- 1Laboratory of Immunobiotechnology, Reference Centre for Lactobacilli (CERELA-CONICET), Tucuman, Argentina
- 2Immunobiotics Research Group, Tucuman, Argentina
- 3Food and Feed Immunology Group, Laboratory of Animal Products Chemistry, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 4Livestock Immunology Unit, International Education and Research Center for Food Agricultural Immunology (CFAI), Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 5Cell Biology Laboratory, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 6Infection Immunology Unit, International Education and Research Center for Food Agricultural Immunology (CFAI), Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
Lactobacillus rhamnosus CRL1505 and Lactobacillus plantarum CRL1506 are immunobiotic strains able to increase protection against viral intestinal infections as demonstrated in animal models and humans. To gain insight into the host–immunobiotic interaction, the transcriptomic response of porcine intestinal epithelial (PIE) cells to the challenge with viral molecular associated pattern poly(I:C) and the changes in the transcriptomic profile induced by the immunobiotics strains CRL1505 and CRL1506 were investigated in this work. By using microarray technology and reverse transcription PCR, we obtained a global overview of the immune genes involved in the innate antiviral immune response in PIE cells. Stimulation of PIE cells with poly(I:C) significantly increased the expression of IFN-α and IFN-β, several interferon-stimulated genes, cytokines, chemokines, adhesion molecules, and genes involved in prostaglandin biosynthesis. It was also determined that lactobacilli differently modulated immune gene expression in poly(I:C)-challenged PIE cells. Most notable changes were found in antiviral factors (IFN-α, IFN-β, NPLR3, OAS1, OASL, MX2, and RNASEL) and cytokines/chemokines (IL-1β, IL-6, CCL4, CCL5, and CXCL10) that were significantly increased in lactobacilli-treated PIE cells. Immunobiotics reduced the expression of IL-15 and RAE1 genes that mediate poly(I:C) inflammatory damage. In addition, lactobacilli treatments increased the expression PLA2G4A, PTGES, and PTGS2 that are involved in prostaglandin E2 biosynthesis. L. rhamnosus CRL1505 and L. plantarum CRL1506 showed quantitative and qualitative differences in their capacities to modulate the innate antiviral immune response in PIE cells, which would explain the higher capacity of the CRL1505 strain when compared to CRL1506 to protect against viral infection and inflammatory damage in vivo. These results provided valuable information for the deeper understanding of the host–immunobiotic interaction and their effect on antiviral immunity. The comprehensive transcriptomic analyses successfully identified a group of genes (IFN-β, RIG1, RNASEL, MX2, A20, IL27, CXCL5, CCL4, PTGES, and PTGER4), which can be used as prospective biomarkers for the screening of new antiviral immunobiotics in PIE cells and for the development of novel functional food and feeds, which may help to prevent viral infections.
Introduction
In the past decade, research has demonstrated that beneficial microbes with the capacity to modulate the mucosal immune system (immunobiotics) are a potential alternative to enhance resistance against viral infections. Immunobiotic lactic acid bacteria (LAB) are able to provide protection against viral infections by modulating innate and adaptive antiviral immunity. Several reports have shown that immunobiotic LAB improve protection against enteric viral infections and shorten the duration of diarrhea, reduce the number of episodes, diminish virus shedding, normalize gut permeability, and increase the production of virus-specific antibodies (1–3). Moreover, it was demonstrated that some immunobiotic strains, when orally administered, are able to increase respiratory defenses and reduce the susceptibility to respiratory viral infections improving virus clearance and diminishing inflammatory-mediated lung tissue damage (4–7).
In developing countries, viral mucosal infections such as bronchitis and diarrhea are the most common infectious diseases in children (8–10). The use of immunobiotics to improve the outcome of those viral infections has been proposed. In this regard, in a randomized controlled trial conducted by Villena et al. (4), the immunobiotic strain Lactobacillus rhamnosus CRL1505 (administered in a yogurt formulation) improved mucosal immunity and reduced the incidence and severity of intestinal and respiratory infection in children. The incidence of infectious events was reduced from 66% in the placebo group to 34% in the group that received the probiotic yogurt. Furthermore, there was also a significant reduction in the occurrence of indicators of disease severity such as fever and the need for antibiotic treatment in children receiving the probiotic yogurt (4). Studies in mice models have proved that orally administered L. rhamnosus CRL1505 improves antiviral immune responses in the intestinal mucosa (local effect) (5, 11) and the respiratory tract (distal effect) (6, 7). Of interest, it was demonstrated that these immunomodulatory capacities are strain specific since other immunobiotic strains such as Lactobacillus plantarum CRL1506 exert only local affects after oral administration (5–7, 11).
The interactions of intestinal epithelial cells (IECs) with luminal antigens and immune cells play a central role in determining the type of immune response triggered by microorganisms in the intestinal mucosa (12, 13). Therefore, by using a previously established porcine intestinal epithelial (PIE) cells that is able to respond to the dsRNA synthetic analog poly(I:C) and are permissive to rotavirus (14, 15), we aimed to evaluate the similarities and differences in the innate antiviral immune response induced by L. rhamnosus CRL1505 and L. plantarum CRL1506. We hypothesized that transcriptomic analyses using microarray technology in PIE cells could provide valuable information to gain insights in the mechanisms involved in the capacity of immunobiotics to modulate the innate antiviral immune response in the gastrointestinal tract and could provide some clues about their ability to stimulate immunity in distal mucosal sites such as the respiratory tract. Therefore, the aim of this study was to investigate the transcriptomic response of PIE cells to the challenge with viral molecular associated pattern poly(I:C) and the changes in that immunotranscriptomic profiles induced by the immunobiotics strains with antiviral capabilities L. rhamnosus CRL1505 and L. plantarum CRL1506. We obtained a global overview of the immune genes involved in the innate antiviral immune response in PIE cells that include type I interferons (IFNs), several IFN-stimulated genes (ISGs), cytokines, chemokines, adhesion molecules, and genes involved in prostaglandin biosynthesis. It was also determined that lactobacilli differently modulated immune gene expression in poly(I:C)-challenged PIE cells by increasing the expression of antiviral factors and cytokines/chemokines and reducing genes involved in poly(I:C)-mediated inflammatory damage. Moreover, the study allowed us to identify a group of genes that could be used as biomarkers for the screening of new antiviral immunobiotics in PIE cells.
Materials and Methods
PIE Cells
PIE cells are intestinal non-transformed cultured cells originally derived from intestinal epithelia isolated from an unsuckled neonatal swine (16). When PIE cells are cultured, they assume a monolayer with a cobblestone and epithelial-like morphology and with close contact between cells (14, 16, 17). PIE cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% fetal calf serum, 100 U/ml streptomycin, and 100 mg/ml penicillin at 37°C in an atmosphere of 5% CO2. PIE cells grow rapidly and are well adapted to culture conditions even without transformation or immortalization (17–19).
Microorganisms
Lactobacillus rhamnosus CRL1505 and L. plantarum CRL1506 belong to CERELA Culture Collection and were originally isolated from goat milk (19). These strains were grown in Man-Rogosa-Sharpe broth at 37°C. For immunomodulatory assays, overnight cultures were harvested by centrifugation, washed three times with sterile PBS, counted in a Petroff-Hausser counting chamber, and resuspended in DMEM until use.
Immunomodulatory Effect of Lactobacilli in PIE Cells
Evaluation of the immunomodulatory activity of L. rhamnosus CRL1505 and L. plantarum CRL1506 was performed using PIE cells as described previously (19). PIE cells were seeded at 3 × 104 cells per well in 12-well type I collagen-coated plates (Sumitomo Bakelite Co., Tokyo, Japan) and cultured for 3 days. After changing medium, lactobacilli (5 × 108 cells/ml) were added, and 48 h later, each well was washed vigorously with medium at least three times to eliminate all stimulants. Then cells were stimulated with poly(I:C) (60 μg/ml) for 3, 6, 12, or 24 h for reverse transcription (RT)-PCR studies or for 12 h for microarray studies.
Microarray Analysis
Total RNA was isolated from lactobacilli-treated and control PIE cells using PureLink RNA Mini Kit (Life Technologies Inc., Gaithersburg, MD, USA) and treated with DNase. RNA integrity of all samples were evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), using the RNA 6000 Nano Kit (20). Complementary DNA synthesis was performed using 200 ng of RNA. Hybridization with Porcine (V2) Gene Expression Microarray (Agilent Technologies) was performed at Hokkaido System Science Co. Scanning and digitization of Microarray were done by Agilent Technologies Microarray Scanner and Agilent Feature Extraction 10.7.3.1, respectively.
Data normalization and expression analysis were performed using GeneSpring software version 13.1 (Agilent Technologies). Significant genes up and downregulated in test samples [those stimulated with poly(I:C) or lactobacilli plus poly(I:C)] with respect to control samples [without poly(I:C) stimulation] were identified. Genes with significant changes in transcript abundance were selected on the basis of two criteria: a t-test P value of less than 0.05, which was considered statistically significant, and a cutoff in transcript abundance of at least twofold. Statistical analysis was conducted using the Limma package from BioConductor in R software (version 3.2.5). Results were expressed as log2 scale (log2 ratio). Genes whose expressions were log2 > 1 and P < 0.05 were annotated using PANTHER 11.1 (pantherdb.org). Genes were further analyzed according to Gene Ontology (GO) classification. Microarray data were submitted to NCBI-GEO under the accession number GSE93225.
Quantitative Expression Analysis by Two-Step Real-time Quantitative PCR (qPCR)
Two-step real-time qPCR was performed to characterize the expression of selected genes in PIE cells. Total RNA was isolated from each PIE cell sample using TRIzol reagent (Invitrogen). All cDNAs were synthesized using a Quantitect RT kit (Qiagen, Tokyo, Japan) according to the manufacturer’s recommendations. Real-time qPCR was carried out using a 7300 real-time PCR system (Applied Biosystems, Warrington, UK) and the Platinum SYBR green qPCR SuperMix uracil-DNA glycosylase with 6-carboxyl-X-rhodamine (Invitrogen). The primers used in this study were described before (19, 20). The PCR cycling conditions were 2 min at 50°C, followed by 2 min at 95°C, and then 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. The reaction mixtures contained 5 µl of sample cDNA and 15 µl of master mix, which included the sense and antisense primers. According to the minimum information for publication of quantitative real-time PCR experiments guidelines, β-actin was used as a housekeeping gene because of its high stability across porcine various tissues (14, 15, 20). Expression of β-actin was used to normalize cDNA levels for differences in total cDNA levels in the samples. The quality of the RNA in all experiments was checked by Agilent 2100 Bioanalyzer, and all samples were determined to be suitable for the qPCR assay considering values of A260/A280 and A260/A230 over 2.0 and the RIN value over 9.0.
Statistical Analysis
Statistical analyses were performed using GLM and REG procedures available in the SAS computer program (SAS, 1994). Comparisons between mean values were carried out using one-way ANOVA and Fisher’s least significant difference test. For these analyses, P values <0.05 were considered significant.
Results
Immunotranscriptomic Changes in PIE Cells after Poly(I:C) Challenge
The transcriptomic response of PIE cells to the challenge with poly(I:C) was first investigated. Microarray analysis was performed in PIE cells 12 h after the stimulation with poly(I:C). When these cells were compared with unchallenged PIE cells, it was found that there were 5,140 transcripts (representing 1,178 unique genes) and 3,359 transcripts (representing 788 unique genes) upregulated and downregulated, respectively (Figures 1A,B).
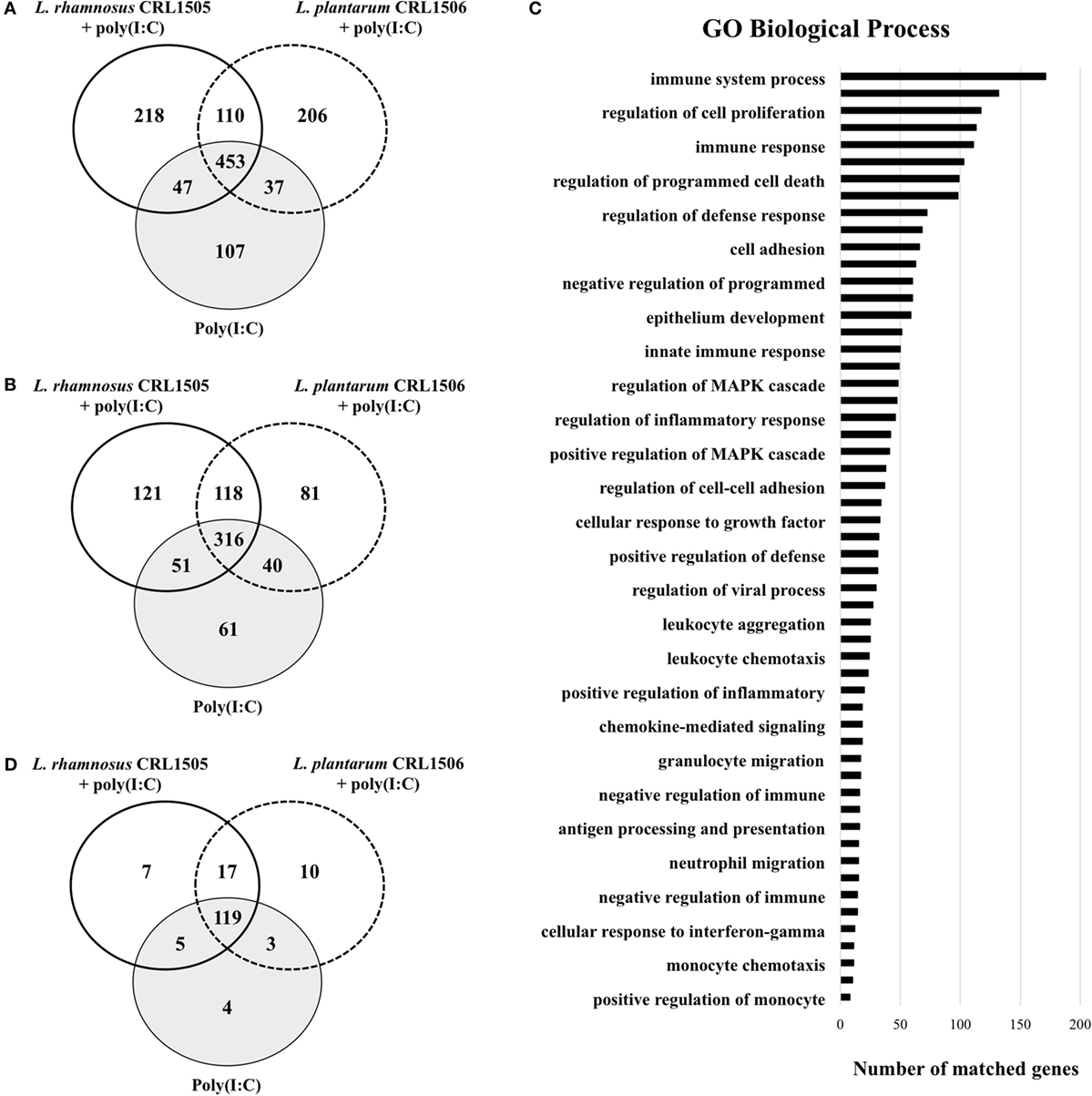
Figure 1. Differentially regulated genes in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C). Non-lactobacilli-treated PIE cells challenged with poly(I:C) were used as controls. The changes in gene expression were evaluated by comparing the three mentioned groups with unchallenged PIE cells. Venn diagrams showing the number of differentially upregulated (A) and downregulated (B) genes for each experimental group. Number of matched genes categorized according to Gene Ontology (GO) database (C). Venn diagram showing the number of differentially regulated genes that are known to have immune-related functions for each experimental group (D).
Of these differentially regulated genes, 165 were assigned to immune-related functions according to GO database (Figures 1C,D; Table S1 in Supplementary Material). Changes in the immunotranscriptome response in PIE cells after poly(I:C) stimulation included genes in the following GO Biological Process pathways: “immune system process,” “regulation of defense response,” “cell adhesion,” “innate immune response,” “regulation of viral process,” “cellular response to interferon-gamma,” and several pathways related to immune cells migration and chemotaxis (Figure 1C).
The most remarkable changes in PIE cells after stimulation with poly(I:C) were found in expression type I IFNs and antiviral factors, cytokines, chemokines, adhesion molecules, and prostaglandins.
A significant increase in the expression of IFN-β and IFN-α was observed in poly(I:C)-challenged PIE cells with fold changes (log2 ratio) of 4.3 and 3.5, respectively (Table S1 in Supplementary Material). Increased expression of the IFN-induced antiviral factors OAS1 (11.2), OASL (10.7), IFIT1 (9.9), IFIT3 (9.1), IFIT2 (8.3), MX1 (7.9), MX2 (6.3), OAS2 (6.3), IFIT5 (3.0), RNASEL (2.2), and RNASE4 (1.9) was also observed. In addition, a significant upregulation of the transcriptional regulators IRF7 (4.6), STAT1 (4.3), IRF1 (3.9), IRF9 (2.5), and STAT2 (2.2) were found in poly(I:C)-challenged PIE cells (Table S1 in Supplementary Material).
The stimulation of PIE cells with poly(I:C) significantly increased the expression of the inflammatory cytokines IL-1α (4.1), IL-6 (4.0), and IL-15 (1.8) (Table S1 in Supplementary Material). There was also a 3.9-fold increase in the expression of the sensor of the inflammasome polymeric complex NPLR3. Chemokines involved in monocyte and T lymphocyte recruitment and activation such as CXCL10 (13.2), CCL5 (8.4), CXCL9 (8.1), CCL4 (7.8), CCL20 (5.9), CCL23 (5.1), CCL28 (2.9), CCL8 (2.3), and CCL2 (2.0) were increased after poly(I:C) stimulation. In addition, we observed a significant upregulation of the chemotactic factors for neutrophils CXCL5 (2.5), CXCL11 (10.3), and CXCL8 (1.2) (Table S1 in Supplementary Material). Moreover, CSF1 (2.9) and CFS2 (2.9) that are factors able to stimulate the growth and differentiation of hematopoietic precursor cells from granulocytes and macrophages were also increased.
An upregulation of genes for adhesion molecules in PIE cells after stimulation with poly(I:C) was observed, including SELE (5.3), VCAM-1 (4.0), SELL (2.6), ICAM-1 (2.2), EPCAM (1.8), and SELP (1.8) (Table S1 in Supplementary Material). There was also a sevenfold increase in the expression of LGALS9 (galectin 9), which is involved in epithelial–lymphocytes interaction.
The microarray analysis revealed increases in the expression of several genes related to prostaglandins biosynthesis in poly(I:C)-challenged PIE cells including PTGS2 (5.0), PTGIR (3.6), PTGIS (1.6), PTGER4 (1.6), and PLA2G4A (1.2). In addition, factors belonging to the complement system were upregulated including C1R (7.5), C1S (5.7), C3 (2.9), and CFB (3.9) (Table S1 in Supplementary Material).
Changes in the expression of some pattern recognition receptors (PRRs) were detected including TLR2 (1.6), and PGLYRP2 (8.1). In addition, we detected changes in the viral innate immune receptors TLR3 (2.8), DDX58 (8.9) also known as retinoic acid inducible gene-I (RIG-I), IFIH1 (5.4) also known as melanoma differentiation associated gene-5 (MDA-5), and PKR (3.2) (Table S1 in Supplementary Material). We also detected increases in the expression of serum amyloid A2 (SAA2) (8.6).
qPCR Analysis of Selected Genes in PIE Cells after Poly(I:C) Challenge
To further evaluate gene expression changes induced by poly(I:C) in PIE cells, qPCR was performed. From the 165 immune and immune-related genes differentially regulated by poly(I:C) (Figure 1D; Table S1 in Supplementary Material), we selected 39 belonging to IFN and IFN-induced antiviral factors, cytokines, chemokines, adhesion molecules, prostaglandins, SAA2, A20, GZMA, LYZ, and trefoil factor 1 (TFF1) to be studied by qPCR. We confirmed that the direction of the changes in gene expression was in agreement with results obtained in the microarray analysis in all the studied genes.
We detected a significant increase in the expression of IFN-β and IFN-α that reached a maximum value on hour 12 after poly(I:C) stimulation (Figure 2). IRF3, RNASEL, MX1, and MX2 showed a peak on hour 12 after poly(I:C) challenge (Figure 2). Similarly, we observed increases in expression of OAS1 and OASL with peaks at hour 24 and in OAS2 with peaks between hours 6 and 12 after the poly(I:C) stimulation.
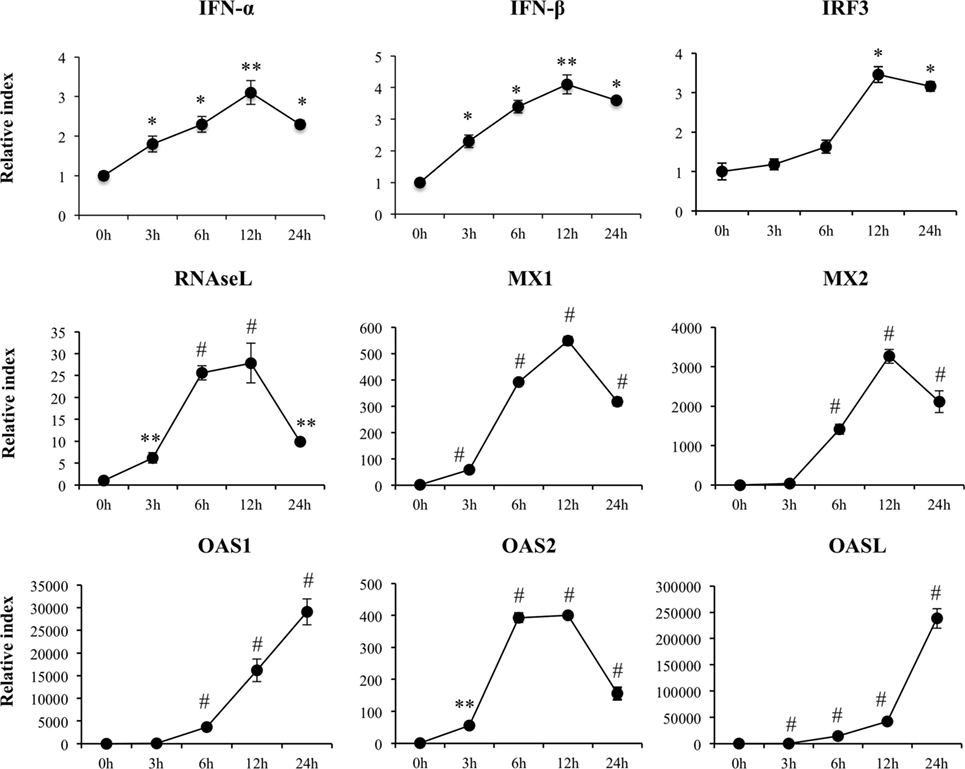
Figure 2. Expression of type I interferons (IFN-β and IFN-α), IFN regulatory factor 3, and IFN-induced antiviral genes in porcine intestinal epithelial (PIE) cells after the challenge with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. The results represent data from three independent experiments. Symbols indicate significant differences when compared to unchallenged control PIE cells (time 0 h) (*P < 0.05, **P < 0.01, #P < 0.001).
A significant increase in expression of CCL4, CCL20, CXCL2, and CXCL5 with peaks on hour 3 after poly(I:C) challenge was also detected (Figure 3). Similarly, we observed increases in expression of CCL8 and CXCL10 with peaks at hour 6 and in CCL11 and CCL5 with peaks at hours 12 and 24, respectively. In addition, expression of CCL23 increased from hour 3 and stayed in the same level between hours 6 and 24 after stimulation of PIE cells (Figure 3). CXCL14 expression was significantly reduced after poly(I:C) challenge and returned to basal levels at hour 24. Poly(I:C) also increased the expression of the adhesion molecules SELE, SELL, ICAM-1, and EPCAM (Figure S1 in Supplementary Material).
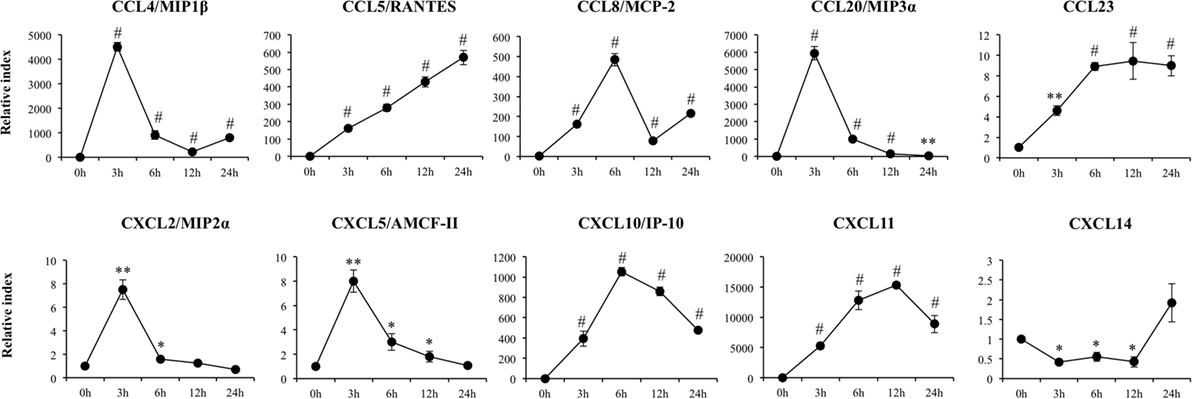
Figure 3. Expression of chemokines genes in porcine intestinal epithelial (PIE) cells after the challenge with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. The results represent data from three independent experiments. Symbols indicate significant differences when compared to unchallenged control PIE cells (time 0 h) (*P < 0.05, **P < 0.01, #P < 0.001).
Increased expression of IL-1β, IL-5, and IL-15 was observed in poly(I:C)-challenged PIE cells (Figure 4) showing all of them their highest values after 6 h of stimulation. Amphiregulin (AREG) was also increased after poly(I:C) challenge with a peak at hour 24. On the contrary, IL-9 expression was significantly reduced between hours 6 and 12 and returned to the basal levels at hour 24 (Figure 4).
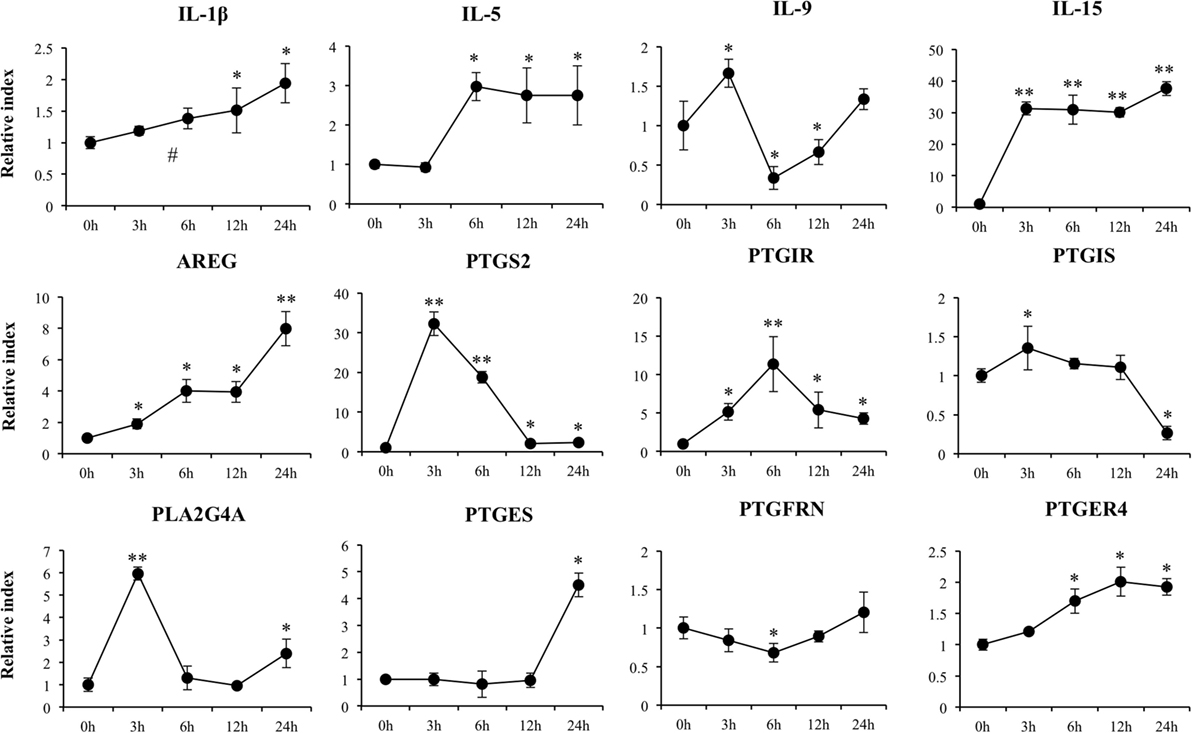
Figure 4. Expression of cytokines and genes involved in prostaglandins biosynthesis in porcine intestinal epithelial (PIE) cells after the challenge with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. The results represent data from three independent experiments. Symbols indicate significant differences when compared to unchallenged control PIE cells (time 0 h) (*P < 0.05, **P < 0.01, #P < 0.001).
We detected a significant increase in the expression of PTGS2, PTGIR, PLA2G4A, PTGES, and PTGER4 (Figure 4). In addition, a slight increase in PTGIS was observed at hour 3, and a significant downregulation occurred at hour 24. A decrease in PTGFRN between hours 3 and 12 was also observed (Figure 4).
Finally, we observed upregulation of GZMA, LYZ, TFF1, and SAA2 with peaks at hours 3, 6, 12, and 24, respectively (Figure S2 in Supplementary Material).
Modulation of Poly(I:C)-Induced Immunotranscriptome Changes in PIE Cells by Immunobiotics
Next, we analyzed microarray data to evaluate the effect of the immunobiotic strains L. rhamnosus CRL1505 and L. plantarum CRL1506 on the immunotranscriptomic response of PIE cells after the challenge with poly(I:C). For that purpose, PIE cells were stimulated with L. rhamnosus CRL1505 or L. plantarum CRL1506 and then challenged with poly(I:C). Comparative analysis of microarray profiles indicated that both CRL1505 and CRL1506 strains differentially modulated the expression of several genes related to the innate antiviral immune response in PIE cells after poly(I:C) stimulation (Table S2 in Supplementary Material).
The Venn diagram analysis was used to find genes that were uniquely and commonly modulated between lactobacilli-treated and control PIE cells (Figure 1D). Of the 165 differentially expressed genes in the Venn diagram analysis, 4 (PPARA, TFF1, STAT3, and DUOX1) were unique to the poly(I:C) challenge. Seven (TNFRSF11B, C5, LOC100127164, PIK3R5, IL27, IL17RC, and IBSP) and 10 (NFIA, CADM4, CDH24, CCR7, IL1RAPL2, TNFAIP8L2, DPEP1, CDH19, BPIFA1, and TNFSF18) unique genes were found in the CRL1505 stimulation plus poly(I:C) challenge and the CRL1506 stimulation plus poly(I:C) challenge groups, respectively. In addition, five genes (RNASE6, PROC, VTN, CCL28, and PLG) were common to CRL1505 treatment plus poly(I:C) and control, whereas three genes (IL23RA, ITGA1, and IL20RB) were common to CRL1506 treatment plus poly(I:C) and control. It was also observed that 119 genes were common to all the 3 treatments (Figure 1D). The cluster analysis in Figure S3 in Supplementary Material depicts the transcriptomic patterns of differentially modulated genes between lactobacilli-treated and control PIE cells. The treatment with CRL1505 plus poly(I:C) clustered closer to the treatment with CRL1506 plus poly(I:C) and both clustered separated from the control.
Closer examination of gene expression revealed differences in several genes sheared by immunobiotic-treated PIE cells and controls (Table S1 in Supplementary Material). Most remarkable differences were found in the genes belonging to IFN and IFN-induced antiviral factors, cytokines, chemokines, and adhesion molecules. Both lactobacilli treatment significantly increased IFN-β, IFN-α, TLR3, OAS1, OASL, MX2, RNASEL, RNASE4, and STAT5A when compared to controls. In addition, stimulation of PIE cells with L. rhamnosus CRL1505 plus poly(I:C) significantly increased the expression levels of IFIT1, IFITM1, DDX58/RIG1, IFIH1/MDA5, IRF7, STAT1, NLRP3, IRF1, STAT2, and IRF2 when compared with PIE cells stimulated only with poly(I:C).
Although expression of IL1A, IL6, IL8, AREG, CXCL10, CCL5, CCL4, CCL20, CCL23, CSF2, CCL3L1, and SELL was upregulated in lactobacilli-treated PIE cells after the challenge with poly(I:C), the increases were significantly higher when compared to control PIE cells without lactobacilli treatment (Table S1 in Supplementary Material). L. rhamnosus CRL1505 plus poly(I:C) also increased the expression levels of VEGFA, IL17RC, CXCL11, CCRL2, CXCL5, CXCL2, SELE, CDHR4, and EPCAM when compared with PIE cells stimulated only with poly(I:C), an effect that was not observed with CRL1506 treatment. Interestingly, IL27 was upregulated only in PIE cells receiving the CRL1505 strain plus poly(I:C). In addition, the expression levels of IL15 and RAE1 were reduced by lactobacilli treatments.
We also observed an increased expression of PLA2G4A, PTGES, and PTGS2 genes in lactobacilli-treated PIE cells after the challenge with poly(I:C) when compared to the control cells, whereas PTGER4 and PTGER2 were diminished in lactobacillus-treated cells (Table S1 in Supplementary Material). L. rhamnosus CRL1505 plus poly(I:C) also increased the expression levels of PTGIR.
Expression of TLR6, MYD88, NCOA1, and NFKB1 was significantly higher in lactobacilli-treated PIE cells after the challenge with poly(I:C) when compared to controls. In addition, the transcripts of other immune and immune-related genes including GZMH, TFF1, LYZ, C1R, CFB, PLG, CFD, SAA2, and NOS2 were higher in lactobacilli-treated PIE cells than controls (Table S1 in Supplementary Material). Stimulation of PIE cells with L. rhamnosus CRL1505 plus poly(I:C) significantly increased the expression levels of C1S, C3, and PLAU when compared with PIE cells stimulated only with poly(I:C).
qPCR Analysis of Selected Genes in PIE Cells after Stimulation with Immunobiotics and Poly(I:C) Challenge
To confirm the changes induced by L. rhamnosus CRL1505 and L. plantarum CRL1506 in the immunotranscriptome response of poly(I:C)-challenged PIE cells, qPCR was performed on selected genes. Genes with or without significant differences between lactobacilli-treated and non-treated PIE cells were chosen. The transcriptional changes evaluated by qPCR indicated a similar overall trend in the transcription.
Both L. rhamnosus CRL1505 and L. plantarum CRL1506 induced a significantly higher expression of IFN-α and IFN-β when compared with control poly(I:C)-challenged PIE cells (Figure 5). In addition, IRF3 and the IFN-induced antiviral factors RNASEL, MX2, OAS1, and OASL were significantly higher in lactobacilli-treated PIE cells than in controls. Furthermore, MX2 expression was higher in PIE cells treated with CRL1505 strain than those treated with CRL1506. Expression of MX1 and OAS2 in lactobacilli-treated PIE cells was not different from the control PIE cells after the challenge with poly(I:C) (Figure 5).
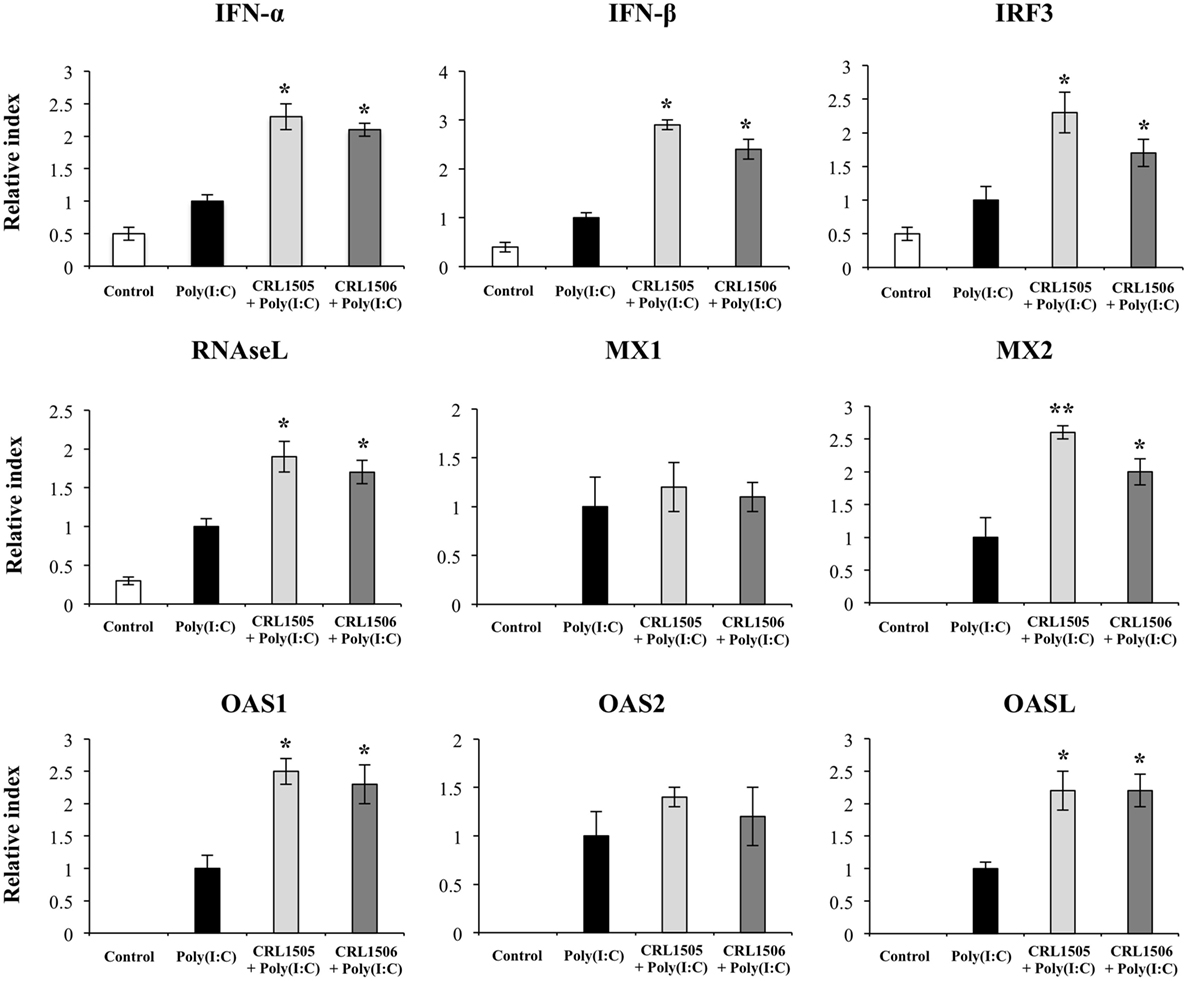
Figure 5. Expression of type I interferons (IFN-β and IFN-α), IFN regulatory factor 3, and IFN-induced antiviral genes in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. Non-lactobacilli-treated PIE cells with or without poly(I:C) challenge were used as controls. The results represent data from three independent experiments. Asterisks indicate significant differences when compared to poly(I:C)-challenged control PIE cells (*P < 0.05, **P < 0.01).
Expression of CCL8 and CXCL14 in lactobacilli-treated PIE cells was not different from the control PIE cells after the challenge with poly(I:C) (Figure 6). In contrast, the levels of CCL23, CXCL8, and SELL were significantly higher in lactobacilli-treated PIE cells when compared to the controls (Figure 6). In addition, both lactobacilli significantly increased the expression of CCL4, CCL5, CCL20, and CXCL10; however, values in L. rhamnosus CRL1505-treated PIE cells were higher than in cells treated with L. plantarum CRL1506. Only L. rhamnosus CRL1505 was able to increase the expression of CXCL2, CXCL5, CXCL11, EPCAM, ICAM-1, and SELE when compared to control PIE cells (Figure 6).
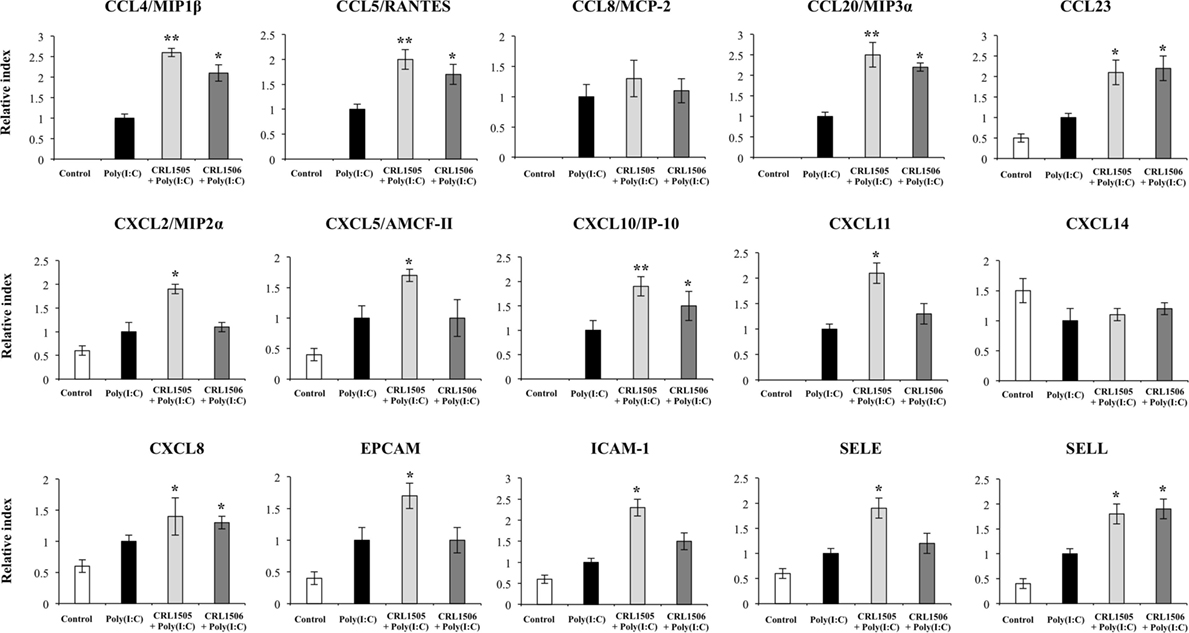
Figure 6. Expression of chemokines and adhesion molecules genes in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. Non-lactobacilli-treated PIE cells with or without poly(I:C) challenge were used as controls. The results represent data from three independent experiments. Asterisks indicate significant differences when compared to poly(I:C)-challenged control PIE cells (*P < 0.05, **P < 0.01).
In agreement with the results from our microarray analysis, both lactobacilli strains were able to increase the expression of IL-1β, IL-6, and AREG and reduce the expression of IL-15 and PTGER4, with no significant differences between them (Figure 7). Moreover, no differences in TGF-β or PTGIS were found between lactobacilli-treated and control PIE cells. Both lactobacilli significantly increased the expression of PLA2G4A, PTGES, and PTGS2; however, values in L. rhamnosus CRL1505-treated PIE cells were higher than in cells treated with L. plantarum CRL1506. In addition, only L. rhamnosus CRL1505 was able to significantly increase the expression of IL-9 and PTGIR when compared to control PIE cells (Figure 7).
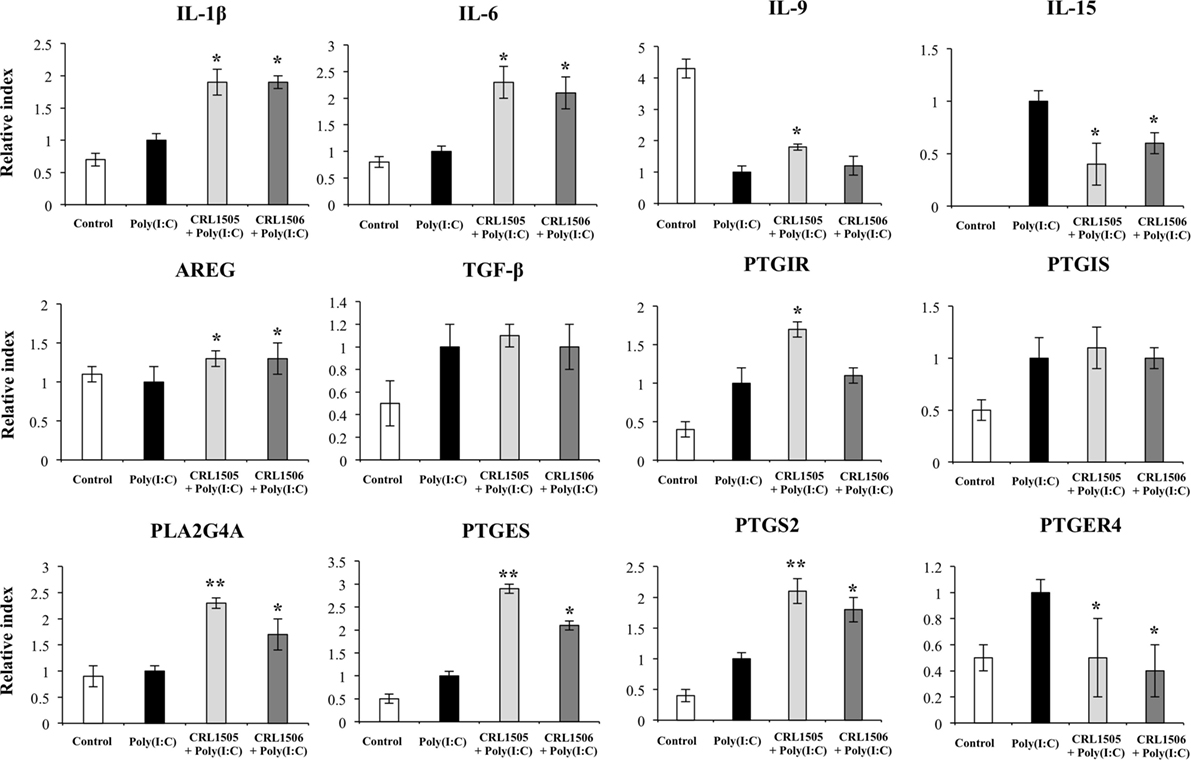
Figure 7. Expression of cytokines and genes involved in prostaglandins biosynthesis in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. Non-lactobacilli-treated PIE cells with or without poly(I:C) challenge were used as controls. The results represent data from three independent experiments. Asterisks indicate significant differences when compared to poly(I:C)-challenged control PIE cells (*P < 0.05, **P < 0.01).
Expression of TLR2 and PGLYRP2 in CRL1505- or CRL1506-tretaed PIE cells was not different from the control PIE cells after the challenge with poly(I:C). In contrast, expression levels of RIG1, TLR3, and TLR6 (Figure 8) were significantly higher in lactobacilli-treated PIE cells when compared to the controls. We also observed that A20 (TNFAIP3) was reduced in lactobacilli-treated PIE cells when compared to the controls (Figure 8). SAA2, GZMA, LYZ, TFF1, and C1R were significantly upregulated in lactobacilli-treated PIE cells when compared to the controls (Figure S4 in Supplementary Material). Only L. rhamnosus CRL1505 was able to significantly increase the expression of C3 when compared to control PIE cells, whereas both lactobacilli reduced the expression of CFB (Figure S4 in Supplementary Material).
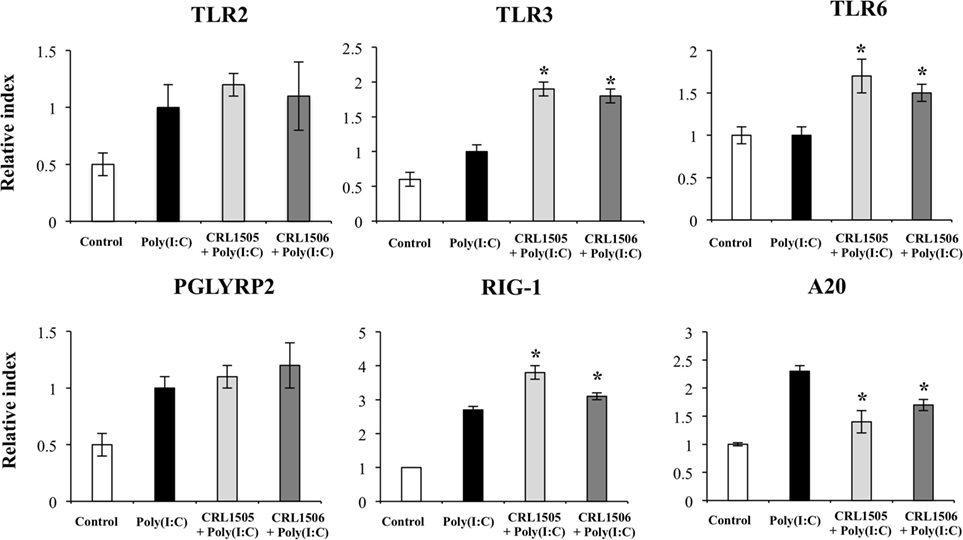
Figure 8. Expression of pattern recognition receptors and A20 genes in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. Non-lactobacilli-treated PIE cells with or without poly(I:C) challenge were used as controls. The results represent data from three independent experiments. Asterisks indicate significant differences when compared to poly(I:C)-challenged control PIE cells (*P < 0.05).
Discussion
It is known that IECs senses viral dsRNA through PRRs including TLR3, RIG-I, and MDA-5. After the recognition of dsRNA by those receptors, cellular signaling cascades are activated to react against viral infection. Antiviral PRRs activation leads to the production of cytokines, chemokines, IFNs, and IFN-regulated gene products that play a key role in establishing an antiviral state for virus clearance and restriction of spread (21).
High-throughput microarray technology has been employed for screening genes involved in the immune responses to enteric virus or poly(I:C) (22, 23). By using a human colon epithelial cell line (HT29 cells), Bagchi et al. (22) evaluated the immunotranscriptomic response of IECs to the challenge with different rotavirus strains. Microarray data revealed a set of commonly differentially regulated genes for the three rotaviruses used in that work. Of interest, several IFN inducible genes (OAS1, MX1, IL18, IITP3, TAP1, and RSAD2) as well as several cytokines and chemokines (CCL5, CXCL10, CXCL11, IL8, and CCL15) were upregulated by rotavirus infection. Later, it was observed that the stimulation of HT29 cells with poly(I:C) enhanced the expression of several genes associated with the dsRNA recognition by PRRs including antiviral factors (IRF1, ISG20, IFIT2, OASL, and STAT5), and proinflammatory cytokines (CSF1, CSF2, IL29, TNF-α, CXCL11, and CLCF1) (23). Those transcriptomic studies indicated that poly(I:C) and rotavirus induce similar innate antiviral immunotranscriptomic responses in IECs.
Previously, the response of PIE cells to poly(I:C) challenge was evaluated, and it was found that MCP-1, IL-8, TNF-α, IL-6, and both IFN-α and IFN-β were upregulated in PIE cells after stimulation (14). The suitability of PIE cells as a model for studying immune signaling pathways after rotavirus infection was also evaluated. Our results showed that PIE cells have functional TLR3, RIG-I, and MDA-5 receptors, which are able to detect rotavirus infection and enhance the expression of IFN-β and the ISGs MxA and RNase L (15), which are important antiviral effectors of IFN pathway. In this study, we corroborated and deepen those findings by using microarray technology and qPCR. We demonstrated that stimulation with poly(I:C) significantly alters gene expression profiles of PIE cells. Of the transcripts differentially modulated by poly(I:C), several were assigned to immune-related functions. Our results showed that the activation of IRF3 and NF-kB pathways in PIE cells by poly(I:C) increased the expression of IFN-α and IFN-β, several ISGs (OAS1, OASL, IFIT1, IFIT3, IFIT2, MX1, MX2, OAS2, IFIT5, RNASEL, and RNASE4), cytokines (IL-1β, IL-5, and IL-15), and chemokines (CCL4, CCL20, CXCL2, CXCL5, CCL8, CXCL10, CCL11, CCL5, and CCL23). Moreover, some adhesion molecules were also significantly upregulated in PIE cells after poly(I:C) stimulation including SELE, SELL, ICAM-1, and EPCAM. In addition, we also observed a significant upregulation of the dsRNA detection sensors TLR3, RIG1, and MDA5. This is in agreement with studies in HT29 cells showing that RIG1 was upregulated by rotavirus infection (22).
These results are in line with the transcriptomics studies mentioned before and indicate that PIE cells are able to mount a complex innate antiviral immune response involving changes needed to induce a mucosal antiviral state and promote the recruitment of inflammatory cells to the intestinal tissue, which are intended to eliminate the viral pathogen (Figure 9A). These features also exhibit that PIE cells are an excellent laboratory tool to study treatments able to favorably modulate the innate antiviral response.
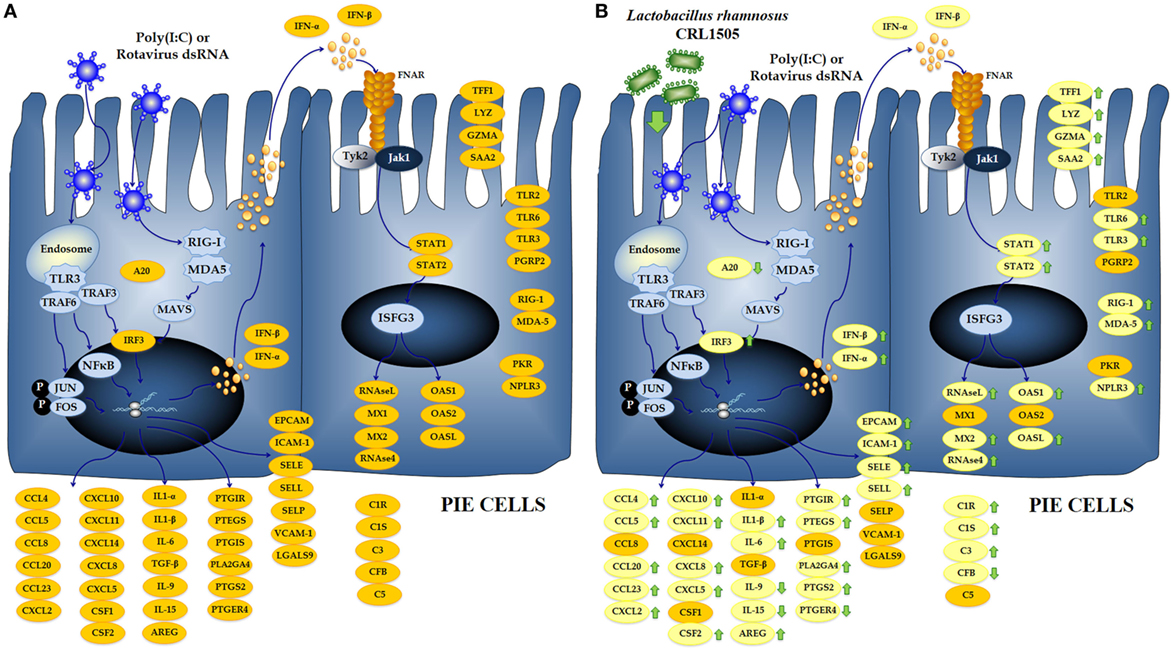
Figure 9. Global overview of the signaling pathways and immune genes differentially regulated in porcine intestinal epithelial (PIE) cells after the challenge with the viral molecular associated pattern poly(I:C) (A). Global overview of the signaling pathways and immune genes differentially regulated in PIE cells treated with Lactobacillus rhamnosus CRL1505 and challenged with poly(I:C) (B).
Several studies have shown that immunobiotics are able to beneficially modulate PRRs-mediated inflammatory response in the gut by regulating the functions of IECs (24). In this regard, our previous studies demonstrated that the immunobiotic strains L. rhamnosus CRL1505 and L. plantarum CRL1506 differentially regulated the expression of IFN-α, IFN-β, MCP-1, IL-8, and IL-6 in PIE cells after TLR3 activation (19). In line with those previous findings, we described here that the treatment of PIE cells with lactobacilli resulted in differential expression of several immune genes in response to the poly(I:C) challenge (Figure 9B), which include not only antiviral factors and cytokines as previously demonstrated but also adhesion molecules, complement factors, enzymes involved in prostaglandin biosynthesis, and PRRs. Most notable changes were found in IFN-α, IFN-β, NPLR3, OAS1, OASL, MX2, RNASEL, and RNASE4 that were significantly increased in lactobacilli-treated PIE cells when compared to the controls. It is known that RNAse L, OAS, MX, and NPLR3 are important factors for the protection of the intestinal mucosa against rotavirus infection (25–27). This finding is of interest because it confirms our previous in vitro (19) and in vivo (11) studies demonstrating the antiviral capacity of L. rhamnosus CRL1505 and L. plantarum CRL1506.
In addition, it was observed that L. rhamnosus CRL1505 and L. plantarum CRL1506 differentially regulated the expression of cytokines, chemokines, and adhesion molecules (Figure 9B). Expression levels of IL-1β, IL-6, SELL, CCL4, CCL5, CCL20, CCL23, CXCL8, and CXCL10 were higher in lactobacilli-treated PIE cells than controls. In addition, ICAM1, EPCAM, CXCL2, CXCL5, and CXCL11 were increased in CRL1505-tretated PIE cells. This pattern of cytokines/chemokines and adhesion molecules gene expression induced by lactobacilli would allow us to predict an improved recruitment and activation of immune cells to the gut mucosa, which could beneficially influence the elimination of the virus. It is also necessary to consider that in several viral infections, the excessive recruitment of inflammatory cells and/or their deregulated activation may contribute to the damage of the infected tissue rather than the resolution of the infection. It was reported that poly(I:C), when administered intraperitoneally to mice, mimics the local intestinal immune response elicited by an enteric viral infection (28, 29). Both purified dsRNA from rotavirus and poly(I:C) are able to induce severe mucosal damage in the gut via TLR3 activation including villous atrophy, mucosal erosion, and gut wall attenuation (28). It was demonstrated that TLR3 activation in IECs by poly(I:C) or rotavirus genomic dsRNA induce the expression of IL-15 and retinoic acid early inducible-1 (RAE1), which mediate epithelial destruction and mucosal injury by interacting with the NKG2D receptor expressed on CD3+NK1.1+CD8αα+ intraepithelial lymphocytes (IELs) (30). Here, we found a significant reduction in the expression of IL-15 and RAE1 in PIE cells treated with lactobacilli. This is in line with our previous work that showed that mice pretreated with immunobiotic lactobacilli responded with reduced levels of TNF-α, IL-15, RAE1, and CD3+NK1.1+CD8αα+ IELs after TLR3 activation with poly(I:C) (11). Those changes significantly diminished the inflammatory damage of the intestinal mucosa.
Our transcriptomic study indicates that other regulatory mechanisms would be improved by lactobacilli to limit the inflammatory damage during intestinal viral infection. A significant upregulation of AREG and TFF1 expression was observed in lactobacilli-treated PIE cells when compared to controls. Recently, it was demonstrated that the mucosal surfaces of lung and intestine are protected from detrimental inflammation by group 2 innate lymphoid cells (ILC2s). Monticelli et al. (31) showed that following activation with IL-33, ILC2s in the gut increased the expression of AREG, limited intestinal inflammation, and decreased disease severity in mice treated with dextran sodium sulfate. Moreover, it was reported that the number of ILC2s increased in the respiratory tract after infection influenza virus and that depletion of those cells induced impaired airway remodeling and altered lung epithelial integrity, diminishing lung function. Notably, these defects were restored by administration of AREG (32). On the other hand, TFF1 is a stable secretory protein expressed in gastrointestinal mucosa that stabilize the mucus layer and affect healing of the epithelium. By using TFF1-knockout mice, it was showed that this factor plays a critical role in maintaining mucosal integrity and regulating the pro-inflammatory response to gastrointestinal pathogens (33). Moreover, a recombinant Lactococcus lactis strain, genetically modified to secrete human TFF1, was able to reduce the severity of mucosal damage in an animal model of oral mucositis (34).
We also observed that poly(I:C) stimulation induced transcriptomic changes in several genes involved in the biosynthesis of prostaglandins and that lactobacilli treatments increased the expression of several of those genes including PLA2G4A, PTGES, and PTGS2. Upregulation of PLA2G4A and PTGES indicates that PIE cells treated with lactobacilli would increase their production of prostaglandin E2 (PGE2). It has been reported that PGE2 regulates immune function in several ways that are able to affect viral pathogenesis. Production of pro-inflammatory cytokines and chemokines by immune cells are inhibited in presence of PGE2, whereas IL-10 is enhanced (35, 36) indicating that PGE2 could have a role in the protective activity of L. rhamnosus CRL1505 and L. plantarum CRL1506 against inflammatory damage. It was also reported that PGE2 inhibits type I IFN production in epithelial and immune cells, thereby causing an increase in virus replication (37). Interestingly, the expression of PGE2 receptors (PTGER4 and PTGER2) was downregulated in PIE cells treated with the immunobiotic strains indicating that cells were protected from this effect of PGE2.
Whether the capacity of L. rhamnosus CRL1505 and L. plantarum CRL1506 to differentially modulate AREG, TFF1, and prostaglandins production is involved in their beneficial effects on intestinal or respiratory viral infections in vivo is an open question, which we propose to address in the near future.
The zinc-finger protein A20 is capable to terminate TLR signaling, which results in inhibition of NF-κB activation and reduction of inflammatory-induced cytotoxicity (38). Saitoh et al. (39) reported that IRF3 activation is suppressed by A20. The A20 protein is able to induce the suppression of the IFN-mediated immune response and IFN-promoter-dependent transcription following engagement of TLR3 by dsRNA. A20 knock down results in enhanced IRF3-dependent transcription triggered by the stimulation of TLR3 or virus infection. Furthermore, it was reported that A20 was upregulated by different rotavirus strains in HT29 cells. Interestingly, the same work demonstrated that the knock down of A20 in IECs by siRNA significantly reduced virus titers indicating that A20 is required for rotavirus infection (39). We have reported previously that two immunobiotic bacteria with antiviral capabilities, Bifidobacterium infantis MCC12 and Bifidobacterium breve MCC1274, significantly reduced the expression of A20 in rotavirus-infected PIE cells (15), which was in line with the capacity of both strains to improve IRF3 activation and IFN-β production. In line with our findings, MacPherson et al. (23) showed that the stimulation of HT29 cells with poly(I:C) alone increased the expression of A20, but the co-stimulation with poly(I:C) and probiotics significantly reduced A20 expression levels. Although our microarray analysis did not show differences between lactobacilli-treated and control PIE cells when the A20 (TNFAIP3) transcript was evaluated, qPCR analysis showed a significant reduction of A20 expression in immunobiotic-treated cells. Therefore, the reduction of A20 in IECs could be a key effect for the antiviral capabilities of immunobiotics.
Lactobacillus rhamnosus CRL1505 and L. plantarum CRL1506 showed quantitative and qualitative differences in their capacities to modulate the innate antiviral immune response in PIE cells. Higher expression levels of the antiviral factors MX2 and IFIT2 were found in CRL1505-treated PIE cells when compared to CRL1506-treated cells. Moreover, some antiviral factors were upregulated only with L. rhamnosus CRL1505 treatment including IFIT1, IFIT3, RIG-1, MDA5, NLRP3, and MSX1. As mentioned before, RIG-1, MDA5, and NLRP3 are important factors in the protection against gastrointestinal virus such as rotavirus. In addition, MSX1 (also known as HOX7) was recently identified as an important modulator of RIG-1-mediated signaling pathway with the ability to induce the activation of the TBK1 kinase and IRF3, increasing the expression of antiviral genes and improving innate antiviral responses (40). Furthermore, L. rhamnosus CRL1505 differentially regulated the expression of proinflamamtory and anti-inflammatory factors in poly(I:C)-challenged PIE cells. Higher expression of CCL4, CCL5, CCL20, and CXCL10 were found in CRL1505-treated PIE cells when compared to CRL1506-treated cells, whereas CXCL2, CXCL5, and CXCL11 were upregulated only with L. rhamnosus CRL1505 treatment, indicating a higher capacity of this strain to induce recruitment of immune cells. It also seems that the CRL1505 strain would have a higher ability to improve the regulation of the inflammatory response. We observed higher expression of PLA2G4A and PTGES that would enhance the production of the anti-inflammatory PGE2. Of interest, microarray analysis showed an increase in the expression of IL-27 in L. rhamnosus CRL1505 treatment, an effect that was not observed in the other experimental groups. IL-27 is a member of IL-12 family of cytokines that is produced mainly by myeloid cell populations, including macrophages, inflammatory monocytes, and dendritic cells, but its production has been reported in endothelial cells and epithelial cells as well (41). This cytokine has important roles in the early regulation of Th1 differentiation and the suppression of cellular activation and production of proinflammatory cytokines (42). It was demonstrated that IL-27 induces IL-10 production from both mouse and human CD4+ and CD8+ T cells and NK cells (43). Moreover, some recent studies reported a role for this cytokine in restricting virus replication (42). These effects would explain the higher capacity of the CRL1505 strain when compared to CRL1506 to protect against viral infection and inflammatory damage (5, 11, 19).
In conclusion, the genome-wide transcriptional profiling performed in this work allowed us to obtain a global overview of the expression patterns of immune and immune-related genes involved in the response of PIE cells to poly(I:C) stimulation. This study also confirmed that L. rhamnosus CRL1505 and L. plantarum CRL1506 differently modulate gene expression in poly(I:C)-challenged PIE cells inducing changes that could help to explain the antiviral activities observed in animal models and clinical trials. These results provided clues for the better understanding the mechanism underlying host–immunobiotic interaction.
The main outcome from the study is that our transcriptomic analysis successfully identified a group of genes (IFN-β, RIG1, RNASEL, MX2, A20, IL27, CXCL5, CCL4, PTGES, and PTGER4), which can be used as prospective biomarkers for the screening of new antiviral immunobiotics in PIE cells. Classically, the selection of potential immunobiotic strains is performed by studying few biomarkers in vitro, and in many cases, the selected strains do not exhibit the same immunomodulatory activity when they are evaluated later in in vivo models. Our preliminary studies indicate that the set of biomarkers found in this work allows an efficient in vitro selection of new strains with antiviral activity in PIE cells, which present antiviral activity when they are evaluated later in animal models. This efficient selection of immunobiotics could improve the development of novel functional food and feeds, which may help to prevent viral infections.
Author Contributions
HA, SA, JV, and HaK designed the study and manuscript writing. LA, HisK, HI, and NS did the laboratory work in the expression and statistical analysis. LA and JV participated in the data analysis of microarray. SS, TN, JV, and HaK contributed to data analysis and interpretation. All the authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This study was supported by a Grant-in-Aid for Scientific Research (B) (2) (No. 24380146, 16H05019), Challenging Exploratory Research (No. 23658216, 26660216, 16 K15028), and Open Partnership Joint Projects of JSPS Bilateral Joint Research Projects from the Japan Society for the Promotion of Science (JSPS) to HaK.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00057/full#supplementary-material.
Figure S1. Expression of adhesion molecules genes in porcine intestinal epithelial (PIE) cells after the challenge with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. The results represent data from three independent experiments. Symbols indicate significant differences when compared to unchallenged control PIE cells (time 0 h) (*P < 0.05, **P < 0.01).
Figure S2. Expression of trefoil factor 1, lysozyme, granzyme, and A20 genes in porcine intestinal epithelial (PIE) cells after the challenge with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. The results represent data from three independent experiments. Symbols indicate significant differences when compared to unchallenged control PIE cells (time 0 h) (*P < 0.05, **P < 0.01, #P < 0.001).
Figure S3. Heat map analysis of the differentially regulated genes in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C). Non-lactobacilli-treated PIE cells challenged with poly(I:C) were used as controls.
Figure S4. Expression of trefoil factor 1, lysozyme, granzyme, serum amyloid A2, and complement system factors genes in porcine intestinal epithelial (PIE) cells treated with immunobiotic Lactobacillus rhamnosus CRL1505 or Lactobacillus plantarum CRL1506 and challenged with the viral molecular associated pattern poly(I:C), analyzed by quantitative PCR. Non-lactobacilli-treated PIE cells with or without poly(I:C) challenge were used as controls. The results represent data from three independent experiments. Asterisks indicate significant differences when compared to poly(I:C)-challenged control PIE cells (*P < 0.05).
References
1. Basu S, Paul DK, Ganguly S, Chatterjee M, Chandra PK. Efficacy of high-dose Lactobacillus rhamnosus GG in controlling acute watery diarrhea in Indian children: a randomized controlled trial. J Clin Gastroenterol (2009) 43:208–13. doi:10.1097/MCG.0b013e31815a5780
2. Liu F, Li G, Wen K, Bui T, Cao D, Zhang Y, et al. Porcine small intestinal epithelial cell line (IPEC-J2) of rotavirus infection as a new model for the study of innate immune responses to rotaviruses and probiotics. Viral Immunol (2010) 23:135–49. doi:10.1089/vim.2009.0088
3. Maragkoudakis PA, Chingwaru W, Gradisnik L, Tsakalidou E, Cencic A. Lactic acid bacteria efficiently protect human and animal intestinal epithelial and immune cells from enteric virus infection. Int J Food Microbiol (2010) 141:S91–7. doi:10.1016/j.ijfoodmicro.2009.12.024
4. Villena J, Salva S, Nuñez M, Corzo J, Tolaba R, Faedda J, et al. Probiotics for everyone! The novel immunobiotic Lactobacillus rhamnosus CRL1505 and the beginning of Social Probiotic Programs in Argentina. Int J Biotechnol Wellness Industries (2012) 1:189–98. doi:10.6000/1927-3037/2012.01.03.05
5. Villena J, Chiba E, Tomosada Y, Salva S, Marranzino G, Kitazawa H, et al. Orally administered Lactobacillus rhamnosus modulates the respiratory immune response triggered by the viral pathogen-associated molecular pattern poly(I:C). BMC Immunol (2012) 13:53. doi:10.1186/1471-2172-13-53
6. Chiba E, Tomosada Y, Vizoso-Pinto MG, Salva S, Takahashi T, Tsukida K, et al. Immunobiotic Lactobacillus rhamnosus improves resistance of infant mice against respiratory syncytial virus infection. Int Immunopharmacol (2013) 17(2):373–82. doi:10.1016/j.intimp.2013.06.024
7. Zelaya H, Tsukida K, Chiba E, Marranzino G, Alvarez S, Kitazawa H, et al. Immunobiotic lactobacilli reduce viral-associated pulmonary damage through the modulation of inflammation–coagulation interactions. Int Immunopharmacol (2014) 19:161–73. doi:10.1016/j.intimp.2013.12.020
8. Bardach A, Ciapponi A, Garcia-Marti S, Glujovsky D, Mazzoni A, Fayad A, et al. Epidemiology of acute otitis media in children of Latin America and the Caribbean: a systematic review and meta-analysis. Int J Pediatr Otorhinolaryngol (2011) 75:1062–70. doi:10.1016/j.ijporl.2011.05.014
9. Edmond K, Scott S, Korczak V, Ward C, Sanderson C, Theodoratou E, et al. Long term sequelae from childhood pneumonia; systematic review and meta-analysis. PLoS One (2012) 7:e31239. doi:10.1371/journal.pone.0031239
10. Gentile A, Bardach A, Ciapponi A, Garcia-Marti S, Aruj P, Glujovsky D, et al. Epidemiology of community-acquired pneumonia in children of Latin America and the Caribbean: a systematic review and meta-analysis. Int J Infect Dis (2012) 16:e5–15. doi:10.1016/j.ijid.2011.09.013
11. Tada A, Zelaya H, Clua P, Salva S, Alvarez S, Kitazawa H, et al. Immunobiotic Lactobacillus strains reduce small intestinal injury induced by intraepithelial lymphocytes after toll-like receptor 3 activation. Inflamm Res (2016) 65:771–83. doi:10.1007/s00011-016-0957-7
12. Parashar UD, Gibson CJ, Bresee JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis (2006) 12:304–6. doi:10.3201/eid1202.050006
13. Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology (2009) 136:1939–51. doi:10.1053/j.gastro.2009.02.076
14. Hosoya S, Villena J, Shimazu T, Tohno M, Fujie H, Chiba E, et al. Immunobiotic lactic acid bacteria beneficially regulate immune response triggered by poly(I:C) in porcine intestinal epithelial cells. Vet Res (2011) 42:111. doi:10.1186/1297-9716-42-111
15. Ishizuka T, Kanmani P, Kobayashi H, Miyazaki A, Soma J, Suda Y, et al. Immunobiotic Bifidobacteria strains modulate rotavirus immune response in porcine intestinal epitheliocytes via pattern recognition receptor signaling. PLoS One (2016) 11:e0152416. doi:10.1371/journal.pone.0152416
16. Moue M, Tohno M, Shimazu T, Kido T, Aso H, Saito T, et al. Toll-like receptor 4 and cytokine expression involved in functional immune response in an originally established porcine intestinal epitheliocyte cell line. Biochim Biophys Acta (2008) 1780:134–44. doi:10.1016/j.bbagen.2007.11.006
17. Shimazu T, Villena J, Tohno M, Fujie H, Hosoya S, Shimosato T, et al. Immunobiotic Lactobacillus jensenii elicits anti-inflammatory activity in porcine intestinal epithelial cells by modulating negative regulators of the toll-like receptor signaling pathway. Infect Immun (2012) 80:276–88. doi:10.1128/IAI.05729-11
18. Tomosada Y, Villena J, Murata K, Chiba E, Shimazu T, Aso H, et al. Immunoregulatory effect of Bifidobacteria strains in porcine intestinal epithelial cells through modulation of ubiquitin-editing enzyme A20 expression. PLoS One (2013) 8:e59259. doi:10.1371/journal.pone.0059259
19. Villena J, Chiba E, Vizoso-Pinto MG, Tomosada Y, Takahashi T, Ishizuka T, et al. Immunobiotic Lactobacillus rhamnosus strains differentially modulate antiviral immune response in porcine intestinal epithelial and antigen presenting cells. BMC Microbiol (2014) 14:126. doi:10.1186/1471-2180-14-126
20. Kobayashi H, Albarracin L, Sato N, Kanmani P, Kober AK, Ikeda-Ohtsubo W, et al. Modulation of porcine intestinal epitheliocytes immunotranscriptome response by Lactobacillus jensenii TL2937. Benef Microbes (2016) 7(5):769–82. doi:10.3920/BM2016.0095
21. Kitazawa H, Villena J. Modulation of respiratory TLR3-antiviral response by probiotic microorganisms: lessons learned from Lactobacillus rhamnosus CRL1505. Front Immunol (2014) 5:201. doi:10.3389/fimmu.2014.00201
22. Bagchi P, Nandi S, Chattopadhyay S, Bhowmick R, Halder UC, Nayak MK, et al. Identification of common human host genes involved in pathogenesis of different rotavirus strains: an attempt to recognize probable antiviral targets. Virus Res (2012) 169:144–53. doi:10.1016/j.virusres.2012.07.021
23. Macpherson C, Audy J, Mathieu O, Tompkins TA. Multistrain probiotic modulation of intestinal epithelial cells’ immune response to a double-stranded RNA ligand, poly(I-C). Appl Environ Microbiol (2014) 80:1692–700. doi:10.1128/AEM.03411-13
24. Villena J, Kitazawa H. Modulation of intestinal TLR4-inflammatory signalling pathways by probiotic microorganisms: lessons learned from Lactobacillus jensenii TL2937. Front Immunol (2014) 4:512. doi:10.3389/fimmu.2013.00512
25. López S, Arias CF. Rotavirus-host cell interactions: an arms race. Curr Opin Virol (2012) 2:389–98. doi:10.1016/j.coviro.2012.05.001
26. Sánchez-Tacuba L, Rojas M, Arias CF, López S. Rotavirus controls activation of the 2’-5’-oligoadenylate synthetase/RNase L pathway using at least two distinct mechanisms. J Virol (2015) 89:12145–53. doi:10.1128/JVI.01874-15
27. Di Fiore IJ, Holloway G, Coulson BS. Innate immune responses to rotavirus infection in macrophages depend on MAVS but involve neither the NLRP3 inflammasome nor JNK and p38 signaling pathways. Virus Res (2015) 208:89–97. doi:10.1016/j.virusres.2015.06.004
28. Zhou R, Wei H, Sun R, Tian Z. Recognition of double-stranded RNA by TLR3 induces severe small intestinal injury in mice. J Immunol (2007) 178:4548–56. doi:10.4049/jimmunol.178.7.4548
29. Araya RE, Jury J, Bondar C, Verdu EF, Chirdo FG. Intraluminal administration of poly I:C causes an enteropathy that is exacerbated by administration of oral dietary antigen. PLoS One (2014) 9:e99236. doi:10.1371/journal.pone.0099236
30. Zhou R, Wei H, Sun R, Zhang J, Tian Z. NKG2D recognition mediates toll-like receptor 3 signaling-induced breakdown of epithelial homeostasis in the small intestines of mice. Proc Natl Acad Sci U S A (2007) 104:7512–5. doi:10.1073/pnas.0700822104
31. Monticelli LA, Osborne LC, Noti M, Tran SV, Zaiss DM, Artis D. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci U S A (2015) 112:10762–7. doi:10.1073/pnas.1509070112
32. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol (2011) 12:1045–1045. doi:10.1031/ni.2131
33. Soutto M, Chen Z, Katsha AM, Romero-Gallo J, Krishna US, Piazuelo MB, et al. Trefoil factor 1 expression suppresses Helicobacter pylori-induced inflammation in gastric carcinogenesis. Cancer (2015) 121:4348–58. doi:10.1002/cncr.29644
34. Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, Vanhoenacker P, Corveleyn S, et al. AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol (2010) 46:564–70. doi:10.1016/j.oraloncology.2010.04.008
35. Kunkel SL, Spengler M, May MA, Spengler R, Larrick J, Remick D. Prostaglandin E2 regulates macrophage-derived tumor necrosis factor gene expression. J Biol Chem (1988) 263:5380–4.
36. Harizi H, Juzan M, Pitard V, Moreau JF, Gualde N. Cyclooxygenase-2-issued prostaglandin e(2) enhances the production of endogenous IL-10, which down-regulates dendritic cell functions. J Immunol (2002) 168:2255–63. doi:10.4049/jimmunol.168.5.2255
37. Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, et al. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity (2014) 40(4):554–68. doi:10.1016/j.immuni.2014.02.013
38. Ning S, Pagano JS. The A20 deubiquitinase activity negatively regulates LMP1 activation of IRF7. J Virol (2010) 84:6130–8. doi:10.1128/JVI.00364-10
39. Saitoh T, Yamamoto M, Miyagishi M, Taira K, Nakanishi M, Fujita T, et al. A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol (2005) 174:1507–12. doi:10.4049/jimmunol.174.3.1507
40. Chen LT, Hu MM, Xu ZS, Liu Y, Shu HB. MSX1 modulates RLR-mediated innate antiviral signaling by facilitating assembly of TBK1-associated complexes. J Immunol (2016) 197:199–207. doi:10.4049/jimmunol.1600039
41. Hall AO, Silver JS, Hunter CA. The immunobiology of IL-27. Adv Immunol (2012) 115:1–44. doi:10.1016/B978-0-12-394299-9.00001-1
42. Duan Y, Jia Y, Wang T, Wang Y, Han X, Liu L. Potent therapeutic target of inflammation, virus and tumor: focus on interleukin-27. Int Immunopharmacol (2015) 26:139–46. doi:10.1016/j.intimp.2015.03.016
Keywords: intestinal epithelial cells, immunotranscriptomic response, TLR3, Lactobacillus rhamnosus CRL1505, Lactobacillus plantarum CRL1506, antiviral response
Citation: Albarracin L, Kobayashi H, Iida H, Sato N, Nochi T, Aso H, Salva S, Alvarez S, Kitazawa H and Villena J (2017) Transcriptomic Analysis of the Innate Antiviral Immune Response in Porcine Intestinal Epithelial Cells: Influence of Immunobiotic Lactobacilli. Front. Immunol. 8:57. doi: 10.3389/fimmu.2017.00057
Received: 15 November 2016; Accepted: 16 January 2017;
Published: 02 February 2017
Edited by:
Juarez Antonio Simões Quaresma, Federal University of Pará, BrazilReviewed by:
Kieran G. Meade, Teagasc – The Irish Agriculture and Food Development Authority, IrelandHaitao Guo, University of North Carolina at Chapel Hill, USA
Copyright: © 2017 Albarracin, Kobayashi, Iida, Sato, Nochi, Aso, Salva, Alvarez, Kitazawa and Villena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haruki Kitazawa, aGFydWtpLmtpdGF6YXdhLmM3QHRvaG9rdS5hYy5qcA==;
Julio Villena, amN2aWxsZW5hQGNlcmVsYS5vcmcuYXI=
†These authors have contributed equally to this work.
 Leonardo Albarracin
Leonardo Albarracin Hisakazu Kobayashi3,4†
Hisakazu Kobayashi3,4† Tomonori Nochi
Tomonori Nochi Hisashi Aso
Hisashi Aso Susana Salva
Susana Salva Susana Alvarez
Susana Alvarez Haruki Kitazawa
Haruki Kitazawa Julio Villena
Julio Villena