- 1Department of Microbiology & Immunology, Faculty of Medicine, Ardabil University of Medical Sciences, Ardabil, Iran
- 2Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Department of Pharmaceutics, Rutgers, The State University of New Jersey, Piscataway, NJ, United States
- 4Medical Oncology Unit—Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Istituto Tumori “Giovanni Paolo II” of Bari, Bari, Italy
- 5Department of Biomedical Sciences and Human Oncology, Department of Internal Medicine and Oncology (DIMO)—University of Bari, Bari, Italy
- 6Department of Immunology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Myeloid-derived suppressor cells (MDSCs) constitute an important component in regulating immune responses in several abnormal physiological conditions such as cancer. Recently, novel regulatory tumor MDSC biology modulating mechanisms, including differentiation, expansion and function, were defined. There is growing evidence that miRNAs and long non-coding RNAs (lncRNA) are involved in modulating transcriptional factors to become complex regulatory networks that regulate the MDSCs in the tumor microenvironment. It is possible that aberrant expression of miRNAs and lncRNA contributes to MDSC biological characteristics under pathophysiological conditions. This review provides an overview on miRNAs and lncRNAs epiregulation of MDSCs development and immunosuppressive functions in cancer.
Introduction
Myeloid-derived suppressor cells (MDSCs) are produced due to aberrant myelopoiesis (1) and represent one of the most pivotal mediators in the orchestration of immunosuppression (2). The normal development and maturation of myeloid linage cells are essential for the promotion of innate immunity, however, under pathological contexts such as chronic inflammation or cancer, normal hematopoiesis is dysregulated, signals that origin from the HSCs niche alter the magnitude and combination of the hematopoietic output, a characteristic of immune regulation known as “emergency” hematopoiesis, needed to provide appropriate supply of both myeloid and lymphoid cells to increased demand (3). In particular, in cancer, modified myelopoiesis generates lineage-restricted hematopoietic progenitors, resulting in the accumulation of myeloid cells with an immature state and more importantly with the characteristic as an immune response suppressive in the bone marrow (4, 5).
Of note, MDSC are considered as not only immature myeloid cells but also matured cells that could be converted to MDSC. Rivoltini et al. reported that culturing monocytes with melanoma-derived exosomes in vitro generates cells which have phenotypic and functional characteristics similar to those exhibited by MDSC (Exo-MDSC). It was indicated that Exo-MDSC have an increased level of mRNA and release of protumourigenic and immunosuppressive cyto/chemokines, decrease the expression of HLA-DR, and suppress T cell proliferation. This hypothesis is well-supported by the evidence that peripheral MDSC from melanoma patients share characteristics of gene-expression and miRNA profile, phenotypic and functional features of Exo-MDSC (6).
MDSCs are accumulated and expand in the tumor microenvironment and in the peripheral lymphoid organs of tumor-bearing hosts (7–9). Several studies indicated the mechanisms of MDSC-mediated immune suppression and its expansion (10, 11). Ongoing investigation has shown epigenetic regulation of MDSCs as a potential approach in attaining this objective. Indeed, epigenetics states all heritable phenotype variations in gene expression (active vs. inactive genes) without any modifications in the underlying DNA sequence. The mentioned epigenetic modifications enable substantial adjustability in the gene expression, rather than just genes to be turned off or on. DNA variation, covalent histone change, and RNA interference are three systems, fundamental in starting and maintaining epigenetic silencing. Epigenetic reprogramming of MDSC's properties results in the alteration of its features, with a consequential rearrangement of the tumor milieu instead of opposing the development and progression of the tumor (12).
More recently, several studies have also unveiled an important regulatory role for miRNAs and long non-coding RNAs (lncRNA) in modulating immune responses and in MDSCs differentiation and functions (13–17). This review will summarize the miRNAs and lncRNAs that contribute to the differentiation, maturation, and immunosuppressive function of MDSCs in cancer. With this goal in mind, this review will summarize miRNAs and lncRNAs that contribute to MDSCs differentiation, maturation, and immunosuppressive function in cancer.
Myeloid-Derived Suppressor Cells
Major studies introduce MDSCs as a heterogeneous population of cells with myeloid origin (18). The heterogeneity features of such cells including their phenotype and functions are highly influenced by the type of tumor and the stage of tumor progression (19). MDSCs are described in animals and humans with malignancy (20, 21). MDSCs in mice are characterized by co-expression of the CD11b (CR3A or integrin αM) and GR1,myeloid-cell lineage differentiation antigen, comprising Ly6C and Ly6G isoforms and divided in two subpopulations polymorphonuclear (PMN)-MDSC (CD11b+Ly6G+Ly6Clo) and monocytic (M)-MDSC (CD11b+Ly6G−Ly6Chi) (22– 24).
According to a new classification by Bronte et al., human MDSCs are generally classified as follows: PMN-MDSC described as CD11b+CD14−CD15+ or CD11b+CD14−CD66b+ and M-MDSC described as HLA-DR−/loCD11b+CD14+ CD15−. Another population also described as early-stage MDSC (eMDSC) and characterized by the HLA-DR− CD33+ Lin− (CD3, CD19, CD56, CD14, and CD15) phenotype, includes mixed cells of MDSC comprising more immature progenitors (25–27). Moreover, Fibrocytic MDSCs (F-MDSCs), a novel MDSC subset, were recently described as CD11b low CD11c low CD33+ Interleukin (IL)-4Ra+ cells with fibrocystic phenotypes (28, 29). In tumor-bearing mice and cancer patients, although PMN-MDSCs are the dominant subsets (70–80%) of the total MDSCs population, this subset demonstrates less immunosuppressive features than M-MDSCs (5, 30).
Several studies suggested that MDSCs represent a “dysfunctional state” of myeloid lineages such as monocytes/neutrophils (31). Dysregulation in myelopoiesis in cancerous and non-cancerous conditions induces MDSC differentiation and expansion and affects the host immune response (22, 32). In the case of cancer, the tumor cells produce different mediators that inhibit the differentiation of mature myeloid cells and, in parallel, stimulate MDSCs expansion, generating an immunosuppressive microenvironment that affects tumor progression (33). These mediators include bombina variegata peptide 8 (Bv8), KIT ligand [stem-cell factor (SCF)], prostaglandin E2 (PGE2), vascular endothelial growth factor (VEGF), FMS-like tyrosine kinase 3 ligand(FLT3L), granulocyte colony-stimulating factor (G-SCF), macrophage colony-stimulating factor(M-CSF), granulocyte-macrophage colony stimulating factor (GM-CSF), IL-1, IL-6, TNF, and IL-10. It was shown that GM-CSF and IL-6 could be the most powerful expanding factors of MDSCs deriving from bone marrow progenitors (11, 34–37). In addition, the immunosuppressive activity of MDSCs requires mediators which induce their activation (38). The most important MDSC activating mediators are the Transforming Growth Factor (TGF-ß), ligands for toll-like receptors, IL-1β, Interferon (IFN)-γ, and IL-4 which are chiefly originated from activated T cells and tumor stromal cells. Most of these mediators promote signaling pathways that converge the Signal transducer and the activator of transcription (STAT)-1, STAT-6, and nuclear factor-κB (NFκB) signaling pathways (4, 39–43).
MDSCs are involved in determining potent immune-negative regulation and tumor-promoting functions in the TME via multiple mechanisms. MDSCs could deplete and capture the amino acids required for T cell activation and proliferation such as L-arginine and L-cysteine, thus generating the reactive oxygen and nitrogen species such as NO, ROS, and peroxynitrite (44–46). Activated M-MDSCs upregulate Arginase-1 (Arg1) and inducible nitric oxide synthase (iNOS), while activated PMN-MDSCs highly express Arg1 and reactive oxygen species (ROS) (47, 48). High expression of Arg1 causes the depletion of L-arginine resources, resulting in T-cell function impairment and inhibition of T cell proliferation via several mechanisms, including translational blockade of CD247 [the ζ chain of the T-cell receptor (TCR)] and decreased production of IFN-γ/IL-2 by T cells (30, 49–52). Arginine is also oxidized by iNOS to produce citrulline and Nitric Oxide (NO) (53). NO, a powerful oxidative modulator, can inhibit T cell proliferation, adhesion, and migration by blocking Jak3 and STAT5 transcription factor activity, inhibiting E-selectin expression on endothelial cells and inducing T-cell apoptosis (54, 55). NO is also changed to the radical peroxynitrite, a reactive nitrogen-oxide species, in the presence of ROS (56). Peroxynitrite alters tyrosine side chains in the TCR-CD8 complex, prevents peptide-MHC-TCR interaction and finally stops T cell responses (57). ROS play a fundamental role in the induction of T cell apoptosis through B cell lymphoma-2 (Bcl-2) downregulation (58). Moreover, by producing indoleamine 2,3 dioxygenase (IDO) and reducing local tryptophan levels, MDSCs can inhibit T cells proliferation (59).
MDSCs also exert immunosuppressive features by skewing anti-tumor immune cells subsets toward immune suppressive counterparts, for example, through the conversion of naïve CD4+ T cells into Tregs and polarization of macrophages toward an alternatively activated macrophages (M2) phenotype (22, 60). As a matter of fact, MDSCs through the production of IL-10 and TGF-β, arginine deprivation and CD40-CD40L interacting can induce Treg cells (61). Chen et al. (62) have demonstrated that TGF-β production leads to Foxp3 gene expression in T cells, which in turn re-polarizes them toward a Treg phenotype with a potent immunosuppressive potential. Moreover, activation of M2 macrophages due to factors like IL-13, IL-10, IL-4, and glucocorticoid, promote tumor growth by secreting high levels of IL-10 and low levels of IL-12. MDSC-polarizes TAMs toward the M2 phenotype via the production of IL-10 in which it suppresses IL-12 and enhances IL-10 production by these cells. These polarized cells create a feedback cycle through MDSCs modification and enhance IL-10 production, which will subsequently facilitate the tumor growth (63).
MDSCs can also down-regulate homing receptors on the surface of T cells, such as L-selectin (CD62L), by expression of aADAM17 (disintegrin and metalloproteinase domain 17) as well as inhibit T cell homing to lymph nodes (52).
Micro RNAs
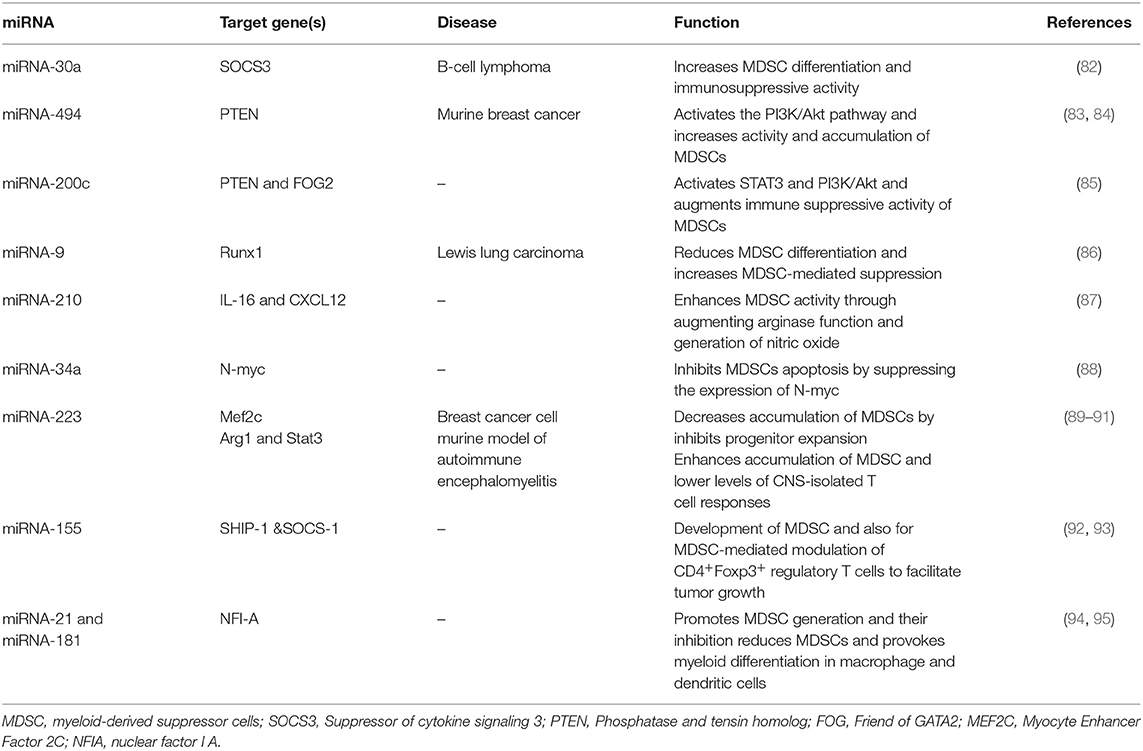
MicroRNAs are small, single-stranded, non-coding RNAs of approximately 22 nucleotides (64). miRNAs control about 30% of human genes (65). They regulate range of physiological processes, such as cell cycle, differentiation, development, and metabolism (66–69), as well as pathological process, such as chronic disease, immuno- or neurodegenerative diseases, and cancer (70–74). MiRNAs are progressively being described as main factors in regulating the immune system. They exert an important role in the epiregulation of the development, the differentiation and the activity of various immune cells, like B and T lymphocytes, dendritic cells, and macrophages (75–78). Moreover, miRNAs show a notable regulatory role in the development, expansion, and function of MDSC (Figure 1) (79). Several studies demonstrated the main role of several miRNA in cancer development. In particular, these molecules have been studied in order to identify potential diagnostic, prognostic or therapeutic targets in several tumors (80, 81). Numerous current researches emphasize the function of certain miRNAs in the MDSCs regulation (Table 1).
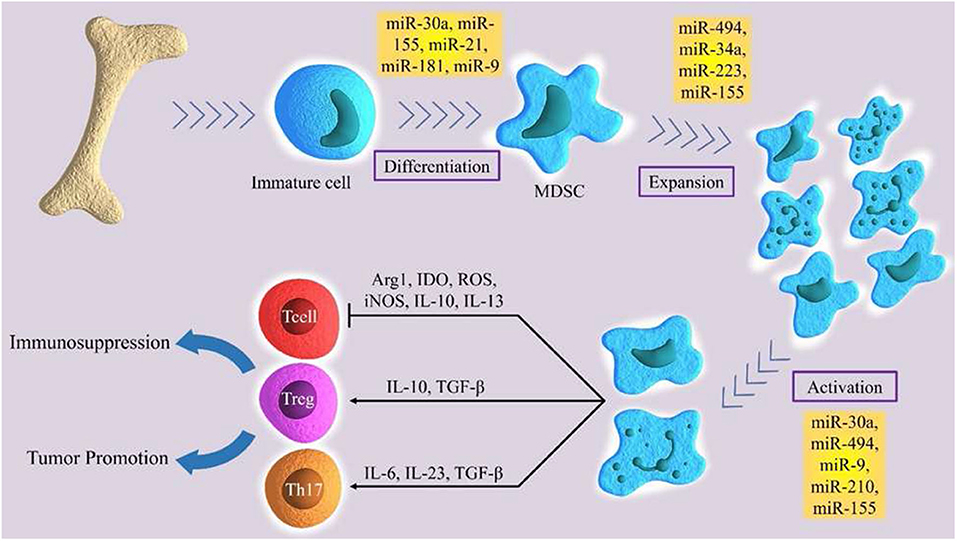
Figure 1. The role of certain miRNAs in regulation of differentiation, expansion and activation of MDSCs.
MicroRNA-30a
A recent study revealed that miR-30a as a member of the miR-30 family increases the differentiation of MDSCs and their immunosuppressive activity through regulating SOCS3 in B-cell lymphoma model mice. Xu et al. (82) detected augmented expression of miR-30a in PMN-MDSCs and M-MDSCs in mice with B cell lymphoma. MicroRNA-30a targets SOCS3, which is a JAK2/STAT3 pathway regulator. SOCS3 negatively regulates MDSCs development and functions by inhibiting STAT3 activation (96). Reduced SOCS3 expression and stimulated JAK2/STAT3 signaling increase MDSCs differentiation and promotes its suppressive function. In both M-MDSCs and PMN-MDSCs, miR-30a is able to promote the expression of suppressive factors, such as arginase-1, IL-10, and ROS, by targeting SOCS3 and stimulating JAK2/STAT3 signaling, thus resulting in tumor growth, which correlates with increased levels of MDSC in the tumor microenvironment, and reduced CD8+ T-cell infiltration in tumors. Furthermore, miR-30a improved the development of tumor by promoting the differentiation and immunosuppressive ability of MDSCs in mice with B-cell lymphoma. MicroRNA-30a antagomir injection in mice demonstrated a considerably improved B-cell lymphoma, while miR-30a agonist injection in mice with B-cell lymphoma stimulated tumor growth (96). These data showed that targeting miR-30a leads to reduced suppression activity of MDSC and lessens the MDSCs numbers, consequently improving anti-tumor response.
MicroRNA-494
MiR-494 is overexpressed by tumor-derived factors and it can regulate the immunosuppressive function of MDSCs (83). Liu et al. reported that tumor cells secrete TGF-β1 which regulates MDSCs activity via stimulation of miR-494 expression; this stimulation is Smad 3 pathway-dependent. An increased miR-494 expression was noticed in MDSCs in tumor-bearing mice compared with those in tumor-free mice. This research found that miR-494 targets phosphatase and tensin homolog (PTEN) and stimulates the PI3K/Akt pathway and controls the accumulation and function of MDSCs. The PTEN downregulation and augmented activation of the PI3K/Akt pathway changed the intrinsic apoptotic/survival signal and also heightened the effectiveness of CXCR4-related recruitment and increased MDSC immunosuppressive function. This study implied that the stimulated PI3K/Akt pathway strongly supports the matrix metalloproteinase (MMPs) expression and results in tumor invasion and metastasis. Additionally, knockdown of miR-494 considerably reversed MDSC function and tumor development and repressed metastasis in an in vivo mouse model of 4T1 mammary carcinoma (84). The data revealed that miR-494 induction by TGF-b1 regulates MDSC accumulation and activity and can be recognized as a possible target in the treatment of cancer.
MicroRNA-155
MicroRNA-155 is a typical miRNA involved in numerous processes in molecular biology, regulating inflammation, hematopoiesis, and immunity (97). Bioinformatics analyses of functional aspects conducted by Robert et al. indicated that multiple miRNAs, including miR-155, are highly expressed in early hematopoietic progenitors and control early stages of hematopoietic differentiation (98). Normal mature B cells, T cells, granulocytes, and monocytes express low levels of miR-155 (99). Moreover, several findings have supported a critical function for miR-155 in concert to MDSCs, emerging as a critical element in the control of their differentiation and function (100). This molecule is markedly elevated in both PMN-MDSCs and M-MDSCs stimulated with GM-CSF and IL-6. Moreover, the expression of miR-155 in MDSCs was controlled by the TGF-β signaling pathway (100). The induction of the miR-155 expression by TGF-β in turn results in the inhibition of Src Homology Inositol Phosphatase (SHIP-1) as a target of miR-155. SHIP-1 is an adaptor protein that negatively controls the survival and proliferation of myeloid cells. During the induction of functional MDSC, down-regulation of this essential molecule promotes STAT3 activation and accordingly induces MDSC expansion (100). Similarly, an experimental study revealed that genetic ablation of miR-155 confers resistance to the growth of transplanted cancer cell in mice tumor models. Moreover, miR-155 is essential for the development of MDSC and also for MDSC-mediated modulation of CD4+Foxp3+ regulatory T cells, indicating that MDSCs are associated with miR-155 to promote tumor growth (92). On the other hand, miR-155 deficiency reduces the immune responses of both dendritic cells and T cells against tumor cells. These discrepancies underscore a context-dependent activity of miR-155 in regulating tumor immunity via distinct subsets of immune cells within the tumor microenvironment (92). Although the above data support a role for miR-155 expression in tumor progression, Wang et al. (93) stated that miR-155 deficiency enhances solid tumor development by augmenting the tumor-supporting properties and recruitment of MDSCs to the tumor milieu. These contradictory observations might be associated with the tumor type and the adjacent microenvironment. Future studies are required to clarify the subject.
MiRNA-34a
Unlike its family members miR-34b/c, miR-34a is expressed in a wide variety of normal human tissues (101). As an example, miR-34a, produced by FoxP3-expressing regulatory T (Treg) cells, has been shown to have a vital role in regulating immune reactions. Yang et al. (102) explored TGF-β signaling activity during hepatocellular carcinoma (HCC) progression, and the results revealed that miR-34a expression is directly regulated by the TGF-β pathway in HCC cell lines. In all HCC cell lines investigated, the stimulation of the TGF-β signaling pathway induced a significant reduction in the expression of miR-34a, leading to the increased production of a miR-34a target gene, the chemokine CCL22, which results in the accumulation of Treg cells which enhance immune escape (102). However, as far as myeloid suppressor cells are concerned, miR-34a resembles a tumor suppressor, since miR-34a-mimic transfected into a murine colorectal carcinoma cell line, CT26 cells, leads to a lower induction of myeloid precursor cells into Gr1+CD11b+ cells, the phenotype of MDSC with immunosuppressive activity, in mice (103). This finding indicates the negative activity of miR-34a in the induction of MDSC. Consistent with the above data on the correlation of TGF-β and miR-34a expression in Treg cell recruitment, miR-34a might control the action of tumor cells, inducing MDSCs via TGF-β and/or IL-10 (103). Moreover, miR-34a repressed MDSC apoptosis by inhibiting N-myc expression, but did not influence MDSCs proliferation. The over-expression of miR-34a results in an increase of MDSCs due to a reduction of MDSCs apoptosis. The proto-oncogene N-myc which is a transcription factor that has a role in activities, including cell growth, differentiation, and apoptosis, is a target of miR-34a in MDSCs (88). As a result, restoring miR-34a expression contemporarily to stopping TGF-β signaling might offer a promising molecular treatment in concert with MDSC inhibition in cancer.
MiRNA-21 and miRNA-181
The miR-181 family has a crucial role in normal myeloid differentiation and adult acute myeloid leukemia (AML) development (104). The miR-181 family consists of four miRs: miR-181a/b/c/d. Overexpression of all of them has been detected in different subtypes of adult AML, suggesting an oncogenic role by the direct inhibition of genes involved in myeloid differentiation such as protein kinase C delta (PRKCD) (104). Also, miR-21 is one of the most studied miRNAs that has attracted particular attention in the myeloid subject area. In particular, miR-21 has been identified as a molecule whose deregulation impairs myelopoiesis (94).
Sepsis is a life-threatening condition characterized by activation of the host's inflammatory pathways. Bacterial sepsis is a scenario where pathological inflammation driving defects in innate immunity can result in bacterial growth. However, less reports describe the regulation of miR21 and miR-181 family expression in tumor-induced immunosuppression by MDSCs, the role of these miRNAs in sepsis-related immunosuppression mediated by MDSCs is well characterized. These investigations provide clues from which we may be able to trace their role in development of MDSCs in the course of human malignancies that resemble some disturbances found in sepsis. In sepsis-induced inflammatory response, the produced miR-21 and miR-181b promotes MDSC generation. Inhibition of this miRNAs decreases the population of MDSCs and significantly enhances late-sepsis survival (95). Importantly, concurrent inhibition of both miR-21 and miR-181b in Gr1+CD11b+ myeloid cells was able to enhance myeloid differentiation into macrophage and dendritic cells and eliminated MDSCs, leading to an enhancement of the innate immunity and improved bacterial clearance. The findings have also linked miR-21 and miR181b with the expression of NFI-A, a transcription factor involved in myeloid differentiation in sepsis (95). These miRNAs upregulate NFI-A, thereby maintaining the myeloid progenitor cells in an immature state (105). This regulatory mechanism underlying the expression of miR-21 and miR-181b in MDSCs under the same pathological condition was further uncovered in a later study. The authors demonstrated that transcriptional regulation of miR-21 and miR-181b is mediated by a combination of factors including STAT3 and C/EBPβ as critical transcription factors in Gr1+CD11b+ myeloid cells in septic mice. STAT3 and C/EBPβ factors bind to miR-21 and miR-181b and promote and activate the expression of these molecules through a pathway that requires IL-6 signaling, implicating phosphorylation on the Rb protein, ultimately leading to MDSC expansion (106). A more recently published study has provided some more details on the mechanisms of miR-21/miR181 functions on MDSC biology in cancer which involves the suppression of the Mixed-lineage leukemia 1 (MLL1) complex that is found to play an important role in regulating hematopoietic stem cell homeostasis. Mechanistically, tumor-secreted factors and GM-CSF + IL-6 signaling activates Stat3 and Cebpβ leading to the expression of miR-21a, miR-21b, and miR-181b. The induction of these microRNAs inhibits the expression of the components of MLL1 complex and thus may play a critical role in PMN-MDSC expansion, activation, and differentiation (107). In addition, miR-21 expresses some common biological functions with the aforementioned miRNA, miR-155, during MDSC induction in tumor bearing mice (100). In this aspect, the cytokine, TGFβ has been shown to promote the induction of MDSC by upregulating the expression of miR-21 and miR-155. MiR-155 and miR-21 synergistically function to promote MDSC stimulation through targeting SHIP-1 and phosphatase and tensin homolog, respectively, leading to STAT3 activation and MDSC expansion.
MicroRNA-200c
Also MiR-200c can regulate the suppressive activity and differentiation of MDSCs. The genomic locus of miR-200c is placed in fragile regions within two chromosomal clusters of miR-200c-141 and miR-200b-200a-429 (108). PTEN, a protein/lipid phosphatase, negatively regulates the PI3K/Akt signaling pathway (109). Friend of Gata 2 (FOG2) is seen to attach to a PI3K regulatory subunit and negatively controls the PI3K/Akt pathway (110). Mei et al. demonstrated that miR-200c enhances the MDSC-mediated suppressive activity by regulating PTEN and FOG2, resulting in activation of STAT3 and PI3K/Akt. They investigated the impact of miR-200c mimics loaded MDSCs on T cell function. CD4+ T or CD8+ T cells were isolated from the spleen, labeled by CFSE and triggered with anti-CD3ε antibody, and then co-cultured with differently modified MDSCs. MDSCs treated with miR-200c mimics indicated significant inhibition on T cell proliferation with a reduction in the proportion of proliferating cells; while inhibitors of miR-200c lessened the MDSC-mediated suppressive activity. These Authors also revealed that GM-CSF has an important role in the stimulation of miR-200c in the microenvironment of tumor. Furthermore, miR-200c stimulated by GM-CSF in the tumor milieu suggests a major function in controlling the expansion and activity of tumor-related MDSCs and can be a possible target for immunomodulation in the immunotherapy of cancer (85).
MicroRNA-9
MicrRNA-9 has been described as an important factor in controlling immune reactions (111), neuronal differentiation (112), posttraumatic stress (113), and different cancers (114, 115). Recently, Tian et al. (86) found the significant regulatory role of miR-9 in differentiation and function of MDSC by targeting Runt-related transcription factor 1 (Runx1). Runx1, as a key target of miR-9, regulates MDSC differentiation and function and is an important transcription factor during the development of hematopoietic stem cells from the hemogenic endothelium (116, 117), which is identified as a main inducer of differentiation. Runx1 controls hematopoietic stem cell differentiation into endothelial cells. It regulates the expression of some myeloid differentiation related genes and supports the differentiation of myeloid, lymphoid, and megakaryocytic lineages (116). The significant role of Runx1 in controlling MDSCs differentiation and function was confirmed by knocking down Runx1. It was found that MDSC differentiation decreased after knocking down Runx1. Moreover, suppressive agents expressed or released through MDSCs, such as arginase, iNOS, and ROS, were highly restored in MDSCs, and the ability of MDSCs to inhibit CD4+ and CD8+ T cells improved after targeting Runx1 (86, 118). These results indicated that miR-9 targeting results in decreased suppressive activity of MDSCs and increases antitumor response, which can thus be considered, with reason, a potential treatment strategy.
MicroRNA-210
MicrRNA-210 is another important miRNA that can boost the tumor-promoting effects of MDSCs. Noman et al. (87) verified that hypoxia via HIF1a stimulates miR-210 in splenic MDSCs and increases the activity of MDSC by promoting Arg-1 expression and regulating IL-16 and CXCL12. Upregulation of miR-210 was sufficient to augment suppression of T cells by MDSC, and targeting miR-210 was enough to reduce MDSC activity against T cells. Thus, miR-210 inhibitor oligonucleotide, as an adjuvant tool, a plus novel developing immunotherapeutic approach might be helpful for improving the immune response in patients with cancer (87).
MicroRNA-223
MicroRNA-223 has a central role in myeloid differentiation and is highly expressed in granulocyte lineage in human hematopoiesis (105). Its expression increasingly rises as granulocytes mature (119). Given its central role in granulocytic differentiation, the augmented expression of miR-223 is apparently able to restore differentiation in leukemic blast cells (89). In the case of MDSCs, the findings proposed a negative function for miR-223 in MDSC development because an overexpression of miR-223 can definitely control the differentiation of myeloid progenitors (90). In a murine model of autoimmune encephalomyelitis, genetic ablation of miR-223 was used to increase understanding of the molecular pathway participated in the pathogenesis of EAE. It was found that miR-223 knockout mice presented a less severe disease characterized by the enhanced accumulation of MDSC and lower levels of CNS-isolated T cell responses compared with control mice (91). A greater suppressive function detected in miR-223−/− MO-MDSCs was associated with an elevated level of Arg1 and Stat3, which are reported as the target genes of miR-223 (91). The downregulation of miR-223 expression by tumor-related factors might enhance the induction and accumulation of MDSCs in the tumor milieu (90). Tumor-associated MDSCs express lower levels of miR-223 than Gr1+CD11b+ cells from the spleens of tumor-free mice. In this process, myeloid ELF1-like factor 2C (MEF2C) was detected as a direct target of miR-223, whose expression induced myeloid progenitor proliferation. MiR-223 activity suppresses the MEF2C gene that inhibits progenitor expansion, leading to decreased accumulation of MDSCs and induce tumor suppression (90).
Long Non-coding RNAs
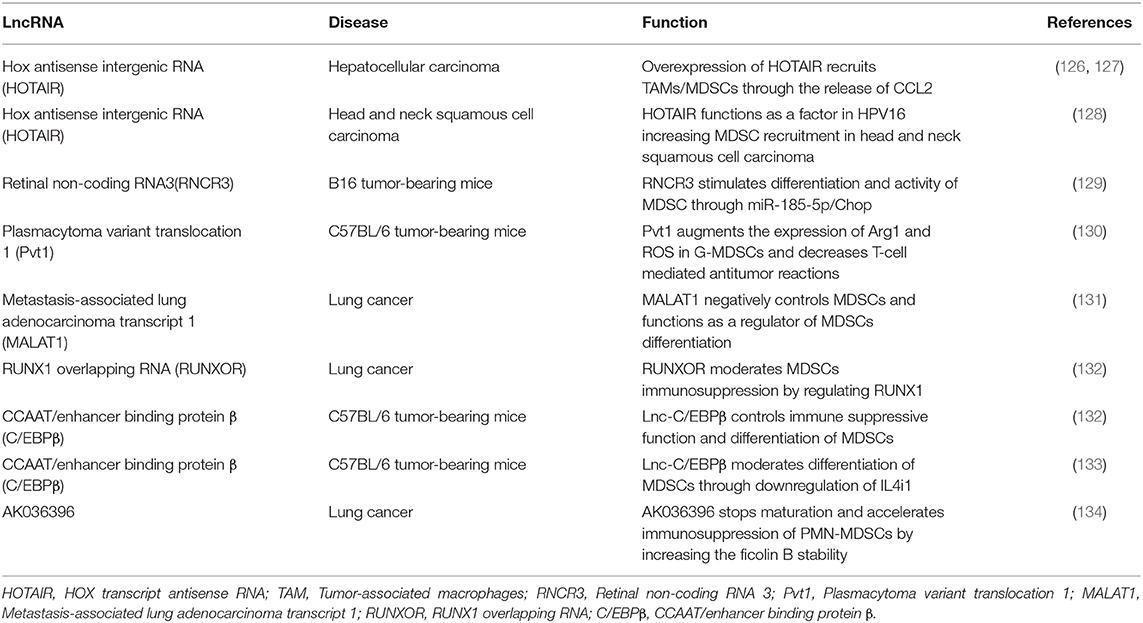
Long non-coding RNAs (lncRNAs) are a large class of non-coding RNAs (ncRNAs) with more than 200 nucleotides in length (120, 121). Initially characterized as “transcriptional noise” in previous RNA sequence researches, lncRNAs are currently considered as efficient RNA factors with properties of shorter length, comprising fewer exons, and low expression levels compared to messenger RNAs (mRNAs) (122–124). lncRNAs are indicated to be overexpressed in immune cells such as monocytes, macrophages, dendritic cells, neutrophils, T cells, B cells and MDSCs during their development, differentiation and activation (125). However, most of researches about lncRNAs controlling cell biology emphasize in their effects on numerous cancer cells, while their significance in MDSC regulation is also worthy of attention (Table 2).
Hox Antisense Intergenic RNA
Hox antisense intergenic RNA (HOTAIR) is a lncRNA and resembles an oncogenic molecule in several cancers. Accumulating evidence proves that HOTAIR has crucial functions in the progression and metastasis of different cancers, such as breast (135), colorectal (136), non-small lung cell (137), and gastric cancer (138). In the case of HCC cell lines, a recent study confirmed that HOTAIR-overexpressing cells release a high level of CCL2 and support macrophage/MDSC proliferation. A report demonstrated that CCL2 is an essential factor for the recruitment of macrophages and the accumulation of MDSC in the tumor milieu (81, 126, 139, 140), signifying that the overexpression of HOTAIR in cancer cells results in TAMs/MDSCs recruitment through CCL2 release, leading to augmented tumor progression and metastasis (127).
In another study, Ma et al. explained the difference in the recruitment of MDSCs seen in human papilloma virus (HPV)-positive head, neck squamous cell carcinoma (HNSCC) and HPV-negative HNSCC by using lncRNA analysis. The results showed significant function and importance of lncRNAs in HPV-associated HNSCC, demonstrating that lncRNA is a possible regulator of MDSCs recruitment in HPV-positive HNSCC (128). The level of MDSCs in HPV-positive HNSCC was notably more than in normal controls, indicating that infection with HPV can support aggregation of MDSCs. Using an array-based method to display the lncRNA expression between HPV-positive HNSCC, HPV-negative HNSCC, and normal oral mucous, 132 various lncRNAs were found in diverse HPV infected conditions of HNSCC. HOTAIR, PROM1, CCAT1, and MUC19 mRNA levels, as detected by qRTPCR, which were reversely related to MDSCs of HPV-related HNSCC. Therefore, lncRNAs resembled a modulatory factor in HPV16 supporting MDSC recruitment in HNSCC (128).
Retinal Non-coding RNA3
Retinal non-coding RNA3 (RNCR3), also named LINC00599 is an intergenic lncRNA and is introduced as a conserved lncRNA in mammals. The proliferation and activity of various cell types can be regulated by RNCR3 (141–143). In a current research, Shang et al. proved the expression of RNCR3 in MDSCs, and it was found that this expression is considerably increased in the inflammatory and tumor microenvironment. Moreover, RNCR3 stimulates the differentiation and activity of MDSC through sponging miR-185-5p so as to produce its target gene named Chop (129). Chop is a sensor of cellular stress, which is stimulated via tumor-related ROS and nitrogen species. Chop acts as a main factor in controlling the accumulation and immunosuppressive activity of MDSCs (144). It is demonstrated that miR-185-5p affected the MDSCs expansion and reversed the impact of RNCR3 on MDSC differentiation and function by targeting Chop. Therefore, this study proposed a RNCR3/miR-185-5p/Chop autologously strengthening complex to support MDSC differentiation and suppressive activity in response to extracellular inflammatory and tumor-related signals (129).
Plasmacytoma Variant Translocation 1
The lncRNA plasmacytoma variant translocation 1 (Pvt1) is intergenic and described as a conserved lncRNA in humans and mice. Upregulation of Pvt1 is detected in numerous cancers, such as melanoma, cervical, gastric, prostate, hepatocellular, esophageal cancers, and AML (145–147). Although lncRNA Pvt1 mechanism in cancer cells is clear, how lncRNA Pvt1 controls function and differentiation of MDSC is not well-known. A recent study indicated that Pvt1 knockdown remarkably stopped the immunosuppressive activity of G-MDSCs in vitro. It is shown that the Pvt1 expression is increased by HIF-1α in G-MDSCs under hypoxia. Pvt1 suppression lessened the Arg1 and ROS in G-MDSCs and increased T-cell-mediated antitumor reactions. The findings of this research demonstrated that Pvt1 targeting may reduce immunosuppression of G-MDSCs, which can be further investigated as a possible therapeutic approach (130).
Metastasis-Associated Lung Adenocarcinoma Transcript 1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a well-recognized lncRNA related to several diseases, including cancer, has increasingly received consideration In 2003, MALAT1 was firstly recognized to be significantly related to the metastasis of early-stage non-small cell lung cancer (NSCLC), and thus MALAT1 was suggested to be a predictive marker for stage I NSCLC (148). MALAT1 is a central lncRNA associated with many biological processes. The vital functions of MALAT1 in gene regulation and the significant impact on the basic activity of cells, including tumor cells, have been established by several researches (149). However, the function of this lncRNA in MDSCs is undefined. A recent research noticed the proportion of MDSCs in patients with lung cancer, the amount of ARG-1 in MDSCs, and the CD8+CTL cells percentage, and they also examined the MALAT1 association with MDSCs in PBMCs and revealed the regulatory activity of MALAT1 on MDSCs stimulation in vitro. Their results indicated a negative link between the MALAT1 level and the MDSCs proportion in PBMCs of lung cancer patients. It is suggested that MALAT1 can have a role in controlling the differentiation of MDSCs. After using MALAT1 siRNA in PBMCs, the proportion of MDSCs was meaningfully augmented as a consequence. These findings indicate that inhibition of MALAT1 promotes the quantity of MDSCs by controlling their differentiation (131).
RUNX1 Overlapping RNA (RUNXOR)
RUNX1 overlapping RNA (RUNXOR) is introduced as an intragenic lncRNA and is about 216 kb in length. RUNXOR regulates the RUNX1 gene, which is introduced as a tumor suppressor and main regulator of hematopoiesis genes. Meanwhile, the effect of lncRNA RUNXOR on the MDSC development is uncertain (150). Tian et al. revealed the overexpression of RUNXOR and downregulation of RUNX1 in patients with lung cancer. Also, they showed that the expression of RUNXOR increases in MDSCs. After knockdown of RUNXOR, the Arg1 expression in MDSCs was downregulated. Moreover, it is indicated that after using siRUNXOR, the expression of RUNX1 is reestablished in MDSCs, and RUNX1 is negatively associated with the MDSCs proportion from patients with lung cancer. These findings presented that RUNXOR can influence the activity of MDSCs by regulation of RUNX1 (132).
CCAAT/Enhancer Binding Protein β
The family of CCAAT/enhancer binding proteins (C/EBPs) transcription factors has vital activities in the expansion and differentiation of several types of cells such as granulocytes. C/EBPβ is informed to be essential for the maturation of monocyte and eosinophil, and currently, for neutrophil differentiation. The C/EBPβ was overexpressed according to the development of granulocytic differentiation (151, 152). LncRNA called lnc-C/EBPβ was described by Gao et al. (132) They confirmed that lnc-C/EBPβ controls numerous transcripts in MDSCs to regulate MDSC differentiation and suppressive activity in inflammatory and tumor milieus. Expression of lnc-C/EBPβ via MDSCs, negatively controls MDSC function, signifying that lnc-C/EBPβ has a negative feedback function in avoiding over-suppression of MDSCs on immune reactions (132). Moreover, it is revealed that lnc-C/EBPβ can increase polymorphonuclear MDSCs (PMN-MDSC) but inhibit the differentiation of monocytic MDSCs (Mo-MDSC). lnc-C/EBPβ can downregulate interleukin 4-induced gene-1 (IL4i1) to affect the MDSC differentiation by attaching with C/EBPβ LIP and WD repeat-containing protein 5 (WDR5) (133).
AK036396
As the largest group of MDSCs, PMN-MDSCs indicate a fundamental role in increasing the immune escape of numerous cancers. Tian et al. recognized that lncRNA AK036396 and its target Ficolin B (Fcnb) were most abundant in PMN-MDSCs, compared with other myeloid cells. They detected the regulatory mechanism of lncRNA AK036396 in the PMN-MDSCs. Downregulation of LncRNA AK036396 decreased Fcnb protein stability in an ubiquitin-proteasome system-dependent mechanism. They revealed that the downregulation of lncRNA AK036396 stimulated the maturation and reduced the suppressive activity of PMN-MDSCs. Moreover, the expression of human M-ficolin, which is an ortholog of mouse Fcnb, was augmented and associated with arginase1 expression (134).
Concluding Remarks
MDSCs are immature immunosuppressive cells that are involved in a wide spectrum of diseases and in particular in cancer. The development of MDSC-targeted approaches to improve immunotherapy appears to be a high priority and might bring new results in the field of immunotherapy. One considerable challenge is the complexity of factors that are found to play a critical role in regulating MDSC function and expansion. Increasing evidence suggests that miRNAs and lncRNAs are implicated in the pathogenesis of diseases or are at least linked to the disease state. Moreover, the aberrantly expressed miRNAs and lncRNAs were significantly associated with MDSC development and function. In addition to intracellular regulation, miRNAs and lncRNAs can show intercellular impacts via exosomes (153). Exosomes are released by diverse cell types, including cells with a myeloid lineage, such as MDSCs. Inadequate evidence on MDSC exosomes reveals that they can exert effects related to immunosuppression and the elevation of tumorigenesis (154). It is presented that exosomes were secreted more plentifully from tumor MDSCs, apparently due to the fact that MDSCs in the TME are in a more stressful environment (155). Furthermore, tumor exosomes have been informed to carry genetic materials and proteins capable of suppressing the immune cells functions and stimulating the activation and expansion of MDSCs in vitro and in vivo (156). It is shown that upregulation of miR-10a and miR-21 in hypoxia-induced glioma-derived exosomes has a powerful impact in MDSC induction. Hypoxia-inducible miR-10a and miR-21 expression in glioma-derived exosomes increased the expansion and suppressive activities of MDSCs. These findings demonstrate that exosomes have a prognostic value, promising positive inputs for new therapeutic directions in the cancer treatment. The promise of targeting miRNAs and lncRNAs, by either antagonizing or restoring function, might provide novel strategies to re-educate immunosuppressive or hyperactive milieu of disease associated with MDSC development, including cancer. Given the heterogeneity of human MDSCs and limitation in clinical application, it is reasonable to predict that more information is urgently needed, and new ideas and questions are proposed for future inquiry.
Author Contributions
ES and ZA: conception, design, and writing. SS, AH, AD, FG, and OB: literature search and writing. NS and BB: editing and correction. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Serafini P, Bronte V. Myeloid-derived suppressor cells in tumor-induced T cell suppression and tolerance. In: Tumor-Induced Immune Suppression. Springer (2008). 222:162–79. doi: 10.3109/08820139.2012.673191
2. De Sanctis F, Bronte V, Ugel S. Tumor-induced myeloid-derived suppressor cells. In: Myeloid Cells in Health and Disease: A Synthesis. (2015) 125:3365–76. doi: 10.1172/JCI80006
3. Sica A, Guarneri V, Gennari A. Myelopoiesis, metabolism and therapy: a crucial crossroads in cancer progression. Cell Stress. (2019) 3:284. doi: 10.15698/cst2019.09.197
4. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. (2009) 9:162–74. doi: 10.1038/nri2506
5. Dai J, El Gazzar M, Li GY, Moorman JP, Yao ZQ. Myeloid-derived suppressor cells: paradoxical roles in infection and immunity. J Innate Immun. (2015) 7:116–26. doi: 10.1159/000368233
6. Huber V, Vallacchi V, Di Guardo L, Cova A, Canevari S, Santinami M, et al. ITOC2–038. Role of exosomes in immune suppression. Eur J Cancer. (2015) 51:S13. doi: 10.1016/j.ejca.2015.01.052
7. Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-derived suppressor cells: critical cells driving immune suppression in the tumor microenvironment. In: Advances in Cancer Research. Elsevier (2015). 128:95–139. doi: 10.1016/bs.acr.2015.04.002
8. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. (2016) 37:208–20. doi: 10.1016/j.it.2016.01.004
9. Ostrand-Rosenberg S, Fenselau C. Myeloid-derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J Immunol. (2018) 200:422–31. doi: 10.4049/jimmunol.1701019
10. Papaspyridonos M, Matei I, Huang Y, Do Rosario Andre M, Brazier-Mitouart H, Waite JC, et al. Id1 suppresses anti-tumour immune responses and promotes tumour progression by impairing myeloid cell maturation. Nat Commun. (2015) 6:6840. doi: 10.1038/ncomms7840
11. Qu P, Wang L-Z, Lin PC. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. (2016) 380:253–6. doi: 10.1016/j.canlet.2015.10.022
12. Zhang C, Wang S, Liu Y, Yang C. Epigenetics in myeloid derived suppressor cells: a sheathed sword towards cancer. Oncotarget. (2016) 7:57452. doi: 10.18632/oncotarget.10767
13. Tian X, Tian J, Tang X, Ma J, Wang S. Long non-coding RNAs in the regulation of myeloid cells. J Hematol Oncol. (2016) 9:99. doi: 10.1186/s13045-016-0333-7
14. Liu SJ. Identification and Functional Characterization of Long Non-Coding RNAs in Development and Disease. San Francisco, CA: University of California (2017).
15. El Gazzar M, McCall CE. MicroRNAs regulatory networks in myeloid lineage development and differentiation: regulators of the regulators. Immunolo Cell Biol. (2012) 90:587–93. doi: 10.1038/icb.2011.74
16. Chen S, Zhang Y, Kuzel TM, Zhang B. Regulating tumor myeloid-derived suppressor cells by microRNAs. Cancer Cell Microenviron. (2015) 2:e637. doi: 10.14800/ccm.637
17. Huber V, Vallacchi V, Fleming V, Hu X, Cova A, Dugo M, et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J Clin Invest. (2018) 128:5505–16. doi: 10.1172/JCI98060
18. Solito S, Pinton L, Mandruzzato S. In Brief: myeloid-derived suppressor cells in cancer. J Pathol. (2017) 242:7–9. doi: 10.1002/path.4876
19. Netherby CS, Abrams SI. Mechanisms overseeing myeloid-derived suppressor cell production in neoplastic disease. Cancer Immunol Immunother. (2017) 66:989–96. doi: 10.1007/s00262-017-1963-5
20. Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. (2015) 125:3356. doi: 10.1172/JCI80005
21. Safarzadeh E, Hashemzadeh S, Duijf PH, Mansoori B, Khaze V, Mohammadi A, et al. Circulating myeloid-derived suppressor cells: an independent prognostic factor in patients with breast cancer. J Cell Physiol. (2019) 234:3515–25. doi: 10.1002/jcp.26896
22. Baniyash M. Myeloid-derived suppressor cells as intruders and targets: clinical implications in cancer therapy. Cancer Immunol Immunother. (2016) 65:857–67. doi: 10.1007/s00262-016-1849-y
23. Ueha S, Shand FH, Matsushima K. Myeloid cell population dynamics in healthy and tumor-bearing mice. Int Immunopharmacol. (2011) 11:783–8. doi: 10.1016/j.intimp.2011.03.003
24. Safarzadeh E, Orangi M, Mohammadi H, Babaie F, Baradaran B. Myeloid-derived suppressor cells: important contributors to tumor progression and metastasis. J Cell Physiol. (2018) 233:3024–36. doi: 10.1002/jcp.26075
25. Talmadge JE, Cole K, Britton H, Dafferner A, Warkentin P. Human myeloid derived suppressor cell (MDSC) subset phenotypes. Am Assoc Immnol. (2017) 198:211.2. doi: 10.1158/1538-7445.AM2017-LB-192
26. Solito S, Marigo I, Pinton L, Damuzzo V, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann NY Acad Sci. (2014) 1319:47–65. doi: 10.1111/nyas.12469
27. Mandruzzato S, Brandau S, Britten CM, Bronte V, Damuzzo V, Gouttefangeas C, et al. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: results from an interim study. Cancer Immunolo Immunother. (2016) 65:161–9. doi: 10.1007/s00262-015-1782-5
28. Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. (2016) 7:12150. doi: 10.1038/ncomms12150
29. Mazza EMC, Zoso A, Mandruzzato S, Bronte V, Serafini P, Inverardi L, et al. Gene expression profiling of human fibrocytic myeloid-derived suppressor cells (f-MDSCs). Genomics data. (2014) 2:389–92. doi: 10.1016/j.gdata.2014.10.018
30. Katoh H, Watanabe M. Myeloid-derived suppressor cells and therapeutic strategies in cancer. Mediat Inflamm. (2015) 2015:159269. doi: 10.1155/2015/159269
31. Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. (2009) 182:4499–506. doi: 10.4049/jimmunol.0802740
32. Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. (2006) 176:284–90. doi: 10.4049/jimmunol.176.1.284
33. Bianchi G, Borgonovo G, Pistoia V, Raffaghello L. Immunosuppressive cells and tumour microenvironment: focus on mesenchymal stem cells and myeloid derived suppressor cells. Histol Histopathol. (2011) 26:941. doi: 10.14670/HH-26.941
34. Meirow Y, Kanterman J, Baniyash M. Paving the road to tumor development and spreading: myeloid-derived suppressor cells are ruling the fate. Front Immunol. (2015) 6:523. doi: 10.3389/fimmu.2015.00523
35. Poschke I, Kiessling R. On the armament and appearances of human myeloid-derived suppressor cells. Clin Immunol. (2012) 144:250–68. doi: 10.1016/j.clim.2012.06.003
36. Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. (2011) 32:19–25. doi: 10.1016/j.it.2010.10.002
37. Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. (2009) 124:2621–33. doi: 10.1002/ijc.24249
38. De Sanctis F, Solito S, Ugel S, Molon B, Bronte V, Marigo I. MDSCs in cancer: conceiving new prognostic and therapeutic targets. Biochim Biophys Acta. (2016) 1865:35–48. doi: 10.1016/j.bbcan.2015.08.001
39. Condamine T, Mastio J, Gabrilovich DI. Transcriptional regulation of myeloid-derived suppressor cells. J Leukocyte Biol. (2015) 98:913–22. doi: 10.1189/jlb.4RI0515-204R
40. Lee JM, Kim EK, Seo H, Jeon I, Chae MJ, Park YJ, et al. Serum amyloid A3 exacerbates cancer by enhancing the suppressive capacity of myeloid-derived suppressor cells via TLR2-dependent STAT3 activation. Eur J Immunol. (2014) 44:1672–84. doi: 10.1002/eji.201343867
41. Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-β1. J Immunol. (2009) 182:240–9. doi: 10.4049/jimmunol.182.1.240
42. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. (2012) 12:253–68. doi: 10.1038/nri3175
43. Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. (2013) 138:105–15. doi: 10.1111/imm.12036
44. Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. (2013) 91:493–502. doi: 10.1038/icb.2013.29
45. Seyfizadeh N. Immune dysfunction induced by myeloid-derived suppressor cells in lymphoma. Cancer Cell Microenviron. (2016) 3:e1111. doi: 10.14800/ccm.1111
46. Fletcher M, Ramirez ME, Sierra RA, Raber P, Thevenot P, Al-Khami AA, et al. l-Arginine depletion blunts antitumor T-cell responses by inducing myeloid-derived suppressor cells. Cancer research. (2015) 75:275–83. doi: 10.1158/0008-5472.CAN-14-1491
47. Movahedi K, Guilliams M, Van Den Bossche J, Van Den Bergh R, Gysemans C, Beschin A, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell–suppressive activity. Blood. (2008) 111:4233–44. doi: 10.1182/blood-2007-07-099226
48. Ostrand-Rosenberg S, Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother. (2010) 59:1593–600. doi: 10.1007/s00262-010-0855-8
49. Youn J-I, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. (2008) 181:5791–802. doi: 10.4049/jimmunol.181.8.5791
50. Ochoa AC, Zea AH, Hernandez C, Rodriguez PC. Arginase prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin Cancer Res. (2007) 13:721–6s. doi: 10.1158/1078-0432.CCR-06-2197
51. Sikalidis AK. Amino acids and immune response: a role for cysteine, glutamine, phenylalanine, tryptophan and arginine in T-cell function and cancer? Pathol Oncol Res. (2015) 21:9–17. doi: 10.1007/s12253-014-9860-0
52. Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. In: Seminars in Cancer Biology. Elsevier (2012) 22:275–81. doi: 10.1016/j.semcancer.2012.01.011
53. Fionda C, Abruzzese MP, Santoni A, Cippitelli M. Immunoregulatory and effector activities of nitric oxide and reactive nitrogen species in cancer. Curr Med Chem. (2016) 23:2618–36. doi: 10.2174/0929867323666160727105101
54. Gehad AE, Lichtman MK, Schmults CD, Teague JE, Calarese AW, Jiang Y, et al. Nitric oxide–producing myeloid-derived suppressor cells inhibit vascular E-selectin expression in human squamous cell carcinomas. J Invest Dermatol. (2012) 132:2642–51. doi: 10.1038/jid.2012.190
55. Bingisser RM, Tilbrook PA, Holt PG, Kees UR. Macrophage-derived nitric oxide regulates T cell activation via reversible disruption of the Jak3/STAT5 signaling pathway. J Immunol. (1998) 160:5729–34.
56. Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. (2007) 6:662. doi: 10.1038/nrd2222
57. Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. (2008) 222:162–79. doi: 10.1111/j.1600-065X.2008.00602.x
58. Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. (2007) 13:828. doi: 10.1038/nm1609
59. Yu J, Du W, Yan F, Wang Y, Li H, Cao S, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. (2013) 190:3783–97. doi: 10.4049/jimmunol.1201449
60. Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, et al. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J Immunol. (2012) 189:5602–11. doi: 10.4049/jimmunol.1201018
61. Chen J, Ye Y, Liu P, Yu W, Wei F, Li H, et al. Suppression of T cells by myeloid-derived suppressor cells in cancer. Hum Immunol. (2017) 78:113–19. doi: 10.1016/j.humimm.2016.12.001
62. Chen W, Jin W, Hardegen N, Lei K-J, Li L, Marinos N, et al. Conversion of peripheral CD4+ CD25− naive T cells to CD4+ CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. (2003) 198:1875–86. doi: 10.1084/jem.20030152
63. Motallebnezhad M, Jadidi-Niaragh F, Qamsari ES, Bagheri S, Gharibi T, Yousefi M. The immunobiology of myeloid-derived suppressor cells in cancer. Tumor Biol. (2016) 37:1387–406. doi: 10.1007/s13277-015-4477-9
64. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509. doi: 10.1038/nrm3838
65. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
66. Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. (2007) 6:2127–32. doi: 10.4161/cc.6.17.4641
67. Harfe BD, MicroRNAs in vertebrate development. Curr Opin Genet Dev. (2005) 15:410–5. doi: 10.1016/j.gde.2005.06.012
68. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. (2001) 294:858–62. doi: 10.1126/science.1065062
69. Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell Cycle. (2006) 5:837–40. doi: 10.4161/cc.5.8.2688
70. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. (2004) 432:226. doi: 10.1038/nature03076
71. Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. (2004) 14:2162–7. doi: 10.1016/j.cub.2004.11.001
72. Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. (2004) 6:1048. doi: 10.1038/ncb1104-1048
73. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. (2005) 435:834. doi: 10.1038/nature03702
74. Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. (2006) 103:2257–61. doi: 10.1073/pnas.0510565103
75. Gracias DT, Katsikis PD. MicroRNAs: key components of immune regulation. In: Crossroads Between Innate and Adaptive Immunity III. Springer. (2011). 780:15–26. doi: 10.1007/978-1-4419-5632-3_2
76. O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. (2010) 10:111. doi: 10.1038/nri2708
77. Lindsay MA. MicroRNAs and the immune response. Trends Immunol. (2008) 29:343–51. doi: 10.1016/j.it.2008.04.004
78. Cao W, Cheng W, Wu W. MicroRNAs reprogram tumor immune response. In: MicroRNA and Cancer. Springer (2018). 1699:67–74. doi: 10.1007/978-1-4939-7435-1_4
79. El-Gazzar M. MicroRNAs as potential regulators of myeloid-derived suppressor cell expansion. Innate Immun. (2014) 20:227–38. doi: 10.1177/1753425913489850
80. Brunetti O, Russo A, Scarpa A, Santini D, Reni M, Bittoni A, et al. MicroRNA in pancreatic adenocarcinoma: predictive/prognostic biomarkers or therapeutic targets? Oncotarget. (2015) 6:23323. doi: 10.18632/oncotarget.4492
81. Gnoni A, Santini D, Scartozzi M, Russo A, Licchetta A, Palmieri V, et al. Hepatocellular carcinoma treatment over sorafenib: epigenetics, microRNAs and microenvironment. Is there a light at the end of the tunnel? Expert Opin Therapeut Targets. (2015) 19:1623–35. doi: 10.1517/14728222.2015.1071354
82. Xu Z, Ji J, Xu J, Li D, Shi G, Liu F, et al. MiR-30a increases MDSC differentiation and immunosuppressive function by targeting SOCS3 in mice with B-cell lymphoma. FEBS J. (2017) 284:2410–24 doi: 10.1111/febs.14133
83. Jiang J, Zhang Y, Yu C, Li Z, Pan Y, Sun C. MicroRNA-492 expression promotes the progression of hepatic cancer by targeting PTEN. Cancer Cell Int. (2014) 14:95. doi: 10.1186/s12935-014-0095-7
84. Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. (2012) 188:5500–10. doi: 10.4049/jimmunol.1103505
85. Mei S, Xin J, Liu Y, Zhang Y, Liang X, Su X, et al. MicroRNA-200c promotes suppressive potential of myeloid-derived suppressor cells by modulating PTEN and FOG2 expression. PLoS ONE. (2015) 10:e0135867. doi: 10.1371/journal.pone.0135867
86. Tian J, Rui K, Tang X, Ma J, Wang Y, Tian X, et al. MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J Immunol. (2015) 195:1301–11. doi: 10.4049/jimmunol.1500209
87. Noman MZ, Janji B, Hu S, Wu JC, Martelli F, Bronte V, et al. Tumor-promoting effects of myeloid-derived suppressor cells are potentiated by hypoxia-induced expression of miR-210. Cancer Res. (2015) 75:3771–87. doi: 10.1158/0008-5472.CAN-15-0405
88. Chen S, Huang A, Chen H, Yang Y, Xia F, Jin L, et al. miR-34a inhibits the apoptosis of MDSCs by suppressing the expression of N-myc. Immunolo Cell Biol. (2016) 94:563–72. doi: 10.1038/icb.2016.11
89. Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. (2007) 12:457–66. doi: 10.1016/j.ccr.2007.09.020
90. Liu Q, Zhang M, Jiang X, Zhang Z, Dai L, Min S, et al. miR-223 suppresses differentiation of tumor-induced CD11b+ Gr1+ myeloid-derived suppressor cells from bone marrow cells. Int J Cancer. (2011) 129:2662–73. doi: 10.1002/ijc.25921
91. Cantoni C, Cignarella F, Ghezzi L, Mikesell B, Bollman B, Berrien-Elliott MM, et al. Mir-223 regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol. (2017) 133:61–77. doi: 10.1007/s00401-016-1621-6
92. Chen S, Wang L, Fan J, Ye C, Dominguez D, Zhang Y, et al. Host miR-155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res. (2015) 75:519–31. doi: 10.1158/0008-5472.CAN-14-2331
93. Wang J, Yu F, Jia X, Iwanowycz S, Wang Y, Huang S, et al. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int J Cancer. (2015) 136:E602–13. doi: 10.1002/ijc.29151
94. Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. (2009) 113:4720–8. doi: 10.1182/blood-2008-11-190215
95. McClure C, Brudecki L, Ferguson DA, Yao ZQ, Moorman JP, Mccall CE, et al. MicroRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect Immun. (2014) 82:3816–25. doi: 10.1128/IAI.01495-14
96. Yu H, Liu Y, Mcfarland BC, Deshane JS, Hurst DR, Ponnazhagan S, et al. SOCS3 deficiency in myeloid cells promotes tumor development: involvement of STAT3 activation and myeloid-derived suppressor cells. Cancer Immunol Res. (2015) 3:727–40. doi: 10.1158/2326-6066.CIR-15-0004
97. Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. (2009) 1792:497–505. doi: 10.1016/j.bbadis.2009.02.013
98. Georgantas RW, Hildreth R, Morisot S, Alder J, Liu C-G. Heimfeld S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. (2007) 104:2750–5. doi: 10.1073/pnas.0610983104
99. Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, Mccoy JP, Sloand EM, et al. Hematopoietic-specific microRNA expression in human cells. Leukemia Res. (2006) 30:643–7. doi: 10.1016/j.leukres.2005.09.001
100. Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang C-Y, et al. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol. (2014) 192:1034–43. doi: 10.4049/jimmunol.1301309
101. Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. (2007) 8:166. doi: 10.1186/1471-2164-8-166
102. Yang P, Li Q-J, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. (2012) 22:291–303. doi: 10.1016/j.ccr.2012.07.023
103. Wang X, Chang X, Zhuo G, Sun M, Yin K. Twist and miR-34a are involved in the generation of tumor-educated myeloid-derived suppressor cells. Int J Mol Sci. (2013) 14:20459–77. doi: 10.3390/ijms141020459
104. Su R, Lin H, Zhang X, Yin X, Ning H, Liu B, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. (2015) 34:3226–39. doi: 10.1038/onc.2014.274
105. Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. (2005) 123:819–31. doi: 10.1016/j.cell.2005.09.023
106. McClure C, Mcpeak MB, Youssef D, Yao ZQ, Mccall CE, El Gazzar M. Stat3 and C/EBPβ synergize to induce miR-21 and miR-181b expression during sepsis. Immunol Cell Biol. (2017) 95:42–55. doi: 10.1038/icb.2016.63
107. Zhang Z, Huang X, Wang E, Huang Y, Yang R. Suppression of Mll1-complex by Stat3/Cebpβ-induced miR-21a/21b/181b maintains the accumulation, homeostasis, and immunosuppressive function of polymorphonuclear myeloid-derived suppressor cells. J Immunol. (2020) 204:3400–15. doi: 10.4049/jimmunol.2000230
108. Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. (2007) 104:16158–63. doi: 10.1073/pnas.0702596104
109. Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. (2006) 441:518. doi: 10.1038/nature04747
110. Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. (2009) 139:1096–108. doi: 10.1016/j.cell.2009.11.020
111. Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. (2009) 106:5282–7. doi: 10.1073/pnas.0810909106
112. Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell–derived neurogenesis. Stem Cells. (2006) 24:857–64. doi: 10.1634/stemcells.2005-0441
113. Zhang Y, Liao Y, Wang D, He Y, Cao D, Zhang F, Dou K. Altered expression levels of miRNAs in serum as sensitive biomarkers for early diagnosis of traumatic injury. J Cell Biochem. (2011) 112:2435–42. doi: 10.1002/jcb.23168
114. Nie K, Gomez M, Landgraf P, Garcia J-F, Liu Y, Tan LH, et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. (2008) 173:242–52. doi: 10.2353/ajpath.2008.080009
115. Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9 a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. (2010) 12:247. doi: 10.1038/ncb2024
116. Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. (2009) 457:887. doi: 10.1038/nature07619
117. Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T, et al. Requirement of Runx1/AML1/PEBP2αB for the generation of haematopoietic cells from endothelial cells. Genes Cells. (2001) 6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x
118. Link KA, Chou FS, Mulloy JC. Core binding factor at the crossroads: determining the fate of the HSC. J Cell Physiol. (2010) 222:50–6. doi: 10.1002/jcp.21950
119. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. (2008) 451:1125–9. doi: 10.1038/nature06607
120. Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. (2001) 2:986–91. doi: 10.1093/embo-reports/kve230
121. Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci Cell Biosci. (2012) 2:37. doi: 10.1186/2045-3701-2-37
122. Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. (2014) 35:408–19. doi: 10.1016/j.it.2014.07.005
123. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. (2013) 154:26–46. doi: 10.1016/j.cell.2013.06.020
124. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
125. Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. (2014) 20:623–31. doi: 10.1016/j.molmed.2014.09.002
126. Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. (2016) 76:5671–82. doi: 10.1158/0008-5472.CAN-16-0144
127. Fujisaka Y, Iwata T, Tamai K, Nakamura M, Mochizuki M, Shibuya R, et al. Long non-coding RNA HOTAIR up-regulates chemokine (C-C motif) ligand 2 and promotes proliferation of macrophages and myeloid-derived suppressor cells in hepatocellular carcinoma cell lines. Oncol Lett. (2018) 15:509–14. doi: 10.3892/ol.2017.7322
128. Ma X, Sheng S, Wu J, Jiang Y, Gao X, Cen X, et al. LncRNAs as an intermediate in HPV16 promoting myeloid-derived suppressor cell recruitment of head and neck squamous cell carcinoma. Oncotarget. (2017) 8:42061. doi: 10.18632/oncotarget.14939
129. Shang W, Tang Z, Gao Y, Qi H, Su X, Zhang Y, et al. LncRNA RNCR3 promotes Chop expression by sponging miR-185–5p during MDSC differentiation. Oncotarget. (2017) 8:111754. doi: 10.18632/oncotarget.22906
130. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer. (2019) 18:61. doi: 10.1186/s12943-019-0978-2
131. Zhou Q, Tang X, Tian X, Tian J, Zhang Y, Ma J, et al. LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. J cancer. (2018) 9:2436. doi: 10.7150/jca.24796
132. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc-C/EBPβ negatively regulates the suppressive function of myeloid-derived suppressor cells. Cancer Immunol Res. (2018) 6:1352–63. doi: 10.1158/2326-6066.CIR-18-0108
133. Gao Y, Shang W, Zhang D, Zhang S, Zhang X, Zhang Y, et al. Lnc-C/EBPβ Modulates differentiation of MDSCs through downregulating IL4i1 with C/EBPβ LIP and WDR5. Front Immunol. (2019) 10:1661. doi: 10.3389/fimmu.2019.01661
134. Tian X, Zheng Y, Yin K, Ma J, Tian J, Zhang Y, et al. LncRNA AK036396 inhibits maturation and accelerates immunosuppression of polymorphonuclear myeloid–derived suppressor cells by enhancing the stability of ficolin B. Cancer Immunol Res. (2020) 8:565–77. doi: 10.1158/2326-6066.CIR-19-0595
135. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071. doi: 10.1038/nature08975
136. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long non-coding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. (2011) 71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021
137. Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. (2013) 436:319–24. doi: 10.1016/j.bbrc.2013.05.101
138. Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE. (2013) 8:e77070. doi: 10.1371/journal.pone.0077070
139. Teng K-Y, Han J, Zhang X, Hsu S-H, He S, Wani NA, et al. Blocking the CCL2–CCR2 axis using CCL2-neutralizing antibody is an effective therapy for hepatocellular cancer in a mouse model. Mol Cancer Therapeut. (2017) 16:312–22. doi: 10.1158/1535-7163.MCT-16-0124
140. Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. (2017) 66:157–67. doi: 10.1136/gutjnl-2015-310514
141. Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. (2010) 11:14. doi: 10.1186/1471-2202-11-14
142. Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, et al. Genomic analysis of mouse retinal development. PLoS Biol. (2004) 2:e247. doi: 10.1371/journal.pbio.0020247
143. Shan K, Jiang Q, Wang X-Q, Yang H, Yao M-D, Liu C, et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. (2016) 7:e2248. doi: 10.1038/cddis.2016.145
144. Thevenot PT, Sierra RA, Raber PL, Al-Khami AA, Trillo-Tinoco J, Zarreii P, et al. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. (2014) 41:389–401. doi: 10.1016/j.immuni.2014.08.015
145. Colombo T, Farina L, Macino G, Paci P. PVT1: a rising star among oncogenic long noncoding RNAs. BioMed Res Int. (2015) 2015:304208. doi: 10.1155/2015/304208
146. Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in cancer: a review and meta-analysis. Clin Chim Acta. (2017) 474:1–7. doi: 10.1016/j.cca.2017.08.038
147. Danza K, Silvestris N, Simone G, Signorile M, Saragoni L, Brunetti O, et al. Role of miR-27a, miR-181a and miR-20b in gastric cancer hypoxia-induced chemoresistance. Cancer Biol Ther. (2016) 17:400–6. doi: 10.1080/15384047.2016.1139244
148. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin β 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. (2003) 22:8031–41. doi: 10.1038/sj.onc.1206928
149. Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: a long non-coding RNA highly associated with human cancers. Oncol Lett. (2018) 16:19–26. doi: 10.3892/ol.2018.8613
150. Wang H, Li W, Guo R, Sun J, Cui J, Wang G, et al. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer. (2014) 135:2783–94. doi: 10.1002/ijc.28922
151. Hirai H, Yokota A, Tamura A, Sato A, Maekawa T. Non-steady-state hematopoiesis regulated by the C/EBP β transcription factor. Cancer Sci. (2015) 106:797–802. doi: 10.1111/cas.12690
152. Satake S, Hirai H, Hayashi Y, Shime N, Tamura A, Yao H, et al. C/EBPβ is involved in the amplification of early granulocyte precursors during candidemia-induced “emergency” granulopoiesis. J Immunol. (2012) 189:4546–55. doi: 10.4049/jimmunol.1103007
153. Mignot G, Chalmin F, Ladoire S, Rébé C, Ghiringhelli F, Xiang X, et al. Tumor exosome-mediated MDSC activation. Am J Pathol. (2011) 178:1403. doi: 10.1016/j.ajpath.2010.11.078
154. Rashid MH, Borin TF, Ara R, Piranlioglu R, Achyut BR, Korkaya H, et al. The critical immunosuppressive effect of MDSC-derived exosomes in the tumor microenvironment. bioRxiv. (2020). doi: 10.1101/2020.03.05.979195
155. Wang Y, Tian J, Tang X, Rui K, Tian X, Ma J, et al. Exosomes released by granulocytic myeloid-derived suppressor cells attenuate DSS-induced colitis in mice. Oncotarget. (2016) 7:15356. doi: 10.18632/oncotarget.7324
Keywords: myeloid-derived suppressor cells, cancer, miRNA, tumor microenvironment, long non-coding RNAs, molecular mechanism
Citation: Safarzadeh E, Asadzadeh Z, Safaei S, Hatefi A, Derakhshani A, Giovannelli F, Brunetti O, Silvestris N and Baradaran B (2020) MicroRNAs and lncRNAs—A New Layer of Myeloid-Derived Suppressor Cells Regulation. Front. Immunol. 11:572323. doi: 10.3389/fimmu.2020.572323
Received: 01 July 2020; Accepted: 28 August 2020;
Published: 02 October 2020.
Edited by:
Fabrizio Mattei, National Institute of Health (ISS), ItalyReviewed by:
Justin Lathia, Case Western Reserve University, United StatesViktor Umansky, German Cancer Research Center (DKFZ), Germany
Copyright © 2020 Safarzadeh, Asadzadeh, Safaei, Hatefi, Derakhshani, Giovannelli, Brunetti, Silvestris and Baradaran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicola Silvestris, n.silvestris@oncologico.bari.it; Behzad Baradaran, baradaranb@tbzmed.ac.ir
†These authors have contributed equally to this work
 Elham Safarzadeh
Elham Safarzadeh Zahra Asadzadeh
Zahra Asadzadeh Sahar Safaei
Sahar Safaei Arash Hatefi
Arash Hatefi Afshin Derakhshani
Afshin Derakhshani Francesco Giovannelli
Francesco Giovannelli Oronzo Brunetti
Oronzo Brunetti Nicola Silvestris
Nicola Silvestris Behzad Baradaran
Behzad Baradaran