- 1Garvan Institute of Medical Research, Sydney, NSW, Australia
- 2Faculty of Medicine, St Vincent’s Clinical School, The University of New South Wales, Sydney, NSW, Australia
In vitro selection technology has transformed the development of therapeutic monoclonal antibodies. Using methods such as phage, ribosome, and yeast display, high affinity binders can be selected from diverse repertoires. Here, we review strategies for the next-generation sequencing (NGS) of phage- and other antibody-display libraries, as well as NGS platforms and analysis tools. Moreover, we discuss recent examples relating to the use of NGS to assess library diversity, clonal enrichment, and affinity maturation.
Introduction
The development of antibody display technology such as phage (1), ribosome (2), yeast (3), and mammalian display (4) has enabled the rapid selection of binders from diverse libraries. These technologies bypass the use of animals and allow for the enrichment of binders within days to weeks. The power of in vitro selection technologies relies on a direct physical link between phenotype (displayed antibody construct) and genotype (antibody variable domain genes), allowing for the identification of binders through sequencing of their encoding genes. Multiple rounds of selection are generally required to identify antigen-specific binders, either by binding to a solid support or through cellular sorting (5). In many cases, later rounds of selections tend to be dominated by a handful of clones, which are then further characterized for affinity. Such clonal dominance can reflect genuine selection for high antigen affinity but might also reflect other properties such as superior expression or display. Consequently, clones with superior affinity may be present at low frequency and may not be readily detectable using traditional screening methods such as ELISA (6).
Recent advances in DNA sequencing technologies and computing power over the last decade has led to a dramatic reduction in the cost of sequencing and has simplified data analyses (7). Although initially developed for genomics applications, such as whole-genome sequencing, transcriptome sequencing, and epigenetics, next-generation sequencing (NGS) technology is now increasingly being applied other fields, including to basic and applied immunology. This includes the sequencing of the paired human heavy and light chain repertoire from isolated naïve (8, 9) and antigen-specific B-cells (10, 11), as well as T-cell receptor (12) and antibody display repertoires (13). While most NGS platforms were originally designed for short reads, technology is evolving rapidly, extending both read length and depth. Here, we review recent advances in NGS technology and key applications to phage display and other in vitro selection technologies.
Strategies for NGS of Antibody Repertoires
Traditionally, antibody display libraries are analyzed by isolation of 102–103 clones in combination with Sanger sequencing (5). Although this approach is sufficient to identify dominant clones after selection, or to broadly validate design objectives, the data obtained represent only a limited snapshot of actual library diversity. By contrast, NGS approaches allow for far-greater insights into library diversity by providing up to 107 sequences (approximately 10,000-fold more sequences than Sanger sequencing).
One of the main challenges in the use of NGS for the analysis of antibody selection systems relates to the size of the encoded genes: the smallest antibody fragments (variable domains) range between 300 and 400 bp in length, while the commonly used scFv and Fab antibody fragment formats range from 700 to 800 bp to over 1,500 bp, respectively. While NGS technologies are particularly well suited for high throughput sequencing of short reads (less than 100 bp), many platforms can nevertheless sequence up to 300–400 bp with reasonable throughput. In particular, Illumina Miseq and Hiseq, 454 GS FLX (instrument discontinued), and Ion Torrent PMG are suited for this task (8, 14, 15); in addition, PacBio sequencing generates particularly long reads at the cost of reduced read numbers (Table 1) (16). Long sequences can also be generated by using paired-end reads: this method is particularly useful for scFv formats, enabling the sequencing of multiple CDR regions of VH and VL domains. In addition to analysis of longer antibody fragment sequences, some studies have focused on sequencing the relatively short VH CDR3 repertoire only (23) [which forms the center of the antigen binding site and is a major determinant of antigen binding (24)].
The use of NGS requires particular attention be paid to sequencing errors (25). DNA amplification inevitably results in polymerase errors, which can be context dependent. Although the error rates of polymerases are generally low (10−5–10−6 per base), errors will inevitably be present in large NGS datasets that encompass billions of bases. In addition, the NGS technologies themselves can be susceptible to the introduction of errors, such as cluster misamplification and base misincorporation, with frequencies ranging from 10−2 (PacBio, Ion Torrent) to 10−3 (Illumina). To help identify PCR and sequencing errors, unique molecular identifiers (UMIs; stretches of 8–10 degenerate DNA bases) can be added to primers during the first two cycles of PCR amplification. Reads that share the same UMI have a high probability of being derived from the same original template. Such reads can then grouped after sequencing and used for error correction (26).
Bioinformatic Tools to Analyze NGS Data
While the analysis of the limited number of clones obtained by Sanger sequencing can be carried out manually, the larger sample size of NGS approaches necessitates the use bioinformatics tools. Following confirmation of the quality of the NGS read data by a tool such as FastQC (27), the data are further processed to clean up reads before analysis of antibody or antibody fragment sequences. The steps undertaken will be highly dependent on the NGS platform utilized and the format of the amplicons but generally will focus on the: removal of adapter sequences [e.g., PRINSEQ (28)], de-multiplexing (if barcodes were used), UMI identification and consensus building, and error correction (26), read quality trimming and filtering [e.g., Trimmomatic (29)] and, if paired-end sequencing was performed, the merging of the read pairs with a program such as PEAR (30). Separate analysis of heavy and light chains may be required for antibody formats such as scFv, where the presence of synthetic linkers can complicate analyses.
Programs such as IMPre (31), IgBLAST (32), IMGT/High V-QUEST (33, 34), and ImmundiveRsity (35), which were originally developed for the analysis of B and T cell receptor repertoires, identify VH and VL germlines as well as VH and VL CDRs. The selection of a tool will depend on the number of NGS reads being analyzed and the computational skill level of the researcher. IgBLAST and IMGT/High V-QUEST are both available as web-based submission systems, with IMGT/High V-QUEST permitting a larger number of reads to be analysis per submission. IMGT/High V-QUEST returns an output format compatible with programs such as Microsoft Excel or OpenOffice, whereas IgBLAST output is text based. The tools use different alignment algorithms, BLAST (IgBLAST) and modified Smith–Waterman (V-QUEST), but both restrict the germline gene repertoires to those defined by the tool’s creators. A stand-alone version of IgBLAST is available, and it has no restriction on the number of input reads, permits the user-defined germline gene databases, provides additional output formats, and can be parallelized on a cluster for processing of large datasets; however, its use does require some command line basics.
Postprocessing of the output of tools such as IgBLAST and IMGT/High V-QUEST is required to generate information about the clone structure within the dataset, and to pair VH and VL domain sequences. Clone structures can be inferred by applying sequence clustering tools, such as CD-hit (36) or UCLUST (37) to CDR3s alone, at either the amino acid or nucleotide sequence level, or to the full-length sequence, to group closely related sequences into “clonal” groups. The choice of parameters will depend on the diversity of the library. Finally, scripts can be used to analyze and summarize the diversity and other compositional characteristics of the library.
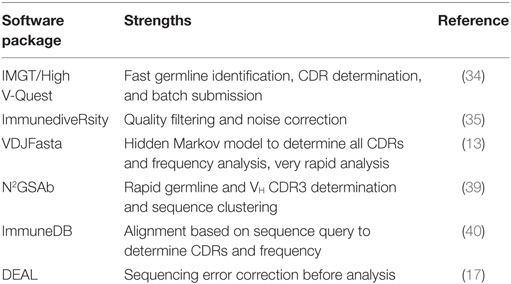
A custom pipeline as described earlier requires a level of informatics skills not always available to researchers, therefore, specialized pipelines for the analysis of recombinant antibody libraries, either naïve or in vitro selected against particular antigens, have been developed. The AbMining Toolbox is particularly suited for identifying VH CDR3, which is determined by using a hidden Markov model (HMM) that captures the conserved sequences upstream and downstream of the CDR3 (38). N2GSAb can rapidly identify germline and VH CDR3 and provides a tool for clustering unique sequences (39). VDJFasta uses an HMM to accurately predict all VH and VL CDRs, as well as the GS linker sequence for scFv fragments, and can generate library diversity plots (13). The ImmuneDB package aligns sequences based on a query sequence, such as a framework region, to delineate CDR regions (40). ImmuneDB also performs mutational and statistical analysis on the sequence library and can construct lineage trees to aid in the interpretation of antigen-selected libraries. More recently, DEAL was developed to better predict library diversity by identifying and correcting sequencing errors (17). In the published example, the library was not generated by PCR but rather by ligation of adapters to avoid any amplification bias and focus on sequencing errors. Reads are clustered using seed sequences of 10–20 bp and analyzed by binary comparison. The clusters are then compared with each read, taking into account the Phred quality score for each base and the error rate of the Phi-X control to identify sequencing errors. A list of software is outlined in Table 2.
It is also possible to outsource the sequencing and/or analysis of antibody libraries to commercial suppliers. Examples include (but are not limited to) CD Genomics and Molecular Cloning Laboratories. Such companies offer a range of options from basic consulting on designing primers for multiplexing and sequencing, to complete analysis from purified DNA or phages.
Application to Design Validation and the Analyses of Naïve Antibody Libraries
When generating antibody display repertoires, either synthetic or derived from immunized animals, it is important to assess the clonal diversity of the library before selection. Several studies have demonstrated the use of NGS to measure diversity to validate the design of displayed libraries. In an early example, Novimmune designed scFv libraries with both synthetic diversity, using degenerated oligonucleotides, and semi-synthetic diversity, from human or rabbit donors (39). Sequencing of VH CDR3 using the Illumina platform revealed that the synthetic libraries had many more unique clones compared with donor-derived libraries, with between 1–16 and 31–69% clonal redundancy, respectively (39). Intriguingly, the extent of clonal redundancy in the donor-derived libraries suggested an upper limit of human VH diversity of around 2–3 × 106 unique clones. This figure correlates with other NGS studies aimed at determining human B-cell diversity (3–9 × 106) (41). In addition, the Novimmune sequencing results also validated the VH CDR3 length distribution in human antibodies, which closely matched that of the IMGT repertoire (39).
A second example, relates to the Ylanthia synthetic antibody library developed by MorphoSys (21). The library was designed to encode a range of VH CDR3 lengths to closely match the natural human antibody repertoire and was analyzed by the Roche 454 sequencing platform (21). The library was found to be composed of about 95% unique clones, and there was no indication of amplification biases during antibody library construction. In addition, the authors used NGS data to validate VH CDR3 diversity and length, as well as VH and VL germline frequencies.
High throughput sequencing approaches are not limited to human sequences, with a recent study assessing the diversity of rabbit (VH and VL) Fab libraries by NGS (Ion PGM) (20). Surprisingly, and unlike human libraries derived from donors, these studies detected very low levels of redundancy within the rabbit libraries, with over 98% of VH clones being of a unique nature (~3 × 109 sequence reads were analyzed).
Next-generation sequencing has also been used to accurately determine library size. A recent study generated a donor-derived VH library for this purpose, which was then sequenced using Illumina adapter ligation (circumventing the need for PCR amplification) (17). Sequencing depth for the VH library exceeded the library size by three-fold suggesting that the diversity was well represented in the NGS output. The authors estimated the minimal functional diversity to be 1.2 × 106 individual unique clones representing just one-fifth of the original number of bacterial clones.
Application to Affinity Maturation and Epitope Mapping
Next-generation sequencing can also be used to guide selection toward high affinity clones. For example, one seminal study employed NGS to guide maturation of an scFv fragment directed against ErbB2 to a final affinity of 25 pM (resulting in a 158-fold improvement over wild type) (18). Guided by structure-based design, individual CDR regions (excluding VL CDR2) were randomized, selected against ErbB2 antigen, and analyzed by NGS before and after panning. This revealed enrichment of novel sequence motifs at diversified CDR positions, with the exception of VH CDR3, which was enriched toward the wild-type motif (suggesting an already optimal sequence). Next, the most frequent CDR substitutions were combined to generate a secondary library (VH CDR3 being reverted to wild type), which was selected against the target. This resulted in improved affinities of between 300 and 25 pM, compared with the wild-type affinity of 4 nM, highlighting the power of this stepwise approach for affinity maturation.
In a further study, deep mutational scanning analysis using NGS was performed on a humanized version of the anti-EGFR monoclonal cetuximab (42). More specifically, independent VH and VL libraries (encoding over 1,000 single amino acid substitutions at 59 different positions—32 in VH and 27 in VL) were selected by mammalian cell display and flow cytometry. Gated populations were analyzed by NGS to identify permissive mutations and to generate a heat map of the antigen binding site. Overall, this strategy identified 67 substitutions that increased affinity, including one mutation with a five-fold KD improvement. Similar strategies can also be used to map epitope surfaces, as exemplified by the interaction of S. aureus toxin with neutralizing antibodies (43).
Conclusion
Next-generation sequencing holds great promise for the development of therapeutic monoclonal antibodies, by allowing unprecedented insights into library diversity and clonal enrichment. Although current NGS platforms were not designed with antibody libraries in mind, the technologies are now at a stage where unique sequence insights into all stages of the selection process can be obtained. Moreover, with ongoing advances in sequencing technology, depth and read length is improving continuously: for instance, the PacBio Sequel system generates approximately seven times more sequences than the previous RS II system but maintains its long-read capability (Pacific Biosciences), while nanopore systems such as the MinIOn (Oxford Nanopore) offer the promise of real-time DNA sequencing in combination with ultra-long reads. We conclude that, with NGS technology evolving at a rapid pace, its importance in the sequence analyses of phage- and other antibody-display libraries is likely to continue to increase.
Author Contributions
RR wrote the manuscript. KJ, DL, and DC edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the Australian National Health and Medical Research Council (APP1090875, APP1148051, APP1113904, APP1113790) and the Australian Research Council (DP16010491).
References
1. Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science (1985) 228:1315–7. doi:10.1126/science.4001944
2. Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A (1997) 94:4937–42. doi:10.1073/pnas.94.10.4937
3. Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol (1997) 15:553–7. doi:10.1038/nbt0697-553
4. Ho M, Nagata S, Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci U S A (2006) 103:9637–42. doi:10.1073/pnas.0603653103
5. Lee CM, Iorno N, Sierro F, Christ D. Selection of human antibody fragments by phage display. Nat Protoc (2007) 2:3001–8. doi:10.1038/nprot.2007.448
6. Rouet R, Lowe D, Dudgeon K, Roome B, Schofield P, Langley D, et al. Expression of high-affinity human antibody fragments in bacteria. Nat Protoc (2012) 7:364–73. doi:10.1038/nprot.2011.448
7. Muir P, Li S, Lou S, Wang D, Spakowicz DJ, Salichos L, et al. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol (2016) 17:53. doi:10.1186/s13059-016-0917-0
8. DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol (2013) 31:166–9. doi:10.1038/nbt.2492
9. DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med (2015) 21:86–91. doi:10.1038/nm.3743
10. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science (2011) 333:1593–602. doi:10.1126/science.1207532
11. Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature (2014) 509:55–62. doi:10.1038/nature13036
12. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood (2009) 114:4099–107. doi:10.1182/blood-2009-04-217604
13. Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A (2009) 106:20216–21. doi:10.1073/pnas.0909775106
14. Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, et al. NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife (2016) 5:e16228. doi:10.7554/eLife.16228
15. Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, et al. By-passing in vitro screening – next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res (2010) 38:e193. doi:10.1093/nar/gkq789
16. Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet (2016) 17:333–51. doi:10.1038/nrg.2016.49
17. Fantini M, Pandolfini L, Lisi S, Chirichella M, Arisi I, Terrigno M, et al. Assessment of antibody library diversity through next generation sequencing and technical error compensation. PLoS One (2017) 12:e0177574. doi:10.1371/journal.pone.0177574
18. Hu D, Hu S, Wan W, Xu M, Du R, Zhao W, et al. Effective optimization of antibody affinity by phage display integrated with high-throughput DNA synthesis and sequencing technologies. PLoS One (2015) 10:e0129125. doi:10.1371/journal.pone.0129125
19. Larman HB, Xu GJ, Pavlova NN, Elledge SJ. Construction of a rationally designed antibody platform for sequencing-assisted selection. Proc Natl Acad Sci U S A (2012) 109:18523–8. doi:10.1073/pnas.1215549109
20. Peng H, Nerreter T, Chang J, Qi J, Li X, Karunadharma P, et al. Mining naive rabbit antibody repertoires by phage display for monoclonal antibodies of therapeutic utility. J Mol Biol (2017) 429(19):2954–73. doi:10.1016/j.jmb.2017.08.003
21. Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs (2013) 5:445–70. doi:10.4161/mabs.24218
22. Vollmers C, Penland L, Kanbar JN, Quake SR. Novel exons and splice variants in the human antibody heavy chain identified by single cell and single molecule sequencing. PLoS One (2015) 10:e0117050. doi:10.1371/journal.pone.0117050
23. Weinstein JA, Jiang N, White RA III, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science (2009) 324:807–10. doi:10.1126/science.1170020
24. Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity (2000) 13:37–45. doi:10.1016/S1074-7613(00)00006-6
25. Fox EJ, Reid-Bayliss KS, Emond MJ, Loeb LA. Accuracy of next generation sequencing platforms. Next Gener Seq Appl (2014) 1. doi:10.4172/jngsa.1000106
26. Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res (2017) 27:491–9. doi:10.1101/gr.209601.116
27. S. Andrews. FastQC: a quality control tool for high throughput sequencing data (2010). Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
28. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics (2011) 27:863–4. doi:10.1093/bioinformatics/btr026
29. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (2014) 30:2114–20. doi:10.1093/bioinformatics/btu170
30. Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate illumina paired-end reAd mergeR. Bioinformatics (2014) 30:614–20. doi:10.1093/bioinformatics/btt593
31. Zhang W, Wang IM, Wang C, Lin L, Chai X, Wu J, et al. IMPre: an accurate and efficient software for prediction of T- and B-cell receptor germline genes and alleles from rearranged repertoire data. Front Immunol (2016) 7:457. doi:10.3389/fimmu.2016.00457
32. Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res (2013) 41:W34–40. doi:10.1093/nar/gkt382
33. Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res (2008) 36:W503–8. doi:10.1093/nar/gkn316
34. Li S, Lefranc MP, Miles JJ, Alamyar E, Giudicelli V, Duroux P, et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun (2013) 4:2333. doi:10.1038/ncomms3333
35. Cortina-Ceballos B, Godoy-Lozano EE, Samano-Sanchez H, Aguilar-Salgado A, Velasco-Herrera Mdel C, Vargas-Chavez C, et al. Reconstructing and mining the B cell repertoire with immunediversity. MAbs (2015) 7:516–24. doi:10.1080/19420862.2015.1026502
36. Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics (2006) 22:1658–9. doi:10.1093/bioinformatics/btl158
37. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (2010) 26:2460–1. doi:10.1093/bioinformatics/btq461
38. D’Angelo S, Glanville J, Ferrara F, Naranjo L, Gleasner CD, Shen X, et al. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. MAbs (2014) 6:160–72. doi:10.4161/mabs.27105
39. Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F, et al. Deep sequencing of phage display libraries to support antibody discovery. Methods (2013) 60:99–110. doi:10.1016/j.ymeth.2013.03.001
40. Rosenfeld AM, Meng W, Luning Prak ET, Hershberg U. ImmuneDB: a system for the analysis and exploration of high-throughput adaptive immune receptor sequencing data. Bioinformatics (2017) 33:292–3. doi:10.1093/bioinformatics/btw593
41. Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One (2011) 6:e22365. doi:10.1371/journal.pone.0022365
42. Forsyth CM, Juan V, Akamatsu Y, DuBridge RB, Doan M, Ivanov AV, et al. Deep mutational scanning of an antibody against epidermal growth factor receptor using mammalian cell display and massively parallel pyrosequencing. MAbs (2013) 5:523–32. doi:10.4161/mabs.24979
Keywords: antibody display technology, next-generation sequencing, phage display, antibody libraries, in vitro selection, antibody therapeutics
Citation: Rouet R, Jackson KJL, Langley DB and Christ D (2018) Next-Generation Sequencing of Antibody Display Repertoires. Front. Immunol. 9:118. doi: 10.3389/fimmu.2018.00118
Received: 27 November 2017; Accepted: 15 January 2018;
Published: 02 February 2018
Edited by:
Prabakaran Ponraj, Intrexon, United StatesReviewed by:
Yang Feng, National Cancer Institute at Frederick, United StatesMasaki Hikida, Kyoto University, Japan
Copyright: © 2018 Rouet, Jackson, Langley and Christ. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Romain Rouet, ci5yb3VldEBnYXJ2YW4ub3JnLmF1
 Romain Rouet
Romain Rouet Katherine J. L. Jackson1
Katherine J. L. Jackson1 Daniel Christ
Daniel Christ