- 1Laboratorio di Immunologia dei Tumori del Sangue (LITS), Centro Interdipartimentale di Biotecnologie Molecolari “Guido Tarone”, Dipartimento di Biotecnologie Molecolari e Scienze della Salute, Università degli Studi di Torino, Torino, Italy
- 2SC Ematologia, AO S.Croce e Carle, Cuneo, Italy
Myeloid derived suppressors cells (MDSC) play major roles in regulating immune homeostasis and immune responses in many conditions, including cancer. MDSC interact with cancer cells within the tumor microenvironment (TME) with direct and indirect mechanisms: production of soluble factors and cytokines, expression of surface inhibitory molecules, metabolic rewiring and exosome release. The two-way relationship between MDSC and tumor cells results in immune evasion and cancer outgrowth. In multiple myeloma (MM), MDSC play a major role in creating protumoral TME conditions. In this minireview, we will discuss the interplay between MDSC and MM TME and the possible strategies to target MDSC.
Introduction
Multiple myeloma (MM) is a paradigm disease in which progression is fueled by intrinsic alterations of myeloma cells and tumor-host interactions in the tumor microenvironment (TME) (1). Disease evolution from monoclonal gammopathy of undetermined significance (MGUS) to smoldering myeloma (SMM), and symptomatic disease is characterized by a progressive increase of myeloma cells associated with co-evolving immunological and metabolic changes making the TME unable to hold the disease in check (1). We and others have shown that immune alterations are already detectable in the very early stage of the disease (2, 3) and that they persist in the remission phase (2). The immune MM TME contexture consists of effector cells (i.e, conventional T cells, unconventional T cells like NKT cells, γδ T cells, NK cells etc), professional suppressor cells [i.e, regulatory T cells (Tregs), regulatory B cells (Bregs), myeloid derived suppressor cells (MDSC)], and cells that are functionally conditioned by the TME and acquire protumoral functions like bone marrow stromal cells (BMSC), endothelial cells, osteoblasts (OB), and osteoclasts (4). Recently, BM-resident neutrophils have also been reported to contribute to the TME-induced suppressive commitment of MM patients (5). Unbalanced distribution of effector and suppressor cells already detectable in MGUS is induced by the progressive accumulation of myeloma cells driven by genetic and epigenetic drivers. The bone marrow (BM), which is where MM originates and propagates, has the capacity to physiologically host around 2-5% polyclonal plasma cells. When myeloma cell infiltration overcomes this threshold, the TME is immunologically and metabolically shaped to support myeloma cell growth, to induce drug resistance, and to suppress immune recognition. MDSC play a major role in the protumoral reset of MM TME.
We have previously shown that MDSC are significantly increased in the BM of MGUS and MM patients: granulocytic/polymorphonuclear MDSC (PMN-MDSC), and not monocytic MDSC (M-MDSC), are responsible for the increase (2). MDSC frequency is very similar in MGUS, MM at diagnosis, and MM in relapse. Unexpectedly, we have found that MDSC frequency is significantly higher in MM in remission (2), indicating that there is no correlation between the proportion of BM myeloma cells and MDSC expansion. Similar data have been reported in mouse models in which MDSC start to accumulate in the TME as early as one week after tumor inoculation when the frequency of myeloma cells is very low (<10%) as in MGUS individuals (6).
Approximately, 20-40% of MDCS express the Programmed Cell Death-Ligand 1+ (PD-L1+) (2) and therefore are very well-suited to engage and suppress immune effector cells like Vγ9Vδ2 cells and NK cells expressing the Programmed Cell Death-1 (PD-1) receptor (2). MDSC are PD-L1+ in MGUS and MM irrespective of the disease stage, including MM in remission when most myeloma cells have been cleared from BM (2). The persistence of PD-L1+ MDSC can hinder the immunomodulatory activity of drugs like bortezomib or lenalidomide after autologous stem cell transplantation.
In conclusion, MDSC play a major role in the establishment of the immune suppressive TME in MM. The aim of this minireview is to discuss the mechanisms exploited by MDSC in cooperation with myeloma cells, professional immune suppressor cells, and other bystander cells to promote myeloma cell growth in the BM of MM patients. We will also discuss possible interventions to dampen the immune suppression operated by MDSC and other suppressor cells to recover the antimyeloma activity of immune effector cells.
MDSC subsets and differentiation
MDSC play a major role in the regulation of immune homeostasis in healthy individuals, and the regulation of immune responses in infectious diseases, autoimmunity, aging, pregnancy, transplantation, and obesity (7). In cancer, the immune suppressive activity of MDSC is exploited by tumor cells to evade immune surveillance and support their survival and accumulation (7).
MDSC are derived from bone marrow hematopoietic stem cells (7). There are two major subsets of MDSC in humans: PMN-MDSC and M-MDSC. The first one are phenotypically and morphologically similar to neutrophils (CD15+ and/or CD66b+), whereas M-MDSC are similar to monocytes (CD14+)(7). More recently, a third subset of phenotypically distinct immature early-MDSC (e-MDSC) has been identified in cancer patients (8). In this review we will use the term MDSC to identify both PMN-MDSC and M-MDSC unless otherwise specified.
MDSC development occurs in two partially overlapping waves (9). The first one is driven by cytokines and soluble factors including granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF), granulocyte colony-stimulating factor (G-CSF), interleukin 6 (IL-6), and vascular endothelial growth factor (VEGF). These cytokines and soluble factors are produced by tumor cells and/or BMSC in the TME and promote MDSC differentiation from hematopoietic progenitor cells via STAT3 and STAT5 activation (10, 11, 12). Mesenchymal stromal cells (MSC) also induce MDSC expansion via the hepatocyte growth factor (HGF), c-Met, and STAT3 phosphorylation (10). The second wave is driven by a different set of cytokines and inflammatory soluble factors like interleukin 13 (IL-13), toll-like receptor (TLR) ligands, and prostaglandin E2 (PGE2) yielding to the functional MDSC activation via the STAT1 and NF-kB pathways (10–12). The TME is highly predisposed to drive the expansion and activation of MDSC at the expense of other myeloid-derived cells like monocytes, macrophages and dendritic cells (DC) (8).
Immuno suppressive MDSC features
The immune suppressive MDSC activity is dependent on: 1) the depletion of essential CD8+ T- cell nutrients in the TME; 2) the production of immune suppressive cytokines and/or soluble factors; 3) the expression of cell surface inhibitory molecules [i.e., (PD-L1)]; 4) the protumoral metabolic TME rewiring at the expense of immune effector cells.
Amino acid depletion
MDSC express the xc- transporter and import cystine, but, unlike DC and macrophages, they are unable to export cysteine because they lack the ASC neutral amino acid transporter (13). Considering the progressive TME invasion by tumor cells and MDSC at the expense of other cells which can supply extracellular cysteine, the TME becomes depleted of cysteine jeopardizing the activation of CD8+T cells that are unable to convert cystine to cysteine to meet their metabolic requirements (13).
MDSC also deplete the TME of tryptophan via the enzyme indoleamine 2, 3-dioxygenase (IDO) (14). T lymphocytes are very susceptible to tryptophan shortage which restrains their proliferative responses by inducing an integrated stress response and the inactivation of the mTOR pathway (15, 16). Tryptophan catabolites can also induce the apoptosis of cytotoxic T cells (17, 18), and the concurrent differentiation of Tregs (16). L-arginine (L-arg) is another essential amino acid which is critical for T-cell immune functions. Arginine metabolism is regulated by the inducible nitric oxide synthase (iNOS) isoenzymes, arginase (Arg 1/2) activity, and proline and polyamines synthesis. MDSC express both iNOS and Arg-1 that induce L-arg depletion in the TME leading to inhibition of CD3-ζ expression in T cells, and induction of apoptosis (7, 9, 19).
Cytokines and soluble factors
The production and release of suppressor cytokines and soluble factors is another mechanism exploited by MDSC to protect tumor cells from immune recognition and killing. Nitric oxide (NO), reactive oxygen species (ROS), peroxynitrite (PNT) (a short-lived product of NO reaction with ROS), interleukin 10 (IL-10), and transforming growth factor-β (TGF-β) are released by MDSC with slightly difference between PMN-MDSC and M-MDSC subsets (7, 9, 20, 21). The hyper-production of ROS and PNT in the TME impairs the ability of CD8+ T cells to bind to peptide–major histocompatibility complexes and to respond to specific peptides (21). NO also hampers the Fc receptor-mediated effector functions of NK cells (22). IL-10 recruits Tregs in the TME and decreases CD8+ T-cell antigen sensitivity by inducing cell surface glycoprotein branching (23). TGF-β is induced by IL-13 (24) and interferon-γ (IFN-γ) (25), and contributes to T-cell suppression through Tregs development (25). Kynurenine is another soluble immune suppressive factor that is generated in the TME as a consequence of tryptophan catabolism by MDSC. Kynurenine can inhibit T-cell and NK cell proliferation and drive the differentiation of naïve T cells into Tregs (16).
Cell surface molecules
The cell surface expression of immune checkpoints ligands (ICP-L) like PD-L1 is another mechanism used by M-MDSC to suppress immune effector cells (2, 7, 9), while PMN-MDSC preferentially exploit the Fas/Fas-ligand pathway to induce T-cell depletion in the TME (26). The V-domain immunoglobulin suppressor of T cell activation (VISTA) is a novel co-inhibitory ligand/receptor highly expressed by MDSC in the TME that suppresses T-cell effector functions and contributes to acquired resistance to PD-1/PD-L1 blockade (27). Lastly, CXCR2 is another cell surface molecule that is critical in mice models and paediatric sarcoma to promote the accumulation of MDSC in the TME and hamper the efficacy of anti-PD-1 treatment (28).
Protumoral metabolic TME rewiring
The TME is a very dynamic ecosystem that is progressively molded by tumor cells to locally create protective conditions to support their growth and resistance to therapy, from conventional chemotherapy to immunotherapy (29, 30). Hypoxia is a major metabolic feature of TME (30), especially in solid tumors, almost always associated with the extracellular acidification induced by lactate accumulation. Tumor cells rewire their metabolism to survive and proliferate in the TME by: 1) increasing glucose and amino acid uptake, glycolytic flux, and lactate production; 2) modifying glutamine metabolism, tricarboxylic acid cycle, and oxidative phosphorylation; 3) increasing the production of mitochondrial ROS; 4) modulating fatty acid synthesis and oxidation (FAO) (30). MDSC partially mimick the metabolic rewiring of tumor cells by adapting their lactate, glucose, and lipid metabolism to the hypoxic and acidic TME conditions (31, 32). As a result, MDSC survive in the TME, contribute to the exacerbation of the protumoral metabolic TME commitment, and maintain unaltered their immune suppressor activity (33–35).
Immune suppressive and metabolic features in MM
MM is a hematologic cancer characterized by the accumulation of malignant plasma cells (myeloma cells) in the BM. Progressive colonization of BM results in a deep remodelling of the BM niche that becomes committed to support myeloma cell growth, immune evasion, and drug resistance (1).
MDSC play a major role in establishing the protumoral TME commitment. We have shown that MDSC accumulation in the BM is already detectable in MGUS, and their expansion persists throughout the entire period of the disease (2), including the remission phase (2). In our hands, PMN-MDSC was the main subpopulation to be expanded in MGUS and MM (2), while other groups have reported the predominance of M-MDSC in MM at diagnosis and in relapse (36, 37). Immunogenomic characterization identified CD11b+CD13+CD16+ cells as the PMN-MDSC subset with strongest capacity to suppress anti-myeloma activity T-cell immune responses (38). MDSC-like suppressive activity is also exhibited by MM neutrophils (5), suggesting that an accurate characterization of MDSC should be based on phenotypic markers, immunosuppressive potential, and transcriptional network.
Development and suppressor functions of MDSC are supported by myeloma cells and bystander cells via direct and indirect mechanisms. Direct mechanisms operated by myeloma cells include: 1) IL-6 production (39, 40) which prevents MDSC differentiation and promotes MDSC accumulation and activation via the STAT3 signaling pathway (41); 2) the induction of Mcl-1, an anti-apoptotic protein sustaining MDSC survival (42); 3) the secretion of galectin-1 that targets CD304 on MDSC and enhances their immune suppressive capacity (43); 4) the production of chemokine ligand 5 (CCL5) and macrophage migration inhibitory factor (MIF) (44). MIF has also been reported to potentiate the immune suppressive activity of MDSC via CD84-mediated PD-L1 upregulation (45); 5) the release of exosomes that promotes MDSC growth and NO production (46)
Bystander cells in the TME also cooperate with myeloma cells in the induction and activation of immune suppressive MDSC via direct mechanisms including: 1) IL-6 release (47, 48); 2) exosome release by BMSC (49); 3) production and release of immune suppressive molecules [i.e. Prostaglandin-Endoperoxide Synthase 2 (PTGS2), TGF-β, Nitric Oxide Synthase 2 (NOS2), IL-10 and IL-6] by MSC and OB (50, 51).
In addition to the direct mechanisms listed above, myeloma cells and bystander cells promote the accumulation and activation of MDSC via indirect mechanisms. An example is the metabolic rewiring operated by myeloma cells and bystander cells that creates an hypoxic and nutrient-depleted TME that promotes the accumulation and activation of MDSC at the expense of immune effector cells (52–54). Lactate over-production shifts MDSC differentiation toward PMN-MDSC (55), which is the subset that we and others have shown to be increased in the peripheral blood (PB) and BM of MM patients (2, 56).
The accumulation and activation of MDSC is beneficial to myeloma cells creating a very effective protumoral loop (3, 57). MDSC facilitate the self-renewal of myeloma stem-cells, enhance their tumorigenic potential via epigenetic regulation (58), and promote myeloma cell survival via AMPK phosphorylation leading to increase β-oxidation, ATP production, and increased NADPH levels (59). MDSC production of S100A9, a calcium-binding protein that promotes the release of TNF-α, IL-6, and IL-10 in autocrine pathway through TLR4 interaction, attracts myeloma cells in the TME (60) and supports myeloma cell growth via the activation of the canonical NFκB pathway (61).
Indirect mechanisms operated by MDSC to support myeloma cells are deprivation of nutrients, production of soluble factors, and the expression of cell surface inhibitory molecules. The common denominator is the impairment of anti-myeloma immune responses. In addition, PMN-MDSC are educated to express angiogenesis-related proteins to support tumor angiogenesis (62).
MDSC upregulate enzymes that contribute to the shortage of amino acids essential for immune effector T cells. Arginase 1 (Arg-1) expression and NO production by MDSC limit the availability of L-Arg needed for effective TCR-mediated signaling (63, 64). MDSC can utilize glutamine for anaplerosis like myeloma cells (65, 66), exacerbating glutamine deprivation in the TME (54).
Several soluble factors and cytokines contribute to the immune suppressor activity of MDSC in the TME, like IL-10, IL-6, TGF-β, CD40-CD40 Ligand, and IFN-γ. These cytokines tip the scales in favor of Tregs (44, 67), whose number is directly correlated with MDSC expansion (56). Lastly, CD38 expression on MDSC (68) contributes to the discontinuous multicellular pathway of adenosine (Ado), an immune suppressive nucleoside highly represented in the TME of MM patients (69).
The expression of immune checkpoint (ICP)/ICP-L contributes to the impairment of anti-myeloma immune responses. We have previously demonstrated that PD-L1 is expressed by BM MDSC in all disease states (2) and can contribute to hold in check anti-myeloma activity of PD1+ effector cells such as T cells, NK cells, and Vγ9Vδ2 T cells. Recently, it has been reported in solid tumors that MDSC can boost the immune suppressive activity of Bregs against T cells via the PD-1/PD-L1 axis (70, 71).
Lastly, MDSC can trans-differentiate into functional osteoclasts (72) to combine immune suppressive functions and direct protumoral functions (73). In mice models, G-MDSC have also been shown to promote angiogenesis (62), another major protumoral TME disruption occurring in human MM (62).
The direct and indirect mechanisms involved in the cross-talk between MDSC, myeloma cells, immune effector, immune suppressor cells, and other bystander cells in the TME of MM patients are shown in Figure 1.
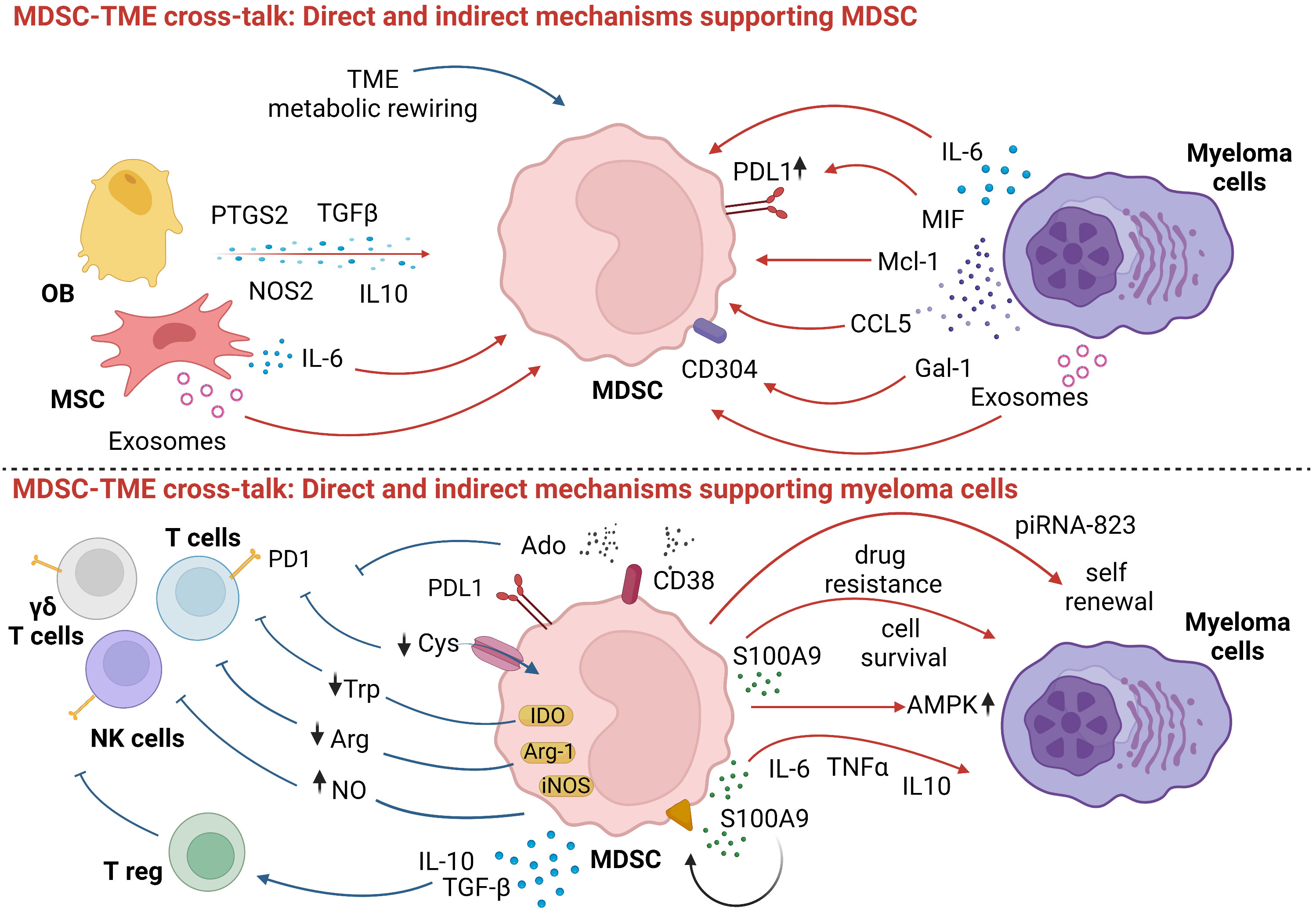
Figure 1 Myeloma cell and surrounding cells promote MDSC differentiation and suppressive functions. In turn, MDSC undermine anti-tumor immune responses and guarantee myeloma cells survival and growth. Red arrows: direct mechanisms; blue arrows: indirect mechanisms. Created by BioRender.com.
Therapeutic interventions
The correlation between the frequency of MDSC and the clinical outcome identifies these cells as potential targets of immune-based therapeutic interventions (74). However, the therapeutic targeting of MDSC is not easy given their multifaceted biological functions and multiple interactions in the TME. Possible strategies are: 1) to restrain their accumulation in the PB and TME; 2) to prevent their functional activation in the TME; 3) to block their protumoral interactions with myeloma cells and bystander cells.
MDSC accumulation can be restrained by immunomodulatory drugs (IMiDs) (44) and proteasome inhibitors (PI) (59). A cereblon (CRBN)-dependent and -independent down-regulation of CCL5 and MIF is a possible mechanism of IMiDs activity on MDSC (44) that can be improved by Arg-1 inhibitors (75). Clinical data confirm the capacity of IMiDs to restrain MDSC in vivo as shown by the decrease of PB MDSC in MM patients treated with pomalidomide, dexamethasone, and daratumumab (76). Daratumumab can also exert a favourable immune modulatory activity in the TME of MM patients by depleting CD38+ MDSC via antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytoxicity (CDC) (68). Data from mice models indicate that demethylating agents like decitabine (DAC), IL-18, and zoledronic acid (ZA) can also affect MDSC survival in the TME (72, 77, 78). ZA is currently used in MM and other solid cancers to prevent osteoclast activation and bone lesions, but this molecule is also endowed with pleiotropic immune modulatory activity (79), including the capacity in murine models to reduce the numbers of MDSC and prevent their differentiation into osteoclasts (72). Targeting CD84 and the CD304-Gal1 axis are other strategies used in myeloma mouse models to restore anti-myeloma T-cell responses by reducing MDSC accumulation and PD-L1 expression (45).
The immune suppressive activity of MM MDSC has also been inhibited in vitro using ABR-238901, a small molecule inhibiting S100A9 interactions (60), and tasquinimod (74). Anti-estrogen therapy may also restrain MDSC suppressive activity, since 17β-estradiol increases their ability to suppress T-cell proliferation (80). iNOS and Arg-1 activities have been down-modulated in mice models with tadalafil (81), a PDE5 inhibitor that has been used with some evidence of clinical efficacy in relapsed/refractory MM patients in combination with lenalidomide (82). Protumoral MDSC cellular interactions in the TME can also be limited by interrupting ICP/ICP-L interactions (2). Daratumumab in combination with the anti-PD1 monoclonal antibody cetrelimab has been reported to decrease the number of circulating MDSC and increase that of CD8+ T cells in the PB of MM patients in relapse (83). In acute myeloid leukemia (AML), knockdown of VISTA, a negative checkpoint regulator in the B7 family, reduced the MDSC-mediated inhibition of T cells (84). Data are not available in MM yet, but VISTA up-regulation is also expected in the BM of MM given the hypoxia and low pH as reported in solid cancer (85).
In conclusion, understanding the mechanisms underlying the immune suppressive activity of MDSC in MM will pave the ground to the therapeutic targeting of these cells to improve the efficacy of immune-based treatments in MM.
Author contributions
CG, FA and MM contributed to the writing of the manuscript, CG designed the figure, MM revised the manuscript.
Funding
This study received funding from the Italian Association for Cancer Research (AIRC) (IG21744 to MM), Associazione Italiana contro le Leucemie-Linfomi e Mielomi ONLUS (AIL) (Sezione di Cuneo “Paolo Rubino”) (MM, FA), and Sanofi (Research-to-Care OncoHematology).
Conflict of interest
MM reports advisory boards for AbbVie, Janssen-Cilag, Sanofi, and research funding from Sanofi.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. García-Ortiz A, Rodríguez-García Y, Encinas J, Maroto-Martín E, Castellano E, Teixidó J, et al. The role of tumor microenvironment in multiple myeloma development and progression. Cancers (2021) 13(2):217. doi: 10.3390/cancers13020217
2. Castella B, Foglietta M, Sciancalepore P, Rigoni M, Coscia M, Griggio V, et al. Anergic bone marrow Vγ9Vδ2 T cells as early and long-lasting markers of PD-1-targetable microenvironment-induced immune suppression in human myeloma. Oncoimmunology. (2015) 4(11):e1047580. doi: 10.1080/2162402X.2015.1047580
3. Zavidij O, Haradhvala NJ, Mouhieddine TH, Sklavenitis-Pistofidis R, Cai S, Reidy M, et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat Cancer (2020) 1(5):493–506. doi: 10.1038/s43018-020-0053-3
4. Leone P, Solimando AG, Malerba E, Fasano R, Buonavoglia A, Pappagallo F, et al. Actors on the scene: Immune cells in the myeloma niche. Front Oncol (2020) 10:599098. doi: 10.3389/fonc.2020.599098
5. Petersson J, Askman S, Pettersson Å, Wichert S, Hellmark T, Johansson ÅCM, et al. Bone marrow neutrophils of multiple myeloma patients exhibit myeloid-derived suppressor cell activity. J Immunol Res (2021) 2021:6344344. doi: 10.1155/2021/6344344
6. Ramachandran IR, Martner A, Pisklakova A, Condamine T, Chase T, Vogl T, et al. Myeloid-derived suppressor cells regulate growth of multiple myeloma by inhibiting T cells in bone marrow. J Immunol (2013) 190(7):3815–23. doi: 10.4049/jimmunol.1203373
7. Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol (2018) 19(2):108–19. doi: 10.1038/s41590-017-0022-x
8. Mortezaee K. Myeloid-derived suppressor cells in cancer immunotherapy-clinical perspectives. Life Sci (2021) 277:119627. doi: 10.1016/j.lfs.2021.119627
9. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol (2021) 21(8):485–98. doi: 10.1038/s41577-020-00490-y
10. Yen BL, Yen ML, Hsu PJ, Liu KJ, Wang CJ, Bai CH, et al. Multipotent human mesenchymal stromal cells mediate expansion of myeloid-derived suppressor cells via hepatocyte growth factor/c-met and STAT3. Stem Cell Rep (2013) 1(2):139–51. doi: 10.1016/j.stemcr.2013.06.006
11. Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol (2011) 32(1):19–25. doi: 10.1016/j.it.2010.10.002
12. Obermajer N, Wong JL, Edwards RP, Odunsi K, Moysich K, Kalinski P. PGE(2)-driven induction and maintenance of cancer-associated myeloid-derived suppressor cells. Immunol Invest (2012) 41(6-7):635–57. doi: 10.3109/08820139.2012.695417
13. Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res (2010) 70(1):68–77. doi: 10.1158/0008-5472.CAN-09-2587
14. Yu J, Wang Y, Yan F, Zhang P, Li H, Zhao H, et al. Noncanonical NF-κB activation mediates STAT3-stimulated IDO upregulation in myeloid-derived suppressor cells in breast cancer. J Immunol (2014) 193(5):2574–86. doi: 10.4049/jimmunol.1400833
15. Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A, et al. T Cell apoptosis by tryptophan catabolism. Cell Death Differ (2002) 9(10):1069–77. doi: 10.1038/sj.cdd.4401073
16. Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol (2006) 176(11):6752–61. doi: 10.4049/jimmunol.176.11.6752
17. Terness P, Bauer TM, Röse L, Dufter C, Watzlik A, Simon H, et al. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J Exp Med (2002) 196(4):447–57. doi: 10.1084/jem.20020052
18. Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol (2010) 185(6):3190–8. doi: 10.4049/jimmunol.0903670
19. Matos A, Carvalho M, Bicho M, Ribeiro R. Arginine and arginases modulate metabolism, tumor microenvironment and prostate cancer progression. Nutrients. (2021) 13(12):4503. doi: 10.3390/nu13124503
20. Raber PL, Thevenot P, Sierra R, Wyczechowska D, Halle D, Ramirez ME, et al. Subpopulations of myeloid-derived suppressor cells impair T cell responses through independent nitric oxide-related pathways. Int J Cancer (2014) 134(12):2853–64. doi: 10.1002/ijc.28622
21. Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, et al. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med (2007) 13(7):828–35. doi: 10.1038/nm1609
22. Stiff A, Trikha P, Mundy-Bosse B, McMichael E, Mace TA, Benner B, et al. Nitric oxide production by myeloid-derived suppressor cells plays a role in impairing fc receptor-mediated natural killer cell function. Clin Cancer Res (2018) 24(8):1891–904. doi: 10.1158/1078-0432.CCR-17-0691
23. Smith LK, Boukhaled GM, Condotta SA, Mazouz S, Guthmiller JJ, Vijay R, et al. Interleukin-10 directly inhibits CD8+ T cell function by enhancing n-glycan branching to decrease antigen sensitivity. Immunity. (2018) 48(2):299–312.e5. doi: 10.1016/j.immuni.2018.01.006
24. Vladimirovna IL, Sosunova E, Nikolaev A, Nenasheva T. Mesenchymal stem cells and myeloid derived suppressor cells: Common traits in immune regulation. J Immunol Res (2016) 2016:7121580. doi: 10.1155/2016/7121580
25. Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, et al. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res (2006) 66(2):1123–31. doi: 10.1158/0008-5472.CAN-05-1299
26. Zhu J, Powis de Tenbossche CG, Cané S, Colau D, van Baren N, Lurquin C, et al. Resistance to cancer immunotherapy mediated by apoptosis of tumor-infiltrating lymphocytes. Nat Commun (2017) 8(1):1404. doi: 10.1038/s41467-017-00784-1
27. Mortezaee K, Majidpoor J, Najafi S. VISTA immune regulatory effects in bypassing cancer immunotherapy: Updated. Life Sci (2022) 310:121083. doi: 10.1016/j.lfs.2022.121083
28. Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med (2014) 6(237):237ra67. doi: 10.1126/scitranslmed.3007974
29. Russell BL, Sooklal SA, Malindisa ST, Daka LJ, Ntwasa M. The tumor microenvironment factors that promote resistance to immune checkpoint blockade therapy. Front Oncol (2021) 11:641428. doi: 10.3389/fonc.2021.641428
30. Belisario DC, Kopecka J, Pasino M, Akman M, De Smaele E, Donadelli M, et al. Hypoxia dictates metabolic rewiring of tumors: Implications for chemoresistance. Cells. (2020) 9(12):2598. doi: 10.3390/cells9122598
31. Jian SL, Chen WW, Su YC, Su YW, Chuang TH, Hsu SC, et al. Glycolysis regulates the expansion of myeloid-derived suppressor cells in tumor-bearing hosts through prevention of ROS-mediated apoptosis. Cell Death Dis (2017) 8(5):e2779. doi: 10.1038/cddis.2017.192
32. Li W, Tanikawa T, Kryczek I, Xia H, Li G, Wu K, et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab (2018) 28(1):87–103.e6. doi: 10.1016/j.cmet.2018.04.022
33. Fu C, Fu Z, Jiang C, Xia C, Zhang Y, Gu X, et al. CD205+ polymorphonuclear myeloid-derived suppressor cells suppress antitumor immunity by overexpressing GLUT3. Cancer Sci (2021) 112(3):1011–25. doi: 10.1111/cas.14783
34. Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, et al. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol Res (2015) 3(11):1236–47. doi: 10.1158/2326-6066.CIR-15-0036
35. Mohammadpour H, MacDonald CR, McCarthy PL, Abrams SI, Repasky EA. β2-adrenergic receptor signaling regulates metabolic pathways critical to myeloid-derived suppressor cell function within the TME. Cell Rep (2021) 37(4):109883. doi: 10.1016/j.celrep.2021.109883
36. Brimnes MK, Vangsted AJ, Knudsen LM, Gimsing P, Gang AO, Johnsen HE, et al. Increased level of both CD4+FOXP3+ regulatory T cells and CD14+HLA-DR⁻/low myeloid-derived suppressor cells and decreased level of dendritic cells in patients with multiple myeloma. Scand J Immunol (2010) 72(6):540–7. doi: 10.1111/j.1365-3083.2010.02463.x
37. Wang Z, Zhang L, Wang H, Xiong S, Li Y, Tao Q, et al. Tumor-induced CD14+HLA-DR (-/low) myeloid-derived suppressor cells correlate with tumor progression and outcome of therapy in multiple myeloma patients. Cancer Immunol Immunother (2015) 64(3):389–99. doi: 10.1007/s00262-014-1646-4
38. Perez C, Botta C, Zabaleta A, Puig N, Cedena MT, Goicoechea I, et al. Immunogenomic identification and characterization of granulocytic myeloid-derived suppressor cells in multiple myeloma. Blood. (2020) 136(2):199–209. doi: 10.1182/blood.2019004537
39. Ara T, Declerck YA. Interleukin-6 in bone metastasis and cancer progression. Eur J Cancer (2010) 46(7):1223–31. doi: 10.1016/j.ejca.2010.02.026
40. Harmer D, Falank C, Reagan MR. Interleukin-6 interweaves the bone marrow microenvironment, bone loss, and multiple myeloma. Front Endocrinol (2019) 9:788. doi: 10.3389/fendo.2018.00788
41. Weber R, Groth C, Lasser S, Arkhypov I, Petrova V, Altevogt P, et al. IL-6 as a major regulator of MDSC activity and possible target for cancer immunotherapy. Cell Immunol (2021) 359:104254. doi: 10.1016/j.cellimm.2020.104254
42. De Veirman K, Van Ginderachter JA, Lub S, De Beule N, Thielemans K, Bautmans I, et al. Multiple myeloma induces mcl-1 expression and survival of myeloid-derived suppressor cells. Oncotarget. (2015) 6(12):10532–47. doi: 10.18632/oncotarget.3300
43. Lim JY, Kim TW, Ryu DB, Park SS, Lee SE, Kim BS, et al. Myeloma-secreted galectin-1 potently interacts with CD304 on monocytic myeloid-derived suppressor cells. Cancer Immunol Res (2021) 9(5):503–13. doi: 10.1158/2326-6066.CIR-20-0663
44. Kuwahara-Ota S, Shimura Y, Steinebach C, Isa R, Yamaguchi J, Nishiyama D, et al. Lenalidomide and pomalidomide potently interfere with induction of myeloid-derived suppressor cells in multiple myeloma. Br J Haematol (2020) 191(5):784–95. doi: 10.1111/bjh.16881
45. Lewinsky H, Gunes EG, David K, Radomir L, Kramer MP, Pellegrino B, et al. CD84 is a regulator of the immunosuppressive microenvironment in multiple myeloma. JCI Insight (2021) 6(4):e141683. doi: 10.1172/jci.insight.141683
46. Wang J, De Veirman K, Faict S, Frassanito MA, Ribatti D, Vacca A, et al. Multiple myeloma exosomes establish a favourable bone marrow microenvironment with enhanced angiogenesis and immunosuppression. J Pathol (2016) 239(2):162–73. doi: 10.1002/path.4712
47. Liu Z, Liu H, Li Y, Shao Q, Chen J, Song J, et al. Multiple myeloma-derived exosomes inhibit osteoblastic differentiation and improve IL-6 secretion of BMSCs from multiple myeloma. J Investig Med (2020) 68(1):45–51. doi: 10.1136/jim-2019-001010
48. Piddock RE, Marlein CR, Abdul-Aziz A, Shafat MS, Auger MJ, Bowles KM, et al. Myeloma-derived macrophage inhibitory factor regulates bone marrow stromal cell-derived IL-6 via c-MYC. J Hematol Oncol (2018) 11(1):66. doi: 10.1186/s13045-018-0614-4
49. Wang J, De Veirman K, De Beule N, Maes K, De Bruyne E, Van Valckenborgh E, et al. The bone marrow microenvironment enhances multiple myeloma progression by exosome-mediated activation of myeloid-derived suppressor cells. Oncotarget. (2015) 6(41):43992–4004. doi: 10.18632/oncotarget.6083
50. Giallongo C, Tibullo D, Parrinello NL, La Cava P, Di Rosa M, Bramanti V, et al. Granulocyte-like myeloid derived suppressor cells (G-MDSC) are increased in multiple myeloma and are driven by dysfunctional mesenchymal stem cells (MSC). Oncotarget. (2016) 7(52):85764–75. doi: 10.18632/oncotarget.7969
51. Xu X, Zhang C, Trotter TN, Gowda PS, Lu Y, Ponnazhagan S, et al. Runx2 deficiency in osteoblasts promotes myeloma progression by altering the bone microenvironment at new bone sites. Cancer Res (2020) 80(5):1036–48. doi: 10.1158/0008-5472.CAN-19-0284
52. Gavriatopoulou M, Paschou SA, Ntanasis-Stathopoulos I, Dimopoulos MA. Metabolic disorders in multiple myeloma. Int J Mol Sci (2021) 22(21):11430. doi: 10.3390/ijms222111430
53. Uckun FM. Overcoming the immunosuppressive tumor microenvironment in multiple myeloma. Cancers (Basel) (2021) 13(9):2018. doi: 10.3390/cancers13092018
54. Castella B, Riganti C, Massaia M. Metabolic approaches to rescue antitumor Vγ9Vδ2 T-cell functions in myeloma. Front Biosci (2020) 25(1):69–105. doi: 10.2741/4795
55. Zhao JL, Ye YC, Gao CC, Wang L, Ren KX, Jiang R, et al. Notch-mediated lactate metabolism regulates MDSC development through the Hes1/MCT2/c-jun axis. Cell Rep (2022) 38(10):110451. doi: 10.1016/j.celrep.2022.110451
56. Favaloro J, Liyadipitiya T, Brown R, Yang S, Suen H, Woodland N, et al. Myeloid derived suppressor cells are numerically, functionally and phenotypically different in patients with multiple myeloma. Leuk Lymphoma (2014) 55(12):2893–900. doi: 10.3109/10428194.2014.904511
57. Görgün GT, Whitehill G, Anderson JL, Hideshima T, Maguire C, Laubach J, et al. Tumor-promoting immune-suppressive myeloid-derived suppressor cells in the multiple myeloma microenvironment in humans. Blood. (2013) 121(15):2975–87. doi: 10.1182/blood-2012-08-448548
58. Ai L, Mu S, Sun C, Fan F, Yan H, Qin Y, et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol Cancer (2019) 18(1):88. doi: 10.1186/s12943-019-1011-5
59. De Veirman K, Menu E, Maes K, De Beule N, De Smedt E, Maes A, et al. Myeloid-derived suppressor cells induce multiple myeloma cell survival by activating the AMPK pathway. Cancer Lett (2019) 442:233–41. doi: 10.1016/j.canlet.2018.11.002
60. De Veirman K, De Beule N, Maes K, Menu E, De Bruyne E, De Raeve H, et al. Extracellular S100A9 protein in bone marrow supports multiple myeloma survival by stimulating angiogenesis and cytokine secretion. Cancer Immunol Res (2017) 5(10):839–46. doi: 10.1158/2326-6066.CIR-17-0192
61. Meng L, Tang Q, Zhao J, Wang Z, Wei L, Wei Q, et al. S100A9 derived from myeloma associated myeloid cells promotes TNFSF13B/TNFRSF13B-dependent proliferation and survival of myeloma cells. Front Oncol (2021) 11:691705. doi: 10.3389/fonc.2021.691705
62. Binsfeld M, Muller J, Lamour V, De Veirman K, De Raeve H, Bellahcène A, et al. Granulocytic myeloid-derived suppressor cells promote angiogenesis in the context of multiple myeloma. Oncotarget. (2016) 7(25):37931–43. doi: 10.18632/oncotarget.9270
63. Grzywa TM, Sosnowska A, Matryba P, Rydzynska Z, Jasinski M, Nowis D, et al. Myeloid cell-derived arginase in cancer immune response. Front Immunol (2020) 11:938. doi: 10.3389/fimmu.2020.00938
64. Van Valckenborgh E, Schouppe E, Movahedi K, De Bruyne E, Menu E, De Baetselier P, et al. Multiple myeloma induces the immunosuppressive capacity of distinct myeloid-derived suppressor cell subpopulations in the bone marrow. Leukemia. (2012) 26(11):2424–8. doi: 10.1038/leu.2012.113
65. Hammami I, Chen J, Murschel F, Bronte V, De Crescenzo G, Jolicoeur M. Immunosuppressive activity enhances central carbon metabolism and bioenergetics in myeloid-derived suppressor cells in vitro models. BMC Cell Biol (2012) 13:18. doi: 10.1186/1471-2121-13-18
66. Wu WC, Sun HW, Chen J, OuYang HY, Yu XJ, Chen HT, et al. Immunosuppressive immature myeloid cell generation is controlled by glutamine metabolism in human cancer. Cancer Immunol Res (2019) 7(10):1605–18. doi: 10.1158/2326-6066.CIR-18-0902
67. Malek E, de Lima M, Letterio JJ, Kim BG, Finke JH, Driscoll JJ, et al. Myeloid-derived suppressor cells: The green light for myeloma immune escape. Blood Rev (2016) 30(5):341–8. doi: 10.1016/j.blre.2016.04.002
68. Krejcik J, Casneuf T, Nijhof IS, Verbist B, Bald J, Plesner T, et al. Daratumumab depletes CD38+ immune regulatory cells, promotes T-cell expansion, and skews T-cell repertoire in multiple myeloma. Blood. (2016) 128(3):384–94. doi: 10.1182/blood-2015-12-687749
69. Horenstein AL, Bracci C, Morandi F, Malavasi F. CD38 in adenosinergic pathways and metabolic re-programming in human multiple myeloma cells: In-tandem insights from basic science to therapy. Front Immunol (2019) 10:760. doi: 10.3389/fimmu.2019.00760
70. Lee-Chang C, Rashidi A, Miska J, Zhang P, Pituch KC, Hou D, et al. Myeloid-derived suppressive cells promote b cell-mediated immunosuppression via transfer of PD-L1 in glioblastoma. Cancer Immunol Res (2019) 7(12):1928–43. doi: 10.1158/2326-6066.CIR-19-0240
71. Liu M L, Wei F, Wang J, Yu W, Shen M, Liu T, et al. Myeloid-derived suppressor cells regulate the immunosuppressive functions of PD-1-PD-L1+ bregs through PD-L1/PI3K/AKT/NF-κB axis in breast cancer. Cell Death Dis (2021) 12(5):465. doi: 10.1038/s41419-021-03745-1
72. Zhuang J, Zhang J, Lwin ST, Edwards JR, Edwards CM, Mundy GR, et al. Osteoclasts in multiple myeloma are derived from gr-1+CD11b+myeloid-derived suppressor cells. PloS One (2012) 7(11):e48871. doi: 10.1371/journal.pone.0048871
73. An G, Acharya C, Feng X, Wen K, Zhong M, Zhang L, et al. Osteoclasts promote immune suppressive microenvironment in multiple myeloma: therapeutic implication. Blood. (2016) 128(12):1590–603. doi: 10.1182/blood-2016-03-707547
74. Fan R, De Beule N, Maes A, De Bruyne E, Menu E, Vanderkerken K, et al. The prognostic value and therapeutic targeting of myeloid-derived suppressor cells in hematological cancers. Front Immunol (2022) 13:1016059. doi: 10.3389/fimmu.2022.1016059
75. Romano A, Parrinello NL, La Cava P, Tibullo D, Giallongo C, Camiolo G, et al. PMN-MDSC and arginase are increased in myeloma and may contribute to resistance to therapy. Expert Rev Mol Diagn (2018) 18(7):675–83. doi: 10.1080/14737159.2018.1470929
76. Pierceall WE, Amatangelo MD, Bahlis NJ, Siegel DS, Rahman A, Van Oekelen O, et al. Immunomodulation in pomalidomide, dexamethasone, and daratumumab-treated patients with Relapsed/Refractory multiple myeloma. Clin Cancer Res (2020) 26(22):5895–902. doi: 10.1158/1078-0432.CCR-20-1781
77. Zhou J, Shen Q, Lin H, Hu L, Li G, Zhang X. Decitabine shows potent anti-myeloma activity by depleting monocytic myeloid-derived suppressor cells in the myeloma microenvironment. J Cancer Res Clin Oncol (2019) 145(2):329–36. doi: 10.1007/s00432-018-2790-6
78. Nakamura K, Kassem S, Cleynen A, Chrétien ML, Guillerey C, Putz EM, et al. Dysregulated IL-18 is a key driver of immunosuppression and a possible therapeutic target in the multiple myeloma microenvironment. Cancer Cell (2018) 33(4):634–648.e5. doi: 10.1016/j.ccell.2018.02.007
79. Castella B, Vitale C, Coscia M, Massaia M. Vγ9Vδ2 T cell-based immunotherapy in hematological malignancies: from bench to bedside. Cell Mol Life Sci (2011) 68(14):2419–32. doi: 10.1007/s00018-011-0704-8
80. Ozerova M, Nefedova Y. Estrogen promotes multiple myeloma through enhancing the immunosuppressive activity of MDSC. Leuk Lymphoma (2019) 60(6):1557–62. doi: 10.1080/10428194.2018.1538511
81. Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med (2006) 203(12):2691–702. doi: 10.1084/jem.20061104
82. Noonan KA, Ghosh N, Rudraraju L, Bui M, Borrello I. Targeting immune suppression with PDE5 inhibition in end-stage multiple myeloma. Cancer Immunol Res (2014) 2(8):725–31. doi: 10.1158/2326-6066.CIR-13-0213
83. Cohen YC, Oriol A, Wu KL, Lavi N, Vlummens P, Jackson C, et al. Daratumumab with cetrelimab, an anti-PD-1 monoclonal antibody, in Relapsed/Refractory multiple myeloma. Clin Lymphoma Myeloma Leuk (2021) 21(1):46–54.e4. doi: 10.1016/j.clml.2020.08.008
84. Wang L, Jia B, Claxton DF, Ehmann WC, Rybka WB, Mineishi S, et al. VISTA is highly expressed on MDSCs and mediates an inhibition of T cell response in patients with AML. Oncoimmunology. (2018) 7(9):e1469594. doi: 10.1080/2162402X.2018.1469594
Keywords: MDSC (myeloid-derived suppressor cell), TME (tumor microenvironment), multiple myeloma, Immunothearpies, immune suppression
Citation: Giannotta C, Autino F and Massaia M (2023) The immune suppressive tumor microenvironment in multiple myeloma: The contribution of myeloid-derived suppressor cells. Front. Immunol. 13:1102471. doi: 10.3389/fimmu.2022.1102471
Received: 18 November 2022; Accepted: 27 December 2022;
Published: 16 January 2023.
Edited by:
Alessandra Romano, University of Catania, ItalyReviewed by:
Markus Munder, Johannes Gutenberg University Mainz, GermanyCopyright © 2023 Giannotta, Autino and Massaia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimo Massaia, bWFzc2ltby5tYXNzYWlhQHVuaXRvLml0
 Claudia Giannotta
Claudia Giannotta Federica Autino
Federica Autino Massimo Massaia
Massimo Massaia