
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 20 January 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1137634
This article is part of the Research Topic Genetic and Epigenetic Regulation of Disease Resistance in Horticultural Plants View all 8 articles
Fusarium wilt, which affects common bean all across the world, is caused by Fusarium oxysporum f. sp. Phaseoli (Fop). It is necessary to have functional genes in response to Fop infection because they might be used to manage disease. As a crucial regulator, TGA-binding transcription factor (TGA) is engaged in the defense mechanism of plants against pathogens. The role of TGA regulators in common bean in response to Fop infection, however, has not been documented. Hence, we performed genome-wide identified and characterized eight TGA genes in common bean. In this study, eight PvTGA genes were distributed on six chromosomes and classified into four subgroups. The PvTGA genes have the same conserved bZIP and DOG1 domains, but there are specific sequence structures in different PvTGAs. Phylogenetic and synteny analysis explained that PvTGA gene has a close genetic relationship with legume TGAs and that PvTGA03 and PvTGA05 may play an important role in evolution. Transcriptome data explained that expression levels of PvTGA genes showed diversity in different tissues. After Fop inoculation, the expression levels of PvTGA03 and PvTGA07 were significantly different between resistant and susceptible genotypes. Under SA treatment, the expression levels of PvTGA03, PvTGA04, PvTGA06, PvTGA07 and PvTGA08 were significantly different. These results imply that PvTGA03 and PvTGA07 play key roles in SA-mediated resistance to Fusarium wilt. Together, these findings advance knowledge of the PvTGA gene family in common bean and will help future studies aimed at reducing Fusarium wilt.
Common bean (Phaseolus vulgaris L.) is an economically important leguminous crop grown worldwide. It is an important source of fiber, proteins, vitamins, and essential micronutrients for human nutrition (Blair et al., 2013; Blair et al., 2016; Suárez-Martínez et al., 2016). The planting area of common bean is second only to soybean and peanut. Common bean is widely cultivated as an important food source in the world, especially in parts of Africa and the South America (Pérez-Vega et al., 2010; Schmutz et al., 2014). The production of common bean is limited by a variety of environmental factors like biotic and abiotic stress (Schwartz et al., 2005). A major fungus disease of common bean is Fusarium wilt, a soil-borne disease that affects a wide variety of crops globally. It is caused by Fusarium oxysporum f. sp. phaseoli (Fop), and has been found and identified in most bean-growing regions of the world. Wet environments or densely planted lands with inadequate crop rotation are especially susceptible to Fusarium wilt (Buruchara and Camacho, 2000). At present, the most effective and environmentally friendly way to control the disease is to discover the resistant genes in common bean, clarify their disease-resistant mechanisms, and apply them to common bean.
Plants have developed a number of barrier defense strategies to shield themselves against invasive pathogens like bacteria, fungi, viruses, and oomycetes (Jones and Dangl, 2006; Spoel and Dong, 2012; Bigeard et al., 2015). Plants activate local defenses that trigger a secondary immune response known as systemic acquired resistance (SAR). SAR provides long-lasting immunity to the distal uninfected tissues by activating various pathogenesis-related genes (Fu and Dong, 2013; Zhou and Zhang, 2020). To date, several molecules have been proposed as mobile signals leading to SAR (Dempsey and Klessig, 2012; Klessig et al., 2018). Salicylic acid (SA) is a phytohormone which plays a role in numerous plant physiological processes in including abiotic or biotic stress regulation. For the initiation of the SAR response and the expression of pathogenesis-related genes (PRs), SA is an essential signaling molecule (Malamy et al., 1990; An and Mou, 2011). The key genes in the synthesis and regulation pathway of salicylic acid have been proved to play an important role in the interaction between common bean and Fusarium oxysporum (Xue et al., 2014; Xue et al., 2021). Therefore, it is of great significance to explore the key genes of common bean resistance to Fusarium wilt mediated by salicylic acid and to clarify their functions for analyzing the molecular mechanism of common bean and Fusarium oxysporum interaction.
Transcription Factors (TF) are proteins that can regulate the transcription of genes by binding to their specific cis-regulatory sequences in the promoter region. TF are crucial in a variety of physiological functions, including metabolic balance, growth, development, and response to adversity stress (Baillo et al., 2019). TGA transcription factors (TGACG motif-binding factor) are members of the Basic Leucine Zipper (bZIP) transcription factor family, which is one of the biggest and most significant TF families. TGA TFs interact with the SA-receptor NPR protein and bind specifically the activation sequence-1 (as-1) motif in the promoter region of PR genes during plants immune response after pathogen attack in Arabidopsis thaliana (Rochon et al., 2006). The first TGA gene cloned in plants was TGA1a, and serves as a crucial point of reference for identifying TGA gene family (Katagiri et al., 1989). More TGA transcription factors were subsequently found in diverse plants (Jakoby et al., 2002; Idrovo Espín et al., 2012). A total of 10 TGA transcription factor genes were identified genome-wide in Arabidopsis (Gatz, 2013). Based on their sequence similarities, the TGAs can be classified into five subgroups (I-V). The two TGAs that most closely resemble tobacco TGA1a are found in Group I: TGA1 and TGA4. TGA2, TGA5, and TGA6 make up Group II; they are connected and have functional redundancy. TGA3 and TGA7 make up Group III, TGA9 and TGA10 make up Group IV, and TGA8 is the solitary member of Group V. Among these identified TGAs, TGA1-TGA7 has been proven to interact with key resistance-related genes such as NPR1, and participate in multiple signal regulation pathways in plants to improve plant biotic and abiotic stress resistance (Johnson et al., 2001; Choi et al., 2004; Thurow et al., 2005; Jiang et al., 2021; Liu et al., 2022). In addition to Arabidopsis and tobacco, TGA has been identified in a variety of plants related to disease resistance, such as the TGA gene in soybean responds to mosaic virus, kiwifruit TGAs plays a role in the process of Pseudomonas syringae pv. Actinidiae (Psa) infection (Jiang et al., 2021; Liu et al., 2022). In Brachypodium distachyon, the TGA-promoted transcription of SA-inducible PR1 is orchestrated by the activator BdNPR2 and the repressor BdNPR1 to enforce immune resistance (Shimizu et al., 2022). However, little is known about TGA gene functions in common bean. At the same time, it is not clear how the TGA family plays a regulatory role in the process of common bean resistance. Therefore, the objectives of this study were to identify members of the TGA gene family in the Phaseolus vulgaris L. genome. The phylogenetic relationships, structural features, chromosomal locations, cis-elements, and expression patterns of PvTGA genes under Fop stress were described and analyzed in order to further investigate the function of the PvTGA gene family in resistance-related functions and the regulatory mechanisms in common bean. The findings offer a theoretical framework for understanding the role of TGA genes in common bean, which may be applied to help create common bean cultivars resistant to Fusarium wilt.
Common bean genotypes BRB130 (Fusarium wilt susceptible) and CAAS260205 (Fusarium wilt resistant) were obtained from the Institute of Crop Sciences of the Chinese Academy of Agricultural Sciences (CAAS) in Beijing, China (Xue et al., 2017; Xue et al., 2021). An aggressive F. oxysporum f. sp. phaseoli (Fop) isolate, FOP-DM01 was used for the experiments as further described by Xue et al. (2012). BRB130 and CAAS260205 plants were inoculated at the fully expanded, “cotyledonary” leaf, seedling development stage using seedlings that had been cultivated in a greenhouse for 10 days using previous described methods (Xue et al., 2017). To explore responses to SA, Fusarium wilt susceptible genotype BRB130 leaves were sprayed for five consecutive days with 2 mM salicylic acid (SA) diluted in Tween 20 (0.02% v/v) using previous described methods (Xue et al., 2014). The plants were grown in a greenhouse with a temperature range of about 22°C to 28°C. For a total of 14 days, all treatments were grown in the same greenhouse with natural sunshine and additional illumination. In the case of the Fop therapy, root tissues were taken at 0, 24, 48, 72, 96, and 120 h post inoculation (hpi), with an additional 6 h collection for the SA treatment. All obtained root tissues were stored at −80°C.
To identify TGA genes in common bean, 10 TGA gene sequences (AtTGA1 (AT5G65210), AtTGA2 (AT5G06950), AtTGA3 (AT1G22070), AtTGA4 (AT5G10030), ATTGA5 (AT5G06960), AtTGA6 (AT3G12250), AtTGA7 (AT1G77920), AtTGA8 (AT1G68640), AtTGA9 (AT1G08320) and AtTGA10 (AT5G06839)) in Arabidopsis were selected to search for candidate TGA genes in common bean. The amino acid sequence of the AtTGA proteins in Arabidopsis was used as queries for a BLAST search for TGAs in the common bean protein database (Phaseolus vulgaris v2.1) (https://phytozome-next.jgi.doe.gov/), and the newly searched proteins whose E-value <10–5 were then used to perform a BLAST search again to determine all the orthologs. All the discovered proteins were submitted to the databases CDD (https://www.ncbi.nlm.nih.gov/cdd), Pfam (http://pfam.xfam.org/), and SMART (http://smart.embl-heidelberg.de/) to check if they included full bZIP and DOG1 domains in order to further confirm TGA proteins. ExPASy (https://web.expasy.org/protparam/) was used to determine the theoretical isoelectric point (pI) and molecular weight (MW) for the retrieved TGA protein sequences. WoLF PSORT (https://www.genscript.com/wolf-psort.html) online software was used for subcellular localization.
To classify PvTGAs, gene cluster analysis was conduction with the common bean and Arabidopsis TGA protein using the maximum-likelihood (ML) method. The MEME program (http://meme-suite.org/) was used to detect conserved motifs of the TGA proteins with the following parameters: the width of the motif ranged from 6 to 50 amino acids, with a maximum of 20 (Bailey et al., 2006). PvTGA gene structure and conserved domain map were drawn using the Gene Structure View program in TBtools based on the download common bean genome annotation files (Phaseolus vulgaris v2.1) (Chen et al., 2019).
To further understand the phylogenetic relationship of PvTGA proteins and other plant species, the phylogenetic tree was constructed. TGA proteins from Arabidopsis thaliana (Cheng et al., 2017), peanut (Arachis hypogaea) (Bertioli et al., 2019), soybean (Glycine max) (Li et al., 2019), grape (Vitis vinifera) (The French–Italian Public Consortium for Grapevine Genome Characterization, 2007), alfalfa (Medicago sativa) (Tang et al., 2014), chickpea (Cicer arietinum) (Varshney et al., 2013), rice (Oryza sativa, Oryza sativa Japonica Group, www.ncbi.nlm.nih.gov), maize (Zea mays, ZmB73_RefGen_v4, www.maizegdb.org) and Sorghum (Sorghum bicolor) (McCormick et al., 2018) were obtained in the same way as common bean. The maximum-likelihood (ML) tree was constructed after the TGA protein sequences were aligned in MEGA-X (https://www.megasoftware.net/) using the MUSCLE method (Kumar et al., 2018).
Gene collinearity and genes involved in duplication were examined using the MCScanX (Wang et al., 2012). Analysis was done on the PEBP homologs between common bean and other plant species. We calculated the homologous genes’ non-synonymous to synonymous mutation rate (Ka/Ks) to look into selection pressure. To evaluate selection pressure, the Ka/Ks Calculator 2.0 program was used to calculate the Ka (non-synonymous substitution rate) and Ks (synonymous substitution rate) values of repeated genes (Wang et al., 2010).
The 2,000 bp upstream area of the start codon (ATG) was extracted and submitted the sequences to the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database to predict the cis-elements in the promoter of PvTGA genes. The distribution of Cis-elements of PvTGA genes was displayed by TBtools (Chen et al., 2019).
The transcriptome data of gene expression in 11 different tissues during the whole growth period of common bean species are from Phytozome 13 genome database including root_10, nodules, root_19, young pods, stem_10, stem_19, green mature buds, leaves, young triloliates, flower buds, and flower. The expression levels in silico were measured using FPKM (expected number of fragments per kilobase of transcript sequence per millions of base pairs sequenced). Using gene expression data in 11 tissues of common bean, the FPKM value of the extracted eight of common bean TGA gene was transformed with log2FPKM. The heatmap program on TBtools platform was used to cluster and draw the heat map.
All root tissues were selected for RNA extraction. Using the Plant Total RNA Extraction Kit (Tiangen Biotech, Beijing, China), total plant RNA was extracted. The Reverse Transcription Kit PrimeScriptTM RT Kit (TaKaRa, Japan) was used to create the reagent cDNA, and the process was based on its guidelines. PrimerBlast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to searching primers for identifying TGA genes with differential expression. The internal control was the Actin11 gene. The SYBR Premix Ex TaqII kit (TliRNaseH Plus) (TaKaRa, Japan) was used to build the reaction system for gene expression analysis, and the ABI7500 was used to detect fluorescence quantitative reactions (Applied Biosystems, United States). The 2−ΔΔCT approach was used to calculate the relative expression analysis (Livak and Schmittgen, 2001).
In this study, 10 AtTGAs in Arabidopsis thaliana were used as a query to identify the TGA genes in the common bean genome, a total of eight putative PvTGA genes were identified. All gene sequences included complete bZIP and DOG1 domains, according to the findings of BLAST analysis. The common bean genome ultimately included eight PvTGA genes, and the relevant genes were renamed PvTGA01–PvTGA08 in accordance with the order of their distribution on different chromosomes. Eight PvTGA genes are located on the six chromosomes of common bean (Figure 1). The full length of the candidate TGA protein sequence, the corresponding gene at the chromosomal location, the molecular weight (MW) of the proteins encoded, subcellular localization and the isoelectric point (pI) were summarized (Table 1). The full length of the eight protein sequences ranges from 351 to 467. The molecular weight (kDa) and theoretical isoelectric points (pI) of putative TGA proteins ranged from 39.72 (PvTGA07) to 51.71 (PvTGA03) kDA and 6.03 (PvTGA05) to 8.53 (PvTGA01), respectively. Additionally, predictions of protein subcellular localization revealed that AcTGA genes were primarily found in cell nuclei.
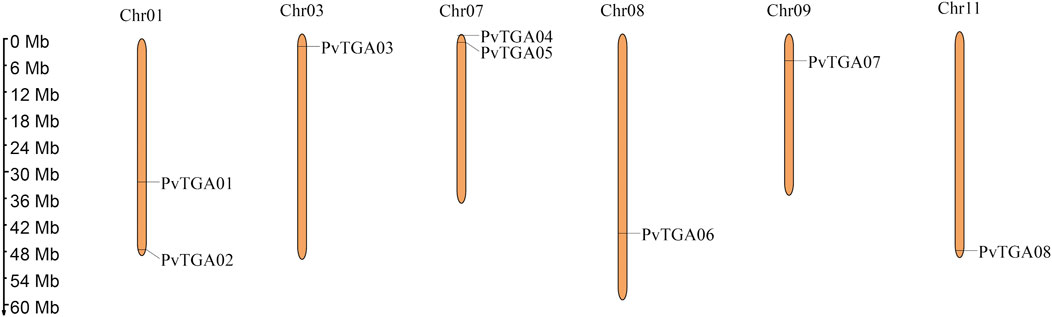
FIGURE 1. Distribution of TGA genes on P. vulgaris chromosome. Chromosome size is indicated by its length. The scale on the left is shown in megabases (Mb).
To further analyze the evolutionary relationship between TGA genes in common bean, Maximum-Likelihood (ML) method was used to construct phylogenetic trees using common bean and Arabidopsis TGA protein sequence. As shown in Figure 2A, 8 PvTGA and 10 AtTGA divided into five subgroups, PvTGAs were distributed in four groups, PvTGA01, PvTGA04, and PvTGA08 belonged to Clade I, PvTGA03 and PvTGA05 to Clade II, PvTGA02 to Clade IV, PvTGA06 and PvTGA07 to Clade V, there was no common bean TGA gene in group III. Using the MEME online program, conserved motifs in the eight PvTGA proteins were found. Nine motifs, numbered from 1 to 10, totaling 11 to 50 amino acids in length, were found in the peanut proteins (Figure 2B; Supplementary Figure S1). The number of motifs contained in TGA protein ranged from six to nine and all of them contained six conserved motifs from Motif 1 to Motif 6. There are different motif distribution characteristics in different clade groups, such as motif 10 only exists in Clade II; Motif 8 and Motif 9 only exist in clade II and V, and Motif 7 is only identified in groups I, III and IV. The different distribution of motifs in different clades may lead to changes in the structure of the TGA genes, which in turn may determine the differentiation of different clade functions. To understand the connection between genomic evolution and functional differentiation, gene structure analysis was crucial. The number of exons in the PvTGA gene family is 9–15. From the evolutionary relationship, the TGA genes with close evolutionary relationship not only have the same number of exons, but also have similar structures (Figure 2C). A bZIP and DOG1 domain was found in every PvTGA protein, and these domains were found in the same relative locations across various sequences, according to a conserved structure study (Figure 2D). These results imply that the PvTGA gene has the same conserved domain in the gene structure, but there are specific sequence structures in different types of TGA.
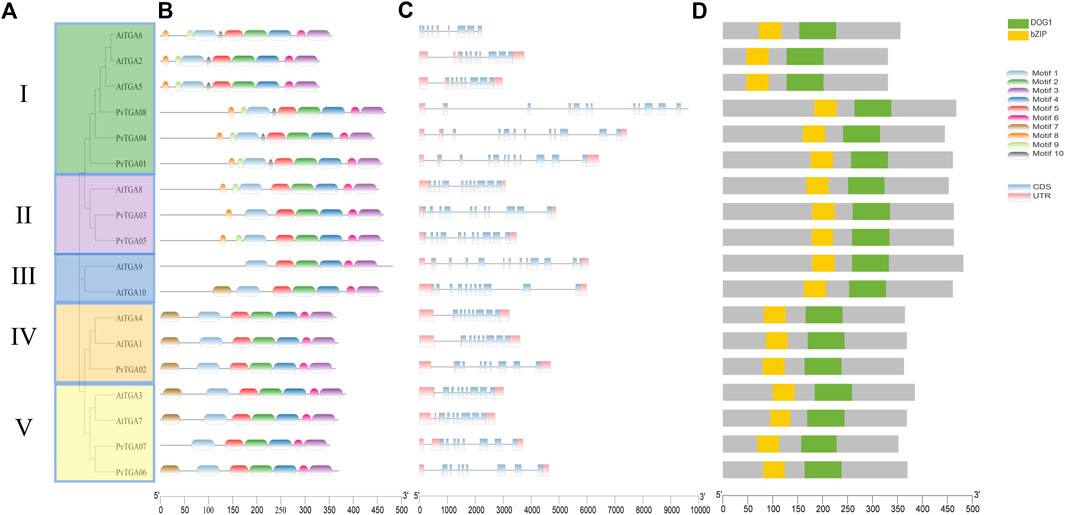
FIGURE 2. Phylogenetic, conserved motifs and gene structure of the TGA genes in P. vulgaris and Arabidopsis. (A) phylogenetic tree of the TGA protein sequence in common bean and Arabidopsis species constructed by the Maximum-Likelihood (ML) method. (B) conserved motifs in PvTGAs using MEME-suite. Various colors represented different motifs. (C) the eight common bean TGA gene structures. (D) comparison of conserved domain among AtTGA and PvTGA.The ruler at the bottom indicates length of the sequences.
Protein homology and cluster analysis were used to build phylogenetic trees of PvTGA genes from various species, and homology relations with other species were identified. In order to further study the evolutionary relationship of TGA family genes in different species, the MEGAX software was used to analyze the relationship between common bean, Arabidopsis, peanut (Arachis hypogaea), soybean (Glycine max), grape (Vitis vinifera), alfalfa (Medicago sativa), chickpea (Cicer arietinum), rice (Oryza sativa), maize (Zea mays) and Sorghum (Sorghum bicolor) TGA protein sequences constructed a phylogenetic tree (Supplementary Table S1). One hundred and thirteen TGA protein sequences were selected for multiple sequence alignment and a phylogenetic tree was constructed (Figure 3). TGAs were divided into five different subgroups, and the eight PvTGAs were assigned to four different groups, namely Group I, Group II, Group IV and V. Among these five subgroups, the number of TGA genes in the group I subgroup was 36. In the second subgroup, there are 18 genes in total, and in subgroup III, there are 26 members in total. In the subgroup IV, a total of 13 genes were included, and the subgroup V contained a total of 14 TGAs. The PvTGAs has a very close genetic distance with other legumes, especially soybean. The PvTGAs Proteins have the highest sequence similarity with those in soybean. Therefore, PvTGAs in common bean may have similar functions to the TGAs in soybean.
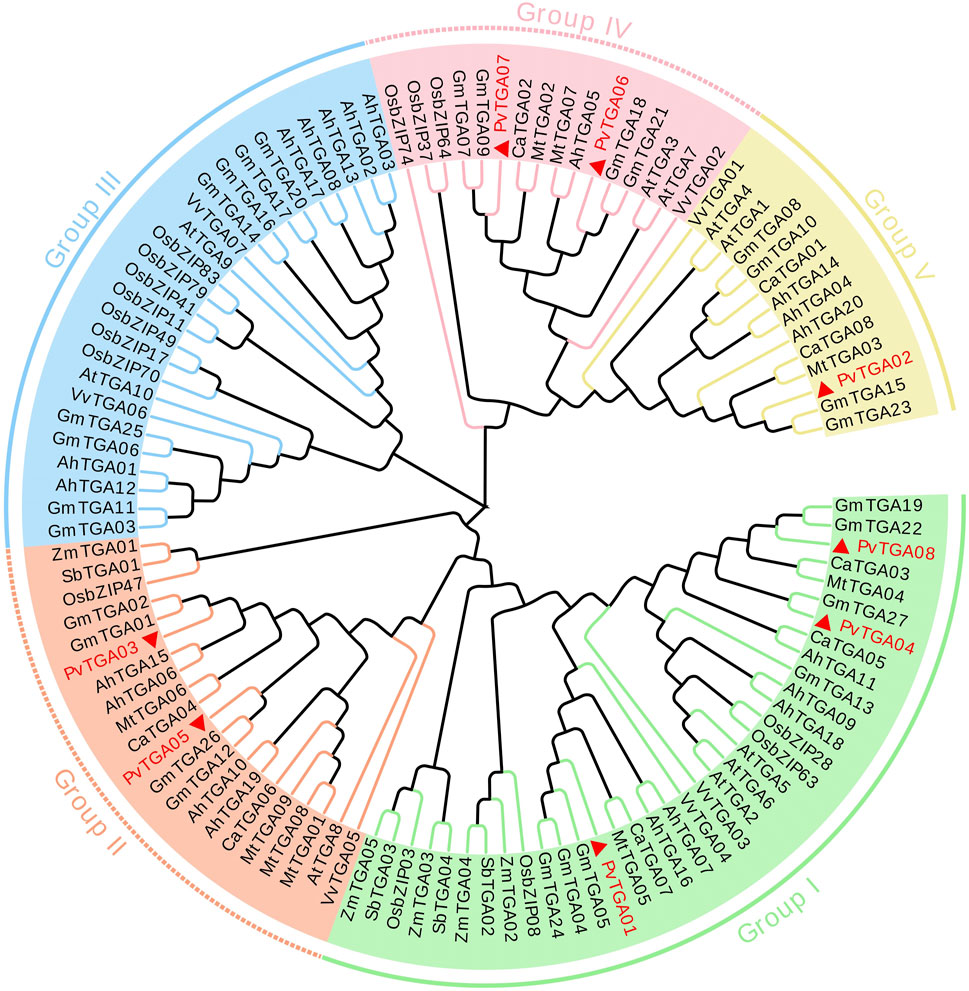
FIGURE 3. Evolutionary relationship analysis of TGA proteins from common bean (Phaseolus vulgaris; Pv). TGA amino acid sequences from the Phaseolus vulgaris, Arabidopsis (At), Arachis hypogaea (Ah), Glycine max (Gm), Cicer arietinum (Ca), rice (Os), corn (Zm), Vitis vinifera (Vv), Medicago truncatula (Mt) and sorghum (Sb) were used to construct a phylogenetic tree. Green line, orange line, blue line, pink line and yellow line represent group I, group II, group III, group IV and group V respectively.
Segmental duplication and tandem repeats were identified in common bean genome. The TGA gene in common bean does not have a tandem repeat event, but PvTGA03 and PvTGA05 are segmental duplications (Figure 4A). To further explore the evolutionary relationship of TGA genes between common bean and other species, the synteny relationship between TGAs and homologues of other species was investigated. Orthologous gene pairs were identified between TGAs of common bean and other plants, including soybean, chickpea, alfalfa, peanut, Arabidopsis, grape, rice, sorghum, and maize (Figure 4B). Common bean and other legumes including soybean, alfalfa, chickpea and peanut have gene pairs of 9, 4, 2, and 5, respectively; dicotyledons grape and Arabidopsis have gene three and one pairs with common bean, and monocots sorghum, rice and maize do not have any gene pairs with common bean. The results showed that TGAs in common bean were genetically closely related to ones in soybean, alfalfa, chickpea, and cultivated peanut, and had distant phylogenetic relationships with Arabidopsis and grapevine. There were no collinear gene pairs of TGAs between common bean and rice, sorghum and maize, suggesting a long-distance relationship between these species and common. PvTGA03 and PvTGA05 in the group II had the most homologous gene pairs with other species (Supplementary Table S2). These two genes co-exist in 6 to 9 gene pairs in common bean and other species, while other TGA genes only have 2 to 7 gene pairs. These results suggest that PvTGA03 and PvTGA05 may play key roles in the TGA gene family during evolution and have important functions.
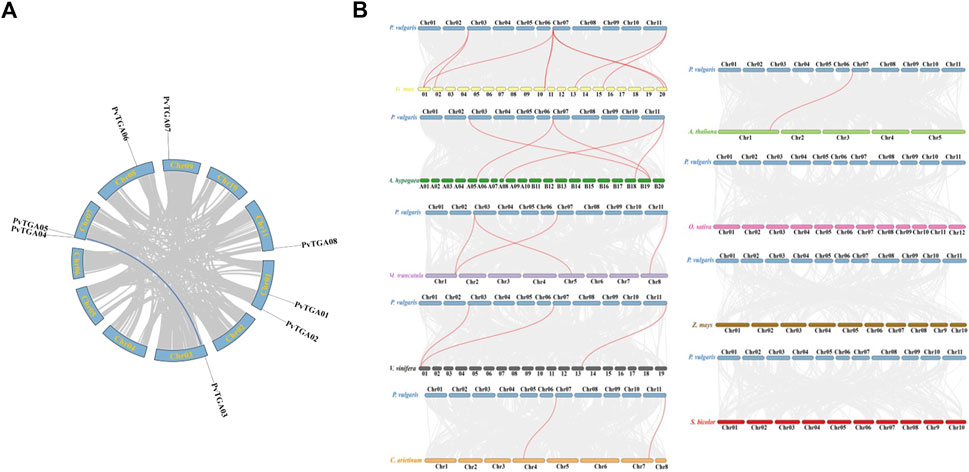
FIGURE 4. Genome scale synteny analysis of Phaseolus vulgaris, Arabidopsis, Glycine max, Vitis vinifera, Medicago truncatula, Cicer arietinum, rice, corn and sorghum TGA genes. (A) syntenic relationship of P. vulgaris TGA genes. All 8 TGA genes are marked according to their chromosome distribution in common bean genome and syntenic gene pairs are connected with blue lines., (B) syntenic pairs of TGA genes between P. vulgaris and other plants. The Phaseolus vulgaris (P. vulgaris), Arabidopsis thaliana (A. thaliana), Arachis hypogaea (A. hypogaea), Glycine max (G. max), Cicer arietinum (C. arietinum), Vitis vinifera (V. vinifera), Medicago truncatula (M. truncatula), Oryzae Sativa (O. Sativa), Zea mays (Z. mays) and Sorghum bicolor (S. bicolor) chromosomes are shown in blue, lightgreen, green, yellow, orange, grey, purple, pink, brown and red bars, respectively. Gray lines in the background indicated the collinear blocks within P. vulgaris and other plant genomes, while the Syntenic TGA gene pairs are linked with red lines.
To analyze the evolutionary selection pressure of TGA genes, we calculated the Ka/Ks ratio of TGA gene pairs. The Ka/Ks ratio of TGA collinear gene pairs between common bean and other species was less than 1, indicating that the TGA gene family was dominated by purifying selection during evolution (Supplementary Table S3).
The promoter activities are essential for controlling how genes operate. The cis-elements in the promoter regions of TGA genes are examined in order to comprehend the genetic processes, metabolic networks, and regulatory mechanisms involved (Figure 5). In total, 164 cis-acting elements of 34 types were predicted to contain potential functions (Supplementary Table S4). It was found 22 cis elements related to growth and development, eight hormone-related cis-elements and four stress-responsive-related elements. There are differences in the number and distribution of cis-elements in each gene (Figure 5A). The PvTGA02 and PvTGA03 promoters contain the most cis-acting elements, with 27, while the PvTGA06 promoter contains the least cis-acting elements, only 15 (Figure 5B). Among the growth and development-related elements, the number of ARE elements on the promoter of PvTGA gene is the largest, followed by Box 4 and GT1-motif; Among the stress-related elements, the number of MBS was the largest, and the number of LTR and CCAAT-box elements was the least; among the hormone-related induction elements, the number of TGACG-motif and CGTCA-motif elements was the largest, and the number of TATC-box was the least. The analysis of cis-acting elements showed that PvTGAs may be involved in plant growth and development and responses to various hormones and stresses.
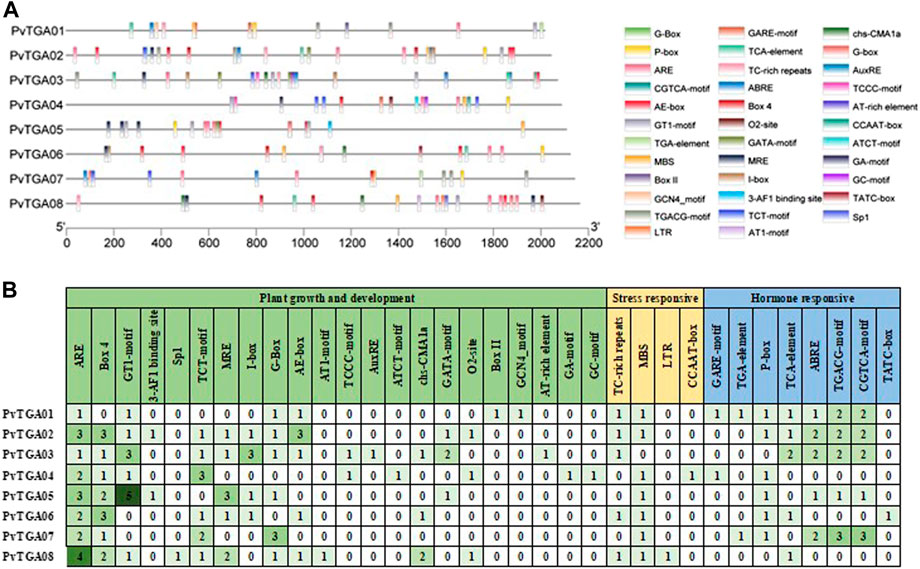
FIGURE 5. Analysis of the cis-acting elements in the promoter regions of 8 TGA genes. (A) the cis-acting elements distribution in 8 TGA genes promoters. Diverse colors were used for representing different cis-elements, as given in on the right side., (B) the names and numbers of cis-acting elements in 8 TGA genes promoters. The heatmap in grid and the color columns indicated the numbers of cis-acting elements.
In this study, we additionally aimed to perform mRNA analysis of TGA genes using publicly available expression data (https://phytozome.jgi.doe.gov). The heatmap, which was derived from mRNA levels, shows the expression variance of identified eight PvTGA genes in different plant tissues (Figure 6). All genes were expressed at low levels in flower buds, young pods, and young trifoliates, while most genes were expressed at high levels in roots. The expression levels of some genes were tissue-specific, for example, PvTGA03 and PvTGA04 were highly expressed in nodules and root 19, while PvTGA1, PvTGA02 and PvTGA08 were highly expressed in flowers and green muture pods. Varying tissues and developmental stages had different PvTGA expression levels, which suggested that these TGA genes were engaged in various developmental and regulatory processes in the common bean.
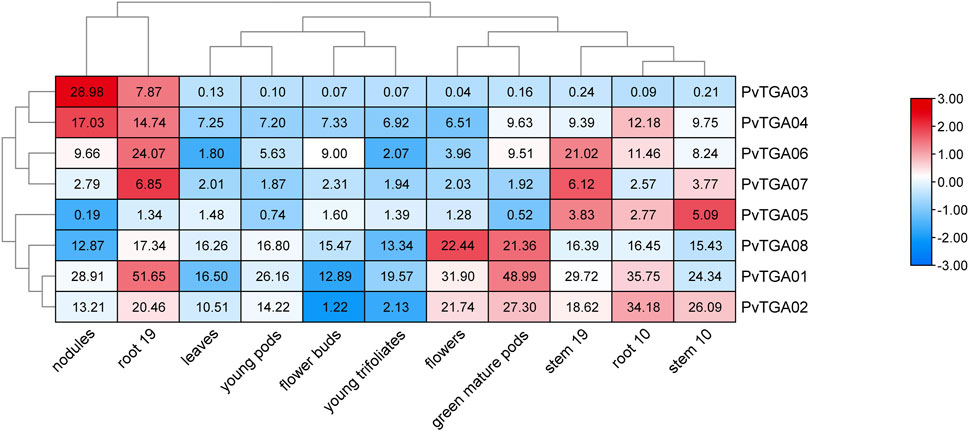
FIGURE 6. Expression profiles of PvTGA genes from common bean in 11 different tissues. Heat-map clustering based on FPKM expression of 8 PvTGA genes in different common bean tissues. Log2-transformed values are used in a color-coded heatmap, with bars representing gene-normalized FPKM (Log2) expression levels, and red for higher expression, blue means low.
The resistant genotype CAAS260205 and the susceptible genotype BRB130 were inoculated with Fop 24 days later, the susceptible genotype BRB130 showed vascular bundle necrosis and caused shoot wilting, while the resistant genotype CAAS260205 did not show vascular bundle necrosis and shoot wilting (Figure 7A). After five consecutive days of exogenous SA, the susceptible genotype BRB130 was inoculated with Fop. Twenty-four days after inoculation, exogenous SA can significantly alleviate the necrosis of vascular bundles in common bean plants and reduce the water loss in the shoot, while BRB130 plants without external application of SA obvious symptoms of wilting appear (Figure 7B).

FIGURE 7. Phenotype of common bean genotype under Fop infection. (A) resistant genotype CAAS260205 and susceptible genotype BRB130 were tested with Fop isolate for 24 days., (B) susceptible genotype BRB130 (left) under five consecutive days of exogenous SA treatment and BRB130 without SA application (right) as control were inoculated with Fop.
Science Fusarium wilt is a typical soil-borne disease, the roots of common plants were collected to detect the expression level of TGA gene. Root tissues were selected for qRT-PCR examination and expression level analysis for eight PvTGA genes in the susceptible (BRB130) and resistant (CAAS260205) genotypes (Figure 8A). After inoculation with Fop, six time points were selected for expression level analysis using qPCR. Specific primers were designed for each PvTGA gene to detect gene expression levels (Supplementary Table S5). The expression levels of eight genes showed significant changes within 120 h after inoculation with Fop (Figure 8A). Between the resistant and susceptible genotypes, PvTGA03 and PvTGA07 showed significant differences in expression levels at all time points except 0 hpi between the resistant and susceptible genotypes. The expression level of PvTGA03 in the resistant genotype CAAS260205 was significantly higher than that in the susceptible genotype BRB130, on the contrary, the expression level of PvTGA07 in the resistant genotype was significantly lower than that in the susceptible genotype. This indicated that these two genes played opposite roles in the response to Fop resistance. Increased expression of PvTGA03 promoted common bean disease resistance, while increased expression of PvTGA07 promoted Fop infection and led to plant disease.
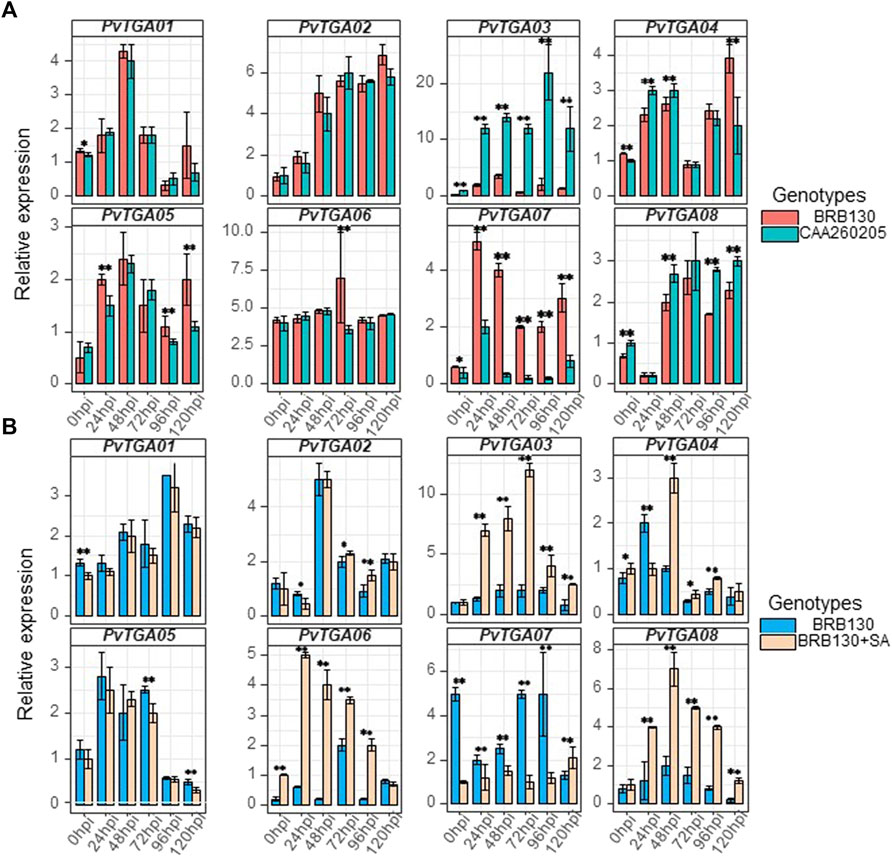
FIGURE 8. Differential expression of eight candidate genes in differernt varieties and processing. (A) relative expression of eight genes in susceptible (BRB130) and resistant (CAA260205) genotype at different time points after inoculation with Fop isolates., (B) relative expression of eight genes in susceptible genotype (BRB130) and sprayed with SA (BRB130 + SA) at different time points after inoculation with Fop isolates. (*, p < 0.05; **, p < 0.01; Student’s t-test).
Previous studies have revealed that TGA protein plays a key role in the salicylic acid-mediated resistance pathway. To explore the molecular mechanism of TGA gene response to salicylic acid in common bean, the expression levels of eight TGA genes were examined under SA treatment using qRT-PCR. After SA treatment of common bean seedlings, some TGA genes did not show significant changes in expression levels, including PvTGA01, PvTGA02 and PvTGA05, while PvTGA03, PvTGA04, PvTGA06, PvTGA07 and PvTGA08 were all showed significant changes in expression after SA treatment (Figure 8B). The disease resistance of common bean plants was enhanced after being treated with SA, and the expression levels of PvTGA03, PvTGA06 and PvTGA08 were increased, while the expression level of PvTGA07 was decreased. Therefore, these genes may be involved in SA-mediated disease resistance.
The defensive response to invading pathogens is all physiological processes in plants that depend on the transcriptional control of gene expression. Through their entire life cycle, plants are controlled by TFs. TFs regulate a wide range of elements of plant growth and development, including flower and root development, and morphological diversity (Suter, 2020; Strader et al., 2022). Therefore, understanding the structure and function of TFs is crucial to comprehending how plant growth and development are regulated. The TGA genes were among the first plant TFs to be identified, and their structure and function have undergone significant research (Pontier et al., 2001; Gatz, 2013). However, related research on the TGA gene family found in common beans is still insufficient. In this study, TGA genes in common bean were identified genome-wide, and gene structure and genetic evolution relationships were explored. The expression pattern of the PvTGA gene was explored including differences in expression levels of different tissues, in resistant and susceptible genotypes, and after exogenous salicylic acid application. This work offers useful data for investigating these molecular processes and advancing common bean resistance breeding.
The TGA protein in common bean are similar to Arabidopsis thaliana, and can be divided into four clade, lacking PvTGA similar to Arabidopsis thaliana subgroup III AtTGA9 and AtTGA10. Exon/intron and motif analysis revealed that the PvTGA members of various groups had considerably diverse gene architectures and lengths of sequences. The sensitivity of gene transcriptional regulation is often correlated with the number of introns. The capacity of the plant to respond to various developmental stages and environmental situations increases with decreasing intron number. The genes without introns or with shorter introns are also one of the active selection propensities of plants in evolution, however, in plants, animals and yeast, introns not only affect transcription to increase the content of mRNA, but also improve the translation efficiency of mRNA (Mattick and Gagen, 2001; Shaul, 2017). PvTGA genes have different annotated intron counts. PvTGAs have a certain amount of introns as a result of long-term evolution. PvTGAs may have different biological activities as a result of their various intron architectures. After conserved domain and motif analysis, it was revealed that all the PvTGAs shared the typical motifs, and motif sequences of each subfamily were relatively similar, indicating that members in each TGA subfamily might have the same function (Kesarwani et al., 2007). All TGA proteins contain key motifs that consist of the bZIP and DOG1 domains, but contain unique motifs in different subgroups. Genes’ functions are determined by their physical and chemical characteristics. PvTGA proteins’ physicochemical characteristics demonstrated their abundance in acidic amino acids (Table 1). The bZIP domain of PvTGA proteins was generally conserved, as shown by protein structure predictions and sequence alignment. Additionally, research has demonstrated that the specificity of DNA and nuclear localization is determined by the alkaline region of the bZIP domain’s interaction with DNA via the fixed nuclear localization signal structure NX7-R/K-X9 (Dröge-Laser et al., 2018). In accordance with TGA transcription factors described in other plants (Idrovo Espín et al., 2012; Xu et al., 2018; Li et al., 2019; Liu et al., 2022), subcellular localization revealed that the PvTGA transcription factors in common bean were mostly found in the cell nucleus, demonstrating a critical function for the PvTGA transcription factors in the nucleus. It was suggested that while the functions of the genes in various branches varied, the functions of the gene family were conserved.
The evolutionary process of orthologous and paralogous genes has an important relationship to the function of gene families (Koonin, 2005; Gabaldón and Koonin, 2013). The 512 Mb genome of common bean contains eight TGA genes, and the identification results based on segmental duplication and tandem repeats show that only PvTGA03 and PvTGA05 are caused by segmental duplication, and no tandem repeats occur among PvTGAs, which may be the TGA gene in the common genome is relatively conservative. The values of all TGA genes in Ka/Ks analysis were less than 1, which also indicated that the PvTGA genes was dominated by purifying selection. Different plant species have different numbers of TGA genes (Idrovo Espín et al., 2012; Xu et al., 2018; Li et al., 2019; Liu et al., 2022). The genome sizes of soybean, chickpea, alfalfa, peanut, Arabidopsis, grape, rice, sorghum, and maize are 967, 525, 408 Mb, 2.5 Gb, 118, 482, 373, 704 Mb, 2.1 Gb, respectively (https://phytozome-next.jgi.doe.gov/). The number of TGA genes identified in different plants is 10 in Arabidopsis, 20 in peanut, 9 in alfalfa, 27 in soybean, 4 in sorghum, 15 in rice, 5 in maize, eight in chickpea, and 7 in grape (Li et al., 2019; Liu et al., 2022). Thus, the genome sizes in these plants are not correlated with the number of TGA genes. Synteny analysis showed that PvTGAs had a closer genetic relationship with legumes, but a farther relationship with monocots, among which the TGA gene in soybean was the closest to the ones in common bean, and had the most orthologous gene pairs. It shows that the TGA genes in common bean and soybean may have similar functions. These results are consistent with the phylogenetic relationship between common bean and the other species. PvTGA03 and PvTGA05 produced by segmental duplication have the most homologous gene pairs in other species, indicating that these two genes may play an important role in the evolution of TGA genes (Fulton et al., 2002).
In this study, CAAS260205, a resistant genotype that highly expresses SA to provide effective SAR against Fop, was used together with the susceptible genotype BRB130 to detect TGA gene responses after inoculation. The results showed that PvTGA03 and PvTGA07 play opposite roles to regulate SA-mediated resistance to Fusarium wilt. And these two genes were highly expressed in the root of common bean, which can provide better sensitivity for the response to Fusarium wilt infection. Interestingly, the soybean TGA genes closest to these two genes in phylogenetic relationship were not found to be stress-responsive to biotic and abiotic stress (Li et al., 2019; Ullah et al., 2019; Jiang et al., 2021). AtTGA8 can control flowering-related morphological traits (Chuang et al., 1999), while PvTGA03 is associated with SA-mediated disease resistance. AtTGA3 interacts with ARR2 and binds to the PR1 promoter under the action of cytokinin (CTK) to improve plant disease resistance, but PvTGA07 plays a negative regulatory role in the process of resistance to Fusarium wilt (Choi et al., 2004; Fang et al., 2017). The role of the same TGA subgroup among different species might be highly diverse (Zhang et al., 2003; Fitzgerald et al., 2005; Thurow et al., 2005). While silencing TGA2.1 in tobacco led to petal-like stamens of TGA2.1, which is important for increasing the resistance to pathogens in silenced rice, knocking out TGA2 in Arabidopsis thaliana resulted in no phenotype of TGA2. Therefore, the anti-Fop response mechanism of TGA mediated by SA may be different from that of other plant TGA genes, and further in-depth functional studies on PvTGA genes are needed to analyze the molecular mechanism.
In summary, the findings of this study lay the groundwork for future investigation into the function of the TGA gene family in the regulation of common bean growth, development, and disease resistance. They also serve as a guide for the potential use of the PvTGA gene family in common bean resistance breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YL, YH, CZ, and RX contributed to conception and design of the study. MF organized the database. WG performed the statistical analysis. YL and YH wrote the first draft of the manuscript. YL, YH, ZL, and CZ wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
The work was supported by Project funded by China Postdoctoral Science Foundation (2021M692237), Free Exploration Project from Special Funding of Central Government’s Guide to Locality for science and technology development. (2022JH6/100100016), National Natural Science Foundation of China (31972962), China Agriculture Research System of MOF and MARA-Food Legumes (CARS-08-Z07), Disciplinary construction project of Liaoning Academy of Agricultural Sciences (2022DD030905), Young and Middle-aged Scientific and Technological Innovative Talents in Shenyang (RC220453).
We thank Professor Jing Wu of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences for supplying the common bean cultivars tested and Professor Zhendong Zhu of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences for providing the aggressive F. oxysporum f. sp. phaseoli (Fop) isolate used in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1137634/full#supplementary-material
An, C., and Mou, Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53 (6), 412–428. doi:10.1111/j.1744-7909.2011.01043.x
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). Meme: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34 (2), W369–W373. doi:10.1093/nar/gkl198
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 10 (10), 771. doi:10.3390/genes10100771
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884. doi:10.1038/s41588-019-0405-z
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8 (4), 521–539. doi:10.1016/j.molp.2014.12.022
Blair, M., Izquierdo, P., Astudillo, C., and Grusak, M. (2013). A legume biofortification quandary: Variability and genetic control of seed coat micronutrient accumulation in common beans. Front. Plant Sci. 4, 275. doi:10.3389/fpls.2013.00275
Blair, M. W., Wu, J., and Wang, S. (2016). Editorial: Food legume diversity and legume research policies. Crop J. 4 (5), 339–343. doi:10.1016/j.cj.2016.09.001
Buruchara, R. A., and Camacho, L. (2000). Common bean reaction to Fusarium oxysporum f. sp. phaseoli, the cause of severe vascular wilt in central Africa. J. Phytopathology 148 (1), 39–45. doi:10.1111/j.1439-0434.2000.tb04622.x
Chen, X., Lu, Q., Liu, H., Zhang, J., Hong, Y., Lan, H., et al. (2019). Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 12 (7), 920–934. doi:10.1016/j.molp.2019.03.005
Cheng, C.-Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., and Town, C. D. (2017). Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804. doi:10.1111/tpj.13415
Choi, J. J., Klosterman, S. J., and Hadwiger, L. A. (2004). A promoter from pea gene DRR206 is suitable to regulate an elicitor-coding gene and develop disease resistance. Phytopathology® 94 (6), 651–660. doi:10.1094/phyto.2004.94.6.651
Chuang, C. F., Running, M. P., Williams, R. W., and Meyerowitz, E. M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes. Dev. 13 (3), 334–344. doi:10.1101/gad.13.3.334
Dempsey, D. M. A., and Klessig, D. F. (2012). SOS – too many signals for systemic acquired resistance? Trends Plant Sci. 17 (9), 538–545. doi:10.1016/j.tplants.2012.05.011
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 45, 36–49. doi:10.1016/j.pbi.2018.05.001
Fang, H., Liu, Z., Long, Y., Liang, Y., Jin, Z., Zhang, L., et al. (2017). The Ca2+/calmodulin2-binding transcription factor TGA3 elevates LCD expression and H2S production to bolster Cr6+ tolerance in Arabidopsis. Plant J. 91 (6), 1038–1050. doi:10.1111/tpj.13627
Fitzgerald, H. A., Canlas, P. E., Chern, M.-S., and Ronald, P. C. (2005). Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae. Plant J. 43 (3), 335–347. doi:10.1111/j.1365-313X.2005.02457.x
Fu, Z., and Dong, X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. plant Biol. 64, 839–863. doi:10.1146/annurev-arplant-042811-105606
Fulton, T. M., Van der Hoeven, R., Eannetta, N. T., and Tanksley, S. D. (2002). Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell. 14 (7), 1457–1467. doi:10.1105/tpc.010479
Gabaldón, T., and Koonin, E. V. (2013). Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 14 (5), 360–366. doi:10.1038/nrg3456
Gatz, C. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant-Microbe Interactions® 26 (2), 151–159. doi:10.1094/mpmi-04-12-0078-ia
Idrovo Espín, F. M., Peraza-Echeverria, S., Fuentes, G., and Santamaría, J. M. (2012). In silico cloning and characterization of the TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiology Biochem. 54, 113–122. doi:10.1016/j.plaphy.2012.02.011
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 (3), 106–111. doi:10.1016/S1360-1385(01)02223-3
Jiang, H., Gu, S., Li, K., and Gai, J. (2021). Two TGA transcription factor members from hyper-susceptible soybean exhibiting significant basal resistance to soybean mosaic virus. Int. J. Mol. Sci. [Online] 22 (21), 11329. doi:10.3390/ijms222111329
Johnson, C., Boden, E., Desai, M., Pascuzzi, P., and Arias, J. (2001). In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 28 (2), 237–243. doi:10.1046/j.1365-313X.2001.01147.x
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi:10.1038/nature05286
Katagiri, F., Lam, E., and Chua, N.-H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340 (6236), 727–730. doi:10.1038/340727a0
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144 (1), 336–346. doi:10.1104/pp.106.095299
Klessig, D. F., Choi, H. W., and Dempsey, D. M. A. (2018). Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant-Microbe Interactions® 31 (9), 871–888. doi:10.1094/mpmi-03-18-0067-cr
Koonin, E. V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39 (1), 309–338. doi:10.1146/annurev.genet.39.073003.114725
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Li, B., Liu, Y., Cui, X.-Y., Fu, J.-D., Zhou, Y.-B., Zheng, W.-J., et al. (2019). Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci. 10, 549. doi:10.3389/fpls.2019.00549
Liu, W., Zhao, C., Liu, L., Huang, D., Ma, C., Li, R., et al. (2022). Genome-wide identification of the TGA gene family in kiwifruit (Actinidia chinensis spp.) and revealing its roles in response to Pseudomonas syringae pv. actinidiae (Psa) infection. Int. J. Biol. Macromol. 222, 101–113. doi:10.1016/j.ijbiomac.2022.09.154
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Malamy, J., Carr, J. P., Klessig, D. F., and Raskin, I. (1990). Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250(4983), 1002–1004. doi:10.1126/science.250.4983.1002
Mattick, J. S., and Gagen, M. J. (2001). The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 18 (9), 1611–1630. doi:10.1093/oxfordjournals.molbev.a003951
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. doi:10.1111/tpj.13781
Pérez-Vega, E., Pañeda, A., Rodríguez-Suárez, C., Campa, A., Giraldez, R., and Ferreira, J. J. (2010). Mapping of QTLs for morpho-agronomic and seed quality traits in a RIL population of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 120 (7), 1367–1380. doi:10.1007/s00122-010-1261-5
Pontier, D., Miao, Z.-H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27 (6), 529–538. doi:10.1046/j.1365-313X.2001.01086.x
Rochon, A., Boyle, P., Wignes, T., Fobert, P. R., and Després, C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell. 18 (12), 3670–3685. doi:10.1105/tpc.106.046953
Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46 (7), 707–713. doi:10.1038/ng.3008
Schwartz, H. F., Steadman, J. R., and Hall, R. (2005). Compendium of bean diseases. 2nd ed. St. Paul, MN: The American phytopathological Society.
Shaul, O. (2017). How introns enhance gene expression. Int. J. Biochem. Cell. Biol. 91, 145–155. doi:10.1016/j.biocel.2017.06.016
Shimizu, K., Suzuki, H., Uemura, T., Nozawa, A., Desaki, Y., Hoshino, R., et al. (2022). Immune gene activation by NPR and TGA transcriptional regulators in the model monocot Brachypodium distachyon. Plant J. 110 (2), 470–481. doi:10.1111/tpj.15681
Spoel, S. H., and Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12 (2), 89–100. doi:10.1038/nri3141
Strader, L., Weijers, D., and Wagner, D. (2022). Plant transcription factors — Being in the right place with the right company. Curr. Opin. Plant Biol. 65, 102136. doi:10.1016/j.pbi.2021.102136
Suárez-Martínez, S. E., Ferriz-Martínez, R. A., Campos-Vega, R., Elton-Puente, J. E., de la Torre Carbot, K., and García-Gasca, T. (2016). Bean seeds: Leading nutraceutical source for human health. CyTA - J. Food 14 (1), 131–137. doi:10.1080/19476337.2015.1063548
Suter, D. M. (2020). Transcription factors and DNA play hide and seek. Trends Cell. Biol. 30 (6), 491–500. doi:10.1016/j.tcb.2020.03.003
Tang, H., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A., Zhou, S., et al. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15, 312. doi:10.1186/1471-2164-15-312
The French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi:10.1038/nature06148
Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolov, K., and Gatz, C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44 (1), 100–113. doi:10.1111/j.1365-313X.2005.02513.x
Ullah, I., Magdy, M., Wang, L., Liu, M., and Li, X. (2019). Genome-wide identification and evolutionary analysis of TGA transcription factors in soybean. Sci. Rep. 9 (1), 11186. doi:10.1038/s41598-019-47316-z
Varshney, R., Song, C., Saxena, R., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246. doi:10.1038/nbt.2491
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics, Proteomics Bioinforma. 8 (1), 77–80. doi:10.1016/S1672-0229(10)60008-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Xu, Z., Zhang, H., Mo, Q., Lü, S., Wang, C., and Ji, W. (2018). Expression analysis of wheat transcription factor TaTGA1 gene responding to infection of powdery mildew. Acta Phytopathol. Sin. 48 (6), 766. doi:10.13926/j.cnki.apps.000189
Xue, R., Feng, M., Chen, J., Ge, W., and Blair, M. W. (2021). A methyl esterase 1 (PvMES1) promotes the salicylic acid pathway and enhances Fusarium wilt resistance in common beans. Theor. Appl. Genet. 134 (8), 2379–2398. doi:10.1007/s00122-021-03830-1
Xue, R. F., Wu, J., Wang, L. F., Blair, M. W., Wang, X. M., De Ge, W., et al. (2014). Salicylic acid enhances resistance to Fusarium oxysporum f. sp. phaseoli in common beans (Phaseolus vulgaris L.). J. Plant Growth Regul. 33 (2), 470–476. doi:10.1007/s00344-013-9376-y
Xue, R., Wu, X., Wang, Y., Zhuang, Y., Chen, J., Wu, J., et al. (2017). Hairy root transgene expression analysis of a secretory peroxidase (PvPOX1) from common bean infected by Fusarium wilt. Plant Sci. 260, 1–7. doi:10.1016/j.plantsci.2017.03.011
Xue, R., Zhu, Z., Huang, Y., Wang, X., Wang, L., and Wang, S. (2012). Quantification of <I>Fusarium oxysporum</I> f. sp. <I>phaseoli</I> Detected by Real-time Quantitative PCR in Different Common Beans Cultivars. Acta Agron. Sin. 38 (5), 791–799. doi:10.3724/sp.j.1006.2012.00791
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell. 15 (11), 2647–2653. doi:10.1105/tpc.014894
Keywords: Common bean, TGA gene family, Gene family, Fusarium oxysporum f. sp. Phaseoli, Fusarium wilt
Citation: Liu Y, Huang Y, Li Z, Feng M, Ge W, Zhong C and Xue R (2023) Genome-wide identification of the TGA genes in common bean (Phaseolus vulgaris) and revealing their functions in response to Fusarium oxysporum f. sp. phaseoli infection. Front. Genet. 14:1137634. doi: 10.3389/fgene.2023.1137634
Received: 04 January 2023; Accepted: 12 January 2023;
Published: 20 January 2023.
Edited by:
Feng Zhang, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Wei Qian, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2023 Liu, Huang, Li, Feng, Ge, Zhong and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Zhong, emhvbmdjaGFvMTEyM0BzeWF1LmVkdS5jbg==; Renfeng Xue, eHVlcmY4MkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.