
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 07 September 2023
Sec. Comparative Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1241934
This article is part of the Research Topic Exploring The Health of Aquatic Organisms Through an Immune Viewpoint View all 5 articles
 Mingming Zhao1
Mingming Zhao1 Zhongyang Lin1
Zhongyang Lin1 Zhihong Zheng1
Zhihong Zheng1 Defu Yao1
Defu Yao1 Shen Yang2
Shen Yang2 Yongzhen Zhao3
Yongzhen Zhao3 Xiuli Chen3
Xiuli Chen3 Jude Juventus Aweya1,2*
Jude Juventus Aweya1,2* Yueling Zhang1*
Yueling Zhang1*Besides dividing the organism’s immune system into adaptive and innate immunity, it has long been thought that only adaptive immunity can establish immune memory. However, many studies have shown that innate immunity can also build immunological memory through epigenetic reprogramming and modifications to resist pathogens’ reinfection, known as trained immunity. This paper reviews the role of mitochondrial metabolism and epigenetic modifications and describes the molecular foundation in the trained immunity of arthropods and mollusks. Mitochondrial metabolism and epigenetic modifications complement each other and play a key role in trained immunity.
The immune system evolved under coevolutionary selection and is the backbone of animal resistance to pathogen attack (1). The immunity of an organism is divided into adaptive immunity and innate immunity. Adaptive immunity evolved independently in vertebrates (2) and is the only one that has memory. However, a growing number of studies have shown that innate immunity can enhance immune responses to secondary infection, which imply that innate immunity has memory (3). However, unlike adaptive immune memory, the memory of innate immunity involves epigenetic modification (4).
In vertebrates, besides adaptive immune memory, innate immune memory or trained immunity has been described (5, 6). The ability of the vertebrate innate immunity to build immunological memory in macrophages was first described in 1986 (7), which seems to result from environmental stress conditions (8–10), and therefore is distinct from classical immunologic memory triggered by the T or B lymphocytes (11, 12) (Figure 1). Many studies on vaccines and pathogens have provided evidence of innate immune memory, such as in SCID mice, which have no T/B lymphocytes, have revealed that Bacille Calmette-Guerin (BCG) could still protect against disseminated candidiasis (13), indicating that some vaccinations and even infections can induce more broad protection against other pathogens through trained immunity mechanisms (5, 12). In vertebrates, trained immunity may improve protection against emerging pathogens and also against future new pandemics (14).
In the last couple of years, trained immunity in mammals has been extensively reviewed (6). Infection and/or vaccine increases the efficiency of immune responses or enhances the resistance to reinfections by related and unrelated pathogens, i.e., offers a cross-protection (15–17). Since the adaptive immune system of vertebrates remembers previous encounters and mounts a robust immune response (18), vertebrates are not ideal models for research into innate immune memory. Recent studies have shown that organisms, such as invertebrates and plants, which lack adaptive immunity, show greater protection from reinfection (19–21). For instance, Anopheles albimanus, Marsupenaeus japonicus, Crassostrea gigas, etc., are reported to build immunological memory to reinfection by the same and different pathogens (22–24).
Among metazoan species that number around 1,162,000, about 1,112,000 (about 95.70%) are invertebrates (25). Given that invertebrates lack lymphocytes and are thought incapable of developing immune memory (26), coupled with the species richness of invertebrates, they are the ideal models for studying innate immune memory. Moreover, many invertebrate species provide steady sources of food globally (27), such as shrimps, scallops, crabs, abalone, etc., are farmed on a largescale (28–31). Thus, understanding how these organisms enhance their immunity, such as via trained immunity, would go a long way toward improving their aquaculture. Therefore, this current review brings together information on trained immunity in invertebrates, especially those that serve as food sources, to understand better how these organisms protect themselves from repeated infections. The concepts and mechanisms from emerging scientific fields will open new avenues for cultivating new species and enhancing disease prevention and treatment of aquaculture animals.
The innate immunity of invertebrates displays some features of an immunological memory (32), which has the same function as the vertebrate adaptive immune system (15, 16). In arthropods and mollusks, trained immunity has been reported in many, including Brine Shrimp (Artemia), the Peruvian scallop (Argopecten purpuratus), Chinese mitten crab (Eriocheir sinensis), Pacific oyster (Crassostrea gigas), kuruma shrimp (Marsupenaeus japonicus), etc. (22, 23, 29, 30, 32). Many studies have shown that the innate immunity in organisms that have or lack adaptive immunity can mount increased resistance to reinfection through innate immunity memory or trained immunity (33–36). For instance, innate immune memory is induced by microbiota to protect mosquitoes against Plasmodium (37), while Bombus terrestris can protect themselves against different pathogens through innate immune memory (38). Similarly, innate immune memory has been identified as an immune defense mechanism in snails (39). In addition to forming memories to the same pathogen, trained immunity helps the host to resist infection by other pathogens, i.e., providing a cross-protection (40) (Figure 2). This phenomenon is because the immune system stays active after the first stimulus, which does not recover to the base level before the next infection (6), indicating that trained immunity also has some specificity (Figure 1). In invertebrates, pattern recognition receptors and/or the genetic diversity of immune molecules and the functional diversity of immune proteins are believed to provide the basis for trained immunity (36, 41–43).
Growing evidence indicates that the molecular mechanisms of trained immunity are epigenetically regulated (17, 44, 45) but not through mechanisms dependent on T and B cell adaptive responses (46). Some mechanisms shown to modulate trained immunity, including histone acetylation, DNA methylation, modulation of microRNA, and noncoding RNA expression (5, 47–49), are defined as epigenetic modifications (50). Innate immune cell reprogramming of metabolic pathways is another basis for forming a trained immunity (51). When innate immune cells are exposed to the first pathogen stimulus, metabolic pathways and epigenetic modifications occur, providing rapid and enhanced immune response upon subsequent pathogen challenge (5, 52). Thus, the trained immunity of arthropods and mollusks could also be affected by epigenetic modifications and metabolic reprogramming, which provides a broader pattern of specificity and immune memory.
During an immune response, host cells’ surface receptors recognize pathogens and transmit the infection signal to the cellular signal transduction pathway to activate target genes’ expression, followed by the release of effector proteins or factors to clear the pathogen. Thus, pathogen recognition initiates the immune response, hence, differences in this recognition induce different immune responses.
Trained immunity has immune memory and primitive specificity to the same pathogen, probably due to receptor diversity (53). The innate immune system’s pattern recognition receptors (PRRs) recognize different microbial species and mediate a broad specificity pattern in vertebrates (53, 54). Similarly, invertebrates have evolved genetic mechanisms capable of producing thousands of different immune proteins from a few genes, which helps them to clear a wide range of pathogens (55). For instance, Tribolium castaneum, Anopheles gambiae, Drosophila melanogaster, and Bombus terrestris, etc. diversify their immune genes’ sequence to enable them to exert a certain degree of specificity to microbial pathogens (38, 56–58).
The ability of invertebrates to discriminate between pathogens is based on a set of PRRs specific for pathogen-associated molecular patterns (PAMPs) of different pathogens (59). Many immune gene families of these PRRs in invertebrates could mediate the non-specific immune response (17, 60, 61), which could also be a form of trained immunity whereby gene expression to synthesize immune proteins is induced by environmental changes. For example, the Down syndrome cell adhesion molecule (Dscam), which is well studied in invertebrates, has been shown to play a role in mounting adaptive-like immunity by specific splicing to produce different immune protein isoforms during pathogens stimulation (62–66). Similarly, C-type lectin-like domain (CTLD) proteins, which perform important tasks in immunity by acting as PRRs (67) and as effector proteins with bactericidal activity (68, 69), are expressed in the genomes of many organisms, including, cephalochordata, echinodermata, insecta, nematoda, cnidaria, porifera, and placozoa (70–76). In penaeid shrimp, such as Litopenaeus vannamei, the C-terminal domain of hemocyanin contains a highly variable genetic sequence that is structurally homologous to immunoglobulin (Ig) and can recognize and bind with bacteria or red blood cells to agglutinate or cause hemolysis (77). The C-terminus of hemocyanin also possesses single nucleotide polymorphisms (SNPs), which is related to shrimp’s resistance to different pathogens (78, 79). In addition, many multigene family proteins in invertebrates, such as npr-1, Sp 185/333 protein, NLRs, TLRs, Caspase gene, and fibrinogen-related proteins (FREPs), are immune effectors and/or modulators of cellular processes involved trained immunity (17, 37, 80–87) (Table 1).
Gene evolution could result from the interaction between hosts and pathogens, indicating that changes in gene expression patterns could be a response to external environmental pressure resulting from long-term evolution. Thus, specific immune pathways and common immune pathways are preserved in the course of evolution. The immune specificity of invertebrates is mainly based on the diversification of somatic gene sequences that encode recognition molecules, effector-enhancement molecules, and other immune molecules (96–98), synonymous with receptor diversification in the adaptive immune system of vertebrates (95). In Arthropods, gene sequence diversity prevents different pathogens from interfering with their immune response (Table 1). The evolutionarily conserved immune system components might explain simple forms of specific immune memory from trained immunity, such as the degree of specific immune reactions of the Toll and Imd pathways (99).
In some invertebrates, macromolecular proteins undergo specific degradation and modifications to allow them to respond to different pathogens. For instance, in response to pathogenic bacteria, the hemocyanin protein of penaeid shrimp degrades into functional peptides to enhance their antimicrobial immunity (100–103) (Table 1). Therefore, arthropods and mollusks do not only encode immune genes but also immune effector molecules with different molecular polymorphisms, which constitute part of their trained immunity (Table 1). Similarly, other invertebrates have diversified their immune gene sequences and repertoire of diversified receptors (Table 1).
The immune system of organism senses and responds to environmental stress, which is high-energy demanding. Trained immunity is associated with many metabolic pathways to increase the ability of immune cells to respond to secondary infections through metabolic reprogramming (104). Environmental cues can also change chromatin structure through epigenetic modification, which could be passed on to the next generation to facilitate adaptation (105). Current studies show that epigenetic modifications, including histone modifications, DNA methylation, chromatin remodeling, and non-coding RNA (106, 107), operate to maintain cell identity (108–110). The mechanism of epigenetic modifications can play an important role in host-pathogen interactions by regulating gene expression (111–116), indicating that metabolic and epigenetic modification are two essential parts of trained immunity.
The cellular immunity of invertebrates mediated by hemocytes consists of inflammatory responses, includes phagocytosis, encapsulation, cytotoxicity, and synthesis or release of microbicidal agents (117). Different immune signals induce different cellular metabolic reorganizations, which are critical for the epigenetic modifications in trained immunity (33, 52, 118). Given that immune responses are high-energy processes, metabolism provides energy to maintain cellular hemostasis and enhances immune cells’ functions (51). For instance, the metabolic product lactate can inhibit the activity of histone deacetylase (HDAC) to increase gene accessibility (119). Moreover, mitochondria are key factors that control many epigenetic enzymes (120). For instance, the activities of alkaline phosphatase, alanine aminotransferase (ALT), phenoloxidase, acid phosphatase (ACP), and lactate dehydrogenase (LDH), peaked at 6-12 h after injection with fungal spores of Spodoptera littoralis, while the highest immune responses and intermediary metabolism occurred 12 h post-injection (121). Similarly, ACP and ALP are the hydrolytic enzymes that mediate the dephosphorylation of nucleotides, proteins, and alkaloids and have been implicated in lipid hydrolysis to provide energy for resistance to external stimuli (122). Thus, metabolic change can promote epigenetic reprogramming under inflammatory stimulation to achieve trained immunity phenotype (5, 33).
During immune stimulation, the metabolic state of cells is modulated to regulate the expression of different genes by retrograde signaling in the mitochondria to promote different cellular functions, such as differentiation, adaptation to challenge, etc. (123, 124). However, under an inappropriate metabolic state, such as due to an effect in the electron transport chain, remedial measures are taken to maintain the production of certain tricarboxylic acid cycle (TCA) intermediates by glutamine-dependent reductive carboxylation (125). The mutual regulation between metabolism and genes expression have been observed in some invertebrates, such as Caenorhabditis elegans, Daphnia pulex, L. vannamei, Argopecten purpuratus, Scylla paramamosain, etc. (29, 126–129).
Mitochondria provide metabolic intermediates and their derived products, e.g., S-adenosyl methionine (SAM) and acetyl-CoA, which drives epigenetic modification, such as histone acetylation by acetyl-CoA (120, 130). Levels of acetyl-CoA affects the activity of histone acetyltransferases (HATs) to regulate gene expression by changing the acetylation of the whole histone (131, 132), which is highly dependent on fatty acid metabolism and glucose availability in mitochondria (131, 133, 134) (Figure 3). Thus, histones acetylation drives the epigenetic control of gene expression through transcriptional programs (135, 136). For instance, exogenous acetate can produce acetyl-CoA to maintain global histone acetylation when acetyl-CoA production by ATP citrate lyase (ACLY) is limited (137). This complementary mechanism relies on two important anaplerotic mechanisms, i.e., the conversion of pyruvate to mitochondrial oxaloacetate by pyruvate decarboxylase and the conversion of glutamate by activation of glutaminolysis and subsequently to α-ketoglutarate (α-KG) (123). However, mitochondria dysfunction induced by exposure to environmental mutagens or pathogen stimulation can suppress mitochondrial oxidative metabolism in invertebrate (138, 139), which could be responsible for the change in gene expression profiles after pathogen challenge (140–142).
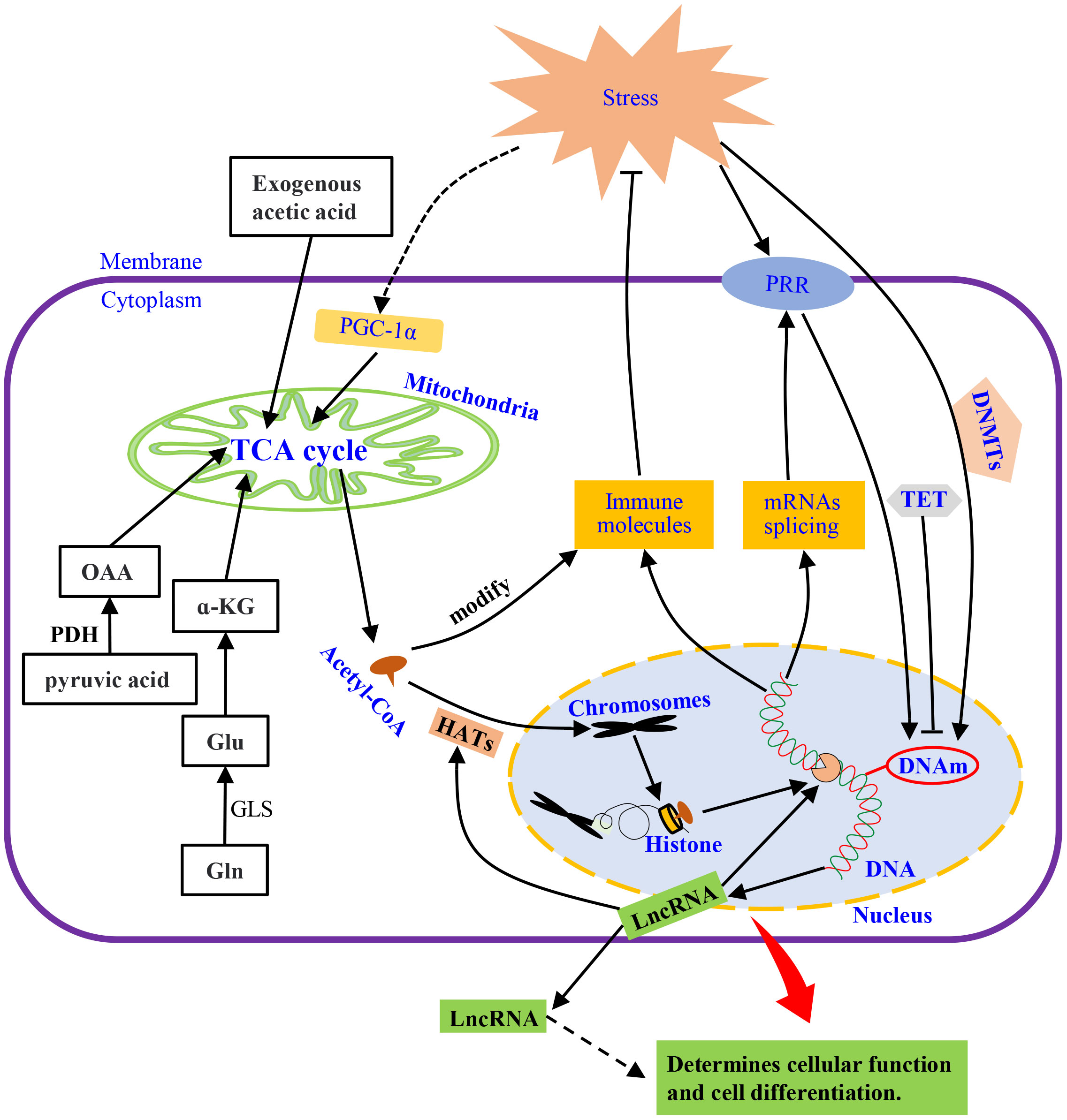
Figure 3 The mechanisms of innate immunity. Under external stimulation, metabolism and membrane receptors directly or indirectly affect DNA methylation, which induces the synthesis of immune molecules by regulating transcription and translation or releases spliced mRNAs. LncRNA is also released to modulate transcription and intercellular information transmission. The external stimuli also activate PGC-a, regulate the TCA cycle, and acetylate chromatin histones through its intermediate product, acetyl-CoA, to promote transcription. Pyruvate, glutamine, or exogenous acetic acid metabolism, supplement the substances needed for the TCA cycle to keep it running. HATs, histone acetyltransferases; α-KG, α-ketoglutarate; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; DNMTs, DNA methyltransferases; TET, Ten-eleven translocation; DNAm, DNA methylation; LncRNAs, Long Non-coding RNAs; OAA, oxaloacetic acid; PDH, pyruvate dehydrogenase; GLS, glutaminase; PRR, pattern recognition receptor; Gln, glutamine; Glu, Glutamate.
An increase in energy metabolism can be regulated by peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) to alter cellular energy demand in different tissues upon activation by stimuli (143, 144). In addition, PGC-1α can coordinate tissue-specific transcription to mediate the plasticity of cells (145) (see Figure 3). These cellular events could be modulated by PGC-1α through steroid receptor coactivator 1 (SRC-1) and CREB-binding protein (CBP) to affect the HAT complex (146). Thus, the influence of immune response through metabolism could affect some aspects of inflammatory or autoimmune diseases.
Nucleosomes are formed when DNA strands are wrapped around eight core histones and then compressed into chromosomes (147). Intrinsically, the compact structure of nucleosomes is repressive regarding unwanted transcription activity. Histone proteins can be modified post-translationally at various residues through methylation, acetylation, phosphorylation, and/or ubiquitination, which changes the chromatin structure (148–150). The increased resistance of Artemia to Vibrio campbellii infection, mediated by increased acetylation levels of histone H3 and H4, could be passed on to the next generation (151). Histone acetylation is a reversible process involving the addition/removal of an acetyl group on lysine residues catalyzed by HAT/histone deacetylase (HDAC) (152, 153) (Figure 3). Although histone deacetylation catalyzed by HDAC tightens the chromatin structure to silence gene expression (153), pathogens infection can disrupt this structure to inhibit host gene expression. For instance, the ICP11 protein of white spot syndrome virus (WSSV) can damage the nucleosome assembly in Penaeus vannamei, by binding to histone proteins to evade the hosts’ immune responses to promote viral replication (154).
Various gene expression changes are induced in the immune system during pathogen infection, which involves regulating epigenetic mechanisms. For instance, DNA methylation, a kind of epigenetic modification, plays an important role in various biological processes (155). DNA methylation is a stable covalent cytosine modification in a cytosine-guanine dinucleotide (CpG) context, which in the genomes of vertebrates, are highly methylated but sparsely methylated in invertebrates (156). This genomic methylation is regulated by DNA methyltransferases (DNMTs), a key mechanism for controlling gene expression in various organisms (157, 158) (Figure 3). The oxidation of 5-methylcytosine (5mC) at CpG dinucleotides to form 5-hydroxymethylcytosine (5-hmC) serves as an epigenetic marker (159) because 5-hmC is enriched at the enhancers of most highly transcribed genes (160). Conversely, the promoters of highly expressed genes that are highly expressed are loss of CpG methylation and form CpG islands (161). Therefore, CpG islands are important for chromosome stability (162, 163).
In Drosophila melanogaster and Aedes aegypti, genomic hypermethylation caused by Wolbachia infection may be associated with the up-regulation of DNA methyltransferases (DNMTs) gene expression (116, 164, 165). Under these conditions, the cytosine-5 of the host insect DNMTs are induced under bacterial infection to affect the expression of antimicrobial peptides (AMPs) (166). In Drosophila, DNMTs are also required for antiviral innate immune responses (167). Since DNA hypermethylation has been linked to transcriptional silencing (163), it is a key and stable mechanism for repressing gene transcription (168), whereas hypomethylation may increase the transcript levels of genes (169). DNA methylation is a reversible process that can be reversed by Ten-eleven translocation enzymes (TET) when gene expression is active (170). Therefore, the interplay between TET proteins and DNMTs controls the DNA methylation landscape (Figure 3).
The methylation of most DNAs in invertebrates is not in the intergenic regions (171, 172) but rather in genomic loci that match small RNAs in gene bodies, which are densely methylated, probably because they regulate the transcription and mRNA splicing of target genes (173). For instance, the DNA methylation in Apis mellifera controls the alternative splicing of mRNA and is involved in gene expression (174, 175). Pathogens may establish successful infection by manipulating the expression of host genes via DNA methylation. In Bombyx mori, cytoplasmic polyhedrosis virus infection may lead to hypermethylation of the p53-2 gene, suppressing its expression to facilitate the proliferation of infected cells (169).
Although histone and DNA methylation are reported widely in most invertebrates, such as arthropods and mollusks, how histone and DNA methylation interact to regulate gene expression and induce trained immunity in invertebrates remains unknown.
Long non-coding RNAs (LncRNAs) are involved in various regulatory functions in animals, including gene regulation at multiple levels, such as at the post-transcriptional levels, enhancers, promoters, and chromatin modification complexes (176, 177). For example, lncRNAs can affect promoter activity or mRNA translation (178) and form miRNA precursors to regulate target gene expression (152). LncRNAs do not only act intracellularly by regulating HDAC, CBP/P300, and HAT but also can be transported to other cells by exosomes (179, 180), which could be one of the mechanisms of epigenetic inheritance across generations (Figure 3). Both lncRNAs (A. aegypti) and microRNAs (C. elegans) show genetic characteristics (181, 182). Thus, in invertebrates, various studies have reported on the regulation of transcriptional activity by non-coding RNAs, such as miRNAs in Galleria mellonella, let-7 miRNA cluster in silkworms (Dazao P50), miRNAs/lncRNAs in A. aegypti, microRNA-8 in Drosophila, miRNA-317 in C. gigas, etc. (183–188).
Adaptive immune memory can last for a long time and respond quickly to reinfection by the same pathogen. Invertebrates such as Artemia display trained immunity with similar features as adaptive immunity (32). The memory of innate immunity could persist for days or almost the entire lifetime, and in some cases across generations (181, 182, 189–191). Trained immune memory induced by inactivated bacterial and viral antigens has been reported in shrimp and crayfish, the main manifestation of which is that the secondary immune response is greatly improved compared with the control (192). For instance, the offspring of Trichoplusia ni from parents raised on a bacteria-rich diet had an increased expression of immune-related genes and immune enzyme activity (193). Similarly, the initiation of transgenerational immunity occurs after the red flour beetle Tribolium castaneum is exposed to heat-killed bacteria (194). Early life microbial exposure improves oyster survival when challenged with the pathogen causing Pacific oyster mortality syndrome (POMS) in both the exposed generation and subsequent generation (195). When challenged with specific bacteria, Artemia could acquire strain-specific immunity that increases resistance against the same strain of bacteria and is transmissible to the progenies of successive generations (196, 197).
From the foregoing, it is clear that trained immunity can also respond specifically to external stimuli and be passed on to the next generation. The transgenerational effects of trained immunity induction in vertebrates have been confirmed (198–200). Moreover, the mechanism of trained immune memory in vertebrates could be mediated by innate immune cells, such as monocytes, macrophages, and natural killer cells (201). Given that invertebrate immunocytes perform the same immune functions as vertebrate macrophages (202), a similar memory mechanism might also be present in invertebrate immunocytes, although the life span of immune-functioning cells in blood is shorter than that of trained immunity. Therefore, more research is needed to further unravel the mechanisms of trans-generational immune priming (TGIP) in invertebrates during trained immunity memory.
Invertebrates lack an adaptive immune system and are an excellent model for studying innate immune defense mechanisms (203). For instance, Drosophila has been used as a valuable insect model to study immune mechanisms of neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease (204, 205).
In aquaculture animals, trained innate immunity has been reported in mollusks, such as oysters and abalone (23, 31, 206, 207). For instance, DNA methylation patterns are found to vary with changes in seasons (especially in temperature) in the oyster Isognomon alatus (208, 209) or changes in ocean acidification and salinity in the Haliotis discus hannai (210, 211). Similarly, the speed of water currents can change DNA methylation patterns, as in the snail Potamopyrgus antipodarum (212, 213). Generally, hypomethylation occurs in different oyster species after infection with toxic algae (214, 215). In the freshwater gastropod Biomphalaria glabrata, Trematode infection induces DNA methylation machinery proteins to impact DNA methylation levels (216, 217). Exposure to air has also been reported to affect innate immunity and DNA methylation in M. japonicus (218). Thus, environmental and biological factors can influence DNA methylation levels in arthropods and mollusks, laying a foundation for the selective breeding of economic species of invertebrates. Although elucidating the epigenetic mechanisms in these species is of great significance to genetic breeding, there is still limited knowledge on pathogen-host interactions, which is one factor limiting trained immunity application for economically important arthropodan and molluscan species.
Prophenoloxidase (ProPO) and transglutaminase (TGase) genes, which play crucial roles in melanization and coagulation, are important constituents of the innate immune system of arthropods that protect the host from invading pathogens (219, 220). For instance, in penaeid shrimp, hemocyanin protein interacts with TGase to modulate its expression, affecting hemolymph clotting (221). Similarly, hemocyanin can be converted into PO-like enzymes in arthropods and mollusks by physical disruption of the structural motifs in the dicopper centers (222), whereas glycosylation modification of hemocyanin or its degradation into functional peptides enhances its antimicrobial activity (223). Despite these findings, it is currently unknown whether the degradation or post-translational modification mechanisms of hemocyanin are regulated by epigenetic inheritance. Thus, further studies would provide better insight into epigenetic reprogramming of the invertebrates’ immune system since such information could be leveraged for designing therapeutic agents for aquaculture invertebrates, such as shrimps, oysters, scallops, etc.
Arthropods and mollusks have evolved to inherit regulatory mechanisms capable of producing thousands of immune proteins from a few genes by epigenetic modifications, allowing them to recognize and eliminate a wide range of pathogens. These organisms select genes through epigenetics to enhance the recognition of pathogens by expressing specific protein receptors and also modify immune molecules, such as hemocyanin, DSCAM, etc., to perform immune functions, whereas mitochondrial metabolism provides the energy and substrates required for epigenetic modifications. The epigenetics of trained immune memories can last long and be passed on to the next generation. Also, arthropods and mollusks activate the trained immunity response to external stimuli, an immune characteristic that can last for a very long time or even into the next generation. Nonetheless, there is still limited knowledge about pathogen-host interactions, an important factor limiting in-depth trained immunity applications in economically important arthropods and mollusks. Therefore, further research on the mechanism of trained immunity would provide vital information for breeding important economic species, optimize the breeding methods, and speed up the breeding process in arthropods and mollusks.
JJA and YLZ conceived the idea. MMZ and JJA performed the literature search, wrote the draft, and revised the paper. YLZ and JJA obtained funding and provided supervision. ZL, ZZ, DY, SY, YZ, and XC provided literature input and suggestions. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Nos. U22A20536, 32073008, and 32273170) and the Natural Science Foundation of Guangdong Province (No. 2022A1515011149).
We thank all members of the Shrimp Disease and Immunology Laboratory for exchanging ideas and for the constructive critique of the manuscript during the drafting.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gerardo NM, Hoang KL, Stoy KS. Evolution of animal immunity in the light of beneficial symbioses. Philos Trans R Soc Lond B Biol Sci (2020) 375(1808):20190601. doi: 10.1098/rstb.2019.0601
2. Orend G, Tucker RP. Did tenascin-C Co-evolve with the general immune system of vertebrates? Front Immunol (2021) 12:663902. doi: 10.3389/fimmu.2021.663902
3. Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature (2021) 596(7872):410–6. doi: 10.1038/s41586-021-03791-x
4. Wimmers F, Donato M, Kuo A, Ashuach T, Gupta S, Li C, et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell. (2021) 184(15):3915–3935.e21. doi: 10.1016/j.cell.2021.05.039
5. Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, et al. Trained immunity: A program of innate immune memory in health and disease. Science (2016) 352(6284):aaf1098. doi: 10.1126/science.aaf1098
6. Divangahi M, Aaby P, Khader SA, Barreiro LB, Bekkering S, Chavakis T, et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat Immunol (2021) 22(1):2–6. doi: 10.1038/s41590-020-00845-6
7. Bistoni F, Vecchiarelli A, Cenci E, Puccetti P, Marconi P, Cassone A. Evidence for macrophage-mediated protection against lethal Candida albicans infection. Infect Immun (1986) 51(2):668–74. doi: 10.1128/iai.51.2.668-674.1986
8. Kaufmann E, Sanz J, Dunn JL, Khan N, Mendonça LE, Pacis A, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell (2018) 172(1-2):176–90. doi: 10.1016/j.cell.2017.12.031
9. Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. (2018) 172(1-2):147–61. doi: 10.1016/j.cell.2017.11.034
10. Christ A, Günther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, et al. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell (2018) 172(1-2):162–75. doi: 10.1016/j.cell.2017.12.013
11. Farber DL, Netea MG, Radbruch A, Rajewsky K, Zinkernagel RM. Immunological memory: lessons from the past and a look to the future. Nat Rev Immunol (2016) 16(2):124–8. doi: 10.1038/nri.2016.13
12. Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe (2011) 9(5):355–61. doi: 10.1016/j.chom.2011.04.006
13. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, et al. Bacille calmette-guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA (2012) 109(43):17537–42. doi: 10.1073/pnas.1202870109
14. Geckin B, Konstantin Föhse F, Domínguez-Andrés J, Netea MG. Trained immunity: implications for vaccination. Curr Opin Immunol (2022) 77:102190. doi: 10.1016/j.coi.2022.102190
15. Kurtz J, Franz K. Innate defence: Evidence for memory in invertebrate immunity. Nature (2003) 425(6953):37–8. doi: 10.1038/425037a
16. Little TJ, Hultmark D, Read AF. Invertebrate immunity and the limits of mechanistic immunology. Nat Immunol (2005) 6(7):651–4. doi: 10.1038/ni1219
17. Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol (2014) 29:1–7. doi: 10.1016/j.coi.2014.02.006
18. Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell (2006) 124(4):815–22. doi: 10.1016/j.cell.2006.02.001
19. Kurtz J. Specific memory within innate immune systems. Trends Immunol (2005) 26(4):186–92. doi: 10.1016/j.it.2005.02.001
20. Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR. Priming for enhanced defense. Annu Rev Phytopathol (2015) 53(1):97. doi: 10.1146/annurev-phyto-080614-120132
21. Gourbal B, Pinaud S, Beckers GJM, van der Meer JWM, Conrath U, Netea MG. Innate immune memory: An evolutionary perspective. Immunol Rev (2018) 283(1):21–40. doi: 10.1111/imr.12647
22. Zang S, Lv LX, Liu CF, Zhang P, Li C, Wang JX. Metabolomic investigation of ultraviolet ray-inactivated white spot syndrome virus-induced trained immunity in. Marsupenaeus japonicus. Front Immunol (2022) 13:885782. doi: 10.3389/fimmu.2022.885782
23. Lafont M, Vergnes A, Vidal-Dupiol J, de Lorgeril J, Gueguen Y, Haffner P, et al. A sustained immune response supports long-term antiviral immune priming in the pacific oyster, Crassostrea gigas. mBio (2020) 11(2):e02777-19. doi: 10.1128/mBio.02777-19
24. Cime-Castillo J, Arts RJW, Vargas-Ponce de León V, Moreno-Torres R, Hernández-Martínez S, Recio-Totoro B, et al. DNA synthesis is activated in mosquitoes and human monocytes during the induction of innate immune memory. Front Immunol (2018) 9:2834. doi: 10.3389/fimmu.2018.02834
25. Bertrand S, Escriva H. The evolution of invertebrate animals. Genes (Basel) (2022) 13(3):454. doi: 10.3390/genes13030454
26. Ali A, Abd El Halim HM. Re-thinking adaptive immunity in the beetles: Evolutionary and functional trajectories of lncRNAs. Genomics (2020) 112(2):1425–36. doi: 10.1016/j.ygeno.2019.08.012
27. Russo V, Songa G, Milani Marin LE, Balzaretti CM, Tedesco DEA. Novel food-based product communication: a neurophysiological study. Nutrients (2020) 12(7):2092. doi: 10.3390/nu12072092
28. Xuan BB, Sandorf ED, Ngoc QTK. Stakeholder perceptions towards sustainable shrimp aquaculture in Vietnam. J Environ Manage (2021) 290:112585. doi: 10.1016/j.jenvman.2021.112585
29. Rojas I, Rivera-Ingraham GA, Cárcamo CB, Jeno K, de la Fuente-Ortega E, Schmitt P, et al. Metabolic cost of the immune response during early ontogeny of the scallop. Argopecten purpuratus. Front Physiol (2021) 12:718467. doi: 10.3389/fphys.2021.718467
30. Zeng Q, Xu Y, Jeppesen E, Gu X, Mao Z, Chen H. Farming practices affect the amino acid profiles of the aquaculture Chinese mitten crab. PeerJ (2021) 9:e11605. doi: 10.7717/peerj.11605
31. Swezey DS, Boles SE, Aquilino KM, Stott HK, Bush D, Whitehead A, et al. Evolved differences in energy metabolism and growth dictate the impacts of ocean acidification on abalone aquaculture. Proc Natl Acad Sci USA (2020) 117(42):26513–9. doi: 10.1073/pnas.2006910117
32. Norouzitallab P, Baruah K, Biswas P, Vanrompay D, Bossier P. Probing the phenomenon of trained immunity in invertebrates during a transgenerational study, using brine shrimp Artemia as a model system. Sci Rep (2016) 6:21166. doi: 10.1038/srep21166
33. Arts RJ, Joosten LA, Netea MG. Immunometabolic circuits in trained immunity. Semin Immunol (2016) 28(5):425–30. doi: 10.1016/j.smim.2016.09.002
34. Yao Y, Jeyanathan M, Haddadi S, Barra NG, Vaseghi-Shanjani M, Damjanovic D, et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell (2018) 175(6):1634–1650.e17. doi: 10.1016/j.cell.2018.09.042
35. Netea MG, Dominguez-Andres J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol (2020) 20(6):375–88. doi: 10.1038/s41577-020-0285-6
36. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and adaptive immune memory: an evolutionary continuum in the host’s response to pathogens. Cell Host Microbe (2019) 25(1):13–26. doi: 10.1016/j.chom.2018.12.006
37. Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles Gambiae mosquitoes. Science (2010) 329(5997):1353–5. doi: 10.1126/science.1190689
38. Sadd BM, Schmid-Hempel P. Insect immunity shows specificity in protection upon secondary pathogen exposure. Curr Biol (2006) 16(12):1206–10. doi: 10.1016/j.cub.2006.04.047
39. Pinaud S, Portela J, Duval D, Nowacki FC, Olive MA, Allienne JF, et al. A shift from cellular to humoral responses contributes to innate immune memory in the vector snail. Biomphalaria glabrata. PloS Pathog (2016) 12(1):e1005361. doi: 10.1371/journal.ppat.1005361
40. Choi JA, Goo J, Yang E, Jung DI, Lee S, Rho S, et al. Cross-protection against MERS-CoV by prime-boost vaccination using viral spike DNA and protein. J Virol (2020) 94(24):e01176-20. doi: 10.1128/jvi.01176-20
41. Pasquier LD. Germline and somatic diversification of immune recognition elements in Metazoa. Immunol Lett (2006) 104(1-2):2–17. doi: 10.1016/j.imlet.2005.11.022
42. Schulenburg H, Boehnisch C, Michiels NK. How do invertebrates generate a highly specific innate immune response? Mol Immunol (2007) 44(13):3338–44. doi: 10.1016/j.molimm.2007.02.019
43. Palmieri B, Vadala M, Palmieri L. Immune memory: an evolutionary perspective. Hum Vaccin Immunother (2021) 17(6):1604–6. doi: 10.1080/21645515.2020.1846396
44. Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol (2010) 11(4):563–76. doi: 10.1111/j.1364-3703.2010.00621.x
45. Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol (2013) 64:839–63. doi: 10.1146/annurev-arplant-042811-105606
46. Kleinnijenhuis J, van Crevel R, Netea MG. Trained immunity: consequences for the heterologous effects of BCG vaccination. Trans R Soc Trop Med Hyg (2015) 109(1):29–35. doi: 10.1093/trstmh/tru168
47. Verhoeven KJ, Jansen JJ, van Dijk PJ, Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol (2010) 185(4):1108–18. doi: 10.1111/j.1469-8137.2009.03121.x
48. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res (2011) 21(3):381–95. doi: 10.1038/cr.2011.22
49. Storz G. An expanding universe of noncoding RNAs. Science (2002) 296(5571):1260–3. doi: 10.1126/science.1072249
50. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science (2014) 345(6204):1251086. doi: 10.1126/science.1251086
51. Sohrabi Y, Godfrey R, Findeisen HM. Altered cellular metabolism drives trained immunity. Trends Endocrinol Metab (2018) 29(9):602–5. doi: 10.1016/j.tem.2018.03.012
52. Mohammadnezhad L, Shekarkar Azgomi M, La Manna MP, Sireci G, Rizzo C, Badami GD, et al. Metabolic reprogramming of innate immune cells as a possible source of new therapeutic approaches in autoimmunity. Cells (2022) 11(10):1663. doi: 10.3390/cells11101663
53. Pees B, Yang W, Zarate-Potes A, Schulenburg H, Dierking K. High innate immune specificity through diversified C-type lectin-like domain proteins in invertebrates. J Innate Immun (2016) 8(2):129–42. doi: 10.1159/000441475
54. Gu Y, Cao J, Zhang X, Gao H, Wang Y, Wang J, et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res (2022) 32(1):24–37. doi: 10.1038/s41422-021-00595-6
55. Ghosh J, Lun CM, Majeske AJ, Sacchi S, Schrankel CS, Smith LC. Invertebrate immune diversity. Dev Comp Immunol (2011) 35(9):0–974. doi: 10.1016/j.dci.2010.12.009
56. Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles Gambiae innate immune system. PloS Biol (2006) 4(7):e229. doi: 10.1371/journal.pbio.0040229
57. Roth O, Sadd BM, Schmid-Hempel P, Kurtz J. Strain-specific priming of resistance in the red flour beetle. Tribolium castaneum. Proc Biol Sci (2009) 276(1654):145–51. doi: 10.1098/rspb.2008.1157
58. Tang C, Kurata S, Fuse N. Genetic dissection of innate immune memory in Drosophila melanogaster. Front Immunol (2022) 13:857707. doi: 10.3389/fimmu.2022.857707
59. Milutinović B, Kurtz J. Immune memory in invertebrates. Semin Immunol (2016) 28(4):328–42. doi: 10.1016/j.smim.2016.05.004
60. Zhang L, Li L, Guo X, Litman GW, Dishaw LJ, Zhang G. Massive expansion and functional divergence of innate immune genes in a protostome. Sci Rep (2015) 5:8693. doi: 10.1038/srep08693
61. McDowell IC, Modak TH, Lane CE, Gomez-Chiarri M. Multi-species protein similarity clustering reveals novel expanded immune gene families in the eastern oyster. Crassostrea virginica. Fish Shellfish Immunol (2016) 53:13–23. doi: 10.1016/j.fsi.2016.03.157
62. Watthanasurorot A, Jiravanichpaisal P, Liu H, Soderhall I, Soderhall K. Retraction: Bacteria-induced Dscam isoforms of the crustacean, Pacifastacus leniusculus. PloS Pathog (2016) 12(5):e1005630. doi: 10.1371/journal.ppat.1005630
63. Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science (2005) 309(5742):1874–8. doi: 10.1126/science.1116887
64. Guichard M, Dainat B, Eynard S, Vignal A, Servin B, Neuditschko M. Two quantitative trait loci are associated with recapping of Varroa destructor-infested brood cells in Apis mellifera. Anim Genet (2022) 53(1):156–60. doi: 10.1111/age.13150
65. Smith PH, Mwangi JM, Afrane YA, Yan G, Obbard DJ, Ranford-Cartwright LC, et al. Alternative splicing of the Anopheles Gambiae Dscam gene in diverse Plasmodium falciparum infections. Malar J (2011) 10:156. doi: 10.1186/1475-2875-10-156
66. Hou S, Li G, Xu B, Dong H, Zhang S, Fu Y, et al. Trans-splicing facilitated by RNA pairing greatly expands sDscam isoform diversity but not homophilic binding specificity. Sci Adv (2022) 8(27):eabn9458. doi: 10.1126/sciadv.abn9458
67. Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol (2014) 16(2):185–94. doi: 10.1111/cmi.12249
68. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science (2011) 334(6053):255–8. doi: 10.1126/science.1209791
69. Li XC, Zhou J, Zhou JF, Wang Y, Ma H, Wang Y, et al. SpBark suppresses bacterial infection by mediating hemocyte phagocytosis in an invertebrate model. Scylla paramamosain. Front Immunol (2019) 10:1992. doi: 10.3389/fimmu.2019.01992
70. Ao J, Ling E, Yu XQ. Drosophila C-type lectins enhance cellular encapsulation. Mol Immunol (2007) 44(10):2541–8. doi: 10.1016/j.molimm.2006.12.024
71. Tanji T, Ohashi-Kobayashi A. Natori S. Participation of a galactose-specific C-type lectin in. Drosophila immunity. Biochem J (2006) 396(1):127–38. doi: 10.1042/bj20051921
72. Watanabe A, Miyazawa S, Kitami M, Tabunoki H, Ueda K, Sato R. Characterization of a novel C-type lectin, Bombyx mori multibinding protein, from the B. mori hemolymph: Mech wide-range microorganism recognition role immunity. J Immunol (2006) 177(7):4594–604. doi: 10.4049/jimmunol.177.7.4594
73. Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev (2004) 198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x
74. Schnitger AK, Yassine H, Kafatos FC, Osta MA. Two C-type lectins cooperate to defend Anopheles Gambiae against Gram-negative bacteria. J Biol Chem (2009) 284(26):17616–24. doi: 10.1074/jbc.M808298200
75. Yu Y, Yu Y, Huang H, Feng K, Pan M, Yuan S, et al. A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J Immunol (2007) 179(12):8425–34. doi: 10.4049/jimmunol.179.12.8425
76. Multerer KA, Smith LC. Two cDNAs from the purple sea urchin, Strongylocentrotus purpuratus, encoding mosaic proteins with domains found in factor H, factor I, and complement components C6 and C7. Immunogenetics (2004) 56(2):89–106. doi: 10.1007/s00251-004-0665-2
77. Zhang YL, Peng B, Li H, Yan F, Wu HK, Zhao XL, et al. C-terminal domain of hemocyanin, a major antimicrobial protein from Litopenaeus vannamei: structural homology with immunoglobulins and molecular diversity. Front Immunol (2017) 8:611. doi: 10.3389/fimmu.2017.00611
78. Zhao X, Guo L, Zhang Y, Liu Y, Zhang X, Lun J, et al. SNPs of hemocyanin C-terminal fragment in shrimp. Litopenaeus vannamei. FEBS Lett (2012) 586(4):403–10. doi: 10.1016/j.febslet.2011.12.038
79. Guo L, Zhao X, Zhang Y, Wang Z, Zhong M, Li S, et al. Evidences of SNPs in the variable region of hemocyanin Ig-like domain in shrimp. Litopenaeus vannamei. Fish Shellfish Immunol (2013) 35(5):1532–8. doi: 10.1016/j.fsi.2013.08.024
80. Mone Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PloS Negl Trop Dis (2010) 4(9):e813. doi: 10.1371/journal.pntd.0000813
81. Zhang T, Qiu L, Sun Z, Wang L, Zhou Z, Liu R, et al. The specifically enhanced cellular immune responses in Pacific oyster (Crassostrea gigas) against secondary challenge with Vibrio splendidus. Dev Comp Immunol (2014) 45(1):141–50. doi: 10.1016/j.dci.2014.02.015
82. Ghosh J, Buckley KM, Nair SV, Raftos DA, Miller C, Majeske AJ, et al. Sp185/333: a novel family of genes and proteins involved in the purple sea urchin immune response. Dev Comp Immunol (2010) 34(3):235–45. doi: 10.1016/j.dci.2009.10.008
83. Ewbank JJ, Pujol N. Local and long-range activation of innate immunity by infection and damage in C. elegans. Curr Opin Immunol (2016) 38:1–7. doi: 10.1016/j.coi.2015.09.005
84. Nakad R, Snoek LB, Yang W, Ellendt S, Schneider F, Mohr TG, et al. Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1. BMC Genomics (2016) 17:280. doi: 10.1186/s12864-016-2603-8
85. Sherman LS, Schrankel CS, Brown KJ, Smith LC. Extraordinary diversity of immune response proteins among sea urchins: nickel-isolated Sp185/333 proteins show broad variations in size and charge. PloS One (2015) 10(9):e0138892. doi: 10.1371/journal.pone.0138892
86. Degnan SM. The surprisingly complex immune gene repertoire of a simple sponge, exemplified by the NLR genes: a capacity for specificity? Dev Comp Immunol (2015) 48(2):269–74. doi: 10.1016/j.dci.2014.07.012
87. Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol (2006) 300(1):349–65. doi: 10.1016/j.ydbio.2006.08.065
88. Brockton V, Henson JH, Raftos DA, Majeske AJ, Kim YO, Smith LC. Localization and diversity of 185/333 proteins from the purple sea urchin–unexpected protein-size range and protein expression in a new coelomocyte type. J Cell Sci (2008) 121(Pt 3):339–48. doi: 10.1242/jcs.012096
89. Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, et al. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol (2008) 25(7):1429–39. doi: 10.1093/molbev/msn087
90. Dong Y, Cirimotich CM, Pike A, Chandra R, Dimopoulos G. Anopheles NF-kappaB-regulated splicing factors direct pathogen-specific repertoires of the hypervariable pattern recognition receptor AgDscam. Cell Host Microbe (2012) 12(4):521–30. doi: 10.1016/j.chom.2012.09.004
91. Zhang Z, Wang F, Chen C, Zheng Z, Aweya JJ, Zhang Y. Glycosylation of hemocyanin in Litopenaeus vannamei is an antibacterial response feature. Immunol Lett (2017) 192:42–7. doi: 10.1016/j.imlet.2017.10.008
92. Yan F, Zhang Y, Jiang R, Zhong M, Hu Z, Du H, et al. Identification and agglutination properties of hemocyanin from the mud crab (Scylla serrata). Fish Shellfish Immunol (2011) 30(1):354–60. doi: 10.1016/j.fsi.2010.11.004
93. Zhang Z, Li R, Aweya JJ, Wang F, Zhong M, Zhang Y. Identification and characterization of glycosylation sites on Litopenaeus vannamei hemocyanin. FEBS Lett (2019) 593(8):820–30. doi: 10.1002/1873-3468.13367
94. Zhao X, Guo L, Lu X, Lu H, Wang F, Zhong M, et al. Evidences of abundant hemocyanin variants in shrimp. Litopenaeus vannamei. Mol Immunol (2016) 77:103–12. doi: 10.1016/j.molimm.2016.07.017
95. Zhi B, Wang L, Wang G, Zhang X. Contribution of the caspase gene sequence diversification to the specifically antiviral defense in invertebrate. PloS One (2011) 6(9):e24955. doi: 10.1371/journal.pone.0024955
96. Oren M, Rosental B, Hawley TS, Kim GY, Agronin J, Reynolds CR, et al. Individual sea urchin Coelomocytes undergo somatic immune gene diversification. Front Immunol (2019) 10:1298. doi: 10.3389/fimmu.2019.01298
97. Johnstun JA, Shankar V, Mokashi SS, Sunkara LT, Ihearahu UE, Lyman RL, et al. Functional diversification, redundancy, and epistasis among paralogs of the drosophila melanogaster obp50a-d gene cluster. Mol Biol Evol (2021) 38(5):2030–44. doi: 10.1093/molbev/msab004
98. Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems–not homogeneous, not simple, not well understood. Immunol Rev (2004) 198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x
99. Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA (1997) 94(26):14614–9. doi: 10.1073/pnas.94.26.14614
100. Wen Y, Zhan S, Huang H, Zhong M, Chen J, You C, et al. Identification and characterization of an 18.4kDa antimicrobial truncation from shrimp. Litopenaeus vannamei hemocyanin upon Vibrio parahaemolyticus infection. Fish Shellfish Immunol (2016) 56:450–8. doi: 10.1016/j.fsi.2016.08.002
101. Zhan S, Aweya JJ, Wang F, Yao D, Zhong M, Chen J, et al. Litopenaeus vannamei attenuates white spot syndrome virus replication by specific antiviral peptides generated from hemocyanin. Dev Comp Immunol (2019) 91:50–61. doi: 10.1016/j.dci.2018.10.005
102. Barberis S QH, Barcia C, Talia JM, Debattista N. Chapter 20-natural food preservatives against microorganisms. In A. M. Grumezescu A. M. Holban (Eds.) Food Saf Preservation (2018) pp:621–58. doi: 10.1016/B978-0-12-814956-0.00020-2
103. Li C, Wang F, Aweya JJ, Yao D, Zheng Z, Huang H, et al. Trypsin of Litopenaeus vannamei is required for the generation of hemocyanin-derived peptides. Dev Comp Immunol (2018) 79:95–104. doi: 10.1016/j.dci.2017.10.015
104. Dominguez-Andres J, Joosten LA, Netea MG. Induction of innate immune memory: the role of cellular metabolism. Curr Opin Immunol (2019) 56:10–6. doi: 10.1016/j.coi.2018.09.001
105. Fallet M, Luquet E, David P, Cosseau C. Epigenetic inheritance and intergenerational effects in mollusks. Gene (2020) 729:144166. doi: 10.1016/j.gene.2019.144166
106. Hu LF. Epigenetic regulation of autophagy. Adv Exp Med Biol (2019) 1206:221–36. doi: 10.1007/978-981-15-0602-4_11
107. Ting HA, de Almeida Nagata D, Rasky AJ, Malinczak CA, Maillard IP, Schaller MA, et al. Notch ligand Delta-like 4 induces epigenetic regulation of Treg cell differentiation and function in viral infection. Mucosal Immunol (2018) 11(5):1524–36. doi: 10.1038/s41385-018-0052-1
108. Titus AJ, Gallimore RM, Salas LA, Christensen BC. Cell-type deconvolution from DNA methylation: a review of recent applications. Hum Mol Genet (2017) 26(R2):R216–24. doi: 10.1093/hmg/ddx275
109. Tran KA, Dillingham CM, Sridharan R. Coordinated removal of repressive epigenetic modifications during induced reversal of cell identity. EMBO J (2019) 38(22):e101681. doi: 10.15252/embj.2019101681
110. Song G, Wang G, Luo X, Cheng Y, Song Q, Wan J, et al. An all-to-all approach to the identification of sequence-specific readers for epigenetic DNA modifications on cytosine. Nat Commun (2021) 12(1):795. doi: 10.1038/s41467-021-20950-w
111. Asgari S. Role of MicroRNAs in insect host-microorganism interactions. Front Physiol (2011) 2:48. doi: 10.3389/fphys.2011.00048
112. Gegner J, Baudach A, Mukherjee K, Halitschke R, Vogel H, Vilcinskas A. Epigenetic mechanisms are involved in sex-specific trans-generational immune priming in the lepidopteran model host Manduca sexta. Front Physiol (2019) 10:137. doi: 10.3389/fphys.2019.00137
113. Morandini AC, Santos CF, Yilmaz O. Role of epigenetics in modulation of immune response at the junction of host-pathogen interaction and danger molecule signaling. Pathog Dis (2016) 74(7):453–63. doi: 10.1093/femspd/ftw082
114. Mukherjee K, Fischer R, Vilcinskas A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front Zool (2012) 9(1):25. doi: 10.1186/1742-9994-9-25
115. Negri I, Jablonka E. Editorial: epigenetics as a deep intimate dialogue between host and symbionts. Front Genet (2016) 7:7. doi: 10.3389/fgene.2016.00007
116. Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in. Aedes aEgypti. Proc Natl Acad Sci USA (2013) 110(25):10276–81. doi: 10.1073/pnas.1303603110
117. Corte CL, Baranzini N, Grimaldi A, Parisi MG. Invertebrate models in innate immunity and tissue remodeling research. Int J Mol Sci (2022) 23(12):204–6. doi: 10.3390/ijms23126843
118. Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol (2014) 32(1):609–34. doi: 10.1146/annurev-immunol-032713-120236
119. Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res (2012) 40(11):4794–803. doi: 10.1093/nar/gks066
120. Matilainen O, Quirós PM, Auwerx J. Mitochondria and epigenetics - crosstalk in homeostasis and stress. Trends Cell Biol (2017) 27(6):453–63. doi: 10.1016/j.tcb.2017.02.004
121. Mirhaghparast SK, Zibaee A, Hajizadeh J. Effects of Beauveria bassiana and Metarhizium anisopliae on cellular immunity and intermediary metabolism of Spodoptera littoralis Boisduval (Lepidoptera: Noctuidae). Invert Surviv J (2013) 10(1):110–9. doi: 10.3201/eid1901.AC1901
122. Zibaee A, Bandani AR, Talaei-Hassanlouei R, Malagoli D. Cellular immune reactions of the sunn pest, Eurygaster integriceps, to the entomopathogenic fungus, Beauveria bassiana and its secondary metabolites. J Insect Sci (2011) 11:138. doi: 10.1673/031.011.13801
123. Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun (2020) 11(1):102. doi: 10.1038/s41467-019-13668-3
124. Chandel NS. Evolution of mitochondria as signaling organelles. Cell Metab (2015) 22:204–6. doi: 10.1016/j.cmet.2015.05.013
125. Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature (2011) 481(7381):385–8. doi: 10.1038/nature10642
126. Yilmaz LS, Li X, Nanda S, Fox B, Schroeder F, Walhout AJ. Modeling tissue-relevant Caenorhabditis elegans metabolism at network, pathway, reaction, and metabolite levels. Mol Syst Biol (2020) 16(10):e9649. doi: 10.15252/msb.20209649
127. Klumpen E, Hoffschröer N, Schwalb A, Gigengack U, Koch M, Paul RJ, et al. Metabolic adjustments during starvation in Daphnia pulex. Comp Biochem Physiol B Biochem Mol Biol (2021) 255:110591. doi: 10.1016/j.cbpb.2021.110591
128. Yen SC, Mao JY, Lin HY, Huang HT, Harroun SG, Nain A, et al. Multifunctional carbonized nanogels to treat lethal acute hepatopancreatic necrosis disease. J Nanobiotechnology (2021) 19(1):448. doi: 10.1186/s12951-021-01194-8
129. Jiang Q, Lu B, Wang G, Ye H. Transcriptional inhibition of sp-IAG by crustacean female sex hormone in the mud crab, Scylla paramamosain. Int J Mol Sci (2020) 21(15):61–74. doi: 10.3390/ijms21155300
130. Weinhouse C. Mitochondrial-epigenetic crosstalk in environmental toxicology. Toxicology (2017) 391:5–17. doi: 10.1016/j.tox.2017.08.008
131. Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab (2014) 20(2):306–19. doi: 10.1016/j.cmet.2014.06.004
132. Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab (2015) 21(3):392–402. doi: 10.1016/j.cmet.2015.02.002
133. Martínez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, et al. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol Cell (2016) 61(2):199–209. doi: 10.1016/j.molcel.2015.12.002
134. McDonnell E, Crown SB, Fox DB, Kitir B, Ilkayeva OR, Olsen CA, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep (2016) 17(6):1463–72. doi: 10.1016/j.celrep.2016.10.012
135. Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol (2015) 33:125–31. doi: 10.1016/j.ceb.2015.02.003
136. Sivanand S, Viney I, Wellen KE. Spatiotemporal control of acetyl-coA metabolism in chromatin regulation. Trends Biochem Sci (2018) 43(1):61–74. doi: 10.1016/j.tibs.2017.11.004
137. Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee JV, et al. ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep (2016) 17(4):1037–52. doi: 10.1016/j.celrep.2016.09.069
138. Wang T, Huang X, Jiang X, Hu M, Huang W, Wang Y. Differential in vivo hemocyte responses to nano titanium dioxide in mussels: Effects of particle size. Aquat Toxicol (2019) 212:28–36. doi: 10.1016/j.aquatox.2019.04.012
139. Tunholi-Alves VM, Tunholi VM, Amaral LS, da Silva Garcia J, Lima MG, DaMatta RA, et al. Alterations in the mitochondrial physiology of Biomphalaria glabrata (Mollusca: Gastropoda) after experimental infection by Angiostrongylus cantonensis (Nematoda: Metastrongylidae). Acta Parasitol (2019) 64(4):693–9. doi: 10.2478/s11686-019-00039-7
140. Guo R, Wang S, Xue R, Cao G, Hu X, Huang M, et al. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl Microbiol Biotechnol (2015) 99(12):5175–87. doi: 10.1007/s00253-015-6634-x
141. Gao K, Deng XY, Qian HY, Qin GX, Hou CX, Guo XJ. Cytoplasmic polyhedrosis virus-induced differential gene expression in two silkworm strains of different susceptibility. Gene (2014) 539(2):230–7. doi: 10.1016/j.gene.2014.01.073
142. Wu P, Wang X, Qin GX, Liu T, Jiang YF, Li MW, et al. Microarray analysis of the gene expression profile in the midgut of silkworm infected with cytoplasmic polyhedrosis virus. Mol Biol Rep (2011) 38(1):333–41. doi: 10.1007/s11033-010-0112-4
143. Kupr B, Handschin C. Complex coordination of cell plasticity by a PGC-1alpha-controlled transcriptional network in skeletal muscle. Front Physiol (2015) 6:325. doi: 10.3389/fphys.2015.00325
144. Martinez-Redondo V, Pettersson AT, Ruas JL. The hitchhiker’s guide to PGC-1alpha isoform structure and biological functions. Diabetologia (2015) 58(9):1969–77. doi: 10.1007/s00125-015-3671-z
145. Kramer AI, Handschin C. How epigenetic modifications drive the expression and mediate the action of PGC-1alpha in the regulation of metabolism. Int J Mol Sci (2019) 20(21):5573. doi: 10.3390/ijms20215449
146. Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science (1999) 286(5443):1368–71. doi: 10.1126/science.286.5443.1368
147. Zhou BR, Bai Y. Chromatin structures condensed by linker histones. Essays Biochem (2019) 63(1):75–87. doi: 10.1042/ebc20180056
148. Wang Y, Yuan Q, Xie L. Histone modifications in aging: The underlying mechanisms and implications. Curr Stem Cell Res Ther (2018) 13(2):125–35. doi: 10.2174/1574888x12666170817141921
149. Molina-Serrano D, Kyriakou D, Kirmizis A. Histone modifications as an intersection between diet and longevity. Front Genet (2019) 10:192. doi: 10.3389/fgene.2019.00192
150. Barnes CE, English DM, Cowley SM. Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem (2019) 63(1):97–107. doi: 10.1042/ebc20180061
151. Norouzitallab P, Baruah K, Vandegehuchte M, Van Stappen G, Catania F, Vanden Bussche J, et al. Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogenetic Artemia model. FASEB J (2014) 28(8):3552–63. doi: 10.1096/fj.14-252049
152. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci (2019) 20(22):: 592–610. doi: 10.3390/ijms20225573
153. Tsai CY, Hsieh SC, Lu CS, Wu TH, Liao HT, Wu CH, et al. Cross-talk between mitochondrial dysfunction-provoked oxidative stress and aberrant noncoding RNA expression in the pathogenesis and pathophysiology of SLE. Int J Mol Sci (2019) 20(20):E3306-15. doi: 10.3390/ijms20205183
154. Encinas-Garcia T, Loreto-Quiroz DL, Mendoza-Cano F, Pena-Rodriguez A, Fimbres-Olivarria D, de la Re-Vega E, et al. White spot syndrome virus down-regulates expression of histones H2A and H4 of Penaeus vannamei to promote viral replication. Dis Aquat Organisms (2019) 137(1):73–9. doi: 10.3354/dao03428
155. Falckenhayn C, Carneiro VC, de Mendonça Amarante A, Schmid K, Hanna K, Kang S, et al. Comprehensive DNA methylation analysis of the Aedes aEgypti genome. Sci Rep (2016) 6:36444. doi: 10.1038/srep36444
156. de Mendoza A, Hatleberg WL, Pang K, Leininger S, Bogdanovic O, Pflueger J, et al. Convergent evolution of a vertebrate-like methylome in a marine sponge. Nat Ecol Evol (2019) 3(10):1464–73. doi: 10.1038/s41559-019-0983-2
157. Sbaraini N, Bellini R, Penteriche AB, Guedes RLM, Garcia AWA, Gerber AL, et al. Genome-wide DNA methylation analysis of Metarhizium anisopliae during tick mimicked infection condition. BMC Genomics (2019) 20(1):836. doi: 10.1186/s12864-019-6220-1
158. Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell (2011) 146(6):866–72. doi: 10.1016/j.cell.2011.08.042
159. Richa R, Sinha RP. Hydroxymethylation of DNA: an epigenetic marker. Excli J (2014) 13:592–610. doi: 10.17877/DE290R-181
160. Tsagaratou A, Aijo T, Lio CW, Yue X, Huang Y, Jacobsen SE, et al. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc Natl Acad Sci USA (2014) 111(32):E3306–15. doi: 10.1073/pnas.1412327111
161. Pal S, Tyler JK. Epigenetics and aging. Sci Adv (2016) 2(7):e1600584. doi: 10.1126/sciadv.1600584
162. Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta (2014) 1839(12):1362–72. doi: 10.1016/j.bbagrm.2014.02.007
163. Schmitz RJ, Lewis ZA, Goll MG. DNA methylation: shared and divergent features across eukaryotes. Trends Genet (2019) 35(11):818–27. doi: 10.1016/j.tig.2019.07.007
164. Bhattacharya T, Newton ILG, Hardy RW. Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PloS Pathog (2017) 13(6):e1006427. doi: 10.1371/journal.ppat.1006427
165. LePage DP, Jernigan KK, Bordenstein SR. The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ (2014) 2:e678. doi: 10.7717/peerj.678
166. Baradaran E, Moharramipour S, Asgari S, Mehrabadi M. Induction of DNA methyltransferase genes in Helicoverpa armigera following injection of pathogenic bacteria modulates expression of antimicrobial peptides and affects bacterial proliferation. J Insect Physiol (2019) 118:103939. doi: 10.1016/j.jinsphys.2019.103939
167. Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, et al. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep (2013) 14(3):269–75. doi: 10.1038/embor.2013.3
168. Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer (2011) 2(6):607–17. doi: 10.1177/1947601910393957
169. Wu P, Jie W, Shang Q, Annan E, Jiang X, Hou C, et al. DNA methylation in silkworm genome may provide insights into epigenetic regulation of response to Bombyx mori cypovirus infection. Sci Rep (2017) 7(1):16013. doi: 10.1038/s41598-017-16357-7
170. Lio CJ, Rao A. TET enzymes and 5hmC in adaptive and innate immune systems. Front Immunol (2019) 10:210. doi: 10.3389/fimmu.2019.00210
171. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet (2008) 9(6):465–76. doi: 10.1038/nrg2341
172. Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science (2010) 330(6004):622–7. doi: 10.1126/science.1190614
173. Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol (2010) 28(5):516–20. doi: 10.1038/nbt.1626
174. Li-Byarlay H, Li Y, Stroud H, Feng S, Newman TC, Kaneda M, et al. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc Natl Acad Sci USA (2013) 110(31):12750–5. doi: 10.1073/pnas.1310735110
175. Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet (2011) 27(4):127–31. doi: 10.1016/j.tig.2011.01.003
176. Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet (2014) 30(8):348–55. doi: 10.1016/j.tig.2014.06.001
177. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by Menzies. Genomics Proteomics Bioinf (2017) 15(3):177–86. doi: 10.1016/j.gpb.2016.12.005
178. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
179. Menzies KJ, Zhang H, Katsyuba E, Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol (2016) 12(1):43–60. doi: 10.1038/nrendo.2015.181
180. Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther (2017) 174:63–78. doi: 10.1016/j.pharmthera.2017.02.020
181. Gabaldón C, Legüe M, Palominos MF, Verdugo L, Gutzwiller F, Calixto A. Intergenerational pathogen-induced diapause in Caenorhabditis elegans is modulated by mir-243. mBio (2020) 11(5):20238. doi: 10.1128/mBio.01950-20
182. Azlan A, Obeidat SM, Yunus MA, Azzam G. Systematic identification and characterization of Aedes aEgypti long noncoding RNAs (lncRNAs). Sci Rep (2019) 9(1):12147. doi: 10.1038/s41598-019-47506-9
183. Mukherjee K, Amsel D, Kalsy M, Billion A, Dobrindt U, Vilcinskas A. MicroRNAs regulate innate immunity against uropathogenic and commensal-like Escherichia coli infections in the surrogate insect model. Galleria mellonella. Sci Rep (2020) 10(1):2570. doi: 10.1038/s41598-020-59407-3
184. Fu Y, Wang Y, Huang Q, Zhao C, Li X, Kan Y, et al. Long noncoding RNA lncR17454 regulates metamorphosis of silkworm through let-7 miRNA cluster. J Insect Sci (2022) 22(3):175. doi: 10.1093/jisesa/ieac028
185. Bishop C, Hussain M, Hugo LE, Asgari S. Analysis of Aedes aEgypti microRNAs in response to wolbachia wAlbB infection and their potential role in mosquito longevity. Sci Rep (2022) 12(1):15245. doi: 10.1038/s41598-022-19574-x
186. Mao W, Zeng Q, She L, Yuan H, Luo Y, Wang R, et al. Wolbachia utilizes lncRNAs to activate the anti-dengue toll pathway and balance reactive oxygen species stress in Aedes aEgypti through a competitive endogenous RNA network. Front Cell Infect Microbiol (2021) 11:823403. doi: 10.3389/fcimb.2021.823403
187. Montigny A, Tavormina P, Duboe C, San Clémente H, Aguilar M, Valenti P, et al. Drosophila primary microRNA-8 encodes a microRNA-encoded peptide acting in parallel of miR-8. Genome Biol (2021) 22(1):118. doi: 10.1186/s13059-021-02345-8
188. Feng D, Li Q, Yu H, Liu S, Kong L, Du S. Integrated analysis of microRNA and mRNA expression profiles in Crassostrea gigas to reveal functional miRNA and miRNA-targets regulating shell pigmentation. Sci Rep (2020) 10(1):20238. doi: 10.1038/s41598-020-77181-0
189. Freitak D, Schmidtberg H, Dickel F, Lochnit G, Vogel H, Vilcinskas A. The maternal transfer of bacteria can mediate trans-generational immune priming in insects. Virulence (2014) 5(4):547–54. doi: 10.4161/viru.28367
190. Tidbury HJ, Pedersen AB, Boots M. Within and transgenerational immune priming in an insect to a DNA virus. Proc Biol Sci (2011) 278(1707):871–6. doi: 10.1098/rspb.2010.1517
191. Nankabirwa V, JK T, PM M, Tylleskar T, Sommerfelt H. Child survival and BCG vaccination: a community based prospective cohort study in Uganda. BMC Public Health (2015) 15:175. doi: 10.1186/s12889-015-1497-8
192. Chang YH, Kumar R, Ng TH, Wang HC. What vaccination studies tell us about immunological memory within the innate immune system of cultured shrimp and crayfish. Dev Comp Immunol (2018) 80:53–66. doi: 10.1016/j.dci.2017.03.003
193. Freitak D, Heckel DG, Vogel H. Dietary-dependent trans-generational immune priming in an insect herbivore. Proc Biol Sci (2009) 276(1667):2617–24. doi: 10.1098/rspb.2009.0323
194. Roth O, Joop G, Eggert H, Hilbert J, Daniel J, Schmid-Hempel P, et al. Paternally derived immune priming for offspring in the red flour beetle. Tribolium castaneum. J Anim Ecol (2010) 79(2):403–13. doi: 10.1111/j.1365-2656.2009.01617.x
195. Fallet M, Montagnani C, Petton B, Dantan L, de Lorgeril J, Comarmond S, et al. Early life microbial exposures shape the Crassostrea gigas immune system for lifelong and intergenerational disease protection. Microbiome (2022) 10(1):85. doi: 10.1186/s40168-022-01280-5
196. Norouzitallab P, Biswas P, Baruah K, Bossier P. Multigenerational immune priming in an invertebrate parthenogenetic. Artemia to pathogenic Vibrio campbellii. Fish Shellfish Immunol (2015) 42(2):426–9. doi: 10.1016/j.fsi.2014.11.029
197. Roy S, Kumar V, Bossier P, Norouzitallab P, Vanrompay D. Phloroglucinol treatment induces transgenerational epigenetic inherited resistance against vibrio infections and thermal stress in a brine shrimp (Artemia franciscana) model. Front Immunol (2019) 10:2745. doi: 10.3389/fimmu.2019.02745
198. Moore RS, Kaletsky R, Murphy CT. Piwi/PRG-1 argonaute and TGF-beta mediate transgenerational learned pathogenic avoidance. Cell (2019) 177(7):1827–1841.e12. doi: 10.1016/j.cell.2019.05.024
199. Berendsen MLT, Oland CB, Bles P, Jensen AKG, Kofoed PE, Whittle H, et al. Maternal priming: bacillus calmette-guerin (BCG) vaccine scarring in mothers enhances the survival of their child with a BCG vaccine scar. J Pediatr Infect Dis Soc (2019) 91(0145-305X):17–25. doi: 10.1093/jpids/piy142
200. Qin Z, Bouteau A, Herbst C, Igyártó BZ. Pre-exposure to mRNA-LNP inhibits adaptive immune responses and alters innate immune fitness in an inheritable fashion. PloS Pathog (2022) 18(9):e1010830. doi: 10.1371/journal.ppat.1010830
201. Hamada A, Torre C, Drancourt M, Ghigo E. Trained immunity carried by non-immune cells. Front Microbiol (2018) 9:3225. doi: 10.3389/fmicb.2018.03225
202. Ottaviani E. Immunocyte: the invertebrate counterpart of the vertebrate macrophage. Invertebr Surviv J (2011) 8(1):1–4. doi: 10.1016/j.trre.2010.10.004
203. Abhyankar V, Kaduskar B, Kamat SS, Deobagkar D, Ratnaparkhi GS. Drosophila DNA/RNA methyltransferase contributes to robust host defense in aging animals by regulating sphingolipid metabolism. J Exp Biol (2018) 221(Pt 22):1248–58. doi: 10.1242/jeb.187989
204. Ji F, Wei J, Luan H, Li M, Cai Z. Study of metabolic disorders associated with BDE-47 exposure in Drosophila model by MS-based metabolomics. Ecotoxicol Environ Saf (2019) 184:109606. doi: 10.1016/j.ecoenv.2019.109606
205. Zhu Y, Cai Q, Zheng X, Liu L, Hua Y, Du B, et al. Aspirin positively contributes to Drosophila intestinal homeostasis and delays aging through targeting imd. Aging Dis (2021) 12(7):1821–34. doi: 10.14336/ad.2020.1008
206. Lafont M, Goncalves P, Guo X, Montagnani C, Raftos D, Green T. Transgenerational plasticity and antiviral immunity in the Pacific oyster (Crassostrea gigas) against Ostreid herpesvirus 1 (OsHV-1). Dev Comp Immunol (2019) 91:17–25. doi: 10.1016/j.dci.2018.09.022
207. Lafont M, Petton B, Vergnes A, Pauletto M, Segarra A, Gourbal B, et al. Long-lasting antiviral innate immune priming in the lophotrochozoan pacific oyster, Crassostrea gigas. Sci Rep (2017) 7(1):13143. doi: 10.1038/s41598-017-13564-0
208. Suarez-Ulloa V, Rivera-Casas C, Michel M, Eirin-Lopez JM. Seasonal DNA methylation variation in the flat tree oyster isognomon alatus from a mangrove ecosystem in north biscayne bay, florida. J Shellfish Res (2019) 38(1):79–88. doi: 10.2983/035.038.0108
209. Wang X, Li A, Wang W, Que H, Zhang G, Li L. DNA methylation mediates differentiation in thermal responses of Pacific oyster (Crassostrea gigas) derived from different tidal levels. Heredity (Edinb) (2021) 126(1):10–22. doi: 10.1038/s41437-020-0351-7
210. Kong N, Liu X, Li J, Mu W, Lian J, Xue Y, et al. Effects of temperature and salinity on survival, growth and DNA methylation of juvenile Pacific abalone. Haliotis discus hannai Ino. Chin J Oceanology Limnology (2017) 35(5):1248–58. doi: 10.1007/s00343-016-5185-z
211. Venkataraman YR, White SJ, Roberts SB. Differential DNA methylation in Pacific oyster reproductive tissue in response to ocean acidification. BMC Genomics (2022) 23(1):556. doi: 10.1186/s12864-022-08781-5
212. Thorson JLM, Smithson M, Beck D, Sadler-Riggleman I, Nilsson E, Dybdahl M, et al. Epigenetics and adaptive phenotypic variation between habitats in an asexual snail. Sci Rep (2017) 7(1):14139. doi: 10.1038/s41598-017-14673-6
213. Huang Z, Xiao Q, Yu F, Gan Y, Lu C, Peng W, et al. Comparative transcriptome and DNA methylation analysis of phenotypic plasticity in the pacific abalone (Haliotis discus hannai). Front Physiol (2021) 12:683499. doi: 10.3389/fphys.2021.683499
214. Farias ND, de Oliveira NFP, da Silva PM. Perkinsus infection is associated with alterations in the level of global DNA methylation of gills and gastrointestinal tract of the oyster Crassostrea gasar. J Invertebr Pathol (2017) 149:76–81. doi: 10.1016/j.jip.2017.08.007
215. Gonzalez-Romero R, Suarez-Ulloa V, Rodriguez-Casariego J, Garcia-Souto D, Diaz G, Smith A, et al. Effects of florida red tides on histone variant expression and DNA methylation in the eastern oyster Crassostrea virginica. Aquat Toxicol (2017) 186:196–204. doi: 10.1016/j.aquatox.2017.03.006
216. Geyer KK, Niazi UH, Duval D, Cosseau C, Tomlinson C, Chalmers IW, et al. The Biomphalaria glabrata DNA methylation machinery displays spatial tissue expression, is differentially active in distinct snail populations and is modulated by interactions with Schistosoma mansoni. PloS Negl Trop Dis (2017) 11(5):e0005246. doi: 10.1371/journal.pntd.0005246
217. Luviano N, Lopez M, Gawehns F, Chaparro C, Arimondo PB, Ivanovic S, et al. The methylome of Biomphalaria glabrata and other mollusks: enduring modification of epigenetic landscape and phenotypic traits by a new DNA methylation inhibitor. Epigenet Chromatin (2021) 14(1):48. doi: 10.1186/s13072-021-00422-7
218. Wang P, Wang J, Su Y, Liu Z, Mao Y. Air exposure affects physiological responses, innate immunity, apoptosis and DNA methylation of kuruma shrimp. Marsupenaeus japonicus. Front Physiol (2020) 11:223. doi: 10.3389/fphys.2020.00223
219. Gao H, Li F, Dong B, Zhang Q, Xiang J. Molecular cloning and characterisation of prophenoloxidase (ProPO) cDNA from Fenneropenaeus chinensis and its transcription injected by Vibrio Anguillarum. Mol Biol Rep (2009) 36(5):1159–66. doi: 10.1007/s11033-008-9292-6
220. Baruah K, Norouzitallab P, Linayati L, Sorgeloos P, Bossier P. Reactive oxygen species generated by a heat shock protein (Hsp) inducing product contributes to Hsp70 production and Hsp70-mediated protective immunity in Artemia franciscana against pathogenic vibrios. Dev Comp Immunol (2014) 46(2):470–9. doi: 10.1016/j.dci.2014.06.004
221. Yao D, Wang Z, Wei M, Zhao X, Aweya JJ, Zhong M, et al. Analysis of Litopenaeus vannamei hemocyanin interacting proteins reveals its role in hemolymph clotting. J Proteomics (2019) 201:57–64. doi: 10.1016/j.jprot.2019.04.013
222. Coates CJ, Decker H. Immunological properties of oxygen-transport proteins: hemoglobin, hemocyanin and hemerythrin. Cell Mol Life Sci (2017) 74(2):293–317. doi: 10.1007/s00018-016-2326-7
Keywords: trained immunity, arthropods, mollusks, metabolism, epigenetic modifications
Citation: Zhao M, Lin Z, Zheng Z, Yao D, Yang S, Zhao Y, Chen X, Aweya JJ and Zhang Y (2023) The mechanisms and factors that induce trained immunity in arthropods and mollusks. Front. Immunol. 14:1241934. doi: 10.3389/fimmu.2023.1241934
Received: 17 June 2023; Accepted: 25 August 2023;
Published: 07 September 2023.
Edited by:
Lingling Wang, Dalian Ocean University, ChinaReviewed by:
Jingguang Wei, South China Agricultural University, ChinaCopyright © 2023 Zhao, Lin, Zheng, Yao, Yang, Zhao, Chen, Aweya and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueling Zhang, emhhbmd5bEBzdHUuZWR1LmNu; Jude Juventus Aweya, amphd2V5YUBqbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.