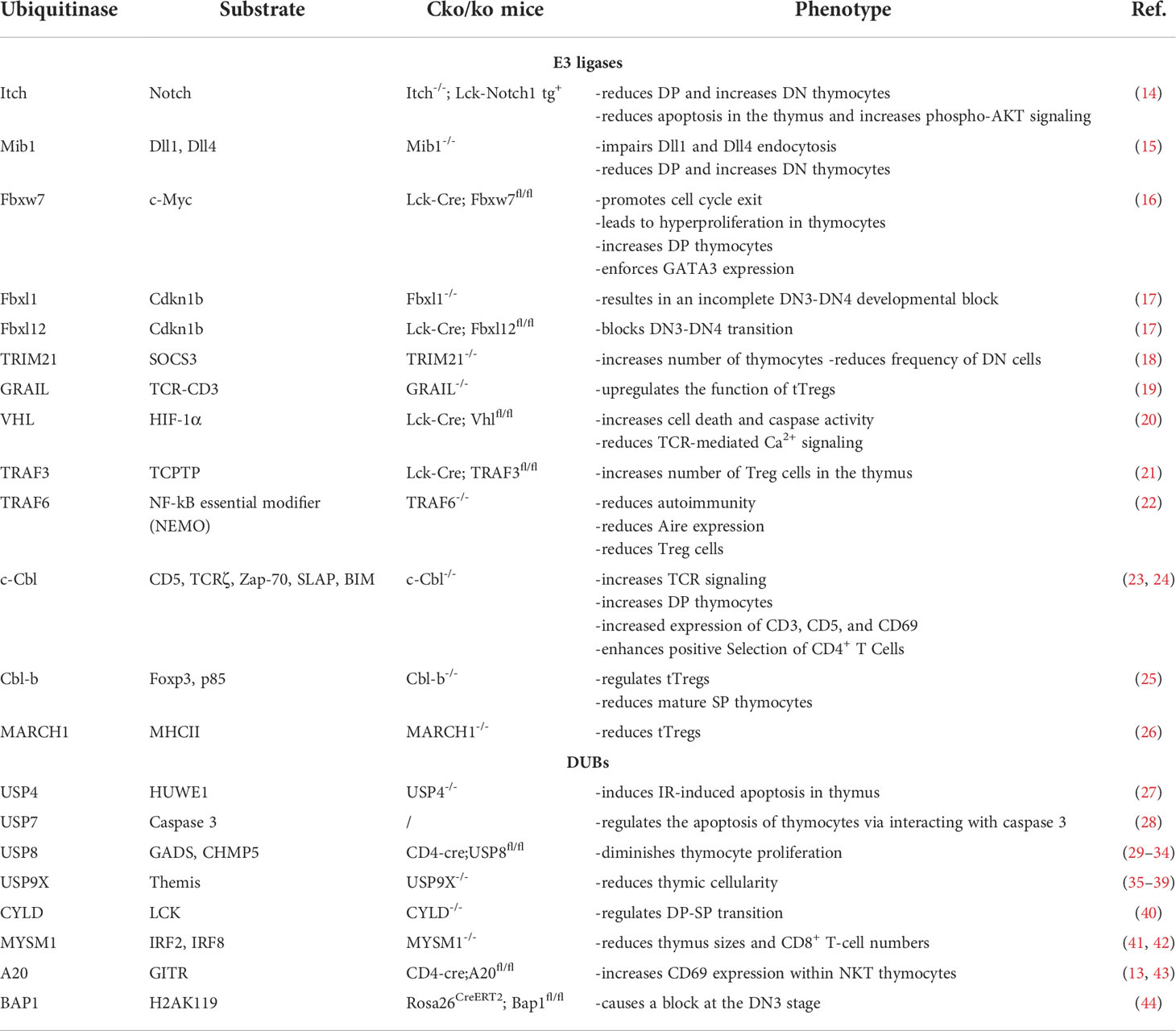
- National Clinical Research Center for Metabolic Diseases, Key Laboratory of Diabetes Immunology, Ministry of Education, and Department of Metabolism and Endocrinology, The Second Xiangya Hospital of Central South University, Changsha, China
As an important form of posttranslational modification, protein ubiquitination regulates a wide variety of biological processes, including different aspects of T cell development and differentiation. During T cell development, thymic seeding progenitor cells (TSPs) in the thymus undergo multistep maturation programs and checkpoints, which are critical to build a functional and tolerant immune system. Currently, a tremendous amount of research has focused on the transcriptional regulation of thymocyte development. However, in the past few years, compelling evidence has revealed that the ubiquitination system also plays a crucial role in the regulation of thymocyte developmental programs. In this review, we summarize recent findings on the molecular mechanisms and cellular pathways that regulate thymocyte ubiquitination and discuss the roles of E3 ligases and deubiquitinating enzymes (DUBs) involved in these processes. Understanding how T cell development is regulated by ubiquitination and deubiquitination will not only enhance our understanding of cell fate determination via gene regulatory networks but also provide potential novel therapeutic strategies for treating autoimmune diseases and cancer.
Introduction
Ubiquitin is a highly conserved protein of 76 amino acids and a versatile posttranslational modifier that is ubiquitously expressed in all eukaryotic cells (1). Protein ubiquitination plays a crucial role in protein homeostasis, thus regulating a vast array of biological processes, such as DNA damage and repair, cell cycle progression, apoptosis and cellular signaling (2, 3). Ubiquitin is added to the protein substrate via a subsequent enzymatic cascade by E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases (4). The specificity of ubiquitination is mainly achieved by E3 ligases, which are responsible for substrate recognition via protein interacting domains and motifs (5). Ubiquitin has seven lysine residues that can be used to assemble polyubiquitin chains: Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63. A substrate can be polyubiquitylated or monoubiquitylated via polyubiquitin chains, and the impact of polyubiquitination on the target protein is greatly dependent on the type of conjugated chain (6). For example, except for Lys63, all six Lys linkages have been implicated in proteasomal degradation, with Lys48 and Lys11 being the predominant type of chains for substrate degradation in cells. Lys63-linked chains are involved in multiple nonproteolytic functions, including activation of NF-κB, DNA damage repair, and regulation of endosomal sorting pathways (7). Ubiquitination is a dynamic and reversible process, and ubiquitination induced by ubiquitin ligases can be counteracted by deubiquitinating enzymes (DUBs) to control the intensity and duration of ubiquitin signaling (8).
The thymus is the primary site for T cell development, thymic seeding progenitor cells (TSPs) arrive at the thymus from the bone marrow and initiate multistep maturation programs and checkpoints comprising lineage commitment, T cell receptor (TCR) gene rearrangement, and positive and negative selection. It is well established that thymocytes mature through ordered progression, including double-negative (CD4–CD8–, DN) stage, double-positive (DP) stage and CD4 or CD8 single-positive (SP) stages (9, 10). In the earlier DN1-3 stages, proliferation and differentiation are mainly driven by Notch signaling and cytokines such as c-kit and IL-7 (11). Then, cells successfully assembled pre-T cell receptor (pre-TCR) complexes will pass β-selection and transition from the DN3 to the DN4 stage. In DP stage, thymocytes undergo positive selection for self-human leukocyte antigen (HLA) recognition under the control of cortical thymic epithelial cells (cTECs) and negative selection to remove strong self-reactive clones based on the interaction with medullary thymic epithelial cells (mTECs) and thymic DCs (tDCs), finally becoming CD4+ SP or CD8+ SP cells (11). “Mature” SP thymocytes exit the thymus to the peripheral lymphoid organs (9).
Ubiquitin signaling modulates a variety of pathways involved in the T cell developmental process primarily through proteolysis-dependent mechanisms, such as Notch, pre-TCR signaling, Signal transducer and activator of transcription 3 (STAT3)-mediated signaling, Wnt signaling, and Nuclear factor κB (NF-κB) pathway (9, 12, 13). Here, we summarize the interplay between the ubiquitination system and T cell developmental programs (Figure 1). Specifically, we highlight the roles of E3 ligases and DUBs involved in these processes as well as the molecular mechanisms and cellular pathways that regulate thymocyte ubiquitination (Table 1).
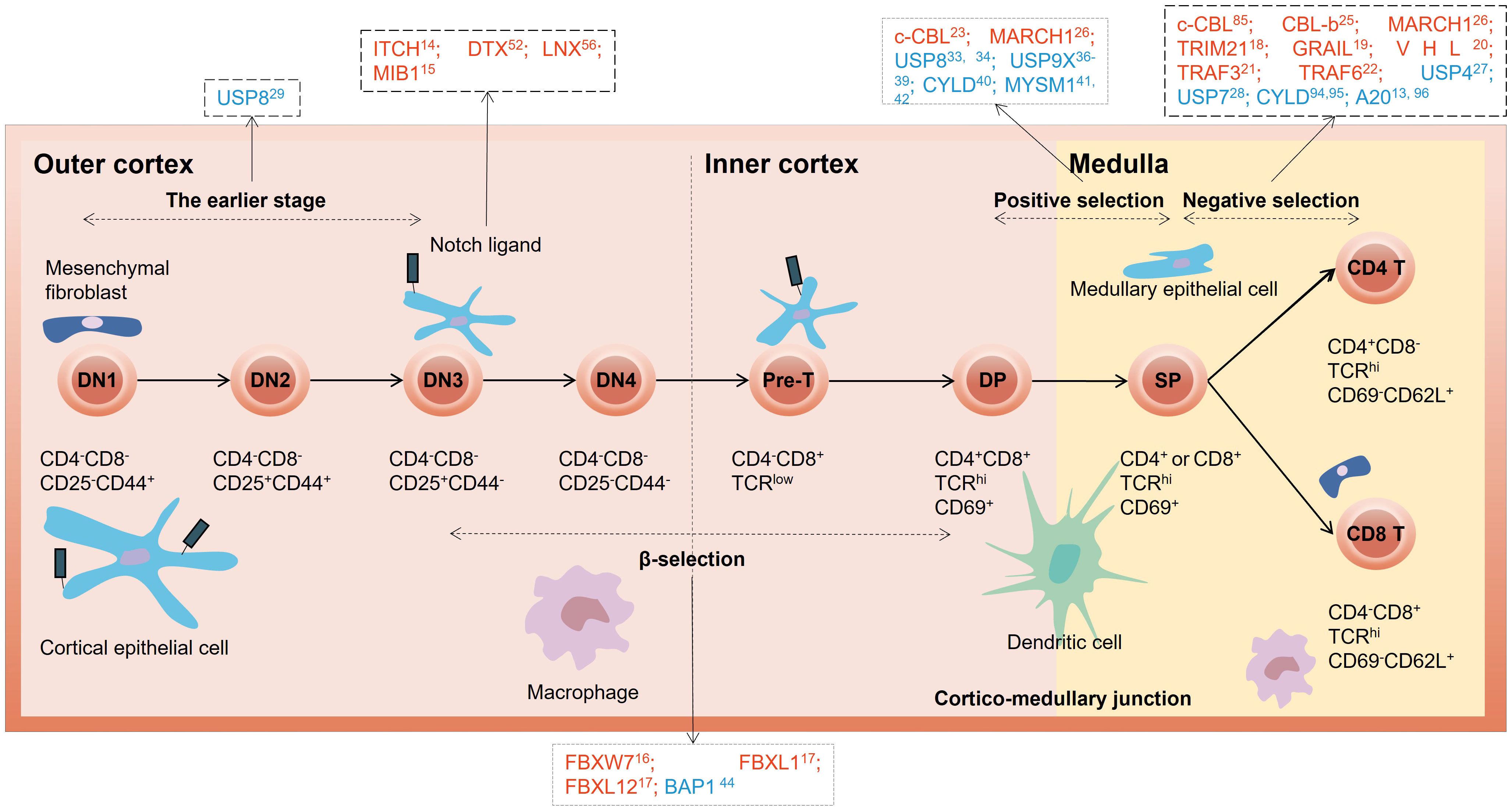
Figure 1 Overview of E3 ubiquitin ligases and DUBs in different stages of thymocyte development. The red letters in the black dotted box represent E3 ubiquitin ligases, and the blue letters represent DUBs. DN, double-negative; DP, double-positive; SP, single-positive.
E3 ubiquitin ligases in T cell development
E3 ligases are crucial components of the Ubiquitin Proteasome System. Several classes of these enzymes have been identified, known as the RING, U-box, HECT and RBR classes (45). As the last component of an enzymatic cascade, E3 ligases determine substrate specificity. Attaching ubiquitin to a protein could have profound effects on the protein’s cellular localization, protein-protein interactions or stability (46). Multiple E3 ligases have been demonstrated to play a role in T cell development.
NOTCH-regulating E3 ligases mainly regulate the early stage of T cell development
Notch signaling has been identified as a key signaling pathway involved in the regulation of T cell development, especially in thymocyte survival, proliferation and differentiation (9, 47–49). E3 ubiquitin ligases that can catalyze the ubiquitylation of Notch include Itch, Ligand of Numb-Protein X (LNX), Deltex (DTX), Mind bomb (Mib) 1, Mib2, Neuralized (Neur) 1, and Neur2 (15). Itch binds to the N-terminal of the Notch intracellular domain via its WW domains and promotes ubiquitination of Notch via K29-linked ubiquitin chains, thus promoting its lysosomal degradation (50). Itch-/- mice with an activated Notch1 transgene in their thymocytes show a reduction of DP and an increase of DN T cells, with a more severe autoimmune phenotype (14). Itch and Notch act in the AKT signaling concurrently in the genesis of autoimmune disease (14). In addition, Itch regulates Notch signaling via interacting with some molecules, such as Numb and DTX. Numb, an adapter protein, was initially identified as a negative regulator of Notch signaling. Numb binds to Itch WW domain and promotes ubiquitination and degradation of Notch1 by Itch (51). DTX, an E3 ligase, has been shown to be an itch homolog that plays a negative role in regulating Notch receptor signaling, and can cooperate with Itch to regulate NOTCH signaling via lysosomal degradation (52). In addition, downregulation of DTX in hematopoietic progenitors promotes T cell development in fetal thymic organ culture and in vivo (53). DTX antagonizes Notch1 signals by inhibiting coactivator recruitment (54) and restores DP thymocyte survival from the glucocorticoid (GC)-induced apoptosis by repressing SRG3 promoter activity (55). LNX can also cause proteasome-dependent degradation of Numb and therefore enhance Notch signaling (56). Mib1 modulates Notch signaling by ubiquitinating the Notch receptors (Dll1 and 4), promoting their endocytosis (57). Reciprocal bone marrow (BM) transplantation experiments revealed that Notch signaling was diminished in the DN thymocytes of Mib1 conditional KO mice (15). Furthermore, knocking down Mib1 in the coculture system causes a delay in T cell growth and a failure of Dll1 endocytosis (15).
SCF complexes play crucial roles in thymic β-selection mediated cell proliferation
The SCF (Skp1-cullin-F-box protein) complex is a well-described multisubunit RING-finger E3 composed of Skp1, Cdc53/cullin, and an F box protein (58). Fbxw7 (F-box and WD-40 domain protein 7)—also known as Fbw7—is an SCF ubiquitin ligase component reported to play a role in thymocyte cell cycle progression by controlling the degradation of c-Myc, c-Jun, cyclin E, and Notch (59). Fbw7 modulates cell cycle progression by controlling c-Myc protein stability, and loss of Fbxw7 leads to hyperproliferation of thymocytes (16). Moreover, the SCF subunits Fbxl1 and Fbxl12, which are transcriptionally induced by Notch and pre-TCR signaling respectively, function identically but additively to promote the degradation of Cdkn1b and proliferation of β-selected thymocytes (17, 60). Deletion of Fbxl1 or Fbxl12 results in an incomplete DN3-DN4 developmental block and a reduced thymus size (17).
TRIM family proteins have crucial roles during negative selection
As RING-type E3 ligases, tripartite motif (TRIM) proteins have been demonstrated to regulate the innate immune response (61, 62). However, recent studies suggest that TRIM21 alters T cell development in the thymus (63). TRIM21−/− mice had an increased number of thymocytes and a reduced frequency of DN cells (18). TRIM21 targets suppressor of cytokine signaling-3 (SOCS3) for proteasomal degradation, thus impairing STAT3 activation in TECs (64). STAT3-mediated signaling has been shown to promote quintessential growth of mTECs (but not cTECs) (12, 65). Double-positive (DP) cells are selected by cTECs to become CD4 or CD8 SP cells (66), while SP thymocytes are further negatively selected in the medulla (67). We can surmise that TRIM21 plays a crucial role during negative selection in the thymus.
GRAIL and VHL regulate T cell development during negative selection
Gene related to anergy in lymphocytes (GRAIL) is a RING-type E3 ligase required for the initiation of CD4+ T cell anergy in vivo. Previous studies considered GRAIL expression patterns in murine CD4+ T cells as a defined anergic phenotype and a negative regulator of the immune response (68, 69). Notably, GRAIL expression is upregulated in tTregs, and its overexpression in DO11.10 T cells convert these cells to a regulatory phenotype (19). Nurieva et al. reported that GRAIL regulates Treg cell function by mediating TCR-CD3 degradation (70). Works are needed to delineate the mechanism(s) of how GRAIL mediates its suppressor activity in the thymus. The von Hippel-Lindau (VHL) is a RING-type E3 ligase that targets hypoxia-inducible factor-1α (HIF-1α) for proteasomal degradation (20). Vhl-deficient mice had a severe reduction in thymus sizes and thymic cellularity due to enhanced caspase 8 activity in the apoptotic pathway, as a result of HIF-1α accumulation (20).
TRAF family proteins regulate T cell development during negative selection
Tumor necrosis factor receptor (TNFR)-associated factor 3 (TRAF3) is a member of the TRAF family of cytoplasmic adaptor proteins and plays a role in modulating IL-2 signaling in T cells. T cell conditional TRAF3 knockout mice resulted in an increased number of Treg cells in the thymus (21) due to more efficient conversion of CD25+ Foxp3– Treg precursors to CD25+ Foxp3+ mature Treg cells (71). TRAF6 is another adaptor E3 ligase that is involved in central tolerance by regulating the development of thymic stroma. TRAF6−/− fetal thymic stroma tissue fails to mediate negative selection (22). Furthermore, specific deletion of TRAF6 in TECs hinders the growth of mTECs (72). Several studies have suggested that TRAF6 regulates the establishment of thymic microenvironments through manipulating RelB (73), RANK (74) and CD40 (75) expression.
Cbl family proteins regulate multiple stages of T cell developmental processes
The Casitas B-lineage lymphoma (Cbl) family of proteins are RING-finger domain containing E3 ubiquitin ligases (76, 77). In mammals, two highly homologous adaptor proteins of the Cbl family, c-Cbl and Cbl-b, are involved in the negative regulation of the immune system (78, 79). Both c-Cbl and Cbl-b contain a highly conserved amino-terminal tyrosine-kinase binding (TKB) domain, a less conserved carboxyl-terminal proline-rich region (PRR) and a RING finger. Through their protein-protein interaction domains, c-Cbl and Cbl-b form multiple complexes together with several signaling molecules to regulate intracellular signaling events (80). The first evidence indicating that Cbl proteins are associated with thymic selection came from experiments showing that thymocytes from c-Cbl-/- mice have increased signaling through the TCR and CD4+ CD8+ DP thymocytes exhibited increased expression of CD3, CD5, and CD69 in the c-Cbl knockout (KO) model (23). Moreover, c-Cbl selectively inhibits thymic-positive selection of CD4 but not CD8 T cells (23). This suggests that the positive selection of thymocytes bearing MHC class II-restricted TCRs is negatively regulated by c-Cbl. Mechanistically, c-Cbl modulates CD4+ T-cell development by promoting TCR-ζ lysosomal degradation. In this model, a transient trimolecular complex of TCRζ-Zap-70-Cbl is formed, and ubiquitin is then shifted from the Cbl-E2 complex to TCRζ (79, 81). In addition to Zap-70, Src-like adaptor protein (SLAP) might also act as a bridge to bond TCRζ and Cbl. In support of this, SLAP-/- mice were shown to have a similar phenotype to c-Cbl-/- mice (82–84). In addition to positive selection, c-Cbl also regulates thymocyte negative selection, probably by ubiquitinating and proteasomal degrading the pro-apoptotic molecule B-cell lymphoma 2-interacting mediator of cell death (BIM) (85). Furthermore, deactivation of c-Cbl reverses T cell developmental detention in SLP-76-deficient mice, in which T cell development is impeded at the DN3 stage (24). In conclusion, the c-Cbl protein modulates multiple stages of T cell developmental processes.
Analyses of Cbl-b KO mice resulted in no similar findings (86). Given that the expression level of Cbl-b in thymocytes is much lower than that of c-Cbl, it would not be surprising. However, Zhao Y et al. reported that Cbl-b, together with Stub1, regulates thymic-derived CD4+ CD25+ regulatory T cells (tTregs) development by targeting Foxp3 for ubiquitination and degradation in the proteasome (25). Moreover, Raberger J et al. reported that the CD4/CD8 developmental profile was noticeably altered and mature SP thymocytes were absent in Vav1-/- or ITK-/- thymocytes (87), and the signaling defects in Vav1-/- or ITK-/- thymocytes can be rescued upon deletion of Cbl-b (87). These results indicate that Cbl-b alters thymus development.
MARCH family E3 ligases modulate the development of tTregs
Membrane-associated RING-CH1 (MARCH1) is an E3 ubiquitin ligase that regulates MHCII ubiquitination (26). Thymocytes and TECs scarcely express MARCH1, while DCs in the thymus express comparatively high levels of MARCH1 (26). MARCH1 deficiency results in an elevated level of MHCII, which leads to a considerable decline in the number of thymic Treg (tTreg) cells but not conventional CD4+ T cells in mice (26). Another E3 ligase, MARCH8, is responsible for MHC II ubiquitination specifically in thymic epithelial cells. In MARCH8-/- mice, TECs express elevated levels of MHC II, but the development of conventional CD4+ T cells or tTreg cells remains unchanged. It is possible that tTreg development does not require MHC II ubiquitination in TECs (88).
DUBs in T cell development
In addition to E3 ligases, the ubiquitin system is also regulated by DUBs. Ubiquitin chains can be removed from the substrate by DUBs, which are essential for the dynamic regulation of the protein ubiquitination process (89, 90). Several DUBs have been identified as regulators in the T cell developmental program.
USP family proteases regulate multiple stages of T cell developmental processes
Ubiquitin-specific proteases (USPs) are the largest subfamily of DUBs and contain more than 100 members (91). Ubiquitin-specific peptidase 4 (USP4) has been shown to inhibit p53 signaling through interacting with and stabilizing ARF-binding protein 1 (ARF-BP1, also known as HUWE1), an E3 ligase for p53 (24). USP4 knockout mice are viable and fertile but exhibit enhanced ionizing radiation (IR)-induced thymocyte apoptosis (27). In addition, USP4, a DUB with dual hydrolyzing activity for K48- and K63-conjugated polyubiquitin chains, interacts with the Nemo like kinase (Nlk) and T-cell factor (TCF) 4, two known components of the Wnt pathway that are essential for cell development (92). USP7 (also known as HAUSP), which is highly expressed in the thymus, also regulates the apoptosis of thymocytes during negative selection via caspase-dependent signaling (28). Likewise, the processing of HAUSP does not occur in caspase 3-deficient thymocytes (28). Ubiquitin-specific protease USP8 is a deubiquitinase involved in the endosomal sorting complex required for transport (ESCRT) system (93). A recent study reported that USP8 is involved in thymocyte maturation and proliferation processes by modulating the Foxo1-IL-7Rα axis (29). Moreover, the amino-terminal SH3BM of USP8 binds with higher affinity to the TCR adaptor GADS in a caspase-dependent manner (30–32). Another study identified USP8 as a deubiquitinase for CHMP5, a component of the ESCRT complex, and uncovered the role of the CHMP5-USP8 complex in regulating thymic positive selection (33, 34). Ubiquitin-specific protease 9X (USP9X) is a member of the peptidase C19 family and encodes a protein similar in structure to ubiquitin-specific proteases. Deletion of Usp9X resulted in an overall reduction in thymic cellularity (35). Mechanistically, USP9X interacts with and stabilizes Themis, an important TCR signaling protein (36), by removing ubiquitin K48-linked chains on Themis upon TCR stimulation, thus affecting thymic positive selection (37–39).
CYLD regulates T cell development during negative selection
Cylindromatosis (CYLD) is a lysine 63-deubiquitinating enzyme that positively regulates TCR signaling by promoting the recruitment of Lck to its substrate, Zap70, in thymocytes (40). CYLD-deficient mice displayed significantly fewer mature CD4+ and CD8+ single-positive thymocytes (40). Previous studies identified CYLD as a switch in T cell development during the transition from double-positive to single-positive thymocytes (40). Furthermore, S. Reissig et al. demonstrated impaired negative selection in the thymus of CYLDex7/8 mice, which overexpresses the naturally occurring CYLD splice variant short CYLD (sCYLD), whereas full-length CYLD (FL-CYLD) is absent (94, 95).
MYSM1, A20 and BAP1 modulate multiple stages of T cell developmental processes
Other types of DUBs involved in T cell development include Myb-like SWIRM and MPN domain containing 1 (MYSM1), A20 and BRCA1-associated protein-1 (BAP1). Conditional ablation of histone H2A deubiquitinase MYSM1 at late stages of thymic development in a mouse model showed a severe reduction in thymus sizes and CD8+ T-cell numbers, indicating the critical role of MYSM1 in the positive selection of CD8+ T cells (41, 42). A20, also known as TNF-α-induced protein 3 (TNFAIP3), regulates tTreg development and maturation by restraining the activation of NF-kB signaling (13, 96). T lineage cell conditional A20 knockout mice showed that tTreg cell compartments are quantitatively enlarged (13). In addition, A20 specifically limits TCR-dependent activation of NKT cells in the thymus (43). BAP1 is a member of the ubiquitin C-terminal hydrolase (UCH) subfamily of DUBs and has been shown to be involved in β-selection mediated cell expansion (44). BAP1 deletion in adult mice led to serious thymic atrophy and loss of cellularity due to defects in cell proliferation (97). Likewise, BAP1 deficiency caused a block at the DN3 stage before the pre-TCR checkpoint by facilitating the ubiquitination of histone H2A at Lys119 (H2AK119) (97).
Conclusion
During the past few years, several lines of evidence have shown that T cell development is regulated at multiple levels; in addition to transcriptional control, posttranslational regulation also plays a crucial role in those processes (9, 98, 99). An increasing number of studies using transgenic mouse models have demonstrated that E3 ubiquitin ligases and DUBs are involved in specific stages of thymocyte maturation by modulating the activity or stability of key proteins during cellular signal transduction cascades (98, 99). Technological advancements in single-cell proteomics, CRISPR/Cas9 mutagenesis and mass cytometry will continue adding valuable findings to this area of research. Future work on the molecular mechanisms of ubiquitination and deubiquitination in T cells will not only enhance our understanding of cell fate determination via gene regulatory networks but also provide potential novel therapeutic strategies for treating autoimmune diseases and cancer.
Author contributions
XLi and BZ: conceptualization and guidance. TZ: writing the original draft. KL: visualization. XLn: provide assistances. ZZ and SL: proofreading. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of China (Grant NO. 82170795, 8207034059) and the Science and Technology Innovation Program of Hunan Province (Grant NO. 2020RC4044).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cruz Walma DA, Chen Z, Bullock AN, Yamada KM. Ubiquitin ligases: Guardians of mammalian development. Nat Rev Mol Cell Biol (2022) 23(5):350–67. doi: 10.1038/s41580-021-00448-5
2. Roberts JZ, Crawford N, Longley DB. The role of ubiquitination in apoptosis and necroptosis. Cell Death Differ (2022) 29(2):272–84. doi: 10.1038/s41418-021-00922-9
3. Oshiumi H. Recent advances and contradictions in the study of the individual roles of ubiquitin ligases that regulate rig-I-Like receptor-mediated antiviral innate immune responses. Front Immunol (2020) 11:1296. doi: 10.3389/fimmu.2020.01296
4. Chang SC, Zhang BX, Ding JL. E2-E3 ubiquitin enzyme pairing - partnership in provoking or mitigating cancers. Biochim Biophys Acta Rev Cancer (2022) 1877(2):188679. doi: 10.1016/j.bbcan.2022.188679
5. Nakasone MA, Majorek KA, Gabrielsen M, Sibbet GJ, Smith BO, Huang DT. Structure of Ube2k-Ub/E3/Polyub reveals mechanisms of K48-linked ub chain extension. Nat Chem Biol (2022) 18(4):422–31. doi: 10.1038/s41589-021-00952-x
6. Donaghy R, Han X, Rozenova K, Lv K, Jiang Q, Doepner M, et al. The brisc deubiquitinating enzyme complex limits hematopoietic stem cell expansion by regulating Jak2 K63-ubiquitination. Blood (2019) 133(14):1560–71. doi: 10.1182/blood-2018-10-877563
7. Rajalingam K, Dikic I. Snapshot: Expanding the ubiquitin code. Cell (2016) 164(5):1074– e1. doi: 10.1016/j.cell.2016.02.019
8. Lange SM, Armstrong LA, Kulathu Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol Cell (2022) 82(1):15–29. doi: 10.1016/j.molcel.2021.10.027
9. Dutta A, Zhao B, Love PE. New insights into tcr B-selection. Trends Immunol (2021) 42(8):735–50. doi: 10.1016/j.it.2021.06.005
10. Liu C, Lan Y, Liu B, Zhang H, Hu H. T Cell development: Old tales retold by single-cell rna sequencing. Trends Immunol (2021) 42(2):165–75. doi: 10.1016/j.it.2020.12.004
11. Fink PJ. Exposing T cell secrets inside and outside the thymus. Annu Rev Immunol (2021) 40:1–14. doi: 10.1146/annurev-immunol-101220-014126
12. Lomada D, Jain M, Bolner M, Reeh KA, Kang R, Reddy MC, et al. Stat3 signaling promotes survival and maintenance of medullary thymic epithelial cells. PloS Genet (2016) 12(1):e1005777. doi: 10.1371/journal.pgen.1005777
13. Fischer JC, Otten V, Kober M, Drees C, Rosenbaum M, Schmickl M, et al. A20 restrains thymic regulatory T cell development. J Immunol (2017) 199(7):2356–65. doi: 10.4049/jimmunol.1602102
14. Matesic LE, Haines DC, Copeland NG, Jenkins NA. Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum Mol Genet (2006) 15(24):3485–97. doi: 10.1093/hmg/ddl425
15. Song R, Kim YW, Koo BK, Jeong HW, Yoon MJ, Yoon KJ, et al. Mind bomb 1 in the lymphopoietic niches is essential for T and marginal zone b cell development. J Exp Med (2008) 205(11):2525–36. doi: 10.1084/jem.20081344
16. Kitagawa K, Shibata K, Matsumoto A, Matsumoto M, Ohhata T, Nakayama KI, et al. Fbw7 targets Gata3 through cyclin-dependent kinase 2-dependent proteolysis and contributes to regulation of T-cell development. Mol Cell Biol (2014) 34(14):2732–44. doi: 10.1128/mcb.01549-13
17. Zhao B, Yoganathan K, Li L, Lee JY, Zúñiga-Pflücker JC, Love PE. Notch and the pre-tcr coordinate thymocyte proliferation by induction of the scf subunits Fbxl1 and Fbxl12. Nat Immunol (2019) 20(10):1381–92. doi: 10.1038/s41590-019-0469-z
18. Gao Y, Liu R, He C, Basile J, Vesterlund M, Wahren-Herlenius M, et al. Socs3 expression by thymic stromal cells is required for normal T cell development. Front Immunol (2021) 12:642173. doi: 10.3389/fimmu.2021.642173
19. MacKenzie DA, Schartner J, Lin J, Timmel A, Jennens-Clough M, Fathman CG, et al. Grail is up-regulated in Cd4+ Cd25+ T regulatory cells and is sufficient for conversion of T cells to a regulatory phenotype. J Biol Chem (2007) 282(13):9696–702. doi: 10.1074/jbc.M604192200
20. Biju MP, Neumann AK, Bensinger SJ, Johnson RS, Turka LA, Haase VH. Vhlh gene deletion induces hif-1-Mediated cell death in thymocytes. Mol Cell Biol (2004) 24(20):9038–47. doi: 10.1128/mcb.24.20.9038-9047.2004
21. Yi Z, Lin WW, Stunz LL, Bishop GA. The adaptor Traf3 restrains the lineage determination of thymic regulatory T cells by modulating signaling Via the receptor for il-2. Nat Immunol (2014) 15(9):866–74. doi: 10.1038/ni.2944
22. Akiyama T, Maeda S, Yamane S, Ogino K, Kasai M, Kajiura F, et al. Dependence of self-tolerance on Traf6-directed development of thymic stroma. Science (2005) 308(5719):248–51. doi: 10.1126/science.1105677
23. Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, et al. Tissue hyperplasia and enhanced T-cell signalling Via zap-70 in c-Cbl-Deficient mice. Mol Cell Biol (1998) 18(8):4872–82. doi: 10.1128/mcb.18.8.4872
24. Chiang YJ, Sommers CL, Jordan MS, Gu H, Samelson LE, Koretzky GA, et al. Inactivation of c-cbl reverses neonatal lethality and T cell developmental arrest of slp-76-Deficient mice. J Exp Med (2004) 200(1):25–34. doi: 10.1084/jem.20040262
25. Zhao Y, Guo H, Qiao G, Zucker M, Langdon WY, Zhang J. E3 ubiquitin ligase cbl-b regulates thymic-derived Cd4+Cd25+ regulatory T cell development by targeting Foxp3 for ubiquitination. J Immunol (2015) 194(4):1639–45. doi: 10.4049/jimmunol.1402434
26. Oh J, Wu N, Baravalle G, Cohn B, Ma J, Lo B, et al. March1-mediated mhcii ubiquitination promotes dendritic cell selection of natural regulatory T cells. J Exp Med (2013) 210(6):1069–77. doi: 10.1084/jem.20122695
27. Zhang X, Berger FG, Yang J, Lu X. Usp4 inhibits P53 through deubiquitinating and stabilizing arf-Bp1. EMBO J (2011) 30(11):2177–89. doi: 10.1038/emboj.2011.125
28. Vugmeyster Y, Borodovsky A, Maurice MM, Maehr R, Furman MH, Ploegh HL. The ubiquitin-proteasome pathway in thymocyte apoptosis: Caspase-dependent processing of the deubiquitinating enzyme Usp7 (Hausp). Mol Immunol (2002) 39(7-8):431–41. doi: 10.1016/s0161-5890(02)00123-2
29. Dufner A, Kisser A, Niendorf S, Basters A, Reissig S, Schönle A, et al. The ubiquitin-specific protease Usp8 is critical for the development and homeostasis of T cells. Nat Immunol (2015) 16(9):950–60. doi: 10.1038/ni.3230
30. Harkiolaki M, Lewitzky M, Gilbert RJ, Jones EY, Bourette RP, Mouchiroud G, et al. Structural basis for Sh3 domain-mediated high-affinity binding between Mona/Gads and slp-76. EMBO J (2003) 22(11):2571–82. doi: 10.1093/emboj/cdg258
31. Kaneko T, Kumasaka T, Ganbe T, Sato T, Miyazawa K, Kitamura N, et al. Structural insight into modest binding of a non-pxxp ligand to the signal transducing adaptor molecule-2 src homology 3 domain. J Biol Chem (2003) 278(48):48162–8. doi: 10.1074/jbc.M306677200
32. Brownlie RJ, Zamoyska R. T Cell receptor signalling networks: Branched, diversified and bounded. Nat Rev Immunol (2013) 13(4):257–69. doi: 10.1038/nri3403
33. Adoro S, Park KH, Bettigole SE, Lis R, Shin HR, Seo H, et al. Post-translational control of T cell development by the escrt protein Chmp5. Nat Immunol (2017) 18(7):780–90. doi: 10.1038/ni.3764
34. Watanabe M, Hatakeyama S. Fine-tuning of thymocyte development by ubiquitination-mediated stability control of the escrt protein Chmp5. Cell Mol Immunol (2017) 14(12):957–9. doi: 10.1038/cmi.2017.91
35. Naik E, Webster JD, DeVoss J, Liu J, Suriben R, Dixit VM. Regulation of proximal T cell receptor signaling and tolerance induction by deubiquitinase Usp9x. J Exp Med (2014) 211(10):1947–55. doi: 10.1084/jem.20140860
36. Paster W, Brockmeyer C, Fu G, Simister PC, de Wet B, Martinez-Riaño A, et al. Grb2-mediated recruitment of themis to lat is essential for thymocyte development. J Immunol (2013) 190(7):3749–56. doi: 10.4049/jimmunol.1203389
37. Fu G, Vallée S, Rybakin V, McGuire MV, Ampudia J, Brockmeyer C, et al. Themis controls thymocyte selection through regulation of T cell antigen receptor-mediated signaling. Nat Immunol (2009) 10(8):848–56. doi: 10.1038/ni.1766
38. Johnson AL, Aravind L, Shulzhenko N, Morgun A, Choi SY, Crockford TL, et al. Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection. Nat Immunol (2009) 10(8):831–9. doi: 10.1038/ni.1769
39. Lesourne R, Uehara S, Lee J, Song KD, Li L, Pinkhasov J, et al. Themis, a T cell-specific protein important for late thymocyte development. Nat Immunol (2009) 10(8):840–7. doi: 10.1038/ni.1768
40. Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, et al. Regulation of T cell development by the deubiquitinating enzyme cyld. Nat Immunol (2006) 7(4):411–7. doi: 10.1038/ni1315
41. Förster M, Boora RK, Petrov JC, Fodil N, Albanese I, Kim J, et al. A role for the histone H2a deubiquitinase Mysm1 in maintenance of Cd8+ T cells. Immunology (2017) 151(1):110–21. doi: 10.1111/imm.12710
42. Gatzka M, Tasdogan A, Hainzl A, Allies G, Maity P, Wilms C, et al. Interplay of H2a deubiquitinase 2a-Dub/Mysm1 and the P19(Arf)/P53 axis in hematopoiesis, early T-cell development and tissue differentiation. Cell Death Differ (2015) 22(9):1451–62. doi: 10.1038/cdd.2014.231
43. Drennan MB, Govindarajan S, Verheugen E, Coquet JM, Staal J, McGuire C, et al. Nkt sublineage specification and survival requires the ubiquitin-modifying enzyme Tnfaip3/A20. J Exp Med (2016) 213(10):1973–81. doi: 10.1084/jem.20151065
44. Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. Bap1 and cancer. Nat Rev Cancer (2013) 13(3):153–9. doi: 10.1038/nrc3459
45. Kannt A, Đikić I. Expanding the arsenal of E3 ubiquitin ligases for proximity-induced protein degradation. Cell Chem Biol (2021) 28(7):1014–31. doi: 10.1016/j.chembiol.2021.04.007
46. Lin AE, Mak TW. The role of E3 ligases in autoimmunity and the regulation of autoreactive T cells. Curr Opin Immunol (2007) 19(6):665–73. doi: 10.1016/j.coi.2007.10.002
47. García-León MJ, Fuentes P, de la Pompa JL, Toribio ML. Dynamic regulation of Notch1 activation and notch ligand expression in human thymus development. Development (2018) 145(16):dev165597. doi: 10.1242/dev.165597
48. Solanki A, Yánez DC, Lau CI, Rowell J, Barbarulo A, Ross S, et al. The transcriptional repressor Bcl6 promotes pre-Tcr-Induced thymocyte differentiation and attenuates Notch1 activation. Development (2020) 147(19):dev192203. doi: 10.1242/dev.192203
49. Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, et al. An activated form of notch influences the choice between Cd4 and Cd8 T cell lineages. Cell (1996) 87(3):483–92. doi: 10.1016/s0092-8674(00)81368-9
50. Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, et al. Recognition and ubiquitination of notch by itch, a hect-type E3 ubiquitin ligase. J Biol Chem (2000) 275(46):35734–7. doi: 10.1074/jbc.M007300200
51. McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem (2003) 278(25):23196–203. doi: 10.1074/jbc.M302827200
52. Chastagner P, Israël A, Brou C. Itch/Aip4 mediates deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep (2006) 7(11):1147–53. doi: 10.1038/sj.embor.7400822
53. Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, et al. Deltex1 redirects lymphoid progenitors to the b cell lineage by antagonizing Notch1. Immunity (2002) 16(2):231–43. doi: 10.1016/s1074-7613(02)00271-6
54. Lehar SM, Bevan MJ. T Cells develop normally in the absence of both Deltex1 and Deltex2. Mol Cell Biol (2006) 26(20):7358–71. doi: 10.1128/mcb.00149-06
55. Jang J, Choi YI, Choi J, Lee KY, Chung H, Jeon SH, et al. Notch1 confers thymocytes a resistance to gc-induced apoptosis through Deltex1 by blocking the recruitment of P300 to the Srg3 promoter. Cell Death Differ (2006) 13(9):1495–505. doi: 10.1038/sj.cdd.4401827
56. Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ. Lnx functions as a ring type E3 ubiquitin ligase that targets the cell fate determinant numb for ubiquitin-dependent degradation. EMBO J (2002) 21(1-2):93–102. doi: 10.1093/emboj/21.1.93
57. Shah DK, Mohtashami M, Zúñiga-Pflücker JC. Role of recycling, Mindbomb1 association, and exclusion from lipid rafts of Δ-like 4 for effective notch signaling to drive T cell development. J Immunol (2012) 189(12):5797–808. doi: 10.4049/jimmunol.1202469
58. Nie L, Perry SS, Zhao Y, Huang J, Kincade PW, Farrar MA, et al. Regulation of lymphocyte development by cell-Type-Specific interpretation of notch signals. Mol Cell Biol (2008) 28(6):2078–90. doi: 10.1128/mcb.00844-07
59. Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alborán IM, Nakayama K, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med (2007) 204(12):2875–88. doi: 10.1084/jem.20062299
60. Kossatz U, Dietrich N, Zender L, Buer J, Manns MP, Malek NP. Skp2-dependent degradation of P27kip1 is essential for cell cycle progression. Genes Dev (2004) 18(21):2602–7. doi: 10.1101/gad.321004
61. Rajsbaum R, García-Sastre A, Versteeg GA. Trimmunity: The roles of the trim E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol (2014) 426(6):1265–84. doi: 10.1016/j.jmb.2013.12.005
62. Espinosa A, Zhou W, Ek M, Hedlund M, Brauner S, Popovic K, et al. The sjogren's syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol (2006) 176(10):6277–85. doi: 10.4049/jimmunol.176.10.6277
63. Espinosa A, Dardalhon V, Brauner S, Ambrosi A, Higgs R, Quintana FJ, et al. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the il-23-Th17 pathway. J Exp Med (2009) 206(8):1661–71. doi: 10.1084/jem.20090585
64. Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (Socs) proteins in regulation of the immune response. Annu Rev Immunol (2004) 22:503–29. doi: 10.1146/annurev.immunol.22.091003.090312
65. Satoh R, Kakugawa K, Yasuda T, Yoshida H, Sibilia M, Katsura Y, et al. Requirement of Stat3 signaling in the postnatal development of thymic medullary epithelial cells. PloS Genet (2016) 12(1):e1005776. doi: 10.1371/journal.pgen.1005776
66. Takahama Y, Ohigashi I, Baik S, Anderson G. Generation of diversity in thymic epithelial cells. Nat Rev Immunol (2017) 17(5):295–305. doi: 10.1038/nri.2017.12
67. Hogquist KA, Xing Y, Hsu FC, Shapiro VS. T Cell adolescence: Maturation events beyond positive selection. J Immunol (2015) 195(4):1351–7. doi: 10.4049/jimmunol.1501050
68. Bandyopadhyay S, Soto-Nieves N, Macián F. Transcriptional regulation of T cell tolerance. Semin Immunol (2007) 19(3):180–7. doi: 10.1016/j.smim.2007.02.006
69. Gómez-Martín D, Díaz-Zamudio M, Alcocer-Varela J. Ubiquitination system and autoimmunity: The bridge towards the modulation of the immune response. Autoimmun Rev (2008) 7(4):284–90. doi: 10.1016/j.autrev.2007.11.026
70. Nurieva RI, Zheng S, Jin W, Chung Y, Zhang Y, Martinez GJ, et al. The E3 ubiquitin ligase grail regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-Cd3 degradation. Immunity (2010) 32(5):670–80. doi: 10.1016/j.immuni.2010.05.002
71. Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity (2008) 28(1):100–11. doi: 10.1016/j.immuni.2007.11.021
72. Bonito AJ, Aloman C, Fiel MI, Danzl NM, Cha S, Weinstein EG, et al. Medullary thymic epithelial cell depletion leads to autoimmune hepatitis. J Clin Invest (2013) 123(8):3510–24. doi: 10.1172/jci65414
73. Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M, et al. Medullary thymic epithelial cells expressing aire represent a unique lineage derived from cells expressing claudin. Nat Immunol (2007) 8(3):304–11. doi: 10.1038/ni1438
74. Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, Hayashi Y, et al. The cytokine rankl produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity (2008) 29(3):438–50. doi: 10.1016/j.immuni.2008.06.018
75. Gray DH, Seach N, Ueno T, Milton MK, Liston A, Lew AM, et al. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood (2006) 108(12):3777–85. doi: 10.1182/blood-2006-02-004531
76. Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol (2004) 22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813
77. Duan L, Reddi AL, Ghosh A, Dimri M, Band H. The cbl family and other ubiquitin ligases: Destructive forces in control of antigen receptor signaling. Immunity (2004) 21(1):7–17. doi: 10.1016/j.immuni.2004.06.012
78. Rao N, Dodge I, Band H. The cbl family of ubiquitin ligases: Critical negative regulators of tyrosine kinase signaling in the immune system. J Leukoc Biol (2002) 71(5):753–63. doi: 10.1189/jlb.71.5.753
79. Liu YC, Gu H. Cbl and cbl-b in T-cell regulation. Trends Immunol (2002) 23(3):140–3. doi: 10.1016/s1471-4906(01)02157-3
80. Thien CB, Langdon WY. Cbl: Many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol (2001) 2(4):294–307. doi: 10.1038/35067100
81. Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in cbl-deficient mice. Proc Natl Acad Sci U.S.A. (1998) 95(26):15547–52. doi: 10.1073/pnas.95.26.15547
82. Dragone LL, Shaw LA, Myers MD, Weiss A. Slap, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol Rev (2009) 232(1):218–28. doi: 10.1111/j.1600-065X.2009.00827.x
83. Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like adaptor protein (Slap) is a negative regulator of T cell receptor signaling. J Exp Med (2000) 191(3):463–74. doi: 10.1084/jem.191.3.463
84. Sosinowski T, Killeen N, Weiss A. The src-like adaptor protein downregulates the T cell receptor on Cd4+Cd8+ thymocytes and regulates positive selection. Immunity (2001) 15(3):457–66. doi: 10.1016/s1074-7613(01)00195-9
85. Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic Bh3-only bcl-2 family member bim. EMBO J (2003) 22(24):6653–64. doi: 10.1093/emboj/cdg635
86. Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of Cd4+ T cells from T cell receptor transgenic mice. J Exp Med (1992) 176(4):1091–8. doi: 10.1084/jem.176.4.1091
87. Raberger J, Boucheron N, Sakaguchi S, Penninger JM, Ellmeier W. Impaired T-cell development in the absence of Vav1 and itk. Eur J Immunol (2008) 38(12):3530–42. doi: 10.1002/eji.200838388
88. Liu H, Wilson KR, Schriek P, Macri C, Blum AB, Francis L, et al. Ubiquitination of mhc class ii is required for development of regulatory but not conventional Cd4+ T cells. J Immunol (2020) 205(5):1207–16. doi: 10.4049/jimmunol.1901328
89. Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell (2005) 123(5):773–86. doi: 10.1016/j.cell.2005.11.007
90. Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem (2009) 78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526
91. Clague MJ, Urbé S, Komander D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol (2019) 20(6):338–52. doi: 10.1038/s41580-019-0099-1
92. Zhao B, Schlesiger C, Masucci MG, Lindsten K. The ubiquitin specific protease 4 (Usp4) is a new player in the wnt signalling pathway. J Cell Mol Med (2009) 13(8b):1886–95. doi: 10.1111/j.1582-4934.2009.00682.x
93. Niendorf S, Oksche A, Kisser A, Löhler J, Prinz M, Schorle H, et al. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol (2007) 27(13):5029–39. doi: 10.1128/mcb.01566-06
94. Reissig S, Hövelmeyer N, Tang Y, Weih D, Nikolaev A, Riemann M, et al. The deubiquitinating enzyme cyld regulates the differentiation and maturation of thymic medullary epithelial cells. Immunol Cell Biol (2015) 93(6):558–66. doi: 10.1038/icb.2014.122
95. Hövelmeyer N, Wunderlich FT, Massoumi R, Jakobsen CG, Song J, Wörns MA, et al. Regulation of b cell homeostasis and activation by the tumor suppressor gene cyld. J Exp Med (2007) 204(11):2615–27. doi: 10.1084/jem.20070318
96. Pujari R, Hunte R, Khan WN, Shembade N. A20-mediated negative regulation of canonical nf-Kb signaling pathway. Immunol Res (2013) 57(1-3):166–71. doi: 10.1007/s12026-013-8463-2
97. Arenzana TL, Lianoglou S, Seki A, Eidenschenk C, Cheung T, Seshasayee D, et al. Tumor suppressor Bap1 is essential for thymic development and proliferative responses of T lymphocytes. Sci Immunol (2018) 3(22):eaal1953. doi: 10.1126/sciimmunol.aal1953
98. Aki D, Li Q, Li H, Liu YC, Lee JH. Immune regulation by protein ubiquitination: Roles of the E3 ligases vhl and itch. Protein Cell (2019) 10(6):395–404. doi: 10.1007/s13238-018-0586-8
Keywords: T cell development, thymocyte, ubiquitination, E3 ubiquitin ligase, deubiquitinating enzyme
Citation: Zhong T, Lei K, Lin X, Xie Z, Luo S, Zhou Z, Zhao B and Li X (2022) Protein ubiquitination in T cell development. Front. Immunol. 13:941962. doi: 10.3389/fimmu.2022.941962
Received: 12 May 2022; Accepted: 11 July 2022;
Published: 04 August 2022.
Edited by:
Jinfang Zhu, National Institute of Allergy and Infectious Diseases (NIH), United StatesCopyright © 2022 Zhong, Lei, Lin, Xie, Luo, Zhou, Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Zhao, binzhao@csu.edu.cn; Xia Li, lixia@csu.edu.cn
†These authors have contributed equally to this work and share first authorship
‡Bin Zhao and Xia Li jointly supervised this work
 Ting Zhong
Ting Zhong