
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 05 July 2023
Sec. Molecular Innate Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1212086
This article is part of the Research Topic Broadening Our View on Nucleic Acid Sensing: Novel Sensors, Signaling Pathways, and involvement in Non-Infectious Diseases View all 9 articles
RNA interference (RNAi) plays pleiotropic roles in animal cells, from the post-transcriptional control of gene expression via the production of micro-RNAs, to the inhibition of RNA virus infection. We discuss here the role of RNAi in regulating the expression of self RNAs, and particularly transposable elements (TEs), which are genomic sequences capable of influencing gene expression and disrupting genome architecture. Dicer proteins act as the entry point of the RNAi pathway by detecting and degrading RNA of TE origin, ultimately leading to TE silencing. RNAi similarly targets cellular RNAs such as repeats transcribed from centrosomes. Dicer proteins are thus nucleic acid sensors that recognize self RNA in the form of double-stranded RNA, and trigger a silencing RNA interference response.
RNA interference (RNAi) is a ubiquitous mechanism of post-transcriptional regulation of gene expression (1). Present in plants, fungi and animals, it relies on the inhibition of messenger RNA translation via the production of micro-RNAs (miRNAs), a family of small RNAs. Irrespective of its function in regulating gene expression, RNAi also carries a role in antiviral immunity (2, 3). The machinery of RNAi is indeed equipped to recognize exogenous viral RNA and target it for cleavage, thereby thwarting infection. If exogenous virus infection represent an obvious threat, cell viability can also be jeopardized by the presence of transposable elements (TEs) in genomic DNA, which have the ability to disrupt gene expression regulation and genomic architecture (4, 5). To avoid such adverse effects, TEs are usually maintained transcriptionally silent via several mechanisms, including RNAi (6, 7). This review discusses the interplay between the machinery of RNAi and endogenous RNAs, including TEs. We focus here on mammals, without detailing the well-documented role of RNAi in controlling TEs present in invertebrates or plants [reviewed in (8)].
Mammalian control of gene expression by the machinery of RNAi depends on the production of primary miRNAs (pri-miRNAs) by genomic transcription, which are cleaved in the nucleus by Drosha, generating precursor miRNAs (pre-miRNAs, Figure 1) (1). After being translocated to the cytoplasm, pre-miRNAs are processed by a protein of the Dicer family, giving rise to mature miRNAs. Incorporated into an RNA-induced silencing complex (RISC), miRNAs guide the interaction of Argonaute (Ago) with cognate mRNAs to inhibit translation (Figure 1). Embryonic lethality of Dicer knock-out mice highlights the physiological importance of this regulatory mechanism (9). The RNAi machinery is additionally involved in the protection against RNA viruses, playing a cornerstone antiviral role in multiple eukaryotes, including plants, worms and insects (3, 10–12). The defensive role of RNAi in mammals and humans has been debated, but recent data indicate that the pathway can be protective in certain cellular contexts. For example, antiviral RNAi protects mammalian stem cells from RNA viruses (13). Viral infection leads to the accumulation of double-stranded RNA (dsRNA) in the cytoplasm, originating from viral replication or from RNA secondary structures within viral genomes. Dicer cleaves dsRNA into small-interfering RNAs (siRNAs), which are small RNAs of approximately 21-23 nucleotides in length (Figure 1). As part of RISC, siRNAs guide the degradation of viral genomes by the endonuclease Ago. Note that, while miRNA-driven RNAi does not canonically function through mRNA cleavage, antiviral RNAi largely depends on the enzymatic activity of mammalian Ago2 (3). Mammals encode a single Dicer gene, which protein product, Dicer, generates miRNAs for transcription regulation. If several studies document human Dicer’s poor ability to target dsRNA for cleavage, the protein has been involved in dsRNA-driven antiviral RNAi in specific conditions (3, 14, 15). An additional isoform, termed antiviral Dicer (aviD), is produced by alternative splicing of mRNAs from the Dicer gene. aviD is expressed in stem cells within tissues and is specialized in targeting viral dsRNA as a starting point for antiviral RNAi (13). Another truncated isoform of Dicer, Dicer°, specific to rodents, is produced in oocytes and targets dsRNA (16).
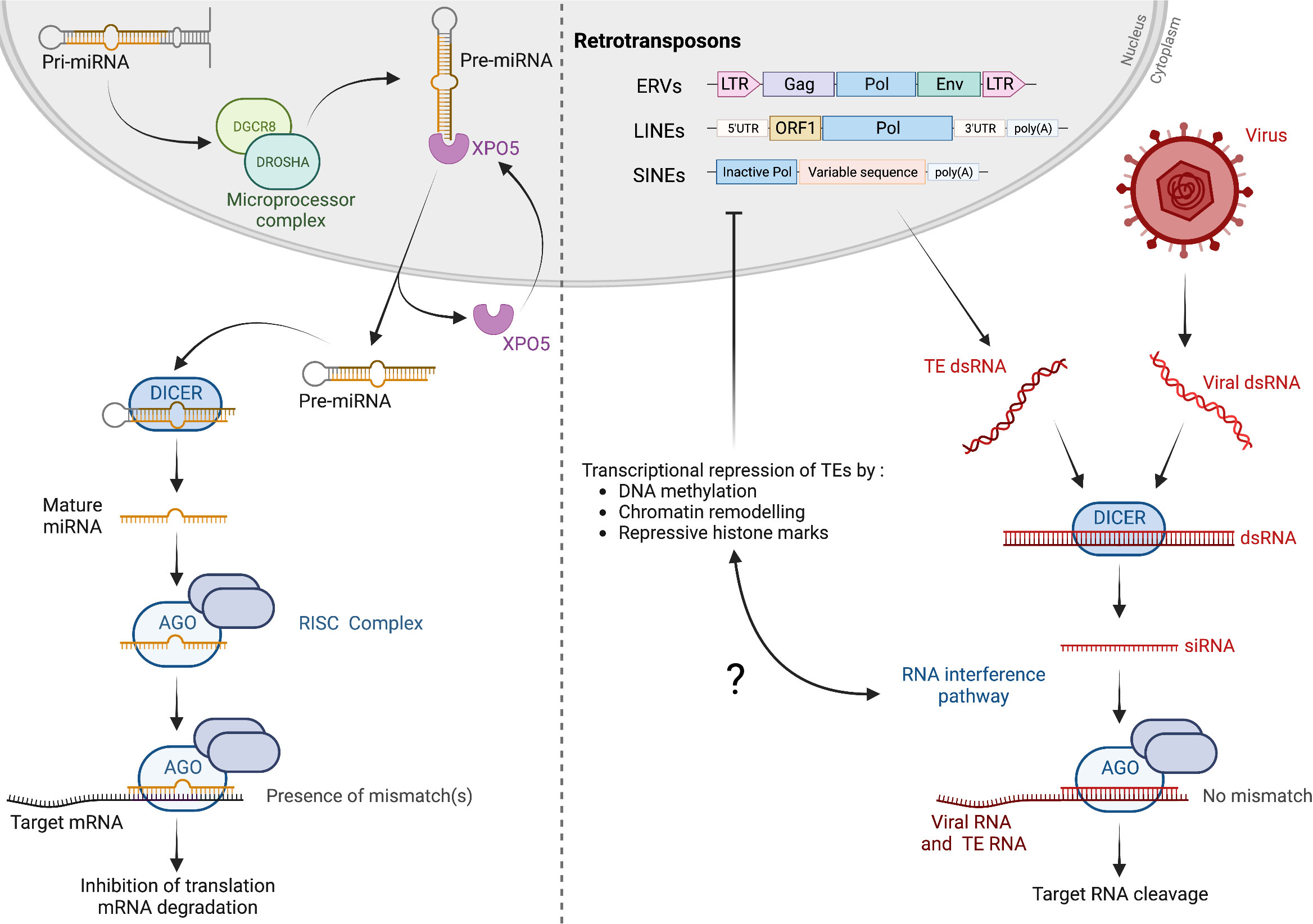
Figure 1 RNA interference in mammals. Left, post-transcriptional regulation of gene expression by RNAi. Pri-miRNAs, produced by genomic transcription, are processed into pre-miRNAs by the Microprocessor complex, composed of Drosha and Dgcr8. After shuffling to the cytoplasm, pre-miRNAs are cleaved by Dicer to generate mature miRNAs of 22 nucleotides in length on average. Loaded into RISC containing the endonuclease Ago, miRNAs guide the sequence-specific interaction with mRNAs to inhibit translation, resulting in mRNA degradation. Mismatch(s) between miRNA and mRNA sequences prevent degradation of mRNAs by Ago2. Right, expression of TEs, or RNA virus infection, lead to the cytosolic accumulation of dsRNA, which is processed by Dicer proteins, giving rise to siRNAs that guide cognate RNA degradation (no mismatch, Ago2 endonuclease is active). TEs are mainly composed of ERVs, LINEs and SINEs. ERVs may encode the ORFs Gag, Pol and Env, delimited by LTRs, while LINEs encode two ORFs. SINEs do not encode any functional ORFs and rely instead on LINE proteins for cycle completion.
An efficient antiviral RNAi response depends on the ability to specifically detect viral RNA, but not cellular RNA. This classical immunology conundrum of discrimination between “self” and “non-self” is achieved by targeting dsRNA, which represents a pathogen-associated molecular pattern (PAMP). In this context, Dicer, aviD and Dicer° display functional similarities with unrelated pattern recognition receptors (PRRs) that detect dsRNA, such as the interferon triggers RIG-I and MDA5 (17). In the absence of virus, formation of dsRNA structures within cellular RNAs is actively curtailed by Adar1, thereby preventing unwanted activation of immunity against “self” (18–21). Recognition of exogenous viral infection through dsRNA thus fits within the binary opposition “self” versus “non-self”. This is not the case when considering TEs, which are sequences –partly of retroviral origin– embedded in the genome. TEs could canonically be considered as “self” because they are encoded by the host, but nonetheless need to be identified and maintained transcriptionally silent.
Following the initial discovery of TEs by Barbara McClintock in 1950, decades of studies documented the quasi universal presence of TEs in organisms from the prokaryote and eukaryote kingdoms (22). TEs can be defined as genomic DNA sequences which, when intact, have the potential to express and re-integrate in the genome (23). While gene-encoding sequences account for 2% of total human genome, TEs constitute almost 50% of genomic DNA (23). Although regrouped under a single appellation, TEs are constituted of unrelated classes of elements, organized by their origin and transposition mechanism [Figure 1 (24)]. Retrotransposons are TEs that perform a “copy and paste” cycle, which starts with TE transcription and RNA translocation in the cytoplasm. TE DNA is subsequently produced via reverse transcription, performed by a TE-encoded enzyme, before integration in genomic DNA. Retrotransposons make up for 90% of mammalian TEs, and are composed of two unrelated groups, identified by the presence or absence of long terminal repeats (LTR) [Figure 1 (24)]. LTR elements originate from ancient events of germline infection by exogenous retroviruses; for this reason, they are coined endogenous retroviruses (ERVs) (25). Non-LTR elements are composed of long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs), with LINEs being the most abundant TEs in mice (approximately 20% of the genome (26). In humans, the LINE-1 (L1) family was documented to be uniquely able to perform a complete cycle, including reverse transcription and integration (25). SINEs, which constitute about 10% of the mouse genome, do not encode an open reading frame, but rather rely on proteins encoded by LINEs. The human genome comports more than a million copies of a hominid-specific SINE termed Alu element, which can be transcribed and form dsRNA in the cytoplasm (27).
Amid the millions of TEs populating the mouse and human genomes, only a small fraction retains the ability to complete a full transposition cycle, which comprise the evolutionary young L1s in humans (4, 28). TE genomic neo-insertions can be a source of genetic innovation, by contributing to the organization of the tridimensional chromatin architecture, as well as by participating to the evolution of gene-regulatory networks (29, 30). Nonetheless, TEs represent a threat for genomic organization, as random TE insertions may occur within coding or regulatory sequences, influencing or disrupting gene expression (30). Indeed, TEs can act as enhancers or promoters for nearby genes, or can induce heterochromatin formation in the vicinity of insertions. In line with TEs’ deleterious effects, deficiencies in TE control have been or may be linked to cancer, neurodegenerative and developmental pathologies (4, 5, 31, 32). Inhibition of TE expression relies on multiple pathways, most of which act at the transcriptional level by inducing the formation of heterochromatin on TE loci. DNA methyltransferases, including Dnmt1, deposit methyl groups on cytosines within TE genomic DNA, providing binding sites for heterochromatin modifiers (6, 33–35). Histone methylation at specific positions, notably on lysine 9 of histone 3 (H3K9me3), represent another pathway of heterochromatin formation (36, 37). The use of a given mechanism of TE inhibition depends on the nature of the family/subfamily of TEs, as well as the cell type and the physiological/pathological context. Prevention of TE expression necessitates the specific recognition of TEs within the diversity of genomic sequences, akin to a distinction between “self” (coding and non-coding genes) and “non-self” (TEs). This partly depends on a family of proteins termed Krüppel-associated box zinc-finger proteins (KRAB-ZFPs) that evolved DNA-binding motifs recognizing TE genomic DNA in a sequence-specific manner (38–40). KRAB-ZFPs interact with cofactor KAP1/TRIM28 and guide the deposition of H3K9me3 on TE DNA by the histone methyltransferase SETDB1 (39, 41, 42). KRAB-driven recognition of TEs depends on the evolution of a TE-specific DNA binding motif, only possible across important evolutionary periods. Consequently, it is tempting to speculate that KRAB-ZFPs may not be able to target the entire spectrum of evolutionary young TEs, raising the question of the means by which such sequences are maintained transcriptionally silent.
Soon after the initial discovery of RNAi by Andrew Fire and Craig Mello in C. elegans, data from the early 2000’s pointed towards a role of the pathway in controlling TEs in embryonic cells (43). Svoboda et al. used siRNAs and dsRNA injection in 1-cell embryos to knock-down Dicer, and demonstrated increased expression of Internal A Particles (IAP) and MERVL, two families of ERVs (44). In cultured human cells, siRNAs mapping to L1s could be detected, which production was linked to the activity of Dicer by a knock-down approach (45). Cumulative evidence similarly points towards a role of RNAi in controlling TE expression in embryonic stem cells (ESCs) (Figure 1). Knocking-out the Dicer gene in mouse ESCs and oocytes correlates with an increased expression of ERVs, LINEs and SINEs (16, 46–52). Additionally, TE expression is repressed by methylation of genomic DNA. Such heterochromatin mark is however erased during early development, upon a wave of demethylation occurring just after fertilization (53). Berrens et al. mimicked the wave of DNA demethylation in mouse ESCs by acutely depleting Dnmt1, leading to transcriptional activation of a limited set of TEs (52). In this context, RNAi participates to TE silencing in mouse ESCs, in compensation for the lack of methylation-driven transcription inhibition. Combining methylation loss and impaired RNAi translates into increased expression of a subset of evolutionary young ERVs such as IAPEz or MERVL (52). RNAi-driven control of TE expression is not solely present in undifferentiated cells such as ESCs. By studying age-related macular degeneration, a condition characterized by a progressive degradation of the retinal pigmented epithelium in aging patients, the Ambati group demonstrated that the phenotype is due to decreased Dicer levels, leading to upregulation of SINE RNAs (B1/B2 SINEs in mice and Alu elements in humans) (27). It is worth noting that the nature of TEs controlled by RNAi varies depending on the cell types and tissues considered. For example, Dicer ablation in cells of the retinal pigmented epithelium translates into increased TEs that seem largely restricted to SINEs, while LINEs and ERVs are upregulated in other cell types (27, 52). This could be related to the cell type-dependent organization of chromatin, linked to differentiation, as well as to other parameters.
In addition to Dicer-driven RNAi, data implicate piwi-interacting RNAs (piRNAs), a class of small RNAs, in the control of TEs (54, 55). The piRNA pathway is a well-documented anti-TE mechanism acting in the germline, through the generation of piRNAs transcribed from piRNA clusters (56). Recently, piRNAs, partly of TE origin, were detected in brain tissues of adult mice. Genetic ablation of the piRNA pathway translates into behavioral deficit, suggesting that it may be an additional small RNA-based mechanism that controls TE in somatic cells (54, 55). The functional importance of such mechanism, as well as the putative role of TE neo-integrations in piRNA clusters, is currently unknown.
Dissection of the molecular mechanism behind RNAi control of TEs suggests that it follows a classical dsRNA-driven RNAi pathway (Figure 1). Knock-down and knock-out approaches targeting the Dicer transcripts/Dicer gene show that protein products of the Dicer gene are essential for the pathway. Upon loss of Dicer in the retinal pigmented epithelium, re-expression of TEs translates into accumulation of cytosolic dsRNA, detected by immunostaining using a specific antibody (27). If the TE sequences that form dsRNA await determination, the existence of TE dsRNA is also highlighted by a body of work showing that PRRs such as MDA5, which detect dsRNA and trigger an interferon response, can be activated upon TE expression in cancer cells (57–59). Formation of TE dsRNA is expected to arise from RNA secondary structures, from transcription of inverted repeats and from bidirectional transcription, when RNAs transcribed from both sense and antisense orientation hybridize (45, 50, 52). dsRNA from TE origin is presumably similar in structure to dsRNA generated upon exogenous infection by RNA viruses. The distinction between “self” and “non-self”, on which relies RNAi-driven control of TEs, thus appears akin to the specific detection of viral infection. Existence of siRNAs from TE origin, lost upon Dicer down-regulation or KO in mouse ESCs and oocytes, points towards a role of product(s) of the Dicer gene in cleaving TE dsRNA (16, 46–50, 52). siRNAs bearing TE sequences can be immunoprecipitated with Ago2, and Ago2 KO leads to increased TE expression, demonstrating that TE control depends on RISC activity (50, 52, 60). This is not universal, as Alu silencing in the retinal pigmented epithelium does not strictly depend on Ago2 (27). Note that, if canonical Dicer is the product of the Dicer gene deemed to be the main actor of dsRNA-driven control of TEs, other Dicer isoforms can participate in TE regulation, such as Dicer° in oocytes (16). Detection of dsRNA structures formed by TE RNA is regulated by Adar1, which prevents their binding by RLRs and the activation of an interferon response (20, 59). Whether Adar1 similarly dampens RNAi silencing of TEs is currently unknown. If data document RNAi control of TE expression through the detection of dsRNA, deviations from this mechanism exist. For example, small RNAs generated by Dicer processing of cellular transfer RNAs can participate to the silencing of certain ERV families (61). Specific miRNAs, such as miR-128 and let-7, regulate the L1 family of LINEs (62, 63). Irrespective of their detailed mechanism of control, inactivation of RNAi-driven TE control translates into increased TE expression, which can have dire functional consequences.
Cytosolic dsRNA resulting from TE expression represents a canonical PAMP, or “non-self” signal, which, in differentiated cells, activates PRRs including MDA5 and the Toll-like receptor 3 (57, 58). Both PRRs’ stimulation results in the activation of innate immunity and production of interferons, potentially leading to autoimmunity. If pathways of interferon activation are functional in differentiated cells, they are severely compromised in stem cells, including ESCs, which could explain why TE re-expression does not translate into interferon-driven cytotoxicity in this context (3, 64). During age macular degeneration, expression of Alus in cells of the retinal pigmented epithelium leads to NLRP3 inflammasome activation and MyD88-dependent apoptosis, ultimately resulting in patient blindness (27, 65). Whether the downregulation of RNAi in pathological contexts could translate into TE re-expression, leading to inflammation and/or genomic instability, remains to be explored.
RNAi-driven control of TEs, relying on dsRNA targeting, mirrors antiviral RNAi thwarting exogenous virus. Even if TEs are part of genomic DNA, one could, in this framework, consider TEs as “non-self”, controlled by the pathway of antiviral RNAi, even if most of the TEs are not from viral origin (ERVs, originating from ancient events of retrovirus integration, constitute around 8% of the human genome, to put in perspective with the 40% of virus-unrelated TEs). The binary distinction between “self” and “non-self” becomes even weaker when considering that a mechanism of RNAi-driven silencing of centromeric repeats has been documented. Centromeric repeats are indeed actively silenced via the RNAi-dependent deposition of heterochromatin marks (16, 66–70). Upon Dicer KO, expression of satellite repeats constitutive of centromeres correlates with major defects in mitosis and meiosis, resulting in impaired spermatogenesis. RNAi regulates expression of centromeric repeats via dicing of satellite dsRNA (69). dsRNA-driven RNAi is thus utilized in various settings, which include –but are not limited to– PAMPs such as TE dsRNA.
There is an established role for dsRNA-driven RNAi in the control of certain families of TEs, which bears strong similarities with the antiviral RNAi pathway acting against exogenous viral infections. In this pathway, Dicer proteins act as PRRs, recognizing PAMPs in the form of dsRNA of TE origin. If such mechanism shows efficiency in controlling TEs, the selective pressures behind its existence remain unclear. Indeed, more robust pathways such as DNA and histone methylation are at play in cells to sturdily prevent the expression of TEs. In that case, why use RNAi at all? Certain situations, such as the initial wave of DNA demethylation during embryonic development, call for compensatory mechanisms. In that case, RNAi fills the space left by the inactivation of the go-to mechanism of TE control. The existence of evolutionary young TEs, not yet targetable by sequence-specific KRAB-ZNFs, may be a second example of RNAi temporarily taking over, although this remains a speculation. Subfamilies of TEs such as IAPEz, ETn and MMERK10C, that are controlled by RNAi upon DNA methylation inhibition, are also the target of KRAB-ZNFs in mouse ESCs (40, 52). Whether RNAi synergizes with other pathways of TE regulation is currently unknown. Outstanding questions of the field include the thorough mapping of TE families controlled by RNAi, which are likely to be cell type and context-dependent, as well as the determination of TE sequences forming dsRNA. The regulation of TE expression participates in various physiological and pathological processes. Increased TE expression is involved, within normal physiology, in the establishment of an immune response against commensal microbiota (71). Cellular senescence, which correlates with aging, is associated with/can be driven by increased TE expression (72–74). Tumors express certain TEs at high levels, which bolsters anti-tumor immunity by triggering an inflammatory response via dsRNA detection, as well as by providing a source of neoantigens for adaptive immunity (75–79). Compounds able to awaken TE expression in tumors and boost immunity thus represent promising anti-cancer therapies (57–59, 80). Whether RNAi is involved in TE regulation in immunity, aging or cancer remains to be explored.
EP and AC wrote the paper. AC designed the figure. All authors contributed to the article and approved the submitted version.
The authors thank members from the Stem Cell Immunity team and members of the Immunity and Cancer Unit from Institut Curie for useful discussions. This work was supported by a grant from the Fondation pour la Recherche Médicale (Amorçage Jeune Équipe ConvAJE202110014409), a grant from the Fondation Chercher Trouver and by funding from Institut Curie. Figure created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. Ding S-W, Han Q, Wang J, Li W-X. Antiviral RNA interference in mammals. Curr Opin Immunol (2018) 54:109–14. doi: 10.1016/j.coi.2018.06.010
3. Anobile DP, Poirier EZ. RNA Interference, an emerging component of antiviral immunity in mammals. Biochem Soc Trans (2023) 2023):BST20220385. doi: 10.1042/BST20220385
4. Garcia-Perez JL, Marchetto MCN, Muotri AR, Coufal NG, Gage FH, O’Shea KS, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet (2007) 16:1569–77. doi: 10.1093/hmg/ddm105
5. Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mobile DNA (2016) 7:9. doi: 10.1186/s13100-016-0065-9
6. Jansz N. DNA Methylation dynamics at transposable elements in mammals. Essays Biochem (2019) 63:677–89. doi: 10.1042/EBC20190039
7. Janssen A, Colmenares SU, Karpen GH. Heterochromatin: guardian of the genome. Annu Rev Cell Dev Biol (2018) 34:265–88. doi: 10.1146/annurev-cellbio-100617-062653
8. Gebert D, Rosenkranz D. RNA-Based regulation of transposon expression: RNAi versus transposons. WIREs RNA (2015) 6:687–708. doi: 10.1002/wrna.1310
9. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet (2003) 35:215–7. doi: 10.1038/ng1253
11. Schuster S, Miesen P, van Rij RP. Antiviral RNAi in insects and mammals: parallels and differences. Viruses (2019) 11:448. doi: 10.3390/v11050448
12. Jin Y, Zhao J-H, Guo H-S. Recent advances in understanding plant antiviral RNAi and viral suppressors of RNAi. Curr Opin Virol (2021) 46:65–72. doi: 10.1016/j.coviro.2020.12.001
13. Poirier EZ, Buck MD, Chakravarty P, Carvalho J, Frederico B, Cardoso A, et al. An isoform of dicer protects mammalian stem cells against multiple RNA viruses. Science (2021) 373:231–6. doi: 10.1126/science.abg2264
14. Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol (2008) 380:237–43. doi: 10.1016/j.jmb.2008.05.005
15. Kennedy EM, Whisnant AW, Kornepati AVR, Marshall JB, Bogerd HP, Cullen BR. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human dicer. Proc Natl Acad Sci (2015) 112:E6945–54. doi: 10.1073/pnas.1513421112
16. Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell (2013) 155:807–16. doi: 10.1016/j.cell.2013.10.001
17. Tan X, Sun L, Chen J, Chen ZJ. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol (2018) 72:447–78. doi: 10.1146/annurev-micro-102215-095605
18. Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, et al. RNA Editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science (2015) 349:1115–20. doi: 10.1126/science.aac7049
19. Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell (2018) 8172(4):811–824.e14. doi: 10.1016/j.cell.2017.12.038
20. Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, et al. Breaching self-tolerance to alu duplex RNA underlies MDA5-mediated inflammation. Cell (2018) 8;172(4):797–810.e13. doi: 10.1016/j.cell.2017.12.016
21. Stok JE, Oosenbrug T, Haar LR, Gravekamp D, Bromley CP, Zelenay S, et al. RNA Sensing via the RIG-i-like receptor LGP2 is essential for the induction of a type I IFN response in ADAR1 deficiency. EMBO J (2022) 41:e109760. doi: 10.15252/embj.2021109760
22. McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U.S.A. (1950) 36:344–55.
23. Kassiotis G. The immunological conundrum of endogenous retroelements. Annu Rev Immunol (2023) 41:annurev–immunol-101721-033341. doi: 10.1146/annurev-immunol-101721-033341
24. Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet (2007) 8:973–82. doi: 10.1038/nrg2165
25. Gagnier L, Belancio VP, Mager DL. Mouse germ line mutations due to retrotransposon insertions. Mob DNA (2019) 10:15. doi: 10.1186/s13100-019-0157-4
26. Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci (2004) 101:14572–9. doi: 10.1073/pnas.0404838101
27. Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. DICER1 deficit induces alu RNA toxicity in age-related macular degeneration. Nature (2011) 471:325–30. doi: 10.1038/nature09830
28. Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature (2009) 460:1127–31. doi: 10.1038/nature08248
29. Choudhary MN, Friedman RZ, Wang JT, Jang HS, Zhuo X, Wang T. Co-Opted transposons help perpetuate conserved higher-order chromosomal structures. Genome Biol (2020) 21:16. doi: 10.1186/s13059-019-1916-8
30. Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet (2017) 18:71–86. doi: 10.1038/nrg.2016.139
31. Tam OH, Ostrow LW, Gale Hammell M. Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mobile DNA (2019) 10:32. doi: 10.1186/s13100-019-0176-1
32. Burns KH. Transposable elements in cancer. Nat Rev Cancer (2017) 17:415–24. doi: 10.1038/nrc.2017.35
33. Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science (2016) 354:909–12. doi: 10.1126/science.aah5143
34. Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature (2004) 431:96–9. doi: 10.1038/nature02886
35. Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet (1997) 13:335–40. doi: 10.1016/s0168-9525(97)01181-5
36. Groh S, Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci (2017) 74:2055–65. doi: 10.1007/s00018-017-2454-8
37. Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, et al. DNA Methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell (2011) 8:676–87. doi: 10.1016/j.stem.2011.04.004
38. Ecco G, Imbeault M, Trono D. KRAB zinc finger proteins. Development (2017) 144:2719–29. doi: 10.1242/dev.132605
39. Wolf G, Yang P, Füchtbauer AC, Füchtbauer E-M, Silva AM, Park C, et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev (2015) 29:538–54. doi: 10.1101/gad.252767.114
40. Imbeault M, Helleboid P-Y, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature (2017) 543:550–4. doi: 10.1038/nature21683
41. Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: targeted silencing of foreign DNA in mammalian genomes by the krüppel-associated box zinc finger protein family. Mobile DNA (2015) 6:17. doi: 10.1186/s13100-015-0050-8
42. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature (2010) 463:237–40. doi: 10.1038/nature08674
43. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature (1998) 391:806–11. doi: 10.1038/35888
44. Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-l and IAP in preimplantation mouse embryos. Dev Biol (2004) 269:276–85. doi: 10.1016/j.ydbio.2004.01.028
45. Yang N, Kazazian HH. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol (2006) 13:763–71. doi: 10.1038/nsmb1141
46. Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev (2008) 22:2773–85. doi: 10.1101/gad.1705308
47. Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA Sequence analysis defines dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci (2007) 104:18097–102. doi: 10.1073/pnas.0709193104
48. Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev (2006) 20:1732–43. doi: 10.1101/gad.1425706
49. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature (2008) 453:534–8. doi: 10.1038/nature06904
50. Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature (2008) 453:539–43. doi: 10.1038/nature06908
51. Bodak M, Cirera-Salinas D, Yu J, Ngondo RP, Ciaudo C. Dicer , a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells. FEBS Open Bio (2017) 7:204–20. doi: 10.1002/2211-5463.12174
52. Berrens RV, Andrews S, Spensberger D, Santos F, Dean W, Gould P, et al. An endosiRNA-based repression mechanism counteracts transposon activation during global DNA demethylation in embryonic stem cells. Cell Stem Cell (2017) 21:694–703.e7. doi: 10.1016/j.stem.2017.10.004
53. Zeng Y, Chen T. DNA Methylation reprogramming during mammalian development. Genes (Basel) (2019) 10:257. doi: 10.3390/genes10040257
54. Nandi S, Chandramohan D, Fioriti L, Melnick AM, Hébert JM, Mason CE, et al. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc Natl Acad Sci USA (2016) 113:12697–702. doi: 10.1073/pnas.1609287113
55. Gasperini C, Tuntevski K, Beatini S, Pelizzoli R, Lo Van A, Mangoni D, et al. Piwil2 (Mili) sustains neurogenesis and prevents cellular senescence in the postnatal hippocampus. EMBO Rep (2023) 24:e53801. doi: 10.15252/embr.202153801
56. Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, et al. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet (2018) 52:131–57. doi: 10.1146/annurev-genet-120417-031441
57. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell (2015) 162(5):974–86. doi: 10.1016/j.cell.2015.07.011
58. Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-Demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell (2015) 162:961–73. doi: 10.1016/j.cell.2015.07.056
59. Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature (2020) 588:169–73. doi: 10.1038/s41586-020-2844-1
60. Petri R, Brattås PL, Sharma Y, Jönsson ME, Pircs K, Bengzon J, et al. LINE-2 transposable elements are a source of functional human microRNAs and target sites. PloS Genet (2019) 15:e1008036. doi: 10.1371/journal.pgen.1008036
61. Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-Retrotransposon control by tRNA-derived small RNAs. Cell (2017) 170:61–71.e11. doi: 10.1016/j.cell.2017.06.013
62. Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, et al. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol (2015) 22:824–31. doi: 10.1038/nsmb.3090
63. Tristán-Ramos P, Rubio-Roldan A, Peris G, Sánchez L, Amador-Cubero S, Viollet S, et al. The tumor suppressor microRNA let-7 inhibits human LINE-1 retrotransposition. Nat Commun (2020) 11:5712. doi: 10.1038/s41467-020-19430-4
64. Wu X, Kwong AC, Rice CM. Antiviral resistance of stem cells. Curr Opin Immunol (2019) 56:50–9. doi: 10.1016/j.coi.2018.10.004
65. Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, et al. DICER1 loss and alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell (2012) 149:847–59. doi: 10.1016/j.cell.2012.03.036
66. Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol (2004) 6:784–91. doi: 10.1038/ncb1155
67. Kanellopoulou C. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev (2005) 19:489–501. doi: 10.1101/gad.1248505
68. Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci (2005) 102:12135–40. doi: 10.1073/pnas.0505479102
69. Yadav RP, Mäkelä J-A, Hyssälä H, Cisneros-Montalvo S, Kotaja N. DICER regulates the expression of major satellite repeat transcripts and meiotic chromosome segregation during spermatogenesis. Nucleic Acids Res (2020) 162(5):974–86. doi: 10.1093/nar/gkaa460
70. Gutbrod MJ, Roche B, Steinberg JI, Lakhani AA, Chang K, Schorn AJ, et al. Dicer promotes genome stability via the bromodomain transcriptional co-activator BRD4. Nat Commun (2022) 13:1001. doi: 10.1038/s41467-022-28554-8
71. Lima-Junior DS, Krishnamurthy SR, Bouladoux N, Collins N, Han S-J, Chen EY, et al. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell (2021) 184:3794–3811.e19. doi: 10.1016/j.cell.2021.05.020
72. De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell (2013) 12:247–56. doi: 10.1111/acel.12047
73. De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature (2019) 566:73–8. doi: 10.1038/s41586-018-0784-9
74. Liu X, Liu Z, Wu Z, Ren J, Fan Y, Sun L, et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell (2023) 186:287–304.e26. doi: 10.1016/j.cell.2022.12.017
75. Kong Y, Rose CM, Cass AA, Williams AG, Darwish M, Lianoglou S, et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat Commun (2019) 10:5228. doi: 10.1038/s41467-019-13035-2
76. Ishak CA, De Carvalho DD. Reactivation of endogenous retroelements in cancer development and therapy. Annu Rev Cancer Biol (2020) 4:159–76. doi: 10.1146/annurev-cancerbio-030419-033525
77. Ng KW, Boumelha J, Enfield KSS, Almagro J, Cha H, Pich O, et al. Antibodies against endogenous retroviruses promote lung cancer immunotherapy. Nature (2023) 616(7957):563–573. doi: 10.1038/s41586-023-05771-9
78. Griffin GK, Wu J, Iracheta-Vellve A, Patti JC, Hsu J, Davis T, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature (2021) 595:309–14. doi: 10.1038/s41586-021-03520-4
79. Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med (2018) 24:1143–50. doi: 10.1038/s41591-018-0116-5
Keywords: RNA interference, mammals, transposable elements, Dicer (Dicer1), epigenetics, pattern recognition receptor (PRR), pathogen-associated molecular pattern (PAMP)
Citation: Cornec A and Poirier EZ (2023) Interplay between RNA interference and transposable elements in mammals. Front. Immunol. 14:1212086. doi: 10.3389/fimmu.2023.1212086
Received: 25 April 2023; Accepted: 20 June 2023;
Published: 05 July 2023.
Edited by:
Lise Chauveau, Institut de Recherche en Infectiologie de Montpellier (IRIM), FranceReviewed by:
Kevin Ng, The Rockefeller University, United StatesCopyright © 2023 Cornec and Poirier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enzo Z. Poirier, ZW56by5wb2lyaWVyQGN1cmllLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.