- Eden Project University Centre, Cornwall College, Bodelva, Cornwall, United Kingdom
Monitoring oomycete populations and communities in bodies of water is vital in developing our understanding of this important group of fungus-like protists that contains many serious pathogens of both crops and wild plants. The methodologies involved in monitoring oomycetes are often presented as a developmental hierarchy, progressing from ‘traditional’ culture-based techniques through immunological techniques and basic PCR to qPCR and metagenomics. Here, techniques are assessed according to the roles they can perform in relation to four stages of the monitoring process: capture, detection and identification, viability determination, and quantification. Possible synergies are then considered for the combined use of different techniques in addressing the various needs relating to different questions asked of monitoring, with an emphasis on the continuing value of cultural and immunodiagnostic procedures. Additionally, the exciting future presented by the ongoing development and improvement of metabarcoding and the use of high throughput sequencing techniques in the measurement and monitoring of oomycete inoculum to determine and mitigate plant disease risks is addressed.
1 Introduction
The Oomycetes are a diverse group of filamentous protists in the clade Stramenopiles. As indicated by their old name, ‘the water molds’, oomycetes generally thrive in aquatic environments, with free water vital for their dispersal and supporting a major part of their asexual lifecycles (Hüberli et al., 2013). Many members of the oomycetes are saprotrophic, but a significant proportion of species are also endophytic and/or parasitic. It is claimed that oomycetes are likely all ‘hard wired’ for parasitism (Beakes et al., 2012) with many early divergent genera in their phylogenetic tree being marine parasites of a diverse range of organisms. In water used for horticulture, members of the orders Peronsporales, Pythiales, and Saprolegniales are of particular importance containing many important plant pathogen genera capable of causing devastating diseases in crops, ornamentals, and native plants (Kamoun, 2009). Phytophthora and Pythium are probably the most notorious oomycete genera and have received the greatest attention. In horticultural practice water used for irrigation, is obtained from a wide range of sources including boreholes and wells, roof-harvested rainwater, water from rivers, lakes and ponds as well as re-cycled hydroponics feed solutions and run-off or tail-water retrieved from production beds (Moorman et al., 2014). The potential risks of contamination of such water sources with plant pathogen inoculum can vary enormously (Moorman et al., 2014) and it is important to monitor water entering and leaving nurseries and gardens for oomycetes to guide management and regulation of disease risks, both in terms of potential crop loss and the threats of greater damage from spread to the wider environment (Jung et al., 2016; Barwell et al., 2020; Green et al., 2021; Dale et al., 2022). The continued development and refinement of measuring and monitoring techniques for oomycetes is essential for building understanding of their lifecycles, dispersal, etiology, and potential environmental impacts. Greater understanding of these factors will help determination and mitigation of disease risks and reduce damage and economic losses. For valid determinations of disease risks, accurate identification, and quantification of inoculum in time and space and in relation to survival, infection, and disease development are vital. The primary oomycete propagules in water are likely sporangia (when caducous), zoospores, and zoospore cysts (Weste, 1983). A great deal is understood about zoospore behavior, especially in the prelude to infection (Judelson and Blanco, 2005; Bassani et al., 2020), although much of this work focusses on interactions within the soil and the rhizosphere and it has often been assumed to be similar in water. However, in water, there are extra factors that are rarely considered concerning zoospore activity in larger bodies of water, where repetitional diplanetism (Drechsler, 1930; Moralejo and Descals, 2011) and zoospore cysts may play an important role in long-distance movement and survival.
Over 400 Phytophthora and Pythium and allied species have now been identified. Only a relatively small proportion of these are readily isolated and regularly seen in water samples (65+ including species of Phytophthora, Pythium, Phytopythium, and Halophytophora: Hwang et al., 2008; Reeser et al., 2011; Hüberli et al., 2013; Huai et al., 2013; Nagel et al., 2013; Zappia et al., 2014; Choudhary et al., 2016; Stamler et al., 2016; Nam and Choi, 2019; Redekar et al., 2019; Riolo et al., 2020). However, it seems likely that many more species are in fact able to spread in bodies of water. During the process of zoospore aggregation prior to root infection, there is evidence of interspecific cooperation (Kong et al., 2010), which may explain why some widespread oomycete pathogens are less frequently detected in water but still appear in root infections. There may be some merit in comparing species mixes in inoculation experiments, which by and large only assess single introductions of single putative pathogens (Hong, 2014).
The techniques developed and used for the monitoring and measurement of oomycetes in water are essentially very similar to, or the same as those developed for other media such as growing substrates, soils or colonised plant tissues. This review refers were possible to published methods applied to test and measure water. Techniques are first outlined in relation to four proposed stages of water assessment. Their varying applicability to water testing scenarios addressing different questions is then considered with the aim of encouraging the use of multiple methods and including culturing techniques where possible.
2 Stages in the process of monitoring oomycetes in water
Effective monitoring and measurement of oomycetes in water can be broken down into four stages: 1) capture, 2) detection and identification, 3) determination of viability, and 4) quantification. Each of these stages and the techniques applicable to them are considered in more detail below and summarized in Figure 1. The capture of detectable oomycete biomass from water is considered as a separate process here because of its high importance in the monitoring of water, even though some capture techniques such as baiting in many circumstances are also effective for detection in their own right which is in contrast to capture approaches like filtration that are purely effective for the physical collection of particles from water. A number of different very sensitive approaches can be used for detection, while relatively recent developments, especially in the field of nucleotide-based chemistries, have greatly expanded the scope for identifications, quantification, and the exploration of contextual information such as wider microbiomes (for example Ruiz Gómez et al., 2019). This exploration is opening up great new opportunities for understanding pathosystems, although such methods still have problems with determinations of viability. There is always room for improvement, but the range of methods now possible means the investigator can develop and tailor an approach to monitoring oomycetes based on the nature of the measurement question(s) being asked (Figure 2).
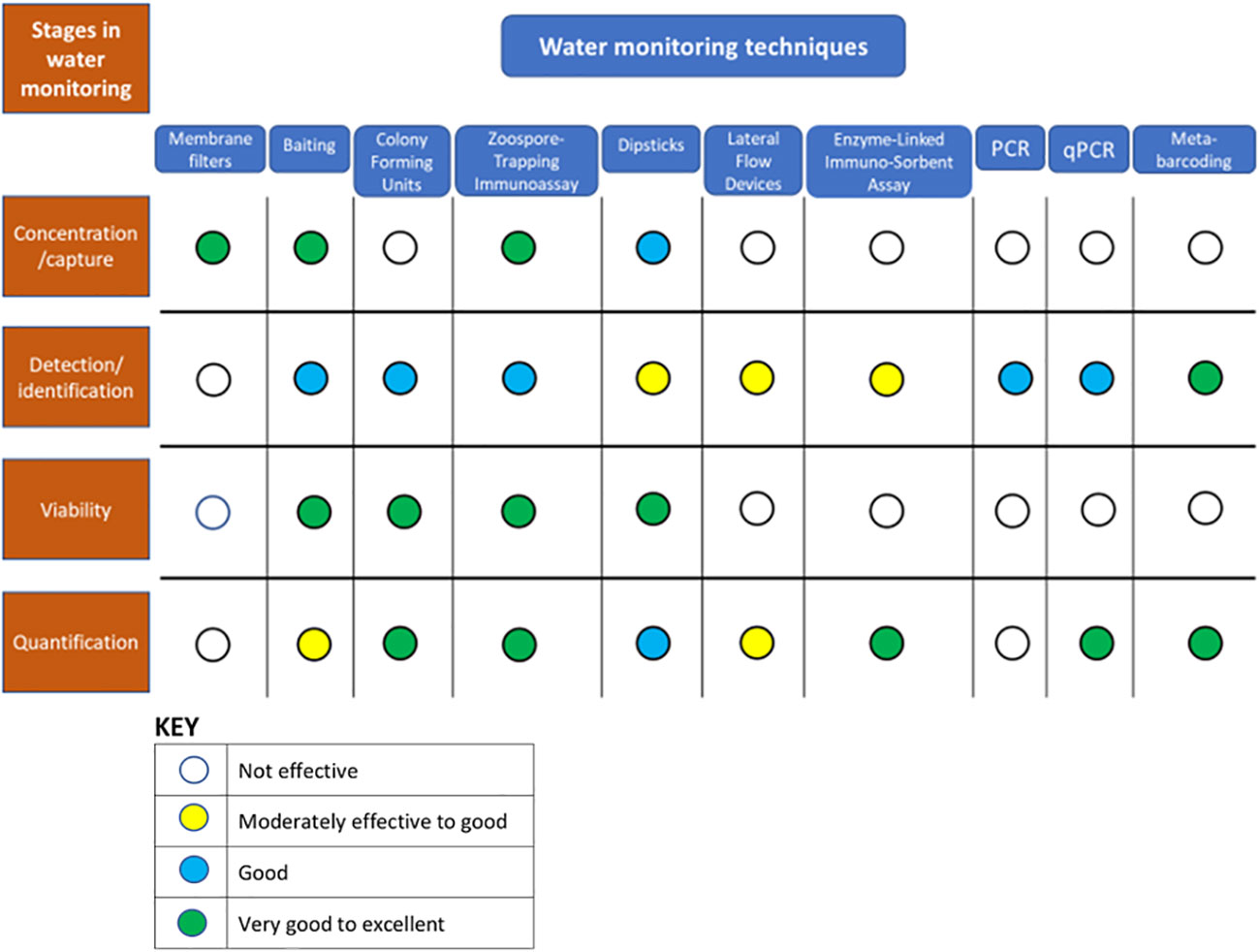
Figure 1 Consideration of the process of monitoring oomycetes in water samples as a series of stages: capture, detection/identification, viability assessment and quantification, and the relative suitability of a range of water monitoring techniques for each of these stages.
2.1 Capture
The central challenge to successful monitoring of aquatic oomycetes is that the amount of target biomass is very small and potentially widely and unevenly dispersed amongst many other taxa and debris in large volumes of water. Effective capture of oomycete biomass from water is essential and is currently achieved largely either by baiting or by filtration methods. Other potential methods include centrifugation and magnetic antibody concentration. Centrifugation has been deployed to extract viable Phytophthora zoospores (most likely as zoospore cysts after centrifugation) from hardy nursery-stock irrigation water (Middleton, 1985). Unwieldy, involving the separation of larger samples into 250 ml aliquots for centrifugation and plating out resuspended pellets onto agar growing media, this method is time-consuming and not efficient for large numbers of samples. In comparison trials, centrifugation was also found to be less effective than other capture methods for molecular detection of Phytophthora in water samples (Scibetta et al., 2012). Immunomagnetic separation, is a process that can be either direct or indirect. Direct immunomagnetic separation deploys target-specific antibodies directly conjugated to super paramagnetic spheres (Cheng et al., 2010; Kennedy and Wakeham, 2013), whereas indirect deploys mouse antibodies raised to the target and then uses an anti-mouse antibody conjugated to magnetic spheres to bind to these. In both cases, the antibodies are introduced to samples where they can bind to target biomass and can then be isolated from the samples for further diagnostic tests by exposure to a magnetic field. Despite showing efficacy in the extraction of cysts of other pathogenic protists from water samples (e.g. Cryptosporidium, Campbell and Smith, 1997, and Giardia lamblia, Bifulco and Schaefer, 1993), this approach has not yet been reported for the successful capture of oomycete propagules from environmental water samples.
2.1.1 Baiting
Since the 1960s, use has been made, primarily of plant tissues, to act as baits to attract oomycete spores out of bodies of water via chemo-attraction and chemotaxis. Many different nuances of the baiting approach exist, and the earliest of these techniques used for Phytophthora species have been comprehensively catalogued by Ribeiro (1978). As they are low in cost, straightforward, and highly adaptable, baiting techniques are still widely developed and successfully used for the capture of oomycetes in water. The baits selected for an assay strongly influence the resulting catch: for example, autoclaved insect parts are likely to attract Saprolegniales and some Pythium species (Sarowar et al., 2013), whereas fresh, whole rhododendron leaves work well for Phytophthora species in temperate streams and hardy nursery-stock irrigation systems (Themann and Werres, 1998; Themann et al., 2002). Green et al. (2020) effectively deployed a mix of different fresh leaf species as baits in a study investigating the range of Phytophthora species present in public parks, whilst in comparative trials in Australia, primarily looking at soils, a multiple bait system used by the Centre for Phytophthora Science and Management was the most effective baiting procedure (Burgess et al., 2021). The age, quality, and physiological state of the plant tissues being used as baits are hugely influential on the results obtained (Themann et al., 2002; Hüberli et al., 2013; Werres et al., 2014), and inclusion of dead tissues within a bait mix can increase the number of species captured (Wielgoss et al., 2009; Aram and Rizzo, 2018; Sarker et al., 2023a). When baiting is deployed in comparative experiments over time, variability can be reduced by using more controllable tissues such as seedlings or seedling parts (Banihashemi and Mitchell, 1975; Erwin and Ribeiro, 1996; Pettitt et al., 1998). Perhaps the ultimate in reproducible baits use just chemical attractants and do not have to rely on maintaining consistency of plant material. This principle was successfully deployed using phenols, alcohols, and amino acids in the development of chemo-attractive dipsticks to detect viable Phytophthora cinnamomi, and later P. nicotianae, zoospores (Cahill and Hardham, 1994a and Cahill and Hardham, 1994b; Gautam et al., 1999). Nevertheless, plant tissue-based baits continue to predominate and be widely and successfully deployed for the capture of oomycetes from water.
Irrespective of the materials used for baits, there are two main approaches to baiting water; in situ and ex situ (Werres et al., 2014). In situ baiting is probably the most frequently deployed and involves placing baits directly within the bodies of water being monitored. Examples include floating bait leaves (Huai et al., 2013; Matsiakh et al., 2016), or fruits (Rodríguez et al., 2021) on pond, stream, or river surfaces for 3-10 days before collection and isolating from lesions. Ex situ baiting, on the other hand, involves the placement of baits in collected and contained samples away from the sampling location, ideally under standardized incubation conditions to encourage zoospore release/re-release and infectivity (Sarker et al., 2021). Redekar et al. (2020) used ex situ baiting as a method for viable propagule capture as part of their sampling routine for metabarcoding analysis of irrigation water at a horticultural nursery in southern California, USA. They placed fresh leaves of Rhododendron catawbiense ‘Grandiflorum’ in 1L water samples and extracted DNA for analysis from lesions formed following incubation for 3 days at 18-22°C. Looking for lesions on harvested baits moves from using baits for capture to the first stages of detection especially of potentially phytopathogenic oomycete species. Only selecting bait lesions runs the risk of bias in favor of detecting only species capable of rapid infection and symptom induction in the bait species used, and when assessed, asymptomatic baits are often found to have captured a wider range of oomycete species (Sarker et al., 2023b). In situ and ex situ baiting of bodies of water differ both temporally and volumetrically. They are temporally different in that ex situ baits test samples taken from locations at specific times that consequentially represent very short time periods (‘freeze frames’). On the other hand, in situ baits might be considered to be sampling their locations for as long as they remain in situ, although their attractiveness and capacity for inoculum capture will likely vary over time depending on their physical condition. Being limited to restricted to pre-set sample volumes, ex situ baiting differs volumetrically to in situ where baits are likely to be capturing inoculum from unconfined and, depending on the movement of the sampled water body, more variable and potentially of much larger volume. In soils, recent work has shown that ex situ baiting of many small samples as opposed to the generally-preferred large pooled samples can increase the diversity of oomycete species captured (Sarker et al., 2023a; Sarker et al., 2023b). This result is thought to be due to the dilution effect of smaller samples reducing the probability of slower-growing or less aggressive species being out competed in bait infection. A similar effect might be observed in ex situ baiting of water samples where it is often assumed that the bulk of inoculum present is motile and infective already and not subject to the limitations of sporulation and release associated with soil. Nevertheless, variation in the aggressiveness of this water-borne inoculum is likely. Additionally, possible bait-induced release of zoospores from caducous sporangia and viable cysts and even the activation of mycelium or oospores in debris, make similar studies with ex situ water baiting worthy of investigation.
In soils, the rates of sporangium formation and zoospore release of different species of Phytophthora have been demonstrated to affect the efficacy of their detection by baits (Sarker et al., 2021). This process might be reflected in water, where inoculum release from infected plants is often in short-term surges (Hallett and Dick, 1981; Pettitt et al., 2015). These inoculum surges are often initiated by changes in the environment such as increased irrigation frequency or by rainfall (Ristaino, 1991; Café-Filho et al., 1995), but can also result from subtle changes in cultural practice, for example sudden reductions in the root zone temperature in hydroponics crops (Kennedy and Pegg, 1990). There are large seasonal variations in the numbers of oomycete propagules seen in irrigation water in the UK (Pettitt et al., 2015), some likely driven by seasonal changes in water temperature (Pettitt and Skjøth, 2016). A fairly consistent double peak in general oomycete CFU counts is seen in clinic samples from UK nurseries, with larger counts in late Spring and in late Summer-Early Autumn and a decline in mid-Summer (Pettitt et al., 2015). This result is in contrast to total filamentous fungal counts and the numbers of Fusarium CFU, which generally reach a single peak in August/September. A similar distribution with distinct peaks of detected CFU in Spring and Autumn was observed in citrus orchard soils (Cacciola and Magnano di San Lio, 2008), and interestingly this represents the annual progress curves of two different species of Phytophthora (P. citrophthora and P. nicotianae).
2.1.2 Filtration
The main alternative to using baits for capture, is to extract oomycete propagules from water by filtration. This approach was initially developed as a precursor to plating viable propagules onto semi-selective agar plates for colony-forming unit (CFU) counts (Ali-Shtayeh et al., 1991; Hong et al., 2002; Pettitt et al., 2002). Filtration to capture dispersed inoculum in water is now also used, often in parallel with baiting, to capture oomycete biomass ready for detection and identification by DNA extraction and nucleotide assays, especially with the increasing use of metabarcoding techniques (Scibetta et al., 2012; Català et al., 2015; Green et al., 2021). To capture viable propagules for colony-plating, membrane filters are used, most frequently of pore size 3 or 5 µm and 47 mm diameter. Once filtration has been completed, membranes can be plated directly onto selective agar media (Davidson et al., 2005; Calvo-Bado et al., 2006; Davidson et al., 2008). Alternatively, the inoculum caught on the filter membrane surface can be released into a small volume of a resuspension medium consisting of 0.09% agar solution (Ali-Shtayeh et al., 1991), ideally containing selective antibiotics at the same concentrations as the plating medium (Pettitt et al., 2002; Büttner et al., 2014). A membrane fabric that readily and reliably releases caught viable propagules is desirable for colony-plating procedures. Hong et al. (2002) investigated a range of membranes and found the most efficient recoveries in terms of filtration speed and colony recoveries were on 5 µm polyvinylidene fluoride (‘Durapore 5’) membranes, although the only cellulose nitrate filters they investigated were of 0.45 µm pore size, which was the size and type originally used by Ali-Shtayeh et al. (1991). This pore size was originally used at Horticulture Research International Efford (UK) but was found to be too fine and too readily blocked for efficient extraction of oomycetes from irrigation water samples. An increase in pore size to 5 µm cellulose nitrate (Whatman International Ltd, Maidstone, UK) gave faster filtration and consistently high colony recoveries of Phytophthora and especially Pythium zoospores (Pettitt et al., 2002). The pore size was later reduced to 3 µm following the findings of Hwang et al. (2008), although later comparisons in 2012 between 3 and 5 µm cellulose nitrate membranes and 3 µm polycarbonate (Cyclopore™ track-etched, Whatman International Ltd, Maidstone, UK) membranes showed no significant differences in colony recoveries from irrigation reservoir samples (Pettitt et al., 2015).
When filtration has been deployed for the capture stage for genomic studies of oomycetes in water, effective use has been made of mixed cellulose ester filters of pore size 1.2 µm to capture target biomass (Scibetta et al., 2012), although again this size has proved too fine for efficient filtration of field samples, and in more recent studies an increased pore size of 5 µm has been used (Mora-Sala et al., 2022). Interestingly, these workers refined their oomycete extractions by using a baiting technique, placing filter membrane halves into longitudinal cuts in apple fruits, a widely used bait, especially for baiting Pythium and Phytophthora species from soil samples and decaying, diseased plant tissues (Campbell, 1949). Cuts containing membrane halves were then sealed with parafilm, and molecular identifications of cultures isolated from the resulting lesions were carried out.
Membrane filtration of water samples taken to the laboratory is normally carried out using bottle-top filter funnels such as the reusable, readily-cleaned, and autoclavable Nalgene™ Polysulfone units (Pettitt, 2016), and passing water through the filter using suction from a vacuum pump. In the field, in the absence of an available electricity supply, this method can still be applied using a hand-held suction pump (Matsiakh et al., 2016; Pettitt et al., 2018). Hand-operated suction pumps are compact and work well, but are tiring to the forearms of the operator after a short time. An alternative approach developed by Scibetta et al. (2012) is to push the water through the filter mounted in an in-line polypropylene filter cartridge using an adapted knapsack sprayer (Cooper Pegler CP3). This method is effective, especially for filtering larger volumes of water (2-5 L), although cleaning the apparatus between samples, particularly when collecting samples for eDNA analysis, is time-consuming and requires a comparatively large amount of oomycete-free water. A refinement of this basic concept was developed during the ‘Phytothreats’ project (Green et al., 2021), using a cyclist’s track pump to apply air pressure to a simple pressure vessel that then drives one or more water sample(s) from their connected collection bottles out through up to 3 parallel filter cartridges. This process greatly reduces potential cross-contamination and the of cleaning needed between samples is reduced to just the filter cartridges and their supply manifold, which can be flushed through with cleaning agent and then rinsed with small volumes of sample water before new filter membranes are mounted.
There can be discrepancies in the numbers and abundance of oomycete species captured by filtration and baiting methods (Sarker et al., 2023a), especially with ex situ baiting which in comparisons, tends to capture fewer species and less biomass than filtration (Pettitt et al., 2002; Redekar et al., 2020). Recent studies looking at assay conditions and sample sizes for in situ baiting of soils are closing this gap (Sarker et al., 2023a; Sarker et al., 2023b), but further studies are required to fully understand the differences seen in water tests.
2.2 Detection and identification
Sensitive techniques for the detection of oomycete propagules have been available for some time (Pettitt et al., 2002; Cooke et al., 2007; O’Brien et al., 2009). However, with the development of new techniques the capacity for rapid, accurate, and precise identifications has increased greatly in the last ten years (Figure 1), especially with the increasing availability of metabarcoding technologies (Burgess et al., 2022). Techniques that can be used for detection and identification fall into three main groups: cultural, immunodiagnostic and nucleotide-based assays.
2.2.1 Culture-based detection and identification
Culture-based detection and identification techniques either consist of plating of captured propagules from baits or directly from membrane filters onto semi-selective antibiotic-amended agar (Tsao, 1970), or of direct observation of signature symptoms and/or sporulation on baits. Depending on the numbers of propagules present in a sample and the condition of the water, culture-based techniques can be highly sensitive with membrane filtration-colony plating potentially resulting in one colony per viable spore (Pettitt et al., 2002). Rapid identifications to genus, either directly from observations of colony morphology on agar plates (O’Brien et al., 2009) or from sporulation on infected baits, are possible (Werres et al., 2014). Culture- plating methods tend to lack specificity and this can be increased by altering the components of the of the mix of fungicides and antibiotics used in the semi-selective medium. Additions of hymexazole to media improves selectivity for many Phytophthora spp. (Tsao, 1983; Erwin and Ribeiro, 1996), but this selectivity often comes at the price of reduced sensitivity as a consequence of inhibitory effects of the antimicrobial mix (Tsao, 1970; Sarker et al., 2020). Nevertheless, isolations and/or further culturing to encourage sporulation and properly observe colony morphology are, more often than not, required to achieve a diagnosis using identification keys, a process requiring patience, practice and skill. Nevertheless, extractions of detected oomycetes can be made from colony picks or from recovered baits for examination and further identification by either immunodiagnostic or nucleotide-based assays.
2.2.2 Immunodiagnostic assays
Baiting techniques can be used as a capture and initial detection step prior to testing by enzyme-linked immunosorbent assays (ELISA) or lateral flow devices (LFDs) (Wedgwood, 2014), whilst immunodiagnostic dipstick assays can carry out the baiting capture step without the need for live plant tissues and on collection can progress straight to a diagnostic staining process (Cahill and Hardham, 1994b; Wilson et al., 2000). Immunodiagnostic assay, both ELISAs and LFDs, have also been used directly on extracts of inoculum captured on membrane filters with some success (Wedgwood, 2014). Trapped propagules can also be subjected to direct immunostaining procedures in situ on a membrane filter and observed by stereo microscope or a hand lens using the zoospore trapping immunoassay (ZTI), a procedure whereby captured, viable zoopsores and cysts are encouraged to germinate in situ prior to immuno-staining (Wakeham et al., 1997; Pettitt et al., 2002). The range of immunodiagnostic procedures of potential use in the testing and monitoring of irrigation water for oomycetes has been more fully reviewed by O’Brien et al. (2009) and more recently by Wakeham and Pettitt (2017).
2.2.3 Nucleotide assays
In recent years, molecular nucleotide assays essentially based around polymerase chain reaction (PCR) have become the preferred method for laboratory-based detection and identification of oomycetes (Schena et al., 2008; Wakeham and Pettitt, 2017: La Spada et al., 2022). For detection of oomycetes in water, PCR tests are used in conjunction with either a filtration or baiting capture technique, and baits have been widely use as a ‘biological amplification’ stage (e.g., Shrestha et al., 2013; Sena et al., 2018: Kunadiya et al., 2019). Over the last 20 years, basic PCR, quantitative PCR and isothermal amplification techniques have been improved, tested, and refined for oomycete detection and identification. Much of this work has focused on Phytophthora because of both the importance of this genus commercially, and the devastating impact of species like P. cinnamomi (Carter, 2004; Shearer et al., 2004; Scott et al., 2013; Hardham and Blackman, 2017) and P. ramorum (Rizzo et al., 2005; Grünwald et al., 2008) on the environment. There are some excellent reviews of these successful, still developing methodologies (Lévesque and DeCock, 2004; Cooke et al., 2007; O’Brien et al., 2009; Robideau et al., 2011; Martin et al., 2012; Schroeder et al., 2013; Magray et al., 2019).
Using DNA sequence data is a precise method of achieving identifications, but there is sometimes a concern about accuracy in that there are many misidentified sequences in public sequence databases, for example for certain species of Phytophthora (Abad et al., 2023). New identifications/diagnoses should be carried out with caution and, where possible, be based on both molecular and morphological characters with close alignment with ex-type sequence data before assigning to species. A good example of this practice is illustrated in the protocol described by Mora-Sala et al. (2022), where new isolates were assigned to a species when the identity was above the 99% cut-off in respect to the ex-type isolates recommended by the IDphy online resource (see Abad et al., 2023).
The large amounts of oomycete genomic data that have been generated, the protocols and PCR primers used to access and collate it, and the identification of so many new taxa (e.g. over 100 new Phytophthora species alone in the last twenty or so years) can be bewildering to the non-specialist. Nevertheless, robust oomycete phylogenies are now building on the early molecular studies of Cooke et al. (2000) and Lévesque and DeCock (2004). These phylogenies consider both morphological and physiological traits as well as nucleotide sequence data. In addition to the originally-used Internal Transcribed Spacer (ITS) region sequence data from other DNA regions including mitochondrially encoded cytochrome oxidase, cox I and II (Martin and Tooley, 2003; Robideau et al., 2011) and β-tubulin (Villa et al., 2006) are now used. Rigorous databases for this information have also been established to guide accurate species identifications for Phytophthora at least (e.g. The Phytophthora Database: http://www.phytophthoradb.org, IDphy: https://idtools.org/phytophthora/).
More recently, increasingly widespread use of metabarcoding technologies has greatly broadened the scope of oomycete community studies and has been successful in a number of studies looking at either oomycete (Redekar et al., 2019; Redekar et al., 2020; Foster et al., 2022) or primarily Phytophthora species (Català et al., 2015; Redondo et al., 2018; Riddell et al., 2020; Green et al., 2021) in water. The wider metagenomic approach also allows broader communities to be considered, meaning that oomycetes in water can be assessed within a broader context (Marčiulynas et al., 2020), an area that needs further exploration. The different ‘next generation’ sequencing (NGS) technologies and the interpretation software used in metabarcoding techniques are beyond the scope of this review. Each method has its own rates of error, and balancing the use of these together with determining the most effective sequence information to read for accurate species identifications is an ongoing, rigorous process of analysis and critique (Riddell et al., 2019), and as more studies are reported, comparisons and improvements can be shared (Burgess et al., 2022). The NGS metabarcoding approach allows the spectrum of species present in samples to be determined and therefore the possible interrelationships to be explored in a previously-unfeasible context. As broad sweeps are possible, samples can be screened for both familiar endemic and novel species all at once. This capacity makes these methods powerful tools for identifying, monitoring, and tracing new and potentially invasive disease threats. Indeed, a large proportion of oomycete studies testing water samples reported so far have, rightly, been primarily focused on biosecurity.
2.3 Viability
Once oomycete propagules have been captured from a water sample, detected and identified, their viability generally needs to be established. The question of propagule viability is of crucial importance in epidemiological studies, and for determinations of immediate disease risks, as well as for assessments of potential management and control measures. Culture plating, including CFU counts and baiting techniques, detect only viable propagules. With appropriate bait selection, an indication of infectivity is also possible (Werres et al., 2014; Garbelotto et al., 2021) and in some cases this can be confirmed by rapid sporulation for example on excised lupine radicals (Pettitt et al., 1998). Dipstick assays also attract viable spores by chemoattraction (Cahill and Hardham, 1994a). The resulting zoospore cysts that form on the dipstick membrane surface can be immediately subjected to a specific immunostaining procedure, or they can be encouraged to germinate on the membrane first in the same way as in zoospore trapping immunoassay (ZTI), thereby reaffirming their viability (Pettitt et al., 2002; Bandte and Pettitt, 2014). ZTI can measure spore viability and give an indication of the predominant inoculum type in water samples by microscopy within 5 hours of sampling (Wakeham et al., 1997). ZTI assays often require pre-filtration to remove debris, and the immunostaining procedure is more prone to interference than on dipsticks, especially when turbid samples are being assessed. ELISA and LFD test strips are not able to discriminate between viable and non-viable material (Wakeham and Pettitt, 2017). Whilst limited success has been achieved by coupling these procedures with a baiting step, there is a danger of false-positive tests as antigenic material has been found to attach to bait tissues in inoculated irrigation water previously subjected to heat, chlorination, or UV sterilization treatments (Wedgwood, 2014). Baits have been used as a biological amplification step in ELISA tests of soils (Yuen et al., 1993), providing both a boost in detectable analytes and an indication of inoculum viability. Unfortunately, these benefits come at the cost of reducing the assay to providing qualitative to semi-quantitative estimates of inoculum concentrations.
CFU counts can be highly sensitive measures of viability (Pettitt et al., 2002) and are especially useful in determining the efficacy of water treatments to eliminate oomycete pathogens. Cayanan et al. (2009) used CFU counts to determine the impact of chlorination (Sodium hypochlorite) treatments on the viability of Phytophthora infestans, P. cactorum and Pythium aphanidermatum zoospores in irrigation water. Despite the sensitivity of CFU plates, they lack specificity as indicated above, sometimes making determinations of target viability difficult. Nevertheless, this lack of specificity can also be useful, for example in monitoring the efficacy of water treatment systems such as chlorination or slow sand filtration when installed on nurseries. In these situations, the populations of target pathogen propagules are likely to be very low or hopefully non-existent, and the spiking of the irrigation system with pathogen inoculum is not an option. Using total oomycete CFU counts on a semi-selective agar can therefore exploit the background populations of saprotrophic zoosporic oomycete species generally common in even the ‘cleanest’ irrigation systems as indicators of water-treatment efficacy (Büttner et al., 2014).
Despite the considerable power of PCR methods for detection and identification, they only detect and amplify pathogen DNA for sequencing and identification and do not differentiate the origin of DNA, or between viable and non-viable tissues (Nielsen et al., 2007; O’Brien et al., 2009; Lau and Botella, 2017). For many applications, this situation is probably of little consequence, as presence of detectable and identifiable DNA indicates presence of the identified species and this information is sufficient. However, in recirculating irrigation water that has been treated to control oomycete propagules, there will likely be many dead pathogen cells and particles of debris present that would not be discerned from viable cells by PCR (Stewart-Wade, 2011), leading to misleading false positive tests. The question as to whether detected DNA represents viable propagules has been directly addressed by two methods: a) the use of the DNA-intercalating dye propidium monoazide (PMA) and b) by extracting RNA and converting it to PCR-measurable cDNA using reverse transcriptase (RT-PCR). PMA cannot enter viable cells and binds with exposed DNA in dead cells by a photo-induced reaction that renders it insoluble and un-extractable (Nocker et al., 2006), while RNA is considerably more labile than DNA and unlikely to remain long in dead tissues (Vettraino et al., 2010; Chimento et al., 2012). Whilst very promising, both methods still present considerable challenges, sometimes producing seemingly contradictory findings. For example, P. ramorum RT-PCR sometimes gave positive tests when culture methods were negative (Chimento et al., 2012), apparently detecting dormant inoculum. However, in P. cinnamomi (Kunadiya et al., 2019), the opposite appeared to be the case, possibly as a result of minimal RNA expression (Rittershaus et al., 2013) in truly dormant tissues. Such differences may always pose problems, and while they are problematic for development of reliable and economic screening procedures, they might possibly be examined together with changes in infectivity of baits and the appearance of CFUs to further our understanding of the cycles of pathogen dormancy and activity.
2.4 Quantification
Once detection, identification, and viability have been established, it is important to establish the sizes of the populations present in the water being monitored to determine species prevalence (Zappia et al., 2014). While there are plenty of data reporting presence/absence of species, there is very little reliable data published on infective propagule numbers and how these relate to potential disease outcomes (Hong, 2014). Of the detection procedures outlined so far, baiting and basic PCR provide only qualitative data on presence/absence of species. Dipstick tests readily give propagule counts and can be calibrated to give quantitative estimations of the numbers of spores in water volumes being tested (Bandte and Pettitt, 2014). The specificity of these tests and of ELISA assays, which can also give good quantitative estimates of target organism biomass (although not of viability), is reliant on the specificity of the antibodies used, most of which are likely to cross-react with some non-target species (O’Brien et al., 2009). The specificity of ELISA can be improved by deploying more than one antibody with different cross-reactivities in a double antibody sandwich (DAS-ELISA, Fang and Ramasamy, 2015), although this is still unlikely to achieve complete species specificity (Pettitt et al., 2018). CFU counts can be very precise but, when used alone, lack specificity and require experience and skill in identifying colony morphology for their interpretation (Hong and Moorman, 2005). Nevertheless, CFU counts have been effectively used in combination with PCR to generate valuable epidemiological data on viable spore numbers (Davidson et al., 2008), and if examined early enough, identification of colony origins can be attempted (Pettitt and Pegg, 1991).
Pathogen biomass can be estimated by measuring the concentration of DNA in a sample using ‘real-time’ or ‘quantitative’ qPCR (Schaad and Frederick, 2002). qPCR assays have been developed for diagnosis and estimating biomass of a wide range of oomycete species (Cooke et al., 2007; Huang et al., 2010; Tuffs and Oidtmann, 2011; Lees et al., 2012; Strand et al., 2012; Mulholland et al., 2013; Li et al., 2014). Once optimized, qPCR assays are fast, sensitive, and have good specificity, although like conventional PCR they cannot discern viable from non-viable DNA. Nonetheless, the technique can be successfully coupled with classical cultural techniques to provide measurement of biomass combined with identifications of the viable species present for example of oomycetes in ornamental nursery-stock (Puertolas et al., 2021). Both PMA and RT treatments mentioned in the viability section above can be used for PMA qPCR or RT qPCR, both of which are promising techniques but still not robust enough to be deployed for the large field sample runs needed for in-depth population monitoring, for example requiring species specific internal assays to calibrate each run (Pettitt et al., 2023).
Metabarcoding techniques have so far been largely used as taxonomic screening tools for oomycetes in water, detecting and identifying species present in samples to determine the presence and distributions of pathogens and potentially invasive species to assess risks to the environment and crops (e.g. Green et al., 2021; Burgess et al., 2022; La Spada et al., 2022; Mora-Sala et al., 2022),. The species abundance data generated by this process can give an indication of relative biomass (Di Muri et al., 2020), but not of propagules’ viability. Also, PCR-based metabarcoding methods can show primer bias depending on the primer sets used (Elbrecht and Leese, 2015; Mendoza et al., 2015; Burgess et al., 2021), possibly affecting the precision of relative abundancy estimates. Baiting has been used alongside metabarcoding methods but more as a complementary sampling strategy and to isolate cultures than to verify viability (La Spada et al., 2022; Mora-Sala et al., 2022). Nevertheless, metabarcoding and other techniques using high throughput sequencing methods ‘have emerged as new paradigms’ (La Spada et al., 2022). In just a few years, these have greatly increased the depth and richness of our understanding of oomycete communities, and are also potentially providing the basis for new phytosanitary testing repertoires (Rossmann et al., 2021).
3 Applicability of techniques to different categories of water monitoring
3.1 In general
With the expanded movements of whole plants and plant parts in world trade since the 1990s, there have been rising concerns about invasive pathogens and an increasing awareness of disease risks to both crops and treasured native plants (Brasier, 2008; Fisher et al., 2012). As mentioned above, the oomycetes already have notoriety as a consequence of several plant disease pandemics as well as many invasive disease problems at more localized scales (Brasier et al., 2022). Water has a key role in infection and spread in many oomycete species, and monitoring oomycetes in water is important at different levels of scale; firstly, at a national or international scale to identify sources and pathways of potential pathogen movement and introduction, and secondly, at a local scale at locations such as horticultural nurseries, gardens, parks, forests, and reserves to identify, manage and control disease risks and disease spread. Oomycete monitoring strategies in water address questions of disease risk, but also can address questions about the basic biology and life cycles of these organisms, which in turn contribute to improved precision of risk prediction and mitigation. Many of the procedures that have been successfully used in monitoring water for oomycetes were initially developed for analysis of soil and plant tissues but inoculum in water tends to be more transient and widely dispersed rather more like airborne inoculum and some aspects of oomycete survival and spread such as cyst formation and polyplanetism have received comparatively very little study. In the first part of this review, the techniques currently used for preparing and analyzing water samples have been considered in relation to a basic framework of monitoring stages: ‘capture’, ‘detection and identification’, ‘viability testing’ and ‘quantification’. This second part of the review looks at some of the key kinds of questions likely to be asked under different monitoring categories or scenarios and how the framework shifts with different emphasis placed on these stages and therefore the possible strengths of particular techniques under these different circumstances. This subtle shift in emphasis attempts consideration of the most appropriate methodologies from those outlined in section 2 above for various scenarios based on efficacy, economy of effort and levels of accuracy and precision needed for the nature and circumstances of the monitoring questions being asked. For example, the question ‘what are the limits of this water treatment’s efficacy?’ will require a quite different approach to ‘does this water source contain potentially infective oomycete species?’. The former question requires high sensitivity of capture, detection and accurate determinations of viability, but not necessarily high species specificity. On the other hand, the latter question requires the ability to detect a wide range of species with high specificity without being immediately concerned with precise estimates of viability. Despite the recent and ongoing exciting developments with metabarcoding and high-throughput sequencing technologies in general, the best results for many applications still come from complementary use of techniques, especially when estimations of viability and the collection of live voucher specimens are needed (Marano et al., 2012; La Spada et al., 2022).
3.2 Water monitoring categories
Four categories of monitoring are considered here, based in each case on what is already known and the specific questions being asked. The first three categories are site-focused (e.g. park or horticultural nursery etc.): 1) water treatment efficacy testing; 2) recognized problem-specific testing (e.g. a recognized and diagnosed recurrent disease) – monitoring epidemics in field situations or within controlled, perhaps inoculated experiments and trials; 3) ‘health checks’ - testing storage tanks, gutters, irrigation lines and similar structures for contamination; 4) surveys – testing water sources from collection ponds, rivers, open reservoirs, and lakes to puddles. This last category of testing could be site-focused, or consist of surveys of many locations, possibly over wide areas. Water testing techniques for oomycetes suit these categories of monitoring to different degrees, and often there are synergies to be gained from combining quite different techniques within a monitoring strategy (Figure 2).
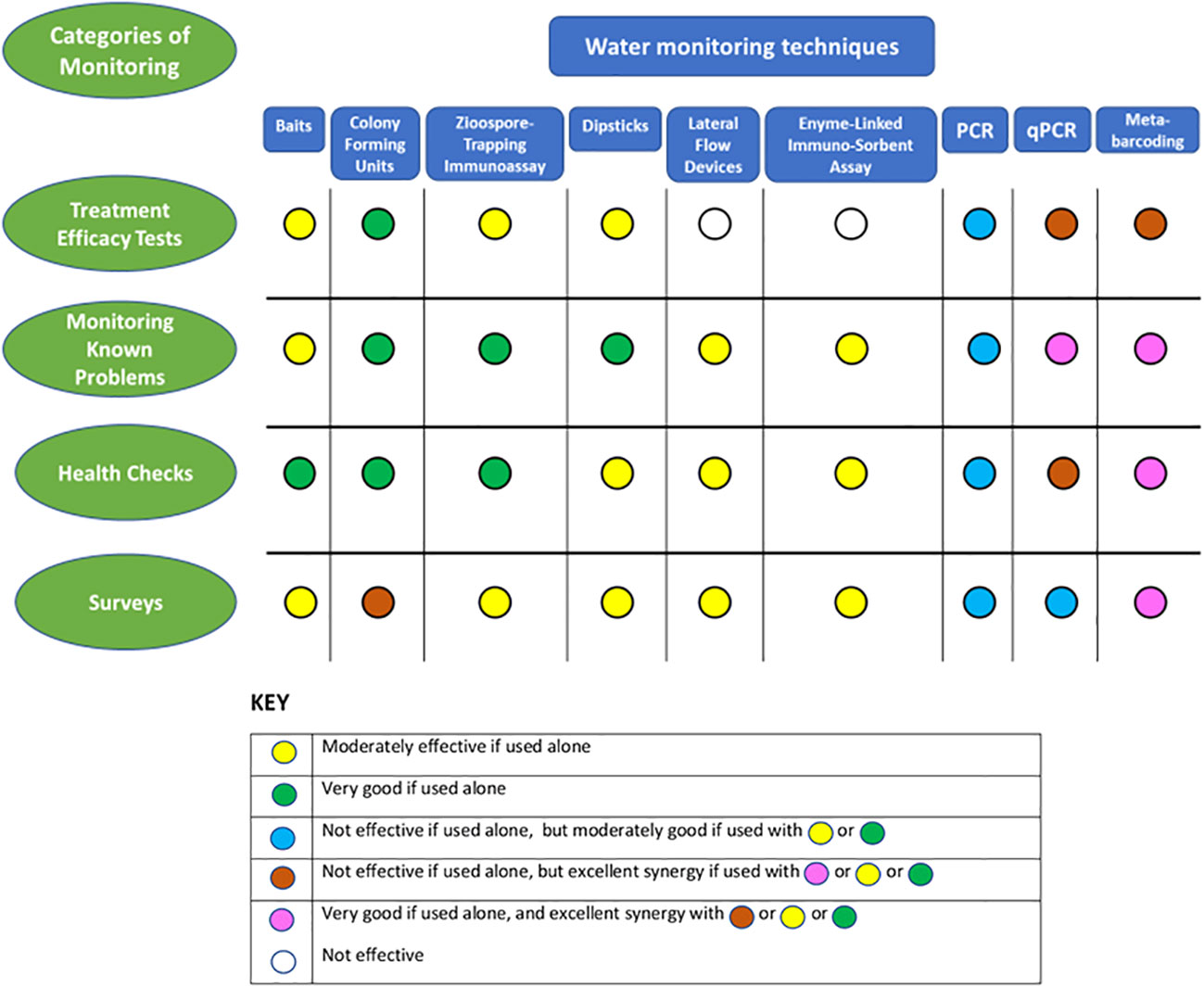
Figure 2 Comparison of the effectiveness of water monitoring techniques for detecting, and evaluating oomycete populations in four different categories of monitoring and an assessment of the possible synergies from their combined use for these individual categories.
3.2.1 Water treatment efficacy testing
For the first category of monitoring, probably the best approach to assessing the efficacy of water treatments in situ is the use of resuspension-colony plating and counting CFU (Büttner et al., 2014), as there is a need for sensitivity but not for a high degree of specificity. Treated and untreated water samples could also be sensitively monitored using ZTI or immuno-dipstick assays using more generic anti-oomycete antibodies than those used by Cahill and Hardham (1994a) or Gautam et al. (1999), (Pettitt et al., 2002).
3.2.2 Local problem-specific testing
The second category of monitoring presents a more complex situation regarding the need for an initial diagnosis for in situ monitoring programs, which would pragmatically still involve a combination of cultural and molecular (metabarcoding) approaches to cast a broad net and minimize presumptive bias (La Spada et al., 2022; Mora-Sala et al., 2022; Sarker et al., 2023a). Depending on the complexity of the diagnosis, it may then be possible to proceed to quantification of inoculum in relation to the specific problem. Again, CFU counts can be used here if colony morphologies are distinctive enough, and ZTI or immuno-dipstick approaches may be workable, depending on the specificity of available antibodies. In many instances in this category of monitoring, it may not be essential to continuously monitor viability. In this case, depending on the availability of suitable primers, qPCR or even multiplex qPCR (Schena et al., 2006) would provide excellent, specific determinations of inoculum concentrations in samples. The assumption here is that this type of monitoring would require large numbers of samples, and currently the costs of using a metagenomics approach for this may prove cost-prohibitive. However, this situation should change as costs will likely reduce with the pace of developments.
3.2.3 ‘Health checks’
The third category of monitoring is generally concerned with sites like horticultural nurseries or gardens that can have complex irrigation rigs (or maybe just gutters for collecting rainwater) that need to be routinely examined for oomycete and other potential pathogens. This is a situation where metabarcoding, combined with either baiting or filtration-colony plating, would provide excellent information, but at the moderate to high intensity of sampling needed, again, might not be economic. Currently, this kind of monitoring is carried out using the same kind of culture-based methodologies as the first category of monitoring, often with the inclusion of generic culture media such as basic potato dextrose agar in addition to oomycete-selective media to crudely assess the ‘background’ microbiota as well as oomycetes (Büttner et al., 2014).
3.2.4 Surveys
The fourth category of monitoring was, for a long time, only feasible using baiting and culturing methods (Erwin and Ribeiro, 1996). These were later supplemented and greatly improved by augmenting morphologically-based identifications with PCR and sequencing (Ghimire et al., 2009; Ghimire et al., 2011; Riolo et al., 2020). This methodology was further adapted for the monitoring of Phytophthora species in water by the extraction and nested PCR procedure of Scibetta et al. (2012) and has subsequently been revolutionized by the use metabarcoding and metagenomics procedures which offer high levels of accuracy, sensitivity and some measure of abundance or relative abundance of detected taxa (Green et al., 2020; Green et al., 2021).
3.3 Future possibilities
There is still much to learn about the ecology and diversity of oomycete species in water and despite continued enormous improvements in monitoring techniques there are still gaps in our knowledge especially relating to prevailing propagule types, their behavior and survival in different types of water environment. Culture-based and immunodiagnostic approaches to monitoring allow some level of propagule assessment and give the most reliable determinations of viability and potential aggressiveness, but lack specificity. DNA-based techniques provide high levels of specificity and with the ongoing improvements in the efficiency and economy of high throughput sequencing technologies, the levels of detection sensitivity and identification specificity and reliability using metagenomic techniques are set to rise. Improvements in analysis of results may also come from software advances, whilst prospecting for addition metagenomic loci to try and improve discrimination of specific taxonomic groups of interest. Also, more basic improvements may well be possible, such as the honing of DNA extraction techniques specifically for oomycetes from water samples or the adaptation of centrifugation techniques used for virus capture (Büttner et al., 2014), possibly to improve effective oomycete capture from more turbid water samples. More conventional methods still also have the potential to be honed for the monitoring of water samples, for example modifications to selective agar media for culture plates are capable of improving selectivity, whilst ongoing studies with soils show possibilities for the improved use of baiting techniques (Sarker et al., 2021; Sarker et al., 2023a; Sarker et al., 2023b).
There are further useful examples for potential future water studies from work in soils, for instance changes in fungal and oomycete community composition have been successfully monitored using a metabarcoding approach (Del Castillo Múnera et al., 2022) which demonstrated significant reductions in the abundance and diversity of oomycete pathogens in the rhizosphere of poinsettia crops as a result of reduced irrigation. Other studies show the power of these techniques: for example, in studies of fungal and oomycete communities in Holm oak soil, Ruiz Gómez et al. (2019) examined some complex interactions and identified a strong correlation between the presence of a taxon of Trichoderma and a decline in the abundance of pathogenic Phytophthora spp.
This exciting work still needs to be complemented by the use of more established approaches, especially cultural techniques. Techniques such as baiting (e.g. La Spada et al., 2022; Mora-Sala et al., 2022; Sarker et al., 2023a), or possibly CFU plates (Davidson et al., 2008), are still key to establishing viability, testing pathogenicity and infectivity, examining morphological characteristics, and obtaining voucher specimens. Nevertheless, metabarcoding/metagenomic studies have obvious application to the monitoring and understanding of oomycete communities in free water, and possibly with the use of autonomous sampling technologies (Searcy et al., 2022; Truelove et al., 2022), a far greater understanding can be achieved of the spread of these specific inocula in time and space. These possibilities show the way for future work with sampling in depth and over time to evaluate and mitigate the risks of disease, and local and international pathogen spread.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Acknowledgments
The author is grateful to Rory Queripel for their very helpful critique and discussion of the manuscript.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abad Z. G., Burgess T. I., Redford A. J., Bienapfl J. C., Mathew R., Srivastava S. K., et al. (2023). IDphy: An international online resource for molecular and morphological identification of Phytophthora. Plant Dis. 4, 987–998. doi: 10.1094/PDIS-02-22-0448-FE
Ali-Shtayeh M. S., MacDonald J. D., Kabashima J. (1991). A method for using commercial ELISA tests to detect zoospores of Phytophthora and Pythium species in irrigation water. Plant Dis. 75, 305–311. doi: 10.1094/PD-75-0305
Aram K., Rizzo D. M. (2018). Distinct trophic specializations affect how Phytophthora ramorum and Clade 6 Phytophthora spp. colonize and persist on Umbellularia californica leaves in streams. Phytopathology 108, 858–869. doi: 10.1094/PHYTO-06-17-0196-R
Bandte M., Pettitt T. R. (2014). “Immunological methods for detection of plant pathogens in irrigation water,” in Biology, detection and management of plant pathogens in irrigation water. Eds. Hong C. X., Moorman G. W., Wohanka W., Büttner C. (St Paul, MN, USA: American Phytopathological Society), 149–160.
Banihashemi Z., Mitchell J. E. (1975). Use of safflower seedlings for the detection and isolation of Phytophthora cactorum from soil and its application to population studies. Phytopathology 65, 1424–1430. doi: 10.1094/Phyto-65-1424
Barwell L. J., Perez-Sierra A., Henricot B., Harris A., Burgess T. I., Hardy G., et al. (2020). Evolutionary trait-based approaches for predicting future global impacts of plant pathogens in the genus Phytophthora. J. Appl. Ecol. 58, 718–730. doi: 10.1111/1365-2664.13820
Bassani I., Larousse M., Tran Q. D., Attard A., Galiana E. (2020). Phytophthora zoospores: From perception of environmental signals to inoculum formation on the host-root surface. Comput. Struct. Biotechnol. J. 18, 3766–3773. doi: 10.1016/j.csbj.2020.10.045
Beakes G. W., Glockling S. L., Sekimoto S. (2012). The evolutionary phylogeny of the oomycete ‘fungi’. Protoplasma 249, 3–19. doi: 10.1007/s00709-011-0269-2
Bifulco J. M., Schaefer F. W. (1993). Antibody-magnetic method for selective concentration of Giardia lamblia cysts from water samples. Appl. Environmental. Microbiol. 59, 772–776. doi: 10.1128/aem.59.3.772-776.1993
Brasier C. M. (2008). The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 57, 792–808. doi: 10.1111/j.1365-3059.2008.01886.x
Brasier C., Scanu B., Cooke D., Jung T. (2022). Phytophthora: an ancient, historic biologically and structurally cohesive and evolutionarily successful generic concept in need of preservation. IMA Fungus 13, 12. doi: 10.1186/s43008-022-00097-z
Burgess T. I., López-Villamor A., Paap T., Williams B., Belhaj R., Crone M., et al. (2021). Towards a best practice methodology for the detection of Phytophthora species in soils. Plant Pathol. 70, 604–614. doi: 10.1111/ppa.13312
Burgess T. I., White D., Sapsford S. J. (2022). Comparison of primers for the detection of phytophthora (and other oomycetes) from environmental samples. J. Fungi 8, 980. doi: 10.3390/jof8090980
Büttner C., Bandte M., Pettitt T. R. (2014). “Filtration and centrifugation for detection of plant pathogens in irrigation water,” in Biology, detection and management of plant pathogens in irrigation water. Eds. Hong C. X., Moorman G. W., Wohanka W., Büttner C. (St Paul, MN, USA: American Phytopathological Society), 139–148.
Cacciola S. O., Magnano di San Lio G. (2008). “Management of citrus diseases caused by Phytophthora spp,” in Integrated management of diseases caused by fungi, phytoplasma and bacteria. Eds. Ciancio A., Mukerjii K. G. (Berlin, Germany: Springer Science & Business Media BV), 61–84.
Café-Filho A. C., Duniway J. M., Davis R. M. (1995). Effects of the frequency of furrow irrigation on root and fruit rots of squash caused by Phytophthora capsici. Plant Dis. 79, 44–48. doi: 10.1094/PD-79-0044
Cahill D. M., Hardham A. R. (1994a). Exploitation of zoospore taxis in the development of a novel dipstick immunoassay for the specific detection of Phytophthora cinnamomi. Phytopathology 84, 193–200. doi: 10.1094/Phyto-84-193
Cahill D. M., Hardham A. R. (1994b). A dipstick immunoassay for the specific detection of Phytophthora cinnamomi in soils. Phytopathology 84, 1284–1292. doi: 10.1094/Phyto-84-1284
Calvo-Bado L. A., Petch G., Parsons N. R., Morgan J. A. W., Pettitt T. R., Whipps J. M. (2006). Microbial community responses associated with the development of oomycete plant pathogens on tomato roots in soilless growing systems. J. Appl. Microbiol. 100, 1194–1207. doi: 10.1111/j.1365-2672.2006.02883.x
Campbell W. A. (1949). A method of isolating Phytophthora cinnamomi directly from soil. Plant Dis. Rep. 33, 134–135.
Campbell A., Smith H. (1997). Immunomagnetic separation of Cryptosporidium oocysts from water samples: round robin comparison of techniques. Water Sci. Technol. 35, 11–12. doi: 10.2166/wst.1997.0766
Carter R. (2004). Arresting Phytophthora Dieback: The biological bulldozer (Sydney, NSW, Australia: Report by WWF Australia and Dieback Consultative Council), 23.
Català S., Pérez-Sierra A., Abad-Campos P. (2015). The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in Northern Spain. PloS One 10, e0119311. doi: 10.1371/journal.pone.0119311
Cayanan D. F., Zhang P., Liu W., Dixon M., Zheng Y. (2009). Efficacy of chlorine in controlling five common plant pathogens. HortScience 44, 157–163. doi: 10.21273/HORTSCI.44.1.157
Cheng W., Tang K., Qi Y., Sheng J., Liu Z. (2010). One-step synthesis of superparamagnetic monodisperse porous Fe3O4 hollow and core-shell spheres. J. Materials Chem. 20, 1799–1805. doi: 10.1039/B919164J
Chimento A., Cacciola S. O., Garbelotto M. (2012). Detection of mRNA by reverse-transcription PCR as an indicator of viability in Phytophthora ramorum. For. Pathol. 42, 14–21. doi: 10.1111/j.1439-0329.2011.00717.x
Choudhary C. E., Burgos-Garay M. L., Moorman G. W., Hong C. (2016). Pythium and Phytopythium species in two Pennsylvania greenhouse irrigation water tanks. Plant Dis. 100, 926–932. doi: 10.1094/PDIS-07-15-0836-RE
Cooke D. E. L., Drenth A., Duncan J. M., Wagels G., Brasier C. M. (2000). A molecular Phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30, 17–32. doi: 10.1006/fgbi.2000.1202
Cooke D. E. L., Schena L., Cacciola S. O. (2007). Tools to detect, identify and monitor Phytophthora species in natural ecosystems. J. Plant Pathol. 89, 13–28.
Dale A. L., Feau N., Berube J. A., Ponchart J., Bilodeau G. L., Hamelin R. C. (2022). Urban environments harbour greater oomycete and Phytophthora diversity, creating a bridgehead for potential new pathogens to natural ecosystems. Environ. DNA 4, 1039–1051. doi: 10.1002/edn3.300
Davidson J. M., Patterson H. A., Rizzo D. M. (2008). Sources of inoculum for Phytophthora ramorum in a redwood forest. Phytopathology 98, 860–866. doi: 10.1094/PHYTO-98-8-0860
Davidson J. M., Wickland A. C., Patterson H. A., Falk K. R., Rizzo D. M. (2005). Transmission of Phytophthora ramorum in mixed evergreen forest in California. Phytopathology 95, 587–596. doi: 10.1094/PHYTO-95-0587
Del Castillo Múnera J., Poret-Petersen A. T., Swett C. L. (2022). Changes in fungal and oomycete community composition following irrigation reductions aimed at increasing water use efficiency in a containerized nursery crop. Phytobiomes J. 6, 247–260. doi: 10.1094/PBIOMES-09-21-0058.R
Di Muri C., Handley L. L., Bean C. W., Li J., Peirson G., Sellers G. S., et al. (2020). Read counts from environmental DNA (eDNA) metabarcoding reflect fish abundance and biomass in drained ponds. Metabarcoding Metagenomics 4, 97–112. doi: 10.3897/mbmg.4.56959
Drechsler C. (1930). Repetitional diplanetism in the genus Phytophthora. J. Agric. Res. 40, 557–573.
Elbrecht V., Leese F. (2015). Can DNA based ecosystem assessments quantify species abundance? Testing primer bias and biomass-sequence relationships with an innovative metabarcoding protocol. PloS One 10, e0130324. doi: 10.1371/journal.pone.0130324
Erwin D. C., Ribeiro O. K. (1996). Phytophthora diseases worldwide (St. Paul, MN, USA: American Phytopathological Society), 562.
Fang Y., Ramasamy R. P. (2015). Review current and prospective methods for plant disease detection. Biosensors 4, 537–561. doi: 10.3390/bios5030537
Fisher M. C., Henk D. A., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Foster Z. S. L., Albornoz F. E., Fieland V. J., Larsen M. M., Jones F. A., Tyler B. M., et al. (2022). A new oomycete metabarcoding method using the rps10 gene. Phytobiomes J. 6, 214–226. doi: 10.1094/PBIOMES-02-22-0009-R
Garbelotto M., Schmidt D., Popenuck T. (2021). Pathogenicity and infectivity of Phytophthora ramorum vary depending on host species, infected plant part, inoculum potential, pathogen genotype, and temperature. Plant Pathol. 70, 287–304. doi: 10.1111/ppa.13297
Gautam Y., Cahill D. M., Hardham A. R. (1999). Development of a quantitative immunodipstick assay for Phytophthora nicotianae. Food Agric. Immunol. 11, 229–242. doi: 10.1080/09540109999753
Ghimire S. R., Richardson P. A., Kong P., Hu J. H., Lea-Cox J. D., Ross D. S., et al. (2011). Distribution and diversity of Phytophthora species in nursery irrigation reservoir adopting water recycling system during winter months. J. Phytopathol. 159, 713–719. doi: 10.1111/j.1439-0434.2011.01831.x
Ghimire S. R., Richardson P. A., Moorman G. W., Lea-Cox J. D., Ross D. S., Hong C. X. (2009). An in-situ baiting bioassay for detecting Phytophthora species in irrigation runoff containment basins. Plant Pathol. 58, 577–583. doi: 10.1111/j.1365-3059.2008.02016.x
Green S., Cooke D. E. L., Dunn M., Barwell L., Purse B., Chapman D. S., et al. (2021). Phyto-Threats: Addressing threats to UK forests and woodlands from Phytophthora; identifying risks of spread in trade and methods for mitigation. Forests 12, 1617. doi: 10.3390/f12121617
Green S., Riddell C. E., Frederickson-Matika D., Armstrong A., Elliot M., Forster J., et al. (2020). Diversity of woody-host infecting Phytophthora species in public parks and botanic gardens as revealed by metabarcoding, and opportunities for mitigation through best practice. Sibbaldia 18, 67–88. doi: 10.23823/Sibbaldia/2020.289
Grünwald N. J., Goss E. M., Press C. M. (2008). Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 9, 729–740. doi: 10.1111/j.1364-3703.2008.00500.x
Hallett I. C., Dick M. W. (1981). Seasonal and diurnal fluctuations of oomycete propagule numbers in the free water of a freshwater lake. J. Ecol. 69, 671–692. doi: 10.2307/2259691
Hardham A. R., Blackman L. M. (2017). Pathogen profile update Phytophthora cinnamomi. Mol. Plant Pathol. 19, 260–285. doi: 10.1111/mpp.12568
Hong C. (2014). “Component analyses of irrigation water in plant disease epidemiology,” in Biology, detection and management of plant pathogens in irrigation water. Eds. Hong C. X., Moorman G. W., Wohanka W., Büttner C. (St Paul, MN, USA: American Phytopathological Society), 111–121.
Hong C. X., Moorman G. W. (2005). Plant pathogens in irrigation water: Challenges and opportunities. Crit. Rev. Plant Sci. 24, 189–208. doi: 10.1080/07352680591005838
Hong C., Richardson P. A., Kong P. (2002). Comparison of membrane filters as a tool for isolating pythiaceous species from irrigation water. Phytopathology 92, 610–616. doi: 10.1094/PHYTO.2002.92.6.610
Huai W.x., Tian G., Hansen E. M., Zhao W.x., Goheen E. M., Grünwald N. J., et al. (2013). Identification of Phytophthora species baited and isolated from forest soil and streams in northwestern Yunnan province, China. For. Pathol. 43, 87–103. doi: 10.1111/efp.12015
Huang J., Wu J., Li C., Xiao C., Wang G. (2010). Detection of Phytophthora nicotianae in soil with real-time quantitative PCR. J. Phytopathol. 158, 15–21. doi: 10.1111/j.1439-0434.2009.01554.x
Hüberli D., St. J. Hardy G. E., White D., Williams N., Burgess T. I. (2013). Fishing for Phytophthora from Western Australia’s waterways: a distribution and diversity survey. Australas. Plant Pathol. 42, 251–260. doi: 10.1007/s13313-012-0195-6
Hwang J., Oak S. W., Jefferes S. N. (2008). “Detecting Phytophthora ramorum and other species of Phytophthora in streams in natural ecosystems using baiting and filtration methods,” in Proceedings of the Sudden Oak Death Third Science Symposium. General Technical Report PSW-GTR-214, technical coordinators. Eds. Frankel S. J., Kliejunas J. T., Palmieri K. M. (Albany, CA, USA: US Department of Agriculture, Forest Service), 55–58.
Judelson H. S., Blanco F. A. (2005). The spores of Phytophthora: weapons of the plant destroyer. Nat. ReviewsMicrobiology 3, 47–58. doi: 10.1038/nrmicro1064
Jung T., Orlikowski L., Henricot B., Abad-Campos P., Aday A. G., Aguín Casal O., et al. (2016). Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 46, 134–163. doi: 10.1111/efp.12239
Kamoun S. (2009). “Plant pathogens: oomycetes (water mold),” in Encyclopedia of microbiology, 3rd Edition. Ed. Schaechter M. (Cambridge, MA, USA: Academic Press), 689–695.
Kennedy R., Pegg G. F. (1990). Phytophthora cryptogea root rot of tomato in rockwool nutrient culture. II. Effect of root zone temperature on infection, sporulation and symptom development. Ann. Appl. Biol. 117, 537–551. doi: 10.1111/j.1744-7348.1990.tb04820.x
Kennedy R., Wakeham A. (2013). “Brassicas: Further development of ‘in field’ tests for resting spores of clubroot and the development of clubroot control based on detection,” in Final report on AHDB project FV349 (Stoneleigh: Agriculture & Horticulture Development Board), 51.
Kong P., Tyler B. M., Richardson P. A., Lee B. W. K., Zhou Z. S., Hong C. (2010). Zoospore interspecific signalling promotes plant infection by Phytophthora. BMC Microbiol. 10, 313. doi: 10.1186/1471-2180-10-313
Kunadiya M. B., Dunstan W. D., White D., Hardy G. E. S. J., Grigg A. H., Burgess T. I. (2019). A qPCR assay for the detection of Phytophthora cinnamomi including an mRNA protocol designed to establish propagule viability in environmental samples. Plant Dis. 103, 2443–2450. doi: 10.1094/PDIS-09-18-1641-RE
La Spada F. L., Cock P. J. A., Randall E., Pane A., Cooke D. E. L., Cacciola S. O. (2022). DNA Metabarcoding and isolation by baiting complement each other in revealing Phytophthora diversity in anthropized and natural ecosystems. J. Fungi 8, 330. doi: 10.3390/jof8040330
Lau H. Y., Botella J. R. (2017). Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02016
Lees A. K., Sullivan L., Lynott J. S., Cullen D. W. (2012). Development of a quantitative real-time PCR assay for Phytophthora infestans and its applicability to leaf, tuber and soil samples. Plant Pathol. 61, 867–876. doi: 10.1111/j.1365-3059.2011.02574.x
Lévesque C. A., DeCock A. W. A. M. (2004). Molecular phylogeny and taxonomy of the genus Pythium. Mycological Res. 108, 1363–1383. doi: 10.1017/S0953756204001431
Li M., Ishiguro Y., Otsubo K., Suzuki H., Tsuji T., Miyake N., et al. (2014). Monitoring by real-time PCR of three water-borne zoosporic Pythium species in potted flower and tomato greenhouses under hydroponic culture systems. Eur. J. Plant Pathol. 140, 229–242. doi: 10.1007/s10658-014-0456-z
Magray A. R., Lone S. A., Ganai B. A., Ahmad F., Dar G. J., Dar J. S., et al. (2019). Comprehensive, classical and molecular characterization methods of Saprolegnia (Oomycota; Stramnipila), an important fungal pathogen of fish. Fungal Biol. Rev. 33, 166–179. doi: 10.1016/j.fbr.2018.12.001
Marano A. V., Gleason F. H., Bärlocher F., Pires-Zottarelli C. L. A., Lilje O., Schmidt S. K., et al. (2012). Quantitative methods for the analysis of zoosporic fungi. J. Microbiological Methods 89, 22–32. doi: 10.1016/j.mimet.2012.02.003
Marčiulynas A., Marčiulyniene D., Lynikienė J., Gedminas A., Vaičiukynė M., Menkis A. (2020). Fungi and oomycetes in the irrigation water of forest nurseries. Forests 11, 459. doi: 10.3390/f11040459
Martin F. N., Abad Z. G., Balci Y., Ivors K. (2012). Identification and detection of Phytophthora: reviewing our progress, identifying our needs. Plant Dis. 96, 1080–1103. doi: 10.1094/PDIS-12-11-1036-FE
Martin F. N., Tooley P. W. (2003). Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95, 269–284. doi: 10.1080/15572536.2004.11833112
Matsiakh I., Oszako T., Kramarets V., Nowakowsk J. A. (2016). Phytophthora and Pythium species detected in rivers of the Polish-Ukrainian border areas. Baltic Forestry 22, 230–238.
Mendoza M. L. Z., Sicheritz-Pontén T., Gilbert M. P. T. (2015). Environmental genes and genomes: understanding the differences and challenges in the approaches and software for their analyses. Briefings Bioinf. 16, 745–758. doi: 10.1093/bib/bbv001
Middleton J. (1985). Potentially pathogenic Phytophthora isolates in irrigation systems. ([Reading, UK]: The University of Reading). [Ph.D. Thesis].
Moorman G. W., Gevens A. J., Granke L. L., Hausbeck M. K., Hendricks K., Roberts P. D., et al. (2014). “Sources and distribution systems of irrigation water and their potential risks for crop health,” in Biology, detection and management of plant pathogens in irrigation water,. Eds. Hong C. X., Moorman G. W., Wohanka W., Büttner C. (St Paul, MN, USA: American Phytopathological Society), 3–11.
Moralejo E., Descals E. (2011). Diplanetism and microcyclic sporulation in Phytophthora ramorum. Forest Pathol. 41, 349–354. doi: 10.111/j.1439-0329.2010.00674.x
Mora-Sala B., León M., Pérez-Sierra A., Abad-Campos P. (2022). New reports of Phytophthora species in plant nurseries in Spain. Pathogens 11, 826. doi: 10.3390/pathogens11080826
Mulholland V., Schlenzig A., MacAskill G. A., Green S. (2013). Development of a quantitative real-time PCR assay for the detection of Phytophthora austrocedrae, an emerging pathogen in Britain. For. Pathol. 43, 513–517. doi: 10.1111/efp.12058
Nagel J. H., Gryzenhout M., Slippers B., Wingfield M. J., Hardy G. E. S. J., Stukely M. J. C., et al. (2013). Characterization of Phytophthora hybrids from ITS clade 6 associated with riparian ecosystems in South Africa and Australia. Fungal Biol. 117, 329–347. doi: 10.1016/j.funbio.2013.03.004
Nam B., Choi Y.-J. (2019). Phytopythium and Pythium species (Oomycota) isolated from freshwater environments of Korea. Mycobiology 47, 261–272. doi: 10.1080/12298093.2019.1625174
Nielsen K. M., Johnsen P. J., Bensasson D., Daffonchio D. (2007). Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6, 37–53. doi: 10.1051/ebr:200703
Nocker A., Cheung C.-Y., Camper A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiological Methods 67, 310–320. doi: 10.1016/j.mimet.2006.04.015
O’Brien P. A., Williams N., St. J Hardy G. E. (2009). Detecting phytophthora. Crit. Rev. Microbiol. 35, 169–181. doi: 10.1080/10408410902831518
Pettitt T. R. (2016). Testing water for plant pathogens. AHDB Factsheet 21/15 (Stoneleigh: Agriculture & Horticulture Development Board), 6. 21 15 Testing water for plant pathogens.pdf (windows.net).
Pettitt T. R., Finlay A. R., Scott M. A., Davies E. M. (1998). Development of a system simulating commercial production conditions for assessing the potential spread of Phytophthora cryptogea root rot of hardy nursery stock in recirculating irrigation water. Ann. Appl. Biol. 132, 61–75. doi: 10.1111/j.1744-7348.1998.tb05185.x
Pettitt T. R., Keane G., John S., Edwards E., Wakeham A. (2018). “Development and testing of single and multiplex diagnostic devices for rapid and precise early detection of oomycete root and collar rot pathogens for disease avoidance, management and control,” in Final report on AHDB project CP 136 (Stoneleigh: Agriculture & Horticulture Development Board), 58.
Pettitt T. R., Lees A., Wood T., Wedgwood E. (2023). “Downy mildew and late blight control,” in Final report on AHDB project CP 184, (Stoneleigh: Agriculture & Horticulture Development Board), 43.
Pettitt T. R., Pegg G. F. (1991). The quantitative estimation of Phytophthora cactorum in infected strawberry tissue. Mycological Res. 95, 233–238. doi: 10.1016/S0953-7562(09)81018-6
Pettitt T. R., Skjøth C. A. (2016). A simple model describes development of early peaks in oomycete zoospore inoculum detected in southern UK outdoors horticultural reservoirs. Acta Agrobotanica 69, 1665. doi: 10.5586/aa.1665
Pettitt T. R., Wakeham A. J., McPherson G. M. (2015). “A desk study to review global knowledge on best practice for oomycete root-rot detection and control,” in Final report on AHDB project CP 126, (Stoneleigh: Agriculture & Horticulture Development Board), 176.
Pettitt T. R., Wakeham A. J., Wainwright M. F., White J. G. (2002). Comparison of serological, culture, and bait methods for detection of Pythium and Phytophthora zoospores in water. Plant Pathol. 51, 720–727. doi: 10.1046/j.1365-3059.2002.00759
Puertolas A., Bonants P. J. M., Boa E., Woodward S. (2021). Application of real-time PCR for the detection and quantification of oomycetes in ornamental nursery stock. J. Fungi 7, 87. doi: 10.3390/jof7020087
Redekar N. R., Bourret T. B., Eberhart J. L., Johnson E., Pitton B. J. L., Haver D. L., et al. (2020). The population of oomycetes in a recycled irrigation water system at a horticultural nursery in southern California. Water Res. 183, 116050. doi: 10.1016/i.watres.2020.116050
Redekar N. R., Eberhart J. L., Parke J. L. (2019). Diversity of Phytophthora, Pythium, and Phytopythium species in recycled irrigation water in a container nursery. Phytobiomes J. 3, 31–45. doi: 10.1094/PBIOMES-10-18-0043-R
Redondo M. A., Boberg J., Stenlid J., Oliva J. (2018). Functional traits associated with the establishment of introduced Phytophthora spp. in Swedish forests. J. Appl. Ecol. 55, 1538–1552. doi: 10.1111/1365-2664.13068
Reeser P. W., Sutton W., Hansen E. M., Remigi P., Adams G. C. (2011). Phytophthora species in forest streams in Oregon and Alaska. Mycologia 103, 22–35. doi: 10.3852/10-013
Riddell C. E., Dun H. F., Elliot M., Armstrong A. C., Clark M., Forster J., et al. (2020). Detection and spread of Phytophthora austrocedri within infected Juniperus communis woodland and diversity of co-associated Phytophthoras as revealed by metabarcoding. For. Pathol. 50, e12602. doi: 10.1111/efp.12602
Riddell C. E., Frederickson-Matika D., Armstrong A. C., Elliot M., Forster J., Hedley P. E., et al. (2019). Metabarcoding reveals a high diversity of woody host-associated Phytophthora spp. in soils at public gardens and amenity woodlands in Britain. PeerJ 7, e6931. doi: 10.7717/peerj.6931
Riolo M., Aloi F., La Spada F., Sciandrello S., Moricca S., Santilli E., et al. (2020). Diversity of Phytophthora communities across different types of Mediterranean vegetation in a nature reserve area. Forests 11, 853. doi: 10.3390/f11080853
Ristaino J. B. (1991). Influence of rainfall, drip irrigation, and inoculum density on the development of Phytophthora root and crown rot epidemics and yield in bell pepper. Phytopathology 81, 922–929. doi: 10.1094/Phyto-81-922
Rittershaus E. S. C., Baek S.-H., Sassetti C. M. (2013). The Normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe 13, 643–651. doi: 10.1016/j.chom.2013.05.012
Rizzo D. M., Garbelotto M., Hansen E. M. (2005). Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 43, 309–335. doi: 10.1146/annurev.phyto.42.040803.140418
Robideau G. P., De Cock A. W. A. M., Coffey M. D., Voglmayr H., Brouwer H., Bala K., et al. (2011). DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 11, 1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x
Rodríguez J. C., Yáñez Juárez M. G., López Orona C. A., Tafoya F. A., López Urquidez G. A., Romero Gómez S. J. (2021). Phytophthora species associated with irrigation water in the Culiacán Valley. Rev. Mexicana Cienc. Agrícolas 12, 473–484. doi: 10.29312/remexca.v12i3.2619
Rossmann S., Lysøe E., Skogen M., Talgø V., Brurberg M. B. (2021). DNA metabarcoding reveals broad presence of plant pathogenic oomycetes in soil from internationally traded plants. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.637068
Ruiz Gómez F. J., Navarro-Cerrillo R. M., Pérez-de-Luque A., Oßwald W., Vannini A., Morales-Rodríguez C. (2019). Assessment of functional and structural changes of soil fungal and oomycete communities in holm oak declined dehesas through metabarcoding analysis. Sci. Rep. 9, 5315. doi: 10.1038/s41598-019-41804-y
Sarker S. R., Burgess T. I., Hardy G. E. S. J., McComb J. (2023a). Closing the gap between the number of Phytophthora species isolated through baiting a soil sample and the number revealed through metabarcoding. Mycological Prog. 23, 39. doi: 10.1007/s11557-023-01892-7
Sarker S. R., McComb J., Burgess T. I., Hardy G. E. S. J. (2020). Antimicrobials in Phytophthora isolation media and the growth of Phytophthora species. Plant Pathol. 69, 1426–1436. doi: 10.1111/ppa.13224
Sarker S. R., McComb J., Burgess T. I., Hardy G. E. S. J. (2021). Timing and abundance of sporangia production and zoospore release influences the recovery of different Phytophthora species by baiting. Fungal Biol. 125, 477–484. doi: 10.1016/j.funbio.2021.01.009
Sarker S. R., McComb J., Hardy G. E. S. J., Burgess T. I. (2023b). Sample volume affects the number of Phytophthora and Phytopythium species detected by soil baiting. Eur. J. Plant Pathol. 166, 303–313. doi: 10.1007/s10658-023-02661-8
Sarowar N. S., van den Berg A. H., McLaggan D., Young M. R., van West P. (2013). Saprolegnia strains isolated from river insects and amphipods are broad spectrum pathogens. Fungal Biol. 117, 752–763. doi: 10.1016/j.funbio.2013.09.002
Schena L., Duncan J. M., Cooke D. E. L. (2008). Development and application of a PCR-based ‘molecular toolbox’ for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathol. 57, 64–75. doi: 10.1111/j.1365-3059.2007.01689.x
Schena L., Hughes K. J. D., Cooke D. E. L. (2006). Detection and quantification of Phytophthora ramorum, P. kernoviae, P. citricola and P. quercina in symptomatic leaves by multiplex real-time PCR. Mol. Plant Pathol. 7, 365–379. doi: 10.1111/j.1364-3703.2006.00345.x
Schroeder K. L., Martin F. N., de Cock A. W. A. M., Lévesque C. A., Spies C. F. J., Okubara P. A., et al. (2013). Molecular detection and quantification of Pythium species: evolving taxonomy, new tools, and challenges. Plant Dis. 97, 4–20. doi: 10.1094/PDIS-03-12-0243-FE
Scibetta S., Schena L., Chimento A., Cacciola S. O., Cooke D. E. L. (2012). A molecular method to assess Phytophthora diversity in environmental samples. J. Microbiological Methods 88, 356–368. doi: 10.1016/j.mimet.2011.12.012
Scott P., Burgess T., Hardy G. (2013). “Globalization and Phytophthora,” in Phytophthora: A global perspective,. Ed. Lamour K. (Wallingford, Oxfordshire, UK & Boston, MA, USA: CABI), 226–232.
Searcy R. T., Boehm A. B., Weinstock C., Preston C. M., Jensen S., Roman B., et al. (2022). High-frequency and long-term observations of eDNA from imperilled salmonids in a coastal stream: temporal dynamics, relationships with environmental factors, and comparisons with conventional observations. Environ. DNA 4, 776–789. doi: 10.1002/edn3.293
Sena K., Dreaden T. J., Crocker E., Barton C. (2018). Detection of Phytophthora cinnamomic in forest soils by PCR on DNA extracted from leaf disc baits. Plant Health Prog. 19, 193–200. doi: 10.1094/PHP-01-18-0004-RS
Shearer B. L., Crane C. E., Cochrane A. (2004). Quantification of the susceptibility of the native flora of the South-West Botanical Province, Western Australia, to Phytophthora cinnamomi. Aust. J. Bot. 52, 435–443. doi: 10.1071/BT03131
Shrestha S. K., Zhou Y., Lamour K. (2013). Oomycetes baited from streams in Tennessee 2010–2012. Mycologia 105, 1516–1523. doi: 10.3852/13-010
Stamler R. A., Sanogo S., Goldberg N. P., Randall J. J. (2016). Phytophthora species in rivers and streams of the Southwestern United States. Appl. Environ. Microbiol. 82, 4696–4704. doi: 10.1128/AEM.01162-16
Stewart-Wade S. M. (2011). Plant pathogens in recycled irrigation water in commercial plant nurseries and greenhouses: their detection and management. Irrigation Sci. 29, 267–297. doi: 10.1007/s00271-011-0285-1
Strand D. A., Jussila J., Viljamaa-Dirks S., Kokko H., Makkonen J., Holst-Jensen A., et al. (2012). Monitoring the spore dynamics of Aphanomyces astaci in the ambient water of latent carrier crayfish. Veterinary Microbiol. 160, 99–107. doi: 10.1016/j.vetmic.2012.05.008
Themann K., Werres S. (1998). Verwendung von Rhododendronblättern zum Nachweis von Phytophthora-Arten in Wurzel- und Bodenproben. Nachrichtenblatt Des. Deutschen Pflenzenschutzdienstes 50, 37–45.
Themann K., Werres S., Diener H. A., Luttmann R. (2002). Comparison of different methods to detect Phytophthora spp. in recycling water from nurseries. J. Plant Pathol. 84, 41–51.
Truelove N. K., Patin N. V., Min M., Pitz K. J., Preston C. M., Yamahara K. M., et al. (2022). Expanding the temporal and spatial scales of environmental DNA research with autonomous sampling. Environ. DNA 4, 972–984. doi: 10.1002/edn3.299
Tsao P. H. (1970). Selective media for isolation of pathogenic fungi. Annu. Rev. Phytopathol. 8, 157–186. doi: 10.1146/annurev.py.08.090170.001105
Tsao P. H. (1983). “Factors affecting isolation and quantification of Phytophthora from soil,” in Phytophthora: its biology, taxonomy, ecology and pathology. Eds. Erwin D. C., Bartnicki-Garcia S., Tsao P. H. (St Paul, MN, USA: American Phytopathological Society), 219–236.
Tuffs S., Oidtmann B. (2011). A comparative study of molecular diagnostic methods designed to detect the crayfish plague pathogen, Aphanomyces astaci. Veterinary Microbiol. 153, 343–353. doi: 10.1016/j.vetmic.2011.06.012
Vettraino A. M., Tomassini A., Vannini A. (2010). Use of mRNA as an indicator of the viability of Phytophthora cambivora. Acta Hortic. 866, 431–434. doi: 10.17660/ActaHortic.2010.866.57
Villa N. O., Kageyama K., Asano T., Suga H. (2006). Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and β-tubulin sequences. Mycologia 98, 410–422. doi: 10.3852/mycologia.98.3.410
Wakeham A. J., Pettitt T. R. (2017). Diagnostic tests and their application in the management of soil- and water-borne oomycete pathogen species. Ann. Appl. Biol. 170, 45–67. doi: 10.1111/aab.12315
Wakeham A. J., Pettitt T. R., White J. G. (1997). A novel method for detection of viable zoospores of Pythium in irrigation systems. Ann. Appl. Biol. 131, 427–435. doi: 10.1111/j.1744-7348.1997.tb05170.x
Wedgwood E. (2014). “Baiting and diagnostic techniques for monitoring Phytophthora spp. and Pythium spp. in irrigation water on ornamental nurseries,” in Final report on HDC project HNS/PO188. (Stoneleigh Park, Warwickshire, UK: Horticultural Development Company), 83.
Werres S., Ghimire S. R., Pettitt T. R. (2014). “Baiting assays for detection of Phytophthora species in irrigation water,” in Biology, detection and management of plant pathogens in irrigation water. Eds. Hong C. X., Moorman G. W., Wohanka W., Büttner C. (St Paul, MN, USA: American Phytopathological Society), 125–138.
Weste G. (1983). “Population dynamics and survival of Phytophthora,” in Phytophthora: its biology, taxonomy, ecology and pathology. Eds. Erwin D. C., Bartnicki-Garcia S., Tsao P. H. (St Paul, MN, USA: American Phytopathological Society), 237–257.
Wielgoss A., Nechwatal J., Bogs C., Mendgen K. (2009). Host plant development, water level and water parameters shape Phragmites australis-associated oomycete communities and determine reed pathogen dynamics in a large lake. FEMS Microbiol. Ecol. 69, 255–265. doi: 10.1111/j.1574-6941.2009.00701.x
Wilson B. A., Aberton J., Cahill D. M. (2000). Relationships between site factors and distribution of Phytophthora cinnamomi in the Eastern Otway Ranges, Victoria. Aust. J. Bot. 48, 247–260. doi: 10.1071/BT98067
Yuen G. Y., Craig M. L., Avila F. (1993). Detection of Pythium ultimum with a species-specific monoclonal antibody. Plant disease. 77, 692–698. doi: 10.1094/PD-77-0692
Keywords: oomycetes, water, detection, propagule-viability, quantification, monitoring
Citation: Pettitt TR (2023) Monitoring oomycetes in water: combinations of methodologies used to answer key monitoring questions. Front. Hortic. 2:1210535. doi: 10.3389/fhort.2023.1210535
Received: 22 April 2023; Accepted: 21 September 2023;
Published: 10 October 2023.
Edited by:
Paul M Severns, University of Georgia, United StatesReviewed by:
Hannah M Rivedal, Agricultural Research Service (USDA), United StatesSarah Sapsford, Murdoch University, Australia
Copyright © 2023 Pettitt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tim R. Pettitt, dGltLnBldHRpdHRAY29ybndhbGwuYWMudWs=
 Tim R. Pettitt
Tim R. Pettitt