/journal.pone.0107903
43. Sebastian S, Gilbert SC. Recombinant modified vaccinia virus Ankara-based malaria vaccines. Expert Rev Vaccines. (2016) 15:91–103. doi: 10.1586/14760584.2016.1106319
44. European Medicines Agency. EMA recommends approval of Imvanex for the prevention of monkeypox disease(2022). Available online at: https://www.ema.europa.eu/en/news/ema-recommends-approval-imvanex-prevention-monkeypox-disease (Accessed January 1, 2023).
45. U.S. Food and Drugs. Vaccines licensed for use in the United States(2022). Available online at: https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states.
46. Goossens M, Pauwels K, Willemarck N, Breyer D. Environmental risk assessment of clinical trials involving modified vaccinia virus ankara (MVA)-based vectors. Curr Gene Ther. (2013) 13:413–20. doi: 10.2174/156652321306140103221941
47. Coulson D, Upton C. Characterization of indels in poxvirus genomes. Virus Genes. (2011) 42:171–7. doi: 10.1007/s11262-010-0560-x
48. Franke A, Pfaff F, Jenckel M, Hoffmann B, Höper D, Antwerpen M, et al. Classification of cowpox viruses into several distinct clades and identification of a novel lineage. Viruses. (2017) 9:1–14. doi: 10.3390/v9060142
49. Diaz-Cánova D, Moens UL, Brinkmann A, Nitsche A, Okeke MI. Genomic sequencing and analysis of a novel human cowpox virus with mosaic sequences from north america and old world orthopoxvirus. Front Microbiol. (2022) 13:868887. doi: 10.3389/FMICB.2022.868887
50. Gao J, Gigante C, Khmaladze E, Liu P, Tang S, Wilkins K, et al. Genome sequences of Akhmeta virus, an early divergent old world orthopoxvirus. Viruses. (2018) 10:1–15. doi: 10.3390/v10050252
51. Gigante CM, Gao J, Tang S, McCollum AM, Wilkins K, Reynolds MG, et al. Genome of alaskapox virus, a novel orthopoxvirus isolated from alaska. Viruses. (2019) 11:1–15. doi: 10.3390/V11080708
52. Gubser C, Hué S, Kellam P, Smith GL. Poxvirus genomes: A phylogenetic analysis. J Gen Virol. (2004) 85:105–17. doi: 10.1099/vir.0.19565-0
53. Qin L, Evans DH. Genome scale patterns of recombination between coinfecting vaccinia viruses. J Virol. (2014) 88:5277–86. doi: 10.1128/jvi.00022-14
54. Qin L, Upton C, Hazes B, Evans DH. Genomic analysis of the vaccinia virus strain variants found in dryvax vaccine. J Virol. (2011) 85:13049. doi: 10.1128/JVI.05779-11
55. Qin L, Favis N, Famulski J, Evans DH. Evolution of and evolutionary relationships between extant vaccinia virus strains. J Virol. (2015) 89:1809. doi: 10.1128/JVI.02797-14
56. Smithson C, Meyer H, Gigante CM, Gao J, Zhao H, Batra D, et al. Two novel poxviruses with unusual genome rearrangements: NY_014 and Murmansk. Virus Genes. (2017) 53:883–97. doi: 10.1007/s11262-017-1501-8
57. Smithson C, Purdy A, Verster AJ, Upton C. Prediction of steps in the evolution of variola virus host range. PloS One. (2014) 9:e91520. doi: 10.1371/JOURNAL.PONE.0091520
58. Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: A mechanism for rapid virus spread. Science (80-). (2010) 327:873–6. doi: 10.1126/science.1183173
59. Laliberte JP, Moss B. A novel mode of poxvirus superinfection exclusion that prevents fusion of the lipid bilayers of viral and cellular membranes. J Virol. (2014) 88:9751–68. doi: 10.1128/jvi.00816-14
60. Campe H, Zimmermann P, Glos K, Bayer M, Bergemann H, Dreweck C, et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg Infect Dis. (2009) 15:777. doi: 10.3201/EID1505.090159
61. Ducournau C, Ferrier-Rembert A, Ferraris O, Joffre A, Favier AL, Flusin O, et al. Concomitant human infections with 2 cowpox virus strains in related cases, France, 2011. Emerg Infect Dis. (2013) 19:1996–9. doi: 10.3201/eid1912.130256
62. Gazzani P, Gach JE, Colmenero I, Martin J, Morton H, Brown K, et al. Fatal disseminated cowpox virus infection in an adolescent renal transplant recipient. Pediatr Nephrol. (2017) 32:533–6. doi: 10.1007/s00467-016-3534-y
63. Andreani J, Arnault JP, Bou Khalil JY, Abrahão J, Tomei E, Vial E, et al. Atypical cowpox virus infection in smallpox-vaccinated patient, France. Emerg Infect Dis. (2019) 25:212–9. doi: 10.3201/eid2502.171433
64. Ferrier A, Frenois-Veyrat G, Schvoerer E, Henard S, Jarjaval F, Drouet I, et al. Fatal cowpox virus infection in human fetus, France, 2017. Emerg Infect Dis. (2021) 27:2570–7. doi: 10.3201/eid2710.204818
65. Krankowska DC, Woźniak PA, Cybula A, Izdebska J, Suchacz M, Samelska K, et al. Cowpox: How dangerous could it be for humans? Case report. Int J Infect Dis. (2021) 104:239–41. doi: 10.1016/j.ijid.2020.12.061
66. Tryland M, Myrmel H, Holtet L, Haukenes G, Traavik T. Clinical cowpox cases in Norway. Scand J Infect Dis. (1998) 30:301–3. doi: 10.1080/00365549850160972
67. Okeke MI, Hansen H, Traavik T. A naturally occurring cowpox virus with an ectromelia virus A-type inclusion protein gene displays atypical A-type inclusions. Infect Genet Evol. (2012) 12:160–8. doi: 10.1016/J.MEEGID.2011.09.017
68. Diaz-Cánova D, Mavian C, Brinkmann A, Nitsche A, Moens U, Okeke MI. Genomic sequencing and phylogenomics of cowpox virus. Viruses. (2022) 14:2134. doi: 10.3390/V14102134
69. WHO. 2022 mpox (Monkeypox) outbreak: global trends(2023). Available online at: https://worldhealthorg.shinyapps.io/mpx_global/.
70. Sutter G, Wyatt LS, Foley PL, Bennink JR, Moss B. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine. (1994) 12:1032–40. doi: 10.1016/0264-410X(94)90341-7
71. Okeke MI, Nilssen I, Moens U, Tryland M, Traavik T. In vitro host range, multiplication and virion forms of recombinant viruses obtained from co-infection in vitro with a vaccinia-vectored influenza vaccine and a naturally occurring cowpox virus isolate. Virol J. (2009) 6:1–13. doi: 10.1186/1743-422X-6-55
72. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:477. doi: 10.1089/CMB.2012.0021
73. Tcherepanov V, Ehlers A, Upton C. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics. (2006) 7:150. doi: 10.1186/1471-2164-7-150
74. Ehlers A, Osborne J, Slack S, Roper RL, Upton C. Poxvirus orthologous clusters (POCs). Bioinformatics. (2002) 18:1544–5. doi: 10.1093/BIOINFORMATICS/18.11.1544
75. Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. (2015) 1:1–5. doi: 10.1093/VE/VEV003
76. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. (1999) 73:160. doi: 10.1128/jvi.73.1.152-160.1999
77. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/MOLBEV/MST010
78. Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. (2000) 16:562–3. doi: 10.1093/BIOINFORMATICS/16.6.562
79. Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. (1999) 265:218–25. doi: 10.1006/VIRO.1999.0056
80. Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. (2005) 21:98–102. doi: 10.1089/AID.2005.21.98
81. Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. (1992) 34:126–9. doi: 10.1007/BF00182389
82. Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc Natl Acad Sci. (2001) 98:13757–62. doi: 10.1073/PNAS.241370698
83. Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. (2000) 16:573–82. doi: 10.1093/BIOINFORMATICS/16.7.573
84. Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. (2007) 176:1035–47. doi: 10.1534/GENETICS.106.068874
85. Sutter G, Moss B. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc Natl Acad Sci U.S.A. (1992) 89:10847–51. doi: 10.1073/pnas.89.22.10847
86. Evans DH, Stuart D, McFadden G. High levels of genetic recombination among cotransfected plasmid DNAs in poxvirus-infected mammalian cells. J Virol. (1988) 62:367–75. doi: 10.1128/jvi.62.2.367-375.1988
87. Willer DO, Mann MJ, Zhang W, Evans DH. Vaccinia virus DNA polymerase promotes DNA pairing and strand-transfer reactions. Virology. (1999) 257:511–23. doi: 10.1006/viro.1999.9705
88. Tolonen N, Doglio L, Schleich S, Krijnse Locker J. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol Biol Cell. (2001) 12:2031–46. doi: 10.1091/mbc.12.7.2031
89. Christen L, Seto J, Niles EG. Superinfection exclusion of vaccinia virus in virus-infected cell cultures. Virology. (1990) 174:35–42. doi: 10.1016/0042-6822(90)90051-R
90. Dobson BM, Procter DJ, Hollett NA, Flesch IEA, Newsome TP, Tscharke DC. Vaccinia virus F5 is required for normal plaque morphology in multiple cell lines but not replication in culture or virulence in mice. Virology. (2014) 456–7:145–56. doi: 10.1016/j.virol.2014.03.020
91. Dobson BM, Tscharke DC. Truncation of gene F5L partially masks rescue of vaccinia virus strain MVA growth on mammalian cells by restricting plaque size. J Gen Virol. (2014) 95:466–71. doi: 10.1099/vir.0.058495-0
92. Morales I, Carbajal MA, Bohn S, Holzer D, Kato SEM, Greco FAB, et al. The vaccinia virus F11L gene product facilitates cell detachment and promotes migration. Traffic. (2008) 9:1283–98. doi: 10.1111/j.1600-0854.2008.00762.x
93. Blasco R, Sisler JR, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. (1993) 67:3325–25. doi: 10.1128/jvi.67.6.3319-3325.1993
94. Roper RL, Wolffe EJ, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. (1998) 72:4192–204. doi: 10.1128/jvi.72.5.4192-4204.1998
95. Zhang W-H, Wilcock D, Smith GL. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J Virol. (2000) 74:11654–62. doi: 10.1128/jvi.74.24.11654-11662.2000
96. Parkinson JE, Smith GL. Vaccinia virus gene A36R encodes a M(r) 43-50 K protein on the surface of extracellular enveloped virus. Virology. (1994) 204:376–90. doi: 10.1006/viro.1994.1542
97. Katz E, Wolffe E, Moss B. Identification of second-site mutations that enhance release and spread of vaccinia virus. J Virol. (2002) 76:11637–44. doi: 10.1128/jvi.76.22.11637-11644.2002
98. Zhou J, Sun XY, Fernando GJP, Frazer IH. The vaccinia virus K2L gene encodes a serine protease inhibitor which inhibits cell-cell fusion. Virology. (1992) 189:678–86. doi: 10.1016/0042-6822(92)90591-C
99. Law KM, Smith GL. A vaccinia serine protease inhibitor which prevents virus-induced cell fusion. J Gen Virol. (1992) 73:678–86. doi: 10.1099/0022-1317-73-3-549
100. Turner PC, Moyer RW. An orthopoxvirus serpinlike gene controls the ability of infected cells to fuse. J Virol. (1992) 66:2076–85. doi: 10.1128/jvi.66.4.2076-2085.1992
101. Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, Rambaut A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol. (2010) 27:2038–51. doi: 10.1093/MOLBEV/MSQ088
102. de Haven BC, Gupta K, Isaacs SN. The vaccinia virus A56 protein: A multifunctional transmembrane glycoprotein that anchors two secreted viral proteins. J Gen Virol. (2011) 92:1971–80. doi: 10.1099/vir.0.030460-0
103. Bratke KA, McLysaght A, Rothenburg S. A survey of host range genes in poxvirus genomes. Infect Genet Evol. (2013) 14:406–25. doi: 10.1016/j.meegid.2012.12.002
104. Panus JF, Smith CA, Ray CA, Smith TD, Patel DD, Pickup DJ. Cowpox virus encodes a fifth member of the tumor necrosis factor receptor family: A soluble, secreted CD30 homologue. Proc Natl Acad Sci U.S.A. (2002) 99:8348–53. doi: 10.1073/pnas.122238599
105. Abrahão JS, Campos RK, De Souza Trindade G, Da Fonseca FG, Ferreira PCP, Kroon EG. Outbreak of severe zoonotic vaccinia virus infection, southeastern Brazil. Emerg Infect Dis. (2015) 21:695. doi: 10.3201/EID2104.140351
106. Kalthan E, Tenguere J, Ndjapou SG, Koyazengbe TA, Mbomba J, Marada RM, et al. Investigation of an outbreak of monkeypox in an area occupied by armed groups, Central African Republic. Médecine Mal Infect. (2018) 48:263–8. doi: 10.1016/J.MEDMAL.2018.02.010
107. Alakunle E, Moens U, Nchinda G, Okeke MI. Monkeypox virus in Nigeria: infection biology, epidemiology, and evolution. Viruses. (2020) 12:1–29. doi: 10.3390/V12111257
108. Cardeti G, Gruber CEM, Eleni C, Carletti F, Castilletti C, Manna G, et al. Fatal outbreak in tonkean macaques caused by possibly novel orthopoxvirus, Italy, january 2015. Emerg Infect Dis. (2017) 23:1941–9. doi: 10.3201/EID2312.162098
109. Springer YP, Hsu CH, Werle ZR, Olson LE, Cooper MP, Castrodale LJ, et al. Novel orthopoxvirus infection in an alaska resident. Clin Infect Dis Off Publ Infect Dis Soc Am. (2017) 64:1737. doi: 10.1093/CID/CIX219
110. Alakunle E, Okeke M. Monkeypox virus: a neglected zoonotic pathogen spreads globally. Nat Rev Microbiol. (2022) 20:507–8. doi: 10.1038/s41579-022-00776-z
111. Naoki O, Arihiro K, Toshiyuki Y, Noriko H, Fumio K, Suyoshi S, et al. The genome landscape of the African Green Monkey kidney-derived vero cell line. DNA Res. (2014) 21:673–83. doi: 10.1093/dnares/dsu029
Keywords: orthopoxvirus, poxvirus, smallpox, superinfection exclusion, recombination
Citation: Diaz-Cánova D, Moens U, Brinkmann A, Nitsche A and Okeke MI (2024) Whole genome sequencing of recombinant viruses obtained from co-infection and superinfection of Vero cells with modified vaccinia virus ankara vectored influenza vaccine and a naturally occurring cowpox virus. Front. Immunol. 15:1277447. doi: 10.3389/fimmu.2024.1277447
Received: 14 August 2023; Accepted: 19 March 2024;
Published: 03 April 2024.
Edited by:
Markus W. Löffler, University of Tübingen, GermanyReviewed by:
David Hugh Evans, University of Alberta, CanadaCopyright © 2024 Diaz-Cánova, Moens, Brinkmann, Nitsche and Okeke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ugo Moens, dWdvLm1vZW5zQHVpdC5ubw==; Malachy Ifeanyi Okeke, bWFsYWNoeS5va2VrZUBhdW4uZWR1Lm5n
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.