
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 04 April 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1152017
Salmonella is an important zoonotic bacterial species and hazardous for the health of human beings and livestock globally. Depending on the host, Salmonella can cause diseases ranging from gastroenteritis to life-threatening systemic infection. In this review, we discuss the effector proteins used by Salmonella to evade or manipulate four different levels of host immune defenses: commensal flora, intestinal epithelial-mucosal barrier, innate and adaptive immunity. At present, Salmonella has evolved a variety of strategies against host defense mechanisms, among which various effector proteins delivered by the secretory systems play a key role. During its passage through the digestive system, Salmonella has to face the intact intestinal epithelial barrier as well as competition with commensal flora. After invasion of host cells, Salmonella manipulates inflammatory pathways, ubiquitination and autophagy processes with the help of effector proteins. Finally, Salmonella evades the adaptive immune system by interfering the migration of dendritic cells and interacting with T and B lymphocytes. In conclusion, Salmonella can manipulate multiple aspects of host defense to promote its replication in the host.
The ability of Salmonella to be transmitted through a fecal-oral route of infection makes it one of the major worldwide public health concerns (1, 2). The intestinal mucus layer provides a barrier against invasion of the epithelium by Salmonella. Commensal microflora in the intestinal mucosal can also promote intestinal stability and prevent pathogens from invading the intestine (3–5). Although the above mentioned barriers play an important role, but some Salmonella can also cross the epithelial barrier and reach the lamina propria where they encounter resident macrophages, dendritic cells (DCs), and intra-epithelial lymphocytes (6). One of their (e.g., resident macrophages, DCs, etc.) most important roles is to eliminate Salmonella by engulfing it and respiratory burst oxidase (RBO) and reactive oxygen species (ROS) (7). In addition, the ubiquitination modification is an enzymatic cascade reaction by which intracellular Salmonella can be labeled with (8). This is one of the pivotal “eat-me” signals that help to initiate the process of autophagy. Finally, ubiquitination can activate the process of inflammation, which also contributes to the resistance to intracellular Salmonella infection (9).
In addition to destroying invading Salmonella, phagocytes create a bridge between innate and acquired immunity by presenting antigens to T cells, thereby enabling the development of long-term immunity (10–12). It has also been suggested that cellular immune responses seem to be more effective in defending against Salmonella infection, and CD4+ Th1 cells have been shown to play a major role (13). This is probably due to the production of IFN-γ and TNF-α (hallmarks of Th1 cell response) that activate macrophages to kill intracellular pathogens (14). Previous studies have suggested that CD8+ T cells participate in secondary, but not primary, bacterial clearance. However, there is also evidence that the CD8+ CTL response plays an important role in resolving primary infection with attenuated Salmonella strain (15). Moreover, the production of anti-Salmonella IgG is essential to enhance phagocytosis in the adaptive immune response (16). In general, the T-cell mediated immune response is vital in host control of Salmonella infection, including primary and subsequent infection clearance (13, 15). Likewise, B cells are also crucial to the maintenance of a proper immune defense by antigen presentation and generation of protective antibodies (17). Thus, both cellular and humoral responses are likely key components of protective immunity.
Despite the presence of various antimicrobial mechanisms in the host, Salmonella have evolved various strategies to overcome these defense mechanisms (18). In Gram-negative bacteria, there are currently known six types of protein secretion systems, identified as type I to type VI secretion systems (T1SS-T6SS), each of which shows a considerable diversity (19, 20). To date, five secretion systems have been described in Salmonella, including the T1SS, T3SS, T4SS, T5SS and T6SS (19). After entering the intestine, Salmonella can use effector proteins secreted by the secretory systems to compete with intestinal flora and establish a colonization advantage (21). Eventually, the bacteria will be absorbed into the host cells, thus promoting the invasion of Salmonella. During this period, T3SS plays an extremely important role (22). Meanwhile, Salmonella resides within a host-derived membrane compartment, so-called Salmonella-containing vacuoles (SCVs) (23). The presence of Salmonella within SCVs avoids the killing effect of the cytoplasmic environment and contributes to its replication. The formation of SCVs can also reduce the presentation of antigenic peptides by DCs or other cell types, thus affecting the adaptive immune response and contributing to the establishment of systemic infection by Salmonella in the later stage (23).
With increasing antibiotic resistance of Salmonella, there is an urgent need to develop novel agents and efficient vaccines for the treatment and control of Salmonella (24). In recent years, it has been found that pathogenicity of bacteria is closely related to various virulence proteins or effector proteins secreted by their own, but these proteins are not essential components for bacterial survival (25). Therefore, the agent or vaccine targeted by this can not only play an essential role in the response to bacterial infection, but also produce less selective pressure on bacteria, thus reducing the possibility of bacterial resistance (26). In summary, the development of vaccines targeting different secretion systems during the Salmonella infection may be an effective strategy to block the spread of the disease.
In this review, we summarize the current knowledge of how pathogenic Salmonella utilize different secretion systems to modulate immune system and facilitate bacterial invasion and colonization. Herein, we describe the various vaccine candidates targeting the secretion systems, with a discussion on their advantages and disadvantages in the context of use scenarios. Through these aspects, we hope to provide a potential novel strategy that may be applied to the development of vaccines against Salmonella.
The intestinal commensal flora plays a critical role in protecting the integrity of the intestinal mucosa (3). Among different secretion systems, T6SS is a contact-dependent secretion device capable of directly injecting effector proteins into other bacteria as well as eukaryotic cells (27). Many Gram-negative enteric pathogens, including Vibrio cholerae, Pseudomonas aeruginosa and Bacteroides fragilis can use its T6SS to defeat other bacteria (28). It suggests that T6SS may help to compete with intestinal flora for effective invasion of the intestinal mucosa. One such secretion system is also encoded in the Salmonella pathogenicity island 19 (SPI-19) present in serotypes Dublin, Weltevreden, Gallinarum and Enteritidis (29). The deletion of SPI-19 in these serotypes significantly affects the survival and colonization of Salmonella in cells and organs, and induces fast bacterial clearance. The SPI-19 deleted strain of S. Dublin also competed significantly weaker than the wild-type strain when co-cultured with strains of pathogenic E. coli, suggesting that this T6SS plays an important role in pathogenicity by killing commensal bacteria in the intestine (30, 31). S. Typhimurium harbors a T6SS encoded in SPI-6, which contributes to the capability of Salmonella to colonize mice (32). Subsequently, it was demonstrated that S. Typhimurium can also use SPI-6 T6SS to kill Klebsiella oxytoca in vitro (21). In a word, these results suggest that T6SS and its effector proteins may be a powerful weapon for Salmonella to compete with other intestinal flora as they breach the intestinal barrier (Figure 1).
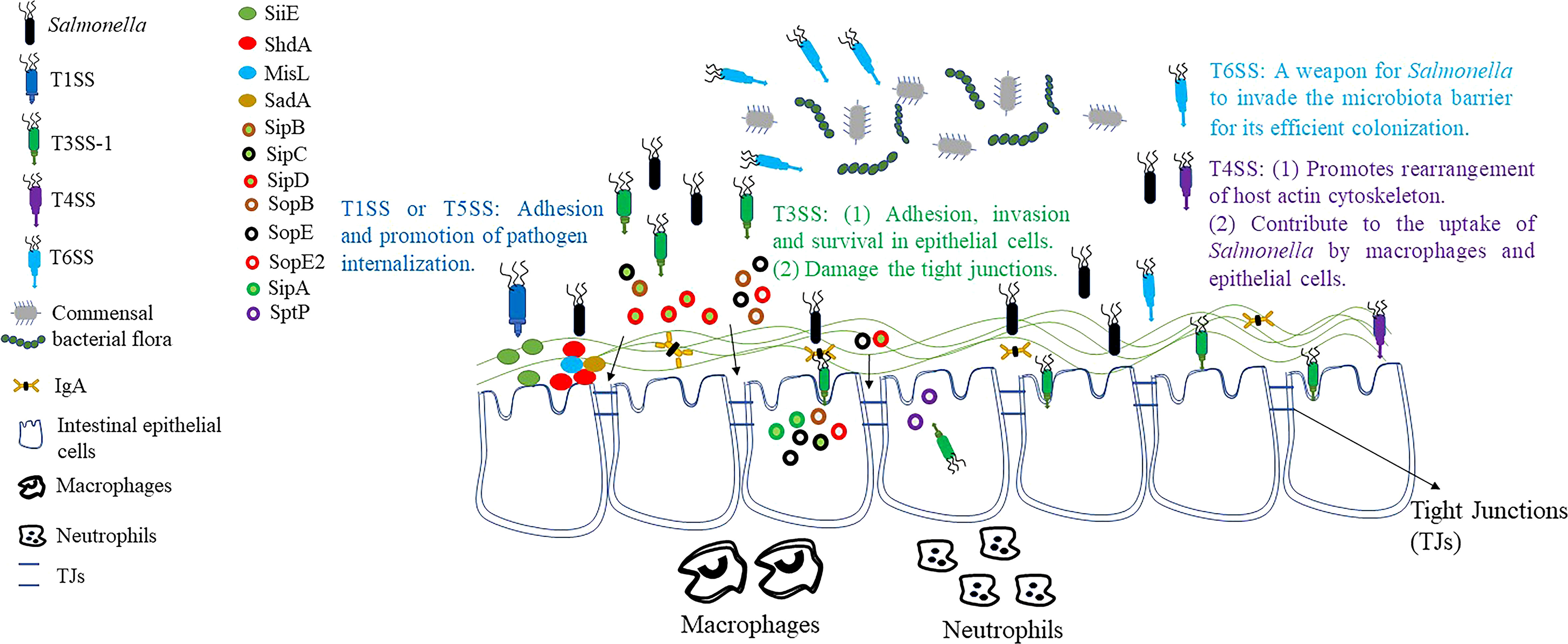
Figure 1 Breakthrough of the intestinal barrier by Salmonella. Before it reaches the intestine, Salmonella utilizes the T6SS to compete with intestinal commensal microorganisms. T1SS and T5SS facilitate the attachment and internalization of Salmonella to IECs. T4SS also contributes to host cytoskeletal rearrangement, which in turn promotes the uptake of Salmonella by macrophages and IECs. Through T3SS-1, Salmonella secretes effector proteins that lead to membrane ruffles, macropinocytosis and bacterial internalization, and ultimately invasion of host cells. At the same time, the effector protein secreted by T3SS-1 helps Salmonella to destroy TJs, which in turn breach the intestinal barrier. However, Salmonella also secretes effector proteins, such as SptP, that restore membrane structure after bacterial internalization and reverse the effects of membrane ruffles caused by other effector proteins.
After breaking through the blockade of the intestinal flora, the adhesion of Salmonella to intestinal epithelial cells (IECs) is a central step in the process of pathogenesis. The initial contact between Salmonella and polarized IECs is established by T1SS (33). This system secretes SiiE, a huge non-fimbrial adhesin that enables the bacterium to adhere the apical surface of host cell. BapA is encoded by SPI-9 and is also secreted by T1SS. It is required for S. Enteritidis to penetrate through IECs, constitutes a first stage in pathogenic processes, and is essential for S. Typhi to adhere to ECs (34, 35).
In addition to the adhesion proteins secreted by the above-mentioned T1SS, the proteins of the T5SS also play an important role in the intestinal invasion of Salmonella (35, 36). In Salmonella, three adhesins of the autotransporter protein family have been characterised earlier. ShdA and MisL are important monomeric adhesins while the putative adhesin SadA is a trimer autotransporter adhesins (TAA) (37–39). The ShdA is the only determining factor known to be required for persistence of S. Typhimurium in the mouse caecum and for efficient and extended shedding of the bacterial with the faeces (40). MisL is an autotransporter protein encoded by SPI-3. The misL mutant colonized poorly in vivo in comparison to the corresponding parental strain, with the bacterial loads recovered significantly lower than those of the wild-type S. Typhimurium strain SL1344 (37, 41). Moreover, expression of MisL enabled S. Typhimurium to bind fibronectin to its cell surface, resulting in adhesion to fibronectin-coated glass slides and in increased invasiveness for epithelial cells. These data indicate that MisL represents a potential extracellular matrix adhesin involved in intestinal colonization. Previous studies have shown that all members of the TAA family are adhesion proteins, and SadA is similar in structure to YadA protein (a member of the TAA family) of Yersinia enterocolitica (35). This result suggested that the SadA is probably also an important mediator of Salmonella adhesion. In addition, expression of SadA led to the agglutination of cell, formation of biofilm, and increased adhesion capability to human IECs (39). In conclusion, these results indicate that the colonization, adhesion and invasion of Salmonella in the intestine may require the cooperation of multiple secretory systems or multiple effector proteins (Figure 1).
Intercellular tight junctions (TJs) connect IECs to form a physical barrier that restricts bacterial pathogen invasion and migration (42). Salmonella has also developed various strategies to destroy TJs (Figure 1). Recent studies discovered that the T3SS-secreted effector proteins SopB, SopE, SopE2, and SipA are responsible for TJs structure and function disruption (43–45). In contrast, AvrA may play an important role in the stabilization of TJs. Previous studies have found that the presence of AvrA stabilizes intestinal TJs as well as normal cellular permeability, and the normal structure of TJs is more severely impaired by AvrA-deficient strains (46). To summarize, AvrA stabilized TJs despite the fact that the other T3SS effector proteins, SopB, SopE, SopE2, and SipA, are reported to disrupt TJs.
After successful adhesion to IECs, Salmonella can promote its internalization by different secretory systems or effector proteins (Figure 1). Currently, there are six effector proteins of Salmonella (SipA, SipC, SopB, SopE, SopE2 and SptP) that can regulate the actin cytoskeleton directly or indirectly (47, 48). Nucleation of microfilaments and their bundling by SipC leads to cytoskeletal rearrangements in cultured cells that result in membrane ruffles below the site of Salmonella attachment (49). Moreover, SipA promotes filament assembly and stabilizes filaments once they have formed (50). Thus, the combined activities of SipC and SipA promote the composition of actin filaments in proximity to attached Salmonella, and stabilize these filaments against disassembly by host regulatory proteins. In addition, the conversion of unbranched filaments into the branched filament networks that drive membrane evagination requires the stimulation of Rac (a member of the Rho family of GTPase, modulate the polymerization of actin) by SopE, as well as the downstream activation of the Arp2/3 (a crucial regulator of the dynamics of the actin cytoskeleton) complex (51). Finally, RhoG (a Rho family small GTPase implicated in cytoskeletal regulation) is indirectly activated by phosphoinositide phosphatase SopB, which aggravates the membrane ruffling phenomenon (52). SopE and SopE2 effector proteins can also lead to actin cytoskeletal rearrangement and pro-inflammatory cytokine expression by activating CDC42 (a member of the Rho family of small GTPases and a key regulator of the actin cytoskeleton) (53, 54). However, both Rac and CDC42 activation were subsequently down-regulated by GAP activity of another effector protein, SptP, which restored the membrane structure after bacterial internalization and reversed the membrane ruffling caused by other effector proteins (55, 56). In recent years, it has been found that SPI-1-deficient S. Typhimurium still has a residual invasion ability, which seems to depend on the outer membrane protein Rck (57, 58). Rck is able to bind to epidermal growth factor receptor (EGFR) and activate Arp2/3 through Rac1 and Akt, which leads to bacterial internalization through the zipper mechanism (58).
The T4SS is used by pathogenic microorganisms to transport macromolecules such as DNA, proteins, and toxins across the host cell. It has been found to be associated with a variety of pathogens, including Legionella spp., Bartonella spp., Brucella spp., Coxiella spp. and Helicobacter pylori (59, 60). For instance, the Helicobacter pylori Cag T4SS has an important role in the pathogenic mechanisms of gastric cancer and peptic ulcer disease (61). However, current studies have shown that only a few serotypes of Salmonella contain T4SS, and due to its particularity, there is little information about its role in Salmonella infection. Furthermore, a previous study found that Salmonella strains with the T4SS were more likely to enter and survive in ECs or macrophages than those without the T4SS (62). Therefore, T4SS may also play an important role in promoting the internalization of Salmonella (Figure 1).
The pro-inflammatory response is a core factor in the pathogenicity of Salmonella (63). On the one hand, S. Typhimurium can initiate intestinal inflammatory responses through the stimulation of innate immune receptors by conserved bacterial products such as lipopolysaccharide (LPS), peptidoglycan or flagellin; on the other hand, it can also bypass these immune receptors to induce inflammation (63). The ability of S. typhimurium to stimulate inflammatory signaling is strictly dependent on SopB, SopE, and SopE2, and the absence of these three effector proteins fails to induce inflammatory signaling (64). Among them, SopE can activate Rac-1 and CDC42 independently, whereas SopE, SopB, and SopE2 can induce CDC42 release from ECs (56, 65). Rac-1 and CDC42 both belong to the Rho family of GTPases, which can lead to the activation of NF-κB and the release of pro-inflammatory cytokines such as IL-1β and IL-23 (66). Alternatively, SopA and SopD can target innate immune inflammatory signals to stimulate inflammation without binding to innate immune receptors, contributing to amplification of the inflammatory response (67, 68). It was also demonstrated that Salmonella can establish an advantage in competition with the commensal flora by activating the inflammatory responses. In conclusion, inflammation alters the balance between the intestinal commensal flora and S. Typhimurium in favor of Salmonella colonization in the host gut (Figure 2A).
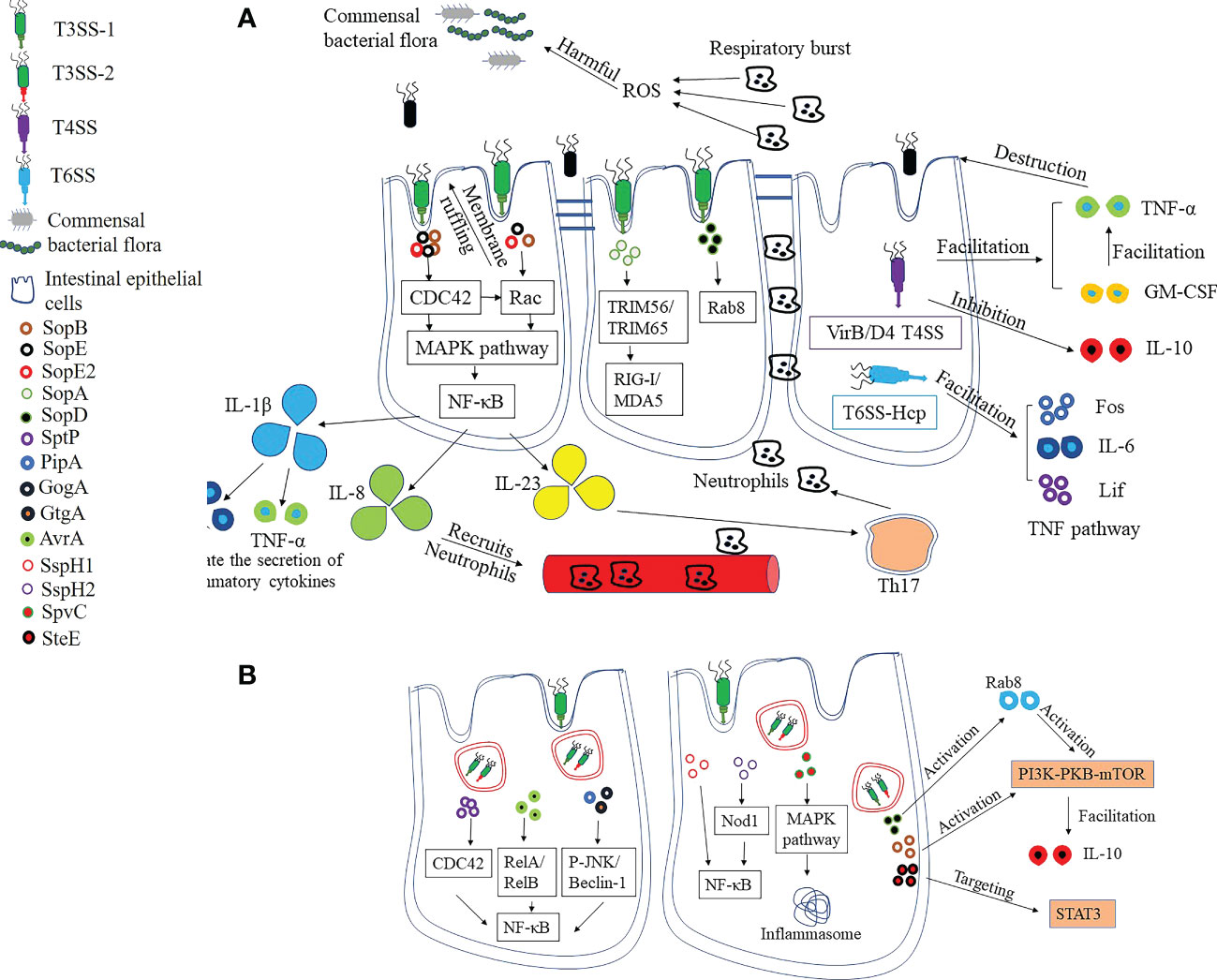
Figure 2 Pro-inflammatory and anti-inflammatory strategies of Salmonella. (A) Pro-inflammatory strategies. The effector proteins SopB, SopE and SopE2 secreted by T3SS are the central components of pro-inflammatory response, which leads to pro-inflammatory cytokine release by facilitating the activation of NF-κB. The production of pro-inflammatory cytokines recruits neutrophils, which contributes to the destruction of intestinal commensal flora and promotes Salmonella colonization. SopA and SopD, meanwhile, target innate immune inflammatory signals to stimulate inflammation and contribute to the amplification of the inflammatory response. T4SS-containing Salmonella can suppress the anti-inflammatory cytokine IL-10 secretion while increasing TNF-α and GM-CSF levels, resulting in intestinal epithelial barrier dysfunction and disruption of integrity. Hcp, one of the structural and effector proteins of T6SS, is involved in the regulation of TNF signaling pathways, including upregulation of Fos, IL-6 and Lif levels. (B) Anti-inflammatory strategies. The excessive inflammatory response leads to epithelial cell death, which in turn affects Salmonella survival. SptP, the effector protein of Salmonella, can inhibit inflammation by limiting the activity of CDC42. There are also some effector proteins that can affect NF-κB activation, which in turn reduces the pro-inflammatory response. In addition, the host mitogen-activated protein kinase (MAPK) is inactivated by SpvC, which also inhibits host autophagy and prevents the formation of inflammasome. SopB and SopD can stimulate the PI3K-PKB-mTOR signaling pathway, respectively, which inhibits the inflammatory response. Finally, the SteE (GogC) protein targets STAT3, a signaling pathway that restores homeostasis after an inflammatory response.
In addition to helping Salmonella compete with normal intestinal flora, the change of pro-inflammatory cytokine profile also helps to change the permeability of ECs, thus promoting the invasion of Salmonella (Figure 2A). Currently, only a few serotypes of Salmonella have been reported to contain T4SS. Compared to strains without T4SS, VirB/D4 T4SS-containing S. enterica Serovar Heidelberg inhibits the secretion of the anti-inflammatory cytokine IL-10 when infecting IECs (69, 70). However, IL-10 deficiency has been linked to increased intestinal permeability, inflammation, and dysfunction, potentially contributing to the successful invasion and persistence of Salmonella in host cells. In the study, it was likewise demonstrated that infection of ECs by T4SS-containing S. enterica Serovar Heidelberg induced elevated levels of TNF-α and GM-CSF, and that changes in the expression of these cytokines may impair epithelial barrier function and thus contribute to bacterial invasion of IECs (69). Hcp has been proposed as a core component and hallmark secreted protein of T6SS, but little is known about the role of Hcp in infection. Expression of Hcp protein in BHK-21 cells by plasmid pEGFP-N1-hcp revealed that it is mainly localized in the cytoplasm and is involved in the regulation of TNF signaling pathways, including up-regulation of Fos, IL-6 and Lif levels, and downregulation of Ccl20, Ccl2 and Map3k8 (71). These findings suggest that T6SS may also contribute to the regulation of inflammation caused by Salmonella, which in turn changes the permeability of ECs.
An excessive inflammatory response will also lead to host cell apoptosis, which in turn puts pressure on the survival of Salmonella. As a result, Salmonella regulates the inflammatory response by expressing effectors that maintain host homeostasis (Figure 2B). Overall, effector proteins of Salmonella antagonize the onset of the inflammatory response through two mechanisms.
In the first mechanism, effector proteins directly antagonize signaling pathways triggered by agonists or pro-inflammatory effectors (63). SptP, for example, can effectively counteract the inflammatory response induced by SopE and SopE2, primarily by suppressing CDC42 activity (72). PipA, GogA, and GtgA were all able to cleave the transcription factors RelA (p65) and RelB of NF-κB, which in turn effectively limited the inflammatory response induced by S. Typhimurium (73). AvrA is also an important anti-inflammatory protein that inhibits the NF-κB pathway by suppressing P-JNK and Beclin-1 molecules (74, 75). SspH1 is mainly localized in the nucleus and down-regulates the production of pro-inflammatory factors by decreasing NF-κB-dependent gene expression (76). The deficiency of SspH2 had no effect on the virulence of Salmonella enterica, but reduced the colonization in vivo and promoted the expression of IL-1, TNF-α, IL-12 and iNOS cytokines, indicating that SspH2 is an important anti-inflammatory effector protein (77). Also, one study has shown that SspH2 is able to interact with Nod1 to regulate inflammation (78). Nod1, an intracellular receptor, is associated with the activation of NF-κB, suggesting that SspH2 exerts its effects also through the downregulation of NF-κB (79). Autophagy relies on the activation of inflammasome as part of the innate immune response and contributes to the host’s defense against Salmonella infection (80). It was shown that the effector protein SpvC of Salmonella can inhibit autophagy and reduce the levels of NLRP3 and NLRC4 (81). Finally, the presence of SpvC exacerbated systemic infection with S. Typhimurium by suppressing fever and intestinal inflammation in mice (82).
There are also some effector proteins that stimulate anti-inflammatory pathways to help maintain host homeostasis, which is the second mechanism by which Salmonella resists the host inflammatory response (Figure 2B). For example, in addition to stimulating the secretion of inflammatory cytokines, SopB activates the PI3K-PKB-mTOR signaling pathway and induces the production of the anti-inflammatory cytokine IL-10 to maintain host cell homeostasis (83). In addition, SopD can also exert anti-inflammatory effects by activating Rab8, which subsequently further stimulates the PI3K-PKB-mTOR signaling pathway (68). Finally, the SteE (GogC) effector protein acts on signal transducer and activator of transcription 3 (STAT3), a signaling pathway for homeostatic restoration after an inflammatory response (84).
The ubiquitin cascade reaction system is also able to regulate protein function, inflammation and immunity, ultimately affecting the innate and adaptive immune response to pathogens (85). Recognition by the ubiquitination system is an important tool for effective identification of intracellular Salmonella, but Salmonella can also evade the host immune response by targeting the ubiquitination pathway with secreted effector proteins (Figure 3).
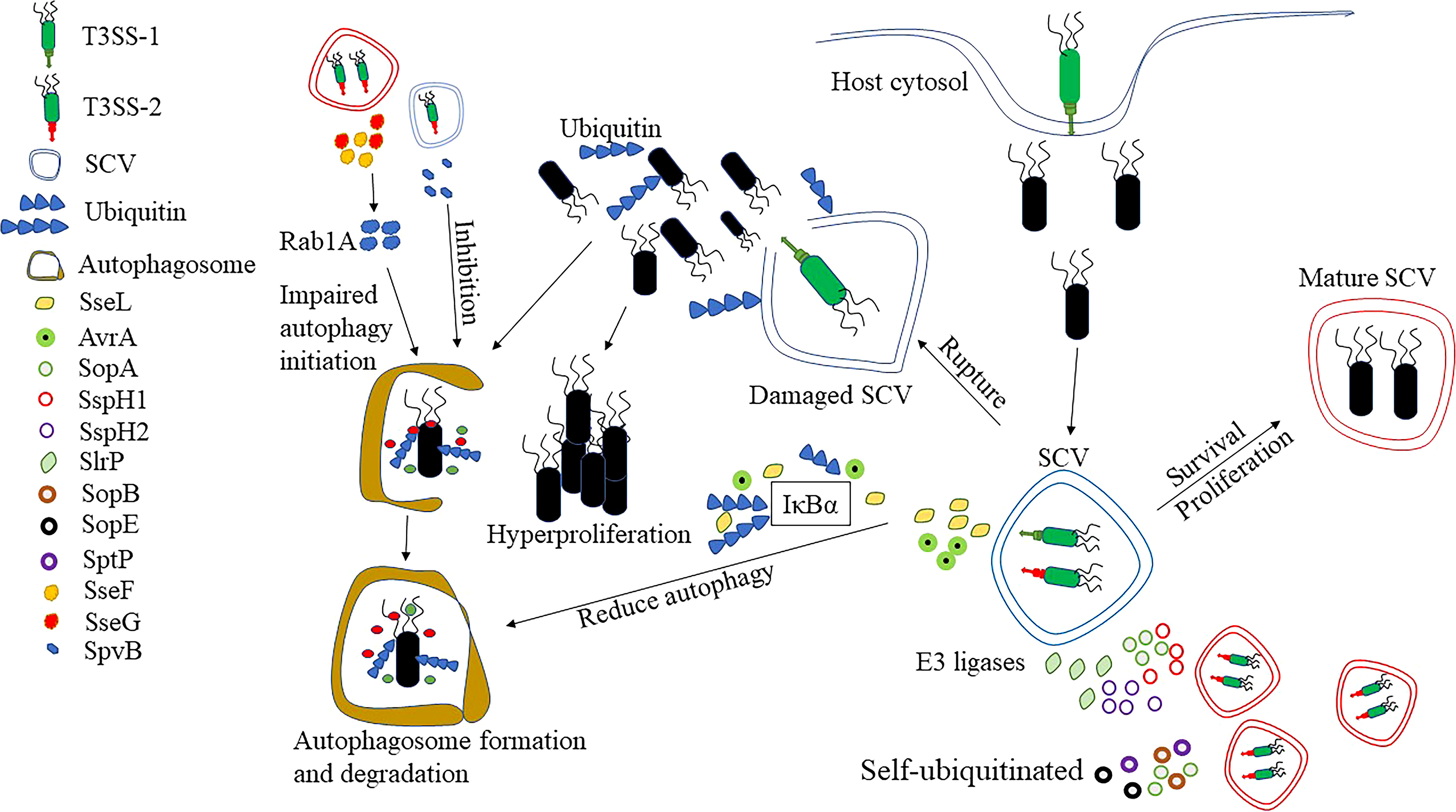
Figure 3 Response of Salmonella to ubiquitination and autophagy. After entering the cytosol, Salmonella can develop a unique ecological niche, SCVs. By the presence of SCVs, Salmonella can avoid recognition by ubiquitination, but studies have shown that the integrity of SCVs can be disrupted by T3SS-1. Some of the Salmonellae escape into the cytoplasm and hyper-proliferate, but there are also some bacteria recognized by ubiquitination. At the same time, ubiquitin also recognizes the damaged SCVs and the bacteria that remain inside the SCVs, and eventually clears the SCVs and bacteria by enzymatic reaction. However, Salmonella can effectively avoid the accumulation of ubiquitin aggregates and aggregate-like induced structures by secreting deubiquitinating enzymes, such as SseL and AvrA, which in turn prevent the recognition of ubiquitin-like modifications by autophagy receptors. There are also effector proteins, such as SopA, SspH1, SspH2 and Slrp that can act as E3 ligases to ubiquitinate host proteins and thus inhibit inflammatory responses. Finally, effector proteins such as SopA, SopB, SopE or SptP can also self-ubiquitinate to influence the antimicrobial response. The disruption of ubiquitination by SseL is also able to reduce the occurrence of autophagy. SseF and SseG, on the other hand, can interact with the small GTPase Rab1A in the host cell to inhibit autophagy initiation. Finally, SpvB depolymerizes actin, inhibiting host cell autophagy, which is also important for Salmonella intracellular survival.
SseL is T3SS effector protein with deubiquitinase activity that effectively avoids the accumulation of ubiquitin aggregates and aggresome-like induced structures, which in turn prevents the recognition of ubiquitin-like modifications by the autophagy receptor proteins (86). It was shown that SseL-deficient strains had significantly more ubiquitination-like aggregates around SCVs compared to wild-type Salmonella, while inducing the onset of autophagy. In addition, SseL affected the ubiquitination and degradation of IκBα, which in turn inhibited NF-κB activation. In contrast, SseL-deficient Salmonella induced a stronger inflammatory response, which may be associated with increased production of NF-κB-dependent cytokines (87). Therefore, as an important effector protein, SseL can further affect the occurrence of autophagy and inflammation through the regulation of ubiquitination modification. AvrA is also an effector protein with deubiquitinating enzyme activity, which effectively dissociates ubiquitin from IκBα and β-catenin (88). Moreover, the normal expression of AvrA in Salmonella helps stabilize IκBα and β-catenin during bacterial-host cell interactions, thereby inhibiting NF-κB signaling and suppressing inflammatory responses.
There is also a class of effector proteins in Salmonella, such as SopA, SspH1, SspH2 and Slrp, all E3 ubiquitin ligases, which are able to ubiquitinate protein substrates, and some are even able to self-ubiquitinate (Figure 3) (89). SopA was found to behave similarly to the mammalian HECT E3 ubiquitin ligase, preferentially recruiting three E2-coupled enzymes, UbcH5a, UbcH5c, and UbcH7E (90). Infection of HeLa cells with wild-type and SopA-deficient Salmonella revealed that the mutants were able to reduce the number of polymorphonuclear leukocytes (PMN) migrating via the epithelium, and thus it was hypothesized that SopA may be involved in regulating Salmonella-induced intestinal inflammation through ubiquitination of bacterial or host proteins. In addition, SspH1 can bind to PKN1 through its leucine-rich repeat (LRR) domain, allowing ubiquitination-proteasomal dependent degradation of PKN1 (91). It has been shown that PKN1 is a potent positive regulator of androgen receptor (AR) signal transduction (92). Also, previous studies have shown that AR can affect the number of neutrophils, macrophage activation and susceptibility to bacteria in mice (93, 94). Thus, the ubiquitination and degradation of PKN1 in cells by SspH1 further leads to the inhibition of AR, with possible effects on a number of cellular functions. As a member of the E3 ubiquitin ligase family, SspH2 can interact with NOD1 and NOD2 to regulate host innate immunity (95). In the study, SspH2 was found to specifically bind to NOD1 and induce ubiquitination modifications, leading to about four-fold higher NOD1 hyperactivation than the original. It also bound to NOD2 in the same way, resulting in a ten-fold increase in activation rate over basal activation. There was also an NF-κb-dependent elevation of IL-8 after SspH2 interacted with NOD1 and NOD2. All these results suggest that SspH2 super-activates NOD1 and NOD2, which in turn increases pro-inflammatory cytokine secretion. Slrp is also an important E3 ubiquitin ligase that binds to thioredoxin (involved in the control of many physiological processes and immune regulation) as well as mediates its ubiquitination, which results in a significant decrease in thioredoxin activity and increased cell death (96).
Another binding target of SlrP was shown to be ERdj3 of the Hsp40/DnaJ family, which plays an important role in the proper folding of proteins (97). However, the binding of SlrP to ERdj3 significantly reduced the interaction of ERdj3 with its substrate. It may lead to the accumulation of unfolded proteins in the endoplasmic reticulum, which eventually leads to cell death. Such an increase in host cell death caused by Salmonella effector proteins may, to some extent, contribute to the escape of bacteria from infected cells and is also necessary to infect new cells as well as to facilitate dissemination.
Finally, Salmonella is also capable of attaching ubiquitin to its own proteins using host E3 ligases, the more important of which are SopA, SopB, SopE or SptP (Figure 3) (89). It has been shown that both SopA and SopE can be degraded by the ubiquitination-proteasome pathway mediated by HsRMA1 (98, 99). And SptP, another representative of self-ubiquitination, can undergo proteasome-dependent degradation after being labeled by ubiquitination (89, 98). SopB, the T3SS effector protein of S. Typhimurium, can diversify its function by targeting to different cellular compartments in a ubiquitin-dependent manner (100). In contradiction to this, the activity of SopB is also down-regulated by ubiquitination mechanisms within the host cell, such as targeting it to lysosomal degradation (101). A further study showed that the E3 ubiquitin ligase TRAF6 is primarily responsible for SopB ubiquitination, which was totally prevented by TRAF6 absence (102). As the analysis of E2 showed, TRAF6-mediated ubiquitination of SopB requires UbcH5c rather than other E2-coupled enzymes in vitro and in vivo. As speculated by the above studies, the corresponding effector proteins minimize the impact on bacterial survival by self-ubiquitination to decrease the ubiquitinating enzymes in the host cells. At the same time, effector proteins are rapidly degraded after they exert their functions, preventing excessive accumulation, which may help to avoid adverse effects on hosts or pathogens.
Autophagy is an essential component of the innate immune system which contributes to the intracellular clearance of Salmonella (103). Upon entrance into the IECs, Salmonella also induces the onset of autophagy. At present, it is believed that Salmonella can induce autophagy through two pathways (Figure 3) (80). The first pathway involves the recognition of Salmonella by ubiquitin (104). The intracellular Salmonella is generally thought to be present within the SCVs, but some can use their T3SS to impair the structural integrity of the SCVs, thereby escaping into the cytoplasm to achieve high replication rates (105). Intracellular Salmonella can be rapidly recognized by the host ubiquitination system, leading to the formation of an intensive layer of ubiquitin chains around the bacteria. Subsequently, ubiquitin-modified Salmonella is identified by autophagy adapters (e. g. NDP52, OPTN and p62). These adapters guide the bacteria to the primary autophagosome by binding to the ubiquitin-modified bacteria through their ubiquitin-binding domains and further interacting with the membrane anchoring protein LC3 of the autophagosome. The second pathway recognizes the damaged SCVs, and upon destruction by T3SS, the entire SCVs, including the Salmonella within it, is degraded by the autophagy pathway (104).
Similarly, Salmonella has also evolved various strategies to evade autophagy, in which T3SS and its secreted effector proteins play a crucial role (Figure 3). After infection of ECs, it was shown that the Salmonella mainly replicated in the SCVs and the ubiquitinated structures were also mainly surrounding the SCVs (106). This seems to be relevant to the effector proteins secreted by T3SS, as the formation of ubiquitinated structures is significantly reduced in cells infected with T3SS-deficient Salmonella. Both SseF and SseG of S. Typhimurium are secreted by T3SS, which impairs the activation of autophagy by directly interacting with Rab1A, a small GTPase in host cell (107). Upon recognition of Salmonella by the ubiquitination machinery, the autophagy receptor p62 is capable of recruiting LC3 that promotes autophagosome formation (106, 108). A co-localization of p62 and LC3 was also found in the ubiquitinated structures induced by Salmonella-infected cells. The deficiency of SseL significantly increased the number of ubiquitinated structures, p62 and LC3, compared to cells infected with wild-type bacteria (86). It was shown that autophagy may be further hindered by SseL through the reduction of ubiquitinated structures around SCVs and the presence of autophagy markers p62 and LC3. Moreover, autophagosome formation was found to be increased in SpvB mutant strains compared to wild-type Salmonella (109). In addition, it was shown that the infection of zebrafish by SpvB mutant strain resulted in increased expression of LC3 and Beclin1, and double membrane-like autophagosome structure also observed, indicating that SpvB can inhibit autophagic activity (110, 111). The polymerization of the actin backbone is involved in the formation of autophagy, and the SpvB effector protein secreted by S. Typhimurium depolymerizes host cell actin (112). Therefore, it is speculated that SpvB may inhibit host cell autophagy by depolymerizing actin.
Antibodies facilitate the uptake of bacteria by phagocytes to prevent infection, and ultimately the destruction of internalized bacteria by phagocytes (16, 113). There are two ways in which antibody exerts its antimicrobial effect. The first one is that pathogens recognized by Fc receptors on macrophages, which is called opsonophagocytosis (113). Alternatively, antibodies binding to the surface of the pathogen can activate the proteins of the complement system (114). The activation of the complement system leads to opsonophagocytosis by binding to complement receptors on phagocytes (115). In addition, other complement elements will recruit phagocytes to the site of infection, and the terminal of complement can directly lyse microorganisms by forming pores in their membranes. However, Salmonella can also disrupt the humoral immune response with the help of the secretory system (Table 1). Bone marrow (BM) is the central tissue for hematopoiesis and immune memory, and serum IgG is mainly produced by IgG-secreting plasma cells in the BM. It has been shown that SiiE, secreted by the Salmonella T1SS, can reduce the persistence of IgG+ plasma cells in the BM to prevent effective humoral immune memory (116). This is the only effector protein that has been reported to interfere with the adaptive immune response in the T1SS.
The activation of CD4+ T cells can effectively target the infection of Salmonella, therefore, if Salmonella wants to establish a systemic infection, it must interfere with the normal biological function of DCs (Table 1). The effector protein secreted by T3SS appears to be more important for the survival of Salmonella inside DCs (Table 1). It was shown that the effector protein SseI secreted by T3SS inhibits normal cell migration of primary macrophages and DCs in vitro, and also inhibits migration of DCs to the mouse spleen in vivo (128). In addition, the researcher found that effector proteins such as SseF, SifA, SspH2, SlrP, and PipB2 seem also to participate in the inhibition of DCs migration, but the exact mechanism needs to be further investigated (134). These evidences suggest that Salmonella can interfere with the normal function of DCs by secreting effector proteins, which in turn impairs the initiation of host adaptive immune response.
Bacterial growth can be hindered by the cytoplasm of ECs and phagocytes. Therefore, the cytosolic environment represents an early selective pressure on Salmonella (23, 137). For survival, the majority of Salmonella is present in a specialized niche SCVs after uptake by cells (23). The presence of SCVs effectively prevents cellular killing of the bacteria and reduces antigen presentation of DCs or other antigen-presenting cells (APCs), which is in turn affects the adaptive immune response. SCVs, on the other hand, frequently binds to lysosomes, causing SCVs rupture and exposing the bacteria to an environment rich in hydrolytic enzymes even if they remain intact. But Salmonella has evolved various ways to both restrain lysosomal binding and maintain SCVs integrity (Table 1), in which SifA plays an important role (120, 121). The absence of the effector protein SifA is capable of causing more than 50% of the SCVs to rupture, leading to the release of the bacteria into the cytosol, which is detrimental to the survival of the bacteria. The cysteine protease GtgE, also secreted by T3SS-1 and T3SS-2, is able to manipulate SCVs transport and prevent the accumulation of Rab29 on SCVs (132). Also, GtgE is able to cleave Rab32 and prevent the fusion of SCVs with lysosomes, contributing to the stabilization of the SCVs membrane (133). In the same way, SopD2 prevents the accumulation of Rab32 to SCVs, thus hindering the endocytosis of host cells (122, 123).
There are also numerous effector proteins secreted by Salmonella to regulate the lipid and protein content of SCVs, which will contribute to the stability of SCVs (Table 1) (138). For example, the SopB effector protein, secreted by T3SS-1, is able to reduce the level of negatively charged lipids on the surface of SCVs, which subsequently leads to the dissociation of many endocytic transporter proteins from SCVs and avoids the occurrence of degradation by lysosomes (117). SopF is a phosphatidylinositol-binding effector protein that binds to a variety of phosphatidylinositols in the protein-lipid overlays after delivery by Salmonella and, furthermore, knockout of the sopF leads to increased cleavage of SCVs, suggesting that it may promote the stability of SCVs (118). SipA, an actin binding protein, is required for effective entrance of Salmonella into host cells, where it can recruit Synaxin8 instead of the host R-SNARE molecule, promote early phagosome fusion with SCVs, avoid maturation of phagosomes to lysosomes, and promote pathogen survival (119, 139). Furthermore, SseJ dissociates cholesterol esters from the phospholipid bilayer in SCVs membranes, which is necessary because increased cholesterol content can affect membrane fluidity, signaling, sorting, and transport (124). In conclusion, as an essential effector protein, SseJ plays an important role in maintaining the stability of SCVs. It was shown that SCVs formed by SseF or SseG-deficient Salmonella undergo irregular movement in the cytoplasm (126). Hence, the prevalence of these two effector proteins in multiple serotypes provides strong evidence that they may contribute to bacterial growth in host cells. Salmonella-induced filaments (SIFs) are a device for nutrient uptake by bacteria within the SCVs. PipB2, an effector protein secreted by both T3SS-1 and T3SS-2, plays an important role in SCVs formation and is also required for the extension of SIFs (135). In combination with SifA, this effector protein serves to facilitate membrane exchange and nutrient delivery by allowing the formation of tubules from SCVs and extension along the microtubule cytoskeleton. SteA also plays an important role in controlling membrane dynamics, and the absence of SteA reduces the ability of Salmonella to form SIFs, increases the aggregation of SCVs, as well as the formation of abnormal vacuoles (136). In conclusion, the presence of SCVs not only contributes to bacterial survival, but also avoids the activation of the adaptive immune response, especially the cellular immune response.
The uptake of bacteria, after processing by DCs, results in the presentation of antigenic peptides to CD4+ T cells via MHC II molecules (140). MHC II molecules play a key role in adaptive immunity by displaying antigenic peptides on the surface of APCs (e.g., DCs) to CD4-restricted T cells, leading to their activation, proliferation and differentiation (141). The infection of DCs by Salmonella exhausts mature MHC II molecules (mMHC II) on the cell surface. A recent study has shown that SteD acts as an important effector protein that can direct the E3 ligase MARCH8 to binding with mMHC II which results in mMHC II ubiquitination and surface exhaustion, ultimately reducing the activation of T cells (129) (Table 1). In addition, inhibition of T-cell activation by SteD was accomplished by reducing the levels of at least three proteins (including MHC II, CD86, and CD97) on the surface of antigen-presenting cells (130, 131). Among them, CD97 mainly stabilizes the immune synapse between DCs and T cells. After degradation by SteD, it eventually inhibits DCs-T cell interactions and reduces T cell activation. Thus, SteD suppresses T-cell immunity through two distinct processes.
T3SS-1 is required for IECs invasion and barrier penetration, and it is an extracellular needle-like device required for effector protein injection into host cells (142). PrgI and SipD are both essential components of the T3SS-1 tip complex (143). Because they are common and highly conserved among all virulent Salmonella species, they may be ideal candidate targets for a broad-spectrum vaccination against Salmonella infection. The levels of immunogenicity induced after immunization of mice with PrgI and SipD proteins alone or in combination by different immunization routes (subcutaneous, intranasal and oral) were investigated and showed that high levels of IgG and IgA titers against both proteins could be induced, where the levels of SipD-specific antibodies were higher (Table 2) (144). In the same study, it was also shown in protective studies that immunization with SipD protein alone or in combination with PrgI protected mice from the lethal challenge by S. Typhimurium with 100×LD50. Furthermore, a study showed that administering SipD protein through intranasal or intragastric routes induced strong IgG (in all immune pathways) and IgA (in intranasal and oral immune pathways) antibody responses and protected mice from lethal challenge by S. Typhimurium or the Shigella spp (Table 2) (145). This is mostly because the structural proteins that constitute T3SS are shared by all pathogenic Salmonella and Shigella spp., particularly the tip protein. Regardless of Salmonella serotype, the two types of T3SS are used to interact with host cells, particularly the tip protein and the first translocation effector protein of Salmonella, both of which are essential for pathogenicity. Based on this, previous vaccine studies fused the tip protein SipD of T3SS-1 to the first translocation effector protein SipB, named S1 protein, and fused the tip protein SseB of T3SS-2 to the first translocation effector protein SseC, named S2 protein, then vaccinated the mice with S1 and S2 alone or in combination (Table 2) (146). Following that, challenge with S. Typhimurium or S. Enteritidis resulted in a 60% survival rate regardless of serotype, indicating that fusion with tip and translocator proteins is a feasible vaccine candidate. However, none of the candidate vaccines elicited an effective mucosal immune response, which may be connected to the immunological route of the protein, as studies have shown that the parenteral route is not a very efficient approach to stimulate IgA production (161–163).
SptP, secreted by T3SS-1, regulates the dynamics of the cytosolic actin backbone and plays an important role in Salmonella invasion (53, 55). C50336 ΔsptP, a sptP-deficient strain, was inoculated in mice, and the humoral and cellular immune responses of the immunized mice were studied afterwards, revealing that the vaccine strain was highly immunogenic and provided 100% protection against S. Enteritidis after challenge (147). It indicates that the deletion of sptP may be a new target for the development of salmonellosis vaccine. Subsequently, live attenuated vaccine strains were constructed by introducing sptP mutations in different S. Enteritidis strains and the protective efficacy was investigated in chickens (164). The results showed that a strong cellular immune response was induced by both lymphocyte proliferation and cytokine assays, and the level of specific IgG antibodies in the immune group was significantly increased, demonstrating the high immunogenicity of the live vaccine. After challenge, it was also able to reduce clinical signs and pathological changes in chickens, with a highest protection rate of 100%. In summary, sptP-deficient Salmonella strains may have good potential for application in both mammals and avian species (Table 2).
By invading through the mucosa and colonizing lymphoid tissues, Salmonella is able to induce a strong mucosal and cellular immune response in the host persistently (165). Based on this, the attenuated Salmonella is often used as a vector to deliver protective antigens of other pathogens (148, 166–168). However, inappropriate attenuation of Salmonella vectors often leads to a severe inflammatory response, which is unacceptable (169). SopB, secreted by T3SS-1, is capable of exacerbating the inflammatory response induced by Salmonella (148). It was shown that sopB deficiency in Salmonella impairs the ability to elicit local inflammatory responses and fluid secretion into the intestinal lumen, but also enhances the immunogenicity of Salmonella as a vector for the presentation of exogenous antigens (148, 168). Subsequently, elevated immunogenicity was demonstrated by the delivery of the Streptococcus pneumoniae surface protein PspA through Salmonella, indicating that the deletion of the SopB contributes to the development of live attenuated vaccines (Table 2). Based on this, our laboratory introduced the sopB deficiency into the vectors of S. Choleraesuis, which we expected to further improve the immunogenicity of vector vaccine. As a result, immunization of the mouse model provided a better protection against either Streptococcus suis, Mycoplasma hyopneumoniae or porcine circovirus infection (166–168).
SsaV is a necessary component of the T3SS-2 secretion apparatus, and the secretion of various effector proteins into the host must be initiated by sensing the neutral pH of the host cytoplasm, and SsaV is the key protein of this transition switch (Table 2) (170). Oral immunization of adult volunteers with S. Typhi or S. Typhimurium (both deficient in aroC and ssaV) showed that ZH9 (candidate strain of S. Typhi) not only had a better security, but also elicited high titers of antibody responses (149). In contrast, immunization of volunteers with WT05 (candidate strain of S. Typhimurium) also elicited high titers of antibody response, but it was shedding in the feces until 23 d. In adult volunteers, the immunogenicity of M01ZH09 (ZH9) immunization with or without carbonate buffer solution was compared separately, demonstrating a well-tolerated with or without carbonate buffer, a mild adverse event after vaccination, and no fever or long-term shedding (150). Furthermore, the vaccine was immunogenic, with more than 88% or 93% of participants in both groups having IgA antibody-secreting cells detectable by ELISPOT, and 81% of participants in both groups producing LPS-specific IgG on day 14. Lymphocyte proliferation and IFN-γ production also showed that the vaccine elicited a strong cellular immune response. A subsequent clinical trial was conducted to determine the tolerability and immunogenicity of a single dose of M01ZH09 (ZH9), which showed that adverse effects were less frequent, the time of fecal shedding was reduced, and the immune response was dose dependent, with the highest dose (5 × 109 CFU) being the most immunogenic (151). Previous studies in healthy adults demonstrated the immunogenicity and acceptable safety of ZH9, 151 children were subsequently recruited in Vietnam in the study, of whom 101 subjects were orally immunized with a single dose of M01ZH09 (ZH9) (152). The results showed that high titers of LPS-specific IgA and IgG could be detected in the serum after immunization, and although no bacteremia was observed, some children experienced adverse reactions, indicating that the vaccine was appropriately immunogenic, but safety should be improved.
The absence of SsaV influences the secretion of effector proteins. However, immunization of mice with the SsaV-deficient Salmonella still elicited O antigen-specific immune responses and improved survival of mice after challenge (171). However, the introduction of the mentioned strains into immunocompromised mice still caused lethal infections, indicating that further attenuation is necessary (Table 2) (171). Therefore, one study has introduced an additional fur deletion into SsaV-deficient Salmonella, a gene that contributes to acid tolerance and iron acquisition (154). The double-deletion strain is safe in immunocompromised mice, while being sufficiently immunogenic to enhance protection against Salmonella. Hha, a nucleoid-associated protein, is able to downregulate the expression of some virulence and invasion-related genes. The introduction of Hha mutation in SsaV-deficient Salmonella has reduced the systemic colonization ability and the adverse effects on immunocompromised hosts (155). Moreover, humoral and cellular immune responses were enhanced after the introduction of Hha mutation. These results suggest that the combination of SsaV-deficient Salmonella with the Hha mutation is a live attenuated candidate vaccine that can be safely used in immunocompromised hosts.
An attractive aspect of live vector vaccines is their ability to stimulate a robust cellular immune response and the efficient delivery of antigen to the MHC I presentation pathway, which is particularly important for CD8+ T cell development (172). It is generally accepted that Salmonella is preferentially present in SCVs upon entry into the cell, and if the escape of Salmonella from SCVs into the cytoplasm can be increased, it may facilitate antigen delivery via the MHC I pathway, thereby enhancing the CTL response (Table 2) (23). Studies have shown that SifA has an important role in maintaining the integrity of SCVs in macrophages and ECs (120). In one study, after the deletion of sifA in AroC-deficient Salmonella, most of the Salmonella was shown to successfully escape into the cytoplasm compared to the AroC-deficient Salmonella only, however, no subsequent increase in MHC I presentation efficiency to the model antigen (Ovalbumin) was detected, nor was an increase in cytotoxic T cell or IFN-γ production levels (156). Another study, however, has shown that the ssaV and aroC double deletion strains induced accelerated maturation of DCs, higher production of TNF-α, IL-12, and IL-1 cytokines, and, most importantly, more efficient antigen presentation than the sifA and aroC deletion strains (153). The above results suggest that enabling the escape of Salmonella from SCVs may not be sufficient to enhance the MHC I presentation efficiency of the antigen. In contrast, the increased efficiency of antigen presentation after ssaV deficiency demonstrates that further screening is needed to investigate whether other effector proteins are involved in interfering with antigen presentation, which may contribute to the development of more effective vaccines targeting CTL.
Activation of the inflammasome contributes to the clearance of intracellular bacteria (Table 2). To construct an effective vaccine targeting the inflammasome, the C-terminus of an E. coli EscI protein was fused to the N-terminal of SspH2, and attenuated Salmonella was used for delivery (157). The strain fused to the C terminus of EscI protein significantly increased IL-1 and IL-18 secretion and cell pyroptosis in mouse intraperitoneal macrophages, while causing less colonization in organs and fewer pathological changes in the spleen and liver than the strain delivered with only the N terminus of SspH2. The fusion protein SspH2-EscI was shown to translocate into macrophages and activate NLRC4 inflammasome, which limits the colonization of Salmonella in the spleen and liver. Subsequently, the vaccine potential of strains delivering the fusion protein SspH2-EscI was investigated, and the results showed that compared to strains delivering only the SspH2 N-terminal or empty plasmids, the colonization of organs was effectively reduced after challenge and the survival rate of mice was improved, indicating that recombinant Salmonella expressing the SspH2-EscI fusion protein enhanced the activation of caspase-1 in macrophages and protected mice from Salmonella challenge (158).
As mentioned previously, attenuated Salmonella strains are an effective vector for delivery of immunogenic proteins of other pathogens (158, 166–168). However, after entering the cells, Salmonella is mainly present in the SCVs, where antigens are mainly presented through MHC II molecules and induce mainly a CD4+ T cell immune response (23, 173). However, CTL induced by CD8+ T cells plays an important role in both viral infections and tumor-associated diseases, so it is also extremely important how Salmonella can be used to elicit a higher CTL response (174, 175). In addition to defects in SCVs stability-related genes that allow Salmonella to escape from SCVs to the cytoplasm, some researchers have also fused heterologous antigens to effector proteins secreted by the T3SS so that the antigens can be delivered to the host cell cytoplasm via the T3SS (Table 2). Previous research has shown that fusion expression of MHC I epitopes of influenza virus nucleoproteins embedded inside SptP proteins can successfully translocate them into the host cytoplasm and induce a CTL response capable of dissolving influenza virus-infected cells (159). As CTL plays a major role in the fight against lymphocytic choroid plexus meningitis virus (LCMV) of mice, the immunogenic protein of LCMV was subsequently embedded inside SptP (159). Oral immunization protected mice from lethal challenge by LCMV, demonstrating that the use of the secreted protein of Salmonella T3SS as a secretory signal can successfully deliver protective antigens of LCMV as well as provoke an effective immune response. It was discovered that delivery of tumor-associated antigens (TAAs) using the above strategy was also extremely effective for the induction of CD8+ T cells. In a study, over 20 effector proteins of SPI-2 were fused with TAAs separately, and it was discovered that fusion of TAAs with SseJ and replacement of the sseJ promoter with that of sifB effectively induced CD8+ T cells with strong antitumor activity (160). It demonstrated that the effector proteins of Salmonella as secretory signals for antigen delivery may be effective cancer vaccine platforms.
It is generally accepted that physical barriers and immune responses are beneficial for host defense against infection because they limit the replication and transmission of pathogens. However, the presence of the immune system is a double-edged sword for Salmonella; on the one hand they limit the replication and systemic spread of Salmonella, and on the other hand, Salmonella can utilize the immune system to compete with the normal flora and establish a systemic infection. The manipulation of Salmonella to these defense mechanisms is mostly related to the secretory systems and the effector proteins presented by these systems. In recent years, the function of some of the effector proteins delivered by the secretory system has been successfully characterized, and their functional analysis has allowed a better understanding of the pathogenesis of Salmonella and deciphering the potential mechanisms by which Salmonella evades the host immune system. However, there are many aspects of secretory systems and effector proteins that are still unknown. It also makes the prevention and control of Salmonella infection difficult because the mechanisms of immune escape are ill-defined. Although the use of antibiotics can effectively control the spread of Salmonella, appearance of multidrug-resistant bacteria is an additional serious threat to public health. Vaccine programs are the most effective and cost-effective measure to prevent and control pathogenic infections. As a zoonosis with numerous serotypes, current vaccines provide protection against only a limited number of serotypes of Salmonella. The presence of T3SS in numerous serotypes makes it an ideal vaccine and drug target, although it is not clear which effector protein induces better protection against Salmonella. Therefore, further studies are necessary to investigate whether the combination of multiple proteins can provide better protection, especially to different Salmonella serotypes, in addition to investigating the vaccine potential of individual effector proteins. Other secretory systems also play an important role in adhesion and invasion, but only a few studies have explored these secretory systems and little is known about their role in the pathogenesis of Salmonella. Therefore, the possibility of various secretory systems as targets for Salmonella vaccine development should also be the focus in subsequent studies.
HS, SW, and QL designed, supervised, and critically revised the manuscript. GZ drafted the manuscript. GZ, QM, and YZ did the reference collection. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant numbers 32172802, 31672516, 32002301, 31172300, 30670079), Jiangsu Province Science and Technology Program Special Fund Project (BZ2022042), the China Postdoctoral Science Foundation (grant number 2019M661953), and Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and supported by the 111 Project D18007. The funding bodies have not been involved in the design of the study as well as the collection, analysis, and interpretation of the data and manuscript writing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Lynch M, Painter J, Woodruff R, Braden C. Surveillance for foodborne-disease outbreaks–united states, 1998-2002. MMWR Surveillance summaries Morbidity mortality weekly Rep Surveillance Summaries / CDC (2006) 55(10):1–42.
2. Sánchez-Vargas FM, Abu-El-Haija MA, Gómez-Duarte OG. Salmonella infections: An update on epidemiology, management, and prevention. Travel Med Infect Dis (2011) 9(6):263–77. doi: 10.1016/j.tmaid.2011.11.001
3. Ahmer BM, Gunn JS. Interaction of Salmonella spp. with the intestinal microbiota. Front Microbiol (2011) 2:101. doi: 10.3389/fmicb.2011.00101
4. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev (2017) 279(1):70–89. doi: 10.1111/imr.12567
5. Sánchez B, Urdaci MC, Margolles A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiol (Reading England) (2010) 156(Pt 11):3232–42. doi: 10.1099/mic.0.044057-0
7. Vazquez-Torres A, Fang FC. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol (2001) 9(1):29–33. doi: 10.1016/S0966-842X(00)01897-7
8. Wang L, Yan J, Niu H, Huang R, Wu S. Autophagy and ubiquitination in Salmonella infection and the related inflammatory responses. Front Cell infection Microbiol (2018) 8:78. doi: 10.3389/fcimb.2018.00078
9. van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hötte K, Pampaloni F, et al. Linear ubiquitination of cytosolic Salmonella typhimurium activates NF-κB and restricts bacterial proliferation. Nat Microbiol (2017) 2:17066. doi: 10.1038/nmicrobiol.2017.66
10. Savina A, Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev (2007) 219(1):143–56. doi: 10.1111/j.1600-065X.2007.00552.x
11. Wick MJ. Monocyte and dendritic cell recruitment and activation during oral Salmonella infection. Immunol Lett (2007) 112(2):68–74. doi: 10.1016/j.imlet.2007.07.007
12. Wick MJ. Innate immune control of Salmonella enterica serovar typhimurium: mechanisms contributing to combating systemic Salmonella infection. J innate Immun (2011) 3(6):543–9. doi: 10.1159/000330771
13. Kurtz JR, Goggins JA, McLachlan JB. Salmonella infection: Interplay between the bacteria and host immune system. Immunol Lett (2017) 190:42–50. doi: 10.1016/j.imlet.2017.07.006
14. Luckheeram RV, Zhou R, Verma AD, Xia B. CD4+ T cells: differentiation and functions. Clin Dev Immunol (2012) 2012:925135. doi: 10.1155/2012/925135
15. Lee S-J, Dunmire S, McSorley SJ. MHC class-i-restricted CD8 T cells play a protective role during primary Salmonella infection. Immunol Lett (2012) 148(2):138–43. doi: 10.1016/j.imlet.2012.10.009
16. Takaya A, Yamamoto T, Tokoyoda K. Humoral immunity vs. Salmonella. Front Immunol (2019) 10:3155. doi: 10.3389/fimmu.2019.03155
17. Cummings LA, Deatherage BL, Cookson BT. Adaptive immune responses during Salmonella infection. EcoSal Plus (2009) 3(2). doi: 10.1128/ecosalplus.8.8.11
18. Nieto PA, Pardo-Roa C, Salazar-Echegarai FJ, Tobar HE, Coronado-Arrázola I, Riedel CA, et al. New insights about excisable pathogenicity islands in salmonella and their contribution to virulence. Microbes Infection (2016) 18(5):302–9. doi: 10.1016/j.micinf.2016.02.001
19. Bao H, Wang S, Zhao J-H, Liu S-L. Salmonella secretion systems: Differential roles in pathogen-host interactions. Microbiological Res (2020) 241:126591. doi: 10.1016/j.micres.2020.126591
20. Wagner C, Hensel M. Adhesive mechanisms of salmonella enterica. Adv Exp Med Biol (2011) 715:17–34. doi: 10.1007/978-94-007-0940-9_2
21. Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, et al. Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci (2016) 113(34):E5044–E51. doi: 10.1073/pnas.1608858113
22. Lou L, Zhang P, Piao R, Wang Y. Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front Cell infection Microbiol (2019) 9:270. doi: 10.3389/fcimb.2019.00270
23. Steele-Mortimer O. The Salmonella-containing vacuole–moving with the times. Curr Opin Microbiol (2008) 11(1):38–45. doi: 10.1016/j.mib.2008.01.002
24. Gawade P, Ghosh P. Genomics driven approach for identification of novel therapeutic targets in salmonella enterica. Gene (2018) 668:211–20. doi: 10.1016/j.gene.2018.05.058
25. Grant AJ, Morgan FJ, McKinley TJ, Foster GL, Maskell DJ, Mastroeni P. Attenuated Salmonella typhimurium lacking the pathogenicity island-2 type 3 secretion system grow to high bacterial numbers inside phagocytes in mice. PloS Pathog (2012) 8(12):e1003070. doi: 10.1371/journal.ppat.1003070
26. Zhang Y, Liu Y, Wang T, Deng X, Chu X. Natural compound sanguinarine chloride targets the type III secretion system of Salmonella enterica serovar typhimurium. Biochem Biophysics Rep (2018) 14:149–54. doi: 10.1016/j.bbrep.2018.04.011
27. Kapitein N, Mogk A. Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Curr Opin Microbiol (2013) 16(1):52–8. doi: 10.1016/j.mib.2012.11.009
28. Sana TG, Lugo KA, Monack DM. T6SS: The bacterial" fight club" in the host gut. PloS Pathog (2017) 13(6):e1006325. doi: 10.1371/journal.ppat.1006325
29. Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics (2009) 10:354. doi: 10.1186/1471-2164-10-354
30. Amaya FA, Blondel CJ, Barros-Infante MF, Rivera D, Moreno-Switt AI, Santiviago CA, et al. Identification of type VI secretion systems effector proteins that contribute to interbacterial competition in Salmonella Dublin. Front Microbiol (2022) 13:811932. doi: 10.3389/fmicb.2022.811932
31. Schroll C, Huang K, Ahmed S, Kristensen BM, Pors SE, Jelsbak L, et al. The SPI-19 encoded type-six secretion-systems (T6SS) of Salmonella enterica serovars gallinarum and Dublin play different roles during infection. Veterinary Microbiol (2019) 230:23–31. doi: 10.1016/j.vetmic.2019.01.006
32. Mulder DT, Cooper CA, Coombes BK. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar typhimurium. Infection Immun (2012) 80(6):1996–2007. doi: 10.1128/IAI.06205-11
33. Gerlach RG, Jäckel D, Stecher B, Wagner C, Lupas A, Hardt WD, et al. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol (2007) 9(7):1834–50. doi: 10.1111/j.1462-5822.2007.00919.x
34. Velásquez JC, Hidalgo AA, Villagra N, Santiviago CA, Mora GC, Fuentes JA. SPI-9 of Salmonella enterica serovar typhi is constituted by an operon positively regulated by RpoS and contributes to adherence to epithelial cells in culture. Microbiol (Reading England) (2016) 162(8):1367–78. doi: 10.1099/mic.0.000319
35. Wagner C, Hensel M. Adhesive mechanisms of Salmonella enterica. Bacterial adhesion. (2011) 715:17–34. doi: 10.1007/978-94-007-0940-9_2
36. Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol (2007) 61:89–112. doi: 10.1146/annurev.micro.61.080706.093233
37. Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Bäumler AJ. Salmonella enterica serotype typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol (2005) 57(1):196–211. doi: 10.1111/j.1365-2958.2005.04666.x
38. Kingsley RA, Abi Ghanem D, Puebla-Osorio N, Keestra AM, Berghman L, Baüumler AJ. Fibronectin binding to the Salmonella enterica serotype typhimurium ShdA autotransporter protein is inhibited by a monoclonal antibody recognizing the A3 repeat. J bacteriology (2004) 186(15):4931–9. doi: 10.1128/JB.186.15.4931-4939.2004
39. Raghunathan D, Wells TJ, Morris FC, Shaw RK, Bobat S, Peters SE, et al. SadA, a trimeric autotransporter from Salmonella enterica serovar typhimurium, can promote biofilm formation and provides limited protection against infection. Infection Immun (2011) 79(11):4342–52. doi: 10.1128/IAI.05592-11
40. Kingsley RA, Van Amsterdam K, Kramer N, Baüumler AJ. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infection Immun (2000) 68(5):2720–7. doi: 10.1128/IAI.68.5.2720-2727.2000
41. Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, et al. Identification of host-specific colonization factors of Salmonella enterica serovar typhimurium. Mol Microbiol (2004) 54(4):994–1010. doi: 10.1111/j.1365-2958.2004.04323.x
42. Paradis T, Bègue H, Basmaciyan L, Dalle F, Bon F. Tight junctions as a key for pathogens invasion in intestinal epithelial cells. Int J Mol Sci (2021) 22(5). doi: 10.3390/ijms22052506
43. Lara-Tejero M, Galán JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infection Immun (2009) 77(7):2635–42. doi: 10.1128/IAI.00077-09
44. Lu L, Walker WA. Pathologic and physiologic interactions of bacteria with the gastrointestinal epithelium. Am J Clin Nutr (2001) 73(6):1124s–30s. doi: 10.1093/ajcn/73.6.1124S
45. Layton AN, Galyov EE. Salmonella-induced enteritis: molecular pathogenesis and therapeutic implications. Expert Rev Mol Med (2007) 9(18):1–17. doi: 10.1017/S1462399407000373
46. Lin Z, Zhang YG, Xia Y, Xu X, Jiao X, Sun J. Salmonella enteritidis effector AvrA stabilizes intestinal tight junctions via the JNK pathway. J Biol Chem (2016) 291(52):26837–49. doi: 10.1074/jbc.M116.757393
47. Ly KT, Casanova JE. Mechanisms of Salmonella entry into host cells. Cell Microbiol (2007) 9(9):2103–11. doi: 10.1111/j.1462-5822.2007.00992.x
48. Schlumberger MC, Hardt W-D. Salmonella type III secretion effectors: pulling the host cell's strings. Curr Opin Microbiol (2006) 9(1):46–54. doi: 10.1016/j.mib.2005.12.006
49. Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive salmonella. EMBO J (1999) 18(18):4926–34. doi: 10.1093/emboj/18.18.4926
50. McGhie EJ, Hayward RD, Koronakis V. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J (2001) 20(9):2131–9. doi: 10.1093/emboj/20.9.2131
51. Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galán JE. S. typhimurium encodes an activator of rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell (1998) 93(5):815–26. doi: 10.1016/s0092-8674(00)81442-7
52. McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol (2009) 12(1):117–24. doi: 10.1016/j.mib.2008.12.001
53. Galan JE, Zhou D. Striking a balance: modulation of the actin cytoskeleton by salmonella. Proc Natl Acad Sci United States America (2000) 97(16):8754–61. doi: 10.1073/pnas.97.16.8754
54. Zhou D, Galán J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infection (2001) 3(14):1293–8. doi: 10.1016/S1286-4579(01)01489-7
55. Fu Y, Galán JEA. salmonella protein antagonizes rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature (1999) 401(6750):293–7. doi: 10.1038/45829
56. Zhou D, Chen LM, Hernandez L, Shears SB, Galán JEA. Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol (2001) 39(2):248–60. doi: 10.1046/j.1365-2958.2001.02230.x
57. Mijouin L, Rosselin M, Bottreau E, Pizarro-Cerda J, Cossart P, Velge P, et al. Salmonella enteritidis rck-mediated invasion requires activation of Rac1, which is dependent on the class I PI 3-kinases-Akt signaling pathway. FASEB J Off Publ Fed Am Societies Exp Biol (2012) 26(4):1569–81. doi: 10.1096/fj.11-189647
58. Wiedemann A, Mijouin L, Ayoub MA, Barilleau E, Canepa S, Teixeira-Gomes AP, et al. Identification of the epidermal growth factor receptor as the receptor for Salmonella rck-dependent invasion. FASEB J Off Publ Fed Am Societies Exp Biol (2016) 30(12):4180–91. doi: 10.1096/fj.201600701R
59. Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol (2006) 9(2):207–17. doi: 10.1016/j.mib.2006.02.008
60. Galán JE, Waksman G. Protein-injection machines in bacteria. Cell (2018) 172(6):1306–18. doi: 10.1016/j.cell.2018.01.034
61. Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, et al. Activation of β-catenin by carcinogenic helicobacter pylori. Proc Natl Acad Sci (2005) 102(30):10646–51. doi: 10.1073/pnas.0504927102
62. Gokulan K, Khare S, Rooney AW, Han J, Lynne AM, Foley SL. Impact of plasmids, including those encodingVirB4/D4 type IV secretion systems, on Salmonella enterica serovar Heidelberg virulence in macrophages and epithelial cells. PloS One (2013) 8(10):e77866. doi: 10.1371/journal.pone.0077866
63. Galán JE. Salmonella typhimurium and inflammation: a pathogen-centric affair. Nat Rev Microbiol (2021) 19(11):716–25. doi: 10.1038/s41579-021-00561-4
64. Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galán JE. Salmonella typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PloS Pathog (2009) 5(8):e1000538. doi: 10.1371/journal.ppat.1000538
65. Müller AJ, Hoffmann C, Galle M, Van Den Broeke A, Heikenwalder M, Falter L, et al. The s. typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe (2009) 6(2):125–36. doi: 10.1016/j.chom.2009.07.007
66. Tong L, Tergaonkar V. Rho protein GTPases and their interactions with NFκB: crossroads of inflammation and matrix biology. Bioscience Rep (2014) 34(3). doi: 10.1042/BSR20140021
67. Kamanova J, Sun H, Lara-Tejero M, Galán JE. The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PloS Pathog (2016) 12(4):e1005552. doi: 10.1371/journal.ppat.1005552
68. Lian H, Jiang K, Tong M, Chen Z, Liu X, Galán JE, et al. The Salmonella effector protein SopD targets Rab8 to positively and negatively modulate the inflammatory response. Nat Microbiol (2021) 6(5):658–71. doi: 10.1038/s41564-021-00866-3
69. Gokulan K, Khare S, Williams K, Foley SL. Transmissible plasmid containing Salmonella enterica Heidelberg isolates modulate cytokine production during early stage of interaction with intestinal epithelial cells. DNA Cell Biol (2016) 35(8):443–53. doi: 10.1089/dna.2015.3142
70. Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflammatory bowel Dis (1999) 5(4):262–70. doi: 10.1097/00054725-199911000-00004
71. Zheng L, Wang S, Ling M, Lv Z, Lin S. Salmonella enteritidis hcp distribute in the cytoplasm and regulate TNF signaling pathway in BHK-21 cells. 3 Biotech (2020) 10(7):1–7. doi: 10.1007/s13205-020-02296-0
72. Lin SL, Le TX, Cowen DS. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting raf activation. Cell Microbiol (2003) 5(4):267–75. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x
73. Sun H, Kamanova J, Lara-Tejero M, Galán JE. A family of Salmonella type III secretion effector proteins selectively targets the NF-κB signaling pathway to preserve host homeostasis. PloS Pathog (2016) 12(3):e1005484. doi: 10.1371/journal.ppat.1005484
74. Yin C, Liu Z, Xian H, Jiao Y, Yuan Y, Li Y, et al. AvrA exerts inhibition of NF-κB pathway in its naïve salmonella serotype through suppression of p-JNK and beclin-1 molecules. Int J Mol Sci (2020) 21(17). doi: 10.3390/ijms21176063
75. Liao AP, Petrof EO, Kuppireddi S, Zhao Y, Xia Y, Claud EC, et al. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PloS One (2008) 3(6):e2369. doi: 10.1371/journal.pone.0002369
76. Haraga A, Miller SIA. Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol (2006) 8(5):837–46. doi: 10.1111/j.1462-5822.2005.00670.x
77. Shappo MOE, Li Q, Lin Z, Hu M, Ren J, Xu Z, et al. SspH2 as anti-inflammatory candidate effector and its contribution in Salmonella enteritidis virulence. Microbial pathogenesis (2020) 142:104041. doi: 10.1016/j.micpath.2020.104041
78. Bhavsar AP, Brown NF, Stoepel J, Wiermer M, Martin DD, Hsu KJ, et al. The Salmonella type III effector SspH2 specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. PloS Pathog (2013) 9(7):e1003518. doi: 10.1371/journal.ppat.1003518
79. Bielig H, Lautz K, Braun PR, Menning M, Machuy N, Brügmann C, et al. The cofilin phosphatase slingshot homolog 1 (SSH1) links NOD1 signaling to actin remodeling. PloS Pathog (2014) 10(9):e1004351. doi: 10.1371/journal.ppat.1004351
80. Wu S, Shen Y, Zhang S, Xiao Y, Shi S. Salmonella interacts with autophagy to offense or defense. Front Microbiol (2020) 11:721. doi: 10.3389/fmicb.2020.00721
81. Zhou L, Li Y, Gao S, Yuan H, Zuo L, Wu C, et al. Salmonella spvC gene inhibits autophagy of host cells and suppresses NLRP3 as well as NLRC4. Front Immunol (2021) 12:639019. doi: 10.3389/fimmu.2021.639019
82. Zuo L, Zhou L, Wu C, Wang Y, Li Y, Huang R, et al. Salmonella spvC gene inhibits pyroptosis and intestinal inflammation to aggravate systemic infection in mice. Front Microbiol (2020) 11:562491. doi: 10.3389/fmicb.2020.562491
83. García-Gil A, Galán-Enríquez CS, Pérez-López A, Nava P, Alpuche-Aranda C, Ortiz-Navarrete V. SopB activates the akt-YAP pathway to promote Salmonella survival within b cells. Virulence (2018) 9(1):1390–402. doi: 10.1080/21505594.2018.1509664
84. Gibbs KD, Washington EJ, Jaslow SL, Bourgeois JS, Foster MW, Guo R, et al. The Salmonella secreted effector SarA/SteE mimics cytokine receptor signaling to activate STAT3. Cell Host Microbe (2020) 27(1):129–39.e4. doi: 10.1016/j.chom.2019.11.012
85. Hu H, Sun SC. Ubiquitin signaling in immune responses. Cell Res (2016) 26(4):457–83. doi: 10.1038/cr.2016.40
86. Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PloS Pathog (2012) 8(6):e1002743. doi: 10.1371/journal.ppat.1002743
87. Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, et al. Salmonella secreted factor l deubiquitinase of Salmonella typhimurium inhibits NF-κB, suppresses IκBα ubiquitination and modulates innate immune responses. J Immunol (2008) 180(7):5045–56. doi: 10.4049/jimmunol.180.7.5045
88. Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol (2007) 171(3):882–92. doi: 10.2353/ajpath.2007.070220
89. Narayanan LA, Edelmann MJ. Ubiquitination as an efficient molecular strategy employed in salmonella infection. Front Immunol (2014) 5:558. doi: 10.3389/fimmu.2014.00558
90. Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol (2006) 62(3):786–93. doi: 10.1111/j.1365-2958.2006.05407.x
91. Keszei AF, Tang X, McCormick C, Zeqiraj E, Rohde JR, Tyers M, et al. Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Mol Cell Biol (2014) 34(3):362–73. doi: 10.1128/MCB.01360-13
92. Metzger E, Müller JM, Ferrari S, Buettner R, Schüle R. A novel inducible transactivation domain in the androgen receptor: implications for PRK in prostate cancer. EMBO J (2003) 22(2):270–80. doi: 10.1093/emboj/cdg023
93. Chuang KH, Altuwaijri S, Li G, Lai JJ, Chu CY, Lai KP, et al. Neutropenia with impaired host defense against microbial infection in mice lacking androgen receptor. J Exp Med (2009) 206(5):1181–99. doi: 10.1084/jem.20082521
94. Lai JJ, Lai KP, Chuang KH, Chang P, Yu IC, Lin WJ, et al. Monocyte/macrophage androgen receptor suppresses cutaneous wound healing in mice by enhancing local TNF-alpha expression. J Clin Invest (2009) 119(12):3739–51. doi: 10.1172/JCI39335
95. Delyea C, Luo S, Dubrule BE, Julien O, Bhavsar AP. NOD1 is super-activated through spatially-selective ubiquitination by the salmonella effector SspH2. BioRxiv (2021) 2021–10. doi: 10.1101/2021.10.08.463692
96. Bernal-Bayard J, Ramos-Morales F. Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J Biol Chem (2009) 284(40):27587–95. doi: 10.1074/jbc.M109.010363
97. Bernal-Bayard J, Cardenal-Muñoz E, Ramos-Morales F. The Salmonella type III secretion effector, salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J Biol Chem (2010) 285(21):16360–8. doi: 10.1074/jbc.M110.100669
98. Kubori T, Galán JE. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell (2003) 115(3):333–42. doi: 10.1016/S0092-8674(03)00849-3
99. Zhang Y, Higashide W, Dai S, Sherman DM, Zhou D. Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. J Biol Chem (2005) 280(46):38682–8. doi: 10.1074/jbc.M506309200
100. Patel JC, Hueffer K, Lam TT, Galán JE. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell (2009) 137(2):283–94. doi: 10.1016/j.cell.2009.01.056
101. Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol (2009) 11(11):1652–70. doi: 10.1111/j.1462-5822.2009.01356.x
102. Ruan HH, Li Y, Zhang XX, Liu Q, Ren H, Zhang KS, et al. Identification of TRAF6 as a ubiquitin ligase engaged in the ubiquitination of SopB, a virulence effector protein secreted by Salmonella typhimurium. Biochem Biophys Res Commun (2014) 447(1):172–7. doi: 10.1016/j.bbrc.2014.03.126
103. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol (2010) 221(1):3–12. doi: 10.1002/path.2697
104. Birmingham CL, Brumell JH. Autophagy recognizes intracellular Salmonella enterica serovar typhimurium in damaged vacuoles. Autophagy (2006) 2(3):156–8. doi: 10.4161/auto.2825
105. Liu W, Jiang Y, Sun J, Geng S, Pan Z, Prinz RA, et al. Activation of TGF-β-activated kinase 1 (TAK1) restricts Salmonella typhimurium growth by inducing AMPK activation and autophagy. Cell Death Dis (2018) 9(5):570. doi: 10.1038/s41419-018-0612-z
106. Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell (2014) 54(2):224–33. doi: 10.1016/j.molcel.2014.03.009
107. Feng ZZ, Jiang AJ, Mao AW, Feng Y, Wang W, Li J, et al. The Salmonella effectors SseF and SseG inhibit Rab1A-mediated autophagy to facilitate intracellular bacterial survival and replication. J Biol Chem (2018) 293(25):9662–73. doi: 10.1074/jbc.M117.811737
108. Ichimura Y, Kumanomidou T, Sou Y-S, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem (2008) 283(33):22847–57. doi: 10.1074/jbc.M802182200
109. Chu Y, Gao S, Wang T, Yan J, Xu G, Li Y, et al. A novel contribution of spvB to pathogenesis of Salmonella typhimurium by inhibiting autophagy in host cells. Oncotarget (2016) 7(7):8295–309. doi: 10.18632/oncotarget.6989
110. Li Y-y, Wang T, Gao S, Xu G-m, Niu H, Huang R, et al. Salmonella plasmid virulence gene spvB enhances bacterial virulence by inhibiting autophagy in a zebrafish infection model. Fish Shellfish Immunol (2016) 49:252–9. doi: 10.1016/j.fsi.2015.12.033
111. Wang L, Li Y, Liu Y, Zuo L, Li Y, Wu S, et al. Salmonella spv locus affects type I interferon response and the chemotaxis of neutrophils via suppressing autophagy. Fish Shellfish Immunol (2019) 87:721–9. doi: 10.1016/j.fsi.2019.02.009
112. Aguilera MO, Berón W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy (2012) 8(11):1590–603. doi: 10.4161/auto.21459
113. Janeway CA Jr., Travers P, Walport M, Shlomchik MJ. The destruction of antibody-coated pathogens via fc receptors. immunobiology: The immune system in health and disease. 5th edition. Garland Science (2001).
114. Daha NA, Banda NK, Roos A, Beurskens FJ, Bakker JM, Daha MR, et al. Complement activation by (auto-) antibodies. Mol Immunol (2011) 48(14):1656–65. doi: 10.1016/j.molimm.2011.04.024
115. Janeway CA Jr., Travers P, Walport M, Shlomchik MJ. The complement system and innate immunity. immunobiology: The immune system in health and disease. 5th edition. Garland Science (2001).
116. Männe C, Takaya A, Yamasaki Y, Mursell M, Hojyo S, Wu TY, et al. Salmonella SiiE prevents an efficient humoral immune memory by interfering with IgG+ plasma cell persistence in the bone marrow. Proc Natl Acad Sci United States America (2019) 116(15):7425–30. doi: 10.1073/pnas.1818242116
117. Bakowski MA, Braun V, Lam GY, Yeung T, Heo WD, Meyer T, et al. The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe (2010) 7(6):453–62. doi: 10.1016/j.chom.2010.05.011
118. Lau N, Haeberle AL, O'Keeffe BJ, Latomanski EA, Celli J, Newton HJ, et al. SopF, a phosphoinositide binding effector, promotes the stability of the nascent Salmonella-containing vacuole. PloS Pathog (2019) 15(7):e1007959. doi: 10.1371/journal.ppat.1007959
119. Singh PK, Kapoor A, Lomash RM, Kumar K, Kamerkar SC, Pucadyil TJ, et al. Salmonella SipA mimics a cognate SNARE for host Syntaxin8 to promote fusion with early endosomes. J Cell Biol (2018) 217(12):4199–214. doi: 10.1083/jcb.201802155
120. Beuzón CR, Méresse S, Unsworth KE, Ruíz-Albert J, Garvis S, Waterman SR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J (2000) 19(13):3235–49. doi: 10.1093/emboj/19.13.3235
121. Sindhwani A, Arya SB, Kaur H, Jagga D, Tuli A, Sharma M. Salmonella exploits the host endolysosomal tethering factor HOPS complex to promote its intravacuolar replication. PloS Pathog (2017) 13(10):e1006700. doi: 10.1371/journal.ppat.1006700
122. Schroeder N, Henry T, de Chastellier C, Zhao W, Guilhon AA, Gorvel JP, et al. The virulence protein SopD2 regulates membrane dynamics of Salmonella-containing vacuoles. PloS Pathog (2010) 6(7):e1001002. doi: 10.1371/journal.ppat.1001002
123. Spanò S, Gao X, Hannemann S, Lara-Tejero M, Galán JE. A bacterial pathogen targets a host rab-family GTPase defense pathway with a GAP. Cell Host Microbe (2016) 19(2):216–26. doi: 10.1016/j.chom.2016.01.004
124. Kolodziejek AM, Miller SI. Salmonella modulation of the phagosome membrane, role of SseJ. Cell Microbiol (2015) 17(3):333–41. doi: 10.1111/cmi.12420
125. Greene AR, Owen KA, Casanova JE. Salmonella typhimurium manipulates macrophage cholesterol homeostasis through the SseJ-mediated suppression of the host cholesterol transport protein ABCA1. Cell Microbiol (2021) 23(8):e13329. doi: 10.1111/cmi.13329
126. Ramsden AE, Mota LJ, Münter S, Shorte SL, Holden DW. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol (2007) 9(10):2517–29. doi: 10.1111/j.1462-5822.2007.00977.x
127. Yu XJ, Liu M, Holden DW. Salmonella effectors SseF and SseG interact with mammalian protein ACBD3 (GCP60) to anchor salmonella-containing vacuoles at the golgi network. mBio (2016) 7(4). doi: 10.1128/mBio.00474-16
128. McLaughlin LM, Govoni GR, Gerke C, Gopinath S, Peng K, Laidlaw G, et al. The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PloS Pathog (2009) 5(11):e1000671. doi: 10.1371/journal.ppat.1000671
129. Bayer-Santos E, Durkin CH, Rigano LA, Kupz A, Alix E, Cerny O, et al. The Salmonella effector SteD mediates MARCH8-dependent ubiquitination of MHC II molecules and inhibits T cell activation. Cell Host Microbe (2016) 20(5):584–95. doi: 10.1016/j.chom.2016.10.007
130. Cerny O, Godlee C, Tocci R, Cross NE, Shi H, Williamson JC, et al. CD97 stabilises the immunological synapse between dendritic cells and T cells and is targeted for degradation by the Salmonella effector SteD. PloS Pathog (2021) 17(7):e1009771. doi: 10.1371/journal.ppat.1009771
131. Godlee C, Cerny O, Liu M, Blundell S, Gallagher AE, Shahin M, et al. The Salmonella transmembrane effector SteD hijacks AP1-mediated vesicular trafficking for delivery to antigen-loading MHCII compartments. PloS Pathog (2022) 18(5):e1010252. doi: 10.1371/journal.ppat.1010252
132. Spanò S, Liu X, Galán JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host salmonella. Proc Natl Acad Sci United States America (2011) 108(45):18418–23. doi: 10.1073/pnas.1111959108
133. Savitskiy S, Wachtel R, Pourjafar-Dehkordi D, Kang HS, Trauschke V, Lamb DC, et al. Proteolysis of Rab32 by Salmonella GtgE induces an inactive GTPase conformation. iScience (2021) 24(1):101940. doi: 10.1016/j.isci.2020.101940
134. McLaughlin LM, Xu H, Carden SE, Fisher S, Reyes M, Heilshorn SC, et al. A microfluidic-based genetic screen to identify microbial virulence factors that inhibit dendritic cell migration. Integr Biol quantitative Biosci nano to macro (2014) 6(4):438–49. doi: 10.1039/C3IB40177D
135. Alberdi L, Vergnes A, Manneville JB, Tembo DL, Fang Z, Zhao Y, et al. Regulation of kinesin-1 activity by the salmonella enterica effectors PipB2 and SifA. J Cell Sci (2020) 133(9). doi: 10.1242/jcs.239863
136. Domingues L, Holden DW, Mota LJ. The Salmonella effector SteA contributes to the control of membrane dynamics of Salmonella-containing vacuoles. Infection Immun (2014) 82(7):2923–34. doi: 10.1128/IAI.01385-13
137. Knodler LA. Salmonella enterica: living a double life in epithelial cells. Curr Opin Microbiol (2015) 23:23–31. doi: 10.1016/j.mib.2014.10.010
138. Creasey EA, Isberg RR. Maintenance of vacuole integrity by bacterial pathogens. Curr Opin Microbiol (2014) 17:46–52. doi: 10.1016/j.mib.2013.11.005
139. Zhou D, Mooseker MS, Galán JE. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc Natl Acad Sci United States America (1999) 96(18):10176–81. doi: 10.1073/pnas.96.18.10176
140. Yrlid U, Svensson M, Kirby A, Wick MJ. Antigen-presenting cells and anti-Salmonella immunity. Microbes Infection (2001) 3(14):1239–48. doi: 10.1016/S1286-4579(01)01484-8
141. Kotsias F, Cebrian I, Alloatti A. Antigen processing and presentation. Int Rev Cell Mol Biol (2019) 348:69–121. doi: 10.1016/bs.ircmb.2019.07.005
142. Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev (2007) 20(4):535–49. doi: 10.1128/CMR.00013-07
143. Rathinavelan T, Lara-Tejero M, Lefebre M, Chatterjee S, McShan AC, Guo DC, et al. NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system. J Mol Biol (2014) 426(16):2958–69. doi: 10.1016/j.jmb.2014.06.009
144. Jneid B, Moreau K, Plaisance M, Rouaix A, Dano J, Simon S. Role of T3SS-1 SipD protein in protecting mice against non-typhoidal Salmonella typhimurium. PloS Negl Trop Dis (2016) 10(12):e0005207. doi: 10.1371/journal.pntd.0005207
145. Jneid B, Rouaix A, Féraudet-Tarisse C, Simon S. SipD and IpaD induce a cross-protection against Shigella and Salmonella infections. PloS Negl Trop Dis (2020) 14(5):e0008326. doi: 10.1371/journal.pntd.0008326
146. Martinez-Becerra FJ, Kumar P, Vishwakarma V, Kim JH, Arizmendi O, Middaugh CR, et al. Characterization and protective efficacy of type III secretion proteins as a broadly protective subunit vaccine against salmonella enterica serotypes. Infection Immun (2018) 86(3). doi: 10.1128/IAI.00473-17
147. Lin Z, Tang P, Jiao Y, Kang X, Li Q, Xu X, et al. Immunogenicity and protective efficacy of a Salmonella enteritidis sptP mutant as a live attenuated vaccine candidate. BMC veterinary Res (2017) 13(1):194. doi: 10.1186/s12917-017-1115-3
148. Li Y, Wang S, Xin W, Scarpellini G, Shi Z, Gunn B, et al. A sopB deletion mutation enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Infection Immun (2008) 76(11):5238–46. doi: 10.1128/IAI.00720-08
149. Hindle Z, Chatfield SN, Phillimore J, Bentley M, Johnson J, Cosgrove CA, et al. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infection Immun (2002) 70(7):3457–67. doi: 10.1128/IAI.70.7.3457-3467.2002
150. Kirkpatrick BD, Tenney KM, Larsson CJ, O'Neill JP, Ventrone C, Bentley M, et al. The novel oral typhoid vaccine M01ZH09 is well tolerated and highly immunogenic in 2 vaccine presentations. J Infect Dis (2005) 192(3):360–6. doi: 10.1086/431605
151. Kirkpatrick BD, McKenzie R, O'Neill JP, Larsson CJ, Bourgeois AL, Shimko J, et al. Evaluation of Salmonella enterica serovar typhi (Ty2 aroC-ssaV-) M01ZH09, with a defined mutation in the Salmonella pathogenicity island 2, as a live, oral typhoid vaccine in human volunteers. Vaccine (2006) 24(2):116–23. doi: 10.1016/j.vaccine.2005.08.008
152. Tran TH, Nguyen TD, Nguyen TT, Ninh TT, Tran NB, Nguyen VM, et al. A randomised trial evaluating the safety and immunogenicity of the novel single oral dose typhoid vaccine M01ZH09 in healthy Vietnamese children. PloS One (2010) 5(7):e11778. doi: 10.1371/journal.pone.0011778
153. Michael A, John J, Meyer B, Pandha H. Activation and genetic modification of human monocyte-derived dendritic cells using attenuated Salmonella typhimurium. TheScientificWorldJournal (2010) 10:393–401. doi: 10.1100/tsw.2010.37
154. Vishwakarma V, Pati NB, Chandel HS, Sahoo SS, Saha B, Suar M. Evaluation of Salmonella enterica serovar typhimurium TTSS-2 deficient fur mutant as safe live-attenuated vaccine candidate for immunocompromised mice. PloS One (2012) 7(12):e52043. doi: 10.1371/journal.pone.0052043
155. Vishwakarma V, Pati NB, Ray S, Das S, Suar M. TTSS2-deficient hha mutant of Salmonella typhimurium exhibits significant systemic attenuation in immunocompromised hosts. Virulence (2014) 5(2):311–20. doi: 10.4161/viru.27605
156. Petrovska L, Aspinall RJ, Barber L, Clare S, Simmons CP, Stratford R, et al. Salmonella enterica serovar typhimurium interaction with dendritic cells: Impact of the sifA gene. Cell Microbiol (2004) 6(11):1071–84. doi: 10.1111/j.1462-5822.2004.00419.x
157. Hu M, Zhao W, Gao W, Li W, Meng C, Yan Q, et al. Recombinant Salmonella expressing SspH2-EscI fusion protein limits its colonization in mice. BMC Immunol (2017) 18(1):21. doi: 10.1186/s12865-017-0203-2
158. Hu M, Zhao W, Li H, Gu J, Yan Q, Zhou X, et al. Immunization with recombinant Salmonella expressing SspH2-EscI protects mice against wild type Salmonella infection. BMC veterinary Res (2018) 14(1):79. doi: 10.1186/s12917-018-1404-5
159. Rüssmann H, Shams H, Poblete F, Fu Y, Galán JE, Donis RO. Delivery of epitopes by the salmonella type III secretion system for vaccine development. Science (1998) 281(5376):565–8. doi: 10.1126/science.281.5376.565
160. Xu X, Hegazy WA, Guo L, Gao X, Courtney AN, Kurbanov S, et al. Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res (2014) 74(21):6260–70. doi: 10.1158/0008-5472.CAN-14-1169
161. Lavelle EC, Ward RW. Mucosal vaccines - fortifying the frontiers. Nat Rev Immunol (2022) 22(4):236–50. doi: 10.1038/s41577-021-00583-2
162. Lycke N. Recent progress in mucosal vaccine development: Potential and limitations. Nat Rev Immunol (2012) 12(8):592–605. doi: 10.1038/nri3251
163. Russell MW, Moldoveanu Z, Ogra PL, Mestecky J. Mucosal immunity in COVID-19: A neglected but critical aspect of SARS-CoV-2 infection. Front Immunol (2020) 11:611337. doi: 10.3389/fimmu.2020.611337
164. Guo Y, Xu Y, Kang X, Gu D, Jiao Y, Meng C, et al. Immunogenic potential and protective efficacy of a sptP deletion mutant of Salmonella enteritidis as a live vaccine for chickens against a lethal challenge. Int J Med Microbiol IJMM (2019) 309(8):151337. doi: 10.1016/j.ijmm.2019.151337
165. Gayet R, Bioley G, Rochereau N, Paul S, Corthésy B. Vaccination against Salmonella infection: the mucosal way. Microbiol Mol Biol Rev MMBR (2017) 81(3). doi: 10.1128/MMBR.00007-17
166. Zhou G, Tian Y, Tian J, Ma Q, Huang S, Li Q, et al. Oral immunization with attenuated Salmonella choleraesuis expressing the P42 and P97 antigens protects mice against Mycoplasma hyopneumoniae challenge. Microbiol Spectr (2022) 10(6):e0236122. doi: 10.1128/spectrum.02361-22
167. Li Q, Lv Y, Li YA, Du Y, Guo W, Chu D, et al. Live attenuated Salmonella enterica serovar choleraesuis vector delivering a conserved surface protein enolase induces high and broad protection against Streptococcus suis serotypes 2, 7, and 9 in mice. Vaccine (2020) 38(44):6904–13. doi: 10.1016/j.vaccine.2020.08.062
168. Li YA, Ji Z, Wang X, Wang S, Shi H. Salmonella enterica serovar choleraesuis vector delivering SaoA antigen confers protection against Streptococcus suis serotypes 2 and 7 in mice and pigs. Veterinary Res (2017) 48(1):89. doi: 10.1186/s13567-017-0494-6
169. Galen JE, Curtiss R. The delicate balance in genetically engineering live vaccines. Vaccine (2014) 32(35):4376–85. doi: 10.1016/j.vaccine.2013.12.026
170. Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T, Yu XJ, et al. Structure of the cytoplasmic domain of SctV (SsaV) from the Salmonella SPI-2 injectisome and implications for a pH sensing mechanism. J Struct Biol (2021) 213(2):107729. doi: 10.1016/j.jsb.2021.107729
171. Periaswamy B, Maier L, Vishwakarma V, Slack E, Kremer M, Andrews-Polymenis HL, et al. Live attenuated s. typhimurium vaccine with improved safety in immuno-compromised mice. PloS One (2012) 7(9):e45433. doi: 10.1371/journal.pone.0045433
172. Jo Wick M, Ljunggren HG. Processing of bacterial antigens for peptide presentation on MHC class I molecules. Immunol Rev (1999) 172(1):153–62. doi: 10.1111/j.1600-065X.1999.tb01363.x
173. Kang HY, Srinivasan J, Curtiss R. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar typhimurium vaccine. Infection Immun (2002) 70(4):1739–49. doi: 10.1128/IAI.70.4.1739-1749.2002
174. Cox MA, Kahan SM, Zajac AJ. Anti-viral CD8 T cells and the cytokines that they love. Virology (2013) 435(1):157–69. doi: 10.1016/j.virol.2012.09.012
Keywords: host defenses, Salmonella infections, secretion systems, effector proteins, vaccine research
Citation: Zhou G, Zhao Y, Ma Q, Li Q, Wang S and Shi H (2023) Manipulation of host immune defenses by effector proteins delivered from multiple secretion systems of Salmonella and its application in vaccine research. Front. Immunol. 14:1152017. doi: 10.3389/fimmu.2023.1152017
Received: 27 January 2023; Accepted: 23 March 2023;
Published: 04 April 2023.
Edited by:
Maria Isabel Colombo, Universidad Nacional de Cuyo, ArgentinaReviewed by:
Shailendra Kumar Verma, La Jolla Institute for Immunology (LJI), United StatesCopyright © 2023 Zhou, Zhao, Ma, Li, Wang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huoying Shi, aHlzaGlAeXp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.