
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 31 August 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1224634
 Nagisa Tokunoh1,2†
Nagisa Tokunoh1,2† Shigeyuki Tamiya2,3†
Shigeyuki Tamiya2,3† Masato Watanabe1
Masato Watanabe1 Toru Okamoto4,5
Toru Okamoto4,5 Jessica Anindita6,7
Jessica Anindita6,7 Hiroki Tanaka6
Hiroki Tanaka6 Chikako Ono5,8
Chikako Ono5,8 Toshiro Hirai2,9,10,11
Toshiro Hirai2,9,10,11 Hidetaka Akita6
Hidetaka Akita6 Yoshiharu Matsuura5,8,11
Yoshiharu Matsuura5,8,11 Yasuo Yoshioka1,2,5,9,10,11,12*
Yasuo Yoshioka1,2,5,9,10,11,12*Introduction: Vaccinations are ideal for reducing the severity of clinical manifestations and secondary complications of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); however, SARS-CoV-2 continues to cause morbidity and mortality worldwide. In contrast to parenteral vaccines such as messenger RNA vaccines, nasal vaccines are expected to be more effective in preventing viral infections in the upper respiratory tract, the primary locus for viral infection and transmission. In this study, we examined the prospects of an inactivated whole-virion (WV) vaccine administered intranasally against SARS-CoV-2.
Methods: Mice were immunized subcutaneously (subcutaneous vaccine) or intranasally (nasal vaccine) with the inactivated WV of SARS-CoV-2 as the antigen.
Results: The spike protein (S)-specific IgA level was found to be higher upon nasal vaccination than after subcutaneous vaccination. The level of S-specific IgG in the serum was also increased by the nasal vaccine, although it was lower than that induced by the subcutaneous vaccine. The nasal vaccine exhibited a stronger defense against viral invasion in the upper respiratory tract than the subcutaneous vaccine and unimmunized control; however, both subcutaneous and nasal vaccines provided protection in the lower respiratory tract. Furthermore, we found that intranasally administered inactivated WV elicited robust production of S-specific IgA in the nasal mucosa and IgG in the blood of mice previously vaccinated with messenger RNA encoding the S protein.
Discussion: Overall, these results suggest that a nasal vaccine containing inactivated WV can be a highly effective means of protection against SARS-CoV-2 infection.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a pandemic with over > 760 million cases and > 6.9 million deaths worldwide as of April 2023, according to World Health Organization reports. SARS-CoV-2 infection leads to a clinical syndrome ranging from mild to severe Coronavirus Disease 2019 (COVID-19), including acute lung injury and acute respiratory disease syndrome (1). SARS-CoV-2 infection is also reported to cause strokes, myocarditis, heart failure, and hyperinflammatory shock syndrome (2–4). High-risk groups, including the elderly and those with chronic respiratory disease, are at an increased risk of severe COVID-19, which is associated with respiratory failure and death (1, 5).
Vaccination is a key strategy to prevent pneumonia, secondary complications, and mortality from SARS-CoV-2 infection at the population level, especially among the elderly and those with chronic respiratory disease (6, 7). To date, injectable vaccines such as messenger RNA (mRNA) vaccines, adenovirus vector-based vaccines, subunit vaccines, and inactivated whole-virion (WV) vaccines against SARS-CoV-2 have been developed and used worldwide (8–11). In particular, mRNA vaccines administered intramuscularly to people worldwide have been effective in reducing disease severity and symptomatic cases (12–14).
Human SARS-CoV-2 infects the upper respiratory tract causing mild disease; however, its progression to the lower respiratory tract leads to pneumonia and serious disease (15, 16). Further, the upper respiratory tract is an important site for the initiation of viral transmission through coughing, sneezing, and talking (15, 16). In the early phase of SARS-CoV-2 infection, the spike (S) protein specifically binds to cellular entry receptors such as angiotensin-converting enzyme 2, which is less expressed in the lungs compared with that in the upper respiratory tract in humans (17–19). Previous reports have shown that during a human challenge with SARS-CoV-2, the virus replicates at significantly higher levels in the nose (20); however, most of the current SARS-CoV-2 vaccines which are administered parenterally via intramuscular injection, fail to induce virus-specific IgA production in the upper respiratory tract, even though they robustly induce production of virus-specific IgG in the blood (21–23). In fact, these vaccines can protect against COVID-19 while providing limited protection against respiratory virus replication and shedding in the upper respiratory tract in non-human primates (24–26). Collectively, these parenteral vaccines are not fully effective in preventing SARS-CoV-2 infection of the upper respiratory tract, though virus-specific IgG in blood may protect against disease exacerbation. Therefore, blocking the initial infection, generation, and shedding of SARS-CoV-2 in the upper respiratory tract is critical for vaccine development.
In contrast to parenteral vaccines, nasally administered vaccines (nasal vaccines) induce the production of antigen-specific IgG and IgA in the blood and respiratory tract, respectively (27, 28). To date, virus-specific IgA in the upper respiratory tract induced by nasal vaccines has shown protection against influenza virus and SARS-CoV-2 infection more efficiently than that with parenteral vaccines (29–34). For example, Hemmi et al. showed that virus-specific IgA induced by a nasal vaccine containing the recombinant S protein from SARS-CoV-2 adjuvanted with CpG oligonucleotides was highly protective against SARS-CoV-2 challenge in the upper respiratory tract of mice (33). In addition, nasal vaccine using the recombinant receptor binding domain of S protein from SARS-CoV-2 adjuvanted with Riboxxim, a Toll-like receptor (TLR)3 ligand, induced robust mucosal IgA antibody in murine models (34). Furthermore, virus-specific T cell responses, particularly respiratory tissue-resident memory CD8+ T cells and CD4+ T cells, induced by nasal vaccine and infection protect against severe disease and mortality induced by influenza virus and SARS-CoV-2 (35–37), although T cell responses induced by the parenteral vaccine are limited to peripheral locations, failing to either access or be maintained at the respiratory mucosa (35). Therefore, a nasal vaccine could be a highly effective means of preventing SARS-CoV-2 infection in the respiratory tract.
Nasal vaccines based on adenovirus vectors and subunit vaccines against SARS-CoV-2 have been developed in preclinical studies and clinical trials (38). Recent research indicates that the intranasal administration of AstraZeneca’s ChAdOx-1 vaccine has poor immunogenicity in humans previously immunized with mRNA vaccines, despite being effective in mice (35, 39). In addition, these vaccines have several limitations. For example, adenovirus vector-based vaccines are ineffective at inducing adenovirus-specific antibodies following the first dose (40). Subunit vaccines require adjuvants to elicit immune responses (41, 42) because intranasal administration of antigens alone does not elicit antigen-specific antibodies. However, no approved adjuvants for nasal vaccines in humans exist, although several adjuvants, such as c-di-GMP, CpG oligonucleotides, poly(I:C), and Riboxxim, have been experimentally implemented in nasal vaccines (34, 43–45).
Inactivated WVs have shown high preventive efficacy, preservation, and transport, and retain sufficient antigenic properties to elicit immune responses (46, 47). Previous reports on influenza viruses have shown that inactivated WV induces strong immune responses due to strong adjuvant activity because inactivated WV contains single-stranded viral RNA (ssRNA), which acts as a ligand for TLR7 (48, 49). In SARS-CoV-2, ssRNA also acts as an activator of TLR7/8 to activate inflammation and immunity (50), indicating strong adjuvant activity of inactivated WV of SARS-CoV-2. To date, parenteral inactivated WVs are available against SARS-CoV-2, and have shown promise in preclinical and clinical studies, leading to approval for their use (51, 52). However, nasal administration of these vaccines remains to be explored. Therefore, in this study, we focused on using the inactivated WV as a nasal vaccine against SARS-CoV-2. We examined the potential of an inactivated WV as an antigen to induce S-specific IgA in the nose and S-specific IgG in blood, as well as to prevent viral infection in both the upper and lower respiratory tracts. We also examined the potential of the nasal inactivated WV as a booster mice previously vaccinated with an mRNA vaccine expressing the S protein.
Specific pathogen-free 10 week-old BALB/c mice were purchased from SLC (Hamamatsu, Japan). The mice were housed in a room with a 12-h:12-h light:dark cycle (lights on, 8:00 am; lights off, 8:00 pm) and unrestricted access to food and water. All animal experiments with or without SARS-CoV-2 were conducted in accordance with the guidelines of Osaka University for the ethical treatment of animals and were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University, Japan (approval numbers: BIKEN-AP-R02-09-0).
The Gamma strain (hCoV-19/Japan/TY7-503/2021) of SARS-CoV-2 was obtained from the National Institute of Infectious Diseases (Tokyo, Japan). The virus was expanded in VeroE6/TMPRSS2 cells (JCRB Cell Bank, Osaka, Japan) and stored at −80 °C until use. VeroE6/TMPRSS2 cells were cultured in DMEM (Nacalai Tesque, Kyoto, Japan) containing 10% fetal calf serum (FCS) and 1% penicillin/streptomycin. Mice were intranasally challenged with MA10, a mouse-adapted SARS-CoV-2. MA10 was generated using a circular polymerase extension reaction as described previously (53). The SARS-CoV-2 NIID strain (2019-nCoV_Japan_TY_WK-5212020) served as the backbone of MA10. MA10 contains seven mutations, which were introduced adaptively into SARS-CoV-2 during serial passages in BALB/c mice (54). Each experiment with live SARS-CoV-2 was performed within a biosafety level 3 facility at Osaka University, adhering to stringent guidelines.
VeroE6/TMPRSS2 cells in 225 cm2 flasks (Corning, Corning, NY, USA) were infected with the Gamma strain of SARS-CoV-2 at 0.01 multiplicity of infection (MOI) at 37 °C. After three days, the cell culture medium was collected via centrifugation at 3000 rpm for 10 min to remove debris. Live SARS-CoV-2 in the supernatant was inactivated using β-propiolactone (Wako, Osaka, Japan) at a concentration of 1:4000 (v/v) at 4 °C for 24 h, then incubated at 37 °C for 2 h to hydrolyze the remaining β-propiolactone. To confirm viral inactivation, we performed a plaque assay before purification. Briefly, VeroE6/TMPRSS2 cell monolayers (1 × 106 cells/well) in 6-well plates (Corning) were incubated with medium as a negative control, live SARS-CoV-2 (diluted 104-fold with medium) as a positive control, or inactivated WV (undiluted) at 37 °C for 2 h. The cells were then rinsed twice with phosphate-buffered saline (PBS) and covered with agar (final 1%; SeaPlaque TM Agarose, Lonza, Basel, Switzerland) in a medium containing 2% FCS and 0.5% penicillin/streptomycin. The plates were then incubated at 37 °C. After three days, the cells were fixed with 10% formalin (Nacalai Tesque) and the agar was removed. The cells were then stained with 1% crystal violet (Tokyo Chemical Industry, Tokyo, Japan) and the plaques were counted. Next, the inactivated WV was purified using sucrose density gradient centrifugation. Briefly, using 20% sucrose, inactivated WV pellets were collected by centrifugation at 32,000 rpm at 4 °C for 2 h. The pellets were resuspended in PBS, and to exclude artefacts, purified through a 15–60% sucrose gradient at 30,000 rpm at 4 °C for 2 h. After centrifugation, 23 fractions (1.5 mL each) were collected from the top to bottom, and the expression of S protein (16–19 fractions) was confirmed by western blotting. Subsequently, to exclude sucrose, 16-19 fractions were diluted with PBS and centrifuged at 30,000 rpm at 4 °C for 3 h. Finally, the pellets were resuspended in PBS and the amount of inactivated WV was quantified using a Pierce protein assay kit (Thermo Fisher Scientific, Hampton, NH, USA) with a bovine serum albumin standard.
To confirm viral inactivation in vivo, mice were treated with SARS-CoV-2 (Gamma strain; 1.7 × 105 Plaque-forming unit; PFU), SARS-CoV-2 MA10 (2 × 105 PFU), or inactivated WV (3 μg or 10 μg) in 20 μL of PBS under anesthesia. The nasal turbinate was harvested and homogenized in DMEM containing 2% FCS and 1% penicillin/streptomycin 3 days after treatment, followed by centrifugation and supernatant collection. To titrate the infectious virus, the supernatant of the nasal turbinate was serially diluted in DMEM containing 2% FCS and 1% penicillin/streptomycin. The plaque assay was performed as described above.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in accordance with previous publications (32). Briefly, purified proteins were mixed in sample buffer solution (Nacalai Tesque) containing 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA) at 1:1 (v/v) ratio and heated at 95 °C for 5 min before being loaded onto a 10% Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad, Hercules, CA, USA). Following electrophoresis, the gels were stained with Coomassie Brilliant Blue (Nacalai Tesque). After gel electrophoresis, protein bands were transferred from the SDS-PAGE gel onto polyvinylidene fluoride membranes (Bio-Rad). After blocking in 5% skim milk (w/v) diluted in distilled water, the transferred membrane was washed thrice with PBST (0.05% Tween-20 in PBS) and incubated with mouse anti-SARS-CoV-2 (2019-nCoV) spike antibody (clone: #42; Sino Biological, Beijing, China) or rabbit anti-SARS membrane protein antibody (polyclonal; Novus Biologicals, Littleton, CO, USA). The membrane was then washed thrice with PBST, and incubated with a rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (Abcam Ltd., Cambridge, UK) or goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Abcam Ltd.), and detected using a ChemiDoc Touch Imaging System (Bio-Rad). The amount of S protein in inactivated WV was determined using western blotting (Supplementary Figure 1B). We used recombinant S protein as standard (from 4.3 ng/well to 69.3 ng/well). After SDS-PAGE, the proteins were transferred onto the membrane for western blotting. The transferred membrane was incubated with rabbit anti-SARS-CoV-2 (2019-nCoV) spike antibody (Polyclonal IgG; Sino Biological). The membrane was then washed thrice with PBST and incubated with a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Abcam Ltd.). The three points (34 ng/well, 68 ng/well, and 136 ng/well) for inactivated WV were averaged. Immunoreactive bands were quantified using Image J software (NIH Image, Bethesda, MD, USA).
The inactivated WV was directly analyzed using a Tecnai G2 20 twin transmission electron microscopy (TEM; FEI Company, Hillsboro, OR, USA) at 200 kV. Carbon-coated copper grids with 250 mesh (STEM Co. Ltd, Tokyo, Japan) were glow-discharged using DII-29020HD (JEOL, Tokyo, Japan). The inactivated WV was fixed in 2% glutaraldehyde overnight at 4 °C. Samples were diluted in PBS to 20 μg/mL immediately before measurement, and 10 μL of the sample was applied to the grids for 5 min. Four-diluted EM stainer (Nisshin EM, Tokyo, Japan) was then used to stain the sample on the grid for 30 min. The specimen was finally gently blotted from the side with filter paper and air-dried before imaging. Images were recorded on an Eagle 2 K × 2 K CCD camera with a defocus of -2 μm at a nominal magnification of 25,000× or 50,000×.
The cDNA of the S protein ectodomain from the Gamma strain of SARS-CoV-2 with a C-terminal hexahistidine tag (His-tag) was cloned into the pcDNA3.1 expression plasmid (Thermo Fisher Scientific). The foldon sequence (GYIPEAPRDGQAYVRKDGEWVLLSTFL) derived from the fibritin of bacteriophage T4 was integrated at the C terminus of the S protein. S protein production was performed in accordance with previous publications (32).
For the transcription of mRNA expressing the S protein of SARS-CoV-2, template pDNA encoding the S protein (Wuhan-Hu-1) was prepared using polymerase chain reaction. The pDNA-spike was linearized using restriction enzymes. After phenol-chloroform extraction and ethanol precipitation, the linearized pDNA was transcribed into mRNA using the MEGAscript™ T7 transcription kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. N1-methylpseudouridine was incorporated into the mRNA by replacing uridine. Residual dsRNA was removed as previously described (55). The 5′ cap was added according to the ScriptCap Cap 1 Capping System protocol (Madison, WI, USA). The 3′ poly(A) tail was added according to the protocol provided with the poly(A) Tailing Kit (Thermo Fisher Scientific). A lipid mixture containing SM-102 (Cayman Chemical Company, Ann Arbor, MI, USA), DMG-PEG2000 (NOF Corporation, Tokyo, Japan), DSPC (NOF Corporation), and cholesterol (Sigma-Aldrich) in an ethanol solution (SM-102/DSPC/cholesterol/DMG-PEG2000 = 50/10/38.5/1.5) were prepared. The mRNA-lipid nanoparticles (LNPs) were produced by mixing the mRNA dissolved in acetic acid/NaOH buffer (pH 5.0) and the lipid mixture in ethanol at an N/P ratio of 5.5 using a NanoAssemblr® instrument (Precision Nanosystems, Vancouver, Canada) at a flow rate ratio of 3:1. The external solution of the mRNA-LNPs was replaced with PBS (Nacalai Tesque) by ultrafiltration using Amicon Ultra-4-100K centrifugal units. The size and zeta potential of mRNA-LNPs were measured using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). The encapsulation efficiency of mRNA was measured using the RibogreenTM reagent (Invitrogen, Carlsbad, CA, USA).
For nasal vaccination, BALB/c mice were administered with inactivated WV (3 μg/mouse) in a total volume of 5 μL, divided equally between both nostrils. Subcutaneous vaccination was achieved via administration of the BALB/c mice at the base of the tail with inactivated WV (3 μg/mouse) combined with Alhydrogel (alum; InvivoGen, San Diego, CA, USA) as an adjuvant (50 μg/mouse) in a total volume of 50 μL. For both routes, the mice were administered on days 0 and 21. Unimmunized mice were used as controls. On day 32, serum, nasal wash, and bronchoalveolar lavage fluid (BALF) samples were collected and stored at -80 °C. The nasal cavity was gently flushed using 400 μL of PBS to obtain nasal wash samples. BALF was obtained via lung lavage with 1.2 mL PBS. In some experiments, mRNA expressing the SARS-CoV-2 Spike (1 μg/mouse) was administered intramuscularly to mice in a total volume of 50 μL. On day 21 after mRNA vaccination, the mice were intranasally administered with inactivated WV (3 μg/mouse) in a total volume of 5 μL, divided equally between both nostrils. On day 28, serum and nasal wash samples were collected.
We determined the levels of S-specific antibodies in the serum, nasal wash, and BALF using enzyme-linked immunosorbent assay (ELISA). To detect SARS-CoV-2-specific total IgG and IgA, ELISA plates (Corning, Corning, NY, USA) were coated overnight at 4°C with S protein of the Gamma strain in PBS (1 μg/mL or 10 μg/mL). ELISA was performed in accordance with previous publications (32). Endpoint titers were determined using the following procedure: The background value was subtracted from the OD value. A subtracted OD value of 0.1 or more was regarded as positive, and the maximum dilution to give a positive result was used as the endpoint titer.
For the neutralization assay, VeroE6/TMPRSS2 cells (1.2 × 104 cells/well) were added to a 96-well half-white plate (Greiner BIO-ONE, Kremsmunster, Austria) and incubated in DMEM containing 10% FCS and 1% penicillin/streptomycin at 37°C for 24 h. On the day after incubation, serum samples and nasal wash samples were evaluated using 50-819,200-fold, or 4-65,536-fold serial dilutions of undiluted solutions. The serial dilutions were mixed 1:1 with pseudotyped viruses, replication-deficient vesicular stomatitis virus (VSV) bearing Gamma spike of SARS-CoV-2, and incubated at 37°C for 1 h. After removing the growth medium from the cells, 50 μL of serum/virus or nasal wash/virus mixture was added to the cells and then incubated at 37°C for 48 h. Following 48 h post-infection, 50 μL of ONE-Glo™ EX Reagent (Promega, Madison, WI, USA) was pipetted into each well and mixed. Luminescence was measured using a microplate reader (CORONA electric, SH-9000, Hitachi High-Tech Corporation, Tokyo, Japan).
To evaluate the percentage of germinal center (GC) B cells in the nasal passage, lymphocytes were collected seven days after the last immunization and analyzed via flow cytometry. The nasal passage lymphocytes were added to a 96-well plate, and incubated with an anti-mouse CD16/CD32 antibody (1:400 dilution; clone:93; BioLegend, San Diego, CA, USA), PE anti-mouse CD45 antibody (1:200 dilution; clone:30-F11; BioLegend), BV421 anti-mouse B220 antibody (1:200 dilution; clone: RA3-6B2; BioLegend), AF647 anti-mouse GL7 antibody (1:200 dilution; clone: GL7; BioLegend), and PE/Cy7 anti-mouse CD95 (Fas) antibody (1:200 dilution; clone: SA367H8; BioLegend) in PBS with 2% FCS, 1 mM EDTA (DOJINDO, Kumamoto, Japan), and 0.05% azide (Wako) for 30 min at 4°C. GC B cells were defined as CD45+ B220+ Fas+ GL7+ cells. Flow cytometry was performed using the CytoFLEX Flow Cytometer (Beckman Coulter, Brea, CA, USA). Kaluza software (Beckman Coulter) was used for analyzing the flow cytometry data.
On day 32 after vaccination, splenocytes (1 × 106 cells/well) were added to a 96-well plate. Subsequently, the cells were stimulated for three days at 37 °C with S protein or left unstimulated (final concentration: 20 μg/mL). Post-incubation, the levels of interferon (IFN)-γ and interleukin (IL)-13 in the supernatants were analyzed using commercial ELISA kits (BioLegend for IFN-γ; R&D Systems, Minneapolis, MN, USA, for IL-13), as per manufacturer’s instructions. Standards supplied with the kits were used to quantify the cytokine levels.
Fourteen days after the last immunization, the mice were challenged intranasally with SARS-CoV-2 MA10 (5 × 104 PFU) in 5 μL of PBS under anesthesia. Unimmunized mice were used as controls. After challenge, the nasal turbinates were harvested and homogenized in 500 μL of DMEM containing 2% FCS and 1% penicillin/streptomycin. Next, 500 μL of DMEM containing 2% FCS and 1% penicillin/streptomycin was added, followed by centrifugation and collection of the supernatant. To titrate the infectious virus, the supernatant of the nasal turbinate was serially diluted in DMEM containing 2% FCS and 1% penicillin/streptomycin. The plaque assay was performed as previously mentioned.
Fourteen days after the last immunization, mice were infected intranasally with SARS-CoV-2 MA10 (5 × 104 PFU or 2 × 105 PFU) in 20 μL of PBS under anesthesia. Unimmunized mice were used as controls. After the infection, the body weight and survival of the mice were observed for eight days. The day of death was defined as the specific day on which the body weight of the mice fell below 75% of their initial weight.
Statistical analyses were performed using Prism software (GraphPad Software, San Diego, CA, USA). All data are presented as mean ± standard deviation (SD). One-way ANOVA was used to determine significance differences, followed by Tukey’s test. P < 0.05 was considered to indicate statistical significance.
To generate inactivated WV, we used a Gamma strain (hCoV-19/Japan/TY7-503/2021) as the seed virus. The virus was propagated in VeroE6 cells expressing transmembrane serine protease 2 (TMPRSS2) and was inactivated using β-propiolactone (Figure 1A). Using a plaque formation assay, we confirmed that β-propiolactone completely inactivated SARS-CoV-2 (Figure 1B). In addition, no live virus was detected in the nasal turbinates of mice intranasally administrated inactivated WV using plaque formation assay (Supplementary Figure 1A). Inactivated WV was purified using sucrose density gradient centrifugation and the purity of the inactivated WV was confirmed using SDS-PAGE (Figure 1C) and western blotting (Figure 1D). SDS-PAGE analysis revealed three major bands. Bands of approximately 150, 50, and 15 kDa indicated S protein, nucleocapsid (N) protein, and membrane (M) protein, respectively (56). Western blot analysis also showed bands for the S and M proteins (Figure 1D). Next, we evaluated the amount of S protein in inactivated WV using western blotting. As a result, we found that 3 μg of inactivated WV contained approximately 2.1 μg of S protein (Supplementary Figure 1B). Furthermore, using transmission electron microscopy (TEM; Figure 1E), we demonstrated that the inactivated WV retained the structure of the virus. These results suggest that inactivated WV could be used as a vaccine antigen.
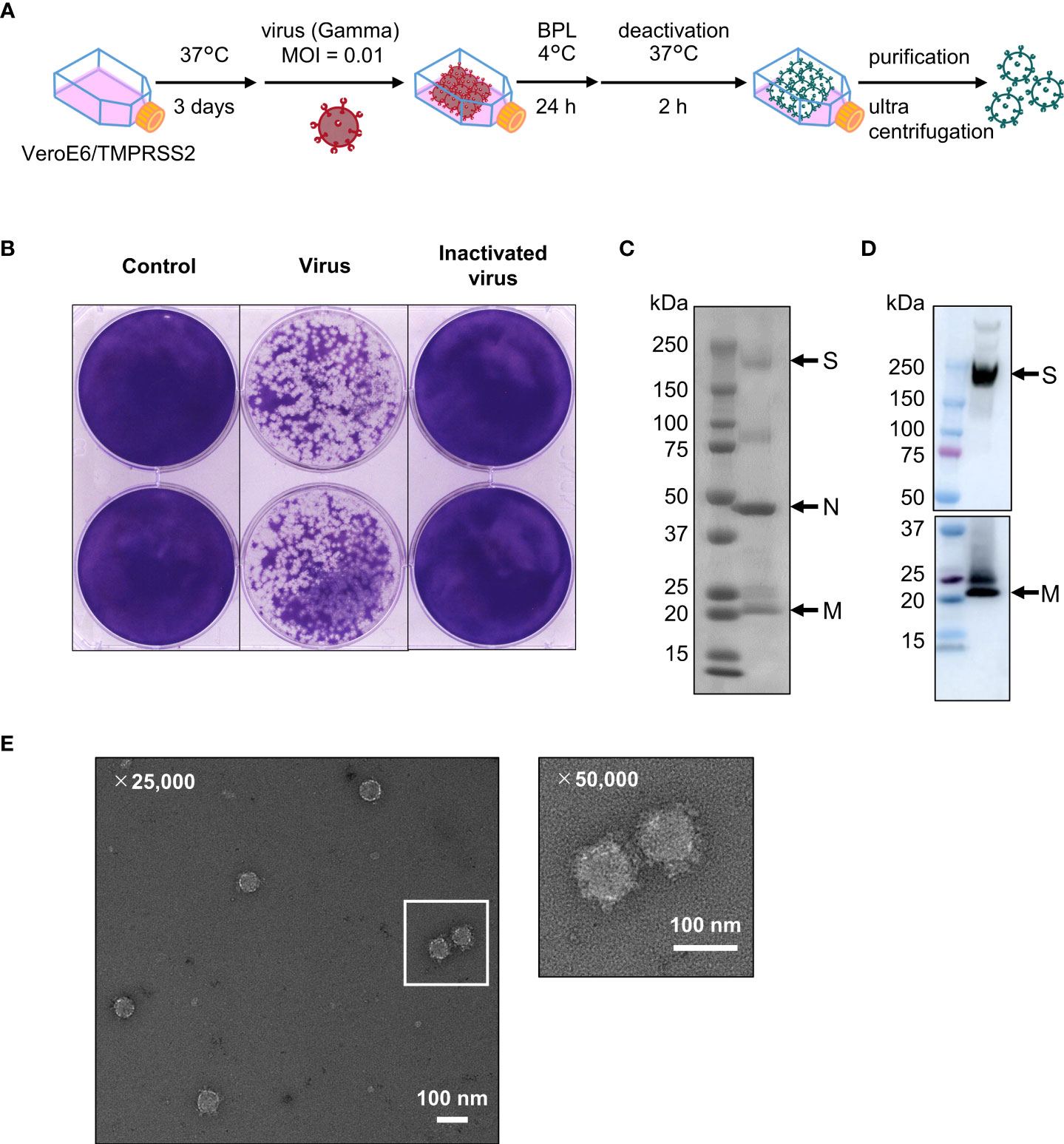
Figure 1 Inactivated whole-virion (WV) vaccine retains the protein composition and the structure of the original virus. (A) Scheme of inactivated WV (Gamma strain) production. (B) VeroE6/TMPRSS2 cells were treated with medium as a negative control, inactivated WV (undiluted) and live virus (diluted 104-fold with medium) at 37 °C for 3 days, followed by plaque counting. (C, D) Inactivated WV were validated using (C) SDS-PAGE; left: marker, right: sample and (D) western blotting; left: marker, right: S protein and M protein. Each experiment was performed in duplicate. (E) TEM micrograph of SARS-CoV-2 virus particles, Scale bar, 100 nm.
To evaluate the functionality of nasal vaccines, mice were immunized intranasally (nasal vaccine) with inactivated WV without an adjuvant or subcutaneously (subcutaneous vaccine) with inactivated WV and alum as an adjuvant. The endpoint titers of S protein-specific IgG and IgA in the serum, bronchoalveolar lavage fluid (BALF), and nasal washes after vaccination were analyzed using ELISA. The endpoint titers of S-specific IgG in the serum of the nasal vaccine group were significantly higher than those in the unimmunized control group but lower than those in the subcutaneous vaccine group (Figure 2A). Subcutaneous vaccines induced significantly higher levels of S-specific IgG in the serum, BALF, and nasal washes than those with nasal vaccines (Figures 2A–C). In contrast, the endpoint titers of S-specific IgA in the serum (Figure 2D) and nasal wash (Figure 2F) were significantly higher in the nasal vaccine group than in the subcutaneous vaccine and unimmunized control groups, although no change was observed in BALF (Figure 2E). To examine neutralizing antibody titer in serum and nasal wash from mice vaccinated with inactivated WV, we used a pseudotyped virus displaying a Gamma spike of SARS-CoV-2. The serum from intranasally and subcutaneously vaccinated mice neutralized the pseudotyped virus, whereas the serum from non-vaccinated mice was ineffective (Figure 2G). In contrast, the nasal wash from intranasally vaccinated mice tended to neutralize compared to that from subcutaneously vaccinated mice and non-vaccinated mice (Figure 2H). Next, to evaluate germinal center (GC) B cells in the nasal passage induced by vaccination, we examined the percentage of GC B cells among total B cells using flow cytometry (Figure 3A; Supplementary Figure 2). Seven days after the last immunization, the percentage of GC B cells among the total B cells was significantly higher in the nasal vaccine group than in the subcutaneous vaccine and unimmunized control groups (Figure 3A). These results indicated that the inactivated WV nasal vaccine strongly induced S-specific IgA in the upper respiratory tract.
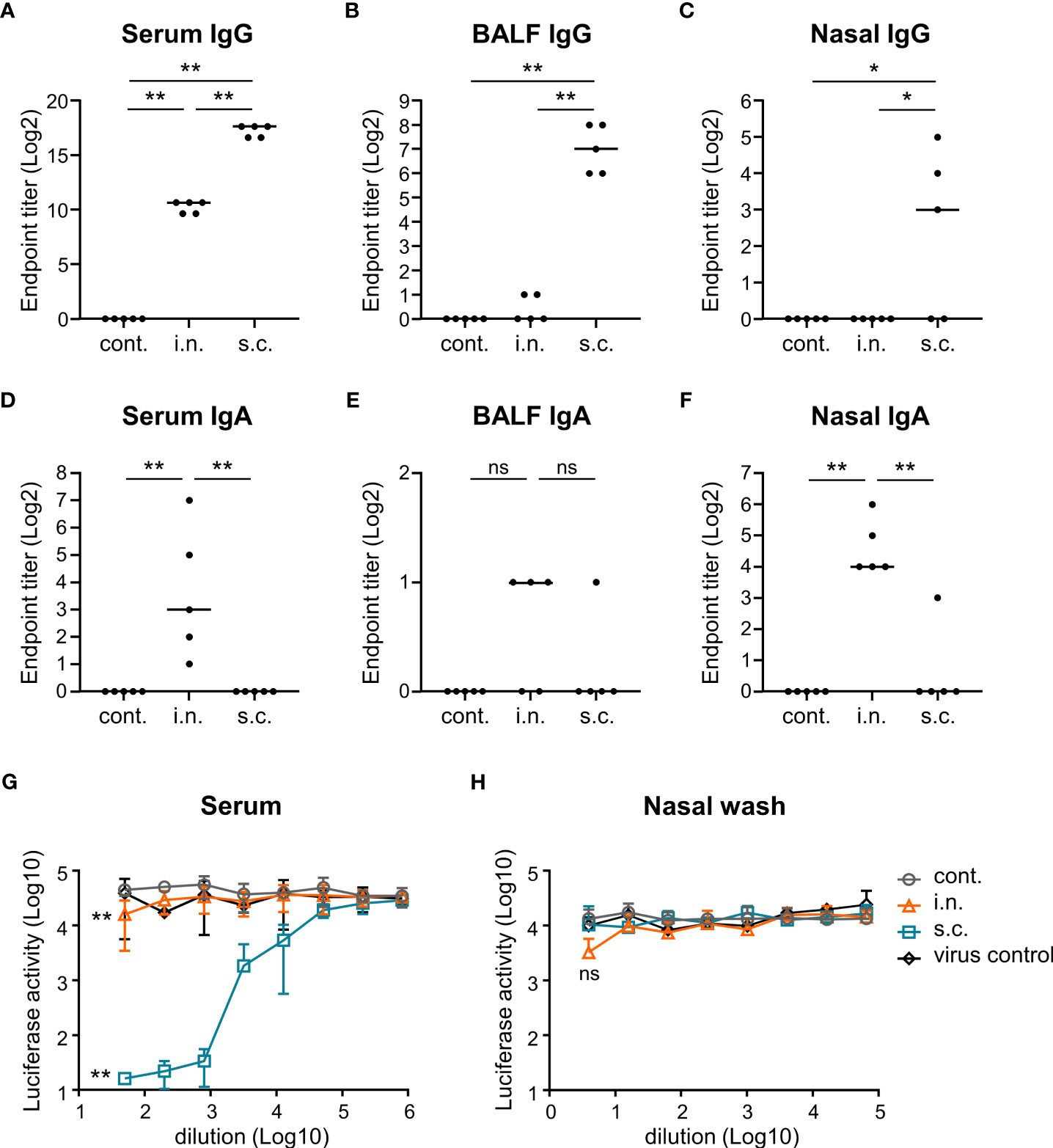
Figure 2 Nasal administration of the inactivated whole-virion (WV) vaccine induces S protein-specific IgG in serum and IgA in nasal wash. (A–H) Mice were immunized twice with inactivated WV intranasally (i.n.) or with inactivated WV and alum subcutaneously (s.c.). Unimmunized mice were used as controls (cont.). (A–F) Endpoint titers of S protein-specific IgG in (A) serum, (B) bronchoalveolar lavage fluid (BALF), and (C) nasal wash, and endpoint titers of S protein-specific IgA in (D) serum, (E) BALF, and (F) nasal wash were evaluated. (G, H) Measurement of neutralization against vesicular stomatitis virus-based pseudotyped viruses displaying Gamma spike of SARS-CoV-2 in (G) serum samples and (H) nasal wash samples. The significance of differences in the (G) 50-fold-diluted serum samples and (H) 4-fold-diluted nasal wash samples was evaluated. The data is presented as mean ± SD. Each experiment was performed in duplicate. (A–H) n = 5 per group. (A–H) *P < 0.05; **P < 0.01 as indicated by Tukey’s test. ns, not statistically significant.
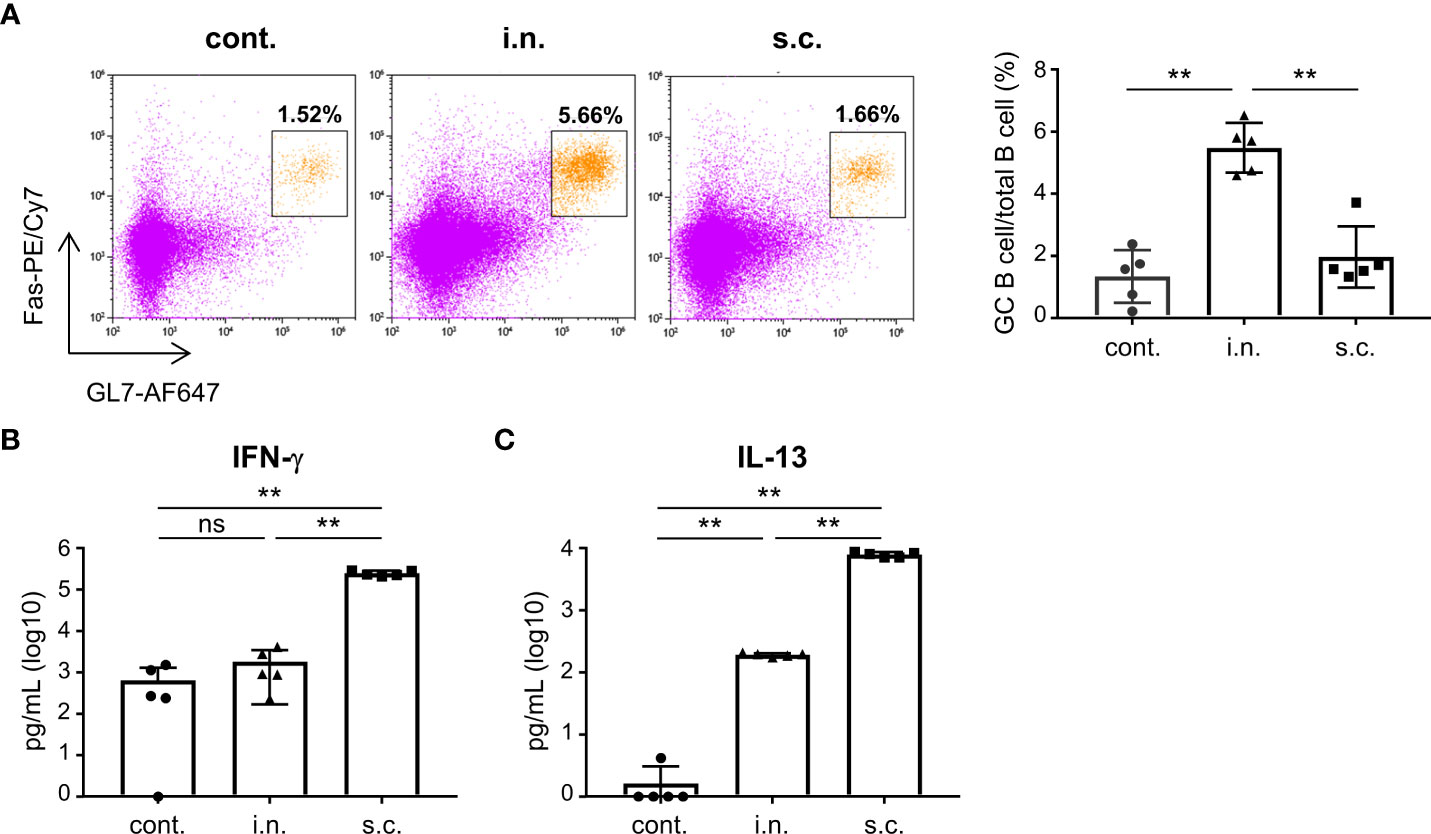
Figure 3 Nasal administration of the inactivated whole-virion (WV) vaccine induces germinal center (GC) B cells in the nasal passage. (A–C) Mice were immunized twice with inactivated whole-virion (WV) vaccine intranasally (i.n.) or with inactivated WV and alum subcutaneously (s.c.). Unimmunized mice were used as controls (cont.). (A) The percentage of germinal center (GC) B cells (CD45+ B220+ Fas+ GL7+) among the total B cells in the nasal passage were measured using flow cytometry. (B, C) Splenocytes were cultured in the presence or absence of S protein for three days, after which the levels of (B) IFN-γ and (C) IL-13 in the supernatant were measured using ELISA. The data is presented as mean ± SD. Each experiment was performed in duplicate. (A–C) n = 5 per group. (A–C) **P < 0.01 as indicated by Tukey’s test.
To examine the SARS-CoV-2-specific CD4+ T cell response induced by vaccination, we examined cytokine production in splenocytes. Splenocytes from vaccinated mice were stimulated with S protein, and the amount of cytokines in the supernatant was analyzed. The levels of IFN-γ and IL-13 (cytokines associated with T helper type 1 and 2 cells, respectively) produced by splenocytes after S protein stimulation were significantly higher in the subcutaneous vaccination group than in the nasal vaccination and unimmunized control groups (Figures 3B, C). In contrast, there was no difference in the level of IFN-γ between the unimmunized control and nasal vaccine groups, whereas the level of IL-13 in the nasal vaccine group was significantly higher than that in the unimmunized controls (Figures 3B, C).
To evaluate the defensive effects of nasal vaccination on the upper respiratory tract, vaccinated mice were challenged intranasally with a mouse-adapted strain of SARS-CoV-2 (SARS-CoV-2 MA10; 5 × 104 PFU) (54) in a total volume of 5 μL. In this model, a plaque assay was used to measure the viral titer in the nasal turbinates as an indicator of vaccination effectiveness. After challenge, the viral titer in the nasal turbinates of the nasal vaccine group was significantly lower than that in the subcutaneous vaccine and unimmunized control groups (Figure 4A). In addition, the virus titer in the nasal turbinates of the subcutaneous vaccine group was significantly lower than that in the unimmunized control group (Figure 4A). Furthermore, to assess the defensive effects of nasal vaccination on the lower respiratory tract, vaccinated mice were challenged intranasally with SARS-CoV-2 MA10 (5 × 104 PFU or 2 × 105 PFU) in a total volume of 20 μL. In this model, body weight loss and survival were measured as indicators of vaccination effectiveness and these parameters were monitored for eight days after infection. At a low-titer challenge (5 × 104 PFU), the unimmunized controls lost weight and some of them died after the challenge, whereas mice in the nasal vaccine and subcutaneous vaccine groups did not (Figures 4B, C). In addition, at a high titer challenge (2 × 105 PFU), the subcutaneous vaccine group completely protected from weight loss and death (Figures 4D, E). The nasal vaccine group was also completely protected from death, even though it showed a body weight decrease (Figures 4D, E). These results suggest that vaccination with intranasally inactivated WV can protect against upper and lower respiratory tract infections caused by SARS-CoV-2.
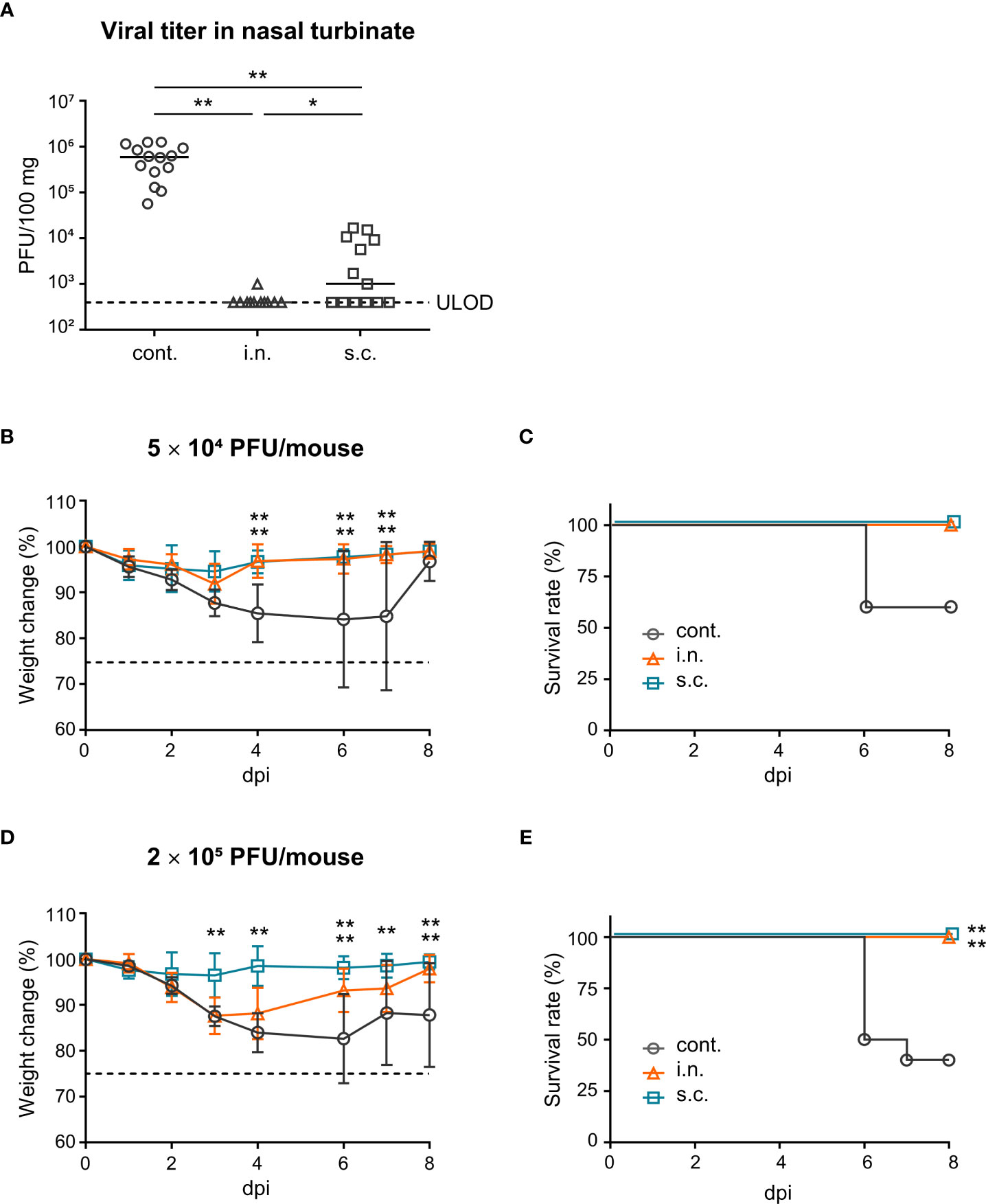
Figure 4 Nasal administration of the inactivated whole-virion (WV) vaccine protects against upper and lower respiratory tract infections caused by SARS-CoV-2. (A–E) Mice were immunized twice with inactivated whole-virion (WV) vaccine intranasally (i.n.) or with inactivated WV and alum subcutaneously (s.c.). Unimmunized mice were used as controls (cont.). (A) Mice were intranasally infected with SARS-CoV-2 MA10 (5 × 104 PFU) in 5 μL of PBS and the SARS-CoV-2 titer in nasal turbinate was assayed using the plaque assay. (B–E) Mice were intranasally infected with SARS-CoV-2 MA10 (B, C; 5 × 104 PFU/mouse, D, E; 2 × 105 PFU/mouse) in 20 μL of PBS; (B, D) body weight loss and (C, E) survival were monitored. The data is presented as the mean ± SD. Each experiment was performed in duplicate. (A) n = 5 per group. (B, C) n = 4–5 per group. (D, E) n = 10 per group. (A, B, D) *P < 0.05; **P < 0.01 as indicated by Tukey’s test. (C, E) **P < 0.01 vs. the control group as indicated by comparing Kaplan–Meier curves using the Log rank test.
Finally, we examined whether systemic priming with an mRNA vaccine, followed by intranasal boosting with the inactivated WV, could enhance antibody responses in the nasal cavity. The mice were divided into four groups (Figure 5A). Group 1 was vaccinated intranasally with inactivated WV for priming with no boosting (prime: i.n., boost: none). Group 2 was vaccinated intranasally with the inactivated WV for both priming and boosting (prime: i.n., boost: i.n.). Group 3 was intramuscularly vaccinated with an mRNA vaccine encoding the S protein for priming and intranasally vaccinated with the inactivated WV for boosting (prime: i.m., boost: i.n.). Group 4 was intramuscularly vaccinated with the mRNA vaccine for both priming and boosting (prime: i.m., boost: i.m.). Seven days after final vaccination, the endpoint titers of S-specific IgA in the nasal mucosa of group 2 were found to be significantly higher than those in groups 1 and 4 (Figure 5B). Further, the endpoint titers of S-specific IgA in nasal mucosa were significantly higher in group 3 than in groups 1 and 4 (Figure 5B). The endpoint titers of S-specific IgG in the serum of group 4 were significantly higher than that of groups 1, 2, and 3 (Figure 5C). Further, the endpoint titers of S-specific IgG in the serum of group 3 were significantly higher than those in group 1 but comparable to those in group 2 (Figure 5C). Our data suggest that inactivated WV can be used as a booster for individuals already administered the mRNA vaccine.
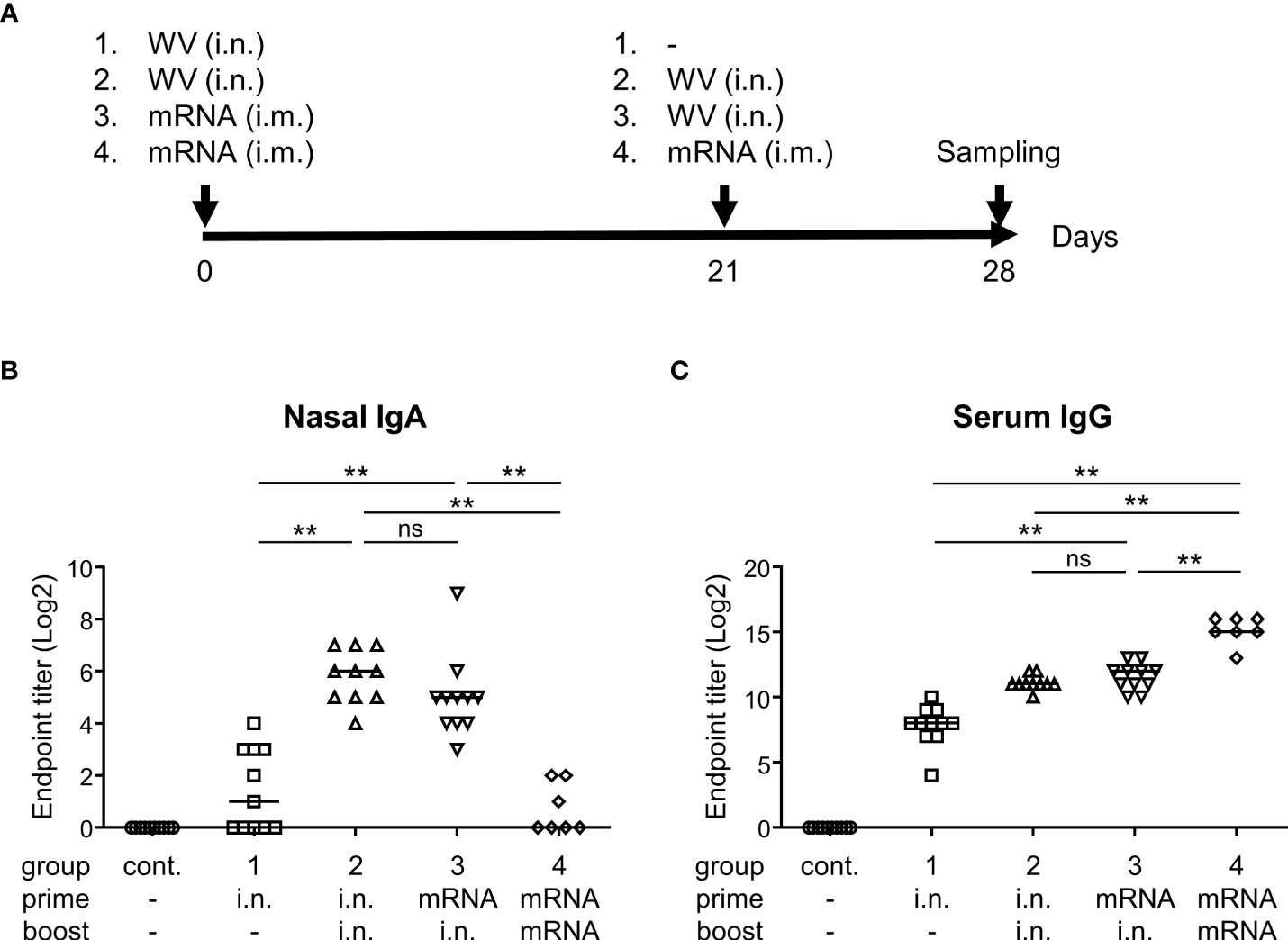
Figure 5 Systemic priming by mRNA vaccine followed by intranasal boosting with inactivated whole-virion (WV) vaccine induces S protein-specific IgG in serum and IgA in nasal wash. (A–C) Mice were immunized intranasally (i.n.) with inactivated WV or intramuscularly (i.m.) with 1 μg of mRNA vaccine encoding SARS-CoV-2 Spike. (A) Graphical illustration of the experimental procedure. (B, C) Endpoint titers of S protein-specific (B) IgA in the nasal wash and (C) IgG in the serum were evaluated. The data is presented as the mean ± SD. Each experiment was performed in duplicate. (B, C) n = 7–11 per group. (B, C) **P < 0.01 as indicated by Tukey’s test. ns, not statistically significant.
In this study, we used the Gamma strain (hCoV-19/Japan/TY7-503/2021) of SARS-CoV-2 as the seed virus to generate inactivated WV. Gamma strains were prevalent when this study was initiated at the end of the year 2020 (57), and we used this strain as the seed virus. We demonstrated that virus particles inactivated using β-propiolactone retained the protein composition on the virus surface and the structure of the virus. In previous reports, using TEM analysis, loss of S protein was observed when Wuhan strains were inactivated (58). In contrast, S proteins have been detected on the surface of inactivated WV in another report (10). Differences from existing reports may be attributed to virus strain and the conditions for ultracentrifugation and inactivation, but the full picture is still unclear. Furthermore, we discovered that 3 μg of inactivated WV in the vaccine contained approximately 2.1 μg of S protein. In general, the amount of antigen used for intranasal vaccines is approximately 1-3 μg (33, 37). Therefore, the amount of inactivated WV used in this study appears to be an appropriate dose. In addition, we demonstrated that the levels of nasal IgA and the percentage of GC B cells in the nasal passage were higher after nasal vaccination than subcutaneous vaccination. In contrast, in the neutralization study using nasal wash, the results showed a slight but not significantly different protective effect compared to the control group due to its poor sensitivity. We believe that these results indicated that nasal IgA may contribute to protection against SARS-CoV-2.
B cells within GC undergo robust proliferation and differentiation processes, such as class switch recombination, somatic mutations, and affinity selection, to produce high-affinity protective antibodies (59–62). B cells within the GC in the nasal passage increased upon nasal administration of inactivated WV, which may be involved in enhancing nasal IgA responses. While we did not assess S-specific B cell responses in GC, nasal vaccination of inactivated WV might induce it in the nasal passage; it may be beneficial to evaluate it in the future. In contrast, we investigated the T-cell response induced by nasal or subcutaneous vaccine. IL-13-secreting cells in the spleen in subcutaneous vaccine with alum as a typical T helper 2 (Th2)-inducing adjuvant were higher than that in nasal vaccine with inactivated WV. When regarding the development of vaccines for SARS-CoV-2, it is important to consider the induction of vaccine-associated enhanced respiratory disease (VAERD), such as lung eosinophilic immunopathology, as a result of the Th2-dominant adaptive immune response (33). Our findings revealed that Th1/Th2 ratio in the nasal vaccine with inactivated WV was lower than that in the subcutaneous vaccine, indicating Th2-dominant immune responses in the nasal vaccine, although the level of IL-13 was significantly lower in the nasal vaccination group than in the subcutaneous vaccination group. While the Th1/Th2 ratio is also important for VAERD induction, strong Th2-dominant immune responses are also required for VAERD induction (63). Therefore, we believe that it is highly unlikely that VAERD would be induced by an inactivated WV nasal vaccine because the Th2 immune responses induced by it are very low.
We demonstrated that the protective efficacy of the nasal vaccine against a viral challenge in the upper respiratory tract was superior to that of the subcutaneous vaccine. This suggests that IgA must be induced in the upper respiratory tract by vaccines to effectively protect against SARS-CoV-2 replication in the airways. In general, influenza virus-specific IgA has stronger cross-protective activity than that of IgG (29, 31, 64, 65). Furthermore, in SARS-CoV-2 infection, virus-specific IgA induced by infection and nasal vaccination were protective against several SARS-CoV-2 variants (33, 66). Further, nasal vaccination against SARS-CoV-2 with adenovirus vector-based vaccines has been shown to reduce viral transmission compared to that with parenteral vaccines in several preclinical studies (67–69). Therefore, a nasal vaccine containing inactivated virus may protect against various strains of SARS-CoV-2 infection and transmission. In contrast, the virus titer in the nasal turbinate from the subcutaneous vaccine group was significantly lower than that in the unimmunized control group, but not as much as that in the nasal vaccine group. The subcutaneous vaccine failed to induce S-specific IgA in the nasal, but strongly elicited S-specific IgG in the nasal mucosa, suggesting that S-specific IgG in the nasal mucosa likely contributes partially to prevent upper respiratory tract infection. The neonatal Fc receptor (FcRn) is reportedly involved in the transport of IgG across cellular barriers (70). Possibly, strongly elicited S-specific IgG in the blood following subcutaneous vaccination may be transported into the nasal cavity via FcRn.
In a lower respiratory tract challenge with a low titer of SARS-CoV-2 MA10, both nasal and subcutaneous vaccines completely protected against weight loss and death. However, the nasal vaccine showed weight loss during a high-titer SARS-CoV-2 MA10 challenge, whereas the subcutaneous vaccine group showed no weight loss. This may be because the nasal vaccine also induces virus-specific IgG in blood, but not as much as the subcutaneous vaccine. However, many pathogens, including SARS-CoV-2 and the influenza virus, cause upper respiratory tract infections in humans. Therefore, protecting the upper respiratory tract using a nasal vaccine is a useful approach for preventing the progression of symptoms to the lower respiratory tract.
To date, mRNA vaccines against the SARS-CoV-2 Wuhan strain have been used worldwide (71). Many people worldwide have been infected with SARS-CoV-2. This indicates that many people already have immunity against SARS-CoV-2. Therefore, we used an mRNA vaccine encoding SARS-CoV-2 Wuhan strain Spike and evaluated the potential use of an inactivated WV as a nasal booster vaccine. A nasal vaccine with inactivated WV increased S-specific nasal IgA and blood IgG levels in mice previously administered the mRNA vaccine. A recent study showed that intranasal vaccination with the S protein induces robust production of S-specific IgA in the respiratory tract by leveraging the pre-existing immunity generated by the mRNA vaccine (37). These findings suggest that systemic priming, such as vaccination or infection, followed by intranasal boosting with inactivated WV, results in enhanced systemic and mucosal immunity, and that inactivated WV can be useful as a booster vaccine to protect against virus transmission and the severity of infection.
Several challenges need to be addressed in the future. We did not assess respiratory tissue-resident T cell responses in this study. In addition, we did not evaluate immune responses to other antigens, such as membrane and nucleocapsid proteins. DNA vaccines expressing the membrane protein partially protect mice from SARS-CoV-2 (72). Furthermore, nucleocapsid protein is a potential target for the development of a new generation of vaccines (73, 74). The inactivated WV nasal vaccine may also induce immune responses to these antigens.
In conclusion, we demonstrated that a nasal vaccine with inactivated WV can be a potential means of preventing future COVID-19 epidemics. At present, sufficient IgG may be present in blood because of vaccine and virus exposure; therefore, the use of inactivated WV as a booster vaccine may be extremely effective.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was approved by the ethical treatment of animals and were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University, Japan. The study was conducted in accordance with the local legislation and institutional requirements.
NT, ST, and YY designed the experiments and interpreted the results. NT, ST, MW, JA, HT, and CO performed the experiments and collected and analyzed the data. TO, TH, HA, and YM provided technical support and conceptual advice. ST and YY drafted the manuscript. YY supervised the study. All authors contributed to the article and approved the submitted version.
This study was supported by grants from the Japan Society for the Promotion of Science (JSPS KAKENHI Grant Numbers: JP17H04009, JP20K21480, and JP20H03404 to YY), Japan Agency for Medical Research and Development (AMED Grant Numbers: 21am0401030h0001, 22am0401030h0002, 22fk0108647h0301, 20pc0101047s0101, and JP223fa627002 to YY), the All-Osaka U Research in “The Nippon Foundation–Osaka University Project for Infectious Disease Project” (to YY), and The Research Foundation for Microbial Diseases of Osaka University (BIKEN).
We thank Mr. Hiroki Uchiyama (The Research Foundation for Microbial Diseases of Osaka University), Dr. Hitomi Sasaki (The Research Foundation for Microbial Diseases of Osaka University), and Dr. Shinya Okamura (The Research Foundation for Microbial Diseases of Osaka University) for the experiments support.
Authors NT, MW, and YY are employees of The Research Foundation for Microbial Diseases of Osaka University.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1224634/full#supplementary-material
Alum, aluminum salts; BALF, bronchoalveolar lavage fluid; c-di-GMP, cyclic diguanylic acid; COVID-19, Coronavirus Disease 2019; CpG, unmethylated cytosine-phosphate-guanine; ELISA, enzyme-linked immunosolvent assay; FCS, fetal calf serum; GC, germinal center; HRP, horseradish peroxidase; IFN, interferon; IL, interleukin; LNP, lipid nanoparticle; MOI, multiplicity of infection; M protein, Membrane protein; mRNA, messenger RNA; mRNA-LNP, LNP containing mRNA; N protein, Nucleocapsid protein; OD, optical density; PFU, plaque-forming unit; Poly(I:C), Polyinosinic-polycytidylic acid sodium salt; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; S protein, Spike protein; TEM, transmission electron microscopy; TMPRSS2, transmembrane serine protease 2; WV, whole-virion vaccine.
1. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet (2020) 395(10229):1054–62. doi: 10.1016/S0140-6736(20)30566-3
2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5
3. Cheung EW, Zachariah P, Gorelik M, Boneparth A, Kernie SG, Orange JS, et al. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City. JAMA (2020) 324(3):294–6. doi: 10.1001/jama.2020.10374
4. Li Y, Li M, Wang M, Zhou Y, Chang J, Xian Y, et al. Acute cerebrovascular disease following COVID-19: a single center, retrospective, observational study. Stroke Vasc Neurol (2020) 5(3):279–84. doi: 10.1136/svn-2020-000431
5. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA (2020) 323(13):1239–42. doi: 10.1001/jama.2020.2648
6. American Geriatrics S. American geriatrics society policy brief: COVID-19 and nursing homes. J Am Geriatr Soc (2020) 68(5):908–11. doi: 10.1111/jgs.16477
7. Zhang J, Litvinova M, Liang Y, Wang Y, Wang W, Zhao S, et al. Changes in contact patterns shape the dynamics of the COVID-19 outbreak in China. Science (2020) 368(6498):1481–6. doi: 10.1126/science.abb8001
8. Lurie N, Saville M, Hatchett R, Halton J. Developing covid-19 vaccines at pandemic speed. N Engl J Med (2020) 382(21):1969–73. doi: 10.1056/NEJMp2005630
9. Dhama K, Sharun K, Tiwari R, Dadar M, Malik YS, Singh KP, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother (2020) 16(6):1232–8. doi: 10.1080/21645515.2020.1735227
10. Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science (2020) 369(6499):77–81. doi: 10.1126/science.abc1932
11. Soleimanpour S, Yaghoubi A. COVID-19 vaccine: where are we now and where should we go? Expert Rev Vaccines (2021) 20(1):23–44. doi: 10.1080/14760584.2021.1875824
12. Shiravi AA, Ardekani A, Sheikhbahaei E, Heshmat-Ghahdarijani K. Cardiovascular complications of SARS-coV-2 vaccines: an overview. Cardiol Ther (2022) 11(1):13–21. doi: 10.1007/s40119-021-00248-0
13. Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature (2020) 586(7830):567–71. doi: 10.1038/s41586-020-2622-0
14. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med (2020) 383(27):2603–15. doi: 10.1056/NEJMoa2034577
15. Liu H, Wang LL, Zhao SJ, Kwak-Kim J, Mor G, Liao AH. Why are pregnant women susceptible to COVID-19? An immunological viewpoint. J Reprod Immunol (2020) 139:103122. doi: 10.1016/j.jri.2020.103122
16. Salzberger B, Buder F, Lampl B, Ehrenstein B, Hitzenbichler F, Holzmann T, et al. Epidemiology of SARS-coV-2. Infection (2021) 49(2):233–9. doi: 10.1007/s15010-020-01531-3
17. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol (2021) 19(3):155–70. doi: 10.1038/s41579-020-00468-6
18. Lee IT, Nakayama T, Wu CT, Goltsev Y, Jiang S, Gall PA, et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun (2020) 11(1):5453. doi: 10.1038/s41467-020-19145-6
19. Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol (2020) 16(7):e9610. doi: 10.15252/msb.20209610
20. Killingley B, Mann AJ, Kalinova M, Boyers A, Goonawardane N, Zhou J, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med (2022) 28(5):1031–41. doi: 10.1038/s41591-022-01780-9
21. Krammer F. SARS-CoV-2 vaccines in development. Nature (2020) 586(7830):516–27. doi: 10.1038/s41586-020-2798-3
22. Sadoff J, Gray G, Vandebosch A, Cardenas V, Shukarev G, Grinsztejn B, et al. Safety and efficacy of single-dose ad26. COV2.S Vaccine against Covid-19. N Engl J Med (2021) 384(23):2187–201. doi: 10.1056/NEJMoa2101544
23. Sheikh-Mohamed S, Isho B, Chao GYC, Zuo M, Cohen C, Lustig Y, et al. Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal Immunol (2022) 15(5):799–808. doi: 10.1038/s41385-022-00511-0
24. Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature (2020) 586(7830):583–8. doi: 10.1038/s41586-020-2607-z
25. Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med (2020) 383(16):1544–55. doi: 10.1056/NEJMoa2024671
26. Bleier BS, Ramanathan M Jr., Lane AP. COVID-19 vaccines may not prevent nasal SARS-coV-2 infection and asymptomatic transmission. Otolaryngol Head Neck Surg (2021) 164(2):305–7. doi: 10.1177/0194599820982633
27. Azegami T, Yuki Y, Kiyono H. Challenges in mucosal vaccines for the control of infectious diseases. Int Immunol (2014) 26(9):517–28. doi: 10.1093/intimm/dxu063
28. Yusuf H, Kett V. Current prospects and future challenges for nasal vaccine delivery. Hum Vaccin Immunother (2017) 13(1):34–45. doi: 10.1080/21645515.2016.1239668
29. Tumpey TM, Renshaw M, Clements JD, Katz JM. Mucosal delivery of inactivated influenza vaccine induces B-cell-dependent heterosubtypic cross-protection against lethal influenza A H5N1 virus infection. J Virol (2001) 75(11):5141–50. doi: 10.1128/JVI.75.11.5141-5150.2001
30. Boyaka PN. Inducing mucosal igA: A challenge for vaccine adjuvants and delivery systems. J Immunol (2017) 199(1):9–16. doi: 10.4049/jimmunol.1601775
31. Maurer MA, Meyer L, Bianchi M, Turner HL, Le NPL, Steck M, et al. Glycosylation of human igA directly inhibits influenza A and other sialic-acid-binding viruses. Cell Rep (2018) 23(1):90–9. doi: 10.1016/j.celrep.2018.03.027
32. Kawai A, Yamamoto Y, Nogimori T, Takeshita K, Yamamoto T, Yoshioka Y. The potential of neuraminidase as an antigen for nasal vaccines to increase cross-protection against influenza viruses. J Virol (2021) 95(20):e0118021. doi: 10.1128/JVI.01180-21
33. Hemmi T, Ainai A, Hashiguchi T, Tobiume M, Kanno T, Iwata-Yoshikawa N, et al. Intranasal vaccination induced cross-protective secretory IgA antibodies against SARS-CoV-2 variants with reducing the potential risk of lung eosinophilic immunopathology. Vaccine (2022) 40(41):5892–903. doi: 10.1016/j.vaccine.2022.08.049
34. Horvath D, Temperton N, Mayora-Neto M, Da Costa K, Cantoni D, Horlacher R, et al. Novel intranasal vaccine targeting SARS-CoV-2 receptor binding domain to mucosal microfold cells and adjuvanted with TLR3 agonist Riboxxim elicits strong antibody and T-cell responses in mice. Sci Rep (2023) 13(1):4648. doi: 10.1038/s41598-023-31198-3
35. Afkhami S, Kang A, Jeyanathan V, Xing Z, Jeyanathan M. Adenoviral-vectored next-generation respiratory mucosal vaccines against COVID-19. Curr Opin Virol (2023) 61:101334. doi: 10.1016/j.coviro.2023.101334
36. Flemming A. Cross-reactive tissue-resident CD8(+) T cells may provide first line of defence against SARS-CoV-2. Nat Rev Immunol (2021) 21(11):693. doi: 10.1038/s41577-021-00638-4
37. Mao T, Israelow B, Pena-Hernandez MA, Suberi A, Zhou L, Luyten S, et al. Unadjuvanted intranasal spike vaccine elicits protective mucosal immunity against sarbecoviruses. Science (2022) 378(6622):eabo2523. doi: 10.1126/science.abo2523
38. Chavda VP, Vora LK, Pandya AK, Patravale VB. Intranasal vaccines for SARS-CoV-2: From challenges to potential in COVID-19 management. Drug Discov Today (2021) 26(11):2619–36. doi: 10.1016/j.drudis.2021.07.021
39. Madhavan M, Ritchie AJ, Aboagye J, Jenkin D, Provstgaad-Morys S, Tarbet I, et al. Tolerability and immunogenicity of an intranasally-administered adenovirus-vectored COVID-19 vaccine: An open-label partially-randomised ascending dose phase I trial. EBioMedicine (2022) 85:104298. doi: 10.1016/j.ebiom.2022.104298
40. Lemke CD, Geary SM, Joshi VB, Salem AK. Antigen-coated poly alpha-hydroxy acid based microparticles for heterologous prime-boost adenovirus based vaccinations. Biomaterials (2013) 34(10):2524–9. doi: 10.1016/j.biomaterials.2012.12.030
41. Casella CR, Mitchell TC. Putting endotoxin to work for us: monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell Mol Life Sci (2008) 65(20):3231–40. doi: 10.1007/s00018-008-8228-6
42. Sa-Nguanmoo N, Namdee K, Khongkow M, Ruktanonchai U, Zhao Y, Liang XJ. Review: Development of SARS-CoV-2 immuno-enhanced COVID-19 vaccines with nano-platform. Nano Res (2022) 15(3):2196–225. doi: 10.1007/s12274-021-3832-y
43. Riese P, Sakthivel P, Trittel S, Guzman CA. Intranasal formulations: promising strategy to deliver vaccines. Expert Opin Drug Deliv (2014) 11(10):1619–34. doi: 10.1517/17425247.2014.931936
44. Takaki H, Ichimiya S, Matsumoto M, Seya T. Mucosal immune response in nasal-associated lymphoid tissue upon intranasal administration by adjuvants. J Innate Immun (2018) 10(5-6):515–21. doi: 10.1159/000489405
45. Ko KH, Cha SB, Lee SH, Bae HS, Ham CS, Lee MG, et al. A novel defined TLR3 agonist as an effective vaccine adjuvant. Front Immunol (2023) 14:1075291. doi: 10.3389/fimmu.2023.1075291
46. Ghasemiyeh P, Mohammadi-Samani S, Firouzabadi N, Dehshahri A, Vazin A. A focused review on technologies, mechanisms, safety, and efficacy of available COVID-19 vaccines. Int Immunopharmacol (2021) 100:108162. doi: 10.1016/j.intimp.2021.108162
47. Meyer CU, Zepp F. Principles in immunology for the design and development of vaccines. Methods Mol Biol (2022) 2410:27–56. doi: 10.1007/978-1-0716-1884-4_2
48. Koyama S, Aoshi T, Tanimoto T, Kumagai Y, Kobiyama K, Tougan T, et al. Plasmacytoid dendritic cells delineate immunogenicity of influenza vaccine subtypes. Sci Transl Med (2010) 2(25):25ra4. doi: 10.1126/scitranslmed.3000759
49. Onodera T, Hosono A, Odagiri T, Tashiro M, Kaminogawa S, Okuno Y, et al. Whole-virion influenza vaccine recalls an early burst of high-affinity memory B cell response through TLR signaling. J Immunol (2016) 196(10):4172–84. doi: 10.4049/jimmunol.1600046
50. Salvi V, Nguyen HO, Sozio F, Schioppa T, Gaudenzi C, Laffranchi M, et al. SARS-CoV-2-associated ssRNAs activate inflammation and immunity via TLR7/8. JCI Insight (2021) 6(18):1–15. doi: 10.1172/jci.insight.150542
51. Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, et al. Development of an inactivated vaccine candidate, BBIBP-corV, with potent protection against SARS-coV-2. Cell (2020) 182(3):713–21 e9. doi: 10.1016/j.cell.2020.06.008
52. Mousavi T, Golpour M, Alizadeh-Navaei R, Mardomi A. The immunogenicity of an inactivated vaccine against SARS-CoV-2 in healthy individuals: A systematic review and meta-analysis. Transpl Immunol (2022) 75:101732. doi: 10.1016/j.trim.2022.101732
53. Torii S, Ono C, Suzuki R, Morioka Y, Anzai I, Fauzyah Y, et al. Establishment of a reverse genetics system for SARS-CoV-2 using circular polymerase extension reaction. Cell Rep (2021) 35(3):109014. doi: 10.1016/j.celrep.2021.109014
54. Leist SR, Dinnon KH 3rd, Schafer A, Tse LV, Okuda K, Hou YJ, et al. A mouse-adapted SARS-coV-2 induces acute lung injury and mortality in standard laboratory mice. Cell (2020) 183(4):1070–85.e12. doi: 10.1016/j.cell.2020.09.050
55. Baiersdorfer M, Boros G, Muramatsu H, Mahiny A, Vlatkovic I, Sahin U, et al. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol Ther Nucleic Acids (2019) 15:26–35. doi: 10.1016/j.omtn.2019.02.018
56. Tok K, Moulahoum H, Ghorbanizamani F, Harmanci D, Balaban Hanoglu S, Durmus C, et al. Simple workflow to repurpose SARS-CoV-2 swab/serum samples for the isolation of cost-effective antibody/antigens for proteotyping applications and diagnosis. Anal Bioanal Chem (2021) 413(29):7251–63. doi: 10.1007/s00216-021-03654-4
57. Oliveira MM, Schemberger MO, Suzukawa AA, Riediger IN, do Carmo Debur M, Becker G, et al. Re-emergence of Gamma-like-II and emergence of Gamma-S:E661D SARS-CoV-2 lineages in the south of Brazil after the 2021 outbreak. Virol J (2021) 18(1):222. doi: 10.1186/s12985-021-01690-1
58. Kordyukova LV, Moiseenko AV, Serebryakova MV, Shuklina MA, Sergeeva MV, Lioznov DA, et al. Structural and immunoreactivity properties of the SARS-coV-2 spike protein upon the development of an inactivated vaccine. Viruses (2023) 15(2):1–15. doi: 10.3390/v15020480
59. Wolniak KL, Shinall SM, Waldschmidt TJ. The germinal center response. Crit Rev Immunol (2004) 24(1):39–65. doi: 10.1615/CritRevImmunol.v24.i1.20
60. Elgueta R, de Vries VC, Noelle RJ. The immortality of humoral immunity. Immunol Rev (2010) 236:139–50. doi: 10.1111/j.1600-065X.2010.00924.x
61. Good-Jacobson KL, Shlomchik MJ. Plasticity and heterogeneity in the generation of memory B cells and long-lived plasma cells: the influence of germinal center interactions and dynamics. J Immunol (2010) 185(6):3117–25. doi: 10.4049/jimmunol.1001155
62. Gatto D, Brink R. The germinal center reaction. J Allergy Clin Immunol (2010) 126(5):898–907. quiz 8-9. doi: 10.1016/j.jaci.2010.09.007
63. Fuentes S, Klenow L, Golding H, Khurana S. Preclinical evaluation of bacterially produced RSV-G protein vaccine: Strong protection against RSV challenge in cotton rat model. Sci Rep (2017) 7:42428. doi: 10.1038/srep42428
64. Takada A, Matsushita S, Ninomiya A, Kawaoka Y, Kida H. Intranasal immunization with forMalin-inactivated virus vaccine induces a broad spectrum of heterosubtypic immunity against influenza A virus infection in mice. Vaccine (2003) 21(23):3212–8. doi: 10.1016/S0264-410X(03)00234-2
65. Okuya K, Yoshida R, Manzoor R, Saito S, Suzuki T, Sasaki M, et al. Potential role of nonneutralizing igA antibodies in cross-protective immunity against influenza A viruses of multiple hemagglutinin subtypes. J Virol (2020) 94(12):1–17. doi: 10.1128/JVI.00408-20
66. Ruiz MJ, Siracusano G, Cottignies-Calamarte A, Tudor D, Real F, Zhu A, et al. Persistent but dysfunctional mucosal SARS-CoV-2-specific IgA and low lung IL-1beta associate with COVID-19 fatal outcome: A cross-sectional analysis. Front Immunol (2022) 13:842468. doi: 10.3389/fimmu.2022.842468
67. van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature (2020) 586(7830):578–82. doi: 10.1038/s41586-020-2608-y
68. Bricker TL, Darling TL, Hassan AO, Harastani HH, Soung A, Jiang X, et al. A single intranasal or intramuscular immunization with chimpanzee adenovirus-vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. Cell Rep (2021) 36(3):109400. doi: 10.1016/j.celrep.2021.109400
69. van Doremalen N, Purushotham JN, Schulz JE, Holbrook MG, Bushmaker T, Carmody A, et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci Transl Med (2021) 13(607):1–15. doi: 10.1126/scitranslmed.abh0755
70. Aaen KH, Anthi AK, Sandlie I, Nilsen J, Mester S, Andersen JT. The neonatal Fc receptor in mucosal immune regulation. Scand J Immunol (2021) 93(2):e13017. doi: 10.1111/sji.13017
71. Chuenkitmongkol S, Solante R, Burhan E, Chariyalertsak S, Chiu NC, Do-Van D, et al. Expert review on global real-world vaccine effectiveness against SARS-CoV-2. Expert Rev Vaccines (2022) 21(9):1255–68. doi: 10.1080/14760584.2022.2092472
72. Chen J, Deng Y, Huang B, Han D, Wang W, Huang M, et al. DNA vaccines expressing the envelope and membrane proteins provide partial protection against SARS-coV-2 in mice. Front Immunol (2022) 13:827605. doi: 10.3389/fimmu.2022.827605
73. Feng W, Xiang Y, Wu L, Chen Z, Li Q, Chen J, et al. Nucleocapsid protein of SARS-CoV-2 is a potential target for developing new generation of vaccine. J Clin Lab Anal (2022) 36(6):e24479. doi: 10.1002/jcla.24479
Keywords: antigen, IgA, inactivated whole-virion, messenger RNA vaccine, nasal vaccine, SARS-CoV-2, upper respiratory tract
Citation: Tokunoh N, Tamiya S, Watanabe M, Okamoto T, Anindita J, Tanaka H, Ono C, Hirai T, Akita H, Matsuura Y and Yoshioka Y (2023) A nasal vaccine with inactivated whole-virion elicits protective mucosal immunity against SARS-CoV-2 in mice. Front. Immunol. 14:1224634. doi: 10.3389/fimmu.2023.1224634
Received: 18 May 2023; Accepted: 14 August 2023;
Published: 31 August 2023.
Edited by:
Mrinmoy Sanyal, Stanford University, United StatesReviewed by:
Akira Ainai, National Institute of Infectious Diseases (NIID), JapanCopyright © 2023 Tokunoh, Tamiya, Watanabe, Okamoto, Anindita, Tanaka, Ono, Hirai, Akita, Matsuura and Yoshioka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuo Yoshioka, y-yoshioka@biken.osaka-u.ac.jp
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.