
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol. , 12 August 2022
Sec. Fundamental Virology
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.980412
 Ben Jackson1
Ben Jackson1 Tiphany Chrun1
Tiphany Chrun1 Kay Childs1
Kay Childs1 Nanchaya Wanasen2
Nanchaya Wanasen2 Jean-Pierre Frossard3
Jean-Pierre Frossard3 Simon P. Graham1
Simon P. Graham1 Julian Seago1*
Julian Seago1*Porcine reproductive and respiratory syndrome virus 1 (PRRSV-1) causes huge economic losses to the European pig industry. PRRSV-1 is divided into 3 subtypes and exhibits considerable antigenic heterogeneity. Due to its high mutation rate, PRRSV-1 is constantly evolving, and highly virulent, particularly subtype 3 strains, are continually emerging. The mechanism(s) underlying PRRSV-1 virulence have not been fully elucidated. In vivo studies have implicated replication kinetics, cell tropism and an enhanced pro-inflammatory cytokine response as potential contributing factors. However, few strains have been directly compared and differences in in vivo study design have hindered comparison, thus limiting our understanding of PRRSV-1 virulence. To address this knowledge gap, we sought to develop a reverse genetics and ex vivo model system, to attempt to identify correlates of PRRSV-1 virulence and attenuation in vitro. Herein we describe the use of primary porcine bone marrow-derived macrophages (BMDM) to investigate the growth kinetics and induced cytokine profiles of the highly virulent SU1-Bel strain, the low virulence 215-06 strain and the attenuated Olot/91 strain. We show that infection of BMDM with virulent PRRSV-1 strains induced higher expression of IL-6 and IL-8 and lower expression of TNF-α when compared with the attenuated strain. In addition, BMDM infected with SU1-Bel secreted significantly more IFN-α than those infected with PRRSV-1 strains of lower virulence. Interestingly, despite inducing less IFN-α than SU1-Bel, Olot/91 induced much higher levels of expression of several interferon-stimulated genes (ISGs), suggesting that Olot/91 may be less able to counteract type I IFN signaling which may contribute to its attenuated phenotype.
Porcine reproductive and respiratory syndrome (PRRS) is caused by two genetically and antigenically distinct species of Beta arterivirus suid (-1 and -2), each containing a single virus member known as PRRS virus 1 (PRRSV-1) or PRRSV-2 respectively (1). Infection with either species is restricted to pigs, where clinical symptoms are dependent on strain virulence, as well as the age, breed, immune and gestational status of the host. Infection in utero induces late term reproductive losses and mortality in weaning pigs, whereas neonatal infection causes respiratory disease (2). Viral persistence and a delayed humoral response create a barrier to effective control, resulting in an estimated US$ 663.91 million yearly loss to the U.S. pig industry (3). Control strategies rely on infection management and vaccination. However, vaccination induces low neutralizing antibody titres and poor heterologous protection, a problem compounded by the antigenic diversity of PRRSV-1 (4, 5). In addition, PRRSV attenuation is not well defined and safety concerns surrounding live vaccine strains persist (6).
PRRSV-1 is divided into three subtypes (1–3). The pan European subtype 1 strains are prominent in Western Europe and also circulate in Asia and North America, whereas subtype 2 and subtype 3 strains are more commonly found in Eastern Europe. Highly virulent strains are found in each subtype but are typically found in subtype 3 (4). The mechanisms contributing to PRRSV-1 virulence are not well defined. Increased viral replication, broader tropism and an enhanced pro-inflammatory response have been suggested mechanisms (7–11). In vivo, pro-inflammatory cytokines IL-1α, IL-1β, IL-6, IL-8, TNF-α, IFN-α, IFN-γ as well as the anti-inflammatory cytokine IL-10 have been shown to be differentially upregulated following infection with PRRSV-1 and/or PRRSV-2 (11–22) Table 1. Unfortunately, it is difficult to assess the importance of cytokine production for PRRSV-1 virulence, as few strains have been compared directly and variations in experimental design may have contributed to differences in the cytokine response observed in these studies. Therefore, to facilitate direct strain comparisons in the future, we sought to develop a reverse genetics and relevant ex vivo system to identify correlates of PRRSV-1 virulence and attenuation in vitro.
In vivo, PRRSV replication is limited to CD163 expressing cells of the monocyte/macrophage lineage (22). PRRSV-2 can adapt to grow in African green monkey cells (MA104 and MARC-145) that express the simian CD163 receptor. Unfortunately, due to increased mutational requirements, reverse genetics is required to facilitate MARC-145 adaption of many PRRSV-1 strains (23). Expression of exogenous porcine CD163 supports productive infection, but whilst secondary cell lines can be used as an initial tool to study replication, they do not represent an ideal system to investigate cytokine responses (24). As a primary target cell, porcine alveolar macrophages (PAM) represent the most relevant system for studying PRRSV-1 induced immune responses in vitro. However, sterile homogeneous PAM are difficult to obtain from heterogenous cell populations in lung lavage and when combined with additional confounding factors such as animal age and health status, introduce unwanted intra-assay variability (25). Both lesions and viral RNA have been detected in the bone marrow (BM) of PRRSV-infected pigs suggesting viral replication in BM cells (26–28). Freshly sorted non-differentiated CD163+ (BM) monocytes and BM pro-genitor matured to macrophages in vitro are permissive to PRRSV infection, with the latter facilitating comparable PRRSV-1 growth kinetics to that observed in PAM (18, 22, 25, 29–33). Since large numbers of bone marrow-derived macrophages (BMDM) can be harvested, cryopreserved and matured as required, we considered them to be a good candidate model cell type to investigate correlates of PRRSV-1 virulence.
To investigate replication kinetics and induced cytokine responses in PRRSV-1 infected BMDM, we developed a reverse genetic system to recover three PRRSV-1 strains; a highly virulent subtype 3 strain (SU1-Bel), a low virulence subtype 1 strain (215–06) and an attenuated subtype 1 strain (ESP-1991-Olot/91, Olot/91) (8, 9, 34). Infection with virulent PRRSV-1 strains induced higher expression of the pro-inflammatory cytokines IL-6 and IL-8 and lower expression of TNF-α compared with the attenuated strain. Differential expression of type I IFN and IFN stimulated genes (ISGs) were also observed in a strain specific manner. These data show that BMDM represent a good model system for studying PRRSV-1 infection in vitro, secreting many of the pro-inflammatory cytokines (IL-6, IL-8, TNF-α, IFN-α) observed following PRRSV infection in vivo. In addition, data presented herein, suggest that differential expression of IFN-α and the differential inhibition of the type I IFN response may contribute to the virulence and attenuation of PRRSV-1.
African green monkey kidney epithelial cells ((MARC-145), American Type Culture Collection, Manassas, US), used for virus preparations of PRRSV-1 strain Olot/91, were cultured in Dulbecco’s modified Eagle’s medium ((DMEM), Thermo Fisher Scientific, Paisley, UK) containing 5 % adult bovine serum ((ABS), Selborne Biological Services Ltd., Alton, UK), 2 mM L-glutamine (Thermo Fisher Scientific), penicillin (100 IU/mL, Merck Life Science UK Ltd., Dorset, UK) and streptomycin (100 µg/mL, Merck). Baby Hamster Kidney ((BHK21), Merck) cells, used for the recovery of recombinant PRRSV-1, were grown in Glasgow’s Modified Eagle’s Medium ((GMEM), Merck) containing 10% ABS, 5% tryptose phosphate broth (Thermo Fisher Scientific), 2 mM L-glutamine, penicillin (100 IU/mL) and streptomycin (100 µg/mL). Porcine bone marrow was obtained mechanically from the leg bones of 8-10 kg PRRSV free pigs and transferred to phosphate buffered saline (PBS) containing 1% ABS, penicillin (100 IU/mL) and streptomycin (100 µg/mL) and agitated at 37°C for 2 h. All additional steps were performed at room temperature (RT). Cells were strained and centrifuged at 280xg for 5 min, washed in 10 mL PBS and centrifuged at 280xg for 5 min, then re-suspended in PBS. 15 mL of Histopaque-1077 (Sigma) was overlayed with 30 mL cell suspension and centrifuged at 1000xg for 30 min. Buffy coats were pooled in 40 mL PBS and centrifuged at 400xg for 5 min. The cell pellet was re-suspended in 5 mL 1x red blood cell lysis buffer (Thermo Fisher Scientific) for 5 min., then mononuclear cells were washed in PBS and centrifuged at 280xg for 5 min. Cell pellets were re-suspended at 1x 107/mL in 90% ABS + 10% dimethyl sulfoxide (Merck) and stored at -80°C. When required, mononuclear cells were re-suspended in RPMI 1640 medium (Thermo Fisher Scientific) containing 10% ABS, 2 mM L-glutamine, penicillin (100 IU/mL), streptomycin (100 µg/mL) and 50 ng/mL recombinant porcine macrophage colony stimulating factor (M-CSF; Bio-Rad, Watford, UK). Mononuclear cells were incubated for 5 days at 37°C/5% CO2 to generate BMDM. Cell maintenance and cellular assays were performed at 37°C/5% CO2 unless otherwise stated.
The nucleotide sequence for 215-06 (AC# OP047897) was provided by the Animal and Plant Health Agency (APHA). Olot/91 (AC# KC862570) and SU1-Bel (AC# KP889243) sequences are available on GenBank. cDNA was ordered from GeneArt gene synthesis (Thermo Fisher Scientific) and cloned into pcDNA3.1 as described in Results Section 3.1. BHK-21 cells grown in 6-well plates were transfected with 3 µg of pcDNA3.1 Olot/91, pcDNA3.1 215-06 or pcDNA3.1 SU1-Bel, using 6 µL Transit-LT1 (Mirus Bio, Madison, US) and incubated for 48 h then subjected to 1x freeze/thaw cycle. P0 supernatants were centrifuged at 1600xg and repeatedly passaged on BMDM or MARC-145 cells to recover recombinant PRRSV-1. In brief, media was removed from confluent T25 flasks of cells and 2 mL P0 added for 2 h, then removed and replaced with respective complete media containing 4% ABS and incubated for 72 h, subjected to 1x freeze/thaw cycle and the process repeated until full cytopathic effect (cpe) was observed.
Confluent BMDM were infected with recombinant 215-06 or SU1-Bel, and MARC-145 were infected with Olot/91. After 72 h incubation, cells were subjected to 1x freeze/thaw cycle and centrifuged at 1600xg for 30 min, then 20 mL of clarified supernatant was layered onto 18 mL of 20% sucrose in PBS and centrifuged at 28 000 rpm for 2.5 h in a SW32 TI rotor using an Optima L-100 XP Ultracentrifuge (Beckman Coulter). Virus pellets were re-suspended in RPMI-1640 media, aliquoted into 1 mL vials and stored at -80°C. All experiments were conducted with P3 virus stocks.
Virus concentrations were determined by Median Tissue Culture Infectious Dose (TCID50) assays. In brief, 96-well plates containing confluent BMDM were infected with 10-fold dilutions of recombinant PRRSV-1 in RPMI-1640 media containing 4% ABS, 2 mM L-glutamine, penicillin (100 IU/ml) and streptomycin (100 µg/mL) and incubated for 72 h. All additional steps were performed at RT unless otherwise stated. Cells were fixed in 4% paraformaldehyde in PBS (Merck), permeabilized with 0.1% Triton-X 100 for 15 min and incubated in blocking buffer (PBS + 10% normal goat serum) for 30 min. Primary SDOW17-A monoclonal antibody (Rural Technologies Inc. (RTI), Brookings, US) was diluted 1:1000 in blocking buffer, added to cells and incubated for 1 h at 4°C. Cells were then washed four times in PBS. Alexa Flour 488 conjugated IgG1 secondary antibody (Thermo Fisher Scientific, #A21121) was diluted 1 in 500 in blocking buffer and added for 1 h, then cells were washed twice in PBS before the addition of 100 µL PBS/well. Infected wells were assessed manually using a Leica DMiL fluorescent microscope and TCID50 calculated by the Spearman–Kärber method (35).
Bone marrow derived mononuclear cells were seeded in 24-well plates at a concentration of 5 x 105 cells/well and cultured for 5 days at 37°C/5% CO2 in RPMI-1640 media containing 10% ABS and 50 ng/mL recombinant porcine CSF-1 (Bio-Rad). Cells were infected (MOI 0.01) with purified recombinant PRRSV-1 in serum free RPMI-1640. After 2 h incubation, media was removed and cells washed twice and replaced with RPMI-1640 containing 4% ABS. Supernatants were collected at 0, 12, 24 and 48 hours post infection (hpi) and titrated on BMDM by TCID50. Experiments were performed on three occasions and all samples from one occasion were titrated on freshly prepared BMDM from a single animal.
Supernatant from infected cells was clarified by centrifugation (10,000xg) and stored at -80°C. Cytokines were quantified using the Mx IFN bioassay and CAT Elisa (Merck, #11363727001) as previously described (36) or by using commercially available ELISA kits to detect, IL-1α (Merck, #ES10RB), IL-1β (R&D systems, Minneapolis, US, #P8000), IL-6 (R&D systems, #P6000B), IL-8 (R&D systems, #PLB00B), IL-10 (R&D systems, #P1000), TNF-α (R&D systems, #PTA00), IFN-α (Merck, #RAB1131), IL-28 (MyBiosource, San Diego, US, #MBS9367259), IL-29 (MyBiosource, #MBS2029742) and IFN-γ (Thermo Fisher Scientific #KSC4021), following the manufacturers’ protocol.
Western blots were performed as previously described (37). Primary antibodies; Anti-RIG-I (Santa Cruz, Dallas, US #sc-376845), Anti-Mx1 (GeneTex, Hsinchu City 300 Taiwan, R.O.C., #GTX110256), Anti-ISG15 (Abcam, Cambridge, UK, #ab233071), Anti-γ-tubulin (Merck, #T6557) were used where indicated. Bound primary antibodies were detected by horseradish peroxidise-conjugated anti-mouse (Promega, Madison, US, #W402B), anti-rabbit (Promega, #W401B), or IRDyeR 800CW anti-mouse (LI-COR, Lincoln, US #926-32210) secondary antibodies as indicated.
Bar graphs were generated using Graphpad Prism® 8 software and represents the mean value of three independent experiments (n = 3). Error bars represent ± standard deviation between these experiments. Samples with equal variance (F-test, p<0.05) were compared using an unpaired student t-test assuming equal variances in Microsoft® excel®. Samples with unequal variance (F-test, p>0.05) were compared using an unpaired student t-test assuming unequal variances in Microsoft® Excel®. The difference between mean values was considered statistically significant when p≤ 0.05 and highly statistically significant when p≤ 0.01.
We aimed to develop an in vitro system to investigate recombinant PRRSV-1 strains of differing virulence. To this end, three PRRSV-1 strains for which full length genomic sequence was available were selected, namely Olot/91, 215-06 and SU1-Bel. Olot/91 PRRSV-1 (AC# KC862570) originated from Spain and is a modified live subtype-1 strain that has been attenuated by passage on MA104 and MARC-145 cells. Strain 215-06 (AC# OP047897) is a low virulence strain that originated from the UK and SU1-Bel (AC# KP889243) is a highly virulent strain from Belarus; both were isolated at the Animal and Plant Health Agency (APHA) and are non-cell culture adapted subtype-1 and -3 strains, respectively (8, 9, 31).
To enable the generation of the selected PRRSV-1 strains as recombinant viruses with defined wild-type genotypes (in the case of 215-06 and SU1-Bel) and to facilitate the future manipulation of viral genomes, a reverse genetic approach was first employed. The PRRSV genome possesses a putative type I 5′ cap structure, so infectious cDNA clones containing each of the selected PRRSV genomes were constructed using the pcDNA3.1 plasmid vector that contains the eukaryotic polymerase II cytomegalovirus (CMV) enhancer-promoter. Each genome was synthesized as 3 fragments of approximately 2800 to 6900 bp with complementary single restriction sites at their termini to facilitate sequential insertion into pcDNA3.1. To facilitate this, some existing restriction sites were removed from the viral genome, whilst others were inserted, by codon alterations using single nucleotide changes that maintained both the integrity of overlapping ORFs and the respective amino acid sequences they encode. These conditions were also maintained when additional codons in the ESP-1991-Olot/91 genome were altered to facilitate the synthesis of fragments. A generic poly(A) tail of 25 was incorporated at the end of each 3’UTR followed by the hepatitis delta virus (HDV) ribozyme to ensure correct 3’ cleavage of the transcripts produced from the cloned cDNA. Where degenerate nucleotides were present in available Olot/91 sequence (AC# KC862570), these were replaced with the specific nucleotide from the Olot/91 sequence published by Lu et al. (2014) (KF203132) which shares approximately 97% identity. The respective number of nucleotides changed in the Olot/91, 215-06 and SU1-Bel genomes was 44, 3, and 4. Following their construction and verification by sequencing across the ligated portion of each fragment, infectious plasmids were used to generate recombinant virus stocks. Although the wild type amino acid sequences were maintained for each recombinant virus, due to the higher number of nucleotide changes that were made to the Olot/91 genome, we compared the growth of wild type and recombinant Olot/91 viruses and showed that they exhibited equal replication kinetics (Figure S1).
We hypothesized that viral infection of the BMDM had stimulated the production of cytokines and that their presence in the virus stocks would influence comparative analyses of the three strains. Indeed, ELISA-based quantification of the SU1-Bel P3 virus stock identified higher concentrations of the pro-inflammatory cytokines IL-1β and IL-8 in comparison to control supernatant. Therefore, to remove such cytokines, virus stocks were purified by sucrose cushion and the absence of IL-1β, IL-6, IL-8, TNF-α and type I and III IFN in each of the viral preparations confirmed by ELISA based cytokine analysis. In addition to utilizing BMDM for the sterile recovery of recombinant virus, we wanted to investigate their potential as an ex vivo system for studying PRRSV-1 growth and virulence. To facilitate this, bone marrow-derived mononuclear cells were isolated and cryopreserved from three animals using established methodology. Purified virus stocks and these cryopreserved mononuclear cells were used in all subsequent experiments.
The correlation between PRRSV-1 growth kinetics and virulence remains unclear. We hypothesized that the enhanced virulence of the PRRSV-1 strain SU1-Bel, could be attributed to enhanced growth rate in certain permissive cell types. Therefore, as an initial step, we compared the growth of SU1-Bel, 215-06 and Olot/91 in BMDM. Cryo-preserved cells from three animals were cultured in the presence of recombinant porcine M-CSF, then infected (MOI 0.01) with the respective PRRSV-1 strains and virus growth monitored over time (Figure 1). When SU1-Bel and 215-06 were compared, higher viral titres were observed for SU1-Bel at 12 and 24 hpi in BMDM from all three animals (12 h: Pig 1 p = 0.062; Pig 2 p = 0.057; Pig 3 p = 0.148 and 24 h: Pig 1 p = 0.039; Pig 2 p = 0.029; Pig 3 p = 0.113). An increase in viral titre at 12 and 24 hpi was also observed when SU1-Bel was compared with the Olot/91 strain (12 h: Pig 1 p = 0.058; Pig 2 p = 0.056; Pig 3 p = 0.144, and 24 h: Pig 1 p = 0.060; Pig 2 p = 0.040; Pig 3 p = 0.105). Despite showing slower growth kinetics, the 215-06 strain reached a similar endpoint titre (48 hpi) to SU1-Bel in BMDM from all three animals (Pig 1: 3.61 x 106 v 2.53 x 106 TCID50/mL, Pig 2: 3.61 x 106 v 3.73 x 106 TCID50/mL and Pig 3: 4.7 x 106 v 5.15 x 106 TCID50/mL, respectively). When compared with the 215-06 strain, Olot/91 showed similar viral titres at 12 and 24 hpi. However, a 0.5~1 log reduction in viral titre was observed at 48 hpi, when Olot/91 was compared with either the SU1-Bel (Pig 1: 1.31 x 106 v 2.53 x 106 TCID50/mL, Pig 2: 2.18 x105 v 3.73x106 TCID50/mL, Pig 3: 9.86 x105 v 5.15x106 TCID50/mL) or 215-06 strains (Pig 1: 1.31 x 106 v 3.61 x 106 TCID50/mL, Pig 2: 2.18 x105 v 3.61 x 106 TCID50/mL, and Pig 3: 9.86 x105 v 4.7 x 106 TCID50/mL), with the difference considered significant in BMDM from one animal (v SU1-Bel at 48 h: Pig 3 p = 0.002, and v 215-06 at 48 h: Pig 3 p = 0.039). Taken together, these data showed that highly virulent SU1-Bel replicated more quickly in BMDM compared to the less virulent 215-06 and attenuated Olot/91, and that Olot/91 reached a lower viral titre compared with the virulent PRRSV-1 strains.
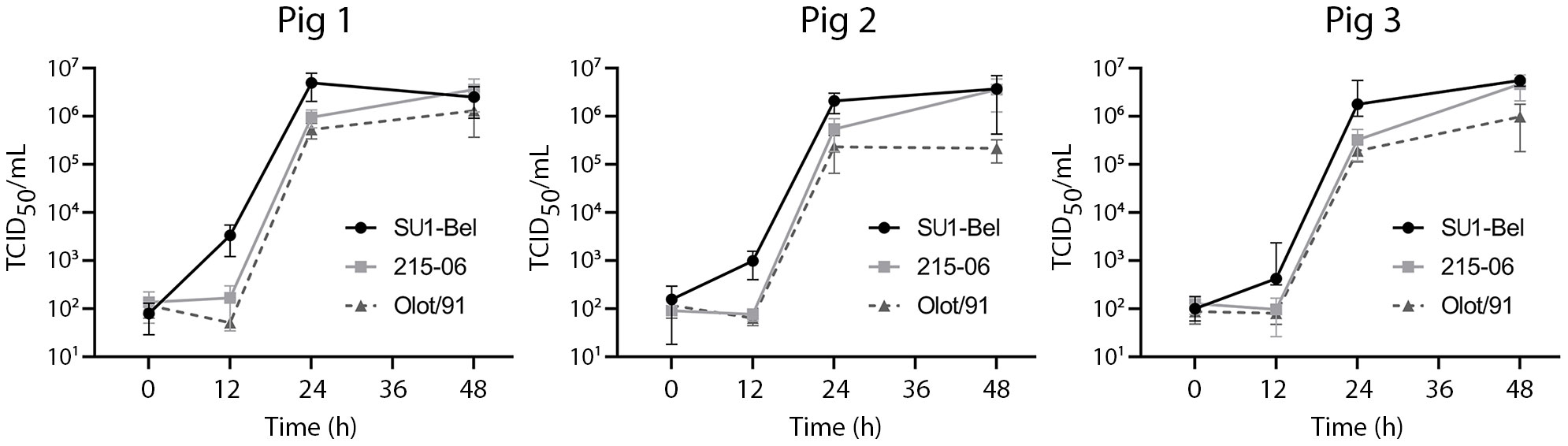
Figure 1 Comparative growth kinetics of PRRSV-1 strains of different virulence in porcine BMDM. Cells from three pigs were infected (MOI 0.01) with either PRRSV-1 strain SU1-Bel, 215-06, or Olot/91 for 48 h. Supernatant was sampled at 0, 12, 24 and 48 hpi and virus titrated by TCID50. Experiments were performed on three separate occasions. Data is presented as the mean +/- standard deviation.
Upregulation of the pro-inflammatory cytokines IL-1α, IL-1β, IL-6, IL-8, TNF-α, IFN-α, IFN-γ, along with the anti-inflammatory cytokine IL-10 have been observed following experimental PRRSV infection in vivo (Table 1). Therefore, we investigated whether PRRSV-1 induced a pro-inflammatory cytokine response in infected porcine BMDM, to explore their potential as an ex vivo model for studying virulence. BMDM were first infected (MOI 0.01) with purified SU1-Bel, 215-06 or Olot/91 and at 48 hpi supernatants were analyzed for IL-1α, IL-1β, IL-6, IL-8, IL-10 and TNF-α by ELISA. When compared with 215-06, infection with SU1-Bel induced significantly higher concentrations of IL-6 (Figure 2A) in BMDM derived from two out of three animals (Pig 1 p = 0.036; Pig 3 p = 0.002). However, the opposite was observed in BMDM from the other animal (Pig 2 p = 0.039). Interestingly, Olot/91 induced significantly lower concentrations of IL-6 in BMDM from two out of three animals when compared with cells infected with either SU1-Bel (Pig 1 p = 0.013; Pig 2 p = 0.01) or 215-06 (Pig 1 p = 0.0002; Pig 2 p = 0.025). In addition to the increased production of IL-6, infection with SU1-Bel or 215-06 also induced significantly higher concentrations of IL-8 (Figure 2B) in BMDM from two out of three animals, when compared with cells infected with Olot/91 (SU1-Bel: Pig 1 p = 0.0006; Pig 2 p = 0.003 and 215-06: Pig 1 p = 0.007; Pig 2 p = 0.0006). Interestingly, strain-specific differences in TNF-α secretion were also observed following infection of BMDM (Figure 2C). Low concentrations of TNF-α were observed following the infection of BMDM with each PRRSV-1 strain, however Olot/91 produced significantly higher levels of TNF-α compared with SU1-Bel (Pig 1 p = 0.04; Pig 2 p = 0.001; Pig 3 p = 0.03) or 215-06 (Pig 1 p = 0.0002; Pig 2 p = 0.006; Pig 3 p = 0.02).
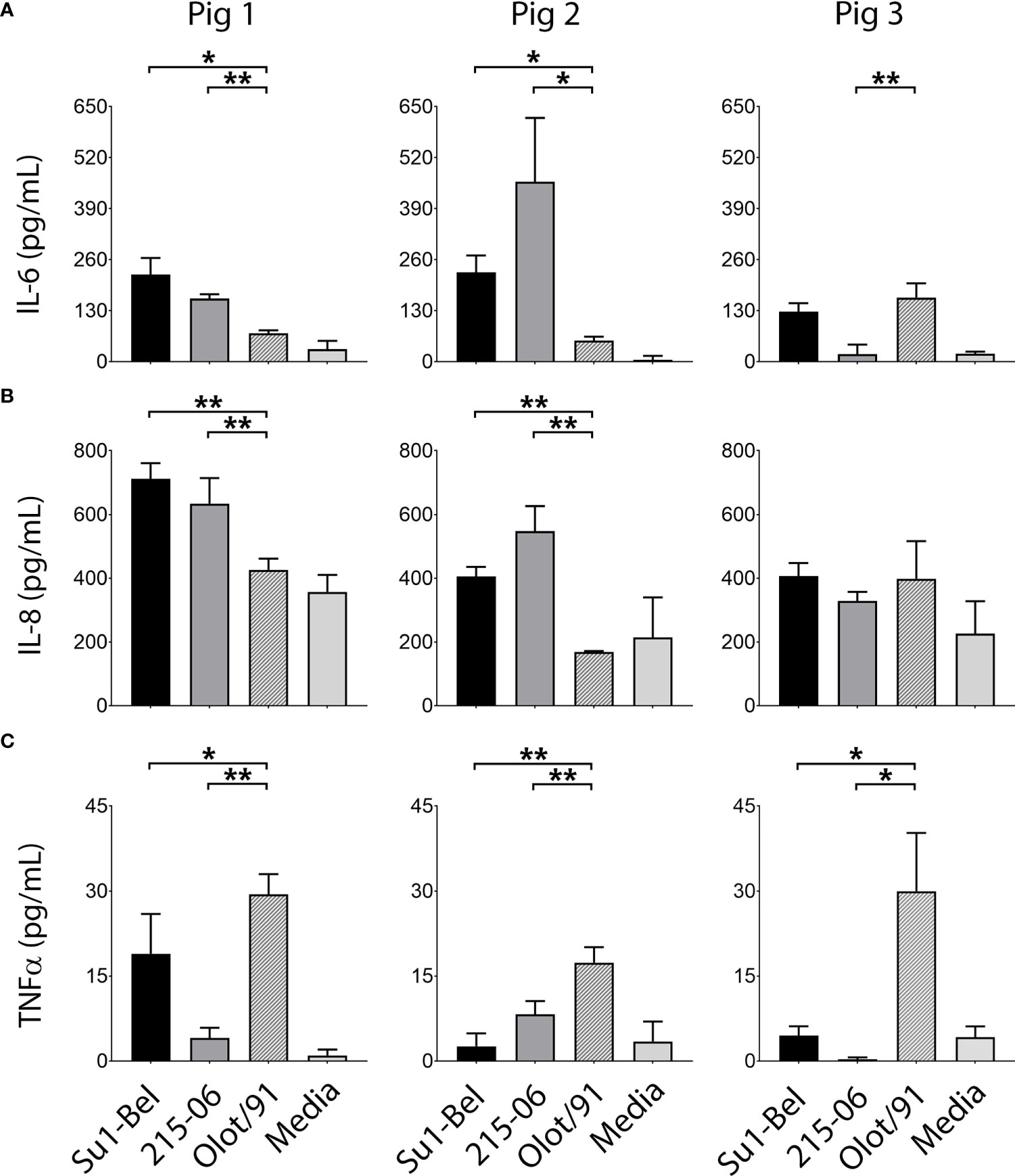
Figure 2 Virulent PRRSV-1 strains induced higher expression of IL-6 and IL-8 and lower expression of TNF-α than the attenuated Olot/91 strain. BMDM from three animals were infected (MOI 0.01) with PRRSV-1 strains of different virulence, SU1-Bel, 215-06 and Olot/91 and at 48 hpi supernatants were analyzed for (A) IL-6, (B) IL-8 and (C) TNF-α by cytokine-specific ELISA. Experiments were performed on three separate occasions. Data is presented as the mean +/- standard deviation (* p < 0.05, ** p < 0.01).
IL-1β was detected in the supernatants of BMDM infected with each PRRSV-1 strain, however the concentration was on the lower limit of detection and considered too variable to draw meaningful conclusions (Figure S2). The pro-inflammatory cytokine IL-1α and the anti-inflammatory cytokine IL-10 were not detected.
To further characterize the cytokine responses of porcine BMDM following infection (MOI 0.01) with the three PRRSV-1 strains, supernatants collected at 48 hpi were analyzed using the Mx-CAT IFN assay (Figure 3A). This assay utilizes the chloramphenicol acetyltransferase (CAT) reporter enzyme under the control of the Mx promoter, which can be stimulated by both type I and type III IFN; it therefore quantifies biologically active type-I and type-III IFNs (38). SU1-Bel infection of BMDM from two out of three animals produced significantly higher concentrations of IFN in comparison to infection with 215-06 (Pig 1; p = 0.00005, Pig 3; p = 0.00008). Similar comparison with Olot/91-infected BMDM revealed significantly higher concentrations of IFN following SU1-Bel infection for all three animals (Pig 1 p = 0.000006; Pig 2 p = 0.002; Pig 3 p = 0.00002). To elucidate the type of IFN produced by these infections, the respective supernatants were subjected to further analyses using type I (IFN-α) and type III IFN (IL-28 and IL-29) specific ELISAs. In agreement with the results of the Mx-CAT assay, a significant increase in IFN-α (Figure 3B) was detected in BMDM from two out of three animals infected with SU1-Bel when compared with cells infected with 215-06 (Pig 1 p = 0.0003; Pig 2 p = 0.25; Pig 3 p = 0.004) and in all BMDM infected with Olot/91 (Pig 1 p = 0.004; Pig 2 p = 0.044; Pig 3 p = 0.014). No significant increase in IL-28 or IL-29 expression was observed in BMDM infected with PRRSV-1 when compared with uninfected controls (Figure S3). These data showed that PRRSV-1 infection induced the production of IFN-α and its expression was enhanced following infection with the highly virulent SU1-Bel strain.
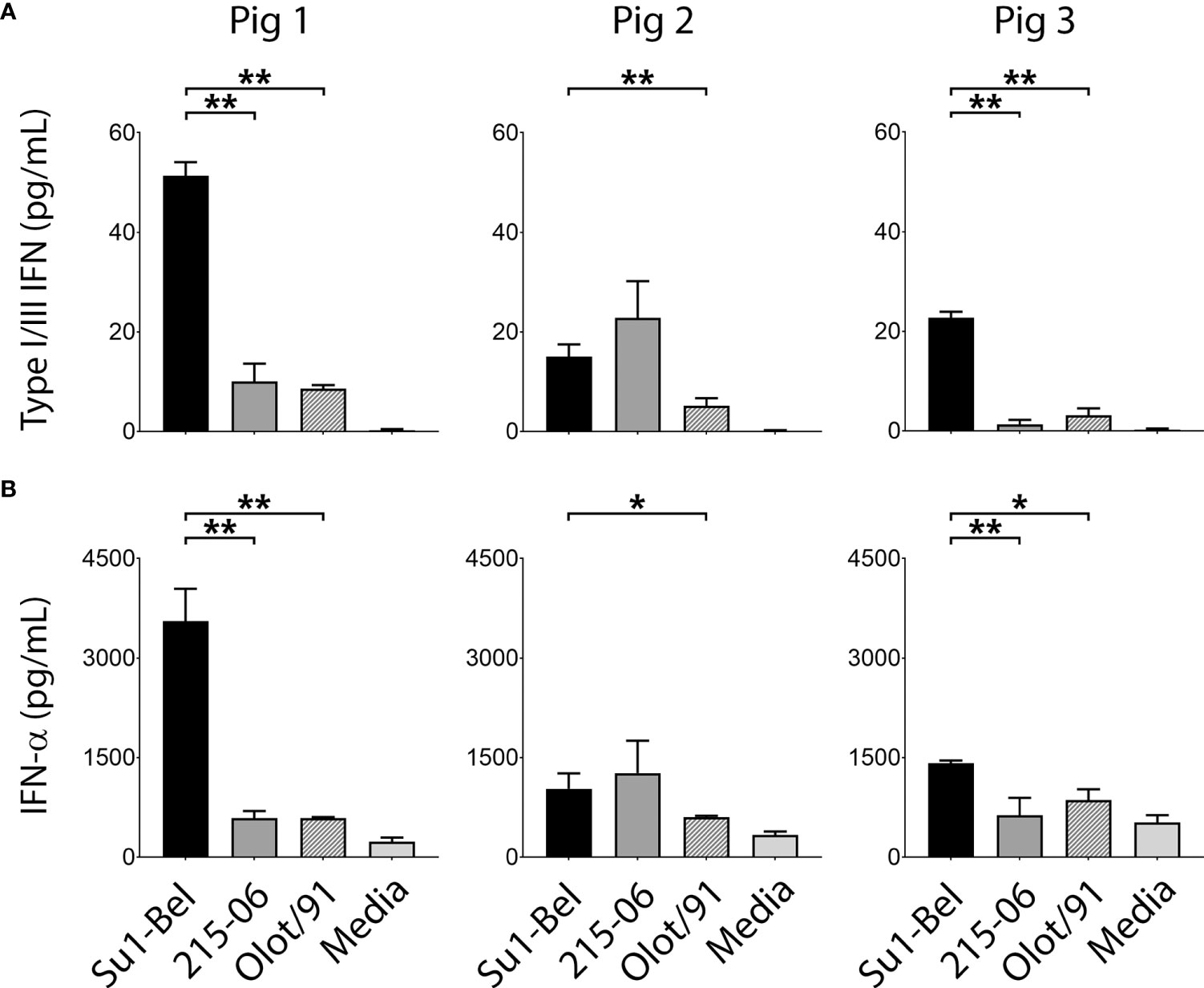
Figure 3 SU1-Bel infected BMDM secrete higher concentrations of IFN-α than BMDM infected with 215-06 or Olot/91. BMDM from three pigs were infected (MOI 0.01) with PRRSV-1 strains SU1-Bel, 215-06 and Olot/91 and at 48 hpi supernatants were analyzed using an (A) Mx-CAT IFN assay or (B) a porcine IFN-α ELISA. Experiments were performed on three separate occasions. Data is presented as the mean +/- standard deviation (*p < 0.05, **p < 0.01).
Having observed that infection with virulent SU1-Bel induced significantly higher expression of IFN-α compared to the less virulent 215-06 and Olot/91 strains, we postulated that this may lead to a higher expression of type I IFN stimulated genes (ISG). We have previously validated the expression of Mx1, RIG-I and ISG15 as markers of ISG expression in immortalized porcine cells (37) and applied this methodology to investigate ISG expression in the porcine BMDM. To facilitate this, BMDM from three animals were infected (MOI 0.01) with either SU1-Bel, 215-06 or Olot/91 and at 48 hpi whole cell lysates were prepared and analyzed for expression of Mx1, RIG-I and ISG15 by Western blot (Figure 4). As expected, Mx1, RIG-I and ISG15 were not observed in the negative control samples (media only) but were induced following stimulation with two different positive control ligands, a TLR3 agonist polyI:polyC (dsRNA) and a RIG-I agonist Sendai virus (SeV). Strikingly, we found that all three ISGs were strongly induced in cells infected with Olot/91, whereas they were only minimally induced or below the limit of detection in cells infected with SU1-Bel or 215-06. These results were consistent in cells from all 3 animals.
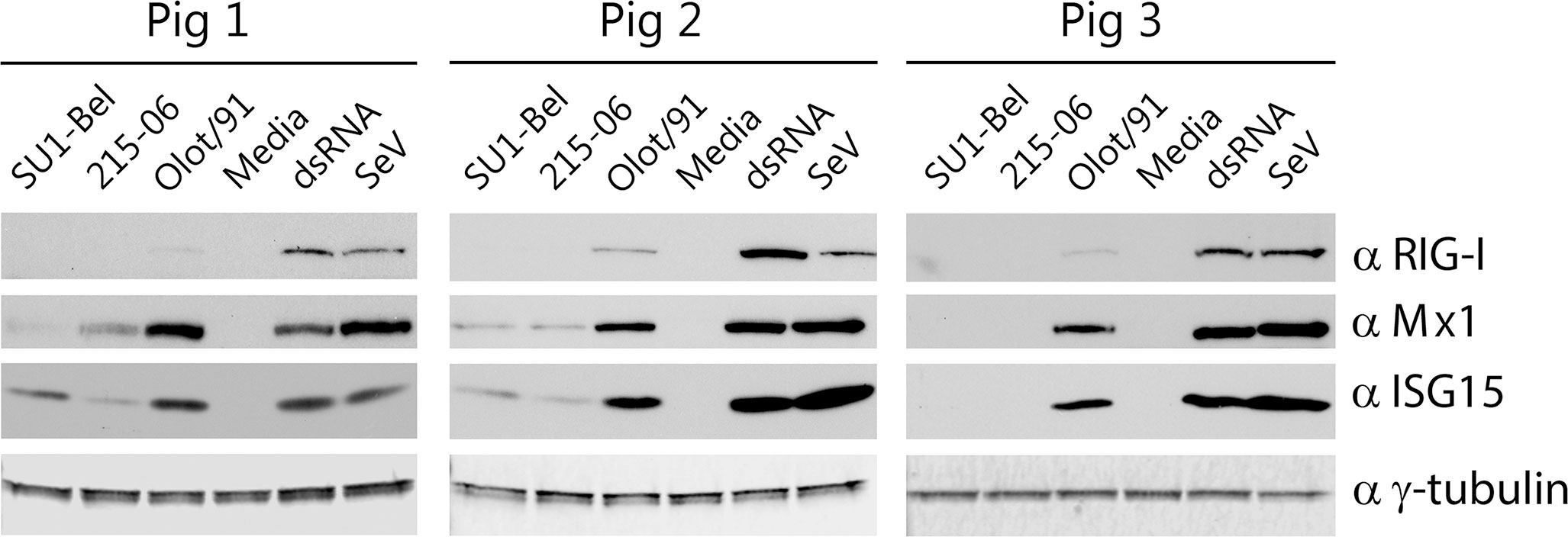
Figure 4 Olot/91 induces higher expression of interferon stimulated genes compared with virulent PRRSV-1 strains. BMDM from three pigs were inoculated with SU1-Bel, 215-06, Olot/91 (MOI 0.01), untreated (media) or stimulated with dsRNA or Sendai Virus for 48 h, then whole cell lysates were analyzed by Western blot using antibodies against RIG-I, Mx1, ISG-15 or γ-tubulin to confirm equal loading.
Interestingly, whole cell lysates prepared from BMDM infected with 215-06 showed a comparatively lower level of ISG15 expression compared with the highly virulent SU1-Bel strain. PAM have been shown to secrete ISG15 in response to infection with a PRRSV-2 vaccine strain (39). Since differential expression of intracellular ISG15 was observed in the whole cell lysates of PRRSV-1 infected BMDM (Figure 4), we also checked the supernatants for secreted ISG15 using a specific ISG15 ELISA. However, ISG15 could not be detected in the supernatants derived from SU1-Bel, 215-06 or Olot/91 infected BMDM.
The mechanism(s) of PRRSV-1 virulence remains unclear as different in vivo studies have suggested enhanced viral growth, wider cellular tropism and differential pro-inflammatory cytokine responses as contributing factors (7–11). Cost as well as practical considerations has prevented more comprehensive strain comparisons in vivo. To address these limitations, we sought to develop a primary porcine ex vivo model system, to attempt to identify correlates of PRRSV-1 virulence and to facilitate wider strain comparison in vitro. Several studies have utilized primary porcine bone marrow-derived monocytes (BMM). When cultured in the presence of M-CSF, BMM develop a macrophage-like phenotype, potent phagocytic activity and express markers associated with blood derived macrophages (18, 25). The majority of these so-called BMDM, express CD163 after 3-4 days of culture and thus support productive PRRSV-1 infection (18, 25). BMDM have been used to investigate the growth and phagocytic suppression of macrophages by virulent PRRSV-1 (40), but to the best of our knowledge a comparative investigation of PRRSV-1 induced cytokine responses has yet to be performed.
For this initial investigation, we used an attenuated, a low and a highly virulent PRRSV-1 strain for comparison. Olot/91 is an attenuated subtype 1 strain. SU1-Bel and 215-06 were selected as their virulence has been compared in several in vivo studies. In each of these studies SU1-Bel was found to be more virulent, inducing higher clinical and gross lung pathology scores and significantly more severe pulmonary and lymphoid lesions (8, 9, 11). Morgan et al. reported that SU1-Bel and 215-06 infected animals had similar viral loads in serum and bronchoalveolar lavage fluid (BALF) (8). In agreement with these in vivo data, we observed no significant difference in the end point titres from SU1-Bel and 215-06 infected BMDM. However, we did observe higher titres at early time points post infection with SU1-Bel and lower end point titres for Olot/91. Two in vivo studies have described significantly higher numbers of PRRSV-1 infected cells in the lung tissue of SU1-Bel infected animals compared with animals infected with 215-06, with the authors suggesting that SU1-Bel may have a wider tropism and/or replicate more efficiently in certain cell types (9, 11). SU1-Bel’s faster growth kinetics in BMDM, may be explained by differences during early infection, including efficiency of virus uptake and early replication which will be investigated in future studies. Whilst our findings require additional strain comparisons, these data appear to suggest that BMDM represent a good primary system for studying PRRSV-1 replication, whereby faster growth kinetics and lower end point titres may be indicative of PRRSV-1 virulence or attenuation, respectively. Recombinant viruses were used in this study to enable the use of more defined populations of PRRSV-1, rather than the quasispecies ‘swarms’ that might be expected from a wild type PRRSV-1 isolate, and to facilitate future identification of the genomic elements that influence cytokine production between strains of differing virulence.
In addition to investigating viral growth, we sought to characterize the PRRSV-1 induced cytokine response in BMDM. We acknowledge that there was variability in the data between animals and care must be taken when drawing conclusions from these experiments. Indeed, this work highlights the need to combine in vivo studies to verify in vitro results and confirm correlates of virulence. Future experiments will involve the evaluation of BMDM infection alongside an immortalized cell line such as the porcine macrophage cell line, ZMAC4 (41). However, it is feasible to suggest that differential secretion of the pro-inflammatory cytokines IL-6, IL-8, TNF-α and IFN-α was observed in a strain dependent manner (Figures 2, 3). To the best of our knowledge, this is the first report that PRRSV-1 infection induces the secretion of IL-1β, IL-6, TNF-α and IFN-α from porcine BMDM. The production of IL-8 from PRRSV-1 infected BMDM has been described previously, but the data reported herein is the first comparative analysis of IL-8 expression following infection with strains of differing virulence. IL-8 is an important pro-inflammatory cytokine that is secreted by macrophages to stimulate cell migration and phagocytosis. The secretion of IL-8 from porcine peripheral blood mononuclear cells (PBMC) and PAM has been demonstrated following PRRSV-1 infection in vitro (12, 17). We found that BMDM secreted IL-8 following infection with all three PRRSV-1 strains. However, whilst similar levels of IL-8 were induced by the virulent strains, significantly lower concentrations of IL-8 were secreted by BMDM infected with the attenuated Olot/91 strain in cells from 2 out of 3 animals (Figure 2). Infection with Olot/91 also induced a weaker IL-6 response in BMDM from 2 out of 3 animals, in comparison with the virulent strains. These observations correspond with experimental PRRSV-1 infection in vivo that showed similar levels of IL-8 in the serum of animals infected with highly virulent (Lena) and virulent (Finistere) strains as well as similar expression of IL-6 in the lung tissue of SU1-Bel and 215-06 infected animals (11, 42). It should be noted that in the work described here, SU1-Bel infection stimulated fairly consistent IL-6 production between BMDM samples (130-220 pg/mL) whereas higher variability was observed following 215-06 infection (20-450 pg/mL). Although the quantities of TNF-α produced were low, BMDM infected with Olot/91 expressed significantly higher levels of TNF-α than cells infected with virulent PRRSV-1 strains. The importance of this observation is unclear as the expression of TNF-α does not appear to correlate with virulence in PBMC in vitro as might be expected (17).
In contrast to the levels of IL-6, IL-8, and TNF-α that were similarly induced by both virulent PRRSV-1 strains, BMDM were found to secrete higher levels of IFN-α following infection with the highly virulent SU1-Bel strain (Figure 3). Whilst this requires in vivo validation, higher IFN-α expression may contribute to the enhanced adaptive immune response and faster viral clearance observed for SU1-Bel relative to 215-06 in vivo (8). Comparatively higher levels of serum IFN-α have been reported for highly virulent PRRSV-2 (43), however there is limited data describing systemic IFN-α during PRRSV-1 infection. PRRSV-1 limits the synthesis of type I IFN in lung tissue and BALF (10, 42), but there is some evidence to suggest higher serum IFN-α may contribute to virulence. Infection with the virulent BSAL/2011 strain induced higher levels of serum IFN-α than the attenuated BS114/2002 strain (44) and infection with the highly virulent Lena strain induced higher serum IFN-α levels than infection with the virulent Finistere and Belgium A strains (7, 42). Since we observed a comparatively higher expression of IFN-α in BMDM, it is plausible to suggest that higher expression of IFN-α in certain cell types may contribute to the virulence of the highly virulent subtype 3 PRRSV-1 strains.
Having observed strain dependent differences in the expression of IFN-α, we thought it necessary to investigate the type I IFN response in PRRSV-1 infected BMDM. We have previously demonstrated that dsRNA and SeV are specific agonists of TLR-3 and RIG-I mediated type I IFN responses in porcine cells, respectively (37). When utilized in BMDM, both ligands induced strong expression of the ISGs, Mx1, ISG15 and RIG-I. Despite infection with Olot/91 inducing lower levels of IFN-α compared to SU1-Bel, and lower or similar levels compared to 215-06, higher levels of all three ISG products were observed. This suggests that Olot/91 both induces and is less able to block type I IFN signaling in comparison to the more virulent strains. It is possible that this inability to inhibit the type I IFN response contributes to the attenuation of Olot/91 in vivo. Ogno et al. have previously shown that highly virulent PRRSV-1 strains are less sensitive to type I IFN treatment in monocyte derived macrophages (MDM) (45). This enhanced resistance could be explained by a more complete inhibition of ISG expression, which may help to explain the increase in viral titres observed by Ogno et al. as well as the increased end-point viral titres observed for the virulent strains employed in this study.
We were unable to detect IL-10 and observed only low levels of IL-1β in PRRSV-1 infected BMDM. These data are in agreement with IL-10 and IL-1β responses observed in PRRSV-1 infected MDM (45). We have previously observed higher levels of IL-1β (108 pg/mL) following sequential passage of SU1-Bel on BMDM. The expression of IL-1β has been shown to be upregulated when MDM are pre-treated with IFN-γ prior to infection with PRRSV-1 (45), therefore pre-treatment of BMDM may prove useful for investigating the PRRSV-1 induced IL-1β response in the future. We were also unable to detect IL-1α or IFN-γ in this study. Despite these limitations, we believe BMDM represent a good model system for studying PRRSV-1 replication and induction of pro-inflammatory cytokines, whereby faster replication and an enhanced production of IFN-α may be indicative of virulence and lower viral yields and expression of Mx1 may indicate an attenuated phenotype.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
All work was approved by The Pirbright Institute Animal Welfare and Ethical Review Body and performed in accordance with the Home Office Guidance on the Operation of the Animals (Scientific Procedures) Act 1986 under the authority of Home Office project license (70/8958).
Conceptualization, JS, BJ, and SG; data curation, BJ; formal analysis, BJ, TC, and JS; funding acquisition, JS; investigation, BJ, TC, and JS; Methodology, JS, BJ, and NW; project administration, JS; resources, JS; supervision, JS; Validation, BJ and TC; visualization, BJ; writing- original draft preparation, BJ and JS; writing- review and editing, JS, BJ, TC, KC, NW, J-PF, and SG. All authors contributed to the article and approved the submitted version.
This work was supported, in whole or in part by BBSRC grants (BBS/E/I/00007034 and BBS/E/I/00007037).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.980412/full#supplementary-material
1. Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, et al. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) (2020) 2020:baaa062. doi: 10.1093/database/baaa062
2. Wensvoort G, de Kluyver EP, Pol JM, Wagenaar F, Moormann RJ, Hulst MM, et al. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: a review of mystery swine disease research at lelystad. Veterinary Microbiol (1992) 33(1-4):185–93. doi: 10.1016/0378-1135(92)90046-V
3. Holtkamp DJ, Kliebenstein JB, Neumann EJ. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on united states pork producers. JSHAP (2013) 21(2):72–84.
4. Stadejek T, Oleksiewicz MB, Scherbakov AV, Timina AM, Krabbe JS, Chabros K, et al. Definition of subtypes in the European genotype of porcine reproductive and respiratory syndrome virus: nucleocapsid characteristics and geographical distribution in Europe. Arch virology (2008) 153(8):1479–88. doi: 10.1007/s00705-008-0146-2
5. Chen N, Li X, Xiao Y, Li S, Zhu J. Characterization of four types of MLV-derived porcine reproductive and respiratory syndrome viruses isolated in unvaccinated pigs from 2016 to 2020. Res Vet Sci (2021) 134:102–11. doi: 10.1016/j.rvsc.2020.12.007
6. Kick AR, Amaral AF, Frias-De-Diego A, Cortes LM, Fogle JE, Crisci E, et al. The local and systemic humoral immune response against homologous and heterologous strains of the type 2 porcine reproductive and respiratory syndrome virus. Front Immunol (2021) 12:637613. doi: 10.3389/fimmu.2021.637613
7. Weesendorp E, Morgan S, Stockhofe-Zurwieden N, Popma-De Graaf DJ, Graham SP, Rebel JM. Comparative analysis of immune responses following experimental infection of pigs with European porcine reproductive and respiratory syndrome virus strains of differing virulence. Veterinary Microbiol (2013) 163(1-2):1–12. doi: 10.1016/j.vetmic.2012.09.013
8. Morgan SB, Graham SP, Salguero FJ, Sanchez Cordon PJ, Mokhtar H, Rebel JM, et al. Increased pathogenicity of European porcine reproductive and respiratory syndrome virus is associated with enhanced adaptive responses and viral clearance. Veterinary Microbiol (2013) 163(1-2):13–22. doi: 10.1016/j.vetmic.2012.11.024
9. Morgan SB, Frossard JP, Pallares FJ, Gough J, Stadejek T, Graham SP, et al. Pathology and virus distribution in the lung and lymphoid tissues of pigs experimentally inoculated with three distinct type 1 PRRS virus isolates of varying pathogenicity. Transbound Emerg Dis (2016) 63(3):285–95. doi: 10.1111/tbed.12272
10. Weesendorp E, Rebel JM, Popma-De Graaf DJ, Fijten HP, Stockhofe-Zurwieden N. Lung pathogenicity of European genotype 3 strain porcine reproductive and respiratory syndrome virus (PRRSV) differs from that of subtype 1 strains. Veterinary Microbiol (2014) 174(1-2):127–38. doi: 10.1016/j.vetmic.2014.09.010
11. Amarilla SP, Gomez-Laguna J, Carrasco L, Rodriguez-Gomez IM, Caridad YOJM, Morgan SB, et al. A comparative study of the local cytokine response in the lungs of pigs experimentally infected with different PRRSV-1 strains: upregulation of IL-1alpha in highly pathogenic strain induced lesions. Vet Immunol Immunopathol (2015) 164(3-4):137–47. doi: 10.1016/j.vetimm.2015.02.003
12. Gimeno M, Darwich L, Diaz I, de la Torre E, Pujols J, Martin M, et al. Cytokine profiles and phenotype regulation of antigen presenting cells by genotype-I porcine reproductive and respiratory syndrome virus isolates. Veterinary Res (2011) 42:9. doi: 10.1186/1297-9716-42-9
13. Liu Y, Shi W, Zhou E, Wang S, Hu S, Cai X, et al. Dynamic changes in inflammatory cytokines in pigs infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Clin Vaccine Immunol CVI (2010) 17(9):1439–45. doi: 10.1128/CVI.00517-09
14. Lunney JK, Benfield DA, Rowland RR. Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res (2010) 154(1-2):1–6. doi: 10.1016/j.virusres.2010.10.009
15. Liu X, Bai J, Jiang C, Song Z, Zhao Y, Nauwynck H, et al. Therapeutic effect of xanthohumol against highly pathogenic porcine reproductive and respiratory syndrome viruses. Veterinary Microbiol (2019) 238:108431. doi: 10.1016/j.vetmic.2019.108431
16. Zhang L, Liu J, Bai J, Wang X, Li Y, Jiang P. Comparative expression of toll-like receptors and inflammatory cytokines in pigs infected with different virulent porcine reproductive and respiratory syndrome virus isolates. Virol J (2013) 10:135. doi: 10.1186/1743-422X-10-135
17. Ferlazzo G, Ruggeri J, Boniotti MB, Guarneri F, Barbieri I, Tonni M, et al. In vitro cytokine responses to virulent PRRS virus strains. Front Vet Sci (2020) 7:335. doi: 10.3389/fvets.2020.00335
18. Fernandez-Caballero T, Alvarez B, Alonso F, Revilla C, Martinez-Lobo J, Prieto C, et al. Interaction of PRRS virus with bone marrow monocyte subsets. Veterinary Microbiol (2018) 219:123–7. doi: 10.1016/j.vetmic.2018.04.027
19. Liu Y, Du Y, Wang H, Du L, Feng WH. Porcine reproductive and respiratory syndrome virus (PRRSV) up-regulates IL-8 expression through TAK-1/JNK/AP-1 pathways. Virology (2017) 506:64–72. doi: 10.1016/j.virol.2017.03.009
20. Barranco I, Gomez-Laguna J, Rodriguez-Gomez IM, Quereda JJ, Salguero FJ, Pallares FJ, et al. Immunohistochemical expression of IL-12, IL-10, IFN-alpha and IFN-gamma in lymphoid organs of porcine reproductive and respiratory syndrome virus-infected pigs. Vet Immunol Immunopathol (2012) 149(3-4):262–71. doi: 10.1016/j.vetimm.2012.07.011
21. Garcia-Nicolas O, Auray G, Sautter CA, Rappe JC, McCullough KC, Ruggli N, et al. Sensing of porcine reproductive and respiratory syndrome virus-infected macrophages by plasmacytoid dendritic cells. Front Microbiol (2016) 7:771. doi: 10.3389/fmicb.2016.00771
22. Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, et al. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PloS pathogens (2017) 13(2):e1006206. doi: 10.1371/journal.ppat.1006206
23. Xie J, Trus I, Oh D, Kvisgaard LK, Rappe JCF, Ruggli N, et al. A triple amino acid substitution at position 88/94/95 in glycoprotein GP2a of type 1 porcine reproductive and respiratory syndrome virus (PRRSV1) is responsible for adaptation to MARC-145 cells. Viruses (2019) 11(1):36. doi: 10.3390/v11010036
24. Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, et al. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J virology (2007) 81(14):7371–9. doi: 10.1128/JVI.00513-07
25. Chaudhuri S, McKenna N, Balce DR, Yates RM. Infection of porcine bone marrow-derived macrophages by porcine respiratory and reproductive syndrome virus impairs phagosomal maturation. J Gen virology (2016) 97(3):669–79. doi: 10.1099/jgv.0.000384
26. Feng W, Laster SM, Tompkins M, Brown T, Xu JS, Altier C, et al. In utero infection by porcine reproductive and respiratory syndrome virus is sufficient to increase susceptibility of piglets to challenge by streptococcus suis type II. J virology (2001) 75(10):4889–95. doi: 10.1128/JVI.75.10.4889-4895.2001
27. Amarilla SP, Gomez-Laguna J, Carrasco L, Rodriguez-Gomez IM, Caridad YOJM, Graham SP, et al. Porcine reproductive and respiratory syndrome type 1 viruses induce hypoplasia of erythroid cells and myeloid cell hyperplasia in the bone marrow of experimentally infected piglets independently of the viral load and virulence. Veterinary Microbiol (2017) 201:126–35. doi: 10.1016/j.vetmic.2016.12.040
28. Wang G, Li L, Yu Y, Tu Y, Tong J, Zhang C, et al. Highly pathogenic porcine reproductive and respiratory syndrome virus infection and induction of apoptosis in bone marrow cells of infected piglets. J Gen virology (2016) 97(6):1356–61. doi: 10.1099/jgv.0.000454
29. Singleton H, Graham SP, Bodman-Smith KB, Frossard JP, Steinbach F. Establishing porcine monocyte-derived macrophage and dendritic cell systems for studying the interaction with PRRSV-1. Front Microbiol (2016) 7:832. doi: 10.3389/fmicb.2016.00832
30. Duan X, Nauwynck HJ, Pensaert MB. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch virology (1997) 142(12):2483–97. doi: 10.1007/s007050050256
31. Teifke JP, Dauber M, Fichtner D, Lenk M, Polster U, Weiland E, et al. Detection of European porcine reproductive and respiratory syndrome virus in porcine alveolar macrophages by two-colour immunofluorescence and in-situ hybridization-immunohistochemistry double labelling. J Comp Pathol (2001) 124(4):238–45. doi: 10.1053/jcpa.2000.0458
32. Bordet E, Blanc F, Tiret M, Crisci E, Bouguyon E, Renson P, et al. Porcine reproductive and respiratory syndrome virus type 1.3 Lena triggers conventional dendritic cells 1 activation and T helper 1 immune response without infecting dendritic cells. Front Immunol (2018) 9:2299. doi: 10.3389/fimmu.2018.02299
33. Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, et al. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol (2010) 91(Pt 7):1659–67. doi: 10.1099/vir.0.020503-0
34. Kvisgaard LK, Hjulsager CK, Kristensen CS, Lauritsen KT, Larsen LE. Genetic and antigenic characterization of complete genomes of type 1 porcine reproductive and respiratory syndrome viruses (PRRSV) isolated in Denmark over a period of 10 years. Virus Res (2013) 178(2):197–205. doi: 10.1016/j.virusres.2013.10.009
35. Hierholzer JC, Killington RA. Virus isolation and quantitation. In: Virology methods manual (1996) Cambridge, Massachusetts: Academic Press, Elsevier. p. 25–46.
36. Fray MD, Mann GE, Charleston B. Validation of an Mx/CAT reporter gene assay for the quantification of bovine type-I interferon. J Immunol Methods (2001) 249(1-2):235–44. doi: 10.1016/S0022-1759(00)00359-8
37. Hardy S, Jackson B, Goodbourn S, Seago J. Classical swine fever virus n(pro) antagonises IRF3 to prevent IFN-independent TLR3 and RIG-i-mediated apoptosis. J Virol (2020) 95(5):e01136–20. doi: 10.1128/JVI.01136-20
38. Reid E, Juleff N, Windsor M, Gubbins S, Roberts L, Morgan S, et al. Type I and III IFNs produced by plasmacytoid dendritic cells in response to a member of the flaviviridae suppress cellular immune responses. J Immunol (2016) 196(10):4214–26. doi: 10.4049/jimmunol.1600049
39. Liu H, Shi B, Zhang Z, Zhao B, Zhao G, Li Y, et al. PRRSV vaccine strain-induced secretion of extracellular ISG15 stimulates porcine alveolar macrophage antiviral response against PRRSV. Viruses (2020) 12(9):1009. doi: 10.3390/v12091009
40. Ruedas-Torres I, Rodriguez-Gomez IM, Sanchez-Carvajal JM, Larenas-Munoz F, Pallares FJ, Carrasco L, et al. The jigsaw of PRRSV virulence. Veterinary Microbiol (2021) 260:109168. doi: 10.1016/j.vetmic.2021.109168
41. Portugal R, Goatley LC, Husmann R, Zuckermann FA, Dixon LK. A porcine macrophage cell line that supports high levels of replication of OURT88/3, an attenuated strain of African swine fever virus. Emerg Microbes Infect (2020) 9(1):1245–53. doi: 10.1080/22221751.2020.1772675
42. Renson P, Rose N, Le Dimna M, Mahe S, Keranflec'h A, Paboeuf F, et al. Dynamic changes in bronchoalveolar macrophages and cytokines during infection of pigs with a highly or low pathogenic genotype 1 PRRSV strain. Veterinary Res (2017) 48(1):15. doi: 10.1186/s13567-017-0420-y
43. Han Z, Liu Y, Wang G, He Y, Hu S, Li Y, et al. Comparative analysis of immune responses in pigs to high and low pathogenic porcine reproductive and respiratory syndrome viruses isolated in China. Transbound Emerg Dis (2015) 62(5):e1–e10. doi: 10.1111/tbed.12190
44. Razzuoli E, Dotti S, Villa R, Martinelli N, Ferrari M, Amadori M eds. Early immune responses to infection by attenuated and non-attenuated, type I PRRS virus strains. In: Proceedings 4th European veterinary immunology workshop. (2012) Edinburgh 23. doi: 10.1016/j.vetmic.2019.03.008
Keywords: PRRSV-1, SU1-Bel, 215-06, Olot/91, bone marrow-derived macrophages, proinflammatory cytokines
Citation: Jackson B, Chrun T, Childs K, Wanasen N, Frossard J-P, Graham SP and Seago J (2022) Analysis of porcine bone marrow-derived macrophage cytokine responses to infection with PRRSV-1 strains of differing virulence. Front. Virol. 2:980412. doi: 10.3389/fviro.2022.980412
Received: 28 June 2022; Accepted: 26 July 2022;
Published: 12 August 2022.
Edited by:
Fernando Bauermann, Oklahoma State University, United StatesReviewed by:
Lauro Velazquez-Salinas, Agricultural Research Service (USDA), United StatesCopyright © 2022 Jackson, Chrun, Childs, Wanasen, Frossard, Graham and Seago. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian Seago, anVsaWFuLnNlYWdvQHBpcmJyaWdodC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.