- 1Department of Public Health, Medical College of Qinghai University, Xining, China
- 2Department of Infection Disease, Qinghai Center for Disease Prevention and Control, Xining, China
- 3The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China
- 4Department of Tuberculosis, Anhui Chest Hospital, Hefei, Anhui, China
The present study was performed to evaluate the association of WNT signaling pathway genes variants with pulmonary tuberculosis (PTB) risk in Chinese Han population. Our study subjects were composed of 452 PTB patients and 465 normal controls, and seventeen SNPs of seven genes in WNT signaling pathway (SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6) were genotyped by SNPscan technique. We found no significant relationship of SFRP1 rs10088390, rs4736958, rs3242, WNT3A rs752107, rs3121310, CTNNB1 rs2293303, rs1798802, rs4135385, WIF-1 rs1026024, rs3782499, DKK-1 rs2241529, rs1569198, LRP5 rs3736228, rs556442, LRP6 rs2302685, rs11054697, rs10743980 polymorphisms with PTB susceptibility. While, WIF-1 rs3782499 variant was associated with susceptibility to PTB under recessive model, and haplotype analysis showed that DKK-1 GA haplotype frequency was significantly increased in PTB patients. The WNT3A rs3121310, CTNNB1 rs2293303 polymorphisms were respectively associated with drug-induced liver injury (DILI), sputum smear-positive in PTB patients. The rs3782499 in WIF-1 gene was related to fever, leukopenia, and the rs1569198 in DKK-1 was linked to sputum smear-positive in PTB patients. In LRP5 gene, rs3736228, rs556442 variants respectively affected the occurrence of DILI, fever, and LRP6 gene rs2302685, rs10743980 variants respectively influenced the development of hypoproteinemia, sputum smear-positive in PTB patients. Our results revealed that WNT signaling pathway genes variation were not associated with the susceptibility to PTB, while WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6 genetic variations might be closely related to the occurrence of several clinical characteristics of PTB patients.
Introduction
Tuberculosis (TB) remains a leading infectious cause of morbidity and mortality caused by infection of Mycobacterium tuberculosis (MTB) in developing countries, with an estimated 9.9 million new cases worldwide in 2021 (1). Epidemiological studies had shown that more than one-third of the global population were infected with MTB, while only 5-10% of infections eventually developed into active TB (2, 3). This suggested that the progression of TB disease was affected environment elements, pathogenic features of MTB, and the host genetic factors (4, 5). Previous association studies had assessed some candidate loci that might be associated with TB susceptibility, including the genes encoding human leukocyte antigen, cytokines and chemokines (6, 7). However, these studies only investigated a few polymorphisms, and explained only part of TB heritability. On the other hand, pathway-based association studies based on the biological functions of related signaling pathway networks were considered to be an effective method to identify the pathogenic genes of complex diseases. Several molecular pathways that regulated immune response, such as JAK/STAT, MAPK, NF-KB signaling, had been the hot area in the studies regarding TB infection and susceptibility (8, 9).
The WNT signaling pathway was considered as an important transduction cascade modulating host immune responses against microbial pathogens, governing ontogeny and homeostatic processes (3). The canonical WNT signaling pathway was activated when WNT proteins bind to LRP5/6-Frizzled receptors and co-receptors, which caused β-catenin co-activating transcription factors to affect the downstream gene expression (10). Recent evidences implicated that the WNT pathway played an etiological role in the pathogenesis of TB. Wu et al. found that the activation of Wnt/β-catenin signaling could induce mycobacteria-infected cell apoptosis and promote the secretion of pro-inflammatory tumor necrosis factor (TNF)-α, interleukin (IL)-6 (11). Another study by Schaale et al. revealed that the mRNA expression of most WNT homologous were significantly decreased during MTB infection, and further suggested that WNT6 was expressed in granulomatous lesions and had a key role in macrophage differentiation and proliferation (12). The specific molecular mechanism of WNT signaling pathway in the pathogenesis of PTB had attracted extensive attention, but it was still poorly understood and needed further exploration.
Genetic variations in WNT signaling pathway genes were proved to be associated with susceptibility to a variety of human diseases, such as infectious diseases and cancer, implying that polymorphisms in the WNT pathway might also affect susceptibility to PTB (3, 13, 14). Hu et al. previously found multiple single nucleotide polymorphisms (SNP) in WNT pathway genes were associated with PTB susceptibility in Western Chinese (3). It was important to note that additional studies were needed to validate these results, however, there were few studies on the epidemiological genetic association between WNT signaling pathway genes and PTB susceptibility. In order to explore the potential roles of WNT pathway genetic polymorphisms in PTB, this study was designed to evaluate the associations of some SNPs from seven key genes in WNT signaling pathway (SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6) with PTB susceptibility in a Chinese population.
Materials and methods
Study participants
In the present study, the PTB patients were selected from the Department of Tuberculosis at Anhui Chest Hospital, and unrelated healthy individuals were recruited from the health examine center as control group subjects. All the subjects were in the same region, and the Han Chinese population. The diagnosis of PTB patients was based on the following criteria: suspicious clinical symptoms, chest radiography, sputum and/or bronchoalveolar lavage fluid MTB culture, microscopy for acid fast bacilli (AFB), and effect of anti-TB treatment. In addition, the exclusion criteria for PTB patients including HIV positive, hepatitis, malignancy, immune deficiency, and other infectious disease. All controls were required to have no history of TB, infectious diseases or malignant neoplasms, negative sputum smear and culture, and normal chest radiographs.
Our study was approved by the Ethics Committee of Anhui Chest Hospital, and followed the Declaration of Helsinki protocols. The written informed and consent was provided by every participant prior to enrollment in the study. Then, the peripheral blood samples, demographic characteristics, and clinical data of study participants were collected. The required clinical information for PTB patients included fever (≥ 37.3°C), drug resistance, drug-induced liver injury (DILI), pulmonary infection, hypoproteinemia (Serum total protein < 60 g/L or albumin < 35 g/L), leukopenia (peripheral blood white blood cell count < 4.0×109/L), sputum smears-positive (the amount of bacteria ≥104/mL), etc.
SNP selection and genotyping
Seven genes in WNT signaling pathway (SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6) were selected for analyses in the present study. The genetic variation information of these genes in Han Chinese people in Beijing was obtained by the Ensembl Genome Browser 85 and CHBS_1000g, and HaploView 4.0 software (Cambridge, MA, USA) was used to select the tag SNPs of each gene. Then, we searched the existing researches regarding the association between these genes and human diseases, and looked for the functional tag SNPs related to human disease. Finally, we selected three SNPs (rs10088390, rs4736958, rs3242) in SFRP1 gene, two SNPs (rs752107, rs3121310) in WNT3A gene, three SNPs (rs2293303, rs1798802, rs4135385) in CTNNB gene, two SNPs (rs1026024, rs3782499) in WIF-1 gene, two SNPs (rs2241529, rs1569198) in DKK-1 gene, two SNPs (rs3736228, rs556442) in LRP5 gene, three SNPs (rs2302685, rs11054697, rs10743980) in LRP6 gene, respectively. Above SNPs satisfied these conditions: minor allele frequency (MAF) ≥ 0.05 in CHB and r2 threshold > 0.8.
We collected the peripheral blood samples from each subject by tubes containing ethylenediaminetetraacetic acid with median cubital vein, and extracted the genomic DNA from peripheral blood by the Flexi Gene-DNA Kit (Qiagen, Valencia, CA). The SNPscan technique was applied for genotyping with the technical support of the Center for Genetic & Genomic Anal-ysis, Genesky Biotechnologies (Inc., Shanghai), and the specific experimental steps could be found in the previous study. Only the subject with 100% success in SNP genotyping were included in the final analysis.
Statistical analysis
All the statistical analysis was performed by SPSS 23.0 (SPSS Inc, IL, USA) in this study. The Hardy-Weinberg equilibrium test of candidate SNPs was evaluated by Chi-square among normal controls. We analyzed the differences of genotype distribution, and allele frequency of all SNPs between the PTB group and control group by Chi-square test, and the associations between these SNPs and the clinical features of PTB patients were also analyzed using Chi-square test. Moreover, logistic regression model was used to calculate the odds ratios (OR) and 95% confidence intervals (CI). The relationship between each SNP and PTB risk in dominant and recessive model was also explored, and haplotype frequencies between PTB group and control group was also estimated by SHEsis (15). A P value < 0.05 was considered as the statistically significant.
Results
Association of WNT signaling pathway genes SNPs with PTB
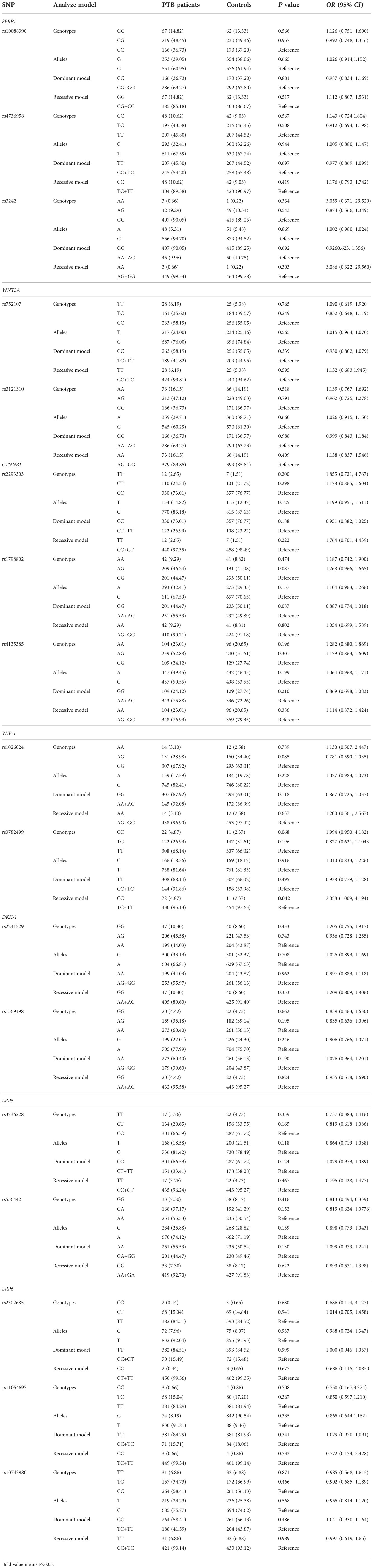
This study included 452 PTB patients (192 females and 260 males) and 465 normal controls (262 females and 203 male), and the mean age of PTB patients and normal controls was 44.88 ± 17.32, 44.12 ± 13.85, respectively. The allele, genotype frequencies of all SNPs in SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6 genes are summarized in Table 1, and the genotype distribution of all SNPs in control met Hardy Weinberg equilibrium.
As illustrated in Table 1, there were no significant differences in allele distribution of SFRP1 rs10088390, rs4736958, rs3242, WNT3A rs752107, rs3121310, CTNNB1 rs2293303, rs1798802, rs4135385, WIF-1 rs1026024, rs3782499, DKK-1 rs2241529, rs1569198, LRP5 rs3736228, rs556442, LRP6 rs2302685, rs11054697, rs10743980 polymorphism between PTB patients and normal controls (all P >0.05). In addition, we did not find any significant association between the genotype distribution of these SNPs and the susceptibility to PTB (all P >0.05). It was worth noting that the increased risk of WIF-1 rs3782499 variant was found to be associated with susceptibility to PTB under recessive model (CC versus TC+TT: P = 0.042).
Association of WNT signaling pathway genes SNPs with several clinical features among with PTB
Among PTB patients, the clinical features of PTB patients were sputum smear-positive (125, 27.65%), pulmonary infection (81, 17.92%), drug resistance (73, 16.15%), fever (69, 15.27%), DILI (64, 14.16%), hypoproteinemia (38, 8.41%), leukopenia (29, 6.42%). We further assessed whether SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6 genes variants affected the occurrence of multiple clinical manifestations in PTB patients (Supplementary Table 1). The WNT3A rs3121310 T allele, CTNNB1 rs2293303 TT genotype frequencies were significantly decreased in PTB patients with DILI, sputum smear-positive, respectively (P = 0.021 P = 0.007). For WIF-1 gene rs3782499, PTB patients carrying the homozygous TT/CC genotype had a highly significant association with increased frequency of fever, and carrying the C allele had a lower frequency of leukopenia (P = 0.032, P = 0.048, respectively).
We also found a high frequency of DKK-1 rs1569198 AG genotype the PTB patients with sputum smear-positive (P = 0.049). In LRP5 gene, the CT genotype frequency of rs3736228 variant was significantly associated with the occurrence of DILI (P = 0.040), and the GA genotype frequency of rs556442 variant were significantly related to the risk of fever (P = 0.025). In addition, the CT genotype frequency of LRP6 gene rs2302685 was significantly decreased with hypoproteinemia (P = 0.049), and the TC genotype frequency of LRP6 gene rs10743980 was significantly increased with sputum smear-positive (P = 0.007).
Haplotype analysis
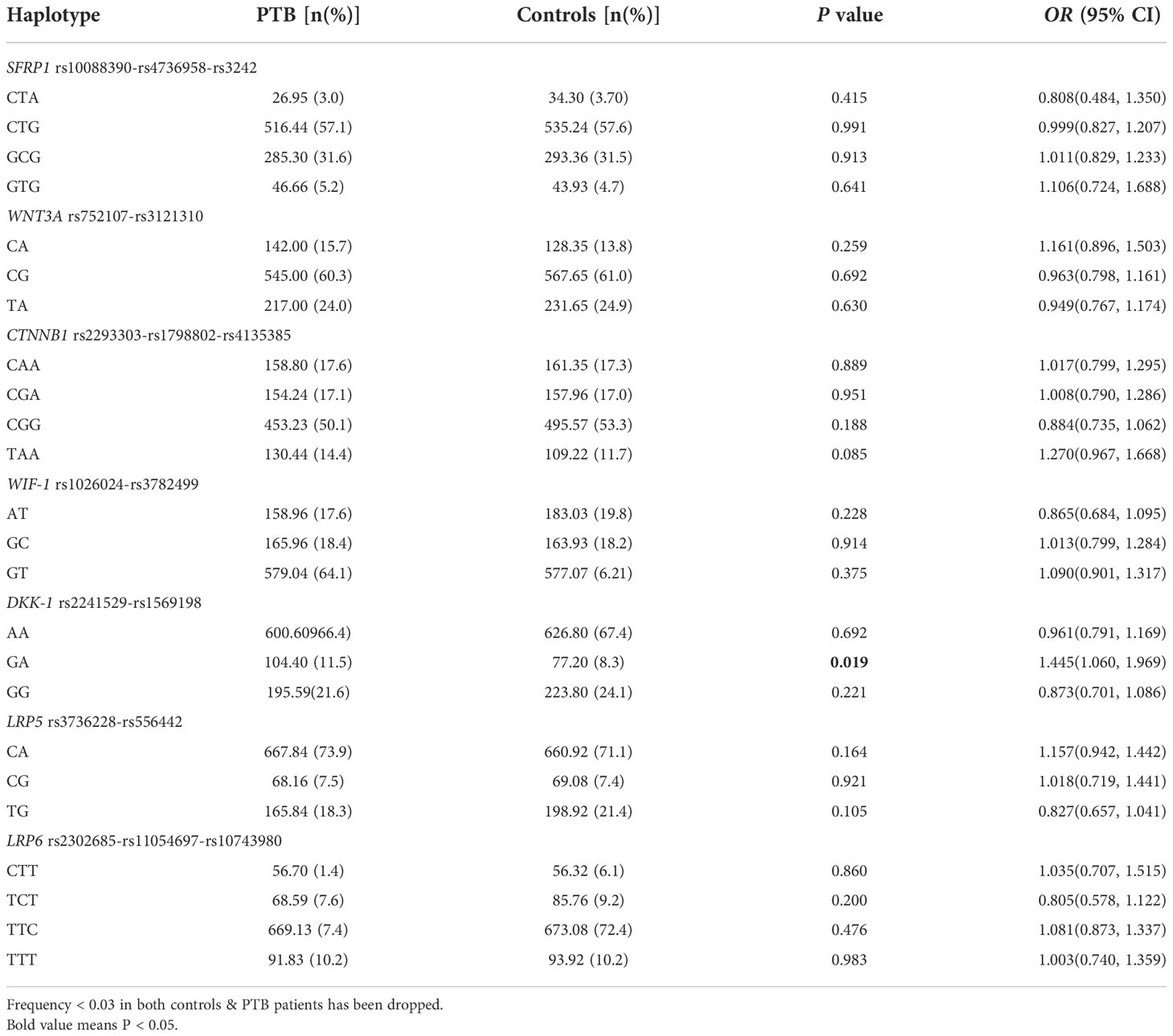
This study firstly detected the haplotypes of SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6 genes by SHEsis software, and then analyzed the potential association between these haplotypes and PTB risk. Four main haplotypes (CTA, CTG, GCG, GTG) for SFRP1, three main haplotypes (CA, CG, TA) for WNT3A, four main haplotypes (CAA, CGA, CGG, TAA) for CTNNB1, three main haplotypes (AT, GC, GT) for WIF-1, three main haplotypes (AA, GA, GG) for DKK-1, three main haplotypes (CA, CG, TG) for LRP5, four main haplotypes (CTT, TCT, TTC, TTT) for LRP6 were detected. The frequency distributions of above haplotypes in PTB group and control group were summarized in Table 2.
Comparing the haplotype frequency between the two groups, we found that DKK-1 GA haplotype frequency was significantly increased in PTB patients than that in controls (OR = 1.445, 95% CI: 1.060-1.969, P = 0.019). However, there was no significant association between other haplotypes frequencies and susceptibility to PTB.
Discussion
Previous studies had shown that the WNT signaling pathway was crucial in bridging innate and adaptive immunity to MTB infection (11, 16, 17), and the mRNA levels of several WNT signaling pathway genes (CTNNB1, SFRP1 and WNT3A) were significantly increased in PTB patients when compared to controls (3). Functional investigation found that CTNNB1 gene variation might significantly decreased the promoter transcriptional activity and associated with decreased CTNNB1 mRNA and phosphorylated β-catenin protein level (3), thereby affecting disease susceptibility. Hence, these genes might play important roles in host immune response to MTB infection. In this study, we comprehensively explored the association between 17 SNPs in WNT signaling pathway genes (SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6) and PTB susceptibility, clinical manifestations, providing clues for in-depth understanding of the pathogenesis of PTB.
β-catenin protein, which was encoded by CTNNB1 gene, played key roles in the coordination of cell adhesion and gene transcription, and aberrant β-catenin was involved in abnormal immune responses to pathogens and disease course, including TB (18, 19). The researchers had also linked CTNNB1 gene variants to susceptibility and prognosis of some cancers (20, 21). SFRP1 was a mature antagonist of WNT signaling pathway and could block the WNT signaling pathway (22). As a novel inflammatory regulator, SFRP1 might play an indispensable role in the pathogenesis of PTB through regulating the balance between anti-inflammatory cytokines and pro-inflammatory cytokines (23). The results of a previous study showed that CTNNB1 rs4135385, SFRP1 rs7832767 variants were associated with PTB risk (24), and another study suggested that SFRP1 rs4736958, rs7832767 were significantly related to PTB susceptibility and could affect the expression levels of some inflammatory markers in PTB patients (25). These results demonstrated a potential role of these variants in the susceptibility to PTB. Therefore, it was reasonable and necessary to screen other PTB related SNPs in WNT signaling pathway. However, our study did not find any correlation between SFRP1 rs10088390, rs4736958, rs3242, as well as CTNNB1 rs2293303, rs1798802, rs4135385 and PTB susceptibility. This inconsistency might be due to the differences in genotyping methods, sample size, etc (25). In addition, Hu et al. investigated the roles of several SNPs in WIF1, WNT3A, WIF-1, DKK-1 genes, and no significant result was found (3). Our results similarly did not support an association between these genetic variants and PTB susceptibility, suggesting that these polymorphisms might not predispose one to this disease. It was worth noting that WIF-1 gene rs3782499 variant was related to PTB risk under recessive model and DKK-1 GA haplotype frequency was significantly increased in PTB patients in our study. These results implied that WIF-1, DKK-1 genes variation might be involved in the pathogenesis of PTB, which needed to be verified with multi-ethnic population, large sample size study. As two key genes in WNT signaling pathway, previous studies had demonstrated the genetic association between LRP5, LRP6 and disease susceptibility (26, 27). Our study was the first to analyze the roles of LRP5 rs3736228, rs556442, LRP6 rs2302685, rs11054697 and rs10743980 polymorphisms in PTB, and no significant association was observed. Despite these irrelevant results, we could not completely rule out the possibility of these genes as putative PTB-related genes due to imperceptible genotype frequencies or slight effects of these polymorphisms. In addition, the effect of WNT signaling genetic variants on the transcription and protein levels of these genes in PTB patients was also needed to be analyzed in future.
Multiple clinical manifestations of PTB patients, such as fever, drug resistance, DILI, and pulmonary infection, etc. might had a certain impact on the rehabilitation and treatment process of patients, and the occurrence of these clinical manifestations was affected by genetic variants (28, 29). Zhao et al. found that PTB patients carrying the CC genotype of lncRNA AC079767.4 rs12477677 variant were highly significantly correlated with lower frequency of fever (29). Another study suggested that the lnc-HNF1B-3:1 rs2542670 polymorphism was associated with a higher risk of thrombocytopenia, leukopenia following medication in PTB patients (30). In this study, we investigated whether all candidate SNPs in WNT signaling pathway genes were associated with the clinical manifestations of PTB. Similarly, we observed that WIF-1 rs3782499, LRP5 rs556442 variants were related to the occurrence of fever, and WNT3A rs3121310, LRP5 rs3736228 polymorphisms had a significant association with the risk of DILI in PTB patients. Moreover, this study demonstrated a significant correction between several SNPs (CTNNB1 rs2293303, DKK-1 rs1569198, LRP6 rs10743980) and sputum smear-positive in PTB patients. The differences between PTB susceptibility related SNPs and clinical phenotypic related gene variation indicated that PTB development and disease manifestations might be independently affected by different gene SNPs, signaling pathways. While the exact molecular mechanism was still unclear, these results supported that gene variations in WNT signaling pathway could influence host defense responses to MTB infection and lead to the development of different clinical manifestations. This was important to accurately understand the progression of clinical manifestations in PTB patients and contributed to making appropriate treatment.
Although the relationship between WNT signaling pathway genes variants and PTB susceptibility, clinical manifestations was studied in detail, there were still some notable limitations in this study. First, our study did not further analyze the gene-environment or gene-gene interaction because of the uncorrelated-PTB risk among these SNPs. Second, we only selected a small number of SNPs for analysis in the whole WNT signaling pathway, and it was still necessary to study other potential and functional SNPs. Third, all the subjects of this study were only from one hospital, which inevitably resulted in a selection bias.
In conclusion, the present study demonstrated that WNT signaling pathway genes variation might not contribute to PTB susceptibility in Chinese Han population, while several SNPs in WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6 genes were related to the risk of several clinical characteristics of PTB patients. For example, WNT3A rs3121310, CTNNB1 rs2293303 were respectively related DILI, sputum smear-positive, and WIF-1 rs3782499, DKK-1 rs1569198 was related to fever, sputum smear-positive. In addition, LRP5 rs3736228, rs556442 were respectively related to DILI, fever, and LRP6 rs2302685, rs10743980 were respectively associated with hypoproteinemia, sputum smear-positive. Replication studies with larger sample size, different ethnic groups were required to verify and explore the genetic mechanisms of WNT signaling pathway in PTB development.
Data availability statement
The data presented in the study are deposited in the dbSNP (1063448). Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Anhui Chest Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
YL, HW, and QH designed the study. QH and T-PZ conducted the experiment. QH and C-CW performed the statistical analyses. HW participated in sample collection. QH drafted the manuscript. Y-GL and C-MZ contributed to the manuscript revision. All authors approved the final submitted version.
Funding
This study was supported by the Qinghai province highend innovative talents thousand talents program (No. 2020-12).
Acknowledgments
We thank Tang Fei of Anhui Chest, Xue-Qian Cai Hospital for their support in the collection of samples for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1011700/full#supplementary-material
References
1. World Health Organization. Global tuberculosis report 2021. Available at: https://www.who.int/tb/publications/global_report/en/.
2. WHO. Guidelines on the management of latent tuberculosis infection (2015). Available at: http://www.who.int/tb/publications/latent-tuberculosisinfection/en (Accessed 20/10/2015).
3. Hu X, Zhou J, Chen X, Zhou Y, Song X, Cai B, et al. Pathway analyses identify novel variants in the WNT signaling pathway associated with tuberculosis in Chinese population. Sci Rep (2016) 6:28530. doi: 10.1038/srep28530
4. Orme IM, Robinson RT, Cooper AM. The balance between protective and pathogenic immune responses in the TB-infected lung. Nat Immunol (2015) 16:57–63. doi: 10.1038/ni.3048
5. Figueira MBA, de Lima DS, Boechat AL, Filho MGDN, Antunes IA, Matsuda JDS, et al. Single-nucleotide variants in the AIM2 - absent in melanoma 2 gene (rs1103577) associated with protection for tuberculosis. Front Immunol (2021) 12:604975. doi: 10.3389/fimmu.2021.604975
6. Liu S, Liu N, Wang H, Zhang X, Yao Y, Zhang S, et al. CCR5 promoter polymorphisms associated with pulmonary tuberculosis in a Chinese han population. Front Immunol (2020) 11:544548. doi: 10.3389/fimmu.2020.544548
7. Liu Q, Chen X, Dai X. The association of cytokine gene polymorphisms with tuberculosis susceptibility in several regional populations. Cytokine (2022) 156:155915. doi: 10.1016/j.cyto.2022.155915
8. Harling K, Adankwah E, Güler A, Afum-Adjei Awuah A, Adu-Amoah L, Mayatepek E, et al. Constitutive STAT3 phosphorylation and IL-6/IL-10 co-expression are associated with impaired T-cell function in tuberculosis patients. Cell Mol Immunol (2019) 16:275–87. doi: 10.1038/cmi.2018.5
9. Sun J, Zhang Q, Yang G, Li Y, Fu Y, Zheng Y, et al. The licorice flavonoid isoliquiritigenin attenuates mycobacterium tuberculosis-induced inflammation through Notch1/NF-κB and MAPK signaling pathways. J Ethnopharmacol (2022) 294:115368. doi: 10.1016/j.jep.2022.115368
10. Akiyama T. Wnt/beta-catenin signaling. Cytokine Growth Factor Rev (2000) 11:273–82. doi: 10.1016/S1359-6101(00)00011-3
11. Wu X, Deng G, Hao X, Li Y, Zeng J, Ma C, et al. A caspase-dependent pathway is involved in wnt/β-catenin signaling promoted apoptosis in bacillus calmette-guerin infected RAW264.7 macrophages. Int J Mol Sci (2014) 15:5045–62. doi: 10.3390/ijms15035045
12. Schaale K, Neumann J, Schneider D, Ehlers S, Reiling N. Wnt signaling in macrophages: augmenting and inhibiting mycobacteria-induced inflammatory responses. Eur J Cell Biol (2011) 90(6–7):553–9. doi: 10.1016/j.ejcb.2010.11.004
13. Liu Y, Qin W, Zhang F, Wang J, Li X, Li S, et al. Association between WNT-1-inducible signaling pathway protein-1 (WISP1) genetic polymorphisms and the risk of gastric cancer in guangxi Chinese. Cancer Cell Int (2021) 21:405. doi: 10.1186/s12935-021-02116-2
14. Chen J, Yin J, Li X, Wang Y, Zheng Y, Qian C, et al. WISP1 polymorphisms contribute to platinum-based chemotherapy toxicity in lung cancer patients. Int J Mol Sci (2014) 15:21011–27. doi: 10.3390/ijms151121011
15. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res (2009) 19:519–23. doi: 10.1038/cr.2009.33
16. Schaale K, Brandenburg J, Kispert A, Leitges M, Ehlers S, Reiling N. Wnt6 is expressed in granulomatous lesions of mycobacterium tuberculosis-infected mice and is involved in macrophage differentiation and proliferation. J Immunol (2013) 191:5182–95. doi: 10.4049/jimmunol.1201819
17. Villaseñor T, Madrid-Paulino E, Maldonado-Bravo R, Urbán-Aragón A, Pérez-Martínez L, Pedraza-Alva G. Mycobacterium tuberculosisActivation of the wnt pathway by: A wnt-wnt situation. Front Immunol (2017) 8:50. doi: 10.3389/fimmu.2017.00050
18. Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, Wang YC, et al. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science (2010) 329:849–53. doi: 10.1126/science.1188510
19. Neumann J, Schaale K, Farhat K, Endermann T, Ulmer AJ, Ehlers S, et al. Frizzled1 is a marker of inflammatory macrophages, and its ligand Wnt3a is involved in reprogramming mycobacterium tuberculosis-infected macrophages. FASEB J (2010) 24:4599–612. doi: 10.1096/fj.10-160994
20. Li Y, Zhang F, Yang D. Comprehensive assessment and meta-analysis of the association between CTNNB1 polymorphisms and cancer risk. Biosci Rep (2017) 37. doi: 10.1042/BSR20171121
21. Butrym A, Rybka J, Łacina P, Gebura K, Frontkiewicz D, Bogunia-Kubik K, et al. Polymorphisms within beta-catenin encoding gene affect multiple myeloma development and treatment. Leuk Res (2015) 39:1462–6. doi: 10.1016/j.leukres.2015.10.007
22. Suzuki H, Watkins DN, Jair KW, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet (2004) 36:417–22. doi: 10.1038/ng1330
23. Barandon L, Casassus F, Leroux L, Moreau C, Allières C, Lamazière JM, et al. Secreted frizzled-related protein-1 improves postinfarction scar formation through a modulation of inflammatory response. Arterioscler Thromb Vasc Biol (2011) 31:e80-7. doi: 10.1161/ATVBAHA.111.232280
24. Hu X, Shang M, Zhou J, Ye Y, Lu X, Tao C, et al. Association of genetic variants in wnt signaling pathway with tuberculosis in Chinese han population. PloS One (2014) 9:e93841. doi: 10.1371/journal.pone.0093841
25. Zhao Z, Peng W, Hu X, Zhang J, Shang M, Zhou J, et al. SFRP1 variations influence susceptibility and immune response to mycobacterium tuberculosis in a Chinese han population. Infect Genet Evol (2016) 37:259–65. doi: 10.1016/j.meegid.2015.11.031
26. Han D, Zhang H, Liu S, Zhuang L, Zhao Z, Ding H, et al. Association between the LRP5 rs556442 gene polymorphism and the risks of NAFLD and CHD in a Chinese han population. BMC Gastroenterol (2022) 22:305. doi: 10.1186/s12876-022-02385-9
27. Guo Q, Lai Y, Chu J, Chen X, Gao M, Sang C, et al. LRP6 polymorphisms is associated with sudden cardiac death in patients with chronic heart failure in the Chinese han population. Front Cardiovasc Med (2021) 8:815595. doi: 10.3389/fcvm.2021.815595
28. Li HM, Wang LJ, Tang F, Pan HF, Zhang TP. Association of leptin and leptin receptor genes variants and pulmonary tuberculosis susceptibility, clinical manifestations in a Chinese population. Microb Pathog (2022) 165:105499. doi: 10.1016/j.micpath.2022.105499
29. Zhao Z, Zhang M, Ying J, Hu X, Zhang J, Zhou Y, et al. Significance of genetic polymorphisms in long non-coding RNA AC079767.4 in tuberculosis susceptibility and clinical phenotype in Western Chinese han population. Sci Rep (2017) 7:965. doi: 10.1038/s41598-017-01163-y
Keywords: pulmonary tuberculosis, infectious disease, Mycobacterium tuberculosis, WNT signaling pathway, single nucleotide polymorphisms
Citation: Huang Q, Wang C-C, Liu Y-G, Zhao C-M, Zhang T-P, Liu Y and Wang H (2022) Clinical relevance of genetic polymorphisms in WNT signaling pathway (SFRP1, WNT3A, CTNNB1, WIF-1, DKK-1, LRP5, LRP6) on pulmonary tuberculosis in a Chinese population. Front. Immunol. 13:1011700. doi: 10.3389/fimmu.2022.1011700
Received: 04 August 2022; Accepted: 21 October 2022;
Published: 07 December 2022.
Edited by:
Jianping Xie, Southwest University, ChinaReviewed by:
Juan José Valdez Alarcón, Universidad Michoacana de San Nicolás de Hidalgo, MexicoWang-Dong Xu, Southwest Medical University, China
Copyright © 2022 Huang, Wang, Liu, Zhao, Zhang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Liu, qhyxyly@aliyun.com; Hua Wang, 1726553540@qq.com
†These authors have contributed equally to this work and share first authorship
 Qian Huang1†
Qian Huang1†