
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 28 October 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1057892
This article is part of the Research Topic Standards in Personalized Phage Therapy: from phage collection to phage production View all 7 articles
 Kosuke Fujimoto1,2
Kosuke Fujimoto1,2 Satoshi Uematsu1,2*
Satoshi Uematsu1,2*Clostridioides difficile is endemic in the intestinal tract of healthy people. However, it is responsible for many healthcare-associated infections, such as nosocomial diarrhea following antibiotic treatment. Importantly, there have been cases of unsuccessful treatment and relapse related to the emergence of highly virulent strains of C. difficile and resistance to antimicrobial agents. Fecal microbiota transplantation (FMT) is considered an effective therapy for recurrent C. difficile infection. However, its safety is of concern because deaths caused by antibiotic-resistant bacterial infections after FMT were reported. Therefore, the development of effective C. difficile-specific treatments is urgently needed. In this review, we summarize the importance of phage therapy against C. difficile, and describe a novel next-generation phage therapy developed using metagenomic data.
The intestinal tract is constantly exposed to commensal microbiota and food antigens. Recently, great advances in intestinal microbial analyses have identified disease-associated alterations of the intestinal environment, such as dysbiosis, and research on the intestinal microbiome is progressing rapidly worldwide. Under dysbiosis, some symbiotic commensal bacteria acquire virulence traits, proliferate, and become directly involved in the development and progression of disease (1). These bacteria are referred to as ‘‘pathobionts,’’ which are distinct from opportunistic pathogens.
Clostridioides difficile (formally Clostridium difficile), a Gram-positive, spore-forming anaerobic bacterium, was first reported as a pathogen in 1978 (2), and is the representative pathobiont of nosocomial diarrhea following antibiotic treatment. C. difficile has three well-characterized toxins (toxin A, toxin B, and binary toxin) at two distinct loci. Whereas toxin A and toxin B are associated with primary virulence factors for C. difficile infection (CDI), binary toxin is involved in the increased severity of CDI. The production of the three toxins was shown to be strain-specific. Interestingly, C. difficile is frequently encountered in the gut microbiota of healthy individuals without any symptoms of disease (3). However, dysbiosis induced by antibiotic usage or malnutrition can initiate CDI with increasing C. difficile toxin concentrations, and C. difficile colonization and biofilm formation in the gut, resulting in severe disease status (4). C. difficile spores are highly resistant to environmental stresses such as high temperature, antibiotics, and disinfectants, which allows C. difficile to spread easily between residents of elderly care facilities (5).
In cases of mild CDI, withdrawing broad spectrum antibiotics such as ampicillin, clindamycin, and third generation cephalosporins is a standard therapy. Also, for cases of severe and recurrent CDI, use of the antibiotics vancomycin and fidaxomicin is an effective treatment (6). Recently, however, FMT has become an alternative therapy (7) and was reported to be successful with a cure rate of approximately 93% in patients with recurrent CDI. This is now accepted as a valid alternative for those failing antibiotic treatment (8). However, two deaths caused by antibiotic-resistant bacterial infections after FMT have been reported (9, 10), suggesting that a modification of FMT or alternative treatments are required to resolve safety concerns about FMT.
Although we are currently facing an unprecedented pandemic caused by severe acute respiratory syndrome-coronavirus-2, we must not forget that the threat of drug-resistant bacteria, including in C. difficile, remains a significant problem. If this trend continues, it is predicted that infections caused by drug-resistant bacteria will be the leading cause of death by 2050 (11, 12). Phage therapy is considered an important development against drug-resistant bacteria, and expectations for phage therapy are growing worldwide (13). This review summarizes recent findings in our understanding of CDI, with a particular focus on phage therapy for C. difficile.
Bacteriophages are viruses that infect bacteria and exhibit bactericidal effects against Gram-positive and Gram-negative bacteria. In 1896, Ernest Hankin discovered “an antiseptic substance” in the water of the Ganges River that killed certain bacteria. Later, in 1915, Frederick Twort discovered “a transparent material” that changed the properties of Staphylococcus aureus, and in 1917 (14), Félix d’Hérelle named an “invisible microbe” that dissolved dysentery bacteria as a “bacteriophage”. Furthermore, d’Hérelle advocated “phage therapy” to treat bacterial infections, and practiced this in humans and animals, including those with dysentery and Vibrio cholerae. d’Hérelle met Giorgi Eliava at the Pasteur Institute, which led to the establishment of several institutes in Tbilisi, Georgia, between 1923 and 1936 with the support of the Soviet Union. One of these, the Eliava Institute, became an important focus for phage research and therapy, and remains the core research institute for phage research today. However, after the discovery of antibiotics, especially penicillin, in 1928 (15) and its subsequent clinical application, phage therapy was abandoned in Western countries. However, during the Cold War, phage therapy was developed as an alternative treatment for infectious diseases in the Soviet Union because of the lack of newly developed antibiotic drugs. Even today, phage cocktails are prescribed and used as an effective treatment for infectious diseases in Russia, Georgia, Poland, and other countries (16). The overuse of antibiotics has led to the emergence of various multidrug-resistant bacteria, which has become a serious medical problem worldwide. Therefore, phage therapy against multidrug-resistant bacteria is beginning to attract attention as a next-generation treatment method.
C. difficile-specific phages have been investigated for the regulation of C. difficile pathogenesis and development of new therapies for CDI. Many C. difficile-specific phages have been identified to date [phiC2 (17, 18), phiC5 (17), phiC6 (17), phiC8 (17), phiCD119 (19), phiCD24-1 (20), phiCD27 (21), phiCD6356 (22), phiCD6365 (22), phiCD38-2 (23), phiMMP01 (24, 25), phiMMP02 (24, 25), phiMMP03 (24, 25), phiMMP04 (24, 25), phiCD24-2 (25), phiCD146 (25), phiCD111 (25), phiCD526 (25), phiCD52 (25), phiCD481-1 (25), phiCD481-2 (25), phiCD505 (25), phiCD506 (25), phiCD508 (25), phiCDHM1 (26), phiCDHM2 (26), phiCDHM3 (26), phiCDHM4 (26), phiCDHM5 (26), phiCDHM6 (26), phiCDHS1 (26), CDKM9 (27), CDKM15 (27), phiSemix9P1 (28), phiCD5763 (29), phiCD5774 (29), phiCD2955 (29), phiCD211/phiCDIF1296T (30), phiHN10 (31), phiHN16-1 (31), phiHN16-2 (31), phiHN50 (31), JD032 (32), and phiCDKH01 (33)] (Supplementary Table S1). The host for all of these phages is C. difficile; however, some were shown to infect multiple C. difficile strains. In addition, the C. difficile-specific phages described above are lysogenic, not lytic, and they are classified in the Myoviridae or Siphoviridae subfamilies of Caudovirales. Because lytic phages against C. difficile should exist, the fact that C. difficile is anaerobic and forms spores makes it difficult to isolate lytic phages experimentally.
The application of phage targeting C. difficile should be considered. Indeed, the efficacy of phage monotherapy against C. difficile has been studied for the past decade (26, 34–37). Unfortunately, none of these studies have led to the development of effective treatments for CDI. Simply identifying and isolating C. difficile-specific phages by conventional methods using ultraviolet or mitomycin C has not led to successful phage therapy against C. difficile (17–23, 31). Therefore, it was considered necessary to develop a new lysis method that takes advantage of the unique properties of phages.
Endolysin, an enzyme encoded by phages, is used by mature phage virions to hydrolyze the bacterial cell wall from the inside. The worldwide increase in antibiotic-resistant bacteria has stimulated research on endolysins as an alternative therapeutic agent. Endolysins can degrade peptidoglycans from the outside, and also have the potential to lyse bacterial biofilms (38). Therefore, the medical applications of endolysin may provide new treatment options for Gram-positive bacterial infections.
Most endolysins derived from phages that infect Gram-positive hosts are modular (39). Their molecular weight is typically 15–40 kDa. They are characterized by one or two (multi-domain) N-terminal enzyme active domains (EAD) often linked to a C-terminal cell wall-binding domain (CBD) by a short flexible linker region. The N-terminal EAD of modular endolysins cleaves various specific peptidoglycan bonds in the host bacterial murein layer, whereas the C-terminal CBD recognizes and binds to different structures in the cell wall to properly anchor the catalytic effect of the EAD (Figure 1) (40). The endolysins of phages that infects Gram-positive host are structurally similar to those of fungal cellulases (41).
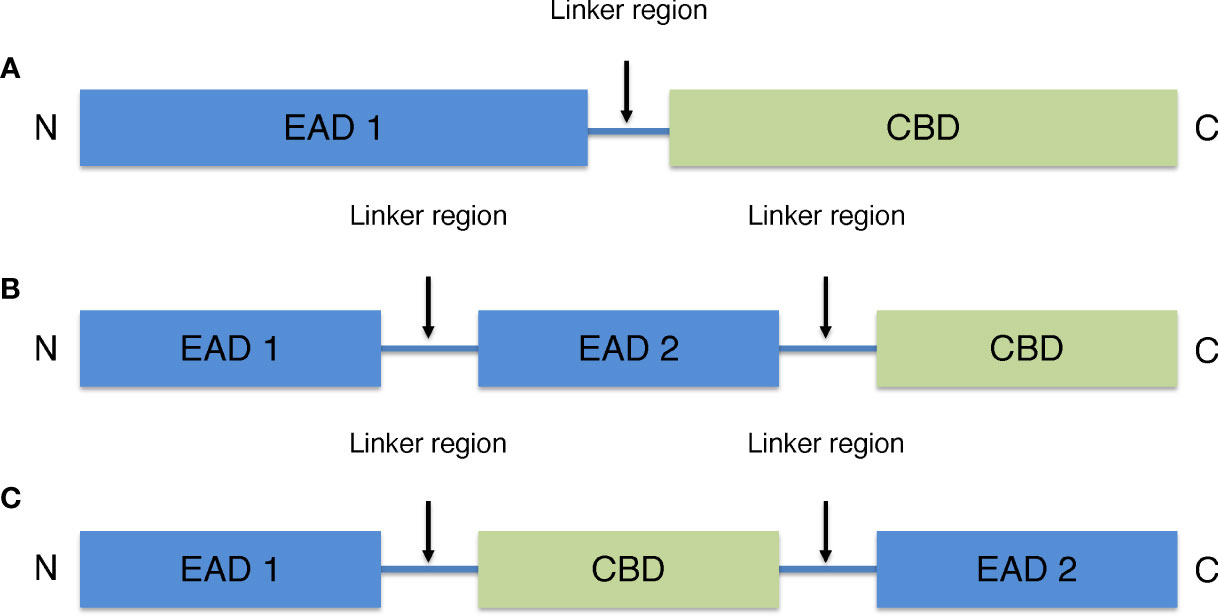
Figure 1 General schematic models of endolysins derived from phages that infects Gram-positive host. (A) Model with one N-terminal EAD and one C-terminal CBD. (B) Multi-domain model with two EAD and one C-terminal CBD. (C) Multi-domain model with one CBD located between two EAD.
There are four types of EAD including amidases, acetylmuramidases, endopeptidases, and glucosaminidases. Amidases break the amide bond between peptide and sugar chains. Acetylmuramidases break the N-acetylmuramoyl β1,4 N-acetylgulcosamine bond of sugar strands. Endopeptidases break the peptide crosslinking in the stem structure of peptidoglycans and glucosaminidases break the N-acetylmuramoyl β1,4 N-acetylmuramine glycosidic bond. Among endolysins, amidases and muramidases are the most common endolysins that target the highly conserved binding of peptidoglycans.
CBD binds to murein ligands and secondary cell wall polymers, which are cell wall components that include teichoic acid and natural polysaccharides of bacteria (42). The main function of CBD is to provide endolysins with host specificity for non-covalent binding to cell membrane ligands. For example, LysM, the most widely reported endolysin, binds to N-acetylglucosamine in the peptidoglycan glycan backbone (43, 44). The CBD motif in the endolysin of many phages that infects Gram-positive host might allow this domain to bind to ligands in the peptidoglycan layer, preventing endolysin diffusion and destroying the nearby host after cell lysis (39). Because EAD do not have specificity for bacterial cell walls, CBD are considered important in defining their specificity for target cells. Although many endolysins retain or increase bacteriolytic activity in the absence of CBD, host specificity is very important when endolysins are used as a treatment for multidrug-resistant bacteria, for example.
As well as other pathogens, the application of phage-derived endolysins targeting C. difficile has been explored [CD27L (21), phyCD (45), CDG (46), CD11 (46), LCD (47), and CWH (48)]. Among these, CD27L derived from phiCD27 was first identified as a C. difficile-specific endolysin. CD27L lyses 30 C. difficile strains, including two strains of the hypervirulent ribotype 027 (21). Importantly, a range of commensal species that inhabited the gastrointestinal tract was insensitive to the endolysin (21, 49), indicating that CD27L is not expected to induce gut dysbiosis. This endolysin might provide a platform for the generation of novel therapeutic agents to overcome C. difficile.
The future practice of phage therapy for CDI is promising. However, it is difficult to identify phages that have C. difficile as a host. Given the versatility of phage therapy for CDI, there is a need for a high-throughput system that can identify phages that infect C. difficile.
Recently, to develop phage therapies to control pathobionts, which are directly involved in the pathogenesis of disease, we established an effective method for obtaining genomic information on host bacteria–phage associations (50, 51). Metagenomic data obtained from the fecal samples of healthy subjects and clinical isolates of C. difficile strains were used to develop a phage therapy specific for C. difficile. The obtained sequencing data were used to search for novel phage-derived endolysins specific for C. difficile. Using the phage genome analysis pipeline, we identified several novel endolysin sequences from the prophage sequence of C. difficile. These endolysins were synthesized and shown to exhibit bacteriolytic activity in vitro and to be effective in a mouse model of CDI (50). This is a practical example of a next-generation phage therapy based on metagenomic information. To isolate phages used in phage therapy, it has been necessary to isolate and culture target bacteria. Phages could not be isolated for non-culturable bacteria in the intestine. Thus, this strategy is not limited to C. difficile and might be applied to various targeted bacteria in the future (Figure 2).
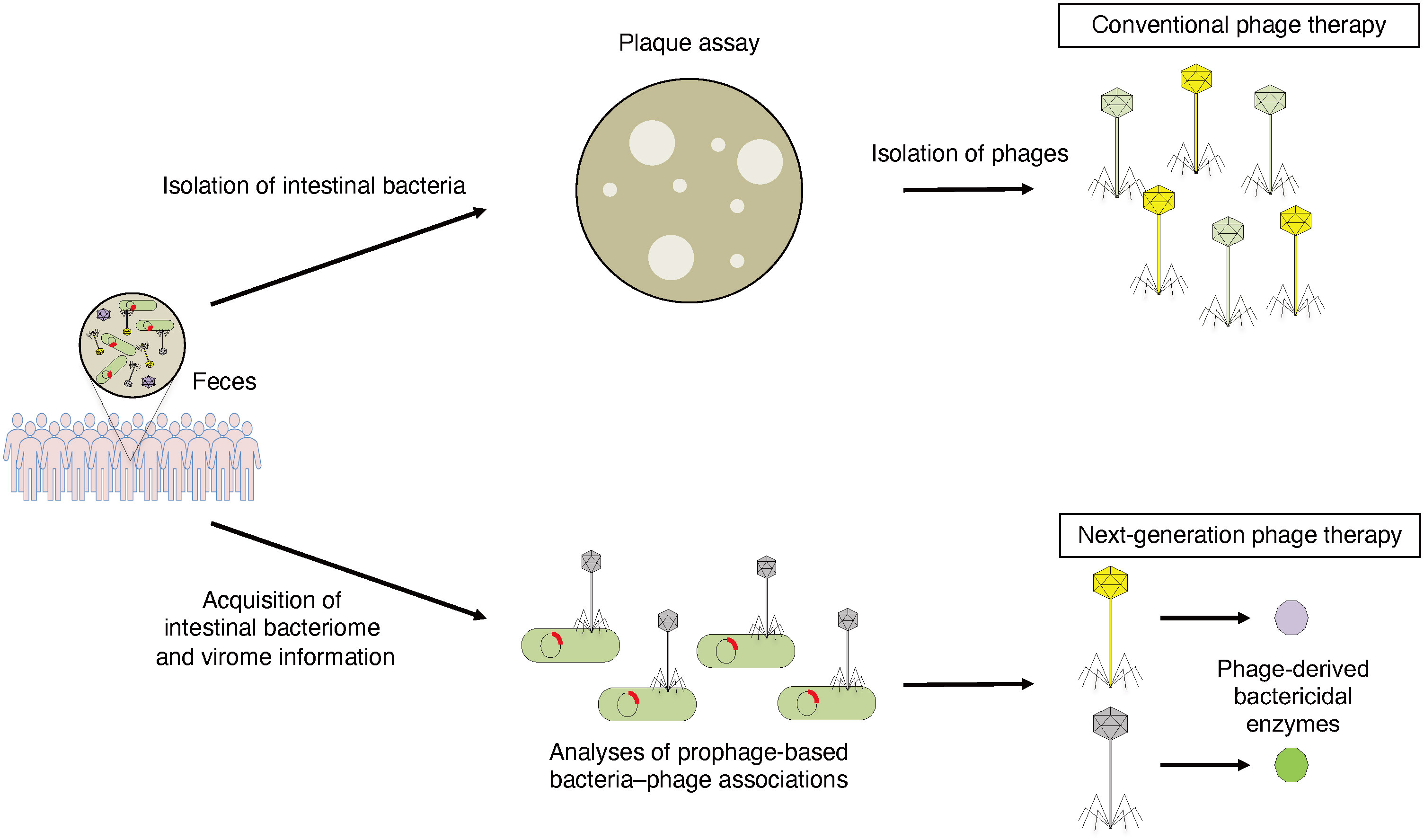
Figure 2 Conventional phage therapy and next-generation phage therapy based on metagenomic information. To isolate phages that can specifically control pathobionts, host bacteria is isolated from human fecal samples and plaque assay is performed. Isolated phages are used for phage therapy (conventional phage therapy). To detect phage-derived antibacterial enzymes that can specifically regulate pathobionts, intestinal bacterial and viral metagenomic information is acquired from human fecal samples. Phage-derived bactericidal enzymes can kill host bacteria specifically (next-generation phage therapy).
Scientific research on bacteriophages has become a hot topic because of the impending problem of multidrug-resistant bacteria. Because the development of antibiotics is not expected to progress as far as expected, it is hoped that phage therapy will become commercially available.
With improvements in genome analysis technology using next-generation sequencing, the analysis of intestinal microbiota has progressed dramatically, and its relationship to disease has gradually become clearer. It is now possible to analyze intestinal phages, which has been difficult in the past, and this is expected to become an extremely powerful analytical tool for the future practice of phage therapy, as well as leading to various industrial applications of phages. Recently, Federici et al. have demonstrated the feasibility of combination phage therapy for pathobionts associated with inflammatory bowel disease (52). It would be an interesting direction to further phage therapies for other intestinal bacteria-mediated diseases including CDI. In the near future, phage science will be developed further by integration with a wide range of fields including medicine, microbiology, bioinformatics, and synthetic biology.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This study was supported by the Japan Agency for Medical Research and Development (AMED) (to KF; 22ae0121048h0001 and to SU; 22fk0108619h0001), and Grant-in-Aid for Early-Career Scientists (to KF; 22K16329) from the JSPS.
We thank K. Ogawa, M. Maeda, and K. Suetsugu for administrative assistance. We thank Edanz Group (https://jp.edanz.com/ac) for editing a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1057892/full#supplementary-material
1. Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science (2012) 336(6086):1325–9. doi: 10.1126/science.1222195
2. Gerding DN. Clostridium difficile 30 years on: What has, or has not, changed and why? Int J Antimicrob Agents (2009) 33 Suppl 1:S2–8. doi: 10.1016/S0924-8579(09)70008-1
3. Adlerberth I, Huang H, Lindberg E, Aberg N, Hesselmar B, Saalman R, et al. Toxin-producing clostridium difficile strains as long-term gut colonizers in healthy infants. J Clin Microbiol (2014) 52(1):173–9. doi: 10.1128/JCM.01701-13
4. Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. Clostridium difficile infection: Toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes (2012) 3(2):121–34. doi: 10.4161/gmic.19399
5. Czepiel J, Drozdz M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, et al. Clostridium difficile infection: Review. Eur J Clin Microbiol Infect Dis (2019) 38(7):1211–21. doi: 10.1007/s10096-019-03539-6
6. Debast SB, Bauer MP, Kuijper EJ, European Society of Clinical M, Infectious D. European Society of clinical microbiology and infectious diseases: Update of the treatment guidance document for clostridium difficile infection. Clin Microbiol Infect (2014) 20 Suppl 2:1–26. doi: 10.1111/1469-0691.12418
7. McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for clostridium difficile infection in adults and children: 2017 update by the infectious diseases society of America (IDSA) and society for healthcare epidemiology of America (SHEA). Clin Infect Dis (2018) 66(7):987–94. doi: 10.1093/cid/ciy149
8. Kassam Z, Hundal R, Marshall JK, Lee CH. Fecal transplant via retention enema for refractory or recurrent clostridium difficile infection. Arch Intern Med (2012) 172(2):191–3. doi: 10.1001/archinte.172.2.191
9. DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, et al. Drug-resistant e. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med (2019) 381(21):2043–50. doi: 10.1056/NEJMoa1910437
10. Grosen AK, Povlsen JV, Lemming LE, Jorgensen SMD, Dahlerup JF, Hvas CL. Faecal microbiota transplantation eradicated extended-spectrum beta-Lactamase-Producing klebsiella pneumoniae from a renal transplant recipient with recurrent urinary tract infections. Case Rep Nephrol Dial (2019) 9(2):102–7. doi: 10.1159/000502336
11. de Kraker ME, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PloS Med (2016) 13(11):e1002184. doi: 10.1371/journal.pmed.1002184
12. Abat C, Fournier PE, Jimeno MT, Rolain JM, Raoult D. Extremely and pandrug-resistant bacteria extra-deaths: Myth or reality? Eur J Clin Microbiol Infect Dis (2018) 37(9):1687–97. doi: 10.1007/s10096-018-3300-0
13. Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe (2019) 25(2):219–32. doi: 10.1016/j.chom.2019.01.014
14. Abedon ST, Thomas-Abedon C, Thomas A, Mazure H. Bacteriophage prehistory: Is or is not hankin, 1896, a phage reference? Bacteriophage (2011) 1(3):174–8. doi: 10.4161/bact.1.3.16591
15. Hutchings MI, Truman AW, Wilkinson B. Antibiotics: Past, present and future. Curr Opin Microbiol (2019) 51:72–80. doi: 10.1016/j.mib.2019.10.008
16. Abedon ST, Garcia P, Mullany P, Aminov R. Editorial: Phage therapy: Past, present and future. Front Microbiol (2017) 8:981. doi: 10.3389/fmicb.2017.00981
17. Goh S, Riley TV, Chang BJ. Isolation and characterization of temperate bacteriophages of clostridium difficile. Appl Environ Microbiol (2005) 71(2):1079–83. doi: 10.1128/AEM.71.2.1079-1083.2005
18. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiol (Reading) (2007) 153(Pt 3):676–85. doi: 10.1099/mic.0.2006/002436-0
19. Govind R, Fralick JA, Rolfe RD. Genomic organization and molecular characterization of clostridium difficile bacteriophage PhiCD119. J Bacteriol (2006) 188(7):2568–77. doi: 10.1128/JB.188.7.2568-2577.2006
20. Fortier LC, Moineau S. Morphological and genetic diversity of temperate phages in clostridium difficile. Appl Environ Microbiol (2007) 73(22):7358–66. doi: 10.1128/AEM.00582-07
21. Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol (2008) 190(20):6734–40. doi: 10.1128/JB.00686-08
22. Horgan M, O'Sullivan O, Coffey A, Fitzgerald GF, van Sinderen D, McAuliffe O, et al. Genome analysis of the clostridium difficile phage PhiCD6356, a temperate phage of the siphoviridae family. Gene (2010) 462(1-2):34–43. doi: 10.1016/j.gene.2010.04.010
23. Sekulovic O, Meessen-Pinard M, Fortier LC. Prophage-stimulated toxin production in clostridium difficile NAP1/027 lysogens. J Bacteriol (2011) 193(11):2726–34. doi: 10.1128/JB.00787-10
24. Meessen-Pinard M, Sekulovic O, Fortier LC. Evidence of in vivo prophage induction during clostridium difficile infection. Appl Environ Microbiol (2012) 78(21):7662–70. doi: 10.1128/AEM.02275-12
25. Sekulovic O, Garneau JR, Neron A, Fortier LC. Characterization of temperate phages infecting clostridium difficile isolates of human and animal origins. Appl Environ Microbiol (2014) 80(8):2555–63. doi: 10.1128/AEM.00237-14
26. Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepinski P, Douce GR, et al. Bacteriophage combinations significantly reduce clostridium difficile growth In vitro and proliferation in vivo. Antimicrob Agents Chemother (2016) 60(2):968–81. doi: 10.1128/AAC.01774-15
27. Rashid SJ, Barylski J, Hargreaves KR, Millard AA, Vinner GK, Clokie MR. Two novel myoviruses from the north of Iraq reveal insights into clostridium difficile phage diversity and biology. Viruses (2016) 8(11). doi: 10.3390/v8110310
28. Riedel T, Wittmann J, Bunk B, Schober I, Sproer C, Gronow S, et al. A clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J Biotechnol (2017) 250:23–8. doi: 10.1016/j.jbiotec.2017.02.017
29. Ramirez-Vargas G, Goh S, Rodriguez C. The novel phages phiCD5763 and phiCD2955 represent two groups of big plasmidial siphoviridae phages of clostridium difficile. Front Microbiol (2018) 9:26. doi: 10.3389/fmicb.2018.00026
30. Garneau JR, Sekulovic O, Dupuy B, Soutourina O, Monot M, Fortier LC. High prevalence and genetic diversity of Large phiCD211 (phiCDIF1296T)-like prophages in clostridioides difficile. Appl Environ Microbiol (2018) 84(3):e02164–17. doi: 10.1128/AEM.02164-17
31. Phothichaisri W, Ounjai P, Phetruen T, Janvilisri T, Khunrae P, Singhakaew S, et al. Characterization of bacteriophages infecting clinical isolates of clostridium difficile. Front Microbiol (2018) 9:1701. doi: 10.3389/fmicb.2018.01701
32. Li T, Zhang Y, Dong K, Kuo CJ, Li C, Zhu YQ, et al. Isolation and characterization of the novel phage JD032 and global transcriptomic response during JD032 infection of clostridioides difficile ribotype 078. mSystems (2020) 5(3):e00017–20. doi: 10.1128/mSystems.00017-20
33. Hinc K, Kabala M, Iwanicki A, Martirosian G, Negri A, Obuchowski M. Complete genome sequence of the newly discovered temperate clostridioides difficile bacteriophage phiCDKH01 of the family siphoviridae. Arch Virol (2021) 166(8):2305–10. doi: 10.1007/s00705-021-05092-0
34. Meader E, Mayer MJ, Gasson MJ, Steverding D, Carding SR, Narbad A. Bacteriophage treatment significantly reduces viable clostridium difficile and prevents toxin production in an in vitro model system. Anaerobe (2010) 16(6):549–54. doi: 10.1016/j.anaerobe.2010.08.006
35. Meader E, Mayer MJ, Steverding D, Carding SR, Narbad A. Evaluation of bacteriophage therapy to control clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe (2013) 22:25–30. doi: 10.1016/j.anaerobe.2013.05.001
36. Nale JY, Chutia M, Carr P, Hickenbotham PT, Clokie MR. 'Get in early'; biofilm and wax moth (Galleria mellonella) models reveal new insights into the therapeutic potential of clostridium difficile bacteriophages. Front Microbiol (2016) 7:1383. doi: 10.3389/fmicb.2016.01383
37. Shan J, Ramachandran A, Thanki AM, Vukusic FBI, Barylski J, Clokie MRJ. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci Rep (2018) 8(1):5091. doi: 10.1038/s41598-018-23418-y
38. Jun SY, Jang IJ, Yoon S, Jang K, Yu KS, Cho JY, et al. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother (2017) 61(6):e02629–16. doi: 10.1128/AAC.02629-16
39. Oliveira H, Sao-Jose C, Azeredo J. Phage-derived peptidoglycan degrading enzymes: Challenges and future prospects for In vivo therapy. Viruses (2018) 10(6):292. doi: 10.3390/v10060292
40. Matamp N, Bhat SG. Phage endolysins as potential antimicrobials against multidrug resistant vibrio alginolyticus and vibrio parahaemolyticus: Current status of research and challenges ahead. Microorganisms (2019) 7(3):84. doi: 10.3390/microorganisms7030084
41. Fischetti VA. Development of phage lysins as novel therapeutics: A historical perspective. Viruses (2018) 10(6):310. doi: 10.3390/v10060310
42. Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol (2012) 7(10):1147–71. doi: 10.2217/fmb.12.97
43. Ohnuma T, Onaga S, Murata K, Taira T, Katoh E. LysM domains from pteris ryukyuensis chitinase-a: A stability study and characterization of the chitin-binding site. J Biol Chem (2008) 283(8):5178–87. doi: 10.1074/jbc.M707156200
44. Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun (2014) 5:4269. doi: 10.1038/ncomms5269
45. Wang Q, Euler CW, Delaune A, Fischetti VA. Using a novel lysin to help control clostridium difficile infections. Antimicrob Agents Chemother (2015) 59(12):7447–57. doi: 10.1128/AAC.01357-15
46. Mehta KK, Paskaleva EE, Wu X, Grover N, Mundra RV, Chen K, et al. Newly identified bacteriolytic enzymes that target a wide range of clinical isolates of clostridium difficile. Biotechnol Bioeng (2016) 113(12):2568–76. doi: 10.1002/bit.26029
47. Peng Z, Wang S, Gide M, Zhu D, Warnakulasuriya L, Patabendige HM, Li C, et al. A novel bacteriophage lysin-human defensin fusion protein is effective in treatment of clostridioides difficile infection in mice. Front Microbiol (2018) 9:3234. doi: 10.3389/fmicb.2018.03234
48. Mondal SI, Akter A, Draper LA, Ross RP, Hill C. Characterization of an endolysin targeting clostridioides difficile that affects spore outgrowth. Int J Mol Sci (2021) 22(11):5690. doi: 10.3390/ijms22115690
49. Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. Structure-based modification of a clostridium difficile-targeting endolysin affects activity and host range. J Bacteriol (2011) 193(19):5477–86. doi: 10.1128/JB.00439-11
50. Fujimoto K, Kimura Y, Shimohigoshi M, Satoh T, Sato S, Tremmel G, et al. Metagenome data on intestinal phage-bacteria associations aids the development of phage therapy against pathobionts. Cell Host Microbe (2020) 28(3):380–9.e9. doi: 10.1016/j.chom.2020.06.005
51. Fujimoto K, Kimura Y, Allegretti JR, Yamamoto M, Zhang YZ, Katayama K, et al. Functional restoration of bacteriomes and viromes by fecal microbiota transplantation. Gastroenterology (2021) 160(6):2089–102.e12. doi: 10.1053/j.gastro.2021.02.013
Keywords: Clostridioides difficile, bacteriophage, endolysin, enzyme active domain, cell wall-binding domain, metagenome
Citation: Fujimoto K and Uematsu S (2022) Phage therapy for Clostridioides difficile infection. Front. Immunol. 13:1057892. doi: 10.3389/fimmu.2022.1057892
Received: 30 September 2022; Accepted: 12 October 2022;
Published: 28 October 2022.
Edited by:
Yoshiyuki Goto, Chiba University, JapanCopyright © 2022 Fujimoto and Uematsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoshi Uematsu, dWVtYXRzdS5zYXRvc2hpQG9tdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.