- Division of Free Radical and Radiation Biology, Department of Radiation Oncology, University of Iowa, Iowa City, IA, United States
Iron-sulfur (Fe-S) clusters are unique, redox-active co-factors ubiquitous throughout cellular metabolism. Fe-S cluster synthesis, trafficking, and coordination result from highly coordinated, evolutionarily conserved biosynthetic processes. The initial Fe-S cluster synthesis occurs within the mitochondria; however, the maturation of Fe-S clusters culminating in their ultimate insertion into appropriate cytosolic/nuclear proteins is coordinated by a late-acting cytosolic iron-sulfur assembly (CIA) complex in the cytosol. Several nuclear proteins involved in DNA replication and repair interact with the CIA complex and contain Fe-S clusters necessary for proper enzymatic activity. Moreover, it is currently hypothesized that the late-acting CIA complex regulates the maintenance of genome integrity and is an integral feature of DNA metabolism. This review describes the late-acting CIA complex and several [4Fe-4S] DNA metabolic enzymes associated with maintaining genome stability.
Introduction
Iron-sulfur (Fe-S) clusters are evolutionarily conserved co-factors ubiquitous throughout biology. Fe-S clusters and Fe-S biogenesis are largely conserved throughout prokaryotic and eukaryotic systems as mammalian Fe-S biogenesis enzymes have many shared features with bacteria that have been proposed to be a central cellular feature passed down from alphaproteobacterium (Freibert et al., 2017). Fe-S clusters exist inside proteins as either a [2Fe-2S]+, [4Fe-4S]2+, or [3Fe-4S]+ clusters. Each cluster type is typically specific to the enzymatic function of either 1) electron transfer, 2) enzyme catalysis, or 3) regulation of biological processes (Saha et al., 2018). Fe-S cluster-containing enzymes control a wide array of cellular functions, most notably mitochondrial respiration by the electron transport chain (complex I, II, and II) (Read et al., 2021). However, Fe-S cluster enzymes and Fe-S metabolism are involved in several other cellular processes including lipid metabolism, protein translation, and DNA replication (Fuss et al., 2015; Mettert and Kiley, 2015; Braymer and Lill, 2017; Crooks et al., 2018; Shi et al., 2021). Thus, synthesizing Fe-S clusters is critical in maintaining global cellular homeostasis, underscored by the number of Fe-S-containing enzymes involved in maintaining genome integrity.
The maintenance of genomic integrity is a critical feature of cellular homeostasis by facilitating stable DNA replication with a low mutational burden and cell survival under stressed conditions. Due to its importance, there exists a highly coordinated DNA metabolic network consisting of multi-faceted DNA polymerases that not only replicate DNA during the S-phase of the cell cycle but also facilitate DNA repair (Fuss et al., 2015; Shi et al., 2021). At each level, these DNA metabolic features can be altered by iron either chemically or metabolically. Thus, iron metabolism should be considered an integral component of DNA metabolism.
Due to its ability to catalyze oxidation reactions through either Fenton chemistry or reactions with molecular oxygen, iron is considered a chemical catalyst for site-specific DNA damage (Wardman and Candeias, 1996; Qian and Buettner, 1999; Kruszewski, 2003). For example, ferrous and ferric iron can enhance both single and double-stranded DNA damage associated with the radiolysis of H2O (Ambroz et al., 2001). However, there are a large number of Fe-S-containing enzymes within the DNA metabolic system (Fuss et al., 2015). Since the discovery that MMS19 coordinates with DNA metabolic enzymes, a foundational and mechanistic link between the late-acting CIA complex and DNA metabolism has been established (Gari et al., 2012; Stehling et al., 2012). Therefore, it can be postulated that Fe-S biogenesis in total plays a critical role in DNA metabolism and the maintenance of genome integrity. In this review, we provide an overview of the insertion of a [4Fe-4S]2+cluster into cytosolic and nuclear apo-proteins via the late-acting CIA complex, describe the role of [4Fe-4S] cluster-containing enzymes in DNA metabolism, and discuss the possible implications of this connection from the systems biology of disease perspective.
Components of the late-acting cytoplasmic iron-sulfur assembly (CIA) complex
The first step of Fe-S biogenesis is de novo [2Fe-2S]+ synthesis that occurs on the inner mitochondrial membrane using ISCU as a scaffold. Following the completion of the [2Fe-2S]+ cluster synthesis on the ISCU scaffold, the co-factor is trafficked to the late-acting CIA complex for insertion into the appropriate apo-proteins. This is a highly coordinated process that encompasses enzymes required for [2Fe-2S]+ synthesis on ISCU (e.g., NFS1) along with [2Fe-2S]+ trafficking/ISCU recycling (e.g., HSC20/HSPA9). Following the formation and trafficking of [4Fe-4S]2+ clusters, insertion into appropriate intra- and extramitochondrial apo-proteins occur in multiple distinct pathways. The primary focus of this review is on the late-acting CIA complex. More extensive detail regarding de novo Fe-S biogenesis can be found in (Petronek et al., 2021).
Extramitochondrial [4Fe-4S]2+ formation and insertion into apo-proteins
Following completion of the de novo [2Fe-2S]+ cluster synthesis on the ISCU scaffold, [4Fe-4S]2+ formation and trafficking are required for insertion into DNA metabolic enzymes. Currently, the formation and trafficking of [4Fe-4S]2+ clusters and their biological implications are an active area of research and many of the connection points require further investigation. Trafficking of [4Fe-4S] clusters to cytosolic and nuclear apo-proteins are carried out by the cytosolic iron-sulfur assembly (CIA) pathway (Figure 1). This process begins with a necessary transfer of a [2Fe-2S]+ cluster from the mitochondria to the cytosol. This initial transfer is facilitated by ABCB7, a transmembrane protein that facilitates the transfer of the cluster out of the mitochondria (Stehling and Lill, 2013). Consistent with its proposed, exclusive role in the maturation of [4Fe-4S] clusters, ABCB7 deletion has little effect on mitochondrial [2Fe-2S]+ protein activity but results in functionally deficient cytosolic and nuclear [4Fe-4S]2+ proteins (Kispal et al., 1999; Pondarré et al., 2006; Miao et al., 2009). It has been proposed that a glutathione-coordinated [2Fe-2S]+ cluster ([2Fe-2S](SG)4) is the natural substrate for ABCB7, and is thus, represents the [2Fe-2S]+ cluster that is utilized by the CIA machinery (Qi et al., 2014; Li and Cowan, 2015) that allows for [4Fe-4S]2+ cluster formation to occur. However, there is still limited data regarding the mechanism of Fe-S transfer out of the mitochondria through ABCB7.
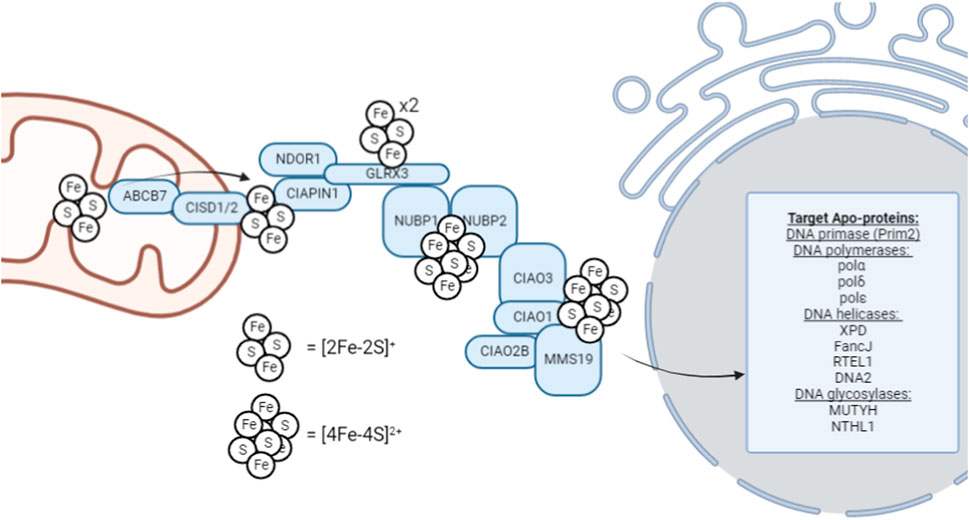
FIGURE 1. Schematic overview of the late-acting, cytosolic iron-sulfur assembly machinery (CIA complex). This schematic represents the general, proposed mechanism of [4Fe-4S]2+ cluster trafficking through the CIA complex for insertion into cytosolic and nuclear [4Fe-4S]2+ enzymes; however, not all factors are required definitively, and the utilization of this pathway can be highly context-dependent. For example, ribonucleotide reductase, critical for S-phase cell cycle entry to begin DNA replication, contains a di-ferric center whose formation is mediated by GLRX3 (Mühlenhoff et al., 2010).
Cytosolic [4Fe-4S]2+ cluster biogenesis in eukaryotes is a complex process that remains an active area of investigation. This process is proposed to be initiated by the outer mitochondrial membrane-bound NEET proteins, which transfer the [2Fe-2S] cluster to the CIA assembly factors for MMS19-mediated insertion into target apo-proteins, however, this hypothesis remains an active area of research. MitoNEET (CISD1) and NAF-1 (CISD2) are [2Fe-2S]+ proteins located on the outer membrane of the mitochondria that aid in the maturation of extramitochondrial Fe-S proteins (Mittler et al., 2019). Both CISD1 and CISD2 can transfer their [2Fe-2S] cluster to anamorsin (CIAPIN1) of the CIA complex; however, their function has not been definitively elucidated (Lipper et al., 2015). The CIAPIN1/NDOR1 complex directly interacts with mitoNEET (CISD1) to reduce the [2Fe-2S]+ cluster (Camponeschi et al., 2017) as the flavorprotein NDOR1 uses an electron from NADPH to reduce the [2Fe-2S]+ cluster (Netz et al., 2010). This reduction step makes the cluster labile and thus provides a [2Fe-2S]+ cluster substrate is available for [4Fe-4S]2+ formation.
[4Fe-4S]2+ cluster formation occurs on the NUBP1-NUBP2 scaffold (Roy et al., 2003; Hausmann et al., 2005; Netz et al., 2012a; Pallesen et al., 2013) where the [2Fe-2S]+ cluster of the CIAPIN1/NDOR1 complex is transferred via GLRX3 to NUBP1-NUBP2 (Camponeschi et al., 2020). GLRX3 can bind two [2Fe-2S]+ clusters and thus, GLRX3-[2Fe-2S]2 can utilize glutathione to transfer the cluster to NUBP1 for [4Fe-4S]2+ cluster formation. NUBP1 and NUBP2 contain conserved cysteine residues at their C-terminal domain to coordinate a bridging [4Fe-4S]2+ cluster (Netz et al., 2012a). However, the [4Fe-4S]2+ cluster formed on the NUBP1-NUBP2 complex is also CIAO3-dependent (Balk et al., 2005). Similarly, CIAO3 is believed to be involved because it contains conserved cysteine motifs at its N- and C-terminal domains for Fe-S binding (Urzica et al., 2009).
Following the formation of a [4Fe-4S]2+ cluster on the NUBP1-NUBP2-CIAO3 complex, it can be transferred to the appropriate apo-proteins by the late-acting CIA complex. This process occurs through MMS19. MMS19 is able to form a complex with CIOA1, CIAO2B, AND CIAO3 to make up the CIA targeting complex (Gari et al., 2012). The completed [4Fe-4S]2+ cluster is hypothesized to transferred to the CIA targeting complex (CIAO1, CIAO2B, and MMS19) by CIAO3 (Kassube and Thomä, 2020), but further data is required to illuminate the role of CIAO3 as a mediator of cluster transfer. CIAO2B and CIAO1 associate with the C-terminus of MMS19 to form a docking site for cytosolic and nuclear apo-proteins, however XPD has been observed to directly interact with MMS19 (Odermatt and Gari, 2017). Interestingly, MMS19 binding prevents CIAO2B proteasomal degradation as an apparent feedback regulatory mechanism to maintain CIA stability (Seki et al., 2013).
When considering the regulation of DNA metabolism, MMS19 is the main connection point as MMS19 serves as a scaffold for the transfer of the completed [4Fe-4S] cluster to the appropriate apo-protein. MMS19 has a docking site that allows it to directly interact with [4Fe-4S]2+ containing DNA metabolic proteins necessary for maintaining genomic stability (Gari et al., 2012). To underscore the importance of MMS19 as a regulatory component of DNA metabolism, MMS19 knockdown has been observed to result in decreased XPD, FANCJ, and DNA polymerase expression. Furthermore, MMS19 depletion strongly decreases the expression of the POLD1 subunit of DNA polymerase δ (Stehling et al., 2012). Thus, it appears that the late-acting CIA-complex, culminating in the insertion of a [4Fe-4S]2+ cluster into nuclear apo-proteins using MMS19 as a scaffold, is a critical regulatory step in ensuring that [4Fe-4S]2+ cluster-containing DNA metabolic enzymes are functional to aid in the maintenance of genomic integrity through DNA replication.
Iron-sulfur clusters in DNA metabolism
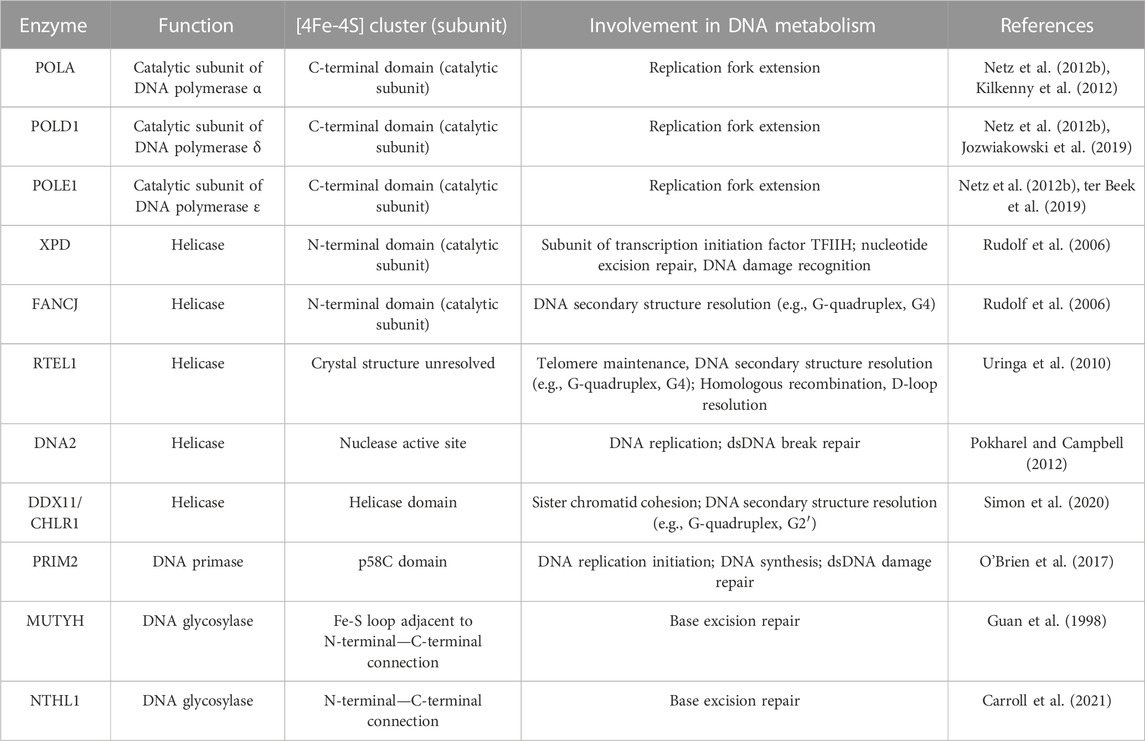
For DNA to be efficiently and accurately passed to progeny cells, high-fidelity DNA replication is required. At each level of maintaining genome integrity, Fe-S-containing enzymes are necessary for this process to occur (Table 1). Mechanistically, it is hypothesized that the [4Fe-4S]2+ clusters serve as electrochemical sensors that can detect electron transport along the DNA backbone. DNA charge transfer occurs when electrons are passed through the pi-stack of base pairs between redox partners (Boal et al., 2009; Slinker et al., 2011). DNA charge transfer can occur in intact double-stranded DNA, but any disruption to the base pair stacking (e.g., DNA base damage, DNA strand break) will disrupt this process. Thus, intact double-stranded DNA can be theoretically considered a wire that allows electrons to move along the strand and impediments to this electron movement will allow for the identification and reparation of damage. When bound to DNA, the [4Fe-4S]2+ cluster contained within proteins have a redox potential of ≈ −200 mV, which allows it to serve as a redox switch by cycling between a [4Fe-4S]2+/3+ oxidation state as electrons move along the double-stranded DNA (Fuss et al., 2015). In this context, the [4Fe-4S] cluster may be critical feature of key DNA metabolic enzymes that can serve as both an electron donor or acceptor, allowing it to function as a redox sensor of DNA damage via cluster oxidation following electron transport along the DNA backbone (Fuss et al., 2015; Arnold et al., 2016; Syed and Tainer, 2019). Furthermore, we describe the function of the various [4Fe-4S] containing enzymes and their role in DNA metabolism.
DNA helicases
For DNA to be replicated, the double-stranded DNA must first be opened by helicases. Helicases are motor proteins that utilize ATP hydrolysis to translocate along and unwind the paired nucleic acids that make up double-stranded DNA. Due to the complexities of double-stranded DNA maintenance, helicases are largely responsible for regulating several different processes of DNA separation (e.g., DNA replication/repair and telomere regulation) (Abdelhaleem, 2010). Thus, helicases are critical features of nucleic acid metabolism and maintaining genome stability. Six superfamilies of helicases are designated based on their amino acid sequence with several containing [4Fe-4S] clusters that are required for their functioning (Singleton et al., 2007). Each of these helicases are linked to the CIA complex through interaction with MMS19 that delivers the completed [4Fe-4S] cluster (Stehling et al., 2012).
DNA2 is an Fe-S containing member of helicase superfamily 1 involved in DNA replication, telomere maintenance, and double-strand break (DSB) repair. Thus, DNA2 serves a central role in maintaining genome stability at multiple phases (Budd et al., 2005; Zheng et al., 2020). Functionally, DNA2 has helicase and nuclease activity, which depend on the presence of a [4Fe-4S] cluster (Pokharel and Campbell, 2012). The [4Fe-4S] cluster of DNA2 is bound to four cysteine residues contained within the nuclease domain, suggesting that the cluster may serve to maintain the structural integrity of the nuclease domain, thus, rendering it essential for facilitating DNA2 binding of broken DNA (Yeeles et al., 2009). A 2020 study by Mariotti et al. (2020) showed that loss of the [4Fe-4S] cluster caused a conformational change in DNA2 resulting in a distortion of the central DNA binding tunnel. This study also showed that oxidation of DNA2 impaired DNA binding in vitro that was reversible by reduction; however, this effect was independent of the presence of the [4Fe-4S] cluster. Thus, DNA2 represents an example of a nuclear enzyme where the [4Fe-4S] protein plays a critical structural role.
Helicase superfamily 2 also encompasses several Fe-S containing members. A commonly recognized Fe-S containing helicase is XPD. XPD is a part of the TFIIH complex that is involved in DNA transcription and unwinding dsDNA for damage verification and initiation of nucleotide excision repair (Houten et al., 2016). Interestingly, a 2014 study by Kuper, et al. showed that the enzymatic activity of XPD within the TFIIH complex is mainly dedicated to DNA damage recognition and resolution, while it primarily functions to maintain the structural integrity of the TFIIH complex during transcription initiation (Kuper et al., 2014). XPD contains a [4Fe-4S] cluster within its catalytic domain that is thought to play a structural role but may be necessary in DNA damage recognition (Wolski et al., 2008; White, 2009). XPD has been observed to interact with the CIA complex and the TFIIH complex in a mutually exclusive fashion suggesting that the [4Fe-4S] cluster of XPD is first inserted in the cytoplasm by the CIA complex before translocation to the nucleus for its association with the TFIIH complex (Vashisht et al., 2015). XPD with deficient Fe-S binding or impaired CIA interaction was unable to join the TFIIH complex and MMS19 deletion causes a depletion of XPD (Kou et al., 2008; Vashisht et al., 2015). XPD can be linked to three separate genetic disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy which can be linked to various mutations in XPD (Taylor et al., 1997). Xeroderma pigmentosum and Cockayne syndrome mutations impair the Fe-S binding domain and impair helicase activity, while trichothiodystrophy mutants have been observed in all four XPD domains to impair the XPD secondary structure likely leading to impaired TFIIH integrity (Fan et al., 2008). Thus, xeroderma pigmentosum and Cockayne syndrome are considered DNA repair related disorders while trichothiodystrophy is due to impaired transcription. Overall, it appears that the insertion of a [4Fe-4S] cluster into XPD is essential for its ability to function within the TFIIH complex to promote efficient DNA damage repair.
Another Fe-S containing superfamily 2 helicase is FANCJ (Brosh and Cantor, 2014). FANCJ is a helicase that functions in double-stranded DNA damage repair through homologous recombination and can resolve DNA secondary structures to promote smooth DNA replication and the avoidance of replication stress (Datta and Brosh, 2019). FANCJ was initially discovered as a result of a physical interaction with the renowned tumor suppressor gene, BRCA1, as it binds at the BRCT motifs of BRCA1 (Cantor et al., 2001). In this seminal report, the FANCJ/BRCA1 complex (initially referred to as BRCA1 interacting C-terminal helicase, BACH1), was shown to be important for the DNA damage response function of BRCA1. In addition to FANCJ, BRCA1 can also associate with a non-Fe-S helicase FANCM, that has been shown to be essential to mitigate replication fork stalling and mediate D-loop dissociations (Gari et al., 2008; Panday et al., 2021). This underscores the importance of the interaction of BRCA1 and the Fancomi family of helicases in the maintenance of genome integrity. Beyond its interaction with BRCA1, FANCJ functions as an ATP-dependent helicase with 5′-3′ specificity that requires an intact [4Fe-4S] cluster, similar to XPD (Rudolf et al., 2006). More specifically, FANCJ is a helicase that can unwind DNA G-quadruplexes ahead of DNA polymerase, which are guanine-rich DNA secondary structures that cause DNA replication stalling and are prone to oxidative damage (London et al., 2008; Wu and Spies, 2016; Lerner and Sale, 2019; Fleming and Burrows, 2021). Also, like XPD, FANCJ has been shown to directly interact with that late acting CIA complex through MMS19 (Wietmarschen et al., 2012) as MMS19 knockdown significantly impairs FANCJ iron insertion to promote genomic instability and sensitivity to DNA damaging agents (Weon et al., 2017). Thus, the insertion of the [4Fe-4S] cluster into FANCJ is a critical step in its ability to preserve genomic integrity through its helicase function.
The third Fe-S containing member of the helicase superfamily 2 is DDX11/CHLR1. Similar to both XPD and FANCJ, DDX11 is an ATP-dependent helicase with 5′-3′ directionality with a preferred single stranded 5′ tail (Hirota and Lahti, 2000; Farina et al., 2008). The helicase function of DDX11 allows it to serve a similar role to FANCJ in the resolution of G-quadruplexes to prevent replication stress (Wu et al., 2012; Bharti et al., 2013; van Schie JJMFaramarz et al., 2020). However, unlike FANCJ, which can efficiently resolve unimolecular (G4) G-quadruplexes (Wu and Spies, 2016), DDX11 efficiently unwinds two stranded anti-parallel (G2′) G-quadruplexes to a much greater extent than G4 structures (Wu et al., 2012). Consistent with its role in maintaining genome integrity, DDX11 depletion has been shown to decrease the amount of single-stranded DNA leading to impaired CHK1 phosphorylation (Simon et al., 2020), a critical step in the DNA damage response pathway, promoting DNA replication stress (Patil et al., 2013; Jegadesan and Branzei, 2021). Importantly, the [4Fe-4S] cluster of DDX11 is indispensable for its functionality in the resolution of DNA secondary structures to prevent replication stress (Simon et al., 2020). DDX11 is primarily recognized for its role in maintaining sister chromatid cohesion as an inactivating mutation results in the cohesinopathy called Warsaw Breakage Syndrome (van der Lelij et al., 2010; Capo-Chichi et al., 2013; Bharti et al., 2014). The DDX11 variant associated with Warsaw Breakage Syndrome (R263Q) cannot bind an Fe-S cluster (Simon et al., 2020). Thus, DDX11 represents a helicase where the [4Fe-4S] cluster is critical to its enzymatic activity.
The final Fe-S containing member of the helicase superfamily 2 is RTEL. RTEL is a [4Fe-4S] cluster critical for maintaining genome stability through telomere maintenance and double-stranded DNA damage repair (Uringa et al., 2010). RTEL has been shown to play a critical role in setting telomere length in mice (Ding et al., 2004) and suppression of RTEL in mouse embryonic fibroblasts results in increased telomere fragility (Sfeir et al., 2009). Moreover, it has been shown that RTEL is required for telomere replication in mouse embryonic fibroblasts. RTEL depletion increases G4 stability at telomeres to prevent telomere replication (Uringa et al., 2012). This is consistent with biochemical data showing that RTEL can resolve G-quadruplexes to prevent replication and promote telomere lengthening throughout the human genome (Wu et al., 2020). Beyond telomere maintenance, RTEL has been shown to play an important role in DNA damage repair (Uringa et al., 2010). RTEL has been shown in C. elegans to regulate homologous recombination and promote genomic stability by resolving D-loops where RTEL mutants showed an increased propensity to accumulate DNA damage (Barber et al., 2008). Currently, the role of the [4Fe-4S] cluster in RTEL (i.e., structural versus functional) remains unclear; however, similar to the other Fe-S containing helicases, RTEL does interact directly with MMS19 (Stehling et al., 2012).
DNA primase and polymerases
Following the opening of a double stranded DNA helix by helicases, replication can occur by DNA polymerases. However, DNA polymerases are unable to initiate the synthesis of a new DNA strand during replication, rather they only extend existing strands and thus, require a primer. DNA primase is the enzyme responsible for synthesizing a short primer for DNA polymerase to use as a template (Frick and Richardson, 2001). Similar to the Fe-S containing helicases, DNA primase acquires a [4Fe-4S] cluster from MMS19 and the functionality of DNA primase is dependent on an intact [4Fe-4S] cluster (Klinge et al., 2007; Stehling et al., 2012). It has been observed that the [4Fe-4S] cluster of DNA primase serves as a redox switch that modulates its DNA binding capacity (Holt et al., 2017). DNA primase is a heterodimer with a small and large subunit with the small subunit being responsible for RNA polymerase activity and the large [4Fe-4S] subunit (PRIM2) being responsible for DNA binding (Agarkar et al., 2011). The Fe-S cluster has a [4Fe-4S]2+ resting state where it is loosely bound to DNA, however, an oxidation of the cluster to a [4Fe-4S]3+ state results in tight DNA binding (O’Brien et al., 2017; O’Brien et al., 2018a; O’Brien et al., 2018b). Thus, it appears that Fe-S mediated DNA charge transfer is an essential feature of DNA replication initiation mediated by PRIM2.
Following the generation of a short primer by DNA primase, DNA is replicated by [4Fe-4S] containing DNA polymerases. In eukaryotic cells, class B family DNA polymerases α, δ, ε, and ζ mediate DNA replication. All three of these polymerases contain a [4Fe-4S] within their C-terminal catalytic subunits (POLA, POLD1, and POLE1, respectively) (Garcia-Diaz and Bebenek, 2007; Shi et al., 2021). It was believed that Zn2+ was the necessary inorganic co-factor for polymerase activity due to the two conserved cysteine residues acting as metal binding motifs in POLA (Evanics et al., 2003; Klinge et al., 2009). However, later structural experiments revealed that all four polymerases coordinate a [4Fe-4S] cluster within the catalytic subunit (Netz et al., 2012b; ter Beek et al., 2019; Suwa et al., 2015; Baranovskiy et al., 2018). Loss of the [4Fe-4S] cluster in POLD1 causes a destabilization of all four enzyme subunits resulting in defective DNA binding and impaired polymerase and exonuclease activities (Jozwiakowski et al., 2019). In yeast, the [4Fe-4S] cluster of DNA polymerase ε is redox active and its polymerase function may be mediated by DNA charge transfer similar to PRIM2 (Jain et al., 2013; Pinto et al., 2021). Consistent with other DNA metabolic enzymes, the assembly of the [4Fe-4S] cluster in the catalytic subunit of DNA polymerases and ultimately their enzymatic activity are mediated by MMS19 of the CIA complex (Gari et al., 2012; Stehling et al., 2012; Han et al., 2015). Therefore, it is apparent that maintenance of high-fidelity DNA replication is largely dependent on the insertion of completed [4Fe-4S] clusters into the appropriate DNA metabolic enzymes by the late acting CIA complex.
DNA glycosylases
While DNA helicases and polymerases aim to perform high-fidelity DNA replication to avoid replication stress, damage to DNA bases can occur through several different chemical modifications including oxidation, alkylation, deamination, and spontaneous hydrolysis (Bauer et al., 2015). The primary enzymatic pathway for the repair of damaged DNA bases is base excision repair (BER), which can occur throughout the cell cycle (Krokan and Bjoras, 2013). BER is initiated by damage recognition by DNA glycosylases which then form AP sites to remove the damaged bases (Jacobs and Schär, 2012; Wallace, 2013). Short patch or long patch base excision repair can occur based on the number of damaged bases. Two DNA glycosylases with [4Fe-4S] clusters have been identified, MUTYH and NTHL1 (Parker et al., 2000; Carroll et al., 2021). Both MUTYH and NTHL1 are mammalian MutY and endonuclease III homologs, E. coli DNA glycosylases that were initially observed coordinate a [4Fe-4S] cluster that mediates their DNA binding and substrate recognition (Kuo et al., 1992; Guan et al., 1998). MUTYH is an adenine-specific glycosylase that removes mismatched adenines from A-G/A-C pairs and can also remove 8-dihydro-8-oxodeoxyguanine (8-oxoG) (McGoldrick et al., 1995). Meanwhile, NTHL1 can excise thymine glycol and oxidize urea lesions (Aspinwall et al., 1997). The [4Fe-4S] cluster of MutY and endonuclease are redox active and serve as an electrochemical sensor to recognize DNA damage through DNA charge transfer (Boal et al., 2005). Thus, it furthers the hypothesis that the [4Fe-4S] clusters of DNA regulatory enzymes serve as conserved sensors of DNA charge transfer to efficiently maintain genomic integrity.
Conclusion and future perspectives
With the current understanding that several [4Fe-4S] cluster enzymes interact with the late-acting CIA complex and play a critical role in DNA metabolism, there is a window of opportunity to accelerate our understanding of how Fe-S biogenesis can regulate metabolic processes (e.g., maintain genomic integrity). As the biomedical community works to understand the various systems that play a role in regulating health and disease, the regulatory role of Fe-S biogenesis remains unclear. However, a wide array of literature suggests that Fe-S biogenesis plays a role in numerous diseases which may result from the dysregulated global metabolic issues that arise from disrupted Fe-S containing enzymes such as those described in this review. For example, mutations associated with the [4Fe-4S] containing helicases have been implicated in the onset of disease as XPD mutations present as xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy (Taylor et al., 1997); FANCJ as Fancomi Anemia (London et al., 2008); DDX11 as Warsaw Breakage Syndrome (van der Lelij et al., 2010; van Schie JJMFaramarz et al., 2020); DNA2 as mitochondrial DNA depletion syndrome (Sun et al., 2022); and RTEL mutations have been associated with familial pulmonary fibrosis (Kannengiesser et al., 2015). Additionally, a majority of these enzymes are associated with cancer development including XPD, FANCJ, DDX11, RTEL, MUTYH, and NTHL1 (Benhamou and Sarasin, 2002; Nicolo et al., 2008; Lubbe et al., 2009; Cantor and Guillemette, 2011; Mazzei et al., 2013; Weren et al., 2015; Yan et al., 2016; Das et al., 2020; Hutchcraft et al., 2021; Magrin et al., 2021; Mahtab et al., 2021).
Following the discovery that MMS19 directly interacts with enzymes that regulate the maintenance of genome integrity and high-fidelity transfer of genetic information including helicases, primase, polymerases, and glycosylases, there is a very clear connection between Fe-S biogenesis through the late-acting CIA complex and DNA metabolism (Gari et al., 2012; Stehling et al., 2012). While the insertion of a complete [4Fe-4S] cluster by MMS19 into these DNA metabolic enzymes represents a critical regulatory step for the maintenance of genomic integrity; it is important to acknowledge the several steps leading to the formation of the completed [4Fe-4S]2+ cluster on MMS19 before the insertion into nuclear enzymes (Petronek et al., 2021). Thus, the transfer of the cluster from the CIA complex through MMS19 to nuclear enzymes represents the penultimate step in a larger process that, if impaired, likely results in genome instability. For example, frataxin loss from de novo [2Fe-2S] cluster synthesis results in Friedrich’s Ataxia and predisposes cells to DNA damage associated with impaired BER, a process that is initiated by [4Fe-4S] containing DNA glycosylases (Haugen et al., 2010; Thierbach et al., 2010; Shen et al., 2016). Thus, it may be imperative to consider the implications of changes in Fe-S biogenesis in totality when investigating global, cellular metabolic alterations in various pathologies associated with genomic instability.
Author contributions
MP and BA both contributed to the writing and editing of this review.
Funding
This work was supported by NIH grants T32 CA078586, P01 CA217797, and the Gateway for Cancer Research grant G-17-1500.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not represent the views of the National Institutes of Health.
References
Abdelhaleem, M. (2010). Helicases: An overview. Methods Mol. Biol. 587, 1–12. doi:10.1007/978-1-60327-355-8_1
Agarkar, V. B., Babayeva, N. D., Pavlov, Y. I., and Tahirov, T. H. (2011). Crystal structure of the C-terminal domain of human DNA primase large subunit: Implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10 (6), 926–931. doi:10.4161/cc.10.6.15010
Ambroz, H., Bradshaw, T., Kemp, T., Kornacka, E., and Przybytniak, G. (2001). Role of iron ions in damage to DNA: Influence of ionising radiation, UV light and H2O2. J. Photochem. Photobiol. A Chem. 142, 9–18. doi:10.1016/s1010-6030(01)00439-7
Arnold, A. R., Grodick, M. A., and Barton, J. K. (2016). DNA charge transport: From chemical principles to the cell. Cell Chem. Biol. 23 (1), 183–197. doi:10.1016/j.chembiol.2015.11.010
Aspinwall, R., Rothwell, D., Roldan Arjona, T., Anselmino, C., Ward, C. J., Cheadle, J. P., et al. (1997). Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl. Acad. Sci. 94, 109–114. doi:10.1073/pnas.94.1.109
Balk, J., Pierik, A. J., Aguilar Netz, D. J., Mühlenhoff, U., Lill, R., and Muhlenhoff, U. (2005). Nar1p, a conserved eukaryotic protein with similarity to Fe-only hydrogenases, functions in cytosolic iron-sulphur protein biogenesis. Biochem. Soc. Trans. 33 (1), 86–89. doi:10.1042/BST0330086
Baranovskiy, A. G., Siebler, H. M., Pavlov, Y. I., and Tahirov, T. H. (2018). “Chapter one - iron–sulfur clusters in DNA polymerases and primases of eukaryotes,” in Methods in enzymology [internet]. Editor S. S. David (Academic Press), 1–20. Available at: https://www.sciencedirect.com/science/article/pii/S0076687917303336.
Barber, L. J., Youds, J. L., Ward, J. D., McIlwraith, M. J., O’Neil, N. J., Petalcorin, M. I. R., et al. (2008). RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135 (2), 261–271. doi:10.1016/j.cell.2008.08.016
Bauer, N. C., Corbett, A. H., and Doetsch, P. W. (2015). The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 43 (21), 10083–10101. doi:10.1093/nar/gkv1136
Benhamou, S., and Sarasin, A. (2002). ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis 17 (6), 463–469. doi:10.1093/mutage/17.6.463
Bharti, S., Khan, I., Banerjee, T., Sommers, J., Wu, Y., and Brosh, R. (2014). Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell. Mol. life Sci. CMLS 71, 2625–2639. doi:10.1007/s00018-014-1569-4
Bharti, S. K., Sommers, J. A., George, F., Kuper, J., Hamon, F., Shin-ya, K., et al. (2013). Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 288 (39), 28217–28229. doi:10.1074/jbc.M113.496463
Boal, A. K., Genereux, J. C., Sontz, P. A., Gralnick, J. A., Newman, D. K., and Barton, J. K. (2009). Redox signaling between DNA repair proteins for efficient lesion detection. Proc. Natl. Acad. Sci. U. S. A. 106 (36), 15237–15242. doi:10.1073/pnas.0908059106
Boal, A. K., Yavin, E., Lukianova, O. A., O’Shea, V. L., David, S. S., and Barton, J. K. (2005). DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry 44 (23), 8397–8407. doi:10.1021/bi047494n
Braymer, J. J., and Lill, R. (2017). Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 292 (31), 12754–12763. doi:10.1074/jbc.R117.787101
Brosh, R., and Cantor, S. (2014). Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi Anemia. Front. Genet. 5, 372. doi:10.3389/fgene.2014.00372
Budd, M. E., Tong, A. H. Y., Polaczek, P., Peng, X., Boone, C., and Campbell, J. L. (2005). A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLOS Genet. 1 (6), e61. doi:10.1371/journal.pgen.0010061
Camponeschi, F., Ciofi-Baffoni, S., and Banci, L. (2017). Anamorsin/Ndor1 complex reduces [2Fe–2S]-MitoNEET via a transient protein–protein interaction. J. Am. Chem. Soc. 139 (28), 9479–9482. doi:10.1021/jacs.7b05003
Camponeschi, F., Prusty, N. R., Heider, S. A. E., Ciofi-Baffoni, S., and Banci, L. (2020). GLRX3 acts as a [2Fe–2S] cluster chaperone in the cytosolic iron–sulfur assembly machinery transferring [2Fe–2S] clusters to NUBP1. J. Am. Chem. Soc. 142 (24), 10794–10805. doi:10.1021/jacs.0c02266
Cantor, S., and Guillemette, S. (2011). Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. Lond. Engl. 7, 253–261. doi:10.2217/fon.10.191
Cantor, S. B., Bell, D. W., Ganesan, S., Kass, E. M., Drapkin, R., Grossman, S., et al. (2001). BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105 (1), 149–160. doi:10.1016/s0092-8674(01)00304-x
Capo-Chichi, J. M., Bharti, S. K., Sommers, J. A., Yammine, T., Chouery, E., Patry, L., et al. (2013). Identification and biochemical characterization of a novel mutation in DDX11 causing Warsaw breakage syndrome. Hum. Mutat. 34 (1), 103–107. doi:10.1002/humu.22226
Carroll, B. L., Zahn, K. E., Hanley, J. P., Wallace, S. S., Dragon, J. A., and Doublié, S. (2021). Caught in motion: Human NTHL1 undergoes interdomain rearrangement necessary for catalysis. Nucleic Acids Res. 49 (22), 13165–13178. doi:10.1093/nar/gkab1162
Crooks, D., Maio, N., Lane, A., Jarnik, M., Higashi, R., Haller, R., et al. (2018), Acute loss of iron–sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J. Biol. Chem. 293, 8297, 8311. doi:10.1074/jbc.ra118.001885
Das, L., Quintana, V. G., and Sweasy, J. B. (2020). NTHL1 in genomic integrity, aging and cancer. DNA Repair 93, 102920. doi:10.1016/j.dnarep.2020.102920
Datta, A., and Brosh, R. (2019). Holding all the cards—how fanconi anemia proteins deal with replication stress and preserve genomic stability. Genes 10, 170. doi:10.3390/genes10020170
Ding, H., Schertzer, M., Wu, X., Gertsenstein, M., Selig, S., Kammori, M., et al. (2004). Regulation of murine telomere length by rtel: An essential gene encoding a helicase-like protein. Cell 117 (7), 873–886. doi:10.1016/j.cell.2004.05.026
Evanics, F., Maurmann, L., Yang, W. W., and Bose, R. N. (2003). Nuclear magnetic resonance structures of the zinc finger domain of human DNA polymerase-alpha. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1651 (1), 163–171. doi:10.1016/s1570-9639(03)00266-8
Fan, L., Fuss, J. O., Cheng, Q. J., Arvai, A. S., Hammel, M., Roberts, V. A., et al. (2008). XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell 133 (5), 789–800. doi:10.1016/j.cell.2008.04.030
Farina, A., Shin, J. H., Kim, D. H., Bermudez, V. P., Kelman, Z., Seo, Y. S., et al. (2008). Studies with the human cohesin establishment factor, ChlR1: Association of ChlR1 with ctf18-RFC and Fen1. J. Biol. Chem. 283 (30), 20925–20936. doi:10.1074/jbc.M802696200
Fleming, A., and Burrows, C. (2021). Oxidative stress-mediated epigenetic regulation by G-quadruplexes. Nar. Cancer 3, zcab038. doi:10.1093/narcan/zcab038
Freibert, S. A., Goldberg, A. V., Hacker, C., Molik, S., Dean, P., Williams, T. A., et al. (2017). Evolutionary conservation and in vitro reconstitution of microsporidian iron-sulfur cluster biosynthesis. Nat. Commun. 8, 13932. doi:10.1038/ncomms13932
Frick, D. N., and Richardson, C. C. (2001). DNA primases. Annu. Rev. Biochem. 70 (1), 39–80. doi:10.1146/annurev.biochem.70.1.39
Fuss, J. O., Tsai, C. L., Ishida, J. P., and Tainer, J. A. (2015). Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim. Biophys. Acta 1853 (6), 1253–1271. doi:10.1016/j.bbamcr.2015.01.018
Garcia-Diaz, M., and Bebenek, K. (2007). Multiple functions of DNA polymerases. Crit. Rev. plant Sci. 26, 105–122. doi:10.1080/07352680701252817
Gari, K., Décaillet, C., Delannoy, M., Wu, L., and Constantinou, A. (2008). Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. U. S. A. 105 (42), 16107–16112. doi:10.1073/pnas.0804777105
Gari, K., León Ortiz, A. M., Borel, V., Flynn, H., Skehel, J. M., and Boulton, S. J. (2012). MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337 (6091), 243–245. doi:10.1126/science.1219664
Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., et al. (1998). MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 5 (12), 1058–1064. doi:10.1038/4168
Han, Y. F., Huang, H. W., Li, L., Cai, T., Chen, S., and He, X. J. (2015). The cytosolic iron-sulfur cluster assembly protein MMS19 regulates transcriptional gene silencing, DNA repair, and flowering time in arabidopsis. PLOS ONE 10 (6), e0129137. doi:10.1371/journal.pone.0129137
Haugen, A. C., Di Prospero, N. A., Parker, J. S., Fannin, R. D., Chou, J., Meyer, J. N., et al. (2010). Altered gene expression and DNA damage in peripheral blood cells from friedreich’s Ataxia patients: Cellular model of pathology. PLOS Genet. 6 (1), e1000812. doi:10.1371/journal.pgen.1000812
Hausmann, A., Aguilar Netz, D. J., Balk, J., Pierik, A. J., Mühlenhoff, U., and Lill, R. (2005). The eukaryotic P loop NTPase Nbp35: An essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U. S. A. 102 (9), 3266–3271. doi:10.1073/pnas.0406447102
Hirota, Y., and Lahti, J. M. (2000). Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 28 (4), 917–924. doi:10.1093/nar/28.4.917
Holt, M. E., Salay, L. E., and Chazin, W. J. (2017). “Chapter twelve - a polymerase with potential: The Fe–S cluster in human DNA primase,” in Methods in enzymology [internet]. Editor S. S. David (Academic Press), 361–390. Available at: https://www.sciencedirect.com/science/article/pii/S007668791730232X.
Houten, B. V., Kuper, J., and Kisker, C. (2016). Role of XPD in cellular functions: To TFIIH and beyond. DNA Repair (Amst) 44, 136–142. doi:10.1016/j.dnarep.2016.05.019
Hutchcraft, M. L., Gallion, H. H., and Kolesar, J. M. (2021). MUTYH as an emerging predictive biomarker in ovarian cancer. Diagnostics 11 (1), 84. doi:10.3390/diagnostics11010084
Jacobs, A. L., and Schär, P. (2012). DNA glycosylases: In DNA repair and beyond. Chromosoma 121 (1), 1–20. doi:10.1007/s00412-011-0347-4
Jain, R., Vanamee, E., Dzikovski, B., Buku, A., Johnson, R., Prakash, L., et al. (2013). An iron–sulfur cluster in the polymerase domain of yeast DNA polymerase ε. J. Mol. Biol., 426.
Jegadesan, N., and Branzei, D. (2021). DDX11 loss causes replication stress and pharmacologically exploitable DNA repair defects. Proc. Natl. Acad. Sci. 118, e2024258118. doi:10.1073/pnas.2024258118
Jozwiakowski, S. K., Kummer, S., and Gari, K. (2019). Human DNA polymerase delta requires an iron–sulfur cluster for high-fidelity DNA synthesis. Life Sci. Alliance 2 (4), e201900321. doi:10.26508/lsa.201900321
Kannengiesser, C., Borie, R., Ménard, C., Réocreux, M., Nitschké, P., Gazal, S., et al. (2015). Heterozygous RTEL1 mutations are associated with familial pulmonary fibrosis. Eur. Respir. J. 46 (2), 474–485. doi:10.1183/09031936.00040115
Kassube, S. A., and Thomä, N. H. (2020). Structural insights into Fe–S protein biogenesis by the CIA targeting complex. Nat. Struct. Mol. Biol. 27 (8), 735–742. doi:10.1038/s41594-020-0454-0
Kilkenny, M. L., De Piccoli, G., Perera, R. L., Labib, K., and Pellegrini, L. (2012). A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J. Biol. Chem. 287 (28), 23740–23747. doi:10.1074/jbc.M112.368951
Kispal, G., Csere, P., Prohl, C., and Lill, R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18 (14), 3981–3989. doi:10.1093/emboj/18.14.3981
Klinge, S., Hirst, J., Maman, J. D., Krude, T., and Pellegrini, L. (2007). An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14 (9), 875–877. doi:10.1038/nsmb1288
Klinge, S., Núñez-Ramírez, R., Llorca, O., and Pellegrini, L. (2009). 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28 (13), 1978–1987. doi:10.1038/emboj.2009.150
Kou, H., Zhou, Y., Gorospe, R. M. C., and Wang, Z. (2008). Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc. Natl. Acad. Sci. U. S. A. 105 (41), 15714–15719. doi:10.1073/pnas.0710736105
Krokan, H., and Bjoras, M. (2013). Base excision repair. Cold Spring Harb. Perspect. Biol. 5 (4), a012583. doi:10.1101/cshperspect.a012583
Kruszewski, M. (2003). Labile iron pool: The main determinant of cellular response to oxidative stress. Mutat. Research/Fundamental Mol. Mech. Mutagen. 531 (1), 81–92. doi:10.1016/j.mrfmmm.2003.08.004
Kuo, C. F., McRee, D. E., Cunningham, R. P., and Tainer, J. A. (1992). Crystallization and crystallographic characterization of the iron-sulfur-containing DNA-repair enzyme endonuclease III from Escherichia coli. J. Mol. Biol. 227 (1), 347–351. doi:10.1016/0022-2836(92)90703-m
Kuper, J., Braun, C., Elias, A., Michels, G., Sauer, F., Schmitt, D. R., et al. (2014). In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol. 12 (9), e1001954. doi:10.1371/journal.pbio.1001954
Lerner, L. K., and Sale, J. E. (2019). Replication of G Quadruplex DNA. Genes 10 (2), 95. doi:10.3390/genes10020095
Li, J., and Cowan, J. A. (2015). Glutathione-coordinated [2Fe-2S] cluster: A viable physiological substrate for mitochondrial ABCB7 transport. Chem. Commun. (Camb) 51 (12), 2253–2255. doi:10.1039/c4cc09175b
Lipper, C. H., Paddock, M. L., Onuchic, J. N., Mittler, R., Nechushtai, R., and Jennings, P. A. (2015). Cancer-related NEET proteins transfer 2Fe-2S clusters to anamorsin, a protein required for cytosolic iron-sulfur cluster biogenesis. PLoS One 10 (10), e0139699. doi:10.1371/journal.pone.0139699
London, T. B. C., Barber, L. J., Mosedale, G., Kelly, G. P., Balasubramanian, S., Hickson, I. D., et al. (2008). FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283 (52), 36132–36139. doi:10.1074/jbc.M808152200
Lubbe, S., Di Bernardo, M. C., Chandler, I., and Houlston, R. (2009). Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J. Clin. Oncol. official J. Am. Soc. Clin. Oncol. 27, 3975–3980. doi:10.1200/JCO.2008.21.6853
Magrin, L., Fanale, D., Brando, C., Fiorino, A., Corsini, L. R., Sciacchitano, R., et al. (2021). POLE, POLD1, and NTHL1: The last but not the least hereditary cancer-predisposing genes. Oncogene 40 (40), 5893–5901. doi:10.1038/s41388-021-01984-2
Mahtab, M., Boavida, A., Santos, D., and Pisani, F. M. (2021). The genome stability maintenance DNA helicase DDX11 and its role in cancer. Genes 12 (3), 395. doi:10.3390/genes12030395
Mariotti, L., Wild, S., Brunoldi, G., Piceni, A., Ceppi, I., Kummer, S., et al. (2020). The iron-sulphur cluster in human DNA2 is required for all biochemical activities of DNA2. Commun. Biol. 3 (1), 322. doi:10.1038/s42003-020-1048-4
Mazzei, F., Viel, A., and Bignami, M. (2013). Role of MUTYH in human cancer. Mutat. Research/Fundamental Mol. Mech. Mutagen. 743–744, 33–43. doi:10.1016/j.mrfmmm.2013.03.003
McGoldrick, J., Yeh, C., Solomon, M., Essigmann, J., and Lu, A. (1995). Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol. Cell Biol. 15 (2), 989–996. doi:10.1128/mcb.15.2.989
Mettert, E. L., and Kiley, P. J. (2015). Fe–S proteins that regulate gene expression. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1853 (6), 1284–1293. doi:10.1016/j.bbamcr.2014.11.018
Miao, R., Kim, H., Koppolu, U. M. K., Ellis, E. A., Scott, R. A., and Lindahl, P. A. (2009). Biophysical characterization of the iron in mitochondria from Atm1p-depleted Saccharomyces cerevisiae. Biochemistry 48 (40), 9556–9568. doi:10.1021/bi901110n
Mittler, R., Darash-Yahana, M., Sohn, Y. S., Bai, F., Song, L., Cabantchik, I. Z., et al. (2019). NEET proteins: A new link between iron metabolism, reactive oxygen species, and cancer. Antioxidants Redox Signal. 30 (8), 1083–1095. doi:10.1089/ars.2018.7502
Mühlenhoff, U., Molik, S., Godoy, J. R., Uzarska, M. A., Richter, N., Seubert, A., et al. (2010). Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12 (4), 373–385. doi:10.1016/j.cmet.2010.08.001
Netz, D. J. A., Pierik, A. J., Stümpfig, M., Bill, E., Sharma, A. K., Pallesen, L. J., et al. (2012). A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation. J. Biol. Chem. 287 (15), 12365–12378. doi:10.1074/jbc.M111.328914
Netz, D. J. A., Stith, C. M., Stümpfig, M., Köpf, G., Vogel, D., Genau, H. M., et al. (2012). Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8 (1), 125–132. doi:10.1038/nchembio.721
Netz, D. J. A., Stümpfig, M., Doré, C., Mühlenhoff, U., Pierik, A. J., and Lill, R. (2010). Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6 (10), 758–765. doi:10.1038/nchembio.432
Nicolo, A., Tancredi, M., Lombardi, G., Flemma, C., Barbuti, S., Cristofano, C., et al. (2008). A novel breast cancer-associated BRIP1 (FANCJ/BACH1) germline mutation impairs protein stability and function. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 14, 4672–4680. doi:10.1158/1078-0432.CCR-08-0087
O’Brien, E., Holt, M. E., Salay, L. E., Chazin, W. J., and Barton, J. K. (2018). Substrate binding regulates redox signaling in human DNA primase. J. Am. Chem. Soc. 140 (49), 17153–17162. doi:10.1021/jacs.8b09914
O’Brien, E., Holt, M. E., Thompson, M. K., Salay, L. E., Ehlinger, A. C., Chazin, W. J., et al. (2017). The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355 (6327), eaag1789. doi:10.1126/science.aag1789
O’Brien, E., Salay, L., Epum, E., Friedman, K., Chazin, W., and Barton, J. (2018). Yeast require redox switching in DNA primase. Proc. Natl. Acad. Sci. 115, 13186–13191. doi:10.1073/pnas.1810715115
Odermatt, D. C., and Gari, K. (2017). The CIA targeting complex is highly regulated and provides two distinct binding sites for client iron-sulfur proteins. Cell Rep. 18 (6), 1434–1443. doi:10.1016/j.celrep.2017.01.037
Pallesen, L. J., Solodovnikova, N., Sharma, A. K., and Walden, W. E. (2013). Interaction with Cfd1 increases the kinetic lability of FeS on the Nbp35 scaffold. J. Biol. Chem. 288 (32), 23358–23367. doi:10.1074/jbc.M113.486878
Panday, A., Willis, N. A., Elango, R., Menghi, F., Duffey, E. E., Liu, E. T., et al. (2021). FANCM regulates repair pathway choice at stalled replication forks. Mol. Cell 81 (11), 2428–2444.e6. doi:10.1016/j.molcel.2021.03.044
Parker, A., Gu, Y., and Lu, A. (2000). Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria. Nucleic acids Res. 28, 3206–3215. doi:10.1093/nar/28.17.3206
Patil, M., Pabla, N., and Dong, Z. (2013). Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell. Mol. Life Sci. 70 (21), 4009–4021. doi:10.1007/s00018-013-1307-3
Petronek, M. S., Spitz, D. R., and Allen, B. G. (2021). Iron–sulfur cluster biogenesis as a critical target in cancer. Antioxidants 10 (9), 1458. doi:10.3390/antiox10091458
Pinto, M. N., ter Beek, J., Ekanger, L. A., Johansson, E., and Barton, J. K. (2021). The [4Fe4S] cluster of yeast DNA polymerase ε is redox active and can undergo DNA-mediated signaling. J. Am. Chem. Soc. 143 (39), 16147–16153. doi:10.1021/jacs.1c07150
Pokharel, S., and Campbell, J. L. (2012). Cross talk between the nuclease and helicase activities of Dna2: Role of an essential iron-sulfur cluster domain. Nucleic Acids Res. 40 (16), 7821–7830. doi:10.1093/nar/gks534
Pondarré, C., Antiochos, B. B., Campagna, D. R., Clarke, S. L., Greer, E. L., Deck, K. M., et al. (2006). The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron–sulfur cluster biogenesis. Hum. Mol. Genet. 15 (6), 953–964. doi:10.1093/hmg/ddl012
Qi, W., Li, J., and Cowan, J. A. (2014). A structural model for glutathione-complexed iron-sulfur cluster as a substrate for ABCB7-type transporters. Chem. Commun. (Camb). 50 (29), 3795–3798. doi:10.1039/c3cc48239a
Qian, S. Y., and Buettner, G. R. (1999). Iron and dioxygen chemistry is an important route to initiation of biological free radical oxidations: An electron paramagnetic resonance spin trapping study. Free Radic. Biol. Med. 26 (11), 1447–1456. doi:10.1016/s0891-5849(99)00002-7
Read, A. D., Bentley, R. E. T., Archer, S. L., and Dunham-Snary, K. J. (2021). Mitochondrial iron–sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox Biol. 47, 02164, 1. doi:10.1016/j.redox.2021.102164
Roy, A., Solodovnikova, N., Nicholson, T., Antholine, W., and Walden, W. E. (2003). A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22 (18), 4826–4835. doi:10.1093/emboj/cdg455
Rudolf, J., Makrantoni, V., Ingledew, W. J., Stark, M. J. R., and White, M. F. (2006). The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23 (6), 801–808. doi:10.1016/j.molcel.2006.07.019
Saha, P. P., Vishwanathan, V., Bankapalli, K., and D’Silva, P. (2018). “Iron-sulfur protein assembly in human cells,” in Reviews of physiology, biochemistry and pharmacology. Editors B. Nilius, P. de Tombe, T. Gudermann, R. Jahn, R. Lill, and O. H. Petersen (Cham: Springer International Publishing), Vol. 174, 25–65. [Internet]. doi:10.1007/112_2017_5
Seki, M., Takeda, Y., Iwai, K., and Tanaka, K. (2013). IOP1 protein is an external component of the human cytosolic iron-sulfur cluster assembly (CIA) machinery and functions in the MMS19 protein-dependent CIA pathway. J. Biol. Chem. 288 (23), 16680–16689. doi:10.1074/jbc.M112.416602
Sfeir, A., Kosiyatrakul, S. T., Hockemeyer, D., MacRae, S. L., Karlseder, J., Schildkraut, C. L., et al. (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138 (1), 90–103. doi:10.1016/j.cell.2009.06.021
Shen, Y., McMackin, M. Z., Shan, Y., Raetz, A., David, S., and Cortopassi, G. (2016). Frataxin deficiency promotes excess microglial DNA damage and inflammation that is rescued by PJ34. PLOS ONE 11 (3), e0151026. doi:10.1371/journal.pone.0151026
Shi, R., Hou, W., Wang, Z. Q., and Xu, X. (2021). Biogenesis of iron–sulfur clusters and their role in DNA metabolism. Front. Cell Dev. Biol. 9, 735678. doi:10.3389/fcell.2021.735678
Simon, A. K., Kummer, S., Wild, S., Lezaja, A., Teloni, F., Jozwiakowski, S. K., et al. (2020). The iron–sulfur helicase DDX11 promotes the generation of single-stranded DNA for CHK1 activation. Life Sci. Alliance 3 (3), e201900547. doi:10.26508/lsa.201900547
Singleton, M. R., Dillingham, M. S., and Wigley, D. B. (2007). Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50. doi:10.1146/annurev.biochem.76.052305.115300
Slinker, J. D., Muren, N. B., Renfrew, S. E., and Barton, J. K. (2011). DNA charge transport over 34 nm. Nat. Chem. 3 (3), 228–233. doi:10.1038/nchem.982
Stehling, O., and Lill, R. (2013). The role of mitochondria in cellular iron-sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 5 (8), a011312. doi:10.1101/cshperspect.a011312
Stehling, O., Vashisht, A. A., Mascarenhas, J., Jonsson, Z. O., Sharma, T., Netz, D. J. A., et al. (2012). MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337 (6091), 195–199. doi:10.1126/science.1219723
Sun, J., Su, W., Deng, J., Qin, Y., Wang, Z., and Liu, Y. (2022). DNA2 mutation causing multisystemic disorder with impaired mitochondrial DNA maintenance. J. Hum. Genet. 67 (12), 691–699. doi:10.1038/s10038-022-01075-4
Suwa, Y., Gu, J., Baranovskiy, A. G., Babayeva, N. D., Pavlov, Y. I., and Tahirov, T. H. (2015). Crystal structure of the human pol α B subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 290 (23), 14328–14337. doi:10.1074/jbc.M115.649954
Syed, A., and Tainer, J. A. (2019). Charge transport communication through DNA by protein Fe–S clusters: How far is not too far? ACS Cent. Sci. 5 (1), 7–9. doi:10.1021/acscentsci.8b00909
Taylor, E., Broughton, B., Botta, E., Stefanini, M., Sarasin, A., Jaspers, N., et al. (1997). Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc. Natl. Acad. Sci. U. S. A. 94 (16), 8658–8663. doi:10.1073/pnas.94.16.8658
ter Beek, J., Parkash, V., Bylund, G. O., Osterman, P., Sauer-Eriksson, A. E., and Johansson, E. (2019). Structural evidence for an essential Fe–S cluster in the catalytic core domain of DNA polymerase ϵ. Nucleic Acids Res. 47 (11), 5712–5722. doi:10.1093/nar/gkz248
Thierbach, R., Drewes, G., Fusser, M., Voigt, A., Kuhlow, D., Blume, U., et al. (2010). The Friedreich’s Ataxia protein frataxin modulates DNA base excision repair in prokaryotes and mammals. Biochem. J. 432, 165–172. doi:10.1042/BJ20101116
Uringa, E. J., Lisaingo, K., Pickett, H., Brind’Amour, J., Rohde, J. H., Zelensky, A., et al. (2012). RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol. Biol. Cell 23, 2782–2792. doi:10.1091/mbc.E12-03-0179
Uringa, E. J., Youds, J., Lisaingo, K., Lansdorp, P., and Boulton, S. (2010). RTEL1: An essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic acids Res. 39, 1647–1655. doi:10.1093/nar/gkq1045
Urzica, E., Pierik, A. J., Mühlenhoff, U., and Lill, R. (2009). Crucial role of conserved cysteine residues in the assembly of two Iron−Sulfur clusters on the CIA protein Nar1. Biochemistry 48 (22), 4946–4958. doi:10.1021/bi900312x
van der Lelij, P., Chrzanowska, K. H., Godthelp, B. C., Rooimans, M. A., Oostra, A. B., Stumm, M., et al. (2010). Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 86 (2), 262–266. doi:10.1016/j.ajhg.2010.01.008
van Schie Jjm, , Faramarz, A., Balk, J. A., Stewart, G. S., Cantelli, E., Oostra, A. B., et al. (2020). Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat. Commun. 11 (1), 4287. doi:10.1038/s41467-020-18066-8
Vashisht, A. A., Yu, C. C., Sharma, T., Ro, K., and Wohlschlegel, J. A. (2015). The association of the xeroderma pigmentosum group D DNA helicase (XPD) with transcription factor IIH is regulated by the cytosolic iron-sulfur cluster assembly pathway. J. Biol. Chem. 290 (22), 14218–14225. doi:10.1074/jbc.M115.650762
Wallace, S. S. (2013). DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 54 (9), 691–704. doi:10.1002/em.21820
Wardman, P., and Candeias, L. P. (1996). Fenton chemistry: An introduction. Radiat. Res. 145 (5), 523–531. doi:10.2307/3579270
Weon, J., Yang, S., and Potts, P. (2017). Cytosolic iron-sulfur assembly is evolutionarily tuned by a cancer-amplified ubiquitin ligase. Mol. Cell, 69.
Weren, R. D. A., Ligtenberg, M. J. L., Kets, C. M., de Voer, R. M., Verwiel, E. T. P., Spruijt, L., et al. (2015). A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 47 (6), 668–671. doi:10.1038/ng.3287
White, M. F. (2009). Structure, function and evolution of the XPD family of iron–sulfur-containing 5′→3′ DNA helicases. Biochem. Soc. Trans. 37 (3), 547–551. doi:10.1042/BST0370547
Wietmarschen, N., Moradian, A., Morin, G., Lansdorp, P., and Uringa, E. J. (2012). The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. J. Biol. Chem., 287.
Wolski, S. C., Kuper, J., Hänzelmann, P., Truglio, J. J., Croteau, D. L., Houten, B. V., et al. (2008). Crystal structure of the FeS cluster–containing nucleotide excision repair helicase XPD. PLOS Biol. 6 (6), e149. doi:10.1371/journal.pbio.0060149
Wu, C. G., and Spies, M. (2016). G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 44 (18), 8742–8753. doi:10.1093/nar/gkw574
Wu, W., Bhowmick, R., Vogel, I., Özer, Ö., Ghisays, F., Thakur, R. S., et al. (2020). RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nat. Struct. Mol. Biol. 27 (5), 424–437. doi:10.1038/s41594-020-0408-6
Wu, Y., Sommers, J. A., Khan, I., de Winter, J. P., and Brosh, R. M. (2012). Biochemical characterization of Warsaw breakage syndrome helicase. J. Biol. Chem. 287 (2), 1007–1021. doi:10.1074/jbc.M111.276022
Yan, S., Xia, R., Jin, T., Chai, F., Yang, H., Li, J., et al. (2016). RTEL1 polymorphisms are associated with lung cancer risk in the Chinese Han population. Oncotarget 7, 70475–70480. doi:10.18632/oncotarget.12297
Yeeles, J. T. P., Cammack, R., and Dillingham, M. S. (2009). An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem. 284 (12), 7746–7755. doi:10.1074/jbc.M808526200
Keywords: iron metabolism, Fe-S biogenesis, genomic integrity, DNA metabolism, CIA complex
Citation: Petronek MS and Allen BG (2023) Maintenance of genome integrity by the late-acting cytoplasmic iron-sulfur assembly (CIA) complex. Front. Genet. 14:1152398. doi: 10.3389/fgene.2023.1152398
Received: 27 January 2023; Accepted: 24 February 2023;
Published: 08 March 2023.
Edited by:
Ashutosh Kumar Pandey, The State University of New Jersey, United StatesReviewed by:
William Walden, University of Illinois at Chicago, United StatesArvind Panday, Harvard Medical School, United States
Copyright © 2023 Petronek and Allen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. S. Petronek, bWljaGFlbC1wZXRyb25la0B1aW93YS5lZHU=
 M. S. Petronek
M. S. Petronek B. G. Allen
B. G. Allen