- 1Inner Mongolia Agricultural University, Hohhot, China
- 2Key Laboratory of Wheat Germplasm Innovation and Utilization Autonomous Region Higher School, Hohhot, China
- 3Key Laboratory of Grassland Resources of the Ministry of Education, Hohhot, China
TCP transcription factors are a unique class of transcription factors that play important roles in alleviating abiotic stresses such as drought and salt. In this study, the whole-genome data of three cultivated varieties, namely, “SFS”, “Sang” and “OT3098v2”, were utilized to identify and analyze the members of the TCP gene family in oats, and their responses to two abiotic stresses, drought and salt, were also investigated. Results showed that there are 83, 65, and 30 non-redundant TCP genes in the three oats, with the highest number of TCP genes specific to the “SFS”, reaching 22 genes. The oat TCP genes can be classified into three subfamilies: PCF, CIN, and CYC/TB1. Most AsTCP genes have important motifs, Motif 1 and Motif 2, which are part of the bHLH domain. Additionally, various cis-acting elements related to hormone response, abiotic stress, light response, and growth and development were found in the promoters of AsTCP genes. The main amplification mechanism of the oat TCP gene family is fragment duplication. Two tandem duplications, AsTCP058/AsTCP059 and AsTCP023/AsTCP025, are stably present in the three oats. The highest number of AsTCP collinear relationships exist in the “SFS” with 89 pairs. After drought and salt stress treatments, significant differences in gene expression were observed among different oat cultivars and treatment periods. Genes that showed significant expression changes under both treatments (AsTCP021, AsTCP033, AsTCP044, AsTCP053, and AsTCP058) may play important roles in oat’s response to abiotic stresses. Notably, AsTCP053 gene was significantly upregulated at 24 h of stress treatment and showed a more sensitive response to salt stress. This study provides insights into the functional characterization of the oat TCP gene family and its molecular mechanisms underlying stress tolerance.
1 Introduction
TCP (Teosinte branched1/Cycloidea/proliferating cell factor) transcription factors are a class of plant-specific proteins believed to have originated originated from algae and bryophytes. These factors were first identified in maize (TB1), cynoglossum (CYC), and rice (PCF1, PCF2) and named after their initials (Braun et al., 2012). Notably, TB1 inhibits the growth and development of lateral branches in maize, while loss of its function promotes lateral branch differentiation (Doebley et al., 1997). In contrast, CYC is involved in the abortion of dorsal stamens and regulates floral symmetry formation in cynoglossum (Luo et al., 1996). PCF is responsible for maintaining chromosome structure and regulating cell cycle progression (Palatnik et al., 2003). These genes share TCP structural domains in their gene structures (Cubas et al., 1999; Kosugi and Ohashi, 1997). The TCP domain is a 59-amino-acid helix-loop-helix (bHLH) structure capable of binding to DNA or facilitating protein-protein interactions (Dhaka et al., 2017; Manassero et al., 2013). Although all TCP genes possess the TCP structural domain, its structure varies among different family members. Based on these differences and their evolutionary relationships, TCP gene familiars can be categorized into two subfamilies: Class I and Class II (Martín-Trillo and Cubas, 2010).
With the identification and characterization of more TCP genes in plants, their functions have been gradually elucidated. These genes play critical roles in regulating plant growth and development. For instance, in A. thaliana, AtTCP14 and AtTCP15 regulate embryo growth during seed germination through the gibberellin signaling pathway (Resentini et al., 2014),while also influencing leaf cell development and internode elongation (Kieffer et al., 2011). In rice, the overexpression of OsPCF7 promotes stem height, root length, and tiller number in transgenic seedlings, while increasing the number of panicles and the proportion of filled grains per plant (Li et al., 2020). Additionally, the cucumber TCP gene CsBRC1 effectively controls lateral shoot growth by repressing the expression of CsPIN3(Junjun et al., 2019). Beyond their role in plant development, TCP genes also play a pivotal role in the adaptation of plants to environmental stresses. Studies have shown that TCP genes enhance stress tolerance through various mechanisms, including the regulation of cellular osmotic pressure (Almeida et al., 2017), signal transduction (Guan et al., 2017; Liu et al., 2020), hormone sensitivity (Ding et al., 2019), and the reduction of reactive oxygen species (ROS) accumulation (Mukhopadhyay and Tyagi, 2015). For instance, the overexpression of PeTCP10 in A. thaliana significantly enhances catalase (CAT) activity, which boosts the plant’s antioxidant capacity and improves its salt tolerance during the nutrient growth period (Xu et al., 2021). However, not all TCP genes contribute positively to stress tolerance. For example, ZmTCP14 in maize promotes the accumulation of ROS, which reduces drought tolerance under drought stress conditions (Jiao et al., 2023). In birch, BpTCP20 enhances salt and drought tolerance by regulating the expression of BpMYB8 and BpIAA5, which reduces the content of ROS and malondialdehyde (MDA) (Li et al., 2024). In wheat, TaTCP21-A negatively regulates cold tolerance by repressing the expression of the cold-responsive gene TaDREB1C (Kankan et al., 2024). Moreover, in upland cotton, GbTCP5 directly activates the expression of GbERD7, GbUBC19, and GbGOLS2, thereby significantly enhancing the plant’s ability to adapt to drought and salt stress (Wang et al., 2023).
Oat, a grain-feeding cash crop (Ju et al., 2022), possesses highly productive and high-quality seeds (Gutierrez-Gonzalez et al., 2013). It is characterized by soft and juicy stems and leaves, and is rich in nutrients (Rasane et al., 2015). Oats have shown remarkable adaptability to various geoclimatic regions and adverse environmental conditions, exhibiting higher resilience compared to other feed crops such as rice and wheat. Additionally, oats can serve as pioneer crops for soil improvement (Han et al., 2014). Among the Oats cultivated today, the most prevalent type is the heterozygous hexaploid species (2n = 6x = 42, AADDCC) known as common Oat (Avena sativa). The common Oat genome is large and complex, and remains one of the least explored genomes and transcriptomes among cereal crops (Rasane et al., 2015). The publication of the oat genome sequences has opened up new possibilities for analyzing gene families on a genome-wide scale. However, there are still numerous gaps in our understanding of the TCP gene family in oats based on the entire gene sequence. Therefore, this study aims to perform a comprehensive analysis of TCP genes identified from different oat genomes and elucidate their response mechanisms to salt stress. The findings of this study will provide novel data and insights for a comprehensive understanding of the molecular mechanism of the oat TCP gene family’s response to adversity stress.
2 Materials and methods
2.1 Experimental materials and stress treatments
Naked oat Nei Avena 6 (NY6) and hulled oat Qing Yin 1 (QY1), provided by the Han Bing Oat Breeding Team at Inner Mongolia Agricultural University, were chosen as the primary experimental materials for this study. Uniformly shaped and sized oat seeds were surface-sterilized by immersing them in 2% NaClO for 5 min, rinsed with sterile water, and placed on filter paper soaked in sterile water. The seeds were then dark-incubated at 16°C in a temperature-controlled incubator until 5 cm shoots and primary roots developed. The seedlings were subsequently transferred to a 96-well hydroponic incubator containing Hoagland’s culture medium and grown for 20 days under day/night temperatures of 22/16°C and a photoperiod of 16/8 h. On the 21st day, seedlings were treated with 20% polyethylene glycol (PEG) 6,000 and 100 mM NaCl, while Hoagland’s solution served as the control. Samples from the treatment groups and control group were collected at 0, 2, 4, 8, 12, and 24 h. After sampling, the tissues were rapidly frozen in liquid nitrogen and stored at −80°C. Each sample included three independent biological replicates and technical replicates for qRT-PCR analysis.
2.2 Screening and characterization of AsTCP gene family
Genomic data of three oat varieties “SFS”, “Sang” and “OT3098v2” used to identify the oat TCP gene family were obtained from the OatBioDB Biology database (http://waoOat.cn/). Among them, “Sang” and “OT3098v2” are skin oats and “SFS” is a naked oat. Genome files and genome annotation files for rice, maize, wheat, A. thaliana, Brachypodium distachyon, A. tauschii, were obtained from the Ensembl Plants database (http://plants.ensembl.org/index.html). The HMM file for the TCP structural domain (PF03634) was derived from the Pfam database (https://pfam.xfam.org), and generated by training and constructing a large number of sequences known to be in this gene family. Using the built-in algorithm of HMMER 3.0 software, the input gene sequences were compared with the HMM model to generate the Score value for match strength and the E-value for statistical significance. According to the screening criteria, genes with E-value below 0.01 were initially labelled as candidate genes (Finn et al., 2011). To improve the identification accuracy, all genes identified by HMMER 3.0 (including a few genes with E-value greater than 0.01) were submitted to the NCBI CDD database (https://www.ncbi.nlm.nih.gov/cdd/) for structural domain validation (Lu et al., 2020). Eventually, genes with TCP structural domains were confirmed as members of the TCP gene family. TCP genes from the “SFS” were designated AsTCP001 to AsTCP083, while genes from the “Sang” and “OT3098v2” retained their respective genomic gene IDs.
2.3 Phylogenetic analysis of AsTCP gene family
DNAman was used to compare selected TCP amino acid sequences and construct a phylogenetic tree for oat TCPs, along with wheat, rice, and A. thaliana using MEGA11, with 1,000 bootstrap replicates. The phylogenetic tree was further refined using the Evolview online tool (https://www.evolgenius.info/evolview/#/treeview) to enhance visualization and clarity (He et al., 2016).
2.4 Analysis of the structure, physicochemical properties and promoter sequence of AsTCP gene
Conserved motifs in the oat TCP gene family were predicted using the MEME suite (https://meme-suite.org/meme/), with the number of predicted motifs set to 10 (Bailey et al., 2009). TCP protein structural domains were identified via the NCBI Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/). Gene structure analysis was conducted using the online tool GSDS 2.0 (http://gsds.cbi.pku.edu.cn/). Visualization of motifs, domains, and gene structures was carried out using TBtools (v2.042) software (Chen et al., 2020). Subcellular localization was predicted using WoLF PSORT (https://wolfpsort.hgc.jp/) and the Molecular Bioinformatics Center (MBC) website (http://cello.life.nctu.edu.tw/) (Horton et al., 2007). Oat TCP family amino acid sequences were analyzed for molecular weight and isoelectric point (pI) using the Expasy website (https://web.expasy.org/compute_pi/) (Artimo et al., 2012). Cis-acting elements within 2 kb upstream of the start codon of oat TCP genes were analyzed using the PlantCARE online tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
2.5 Chromosomal localization, covariance and interaction network analysis of AsTCP gene
Chromosomal localization of oat TCP genes was performed using TBtools software (Chen et al., 2020). Nonsynonymous (Ka) and synonymous (Ks) substitution rates were calculated for oat TCP genes using TBtools, and the Ka/Ks ratio was computed to assess evolutionary pressures influencing gene trends (>1 indicates positive selection, = 1 neutral selection, <1 purifying selection) (Chen et al., 2020). Duplication events of AsTCP genes were analyzed using the Multicollinearity Scanning Toolkit (MCScanX) with default parameters (Wang et al., 2012). The Dual Synteny Plotter tool within TBtools was employed to visualize synteny relationships of oat TCP genes with those from rice, maize, wheat, two-spike phragmites, and knapweed genomes (Chen et al., 2020). STRING (https://cn.string-db.org/) was employed to predict interacting proteins using Arabidopsis as the reference species. Additionally, psRNATarget (https://www.zhaolab.org/psRNATarget/analysis) was used to predict the miRNAs targeting AsTCP proteins (Dai et al., 2018). All results were visualized using Cytoscape 3.10.0 software.
2.6 Transcriptome data analysis
Transcriptome data related to silicon-mediated drought stress alleviation and salt stress in oats were sourced from the public NCBI (https://www.ncbi.nlm.nih.gov/sra/?term=Oat) (data number SRP237902, SRP093940) (Bray et al., 2016). FPKM values (log2 transformed) were used to analyze the expression of TCP family genes under the 2 treatments, and heatmaps were drawn using the Heatmap program of TBtools (Chen et al., 2020).
2.7 Real-time fluorescent quantitative PCR assay
Total RNA extraction from plants was performed using the Transzol Up Plus kit from Beijing All Style Gold. The first strand of cDNA was synthesized via reverse transcription, following the instructions provided with the PrimeScript RT kit from Takara. Quantitative PCR primers for AsTCP021, AsTCP025, AsTCP033, AsTCP044, AsTCP053, and AsTCP058 were designed using Primer 5.0 software (Supplementary Table S1) and synthesized by Beijing Liuhe Huada Gene Science and Technology Co. Oat β-Actin was used as the internal reference gene (Zhang, 2023). The expression level of each AsTCP gene was quantified using the 2-(ΔΔCt) method. The PCR reaction conditions were as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 20 s, and extension at 72°C for 20 s.
3 Results
3.1 Identification of members of AsTCP gene family
Using the genomic data of oat cultivars “SFS”, “Sang”, and “OT3098v2”, we identified 83, 65, and 30 members of the AsTCP gene family, respectively (Supplementary Tables S2, S3). In “SFS” and “Sang”, the amino acid sequences of 34 AsTCPs showed 100% identity, and those of 13 AsTCPs had over 90% identity. In “SFS” and “OT3098v2”, the amino acid sequences of 16 AsTCPs exhibited 100% identity, and those of 5 AsTCPs had over 90% identity. In “Sang” and “OT3098v2”, the amino acid sequences of 16 AsTCPs presented 100% identity, and those of 6 AsTCPs had over 90% identity. The “OT3098v2” contained three unique AsTCP genes (AVESA.00001b.r1.3Cg0000521.1, AVESA.00001b.r1.5Ag0002232.1, AVESA.00001b.r1.2Dg0002764.1). The “Sang” had four unique AsTCP genes (AVESA.00010b.r2.2DG0346510.1, AVESA.00010b.r2.2DG0402990.1, AVESA.00010b.r2.6DG1166000.1, and AVESA.00010b.r2.7AG1210110.1). The “SFS” had 22 unique AsTCP genes (AsTCP020 to AsTCP022, AsTCP033, AsTCP035, AsTCP040, AsTCP045, AsTCP056, AsTCP064, AsTCP065, AsTCP068, AsTCP070, AsTCP072, AsTCP075 to AsTCP083). Analysis of the amino acid sequence lengths of AsTCP genes in the “SFS” showed substantial variation, ranging from 71 to 609 amino acids (aa). The shortest protein, AsTCP081, comprised 71 aa, whereas the longest protein, AsTCP037, comprised 609 aa. The molecular weights of the 83 AsTCP proteins varied from 7954.97 to 64420.89 Da, and their isoelectric points (pI) ranged from 4.3 to 10.78 (Supplementary Table S4). Subcellular localization analysis indicated that 72 (86.75%) of the AsTCP proteins were localized in the nucleus. The remaining proteins were distributed as follows: five in the extracellular region, two at the plasma membrane, two in the cytoplasm, one in the chloroplast, and one in the mitochondrion (Supplementary Table S4).
3.2 Phylogeny and classification of the oat AsTCP gene family
Phylogenetic analysis of TCP proteins from oat, A. thaliana, rice, and wheat revealed that the 83 AsTCP proteins encoded by the “SFS” could be classified into three subfamilies: class I PCF, class II CYC/TB1, and CIN. Specifically, 40 proteins were categorized under the PCF subfamily, 13 under CYC/TB1, and 30 under CIN (Supplementary Figure S1A). In the “Sang”, 37 AsTCP proteins were classified as PCF, 10 as CYC/TB1, and 18 as CIN (Supplementary Figure S1B). Similarly, the “OT3098v2” showed 16 proteins in the PCF subfamily, 3 in CYC/TB1, and 11 in CIN (Supplementary Figure S1C). In the evolutionary relationship of TCP genes among oat, wheat, rice and Arabidopsis, the TCP genes in oat have a closer phylogenetic relationship with the TCP genes in wheat. A total of 14 AsTCP genes were found in the hulled and naked oat varieties “SFS”, “Sang” and “OT3098v2”, and the amino acid sequences encoded by them showed complete consistency (Supplementary Tables S2, S3). Within the oat genome, there are also different evolutionary relationships among members of the TCP gene family. Most oat TCP genes, such as AsTCP024/AsTCP023/AsTCP025, which are homologous genes distributed in subgroups A/C/D, are the closest in evolution, and these three genes are all distributed in the three oat genomes. A small number of oat TCP genes, such as AsTCP015/AsTCP017/AsTCP018 show a closer relationship to AsTCP021/AsTCP020/AsTCP022, AsTCP001/AsTCP003/AsTCP002 with AsTCP009/AsTCP008/AsTCP010, AsTCP043/AsTCP035 with AsTCP049/AsTCP048/AsTCP050, and AsTCP013/AsTCP012 with AsTCP014/AsTCP005. These types of partially homologous gene families have similar evolutionary relationships (Supplementary Figure S1; Supplementary Table S2).
Analysis of amino acid sequences revealed that 65, 59, and 27 AsTCP proteins in the “SFS”, “Sang”, and “OT3098v2”, respectively, possessed a complete helix-loop-helix (bHLH) structure, indicating a high conservation of the TCP structural domain in oats. Notably, the basic region within the bHLH of CYC/TB1 and CIN subfamilies contained a bidirectional nuclear localization signal (NLS), crucial for protein translocation to the nucleus. In contrast, the PCF subfamily exhibited a partial NLS in its basic region (Supplementary Figure S2A). These sequence differences likely contribute to the observed conservation pattern within the PCF subfamily, and suggest functional divergence between the subfamilies. Furthermore, variations were observed in the basic region of the bHLH domain. The CYC/TB1 and CIN subfamilies have four additional amino acids compared to the PCF subfamily. Specific AsTCP proteins, such as AsTCP69 and AsTCP71 in both “SFS” and “Sang”, exhibited amino acid deletions in the basic region (Supplementary Figures S2A, B). Additionally, 16, 3, and 3 AsTCP proteins in “SFS”, “Sang”, and “OT3098v2”, respectively, showed deletions spanning the entire bHLH domain (Supplementary Figure S3).
3.3 AsTCP conserved motifs, structural domains and gene structure
The phylogenetic tree results show that the 83 AsTCP genes are divided into three subfamilies: PCF, CIN, and CYC/TB1 (Figure 1A). Conservative base sequencing analysis identified 10 motifs present in the 83 AsTCP proteins (Supplementary Table S5). Several TCP genes exhibited deletions within the bHLH structural domain: AsTCP051, AsTCP067, AsTCP072, AsTCP077, AsTCP080, AsTCP078, and AsTCP081 lacked Motif 1 and Motif 2. While AsTCP070, AsTCP073, and AsTCP074 were missing Motif 1 (Figure 1B). Similarly, the AsTCP gene of “Sang” (AVESA.00010b.r2.5CG0872470.1, AVESA.00010b.r2.6CG1108510.1) and the AsTCP gene of “OT3098v2” (AVESA.001b.r1.2Dg0002764.1, AVESA.001b.r1.5Ag0002232.1,AVESA.00001b.r1.3Cg0000521.1) is also in a similar situation (Supplementary Figures S4, S5). Genes within the same subfamily exhibited similar motif compositions, indicating conserved motif types and distributions among closely related genes. Motif 4 is specific to the Class II (CYC/TB1 and CIN subfamilies), Motif 8 is specific to the CIN subfamily, and Motif 9 is specific to the PCF subfamily. In addition to the TCP structural domain, AsTCP061, AsTCP073, and AsTCP074 also contained two additional structural domains, flgK and HAD (Figure 1C). Gene structure analysis revealed that among the 18 AsTCP genes analyzed, intron numbers ranged from 1 to 4, with 78% of genes being intronless. Notably, the CIN subfamily exhibited the highest intron count among AsTCP genes (Figure 1D). Intronless AsTCP genes accounted for 65% and 40% of the total in the “Sang” and “OT3098v2”, respectively, significantly influencing the untranslated region (UTR) annotations of AsTCP genes (Supplementary Figures S4, S5). Additionally, differences in intron lengths within genes of the same subfamily contributed to significant variations in gene lengths.
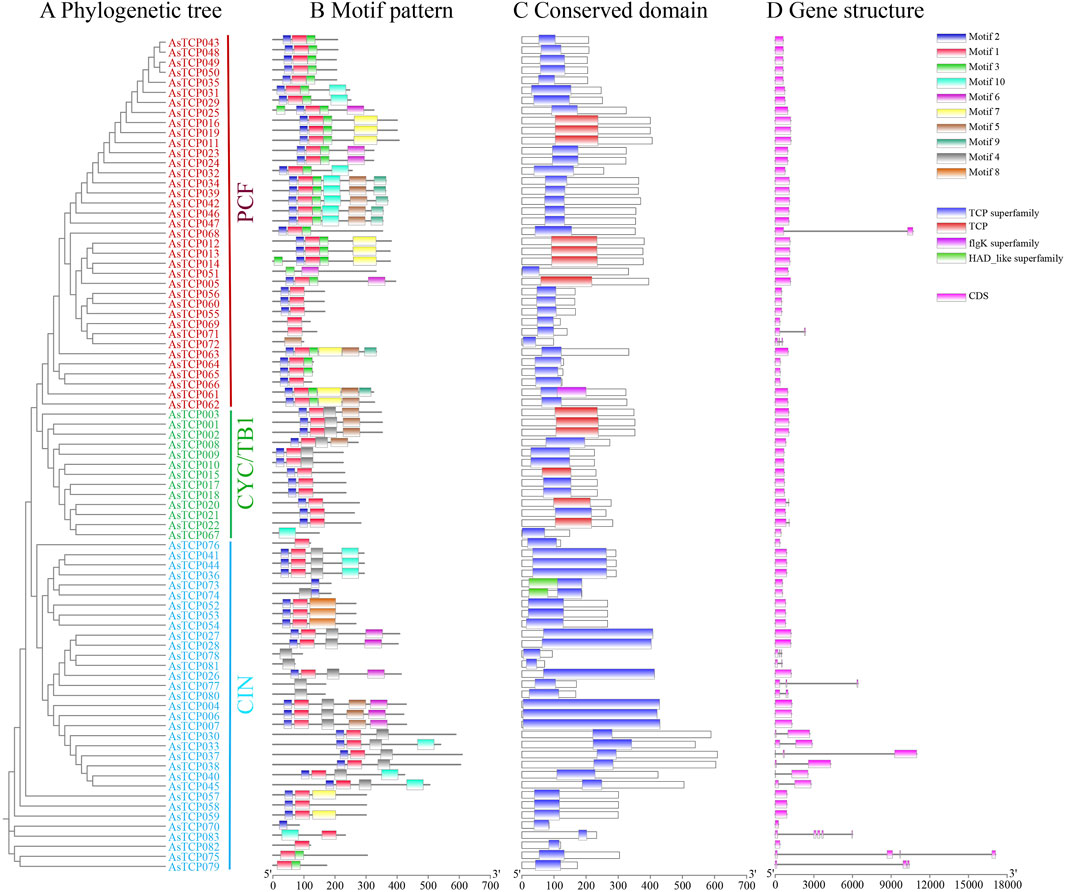
Figure 1. Phylogenetic analysis, motif patterns, conserved domains, and gene structure of TCP genes in the “SFS”. (A) Neighbor-Joining tree of oat TCP proteins; (B) Motif patterns, with motifs numbered 1-10 and represented by different colored boxes; (C) Conserved domains identified in the oat TCP proteins; (D) Gene structure representation, where CDS and introns are represented with pink boxes and black lines respectively.
3.4 Prediction of cis-acting elements in the promoter of AsTCP gene family
A total of 4 types of elements, including 51 different cis-acting elements, were discovered in the promoter region of the AsTCP gene in “SFS” (Supplementary Table S6). 19 hormone response elements were widely distributed in the promoter regions of the AsTCPs (Figure 2), with the highest number of ABREs attributed to abscisic acid response elements, accounting for about 21% of the total number of hormone elements. In addition, the promoter regions harbored 14 types of stress-responsive elements, including those responsive to drought, endosperm-specific expression, low-temperature, cell cycle regulation, meristem expression, maize protein metabolism regulation, and defense stress responses (Figure 2). MYC and MYB elements, crucial for environmental adaptation, were particularly abundant, accounting for approximately 23.7% and 23.2% of total stress-responsive elements, respectively. 13 types of light-responsive elements were also identified, with G-box elements (Figure 2), which can bind MYC proteins, being the most prevalent at about 34.2% of all light-responsive elements. Furthermore, 5 types of physiological response elements were found, widely distributed across AsTCP gene promoters (Figure 2). The CCGTCC motif was the most abundant, comprising approximately 30.6% of all physiological response elements. The types of elements in the promoter regions of the AsTCP gene in “SFS”, “Sang” and “OT3098v2” are similar (Supplementary Figures S6– S8).
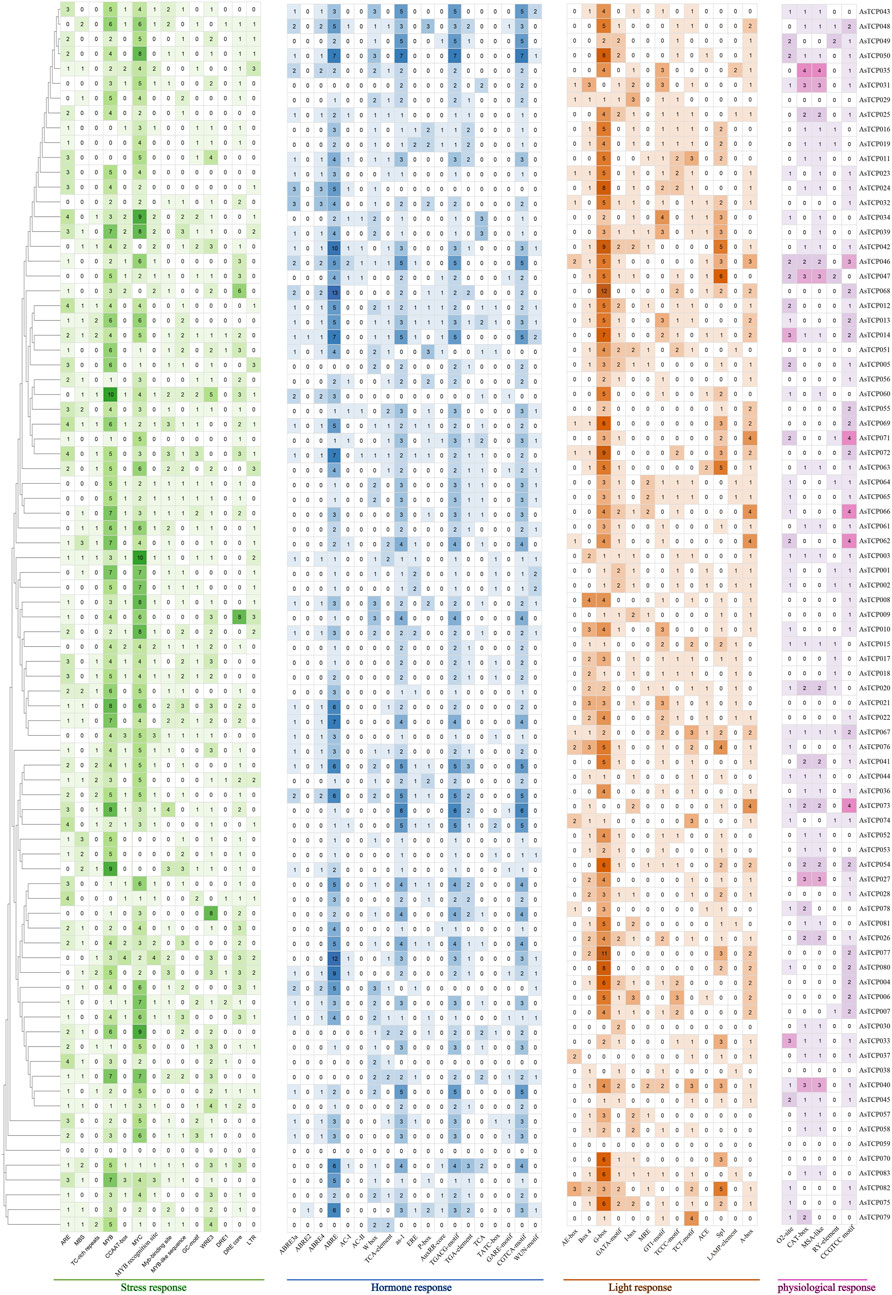
Figure 2. Cis-acting elements on promoters of oat TCP genes. ARE, cis-acting regulatory element essential for the anaerobic induction; MBS, MYB binding site involved in drought-inducibility; TC-rich repeats, cis-acting element involved in defense and stress responsiveness; CCAAT-box, MYBHv1 binding site; GC-motif, enhancer-like element involved in anoxic specific inducibility; LTR, cis-acting element involved in low-temperature responsiveness; ABRE, cis-acting element involved in the abscisic acid responsiveness; TCA-element, cis-acting element involved in salicylic acid responsiveness; P-box and GARE-motif, gibberellin-responsive element; AuxRR-core, cis-acting regulatory element involved in auxin responsiveness; TGACG-motif, cis-acting regulatory element involved in the MeJA-responsiveness; TGA-element, auxin-responsive element; TATC-box, cis-acting element involved in gibberellin-responsiveness; WUN-motif, wound-responsive element; AE-box, part of a module for light response; Box 4, part of a conserved DNA module involved in light responsiveness; G-box, cis-acting regulatory element involved in light responsiveness; GATA-motif, I-box, TCCC-motif, TCT-motif, and LAMP-element, part of a light responsive element; MRE, MYB binding site involved in light responsiveness; GT1-motif and Sp1, light responsive element; ACE, cis-acting element involved in light responsiveness; A-box, cis-acting regulatory element/sequence conserved in alpha-amylase promoters; O2-site, cis-acting regulatory element involved in zein metabolism regulation; CAT-box, cis-acting regulatory element related to meristem expression; MSA-like, cis-acting element involved in cell cycle regulation; RY-element, cis-acting regulatory element involved in seed-specific regulation.
3.5 Comparative analysis of chromosome localization and covariance
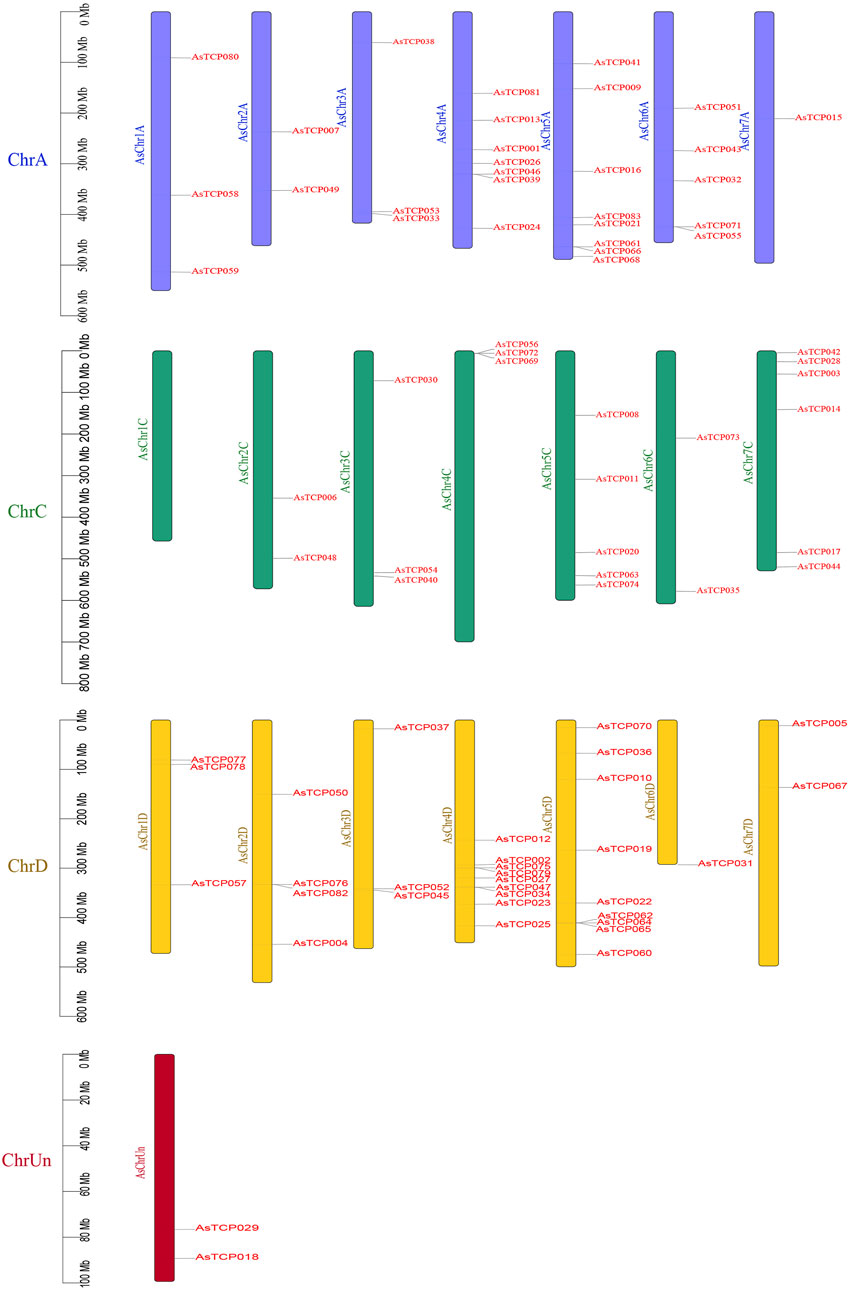
In the “SFS”, 21 AsTCP genes were localized on chromosome A, 29 on chromosome C, 31 on chromosome D, and 2 on Un (Figure 3). In the “Sang”, 22 AsTCP genes were localized on chromosome A, 16 on chromosome C, 23 on chromosome D, and 4 on Un (Supplementary Figure S9A). For the “OT3098v2”, 12 AsTCP genes were localized on chromosome A, 5 on chromosome C, 11 on chromosome D, and 2 on Un (Supplementary Figure S9B). Generally, the distribution of AsTCP genes across the A, C, and D subgenomes was relatively even, with the highest number of AsTCP genes found on subgenomes 4D and 5D, each containing 9 genes. Notably, only one gene was found on subgenome 6D, and none were present on 1C.
Gene duplication event analysis revealed 89, 73, and 20 homologous pairs of AsTCP gene family members in the “SFS”, “Sang”, and “OT3098v2”, respectively. Among these, two pairs (AsTCP058/AsTCP059 and AsTCP023/AsTCP025) were identified as tandem duplications across all three genomes, while the remaining 87, 71, and 18 pairs were segmental duplications (Figure 4; Supplementary Figure S10). Notably, 89 homologous gene pairs in the “SFS” exhibited Ka/Ks < 1, indicating that these genes were primarily subject to purifying selection, with no pairs evolving under strong positive selection (Ka/Ks > 1) post-duplication (Supplementary Table S7). These findings suggest that oat TCP genes underwent both segmental and tandem duplications, with segmental duplication being the predominant mode, and two pairs of tandem duplications remaining stable across the three oat genomes.
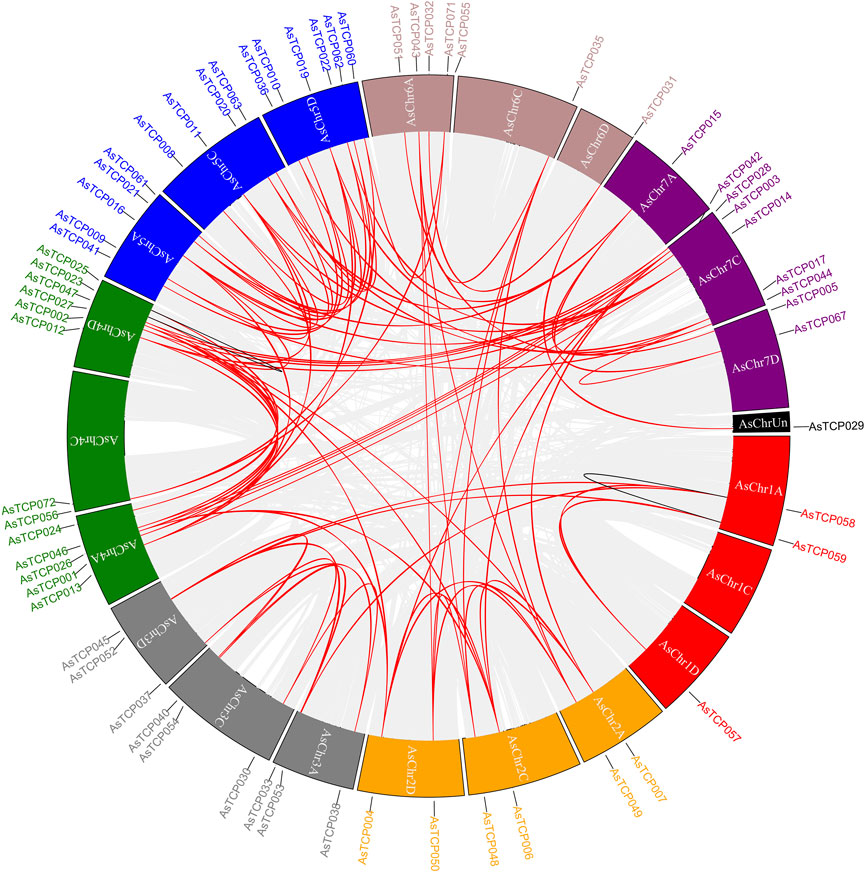
Figure 4. Synteny analysis of AsTCP genes in oat. Red colored lines indicate segmental duplication gene pairs, black lines indicate tandem duplication pairs. Chromosomes 1 red, 2 orange, 3 gray, 4 green, 5 blue, 6 maroon, 7 purple, Un black.
Comparative collinearity analysis between oat (“SFS”) and other species, including A. thaliana, rice, Brachypodium, Setaria, maize, and wheat, revealed the highest TCP gene homology with wheat, comprising 175 collinear pairs between 54 AsTCPs and 54 TaTCPs. Conversely, the lowest homology was observed with A. thaliana, where 14 AsTCPs exhibited collinearity with 4 AtTCPs, resulting in 15 collinear pairs (Figure 5; Supplementary Table S8). 42 AsTCP genes in the oat genome showed collinearity with TCP genes in all other species, excluding Arabidopsis. 9 out of the 12 AsTCP genes collinear with Arabidopsis were also part of these 42 genes (Supplementary Table S8). These duplicated AsTCP genes, exhibiting collinearity with other species, likely participated more frequently in gene duplication events, playing significant roles in the evolutionary trajectory of oats.

Figure 5. Synteny analysis of AsTCP genes between oat and six representative plant species. The gray lines show colinear blocks in the genomes of oat with other plants, while the red line highlights the colinear TCP pairs. The species name with the prefixes “A.sativa”, “A.thaliana”“O. sativa” ‘B.distachyon”, “A.tauschii”, “Z.mays” and “T. aestivum” indicate Avena sativa, Arabidopsis thaliana, Oryza sativa, Brachypodium distachyon, Aegilops tauschii, Zeamays, and Triticum aestivum, respectively.
3.6 Protein interaction network and miRNA prediction of AsTCP gene
The predicted protein interaction relationships of AsTCP genes as well as miRNA realisation results were visualised by Cytoscape software. The results showed that 35 AsTCP proteins were predicted to have interaction relationships with other proteins, among which, 8 proteins, AsTCP005, AsTCP016, AsTCP019, AsTCP030, AsTCP037, AsTCP038, and AsTCP051, had a higher number of interacting proteins. Among those interacting proteins, SAUR65, SRFR1, APRR1, SAP11, and CYCB1-1 have higher chance to interact with these 8 AsTCP proteins as mentioned above (Figure 6A). When predicting the upstream regulator miRNAs of AsTCP genes, 197 upstream regulator miRNAs were found for 15 AsTCP genes, including AsTCP004, AsTCP006, AsTCP007, AsTCP026, and AsTCP027. When the expectation value was less than 2.5, all the upstream regulatory factors were miR319 (Figure 6B; Supplementary Table S9). Therefore, miR319 may play an important regulatory role in the AsTCP gene family.
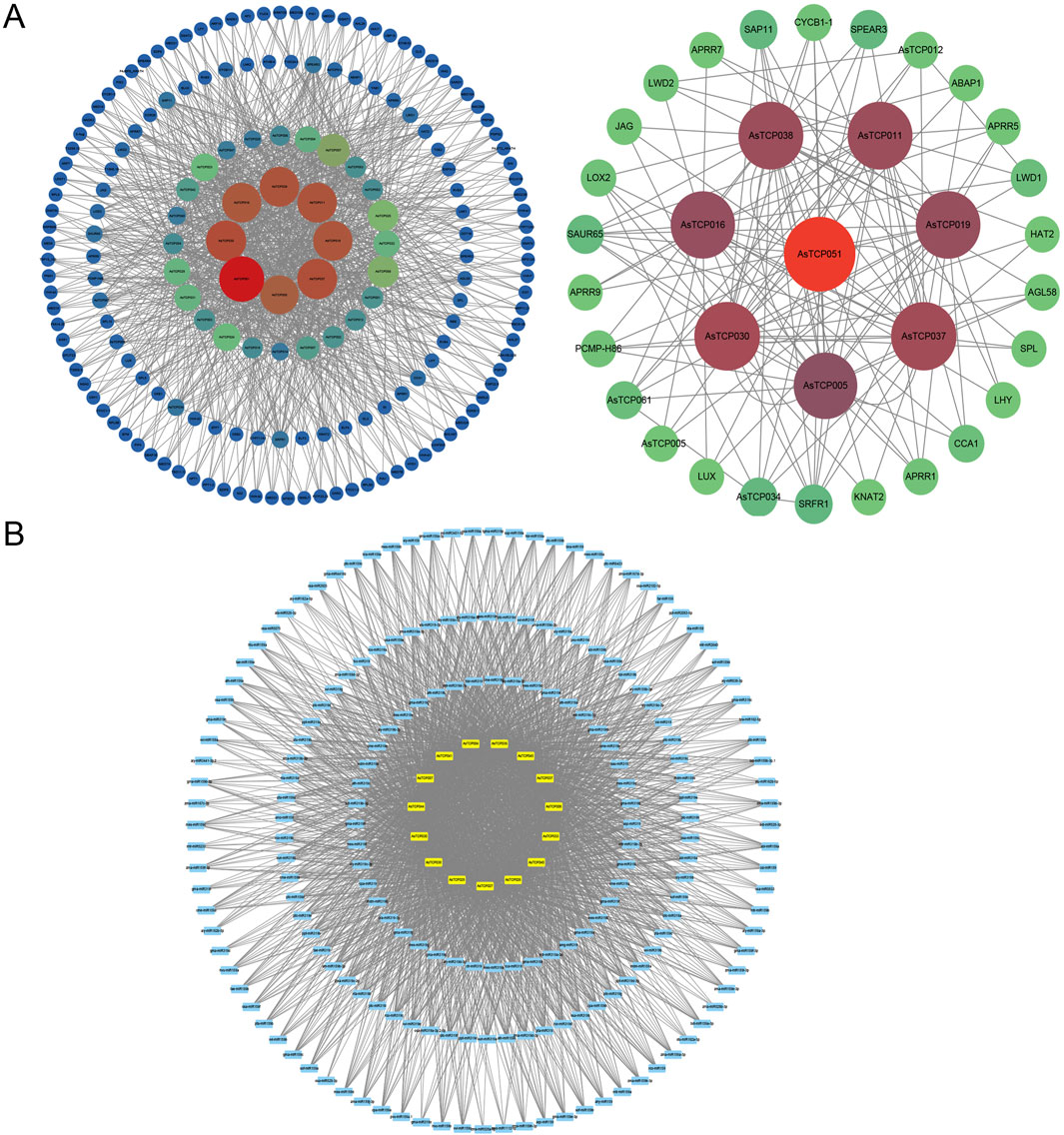
Figure 6. Protein-Protein Interaction Network and miRNA Prediction for AsTCP Genes. (A) The protein interaction network based on the Arabidopsis homologue AsTCP protein; (B) The relationship between members of the AsTCP gene family and miRNAs.
3.7 Expression profiles of AsTCP genes under abiotic stresses
Given the critical role of the TCP gene family in regulating plant stress resistance, the expression patterns of 83 AsTCP genes under silicon-mediated drought stress alleviation and salt stress treatments were analyzed (Supplementary Tables S10, S11). 83 AsTCP genes (excluding AsTCP068, AsTCP020, AsTCP076, AsTCP078, AsTCP077, AsTCP082, AsTCP075, AsTCP079) had significant responses to drought and salt stresses (Figure 7). Under drought treatment, AsTCP005, AsTCP010, AsTCP011, AsTCP021, AsTCP053, AsTCP056, AsTCP065, AsTCP069, and AsTCP072 were significantly downregulated, whereas in the control group, these genes were generally significantly upregulated. On the other hand, AsTCP007, AsTCP033, AsTCP044, AsTCP045, AsTCP071, AsTCP073, and AsTCP080 were significantly upregulated under drought treatment but significantly downregulated silicon-mediated drought stress alleviation. These genes generally exhibited low expression levels in the control group (Figure 7A). These results suggest that the aforementioned genes may play key roles in responding to drought stress. Under short-term salt stress, different oat varieties exhibited varying sensitivities to salt stress. There were significant differences in the timing and expression patterns of some AsTCP genes between the two cultivars, Huazao-2 and Hanyou-5. In addition, the majority of AsTCPs were significantly upregulated at 8 h and 12 h of salt stress (Figure 7B).
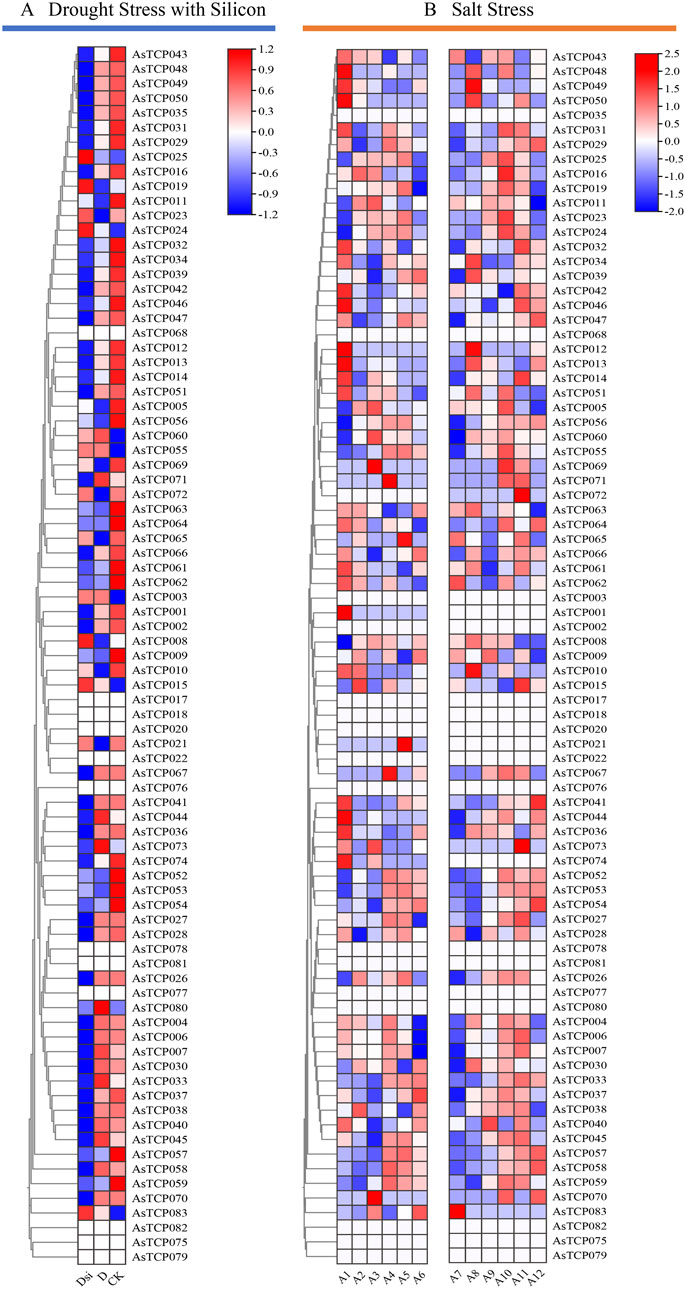
Figure 7. The expression patterns of 83 AsTCP genes stress levels are based on RNA-seq data.(A) Analysis of TCP gene expression induced by exogenous silicon addition Under drought conditions,D (Drought stress),Dsi (Adding exogenous silicon under drought stress); (B) Expression analysis of TCP genes was conducted on Huazao-2 under salt stress at 0 h (A1), 2 h (A2), 4 h (A3), 8 h (A4), 12 h (A5), and 24 h (A6), and on Hanyou-5 at 0 h (A7), 2 h (A8), 4 h (A9), 8 h (A10), 12 h (A11), and 24 h (A12) under salt stress.
3.8 Expression patterns of AsTCP genes in abiotic stresses
To investigate the expression levels of AsTCP genes in different oat varieties under drought stress (20% PEG6000) and salt stress (100 mM NaCl), six stress-responsive genes were selected from the oat TCP expression profile for qRT-PCR analysis. Among them, AsTCP021 and AsTCP033 were identified in the “SFS”, AsTCP044 and AsTCP053 were identified in both the “SFS” and “Sang”, while AsTCP025 and AsTCP058 were identified in the “SFS”, “Sang” and “OT3098v2”. In NY6, AsTCP044 and AsTCP058 were significantly upregulated at 4 h under NaCl treatment, whereas AsTCP021, AsTCP033, and AsTCP053 were significantly downregulated at 4 h (Figure 8A). In QY1, AsTCP021, AsTCP033, and AsTCP053 were significantly upregulated at 24 h under NaCl treatment, while AsTCP033, AsTCP044, and AsTCP058 were significantly downregulated at 0 h (Figure 8B). Comparison with transcriptome results from Huazao-2 and Hanyou-5 revealed that AsTCP025, AsTCP033, AsTCP044, and AsTCP058 displayed similar expression trends at 0 h and 2 h across the four varieties. Furthermore, AsTCP053 showed consistent expression trends in the four varieties, being downregulated at 0 h, 2 h, and 4 h, and upregulated at 8 h, 12 h, and 24 h, except for QY1, where it was upregulated at 2 h. In NY6, AsTCP021, AsTCP025, and AsTCP053 were significantly upregulated at 24 h under PEG treatment, while AsTCP021, AsTCP025, AsTCP033, and AsTCP053 were significantly downregulated at 4 h (Figure 8A). In QY1, AsTCP033, AsTCP044, and AsTCP053 were significantly upregulated at 24 h under PEG treatment, while AsTCP021 and AsTCP033 were significantly downregulated at 0 h (Figure 8B). The qRT-PCR results for AsTCP021 and AsTCP053 in NY6 under drought stress from 0 h to 12 h were largely consistent with transcriptome results, both showing a trend of downregulation.
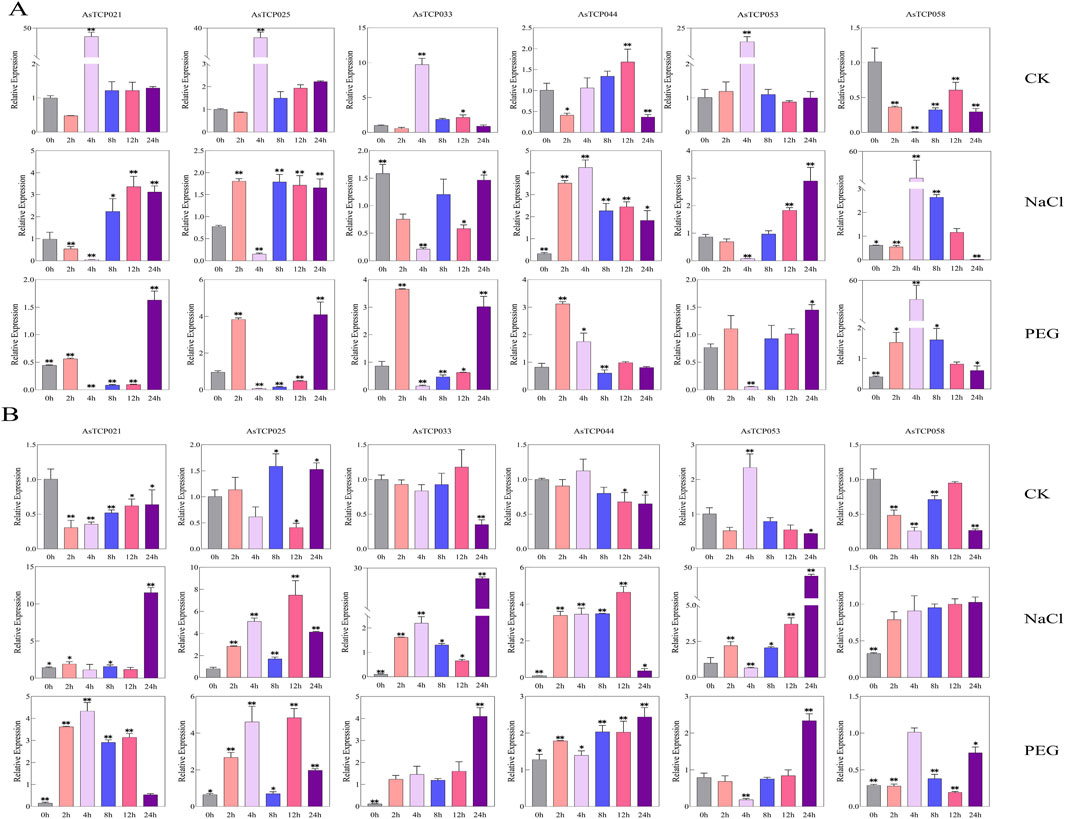
Figure 8. Expression of 6 AsTCP Genes Under Untreated (CK), Drought (PEG6000), and Salt (100 mM NaCl) Conditions. (A) Expression of six AsTCP genes in NY6; (B) Expression of six AsTCP genes in QY1. Explanations: CK Group: AsTCP genes expression with 0 h as the control, NaCl and PEG Groups. AsTCP genes expression with the corresponding CK time point as the control. Significant differences between treatment and control groups are indicated by * (LSD test, p < 0.05) and ** (p < 0.01).
In both NY6 and QY1, the six AsTCP genes exhibited significant responses to both salt and drought stress, with the most notable changes observed at 0 h, 4 h, and 24 h. At 0 h under drought and salt stress, AsTCP058 was significantly downregulated in both varieties. At 4 h, AsTCP044 was significantly upregulated in both varieties, while AsTCP053 was significantly downregulated. At 24 h, AsTCP025, AsTCP033, and AsTCP053 were significantly upregulated in both varieties. Additionally, in the QY1 variety, AsTCP033 and AsTCP053 showed the most significant upregulation at 24 h under both drought and salt stress. The expression patterns of different genes at different time points varied significantly between the two varieties, particularly at 24 h, where the notable upregulation of some genes in QY1 reflected a stronger response to stress. These findings indicate that varietal differences in stress sensitivity significantly influence the expression patterns of these genes.
4 Discussion
TCP gene family members have been identified in various grass species, including 66 in wheat (Zhao et al., 2018), 46 in maize (Ding et al., 2019), 24 in Arabidopsis, and 28 in rice (Yao et al., 2007), as well as 20 in sorghum (Lei et al., 2021), 42 in willowherb (Huo et al., 2019), and 22 in orchardgrass (Wang C. et al., 2023). In this study, we identified 83, 65, and 30 TCP genes in the three oat genomes, respectively. Each of these AsTCP genes was specific to their corresponding oat genomes (Supplementary Table S2). As oats are heterozygous hexaploids, the presence of multiple gene copies within their genomes can be attributed to extensive replication and diversification during evolution (Song et al., 2015). The differences in the number of TCP genes among the three oat genomes may be attributed to several factors. Firstly, with the advancement of sequencing technologies and the availability of reference oat genomes, the genome assembly techniques and methods used in later sequencing projects have become more refined, allowing for greater sequencing depth. Additionally, different gene annotation methods and parameters can lead to variations in the number of identified genes (Tortuero et al., 2021). Secondly, transcription factors exist in various subtypes or alternative splicing forms, which may be identified as distinct genes in different genomes. Moreover, transcription factor families evolve rapidly, and events such as gene duplication, loss, or variation in different genomes can result in significant differences in gene copy numbers within these families (Adams and Wendel, 2005; Wang et al., 2018). Despite these differences, the AsTCP genes were classified into three subclasses: Class I (PCF), Class II (CIN), and (CYC/TB1) (Supplementary Figure S1). The high similarity between oat TCP genes and those in Arabidopsis, rice, and wheat suggests that TCP genes are highly conserved in plants (Martín-Trillo and Cubas, 2010). This further indicates that the AsTCP genes in oats evolved from a common ancestor within the Gramineae family, undergoing different modes of divergence in different lineages. In our study, we found that most of the TCP genes with amino acid deletions within the bHLH structure lacked Motif 1 and Motif 2, which are presumed to be key components of the TCP structural domain. Additionally, we observed that most oat TCP genes lacked intronic structures (Figure 1D and Supplementary Figures S4, S5). This may be related to the fact that intronless genes are often derived from horizontal gene transfer of intronless ancient prokaryotes, replication of existing intronless genes, or retrotranscription of intron-containing genes (Zou et al., 2011). Moreover, differences in conserved motifs and gene structures among subfamilies within the oat AsTCP gene family likely contribute to its functional diversity.
Members of the oat TCP family exhibit extensive variation in amino acid sequences, isoelectric points, relative molecular masses, and exon numbers, indicating their structural complexity and functional diversity (Xiao et al., 2018). Subcellular localization analysis revealed that most oat TCP proteins are localized in the nucleus and are also present in the extracellular, plasma membrane, cytoplasm, chloroplasts, and mitochondria (Supplementary Table S4). This distribution pattern may be associated with the broad range of roles that TCP genes play in plant growth and development. Transcription factors (TFs) specifically bind to cis-acting elements to regulate the expression of target genes (Zhao et al., 2021), and the diversity of these cis-acting sites determines the regulatory functions of TFs(Lei et al., 2021). Our analysis of promoter cis-acting elements found a variety of elements involved in hormone response, light response, and stress response were widely distributed in the promoters of the 83 AsTCP genes (Figure 2). This further highlights the involvement of TCP family genes in various biological processes, including photosynthesis, hormone regulation, growth and development, and stress response in plants (Ling et al., 2020; Ren et al., 2021; Xiong et al., 2022). In the oat genome, TCP genes exhibit a widespread phenomenon of multiple copies, such as AsTCP024/AsTCP023/AsTCP025, which are distributed in the A/C/D subgenomes. They demonstrate the closest evolutionary relationship among the three oat subgenomes. Additionally, AsTCP001/AsTCP002 show a more similar evolutionary relationship to AsTCP009/AsTCP008/AsTCP010, as well as AsTCP013/AsTCP012 to AsTCP014/AsTCP015. These genes are distributed in both the “SFS” and “Sang” (Supplementary Figure S1; Supplementary Table S2). These closely related genes in evolutionary terms likely share more similar sequences and structures, potentially indicating higher functional similarity (Peterson et al., 2010). Plants have undergone large-scale chromosome doubling events during evolution (Flagel and Wendel, 2009), and each replication or doubling of the genome leaves traces of loss, transfer, and recombination on the chromosomes (Peng et al., 2022). Our study revealed a direct correlation between the distribution of TCP genes in the three oat genomes and the lengths of the chromosomes, with none of the being located on chromosome 1C. Gene duplication analysis indicated that segmental duplications predominated in the oat TCP gene family, with two pairs of tandem duplication events consistently present in all three genomes (Figure 4 and Supplementary Figure S10). These findings suggest that segmental duplications contribute to the amplification and evolution of the oat TCP gene family. Interestingly, segmental duplications are also prevalent in Arabidopsis and rice, suggesting a common mechanism for TCP gene duplication in plant genomes (Liu et al., 2022). Genome-wide covariance analysis demonstrated that oats had the highest number of covariant pairs with wheat and the fewest with Arabidopsis (Figure 5). This difference may be attributed to the genomic characteristics and evolution of oats, being a homozygous hexaploid plant that shares a closer evolutionary origin with wheat. Arabidopsis thaliana, a diploid dicotyledonous plant, evolved from a common ancestor shared with gramineous plants. However, the evolution of monocots predates that of dicots, and TCP genes may have undergone different modes of divergence between these two plant groups.
Oats are known for their high adaptability to harsh environments, and a gene’s function can often be inferred from its expression profile (Jewiss, 1972). Our analysis revealed that more than half of the AsTCP genes are involved in the response of oats to drought and salt stress (Figure 7). Similar results have been reported in canola (Brassica napus) (Wen et al., 2021), cotton (Yin et al., 2018), and orchardgrass (Huo et al., 2019). Studies have shown that TCP genes are involved in plant defense against abiotic stresses by regulating the expression of downstream genes. For example, rice OsPCF2 regulates the downstream OsNHX1 genes to enhance salt and drought tolerance (Almeida et al., 2017), whereas bamboo PeTCP10 binds to the downstream BT gene to regulate drought tolerance (Liu et al., 2020). In our study, we observed temporal differences in the response of TCP genes to salt and drought stress in two oat varieties, with variations in expression patterns and degrees of response (Figure 7B). The same conclusions were drawn from the expression profiles of Huazao-2 and Hanyou-5 under salt stress. Specifically, in QY1, the expression of AsTCP044, AsTCP058, and AsTCP033 was significantly downregulated at 0 h and 4 h after NaCl treatment (Figure 8B). In NY6, their expression was significantly downregulated at 0 h and 4 h but upregulated at 0 h after NaCl treatment (Figure 8A). Similarly, after PEG treatment, AsTCP021 and AsTCP053 were significantly downregulated and upregulated at 4 h and 24 h in NY6(Figure 8A), but downregulated at 0 h in QY1 (Figure 8B). These differences in expression may be attributed to the varying sensitivities of different oat varieties to the stress environment. Furthermore, at 0 h, 4 h, and 24 h of both stress treatments, the expression pattern of the AsTCP053 gene was consistent in the two varieties, but there were differences in the degree of response to stress between them at 4 h and 24 h. Notably, at 24 h of drought and salt stress, the upregulation of the AsTCP053 gene was more prominent in QY1 compared to NY6 Particularly under salt stress at 24 h, the upregulation of AsTCP053 in QY1 was remarkably significant. Our study revealed that the expression of the AsTCP053 gene exhibited an upregulation trend at 8 h, 12 h, and 24 h under salt stress treatment across four varieties, with expression levels gradually increasing over time. Prediction analysis indicated that the promoter region of AsTCP053 is enriched with various cis-acting elements, including MYB and ARE. Under salt stress conditions, ARE element may interact with Nrf2-like transcription factors to activate genes related to antioxidant synthesis, thereby scavenging ROS accumulation (Zgorzynska et al., 2021). Meanwhile, the MYB element may function through the ABA signaling pathway by cooperating with MYB transcription factors to further activate downstream stress-responsive genes (Bo et al., 2015). The coordinated regulation of antioxidant and stress-responsive mechanisms mediated by ARE and MYB elements likely constitutes the key molecular basis for the dynamic regulation of AsTCP053 under salt stress conditions. We hypothesize that the AsTCP053 gene is a key regulator in oat response to salt stress, but currently, there is a lack of reports on its downstream genes in the salt stress pathway. We will focus on the exploration of the downstream gene functions of AsTCP053 in our future research.
5 Conclusion
We analyzed the genome data of oats “SFS”, “Sang” and “OT3098v2” to identify TCP genes and perform bioinformatics analysis on them. Additionally, we selected six representative AsTCP genes to examine their expression patterns in hulled and hulless oats under drought and salt stress treatments.
1. In the genomes of the three oat varieties “SFS”, “Sang” and “OT3098v2”, we identified 83, 65, and 30 TCP genes, respectively, with some genes being unique to specific genomes.
2. The bHLH domain of some AsTCPs showed varying degrees of deletion, with Motif 1 and Motif 2 being key components of the bHLH domain. Only a small proportion of AsTCP genes in oats contain introns.
3. Segmental duplication is a common mechanism driving the expansion of the AsTCP gene family, with two pairs of tandemly duplicated TCP genes consistently present in the three oat genomes.
4. The genes AsTCP021, AsTCP025, AsTCP033, AsTCP044, AsTCP053, and AsTCP058 are involved in the oat response to drought and salt stress, with significant gene expression observed at 0 h, 4 h, and 24 h. Due to differences in variety sensitivity to stress, the response time and gene expression patterns varied between the two oat varieties under the same stress conditions
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
JN: Methodology, Writing–review and editing, Data curation, Visualization, Writing–original draft. HZ: Funding acquisition, Methodology, Validation, Writing–original draft, Writing–review and editing. XG: Data curation, Supervision, Writing–review and editing. TZ: Methodology, Project administration, Supervision, Validation, Visualization, Writing–review and editing. BH: Data curation, Project administration, Supervision, Validation, Writing–review and editing. HL: Project administration, Supervision, Validation, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the First-class Academic Subjects Special Research Project of the Education Department of Inner Mongolia Autonomous Region (YLXKZX-NND-031), the National Natural Science Foundation of China (32360025) and the Central Guided Local Science and Technology Development Funds (2022ZY0069). This study was also supported by the Water Conservancy Science and Technology Project of Inner Mongolia Autonomous Region (NSK202405) and the Natural Science Foundation of Inner Mongolia Autonomous Region (2021MS03094).
Acknowledgments
We thank Yongwang Dong, Qidi Zhang, and Simeng Cai for their valuable help in sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1533562/full#supplementary-material
References
Adams, K. L., and Wendel, J. F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 (2), 135–141. doi:10.1016/j.pbi.2005.01.001
Almeida, D. M., Gregorio, G. B., Oliveira, M. M., and Saibo, N. J. (2017). Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 93, 61–77. doi:10.1007/s11103-016-0547-7
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic acids Res. 40 (W1), W597–W603. doi:10.1093/nar/gks400
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic acids Res. 37 (Suppl. l_2), W202–W208. doi:10.1093/nar/gkp335
Bo, L., Wengzheng, Z., Chunyu, L., and Feng, M. (2015). The MYB family transcription factor OsMYB84 participates in salt stress response through ABA signaling pathway. J. Fudan Univ. Nat. Sci. Ed. (5), 10. doi:10.15943/j.cnki.fdxb-jns.2015.05.007
Braun, N., de Saint Germain, A., Pillot, J.-P., Boutet-Mercey, S., Dalmais, M., Antoniadi, I., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant physiol. 158 (1), 225–238. doi:10.1104/pp.111.182725
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34 (5), 525–527. doi:10.1038/nbt.3519
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18 (2), 215–222. doi:10.1046/j.1365-313x.1999.00444.x
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic acids Res. 46 (W1), W49–W54. doi:10.1093/nar/gky316
Dhaka, N., Bhardwaj, V., Sharma, M. K., and Sharma, R. (2017). Evolving tale of TCPs: new paradigms and old lacunae. Front. plant Sci. 8, 479. doi:10.3389/fpls.2017.00479
Ding, S., Cai, Z., Du, H., and Wang, H. (2019). Genome-wide analysis of TCP family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 20 (11), 2762. doi:10.3390/ijms20112762
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 (6624), 485–488. doi:10.1038/386485a0
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic acids Res. 39 (Suppl. l_2), W29–W37. doi:10.1093/nar/gkr367
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 (3), 557–564. doi:10.1111/j.1469-8137.2009.02923.x
Guan, P., Ripoll, J.-J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U. S. A. 114 (9), 2419–2424. doi:10.1073/pnas.1615676114
Gutierrez-Gonzalez, J. J., Tu, Z. J., and Garvin, D. F. (2013). Analysis and annotation of the hexaploid oat seed transcriptome. BMC genomics 14 (1), 1–17. doi:10.1186/1471-2164-14-471
Han, L., Eneji, A. E., Steinberger, Y., Wang, W., Yu, S., Liu, H., et al. (2014). Comparative biomass production of six oat varieties in a saline soil ecology. Commun. soil Sci. plant analysis 45 (19), 2552–2564. doi:10.1080/00103624.2014.912299
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W.-H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic acids Res. 44 (W1), W236–W241. doi:10.1093/nar/gkw370
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic acids Res. 35 (Suppl. l_2), W585–W587. doi:10.1093/nar/gkm259
Huo, Y., Xiong, W., Su, K., Li, Y., Yang, Y., Fu, C., et al. (2019). Genome-wide analysis of the TCP gene family in switchgrass (Panicum virgatum L.). Int. J. genomics 2019 (1), 8514928. doi:10.1155/2019/8514928
Jewiss, O. (1972). Tillering in grasses—its significance and control. Grass Forage Sci. 27 (2), 65–82. doi:10.1111/j.1365-2494.1972.tb00689.x
Jiao, P., Liu, T., Zhao, C., Fei, J., Guan, S., and Ma, Y. (2023). ZmTCP14, a TCP transcription factor, modulates drought stress response in Zea mays L. Environ. Exp. Bot. 208, 105232. doi:10.1016/j.envexpbot.2023.105232
Ju, Z., Liu, K., Zhao, G., Ma, X., and Jia, Z. (2022). Nitrogen fertilizer and sowing density affect flag leaf photosynthetic characteristics, grain yield, and yield components of oat in a semiarid region of northwest China. Agronomy 12 (9), 2108. doi:10.3390/agronomy12092108
Junjun, S., Yaqi, Z., Danfeng, Z., Wang, W., Song, R., Gu, R., et al. (2019). CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. U. S. A. 116 (34), 17105–17114. doi:10.1073/pnas.1907968116
Kankan, P., Zhipeng, R., Shengnan, W., Yu, T., Shuo, N., Xuan, M., et al. (2024). TaTCP21-A negatively regulates wheat cold tolerance via repressing expression of TaDREB1C. Plant Physiology Biochem. 219, 109353. doi:10.1016/j.plaphy.2024.109353
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. Cell and Mol. Biol. 68 (1), 147–158. doi:10.1111/j.1365-313X.2011.04674.x
Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 (9), 1607–1619. doi:10.1105/tpc.9.9.1607
Lei, P., Wei, X., Gao, R., Huo, F., Nie, X., Tong, W., et al. (2021). Genome-wide identification of PYL gene family in wheat: evolution, expression and 3D structure analysis. Genomics 113 (2), 854–866. doi:10.1016/j.ygeno.2020.12.017
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Li, W., Chen, G., Xiao, G., Zhu, S., Hu, T., Zhu, P., et al. (2020). Overexpression of TCP transcription factor OsPCF7 improves agronomic trait in rice. Mol. Breed. 40 (5), 48. doi:10.1007/s11032-020-01129-5
Li, Y., Shi, Y., Wang, B., Li, F., An, L., Jiang, J., et al. (2024). The overexpression of the BpTCP20 gene enhances cell proliferation and improves tolerance to drought and salt stress in Betula platyphylla. Industrial Crops and Prod. 214, 118521. doi:10.1016/j.indcrop.2024.118521
Ling, L., Zhang, W., An, Y., Du, B., Wang, D., and Guo, C. (2020). Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. and Integr. genomics 20, 537–550. doi:10.1007/s10142-020-00733-0
Liu, D.-H., Luo, Y., Han, H., Liu, Y.-Z., Alam, S. M., Zhao, H.-X., et al. (2022). Genome-wide analysis of citrus TCP transcription factors and their responses to abiotic stresses. BMC plant Biol. 22 (1), 325. doi:10.1186/s12870-022-03709-3
Liu, H., Gao, Y., Wu, M., Shi, Y., Wang, H., Wu, L., et al. (2020). TCP10, a TCP transcription factor in moso bamboo (Phyllostachys edulis), confers drought tolerance to transgenic plants. Environ. Exp. Bot. 172, 104002. doi:10.1016/j.envexpbot.2020.104002
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic acids Res. 48 (D1), D265–D268. doi:10.1093/nar/gkz991
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 (6603), 794–799. doi:10.1038/383794a0
Manassero, N. G. U., Viola, I. L., Welchen, E., and Gonzalez, D. H. (2013). TCP transcription factors: architectures of plant form. Biomol. concepts 4 (2), 111–127. doi:10.1515/bmc-2012-0051
Martín-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends plant Sci. 15 (1), 31–39. doi:10.1016/j.tplants.2009.11.003
Mukhopadhyay, P., and Tyagi, A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulatingABI4-mediated pathways. Sci. Rep. 5 (1), 9998. doi:10.1038/srep09998
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 (6955), 257–263. doi:10.1038/nature01958
Peng, Y., Yan, H., Guo, L., Deng, C., Wang, C., Wang, Y., et al. (2022). Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 54 (8), 1248–1258. doi:10.1038/s41588-022-01127-7
Peterson, M. E., Chen, F., Saven, J. G., Roos, D. S., and Sali, A. (2010). Evolutionary constraints on structural similarity in orthologs and paralogs. Protn ence 18 (6), 1306–1315. doi:10.1002/pro.143
Rasane, P., Jha, A., Sabikhi, L., Kumar, A., and Unnikrishnan, V. (2015). Nutritional advantages of oats and opportunities for its processing as value added foods-a review. J. food Sci. Technol. 52, 662–675. doi:10.1007/s13197-013-1072-1
Ren, L., Wu, H., Zhang, T., Ge, X., Wang, T., Zhou, W., et al. (2021). Genome-wide identification of TCP transcription factors family in sweet potato reveals significant roles of miR319-targeted TCPs in leaf anatomical morphology. Front. Plant Sci. 12, 686698. doi:10.3389/fpls.2021.686698
Resentini, F., Felipo-Benavent, A., Colombo, L., Blázquez, M. A., and Masiero, S. (2014). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8 (3), 482–485. doi:10.1016/j.molp.2014.11.018
Song, J., Gao, Z., Huo, X., Sun, H., Xu, Y., Shi, T., et al. (2015). Genome-wide identification of the auxin response factor (ARF) gene family and expression analysis of its role associated with pistil development in Japanese apricot (Prunus mume Sieb. et Zucc). Acta Physiol. Plant. 37, 1–13. doi:10.1007/s11738-015-1882-z
Tortuero, E. G., Krishnamurthi, R., Allison, H. E., and James, C. (2021). Comparative analysis of gene prediction tools for viral genome annotation. bioRxiv 12. doi:10.1101/2021.12.11.472104
Wang, C., Feng, G., Xu, X., Huang, L., Nie, G., Li, D., et al. (2023a). Genome-wide identification, characterization, and expression of TCP genes family in orchardgrass. Genes 14 (4), 925. doi:10.3390/genes14040925
Wang, P., Moore, B. M., Panchy, N. L., Meng, F., Lehti-Shiu, M. D., and Shiu, S.-H. (2018). Factors influencing gene family size variation among related species in a plant family, Solanaceae. Genome Biol. Evol. 10 (10), 2596–2613. doi:10.1093/gbe/evy193
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wang, Y., Yu, Y., Wan, H., and Ni, Z. (2023). Sea-island cotton (Gossypium barbadense L.) GbTCP5 improves plant adaptation to drought and salt stress by directly activating GbERD7, GbUBC19, and GbGOLS2 expression. Industrial Crops and Prod. 203, 117209. doi:10.1016/j.indcrop.2023.117209
Wen, Y., Raza, A., Chu, W., Zou, X., Cheng, H., Hu, Q., et al. (2021). Comprehensive in silico characterization and expression profiling of TCP gene family in rapeseed. Front. Genet. 12, 794297. doi:10.3389/fgene.2021.794297
Xiao, X., Feng, X., Gao, X., Zou, H., Li, K., and Jin, H. (2018). Genome-wide identification and analysis of TCP gene in potato. Mol. Plant Breed. 16, 7933–7938.
Xiong, W., Zhao, Y., Gao, H., Li, Y., Tang, W., Ma, L., et al. (2022). Genomic characterization and expression analysis of TCP transcription factors in Setaria italica and Setaria viridis. Plant Signal. and Behav. 17 (1), 2075158. doi:10.1080/15592324.2022.2075158
Xu, Y., Liu, H., Gao, Y., Xiong, R., Wu, M., Zhang, K., et al. (2021). The TCP transcription factor PeTCP10 modulates salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 40 (10), 1971–1987. doi:10.1007/s00299-021-02765-7
Yao, X., Ma, H., Wang, J., and Zhang, D. (2007). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49 (6), 885–897. doi:10.1111/j.1744-7909.2007.00509.x
Yin, Z., Li, Y., Zhu, W., Fu, X., Han, X., Wang, J., et al. (2018). Identification, characterization, and expression patterns of TCP genes and microRNA319 in cotton. Int. J. Mol. Sci. 19 (11), 3655. doi:10.3390/ijms19113655
Zgorzynska, E., Dziedzic, B., and Walczewska, A. (2021). An overview of the Nrf2/ARE pathway and its role in neurodegenerative diseases. Int. J. Mol. Sci. 22 (17), 9592. doi:10.3390/ijms22179592
Zhang, T. (2023). Identification of AsSPL gene family members in the oat genome and analysis of their expression patterns at the tillering stage in naked oat Master's thesis. Inner Mongolia Agricultural University. doi:10.27229/d.cnki.gnmnu.2023.000466
Zhao, J., Zhai, Z., Li, Y., Geng, S., Song, G., Guan, J., et al. (2018). Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. plant Sci. 9, 1282. doi:10.3389/fpls.2018.01282
Zhao, X., Yang, J., Li, G., Sun, Z., Hu, S., Chen, Y., et al. (2021). Genome-wide identification and comparative analysis of the WRKY gene family in aquatic plants and their response to abiotic stresses in giant duckweed (Spirodela polyrhiza). Genomics 113 (4), 1761–1777. doi:10.1016/j.ygeno.2021.03.035
Keywords: oat, TCP gene family, genome-wide identification, abiotic stress, qRT-PCR
Citation: Nie J, Zhao H, Guo X, Zhang T, Han B and Liu H (2025) Genome-wide identification of oat TCP gene family and expression patterns under abiotic stress. Front. Genet. 16:1533562. doi: 10.3389/fgene.2025.1533562
Received: 26 November 2024; Accepted: 17 January 2025;
Published: 04 February 2025.
Edited by:
Feng Li, National Agriculture and Food Research Organization (NARO), JapanReviewed by:
Chunjie Fan, Chinese Academy of Forestry, ChinaShujun Wei, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Nie, Zhao, Guo, Zhang, Han and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Zhao, aGJ6aGFvMDRAMTYzLmNvbQ==
 Jiaming Nie
Jiaming Nie Hongbin Zhao
Hongbin Zhao Xiaodong Guo1,2
Xiaodong Guo1,2