- 1Department of Evolutionary Ecology and Genetics, Zoological Institute, Christian-Albrechts-Universität zu Kiel, Kiel, Germany
- 2RD3 Marine Symbioses, GEOMAR Helmholtz Centre for Ocean Research, Kiel, Germany
Multicellular organisms live in close association with a plethora of microorganism, which have a profound effect on multiple host functions. As such, the microbiota and its host form an intimate functional entity, termed the metaorganism or holobiont. But how does the metaorganism communicate? Which receptors recognize microbial signals, mediate the effect of the microbiota on host physiology or regulate microbiota composition and homeostasis? In this review we provide an overview on the function of different receptor classes in animal host-microbiota communication. We put a special focus on invertebrate hosts, including both traditional invertebrate models such as Drosophila melanogaster and Caenorhabditis elegans and “non-model” invertebrates in microbiota research. Finally, we highlight the potential of invertebrate systems in studying mechanism of host-microbiota interactions.
Introduction
A constantly and rapidly growing body of evidence supports the critical impact of the microbiota on multiple host functions, as diverse as digestion, development, metabolism, immune defenses, and behavior. In fact, almost every host process seems to be affected by the microbiota. As a consequence, hosts and their microbiota form an intimate functional entity, termed the “metaorganism” (1) or “holobiont” (2, 3). Metaorganism research aims at moving from correlation to causality, i.e., to understand how the microbiota shapes organism health and how microbiota and host activities emerge into metaorganism functions that also impact broader communities and ecosystems (4, 5). Moreover, the microbiota has become the target for novel therapies seeking to enhance health (6, 7), productivity (8), or even favor acclimation to new environmental conditions [e.g., (9)]. However, understanding the underlying mechanisms of host-microbiota interactions is key to translate metaorganism research into effective therapies and managing strategies.
Host-microbiota interactions are based on the exchange of information, which from the host point of view comes down to microbial signal—host receptor interactions. One group of host receptors that seem to play a crucial role in host-microbiota communication are the pattern recognition receptors (PRRs) of the innate immune system, such as Toll-like receptors (TLRs), NOD-like receptors (NLRs), and C-type lectin receptors (CTLRs) (10–12). PRRs recognize microbial molecules that are essential for microbes but absent in eukaryotic organisms, such as the cell surface molecules lipopolysaccharide (LPS), peptidoglycan (PGN), or flagellin (13, 14). As these so-called microbial-associated molecular patterns are produced by pathogenic as well as commensal bacteria, it was repeatedly suggested that PRRs, in addition to their crucial role in regulating defense responses to pathogens, may have evolved to communicate with commensal microbes [e.g., (10)]. Indeed, PRRs like TLRs and NLRs have been shown to mediate the effect of the microbiota on the immune system of the host [reviewed in (15, 16)]. Two other groups of host receptors that function in recognition of microbiota-derived signals are the G-protein coupled receptors (GPCRs) and peptidoglycan recognition proteins (PGRPs). GPCRs recognize bacteria-derived molecules, such as signal peptides or short chain fatty acids (17, 18).
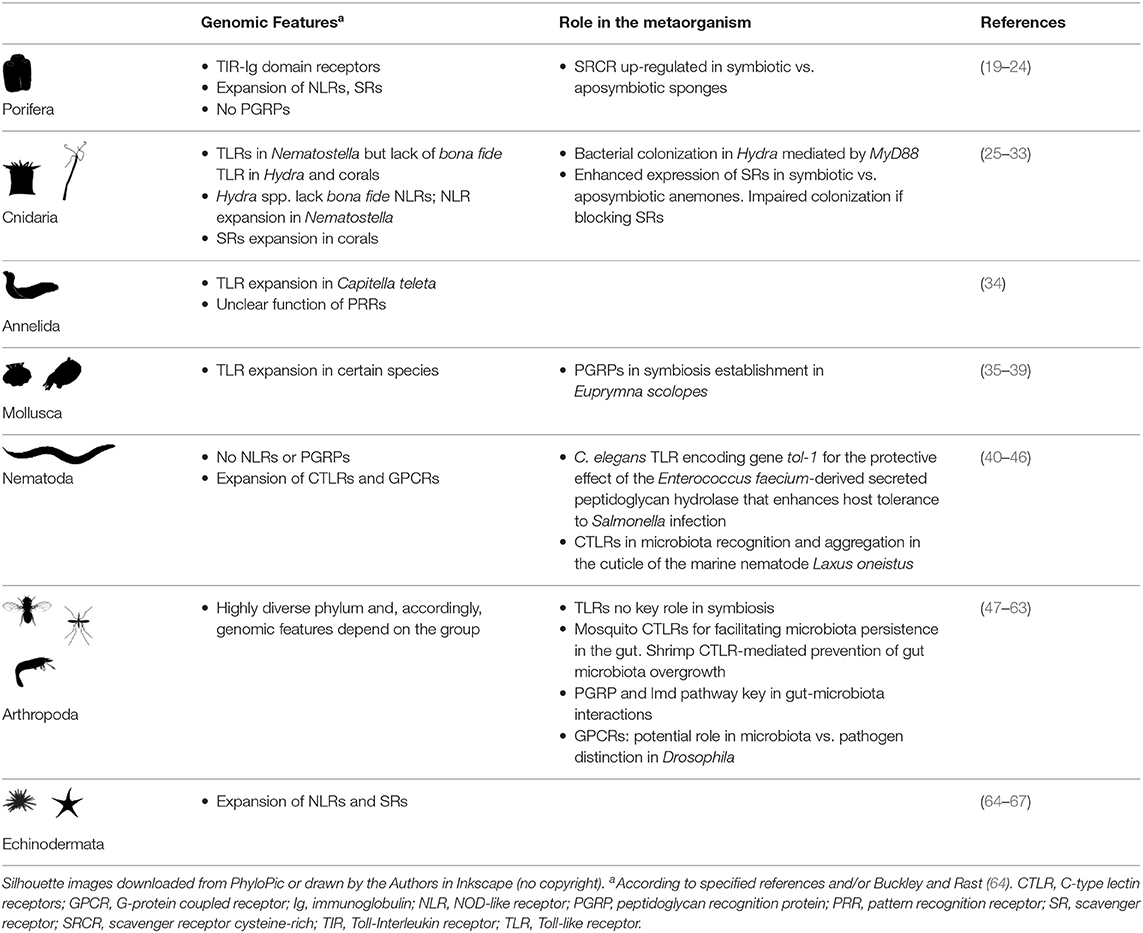
In this review we summarize what is known about invertebrate host-microbe communication, with a particular focus on the role of the above-mentioned receptor classes, for which evidence of an involvement in host-microbiota interactions is available. For each receptor class, we will first give a brief overview and present examples of their function in mediating microbiota-host interactions in humans and mice. We will then review experimental evidence for a role of these receptors in microbiota-host interactions in invertebrates (Table 1). We focus not only on classical model organisms like the fruit fly Drosophila melanogaster or the nematode Caenorhabditis elegans, but also summarize evidence from diverse other taxonomic groups. We specify if evidence comes from genomic data, differential gene expression analysis or functional analysis. Finally, we will discuss the potential of invertebrate systems for the study of host-microbiota communication.
Toll-like Receptors (TLRs)
TLRs are transmembrane receptors with several extracellular leucine-rich repeat (LRR) motifs and an intracellular Toll/interleukin-1 receptor (TIR) domain. The extracellular LRR motifs of TLRs can bind a wide range of microbe-derived signals (e.g., LPS, flagellin, PGN, lipoteichoic acid), but also endogenous ligands derived from damaged cells such as the extracellular matrix molecules fibronectin and biglycan [reviewed in (68)]. TLR stimulation ultimately leads to the nuclear translocation of the transcription factors NF-κB or c-Jun and subsequently to the production of inflammatory cytokines or antimicrobial peptides (AMPs). In addition to NF-κB signaling, TLR receptors can activate mitogen-activated protein kinase (MAPK) and interferon regulatory factor signaling cascades [reviewed in: (69, 70)]. Extensive reviews on the evolution of TLRs, their ligands and downstream signaling cascade can be found in Brennan and Gilmore (71) and Nie et al. (72).
Human and Mouse
The role of TLRs in microbial recognition is well-studied, particularly in the context of pathogens. But in 2004 the group of Ruslan Medzhitov proposed two distinct TLR functions—host defense against infection through recognition of pathogens and control of intestinal homeostasis through recognition of commensal bacteria: TLR-2-, TLR-4-, and MyD88-deficient mice showed increased susceptibility to intestinal injury induced by administration of dextran sulfate sodium (DSS) (73). Interestingly, germ-free mice were also highly susceptible to DSS-induced intestinal injury but could be rescued by administration of LPS. The authors concluded that the protection against intestinal injury occurred through recognition of commensal products by TLRs (73). This study was the first to describe a function of TLRs in the crosstalk between host innate immunity and the microbiota and thus in the control of intestinal homeostasis. Since then several studies have provided evidence of the involvement of TLRs in immunomodulation by the microbiota. For example, microbiota-derived polysaccharide A signals through TLR2 to suppress TH17 responses (74) and TLR 2 is required to sense outer membrane vesicle-associated polysaccharide from Bacteroides fragilis (75).
It thus seems that TLRs and also other PRRs are sentinels of microbial colonization by microbes in general, not only by pathogens. This makes sense as the microbial ligands of these receptors are not only produced by pathogens, but also commensal or beneficial bacteria. The challenge for the host is to detect microbial signals and interpret them in the appropriate context, preventing over-activation of defense responses and thus tolerating beneficial microbes while controlling overgrowth and responding appropriately to pathogens or opportunists. Vatanen et al. showed that the different members of the human gut microbiome present different LPS immunogenicity, as measured by TLR4 and NF-kB activation (76). D'Hennezel et al. showed that LPS of gut commensals of the order Bacteroidales silences TLR4 signaling and they proposed that this immunoinhibitory activity may sustain the tolerance of microbes in the gut (77). Decreased apical surface expression of TLRs and spatial segregation of host cells and commensal bacteria by mucus layers also prevent over-activation of TLR signaling (78, 79). The spatial segregation of microbiota and host epithelium depends on the regulatory feedback loop that senses bacterial colonizers via TLR-signaling and the activation of the expression of the antibacterial lectin RegIIIγ (80).
In addition, TLRs seem to play an important role in shaping and regulating the intestinal microbiota, as suggested by several studies analyzing the effect of TLR deficiency on microbial composition (81, 82). However, knock-out and wildtype mice used in these studies were separately maintained over multiple generations and a subsequent study comparing wild type and TLR-deficient littermate control mice, in which offspring resulted from crosses of mice heterozygous for each depletion identified no significant changes in the intestinal microbiota composition (83). The role of TLRs in regulating microbiota composition thus remains controversial.
Together, TLRs and TLR signaling play a crucial role in the recognition of both pathogenic and commensal bacteria and in the reciprocal interaction between the immune system and the intestinal microbiota in human and mice. It is unclear in how far TLRs directly mediate regulation of gut microbial composition, but it appears that balanced TLR signaling is most important to maintain intestinal homeostasis.
Invertebrates
The first Toll receptor was discovered in D. melanogaster. Drosophila Toll is expressed in many tissues in a complex spatial and temporal pattern (84) and was initially identified as essential in Drosophila early embryonic development (85). It was subsequently found that in adult flies Toll signaling mediates defense responses against bacterial and fungal pathogens by regulating, among others, the expression of the antifungal peptide drosomycin in the fat body (47, 86). Only one Toll homolog, termed TOL-1, was identified in C. elegans but the worm lacks central proteins of the canonical TLR-signaling cascade such as NF-kB (48). TOL-1 is expressed in neurons and not required for C. elegans resistance to a number of pathogens, but for development (48) and also for development of chemosensory neurons that function in microbe sensing (87). C. elegans tol-1 mutants are thus defective in pathogen avoidance behavior (87, 88). The first functional study of TLR signaling in a non-bilaterian animal demonstrated that recognition of bacteria and its contribution to defense is an ancestral function of TLRs (25). In the anthozoan Nematostella vectensis (phylum Cnidaria) a single TLR receptor could be identified (89) that is expressed in cnidocytes, stimulatedby flagellin in vitro and is involved in the recognition of the pathogen Vibrio coralliilyticus (26). In Hydra (phylum Cnidaria), however, conventional TLRs are absent; yet, they express a LRR domain protein that interacts with a TIR domain-containing protein and recognizes bacterial flagellin in vitro (90). In sponges (phylum Porifera), the function of TLR signaling is unknown. However, transcriptomic and genomic analyses of different sponge species from four different classes identified all essential genes involved in TLR signaling, but no conventional TLR (19–21, 91–93). Instead, sponges contain a receptor class with a TIR domain homolog of the TIR-domain of vertebrate TLRs, combined with extracellular immunoglobulin domains rather than LRR motifs. Components of the TLR pathway such as MyD88 were activated in response to microbial signals in some sponge species (94, 95). The role of these Poriferan receptors and the TLR pathway in bacterial recognition thus remains to be probed (21).
While the function of TLRs in pathogen recognition and immune defense has been first demonstrated in an invertebrate, the role of TLRs in the communication between the microbiota and invertebrate hosts is less clearly defined. Intriguingly, The Toll pathway does not play a role in regulating gut homeostasis in Drosophila. Instead, flies rely on the immune deficiency (Imd) pathway, which is activated by peptidoglycan recognition proteins (PGRPs, see section below) for host-microbiota crosstalk (96). There are only two studies reporting experimental evidence of the involvement of invertebrate TLRs in mediating host-microbiota interactions, one in Hydra and one in C. elegans. Analysis of MyD88-deficient Hydra polyps revealed that TLR-signaling affects microbiome resilience after antibiotic disturbance (25). The only C. elegans TLR encoding gene tol-1 was found to be required for the protective effect of the Enterococcus faecium-derived secreted peptidoglycan hydrolase SagA that enhances C. elegans tolerance to Salmonella infection (40). SagA remodels the peptidoglycan and generates muramyl-peptide fragments, which protect wildtype, but not tol-1 mutant worms from Salmonella pathogenesis (40). However, it remains unclear how exactly tol-1 is involved in SagA-mediated protection.
In summary, TLR signaling in invertebrates is functionally studied only in few model systems, yet recognition of bacteria seems to be an ancestral function of TLR signaling. Several studies in cnidarians, mollusks, and annelids have shown that bacterial recognition modulates the expression of genes encoding TLR pathway components [reviewed in (71)]. Further studies are needed to explore the potential function of TLR signaling in invertebrate host-microbe communication.
NOD-like Receptors (NLRs)
Also known as nucleotide binding oligomerization domain (NOD)-like receptors, the standard nomenclature for the family has been designated “nucleotide-binding domain and leucine-rich repeat containing” receptors (NLRs), in order to emphasize the presence of these two conserved domains in bona fide NLRs (97): a nucleotide-binding domain denoted NACHT and a C-terminal LRR domain. The architecture of metazoan NLRs usually includes a third N-terminal domain, mainly a Death domain, a CARD domain or a Pyrin domain [reviewed in (97)]. NLRs exemplify how investigating different animal groups broadens our knowledge on the evolution of the immune system. Because D. melanogaster and C. elegans lack NLRs, it was long thought this family had its origin in teleost fish. The availability of genomes and transcriptomes from non-model invertebrates, such as the publication of the sea urchin Strongylocentrotus purpuratus genome (98) and later the sponge Amphimedon queenslandica genome (20), revealed the ancient origin of animal NLRs (22). The evolutionary trajectory of animal NLRs is complex and diverse, with remarkable losses and expansions. Expansions of NLRs occurred in the sponge A. queenslandica (135 genes), the sea urchin S. purpuratus (203 genes) (22), the coral Acropora digitifera (66 genes, (99)), the lancelet Branchiostoma floridae [92 genes, as calculated by (64)], and in the zebra fish Danio rerio [>250 genes, (100)]. In contrast, the cnidarian Hydra magnipapillata, the ctenophore Mnemiopsis sp., the nematode C. elegans, the urochordate Okopleura dioica, and various arthropods lack bona fide NLRs (22). Some of these animals, such as Hydra, harbor instead a diverse repertoire of proteins containing a nucleotide binding domain that could potentially have a similar role as bona fide NLRs (27).
Human and Mouse
Most of our knowledge about NLR signaling pathways and function in animal-microbe interactions comes from research on mice and human cell lines [reviewed in (101)]. NLRs recognize microbes through the C-terminal LRR domain by direct binding a diverse range of ligands that include LPS, PGN, and small bacterial peptides [e.g., (102, 103)]. Microbial recognition can also happened indirectly; for example, in HEK293 cells, the exposure to Salmonella-derived proteins activated the small Rho-GTPases such as RAC1 and the active GTP-bound state induced NOD1-dependent signaling (104). The N-terminal domain engages in protein-protein interactions and triggers downstream signaling cascades. Mammalian NLRs, such as NOD1 and NOD2, mainly activate NF-κB, but they can also induce MAPK signaling cascades (e.g. JNKs, ERKs, p38) [reviewed in: (101, 105)]. Other NLRs form multimeric complexes known as “inflammasomes” that consists of one or several NLRs, an adapter protein containing a CARD domain, and a caspase as effector [reviewed in (106)]. NLR activation and subsequent signaling result in regulation of reactive oxygen species formation, secretion of cytokines, production of AMPs, as well as apoptosis [reviewed in: (105, 107)].
NLRs have received great attention in the context of human-gut microbiota interaction because mutations in NLR genes were repeatedly associated with chronic inflammatory diseases (e.g., Nod2 mutation as a risk factor for Crohn's disease) [reviewed in (101, 108)]. Impairment of Nod2 and Nlrp 6-inflammasome genes in a mouse model correlated with dysbiosis, suggesting that these receptors could regulate gut microbiota composition [e.g., (109, 110)]. However, other studies could not detect differences in microbiota composition between Nod1, Nod2, or Nlrp6 deficient mice and wild type mice, when controlling for mouse breeding and housing effects [e.g., (111–113)].
In a different approach, Schieber et al. (114) showed that the commensal gut bacterium E. coli O21:H+ act via the Nlrc4 inflammasome to promote disease tolerance as defense during infection and tissue inflammation. Upon infection by Burkholderia thailandesis (a model for pneumonic infection which provokes wasting of skeletal muscle but does not compromise intestinal barrier), the commensal E. coli O21:H+ translocates into the white adipose tissue and activates the Nlrc4 inflammasome to induce sustained insulin-like growth factor 1 signaling in the skeletal muscle, resulting in prevention of muscle loss (114). Thus, the location of the microbial signal is also relevant for determining host response. Recently, Kim et al. (115) showed that the probiotic effect of E. faecium against Clostridium difficile pathogenesis in mice depends on activation of NOD2. NOD2 is activated by small muropeptides that are generated by the E. faecium-derived peptidoglycan hydrolase SagA. These studies provide evidence of key interactions between NLRs and commensal bacteria and the consequences for host health.
Thus, mouse NLRs detect microbial signals derived from commensal and pathogenic bacteria, but the direct link between these NLRs and the gut microbiota composition remains highly debated and needs to be resolved by future studies.
Invertebrates
To what extent do the NLR functions described in mammals apply to invertebrates? The expansion of the NLR family and genetic diversity of the recognition domain (i.e., high polymorphism of C-terminal LRRs) is considered an indication for specific recognition of diverse microbial ligands (116). However, only a handful of studies provide evidence of a potential role of NLRs in invertebrate host-microbiota interactions, and all are based on gene expression analysis. Sponges expressed a high diversity of NLRs and, recently, the enhanced expression of NLRs in response to PGN and LPS was reported (21). In the medicinal leech Hirudo medicinalis (phylum Annelida), a LRR-domain containing protein with sequence similarity to vertebrate NLRs was identified and its expression in nerve chords was enhanced upon exposure to heat killed Micrococcus nishinomiyaensis, as well as to E. coli LPS, lipoteichoic acid, and muramyl dipeptide (34). Finally, a non-conventional Hydra NLR protein, mainly expressed in the endoderm of this animal, responded to LPS and flagellin stimulation and recruited an effector caspase in a heterologous expression system (27). Thus, the expansion of NLRs in certain groups and these first results suggest that invertebrate NLRs are good candidates to mediate host-microbiota crosstalk.
C-type Lectin Receptors (CTLRs)
The C-type lectin-like domain family contains secreted as well as transmembrane proteins that are highly diverse regarding their overall domain architecture, but all share primary and secondary structural homology in their carbohydrate recognition domain [reviewed in: (117, 118)]. The first described members of this family indeed bound carbohydrates in a calcium-dependent (C-type) manner, and were thus bona fide lectins. However, the carbohydrate recognition domain was subsequently also identified in proteins that did not bind carbohydrates, but other ligands such as proteins and lipids, and also did not require calcium for binding. The term C-type lectin-like domain (CTLD) was thus introduced to reflect the structural similarity to the carbohydrate recognition domain of prototype C-type lectins without implying common function (119). CTLD genes are abundant in metazoan genomes and constitute highly diverse and expanded gene families. The human genome encodes 100 CTLD genes, 132 CTLD genes are encoded in the mice genome, 56 CTLD genes in D. melanogaster, 283 CTLD genes in C. elegans, 67 CTLD genes in Nematostella vectensis, and 2 CTLD genes in the sponge A. queenslandica [reviewed in (49)].
Human and Mouse
CTLD proteins perform multiple functions in human and mouse immune defense and are commonly called C-type lectin receptors (CTLRs). CTLRs function as PRRs that bind glycans, such as mannose, fucose, and N-acetylgalactosamine residues on the surface of pathogens, including bacteria, fungi, viruses, and parasites, and regulate innate and adaptive immune responses. Other CTLRs bind endogenous ligands (self-antigens) and thus play an important role in immune homeostasis [for a review of CTLR functions in the human/mouse immune system see (120, 121)]. The high diversity of glycosylated bacterial proteins or lipids attached to the cell surface or to secreted molecules constitute ideal ligands to establish specific interactions with the host (122).
CTLRs are crucial for the recognition of bacterial glycoconjugates in the context of pathogen infection and there is some evidence for CTLR ligands from commensal bacteria. For example, the beneficial human gut bacterium Lactobacillus acidophilus produces the glycosylated surface layer A protein, which modulates human dendritic cell and T-cell responses via the interaction with the CTLR dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) (123). Similarly, Lactobacillus rhamnosus produces a glycosylated adhesive heterotrimeric pili, which interacts with DC-SIGN resulting in modulation of the cytokine response of human dendritic cells (124). These two Lactobacillus glycoproteins are the as yet only identified CTLR ligands from commensal bacteria. Furthermore, two studies provide evidence of an interaction between CTLRs and commensal bacteria and its impact on gut homeostasis in experimental murine colitis models. The closest murine homolog of DC-SIGN, the specific intracellular adhesion molecule-3 grabbing non-integrin homolog-related 3 (SIGNR3), interacted with L. acidophilus surface layer protein A contributing to the maintenance of healthy gastrointestinal microbiota, protection of the gut mucosal barrier function, and mitigation of colitis (125). Hütter and colleagues showed that the two CTLRs, macrophage C-type lectin (MCL) and dendritic cell immunoreceptor (DCIR), bind commensal intestinal bacteria in vitro (126). However, MCL−/− and DCIR−/− knock-out mice showed only a slight increase in inflammation in a DSS murine colitis model and the role of MCL and DCIR in regulating gut homeostasis thus remains unclear. Interestingly, recognition of commensal fungi by CTLRs also seems to support intestinal immune homeostasis. Dectin-1, a major PRR in antifungal immunity, and SIGNR3 regulated the host response to commensal fungi and influenced immune homeostasis in a DSS murine colitis model (127, 128). Moreover, a polymorphism of the Dectin-1/CLEC7A gene in humans was associated with severe ulcerative colitis (127). Together, these studies demonstrate an emerging role of vertebrate CTLD proteins in microbiota-host interactions, including not only bacteria but also fungi.
Invertebrates
In invertebrates, CTLD genes contribute to immune responses in a variety of different taxa, including insects, crustaceans, and nematodes [reviewed in (49)]. Insect and crustacean CTLD proteins are involved in cellular immune responses, such as hemocyte nodule formation, encapsulation, melanization, and activation of phagocytosis, and in the direct elimination of pathogens by exhibiting antimicrobial activity [reviewed in (49)]. In C. elegans, CTLD genes are mainly expressed in the intestine and can mediate both physiological, as well as behavioral immune responses (41). Invertebrate CTLD proteins were thus suggested to function as PRRs. We however know almost nothing about the downstream signaling pathways that are activated by invertebrate CTLD proteins. The one exception is a study on the CTLD proteins FcLec4 from the kuruma shrimp Marsupenaeus japonicas, which binds β-integrin in the membrane of hemocytes to promote phagocytosis (50).
Three studies linked CTLD protein function to host colonization by microbiota bacteria in invertebrates (Figure 1). In the mosquito Aedes aegypti the CTLD proteins mosGCTL-29 and mosGCTL-32 seem to facilitate colonization and persistence of microbiota bacteria in the gut: mosGCTL-29 and mosGCTL-32 bind E. coli cells and protect E. coli and microbiota bacteria against AMP activity by coating the bacterial surface. Silencing the expression of mosGCTL-29 and mosGCTL-32 by RNAi or blocking the CTLD proteins by feeding mosquitos mosGCTL antisera, reduced bacterial colonization (51). In contrast, the CTLD protein MjHeCL from the kuruma shrimp M. japonicas controls the hemolymph microbiota by inhibiting bacterial proliferation (52). Silencing the expression of MjHeCL by RNAi increased proliferation of the hemolymph microbiota. MjHeCL was shown to bind to several hemolymph microbiota isolates and to be required for expression of certain AMPs in M. japonicas hemocytes. It was thus suggested that MjHeCL plays a role in restricting the growth of the hemolymph microbiota by regulating AMP expression (52). Finally, a CTLD protein mediates symbiont attachment in the marine nematode Laxus oneistus. The L. oneistus cuticle is covered by a monolayer of a single phylotype of sulfur-oxidizing bacteria. The mannose-specific CTLD protein Mermaid, which is similar to human DC-SIGN, is secreted onto the cuticle and mediates symbiont aggregation and attachment to the worm (42). Moreover, different Mermaid isoforms serve to discriminate different bacterial symbionts and were thus suggested to be involved in the specific recruitment of symbionts (43).
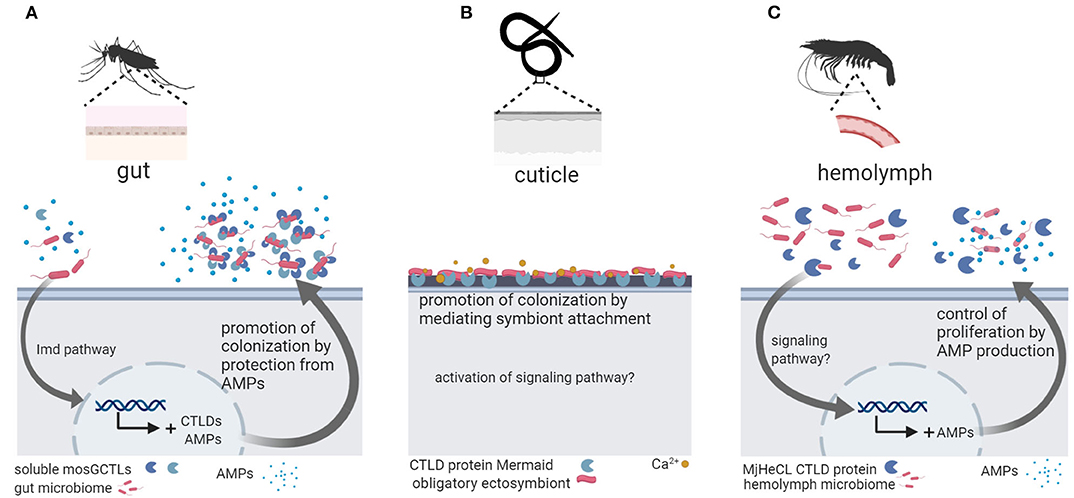
Figure 1. Invertebrate C-type lectin-like domain (CTLD) proteins function in host colonization by the microbiota. (A) In the mosquito Aedes aegypti, gut microbiota bacteria are coated by soluble mosCTLs and, in this way, protected from antimicrobial peptides (AMPs) secreted by the host, promoting colonization and persistence in the gut (51). (B) In the marine nematode Laxus oneistus, the calcium-dependent CTLD protein Mermaid mediates the agglutination and attachment of the obligatory ectosymbiont, a sulfur-oxidizing bacterium that forms a monolayer in the cuticle. If symbiont attachment via Mermaid leads to activation of intracellular signaling remains unknown. (C) In the kuruma shrimp Marsupenaeus japonicas, the CTLD protein MjHeCL binds microbiota bacteria in the hemolymph and activates AMP production to prevent bacterial overgrowth. Figure created with Biorender.com. Silhouette image for L. oneistus was downloaded from PhyloPic (no copyright).
Taken together, future investigations in other animal groups will help understanding the role of invertebrate CTLD proteins in host-microbiota interactions.
Peptidoglycan Recognition Proteins (PGRPs)
PGRPs (also known as PGLYRPs in mammals) are a family of homologous receptors characterized by the presence of at least one PGRP domain with structure and sequence similarity to bacteriophage type 2 amidases [reviewed in (53)]. PGRPs are secreted, transmembrane, or intracellular proteins that bind and often also hydrolyze PGN. However, certain PGRPs can also bind other molecules such as LPS and lipoteichoic acid [reviewed in (53)]. PGRPs are present in many animals groups, from insects to mammals; yet absent from the genomes of representatives for other animal phyla such as the sponge A. queenslandica, the cnidarian N. vectensis, the nematode C. elegans, or the crustacean Daphnia pulex [reviewed in (64)]. In vertebrates, the suite of PGRPs is rather small (e.g., 4 PGRPs in mammals and in zebrafish). In zebrafish, PGRPs are expressed in skin, gills, liver, intestine, and pancreas (129) and have been implicated in immune defenses [reviewed in (53)]. In mammals, all 4 PGRPs (PGLYRP1-PGLYRP4) recognize PGN [reviewed in (53)], but may also recognize other ligands, including LPS [reviewed in (130)].
Human and Mouse
Mammalian PGRPs have antibacterial activity and assist in macrophage activation during immune responses [reviewed in (130)]. They are expressed in different tissues, including the gastrointestinal tract [reviewed in (131)]. Saha et al. (132) investigated the role of PGRPs in the gut by analyzing DSS-induced colitis in mice that were deficient for individual pglyrp genes. All PGRP deficient mice were more susceptible to colitis and showed significant changes in gut microbiota composition (as bacterial abundances quantified by qPCR). Interestingly, deficiency in each pglyrp induced different changes, suggesting specific functions of these genes. When transferred into germ-free mice, stools from pglyrp-deficient mice produced higher inflammatory activity and increased sensitivity to colitis in the receiving mice than stools from wild type mice. Therefore, PGRPs might play a protective role in the gut by preventing inflammation upon damage, possibly, by directly regulating gut microbiota composition.
Invertebrates
In insects, PGRPs play a key role in pathogen defense [reviewed in (53)]. In the sea star Asterias rubens, two secreted PGRPs have been characterized, PGRP-S1a, present in the coelomic plasma, and PGRP-S2a, expressed in phagocytes. PGRP-S2a opsonized Micrococcus luteus, increasing their phagocytosis (65). Importantly, accumulating experimental evidence supports a crucial role of PGRPs in fly and mosquito host-microbiota crosstalk and in the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri (Figure 2).
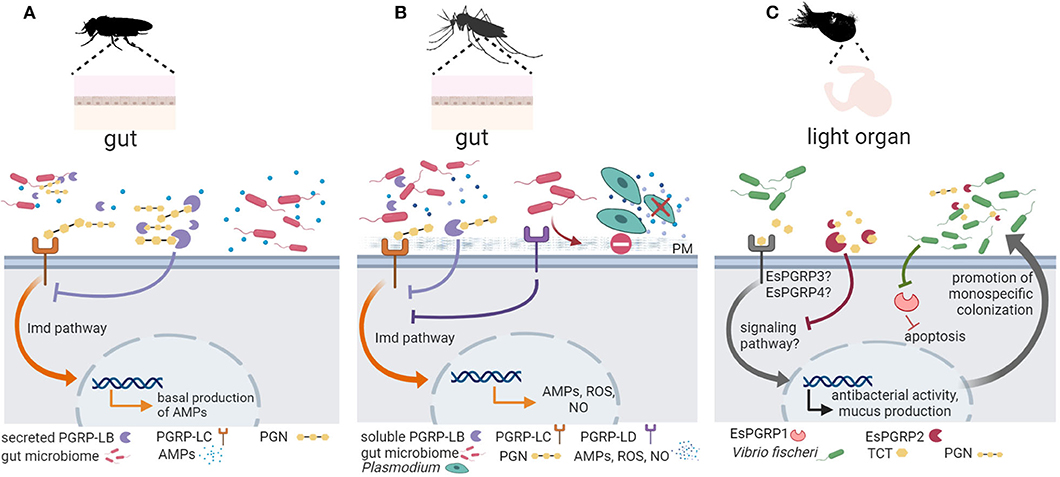
Figure 2. Invertebrate PGRP function in symbiont colonization and symbiosis homeostasis. (A) In the fruit fly Drosophila melanogaster, peptidoglycan recognition proteins (PGRPs) are important for keeping the balance between activating an immune response to pathogenic bacteria and preserving the beneficial microbiota [reviewed in (57)]. Peptidoglycan (PGN) derived from gut microbiota activates basal antimicrobial peptide (AMP) production via the PGRP receptor PGRP-LC (shown in orange) and the immune deficiency (Imd) signaling pathway. Secreted PGRP-LB (shown in purple) hydrolyzes excessive PGN downregulating antimicrobial defenses to promote host tolerance toward the microbiota (58). (B) In mosquitos, PGRPs mediate gut homeostasis and symbiosis. Similar to its function in D. melanogaster, PGRP-LC (shown in orange) detects microbiota derived PGN and activates the Imd pathway to promote immune effectors (AMPs, reactive oxygen species (ROS), nitric oxide (NO), whereas a soluble PGRP (PGRP-LB) (shown in purple) hydrolyzes excessive PGN to promote tolerance [see (53) and references therein]. In addition, the receptor PGRP-LD (shown in dark purple) also protects the gut microbiota by dampening immune activities, and this action is key to maintaining the integrity of the peritrophic matrix (PM), which acts as barrier against parasitic infection by Plasmodium (61). (C) In the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri, the symbiont PGN-derived tracheal cytotoxin (TCT) activates mucus production to promote colonization. As symbiont colonization progresses, EsPGRP1 is silenced to induce apoptosis and rearrangement of the host light organ. The amidase activity of EsPGRP2 detoxifies TCT to promote symbiont tolerance. Figure created with Biorender.com. Silhouette image for Drosophila was downloaded from PhyloPic (no copyright).
In the fly, recognition of PGN through PGRPs activates the Imd pathway, which is in addition to the Toll pathway the major AMP-regulating signaling pathway in Drosophila (54, 55). While the Toll pathway only regulates AMP expression in the body cavity, the Imd pathway controls AMP expression in the body cavity and in the gut. The expression of AMPs in the gut is strongly reduced in axenic flies, which indicates that the gut microbiota activates the Imd pathway, leading to a basal expression of AMPs in the presence of commensal bacteria (56, 133). D. melanogaster has 13 PGPR genes that encode more than 20 transmembrane, cytoplasmic or soluble proteins, which act in different immune tissues such as the fat body, hemocytes, and barrier epithelia in the gut and trachea (53). While some PGRPs function as recognition proteins and activate antimicrobial responses, other PGRPs function as inhibitory receptors or PGN-hydrolyzing amidases that downregulate antimicrobial defenses. Drosophila PGRPs thus seem to play an important role in keeping the balance between activating an immune response to pathogenic bacteria and preserving the beneficial microbiota [reviewed in (57)]. For example, secreted and cytosolic isoforms of the PGN-cleaving amidase PGRP-LB control the level of extracellular PGN in the gut lumen and of intracellular PGN inside enterocytes, respectively, to prevent constitutive activation of NF-κB in response to microbiota bacteria (58). Moreover, the extracellular receptor PGRP-SD, which is important for the detection of bacterial pathogens in Drosophila (59), also affect microbiota composition: The intestinal microbiota of PRGP-SD knock-out flies showed an increased abundance of Lactobacillus plantarum. The excessive proliferation of L. plantarum derived in excessive levels of metabolite lactic acid and resulted in the generation of reactive oxygen species, which in turn promoted intestinal damage and increased proliferation of intestinal stem cells, and dysplasia (60).
In mosquitos, there is strong support for the role of PGRPs in regulating gut homeostasis and promoting symbiosis. Song et al. (61) showed that knockdown of Aedes stephensi PGRP-LD activated immune responses and changed the abundance and spatial distribution of the gut microbiota. Reduction of the microbiota in PGRP-LD knock down mosquitoes and in antibiotic-treated mosquitos compromised the peritrophic matrix, which represents the physical barrier of mosquito midgut epithelium and its luminal contents (61). Thus, PGRP-LD dampens immune responses to protect gut microbiota, which in turn maintains the structural integrity of the peritrophic matrix. In the mosquito Aedes aegypti, symbiosis with Wolbachia activates PGRP-LE, which is a receptor of the Imd pathway (134). In independent experiments using genetic tools to silence PGRP-LE or components and regulators of Imd and Toll pathways, the authors provide strong evidence of the role of PGRP-LE and downstream signaling in controlling symbiont load in A. aegypti (134).
In the Hawaiian bobtail squid Euprymna scolopes, recognition of LPS, PGN, and PGN derivate tracheal cytotoxin (TCT) is essential for the highly specific colonization of the E. scolopes light organ by Vibrio fischeri [reviewed in (14)]. PGN triggers mucus production, which facilitates V. fischeri aggregation, and later LPS and TCT induce apoptosis and epithelium regression for the formation of the light organ. Ligand assays are missing, but PGRP expression profiles agree with their role in mediating the establishment of the symbiosis in the squid juvenile. For example, the E. scolopes PGRP EsPGRP1 is expressed in the nucleus of epithelial cells but TCT alters its localization during the inducement of light organ morphogenesis (35). This effect disappears when the host encounters a V. fischeri mutant that is defective in the release of TCT (35). Another E. scolopes PGRP, EsPGRP2, degrades TCT via amidase activity, reducing its toxicity (36) and, thus, authors suggests that EsPGRP2 may help dampen the response to this powerful toxin during periods when symbiont concentrations are high and promote tolerance (36). EsPGRP5 is also predicted to present amidase activity and is highly expressed in the hemocytes (37), which are E. scolopes immune cells. EsPGRP5 gene expression is altered in hemocytes from symbiotic vs. aposymbiotic hosts (37).
To summarize, PGRPs play a vital role in maintaining intestinal homeostasis in insects and in establishing the squid-Vibrio symbiosis, two distantly-related animal groups. The function of PGRPs in host-microbiota interactions in other invertebrates remains to be investigated.
Scavenger Receptors (SRs)
SRs were first characterized based on their capacity to bind altered low-density lipoproteins (135). Since then, additional ligands have been identified, such as LPS, β-glucan, maleylated bovine serum albumin, and viruses [reviewed in (136)]. Despite similar ligand affinities, SRs comprise both secreted and transmembrane proteins that are structurally very heterogeneous. The high complexity of protein domain architectures reveals the lack of (or little) homology within this superfamily [reviewed in (136)]. Some of the domains reported in SRs include: the scavenger receptor cysteine-rich (SRCR) domains (class A and I), collagen (class A), CD36 (class B), or the epidermal growth factor (EGF) and EGF-like domains (class F and H). The exact binding capacity and downstream signaling vary between different SRs.
Human and Mouse
Our understanding of the functions and signaling mediated by SRs relies mainly on results from mammals, where they play a role in immunity (e.g., inducing bacterial clearance) but also in homeostasis by regulating lipid transport [reviewed in (136)]. However, they have been little studied in the context of host-microbiota crosstalk. CD36 domain-containing SRs are found in all animals, from sponges to humans (28, 137, 138). They conform the SR-B class and probably the most studied one is the vertebrate receptors CD36 [reviewed in (136)]. CD36 binds diacyl fatty acids of microbial cells, acting as a PRR (139, 140). Moreover, CD36 mediates the recognition of apoptotic cells via the detection of modified lipids and thrombospondin-1 and is also involved in lipid transport (141). The signaling pathways activated by CD36 vary depending on the ligand. For example, binding thrompospondin-1 yields actin rearrangement and pro-apoptotic signals through MAPKs such as p38 or JNK [reviewed in (136)], whereas bacteria (139) and endogenous modified lipoproteins (142) induce inflammatory responses. In mice, CD36 function in host-microbiota crosstalk is mainly indirect and relies on its role in lipid metabolism, as the microbiota influences host lipid content and these changes affect CD36 gene expression levels [e.g., (143, 144)].
Invertebrates
In invertebrates, CD36 domain-containing SRs seem to be involved in the response to bacterial challenge in diverse animal groups. For example, the SR-B family member MjSR-B1 in the kuruma shrimp Marsupenaue japonicus is mainly expressed in hemocytes, hepatopancreas, and heart (62). MjSR-B1 knockdown impairs bacteria agglutination, phagocytosis and expression of antimicrobial peptides upon exposure to the pathogens Vibrio anguillarum and Staphyloccoccus aureus, whereas overexpression of MjSR-B1 has the opposite effect (62). In the octopus Octopus ocellatus, OoSR-B localizes to the surface of hemocytes and stimulates phagocytosis upon exposure to V. anguillarum and to Micrococcus luteus (38). Knockdown of OoSR-B impaired the transcription of TLRs and downstream components of the cascade (i.e., MyD88 and TRAF6), suggesting that the interaction of OoSR-B and TLRs is key in the response to bacteria (38). A CD36-like gene is strongly upregulated in symbiotic vs. aposymbiotic (i.e., dinoflagellate-free) sea anemones Aiptasia sp. and Anthopleura elegantissima (phylum Cnidaria) (29, 30); however, exact role cnidarian CD36 domain-containing SRs play in cnidarian-dinoflagellate symbiosis is unknown.
In invertebrates, the group of SRCRs has received special attention due to their high diversity in the genome of several animal groups. In contrast to mammals (145), invertebrate SRCR protein architectures are highly diverse: SRCR domains are reported as single domains, arranged in tandem and/or in combination with other conserved domains such as immunoglobulin domains, collagen, low density lipoprotein receptor, and fibronectin type III [e.g., (146–148)]. In terms of total number of SRCR domain and SRCR genes, some invertebrates showed similar or even lower diversification than mammals (e.g., 3 SRCR domains in C. elegans, 22 SRCR domains in Ciona intestinalis) (64). However, sea urchins, sponges and in less order of magnitude, lancelets (amphioxus) showed significant expansions of the SRCR gene family, with hundreds of genes reported in their genomes (64).
A handful of studies have explored the function of invertebrate SRCRs in immunity, mainly through gene expression analyses. In sponges, one SRCR gene was up-regulated in the Mediterranean species Aplysina aerophoba in response to LPS and PGN (21). Also, a diverse array of SRCRs is activated in juveniles of the sponge Amphimedon queenslandica when exposed to their own microbiota as well as to seawater bacteria (95). Interestingly, the expression of SRCR-containing genes is enriched in the choanocytes of adult A. queenslandica, which are phagocytic cells that are in direct contact with the external environment (149). In adults of the purple sea urchin S. purpuratus SRCRs are specifically expressed in coelomocytes, the sea urchin immune cells (150). The expression profiles were individual specific and fluctuated over time (e.g., their expression levels can fluctuate up to 10-fold in 1 week) (147). In S. purpuratus larvae, the SRCR gene srcr142 was expressed in pigment cells, phagocytic cells and also in the amoeboid cells, which are the specialized immune cells in the larvae (66). Moreover, srcr142 expression was enhanced upon exposure of larvae to the seawater bacterium Vibrio diazotrophicus (66). In the sea star Asterina pectinifera, a secreted SRCR protein (ApSRCR1) is highly expressed in coelomocytes and mediates bacterial binding and clearance (67). In several coral species, the dmbt1 gene, annotated based on protein sequence similarity, is differentially-expressed in response to LPS stimulation (31), during disease (151), and upon exposure to Vibrio bacterium (Vibrio owensii and V. diazotrophicus) (32). The scallop SRCR-containing gene CfSR was up-regulated upon stimulation with different microbial-associated molecular patterns (i.e., LPS, PGN, ß-glucan) and the protein was expressed at the surface of hemocytes (39). In this case, binding assays on the purified Cf SR protein showed its affinity for acetylated low density lipoprotein, dextran sulfate, LPS, PGN, zymosan, and mannan (39), which confirmed its function as PRR.
An additional study points to the potential role of SRCRs in regulating host colonization by the microbiota in sponges. Steindler et al. (23) identified a SRCR-containing gene as potential mediator in the establishment of the intracellular cyanobacterial symbiont in the Mediterranean sponge Petrosia ficiformes. Specimens of P. ficiformis become naturally aposymbiotic (loss of cyanobacteria) when growing in dark caves. In comparison to aposymbiotic specimens, symbiotic sponges (harboring cyanobacteria) living at a short distance in illuminated areas showed elevated expression of a gene containing a SRCR domain (23). The alignment of the predicted protein revealed similarity with the SRCR-containing protein derived from other sponge species, Geodia cydonium, as well as the sea urchin S. purpuratus and the zebrafish Danio rerio. However, further studies are necessary to unequivocally confirm the role of SRCR in symbiosis establishment in sponges.
Interestingly, Neubauer et al. (28) impaired the ability of the sea anemone Aiptasia pallida to acquire the endosymbiotic dinoflagellate Symbiodium by pre-incubating aposymbiotic anemones with the SR ligand fucoidan, which blocks the binding sites of SRs. Although the class of the blocked SR(s) was not identified, the experiment supports the role of SRs in the establishment of anemone-dinoflagellate symbiosis (28).
In summary, the great diversity of scavenger receptors and their role in invertebrate response to microorganisms, with preliminary evidence of a role in symbiosis establishment, makes this PRR group an interesting target for future studies on host-microbiota crosstalk.
G Protein-coupled Receptors (GPCRs)
GPCRs are central for the perception of external stimuli and the transduction of the signal to the cellular cytoplasm and thus vital for the connection of the cell and organism to its environment. The family of GPCRs represents the largest receptor family in animals (152). The human genome contains over 800 GPCR-encoding genes, the mice genome comprises over 1,300 GPCRs and zebrafish genome includes over 700 GPCRs (153). In invertebrate genomes, 116 GPCRs were found in D. melanogaster (154), over 1,000 GPCRs in C. elegans (155), and 220 GPCRs in the sponge Amphimedon queenslandica (156). All GPCRs share a common structure and the general mechanism of activation [reviewed in (157)]: They are characterized by a conserved signature motif consisting of seven transmembrane (7TM) spanning helix domains. The extracellular N-terminal domain or the extracellular loops joining the 7TM domains mediate receptor activation through ligand binding, which causes conformational changes in the 7TM domain and in turn activates the cytoplasmic C-terminal domain. The activated C-terminal domain starts the intracellular signaling cascades typically through coupling to heterotrimeric guanine nucleotide-binding regulatory proteins (G proteins), but also through G protein-independent pathways mainly via G protein-coupled receptor kinases (GRKs) and arrestin [reviewed in: (158, 159)]. GPCR ligands are as diverse as photons, Ca2+, proteins, hormones, drugs, odorants, and small molecules such as amino acid residues, nucleotides, and peptides. GPCRs are thus involved in a multitude of different physiological functions, such as development, reproduction, metabolism, neuronal function, taste, smell, cell adhesion, immune function, and the regulation of feeding (160, 161).
Human and Mouse
In vertebrates, GPCRs play an important role in the crosstalk between microbes and the host. In vertebrate immunity GPCRs function next to TLRs and NLRs in the recognition of microbial-associated molecular patterns [reviewed in (162)]. Cells of the innate and adaptive immune system abundantly express GPCRs, which regulate the immune response and cell migration by sensing both endogenous signals (e.g., chemokines and other inflammatory factors such as activated complement) and bacterial-derived signals [e.g., formyl peptides or short chain carboxylic acids (17, 18)]. GPCRs bind products of bacterial fermentation, in particular the short-chain fatty acids (SCFAs), induce migration and activate innate immune responses in human and mouse leukocytes [reviewed in (163)]. Besides, these GPCRs mediate the activation of AMP production by bacterial SCFAs in intestinal epithelial cells (164). In addition to their function and expression on immune cells, certain GPCRs (e.g., GPCR 41 and 43) are also constantly expressed on intestinal epithelial cells, where they sense bacterial-derived metabolites. In recent years it became more and more clear that GPCRs represent a crucial point of contact between the vertebrate host and commensal bacteria in the gut and, consequently, act as key players in the maintenance of gut homeostasis. Their central role in the recognition of both host- and microbiota-derived metabolites was recently reviewed in Husted et al. (165) and the list of GPCR-metabolite interactions is constantly expanding [e.g., (166, 167)]. Interestingly, commensal bacteria produce GPCR ligands that mimic human signaling molecules: Human microbiome-encoded N-acyl amides are structurally similar to endogenous GPCR-active lipids and can activate GPCRs that regulate gastrointestinal tract physiology (168). Moreover, GPCRs on entero-endocrine cells sense microbial metabolites such as SCFAs and seem to mediate the effect of the microbiota on the secretion of hormones and thus on several aspects of host physiology, including behavior [reviewed in (169)].
Thus, in human and mouse, it is well-established that GPCRs detect microbial-derived signals and that the microbiota can affect host physiology through GPCRs.
Invertebrates
Evidence of the involvement of GPCRs in invertebrate immunity mainly comes from the two model organisms C. elegans and D. melanogaster [reviewed in (170)]. As an example, in C. elegans, the GPCR DCAR-1 was identified as an epidermal DAMP receptor. DCAR-1 responds to the endogenous ligand hydroxyphenyllactic acid, a tyrosine derivate that accumulates in the epidermis upon wounding or fungal infection (45). A Gα protein-PLCβ-DAG-PKC pathway, which then converts onto a p38 MAPK signaling cascades and a STAT-like transcription factor, acts downstream of DCAR-1 to activate the expression of AMPs (44, 45, 171, 172). Moreover, several C. elegans GPCRs and GPCR signaling were implicated in the regulation of pathogen avoidance behavior and the expression of intestinal immune defense genes [e.g., (46, 173–178)]. Two Pseudomonas aeruginosa secondary metabolites, phenazine-1-carboxamide and pyochelin, activate GPCR signaling in chemosensory neurons, which in turns promotes pathogen avoidance behavior (177). C. elegans GPCRs thus present a potential link between the nervous system and immune defenses, it is however as yet unclear if they directly respond to microbial-derived stimuli or to endogenous ligands.
In D. melanogaster, GPCRs play a potential role in the activation of dual oxidase (DUOX) enzymes that produce reactive oxygen species, which act as important antimicrobial effectors for the control of pathogens (179). Moreover, the community of commensal bacteria in the gut of DUOX knockdown flies is modified (180). However, DUOX-dependent gut immunity is mainly triggered by opportunistic pathogens, but not by commensal bacteria (63) and DUOX may thus play a role in the discrimination between symbionts and pathogens (181). Genetic and biochemical analyses revealed that DUOX activity and reactive oxygen species production is triggered by the recognition of pathogen-derived uracil (63), and require a Gαq-PLCβ-Ca2+ pathway (180). This indicates that DUOX is activated by an upstream GPCR, but the identification of this GPCR remains a challenge for the future.
In other animal models, GPCRs and GPCR signaling components were identified as potential mediators of host-microbe interactions by -omic approaches. For example, GPCR signaling might play a role in the establishment of cnidarian-dinoflagellate endosymbiosis: Transcriptomic and metabolomic analyses of the sea anemone Aiptasia pallida response to different endosymbiont Symbiodinium species revealed GPCR signaling as one of the major differentially enriched processes (182). In addition, proteomic analysis identified that GPCRs are present in the membranes of symbiosomes, the intracellular vacuoles containing microalgae in a cnidaria-dinoflagellate endosymbiosis (183). Together, studies on different animal models suggest that GPCRs are potential mediators of host-microbe interactions also in invertebrates.
Perspectives
The metaorganism can only function as entity when host and microbiota efficiently communicate with each other. Here, we presented our current understanding on the role of PRRs and GPCRs in maintaining host-microbiota interactions within the metaorganism throughout different animal groups (Table 1). Accumulating evidence suggests that TLRs, NLRs, CTLDs, and GPCRs are crucial mediators in host-microbiota communication in mammals. However, it remains unclear if this function of these PRRs is conserved throughout the animal kingdom, as is their function in pathogen recognition. Research on PGRPs in diverse animal groups provides strong support of their conserved role in mediating host-microbiota interactions. For mammalian SRs, their interaction with the microbiota seems to be indirect as modulators of host metabolism. In contrast, the work on the sea anemone Aiptasia point to a role of SRs in establishment of the symbiosis with Symbiodinium and this may be also the case in other animal groups, such as sponges.
Research on animal receptors ranges from comparative genomic studies, differential gene expression or metabolic analysis, biochemical characterization of ligands and receptors and -in a few animal models- functional validation by silencing of target genes and investigation of germ-free vs. conventionally raised animals. In this way, we are advancing our knowledge on the receptors mediating host-microbiota communication but open questions remain. In most cases, the microbiota-derived molecules that the receptors recognize and the signaling pathways that they trigger remain elusive. Moreover, synergies among receptors, which may be key to determine specific responses, are not taken into consideration (184). Understanding these interactions and the downstream signaling upon encounter with different microbes is key to reveal the biological role of PRRs.
Exploring host-microbiota interactions in less complex invertebrate organisms may help to decipher the molecular language used in host-microbiota communication (185). The two most intensively studied invertebrate model organisms are the fruit fly D. melanogaster and the nematode C. elegans. D. melanogaster-gut microbiome research started several years ago with the focus on the effect of gut microbiome on host nutrition [reviewed in (186)], as well as other host functions such as larval growth (187, 188), mate selection (189), and behavior (190, 191). C. elegans is only beginning to emerge as model system for metaorganism research (192), but several recent studies have already strengthen the big potential of C. elegans to study molecular mechanisms underlying host-microbiota interactions (193–195), including microbiota-mediated protection against pathogens (196). C. elegans and D. melanogaster both offer experimental advantages for e.g., using large scale genetic screens as tools for receptor identification in host-microbiota interactions, as they combine genetic amenability, low cost, and undemanding culture conditions. They provide the ability to screen knockout mutants or animals, in which genes are individually knockdown by RNAi, for the effect of gene inactivation on microbiota-dependent phenotypes (such as the above mentioned larval growth or microbiota-mediated protection against pathogen infection). These forward genetic screens can be unbiased whole genome screens and may thus offer the potential to discover novel receptors, or targeted on a certain known receptor group, such as GPCRs.
Obviously, not all invertebrates are genetically tractable, have a simple microbiota, and culturable symbionts. However, the study of diverse animal hosts has the potential to bring completely new insights into animal-microbe interactions (197, 198). The early branching metazoan Hydra harbors a relatively simple microbiome and offers functional tractability of host and bacteria, in addition to the accessibility of a germfree model. The Hydra system helped in deciphering the role of the immune system and the nervous system in shaping the microbiome (199–201) and opening new venues of research, like the role of quorum quenching in host-symbiont interactions (202). The sea anemone Aiptasia is arising as a model underlying molecular processes in cnidarian-dinoflagellate symbiosis, which promises to set the basis for understanding coral bleaching (182, 203, 204). Finally, the one-on-one symbiosis between the Hawaiian bobtail squid E. scolopes and the bioluminescent bacterium V. fischeri has served as beautiful model for studying symbiont colonization and the genetic basis of host-microbe “conversations.” Discoveries such as the release of LPS and PGN signals by V. fischeri that trigger the maturation of the host light organ (13) were not only relevant to understanding this specialized and intimate symbiosis, but also broadened our general perspective on host-microbe interactions, changing the focus from pathogenesis to beneficial symbiosis. In this sense, the Vibrio-squid system has been key in fostering the field of metaorganism research.
The study of various invertebrate hosts thus expands the evolutionary view on host-microbiota interactions and provides a high diversity of contexts to interrogate the underlying principles and mechanisms of the metaorganism. For example, as early-diverging animals, sponges offer a window into fundamental processes that have allowed animals to evolve in a microbial world. Despite current low tractability, the diversity of this group allows us to interrogate host-microbiota interactions by comparing different groups; for example the effect of harboring a high dense vs. low dense microbial communities on host immune strategies (21). And even in these complex holobionts, surprising mechanisms of interkingdom interactions have been revealed, like that of “ankyphages”: phage-derived proteins which facilitate the persistence of their host bacteria within the animal host (205). Growing genomic information combined with the amenability of the system to lab manipulation are also bringing echinoderm larva as a promising model for host-microbiota interactions (206, 207). In other invertebrates, metaorganism research is directly link to economic activities. For example, the causes of mortalities in the Pacific oyster Crassostrea gigas can only be explained from a holistic approach that addresses immunity-microbe-environment interactions (208). The development of sequencing, imaging, modeling, and genome editing technologies is opening up the mechanisms involved in a priori more cryptic animal-microbe interactions.
The study of invertebrate-microbiota interactions broadens the metaorganism concept and provides the wide lens and evolutionary perspective that is required to disentangle core mechanisms of host-microbe interaction as well as the underlying regulatory principles.
Author Contributions
KD and LP conceived and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was funded by the German Science Foundation DFG (Collaborative Research Center CRC1182 Origin and Function of Metaorganisms, Project A1.2 to KD and Project IMMUBASE to LP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the CRC1182-Origin and Function of Metaorganisms for fostering the exchange of ideas and in particular Sebastian Fraune (Düsseldorf University) for critical discussion of the manuscript and encouragement.
References
1. Bosch TCG, McFall-Ngai MJ. Metaorganisms as the new frontier. Zoology. (2011) 114:185–90. doi: 10.1016/j.zool.2011.04.001
2. Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. (2002) 243:1–10. doi: 10.3354/meps243001
3. Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. (2006) 8:2068–73. doi: 10.1111/j.1462-2920.2006.01148.x
4. McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. (2013) 110:3229–36. doi: 10.1073/pnas.1218525110
5. Pita L, Rix L, Slaby BM, Franke A, Hentschel U. The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome. (2018) 6:46. doi: 10.1186/s40168-018-0428-1
6. Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol. (2013) 108:500–8. doi: 10.1038/ajg.2013.59
7. Scott KP, Jean-Michel A, Midtvedt T, Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis. (2015) 26:2235. doi: 10.3402/mehd.v26.25877
8. Parnell JJ, Berka R, Young HA, Sturino JM, Kang Y, Barnhart DM, et al. From the lab to the farm: an industrial perspective of plant beneficial microorganisms. Front Plant Sci. (2016) 7:1110. doi: 10.3389/fpls.2016.01110
9. Rosado PM, Leite DCA, Duarte GAS, Chaloub RM, Jospin G, Nunes da Rocha U, et al. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. (2018) 13:921–36. doi: 10.1038/s41396-018-0323-6
10. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. (2013) 14:668–75. doi: 10.1038/ni.2635
11. Brown RL, Clarke TB. The regulation of host defences to infection by the microbiota. Immunology. (2017) 150:1–6. doi: 10.1111/imm.12634
12. Rosenstiel P, Philipp EE, Schreiber S, Bosch TC. Evolution and function of innate immune receptors–insights from marine invertebrates. J Innate Immun. (2009) 1:291–300. doi: 10.1159/000211193
13. Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. (2004) 306:1186–8. doi: 10.1126/science.1102218
14. Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. (2012) 10:815–27. doi: 10.1038/nrmicro2894
15. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
16. Skelly AN, Sato Y, Kearney S, Honda K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol. (2019) 19:305–23. doi: 10.1038/s41577-019-0144-5
17. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. (2003) 278:11312–9. doi: 10.1074/jbc.M211609200
18. Bufe B, Zufall F. The sensing of bacteria: emerging principles for the detection of signal sequences by formyl peptide receptors. Biomol Concepts. (2016) 7:205–14. doi: 10.1515/bmc-2016-0013
19. Gauthier MEA, Du Pasquier L, Degnan BM. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol Dev. (2010) 12:519–33. doi: 10.1111/j.1525-142X.2010.00436.x
20. Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. (2010) 466:720–6. doi: 10.1038/nature09201
21. Pita L, Hoeppner MP, Ribes M, Hentschel U. Differential expression of immune receptors in two marine sponges upon exposure to microbial- associated molecular patterns. Sci Rep. (2018) 8:16081. doi: 10.1038/s41598-018-34330-w
22. Yuen B, Bayes JM, Degnan SM. The characterization of sponge NLRs provides insight into the origin and evolution of this innate immune gene family in animals. Mol Biol Evol. (2014) 31:106–20. doi: 10.1093/molbev/mst174
23. Steindler L, Schuster S, Ilan M, Avni A, Cerrano C, Beer S. Differential gene expression in a marine sponge in relation to its symbiotic state. Mar Biotechnol. (2007) 9:543–9. doi: 10.1007/s10126-007-9024-2
24. Wiens M, Korzhev M, Perovic-Ottstadt S, Luthringer B, Brandt D, Klein S, et al. Toll-like receptors are part of the innate immune defense system of sponges (demospongiae: Porifera). Mol Biol Evol. (2007) 24:792–804. doi: 10.1093/molbev/msl208
25. Franzenburg S, Fraune S, Künzel S, Baines JF, Domazet-lo T, Bosch TCG. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA. (2012) 109:19374–9. doi: 10.1073/pnas.1213110109
26. Brennan JJ, Messerschmidt JL, Williams LM, Matthews BJ, Reynoso M, Gilmore TD. Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-κB signal transduction, and development. Proc Natl Acad Sci USA. (2017) 114:E10122–31. doi: 10.1073/pnas.1711530114
27. Lange C, Hemmrich G, Klostermeier UC, López-Quintero JA, Miller DJ, Rahn T, et al. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. (2011) 28:1687–702. doi: 10.1093/molbev/msq349
28. Neubauer EF, Poole AZ, Weis VM, Davy SK. The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ. (2016) 4:e2692. doi: 10.7717/peerj.2692
29. Rodriguez-Lanetty M, Phillips WS, Weis VM. Transcriptome analysis of a cnidarian-dinoflagellate mutualism reveals complex modulation of host gene expression. BMC Genomics. (2006) 7:23. doi: 10.1186/1471-2164-7-23
30. Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, Pringle JR. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. Genes Genomes Genet. (2014) 4:277–95. doi: 10.1534/g3.113.009084
31. Fuess LE, Pinzón CJH, Weil E, Mydlarz LD. Associations between transcriptional changes and protein phenotypes provide insights into immune regulation in corals. Dev. Comp. Immunol. (2016) 62:17–28. doi: 10.1016/j.dci.2016.04.017
32. Wright RM, Kenkel CD, Dunn CE, Shilling EN, Bay LK, Matz MV. Intraspecific differences in molecular stress responses and coral pathobiome contribute to mortality under bacterial challenge in Acropora millepora. Sci Rep. (2017) 7:1–13. doi: 10.1038/s41598-017-02685-1
33. Mansfield KM, Carter NM, Nguyen L, Cleves PA, Alshanbayeva A, Williams LM, et al. Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci Rep. (2017) 7:16025. doi: 10.1038/s41598-017-16168-w
34. Cuvillier-Hot V, Boidin-Wichlacz C, Slomianny C, Salzet M, Tasiemski A. Characterization and immune function of two intracellular sensors, HmTLR1 and HmNLR, in the injured CNS of an invertebrate. Dev Comp Immunol. (2011) 35:214–26. doi: 10.1016/j.dci.2010.09.011
35. Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, et al. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol. (2009) 11:1114–27. doi: 10.1111/j.1462-5822.2009.01315.x
36. Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. (2010) 12:2190–203. doi: 10.1111/j.1462-2920.2009.02121.x
37. Collins AJ, Schleicher TR, Rader BA, Nyholm SV, Weis V, State O. Understanding the role of host hemocytes in a squid/Vibrio symbiosis using transcriptomics and proteomics. Front Immunol. (2012) 3:91. doi: 10.3389/fimmu.2012.00091
38. Wei X, Zhao T, Ai K, Li H, Jiang X, Li C, et al. Role of scavenger receptor from Octopus ocellatus as a co-receptor of Toll-like receptor in initiation of TLR-NF-κB signaling during anti-bacterial response. Dev Comp Immunol. (2018) 84:14–27. doi: 10.1016/j.dci.2018.01.023
39. Liu L, Yang J, Qiu L, Wang L, Zhang H, Wang M, et al. A novel scavenger receptor-cysteine-rich (SRCR) domain containing scavenger receptor identified from mollusk mediated PAMP recognition and binding. Dev Comp Immunol. (2011) 35:227–39. doi: 10.1016/j.dci.2010.09.010
40. Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, et al. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science. (2016) 353:1434–7. doi: 10.1126/science.aaf3552
41. Pees B, Kloock A, Nakad R, Barbosa C, Dierking K. Enhanced behavioral immune defenses in a C. elegans C-type lectin-like domain gene mutant. Dev Comp Immunol. (2017) 74:237–42. doi: 10.1016/j.dci.2017.04.021
42. Bulgheresi S, Schabussova I, Chen T, Mullin NP, Maizels RM, Ott JA. A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl Environ Microbiol. (2006) 72:2950–6. doi: 10.1128/AEM.72.4.2950-2956.2006
43. Bulgheresi S, Gruber-Vodicka HR, Heindl NR, Dirks U, Kostadinova M, Breiteneder H, et al. Sequence variability of the pattern recognition receptor Mermaid mediates specificity of marine nematode symbioses. ISME J. (2011) 5:986–98. doi: 10.1038/ismej.2010.198
44. Dierking K, Polanowska J, Omi S, Engelmann I, Gut M, Lembo F, et al. Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe. (2011) 9:425–35. doi: 10.1016/j.chom.2011.04.011
45. Zugasti O, Bose N, Squiban B, Belougne J, Kurz CL, Schroeder FC, et al. Activation of a G protein-coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nat Immunol. (2014) 15:833–8. doi: 10.1038/ni.2957
46. Cao X, Kajino-Sakamoto R, Doss A, Aballay A. Distinct roles of sensory neurons in mediating pathogen avoidance and neuropeptide-dependent immune regulation. Cell Rep. (2017) 21:1442–51. doi: 10.1016/j.celrep.2017.10.050
47. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. (1996) 86:973–83. doi: 10.1016/S0092-8674(00)80172-5
48. Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. (2001) 11:809–21. doi: 10.1016/s0960-9822(01)00241-x
49. Pees B, Yang W, Hinrich AZ. High innate immune specificity through diversified C-Type Lectin-like Domain Proteins in invertebrates. J Innate Immun. (2016) 8:129–42. doi: 10.1159/000441475
50. Wang XW, Zhao XF, Wang JX. C-type lectin binds to β-integrin to promote hemocytic phagocytosis in an invertebrate. J Biol Chem. (2014) 289:2405–14. doi: 10.1074/jbc.M113.528885
51. Pang X, Xiao X, Liu Y, Zhang R, Liu J, Liu Q, et al. Mosquito C-type lectins maintain gut microbiome homeostasis. Nat Microbiol. (2016) 1:16023. doi: 10.1038/nmicrobiol.2016.23
52. Wang XW, Xu JD, Zhao XF, Vasta GR, Wang JX. A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J Biol Chem. (2014) 289:11779–90. doi: 10.1074/jbc.M114.552307
53. Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol. (2011) 11:837–51. doi: 10.1038/nri3089
54. Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, Charroux B. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe. (2012) 12:153–65. doi: 10.1016/J.CHOM.2012.06.002
55. Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of Gram-Negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol. (2012) 189:1886–97. doi: 10.4049/jimmunol.1201022
56. Ryu J-H, Kim S-H, Lee H-Y, Bai JY, Nam Y-D, Bae J-W, et al. Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science. (2008) 319:777–82. doi: 10.1126/science.1149357
57. Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster — from microbial recognition to whole- organism physiology. Nat Rev Immunol. (2014) 14:796–810. doi: 10.1038/nri3763
58. Charroux B, Capo F, Kurz CL, Peslier S, Chaduli D, Viallat-Lieutaud A, et al. Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe. (2018) 23:215–28.e4. doi: 10.1016/j.chom.2017.12.007
59. Iatsenko I, Kondo S, Mengin-Lecreulx D, Lemaitre B. PGRP-SD, an extracellular Pattern-Recognition Receptor, enhances peptidoglycan-mediated activation of the Drosophila Imd pathway. Immunity. (2016) 45:1013–23. doi: 10.1016/j.immuni.2016.10.029
60. Iatsenko I, Boquete J-P, Lemaitre B. Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity. (2018) 49:929–42.e5. doi: 10.1016/j.immuni.2018.09.017
61. Song X, Wang M, Dong L, Zhu H, Wang J. PGRP-LD mediates A. stephensi vector competency by regulating homeostasis of microbiota-induced peritrophic matrix synthesis. PLoS Pathog. (2018) 14:e1006899. doi: 10.1371/journal.ppat.1006899
62. Bi W-J, Li D-X, Xu Y-H, Xu S, Li J, Zhao X-F, et al. Scavenger receptor B protects shrimp from bacteria by enhancing phagocytosis and regulating expression of antimicrobial peptides. Dev Comp Immunol. (2015) 51:10–21. doi: 10.1016/j.dci.2015.02.001
63. Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, et al. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell. (2013) 153:797–811. doi: 10.1016/j.cell.2013.04.009
64. Buckley KM, Rast JP. Diversity of animal immune receptors and the origins of recognition complexity in the deuterostomes. Dev Comp Immunol. (2015) 49:179–89. doi: 10.1016/j.dci.2014.10.013
65. Coteur G, Mellroth P, Lefortery C, De Gillan D, Dubois P, Communi D, et al. Peptidoglycan recognition proteins with amidase activity in early deuterostomes (Echinodermata). Dev Comp Immunol. (2007) 31:790–804. doi: 10.1016/j.dci.2006.11.006
66. Ho ECH, Buckley KM, Schrankel CS, Schuh NW, Hibino T, Solek CM, et al. Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol Cell Biol. (2016) 94:861–74. doi: 10.1038/icb.2016.51
67. Furukawa R, Matsumoto M, Kaneko H. Characterization of a scavenger receptor cysteine-rich-domain-containing protein of the starfish, Asterina pectinifera: ApSRCR1 acts as an opsonin in the larval and adult innate immune systems. Dev Comp Immunol. (2012) 36:51–61. doi: 10.1016/j.dci.2011.06.005
68. Yu L, Wang L, Chen S. Endogenous toll-like receptor ligands and their biological significance. J Cell Mol Med. (2010) 14:2592–603. doi: 10.1111/j.1582-4934.2010.01127.x
69. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
70. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
71. Brennan JJ, Gilmore TD. Evolutionary origins of Toll-like receptor signaling. Mol Biol Evol. (2018) 35:1576–87. doi: 10.1093/molbev/msy050
72. Nie L, Cai S-Y, Shao J-Z, Chen J. Toll-Like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
73. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. (2004) 118:229–41. doi: 10.1016/j.cell.2004.07.002
74. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. (2011) 332:974–7. doi: 10.1126/science.1206095
75. Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. (2012) 12:509–20. doi: 10.1016/j.chom.2012.08.004
76. Vatanen T, Kostic AD, D'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. (2016) 165:842–53. doi: 10.1016/j.cell.2016.04.007
77. Hennezel E, Abubucker S, Murphy LO, Cullen TW. Total lipopolysaccharide from the human gut microbiome silences Toll-like receptor signaling. mSystems. (2017) 2:e00046-17. doi: 10.1128/mSystems.00046-17
78. Otte J-M, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. (2004) 126:1054–70. doi: 10.1053/j.gastro.2004.01.007
79. Cario E, Podolsky DK. Intestinal epithelial TOLLerance versus inTOLLerance of commensals. Mol Immunol. (2005) 42:887–93. doi: 10.1016/j.molimm.2004.12.002
80. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIII promotes the spatial segregation of microbiota and host in the intestine. Science. (2011) 334:255–8. doi: 10.1126/science.1209791
81. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. (2008) 455:1109–13. doi: 10.1038/nature07336
82. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. (2010) 328:228–31. doi: 10.1126/science.1179721
83. Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. (2012) 209:1445–56. doi: 10.1084/jem.20120504
84. Gerttula S, Jin YS, Anderson KV. Zygotic expression and activity of the Drosophila Toll gene, a gene required maternally for embryonic dorsal-ventral pattern formation. Genetics. (1988) 119:123–33
85. Anderson KV, Jürgens G, Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. (1985) 42:779–89.
86. Rosetto M, Engström Y, Baldari CT, Telford JL, Hultmark D. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem Biophys Res Commun. (1995) 209:111–6. doi: 10.1006/bbrc.1995.1477
87. Brandt JP, Ringstad N. Toll-like receptor signaling promotes development and function of sensory neurons required for a C. elegans pathogen-avoidance behavior. Curr Biol. (2015) 25:2228–37. doi: 10.1016/j.cub.2015.07.037
88. Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA. (2007) 104:2295–300. doi: 10.1073/pnas.0610281104
89. Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, et al. The innate immune repertoire in cnidaria–ancestral complexity and stochastic gene loss. Genome Biol. (2007) 8:R59. doi: 10.1186/gb-2007-8-4-r59
90. Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. (2009) 33:559–69. doi: 10.1016/j.dci.2008.10.004
91. Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. (2014) 31:1102–20. doi: 10.1093/molbev/msu057
92. Ryu T, Seridi L, Moitinho-Silva L, Oates M, Liew YJ, Mavromatis C, et al. Hologenome analysis of two marine sponges with different microbiomes. BMC Genomics. (2016) 17:158. doi: 10.1186/s12864-016-2501-0
93. Germer J, Cerveau N, Jackson DJ. The holo-transcriptome of a calcified early branching metazoan. Front Mar Sci. (2017) 4:81. doi: 10.3389/fmars.2017.00081
94. Wiens M, Korzhev M, Krasko A, Thakur NL, Perović-Ottstadt S, Breter HJ, et al. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway: induction of a perforin-like molecule. J Biol Chem. (2005) 280:27949–59. doi: 10.1074/jbc.M504049200
95. Yuen B. Deciphering the genomic toolkit underlying animal-bacteria interactions – insights through the demosponge Amphimedon queenslandica (Ph.D. thesis). The University of Queensland, Brisbane, QLD, Australia (2016). doi: 10.14264/uql.2017.39
96. Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio. (2014) 5:1–13. doi: 10.1128/mBio.01117-14
97. Ting JPY, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity. (2008) 28:285–7. doi: 10.1016/j.immuni.2008.02.005
98. Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. (2006) 314:952–6. doi: 10.1126/science.1134301
99. Hamada M, Shoguchi E, Shinzato C, Kawashima T, Miller DJ, Satoh N. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol Biol Evol. (2013) 30:167–76. doi: 10.1093/molbev/mss213
100. Howe K, Schiffer PH, Zielinski J, Wiehe T, Laird GK, Marioni JC, et al. Structure and evolutionary history of a large family of NLR proteins in the zebrafish. Open Biol. (2016) 6:160009. doi: 10.1098/rsob.160009
101. Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: versatile cytosolic sentinels. Physiol Rev. (2015) 95:149–78. doi: 10.1152/physrev.00009.2014
102. Laroui H, Yan Y, Narui Y, Ingersoll SA, Ayyadurai S, Charania MA, et al. L-Ala-gamma-D-Glu-meso-diaminopimelic acid (DAP) interacts directly with leucine-rich region domain of nucleotide-binding oligomerization domain 1, increasing phosphorylation activity of receptor-interacting serine/threonine- protein kinase 2 and its inte. J Biol Chem. (2011) 286:31003–13. doi: 10.1074/jbc.M111.257501
103. Mo J, Boyle JP, Howard CB, Monie TP, Davis BK, Duncan JA. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J Biol Chem. (2012) 287:23057–67. doi: 10.1074/jbc.M112.344283
104. Keestra AM, Winter MG, Auburger JJ, Fräßle SP, Xavier MN, Winter SE, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. (2013) 496:233–7. doi: 10.1038/nature12025
105. Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: Regulators of inflammation in health and disease. Nat Rev Immunol. (2014) 14:9–23. doi: 10.1038/nri3565
106. Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. (2009) 27:229–65. doi: 10.1146/annurev.immunol.021908.132715
107. Lipinski S, Rosenstiel P. Debug your bugs - how NLRs shape intestinal host-microbe interactions. Front Immunol. (2013) 4:479. doi: 10.3389/fimmu.2013.00479
108. Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol. (2011) 12:121–8. doi: 10.1038/ni.1985
109. Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. (2013) 123:700–11. doi: 10.1172/JCI62236
110. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. (2011) 145:745–57. doi: 10.1016/j.cell.2011.04.022
111. Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, et al. The NLR protein NLRP6 does not impact gut microbiota composition. Cell Rep. (2017) 21:3653–61. doi: 10.1016/j.celrep.2017.12.026
112. Mamantopoulos M, Ronchi F, Van Hauwermeiren F, Vieira-Silva S, Yilmaz B, Martens L, et al. Nlrp6- and ASC-dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity. (2017) 47:339–348.e4. doi: 10.1016/j.immuni.2017.07.011
113. Robertson SJ, Yu Zhou J, Geddes K, Rubino SJ, Ho Cho J, Girardin SE, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. (2013) 4: 222–31. doi: 10.4161/gmic.24373
114. Schieber AMP, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, et al. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science. (2015) 350:558–63. doi: 10.1126/science.aac6468
115. Kim B, Wang YC, Hespen CW, Espinosa J, Salje J, Rangan KJ, et al. Enterococcus faecium secreted antigen a generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife. (2019) 8:1–25. doi: 10.7554/eLife.45343
116. Degnan SM. The surprisingly complex immune gene repertoire of a simple sponge, exemplified by the NLR genes: a capacity for specificity? Dev Comp Immunol. (2015) 48:269–74. doi: 10.1016/j.dci.2014.07.012
117. Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. (2005) 272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x
118. Cummings RD, McEver RP. C-type lectins. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology, 2nd Edn. La Jolla, CA: Cold Spring Harbor Laboratory Press (2009). p. 1857–929
119. Drickamer K. C-type lectin-like domains. Curr Opin Struct Biol. (1999) 9:585–90. doi: 10.1016/S0959-440X(99)00009-3
120. Drickamer K, Taylor ME. Recent insights into structures and functions of C-type lectins in the immune system. Curr Opin Struct Biol. (2015) 34:26–34. doi: 10.1016/j.sbi.2015.06.003
121. Mayer S, Raulf MK, Lepenies B. C-type lectins: their network and roles in pathogen recognition and immunity. Histochem Cell Biol. (2017) 147:223–37. doi: 10.1007/s00418-016-1523-7
122. Tytgat HLP, de Vos WM. Sugar coating the envelope: glycoconjugates for microbe–host crosstalk. Trends Microbiol. (2016) 24:853–61. doi: 10.1016/j.tim.2016.06.004
123. Konstantinov SR, Smidt H, de Vos WM, Bruijns SCM, Singh SK, Valence F, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. (2008) 105:19474–9. doi: 10.1073/pnas.0810305105
124. Tytgat HLP, van Teijlingen NH, Sullan RMA, Douillard FP, Rasinkangas P, Messing M, et al. Probiotic gut microbiota isolate interacts with dendritic cells via glycosylated geterotrimeric pili. PLoS ONE. (2016) 11:e0151824. doi: 10.1371/journal.pone.0151824
125. Lightfoot YL, Selle K, Yang T, Goh YJ, Sahay B, Zadeh M, et al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. (2015) 34:881–95. doi: 10.15252/embj.201490296
126. Hütter J, Eriksson M, Johannssen T, Klopfleisch R, Von Smolinski D, Gruber AD, et al. Role of the C-Type lectin receptors MCL and DCIR in experimental colitis. PLoS ONE. (2014) 9:e103281. doi: 10.1371/journal.pone.0103281
127. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science. (2012) 336:1314–7. doi: 10.1126/science.1221789
128. Eriksson M, Johannssen T, von Smolinski D, Gruber AD, Seeberger PH, Lepenies B. The C-type lectin receptor SIGNR3 binds to fungi present in commensal microbiota and influences immune regulation in experimental colitis. Front Immunol. (2013) 4:196. doi: 10.3389/fimmu.2013.00196
129. Li X, Wang S, Qi J, Echtenkamp SF, Chatterjee R, Wang M, et al. Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immunity. (2007) 27:518–29. doi: 10.1016/j.immuni.2007.07.020
130. Wolf AJ, Underhill DM. Peptidoglycan recognition by the innate immune system. Nat Rev Immunol. (2018) 18:243–54. doi: 10.1038/nri.2017.136
131. Dziarski R, Gupta D. Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun. (2010) 16:168–74. doi: 10.1177/1753425910366059
132. Saha S, Jing X, Park SY, Wang S, Li X, Gupta D, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-γ. Cell Host Microbe. (2010) 8:147–62. doi: 10.1016/j.chom.2010.07.005
133. Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. (2008) 4:147–58. doi: 10.1016/j.chom.2008.07.004
134. Pan X, Pike A, Joshi D, Bian G, McFadden MJ, Lu P, et al. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. (2018) 12:277–88. doi: 10.1038/ismej.2017.174
135. Brown M, Goldstein J. Receptor-mediated endocytosis: insights from the lipoprotein receptor system. Proc Natl Acad Sci USA. (1979) 76:3330–7. doi: 10.1073/pnas.76.7.3330
136. Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol. (2013) 13:621–34. doi: 10.1038/nri3515
137. Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RAB. Croquemort, a novel drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. (1996) 4:431–43. doi: 10.1016/S1074-7613(00)80410-0
138. Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. (2006) 300:349–65. doi: 10.1016/j.ydbio.2006.08.065
139. Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, et al. CD36 is a sensor of diacylglycerides. Nature. (2005) 433:523–7. doi: 10.1038/nature03253
140. Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. (2005) 170:477–85. doi: 10.1083/jcb.200501113
141. Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. (2001) 108:785–91. doi: 10.1172/JCI200114006.CD36
142. Stewart CR, Stuart LM, Wilkinson K, Van Gils JM, Deng J, Halle A, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. (2010) 11:155–61. doi: 10.1038/ni.1836
143. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. (2011) 472:57–65. doi: 10.1038/nature09922
144. Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. (2014) 158:705–21. doi: 10.1016/j.cell.2014.05.052
145. Zani I, Stephen S, Mughal N, Russell D, Homer-Vanniasinkam S, Wheatcroft S, et al. Scavenger receptor structure and function in health and disease. Cells. (2015) 4:178–201. doi: 10.3390/cells4020178
146. Guzman C, Conaco C. Comparative transcriptome analysis reveals insights into the streamlined genomes of haplosclerid demosponges. Sci Rep. (2016) 6:18774. doi: 10.1038/srep18774
147. Pancer Z. Dynamic expression of multiple scavenger receptor cysteine-rich genes in coelomocytes of the purple sea urchin. Proc Natl Acad Sci USA. (2000) 97:13156–61. doi: 10.1073/pnas.230096397
148. Wippler J, Kleiner M, Lott C, Gruhl A, Abraham PE, Giannone RJ, et al. Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis. BMC Genomics. (2016) 17:942. doi: 10.1186/s12864-016-3293-y
149. Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, Mukamel Z, et al. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol. (2018) 2:1176–88. doi: 10.1038/s41559-018-0575-6
150. Pancer Z, Rast JP, Davidson EH. Origins of immunity: transcription factors and homologues of effector genes of the vertebrate immune system expressed in sea urchin coelomocytes. Immunogenetics. (1999) 49:773–86. doi: 10.1007/s002510050551
151. Fuess LE, Mann WT, Jinks LR, Brinkhuis V, Mydlarz LD. Transcriptional analyses provide new insight into the late-stage immune response of a diseased Caribbean coral. R Soc Open Sci. (2018) 5:172062. doi: 10.1098/rsos.172062
152. Krishnan A, Almén MS, Fredriksson R, Schiöth HB. The origin of GPCRs: Identification of mammalian like rhodopsin, adhesion, glutamate and frizzled GPCRs in fungi. PLoS ONE. (2012) 7:e29817. doi: 10.1371/journal.pone.0029817
153. Fredriksson R, Schiöth HB. The repertoire of G-Protein-Coupled Receptors in fully sequenced genomes. Mol Pharmacol. (2005) 67:1414–25. doi: 10.1124/mol.104.009001.sequenced
154. Hanlon CD, Andrew DJ. Outside-in signaling - a brief review of GPCR signaling with a focus on the Drosophila GPCR family. J Cell Sci. (2015) 128:3533–42. doi: 10.1242/jcs.175158
156. Krishnan A, Dnyansagar R, Almén MS, Williams MJ, Fredriksson R, Manoj N, et al. The GPCR repertoire in the demosponge Amphimedon queenslandica: insights into the GPCR system at the early divergence of animals. BMC Evol Biol. (2014) 14:270. doi: 10.1186/s12862-014-0270-4
157. Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. (2002) 3:639–50. doi: 10.1038/nrm908
158. Bockaert J, Pin JP, Zuker C. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. (1999) 18:1723–9. doi: 10.1093/emboj/18.7.1723
159. De Mendoza A, Sebé-Pedrós A, Ruiz-Trillo I. The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biol Evol. (2014) 6:606–19. doi: 10.1093/gbe/evu038
160. Cardoso JCR, Félix RC, Fonseca VG, Power DM. Feeding and the rhodopsin family G-protein coupled receptors in nematodes and arthropods. Front Endocrinol. (2012) 3:157. doi: 10.3389/fendo.2012.00157
161. Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-Protein-Coupled Receptors. Nature. (2014) 459:356–63. doi: 10.1038/nature08144
162. Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. (2010) 28:623–67. doi: 10.1146/annurev-immunol-030409-101330
163. Bloes DA, Kretschmer D, Peschel A. Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat Rev Microbiol. (2015) 13:95–104. doi: 10.1038/nrmicro3390
164. Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S, et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. (2018) 11:752–62. doi: 10.1038/mi.2017.118
165. Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW. GPCR-mediated signaling of metabolites. Cell Metab. (2017) 25:777–96. doi: 10.1016/j.cmet.2017.03.008
166. Chen H, Nwe P-K, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. (2019) 117:1217–31. doi: 10.1016/j.cell.2019.03.036
167. Colosimo DA, Kohn JA, Luo PM, Piscotta FJ, Han SM, Pickard AJ, et al. Mapping interactions of microbial metabolites with human G-Protein-Coupled Receptors. Cell Host Microbe. (2019) 26:273–82.e7. doi: 10.1016/j.chom.2019.07.002
168. Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. (2017) 549:48–53. doi: 10.1038/nature23874
169. Mazzoli R, Pessione E. The neuro-endocrinological role of microbial glutamate and GABA signaling. Front Microbiol. (2016) 7:1934. doi: 10.3389/fmicb.2016.01934
170. Reboul J, Ewbank JJ. GPCRs in invertebrate innate immunity. Biochem Pharmacol. (2016) 114:82–7. doi: 10.1016/j.bcp.2016.05.015
171. Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, et al. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. (2008) 18:481–9. doi: 10.1016/j.cub.2008.02.079
172. Ziegler K, Kurz CL, Cypowyj S, Couillault C, Pophillat M, Pujol N, et al. Antifungal innate immunity in C. elegans: PKCδ links G Protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe. (2009) 5:341–52. doi: 10.1016/j.chom.2009.03.006
173. Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. (2008) 322:460–4. doi: 10.1126/science.1163673
174. Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. (2009) 323:382–4. doi: 10.1126/science.1166527
175. Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating non- canonical unfolded protein response genes. Science. (2011) 332:729–32. doi: 10.1126/science.1203411.Neuronal
176. Anderson A, Laurenson-Schafer H, Partridge FA, Hodgkin J, McMullan R. Serotonergic chemosensory neurons modify the C. elegans immune response by regulating G-Protein signaling in epithelial cells. PLoS Pathog. (2013) 9:e1003787. doi: 10.1371/journal.ppat.1003787
177. Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. (2014) 159:267–80. doi: 10.1016/j.cell.2014.09.011
178. Labed SA, Wani KA, Jagadeesan S, Hakkim A, Najibi M, Irazoqui JE. Intestinal epithelial Wnt signaling mediates acetylcholine-triggered host defense against infection. Immunity. (2018) 48:963–978.e4. doi: 10.1016/j.immuni.2018.04.017
179. Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. (2005) 310:847–50. doi: 10.1126/science.1117311
180. Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, et al. Regulation of DUOX by the Gαq-phospholipase Cβ-Ca2+ pathway in Drosophila gut immunity. Dev Cell. (2009) 16:386–97. doi: 10.1016/j.devcel.2008.12.015
181. Kim S-H, Lee W-J. Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front Cell Infect Microbiol. (2014) 3:116. doi: 10.3389/fcimb.2013.00116
182. Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, et al. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci USA. (2017) 114:201710733. doi: 10.1073/pnas.1710733114
183. Peng S-E, Wang Y-B, Wang L-H, Chen W-NU, Lu C-Y, Fang L-S, et al. Proteomic analysis of symbiosome membranes in Cnidaria-dinoflagellate endosymbiosis. Proteomics. (2010) 10:1002–16. doi: 10.1002/pmic.200900595
184. Schulenburg H, Boehnisch C, Michiels NK. How do invertebrates generate a highly specific innate immune response? Mol Immunol. (2007) 44:3338–44. doi: 10.1016/j.molimm.2007.02.019
185. Douglas AE. Simple animal models for microbiome research. Nat Rev Microbiol. (2019) 17:764–75. doi: 10.1038/s41579-019-0242-1
186. Douglas AE. The Drosophila model for microbiome research. Lab Anim. (2018) 47:157–64. doi: 10.1038/s41684-018-0065-0
187. Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. (2011) 14:403–14. doi: 10.1016/j.cmet.2011.07.012
188. Bozonnet L, Daniel C, Schwarzer M, Strigini M, Matos R, Leulier F, et al. Drosophila perpetuates nutritional mutualism by promoting the fitness of its intestinal symbiont Lactobacillus plantarum. Cell Metab. (2017) 27:362–77.e8. doi: 10.1016/j.cmet.2017.11.011
189. Sharon G, Segal D, Ringo J, Hefetz A, Zilber-Rosenberg I, Rosenbeg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA. (2010) 107:20051–6. doi: 10.1073/pnas.1302980110
190. Fischer C, Trautman EP, Crawford JM, Stabb EV, Handelsman J, Broderick NA. Metabolite exchange between microbiome members produces compounds that influence drosophila behavior. Elife. (2017) 6:1–25. doi: 10.7554/eLife.18855
191. Schretter CE, Vielmetter J, Bartos I, Marka Z, Marka S, Argade S, et al. A gut microbial factor modulates locomotor behaviour in Drosophila. Nature. (2018) 563:402–6. doi: 10.1038/s41586-018-0634-9
192. Zhang F, Berg M, Dierking K, Félix MA, Shapira M, Samuel BS, et al. Caenorhabditis elegans as a model for microbiome research. Front Microbiol. (2017) 8:485. doi: 10.3389/fmicb.2017.00485
193. Berg M, Monnin D, Cho J, Nelson L, Crits-Christoph A, Shapira M. TGFβ/BMP immune signaling affects abundance and function of C. elegans gut commensals. Nat Commun. (2019) 10:604. doi: 10.1038/s41467-019-08379-8
194. Yang W, Petersen C, Pees B, Zimmermann J, Waschina S, Dirksen P, et al. The inducible response of the nematode Caenorhabditis elegans to members of its natural microbiota across development and adult life. Front Microbiol. (2019) 10:1793. doi: 10.3389/fmicb.2019.01793
195. Zimmermann J, Obeng N, Yang W, Pees B, Petersen C, Waschina S, et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. ISME J. (2019) 14:26–38. doi: 10.1038/s41396-019-0504-y
196. Kissoyan KAB, Drechsler M, Stange EL, Zimmermann J, Kaleta C, Bode HB, et al. Natural C. elegans microbiota protects against infection via production of a cyclic lipopeptide of the viscosin group. Curr Biol. (2019) 29:1030–7.e5. doi: 10.1016/j.cub.2019.01.050
197. Petersen JM, Osvatic J. Microbiomes In Natura: importance of invertebrates in understanding the natural variety of animal-microbe interactions. mSystems. (2018) 3:1–7. doi: 10.1128/msystems.00179-17
198. Bosch TCG, Guillemin K, McFall-Ngai M. Evolutionary “experiments” in Symbiosis: the study of model animals provides insights into the mechanisms underlying the diversity of host–microbe interactions. Bioessays. (2019) 41:1800256. doi: 10.1002/bies.201800256
199. Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci USA. (2013) 110:E3730–8. doi: 10.1073/pnas.1304960110
200. Bosch TCG. Rethinking the role of immunity: lessons from Hydra. Trends Immunol. (2014) 35:495–502. doi: 10.1016/j.it.2014.07.008
201. Augustin R, Schröder K, Murillo Rincón AP, Fraune S, Anton-Erxleben F, Herbst E-M, et al. A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat Commun. (2017) 8:698. doi: 10.1038/s41467-017-00625-1
202. Pietschke C, Treitz C, Forêt S, Schultze A, Künzel S, Tholey A, et al. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc Natl Acad Sci USA. (2017) 114:E8488–97. doi: 10.1073/pnas.1706879114
203. Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci USA. (2015) 112:11893–8. doi: 10.1073/pnas.1513318112
204. Neff EP. The quest for an animal model of coral health and disease. Lab Anim. (2020) 49:37–41. doi: 10.1038/s41684-019-0467-7
205. Jahn MT, Arkhipova K, Markert SM, Stigloher C, Lachnit T, Pita L, et al. A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host Microbe. (2019) 26:542–50.e5. doi: 10.1016/j.chom.2019.08.019
206. Buckley KM, Rast JP. An organismal model for gene regulatory networks in the gut-associated immune response. Front Immunol. (2017) 8:1297. doi: 10.3389/fimmu.2017.01297
207. Carrier TJ, Reitzel AM. Convergent shifts in host-associated microbial communities across environmentally elicited phenotypes. Nat Commun. (2018) 9:952. doi: 10.1038/s41467-018-03383-w
Keywords: innate immunity, pattern recognition receptors, holobiont, microbiome, invertebrate
Citation: Dierking K and Pita L (2020) Receptors Mediating Host-Microbiota Communication in the Metaorganism: The Invertebrate Perspective. Front. Immunol. 11:1251. doi: 10.3389/fimmu.2020.01251
Received: 12 February 2020; Accepted: 18 May 2020;
Published: 16 June 2020.
Edited by:
Shoichiro Kurata, Tohoku University, JapanReviewed by:
Akira Goto, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceMaria Forlenza, Wageningen University and Research, Netherlands
Copyright © 2020 Dierking and Pita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katja Dierking, a2RpZXJraW5nJiN4MDAwNDA7em9vbG9naWUudW5pLWtpZWwuZGU=; Lucía Pita, bHBpdGEmI3gwMDA0MDtnZW9tYXIuZGU=
 Katja Dierking
Katja Dierking Lucía Pita
Lucía Pita