- 1Institute of Hematology, School of Medicine, Jinan University, Guangzhou, China
- 2Key Laboratory for Regenerative Medicine of Ministry of Education, Jinan University, Guangzhou, China
- 3Department of Pathology, School of Medicine, Jinan University, Guangzhou, China
- 4Key Laboratory of Viral Pathogenesis and Infection Prevention and Control, Jinan University, Guangzhou, China
Acute myeloid leukemia (AML) is a malignant clonal tumor originating from immature myeloid hematopoietic cells in the bone marrow with rapid progression and poor prognosis. Therefore, an in-depth exploration of the pathogenesis of AML can provide new ideas for the treatment of AML. In recent years, it has been found that exosomes play an important role in the pathogenesis of AML. Exosomes are membrane-bound extracellular vesicles (EVs) that transfer signaling molecules and have attracted a large amount of attention, which are key mediators of intercellular communication. Extracellular vesicles not only affect AML cells and normal hematopoietic cells but also have an impact on the bone marrow microenvironment and immune escape, thereby promoting the progression of AML and leading to refractory relapse. It is worth noting that exosomes and the various molecules they contain are expected to become the new markers for disease monitoring and prognosis of AML, and may also function as drug carriers and vaccines to enhance the treatment of leukemia. In this review, we mainly summarize to reveal the role of exosomes in AML pathogenesis, which helps us elucidate the application of exosomes in AML diagnosis and treatment.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematological malignancy that affects adults. It is characterized by the abnormal proliferation and differentiation of immature myeloblasts, which accumulate in the bone marrow (BM) and peripheral blood and impair normal hematopoiesis. Although 50% of patients with AML can achieve complete remission after induction chemotherapy and post-remission treatment, more than 20% of patients with AML remain unresponsive and refractory (1). The precise treatment of AML is impeded by the disease’s aggressive and heterogeneous nature, which is characterized by genetic abnormalities, extensive epigenetic changes, and abnormal tumor microenvironment (TME) (2, 3). Therefore, understanding of the pathogenesis of AML must be improved, and novel biomarkers for diagnosis for its diagnosis and prognosis must be developed.
Exosomes, also known as intraluminal vesicles, are 30-100nm extracellular vesicles (EVs) secreted by various cells, including tumor cells, into the body fluids, blood, urine, semen, saliva, breast milk, amniotic fluid, ascitic fluid, cerebrospinal fluid, and bile (4). The circulatory system serves as the primary medium for exosomes to perform their long-distance communication function (5, 6). Tumor-derived exosomes (TEXs) induce vascular leakage, inflammation, and BM progenitor recruitment during pre-metastatic niche formation and metastasis (7), and finally induce tumor growth and metastasis, affecting tumor progression and prognosis (8).
TEXs in TME transport a substantial amount of genetic material from maternal tumor cells (9). By regulating the physiology of recipient cells, including signaling to tumor and stromal cells, exosomes secreted into the extracellular environment can reshape the TME and promote tumor growth. Exosomes are crucial components in tumorigenesis and tumor proliferation, angiogenesis, invasion, and metastasis (10). This intercellular communication influences cells of various lineages remotely or in situ. Exosomal communication involving immune cells can induce intricate cellular modifications and considerably influence the course of cancer progression by eliciting an immune response. Recent evidence suggests that exosomes greatly influence cell-to-cell and cell-to-environment communication in AML (11). Exosomes are essential for the progression of leukemia and facilitate the survival and chemoresistance of leukemic cells by transferring their molecular cargo (12, 13). Therefore, TEXs are essential to the evaluation of the effects of AML disease activity, severity, and treatment response. In this review, we briefly describe the production of exosomes and how vesicles mediate cellular communication, and then explore the potential use of exosomes in AML diagnosis and treatment.
Origin and mechanisms of exosomes
Approximately 50 years ago, scientists observed that cells in culture fluid “shed” small vesicles of unknown function -called exosomes. Previously, “waste” produced by cellular physiological metabolism. Owing to the development of high-throughput proteomics and genomics, exosomes have been demonstrated to be involved in intercellular communication in living organisms.
Exosome biogenesis
Exosome biogenesis is a multistep process involving several pathways. First, multivesicular bodies (MVBs) are generated through two stages of inward membrane budding. The invagination of the cell membrane generates early endosomes, from which exosomes bud inward and late endosomes or MVBs are formed (14). Then, MVBs fuse with the plasma membrane through exocytosis and release exosomes from the cells in tubular vesicles (15). The mechanisms underlying MVB formation including endosomal sorting complex required for transport (ESCRT) pathway, and tetraspanin-dependent pathway (16, 17). ESCRT consists of four subunits (ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III) and related molecules (VPS4, VTA1, and ALIX). ESCRT-0 complex initiates the ESCRT pathway through its subunit hepatocyte growth factor-regulated tyrosine kinase substrate, which not only recognizes ubiquitinated proteins and binds to phosphoinositide in the endosomal membrane but also recruits ESCRT-I by binding its TSG101 subunit (18–22). Then, ESCRT-I, and -II promote membrane endosome invagination, ESCRT-III and VPS4 drive the abscission of vesicles from the membrane, and exosomes are generated (23–29). However, the ESCRT system is not the sole pathway for regulating exosome formation. Several tetraspanins, such as CD63, CD81, and CD9, can sequester multiple proteins and form tetraspanin-enriched exosomes (30, 31). Apart from these pathways, other regulators have been identified, including syntenin, ceramide activation via neutral sphingomyelinase, and lipid-raft formation (32–34).
Secretion of exosome
The mechanism by which MVBs are delivered to the plasma membrane is still not fully understood. Nevertheless, research has demonstrated that the process is controlled by small GTPase molecules that interact with cytoskeletal proteins, and cortactin and ALIX play roles in the intracellular distribution of MVBs (35–37). MVBs fuse with the plasma membrane through a series of proteins, including the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE). Vesicle-associated membrane proteins bind to the plasma membrane proteins syntaxin and SNAP and then trans-SNARE complexes are formed, which provide the necessary force for the movement of MVBs toward the plasma membrane (38). Finally, other SNARE proteins promote the fusion of MVBs and exosome (38–40).
Exosome uptake
After secretion, exosomes with cargoes are released into the extracellular environment. The membranes of exosomes can protect biomacromolecules that exist stably in the body fluid. Therefore, peripheral blood is the main environment in which exosomes perform long-distance communication functions (5, 6). In vivo, exosomes can be assimilated by target cells through the direct fusion of membranes, ligand-receptor interactions, or endocytosis. First, exosomes can directly activate receptors on the surfaces of target cells through protein molecules on the surfaces or lipid ligands, generating signaling complexes and activating intracellular signaling pathways (41). Second, in the extracellular matrix, exosomes release intracellular substances that act as ligands to bind to receptors on the cell membrane, thereby activating intracellular signaling pathways (42). Third, exosomes encode essential integrin molecules and fuse with the plasma membranes of target cells or are endocytosed directly into the cells and release nonspecific proteins, noncoding RNA, and nucleic acids (43). Biomolecules transferred by exosomes can alter the phenotypes and functions of recipient cells by altering gene expression and are involved in many physiological and pathological processes in recipient and donor cells (44, 45). Figure 1 shows the relevant mechanism.
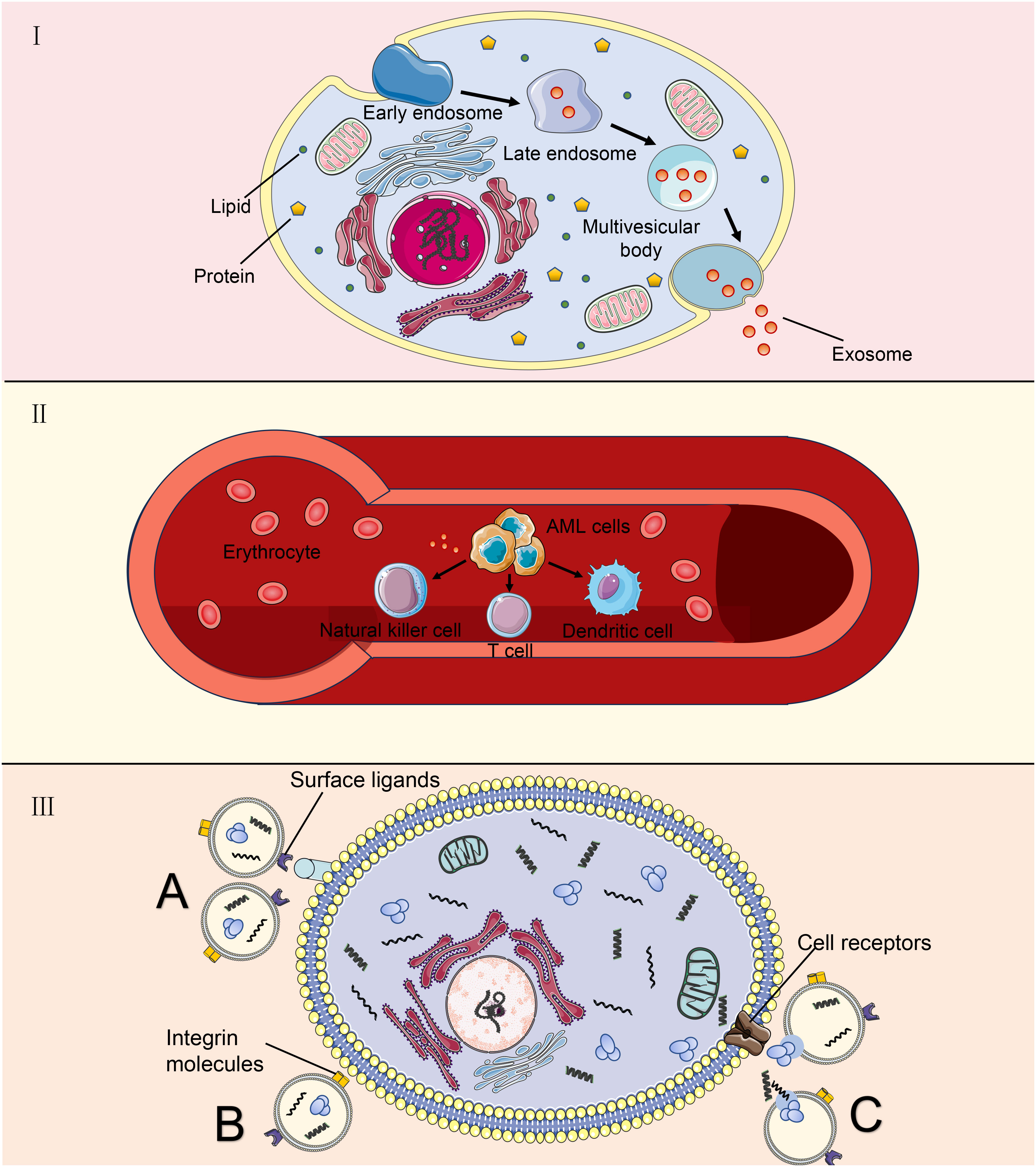
Figure 1 Exosomes derived from AML cells are used as target cells. (A) AML cells undergo a process from tubular vesicles (early intracellular bodies) to late intracellular bodies to multivesicular bodies, and finally, they release exosomes into the extracellular space through fusion with the plasma membrane. (B) AML-derived exosomes play a long-distance communication role mainly through peripheral blood, which can affect some immune cells; (C) Exosomes communicate with target cells. Exosomes can directly activate receptors on the surface of target cells through surface ligands. Integrin molecules on the membrane of exosomal cells directly fuse with the plasma membrane of target cells or endocytosis enters the cell. Outside the cell, exosomes release intracellular substances that bind to receptors on the cell membrane.
Content of exosome
Recent data obtained from the Exosome Database reveal that exosomes comprise 1116 lipids, 9769 proteins, 3408 mRNAs, and 2838 microRNAs (miRNAs). The lipid content of exosomes consists of cholesterol, sphingomyelin, ceramide, phosphatidylserine, lysophosphatidic acid, and prostaglandins, which are important for the mechanistic and biophysical aspects of bilayer formation, curvature, and fluidity and affect membrane fusion (46). Proteins in exosomes include tetraspanins, which are membrane transport and fusion proteins on the surfaces of exosomes and act as specific markers. They include specific proteins that are excellent markers for exosome recognition, heat shock proteins (HSP-60, HSP-70, and HSP-90), chaperone proteins, adhesion proteins, MHC (e.g., MHC I and MHC II, which are evolved in antigen presentation), cytoskeletal proteins, multivesicular body synthesis proteins, and lipid-associated proteins (47). In addition, AML-derived exosomes contain the tumor antigens CD33, CD34, and CD117 (48). Exosomes express the adhesion molecules ICAM-1 and integrins, which mediate the interaction and binding of exosomal membranes to receptor cells for cargo delivery (49). Nucleic acids, including DNA, mRNA, and noncoding RNA, are associated with the detection of cancer-associated mutations in serum exosomes (50). Thus, exosome-specific nucleic acids and proteins are crucial for identifying biomarkers of serum exosomes associated with tumor gene mutations and predicting tumor development and prognosis (51).
AML-derived exosomes cause the dysfunction of immune cells
Exosomes originating from AML induce T-cell differentiation towards a pro-tumor phenotype
The efficacy of tumor immunotherapy is restricted by tumor cells evading host immune system surveillance and downregulating the function of immune cells, especially antitumor effector cells, including CD8+ T and CD4+ T cells, natural killer cells (NK), and dendritic cells (DCs) (52, 53). Immune cell dysfunction is a common feature of AML. AML-derived exosomes are key mediators in the TME and function as immunosuppressants, enabling AML cells to evade immune surveillance (12). Exosomes isolated from the plasma of AML patients are loaded with leukemia-associated antigens and inhibitory molecules, which can disrupt the functions of immune cells used in adoptive cell therapy, thereby limiting the expected therapeutic effect of adoptive cell therapy and resulting in immune dysfunction (54). Human TEXs induce apoptosis in activated CD8+ T cells, promote the expansion and function of regulatory T (Treg) cells, and thus promote tumor evasion. The proliferation of activated CD8+ T cells is inhibited by co-cultivation with TEX, but TEXs increase the proportion of activated CD4+ T cells. Additionally, TEXs promote Treg cell expansion and transport transforming growth factor β (TGFβ) and IL-10, which promote the conversion of T cells into Treg cells (55). Treg cells constitute a subpopulation of T cells, mainly CD4+CD25+ cells, and are classified according to their origin. Elevated levels of Treg cells in peripheral blood are associated with poor outcomes in patients with AML (56). Pando et al. investigated the effects of AML-derived EVs on T cell subsets by an in vitro approach to study the effects of EVs derived from the human AML cell line MOLM-14 cells on CD4+, CD4+CD39+, and CD8+ T cell subsets from healthy individuals; the results showed that tumor-derived EVs modulate T cell responses by upregulating immune processes, such as immunosuppression and oncogenic gene expression (57).
AML-derived exosomes downregulate the natural killer receptor of NK cells
NK cells are major innate immune cells in the bloodstream and target tumor cells. In AML, the ability of NK cells to eliminate leukemic cells is dependent on the predominance of activation signals. Weak activation signals among NK cells lead to their inability to exert their cytotoxicity and render them unresponsive to leukemic cells (58). The low expression of the activation receptor natural killer group 2D (NKG2D) of NK cells in patients with AML results in a decline in NK cell activity and the inhibition of its killing function (59). Szczepanski et al. examined serum exosomes from 19 patients with AML and 14 healthy controls and found that serum exosomes from patients with AML disrupted NK cell activation by downregulating the expression of NKG2D; this effect reduced the toxicity of NK cells to tumor cells, but interleukin 15 counteracted this inhibitory effect (60). AML-derived exosomes reduced the cytolytic activity of normal NK cells by downregulating NKG2D receptor expression and inducing Smad phosphorylation in NK cells (61). Hong et al. isolated exosomes from the plasma of the AML human-derived tissue xenograft model they developed; they observed that the expression levels of surface markers in the exosomes were similar to those in the exosomes from patients with AML. The AML-derived exosomes that carried immunosuppressive ligands activated on human NK cell or CD8+ T cell receptors, leading to their dysfunction (62).
AML-derived exosomes inhibit the direct and indirect anti-tumor effects of DCs
DCs are major antigen-presenting cells and play an important role in innate immunity. However, DCs generated in the presence of TEX under express costimulatory molecules and produce suppressive cytokines, thus inducing the dose-dependent suppression of T cell proliferation and antitumor cytotoxicity (63). In the context of AML, type I interferons produced by plasmacytoid DCs can clear AML cells. This finding suggests that DCs eliminate AML cells. Benites et al. used exosomes or lysates derived from the leukemia K562 cell line as antigen sources of DC pulses, which initiated the maturation of DCs into a cytotoxic phenotype and markedly enhanced the cleavage of target cells; conversely, when the serum exosomes of patients with AML were used as the pulse sources, opposite effects were observed, which may have induced the immune tolerance of DCs. Considering these contrasting effects can contribute to the mitigation of in vivo immune tumor evasion mechanisms (64). In summary, AML-derived exosomes transport substances that induce dysfunction in immune cells and exert a suppressive effect on the immune system (Figure 2).
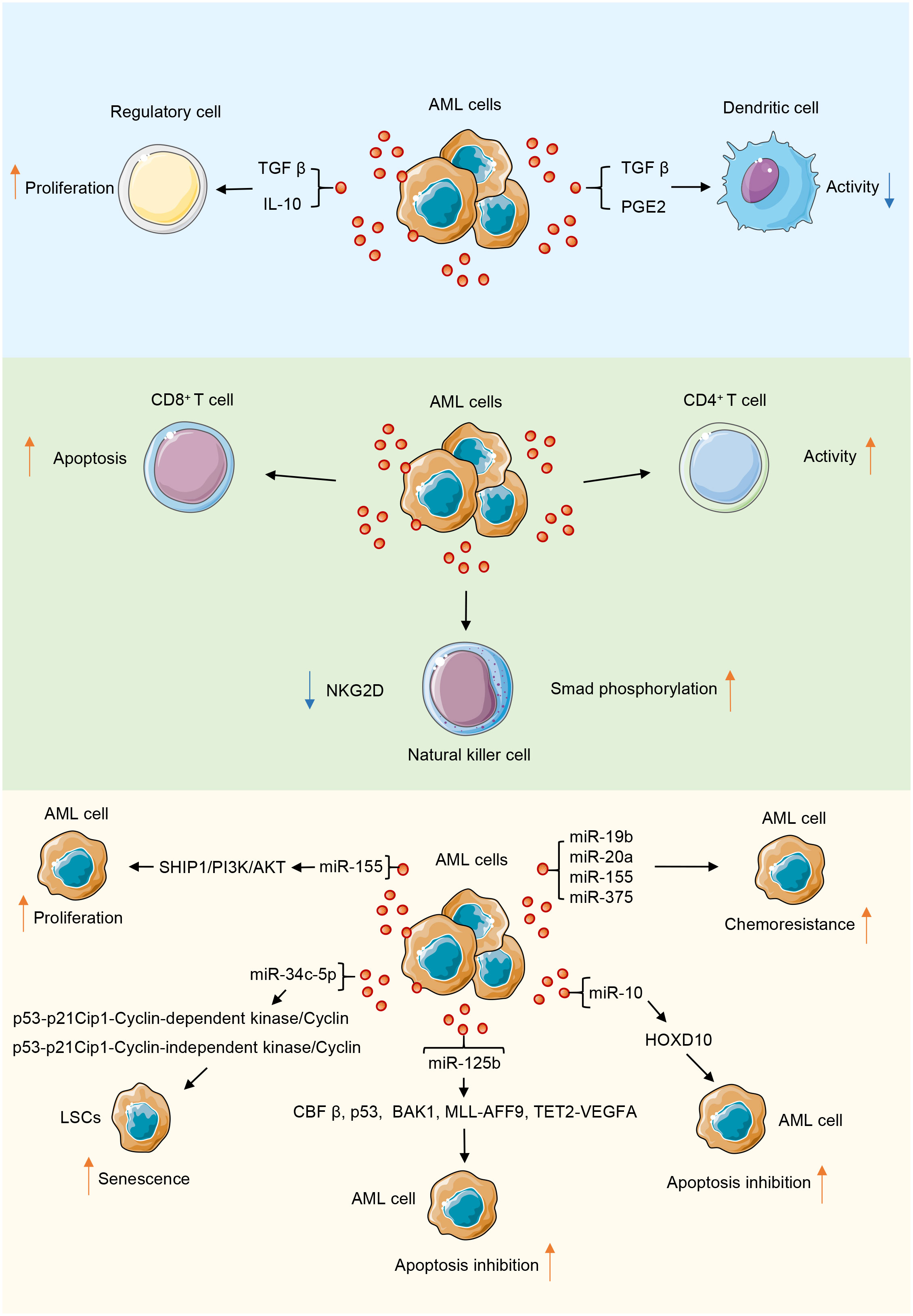
Figure 2 AML-derived exosomes play a role in promoting or inhibiting tumor progression through their contents in the tumor microenvironment. During the interaction of AML-derived exosomes with immune cells, their contents mainly inhibit the function of immune cells.
AML-derived exosomes related to AML progress
Exosomes participate in BM microenvironment reconstitution
The leukemic microenvironment is a complex and heterogeneous ecological niche composed of various cells, including leukemic, immune, mesenchymal stem, and endothelial cells. The interaction of tumor cells with the microenvironment and tumor stem cells in the BM promotes the relapse of leukemia and metastasis to lymphoid tissues (65). Exosomes are important for the induction of immune responses in a pro-tumor microenvironment and for tumor progression and survival. They promote tumor survival by remodeling the extracellular matrix and inducing angiogenesis and tumor cell proliferation (66). AML can reconstitute the BM microenvironment to one that promotes the growth of leukemic cells but inhibits normal hematopoietic function by secreting exosomes. Exosomes released by AML cells upregulate DKK1 in BM mesenchymal stromal cells and thereby inhibit normal hematopoiesis through the WNT signaling pathway, and AML-derived exosomes stimulate vascular endothelial growth factor (VEGF) signaling in human umbilical vein endothelial cells (HUVECs) by transferring angiogenic factors or proteins and miRNAs, which form vascular tubular structures that promote tumor growth (4, 67). Some studies have confirmed that exosomal miRNAs secreted by AML cells contribute to the progression of AML by altering the expression of downstream genes (68). Point mutation inactivation and reduced SHIP1 gene activity have been observed in patients with AML, and the miR-155-mediated suppression of SHIP1 expression is involved in the pathogenesis of AML. The miR-155/SHIP1/PI3K/AKT signaling pathway potentially has a tumor-suppressive function in the pathogenesis of AML (69). miR-155 is upregulated in FLT3-ITD-associated AML and targets the myeloid transcription factor PU.1. The knockdown of miR-155 inhibits proliferation of FLT3-ITD-associated leukemic cells and induces their apoptosis (70). miR-34c-5p is a core miRNA in pathways regulating aging. It is expressed through the p53-p21Cip1-cyclin-dependent kinase (CDK)/cyclin or the p53-independent CDK/cyclin pathway (p53-p21Cip1-CDK/cyclin or p53-independent CDK/cyclin pathway (p53-p21Cip1-CDK/cyclin or p53-independent CDK/cyclin pathways) and promotes leukemia stem cells senescence. However, miR-34c-5p is downregulated in AML (excluding APL) stem cells; poor prognosis and poor therapeutic effect are clinical manifestations of this outcome (71).
The role of exosomes in the apoptosis of AML
Apoptosis is one of the key mechanisms affecting the survival of AML cells, and the dysregulation of apoptosis may lead to the chemoresistance of AML cells and disease relapse (72). Exosomes carry many complex cargoes, which can serve as the key mediators of intercellular communication and regulate cell proliferation (73). Exosomal miRNAs enter body fluids through autocrine secretion and create a microenvironment in malignant regulatory pathways that facilitate the growth of AML cells by cross talk with other cells, thereby promoting leukemic cell survival, proliferation, and migratory infiltration (74). AML cells highly resistant to apoptosis can affect the expression of apoptosis-related proteins in chemo-sensitive cells. Jiang et al. showed that exosomes secreted by AML cells are enriched in miR-125b (75). The mechanisms by which miR-125b affects apoptosis in AML cells are as follows: First, miR-125b partly targets core binding factor β (CBFβ) and blocks apoptosis by downregulating multiple genes involved in the p53 pathway (76). Second, it inhibits apoptosis and promotes cell proliferation by affecting brassinosteroid-insensitive 1-associated receptor kinase 1 (BAK1) expression (77). Third, miR-125b facilitates the progression of leukemia by promoting the expression of oncogenic MLL-AFF9 in vivo, and it upregulates VEGFA, providing conditions conducive to the expansion of leukemic cells. This process involves carcinogenic miRNAs mediating noncellular endogenous leukemia and promoting the miR-125b-TET2-VEGFA pathway. Fourth, caudal-related homeobox transcription factor 2 (CDX2) binds to the promoter region of the miR-125b gene and activates the expression of miR-125b in malignant myeloid cells, and the generated miR-125b inhibits the translation of CBFβ, thereby inhibiting the differentiation of myeloid cells in granulocyte lineaments and promoting the occurrence of leukemia (78). Exosomes in the sera of patients with AML are enriched in miR-10, and miR-10b can inhibit apoptosis and homeobox D10 expression in AML cells by directly targeting homeobox D10 (79).
Exosomes are involved in the development of drug resistance in AML
Co-culturing of exosomes with multi-drug-resistant AML cell line with chemo-sensitive HL-60 may cause chemo-resistance because of the transfer of miR-19b and miR-20a to the exosomes; thus, exosomes can make chemo-sensitive cells resistant to chemotherapy (80). Chen et al. demonstrated that exosomes secreted by the AML cell KG1α can drive BM stromal cells to produce IL-8, which can inhibit the chemotherapy-induced apoptosis of AML cells (81). Moreover, an exosome-mediated communication mechanism may impede drug therapy. Hekmatirad et al. found that U937 cells (an AML cell line) increase their resistance to the cytotoxic effects of doxorubicin (PLD) in pegylated liposomes through exosome-mediated drug expulsion (82). Another study that investigated the chemoresistance of AML-BMSC exosomes showed that miR-155 and miR-375 in exosomes derived from AML cells are responsible for chemoresistance to chemotherapeutic drugs cytarabine and AC220; the possible mechanism is the miRNA-induced downregulation of the promoters of apoptosis or cell differentiation under the guidance; free leukemic cells become independent of the kinase pathway through this mechanism (83).
Application of exosomes in the diagnosis and prognosis of AML
Exosomes as biomarkers of tumors have attracted considerable interest (84). They are present in various body fluids and easy to isolate and can be extracted from a small amount of serum (85). Moreover, they have a unique molecular profile (61). AML-secreted exosomal miRNAs are involved in the progression of AML and can be used as entry points for AML treatment (86, 87).
AML might reflect unique miRNA profiles. Compared with the sera of healthy individuals, the sera of patients with tumors contain a large number of exosomes and specific pathogenic information molecules of parental cell origin, which represent the biological behavior of parental cells (88). For example, miRNAs, are important cargoes carried by exosomes because they act in tumor tissues through targeted molecules. miRNAs are important biomarkers of tumor development and prognosis and are protected by exosomal surface membranes with highly conserved sequences. These membranes are stable under extreme conditions and can prevent miRNAs from being released into the circulation (89). miRNAs have potential use in the diagnosis of multiple diseases (90). Exosomal miRNA can be collected from 20 μL of serum and can be used as an ideal molecular marker for the targeted diagnosis and prognosis of leukemia (91). Serum exosomal miR-10b is an independent prognostic factor for overall survival in AML patients. miR-10b expression levels are elevated in the sera of patients with AML, and its expression levels are strongly correlated with poor prognosis, and miR-10b level considerably increases in patients with AML (92–94). Therefore, serum exosomal miR-10b is a potential diagnostic and prognostic marker for AML. The expression levels of miR-146a/b, miR-181a/b/d, miR-130a, miR-663, and miR-135b were high in M1, whereas those of miR-21, miR-193a, and miR-370 were high in M5 (95). In addition, miR-155 is downregulated in peripheral blood mononuclear cells from patients with multi-drug-resistant AML and adriamycin-resistant AML cell lines and showed a positive correlation in these patients. miR-155 can be used as a monitoring indicator for drug resistance and micro-residual focus with high sensitivity (96).
As mentioned above, numerous biomarkers can demonstrate powerful uniqueness in the diagnostic prediction of AML (Table 1). AML-derived exosomes are rich in CD33, CD34, and CD117, and their overall protein content is significantly higher than that of healthy controls; the content of some proteins, such as TGF-β1, decreases at the initial diagnosis and effective treatment of AML and can thus be used for detecting leukemia relapse and drug resistance status (97). Plasma exosomal lncRNAs are potential cell-free indicators for the diagnosis and therapeutic monitoring of AML and offer novel and cutting-edge concepts for the liquid biopsy of hematologic cancers (98). Bernardi et al. first used the commercially available CE-IVD-based kits for exosome-enrichment methods to investigate leukemic sources and exosomal dsDNA target resequencing for adult AML marker detection; they performed next-generation sequencing analysis of exosome-derived dsDNA isolated from 14 adult patients with AML and identified the optimal amount of exosomal dsDNA as a potential AML biomarker for liquid biopsies; they found exosomal dsDNA can be developed as a tool that can facilitate the monitoring of AML progression and the early diagnosis of relapse after allogeneic hematopoietic stem cell transplantation (99). In summary, exosomes may offer a novel perspective on AML diagnosis and treatment response (Table 2).
Use of exosomes in the treatment of AML
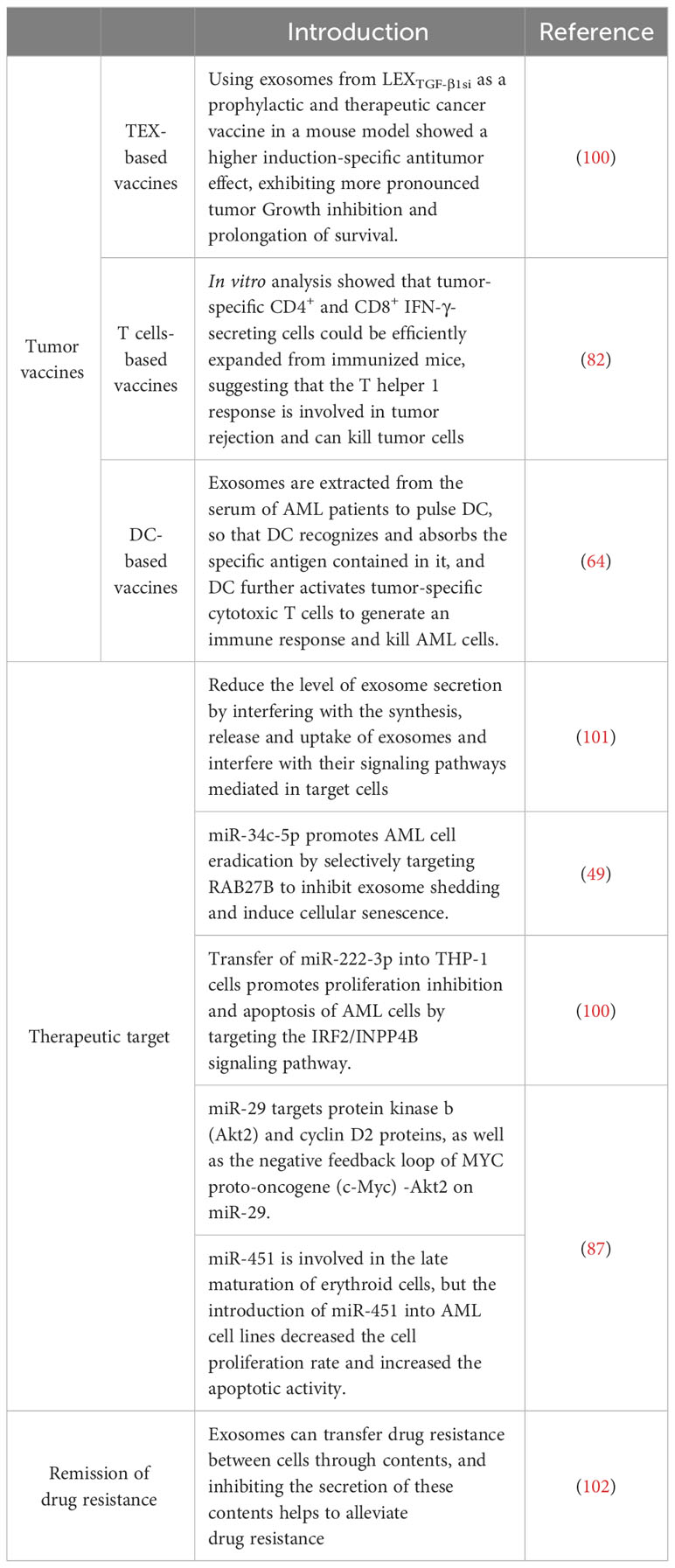
Currently, patients with refractory/recurrent AML experience an aggressive clinical course and have poor prognoses. Therefore, complementary alternative therapies are urgently needed to improve conventional treatments and increase survival rate (103). Blocking exosome-induced production, secretion, and reprogramming has emerged as a novel approach to treating AML and other types of leukemia. This exosome-targeted therapy may be financially beneficial for elderly patients with AML or patients with AML who cannot tolerate strongly induced chemotherapy (101, 104). Therefore, exosome-based immunotherapy has attracted considerable interest. In T-cell lymphoma mice, it effectively eliminated minimal residual disease and prolonged disease-free survival (100). TEX-carrying tumor-associated antigens are potential cell-free tumor treatments for the specific eradication of minimal residual leukemic cells (105). Huang et al. found that lentiviral shRNA– silenced TGF-β1 expression in parental cells of mouse acute lymphoblastic leukemia cell lines and exosomes from TGF-β1-silenced leukemia cells (LEXTGF-β1si) had a higher induction-specific antitumor effect than unmodified leukemia cell–derived exosomes. In mouse models, LEXTGF-β1si inhibited tumor growth and prolonged survival as a prophylactic and therapeutic cancer drug (102). This finding suggests that exosomal immune vaccines hold promise for the treatment of leukemia through immunotherapy.
As actionable drug-resistant mediators, exosomes may play a key role in current and future AML treatment. Hekmatirad et al. found that tumor cells can excrete drugs through exosomes and result in resistance and that inhibiting exosome release with GW4869 increases the sensitivity of U937 cells to PLD Therefore, the use of exosome inhibitors is a potential strategy for increasing the sensitivity of cancer cells to treatment. Inhibitors that pharmacologically inhibit exosome release (such as neticonazole, ketotifen, cannabidiol, and GW4869) are effective. Mortality and morbidity in AML are associated with frequent cytopenia, and exosomes may be involved in the suppression of normal hematopoiesis in leukemia (82). Namburi et al. found that exosomes isolated from the plasma of AML patients carry abundant dipeptidyl peptidase 4 (DPP4) and inhibit the differentiation of normal hematopoietic progenitor cells in vitro; pharmacologically inhibiting DPP4 reverses exosome-mediated colony formation; therefore, reversing the negative effects of DPP4 exosomes, improving platelet and neutrophil counts, and restoring BM function in patients are promising treatment approaches for AML; however, many regulatory proteins in exosomes contain DPP4 truncation sites and may have different induction, enhancement, or inhibition effects on hematopoiesis (106).
The systematic design of drug delivery vehicles can address many issues, such as low water solubility, poor biocompatibility, rapid metabolism in vivo, easy accumulation in nonpathological tissues, and poor ability to penetrate the membranes of some drugs (107). Exosomes as drug delivery vehicles are mainly dependent on their unique natural physicochemical properties, including phospholipid membrane structure that protects them from destruction by nucleases and proteases, high stability in blood, and long blood half-life (108). Nanoscale and lipid bilayer membranes prevents their removal by mononuclear phagocytes and reduce immunogenicity (especially of autologous cell origin), resulting in low cytotoxicity. The specific lipid and protein composition makes them highly stable in body fluids and enables them to readily fuse with target cells, rendering them chemotactic for specific target cells. Exosomes possess distinctive membrane structures that enables them to easily cross the biofilm barrier and act as carriers through specific delivery modes and modifications (109). These advantageous features of exosomes as drug carriers render them highly attractive for precision medicine. Bellavia et al. used HEK293T cells to express Lamp2b, an exosomal protein fused to an IL-3 fragment, and showed that IL3-Lamp2b exosomes loaded with imatinib targeted chronic myelogenous leukemic cells and inhibited cancer cell growth in vitro and in vivo (110). Kim et al. used exosomes to deliver paclitaxel or doxorubicin to mitigate multidrug resistance in lung cancer (111).
However, exosomes as drug carriers for AML treatment are currently underdeveloped.
Conclusion
During the progression of AML, exosomes secreted by AML cells can promote the development of AML by affecting the proliferation and apoptosis of AML cells, regulating the BM microenvironment, affecting angiogenesis, and inhibiting hematopoiesis. Therefore, according to the characteristics of AML cell-derived exosomes, exosomes can also be used as biomarkers of AML prognosis, preparing vaccines and drug carriers.
Author contributions
WDW: Writing – original draft. XFW: Writing – original draft. RY: Visualization, Writing – review & editing. JMZ: Validation, Writing – review & editing. YQL: Writing – review & editing. XLW: Writing – review & editing. LX: Writing – review & editing. ZYJ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Science and Technology Program of Guangzhou City (202201010164), the National Natural Science Foundation of China (81800143, 81770150, 81200388, and 82170220), the Natural Science Foundation of Guangdong Province (2020A1515010817, 2022A1515010313, and 2023A1515030271), the National Innovation and Entrepreneurship Training Program for Undergraduates (202310559054), Guangdong’s Innovation and Entrepreneurship Training Program for Undergraduates (202010559081 and 202210559083), and Guangdong College Students’ Scientific and Technological Innovation (CX23304).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol (2011) 29(5):487–94. doi: 10.1200/JCO.2010.30.1820
2. Cheng Y, Wang X, Qi P, Liu C, Wang S, Wan Q, et al. Tumor microenvironmental competitive endogenous RNA network and immune cells act as robust prognostic predictor of acute myeloid leukemia. Front Oncol (2021) 11:584884. doi: 10.3389/fonc.2021.584884
3. Tang BJ, Sun B, Chen L, Xiao J, Huang ST, Xu P. The landscape of exosome-derived non-coding RNA in leukemia. Front Pharmacol (2022) 13:912303. doi: 10.3389/fphar.2022.912303
4. Mirfakhraie R, Noorazar L, Mohammadian M, Hajifathali A, Gholizadeh M, Salimi M, et al. Treatment failure in acute myeloid leukemia: focus on the role of extracellular vesicles. Leukemia Res (2022) 112:106751. doi: 10.1016/j.leukres.2021.106751
5. Keller S, Ridinger J, Rupp A-K, Janssen JWG, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Trans Med (2011) 9:86. doi: 10.1186/1479-5876-9-86
6. Huang F, Wan J, Hu W, Hao S. Enhancement of anti-leukemia immunity by leukemia–derived exosomes via downregulation of TGF-β1 expression. Cell Physiol Biochem (2017) 44(1):240–54. doi: 10.1159/000484677
7. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Mark MT, et al. Tumour exosome integrins determine organotropic metastasis. Nat (2015) 527(7578):329–35. doi: 10.1038/nature15756
8. Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev (2013) 32(3-4):623–42. doi: 10.1007/s10555-013-9441-9
9. Meng W, Hao Y, He C, Li L, Zhu G. Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer (2019) 18(1):57. doi: 10.1186/s12943-019-0982-6
10. Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol (2018) 8:66. doi: 10.3389/fonc.2018.00066
11. Wolf-Dennen K, Kleinerman ES. Exosomes: dynamic mediators of extracellular communication in the tumor microenvironment. Adv Exp Med Biol (2020) 1258:189–97. doi: 10.1007/978-3-030-43085-6_13
12. Amin AH, Al Sharifi LM, Kakhharov AJ, Opulencia MJC, Alsaikhan F, Bokov DO, et al. Role of Acute Myeloid Leukemia (AML)-Derived exosomes in tumor progression and survival. Biomed Pharmacother (2022) 150:113009. doi: 10.1016/j.biopha.2022.113009
13. Kumar B, Garcia M, Murakami JL, Chen C-C. Exosome-mediated microenvironment dysregulation in leukemia. Biochim Et Biophys Acta-Molecular Cell Res (2016) 1863(3):464–70. doi: 10.1016/j.bbamcr.2015.09.017
14. Zuo Z, Hong CS, Muller L, Boyiadzis M, Whiteside TL. Isolation and characterization of CD34+ Blast-derived exosomes in acute myeloid leukemia. PloS One (2014) 9(8):e103310. doi: 10.1371/journal.pone.0103310
15. Stahl PD, Raposo G. Extracellular vesicles: exosomes and microvesicles, integrators of homeostasis. Physiol (2019) 34(3):169–77. doi: 10.1152/physiol.00045.2018
16. Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol (2008) 20(1):4–11. doi: 10.1016/j.ceb.2007.12.002
17. Rädler J, Gupta D, Zickler A, Andaloussi SE. Exploiting the biogenesis of extracellular vesicles for bioengineering and therapeutic cargo loading. Mol Ther J Am Soc Gene Ther (2023) 31(5):1231–50. doi: 10.1016/j.ymthe.2023.02.013
18. Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol (2002) 4(7):534–9. doi: 10.1038/ncb815
19. Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol (2002) 4(5):394–8. doi: 10.1038/ncb791
20. Wegner CS, Schink KO, Stenmark H, Brech A. Monitoring phosphatidylinositol 3-phosphate in multivesicular endosome biogenesis. Methods enzymol (2014) 534:3–23. doi: 10.1016/B978-0-12-397926-1.00001-9
21. Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol (2003) 162(3):435–42. doi: 10.1083/jcb.200302131
22. Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci United States America (2003) 100(13):7626–31. doi: 10.1073/pnas.0932599100
23. Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell (2006) 125(1):99–111. doi: 10.1016/j.cell.2006.01.047
24. Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Emr SD, et al. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J (2007) 26(2):600–12. doi: 10.1038/sj.emboj.7601501
25. Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell (2008) 15(4):578–89. doi: 10.1016/j.devcel.2008.08.013
26. Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nat (2009) 458(7235):172–7. doi: 10.1038/nature07836
27. Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, et al. Helical structures of ESCRT-III are disassembled by VPS4. Sci (New York NY) (2008) 321(5894):1354–7. doi: 10.1126/science.1161070
28. Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell (2009) 136(1):97–109. doi: 10.1016/j.cell.2008.11.013
29. Jackson CE, Scruggs BS, Schaffer JE, Hanson PI. Effects of inhibiting VPS4 support a general role for ESCRTs in extracellular vesicle biogenesis. Biophys J (2017) 113(6):1342–52. doi: 10.1016/j.bpj.2017.05.032
30. Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem (2013) 288(17):11649–61. doi: 10.1074/jbc.M112.445304
31. Jankovičová J, Sečová P, Michalková K, Antalíková J. Tetraspanins, more than markers of extracellular vesicles in reproduction. Int J Mol Sci (2020) 21(20):7568. doi: 10.3390/ijms21207568
32. Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res (2021) 31(2):157–77. doi: 10.1038/s41422-020-00409-1
33. Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol (2010) 190(6):1079–91. doi: 10.1083/jcb.201002049
34. Arya SB, Chen S, Jordan-Javed F, Parent CA. Ceramide-rich microdomains facilitate nuclear envelope budding for non-conventional exosome formation. Nat Cell Biol (2022) 24(7):1019–28. doi: 10.1038/s41556-022-00934-8
35. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun (2011) 2:282. doi: 10.1038/ncomms1285
36. Sinha S, Hoshino D, Hong NH, Kirkbride KC, Grega-Larson NE, Seiki M, et al. Cortactin promotes exosome secretion by controlling branched actin dynamics. J Cell Biol (2016) 214(2):197–213. doi: 10.1083/jcb.201601025
37. Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J Cell science (2005) 118(Pt 12):2625–35. doi: 10.1242/jcs.02382
38. Han J, Pluhackova K, Böckmann RA. The multifaceted role of SNARE proteins in membrane fusion. Front Physiol (2017) 8:5. doi: 10.3389/fphys.2017.00005
39. Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol (2012) 14(10):1036–45. doi: 10.1038/ncb2574
40. Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun (2017) 8:14041. doi: 10.1038/ncomms14041
41. Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Science (2011) 124(3):447–58. doi: 10.1242/jcs.074088
42. Feng D, Zhao W-L, Ye Y-Y, Bai X-C, Liu R-Q, Chang L-F, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic (2010) 11(5):675–87. doi: 10.1111/j.1600-0854.2010.01041.x
43. Morelli AE, Larregina AT, Shufesky WJ, Sullivan MLG, Stolz DB, Papworth GD, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood (2004) 104(10):3257–66. doi: 10.1182/blood-2004-03-0824
44. Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol (2015) 77(1):13–27. doi: 10.1146/annurev-physiol-021014-071641
45. Yáñez-Mó M, Siljander PRM, Andreu Z, Bedina Zavec A, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracellular Vesicles (2015) 4(1):27066. doi: 10.3402/jev.v4.27066
46. Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol (2017) 13(12):731–49. doi: 10.1038/nrneph.2017.148
47. Singh N, Huang L, Wang DB, Shao N, Zhang XE. Simultaneous detection of a cluster of differentiation markers on leukemia-derived exosomes by multiplex immuno-polymerase chain reaction via capillary electrophoresis analysis. Analytical Chem (2020) 92(15):10569–77. doi: 10.1021/acs.analchem.0c01464
48. Kalra H, Drummen G, Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci (2016) 17(2):170. doi: 10.3390/ijms17020170
49. Chaput N, Taieb J, Schartz NEC, Andre F, Angevin E, Zitvogel L. Exosome-based immunotherapy. Cancer Immunol Immunother (2004) 53(3):234–9. doi: 10.1007/s00262-003-0472-x
50. Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J Cell Physiol (2018) 233(9):6370–80. doi: 10.1002/jcp.26481
51. Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol (2020) 13(1):152. doi: 10.1186/s13045-020-00987-y
52. Whiteside T. Immune suppression in cancer: Effects on immune cells, mechanisms and future therapeutic intervention. Semin Cancer Biol (2006) 16(1):3–15. doi: 10.1016/j.semcancer.2005.07.008
53. Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J Immunol (2009) 183(6):3720–30. doi: 10.4049/jimmunol.0900970
54. Hong C-S, Sharma P, Yerneni SS, Simms P, Jackson EK, Whiteside TL, et al. Circulating exosomes carrying an immunosuppressive cargo interfere with cellular immunotherapy in acute myeloid leukemia. Sci Rep (2017) 7(1):14684. doi: 10.1038/s41598-017-14661-w
55. Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PloS One (2010) 5(7):e11469. doi: 10.1371/journal.pone.0011469
56. Zhang SH, Han YX, Wu JB, Yu K, Bi LX, Zhuang Y, et al. Elevated frequencies of CD4(+)CD25(+)CD127(lo) regulatory T cells is associated to poor prognosis in patients with acute myeloid leukemia. Int J Cancer (2011) 129(6):1373–81. doi: 10.1002/ijc.25791
57. Pando A, Schorl C, Fast LD, Reagan JL. Tumor derived extracellular vesicles modulate gene expression in T cells. Gene (2023) 850:146920. doi: 10.1016/j.gene.2022.146920
58. Lion E, Willemen Y, Berneman ZN, Van Tendeloo VFI, Smits ELJ. Natural killer cell immune escape in acute myeloid leukemia. Leukemia (2012) 26(9):2019–26. doi: 10.1038/leu.2012.87
59. Venton G, Labiad Y, Colle J, Fino A, Afridi S, Torres M, et al. Natural killer cells in acute myeloid leukemia patients: from phenotype to transcriptomic analysis. Immunologic Res (2016) 64(5-6):1225–36. doi: 10.1007/s12026-016-8848-0
60. Hong C-S, Muller L, Whiteside TL, Boyiadzis M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front Immunol (2014) 5:160. doi: 10.3389/fimmu.2014.00160
61. Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor- 1. Haematologica (2011) 96(9):1302–9. doi: 10.3324/haematol.2010.039743
62. Hong C-S, Danet-Desnoyers G, Shan X, Sharma P, Whiteside TL, Boyiadzis M. Human acute myeloid leukemia blast-derived exosomes in patient-derived xenograft mice mediate immune suppression. Exp Hematol (2019) 76:60–6. doi: 10.1016/j.exphem.2019.07.005
63. Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, et al. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res (2006) 66(18):9290–8. doi: 10.1158/0008-5472.CAN-06-1819
64. Benites BD, Duarte ADS, Longhini ALF, Santos I, Alvarez MC, Ribeiro LND, et al. Exosomes in the serum of Acute Myeloid Leukemia patients induce dendritic cell tolerance: Implications for immunotherapy. Vaccine (2019) 37(11):1377–83. doi: 10.1016/j.vaccine.2019.01.079
65. Deng W, Wang L, Pan M, Zheng J. The regulatory role of exosomes in leukemia and their clinical significance. J Int Med Res (2020) 48(8):300060520950135. doi: 10.1177/0300060520950135
66. Kumar B, Garcia M, Weng L, Jung X, Murakami JL, Hu X, et al. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia (2017) 32(3):575–87. doi: 10.1038/leu.2017.259
67. Miyamoto KN, Bonatto D. Circulating cells and exosomes in acute myelogenous leukemia and their role in disease progression and survival. Clin Immunol (2020) 217:108489. doi: 10.1016/j.clim.2020.108489
68. Pei Y, Zhang H, Lu K, Tang X, Li J, Zhang E, et al. Circular RNA circRNA_0067934 promotes glioma development by modulating the microRNA miR-7/ Wnt/β-catenin axis. Bioengineered (2022) 13(3):5792–802. doi: 10.1080/21655979.2022.2033382
69. Xue H, Hua LM, Guo M, Luo JM. SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol Rep (2014) 32(5):2253–9. doi: 10.3892/or.2014.3435
70. Gerloff D, Grundler R, Wurm AA, Bräuer-Hartmann D, Katzerke C, Hartmann JU, et al. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia (2014) 29(3):535–47. doi: 10.1038/leu.2014.231
71. Peng DY, Wang HF, Li L, Ma X, Chen Y, Zhou H, et al. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia (2018) 32(5):1180–8. doi: 10.1038/s41375-018-0015-2
72. Sharawat SK, Bakhshi R, Vishnubhatla S, Gupta R, Bakhshi S. BAX/BCL2RMFI ratio predicts better induction response in pediatric patients with acute myeloid leukemia. Pediatr Blood Cancer (2013) 60(8):E63–E6. doi: 10.1002/pbc.24518
73. Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci United States America (2008) 105(40):15535–40. doi: 10.1073/pnas.0808266105
74. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol (2009) 27(34):5848–56. doi: 10.1200/JCO.2009.24.0317
75. Jiang LJ, Deng TR, Wang D, Xiao Y. Elevated serum exosomal miR-125b level as a potential marker for poor prognosis in intermediate-risk acute myeloid leukemia. Acta Haematologica (2018) 140(3):183–92. doi: 10.1159/000491584
76. Bousquet M, Nguyen D, Chen C, Shields L, Lodish HF. MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica (2012) 97(11):1713–21. doi: 10.3324/haematol.2011.061515
77. Liu J, Guo B, Chen Z, Wang N, Iacovino M, Cheng J, et al. miR-125b promotes MLL-AF9–driven murine acute myeloid leukemia involving a VEGFA-mediated non–cell-intrinsic mechanism. Blood (2017) 129(11):1491–502. doi: 10.1182/blood-2016-06-721027
78. Lin K-Y, Zhang X-J, Feng D-D, Zhang H, Zeng C-W, Han B-W, et al. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic Malignancies. J Biol Chem (2011) 286(44):38253–63. doi: 10.1074/jbc.M111.269670
79. Wang CJ, Zou H, Feng GF. MiR-10b regulates the proliferation and apoptosis of pediatric acute myeloid leukemia through targeting HOXD10. Eur Rev Med Pharmacol Sci (2018) 22(21):7371–8. doi: 10.26355/eurrev_201811_16275
80. Bouvy C, Wannez A, Laloy J, Chatelain C, Dogné J-M. Transfer of multidrug resistance among acute myeloid leukemia cells via extracellular vesicles and their microRNA cargo. Leukemia Res (2017) 62:70–6. doi: 10.1016/j.leukres.2017.09.014
81. Chen T, Zhang G, Kong L, Xu S, Wang Y, Dong M. Leukemia-derived exosomes induced IL-8 production in bone marrow stromal cells to protect the leukemia cells against chemotherapy. Life Sci (2019) 221:187–95. doi: 10.1016/j.lfs.2019.02.003
82. Hekmatirad S, Moloudizargari M, Moghadamnia AA, Kazemi S, Mohammadnia-Afrouzi M, Baeeri M, et al. Inhibition of exosome release sensitizes U937 cells to PEGylated liposomal doxorubicin. Front Immunol (2021) 12:692654. doi: 10.3389/fimmu.2021.692654
83. Viola S, Traer E, Huan J, Hornick NI, Tyner JW, Agarwal A, et al. Alterations in acute myeloid leukaemia bone marrow stromal cell exosome content coincide with gains in tyrosine kinase inhibitor resistance. Br J Haematol (2016) 172(6):983–6. doi: 10.1111/bjh.13551
84. Makler A, Asghar W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert Rev Mol Diagnostics (2020) 20(4):387–400. doi: 10.1080/14737159.2020.1731308
85. Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep (2015) 5:11295. doi: 10.1038/srep11295
86. Fletcher D, Brown E, Javadala J, Uysal-Onganer P, Guinn B. microRNA expression in acute myeloid leukaemia: New targets for therapy? eJHaem (2022) 3(3):596–608. doi: 10.1002/jha2.441
87. Buhagiar A, Borg J, Ayers D. Overview of current microRNA biomarker signatures as potential diagnostic tools for leukaemic conditions. Non-Coding RNA Res (2020) 5(1):22–6. doi: 10.1016/j.ncrna.2020.02.001
88. Tickner JA, Richard DJ, O'Byrne KJEV. Microvesicles/microRNAs and stem cells in cancer. Adv Exp Med Biol (2018) 1056:123–35. doi: 10.1007/978-3-319-74470-4_8
89. He Y, Lin J, Kong D, Huang M, Xu C, Kim T-K, et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem (2015) 61(9):1138–55. doi: 10.1373/clinchem.2015.241190
90. Ni L, Tang C, Wang Y, Wan J, Charles MG, Zhang Z, et al. Construction of a miRNA-based nomogram model to predict the prognosis of endometrial cancer. J personalized Med (2022) 12(7):1154. doi: 10.3390/jpm12071154
91. Abdelhamed S, Butler JT, Jung S, Chen D-W, Jenkins G, Gao L, et al. Rational biomarker development for the early and minimally invasive monitoring of AML. Blood Advances (2021) 5(21):4515–20. doi: 10.1182/bloodadvances.2021004621
92. Fang ZG, Wang XZ, Wu JY, Xiao RZ, Liu JJ. High serum extracellular vesicle miR-10b expression predicts poor prognosis in patients with acute myeloid leukemia. Cancer Biomarkers (2020) 27(1):1–9. doi: 10.3233/CBM-190211
93. Zou Q, Tan S, Yang ZL, Wang J, Xian JR, Zhang SS, et al. The human nucleophosmin 1 mutation A inhibits myeloid differentiation of leukemia cells by modulating miR-10b. Oncotarget (2016) 7(44):71477–90. doi: 10.18632/oncotarget.12216
94. Bi L, Sun L, Jin Z, Zhang S, Shen Z. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol letters (2018) 15(4):5611–9. doi: 10.3892/ol.2018.8050
95. Lutherborrow M, Bryant A, Jayaswal V, Agapiou D, Palma C, Yang YH, et al. Expression profiling of cytogenetically normal acute myeloid leukemia identifies microRNAs that target genes involved in monocytic differentiation. Am J Hematol (2011) 86(1):2–11. doi: 10.1002/ajh.21864
96. Yu YN, Kou DQ, Liu B, Huang YR, Li SD, Qi Y, et al. LncRNA MEG3 contributes to drug resistance in acute myeloid leukemia by positively regulating ALG9 through sponging miR-155. Int J Lab Hematol (2020) 42(4):464–72. doi: 10.1111/ijlh.13225
97. Boyiadzis M, Whiteside TL. Plasma-derived exosomes in acute myeloid leukemia for detection of minimal residual disease: are we ready? Expert Rev Mol Diagnostics (2016) 16(6):623–9. doi: 10.1080/14737159.2016.1174578
98. Xiao QL, Lin C, Peng MX, Ren J, Jing YP, Lei L, et al. Circulating plasma exosomal long non-coding RNAs LINC00265, LINC00467, UCA1, and SNHG1 as biomarkers for diagnosis and treatment monitoring of acute myeloid leukemia. Front Oncol (2022) 12:1033143. doi: 10.3389/fonc.2022.1033143
99. Bernardi S, Farina M, Bosio K, Di Lucanardo A, Leoni A, Re F, et al. Feasibility of leukemia-derived exosome enrichment and co-isolated dsDNA sequencing in acute myeloid leukemia patients: A proof of concept for new leukemia biomarkers detection. Cancers (2022) 14(18):4504. doi: 10.3390/cancers14184504
100. Menay F, Herschlik L, De Toro J, Cocozza F, Tsacalian R, Jose Gravisaco M, et al. Exosomes isolated from ascites of T-cell lymphoma-bearing mice expressing surface CD24 and HSP-90 induce a tumor-specific immune response. Front Immunol (2017) 8:286. doi: 10.3389/fimmu.2017.00286
101. Zhang F, Lu Y, Wang M, Zhu J, Li J, Zhang P, et al. Exosomes derived from human bone marrow mesenchymal stem cells transfer miR-222-3p to suppress acute myeloid leukemia cell proliferation by targeting IRF2/INPP4B. Mol Cell probes (2020) 51:101513. doi: 10.1016/j.mcp.2020.101513
102. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci (2018) 75(2):193–208. doi: 10.1007/s00018-017-2595-9
103. Zeichner SB, Gleason S, Antun AG, Langston A, Heffner LT Jr., Kota VK, et al. Survival of patients diagnosed with primary refractory and relapsed acute myeloid leukemia from 2008-2012: A single institution experience. Blood (2015) 126(23):4955. doi: 10.1182/blood.V126.23.4955.4955
104. Rashed MH, Bayraktar E, Helal GK, Abd-Ellah MF, Amero P, Chavez-Reyes A, et al. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci (2017) 18(3):538. doi: 10.3390/ijms18030538
105. Whiteside Theresa L. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes). Biochem Soc Trans (2013) 41(1):245–51. doi: 10.1042/BST20120265
106. Namburi S, Broxmeyer HE, Hong CS, Whiteside TL, Boyiadzis M. DPP4(+) exosomes in AML patients' plasma suppress proliferation of hematopoietic progenitor cells. Leukemia (2021) 35(7):1925–32. doi: 10.1038/s41375-020-01047-7
107. Peng H, Ji WH, Zhao RC, Yang J, Lu ZG, Li Y, et al. Exosome: a significant nano-scale drug delivery carrier. J Materials Chem B (2020) 8(34):7591–608. doi: 10.1039/D0TB01499K
108. Yong TY, Zhang XQ, Bie NN, Zhang HB, Zhang XT, Li FY, et al. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nat Commun (2019) 10(1):3838. doi: 10.1038/s41467-019-11718-4
109. Bunggulawa EJ, Wang W, Yin TY, Wang N, Durkan C, Wang YZ, et al. Recent advancements in the use of exosomes as drug delivery systems. J Nanobiotechnol (2018) 16(1):81. doi: 10.1186/s12951-018-0403-9
110. Bellavia D, Raimondo S, Calabrese G, Forte S, Cristaldi M, Patinella A, et al. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics (2017) 7(5):1333–45. doi: 10.7150/thno.17092
Keywords: acute myeloid leukemia, exosomes, prognosis, biomarker, immune evasion
Citation: Wang W, Wu X, Zheng J, Yin R, Li Y, Wu X, Xu L and Jin Z (2024) Utilizing exosomes as sparking clinical biomarkers and therapeutic response in acute myeloid leukemia. Front. Immunol. 14:1315453. doi: 10.3389/fimmu.2023.1315453
Received: 10 October 2023; Accepted: 29 December 2023;
Published: 16 January 2024.
Edited by:
Xin He, City of Hope National Medical Center, United StatesReviewed by:
Kuo Chen, First Affiliated Hospital of Zhengzhou University, ChinaConcha Herrera, Hospital Reina Sofía de Córdoba, Spain
Copyright © 2024 Wang, Wu, Zheng, Yin, Li, Wu, Xu and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuli Wu, c2l1bGllckAxNjMuY29t; Ling Xu, bGluZ3h1MTE0QDE2My5jb20=; Zhenyi Jin, amluemhlbnlpam51QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Wandi Wang
Wandi Wang Xiaofang Wu1†
Xiaofang Wu1† Yangqiu Li
Yangqiu Li Xiuli Wu
Xiuli Wu Ling Xu
Ling Xu Zhenyi Jin
Zhenyi Jin