
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 28 September 2023
Sec. Inflammation
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1189036
This article is part of the Research Topic Pathogenetic mechanism and therapeutic target for inflammation in autoimmune disease View all 22 articles
Alterations in the composition or function of the gut microbiota are associated with the etiology of human diseases. Drug-microbiota interactions can affect drug bioavailability, effectiveness, and toxicity through various routes. For instance, the direct effect of microbial enzymes on drugs can either boost or diminish their efficacy. Thus, considering its wide range of metabolic capabilities, the gut microbiota is a promising target for pharmacological modulation. Furthermore, drugs can alter the microbiota and the mechanisms by which they interact with their host. Individual variances in microbial profiles can also contribute to the different host responses to various drugs. However, the influence of interactions between the gut microbiota and drugs on treatment efficacy remains poorly elucidated. In this review, we will discuss the impact of microbiota dysbiosis in the pathogenesis of rheumatoid arthritis (RA), and we will attempt to elucidate the crosstalk between the gut microbiota and disease-modifying anti-rheumatic drugs (DMARDs), with an emphasis on how drug-microbiota interactions affect the treatment efficacy in RA. We speculate that improved knowledge of these critical interactions will facilitate the development of novel therapeutic options that use microbial markers for predicting or optimizing treatment outcomes.
Rheumatoid arthritis (RA) is one of the most common immune-mediated disorders. Its primary manifestation is inflammatory arthritis, characterized by symmetric, polyarticular pain and swelling, frequently affecting peripheral joints (1). Despite recent significant advances in RA therapy, overall remission remains unsatisfactory, and once bone or joint degradation has started it is virtually impossible to reverse (2). The poorly defined RA pathogenetic mechanisms may be related to the intricate relationship between genetic, environmental, and immunological responses, resulting in immune system dysregulation and lack of autoimmune tolerance.
Over the past years, a growing number of evidence from epidemiological and translational research suggests that interactions between mucosal sites and dysbiotic microbiota may play a causal role in RA development (3, 4). Notably, altered composition of microbial flora has been observed in the preclinical stages of RA patients. Furthermore, the gut contains more innate and adaptive immune cells than any other organ in the body (5, 6). The intricate linkages between altered gut microbiota and an immune system genetically predisposed to autoimmunity may be the basis of inflammatory arthritis. These changes in the gut microbiota may result from a systemic inflammation that affects the gut, or from an interplay between the environment and the innate immune system in individuals genetically prone to RA (7). Nevertheless, studies in early-stage RA patients and results from murine models indicate that these changes may predate the onset of arthritis, and constitute a veiled trigger of systemic inflammation (3).
The emergence of new innovative technologies has facilitated the investigation of the gut microbiota, which is pivotal in determining the clinical features and therapeutic responses of RA patients (8). Great advances have been made in relation to microbial diversity and the investigation of the bacterial metagenome in RA by sequencing analysis of the bacterial 16s ribosomal RNA (rRNA) and metagenomics (8, 9). In particular, a wide range of bacterial species associated with the RA clinical presentation spectrum has been identified in RA patients (10). Additionally, cohort studies have aided in establishing a link between gut microbiota and therapeutic response variability in RA (3, 10). Therefore, understanding the mechanisms of the effects disease-modifying anti-rheumatic drugs (DMARDs) exert on the gut microbiota, the consequences of gut dysbiosis in regulating treatment efficacy, and the approaches to restore microbial symbiosis in RA is crucial.
Animal studies have demonstrated that alterations in the gut microbiota can affect local and systemic immunity, thereby resulting in joint inflammation (11, 12) (Figure 1). Antibiotics treatment has been shown to worsen arthritis in collagen-induced arthritis (CIA) mice, and to elevate levels of proinflammatory cytokines such as interleukin (IL)-6, interferon (IFN)-gamma, and IL-17 (13). Desulfovibrio, Prevotella, Parabacteroides, Odoribacter, Acetatifactor, Blautia, Coprococcus, and Ruminococcus genera were abundant, and levels of serum IL-17 and splenic CD4+ Th17 cells were elevated in arthritic mice, suggesting that the gut microbiota composition differs between CIA-susceptible and CIA-resistant mice (13). IL-17 and IL-1β production, and toll like receptor-2 (TLR2) and TLR4 activation, are increased in IL-1RA knockout mice, whereas these inflammatory responses are suppressed under germ-free conditions, attenuating arthritis development (14–16). Nevertheless, the clinical scores of these IL-RA knockout germ-free mice improved upon colonization with Bifidobacterium bifidum compared with those maintained under standard conditions (15). Maeda et al. discovered that transferring gut microbiota of RA patients to germ-free arthritis-prone SKG mice led to an increase in Th17 cells and severe arthritis. Further evidence revealed that co-culturing SKG dendritic cells with Prevotella copri resulted in activation of autoreactive cells in the gut and exacerbated joint inflammation in response to RA autoantigens (17). Pianta et al. reported that CD4+ T cells recognizing the autoantigens Filamin A and N-acetylglucosamine-6-sulfatase also recognized similar sequences from Prevotella, Butyricimonas, and Parabacteroides (18). Interestingly, they also identified antibodies against P. copri in new-onset RA patients, but not in healthy adults (18). Moreover, P. copri-specific IgA responses were strongly associated with serum concentrations of Th1- and Th17-related cytokines. This indicates that the immune response to elevated P. copri in the gut may be pivotal in initiating RA. Together, these findings suggest that the microbiota may function as a molecular mimic that triggers autoimmune responses in animal models and RA patients.
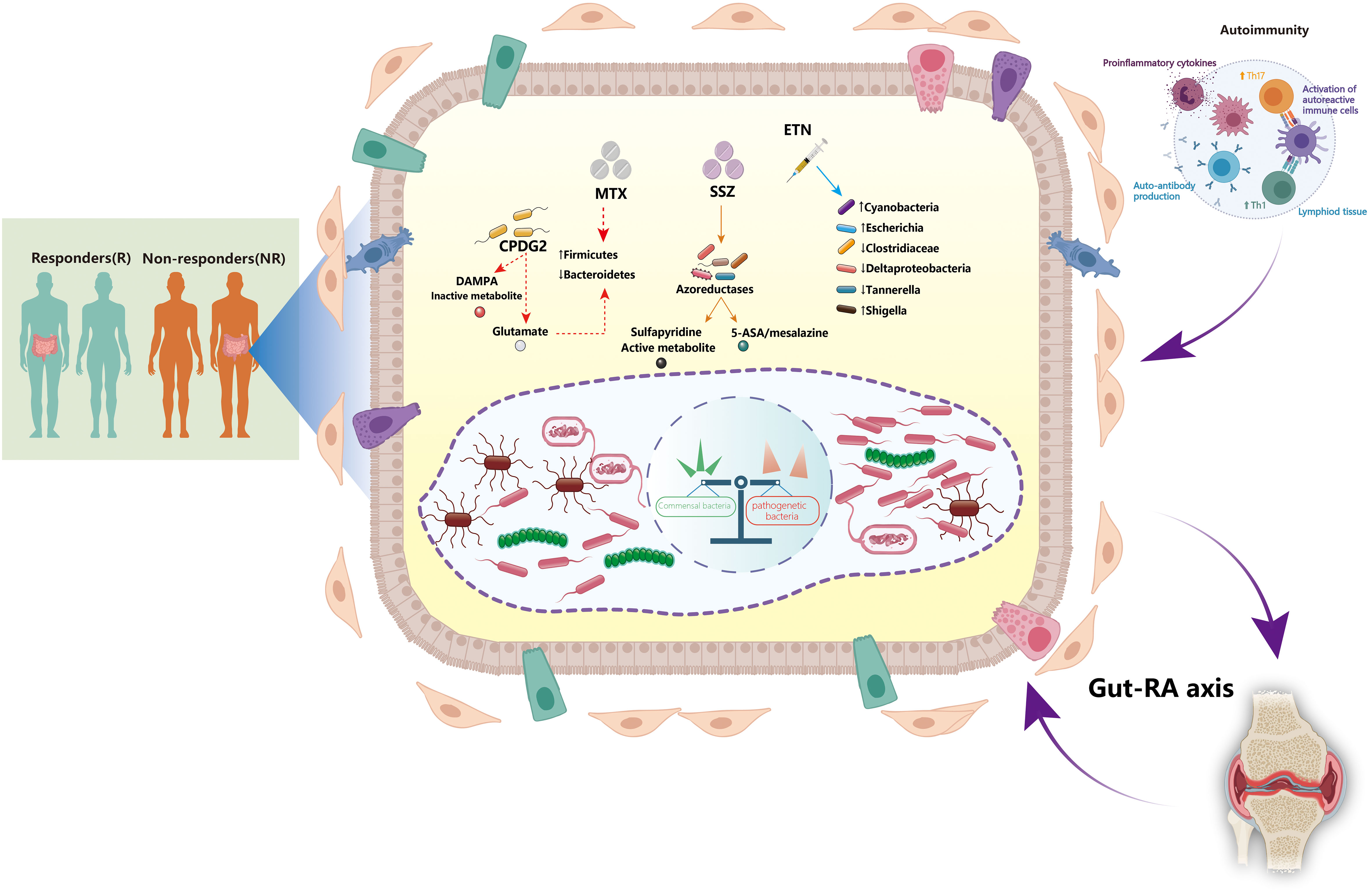
Figure 1 Interactions between the gut microbiota and disease-modifying anti-rheumatic drugs: implications for treatment efficacy in rheumatoid arthritis. The gut microbiota plays a critical role in the pathogenesis of rheumatoid arthritis (RA). The dynamic gut microbiota composition helps regulating host autoimmunity. Expansion of some opportunistic commensal bacteria may influence RA development by modifying the host’s microbiome, metabolic profile, and immune responses. The microbiota and its metabolite-associated signals are responsible for the activation and function of different immune cells. Autoreactive cells (e.g., Th1 and Th17 cells) are activated in lymphoid tissues, leading to inflammatory cytokine responses and production of autoantibodies. Certain disease-modifying anti-rheumatic drugs (DMARDs), such as methotrexate (MTX), sulfasalazine (SSZ), and etanercept (ETN), can directly affect the growth of gut microbiota. Furthermore, alteration of the gut microbiota may also contribute to the treatment efficacy of DMARDs by various mechanisms. CPDG2, carboxypeptidase-G2; DAMPA, 2,4-diamino-N(10)-methylpteroic acid; ETN, etanercept; MTX, methotrexate; SSZ, sulfasalazine; Th1, T helper 1 cells; Th17, T helper 17 cells.
Chiang et al. used metagenomic analysis to investigate the gut microbiota of RA patients with variable clinical characteristics, and observed that those with positive anti-cyclic citrullinated peptide antibodies had reduced microbial diversity and higher abundance of the Blautia, Akkermansia, and Clostridiales genera (19). A functional link between genes and the arginine deiminase enzyme was also discovered, suggesting a significant role in RA onset (20). This raises the possibility that some gut bacteria may be responsible for protein citrullination and may contribute to the abnormal autoimmunity in RA. The discovery of 21 citrullinated peptides in colon tissues of RA patients suggests that the colon contents create a tolerance site owing to loss of intestinal integrity. The presence of citrullinated antigens may trigger and perpetuate immune responses in RA (21). Patricia et al. investigated the relationship between gut microbiota and inflammatory activities in RA patients (22) and reported that the gut microbiota composition varies with RA disease activities. Despite notable variances across participants, Collinsella was strongly associated with cumulative inflammatory burden in RA patients, which is consistent with previous observations (20, 23). Cheng et al. conducted a large-scale cohort study and demonstrated dynamic changes in the gut microbiota and plasma metabolome, as well as their persistent involvement in RA pathogenesis throughout the four distinct RA stages (24). More importantly, they presented solid evidence confirming that microbial invasion of the joint synovial fluid occurs in the fourth stage of RA. Thus, a stage-specific intervention of microbial dysbiosis and metabolic disorders is warranted for improved RA prognosis and prevention.
DMARDs are encouraged by international clinical practice guidelines for the therapeutic management of RA (25). These can be administered as a single agent or in combination with other DMARDs, such as methotrexate (MTX), sulfasalazine (SSZ), and leflunomide (LEF). Corticosteroids (Cs) can be either gradually or intermittently administered in conjunction with DMARDs (25). Moreover, chloroquine (CLQ) and hydroxychloroquine (HCQ) are also recommended treatments (26, 27). Treat-to-target treatment (T2T) has recently been proposed as the optimum therapy to achieve clinical remission in RA patients by the rational use of medications. T2T comprises the combined use of MTX + SSZ + HCQ (28). When traditional medications fail to alleviate RA symptoms, clinicians may use biologic DMARDs (bDMARDs) or targeted synthetic DMARDs (tsDMARDs) (1, 29, 30). Currently, conventional synthetic DMARDs (csDMARDs) remain the gold standard for treating RA worldwide.
Despite the recent introduction of various effective therapies, low-dose MTX remains the cornerstone drug for RA treatment and is widely accepted to promote the efficacy of biologics (31, 32). There are three distinct metabolic routes for MTX (33, 34) (Figure 1). First, 2,4-diamino-N (10)-methylpteroic acid (DAMPA) is generated as a metabolite of MTX by gut bacteria; carboxypeptidase-G2 (CPDG2) is a bacterial enzyme that forms non-toxic metabolites such DAMPA and glutamate by hydrolyzing MTX (35, 36). Pseudomonas species catalyze the production of glutamate through CPDG2 from MTX in vitro, suggesting that gut bacteria may govern the availability of active metabolites of drugs as well as control their operational mechanisms (37). Second, the liver’s metabolic pathway is responsible for the biotransformation of MTX to 7-OH-MTX (38). 7-OH-MTX inhibits the dihydrofolate reductase (DHFR) enzyme. DHFR is also present in the gut microbiota, suggesting it may affect the metabolism of drugs, and vice versa, resulting in a reciprocal interaction between the drug and microbial metabolism (34, 38). Third, MTX is converted into polyglutamate within the cells. As it comprises the primary mechanism of immunomodulation, this route is prioritized over others (39, 40). Hence, the main active form of MTX relies on gut homeostasis and intestinal barrier integrity.
Gut microbiota can also indirectly regulate pharmacological metabolism by preserving the integrity of the intestinal barrier. Microbial dysbiosis affects microbial diversity and impacts translocation, immunomodulation, metabolism, and enzymatic degradation in the gut (8, 41). High MTX dosages exert anti-bacterial effects, leading to decreased Bacteroidetes and increased Firmicutes abundance in the host (42). The abundance of Bacteroides fragilis, but not that of Lactobacillales, is decreased after MTX administration in mouse models (43, 44). Furthermore, the effects of SSZ on the gut microbiota have also been confirmed despite its use as a monotherapy or in conjunction with MTX (25). Reduction of SSZ into sulfapyridine and 5-aminosalicylic acid (5-ASA/mesalazine) facilitates its metabolism by the gut microbiota through azoreductase-mediated chemical reactions (45). Most 5-ASA is stored in the colon, recirculated via the enterohepatic system, and eliminated in the feces (46). Meanwhile, the anti-inflammatory effects of 5-ASA might be neutralized by microbial arylamine of N-acetyltransferases (NATs) (47). Sulfapyridine can be metabolized in the liver by acetylation by the arylamine NAT-2, hydroxylation, and glucuronidation, all of which contribute to its anti-microbial activity (48–50). Based on the aforementioned factors, the abundance of bacteria belonging to Bifidobacterium, Lactobacillus, Enterococcus, Clostridium, Eubacterium, and Bacteroides genera, which generate azoreductases, as well as those that generate NATs, may have a significant effect on the parameters that define response to RA therapy (51–53).
bDMARDs are routinely prescribed as an alternative therapy for RA patients who do not respond well to conventional DMARDs. bDMARDs successfully delay RA progression, alleviate symptoms, and improve the overall quality of life by targeting specific proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α), IL-1, and IL-6 (1). As a TNF-α inhibitor, etanercept has a favorable impact on the gut microbiota. Etanercept administration in RA patients was related to a decline in the amount of Clostridiaceae and Deltaproteobacteria and an increased abundance of Cyanobacteria (54). In CIA mice, etanercept treatment decreased diversity and richness of the microbial community; notably, Escherichia and Shigella became more prevalent, whereas Clostridium XIVa, Tannerella, and Lactobacillus became less common (55). However, our understanding of the impact of biologics on the gut microbiome remains limited. Further research is required to address these knowledge gaps by establishing the mechanisms by which bDMARDs influence the intestinal microbiota diversity in RA.
Traditional Chinese medicine (TCM)-based treatments have considerable therapeutic efficacy and cause few side effects in RA (56, 57). Moreover, the potential success of TCM in treating RA may be partially attributed to its ability to alter the composition of the gut microbes (58, 59). Qingluo Tongbi decoction limits inflammatory responses controlled by the gut microbiota and effectively treats arthritis in AIA mice (60). Total paeony glucosides significantly improved microbial taxonomic diversity and increased the relative abundance of some preferable commensal bacteria in CIA rats (61). Xie et al. recently found that the protective effects of ASPS are mediated through the fecal microbiota and inhibited by a concomitant antibiotic cocktail, indicating that gut microbiota may be associated with ASPS (62).
Tripterygium wilfordii, a classical Chinese herbal medicine, is commonly used to treat RA in China. It can reduce inflammation and bone damage in RA through various approaches (63). Intestinal microbes, such as Holdemania fliformis and Bacteroides, are particularly abundant in RA patients; however, the abundance of these bacteria reduced substantially after treatment with glycosides from T. wilfordii. Furthermore, the microbiome of RA patients treated with MTX and T. wilfordii is abundant in Prevotella intermedia compared with those treated with T. wilfordii or MTX alone (64). Another independent study revealed that during incubation with Tripterygium glycosides and their active components, the gut microbiota produced various metabolites in the tryptophan (Trp) and phenylalanine (Phe) pathways, including two potentially favorable Trp metabolites: indole propionic acid and indole acetic acid (65). Taken together, these findings suggest that Tripterygium wilfordii can impact the host-microbiome composition and modulate metabolite production, underlining their considerable therapeutic potential in RA.
To date, the pathogenesis of RA remains obscure and conventional therapy either yields inadequate clinical efficacy or has severe adverse events. For example, up to 50% of RA patients who received MTX treatment could not acquire a clinically satisfactory outcome (66, 67); this inadequacy may be attributed to gut microbiota dysbiosis, suggesting that drug metabolism is closely associated with the gut microbiota.
Treatment with DMARDs affects the gut microbiota composition; furthermore, a feedback loop may exist between DMARDs and their effects (8) (Figure 1). Significantly, the Firmicutes and Bacteroidetes phyla could increase owing to RA treatment, which is a desired outcome in this respect (42, 68, 69). Dysbiosis can be caused by either the intrinsic ability to consume a xenobiotic or by extrinsic variables such as drug combination, prescription dosage, or treatment duration. Hence, further investigation into this area is required for the development of optimum RA treatment strategies. Increased abundance of Clostridium perfringes in the gut microbiota was observed in an RA patient cohort nonresponsive to standard drug treatment (70). In contrast, the amount of Clostridium perfringens was reduced in RA patients who responded well to drug treatment (70, 71). These observations suggest that the microbiota may influence clinical responses to RA treatment. SSZ administration is also associated with decreased E. coli and Bacteroides abundance, which highlights its anti-bacterial properties (72). Moreover, nonresponse to RA therapy has been linked to MTX consumption, and is currently attributed to increased abundance of Clostridia and decreased abundance of Bacteroidia (68, 73–75). Certain Bacteroides species may be more vulnerable or resistant to csDMARDs, and Bacteroides have been demonstrated to possess anti-microbial resistance genes (76, 77).
Variability in DMARDs responses is associated with the presence of bacterial enzymes that catalyze their metabolism. Overexposure to a protein with DHFR activity restores sensitivity to intracellular MTX in strains of medication-resistant Escherichia coli. MTX may share compatibility with bacterial DHFR, as in the case of Escherichia coli and Lactobacillus casei (78–80). Consequently, MTX is deposited in cells harboring mutations in acrA or tolC, which have rendered the efflux pump resistant to multiple AcrAB treatments that rely on tolC inactivity (81). Furthermore, MTX can be converted into MTX polyglutamate by the gut flora (44). Some patients may not respond to the initial strategy of orally administered medicine because the production pathway of tetrahydrofolate reductase, dictated by intestinal metagenomes such as Bacteroides, might compete with the DHFR and MTX metabolism in the host, thereby disturbing the anti-inflammatory effects of MTX (68).
In addition to discovering the close association between treatment efficacy and gut microbiota dysbiosis, several studies have comprehensively explored the underlying mechanisms, including immune regulation and metabolic modulation. Herein, we have exemplified the role of MTX, the anchor drug of RA treatment, to illustrate the interactions between gut microbiota and treatment responses. Nayak et al. reported that MTX significantly modifies the human gut microbiome. Despite differing drug susceptibility between strains, the action mechanism against DHFR is seemingly conserved in human and bacterial cells (42). The gut microbiota of RA patients responded differently to MTX treatment in terms of alterations in bacterial taxa and abundance in gene families. Immune activation was suppressed after transplanting post-treatment samples into germ-free mice exposed to inflammatory triggers, permitting the detection of MTX-modulated bacterial taxa associated with intestinal and splenic immune cells (42). Artacho et al. revealed a significant correlation between the abundance of gut bacterial taxa and their genes, particularly orthologs involved in methotrexate and purine metabolism, with clinical responses (82). Additionally, they created a microbiome-based model that predicts MTX non-response in a different set of patients. Intriguingly, clinical response was strongly associated with MTX levels remaining after ex vivo incubation with distal gut samples from pre-treatment RA patients, implying a direct impact of the gut microbiota on MTX metabolism and therapeutic efficacy. More recently, using machine learning Han et al. discovered that the composition of genes involved in MTX metabolism differed significantly between the response and non-response groups (83). These genes were predominantly related to Firmicutes and Bacteroidetes. Furthermore, they demonstrated that the catabolic ability of the drug in the gut microbiota is closely associated with the response mechanism to MTX in RA patients, and proposed that metabolic capability is a critical component in determining the host response to MTX.
The gut microbiota has been extensively studied over the last decades, and its importance in health and disease states has been established. Gut microbiota influence almost every biological process within the host, and microbiota dysbiosis is associated with compromised immunological tolerance and RA development. Moreover, alterations in the gut microbiota have been linked to RA disease activity, even before clinical arthritis onset. Analyzing the gut microbiota has provided novel insights into variables that promote or limit the sensitivity to disease, and has become a feasible method for predicting and reducing RA occurrence. The human gut microbiota and its enzymatic products can also directly and/or indirectly influence drug bioavailability, clinical efficacy, and toxicity. Conversely, certain medications and active ingredients can influence the immune system by modifying the gut microbiota composition, thus strengthening the host defenses. Despite significant abnormalities in specific microbial communities being associated with RA progression, several critical aspects must be addressed to facilitate development of gut microbiota-targeted treatments. First, future studies need to establish the causes of dysbiosis and to determine exactly how and when gut dysbiosis influences RA development. Second, the association of the disease with altered microbial composition and the mechanistic pathways influencing RA development must be elucidated, to acquire effective diagnostic, prognostic, and therapeutic targets. Third, it is necessary to understand the mechanism of the effects DMARDs exert on the gut microbiota and the implications of gut dysbiosis on the modulation of treatment response, in order to optimize therapeutic strategies that restore microbial symbiosis in RA patients. Collectively, further research into these unresolved topics would help promote treatment efficacy, reduce toxicity risk, and improve RA clinical outcomes.
JF conceived and wrote the manuscript. TJ collected the references. DH supervised the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Nature Science Foundation of China (82104634).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gravallese EM, Firestein GS. Rheumatoid arthritis - common origins, divergent mechanisms. N Engl J Med (2023) 388(6):529–42. doi: 10.1056/NEJMra2103726
2. Wijbrandts CA, Tak PP. Prediction of response to targeted treatment in rheumatoid arthritis. Mayo Clin Proc (2017) 92(7):1129–43. doi: 10.1016/j.mayocp.2017.05.009
3. Zaiss MM, Joyce Wu HJ, Mauro D, Schett G, Ciccia F. The gut-joint axis in rheumatoid arthritis. Nat Rev Rheumatol (2021) 17(4):224–37. doi: 10.1038/s41584-021-00585-3
4. Miyauchi E, Shimokawa C, Steimle A, Desai MS, Ohno H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat Rev Immunol (2023) 23(1):9–23. doi: 10.1038/s41577-022-00727-y
5. Zhao T, Wei Y, Zhu Y, Xie Z, Hai Q, Li Z, et al. Gut microbiota and rheumatoid arthritis: from pathogenesis to novel therapeutic opportunities. Front Immunol (2022) 13:1007165. doi: 10.3389/fimmu.2022.1007165
6. Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol (2020) 11:282. doi: 10.3389/fimmu.2020.00282
7. Garabatos N, Santamaria P. Gut microbial antigenic mimicry in autoimmunity. Front Immunol (2022) 13:873607. doi: 10.3389/fimmu.2022.873607
8. Zaragoza-Garcia O, Castro-Alarcon N, Perez-Rubio G, Guzman-Guzman IP. Dmards-gut microbiota feedback: implications in the response to therapy. Biomolecules (2020) 10(11):1479. doi: 10.3390/biom10111479
9. Xu X, Wang M, Wang Z, Chen Q, Chen X, Xu Y, et al. The bridge of the gut-joint axis: gut microbial metabolites in rheumatoid arthritis. Front Immunol (2022) 13:1007610. doi: 10.3389/fimmu.2022.1007610
10. Romero-Figueroa MDS, Ramirez-Duran N, Montiel-Jarquin AJ, Horta-Baas G. Gut-joint axis: gut dysbiosis can contribute to the onset of rheumatoid arthritis via multiple pathways. Front Cell Infect Microbiol (2023) 13:1092118. doi: 10.3389/fcimb.2023.1092118
11. Brusca SB, Abramson SB, Scher JU. Microbiome and mucosal inflammation as extra-articular triggers for rheumatoid arthritis and autoimmunity. Curr Opin Rheumatol (2014) 26(1):101–7. doi: 10.1097/BOR.0000000000000008
12. Dorozynska I, Majewska-Szczepanik M, Marcinska K, Szczepanik M. Partial depletion of natural gut flora by antibiotic aggravates collagen induced arthritis (Cia) in mice. Pharmacol Rep (2014) 66(2):250–5. doi: 10.1016/j.pharep.2013.09.007
13. Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci Rep (2016) 6:30594. doi: 10.1038/srep30594
14. Horai R, Saijo S, Tanioka H, Nakae S, Sudo K, Okahara A, et al. Development of chronic inflammatory arthropathy resembling rheumatoid arthritis in interleukin 1 receptor antagonist-deficient mice. J Exp Med (2000) 191(2):313–20. doi: 10.1084/jem.191.2.313
15. Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, et al. Stimulation of tlr2 and tlr4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest (2008) 118(1):205–16. doi: 10.1172/JCI32639
16. Rogier R, Koenders MI, Abdollahi-Roodsaz S. Toll-like receptor mediated modulation of T cell response by commensal intestinal microbiota as a trigger for autoimmune arthritis. J Immunol Res (2015) 2015:527696. doi: 10.1155/2015/527696
17. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol (2016) 68(11):2646–61. doi: 10.1002/art.39783
18. Pianta A, Arvikar SL, Strle K, Drouin EE, Wang Q, Costello CE, et al. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J Clin Invest (2017) 127(8):2946–56. doi: 10.1172/JCI93450
19. Chiang HI, Li JR, Liu CC, Liu PY, Chen HH, Chen YM, et al. An association of gut microbiota with different phenotypes in chinese patients with rheumatoid arthritis. J Clin Med (2019) 8(11):1770. doi: 10.3390/jcm8111770
20. Mena-Vazquez N, Ruiz-Limon P, Moreno-Indias I, Manrique-Arija S, Tinahones FJ, Fernandez-Nebro A. Expansion of rare and harmful lineages is associated with established rheumatoid arthritis. J Clin Med (2020) 9(4):1044. doi: 10.3390/jcm9041044
21. Bennike TB, Ellingsen T, Glerup H, Bonderup OK, Carlsen TG, Meyer MK, et al. Proteome analysis of rheumatoid arthritis gut mucosa. J Proteome Res (2017) 16(1):346–54. doi: 10.1021/acs.jproteome.6b00598
22. Ruiz-Limon P, Mena-Vazquez N, Moreno-Indias I, Manrique-Arija S, Lisbona-Montanez JM, Cano-Garcia L, et al. Collinsella is associated with cumulative inflammatory burden in an established rheumatoid arthritis cohort. BioMed Pharmacother (2022) 153:113518. doi: 10.1016/j.biopha.2022.113518
23. Jeong Y, Kim JW, You HJ, Park SJ, Lee J, Ju JH, et al. Gut microbial composition and function are altered in patients with early rheumatoid arthritis. J Clin Med (2019) 8(5):693. doi: 10.3390/jcm8050693
24. Cheng M, Zhao Y, Cui Y, Zhong C, Zha Y, Li S, et al. Stage-specific roles of microbial dysbiosis and metabolic disorders in rheumatoid arthritis. Ann Rheum Dis (2022) 81(12):1669–77. doi: 10.1136/ard-2022-222871
25. Smolen JS, Landewe R, Bijlsma J, Burmester G, Chatzidionysiou K, Dougados M, et al. Eular recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann Rheum Dis (2017) 76(6):960–77. doi: 10.1136/annrheumdis-2016-210715
26. Avina-Zubieta JA, Galindo-Rodriguez G, Newman S, Suarez-Almazor ME, Russell AS. Long-term effectiveness of antimalarial drugs in rheumatic diseases. Ann Rheum Dis (1998) 57(10):582–7. doi: 10.1136/ard.57.10.582
27. Cardiel MH, Pons-Estel BA, Sacnun MP, Wojdyla D, Saurit V, Marcos JC, et al. Treatment of early rheumatoid arthritis in a multinational inception cohort of latin american patients: the gladar experience. J Clin Rheumatol (2012) 18(7):327–35. doi: 10.1097/RHU.0b013e31826d6610
28. Smolen JS. Treat to target in rheumatology: A historical account on occasion of the 10th anniversary. Rheum Dis Clin North Am (2019) 45(4):477–85. doi: 10.1016/j.rdc.2019.07.001
29. Drosos AA, Pelechas E, Voulgari PV. Treatment strategies are more important than drugs in the management of rheumatoid arthritis. Clin Rheumatol (2020) 39(4):1363–8. doi: 10.1007/s10067-020-05001-x
30. Smolen JS, Landewe RBM, Bijlsma JWJ, Burmester GR, Dougados M, Kerschbaumer A, et al. Eular recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis (2020) 79(6):685–99. doi: 10.1136/annrheumdis-2019-216655
31. Visser K, van der Heijde D. Optimal dosage and route of administration of methotrexate in rheumatoid arthritis: A systematic review of the literature. Ann Rheum Dis (2009) 68(7):1094–9. doi: 10.1136/ard.2008.092668
32. Singh JA, Saag KG, Bridges SL Jr., Akl EA, Bannuru RR, Sullivan MC, et al. 2015 American college of rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res (Hoboken) (2016) 68(1):1–25. doi: 10.1002/acr.22783
33. Bleyer WA. The clinical pharmacology of methotrexate: new applications of an old drug. Cancer (1978) 41(1):36–51. doi: 10.1002/1097-0142(197801)41:1<36::aid-cncr2820410108>3.0.co;2-i
34. Cronstein BN. Molecular therapeutics. Methotrexate and its mechanism of action. Arthritis Rheum (1996) 39(12):1951–60. doi: 10.1002/art.1780391203
35. Buchen S, Ngampolo D, Melton RG, Hasan C, Zoubek A, Henze G, et al. Carboxypeptidase G2 rescue in patients with methotrexate intoxication and renal failure. Br J Cancer (2005) 92(3):480–7. doi: 10.1038/sj.bjc.6602337
36. Ramsey LB, Balis FM, O'Brien MM, Schmiegelow K, Pauley JL, Bleyer A, et al. Consensus guideline for use of glucarpidase in patients with high-dose methotrexate induced acute kidney injury and delayed methotrexate clearance. Oncologist (2018) 23(1):52–61. doi: 10.1634/theoncologist.2017-0243
37. Levy CC, Goldman P. The enzymatic hydrolysis of methotrexate and folic acid. J Biol Chem (1967) 242(12):2933–8. doi: 10.1016/S0021-9258(18)99594-3
38. Seideman P, Beck O, Eksborg S, Wennberg M. The pharmacokinetics of methotrexate and its 7-hydroxy metabolite in patients with rheumatoid arthritis. Br J Clin Pharmacol (1993) 35(4):409–12. doi: 10.1111/j.1365-2125.1993.tb04158.x
39. Grim J, Chladek J, Martinkova J. Pharmacokinetics and pharmacodynamics of methotrexate in non-neoplastic diseases. Clin Pharmacokinet (2003) 42(2):139–51. doi: 10.2165/00003088-200342020-00003
40. Treon SP, Chabner BA. Concepts in use of high-dose methotrexate therapy. Clin Chem (1996) 42(8 Pt 2):1322–9. doi: 10.1093/clinchem/42.8.1322
41. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol (2017) 14(6):356–65. doi: 10.1038/nrgastro.2017.20
42. Nayak RR, Alexander M, Deshpande I, Stapleton-Gray K, Rimal B, Patterson AD, et al. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host Microbe (2021) 29(3):362–77 e11. doi: 10.1016/j.chom.2020.12.008
43. Zhou B, Xia X, Wang P, Chen S, Yu C, Huang R, et al. Induction and amelioration of methotrexate-induced gastrointestinal toxicity are related to immune response and gut microbiota. EBioMedicine (2018) 33:122–33. doi: 10.1016/j.ebiom.2018.06.029
44. Tang D, Zeng T, Wang Y, Cui H, Wu J, Zou B, et al. Dietary restriction increases protective gut bacteria to rescue lethal methotrexate-induced intestinal toxicity. Gut Microbes (2020) 12(1):1714401. doi: 10.1080/19490976.2020.1714401
45. Haiser HJ, Turnbaugh PJ. Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res (2013) 69(1):21–31. doi: 10.1016/j.phrs.2012.07.009
46. Azadkhan AK, Truelove SC, Aronson JK. The disposition and metabolism of sulphasalazine (Salicylazosulphapyridine) in man. Br J Clin Pharmacol (1982) 13(4):523–8. doi: 10.1111/j.1365-2125.1982.tb01415.x
47. Delomenie C, Fouix S, Longuemaux S, Brahimi N, Bizet C, Picard B, et al. Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: evidence for highly selective acetylation of 5-aminosalicylic acid. J Bacteriol (2001) 183(11):3417–27. doi: 10.1128/JB.183.11.3417-3427.2001
48. Bishop JB, Witt KL, Gulati DK, MacGregor JT. Evaluation of the mutagenicity of the anti-inflammatory drug salicylazosulfapyridine (Sasp). Mutagenesis (1990) 5(6):549–54. doi: 10.1093/mutage/5.6.549
49. Das KM, Eastwood MA, McManus JP, Sircus W. Adverse reactions during salicylazosulfapyridine therapy and the relation with drug metabolism and acetylator phenotype. N Engl J Med (1973) 289(10):491–5. doi: 10.1056/NEJM197309062891001
50. Pullar T, Capell HA. Variables affecting efficacy and toxicity of sulphasalazine in rheumatoid arthritis. A review. Drugs (1986) 32(Suppl 1):54–7. doi: 10.2165/00003495-198600321-00011
51. Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther (1972) 181(3):555–62.
52. Rafii F, Franklin W, Cerniglia CE. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol (1990) 56(7):2146–51. doi: 10.1128/aem.56.7.2146-2151.1990
53. Rafii F, Cerniglia CE. Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ Health Perspect (1995) 103 Suppl 5(Suppl 5):17–9. doi: 10.1289/ehp.95103s417
54. Picchianti-Diamanti A, Panebianco C, Salemi S, Sorgi ML, Di Rosa R, Tropea A, et al. Analysis of gut microbiota in rheumatoid arthritis patients: disease-related dysbiosis and modifications induced by etanercept. Int J Mol Sci (2018) 19(10):2938. doi: 10.3390/ijms19102938
55. Wang B, He Y, Tang J, Ou Q, Lin J. Alteration of the gut microbiota in tumor necrosis factor-alpha antagonist-treated collagen-induced arthritis mice. Int J Rheum Dis (2020) 23(4):472–9. doi: 10.1111/1756-185X.13802
56. Mei L, Gao K, He X, Jakobsson PJ, Huang R. Editorial: disease-modifying antirheumatic drugs: approaches and lessons learned from traditional medicine. Front Pharmacol (2023) 14:1135803. doi: 10.3389/fphar.2023.1135803
57. Jakobsson PJ, Robertson L, Welzel J, Zhang M, Zhihua Y, Kaixin G, et al. Where traditional chinese medicine meets western medicine in the prevention of rheumatoid arthritis. J Intern Med (2022) 292(5):745–63. doi: 10.1111/joim.13537
58. Wang Y, Chen S, Du K, Liang C, Wang S, Owusu Boadi E, et al. Traditional herbal medicine: therapeutic potential in rheumatoid arthritis. J Ethnopharmacol (2021) 279:114368. doi: 10.1016/j.jep.2021.114368
59. Lu Q, Xu J, Jiang H, Wei Q, Huang R, Huang G. The bone-protective mechanisms of active components from tcm drugs in rheumatoid arthritis treatment. Front Pharmacol (2022) 13:1000865. doi: 10.3389/fphar.2022.1000865
60. Huang Y, Li M, Zhou L, Xu D, Qian F, Zhang J, et al. Effects of qingluo tongbi decoction on gut flora of rats with adjuvant-induced arthritis and the underlying mechanism. Evid Based Complement Alternat Med (2019) 2019:6308021. doi: 10.1155/2019/6308021
61. Peng J, Lu X, Xie K, Xu Y, He R, Guo L, et al. Dynamic alterations in the gut microbiota of collagen-induced arthritis rats following the prolonged administration of total glucosides of paeony. Front Cell Infect Microbiol (2019) 9:204. doi: 10.3389/fcimb.2019.00204
62. Liu A, Zhang M, Wu Y, Zhang C, Zhang Q, Su X, et al. Asps exhibits anti-rheumatic effects by reprogramming gut microbiota and increasing serum gamma-glutamylcysteine level. Adv Sci (Weinh) (2023) 10(3):e2205645. doi: 10.1002/advs.202205645
63. Fan D, Guo Q, Shen J, Zheng K, Lu C, Zhang G, et al. The effect of triptolide in rheumatoid arthritis: from basic research towards clinical translation. Int J Mol Sci (2018) 19(2):376. doi: 10.3390/ijms19020376
64. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med (2015) 21(8):895–905. doi: 10.1038/nm.3914
65. Xu H, Pan LB, Yu H, Han P, Fu J, Zhang ZW, et al. Gut microbiota-derived metabolites in inflammatory diseases based on targeted metabolomics. Front Pharmacol (2022) 13:919181. doi: 10.3389/fphar.2022.919181
66. Bello AE, Perkins EL, Jay R, Efthimiou P. Recommendations for optimizing methotrexate treatment for patients with rheumatoid arthritis. Open Access Rheumatol (2017) 9:67–79. doi: 10.2147/OARRR.S131668
67. Yamakawa H, Ogura T, Kameda H, Kishaba T, Iwasawa T, Takemura T, et al. Decision-making strategy for the treatment of rheumatoid arthritis-associated interstitial lung disease (Ra-ild). J Clin Med (2021) 10(17):3806. doi: 10.3390/jcm10173806
68. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
69. Hammad DBM, Hider SL, Liyanapathirana VC, Tonge DP. Molecular characterization of circulating microbiome signatures in rheumatoid arthritis. Front Cell Infect Microbiol (2019) 9:440. doi: 10.3389/fcimb.2019.00440
70. Neumann VC, Shinebaum R, Cooke EM, Wright V. Effects of sulphasalazine on faecal flora in patients with rheumatoid arthritis: A comparison with penicillamine. Br J Rheumatol (1987) 26(5):334–7. doi: 10.1093/rheumatology/26.5.334
71. Bradley SM, Neumann VC, Barr K, Troughton PR, Astbury C, Bird HA, et al. Sequential study of bacterial antibody levels and faecal flora in rheumatoid arthritis patients taking sulphasalazine. Br J Rheumatol (1993) 32(8):683–8. doi: 10.1093/rheumatology/32.8.683
72. Kanerud L, Scheynius A, Nord CE, Hafstrom I. Effect of sulphasalazine on gastrointestinal microflora and on mucosal heat shock protein expression in patients with rheumatoid arthritis. Br J Rheumatol (1994) 33(11):1039–48. doi: 10.1093/rheumatology/33.11.1039
73. Rodrigues GSP, Cayres LCF, Goncalves FP, Takaoka NNC, Lengert AH, Tansini A, et al. Detection of increased relative expression units of bacteroides and prevotella, and decreased clostridium leptum in stool samples from Brazilian rheumatoid arthritis patients: A pilot study. Microorganisms (2019) 7(10):413. doi: 10.3390/microorganisms7100413
74. Lee JY, Mannaa M, Kim Y, Kim J, Kim GT, Seo YS. Comparative analysis of fecal microbiota composition between rheumatoid arthritis and osteoarthritis patients. Genes (Basel) (2019) 10(10):748. doi: 10.3390/genes10100748
75. Qiao J, Zhang SX, Chang MJ, Cheng T, Zhang JQ, Zhao R, et al. Specific enterotype of gut microbiota predicted clinical effect of methotrexate in patients with rheumatoid arthritis. Rheumatol (Oxford) (2023) 62(3):1087–96. doi: 10.1093/rheumatology/keac458
76. Kouhsari E, Mohammadzadeh N, Kashanizadeh MG, Saghafi MM, Hallajzadeh M, Fattahi A, et al. Antimicrobial resistance, prevalence of resistance genes, and molecular characterization in intestinal bacteroides fragilis group isolates. APMIS (2019) 127(6):454–61. doi: 10.1111/apm.12943
77. Vishwanath S, Shenoy PA, Chawla K. Antimicrobial resistance profile and nim gene detection among bacteroides fragilis group isolates in a university hospital in south India. J Glob Infect Dis (2019) 11(2):59–62. doi: 10.4103/jgid.jgid_116_18
78. Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J. Crystal structures of escherichia coli and lactobacillus casei dihydrofolate reductase refined at 1.7 a resolution. I. General features and binding of methotrexate. J Biol Chem (1982) 257(22):13650–62.
79. Filman DJ, Bolin JT, Matthews DA, Kraut J. Crystal structures of escherichia coli and lactobacillus casei dihydrofolate reductase refined at 1.7 a resolution. Ii. Environment of bound nadph and implications for catalysis. J Biol Chem (1982) 257(22):13663–72.
80. Gargaro AR, Soteriou A, Frenkiel TA, Bauer CJ, Birdsall B, Polshakov VI, et al. The solution structure of the complex of lactobacillus casei dihydrofolate reductase with methotrexate. J Mol Biol (1998) 277(1):119–34. doi: 10.1006/jmbi.1997.1560
81. Kopytek SJ, Dyer JC, Knapp GS, Hu JC. Resistance to methotrexate due to acrab-dependent export from escherichia coli. Antimicrob Agents Chemother (2000) 44(11):3210–2. doi: 10.1128/AAC.44.11.3210-3212.2000
82. Artacho A, Isaac S, Nayak R, Flor-Duro A, Alexander M, Koo I, et al. The pretreatment gut microbiome is associated with lack of response to methotrexate in new-onset rheumatoid arthritis. Arthritis Rheumatol (2021) 73(6):931–42. doi: 10.1002/art.41622
Keywords: gut microbiota, dysbiosis, rheumatoid arthritis, immune response, DMARDs
Citation: Fan J, Jiang T and He D (2023) Advances in the implications of the gut microbiota on the treatment efficacy of disease-modifying anti-rheumatic drugs in rheumatoid arthritis. Front. Immunol. 14:1189036. doi: 10.3389/fimmu.2023.1189036
Received: 18 March 2023; Accepted: 15 September 2023;
Published: 28 September 2023.
Edited by:
Jian Gao, Shanghai Children’s Medical Center, ChinaReviewed by:
Dongdong Qin, Yunnan University of Chinese Medicine, ChinaCopyright © 2023 Fan, Jiang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongyi He, aGVkb25neWkxOTY3QHNodXRjbS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.