- 1Center for Molecular and Cellular Biosciences, The University of Southern Mississippi, Hattiesburg, MS, United States
- 2Department of Microbiology and Immunology, University of Arkansas for Medical Sciences, Little Rock, AR, United States
Staphylococcus aureus is an important human pathogen that can infect almost every organ system, resulting in a high incidence of morbidity and mortality. The msaABCR operon is an important regulator of several staphylococcal phenotypes, including biofilm development, cell wall crosslinking, antibiotic resistance, oxidative stress, and acute and chronic implant-associated osteomyelitis. Our previous study showed that, by modulating murein hydrolase activity, the msaABCR operon negatively regulates the proteases that govern cell death. Here, we report further elucidation of the mechanism of cell death, which is regulated by the msaABCR operon at the molecular level in the USA300 LAC strain. We showed that deletion of msaABCR enhances weak-acid-dependent cell death, because, in the biofilm microenvironment, this mutant strain consumes glucose and produces acetate and acetoin at higher rates than wild-type USA300 LAC strain. We proposed the increased intracellular acidification leads to increased cell death. MsaB, a dual-function transcription factor and RNA chaperone, is a negative regulator of the cidR regulon, which has been shown to play an important role in overflow metabolism and programmed cell death during biofilm development in S. aureus. We found that MsaB binds directly to the cidR promoter, which represses expression of the cidR regulon and prevents transcription of the cidABC and alsSD operons. In addition, we observed that pyruvate induced expression of the msaABCR operon (MsaB). The results reported here have enabled us to decipher the role of the msaABCR operon in staphylococcal metabolic adaption during biofilm development.
Introduction
Staphylococcus aureus is a human pathogen that significantly impacts both community and health care settings, causing a wide spectrum of community-acquired and nosocomial infections, respectively. A growing concern for treatment of S. aureus is the increase in its formation of biofilms within host tissues and on the surfaces of implanted medical devices. These biofilms confer protection against antibiotics, stimulate resistance against clearance by the host immune response, and support spread from the infection site (Davis et al., 2008; Otto, 2018), leading to chronic wound infections and systemic diseases, such as bacteremia, osteomyelitis, and endocarditis (Petti and Fowler, 2002; Tong et al., 2015; Dayan et al., 2016). Biofilms are structured bacterial communities in which bacteria stick together or to a surface by self-produced, sticky polymeric molecules, including polysaccharides (e.g., polysaccharide intercellular adhesin), extracellular DNA (eDNA), and proteins (Otto, 2018), as well as others. It is expected that elucidating the mechanisms of formation and maintenance of bacterial biofilms will help identify new strategies for inhibiting their production.
Programmed cell death (PCD) is a phenomenon by which some cells undergo a suicidal mechanism for the benefit of the whole organism and is well defined in eukaryotes (Rice and Bayles, 2003; Bayles, 2007). The concept of PCD has been extended to prokaryotes that develop multicellular communities, such as bacterial biofilms, with a particular focus on production of eDNA and pronounced death and lysis during bacterial biofilm development (Rice and Bayles, 2003; Bayles, 2007; Moormeier and Bayles, 2017). The main purpose of bacterial PCD in this context is hypothesized to be the release of genomic DNA, proteins, and polysaccharides that serve as constituents of the biofilm matrix. However, this process is a finely balanced phenomenon, as several bacterial mutants defective in lysis and the release of eDNA are poor biofilm formers (Rice et al., 2003, 2005; Patton et al., 2005; Yang et al., 2006; Bayles, 2007; Thomas et al., 2014; Chaudhari et al., 2016; Windham et al., 2016). The LysR-type transcriptional regulator, CidR, is a transcriptional activator of the cidABC, lrgAB, and alsSD operons (i.e., cidR regulon) and has been shown to play an important role in autolysis and cell death during biofilm formation (Rice et al., 2003, 2005; Patton et al., 2005; Yang et al., 2006; Thomas et al., 2014; Chaudhari et al., 2016; Windham et al., 2016; Sadykov et al., 2019).
In addition to the cidA–lrgA holin–antiholin system, other genes, such as cidC and alsSD, regulate PCD by modulating overflow metabolism in S. aureus (Patton et al., 2005; Yang et al., 2006; Thomas et al., 2014; Chaudhari et al., 2016; Sadykov et al., 2019). When grown under aerobic conditions, most of the carbon (including glucose) is directed to substrate-level phosphorylation that is catalyzed by the Pta–AckA pathway, which is important for yielding secondary energy, producing acetate when activity of the TCA cycle is limited by repression of carbon catabolites (known as the Crabtree effect; Sadykov et al., 2013; Marshall et al., 2016). When glucose is completely exhausted in the medium, S. aureus generates the energy required for classic diauxic growth by reusing acetate that was excreted via the TCA cycle (Somerville et al., 2002, 2003; Thomas et al., 2014). Overflow metabolism is a wasteful strategy for catabolizing excess nutrients through incomplete oxidation, even under aerobic conditions, because it produces fewer energy metabolites per glucose molecule than the energy-efficient respiration pathway (Thomas et al., 2014). S. aureus overflow metabolism involves catabolism of excess pyruvate that is produced by increased glycolytic flux that exceeds its TCA flux. This results in the formation of acetate via the AckA–Pta and CidC pathways, of acetoin via the AlsSD pathway, and of 2,3-butanediol via the ButA pathway (Patton et al., 2005; Yang et al., 2006; Sadykov et al., 2013; Thomas et al., 2014; Chaudhari et al., 2016; Marshall et al., 2016; Zhang et al., 2017).
Two genes of the CidR regulon—cidC (encoding pyruvate:menaquinone oxidoreductase) and alsSD (encoding α-acetolactate synthetase/decarboxylase)—have enzymatic products that use the same substrate—pyruvate—and produce acetate and acetoin, respectively (Patton et al., 2005; Yang et al., 2006; Thomas et al., 2014; Zhang et al., 2017). Acetate and acetoin metabolites affect cell death antithetically, suggesting an intimate relationship between pyruvate metabolism and cell death. cidC-encoded pyruvate oxidase promotes cell death during stationary phase by increasing acidification of the growth medium via production of acetate (Thomas et al., 2014). The alsSD operon encodes α-acetolactate synthase (AlsS) and α-acetolactate decarboxylase (AlsD), which catabolize pyruvate to acetoin, which can be processed by acetoin reductase (ButA) to create 2,3-butanediol (Yang et al., 2006; Thomas et al., 2014; Chaudhari et al., 2016). The synthesis of acetoin and 2,3-butanediol (both neutral metabolites) redirects pyruvate catabolism away from the cidC pathway and actively consumes protons, promoting a neutral environment (Thomas et al., 2014).
While CidR is the main transcriptional activator of the cidABC and alsSD operons, other regulators, such as SrrAB and CcpA, also are involved in the complex regulatory network of these operons (Patton et al., 2005, 2006; Yang et al., 2006; Chaudhari et al., 2016; Windham et al., 2016; Sadykov et al., 2019). The SrrAB two-component system, which responds to nitric oxide stress and oxygen availability, appears to repress expression of the cidABC operon (Kinkel et al., 2013; Windham et al., 2016). Furthermore, in agreement with the facts that excess glucose is needed to induce expression of the cidABC operon and that CcpA is a master regulator of carbohydrate metabolism, both CcpA and CidR are required to fully induce expression of cidABC and alsSD (Seidl et al., 2009; Sadykov et al., 2011, 2019).
The msaABCR operon in S. aureus previously was found to regulate expression of global regulator sarA, as well as biofilm development, virulence, antibiotic resistance, persistence, and chronic implant-associated osteomyelitis (Sahukhal and Elasri, 2014; Samanta and Elasri, 2014; Sahukhal et al., 2015, 2017, 2020; Batte et al., 2018; GC et al., 2019; Pandey et al., 2019; Rom et al., 2020). The four-gene operon is composed of msaA, msaB, msaC, and the antisense RNA msaR (Sahukhal and Elasri, 2014). msaA, msaC, and msaR are noncoding RNAs and are thought to regulate expression of MsaB. msaB is the only protein-coding gene of the msaABCR operon; it encodes the MsaB protein, which acts as a transcription factor and as an RNA chaperone (Sahukhal and Elasri, 2014; Batte et al., 2016, 2018; Pandey et al., 2019). Studies from our laboratory have shown that deletion of the msaABCR operon results in decreased minimum inhibitory concentration for antibiotics that target the cell wall (e.g., vancomycin and certain β-lactams) in vancomycin-intermediate S. aureus (VISA) strains (Mu50, HIP6297, and LIM2) and in a methicillin-resistant S. aureus (MRSA) strain (USA300 LAC; Samanta and Elasri, 2014; Sahukhal et al., 2017; GC et al., 2019). We also showed that deletion of the msaABCR operon in S. aureus cells results in a significant decrease in biofilm thickness and in a significant increase in cell death, relative to the wild-type strain (Sahukhal et al., 2015). Notably, the biofilm defect in the ∆msaABCR mutant resulted from uncontrolled cell death (Sahukhal et al., 2015). Furthermore, we demonstrated that the msaABCR operon represses expression of genes encoding four extracellular proteases (Aur, Scp, Ssp., and Spl) in different growth phases (Sahukhal and Elasri, 2014; Sahukhal et al., 2015). Serine protease Ssp., the cysteine proteases, and other proteases are involved in processing AtlA, a major murein hydrolase, thereby regulating murein hydrolase activity and cell death during biofilm development (Rice et al., 2001; Thomas et al., 2008; Chen et al., 2013; Sahukhal et al., 2015).
These findings have led to the present study, in which we further elucidated the interrelation between the msaABCR operon (MsaB) and the CidR regulon in regulating pyruvate homeostasis, overflow metabolism, and programmed cell death during biofilm development.
Materials and Methods
Bacteria and Growth Conditions
Experiments were conducted with USA300 LAC, a clinically significant community-acquired MRSA strain. The detailed lists of all the strains and mutant strains used in this study are listed in Supplementary Table 1. The allelic replacement method was used to generate msaB and msaABCR-deletion mutants in USA300 LAC (Bae and Schneewind, 2006; Elbarasi, 2014; Samanta and Elasri, 2014). For trans-complementation, the msaABCR region was cloned into pCN34, a low-copy vector that was modified to replace the kanamycin selectable marker with a chloramphenicol-resistance marker, as described previously (Samanta and Elasri, 2014). The cidC:Tn, CidR:Tn, ∆msaABCR/cidC:Tn, and ∆msaABCR/cidC:Tn mutants were created by transducing, via bacteriophage Φ11, cidC:Tn and cidR:Tn (bursa aurealis transposon mutants, obtained from Nebraska Transposon Mutant Library) to USA300 LAC wild-type strain and ∆msaABCR mutant strains. S. aureus strains were grown in tryptic soy agar (TSA) or tryptic soy broth (TSB). Overnight bacterial cultures were prepared by inoculating cells from frozen culture stocks into culture tubes containing 5 ml of freshly prepared TSB or Mueller–Hinton broth and incubating at 37°C with continuous shaking (225 rpm). The overnight cultures were diluted 1:10 in fresh medium, incubated for 2 h, and then normalized to 0.05 optical density at 600 nm (OD600) for use as the starting cultures for experiments.
Biofilm Assays
Biofilm assays were performed according to the original protocol outlined by Sambanthamoorthy et al., with modifications added by Sahukhal and Elasri (Sambanthamoorthy et al., 2008; Sahukhal and Elasri, 2014). First, we pre-coated each well of the microtiter plates with 20% human plasma and incubated the plates overnight at 4°C. Next, each well was inoculated with 2 ml of starter culture (prepared as described above, “Bacteria and growth conditions”). The biofilm medium for this assay was prepared by mixing TSB with 3% NaCl and 0.25% glucose; for excess glucose conditions, an additional 0.5% glucose was added to the appropriate wells. We also added vancomycin (0.2 μg/ml) when necessary. The microtiter plates were incubated at 37°C for 24 h with shaking at 150 rpm. After 24 h, we washed each well with sterile phosphate-buffered saline (PBS), stained the resulting biofilms with crystal violet dye, and eluted with 5% acetic acid. To measure the amount of biofilm, we used a spectrophotometer to quantitate absorbance at 595 nm. The assays were performed in experimental duplicates for USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains; biofilm assays were performed three times. Biofilm formation values were calculated as the percent activity relative to USA300 LAC (wild type), which was set as 100%; mean values and standard deviations of each growth condition then were calculated.
Growth Curves and CFU Count
To determine the effects of excess glucose on cell death, the survival of different strains was monitored for 5 days (120 h). Wild type, ∆msaABCR mutant, and complementation strains of USA300 LAC were grown aerobically in TSB with 50 mM glucose (TSB-50 mM glucose) and, when indicated, buffered with morpholinepropanesulfonic acid (MOPS) buffer (50 mM, pH 7.3). Cultures were incubated for 120 h at 37°C with shaking at 220 rpm, and samples were collected at 24-h intervals. To measure the colony-forming units (CFUs) of each sample, cultures were serially diluted up to the appropriate dilutions, and 100-μl aliquots were placed on TSA plates, which then were incubated overnight at 37°C. To verify the consistency of these results, cell death assays for each sample were performed in triplicate and were repeated at least three times. The CFUs for each sample were averaged to obtain mean values, and the standard deviation was calculated.
Metabolite Analyses
For these analyses, bacterial growth was allowed to proceed (at 37°C and 225 rpm) in flasks containing TSB-50 mM glucose at a 1:10 flask-to-volume ratio. Metabolite excretion profiles were determined from culture supernatants that were harvested at the indicated times and under the indicated growth conditions after incubation. Glucose and acetate were measured with commercial kits (R-Biopharm), according to the manufacturer’s instructions.
Acetoin assays were performed as previously described (Nicholson, 2008). Briefly, 300 μl of supernatant was mixed with 210 μl of 0.5% creatine, 300 μl of 5% α-napthol, and 300 μl of 40% KOH. Each sample was incubated for 15–30 min. OD560 was measured and used to determine the concentration of acetoin.
RNA Extraction, Reverse Transcription, and Quantitative Reverse-Transcription PCR
Starting cultures were grown in the specified growth medium for 5 h, and the cells were harvested by centrifugation. The bacterial pellet was treated with RNAprotect bacterial reagent (Qiagen), and the total RNA was extracted as described previously (Samanta and Elasri, 2014; Sahukhal et al., 2015). Relative gene expression was quantified with real-time PCR analysis of cDNA prepared from the total RNA samples. All experiments used housekeeping gene gyrB as an internal control (Goerke et al., 2000; Samanta and Elasri, 2014; Sahukhal et al., 2015). The relative fold-change in gene expression was calculated with the ∆CT method. All primers used for qRT-PCR are listed in Supplementary Table 2.
Expression and Purification of MsaB
MsaB protein was cloned into pCN51, an inducible expression vector that, when induced, produces the protein with 6 × His (MsaBhis) at the C-terminus. The construct was transformed into E. coli strain DH5α, and the plasmids isolated from these cells were used to transduce competent cells of the restriction-deficient S. aureus RN4220 strain. The plasmid then was moved to the USA300 LAC ∆msaABCR mutant through generalized transduction with bacteriophage ϕ11, as described previously (Pandey et al., 2019). To induce expression of the 6 × His–MsaB fusion protein (i.e., MsaBhis) in the ∆msaABCR mutant of the USA300 LAC strain, 20 μM cadmium chloride (CdCl2) was added to cell cultures during exponential growth phase; cell cultures were incubated an additional 4 h with shaking. The cells then were pelleted, resuspended in PBS (pH 7.4) with a protease inhibitor cocktail, and lysed by bead beating followed by sonication. The cell lysate was centrifuged at 10,000×g for 30 min to remove cell debris, and MsaBhis was purified from the clear lysate with a nickel column (HisPur Ni-nitrilotriacetic acid [Ni-NTA] resin; Thermo Scientific).
Electrophoretic Mobility Shift Assay
The ability of MsaB to bind the cidR promoter was determined according to electrophoretic mobility shift, as described previously (Pandey et al., 2019). Briefly, the 5′-biotinylated PcidR-Electrophoretic Mobility Shift assay (EMSA) forward primers and reverse primers were used to amplify the promoter region. The PCR product (250 μl) was loaded onto a 2% agarose gel and separated by electrophoresis, and the corresponding PcidR fragment was extracted from the gel (Wizard SV Gel and PCR Cleanup System, Promega). The concentration and quality of the purified DNA product were determined with a NanoDrop spectrophotometer (Thermo Scientific). To study the ability MsaB to bind to the CidABC and alsSD promoters, the 5′-biotinylated duplex oligonucleotide sequence of the promoter regions of these genes was used, as in previous studies (Sadykov et al., 2019). For EMSA experiments, the LightShift Chemiluminescent EMSA kit (Pierce) was used according the manufacturer’s protocol. The binding reaction mixture (20 μl) contained ultrapure water, 1X binding buffer, 50 ng μl− 1 poly (dI-dC), 2.5% (vol/vol) glycerol, 5 mM MgCl2, and 5′-biotin-labeled DNA probe; increasing concentrations of MsaBhis protein and unlabeled specific probe, when required, were added. The reaction mixture was incubated at room temperature for 20 min and separated by electrophoresis (1 h, 100 V) in a pre-run 5% Tris-borate-EDTA (TBE) gel. The samples in the gel were then transferred to a nylon membrane (1 h, 4°C), cross-linked in a UV crosslinker, and processed for detection. The blots were developed and visualized with the kit detection module, according to the manufacturer’s protocol, and were imaged with the ChemiDoc system (Bio-Rad).
Murein Hydrolase Assay
Starter culture was incubated for 16 h at 37°C with shaking at 250 rpm. The culture supernatants were collected by centrifugation and were concentrated (Centricon-3 concentrator; Millipore) approximately 10-fold. Protein concentrations of the extracellular supernatants were determined with the BCA protein assay kit (Life Technologies), according to the manufacturer’s recommendations. Quantitative cell wall hydrolysis assays were performed as described previously (Sahukhal et al., 2015).
Western Blotting of MsaBhis
To quantify the amount of MsaBhis produced under different growth conditions, Western blotting was performed with whole-cell lysates, as described previously (Batte et al., 2016). The USA300 LAC msaABhisCR complementation strain was used for this study. It was generated by introducing the pCN34–msaABhisCR operon construct (with His-tagged MsaB at the 5′ end) into USA300 LAC ∆msaABCR. The USA300 LAC msaABhisCR complementation cells were grown in TSB without glucose, TSB-50 mM glucose, and TSB without glucose supplemented with 2% pyruvate. Cells were grown for 5 h, pelleted by centrifugation, washed with PBS, resuspended in lysis buffer (PBS with a protease inhibitor), and lysed with a Fastprep instrument with glass beads. The clear supernatants were obtained after centrifugation of the whole-cell lysates to remove debris. Proteins from different growth conditions were quantified with the BCA method (Pierce BCA protein assay kit; Life Technologies), and 25 μg of proteins were separated with SDS-polyacrylamide gel electrophoresis. Proteins in the gels were transferred to polyvinylidene difluoride (PVDF) membrane by blotting, blots were blocked wit 5% non-fat milk, and MsaBhis was detected in the blots with an anti-His antibody and a peroxidase-conjugated secondary antibody.
Statistical Analysis
All statistical analyses to test for significant differences in this study were done with OriginPro software (Origin Lab). A statistical significance level of p < 0.05 was used as the cutoff for significance in performing statistical analyses between the strains. Student’s t-test (unpaired) or one-way analysis of variance (ANOVA) were used to compare the results from wild-type strains with those from mutant or complementation strains (*p < 0.05, **p < 0.005, ***p < 0.0005).
Results
The msaABCR Operon Represses Acetate-Mediated Cell Death During Biofilm Formation
We performed biofilm assays under conditions of excess glucose (0.5%) and/or a sub-inhibitory concentration of vancomycin in the USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains. The USA300 LAC strain showed significantly increased biofilm production under the supplementation conditions tested (p < 0.005); however, under these same conditions, biofilm production was not induced to significant level in the ∆msaABCR mutant (Figure 1). These observations suggest that the msaABCR operon plays an important role in biofilm formation in the presence of excess glucose, and/or subinhibitory concentration of vancomycin.
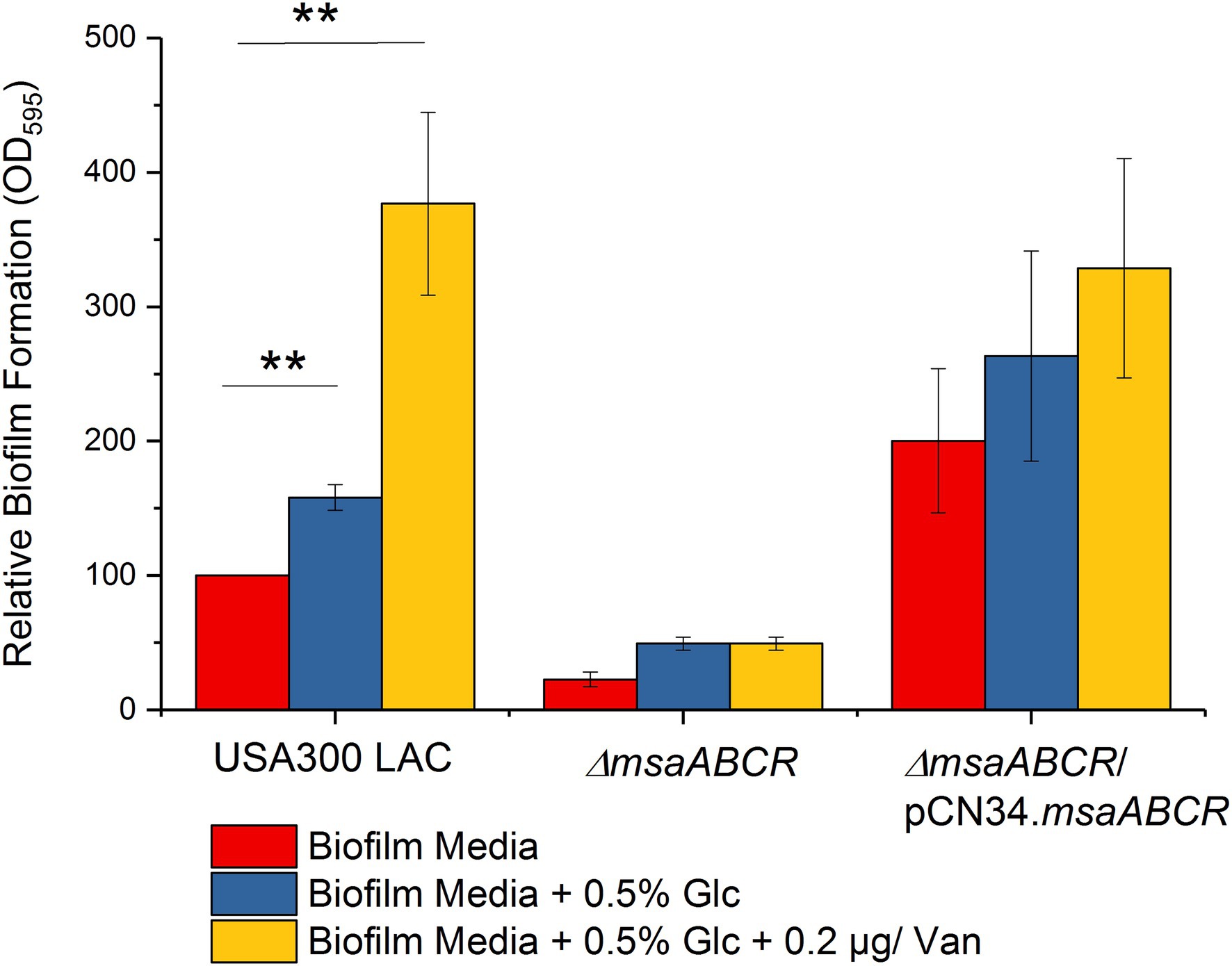
Figure 1. Biofilm formation by the USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains. Biofilm formation measured in biofilm medium, biofilm medium supplemented with excess glucose (0.5%), and biofilm medium supplemented with excess glucose (0.5%) and a sub-inhibitory concentration of vancomycin (0.2 μg/ml). Values are shown as the percent activity relative to the wild-type USA300 LAC, which was set at 100%. Each vertical bar represents the mean of three independent experiments. Error bars represent the standard error (SE). One-way ANOVA was used to compare the results for wild-type, ∆msaABCR mutant, and complementation strains in different growth media (**p < 0.005).
Previously, we showed that a phenotype of increased cell death was associated with the ∆msaABCR mutation under conditions for both planktonic and biofilm growth (Samanta and Elasri, 2014; Sahukhal et al., 2015; GC et al., 2019). Other studies reported that aerobic growth of S. aureus in TSB-50 mM glucose impairs stationary-phase survival of S. aureus, due to accumulation of acetate derived from the incomplete catabolism of glucose, thus potentiating cell death (Thomas et al., 2014; Augagneur et al., 2020). Therefore, we monitored survival of the USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains during stationary phase when cultured in TSB supplemented with 14 or 50 mM glucose. In TSB with 14 mM supplemental glucose, all test strains, including the ∆msaABCR mutant, demonstrated uncompromised survival during stationary phase over a period of 5 days (Figure 2A). However, in TSB-50 mM glucose, survival of the ∆msaABCR mutant, relative to that of the USA300 LAC and complementation strains, significantly decreased over a period of 5 days (Figure 2B). Based on these results, we hypothesized that intracellular acidification might be responsible for the reduced survival seen in the ∆msaABCR mutant. To assess whether the reduced cell survival of the ∆msaABCR mutant grown in excess glucose was indeed caused by intracellular acidification, we performed a cell viability assay in TSB-50 mM glucose and buffered with 50 mM morpholinepropanesulfonic acid (MOPS, pH 7.3). Under these conditions, survival of the ∆msaABCR mutant improved and reverted to the level of USA300 LAC (Figure 2C). Together, these results suggest that the observed increase in cell death of the ∆msaABCR mutant strain is due to cellular acidification.
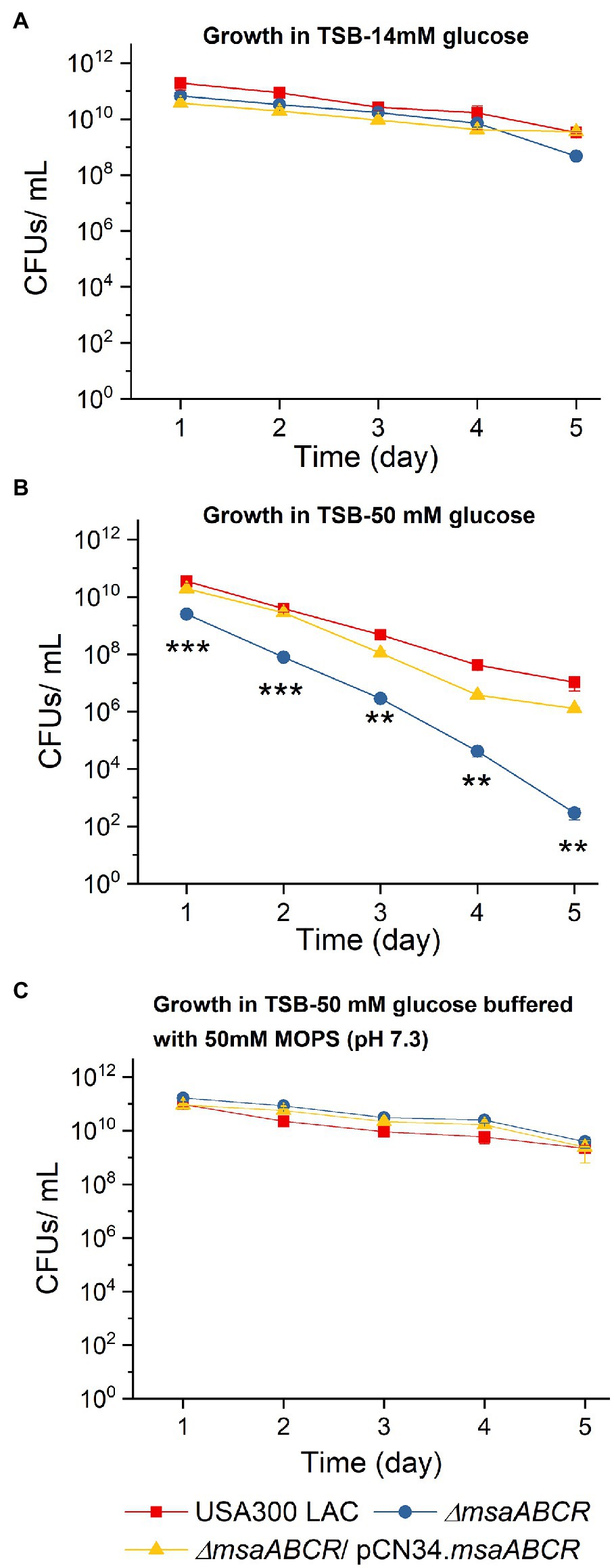
Figure 2. Survival of the ∆msaABCR mutant in stationary phase. Cell viability of Staphylococcus aureus USA300 LAC (wild type), ∆msaABCR mutant, and complementation strain was monitored every 24 h over a period of 5 days in TSB plus 14 mM glucose (A), TSB-50 mM glucose (B), and TSB-50 mM glucose buffered with MOPS (pH 7.3; C). Each data point represents the mean of three independent experiments. Error bars represent the SE. One-way ANOVA was used to compare the results for wild type with those of its mutant and complementation strains (**p < 0.005, ***p < 0.0005).
Studies have shown that when S. aureus is grown aerobically under excess glucose conditions, pyruvate is catabolized to acetate via the AckA–Pta and CidC pathways (Patton et al., 2005; Yang et al., 2006; Thomas et al., 2014; Chaudhari et al., 2016; Zhang et al., 2017). To examine how the msaABCR operon is involved in glucose catabolism to acetate, we measured glucose utilization and acetate production in all three test strains by growing the cells for 24 h in TSB-50 mM glucose. OD600 of all test strains was similar for the first 12 h, but OD600 the ∆msaABCR mutant was significantly reduced after 24 h of growth (Figure 3A), most likely due to increased lysis of ∆msaABCR mutant cells. When we measured the pH of the supernatants collected from cell cultures, we observed that all strains underwent similar changes in pH over 24 h (Figure 3B). Measurements of glucose and acetate kinetics showed that the ∆msaABCR mutant consumed glucose and produced acetate significantly faster after late exponential growth phase (4 h) than the USA300 LAC (wild type) and complementation strains (Figures 3C,D). The ∆msaABCR mutant consumed the most glucose before 12 h of growth and had minimal glucose (~2 mM) remaining in the medium at 12 h; however, the wild type and complementation strains maintained ~4 mM glucose in the medium, even after 16 h of growth (Figure 3C). Acetate levels for all the strains remained almost constant from 16 to 24 h, suggesting that none of the strains could reuse acetate when grown in excess glucose, even after glucose was depleted from the culture medium (Figure 3D). Therefore, these strains did not undergo classic diauxic growth in an acidic growth environment (Figure 3A).
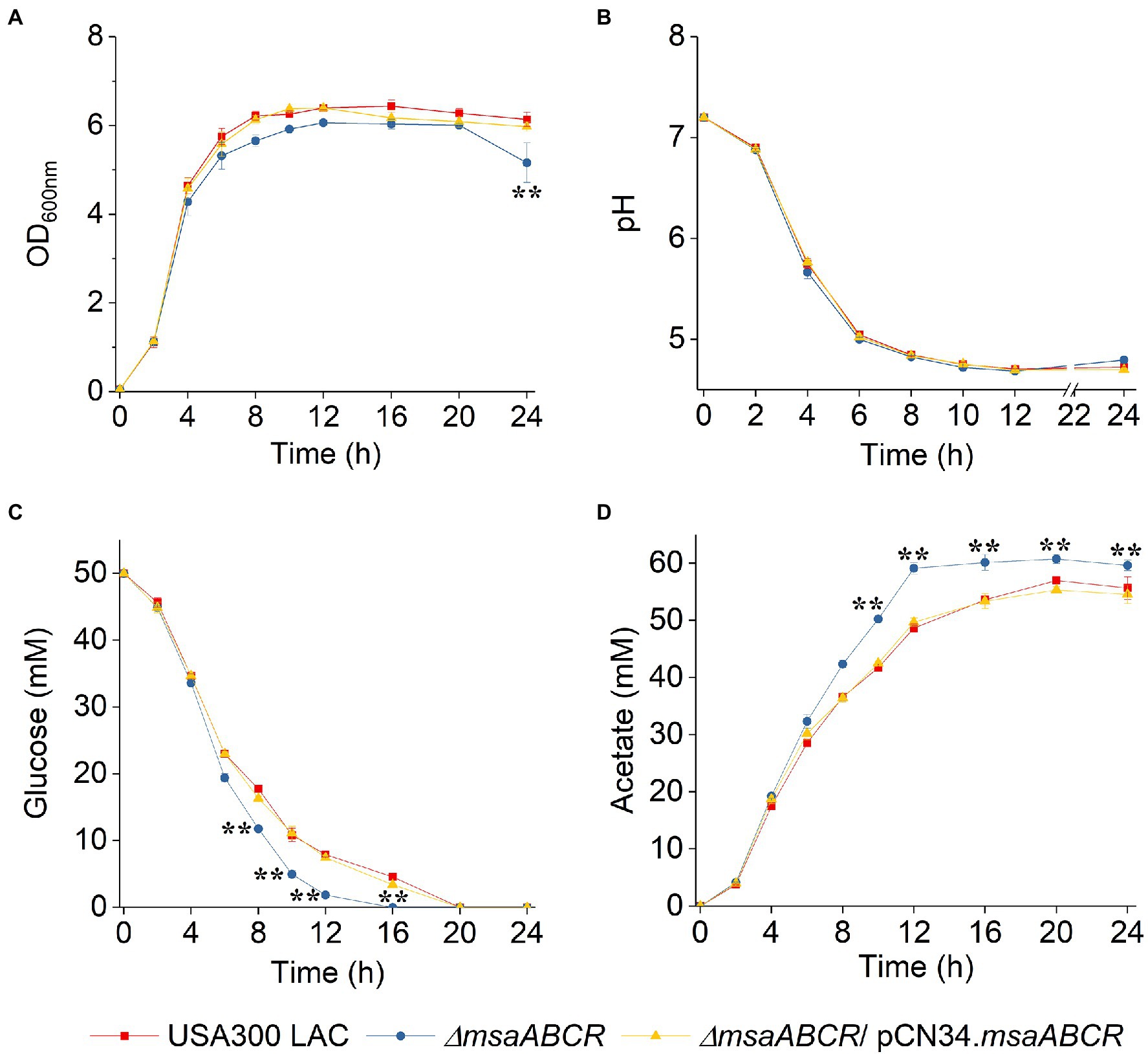
Figure 3. Growth curve, pH kinetics, glucose kinetics, and acetate kinetics of the USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains grown aerobically in TSB-50 mM glucose. The OD600nm (A) and pH (B) of the culture medium were determined at the indicated times. Temporal depletion of glucose from (C) or accumulation of acetic acid in (D) the culture media of the indicated strains. Glucose and acetate concentrations in culture supernatants were measured at the indicated from 0 to 24 h of growth in TSB-50 mM glucose. Each data point represents the mean of three independent experiments. Error bars represent the SE. One-way ANOVA was used to compare the results for wild type with those of the mutant and complementation strains (**p < 0.005).
Because we observed the effects of msaABCR deletion after late exponential phase (after 4 h) and the growth medium pH reached ~4.8 at 8 h (Figure 3B), the period of 4–8 h was chosen for measuring the rates of glucose consumption and acetate production. These rates were measured as the difference in acetate or glucose levels in the growth medium from 4 to 8 h of growth per unit time per OD600 at 8 h. When we measured the rate of glucose consumption and acetate production in this way, we observed that the ∆msaABCR mutant had a significantly (p < 0.0005) higher rate of glucose consumption (Figure 4A) and a significantly (p < 0.005) higher rate of acetate production (Figure 4B) than the USA300 LAC (wild type) and complementation strains. In an acidic environment (pH ~ 4.76 = pKa of acetate), the equilibrium between acetate and acetic acid shifts toward the neutral form—acetic acid. Neutral acetic acid then can pass freely through the cell membrane, which is thought to be the cause of intracellular acidification (Thomas et al., 2014). Thus, increased acetate production in the ∆msaABCR mutant under lower pH conditions (close to pH 4.8), suggests an increase in neutral acetic acid, which can diffuse across the membrane and cause intracellular acidification in the mutant compared with the USA300 LAC (wild type) and complementation strains. Thus, we hypothesized that the decreased viability of the ∆msaABCR mutant during stationary phase (24–120 h) in TSB-50 mM glucose (Figure 2B) is due to increased intracellular acidification and its pleiotropic effects.
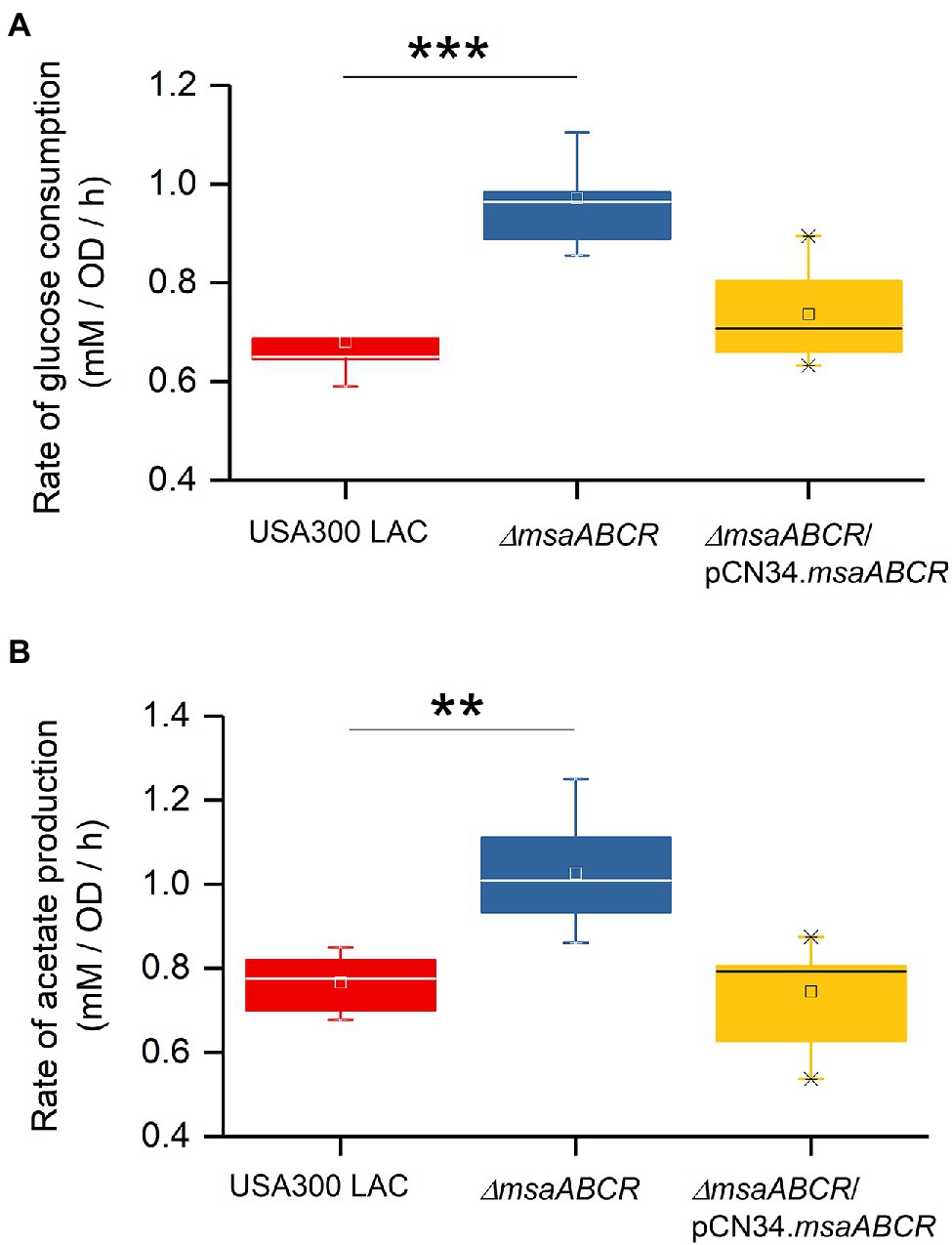
Figure 4. Rate of glucose consumption and production of acetate under TSB-50 mM growth conditions. The rate of glucose consumption was measured as the concentration of glucose (mM) consumed from 4 to 8 h of growth per OD600 at the 8-h time point per unit time (h; A). The rate of acetate production was measured as the concentration of acetate (mM) produced from 4 to 8 h of growth per OD600 at the 8-h time point per unit time (h) in Staphylococcus aureus USA300 LAC (wild type) and the ∆msaABCR mutant strains (B). Glucose and acetate concentration in culture supernatants were measured at the indicated growth times in TSB-50 mM glucose. The box indicates Q1 and Q3 values; the solid line represents Q2. One way ANOVA was used to compare the results for wild type with those of the mutant and complementation strains (**p < 0.005, ***p < 0.0005).
To assess whether the observed increases in consumption of glucose and production of acetate by the ∆msaABCR mutant are influenced by pH in the culture medium, we measured glucose consumption and acetate production over the first 24 h when cells were grown in TSB-50 mM glucose buffered with 50 mM MOPS (pH 7.3; Figure 5). As was observed in TSB-50 mM glucose without MOPS buffer (Figures 3C,D), after late exponential growth phase (4 h), glucose consumption and acetate production in TSB-50 mM glucose with MOPS buffer (pH 7.3) were significantly higher for the ∆msaABCR mutant than for the USA300 LAC (wild type) and complementation strains (Figures 5B,C). This significantly increased difference for the ∆msaABCR mutant is supported by the calculated rates of glucose consumption and acetate production during the period of 4–8 h of growth (Figure 6). We also confirmed a previous observation that MOPS buffer does not allow pH in the culture medium to drop below 5.5, even while all the strains were producing ~50 mM acetate (data not shown; Thomas et al., 2014). This observation suggests that acetate produced under conditions of excess glucose and buffered with MOPS produces significantly fewer neutral acetic acid molecules (as pH > 4.8) that could passively breach the S. aureus cell membrane and cause intracellular acidification. The improved survival of the ∆msaABCR mutant in TSB-50 mM glucose buffered with MOPS to a pH of 7.3 is most likely due to the inability of acetate (pKa = 4.8) to permeate cells and acidify the cytoplasm under relatively neutral conditions. In addition, we found that, under MOPS-buffered conditions, all test strains were able to transport acetate inside cells and reuse acetate after glucose was depleted from the medium, which was indicated by decreased acetate levels between the 12- and 24-h measurements (Figure 5C). Interestingly, the ∆msaABCR mutant reused acetate significantly faster than the USA300 LAC (wild type) and complementation strains (Figure 5C), and, as a result, the mutant strain’s growth was significantly higher at the 24-h time point (p < 0.0005, Figure 5A).
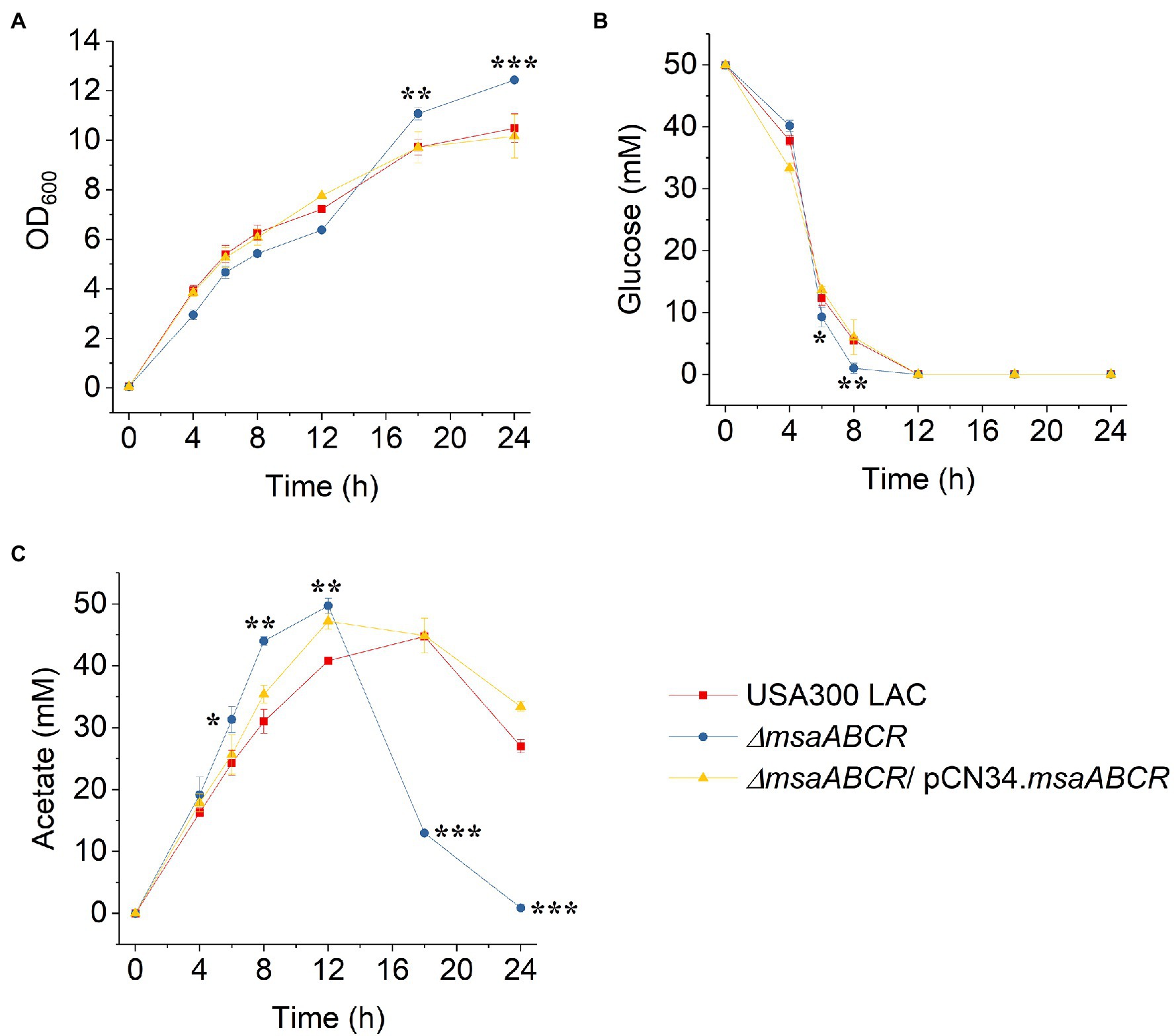
Figure 5. Growth and kinetics of glucose and acetate for the USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains grown aerobically in TSB-50 mM glucose buffered with MOPS (50 mM, pH 7.3). The OD600 of the culture medium was determined at the indicated times (A). Temporal depletion of glucose from (B) or accumulation of acetic acid and reuse in (C) the culture media of the indicated strains. Glucose and acetate concentrations in culture supernatants were measured at the indicated time of culture from 0 to 24 h of growth in TSB-50 mM glucose buffered with 50 mM MOPS, pH 7.3. Each data point represents the mean of three independent experiments. Error bars represent the SE. One-way ANOVA was used to compare the results for wild type with those of the mutant and complementation strains (*p < 0.05, **p < 0.005, ***p < 0.0005).
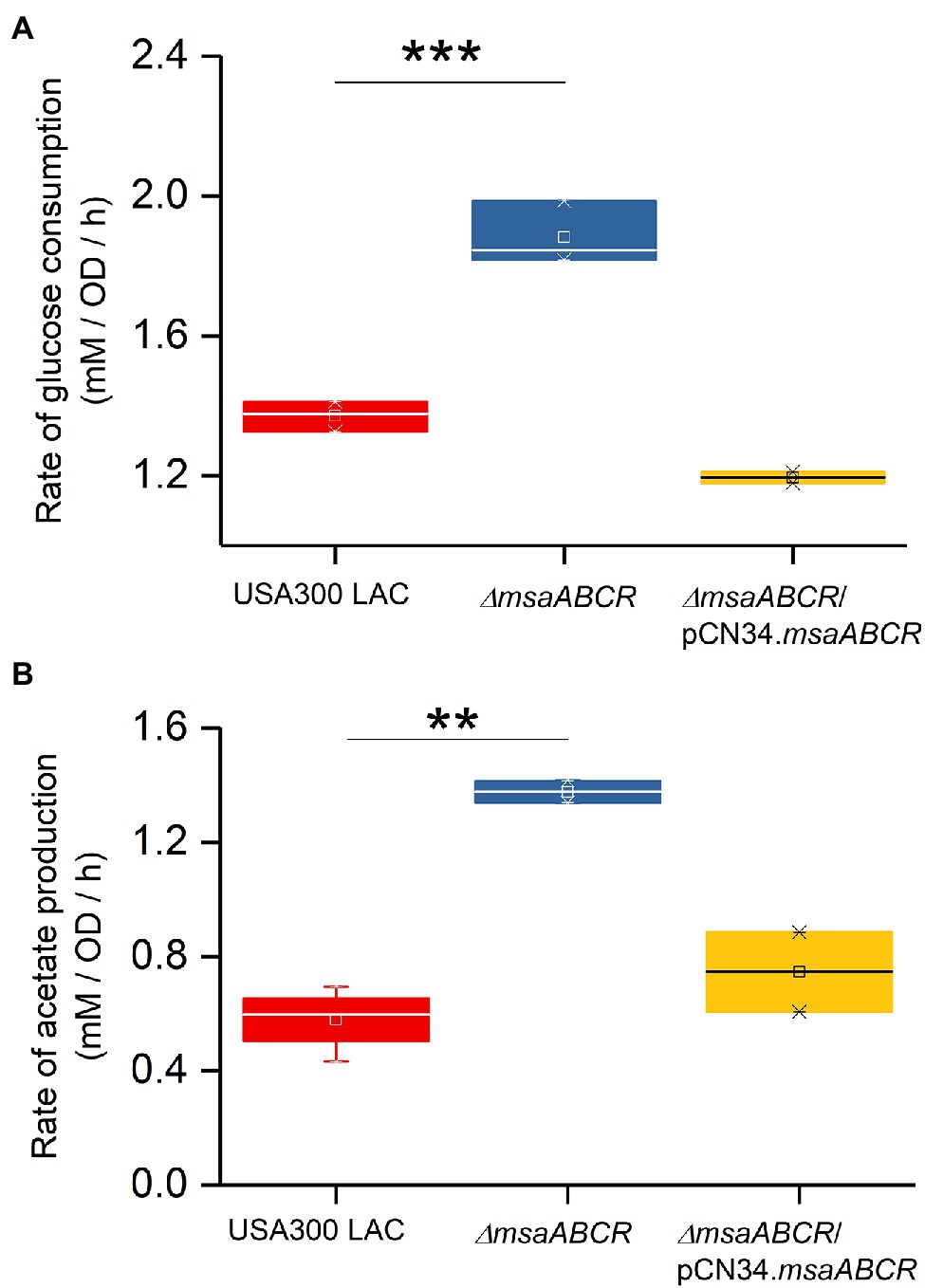
Figure 6. Rates of glucose consumption and acetate production when grown aerobically in TSB-50 mM glucose buffered with MOPS (50 mM, pH 7.3). The rate of glucose consumption was measured as glucose concentration (mM) consumed from 4 to 8 h of growth per OD600 at the 8-h time point per unit time (h; A). The rate of acetate production was measured as acetate concentration (mM) produced from 4 to 8 h of growth per OD600 at the 8-h time point per unit time (h; B) in the indicated Staphylococcus aureus strains. The box indicates Q1 and Q3 values; the solid line represents Q2. One-way ANOVA was used to compare the results for wild type with the mutant and complementation strains (**p < 0.005, ***p < 0.0005).
Overall, the ∆msaABCR mutant consumed glucose and produced acetate at significantly faster rates than the USA300 LAC (wild type) and complementation strains did. Therefore, we conclude that one function of the msaABCR operon is to repress consumption of glucose and catabolism of pyruvate to acetate during or after late exponential growth phase.
The msaABCR Operon Represses Expression of the CidR Regulon to Repress cidC-Mediated, Acetate-Dependent Cell Death
Several studies have reported that cidR, cidABC, lrgAB, pta–ack, and alsSD play major roles in the pathways involved in PCD and overflow metabolism (Patton et al., 2005; Yang et al., 2006; Sadykov et al., 2013, 2019; Thomas et al., 2014; Chaudhari et al., 2016; Zhang et al., 2017). Therefore, we measured with qRT-PCR the relative expression of these genes in the ∆msaABCR mutant cultured in TSB-50 mM glucose during late exponential growth phase (after 5 h of growth). Results showed that the relative expression of genes controlled by the CidR regulon (cidR, cidA, cidC, lrgA, and alsS) was higher in the ∆msaABCR mutant than in the USA300 LAC (wild type) strain (Table 1); however, expression of pta, ack, acsA, and pykA genes was not altered in the ∆msaABCR mutant (Table 1). These results suggest that the msaABCR operon represses expression of the genes of the cidR regulon, including those of the cidABC, alsSD, and lrgAB operons. When we compared expression of CidR regulon genes (cidR, cidA, and alsS) by the ∆msaABCR mutant with expression by the USA300 LAC (wild type) strain during late exponential growth phase in TSB without glucose, we observed a statistically insignificant increase (<2-fold) in expression of the genes but not to the same extent as when the cultures were grown in TSB-50 mM glucose (Supplementary Table 3). These results suggest that glucose or its metabolites are involved in msaABCR-mediated regulation of the CidR regulon when excess glucose is present as an environmental stressor. Among the two pathways for acetate production in S. aureus, the pta–ackA pathway appeared to be unaffected by deletion of the msaABCR operon; this was evident as no change in expression of the pta and ackA genes. Because we observed increased expression of cidC in the ∆msaABCR mutant, we examined how cidC-mediated acetate production is involved in survival of the ∆msaABCR mutant. We introduced a cidC transposon mutation in the ∆msaABCR mutant and measured survival of the ∆msaABCR/cidC:Tn double mutant during stationary phase under excess-glucose conditions (TSB-50 mM glucose; Figure 7A). Survival (assessed as CFU/ml) of the ∆msaABCR/cidC:Tn and cidC:Tn mutants were significantly higher than USA300 LAC (wild type; p < 0.0005, Figure 7A; Supplementary Figure 1A). The rate of glucose consumption of ∆msaABCR/cidC:Tn was similar to ∆msaABCR mutant (Figures 7B; Supplementary Figure 1B). However, the rate of acetate production was comparable to USA300 LAC but significantly less compared to ∆msaABCR mutant (Figures 7C; Supplementary Figure 1C). We also measured the acetoin production by all test strains (Figure 7D). We observed significant increase in acetoin production in the ∆msaABCR mutant (Figure 7D). These observations suggest that CidC-mediated acetate production contributed to the increased cell death in the ∆msaABCR mutant. Therefore, we speculated that increased expression of cidC contributes to increased cell death in the ∆msaABCR mutant.
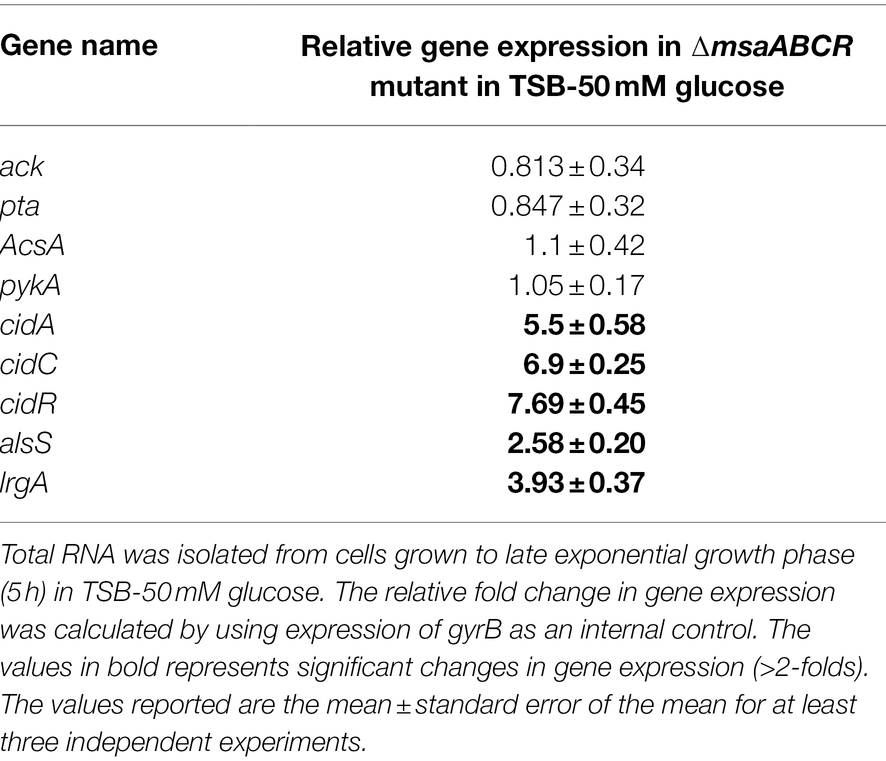
Table 1. Measurement of gene expression involved in overflow metabolism in the ∆msaABCR mutant compared with the wild-type USA300 LAC strain.
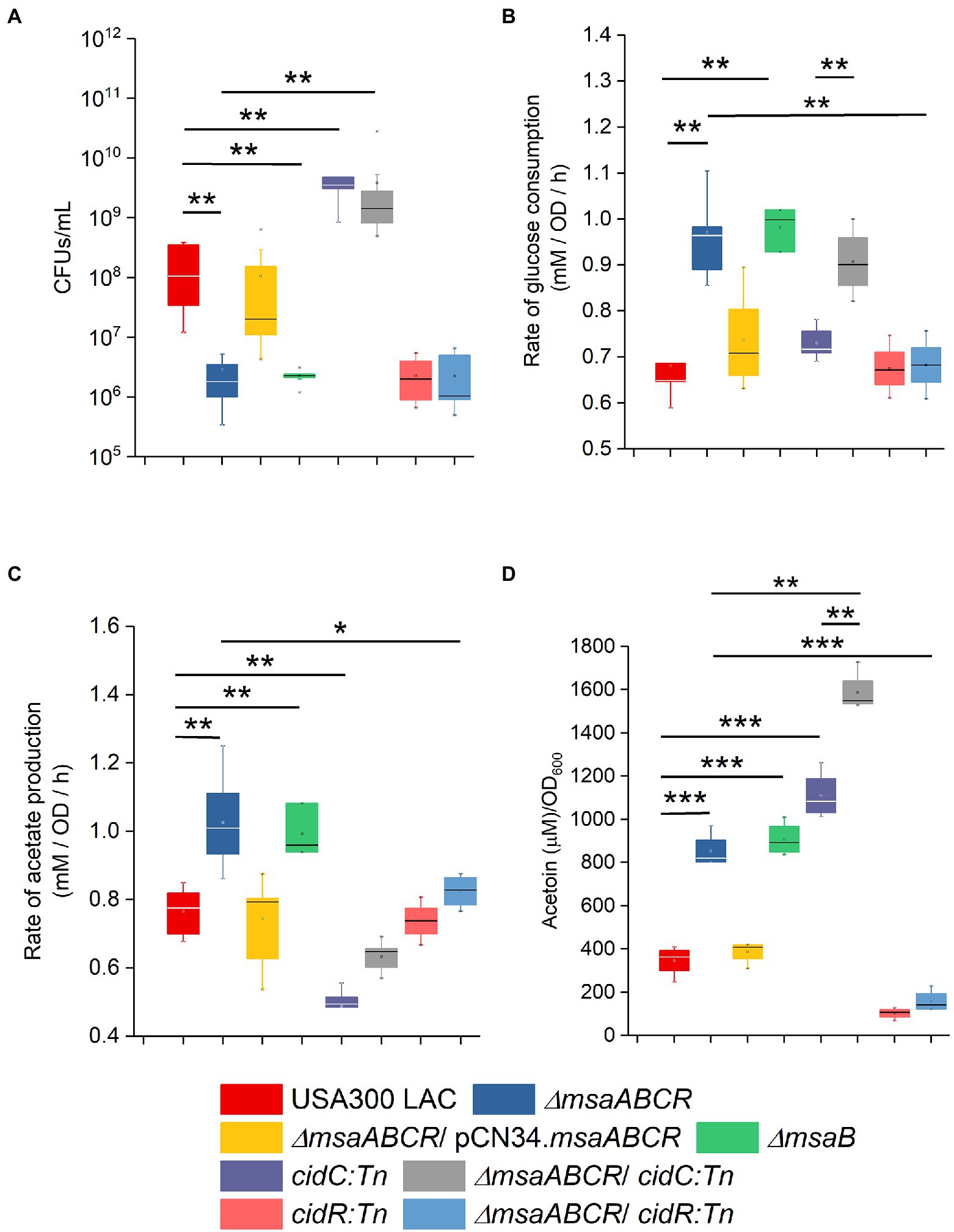
Figure 7. Stationary-phase survival, rate of glucose consumption, rate of acetate production, and rate of acetoin production by the indicated mutants. Cell viability of Staphylococcus aureus USA300 LAC (wild type), ∆msaABCR mutant, ∆msaB mutant, cidC:Tn mutant, ∆msaABCR/cidC:Tn mutant, cidR:Tn mutant, and ∆msaABCR/cidR:Tn mutant strains were monitored after 72 h of growth (A). Rate of glucose consumption for all the strains was measured as the concentration of glucose (mM) consumed from 4 to 8 h of growth per OD600 at the 8-h time point per unit time (h; B). Rate of acetate production was measured as the concentration of acetate (mM) produced from 4 to 8 h of growth per OD600 at the 8-h time point per unit time (h; C). Glucose and acetate concentrations in culture supernatants were measured at the indicated growth times in TSB-50 mM glucose. Temporal accumulation of acetoin in the culture media of the indicated strains at the 12-h time point (D). All the strains were grown aerobically in TSB-50 mM glucose for these experiments. The box indicates Q1 and Q3 values; the solid line represents Q2. Oneway ANOVA was used to compare the results for the two different strains (*p < 0.05, **p < 0.005, ***p < 0.0005).
MsaB Regulates the CidR Regulon to Repress Overflow Metabolism
Another overflow metabolic pathway regulated by CidR is AlsSD. Because we observed increased expression of the alsS gene in the ∆msaABCR mutant (Table 1), we also assessed whether the msaABCR deletion affects pyruvate catabolism via the AlsSD pathway. For this, we measured acetoin production in cells grown in TSB-50 mM glucose for 12 h. Results showed that the ∆msaABCR mutant produced significantly more (~3-fold greater) acetoin in the culture medium than the USA300 LAC and complementation strains (Figure 7D). Therefore, increased expression of two metabolic genes, cidC and alsS, in the ∆msaABCR mutant, relative to expression in the USA300 LAC (wild type) strain, also was correlated with the respective increases in acetate (Figures 3D, 7C,) and acetoin (Figure 7D) production that were observed during the post-exponential growth phase. Furthermore, the ∆msaABCR/cidC:Tn mutant showed significantly higher acetoin production compared to other test strains (Figure 7D) explained why ∆msaABCR/cidC:Tn survived much better than other test strains (Figure 7A). All these observations suggest that the msaABCR operon plays an important role in repressing overflow metabolism.
MsaB is the only protein translated from the msaABCR operon that acts both as a transcription factor and as an RNA chaperone (Sahukhal and Elasri, 2014; Batte et al., 2016, 2018; Pandey et al., 2019); the functions of the other ncRNAs in the msaABCR operon remain unknown. Therefore, we next examined the contributions of the msaB gene in stationary-phase cell death and overflow metabolism. We confirmed the survival, the rate of glucose consumption, the rate of acetate production, and acetoin production in the ∆msaB mutant of USA300 LAC (Figure 7; Supplementary Figure 2). The ∆msaB mutant showed phenotypes similar to those of the ∆msaABCR mutant thus suggesting that MsaB protein is essential for these phenotypes. These observations also confirm our previous findings that ∆msaABCR and ∆msaB mutants have similar phenotypes (Sahukhal and Elasri, 2014; Batte et al., 2016, 2018; Pandey et al., 2019).
We used EMSA to investigate whether MsaB binds to the promoter region of cidR, cidABC, and alsSD, which are genes important for regulating pathways involved in PCD and overflow metabolism. We prepared purified MsaB protein and examined the gel shift of the protein–DNA complex with the 5′-biotinylated, amplified promoter region for each of the three target genes. The presence of a shifted band as a result of MsaB binding to the 5′-biotinylated promoter regions was observed only with the promoter region of the cidR gene (Figure 8) and not with the promoter regions of the cidABC (Supplementary Figure 3) or alsSD (Supplementary Figure 4) genes. Therefore, MsaB could act as a direct transcriptional regulator of the cidR gene. To determine the specificity of this binding, a 100-fold excess of unlabeled PcidR probe was added to the reaction mixture. Addition of this competitive probe resulted in an almost complete elimination of the band corresponding to the labeled PcidR–MsaB complex, indicating that the non-labeled specific probe competed with the labeled probe for the limited amount of MsaB (Figure 8). These results indicate that the MsaB protein binds to the cidR promoter and that recognition of the promoter is specific. Therefore, specific binding of MsaB protein with the cidR promoter and increased expression of the cidR gene in the ∆msaABCR mutant under excess-glucose conditions suggests that MsaB may be a transcriptional repressor of the cidR gene under these conditions.
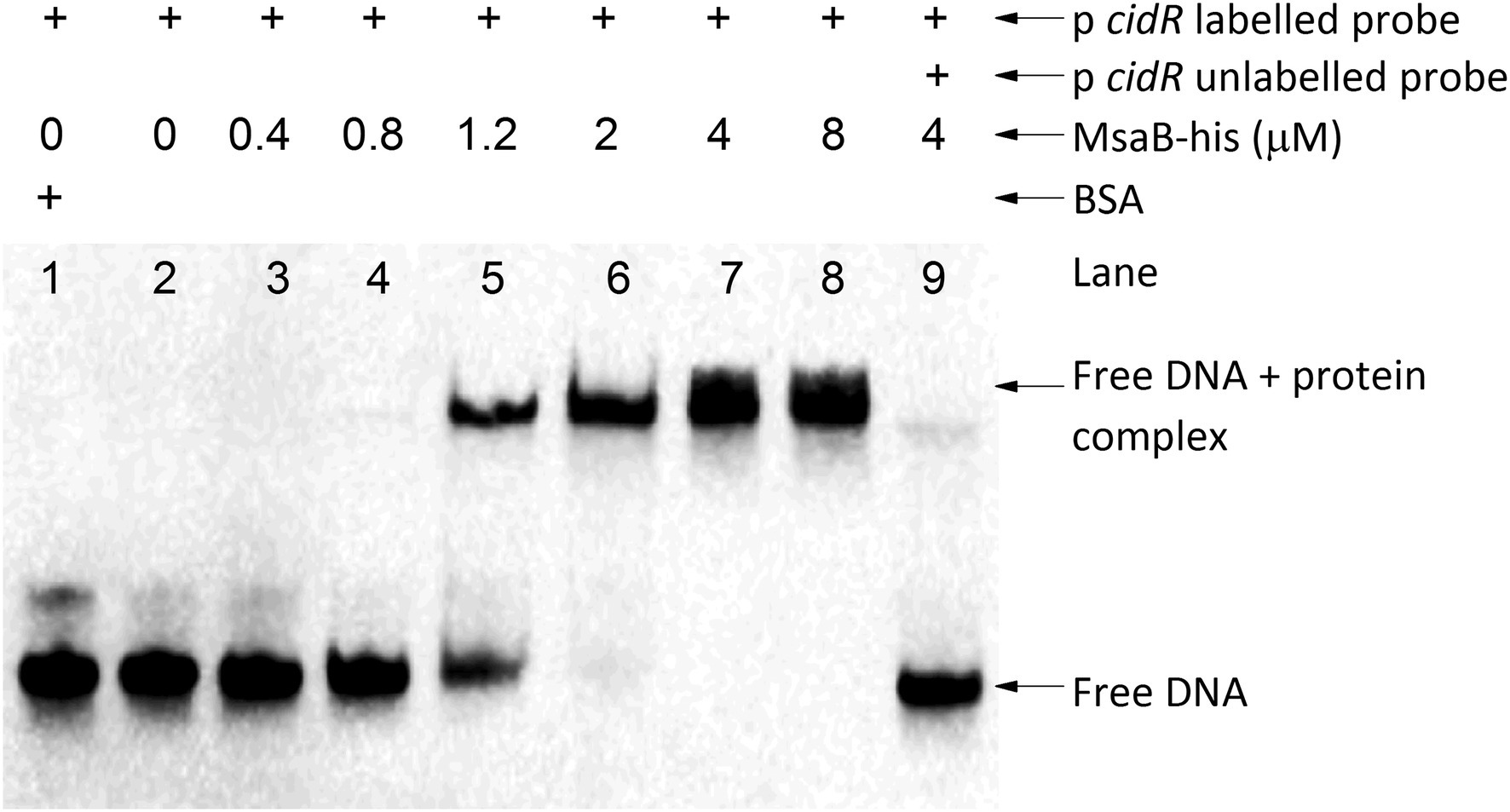
Figure 8. MsaB binds to the cidR promoter. EMSA with the cidR promoter. The shifted DNA–protein complex (PcidR–MsaBhis complex) is shown after an increased concentration of MsaBhis was incubated with a biotin-labeled cidR promoter target (labeled probe). The gel shift was reversed when a 100-fold excess concentration of unlabeled cidR promoter probe (unlabeled probe) was added to the reaction mixture, demonstrating that the label did not bind nonspecifically to MsaBhis.
To examine whether the cell death and overflow metabolism phenotypes observed for the ∆msaABCR mutant are due to MsaB affecting the cidR regulator, we created the ∆msaABCR/cidR:Tn mutant. By comparing the ∆msaABCR/cidR:Tn mutant with the wild-type, ∆msaABCR mutant, and cidR:Tn mutant strains, we assessed the following phenotypes: stationary-phase survival after 72 h of growth, glucose and acetate kinetics over 24 h, rates of glucose consumption and acetate generation after late exponential growth phase, and acetoin production after 12 h of growth (Figures 7A–D; Supplementary Figure 5). As observed in previous studies (Chaudhari et al., 2016), deletion of cidR caused a significant decrease in survival, relative to the wild-type strain, after 72 h of growth (Figure 7A). The ∆msaABCR/cidR:Tn mutant showed survival similar to that of the cidR:Tn-only mutant (Figure 7A). Deletion of cidR did not affect rates of glucose consumption and acetate production, relative to rates for the wild-type strain (Figures 7B,C; Supplementary Figure 5). However, deletion of cidR in the ∆msaABCR mutant resulted in rates of glucose consumption and acetate production that were similar to those of the wild-type and cidR mutant strains (Figures 7B,C). In addition, acetoin production was significantly decreased with deletion of cidR (Figure 7D), as shown in previous studies (Yang et al., 2006; Chaudhari et al., 2016). Deletion of cidR in the ∆msaABCR mutant also significantly decreased acetoin production, relative to the ∆msaABCR mutant (Figure 7D), and the ∆msaABCR/cidR:Tn mutant produced acetoin at a level similar to that of the cidR:Tn mutant (Figure 7D). These results suggest that the cell death and overflow metabolism phenotypes observed in the ∆msaABCR mutant are due to effects of MsaB on the cidR regulator.
CidR increases murein hydrolase activity, via an unknown mechanism, when cells are grown under excess-glucose conditions (Yang et al., 2006), and the ∆msaABCR mutant also represses this activity (Samanta and Elasri, 2014; Sahukhal et al., 2015). To examine whether increased murein hydrolase activity in the ∆msaABCR mutant is due to the effects of MsaB on the cidR regulator, we measured this activity in the ∆msaABCR/cidR:Tn mutant and compared it with that of the USA300 LAC, ∆msaABCR mutant, and cidR:Tn mutant strains. The cidR:Tn mutant showed significantly decreased murein hydrolase activity, confirming the previous study (Yang et al., 2006). However, this activity in the ∆msaABCR/cidR:Tn mutant was not completely restored to either wild type or to the cidR:Tn mutant activity level; rather, in the ∆msaABCR/cidR:Tn mutant, murein hydrolase activity remained significantly lower than that of the ∆msaABCR mutant (Figure 9). These observations suggest that cidR is not the sole regulator contributing to the murein hydrolase phenotype in the ∆msaABCR mutant.
Figure 9. Quantitative murein hydrolase assay. Extracellular proteins (100 μg) extracted from 16-h cultures of USA300 LAC, ∆msaABCR mutant, cidR:Tn mutant, and ∆msaABCR/cidR:Tn mutant strains grown in TSB-50 mM glucose were each added to a 1-ml M. luteus cell suspension containing 1 mg/ml glucose. Murein hydrolase activity of each sample was measured as a decline in OD600 of the cell suspension every 30 min over 3 h. Each data point represents the mean of three independent experiments. Error bars represent the SE.
Pyruvate Induces Expression of the msaABCR Operon to Repress Growth and Pyruvate Catabolism Under Conditions of Excess-Pyruvate During the Post-exponential Growth Phase
We observed a significant increase in expression (4.7-fold) of the msaB gene when the USA300 LAC (wild type) strain was grown in TSB-50 mM glucose, compared with TSB without glucose (Supplementary Table 4). We also confirmed previous reports of activation of the CidR regulon with glucose (Supplementary Table 4; Rice et al., 2005; Yang et al., 2006; Thomas et al., 2014). It has been speculated that, during overflow metabolism, when S. aureus cells are grown in medium with excess glucose, carbon catabolite repression may result in accumulation of intracellular pyruvate from glucose catabolism (Rice and Bayles, 2008; Sadykov and Bayles, 2012).
To determine whether the expression of msaABCR that is induced by excess glucose occurs via its catabolism to produce excess intracellular pyruvate, we used qRT-PCR to measure the relative expression of genes in the msaABCR operon (msaA, msaB, msaC) in the USA300 LAC strain after 5 h of growth in TSB without glucose supplemented with 2% pyruvate relative to the strain grown in TSB without glucose and pyruvate (i.e., control medium). Results showed that expression of the msaABCR operon genes is significantly higher when grown in TSB without glucose supplemented with 2% pyruvate compared with control medium, suggesting that pyruvate induces expression of the msaABCR operon (Table 2). We also measured MsaB production in USA300 LAC cells grown under the following conditions: TSB without glucose (control), TSB without glucose supplemented with 2% pyruvate, and TSB-50 mM glucose. We observed that MsaB production in cells grown in TSB-50 mM glucose and in TSB without glucose supplemented with 2% pyruvate was significantly higher than in cells grown in TSB without glucose and pyruvate (Figure 10). This observation is consistent with increased expression of the msaB gene that occurs when cells were grown under the same conditions (Table 2 Supplementary Table 4). These results show that both glucose and pyruvate induce expression of the msaABCR operon and production of MsaB in the USA300 LAC strain.
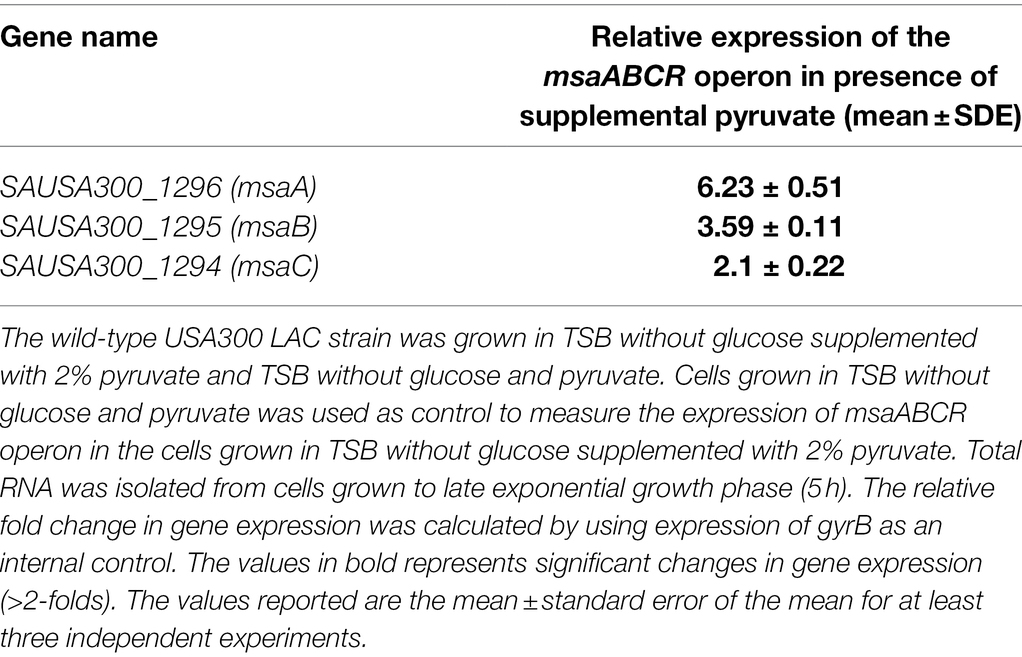
Table 2. Expression of the msaABCR operon in the wild-type USA300 LAC strain in presence of supplemental pyruvate in TSB media.
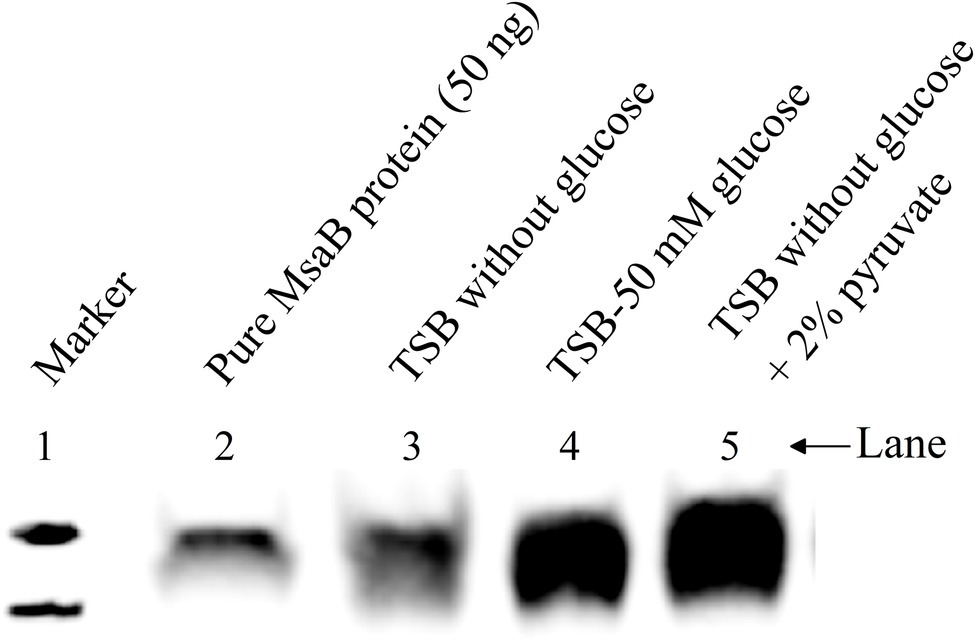
Figure 10. Western blot to show the MsaBhis production in USA300 LAC msaABhisCR complementation cells under different growth conditions. Cells were grown in different growth conditions like TSB without glucose, TSB-50 mM glucose, and TSB without glucose supplemented with 2% pyruvate. Cells were harvested and cell lysate were prepared. From the crude cell lysate, 25 μg of total protein was loaded in each lane. These results are representative of three independent experiments for each sample set.
Because we observed that pyruvate induces msaABCR expression and MsaB production, we evaluated the ∆msaABCR mutant strain’s growth and ability to catabolize pyruvate to acetate when grown in TSB with 1% pyruvate for 5 h (Figure 11). For the USA300 LAC strain, growth (measured at OD600nm) in TSB was not statistically different from that in TSB with 1% pyruvate. Growth of the ∆msaABCR mutant in TSB was slightly lower than that of the USA300 LAC (wild type) strain. However, the ∆msaABCR mutant’s growth was significantly increased in TSB with 1% pyruvate, relative to its growth in TSB (p < 0.05) and were comparable to the growth of USA300 LAC under both TSB, and TSB with 1% pyruvate (Figure 11A). We also measured the amount of acetate (i.e., the product of pyruvate catabolism) produced under the same conditions. All the test strains produced same levels of acetate when grown in TSB medium. Addition of pyruvate to the medium (TSB with 1% pyruvate) invariably increased acetate production of all strains tested (Figure 11B). The ∆msaABCR mutant produced even more acetate than the USA300 LAC (wild type, p < 0.005; Figure 11B), however reversed the growth phenotype back to wild-type USA300 LAC level. Thus, pyruvate corrected the cell death phenotype without reducing acetate levels, thus suggesting that reduced fitness in the ∆msaABCR mutant may be partly caused by depletion of pyruvate due to aberrant expression of cidABC and alsSD operons. These observations suggest that pyruvate plays a significant role in programmed cell death. However, additional investigation is required to study the role of pyruvate in cell death phenomenon.
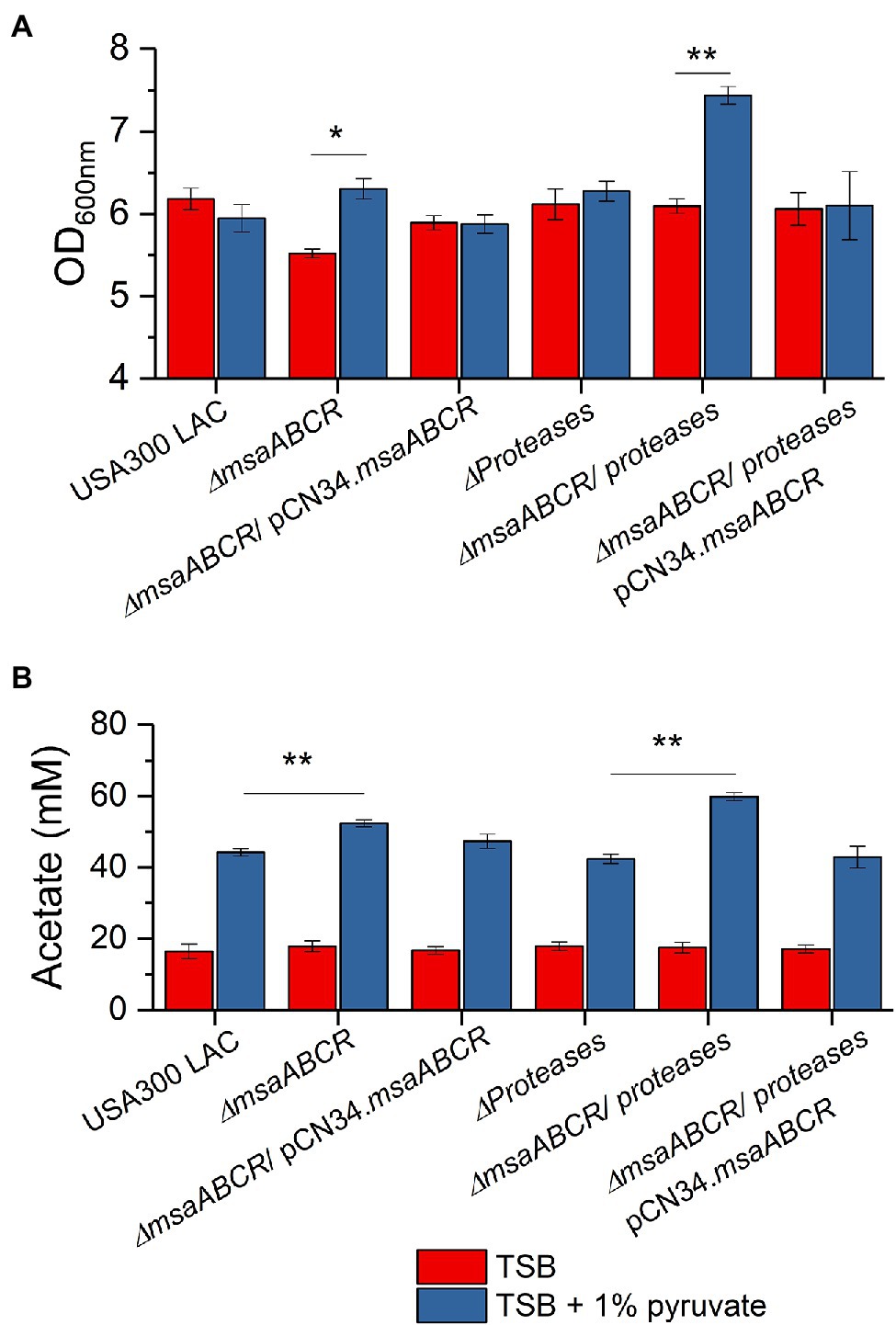
Figure 11. The OD600 measurement and acetate accumulation by the USA300 LAC (wild type), ∆msaABCR mutant, and complementation strains grown in TSB with 1% pyruvate. OD600 (A) and temporal accumulation of acetic acid (B) in the culture media of the indicated strains when grown in the indicated growth media in late exponential growth phase (5 h). Each vertical bar represents the mean of three independent experiments. Error bars represent the SE. Student’s t-test (unpaired) was used to compare the results for the two test groups (*p < 0.05, **p < 0.005).
Previously, our study showed that the ∆msaABCR mutant has increased protease activity, which modulates murein hydrolase activity (Sahukhal et al., 2015), and other studies have showed that increased concentrations of intracellular pyruvate induce extracellular proteases (Harper et al., 2018). In order to discern the effects of increased protease activity in the ∆msaABCR mutant, we used our currently tested strains combined with an all-protease-knockout strain—LAC all-protease mutant, ∆msaABCR/all-protease double mutant, and ∆msaABCR/all-protease double mutant complementation strains—and studied their growth in TSB with 1% pyruvate. We observed that growth of the ∆msaABCR/all-protease mutant was significantly increased in TSB with 1% pyruvate than in TSB (p < 0.005), but growth of the LAC all-protease mutant was the same in these two conditions (Figure 11A). In addition, when grown in TSB with 1% pyruvate, the ∆msaABCR/all-protease mutant produced significantly more acetate than the all-protease mutant (p < 0.005) and complementation strains (Figure 11B). These results show that, when cells are cultured in excess-pyruvate conditions, induces production of MsaB. Increased MsaB thus might have important role in repressing expression of cidABC and alsSD operons via cidC under increased intracellular pyruvate condition.
Discussion
In this study, we investigated the role of the msaABCR operon in regulation of the CidR regulon, and we defined the mechanism of cell death as regulated by the msaABCR operon in the USA300 LAC strain. The results showed that the ∆msaABCR mutant has increased CidC-mediated, weak-acid–dependent cell death. We observed an increased rate of glucose consumption and of acetate and acetoin production in the ∆msaABCR mutant, relative to the USA300 LAC (wild type) strain, under excess-glucose conditions. We showed that msaABCR represses expression of the cidABC and alsSD operons, which repress pyruvate catabolism to acetate and acetoin, respectively. Our results suggest that MsaB represses overflow metabolism via cidR by binding to its promoter region. Pyruvate also was shown to induce expression of the msaABCR operon. Our findings further clarify the role of the msaABCR operon in staphylococcal metabolic adaption under excess glucose condition.
Previous studies have shown that addition of glucose or antibiotics, such as vancomycin, induces biofilm formation in S. aureus (Ferreira et al., 2012; You et al., 2014; Hsu et al., 2015; He et al., 2017). Consistent with previous studies, we observed induced biofilm formation in the USA300 LAC (wild type) strain with excess glucose and/or subinhibitory concentrations of vancomycin (Hsu et al., 2011, 2015; Mirani and Jamil, 2011; He et al., 2017). However, these conditions failed to induce biofilm formation in the ∆msaABCR mutant (Figure 1), indicating that the msaABCR operon plays a role in vancomycin- and glucose-induced biofilm formation. Few studies have shown that cidA mediates induction of biofilm development under these conditions (Hsu et al., 2011, 2015). In addition, release of eDNA via cidA-mediated cell death is a major contributor to vancomycin-enhanced biofilm formation (Hsu et al., 2011, 2015; Mirani and Jamil, 2011). Defective biofilm development in the ∆msaABCR mutant under normal conditions and conditions of excess glucose or subinhibitory antibiotic stress, as well as our previous report of its reduced biofilm formation in bone implants in a rat osteomyelitis model, suggest that the msaABCR operon is essential for establishing staphylococcal biofilm infections (Sahukhal et al., 2015, 2020). Previously, we showed that deletion of the msaABCR operon results in a defective biofilm phenotype, with significantly increased dead cell biomass, decreased live cell biomass, and decreased biofilm thickness (Sahukhal and Elasri, 2014; Sahukhal et al., 2015). The process of cell death is essential for adequate biofilm formation, but proper biofilm development requires maintenance of a homeostatic balance between the dying and growing cells (Bayles, 2007; Rice and Bayles, 2008; Sadykov and Bayles, 2012; Thomas et al., 2014; Windham et al., 2016). Thus, we hypothesized that the msaABCR operon plays an important role in maintaining a balance between cell death and cell growth during normal, excess glucose and vancomycin induced biofilm development. It has been shown that the expression dynamics of the major genes involved in cell death, such as cidR, cidABC, and alsSD, that are observed in staphylococcal biofilm microcolonies are mimicked during planktonic growth in TSB-50 mM glucose (Thomas et al., 2014). Therefore, to better understand the mechanism of programmed cell death during biofilm formation, stationary-phase survival (Figure 2), overflow metabolism (Figures 3–7), and transcription of genes regulated by the CidR regulon (Table 1) by msaABCR operon, we performed these experiments in TSB-50 mM glucose.
Previous studies have proposed that acetic acid (produced during glucose catabolism) accumulates in biofilm microcolonies, creating acidic microenvironments due to low diffusion within the biofilm structure. In an environment with a pH close to the pKa of acetate (~4.8), acetate is protonated to its neutral acetic acid form, which can easily diffuse through a subpopulation of cells in microcolonies. Increased localized accumulation of acetic acid within these microcolonies then increases intracellular acidification, which can lead to unfolding or misfolding of proteins and reduce the functionality of the electron transport chain, which can, in turn, result in increased production of reactive oxygen species (ROS) and, ultimately, cause cell death in S. aureus (Bayles, 2007; Sadykov and Bayles, 2012; Thomas et al., 2014; Chaudhari et al., 2016). The present study showed that the ∆msaABCR mutant consumes glucose and produces acetate at significantly higher rates than the wild-type USA300 LAC strain during overflow metabolism (Figures 3, 4), so we hypothesized that increased acetate production and decreased extracellular pH increases cell death in the ∆msaABCR mutant. This hypothesis is supported by our observation that under MOPS-buffered condition, the increased cell death phenotype of ΔmsaABCR mutant reverted to wild-type level without affecting the rate of glucose consumption and acetate production (Figures 5B,C). The MOPS buffer does not allow pH in the culture medium to drop below 5.5, despite the ΔmsaABCR mutant produced ~50 mM acetate at 12-h time point. Under MOPS-buffered conditions, all test strains were able to reutilize acetate as secondary source of carbon after glucose was depleted from the medium (Figure 5C). Interestingly, the ΔmsaABCR mutant reutilized acetate significantly faster than the USA300 LAC (wild type) and complementation strains (Figure 5C). In our previous study, we showed that msaABCR operon represses TCA cycle (Pandey et al., 2021). Increased reutilization of acetate in ΔmsaABCR mutant is most likely due to increased TCA activity. However, further studies are needed to investigate the role of msaABCR operon in acetate reutilization mechanism.
The ∆msaABCR/cidC:Tn mutant produced a significantly lower amount of acetate than the ∆msaABCR mutant. Stationary-phase survival of the ∆msaABCR mutant also was reversed to the USA300 LAC (wild type) level when we introduced the cidC mutation into this mutant (Figure 7). These observations also suggest that increased acetate production via the CidC pathway contributes to increased weak-acid-dependent cell death in the ∆msaABCR mutant. Previously, our lab found the ∆msaABCR mutant to be defective in its oxidative stress response (Pandey et al., 2019). Therefore, we hypothesized that the inability of the ∆msaABCR mutant to cope with acetate-mediated ROS oxidative stress may also contribute to increased cell death in this mutant. Interestingly, mutation of cidC in the ∆msaABCR mutant reduced acetate production but did not reduce the rate of glucose consumption compared with the ∆msaABCR mutant without mutation (Figure 7B). However, the ∆msaABCR/cidC:Tn double mutant produced a significantly greater amount of acetoin than the cidC mutant. This suggests that the excess pyruvate (resulting from excess glucose consumption) was funneled through the AlsSD pathway to produce acetoin when acetate formation via the CidC pathway was blocked (by mutation of the cidC gene) in the ∆msaABCR/cidC:Tn mutant (Figure 7D). This finding indicates a further role for the msaABCR operon in repression of pyruvate catabolism and maintenance of pyruvate homeostasis.
In this study, we also showed that MsaB binds to the cidR gene promoter region, suggesting that it act as a transcriptional regulator of cidR (Figure 8). CidR is a transcriptional activator of two operons, cidABC and alsSD, that display pro- and anti-death functions, respectively (Yang et al., 2006; Thomas et al., 2014; Chaudhari et al., 2016; Sadykov et al., 2019). These two operons have been shown to play important roles in catabolism of pyruvate. The alsSD operon encodes acetolactate synthase and acetolactate decarboxylase, which are required for converting pyruvate to the neutral byproduct acetoin, whereas cidC encodes pyruvate:menaquinone oxidoreductase for decarboxylation of pyruvate to form acetate (Yang et al., 2006; Thomas et al., 2014; Chaudhari et al., 2016; Sadykov et al., 2019). Several studies suggest that the metabolic activities of the alsSD and cidC gene products play important roles in determining the direction of carbon flux at the pyruvate node, thereby determining cell fate during aerobic growth in excess glucose (Thomas et al., 2014; Chaudhari et al., 2016; Sadykov et al., 2019). The cidABC operon previously was shown to be transcribed into two different overlapping transcripts, cidABC and cidBC, depending on growth conditions (Rice et al., 2004). Sigma factor B (SigB) was found to activate the smaller cidBC transcript, produced during exponential growth phase (Rice et al., 2004), while the LTTR family transcriptional regulator, CidR, induces the full-length cidABC transcript during late exponential growth phase after acidification of the culture medium occurs due to increased acetate when cells are grown under excess-glucose conditions (Yang et al., 2006; Chaudhari et al., 2016). In our study, deletion of msaABCR resulted in increased cidR expression, increased cidABC and alsSD transcription, and increased acetate and acetoin production during the post-exponential phase. However, expression of the pta and ackA genes was not altered in the ∆msaABCR mutant (Table 1). Previous studies have reported that deletion of the msaABCR operon results in decreased sigB expression during exponential growth phase (Sahukhal and Elasri, 2014). All these results suggest that increased acetate and acetoin production in the ∆msaABCR mutant most likely is due to increased cidR expression that leads to the aberrant expression of cidABC and alsSD. Interestingly, deletion of cidR in the ∆msaABCR mutant reversed the rate of glucose consumption, acetate and acetoin production, and stationary-phase survival comparable to wild type and/or cidR mutant strains (Figures 7A–D; Supplementary Figure 5). These observations suggest that the observed cell death and overflow metabolism phenotypes in the ∆msaABCR mutant occur via the effects of MsaB on the cidR regulator.
Studies have shown that an acidic pH within microcolonies can activate the CidR regulon in a subpopulation of cells (Thomas et al., 2014). CidR activation causes increased murein hydrolase activity and cidABC activation, leading to a feed-forward loop in which toxic levels of acetate are reached as a result of increased CidC activity, ultimately resulting in cell death with the exhaustion of repair mechanisms (Patton et al., 2005; Thomas et al., 2014; Chaudhari et al., 2016; Marshall et al., 2016). Thus, regulation of cidR expression plays an important role in determining the fate of staphylococcal cells. Our results suggest that the msaABCR operon, through MsaB binding to the cidR promoter (Figure 8), is involved in controlling cidR expression to limit autolysis and acetate-dependent potentiation of cell death.
In our previous study, we showed that the ∆msaABCR mutant has increased expression of cell-wall–bound and extracellular murein hydrolases, and this activity was further increased by enhanced activity of extracellular proteases (Sahukhal et al., 2015; GC et al., 2019). We also showed that deletion of all proteases in the ∆msaABCR mutant decreased dead cells in biofilms of the ∆msaABCR mutant strain (Sahukhal et al., 2015; GC et al., 2019). CidR has been found to activate murein hydrolase activity, via an unknown mechanism, in cells grown in excess-glucose conditions (Yang et al., 2006). This activity decreased as a result of deletion of cidR in the ∆msaABCR mutant, relative to activity in the ∆msaABCR mutant (Figure 9). However, the ∆msaABCR/cidR:Tn mutant had greater murein hydrolase activity than the cidR:Tn-only mutant (Figure 9). These observations suggest that CidR-mediated murein hydrolase activity partially contributes to the increased activity in the ∆msaABCR mutant and suggest the existence of another mechanism that involves the msaABCR operon in repression of this activity.
During overflow metabolism, repression of carbon catabolites may result in the accumulation of intracellular pyruvate (Rice and Bayles, 2008; Sadykov and Bayles, 2012). Several studies have speculated that pyruvate is an inducer molecule for activation of cidR expression when cells are grown in excess glucose under acidic conditions (Thomas et al., 2014; Sadykov et al., 2019). Thus, pyruvate is the central metabolite directing overflow metabolism. Because pyruvate lies at the junction of several essential pathways in S. aureus cells, tight control of pyruvate homeostasis and its fate is crucial for metabolic adaptability in S. aureus. In this study, we showed that pyruvate induces expression of the msaABCR operon and subsequent production of MsaB. We also showed that MsaB indirectly represses pathways of pyruvate catabolismby repressing CidR regulon (cidC and alsSD) via MsaB binding to the cidR promoter. Therefore, these results suggest that the msaABCR operon (MsaB) might respond to intracellular pyruvate concentration, which would allow it to direct optimal expression of cidR to regulate programmed cell death during biofilm development and pyruvate catabolism, thereby maintaining pyruvate homeostasis. However, further studies are needed to determine whether MsaB senses pyruvate directly or indirectly. Several transcriptional regulators, including AgrAC, SaeRS, and ArlRS, have been found to be essential for pyruvate-mediated virulence regulation (Harper et al., 2018). Thus, we also cannot rule out the involvement of other transcriptional regulators and systems that are controlled by changes in glycolytic flux in pyruvate sensing by MsaB protein. Therefore, a detailed analysis of intracellular pyruvate sensing by MsaB requires further study.
In conclusion, this report suggests that the msaABCR operon regulates the process of weak-acid-dependent cell death in S. aureus (Figure 12). This study also showed that the msaABCR operon directly represses the cidR regulon to repress pyruvate catabolism during overflow metabolism. Our new findings help decipher the role of the msaABCR operon in staphylococcal overflow metabolic adaption during biofilm development, as well as its roles in pyruvate catabolism and maintenance of pyruvate homeostasis (Figure 12). Going forward, we will seek to further define the mechanism by which the msaABCR operon senses the central metabolite, pyruvate. Continued research is crucial for understanding S. aureus metabolic adaptation during chronic infection and for developing alternative treatments to combat the current rise in highly virulent, antibiotic-resistant strains in clinical infections.
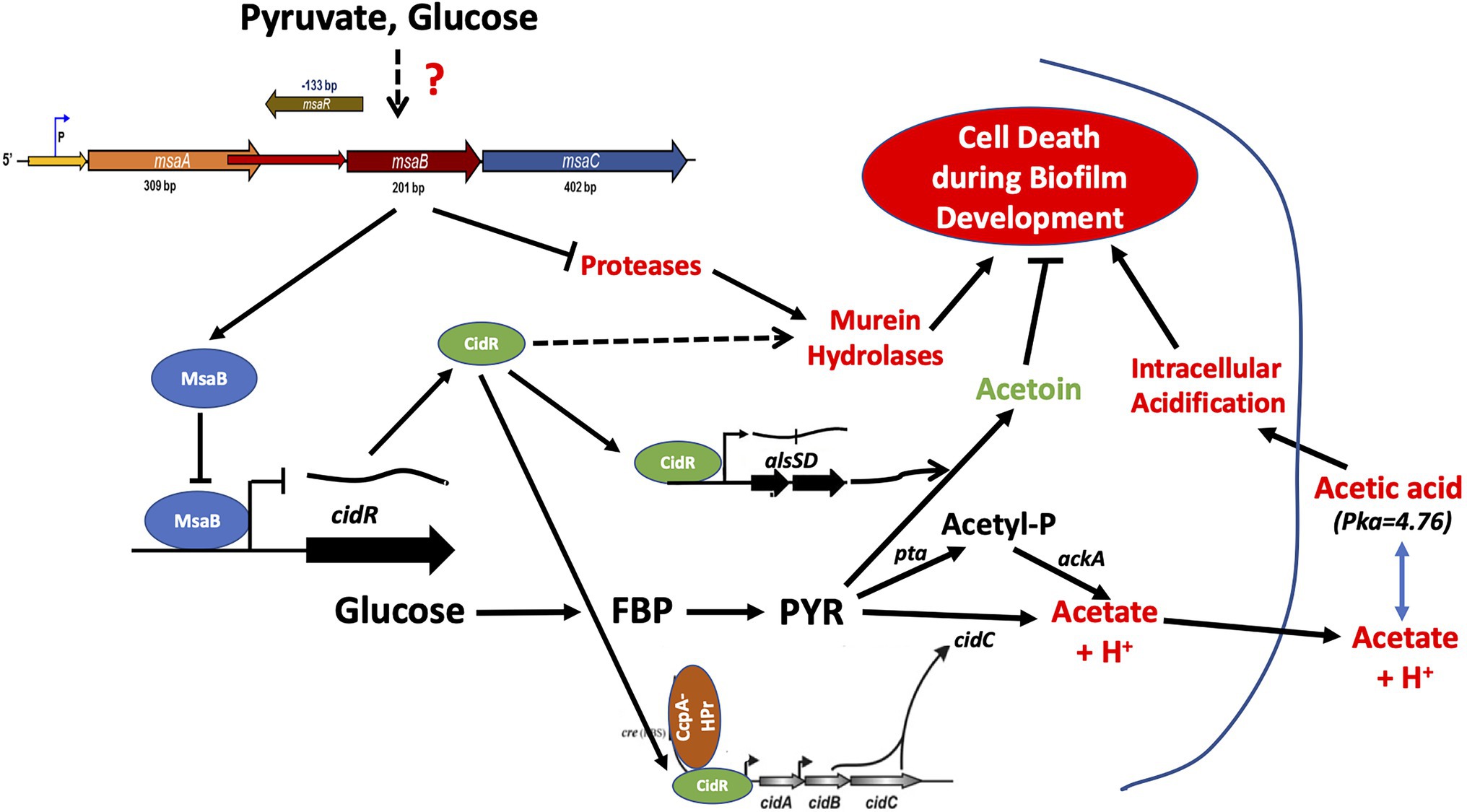
Figure 12. MsaB regulatory pathway during staphylococcal overflow metabolism. Pyruvate induces MsaB production, which, in turn, represses pyruvate catabolic pathways via the cidC and alsSD pathways by repressing CidR activity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
GSS and MOE supervised the project. BGC performed the experiments. All authors designed the project, wrote the manuscript, and read and approved the final manuscript.
Funding
This work was partially supported by the Mississippi INBRE, funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103476 and by startup funding (Z1-50328-01) from the University of Arkansas for Medical Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Lindsey Shaw, Mark Smeltzer, and the ATCC for sharing Staphylococcus aureus strains and mutants. We appreciate BEI Resources (Manassas, VA) for providing Network on Antimicrobial Resistance in S. aureus (NARSA) isolates used in this study. We are also thankful to Shanti Pandey and Sarah E. Polley for their contributions to this study. The manuscript was edited by the Science Communication Group at the University of Arkansas for Medical Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.914512/full#supplementary-material
References
Augagneur, Y., King, A. N., Germain-Amiot, N., Sassi, M., Fitzgerald, J. W., Sahukhal, G. S., et al. (2020). Analysis of the CodY RNome reveals RsaD as a stress-responsive riboregulator of overflow metabolism in Staphylococcus aureus. Mol. Microbiol. 113, 309–325. doi: 10.1111/mmi.14418
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Batte, J. L., Sahukhal, G. S., and Elasri, M. O. (2018). MsaB and CodY interact to regulate Staphylococcus aureus capsule in a nutrient-dependent manner. J. Bacteriol. 200:e00294-18. doi: 10.1128/JB.00294-18
Batte, J. L., Samanta, D., and Elasri, M. O. (2016). MsaB activates capsule production at the transcription level in Staphylococcus aureus. Microbiology 162, 575–589. doi: 10.1099/mic.0.000243
Bayles, K. W. (2007). The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5, 721–726. doi: 10.1038/nrmicro1743
Chaudhari, S. S., Thomas, V. C., Sadykov, M. R., Bose, J. L., Ahn, D. J., Zimmerman, M. C., et al. (2016). The LysR-type transcriptional regulator, CidR, regulates stationary phase cell death in Staphylococcus aureus. Mol. Microbiol. 101, 942–953. doi: 10.1111/mmi.13433
Chen, C., Krishnan, V., Macon, K., Manne, K., Narayana, S. V. L., and Schneewind, O. (2013). Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 288, 29440–29452. doi: 10.1074/jbc.M113.502039
Davis, S. C., Ricotti, C., Cazzaniga, A., Welsh, E., Eaglstein, W. H., and Mertz, P. M. (2008). Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 16, 23–29. doi: 10.1111/j.1524-475X.2007.00303.x
Dayan, G. H., Mohamed, N., Scully, I. L., Cooper, D., Begier, E., Eiden, J., et al. (2016). Staphylococcus aureus: the current state of disease, pathophysiology and strategies for prevention. Expert Rev. Vaccines 15, 1373–1392. doi: 10.1080/14760584.2016.1179583
Elbarasi, A. (2014). “Identification and Characterization of msaB gene Involved in Biofilm Formation and Virulence in Staphylococcus aureus”. master’s theses. 49. Available at: https://aquila.usm.edu/masters_theses/49
Ferreira, F. A., Souza, R. R., Bonelli, R. R., Americo, M. A., Fracalanzza, S. E., and Figueiredo, A. M. (2012). Comparison of in vitro and in vivo systems to study Ica-independent Staphylococcus aureus biofilms. J. Microbiol. Methods 88, 393–398. doi: 10.1016/j.mimet.2012.01.007
GC, B., Sahukhal, G. S., and Elasri, M. O. (2019). Role of the msaABCR operon in cell wall biosynthesis, autolysis, integrity, and antibiotic resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 63:e00680-00619. doi: 10.1128/AAC.00680-19
Goerke, C., Campana, S., Bayer, M. G., Doring, G., Botzenhart, K., and Wolz, C. (2000). Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68, 1304–1311. doi: 10.1128/IAI.68.3.1304-1311.2000
Harper, L., Balasubramanian, D., Ohneck, E. A., Sause, W. E., Chapman, J., Mejia-Sosa, B., et al. (2018). Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. MBio 9:e02272-17. doi: 10.1128/mBio.02272-17
He, X., Yuan, F., Lu, F., Yin, Y., and Cao, J. (2017). Vancomycin-induced biofilm formation by methicillin-resistant Staphylococcus aureus is associated with the secretion of membrane vesicles. Microb. Pathog. 110, 225–231. doi: 10.1016/j.micpath.2017.07.004
Hsu, C. Y., Lin, M. H., Chen, C. C., Chien, S. C., Cheng, Y. H., Su, I. N., et al. (2011). Vancomycin promotes the bacterial autolysis, release of extracellular DNA, and biofilm formation in vancomycin-non-susceptible Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 63, 236–247. doi: 10.1111/j.1574-695X.2011.00846.x
Hsu, C. Y., Shu, J. C., Lin, M. H., Chong, K. Y., Chen, C. C., Wen, S. M., et al. (2015). High glucose concentration promotes Vancomycin-enhanced biofilm formation of Vancomycin-non-susceptible Staphylococcus aureus in diabetic mice. PLoS One 10:e0134852. doi: 10.1371/journal.pone.0134852
Kinkel, T. L., Roux, C. M., Dunman, P. M., and Fang, F. C. (2013). The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. MBio 4, e00696–e00613. doi: 10.1128/mBio.00696-13
Marshall, D. D., Sadykov, M. R., Thomas, V. C., Bayles, K. W., and Powers, R. (2016). Redox imbalance underlies the fitness defect associated with inactivation of the Pta-AckA pathway in Staphylococcus aureus. J. Proteome Res. 15, 1205–1212. doi: 10.1021/acs.jproteome.5b01089
Mirani, Z. A., and Jamil, N. (2011). Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J. Basic Microbiol. 51, 191–195. doi: 10.1002/jobm.201000221
Moormeier, D. E., and Bayles, K. W. (2017). Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 104, 365–376. doi: 10.1111/mmi.13634
Nicholson, W. L. (2008). The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74, 6832–6838. doi: 10.1128/AEM.00881-08
Otto, M. (2018). Staphylococcal biofilms. Microbiol. Spectr. 6, 6–4. doi: 10.1128/microbiolspec.GPP3-0023-2018
Pandey, S., Sahukhal, G. S., and Elasri, M. O. (2019). The msaABCR operon regulates the response to oxidative stress in Staphylococcus aureus. J. Bacteriol. 201:e00417-19. doi: 10.1128/JB.00417-19
Pandey, S., Sahukhal, G. S., and Elasri, M. O. (2021). The msaABCR Operon Regulates Persister Formation by Modulating Energy Metabolism in Staphylococcus aureus. Front. Microbiol. 12:657753. doi: 10.3389/fmicb.2021.657753
Patton, T. G., Rice, K. C., Foster, M. K., and Bayles, K. W. (2005). The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 56, 1664–1674. doi: 10.1111/j.1365-2958.2005.04653.x
Patton, T. G., Yang, S. J., and Bayles, K. W. (2006). The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 59, 1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x
Petti, C. A., and Fowler, V. G. Jr. (2002). Staphylococcus aureus bacteremia and endocarditis. Infect. Dis. Clin. North Am. 16, 413–435. doi: 10.1016/S0891-5520(01)00003-4
Rice, K. C., and Bayles, K. W. (2003). Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50, 729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x
Rice, K. C., and Bayles, K. W. (2008). Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72, 85–109. doi: 10.1128/MMBR.00030-07
Rice, K. C., Firek, B. A., Nelson, J. B., Yang, S. J., Patton, T. G., and Bayles, K. W. (2003). The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185, 2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003
Rice, K. C., Nelson, J. B., Patton, T. G., Yang, S. J., and Bayles, K. W. (2005). Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187, 813–821. doi: 10.1128/JB.187.3.813-821.2005
Rice, K. C., Patton, T., Yang, S. J., Dumoulin, A., Bischoff, M., and Bayles, K. W. (2004). Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 186, 3029–3037. doi: 10.1128/JB.186.10.3029-3037.2004
Rice, K., Peralta, R., Bast, D., De Azavedo, J., and Mcgavin, M. J. (2001). Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69, 159–169. doi: 10.1128/IAI.69.1.159-169.2001
Rom, J. S., Ramirez, A. M., Beenken, K. E., Sahukhal, G. S., Elasri, M. O., and Smeltzer, M. S. (2020). The impacts of msaABCR on sarA-associated phenotypes are different in divergent clinical isolates of Staphylococcus aureus. Infect. Immun. 8:e00530-19. doi: 10.1128/IAI.00530-19
Sadykov, M. R., and Bayles, K. W. (2012). The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr. Opin. Microbiol. 15, 211–215. doi: 10.1016/j.mib.2011.12.010
Sadykov, M. R., Hartmann, T., Mattes, T. A., Hiatt, M., Jann, N. J., Zhu, Y., et al. (2011). CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 157, 3458–3468. doi: 10.1099/mic.0.051243-0
Sadykov, M. R., Thomas, V. C., Marshall, D. D., Wenstrom, C. J., Moormeier, D. E., Widhelm, T. J., et al. (2013). Inactivation of the Pta-AckA pathway causes cell death in Staphylococcus aureus. J. Bacteriol. 195, 3035–3044. doi: 10.1128/JB.00042-13
Sadykov, M. R., Windham, I. H., Widhelm, T. J., Yajjala, V. K., Watson, S. M., Endres, J. L., et al. (2019). CidR and CcpA synergistically regulate Staphylococcus aureus cidABC expression. J. Bacteriol. 201, e00371–e00379. doi: 10.1128/JB.00371-19
Sahukhal, G. S., Batte, J. L., and Elasri, M. O. (2015). msaABCR operon positively regulates biofilm development by repressing proteases and autolysis in Staphylococcus aureus. FEMS Microbiol. Lett. 362, 1–10. doi: 10.1093/femsle/fnv006
Sahukhal, G. S., and Elasri, M. O. (2014). Identification and characterization of an operon, msaABCR, that controls virulence and biofilm development in Staphylococcus aureus. BMC Microbiol. 14:154. doi: 10.1186/1471-2180-14-154
Sahukhal, G. S., Pandey, S., and Elasri, M. O. (2017). msaABCR operon is involved in persister cell formation in Staphylococcus aureus. BMC Microbiol. 17:218. doi: 10.1186/s12866-017-1129-9
Sahukhal, G. S., Tucci, M., Benghuzzi, H., Wilson, G., and Elasri, M. O. (2020). The role of the msaABCR operon in implant-associated chronic osteomyelitis in Staphylococcus aureus USA300 LAC. BMC Microbiol. 20:324. doi: 10.1186/s12866-020-01964-8
Samanta, D., and Elasri, M. O. (2014). The msaABCR operon regulates resistance in vancomycin-intermediate Staphylococcus aureus strains. Antimicrob. Agents Chemother. 58, 6685–6695. doi: 10.1128/AAC.03280-14
Sambanthamoorthy, K., Schwartz, A., Nagarajan, V., and Elasri, M. O. (2008). The role of msa in Staphylococcus aureus biofilm formation. BMC Microbiol. 8:221. doi: 10.1186/1471-2180-8-221
Seidl, K., Muller, S., Francois, P., Kriebitzsch, C., Schrenzel, J., Engelmann, S., et al. (2009). Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9:95. doi: 10.1186/1471-2180-9-95
Somerville, G. A., Chaussee, M. S., Morgan, C. I., Fitzgerald, J. R., Dorward, D. W., Reitzer, L. J., et al. (2002). Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70, 6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002
Somerville, G. A., Said-Salim, B., Wickman, J. M., Raffel, S. J., Kreiswirth, B. N., and Musser, J. M. (2003). Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 71, 4724–4732. doi: 10.1128/IAI.71.8.4724-4732.2003
Thomas, V. C., Sadykov, M. R., Chaudhari, S. S., Jones, J., Endres, J. L., Widhelm, T. J., et al. (2014). A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 10:e1004205. doi: 10.1371/journal.ppat.1004205
Thomas, V. C., Thurlow, L. R., Boyle, D., and Hancock, L. E. (2008). Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190, 5690–5698. doi: 10.1128/JB.00314-08
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Windham, I. H., Chaudhari, S. S., Bose, J. L., Thomas, V. C., and Bayles, K. W. (2016). SrrAB modulates Staphylococcus aureus cell death through regulation of cidABC transcription. J. Bacteriol. 198, 1114–1122. doi: 10.1128/JB.00954-15
Yang, S. J., Dunman, P. M., Projan, S. J., and Bayles, K. W. (2006). Characterization of the Staphylococcus aureus CidR regulon: elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol. Microbiol. 60, 458–468. doi: 10.1111/j.1365-2958.2006.05105.x
You, Y., Xue, T., Cao, L., Zhao, L., Sun, H., and Sun, B. (2014). Staphylococcus aureus glucose-induced biofilm accessory proteins, GbaAB, influence biofilm formation in a PIA-dependent manner. Int. J. Med. Microbiol. 304, 603–612. doi: 10.1016/j.ijmm.2014.04.003
Keywords: Staphylococcus aureus, msaABCR operon, overflow metabolism, pyruvate catabolism, cidR regulon, programmed cell death, biofilm formation
Citation: G C B, Sahukhal GS and Elasri MO (2022) Delineating the Role of the msaABCR Operon in Staphylococcal Overflow Metabolism. Front. Microbiol. 13:914512. doi: 10.3389/fmicb.2022.914512
Edited by:
Michal Letek, Universidad de León, SpainReviewed by:
Vinai Chittezham Thomas, University of Nebraska Medical Center, United StatesVolker Behrends, University of Roehampton London, United Kingdom
Lin Zeng, University of Florida, United States
Copyright © 2022 G C, Sahukhal and Elasri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gyan S. Sahukhal, Z3NzYWh1a2hhbEB1YW1zLmVkdQ==
 Bibek G C
Bibek G C Gyan S. Sahukhal
Gyan S. Sahukhal Mohamed O. Elasri2
Mohamed O. Elasri2