
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 October 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1002616
This article is part of the Research Topic Research on the Mechanism of Pathogenesis and Immune Evasion of ASFV View all 7 articles
 Changjie Lv1,2,3
Changjie Lv1,2,3 Qiang Zhang4
Qiang Zhang4 Li Zhao5
Li Zhao5 Jingyu Yang5
Jingyu Yang5 Zhong Zou1,2
Zhong Zou1,2 Ya Zhao1,3
Ya Zhao1,3 Chengfei Li1,3
Chengfei Li1,3 Xiaomei Sun1,3
Xiaomei Sun1,3 Xian Lin6*
Xian Lin6* Meilin Jin1,2,3*
Meilin Jin1,2,3*African swine fever (ASF) is the most dangerous pig disease, and causes enormous economic losses in the global pig industry. However, the mechanisms of ASF virus (ASFV) infection remains largely unclear. Hence, this study investigated the host response mechanisms to ASFV infection. We analyzed the differentially expressed proteins (DEPs) between serum samples from ASFV-infected and uninfected pigs using quantitative proteomics. Setting the p-value < 0.05 and |log2 (fold change)| > 1.5, we identified 173 DEPs, comprising 57 upregulated and 116 downregulated proteins, which belonged to various biological processes and pathways based on the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses. The enriched pathways include immune responses, metabolism, and inflammation signaling pathways. Western blot analysis validated the DEPs identified using quantitative proteomics. Furthermore, our proteomics data showed that C1QTNF3 regulated the inflammatory signaling pathway. C1QTNF3 knockdown led to the upregulation of pro-inflammatory factors IL-1β, IL-8, and IL-6, thus inhibiting ASFV replication. These results indicated that C1QTNF3 was critical for ASFV infection. In conclusion, this study revealed the molecular mechanisms underlying the host-ASFV interaction, which may contribute to the development of novel antiviral strategies against ASFV infection in the future.
African swine fever (ASF) is one of the most severe infectious diseases of pigs worldwide with its virulent strain mortality rate of up to 100% (1, 2), and causes enormous economic loss. ASF virus (ASFV) is the only known DNA arbovirus, and it is a sole member of the Asfarviridae family and Asfivirus genus (3). ASFV contains a 170–190 kb genome that encodes 151–167 genes depending on the strain (4). ASFVs are divided into 24 genotypes based on the B646L gene-encoding capsid protein p72 (5). To date, there have been no effective vaccines or antiviral drugs to control ASFVs in domesticated pigs and wild boars (6, 7). Currently, the prevention and control of ASF mainly depend on rigorous import policies and biosecurity measures, including disinfection, culling of infected animals, and early detection.
ASF was first reported in Kenya in 1921, followed by its discovery in other African countries. ASF was reported in Europe in the 1950s (8). Later in 2007, there was an outbreak of genotype II ASFV in Georgia and Armenia. In China, ASF was first discovered in 2018, followed by outbreaks in Mongolia and Vietnam in 2019 (9). In recent years, various naturally-mutated, more transmissible, and more virulent ASFV strains have emerged, which poses a great challenge to the prevention and control of the virus (10, 11). Therefore, it is necessary to elucidate the host–virus interaction mechanisms of ASFV. A genome-wide transcriptomic analysis of ASFV-infected primary porcine alveolar macrophages (PAMs) revealed that multiple pathways are involved in the responses against ASFV, including the host immune response and metabolic processes (12). Proteomics of ASFV-infected PAMs also show that immune system response, complement and coagulation cascade, and metabolic processes are crucial pathways during the infection (13). Furthermore, transcriptomics of the different tissues infected by ASFV can reveal the activation of their homologous pathways (14). Although such studies have found numerous critical proteins and pathways, the molecular mechanisms of host-virus interaction remain largely unknown.
Quantitative proteomic analysis of serum samples has been used to investigate the host response to several viruses, including the chikungunya virus, human immunodeficiency virus, porcine epidemic diarrhea virus, and classical swine fever virus. Studies revealed that antiviral factors and immune- and inflammation-related pathways are vital for the survival or protection of hosts (15–18). The proteomic analysis of serum samples may contribute to exploring the changes in host proteins during viral infection. Therefore, this study aimed to reveal the host-virus interaction mechanisms after ASFV infection using quantitative proteomic analysis of ASFV-infected serum samples.
We analyzed the quantifiable proteins from the serum samples of ASFV-infected pigs using tandem mass tagging (TMT)-based quantitative proteomics. Comparing infected with uninfected samples, we found that the differentially expressed proteins (DEPs) were mainly enriched in the immune-, inflammation-, and metabolism-related pathways. Of these altered proteins, we focused on inflammation-related C1Q and tumor necrosis factor-related protein 3 (C1QTNF3, also called CORS26, CTRP3, or cartducin), which is a member of the C1q/TNF-related protein-family (19, 20). C1QTNF3 has four domains, namely the N-terminal signal peptide, C-terminal C1Q globular domain, variable domain, and collagen repeat (20). Of these four domains, C1QTNF3 exerts functions mainly based on the C1Q globular domain. C1QTNF3 plays an important regulatory role in cartilage development, inflammatory response, cardiovascular disease, adipocyte differentiation, and metabolism (21–24). C1QTNF3 inhibits LPS-induced inflammation via PPAR-γ and TLR4-mediated pathways (25). ASFV infection also induces the expression of inflammatory cytokines through the TLRs/MyD88 pathway (26). C1QTNF3 might affect ASFV proliferation through the inflammatory signaling pathway. In addition, we confirmed that the downregulation of C1QTNF3 by siRNA increased ASFV-induced inflammatory cytokine production in PAMs, thereby inhibiting ASFV proliferation. Our findings provide the first insights into the host response to ASFV infection using serum samples, and lay a solid foundation for further research on the host-virus interaction of ASFV.
PAMs were obtained from 5-week-old specific pathogen-free pigs using bronchoalveolar lavage as previously described (27). Porcine kidney-15 (PK-15) cells were obtained from our laboratory. PAMs and PK-15 cells were grown in RPMI 1640 culture medium (Gibco, USA) and Dulbecco’s modified Eagle medium (Gibco, USA), respectively, supplemented with 10% fetal bovine serum (Gibco, USA), 100 μg/mL streptomycin, and 100 IU/mL penicillin at 37°C in 5% CO2. The virulent genotype II ASFV was previously isolated from wild boar and kept in a biosafety level 3 laboratory for in vitro experiments, and the genome has been uploaded to the National Center for Biotechnology Information database (GenBank: OM161110). The virus was proliferated in PAMs and stored at –80°C until used. Swine serum samples of ASFV-infected and uninfected pigs from the same pig farm were stored in our laboratory.
The recombinant porcine IL-1β and IL-6 proteins were purchased from Sino Biological (China), the recombinant porcine IL-8 proteins were purchased from Abcam (UK), and dsDNA naked Poly (dA:dT) was purchased from In vivoGen (USA). The primary antibodies used in this study were rabbit anti-PSMC3 (1:1000, Cell Signaling Technology, USA), rabbit anti-SLC38A2 (1:1000, Invitrogen, USA), rabbit anti-EEF1G (1:1000, Invitrogen, USA), rabbit anti-MMP8 (1:1000, Cell Signaling Technology, USA), rabbit anti-C1QTNF3 (1:1500, Bioss, China), and mouse anti-GAPDH (1:8000, Proteintech, China). The secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10000, Proteintech, China) and goat anti-mouse IgG (1:10000, Proteintech, China).
The cellular debris and impurities in the 100-μL serum sample were removed using centrifugation at 4 °C at 12,000 × g for 10 min. Subsequently, in a new 1.5-mL centrifuge tube, the supernatant was transferred and the protein concentration was determined with the PierceTM BCA protein assay kit (Thermo Fisher Scientific, USA), according to the manufacturer’s instructions.
For trypsin digestion, the protein solution was deoxidized with 5 mM dithiothreitol at 56 °C for 30 min, and alkylated with 11 mM iodoacetamide for 15 min in the dark at 37°C. Then, the protein sample was diluted by adding 100 mM tetraethylammonium bromide, leading to the final concentration of urea not being more than 2 M. Finally, trypsin was added at 1:50 mass ratio overnight for the first digestion, followed by the secondary digestion at 1:100 mass ratio for 4 h.
The different groups of peptides produced after trypsin digestion were labeled using the TMT kit (Thermo Fisher, USA), according to the manufacturer’s instructions. Briefly, the TMT reagent was thawed and dissolved in acetonitrile. The peptide mixtures were then incubated for 2 h at 37°C, desalted, and dried using vacuum centrifugation.
The labeled peptides were fractionated using high pH reverse-phase high-performance liquid chromatography with the Thermo Betasil C18 column (10 mm ID, 5 μm particles, and 250 mm in length), according to the manufacturer’s instructions. The tryptic peptides in 0.1% formic acid (solvent A) were dissolved and directly loaded onto a homemade reversed-phase analytical column (75 μm, 15 cm length) (Thermo Fisher Scientific, USA). The gradient comprised of an increasing percentage of solvent B (0.1% formic acid in 98% acetonitrile) over 26 min from 6%–23%, 23%–35% in the next 12 min, 35%–80% in the next 3 min, and then kept at 80% for the last 3 min; all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 ultra-performance liquid chromatography (UPLC) system (Thermo Fisher, USA).
The resulting peptides were subjected to a nanospray ionization source followed by tandem mass spectrometry (MS/MS) using the Q Exactive™ Plus Orbitrap mass spectrometer (Thermo Fisher, USA) coupled online to the UPLC system. The electrospray voltage was set as 2.0 kV. A 350–1800 m/z MS was used for the entire scan range. The Orbitrap was used to detect intact peptides at a resolution of 70,000. We then selected peptides for MS/MS using 28% normalized collision energy, and fragments were detected in the Orbitrap at a resolution of 17,500.The automatic gain control was set at 5E4 for MS/MS and the fixed first mass was set as 100 m/z. A data-dependent procedure alternated between one MS scan and 20 MS/MS scans with a dynamic exclusion time of 15 s.
The MS/MS data were analyzed using the Maxquant software (version 1.5.2.8). Concatenated with the reverse decoy database, the tandem mass spectra were searched against the UniProt database of Sus scrofa (pig). Trypsin/P was specified as the cleavage enzyme allowing up to two missing cleavages. For the initial search, the mass tolerance was set as 20 ppm for precursor ions and 5 ppm in the mass search, and the mass tolerance for fragment ions was set as 0.02 Da. Oxidation of methionine was specified as a variable modification and the modification of cysteine with carbamidomethyl was specified as a fixed modification. The false discovery rate was modulated to < 1% and the minimum score for modified peptides was set as > 40.
To further explore the internal relationships between the DEPs, an enrichment analysis was performed. The Gene Ontology (GO) annotation was analyzed from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/) for three categories—biological process, molecular function, and cellular component. The Clusters of Orthologous Groups of protein/Eukaryotic Orthologous Groups of proteins (COG/KOG) analysis was performed using an online database (http://ftp.ncbi.nih.gov/pub/COG/KOG/kyva).
To confirm the enrichment of DEPs, the two-tailed Fisher’s exact test was used to identify enriched pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG). The whole quantified protein annotations were used as background data.
For western blot analysis, the proteins of cells or serum samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose filter membrane (GE Technology, USA) in the transfer buffer. The membrane was blocked with 1% bovine serum albumin at 37°C for 1 h. Subsequently, the membrane was incubated with a primary antibody at 37°C for 2 h followed by three washes with tris buffered saline plus Tween 20 (TBST). Finally, the membrane was incubated with the corresponding secondary antibody at 37°C for 1 h and washed three times with TBST. The specific bands on the membrane were visualized using the western blot ECL reagent (Advansta, USA).
A commercially available ASFV VP72 antibody blocking ELISA kit (INgezim 11.PPA.K3, Madrid, Spain) was used to detect antibodies of six serum samples. Cell supernatants were collected, and porcine IL-6, IL-1β, and IL-8 were detected using porcine IL-6, IL-1β, and IL-8 ELISA kit (Solarbio). The measured value was calculated based on a standard curve according to the manufacturer’s instructions.
To assess the change in cytokines of PAMs during ASFV infection, the real-time quantitative polymerase chain reaction (RT-qPCR) was performed using the primers listed in Table S1. Briefly, the total RNA of PAMs was extracted using an RNA extraction kit (Magen, Guangzhou, China), followed by reverse transcription to cDNA using HiScript Reverse Transcriptase (Vazyme, Nanjing, China). PCRs were conducted in a QuantStudio™ 6 Flex system (Life Technologies, USA) as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. The 2-ΔΔCt quantification method was used to analyze the qPCR results.
We performed the hemadsorption (HAD) assay to detect the titer of ASFV as previously described (28), with minor modifications. Briefly, PAMs were seeded in 96-well plates. The virus samples were diluted in a continuous 10-fold dilution and then pipetted into the wells of the 96-well plates with eight replicates in each gradient. Next, 30 µL of 1% swine red blood cells was added to each well after 3 days of cultivation, and a rosette formation representing hemadsorption was observed around the infected cells. The hemadsorption was observed until 7 days postinfection and 50% HAD dose (HAD50) was calculated using the Reed–Muench method (29).
The siRNA of porcine C1QTNF3 or the negative control was synthesized by JTS scientific (Wuhan, China). The compositive siRNA was dissolved in sterile diethylpyrocarbonate-treated water and transfected into PAMs at a final concentration of 60 nM, using the Lipofectamine™ RNAiMAX Transfection Reagent (Thermo Fisher, USA), according to the manufacturer’s instructions. The siRNA sequences are listed in Table S2.
The full-length sequence of the porcine C1QTNF3 was cloned into a pCAGGS-HA vector to construct the recombinant plasmid C1QTNF3-HA. The constructed plasmid was analyzed and verified using DNA sequencing. PAMs and PK-15 cells were transfected with the recombinant plasmid C1QTNF3-HA using the jetPRIME® transfection reagent (Polyplus-transfection, France) according to the manufacturer’s instructions.
All live ASFV experiments were performed in an animal biosafety level 3 (ABSL-3) laboratory according to the relevant laws and regulations.
All data were analyzed for statistical significance using the Student’s t-test. Significant differences were affirmed when p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
The schematic of the experimental workflow is shown in Figure 1A. Serum samples of healthy pigs (n = 3) and ASFV-infected pigs (n = 3) were collected from the same pig farm. Pigs infected with ASFV rather than healthy pigs showed symptoms of the disease, including staggering gait, anorexia, depression, and cough. The pigs were approximately 110 days old. Six serum samples (one from each pig) were analyzed using the OIE-recommended INgezim ASFV antibody blocking ELISA kit according to the manufacturer’s instructions, and three negative and three positive results were obtained (Figure 1B). The viral titers of the six serum samples were also analyzed using the HAD assay (Figure 1C). The viral titers of the positive serum samples were less than 102 HAD50/mL (Figure 1D), which is consistent with a previous report that viral titers were less than 102 HAD50/mL in the blood in the early stage of infection (30). The proteomic analysis yielded 232,596 raw data (Figure 1E). The raw spectra were further searched in the protein database, and 26,501 clean spectra were obtained (Figure 1E). Ultimately, 945 quantifiable proteins were mapped to the pig reference genome. Based on the thresholds of p-value < 0.05 and |log2 (fold change)| > 1.5, 173 DEPs were identified from the swine serum samples by comparing the ASFV versus non-ASFV infection groups, of which 57 were upregulated and 116 were downregulated DEPs (Figures 1F, G). C1QTNF3 is marked in Figure 1F. The identified DEPs are presented in Supplementary Table S3.
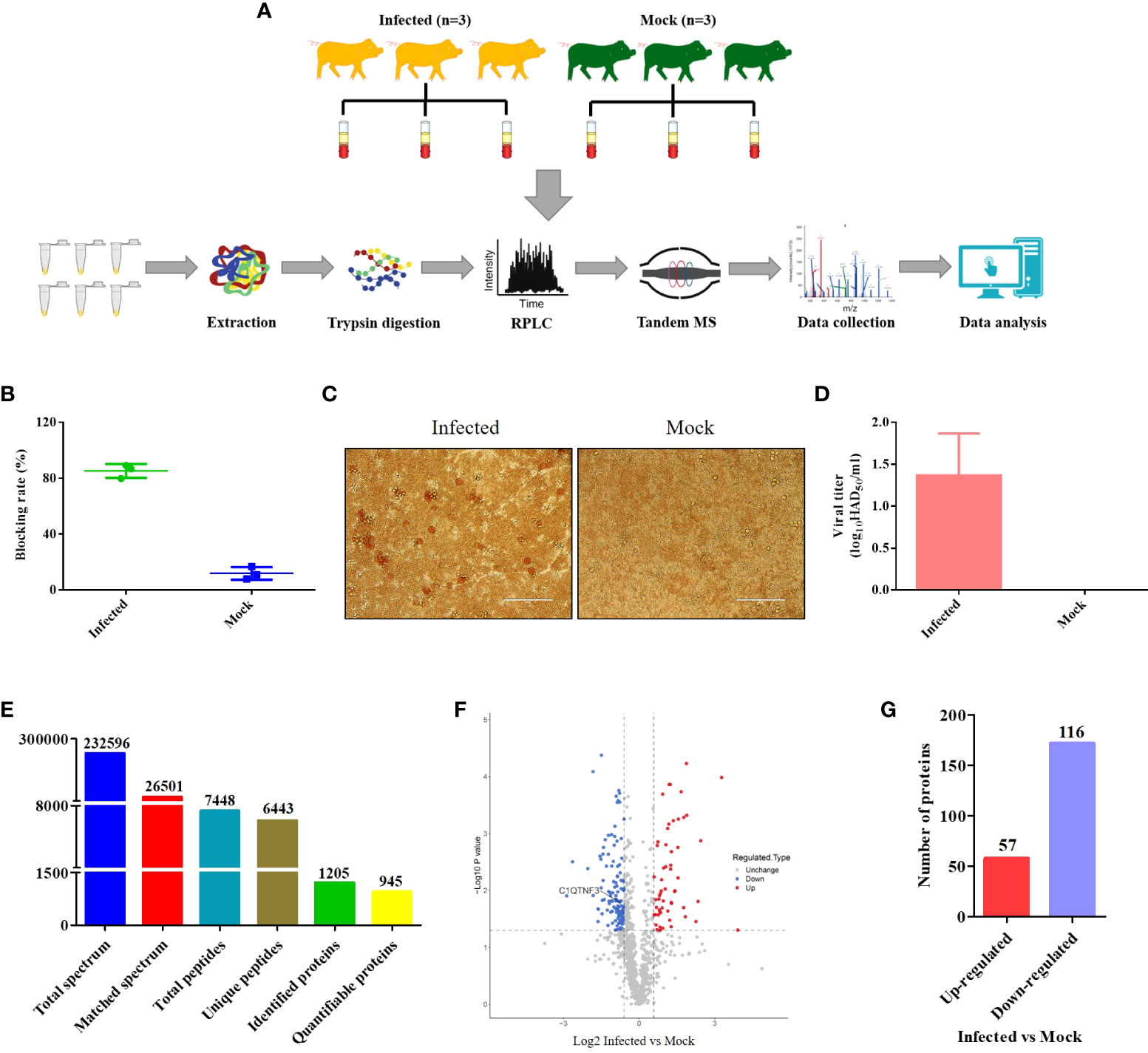
Figure 1 Schematic of the experimental method and data on the differentially expressed proteins (DEPs) identified between the ASFV-infected and uninfected serum samples using proteomics. (A) The serum samples of three ASFV-infected and uninfected pigs were collected from the same pig farm and subjected to quantitative proteomics through tandem mass spectrometry technology, high-performance liquid chromatography, and mass spectrometry. (B) Antibody, (C, D) Titer of the African swine fever virus was measured between the ASFV-infected and uninfected serum samples using ELISA and the HAD assay, respectively. (E) Summary of the quantitative proteomics data of swine serum samples. (F) Volcano map of DEPs between the ASFV-infected and uninfected serum samples. (G) Number of DEPs between the ASFV-infected and uninfected serum samples.
The 173 DEPs were functionally annotated using GO enrichment, KEGG pathway analysis, and COG/KOG analysis. The subcellular localization results showed that the largest number of the upregulated (45.61%) and downregulated proteins (43.97%) were distributed in the extracellular matrix (Figures 2A, B). In addition, 15.79% of the upregulated proteins were located in the nucleus and 29.31% of the downregulated proteins were located in the cytoplasm (Figures 2A, B).
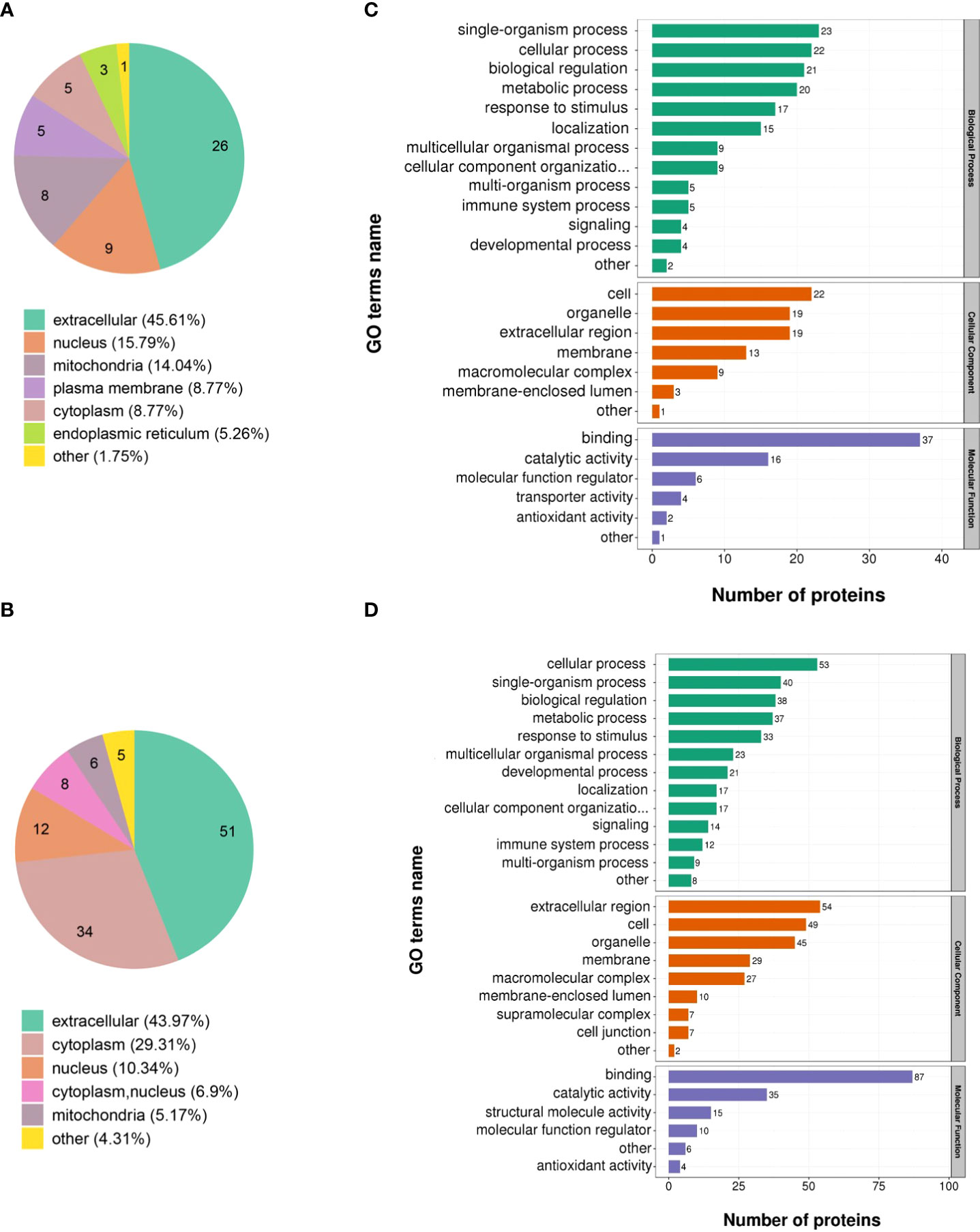
Figure 2 Functional analysis of the DEPs identified through quantitative proteomics in ASFV-infected versus uninfected samples. Subcellular structural analysis of (A) upregulated DEPs and (B) downregulated DEPs. GO enrichment analysis of (C) upregulated DEPs and (D) downregulated DEPs.
GO annotation categorized the 173 DEPs based on biological processes, cellular components, and molecular function. The upregulated proteins were classified into 26 functional groups. Among them, biological processes possessed 13 GO terms, and the most representative were “single-organism process” and “cellular process.” Cellular components possessed 7 GO terms, and the most representative were “cell” and “organelle.” Moreover, molecular function possessed 6 GO terms, and the most representative terms were “binding” and “catalytic activity” (Figure 2C). Furthermore, the downregulated proteins were annotated to 28 GO terms. Among them, 13 GO terms represented biological processes, and the most representative were “cellular process” and “single-organism process.” The 9 and 6 GO terms belonged to the cellular component and molecular function, respectively, and the most representative cellular component GO terms were “extracellular region” and “cell” while the most representative molecular function GO terms were “binding” and “catalytic activity” (Figure 2D).
The KEGG pathway enrichment analysis was used based on the upregulated and downregulated proteins. The results show that the upregulated proteins were enriched in the pantothenate and CoA biosynthesis, adipocytokine signaling pathway, viral myocarditis, and PPAR signaling pathway (Figure 3A). Correspondingly, the downregulated proteins were enriched in oocyte meiosis, lysine degradation, cell cycle, hepatitis C infection, alcoholism, T cell receptor signaling pathway, osteoclast differentiation, viral carcinogenesis, Hippo signaling pathway, and gap junction (Figure 3B).
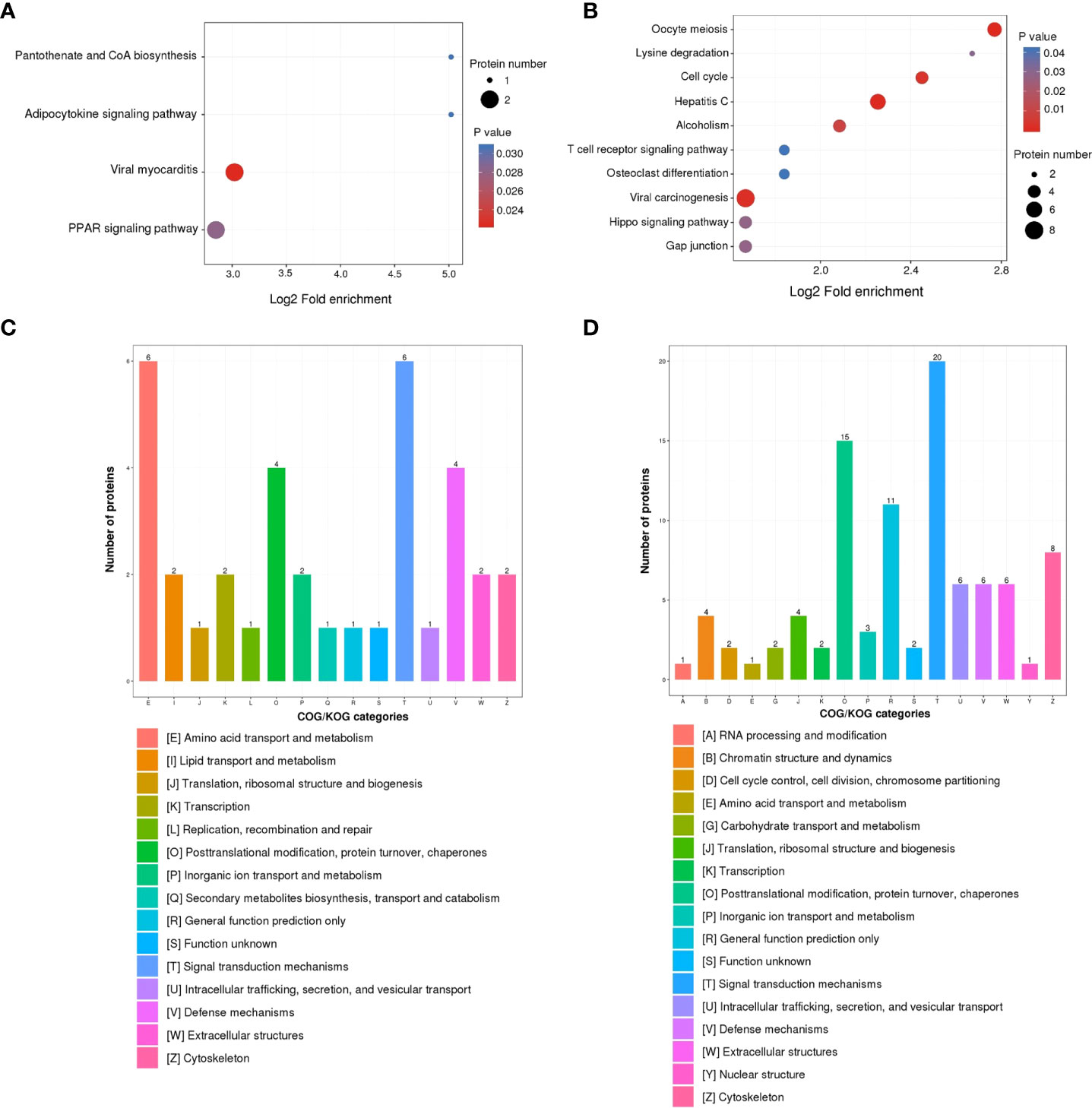
Figure 3 KEGG pathway and COG/KOG functional classification analysis of DEPs in ASFV-infected versus uninfected samples using quantitative proteomics. KEGG pathway analysis of (A) upregulated DEPs and (B) downregulated DEPs. COG/KOG functional classification analysis of (C) upregulated DEPs and (D) downregulated DEPs.
To better analyze the DEPs, the COG/KOG analysis was applied. Among the upregulated proteins, the largest proportion of proteins belonged to the categories of amino acid transport, metabolism, and signal transduction mechanisms; followed by posttranslational modification, protein turnover, chaperones, and defense mechanisms (Figure 3C). Furthermore, the downregulated proteins were in the categories of signal transduction mechanisms, posttranslational modification, protein turnover, and chaperones (Figure 3D).
Western blot analysis was used to validate some of the DEPs identified in the LC-MS/MS analysis. To more accurately evaluate the results of LC-MS/MS, we did not choose proteins with the highest or lowest levels, but randomly selected DEPs—PSMC3, SLC38A2, EEF1G, MMP8, and C1QTNF3 (Figure S1). We found that the levels of PSMC3 and SLC38A2 were higher, whereas the levels of EEF1G, MMP8, and C1QTNF3 were lower in the ASFV-infected serum samples than in the mock group, which was consistent with the results of the quantitative proteomics, indicating the reliability of the LC-MS/MS results.
We also screened DEPs that were clustered in the inflammatory response from the quantitative proteomics. We found 11 inflammation-related DEPs, comprising four upregulated DEPs (HP, ADIPOQ, VNN1, and LOC100517145) and seven downregulated DEPs (PRDX3, C1QTNF3, CHI3L1, RPS3, CALR, MMP8, and LTF) in our study. The GO and KEGG analyses showed that HP and VNN1 were enriched in the acute inflammatory response signaling pathway; ADIPPQ and C1QTNF3 were clustered in the negative regulation of inflammatory response signaling pathway; LOC100517145 and CHI3L1 were clustered in the inflammatory response signaling pathway; PRDX3, RPS3, and LTF were enriched in the positive regulation of NF-κB transcription factor activity signaling pathway; CALR and MMP8 were enriched in the positive regulation of NIK/NF-κB signaling. The inflammation-related DEPs are listed in Table S4.
The complement component is one of the important parts of serum samples, and regulates multiple signaling pathways, including inflammatory response (31). Therefore, we investigated the role of complement-related proteins in these inflammation-related DEPs, and found that among the 11 DEPs, C1QTNF3 is a member of the complement C1q/TNF-related protein family. Previous research has shown that ASFV infection could cause an imbalanced pro- and anti-inflammatory response (32). From the proteomics data, C1QTNF3 decreased by 0.591 times in ASFV-infected cells compared with that in the control. Previous research has indicated that C1QTNF3 regulates the inflammatory signaling pathway (33), and may be critical for ASFV-induced inflammatory response. Hence, we explored whether ASFV replication affects C1QTNF3 expression in vitro. PAMs were infected with ASFV at a multiplicity of infection (MOI) of 0.1, and the cell samples were collected 24 h, 48 h, and 72 h postinfection (hpi) to detect the expression of C1QTNF3 and other inflammatory factors. The results show that the expression of C1QTNF3 decreased gradually with infection time, which was 69% lower in the infected group than in the uninfected group at 72 hpi (Figure 4A). This result was consistent with that of the western blot analysis, and ASFV p72 proteins increased gradually with infection time (Figure 4B). The mRNA levels of the inflammatory factors, namely IL-1β, IL-6, and IL-8, increased gradually with the extended infection time (Figures 4C-E). Compared with the ASFV uninfected group, the levels of IL-1β, IL-6, and IL-8 in the ASFV-infected PAMs group increased by 26.9-, 4.86-, and 15.4-fold at 72 hpi, respectively. Simultaneously, the protein levels of the inflammatory factors, including IL-1β, IL-6, and IL-8, were upregulated comparing the ASFV-infected to uninfected with infection time (Figures 4F-H). These results show that the inflammatory responses were activated, and the expression of C1QTNF3 was inhibited in PAMs after ASFV infection.
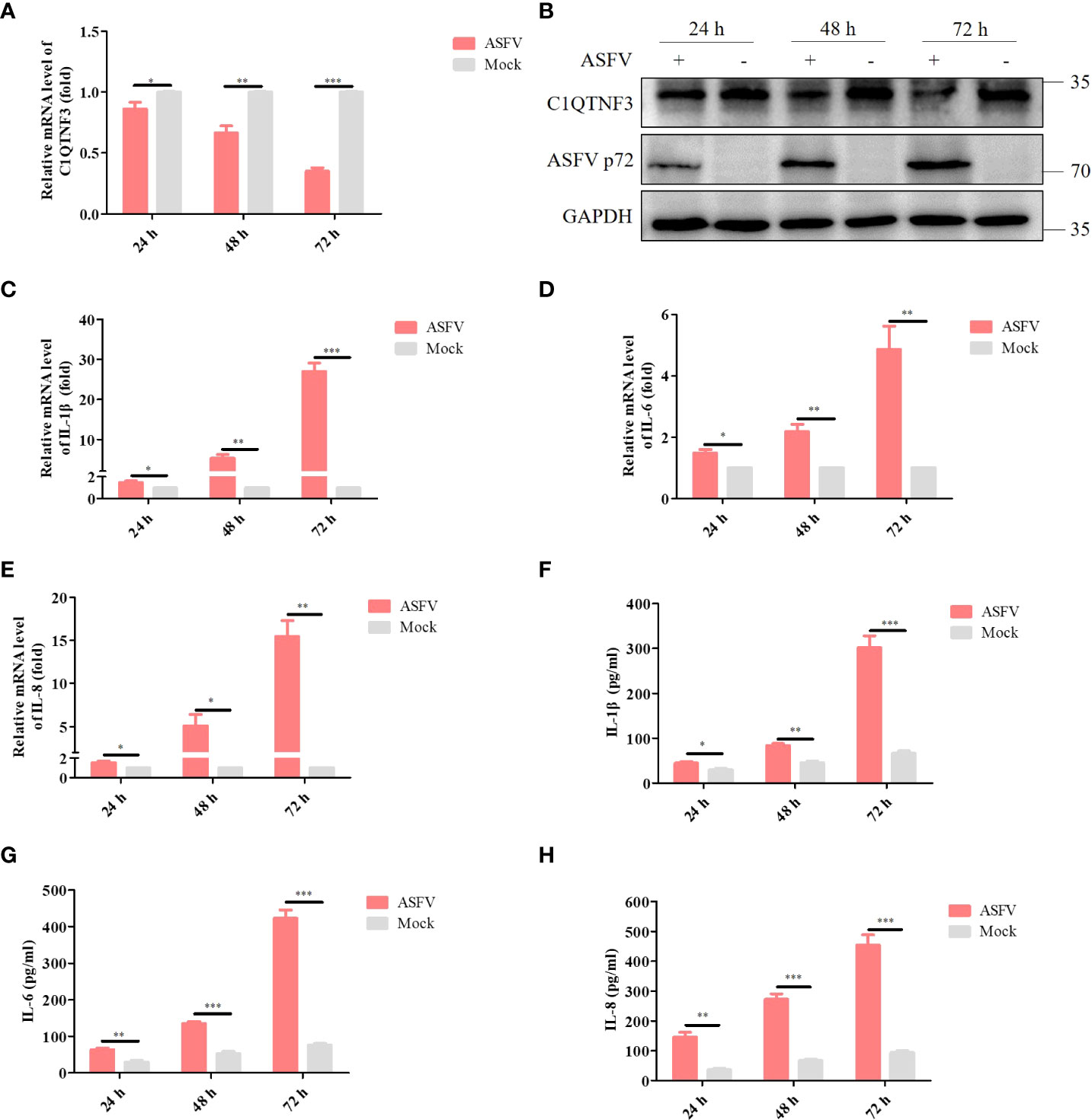
Figure 4 Expression of C1QTNF3 and pro-inflammatory factors IL-1β, IL-6, and IL-8 in ASFV-infected and mock-infected PAMs at 24, 48, and 72 h post-infection. The mRNA levels of (A) C1QTNF3, (C) IL-1β, (D) IL-6, and (E) IL-8 in ASFV-infected and mock-infected PAMs. The GAPDH gene was used as the internal control for normalization. The 2-ΔΔCt method was used to analyze the results of qPCR. The protein levels of (B) C1QTNF3, ASFV p72 (F) IL-1β, (G) IL-6, and (H) IL-8 in ASFV-infected and mock-infected PAMs p <0.05 (*), p < 0.01 (**), and p < 0.001 (***).
To determine whether C1QTNF3 affected the inflammatory response signaling pathway upon ASFV infection, we transfected siRNAs to downregulate the C1QTNF3 gene in PAMs. Western blot analysis results showed that among the several siRNAs, the siRNA #1 demonstrated the best inhibition efficiency (Figure 5A), which was consistent with the analysis of the relative protein band intensity using ImageJ software (Figure 5B). Hence, we chose the siRNA #1 for the subsequent experiments. Upon the downregulation of the C1QTNF3 gene by siRNA in PAMs 24 h before ASFV infection, the mRNA levels of IL-8, IL-1β, and IL-6 were upregulated in the C1QTNF3 downregulated cells compared with that in the siNC-treated cells at 48 hpi (Figures 5D-F). Compared with the siNC-treated cells followed by ASFV infection, the levels of IL-6, IL-1β, and IL-8 in the C1QTNF3 downregulated cells followed by ASFV infection increased by 1.5-, 2.4-, and 1.76-fold, respectively. The protein levels of IL-8, IL-1β, and IL-6 were also detected using ELISA (Figures 5H-J). The results were consistent with the qPCR results. In addition, the ASFV titer was lower in the ASFV-infected PAMs with C1QTNF3 downregulation by siRNA than in the siNC-treated PAMs (Figure 5C). To explore the role of inflammatory cytokines during ASFV proliferation, the cytotoxicity of porcine IL-1β, IL-6, and IL-8 proteins was measured in the PAMs using the CCK-8 assay according to the manufacturer’s instruction. When the concentrations of IL-1β, IL-6, and IL-8 proteins were less than 300 ng/mL, no cytotoxic effects were detected after 24 h or 48 h (Figure S2). Therefore, 50 ng/mL porcine IL-1β, IL-6, and IL-8 proteins were incubated with the PAMs for 24 h, followed by infection with 0.1 MOI ASFV for 48 h. The ASFV titer of the cell supernatants was measured using the HAD assay. The ASFV titer decreased in the PAMs treated with porcine IL-1β, IL-6, and IL-8 proteins compared with that of the untreated cells (Figure 5G). Thus, the downregulation of C1QTNF3 could lead to the upregulation of ASFV-induced inflammatory cytokines, and the expression of specific inflammatory cytokines can inhibit ASFV proliferation.
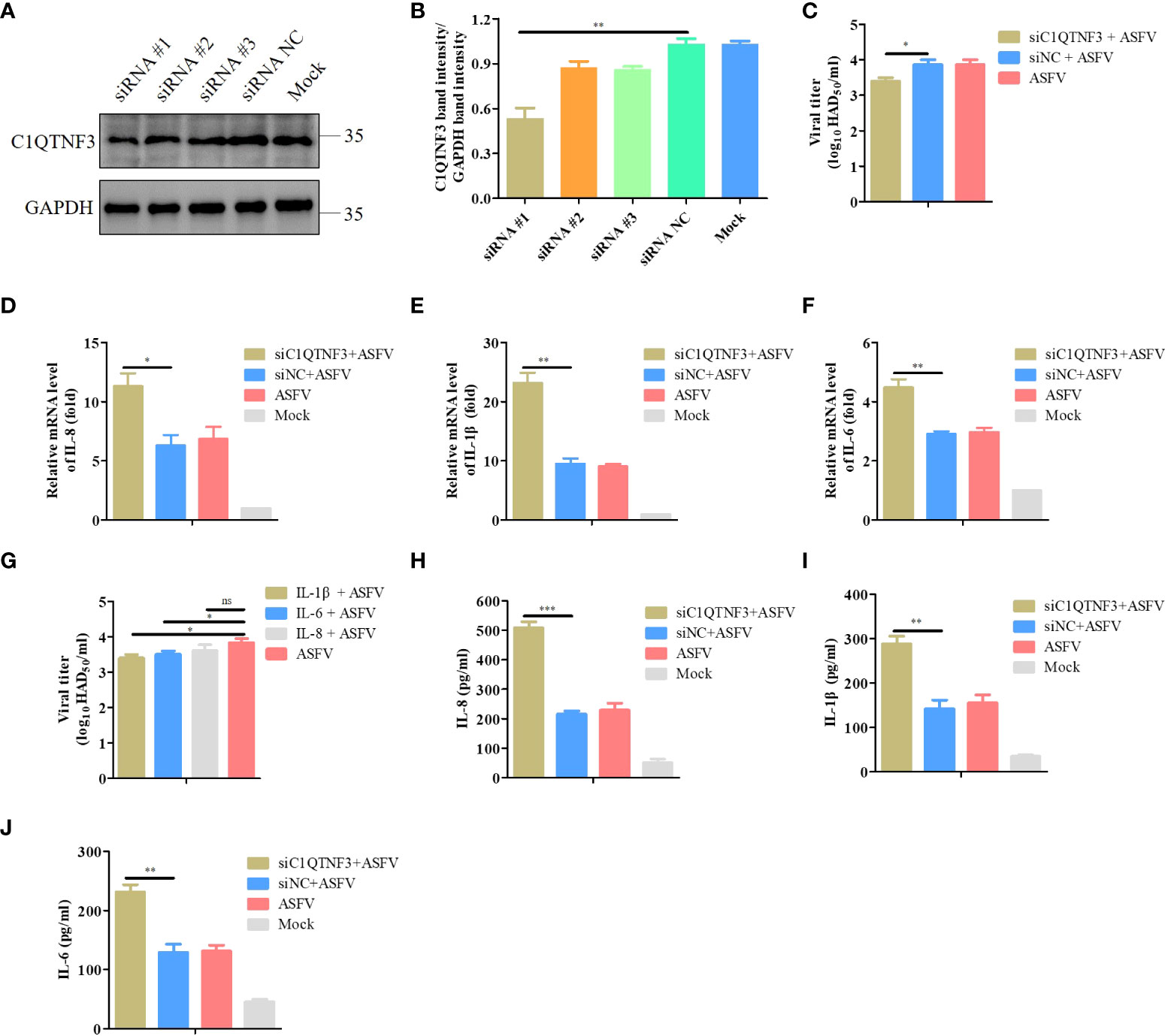
Figure 5 Upregulated pro-inflammatory cytokines inhibit ASFV infection in PAMs. (A, B) Detection of C1QTNF3 siRNA knockdown efficiency using western blot analysis. (C) C1QTNF3 knockdown in PAMs inhibits ASFV replication as detected using the HAD assay. The mRNA levels of (D) IL-8, (E) IL-1β, and (F) IL-6 upon C1QTNF3 knockdown in PAMs infected with ASFV. The GAPDH gene was used as the internal control for normalization. The 2-ΔΔCt method was used to analyze the results of qPCR. (G) Porcine IL-1β, IL-6, and IL-8 proteins inhibit ASFV replication, as detected using the HAD assay. The protein levels of (H) IL-8, (I) IL-1β, and (J) IL-6 upon C1QTNF3 knockdown in PAMs infected with ASFV p <0.05 (*), p < 0.01 (**), and p < 0.001 (***), ns means no difference.
We explored the inflammatory response signaling pathway regulated by C1QTNF3 after ASFV infection in PAMs. PAMs were transfected with the C1QTNF3-HA plasmid for 24 h, followed by infection with ASFV for 48 h. The protein level of C1QTNF3 was detected using western blot analysis (Figure 6A), and the band intensity was analyzed using ImageJ (Figure 6B), which confirmed that C1QTNF3 was overexpressed in the PAMs. Comparing the C1QTNF3 overexpression group with the control group, the mRNA levels of IL-1β, IL-8, and IL-6 were reduced (Figures 6D-F). The protein levels of IL-8, IL-6, and IL-1β also showed concordant results (Figures 6G-I). The results indicate that the ASFV titer was higher in the supernatant of the ASFV-infected PAMs with C1QTNF3 overexpression than in the control group (Figure 6C). Therefore, C1QTNF3 overexpression could cause the downregulation of inflammatory cytokines after ASFV infection and further promoted viral proliferation.
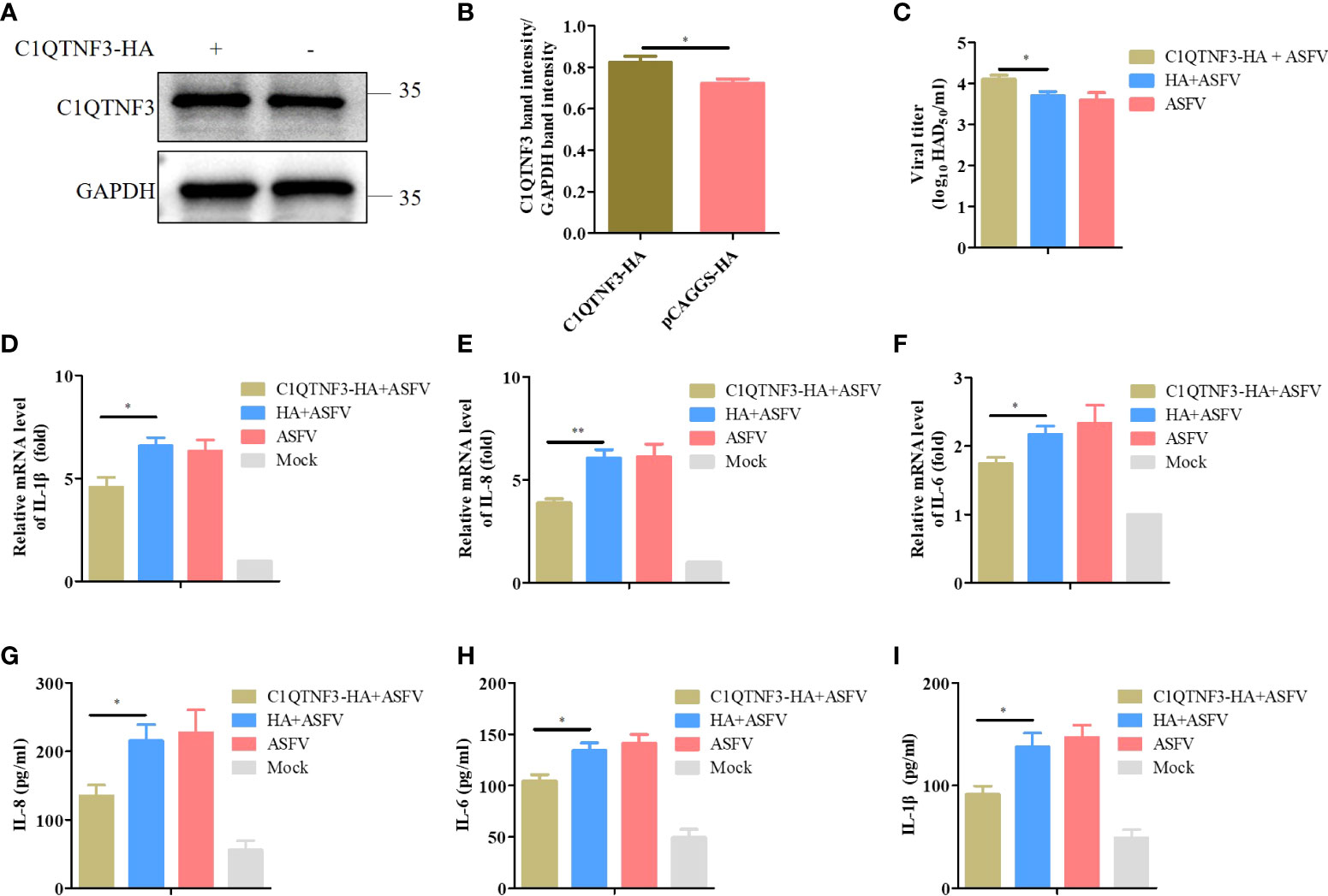
Figure 6 Downregulated pro-inflammatory cytokines increase ASFV infection in PAMs. (A, B) Overexpression of HA-tagged C1QTNF3 proteins was detected using western blot analysis. (C) C1QTNF3 overexpression in PAMs increases ASFV replication as detected using the HAD assay. The mRNA levels of (D) IL-1β, (E) IL-8, and (F) IL-6 upon C1QTNF3 overexpression in PAMs infected with ASFV. The GAPDH gene was used as the internal control for normalization. The 2-ΔΔCt method was used to analyze the results of qPCR. The protein levels of (G) IL-8, (H) IL-6, and (I) IL-1β upon C1QTNF3 overexpression in PAMs infected with ASFV p <0.05 (*), p < 0.01 (**).
Considering the low transfection efficiency of PAMs, PK-15 cells were employed to assess the role of C1QTNF3 overexpression in the inflammatory signaling pathway. ASFV cannot infect PK-15 cells, and thus poly (dA:dT) double-stranded DNA fragments were selected as the inflammatory stimulus for these cells. The PK-15 cells were transfected with the C1QTNF3-HA plasmid for 24 h, followed by transfection with poly (dA:dT) for 48 h. The antibody of porcine endogenous C1QTNF3 was used to detect the protein level of overexpression of HA-tagged C1QTNF3 in the PK-15 cells using western blot analysis (Figure 7A), and the relative protein band intensity was analyzed using ImageJ software (Figure 7B), which show that C1QTNF3 was overexpressed in the PK-15 cells. Additionally, the mRNA level of TLR9 was upregulated comparing PK-15 cell transfection with poly (dA:dT) to the control cells, which confirmed that the inflammatory signaling pathway was activated (Figure S3). The mRNA levels of IL-6 and IL-8 were lower in cells with C1QTNF3 overexpression than in the control group (Figures 7C, D). The same results were also obtained for the protein levels of IL-8 and IL-6 (Figures 7E, F). Hence, it can be concluded that C1QTNF3 was highly likely to be a broad-spectrum anti-inflammatory mediator and is negatively associated with the expression of pro-inflammatory cytokines in an inflammatory reaction.
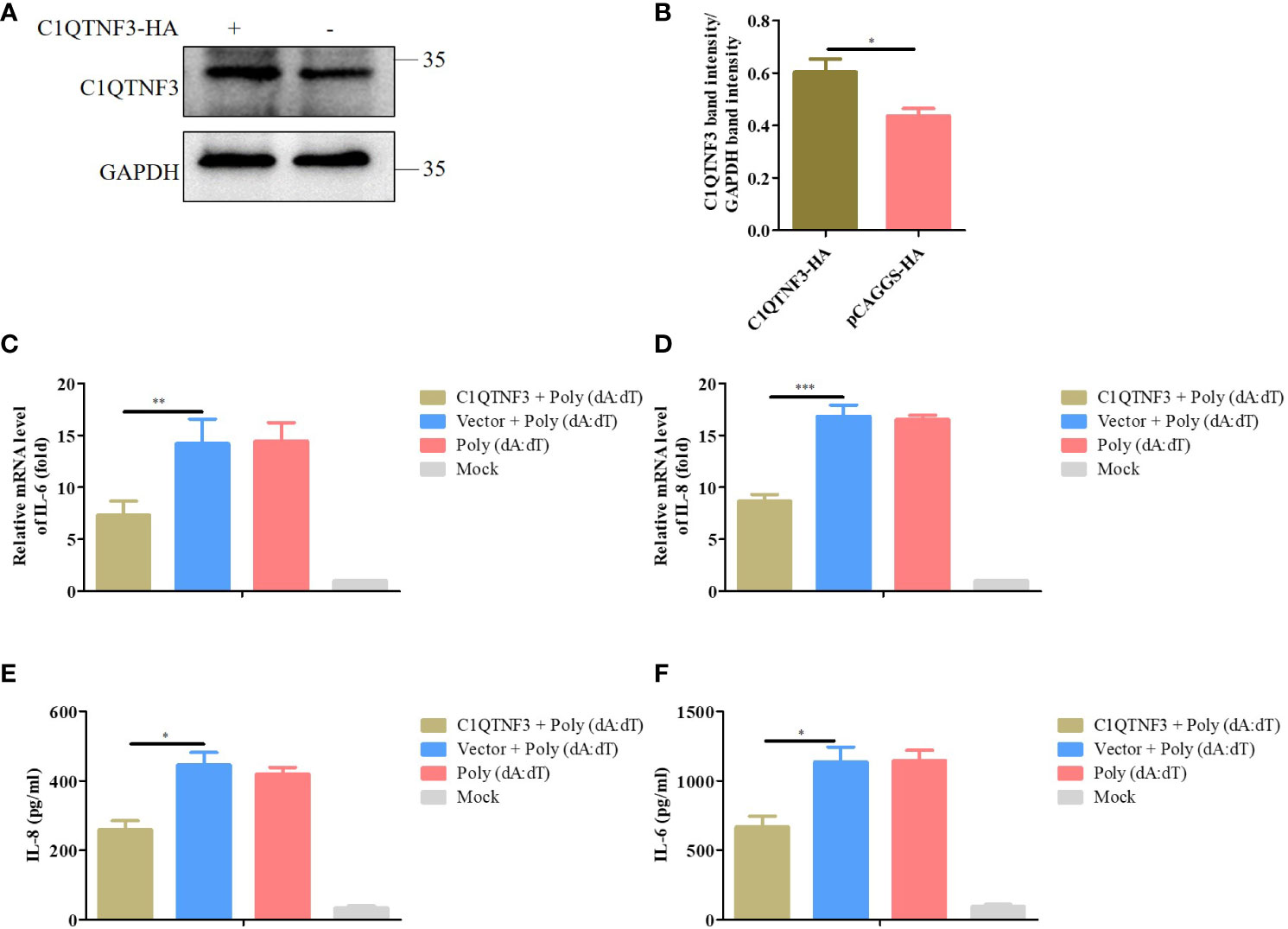
Figure 7 Overexpression C1QTNF3 inhibits poly (dA:dT)-induced inflammatory cytokine production in PK-15 cells. (A, B) Overexpression of HA-tagged C1QTNF3 proteins was detected using western blot analysis. The mRNA levels of (C) IL-6 and (D) IL-8 in PK-15 cells with C1QTNF3 overexpression for 24 h followed by poly (dA:dT) stimulation for 48 h. The GAPDH gene was used as the internal control for normalization. The 2-ΔΔCt method was used to analyze the results of qPCR. The protein levels of (E) IL-8 and (F) IL-6 in PK-15 cells with C1QTNF3 overexpression for 24 h followed by poly (dA:dT) stimulation for 48 h p <0.05 (*), p < 0.01 (**), and p < 0.001 (***).
We explored the effect of the knockdown of C1QTNF3 in poly (dA:dT)-induced expression of inflammatory cytokines. PK-15 cells were transfected with siRNA #1 for 24 h, followed by transfection with poly (dA:dT) for 48 h. The antibody of porcine endogenous C1QTNF3 was used to detect the protein level of C1QTNF3 in the PK-15 cells using western blot analysis (Figure 8A), and the relative protein band intensity was analyzed using ImageJ software (Figure 8B), which indicates that C1QTNF3 was downregulated in the PK-15 cells. The mRNA levels of IL-6 and IL-8 were higher in the PK-15 cells with C1QTNF3 downregulation by siRNA #1 than in the siNC-treated PK-15 cells (Figures 8C, D). The same results were obtained for the protein levels of IL-8 and IL-6 (Figures 8E, F). Therefore, these results further indicate that the expression of C1QTNF3 was negatively correlated with that of the inflammatory cytokines.
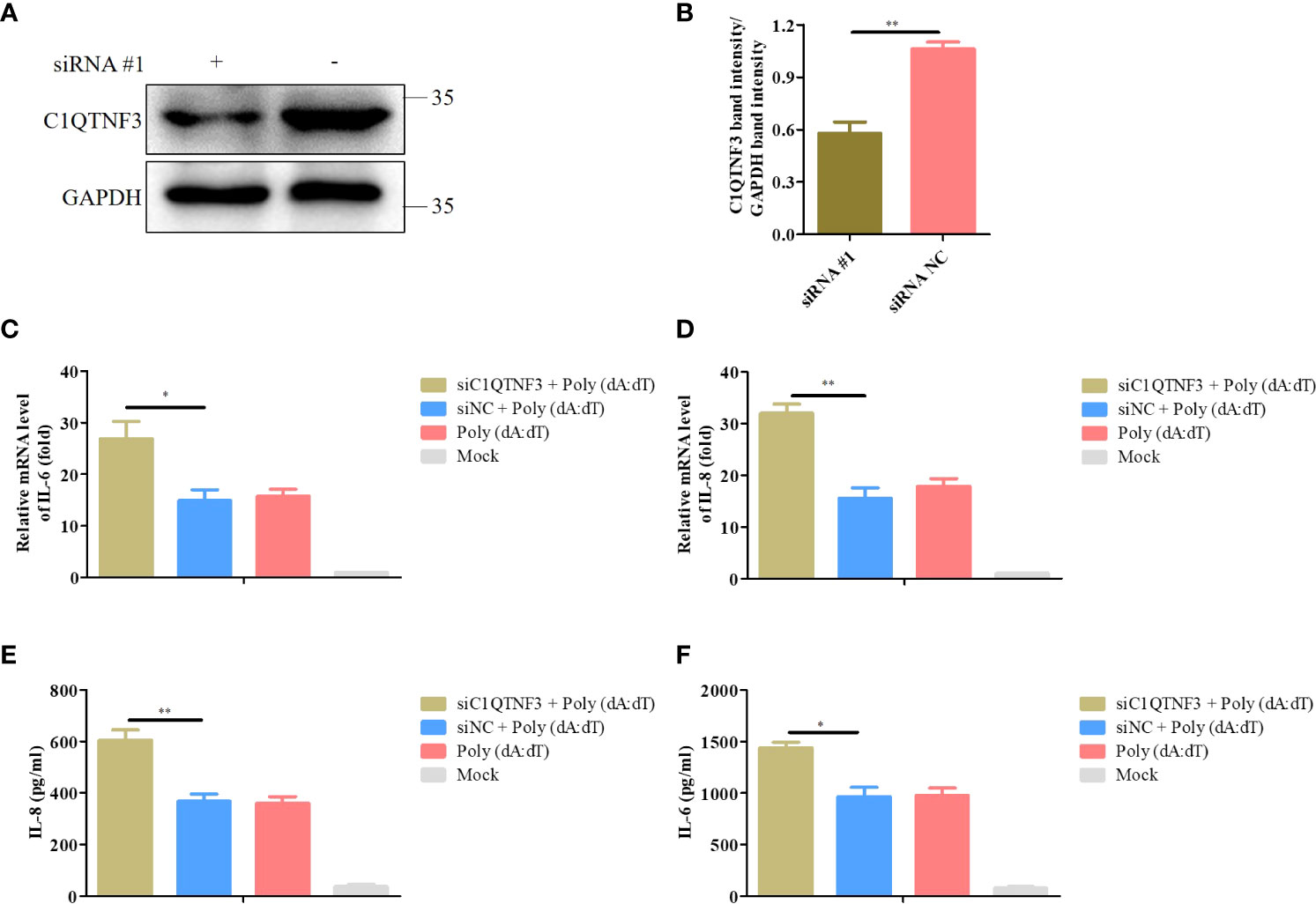
Figure 8 Knockdown of C1QTNF3 increases poly (dA:dT)-induced inflammatory cytokine production in PK-15 cells. (A, B) Detection of C1QTNF3 siRNA knockdown efficiency using western blot analysis. The mRNA levels of (C) IL-6 and (D) IL-8 in PK-15 cells with C1QTNF3 downregulation for 24 h followed by poly (dA:dT) stimulation for 48 h. The GAPDH gene was used as the internal control for normalization. The 2-ΔΔCt method was used to analyze the results of qPCR. The protein levels of (E) IL-8 and (F) IL-6 in PK-15 cells with C1QTNF3 downregulation for 24 h followed by poly (dA:dT) stimulation for 48 h p <0.05 (*), p < 0.01 (**).
A virus hijacks the host’s transcription and translation to complete its replication (34, 35). Simultaneously, the host cells develop several antiviral mechanisms after sensing the viral genetic material (36, 37). If there is an imbalance between the viral infection and host regulation, it may lead to cell death or virus cleaning. Therefore, exploring the host-virus interaction is critical for understanding the pathogenic mechanisms of the virus. In this study, to unravel the mechanisms of the host’s response to ASFV infection, we compared the serum protein level profiles between ASFV-infected samples and the uninfected. Upon label-free LC-MS/MS quantification, 173 DEPs were found between the two groups. These DEPs were involved in several biological processes, comprising the immune system, metabolism, and inflammation. Inflammatory response plays an important role in ASFV proliferation. The serum samples show the changes in multiple inflammatory cytokines after ASFV infection (32). Therefore, the proteomic analysis of serum samples can better reveal the inflammatory response after ASFV infection than other omics. However, the weakness of proteomics of serum samples is that the DEPs are mainly located in the extracellular region compared with the omics of cell or tissue samples, which leads to numerous intracellular proteins not being detected (13, 14). In the present study, C1QTNF3 was downregulated after ASFV infection, which increased the ASFV-induced inflammatory response, inhibiting ASFV replication.
Our data showed that natural and acquired immune responses were activated during ASFV infection (Figures 2C, D). ASFV has been reported to encode various proteins to repress the host’s immunity and complete its propagation. For instance, ASFV Armenia/07 inhibits the cGAS-STING pathway by suppressing STING activation, thereby restraining type I interferons and interferon-stimulated gene expression (38). ASFV E120R interferes with the recruitment of IRF3 to TANK-binding kinase 1, which suppresses IRF3 phosphorylation, decreasing interferon expression (39). Previous proteomic and transcriptomic studies have shown that the metabolic pathways are activated in ASFV-infected PAMs (12, 13). Fatty acid synthesis is essential for the red-spotted grouper nervous necrosis virus to complete its life cycle (40). In this study, we found that the upregulated proteins were involved in regulating fatty acid metabolism after ASFV infection. However, the metabolic pathway involved in ASFV infection remains to be further investigated, which will contribute to understanding the pathogenic mechanisms of ASFV.
ASFV infection activates an inflammatory response in vivo and in vitro (32, 41), which was corroborated by our KEGG analysis result that DEPs were enriched in the inflammatory signaling pathways (Figure 3A). The balance of inflammatory factor expression is vital for the host’s response to ASFV infection. C1QTNF3, as a multifunctional protein, is an anti-inflammatory mediator, and it negatively regulates the pro-inflammatory cytokines TNF-α, IL-6, and C-reactive protein (33). This ability may explain our finding that the expression of C1QTNF3 was negatively correlated with that of the inflammatory cytokines IL-6, IL-8, and IL-1β after ASFV infection (Figure 4). One previous study showed that miR-495-targeted C1QTNF3 inhibits hepatocellular carcinoma cell growth (42). In this study, the knockdown of the C1QTNF3 gene in ASFV-infected PAMs resulted in the increased expression of inflammatory cytokines, thereby inhibiting viral infection (Figure 5). Furthermore, we found that porcine IL-1β, IL-8, and IL-6 proteins repressed ASFV replication (Figure 5G). However, how C1QTNF3 regulates the inflammation process during ASFV infection needs further study.
Previous microarray analysis has identified C1QTNF3 as a downstream molecule of HIF-1α, and C1QTNF3 exerts anticatabolic effects by suppressing NF-κB (43). The inflammatory response is reported to resist viral proliferation; for instance, ASFV MGF505-7R inhibits NF-κB activation and NLRP3 inflammasome assembly, thus reducing IL-1β production, finally promoting viral replication (26). ASFV-encoded early viral protein UBCv1 can block p65 nuclear translocation, inhibiting the inflammatory response (44). I226L, A151R, NP419L, and QP383R also inhibit the inflammatory response in an iGLuc reporter system (45). Moreover, the human cytomegalovirus-encoded protein IE86 impairs the synthesis of the IL-1β transcript and the stability of the immature protein, thus leading to immune evasion through the regulation of inflammasome activation (46). We found that in ASFV-infected PAMs, the expression of inflammatory cytokines was upregulated, and the C1QTNF3 expression was downregulated. Overexpression of C1QTNF3 inhibited inflammatory cytokine production after ASFV infection, in turn enhancing viral proliferation (Figure 6). However, whether the interaction between C1QTNF3 and ASFV-encoded proteins inhibiting the inflammatory response regulates inflammation and viral proliferation deserves further investigation. In addition, the overexpression of C1QTNF3 could inhibit the production of poly (dA:dT)-induced inflammatory cytokines (Figure 7). Therefore, C1QTNF3 may be a broad-spectrum anti-inflammatory mediator and a potential drug to reduce an inflammatory reaction.
In conclusion, this study revealed the ASFV-host interactions in ASFV-infected and uninfected serum samples through quantitative proteomics. We also found that reducing C1QTNF3 expression promotes the production of ASFV infection-induced inflammatory cytokines, thereby inhibiting viral replication. Therefore, this study provides insights into potential antiviral targets in the pathogenic process of ASFV infection.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org, accession ID: PXD035545.
The animal study was reviewed and approved by Research Ethics Committee, Huazhong Agricultural University, Hubei, China.
CJL, MJ, and XL contributed to conception and design of the study. QZ and ZZ organized the database. CFL and XS performed the statistical analysis. CJL wrote the first draft of the manuscript. LZ, JY, and YZ wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This study was supported by the National Key R&D Program of China (Approval No: 2021YFD1801405) and Key technologies for ASFV control from the Department of Science and Technology of Hubei province, China (Approval No: 2019ABA08).
We would like to thank the National Key R&D Program of China, the Department of Science and Technology of Hubei province, and all the members of the animal biosafety level 3 (ABSL-3) laboratory of Huazhong Agricultural University.
Authors CL, ZZ and MJ were employed by company Wuhan Keqian Biology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1002616/full#supplementary-material
1. Dunn LEM, Ivens A, Netherton CL, Chapman DAG, Beard PM. Identification of a functional small noncoding RNA of African swine fever virus. J Virol (2020) 94(21):e01515–20. doi: 10.1128/JVI.01515-20
2. Borca MV, Ramirez-Medina E, Silva E, Vuono E, Rai A, Pruitt S, et al. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. J Virol (2020) 94(7):e02017–19. doi: 10.1128/JVI.02017-19
3. Cackett G, Matelska D, Sykora M, Portugal R, Malecki M, Bahler J, et al. The African swine fever virus transcriptome. J Virol (2020) 94(9):e00119–20. doi: 10.1128/JVI.00119-20
4. Wang A, Jia R, Liu Y, Zhou J, Qi Y, Chen Y, et al. Development of a novel quantitative real-time PCR assay with lyophilized powder reagent to detect African swine fever virus in blood samples of domestic pigs in China. Transboundary emerging Dis (2020) 67(1):284–97. doi: 10.1111/tbed.13350
5. Achenbach JE, Gallardo C, Nieto-Pelegrin E, Rivera-Arroyo B, Degefa-Negi T, Arias M, et al. Identification of a new genotype of African swine fever virus in domestic pigs from Ethiopia. Transboundary emerging Dis (2017) 64(5):1393–404. doi: 10.1111/tbed.12511
6. Urbano AC, Ferreira F. Role of the DNA-binding protein pA104R in ASFV genome packaging and as a novel target for vaccine and drug development. Vaccines (2020) 8(4):585. doi: 10.3390/vaccines8040585
7. Lv C, Zhao Y, Jiang L, Zhao L, Wu C, Hui X, et al. Development of a dual ELISA for the detection of CD2v-unexpressed lower-virulence mutational ASFV. Life (2021) 11(11):1214. doi: 10.3390/life11111214
8. Wang N, Zhao D, Wang J, Zhang Y, Wang M, Gao Y, et al. Architecture of African swine fever virus and implications for viral assembly. Science (2019) 366(2019):640–4. doi: 10.1126/science.aaz1439
9. Zhao D, Liu R, Zhang X, Li F, Wang J, Zhang J, et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerging Microbes infections (2019) 8(1):438–47. doi: 10.1080/22221751.2019.1590128
10. Sun E, Zhang Z, Wang Z, He X, Zhang X, Wang L, et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci China Life Sci (2021) 64(5):752–65. doi: 10.1007/s11427-021-1904-4
11. Gallardo C, Soler A, Rodze I, Nieto R, Cano-Gomez C, Fernandez-Pinero J, et al. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transboundary emerging Dis (2019) 66(3):1399–404. doi: 10.1111/tbed.13132
12. Ju X, Li F, Li J, Wu C, Xiang G, Zhao X, et al. Genome-wide transcriptomic analysis of highly virulent African swine fever virus infection reveals complex and unique virus host interaction. Vet Microbiol (2021) 261:109211. doi: 10.1016/j.vetmic.2021.109211
13. Ai Q, Lin X, Xie H, Li B, Liao M, Fan H. Proteome analysis in PAM cells reveals that African swine fever virus can regulate the level of intracellular polyamines to facilitate its own replication through ARG1. Viruses (2021) 13(7):1236. doi: 10.3390/v13071236
14. Fan W, Cao Y, Jiao P, Yu P, Zhang H, Chen T, et al. Synergistic effect of the responses of different tissues against African swine fever virus. Transboundary emerging Dis (2021) 69(4):e204–15. doi: 10.1111/tbed.14283
15. Chen J, Jin L, Yan M, Yang Z, Wang H, Geng S, et al. Serum exosomes from newborn piglets restrict porcine epidemic diarrhea virus infection. J Proteome Res (2019) 18(5):1939–47. doi: 10.1021/acs.jproteome.9b00195
16. Chen Y, Huang A, Ao W, Wang Z, Yuan J, Song Q, et al. Proteomic analysis of serum proteins from HIV/AIDS patients with talaromyces marneffei infection by TMT labeling-based quantitative proteomics. Clin Proteomics (2018) 15:40. doi: 10.1186/s12014-018-9219-8
17. Sun JF, Shi ZX, Guo HC, Li S, Tu CC. Proteomic analysis of swine serum following highly virulent classical swine fever virus infection. Virol J (2011) 8:107. doi: 10.1186/1743-422X-8-107
18. Puttamallesh VN, Sreenivasamurthy SK, Singh PK, Harsha HC, Ganjiwale A, Broor S, et al. Proteomic profiling of serum samples from chikungunya-infected patients provides insights into host response. Clin Proteomics (2013) 10(1):14. doi: 10.1186/1559-0275-10-14
19. Micallef P, Vujicic M, Wu Y, Peris E, Wang Y, Chanclon B, et al. C1QTNF3 is upregulated during subcutaneous adipose tissue remodeling and stimulates macrophage chemotaxis and M1-like polarization. Front Immunol (2022) 13:914956. doi: 10.3389/fimmu.2022.914956
20. Maeda T, Abe M, Kurisu K, Jikko A, Furukawa S. Molecular cloning and characterization of a novel gene, CORS26, encoding a putative secretory protein and its possible involvement in skeletal development. J Biol Chem (2001) 276(5):3628–34. doi: 10.1074/jbc.M007898200
21. Maeda T, Jikko A, Abe M, Yokohama-Tamaki T, Akiyama H, Furukawa S, et al. Cartducin, a paralog of Acrp30/adiponectin, is induced during chondrogenic differentiation and promotes proliferation of chondrogenic precursors and chondrocytes. J Cell Physiol (2006) 206(2):537–44. doi: 10.1002/jcp.20493
22. Otani M, Kogo M, Furukawa S, Wakisaka S, Maeda T. The adiponectin paralog C1q/TNF-related protein 3 (CTRP3) stimulates testosterone production through the cAMP/PKA signaling pathway. Cytokine (2012) 58(2):238–44. doi: 10.1016/j.cyto.2012.01.018
23. Weigert J, Neumeier M, Schaffler A, Fleck M, Scholmerich J, Schutz C, et al. The adiponectin paralog CORS-26 has anti-inflammatory properties and is produced by human monocytic cells. FEBS Lett (2005) 579(25):5565–70. doi: 10.1016/j.febslet.2005.09.022
24. Deng W, Li C, Zhang Y, Zhao J, Yang M, Tian M, et al. Serum C1q/TNF-related protein-3 (CTRP3) levels are decreased in obesity and hypertension and are negatively correlated with parameters of insulin resistance. Diabetol Metab syndrome (2015) 7:33. doi: 10.1186/s13098-015-0029-0
25. Lin J, Liu Qi, Zhang H, Huang X, Zhang R, Chen S, et al. C1q tumor necrosis factor related protein 3 protects macrophages against LPS induced lipid accumulation, inflammation and phenotype transition via PPARγ and TLR4 mediated pathways. Oncotarget (2017) 8(47):82541–57. doi: 10.18632/oncotarget.19657
26. Li J, Song J, Kang L, Huang L, Zhou S, Hu L, et al. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PloS Pathog (2021) 17(7):e1009733. doi: 10.1371/journal.ppat.1009733
27. C AL, Santaren JF, Viruela E. Production and titration of African swine fever virus in porcine alveolar macrophages. J virol Methods (1982) 3(1982):303–10. doi: 10.1016/0166-0934(82)90034-9
28. Malmquist Wa, Hay D. Hemadsorption and cytopathic effect produced by African swine fever virus in swine bone marrow and buffy coat cultures. Am J vet Res (1960) 21:104–8. doi: 10.1080/01944365908978338
29. Reed LJ HM. A simple method of estimaing fifty percent endpoints. Am J Epidemiol (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
30. Guinat C, Reis AL, Netherton CL, Goatley L, Pfeiffer DU, Dixon L. Dynamics of African swine fever virus shedding and excretion in domestic pigs infected by intramuscular inoculation and contact transmission. Vet Res (2014) 45(1):93. doi: 10.1186/s13567-014-0093-8
31. Giang J, Seelen MAJ, van Doorn MBA, Rissmann R, Prens EP, Damman J. Complement activation in inflammatory skin diseases. Front Immunol (2018) 9:639. doi: 10.3389/fimmu.2018.00639
32. Wang S, Zhang J, Zhang Y, Yang J, Wang L, Qi Y, et al. Cytokine storm in domestic pigs induced by infection of virulent African swine fever virus. Front vet Sci (2020) 7:601641. doi: 10.3389/fvets.2020.601641
33. Li Y, Wright GL, Peterson JM. C1q/TNF-related protein 3 (CTRP3) function and regulation. Compr Physiol (2017) 7(3):863–78. doi: 10.1002/cphy.c160044
34. Perrin-Cocon L, Diaz O, Jacquemin C, Barthel V, Ogire E, Ramiere C, et al. The current landscape of coronavirus-host protein-protein interactions. J Trans Med (2020) 18(1):319. doi: 10.1186/s12967-020-02480-z
35. Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang Ke, Cohen P, Patel RS, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. PNAS (2020) 117(45):28344–54. doi: 10.1073/pnas.2016650117/-/DCSupplemental
36. Zhang X, Bai XC, Chen ZJ. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity (2020) 53(1):43–53. doi: 10.1016/j.immuni.2020.05.013
37. Qiao Y, Zhu S, Deng S, Zou SS, Gao B, Zang G, et al. Human cancer cells sense cytosolic nucleic acids through the RIG-I-MAVS pathway and cGAS-STING pathway. Front Cell Dev Biol (2020) 8:606001. doi: 10.3389/fcell.2020.606001
38. Garcia-Belmonte R, Perez-Nunez D, Pittau M, Richt JA, Revilla Y. African Swine fever virus Armenia/07 virulent strain controls interferon beta production through the cGAS-STING pathway. J Virol (2019) 93(12):e02298–18. doi: 10.1128/JVI.02298-18
39. Liu H, Zhu Z, Feng T, Ma Z, Xue Q, Wu P, et al. African Swine fever virus E120R protein inhibits interferon beta production by interacting with IRF3 to block its activation. J Virol (2021) 95(18):e00824–21. doi: 10.1128/JVI.00824-21
40. Huang Y, Zhang Y, Zheng J, Wang L, Qin Q, Huang X. Metabolic profiles of fish nodavirus infection in vitro: RGNNV induced and exploited cellular fatty acid synthesis for virus infection. Cell Microbiol (2020) 22(9):e13216. doi: 10.1111/cmi.13216
41. Zhu JJ, Ramanathan P, Bishop EA, O'Donnell V, Gladue DP, Borca MV. Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PloS One (2019) 14(11):e0223955. doi: 10.1371/journal.pone.0223955
42. Zhang R, Guo C, Liu T, Li W, Chen X. MicroRNA miR-495 regulates the development of hepatocellular carcinoma by targeting C1q/tumor necrosis factor-related protein-3 (CTRP3). Bioengineered (2021) 12(1):6902–12. doi: 10.1080/21655979.2021.1973878
43. Okada K, Mori D, Makii Y, Nakamoto H, Murahashi Y, Yano F, et al. Hypoxia-inducible factor-1 alpha maintains mouse articular cartilage through suppression of NF-kappaB signaling. Sci Rep (2020) 10(1):5425. doi: 10.1038/s41598-020-62463-4
44. Barrado-Gil L, Del Puerto A, Galindo I, Cuesta-Geijo MA, Garcia-Dorival I, de Motes CM, et al. African Swine fever virus ubiquitin-conjugating enzyme is an immunomodulator targeting NF-kappaB activation. Viruses (2021) 13(6):1160. doi: 10.3390/v13061160
45. Song J, Li K, Li T, Zhao G, Zhou S, Li H, et al. Screening of PRRSV- and ASFV-encoded proteins involved in the inflammatory response using a porcine iGLuc reporter. J virol Methods (2020) 285:113958. doi: 10.1016/j.jviromet.2020.113958
46. Botto S, Abraham J, Mizuno N, Pryke K, Gall B, Landais I, et al. Human cytomegalovirus immediate early 86-kDa protein blocks transcription and induces degradation of the immature interleukin-1beta protein during virion-mediated activation of the AIM2 inflammasome. mBio (2019) 10(1):e02510–18. doi: 10.1128/mBio.02510-18
Keywords: African swine fever virus, quantitative proteomics, C1QTNF3, inflammatory process, host-virus infection
Citation: Lv C, Zhang Q, Zhao L, Yang J, Zou Z, Zhao Y, Li C, Sun X, Lin X and Jin M (2022) African swine fever virus infection activates inflammatory responses through downregulation of the anti-inflammatory molecule C1QTNF3. Front. Immunol. 13:1002616. doi: 10.3389/fimmu.2022.1002616
Received: 25 July 2022; Accepted: 26 September 2022;
Published: 12 October 2022.
Edited by:
Li Huang, Harbin Veterinary Research Institute (CAAS), ChinaReviewed by:
Bin Li, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaCopyright © 2022 Lv, Zhang, Zhao, Yang, Zou, Zhao, Li, Sun, Lin and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian Lin, linxian1221@163.com; Meilin Jin, jml8328@126.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.