
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 03 February 2023
Sec. Inflammation
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1084636
Macrophages have a wide variety of roles in physiological and pathological conditions, making them promising diagnostic and therapeutic targets in diseases, especially metabolic disorders, which have attracted considerable attention in recent years. Owing to their heterogeneity and polarization, the phenotypes and functions of macrophages related to metabolic disorders are diverse and complicated. In the past three decades, the rapid progress of macrophage research has benefited from the emergence of specific molecular markers to delineate different phenotypes of macrophages and elucidate their role in metabolic disorders. In this review, we analyze the functions and applications of commonly used and novel markers of macrophages related to metabolic disorders, facilitating the better use of these macrophage markers in metabolic disorder research.
In the last three decades, rapidly evolving technology has greatly expanded and deepened our understanding of the characteristics of macrophages. (a) Macrophages are professional phagocytes present in virtually every tissue under homeostatic physiological conditions (1). (b) Macrophages not only have important immunomodulatory functions, initiating the innate response and inflammation, but also maintain tissue homeostasis and repair (2). (c) The characteristics of macrophages are heterogeneity and plasticity, and they can be phenotypically polarized by surrounding micro-environmental stimuli (2, 3). (d) Macrophages have three distinct precursors: yolk sac (YS) macrophages, fetal liver (FL) monocytes, and bone-marrow-derived monocytes, which can be divided into two groups: tissue-resident macrophages (originating from YS and FL) and monocyte-derived macrophages (derived from bone marrow-derived monocytes) (4–6).
To study macrophage heterogeneity and plasticity, the M1/M2 dichotomy has been developed for 20 years, which offers a useful framework (7). (a) M1 macrophages play a proinflammatory role by upregulating inducible nitric oxide synthase (iNOS) to produce reactive oxygen species (ROS) and reactive nitrogen species (RNS) for activating glycolysis, fatty acid synthesis (FAS), and the pentose phosphate pathway (PPP); the M1 phenotype also suppresses the tricarboxylic acid (TCA) cycle and mitochondrial oxidative phosphorylation (OXPHOS), which promote the inflammatory response and phagocytosis. (b) M2 macrophages play an anti-inflammatory role by upregulating arginase-1 (Arg-1) to produce ornithine and urea to enhance OXPHOS, FAS, and glutamine metabolism; they also suppress PPP, thereby promoting the anti-inflammatory response and tissue repair (8, 9). However, the M1/M2 dichotomy is too simple to explain complex macrophages with various phenotypes and activation statuses in different tissues (10). Recently, cytometry and single-cell RNA sequencing have facilitated the development of macrophage marker biology. Consequently, identifying phenotypes using markers and exploring their relationship with macrophage metabolism are key points in the study of macrophages.
Metabolic diseases are noncommunicable diseases characterized by disorders of blood pressure, glucose, and lipid levels, including obesity, diabetes, hypertension, and neurodegenerative diseases (11, 12). Macrophages are important for the maintenance of homeostasis and play a profound role in the pathological state of metabolic diseases (13). (a) Obesity and diabetes: unique metabolic activation of adipose tissue macrophages (ATMs) in obesity and diabetes increases OXPHOS and glycolysis, which may be therapeutic targets to alleviate inflammation and insulin resistance (14). (b) Neurological disorders: microglia stimulated by pathological signals reprogram their metabolic pathways, such as increasing glycolysis, iron accumulation, and decreasing mitochondrial respiration, to influence neuronal functionality and survival in the brain to regulate neurological disorders (15, 16). (c) Cardiometabolic disorders: dysregulation of polarization between proinflammatory macrophages (PIMs) and anti-inflammatory macrophages (AIMs) promotes excessive inflammation and cardiac injury, resulting in cardiometabolic diseases; therefore, exploring cardiac macrophages in polarization mechanisms and their interaction with other cardiac cells is vital for future research (17).
The identification of macrophages in metabolic diseases is beneficial for the development of macrophage marker biology. In this review, we selected classical pan-markers of macrophages (F4/80 and CD68), conventional markers of macrophages in inflammatory states (iNOS and Arg-1), markers of macrophages in different tissues (C-X3-C motif chemokine receptor 1 (CX3CR1), CC chemokine receptor 2 (CCR2), lymphatic vessel hyaluronan receptor1 (Lyve1), and major histocompatibility complex class II (MHCII)), and novel markers emerging in recent years (CD9 and triggering receptor expressed on myeloid cells 2 (TREM2)), expanding on their structure and location (Figure 1), biological function (Table 1), and application in research.
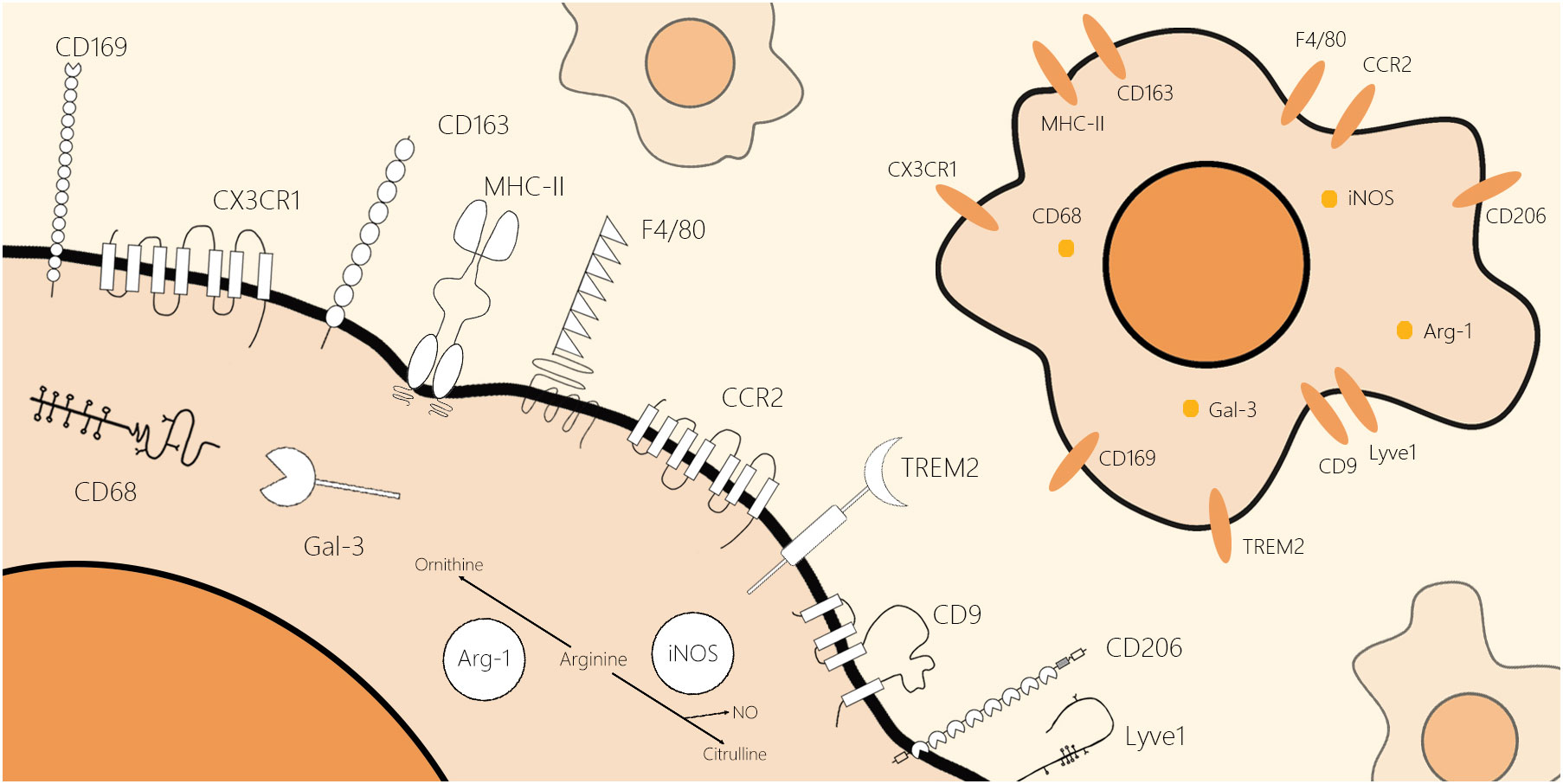
Figure 1 Schematic molecular structure and location of classical and novel markers of the macrophages. These markers of macrophages can be roughly divided into two categories by their location: cell surface markers and intracellular markers. F4/80, CCR2, CD169, CX3CR1, CD206, CD163, Lyve1, CD9, TREM2, and MHCII are macrophage markers located on the cell membrane. Moreover, CD68, iNOS, Arg-1, and Gal-3 are macrophage markers located inside the cell. All schematics of markers are based on their two-dimensional molecular structure.
Obesity-induced adipose tissue hypoxia promotes macrophage switching into PIMs with overexpression of iNOS (39–41). Cluster of Differentiation 9 (CD9) and TREM2 are two macrophage markers in the white adipose tissue of obese patients. Human adipocyte-related macrophages are labeled with Cluster of Differentiation 68 (CD68) and CD9 (42–44). The pan-marker CD68, proinflammatory marker iNOS, and novel markers TREM2 and CD9 are widely used markers in macrophages that are strongly associated with obesity and diabetes.
CD68, also known as macrosialin, is a highly glycosylated transmembrane protein that is deemed a member of the scavenger receptor family because of its significant role in binding oxidized low-density lipoprotein (ox-LDL) on the macrophage cell surface (45).
CD68 is strongly expressed by the mononuclear phagocyte system, including macrophages, resulting in its use as a pan-macrophage marker (4, 46). CD68 is regarded as a marker of ATMs and has been shown to be the strongest predictor of insulin-resistant obesity (47). CD68+ ATMs in obese mice increased and presented a proinflammatory phenotype, which produced inflammatory cytokines, regulated glycolysis, and cleared lipids and dead adipocytes (48, 49). Similarly, CD68+ pancreatic islet macrophages (PLMs) are increased in diabetic patients, and their accumulation is associated with the pathogenesis of type 2 diabetes mellitus (T2DM) in humans, such as amyloid deposition (50, 51). CD68+ PLMs promote the compensatory proliferation of β cells and reduce glucose-stimulated insulin secretion (50, 52). In summary, CD68 is a commonly used pan-marker of tissue macrophages related to glucolipid metabolic disorder diseases, especially ATM- and PLM-mediated uptake of modified lipoproteins.
iNOS is widely used as a marker of PIMs because PIMs have a 30-fold higher expression of iNOS than AIMs (53). NO produced by iNOS has a significant effect on the glucolipid metabolic response. First, iNOS-derived NO in activated resident macrophages could destroy the antilipolytic effect of insulin in adipocytes, resulting in accelerated triglyceride lipolysis in adipose tissue. Second, iNOS participates in the metabolic reprogramming of immune cells to promote aerobic glycolysis in PIMs to strengthen the inflammatory response (54).
The metabolism of arginine is important in regulating macrophage polarization between pro- and anti-inflammatory subtypes because arginine is a substrate of iNOS in PIMs; in contrast, Arg-1 triggers the anti-inflammatory macrophage phenotype, competing with iNOS for arginine (55). Therefore, the future task of macrophage polarization research is to dissect the relationship between pro- and anti-inflammatory phenotypes and metabolism with respect to iNOS-derived NO.
Previously, the number of PLMs in T2DM patients with islet amyloid deposition expressing CD68 and iNOS increased, which was associated with lesion progression (56). Recent studies have shown that the downregulation of proinflammatory iNOS expression in macrophages is positively associated with improved glucose intolerance and insulin resistance after treatment (57, 58). Therefore, iNOS+ PIMs in obese and T2DM patients have a potent phagocytosing capacity through NO production; thus, inhibiting their formation may be a useful treatment strategy.
CD9, also called human lymphohematopoietic progenitor cell surface antigen P24, was first identified by Kersey when combining monoclonal antibodies with acute lymphoblastic leukemia cells (59). CD9 has multiple biological functions that are involved in many vital physiological and pathological processes, such as neuroectodermal growth, myotubular formatting, and the incidence and transfer of tumors (60).
CD9 can restrict the activation of macrophages in inflammatory responses, so it is recognized as a marker for anti-inflammatory monocytes and macrophages (61, 62). However, recent studies on metabolic disorders have suggested that CD9 and TREM2 are markers of lipid-associated macrophages (LAMs) that produce proinflammatory cytokines in humans (43). In white adipose tissue, CD9+TREM2+ macrophages correlated with the severity of inflammation and influenced obesity pathology (42). In the aging brain, CD9+TREM2+ macrophages containing lipid droplets may play a pathogenic role in neurodegenerative diseases (63). Taken together, CD9 is involved in the macrophage response to lipids, and infiltrating CD9+ LAMs exacerbate metabolic diseases.
TREM2 is a lipid-sensing extracellular receptor expressed by the myeloid lineage (64, 65). TREM2 has been implicated in various biological processes, including maturation, activation, survival of cells, and regulation of inflammatory responses (65). TREM2 facilitates phagocytosis and transcription of anti-inflammatory cytokines to inhibit the production of inflammatory cytokines (66).
TREM2 was first found to be expressed on the surface of monocyte-derived dendritic cells (DCs) in humans, and it is expressed on the myeloid lineage, including macrophages (65, 67). It is expressed by a small subset of physiological tissue macrophages, such as microglia and ATMs (68). TREM2+ macrophages are good at lipolysis and are enriched in atherosclerotic lesions (69). TREM2 expressed by macrophages inhibits the development of metabolic disorders by facilitating cell death of prone adipocytes (43). Overall, TREM2 participates in the transmission of an inhibitory signal that reduces the inflammatory response, and TREM2+ macrophages are associated with lipid metabolism by suppressing lipid peroxidation and ROS to prevent systemic metabolic dysregulation.
Metabolic disorders trigger gut microbiota dysbiosis and low-grade systemic inflammation, leading to blood–brain barrier (BBB) dysfunction. All circulating immune cells and molecules infiltrate the brain because of increased BBB permeability, resulting in neuroinflammation and amyloid imbalance (70–72). The innate immune cells involved in this process are mainly microglia, whose classical markers are F4/80 and CX3CR1 (73–75). Recent studies have shown that M2 macrophages provide neuroprotective and regenerative effects, the targeting of which might be promising for treating chronic neurological diseases (76). In conclusion, F4/80 and CX3CR1-expressing macrophages play important regulatory roles in neuroinflammation, which polarizes to the Arg1+ M2 phenotype, contributing to the treatment of neurological disorders.
F4/80 has been extensively used in the identification and study of murine macrophages under physiological and pathological conditions since 1981, and it has greatly boosted research on macrophages (77). Extracellular epithelial growth factor (EGF) module containing mucin-like hormone receptor 1 (EMR1), the homology of F4/80 in humans, is a glycoprotein with an EGF-like domain and a seven-transmembrane motif (TM7) (78, 79). Although human EMR1 and murine F4/80 share a similar structure, EMR1 is not expressed in human macrophages but is highly expressed in eosinophils, whereas F4/80 is a well-known marker of mouse macrophages and microglia (18, 80).
F4/80 is restricted to murine macrophages in almost all tissues, including the liver, splenic red pulp, adrenal glands, and central nervous system; it may also be implicated in the generation of efferent CD8+ Treg cells required for inducing peripheral immune tolerance (18, 19). Nevertheless, it was later proven that apart from macrophages, F4/80 is also expressed in other myeloid cells, such as DCs (81, 82). This evidence indicates that cell specificity and limitations of F4/80 expression should be considered when labeling macrophages and microglia with F4/80.
CX3CR1 is a receptor for C-X3-C motif chemokine ligand 1 (CX3CL1) (83). CX3CL1-CX3CR1 has been shown to be a novel regulator of leukocyte transportation with adhesive and chemotactic functions; it is also a connecting bridge between neuronal cells and microglia (84). The CX3CL1-CX3CR1 axis in microglia internalizes and degrades amyloid-β deposits and Tau aggregates, which influence the development of Alzheimer’s disease (AD) (85). Hence, regulating CX3CL1 to bind microglia-expressing CX3CR1 may be a possible therapy for disease progression in the central nervous system.
CX3CR1 is widely expressed by various cells belonging to the macrophage lineage and is involved in macrophage development. For example, in the central nervous system, CX3CR1 is predominantly expressed by microglia and neurons and is engaged in activated microglia recruitment to inflammatory sites following ischemia (86). In recent years, it has served as a marker for intestinal macrophages, microglia, and patrolling monocytes (34). For instance, activated microglia were shown to have high CX3CR1 and/or MHCII expression (87). CX3CR1 can serve as a marker of tissue-resident macrophages that migrate from monocytes, especially in the brain and intestine, as well as in pathological conditions that promote inflammatory macrophages. CX3CR1+ microglia perform different activations to change mitochondrial dynamics and switch between OXPHOS and glycolysis, which directly affects neurological disorders.
Arg-1 is a member of the ureohydrolase family of enzymes that catalyze the hydrolysis of l-arginine to urea and l-ornithine (88). The physiological functions of Arg-1 in healthy conditions include detoxification of ammonia in urea, neuroprotection, and wound healing (88, 89).
Arg-1 is a well-known marker of M2 macrophages with anti-inflammatory properties that maintain tissue homeostasis and resolve inflammation (90). A recent study has shown that promoting the polarization of microglia toward the Arg-1+ M2 phenotype to enhance Aβ-induced neurite atrophy and neuronal regeneration might be a therapeutic approach in the treatment of AD (91). Arg-1+ microglia are mainly involved in neuroprotection, and it is necessary to develop new potential drugs for neurodegenerative diseases that modulate M2 microglial polarization.
Metabolic disorders are major risk factors for cardiovascular disease (CVD), and metabolic syndrome increases the risk of CVD twofold (92, 93). Cardiac macrophages are identified by MHCII and CCR2, which are involved in maintaining homeostasis, immune surveillance, angiogenesis, injury repair, and assisting atrioventricular conduction (94). Recent studies have used Lyve1 and MHCII to label macrophages near blood vessels, which are involved in suppressing inflammation and fibrosis (95). Taken together, CCR2, MHCII, and Lyve1 are commonly used markers of macrophages in the cardiac system.
The CCR2 is a chemokine receptor, also known as the CCL2 receptor, of the monocyte chemotactic protein (MCP) family, which has a high affinity for binding (96). CCR2 engages in monocyte extravasation, adherence, and migration into inflammatory tissues, where they differentiate into macrophages (97). CCR2, antagonized by pharmacological action, can upregulate insulin sensitivity to cure obesity in mice (98). Hence, future studies on the pharmacological inhibition of CCR2 may offer guidance for therapeutic approaches in inflammatory diseases.
CCR2 has been used to label tissue macrophages originating from monocytes (99). It controls the migration of monocytes and macrophages, which play a vital role in various diseases, especially cardiometabolic disease (100). There are two distinct subsets of cardiac macrophages divided by CCR2 expression of different origins: YS-derived CCR2 macrophages and monocyte-derived CCR2+ macrophages (24). Over time, CCR2− macrophages are progressively replaced by CCR2+ macrophages via circulation, which presents an anti-inflammatory phenotype and plays a cardioprotective role (24). Tissue-resident CCR2+ macrophages are vital regulators of monocyte movement, inflammation, cardiac pacemaking, and electrical propagation (101). Recent studies have expounded that the inhibition of CCR2 affects the polarization of macrophages, and the inhibition of CCL2 binding to CCR2 upregulates the expression of proinflammatory genes (102, 103). Thus, the expression and functional regulation of CCR2 in macrophages to affect inflammatory diseases need to be further investigated.
MHCII is a glycoprotein involved in the generation of immune responses. Its main function is to present peptide fragments from antigens to T cells to initiate an immune response (104). Moreover, MHCII also activates intracellular signaling pathways as a signaling receptor, which leads to the apoptosis of antigen-presenting cells (APCs), resulting in the termination of immune responses (105).
MHCII is expressed by innate immune cells, particularly APCs such as the monocyte-macrophage lineage (106). Macrophages expressing MHCII are activated by various inflammatory agents. IFN-γ activates macrophages to PIMs by upregulating the expression of MHCII and CD86 (20, 107). M2a macrophages induced by IL-4 are associated with increased expression of MHCII (21). IL-4 and IFN-γ both enhance MHCII expression in macrophages while also affecting macrophage polarization into different subtypes. Therefore, MHCII can be used as a pro- and anti-inflammatory marker for macrophages.
Recent studies have utilized the expression of MHCII to classify different functional macrophage subsets in metabolic diseases. In murine atherosclerosis, macrophages are subclassified into at least five subsets based on MHCII and CCR2 expression (108). In the murine heart, MHCIIhi macrophages are most prevalent under physiological conditions, while MHCIIlow macrophages become the major macrophages at the early stage of myocardial infarction because MHCII expression on macrophages is transiently modulated by ischemia-reperfusion injury (109). In conclusion, macrophages with high MHCII expression play key roles in innate and adaptive immune responses and can polarize into different subtypes, thereby regulating inflammation to treat metabolic diseases by antigen presentation and inducing apoptosis.
Lyve1 is a receptor of the extracellular matrix glycosaminoglycan hyaluronan located in the lymphatic endothelium (110). Lyve1 mediates the docking and transit of leukocytes, including macrophages, to influence inflammation and regulate the movement of hyaluronan, which enters peripheral lymphatics for immune activation (111–113).
Chakarov (95) demonstrated that monocyte-derived tissue macrophages could be separated into two subsets: Lyve1lowMHCIIhigh subsets adjacent to nerve bundles and fibers and Lyve1highMHCIIlow subsets near blood vessels—the latter of which played a role in inflammation influencing lung and heart fibrosis. Lim et al. (114) proved that Lyve1 expressed on perivascular macrophages interacts with hyaluronan on smooth muscle cells to protect against arterial stiffness. In conclusion, Lyve1 is mainly restricted to lymphatic endothelia and is also expressed in the liver, spleen, and lungs (111, 115, 116). Furthermore, it is expressed in a rare anti-inflammatory macrophage subset with a potent endothelial progenitor appearing in inflammatory and tumor sites (116–118). Lyve1+ macrophages interact with hyaluronan, influencing endothelial junctional retraction and proliferation. Future research should investigate the relationship between macrophages in the lymphatic and vascular systems and Lyve1.
Since these markers are expressed but not restricted to certain tissues, macrophages identified by these markers distribute in virtually all tissues related to metabolic disorders. For instance, TREM2+ macrophages resided in different tissues during obesity, including adipose tissue and liver, with different phenotypes, such as LAMs, non-alcoholic steatohepatitis-associated macrophages, and scar-associated macrophages (119). Identifying various macrophage phenotypes requires the combination of multiple markers, which helps to visualize macrophage heterogeneity and plasticity in affecting the metabolic microenvironment of tissues. For example, glucose uptake and utilization are upregulated in cardiac macrophages (classified by TIMD4, Lyve1, MHCII, and CCR2), and free fatty acids promote dysregulation of polarization, resulting in excessive inflammation, activation of myofibroblast, and apoptosis of cardiomyocytes during the metabolic disorders (120, 121). This evidence proves macrophages identified by various markers in metabolic tissues may be therapeutic targets to alleviate inflammation and insulin resistance.
Macrophages are critical immune cells located in various tissues and are polarized to various phenotypes depending on the tissue microenvironment. Macrophage polarization influences metabolism by regulating inflammation in metabolic diseases. F4/80 is a pan-marker of mouse macrophages and microglia, whereas CD68 is a pan-marker of human and mouse macrophages. iNOS has been identified as a proinflammatory marker, Arg-1 and CD9 are anti-inflammatory markers, and MHCII is a marker of both states. CD9 and TREM2 are novel markers that are associated with glucose and lipid metabolism, respectively. CX3CR1 is expressed on microglia, Lyve1 on macrophages of the vasculature, and CCR2 and MHCII on cardiac macrophages. Applying a combination of origin, recruitment dynamics, physiological and pathological functions, and marker expression to defined macrophage phenotypes may be a new approach to investigating macrophages in metabolic disorders.
QW: conceptualization, writing–Original draft preparation, and visualization. LW: conceptualization, supervision, and project administration. YD: writing—original draft preparation. QY: resources. AZ: investigation. All authors contributed to the article and approved the submitted version.
This work was supported by the Basic and Applied Basic Research Project of Guangdong Province of China (2020A1515010155), the “Innovation and Strengthening University Project” Subsidized Project of the Guangdong Pharmaceutical University (2018KTSCX112), and the Ph.D. Startup Fund of the Natural Science Foundation of Guangdong Province (2018A030310403).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol (2020) 15:123–47. doi: 10.1146/annurev-pathmechdis-012418-012718
2. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature (2013) 496(7446):445–55. doi: 10.1038/nature12034
3. Wang LX, Zhang SX, Wu HJ, Rong XL, Guo J. M2b macrophage polarization and its roles in diseases. J Leukoc Biol (2019) 106(2):345–58. doi: 10.1002/JLB.3RU1018-378RR
4. Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, Bobryshev YV. CD68/macrosialin: not just a histochemical marker. Lab Invest (2017) 97(1):4–13. doi: 10.1038/labinvest.2016.116
5. Bleriot C, Chakarov S, Ginhoux F. Determinants of resident tissue macrophage identity and function. Immunity. (2020) 52(6):957–70. doi: 10.1016/j.immuni.2020.05.014
6. Gentek R, Molawi K, Sieweke MH. Tissue macrophage identity and self-renewal. Immunol Rev (2014) 262(1):56–73. doi: 10.1111/imr.12224
7. Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol (2000) 164(12):6166–73. doi: 10.4049/jimmunol.164.12.6166
8. Liu Y, Xu R, Gu H, Zhang E, Qu J, Cao W, et al. Metabolic reprogramming in macrophage responses. Biomark Res (2021) 9(1):1. doi: 10.1186/s40364-020-00251-y
9. Perez S, Rius-Perez S. Macrophage polarization and reprogramming in acute inflammation: A redox perspective. Antioxidants (Basel) (2022) 11(7):1394. doi: 10.3390/antiox11071394
10. Nahrendorf M, Swirski FK. Abandoning M1/M2 for a network model of macrophage function. Circ Res (2016) 119(3):414–7. doi: 10.1161/CIRCRESAHA.116.309194
11. Castro-Barquero S, Ruiz-Leon AM, Sierra-Perez M, Estruch R, Casas R. Dietary strategies for metabolic syndrome: A comprehensive review. Nutrients (2020) 12(10):2983. doi: 10.3390/nu12102983
12. Bishehsari F, Voigt RM, Keshavarzian A. Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat Rev Endocrinol (2020) 16(12):731–9. doi: 10.1038/s41574-020-00427-4
13. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol (2011) 11(11):723–37. doi: 10.1038/nri3073
14. Boutens L, Hooiveld GJ, Dhingra S, Cramer RA, Netea MG, Stienstra R. Unique metabolic activation of adipose tissue macrophages in obesity promotes inflammatory responses. Diabetologia. (2018) 61(4):942–53. doi: 10.1007/s00125-017-4526-6
15. Bernier LP, York EM, MacVicar BA. Immunometabolism in the brain: How metabolism shapes microglial function. Trends Neurosci (2020) 43(11):854–69. doi: 10.1016/j.tins.2020.08.008
16. Lynch MA. Can the emerging field of immunometabolism provide insights into neuroinflammation? Prog Neurobiol (2020) 184:101719. doi: 10.1016/j.pneurobio.2019.101719
17. Mouton AJ, Li X, Hall ME, Hall JE. Obesity, hypertension, and cardiac dysfunction: Novel roles of immunometabolism in macrophage activation and inflammation. Circ Res (2020) 126(6):789–806. doi: 10.1161/CIRCRESAHA.119.312321
18. Gordon S, Hamann J, Lin HH, Stacey M. F4/80 and the related adhesion-GPCRs. Eur J Immunol (2011) 41(9):2472–6. doi: 10.1002/eji.201141715
19. Kinoshita M, Uchida T, Sato A, Nakashima M, Nakashima H, Shono S, et al. Characterization of two F4/80-positive kupffer cell subsets by their function and phenotype in mice. J Hepatol (2010) 53(5):903–10.
20. Mizuno N, Yanagawa Y. Tofacitinib enhances interferon-gamma-induced expression of major histocompatibility complex class II in macrophages. Eur J Pharmacol (2022) 915:174564.
21. Klopfleisch R. Macrophage reaction against biomaterials in the mouse model - phenotypes, functions and markers. Acta Biomater (2016) 43:3–13. doi: 10.1016/j.actbio.2016.07.003
22. Rath M, Muller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front Immunol (2014) 5:532. doi: 10.3389/fimmu.2014.00532
23. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol (2004) 25(12):677–86. doi: 10.1016/j.it.2004.09.015
24. Yap J, Cabrera-Fuentes HA, Irei J, Hausenloy DJ, Boisvert WA. Role of macrophages in cardioprotection. Int J Mol Sci (2019) 20(10). doi: 10.3390/ijms20102474
25. Fantuzzi L, Borghi P, Ciolli V, Pavlakis G, Belardelli F, Gessani S. Loss of CCR2 expression and functional response to monocyte chemotactic protein (MCP-1) during the differentiation of human monocytes: Role of secreted MCP-1 in the regulation of the chemotactic response. Blood (1999) 94(3):875–83. doi: 10.1182/blood.V94.3.875.415k28_875_883
26. Van Gorp H, Delputte PL, Nauwynck HJ. Scavenger receptor CD163, a jack-of-all-trades and potential target for cell-directed therapy. Mol Immunol (2010) 47(7-8):1650–60. doi: 10.1016/j.molimm.2010.02.008
27. Vidyarthi A, Agnihotri T, Khan N, Singh S, Tewari MK, Radotra BD, et al. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother (2019) 68(12):1995–2004. doi: 10.1007/s00262-019-02423-8
28. O'Neill AS, van den Berg TK, Mullen GE. Sialoadhesin - a macrophage-restricted marker of immunoregulation and inflammation. Immunology (2013) 138(3):198–207. doi: 10.1111/imm.12042
29. Hiemstra IH, Beijer MR, Veninga H, Vrijland K, Borg EG, Olivier BJ, et al. The identification and developmental requirements of colonic CD169(+) macrophages. Immunology (2014) 142(2):269–78. doi: 10.1111/imm.12251
30. Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm (2015) 2015:816460. doi: 10.1155/2015/816460
31. Rey-Giraud F, Hafner M, Ries CH. In vitro generation of monocyte-derived macrophages under serum-free conditions improves their tumor promoting functions. PLos One (2012) 7(8):e42656. doi: 10.1371/journal.pone.0042656
32. Ikarashi M, Nakashima H, Kinoshita M, Sato A, Nakashima M, Miyazaki H, et al. Distinct development and functions of resident and recruited liver kupffer cells/macrophages. J Leukoc Biol (2013) 94(6):1325–36. doi: 10.1189/jlb.0313144
33. Suzuki M, Tachibana I, Takeda Y, He P, Minami S, Iwasaki T, et al. Tetraspanin CD9 negatively regulates lipopolysaccharide-induced macrophage activation and lung inflammation. J Immunol (2009) 182(10):6485–93. doi: 10.4049/jimmunol.0802797
34. Zaid A, Tharmarajah K, Mostafavi H, Freitas JR, Sheng KC, Foo SS, et al. Modulation of monocyte-driven myositis in alphavirus infection reveals a role for CX3CR1(+) macrophages in tissue repair. mBio (2020) 11(2). doi: 10.1128/mBio.03353-19
35. Wehrhan F, Buttner-Herold M, Distel L, Ries J, Moebius P, Preidl R, et al. Galectin 3 expression in regional lymph nodes and lymph node metastases of oral squamous cell carcinomas. BMC Cancer (2018) 18(1):823. doi: 10.1186/s12885-018-4726-6
36. Lu G, Zhang R, Geng S, Peng L, Jayaraman P, Chen C, et al. Myeloid cell-derived inducible nitric oxide synthase suppresses M1 macrophage polarization. Nat Commun (2015) 6:6676. doi: 10.1038/ncomms7676
37. Lee LK, Ghorbanian Y, Wang W, Wang Y, Kim YJ, Weissman IL, et al. LYVE1 marks the divergence of yolk sac definitive hemogenic endothelium from the primitive erythroid lineage. Cell Rep (2016) 17(9):2286–98. doi: 10.1016/j.celrep.2016.10.080
38. Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol (2006) 177(6):3520–4. doi: 10.4049/jimmunol.177.6.3520
39. Fujisaka S, Usui I, Ikutani M, Aminuddin A, Takikawa A, Tsuneyama K, et al. Adipose tissue hypoxia induces inflammatory M1 polarity of macrophages in an HIF-1alpha-dependent and HIF-1alpha-independent manner in obese mice. Diabetologia. (2013) 56(6):1403–12. doi: 10.1007/s00125-013-2885-1
40. Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes (2007) 56(1):16–23. doi: 10.2337/db06-1076
41. Boutens L, Stienstra R. Adipose tissue macrophages: going off track during obesity. Diabetologia (2016) 59(5):879–94. doi: 10.1007/s00125-016-3904-9
42. Olona A, Mukhopadhyay S, Hateley C, Martinez FO, Gordon S, Behmoaras J. Adipoclast: A multinucleated fat-eating macrophage. BMC Biol (2021) 19(1):246. doi: 10.1186/s12915-021-01181-3
43. Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell (2019) 178(3):686–98 e14. doi: 10.1016/j.cell.2019.05.054
44. Vijay J, Gauthier MF, Biswell RL, Louiselle DA, Johnston JJ, Cheung WA, et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat Metab (2020) 2(1):97–109.
45. Ramprasad MP, Fischer W, Witztum JL, Sambrano GR, Quehenberger O, Steinberg D. The 94- to 97-kDa mouse macrophage membrane protein that recognizes oxidized low density lipoprotein and phosphatidylserine-rich liposomes is identical to macrosialin, the mouse homologue of human CD68. Proc Natl Acad Sci USA (1995) 92(21):9580–4. doi: 10.1073/pnas.92.21.9580
46. Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood (1993) 81(6):1607–13. doi: 10.1182/blood.V81.6.1607.1607
47. Haase J, Weyer U, Immig K, Kloting N, Bluher M, Eilers J, et al. Local proliferation of macrophages in adipose tissue during obesity-induced inflammation. Diabetologia (2014) 57(3):562–71. doi: 10.1007/s00125-013-3139-y
48. Russo L, Lumeng CN. Properties and functions of adipose tissue macrophages in obesity. Immunology (2018) 155(4):407–17. doi: 10.1111/imm.13002
49. Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest (2003) 112(12):1796–808. doi: 10.1172/JCI200319246
50. Ying W, Lee YS, Dong Y, Seidman JS, Yang M, Isaac R, et al. Expansion of islet-resident macrophages leads to inflammation affecting beta cell proliferation and function in obesity. Cell Metab (2019) 29(2):457–74.e5. doi: 10.1016/j.cmet.2018.12.003
51. Ehses JA, Perren A, Eppler E, Ribaux P, Pospisilik JA, Maor-Cahn R, et al. Increased number of islet-associated macrophages in type 2 diabetes. Diabetes. (2007) 56(9):2356–70. doi: 10.2337/db06-1650
52. Donath MY, Boni-Schnetzler M, Ellingsgaard H, Halban PA, Ehses JA. Cytokine production by islets in health and diabetes: cellular origin, regulation and function. Trends Endocrinol Metab (2010) 21(5):261–7. doi: 10.1016/j.tem.2009.12.010
53. Kapralov AA, Yang Q, Dar HH, Tyurina YY, Anthonymuthu TS, Kim R, et al. Redox lipid reprogramming commands susceptibility of macrophages and microglia to ferroptotic death. Nat Chem Biol (2020) 16(3):278–90. doi: 10.1038/s41589-019-0462-8
54. Anavi S, Tirosh O. iNOS as a metabolic enzyme under stress conditions. Free Radic Biol Med (2020) 146:16–35. doi: 10.1016/j.freeradbiomed.2019.10.411
55. Nath N, Kashfi K. Tumor associated macrophages and 'NO'. Biochem Pharmacol (2020) 176:113899. doi: 10.1016/j.bcp.2020.113899
56. Kamata K, Mizukami H, Inaba W, Tsuboi K, Tateishi Y, Yoshida T, et al. Islet amyloid with macrophage migration correlates with augmented beta-cell deficits in type 2 diabetic patients. Amyloid (2014) 21(3):191–201. doi: 10.3109/13506129.2014.937857
57. Luo D, Guo Y, Cheng Y, Zhao J, Wang Y, Rong J. Natural product celastrol suppressed macrophage M1 polarization against inflammation in diet-induced obese mice via regulating Nrf2/HO-1, MAP kinase and NF-kappaB pathways. Aging (Albany NY). (2017) 9(10):2069–82. doi: 10.18632/aging.101302
58. Yu S, Cheng Y, Zhang L, Yin Y, Xue J, Li B, et al. Treatment with adipose tissue-derived mesenchymal stem cells exerts anti-diabetic effects, improves long-term complications, and attenuates inflammation in type 2 diabetic rats. Stem Cell Res Ther (2019) 10(1):333. doi: 10.1186/s13287-019-1474-8
59. Kersey JH, LeBien TW, Abramson CS, Newman R, Sutherland R, Greaves M. P-24: a human leukemia-associated and lymphohemopoietic progenitor cell surface structure identified with monoclonal antibody. J Exp Med (1981) 153(3):726–31. doi: 10.1084/jem.153.3.726
60. CS P, Shetty SS, Nalilu SK, Shetty PK, Patil P. Tetraspanin CD9: A friend or foe of head and neck cancer (Review). Oncol Rep (2022) 47(5). doi: 10.3892/or.2022.8299
61. Brosseau C, Colas L, Magnan A, Brouard S. CD9 tetraspanin: A new pathway for the regulation of inflammation? Front Immunol (2018) 9:2316. doi: 10.3389/fimmu.2018.02316
62. Tippett E, Cameron PU, Marsh M, Crowe SM. Characterization of tetraspanins CD9, CD53, CD63, and CD81 in monocytes and macrophages in HIV-1 infection. J Leukoc Biol (2013) 93(6):913–20. doi: 10.1189/jlb.0812391
63. Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci (2020) 23(2):194–208. doi: 10.1038/s41593-019-0566-1
64. Kober DL, Brett TJ. TREM2-ligand interactions in health and disease. J Mol Biol (2017) 429(11):1607–29. doi: 10.1016/j.jmb.2017.04.004
65. Paradowska-Gorycka A, Jurkowska M. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Hum Immunol (2013) 74(6):730–7. doi: 10.1016/j.humimm.2013.02.003
66. Neumann H, Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol (2007) 184(1-2):92–9. doi: 10.1016/j.jneuroim.2006.11.032
67. Colonna M, Turnbull I, Klesney-Tait J. The enigmatic function of TREM-2 in osteoclastogenesis. Adv Exp Med Biol (2007) 602:97–105. doi: 10.1007/978-0-387-72009-8_13
68. Deczkowska A, Weiner A, Amit I. The physiology, pathology, and potential therapeutic applications of the TREM2 signaling pathway. Cell (2020) 181(6):1207–17. doi: 10.1016/j.cell.2020.05.003
69. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, et al. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res (2018) 122(12):1661–74. doi: 10.1161/CIRCRESAHA.117.312509
70. Curley S, Gall J, Byrne R, Yvan-Charvet L, McGillicuddy FC. Metabolic inflammation in obesity-At the crossroads between fatty acid and cholesterol metabolism. Mol Nutr Food Res (2021) 65(1):e1900482. doi: 10.1002/mnfr.201900482
71. He JT, Zhao X, Xu L, Mao CY. Vascular risk factors and alzheimer's disease: Blood-brain barrier disruption, metabolic syndromes, and molecular links. J Alzheimers Dis (2020) 73(1):39–58. doi: 10.3233/JAD-190764
72. Sweeney MD, Zhao Z, Montagne A, Nelson AR, Zlokovic BV. Blood-brain barrier: From physiology to disease and back. Physiol Rev (2019) 99(1):21–78. doi: 10.1152/physrev.00050.2017
73. Leng F, Edison P. Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat Rev Neurol (2021) 17(3):157–72. doi: 10.1038/s41582-020-00435-y
74. Harrison JK, Jiang Y, Chen S, Xia Y, Maciejewski D, McNamara RK, et al. Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia. Proc Natl Acad Sci USA (1998) 95(18):10896–901. doi: 10.1073/pnas.95.18.10896
75. Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res (1999) 117(2):145–52. doi: 10.1016/S0165-3806(99)00113-3
76. Song GJ, Suk K. Pharmacological modulation of functional phenotypes of microglia in neurodegenerative diseases. Front Aging Neurosci (2017) 9:139. doi: 10.3389/fnagi.2017.00139
77. Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol (1981) 11(10):805–15. doi: 10.1002/eji.1830111013
78. Dos Anjos Cassado A. F4/80 as a major macrophage marker: The case of the peritoneum and spleen. Results Probl Cell Differ (2017) 62:161–79. doi: 10.1007/978-3-319-54090-0_7
79. Ruytinx P, Proost P, Van Damme J, Struyf S. Chemokine-induced macrophage polarization in inflammatory conditions. Front Immunol (2018) 9:1930. doi: 10.3389/fimmu.2018.01930
80. McKnight AJ, Macfarlane AJ, Dri P, Turley L, Willis AC, Gordon S. Molecular cloning of F4/80, a murine macrophage-restricted cell surface glycoprotein with homology to the G-protein-linked transmembrane 7 hormone receptor family. J Biol Chem (1996) 271(1):486–9. doi: 10.1074/jbc.271.1.486
81. Hamann J, Koning N, Pouwels W, Ulfman LH, van Eijk M, Stacey M, et al. EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. Eur J Immunol (2007) 37(10):2797–802. doi: 10.1002/eji.200737553
82. van den Berg TK, Kraal G. A function for the macrophage F4/80 molecule in tolerance induction. Trends Immunol (2005) 26(10):506–9.
83. Trinchieri G. Critical role for CX(3)CR1(+) mononuclear phagocytes in intestinal homeostasis. J Exp Med (2014) 211(8):1500–1. doi: 10.1084/jem.2118insight2
84. Nishimura M, Umehara H, Nakayama T, Yoneda O, Hieshima K, Kakizaki M, et al. Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme b+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J Immunol (2002) 168(12):6173–80. doi: 10.4049/jimmunol.168.12.6173
85. Chidambaram H, Das R, Chinnathambi S. Interaction of tau with the chemokine receptor, CX3CR1 and its effect on microglial activation, migration and proliferation. Cell Biosci (2020) 10:109. doi: 10.1186/s13578-020-00474-4
86. Sheridan GK, Murphy KJ. Neuron-glia crosstalk in health and disease: fractalkine and CX3CR1 take centre stage. Open Biol (2013) 3(12):130181. doi: 10.1098/rsob.130181
87. Villapol S, Faivre V, Joshi P, Moretti R, Besson VC, Charriaut-Marlangue C. Early sex differences in the immune-inflammatory responses to neonatal ischemic stroke. Int J Mol Sci (2019) 20(15). doi: 10.3390/ijms20153809
88. Narayanan SP, Rojas M, Suwanpradid J, Toque HA, Caldwell RW, Caldwell RB. Arginase in retinopathy. Prog Retin Eye Res (2013) 36:260–80. doi: 10.1016/j.preteyeres.2013.06.002
89. da Cunha Colombo Tiveron LR, da Silva IR, da Silva MV, Peixoto AB, Rodrigues DBR, Rodrigues V Jr. High in situ mRNA levels of IL-22, TFG-beta, and ARG-1 in keloid scars. Immunobiology (2018) 223(12):812–7. doi: 10.1016/j.imbio.2018.08.010
90. Subramaniam SR, Federoff HJ. Targeting microglial activation states as a therapeutic avenue in parkinson's disease. Front Aging Neurosci (2017) 9:176. doi: 10.3389/fnagi.2017.00176
91. Yang Z, Liu B, Yang LE, Zhang C. Platycodigenin as potential drug candidate for alzheimer's disease via modulating microglial polarization and neurite regeneration. Molecules (2019) 24(18). doi: 10.3390/molecules24183207
92. Wiebe N, Stenvinkel P, Tonelli M. Associations of chronic inflammation, insulin resistance, and severe obesity with mortality, myocardial infarction, cancer, and chronic pulmonary disease. JAMA Netw Open (2019) 2(8):e1910456. doi: 10.1001/jamanetworkopen.2019.10456
93. Mottillo S, Filion KB, Genest J, Joseph L, Pilote L, Poirier P, et al. The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. J Am Coll Cardiol (2010) 56(14):1113–32. doi: 10.1016/j.jacc.2010.05.034
94. Wang Z, Koenig AL, Lavine KJ, Apte RS. Macrophage plasticity and function in the eye and heart. Trends Immunol (2019) 40(9):825–41. doi: 10.1016/j.it.2019.07.002
95. Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science (2019) 363(6432). doi: 10.1126/science.aau0964
96. Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International union of basic and clinical pharmacology. [corrected]. LXXXIX. update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev (2014) 66(1):1–79.
97. Dunay IR, Damatta RA, Fux B, Presti R, Greco S, Colonna M, et al. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen toxoplasma gondii. Immunity. (2008) 29(2):306–17. doi: 10.1016/j.immuni.2008.05.019
98. Niu J, Kolattukudy PE. Role of MCP-1 in cardiovascular disease: molecular mechanisms and clinical implications. Clin Sci (Lond) (2009) 117(3):95–109. doi: 10.1042/CS20080581
99. Mildner A, Mack M, Schmidt H, Bruck W, Djukic M, Zabel MD, et al. CCR2+Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain (2009) 132(Pt 9):2487–500. doi: 10.1093/brain/awp144
100. Huma ZE, Sanchez J, Lim HD, Bridgford JL, Huang C, Parker BJ, et al. Key determinants of selective binding and activation by the monocyte chemoattractant proteins at the chemokine receptor CCR2. Sci Signal (2016) 10(480):eaai8529. doi: 10.1126/scisignal.aai8529
101. Bajpai G, Bredemeyer A, Li W, Zaitsev K, Koenig AL, Lokshina I, et al. Tissue resident CCR2- and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circ Res (2019) 124(2):263–78. doi: 10.1161/CIRCRESAHA.118.314028
102. Sierra-Filardi E, Nieto C, Dominguez-Soto A, Barroso R, Sanchez-Mateos P, Puig-Kroger A, et al. CCL2 shapes macrophage polarization by GM-CSF and m-CSF: identification of CCL2/CCR2-dependent gene expression profile. J Immunol (2014) 192(8):3858–67. doi: 10.4049/jimmunol.1302821
103. Deci MB, Ferguson SW, Scatigno SL, Nguyen J. Modulating macrophage polarization through CCR2 inhibition and multivalent engagement. Mol Pharm (2018) 15(7):2721–31. doi: 10.1021/acs.molpharmaceut.8b00237
104. Wu Y, Zhang N, Hashimoto K, Xia C, Dijkstra JM. Structural comparison between MHC classes I and II; in evolution, a class-II-Like molecule probably came first. Front Immunol (2021) 12:621153. doi: 10.3389/fimmu.2021.621153
105. Holling TM, Schooten E, van Den Elsen PJ. Function and regulation of MHC class II molecules in T-lymphocytes: of mice and men. Hum Immunol (2004) 65(4):282–90. doi: 10.1016/j.humimm.2004.01.005
106. Landsverk OJ, Bakke O, Gregers TF. MHC II and the endocytic pathway: regulation by invariant chain. Scand J Immunol (2009) 70(3):184–93. doi: 10.1111/j.1365-3083.2009.02301.x
107. Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol (2018) 233(9):6425–40. doi: 10.1002/jcp.26429
108. Cole JE, Park I, Ahern DJ, Kassiteridi C, Danso Abeam D, Goddard ME, et al. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc Res (2018) 114(10):1360–71. doi: 10.1093/cvr/cvy109
109. Weinberger T, Rauber S, Schneider V, Messerer D, Fischer M, Rudi WS, et al. Differential MHC-II expression and phagocytic functions of embryo-derived cardiac macrophages in the course of myocardial infarction in mice. Eur J Immunol (2021) 51(1):250–2. doi: 10.1002/eji.202048560
110. Stanly TA, Fritzsche M, Banerji S, Shrestha D, Schneider F, Eggeling C, et al. The cortical actin network regulates avidity-dependent binding of hyaluronan by the lymphatic vessel endothelial receptor LYVE-1. J Biol Chem (2020) 295(15):5036–50. doi: 10.1074/jbc.RA119.011992
111. Jackson DG. Hyaluronan in the lymphatics: The key role of the hyaluronan receptor LYVE-1 in leucocyte trafficking. Matrix Biol (2019) 78-79:219–35. doi: 10.1016/j.matbio.2018.02.001
112. Johnson LA, Banerji S, Lawrance W, Gileadi U, Prota G, Holder KA, et al. Dendritic cells enter lymph vessels by hyaluronan-mediated docking to the endothelial receptor LYVE-1. Nat Immunol (2017) 18(7):762–70. doi: 10.1038/ni.3750
113. Vieira JM, Norman S, Villa Del Campo C, Cahill TJ, Barnette DN, Gunadasa-Rohling M, et al. The cardiac lymphatic system stimulates resolution of inflammation following myocardial infarction. J Clin Invest (2018) 128(8):3402–12. doi: 10.1172/JCI97192
114. Lim HY, Lim SY, Tan CK, Thiam CH, Goh CC, Carbajo D, et al. Hyaluronan receptor LYVE-1-Expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen. Immunity (2018) 49(2):326–41 e7. doi: 10.1016/j.immuni.2018.06.008
115. Martens JH, Kzhyshkowska J, Falkowski-Hansen M, Schledzewski K, Gratchev A, Mansmann U, et al. Differential expression of a gene signature for scavenger/lectin receptors by endothelial cells and macrophages in human lymph node sinuses, the primary sites of regional metastasis. J Pathol (2006) 208(4):574–89. doi: 10.1002/path.1921
116. Novak N, Bieber T, Hoffmann M, Folster-Holst R, Homey B, Werfel T, et al. Efficacy and safety of subcutaneous allergen-specific immunotherapy with depigmented polymerized mite extract in atopic dermatitis. J Allergy Clin Immunol (2012) 130(4):925–31 e4.
117. Gordon EJ, Gale NW, Harvey NL. Expression of the hyaluronan receptor LYVE-1 is not restricted to the lymphatic vasculature; LYVE-1 is also expressed on embryonic blood vessels. Dev Dyn (2008) 237(7):1901–9.
118. Schledzewski K, Falkowski M, Moldenhauer G, Metharom P, Kzhyshkowska J, Ganss R, et al. Lymphatic endothelium-specific hyaluronan receptor LYVE-1 is expressed by stabilin-1+, F4/80+, CD11b+ macrophages in malignant tumours and wound healing tissue in vivo and in bone marrow cultures in vitro: implications for the assessment of lymphangiogenesis. J Pathol (2006) 209(1):67–77.
119. Hendrikx T, Porsch F, Kiss MG, Rajcic D, Papac-Milicevic N, Hoebinger C, et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2(+) macrophages that localize to fibrotic areas and limit NASH. J Hepatol (2022) 77(5):1373–85.
120. Lafuse WP, Wozniak DJ, Rajaram MVS. Role of cardiac macrophages on cardiac inflammation, fibrosis and tissue repair. Cells (2020) 10(1).
Keywords: macrophage, molecular marker, metabolic disease, cell marker, metabolic disorders
Citation: Wei Q, Deng Y, Yang Q, Zhan A and Wang L (2023) The markers to delineate different phenotypes of macrophages related to metabolic disorders. Front. Immunol. 14:1084636. doi: 10.3389/fimmu.2023.1084636
Received: 31 October 2022; Accepted: 17 January 2023;
Published: 03 February 2023.
Edited by:
Aline Bozec, University of Erlangen Nuremberg, GermanyCopyright © 2023 Wei, Deng, Yang, Zhan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lexun Wang, d2FuZ2xleHVuMTIzNDU2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.