- 1Department of Field Crops, Ege University, Izmir, Turkey
- 2Agriculture and Agri-Food Canada, Lethbridge, AB, Canada
- 3Turkey-ICARDA Regional Cereal Rust Research Center (RCRRC), Izmir, Turkey
- 4Agean Agricultural Research Institute, Regional Cereal Rust Research Center (RCRRC), Izmir, Turkey
- 5International Maize and Wheat Improvement Center (IWWIP-Turkey), Ankara, Turkey
Stripe rust caused by Puccinia striiformis Westend. f. sp. tritici. is a major bread wheat disease worldwide with yield losses of up to 100% under severe disease pressure. The deployment of resistant cultivars with adult plant resistance to the disease provides a long-term solution to stripe rust of wheat. An advanced line from the International Winter Wheat Improvement Program (IWWIP) 130675 (Avd/Vee#1//1-27-6275/Cf 1770/3/MV171-C-17466) showed a high level of adult plant resistance to stripe rust in the field. To identify the adult plant resistance genes in this elite line, a mapping population of 190 doubled haploid (DH) lines was developed from a cross between line 130675 and the universal stripe rust-susceptible variety Avocet S. The DH population was evaluated at precision wheat stripe rust phenotyping platform, in Izmir during 2019, 2020, and 2021 cropping seasons under artificial inoculations. Composite interval mapping (CIM) identified two stable QTLs QYr.rcrrc-3B.1, and QYr.rcrrc-3B.2, which were detected in multiple years. In addition to these two QTLs, five more QTLs, QYr.rcrrc-1B, QYr.rcrrc-2A, QYr.rcrrc-3A, QYr.rcrrc-5A, and QYr.rcrrc-7D, were identified, which were specific to the cropping year (environment). All QTLs were derived from the resistant parent, except QYr.rcrrc-3A. The significant QTLs explained 3.4–20.6% of the phenotypic variance. SNP markers flanking the QTL regions can be amenable to marker-assisted selection. The best DH lines with high yield, end-use quality, and stripe rust resistance can be used for further selection for improved germplasm. SNP markers flanking the QTL regions can aid in identifying such lines.
Introduction
Wheat stripe (yellow) rust, caused by Puccinia striiformis Westend. f. sp. tritici (Pst), is one of the most important and devastating diseases of common wheat (Triticum aestivum L.) around the world (Hovmøller et al., 2011). It remains a significant threat to wheat yield loss, and under severe disease pressure, yield losses of up to 100% are observed (Ali et al., 2014). Stripe rust was historically considered a disease of wheat-growing areas with cool temperatures; however, with the emergence of adapted races to high temperatures and more aggressive races, the disease is now spreading to areas where it was previously considered unfavorable (de Vallavieille-Pope et al., 2012; Muleta et al., 2017; Godoy et al., 2018). Today, the new pathotypes of stripe rust are prevalent from Europe to Australia, Asia, and America and as a result threatens the wheat production on a global scale (Ali et al., 2014).
The continuous occurrence of new stripe rust races requires the identification of new sources of resistance and the deployment of resistant varieties in a timely manner. The conventional approaches for controlling stripe rust include cultural practices like early sowing and crop rotation to avoid infection during the disease infestation period (Boyd 2005). Additionally, fungicide application is also an effective way of controlling stripe rust; however, it is not the most economical and recommended method (Brar et al., 2018). The most effective strategy to control stripe rust outbreaks is the exploitation of genetic resistance and pyramiding of multiple minor and major stripe rust resistance genes conferring seedling and adult plant resistance (APR) (Chen et al., 2014; Tadesse et al., 2014; Muleta et al., 2017; Cobo et al., 2018). Most breeding programs in the world rely on two types of genetic resistance based on major and minor genes (Chen et al., 2014). Genetic resistance due to major genes is termed as a seedling and/or all-stage resistance and is often race-specific and based on the gene for gene hypothesis and is effective throughout a plant’s life (Burdon et al., 2014; Tehseen et al., 2021). However, such resistance in commercial wheat cultivars is often short-lived and is overcome by new races of stripe rust pathogens virulent on the major resistance gene (Boyd 2005; Ellis et al., 2014; Hulbert and Pumphrey 2014), whereas the minor gene resistance is often not expressed until in the later stages of plant life and is commonly referred to as horizontal or adult plant stage resistance (Steele et al., 2001; Boyd 2005). Therefore, many wheat breeding programs consider pyramiding of both seedling and APR genes for enhancing the durability of resistance to multiple prevalent races of stripe rust, hence minimizing yield losses. Due to new emerging races of the stripe rust pathogen virulent to numerous seedling or race-specific genes, the best strategy would be to stack multiple non-race-specific or APR genes in combinations for durable stripe rust resistance (Rajaram 2015). Therefore, although the characterization of seedling resistance genes from highly resistant lines is crucial, the elite breeding lines with multiple adult plant QTLs having high to moderate resistance levels should be considered more important. Elite breeding lines having higher agronomical, biotic, and abiotic stress resistance and end-use quality traits tend to be the ideal candidates for gene mapping as they can be readily used in the ongoing breeding programs.
The bread wheat has a very large genome size; additionally, the allopolyploidy further hampers the progress of mapping new quantitative trait loci (QTLs) and as a result slows down the breeding process (Liu et al., 2021). The whole genome of the common wheat cultivar Chinese Spring was completed 14 years later than some of the other gramineous crops such as rice; thus, it made genetic association comparisons at the whole genome level more complex than other crops (Yu et al., 2002; Wang et al., 2018). Recently, with advances in wheat genome sequencing, multiple versions of the annotated wheat genome have been published consequently accelerating forward genetic research (Clavijo et al., 2017; Zimin et al., 2017; Appels et al., 2018). Today, due to high-throughput sequencing platforms, the development of a large number of high-quality markers is possible, thus facilitating more efficient mapping techniques to analyze a large number of traits across different treatments and environments and opening new opportunities in wheat breeding for biotic and abiotic studies (Rimbert et al., 2018). The International Centre for Agricultural Research in Dry Areas (ICARDA) and the International Maize and Wheat Improvement Centre (CIMMYT) have both played pivotal roles in the development of high-yielding, abiotic stress-tolerant, disease-resistant, higher end-use quality, and widely adaptive global wheat germplasm (Wu et al., 2021).
An improved wheat line 130675 from the International Winter Wheat Breeding Program (IWWIP) (Avd/Vee#1//1-27-6275/Cf 1770/3/MV171-C-17466) selected from the Facultative and Winter Wheat Observation Nursery (FAWWON 2013-2014) possesses several desirable traits, including yield and early maturity, and showed APR to stripe rust in multiple field trials in Turkey. However, it was susceptible to PstS2 and Warrior races at the seedling stage, indicating typical APR for both races. The resistance to stripe rust of the wheat line 130675 has not been characterized. Therefore, the current study aimed to map and characterize adult plant stripe rust resistance loci in the doubled haploid (DH) population derived from a cross between wheat line 130675 and universal stripe rust-susceptible variety Avocet S.
Material and Methods
Plant Material and Pathogen
The panel of 190 DH lines from the cross of an improved IWWIP line 130675 (Avd/Vee#1//1-27-6275/Cf 1770/3/MV171-C-17466) and Avocet S (AvS) were evaluated for adult plant stripe rust resistance. The DH lines derived from the F1 generation (F1DH) were developed using the wheat maize hybridization protocol (Sadasivaiah et al., 1999). The parents were selected due their diverse genetic backgrounds and different levels of stripe rust resistance. The stripe rust isolates PstS2 and Warrior (PstS7) were used in artificial field inoculations, and both belonged to PstS2v27 and PstS7vWarrior lineages, and the virulence/avirulence formula of the two races are given in Table 1.
Field Adult Plant Resistance Assessment
The field experiments were carried out at the precision wheat stripe rust phenotyping platform, Regional Cereal Rust Research Center (RCRRC), Izmir, Turkey, during the cropping seasons 2019, 2020, and 2021. The experiment was laid out as an augmented design with un-replicated test entries and repeated check rows in 12 blocks. Each block contained 16 test entries and seven checks. Thirty seeds from each accession were planted in a 1-m row with 30-cm spacing between the rows. To ensure sufficient inoculum production for disease infection, a mixture of the universally susceptible varieties “Morocco,” “Seri 82,” and “Avocet S” along with the locally susceptible varieties “Bolani,” “Basribey” (also derived from the CIMMYT cross “Kauz”), and “Cumhuriyet 75,” “Kunduru,” “Kasifbey,” and “Gonen” were planted as spreader after every 20 rows, as well as spreader rows bordering the nurseries. The experiments were managed as per the standard local agronomic practices during the crop season.
PstS2 and Warrior (PstS7) pathotypes of stripe rust preserved at RCRRC were multiplied using susceptible variety AvS, and the freshly collected urediniospores were used for field inoculations. The DH panel along with the spreader rows bordering the experiment was artificially sprayed with a mixture of the two races in talcum powder using a backpack sprayer at the seedling, tillering, and booting stages. The field was irrigated through a mist irrigation system.
Field scoring started when disease severity reached 100% on the susceptible checks, “Morocco” and AvS. Adult plant responses were recorded three times at 10-day intervals for the major infection types resistant (R), moderate resistant (MR), moderate (M), moderate susceptible (MS), and susceptible (S) (Roelfs et al., 1992), and the disease severities (0-100%) following the modified Cobb’s scale (Peterson et al., 1948). All the three recordings were averaged, and the coefficients of infection (CI) were calculated. The CIs were calculated by multiplying the constant values of the infection types and disease severity. The constant values of infection types were used as R = 0.2, MR = 0.4, M = 0.6, MS = 0.8, and S = 1 (Saari and Wilcoxson 1974; Stubbs et al., 1986).
DNA Extraction and Genotyping
Genomic DNA was extracted from fresh leaves collected from three individual 10-day-old seedlings using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987). The seedling leaves were collected in labeled Eppendorf tubes and stored in an Ultra freezer at −80°C for subsequent DNA extraction. The leaf samples were grounded using a mortar in liquid nitrogen until a fine powder was obtained, and 0.1 g of the powdered leaf samples were used for DNA extraction using the CTAB method (Doyle and Doyle 1987). The extracted DNA was dissolved in 100 µl Tris–EDTA (TE) buffer. The samples were analyzed on 1% agarose gel for the purity test and quantified with a biophotometer (BioPhotometer, Eppendorf). The DNA samples were then kept at −80°C. The extracted DNA samples of the DH panel and two parental lines were sent to Diversity Arrays Technology Pty Ltd. (Canberra, Australia, http://www.DiversityArrays.com/) for genotyping. The genotypic data obtained for 172 DH lines including parents were filtered, and markers with > 10% missing data and < 0.1% minor allele frequency were eliminated and not used in the subsequent analysis.
Statistical Analysis
Descriptive statistics and analysis of variance (ANOVA) were performed using the R package “AugmentedRCBD”. Broad-sense heritability was estimated as the ratio of genetic variance (σ2g) to phenotypic variance (σ2g + σ2ε), where σ2ε represents error variance and is represented as follows:
Linkage Map Construction and QTL Mapping
The marker genetic data were used to construct the linkage map using the software QTL Ici-Mapping software V4.2. The Kosambi function was used to calculate the genetic distances between the markers (Kosambi 1944). The stripe rust resistance QTLs were estimated in the DH population based on the CI of the 3 years. The composite interval mapping (CIM) method was used for the detection of QTL using QTL Ici-Mapping software V4.2. The threshold value for the logarithm of odds (LOD) score was calculated after running a permutation test of 1,000 runs and was 2.1, 2.0, and 2.4 for 2019, 2020, and 2021 experiments, respectively, with a walking step of 1 cM (Van Ooijen 1999). The QTLs were also reported significant at a threshold of 2.0 if found in multiple years. The effects of QTLs were calculated as the proportion of phenotypic variance explained by the QTL. The genomic locations of the significant QTL were indicated using the software Map Chart V2.3.
Gene Annotation
The candidate genes with their putative proteins/enzymes were predicted within the interval of 500 kb upstream and downstream from the closest significant markers using Ensembl, a plant database available at http://plants.ensembl.org/Triticum_aestivum/Info/Index, and the International Wheat Genome Sequencing Consortium (IWGSC) RefSeq v1.1 annotations (Appels et al., 2018) available at https://wheat-urgi.versailles.inra.fr/Seq-Repository/Annotations. The nearby genes in the linkage regions of significant markers with putative functions that could be related to the trait were selected as candidates.
Results
Field Assessment of Resistance
In adult plant assessment, the estimates of genetic variance identified significant differences among the DH lines (Table 2). A small variation was observed in the disease severity scores of the tested accessions during the 3 years as in 2021, and the data were more skewed toward the resistance side (Figure 1). Overall, during the 3 years 34.9, 36.9, and 47.92% of the DH lines showed resistance response. The mean values of CI for 2019 and 2020 were 34.5 and 37, respectively, whereas in 2021, the mean value dropped to 21.6. The mean values for the parents ranged from 0.2 to 20 for the resistant parent, while 79 to 90 for the susceptible parent. The broad-sense heritability was 87.04, 91.14, and 77.18 for 2019, 2020, and 2021, respectively. Significant positive correlations were found between the 3 years of field data. The highest correlation (0.69) was found between the CIs from 2019 and 2020, whereas the lowest CI correlation (0.38) was observed between 2019 and 2021 (Figure 1).
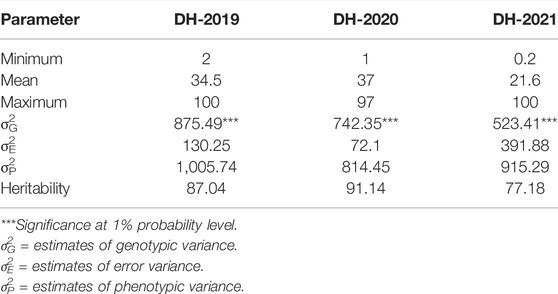
TABLE 2. Basic statistics of adult plant response of bread wheat DH lines against PstS2 and Warrior pathotypes of stripe rust, estimates of variance components, and broad-sense heritability.
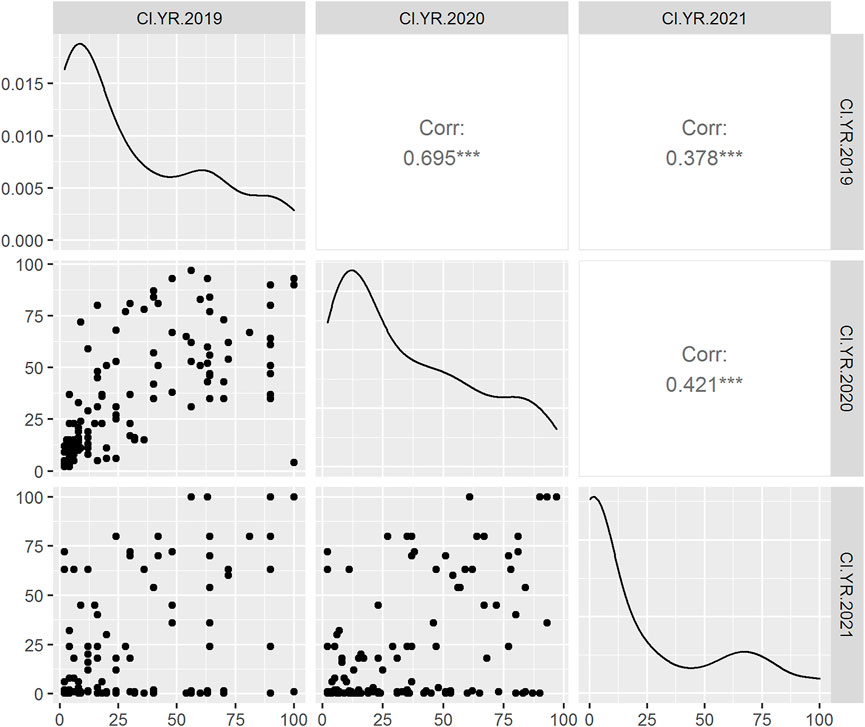
FIGURE 1. Scatter plot (lower triangle) with the distribution of phenotypic data during 2019, 2020, and 2021 field years from left to right anticlockwise, respectively; density plot (diagonal line) and Pearson correlation analysis (upper triangle) between the 3 years of DH population in field condition. The X-axis and Y-axis represent the stripe rust coefficient of infection (CI).
Linkage Map and Identification of QTLs for Adult Plant Resistance to Stripe Rust in the DH Population
After filtering for quality parameters such as missing data and segregation distortion, a set of 590 skeleton SNP markers were used to construct a linkage map for the 130675 × AvS DH population. The markers covered the whole genome and were divided into 28 linkage groups, marker order in the linkage group was generally in agreement with the published consensus map (Li et al., 2015). Genomes A, B, and D had 244 (41.33%), 237 (40.17%), and 109 (18.47%) markers, respectively, and the total map length was 2,232 cm. Composite interval mapping identified 10 QTLs in 3 years on seven genomic regions across the genome for resistance to yellow rust (Yr) at the adult plant growth stage; the QTLs’ were named QYr.rcrrc.1B, QYr.rcrrc.2A, QYr.rcrrc.3A, QYr.rcrrc.3B.1, QYr.rcrrc.3B.2, QYr.rcrrc.5A, and QYr.rcrrc.7D. Out of these 10 QTLs, three were detected in the 2019 and 2021 field years, while four were detected in 2020. The QTLs were detected on seven genomic regions in chromosomes 1B, 2A, 3A, 3B, 5A, and 7D (Figure 2).

FIGURE 2. Segments of genetic linkage maps of QTL conferring adult plant stripe rust resistance. Single-nucleotide polymorphism (SNP) markers are shown on the left and their genetic positions (cM) are on the right of chromosomes. The region containing the QTL is indicated by a vertical bar on the right and followed by the name of the QTL. The markers in red are associated with the QTL.
The phenotypic variance explained by an individual QTL ranged from 3.4 to 20.6%. Two stable QTLs on chromosome 3B, that is, QYr.rcrrc.3B.1 and QYr.rcrrc.3B.2 were detected in multiple years and contributed 5.2–19.8% toward phenotypic variation. The QTL that explained a phenotypic variance of more than 10% was considered as a major QTL. All QTLs were contributed by the resistant parent 130675 except one on chromosome 3A, which was contributed by the susceptible parent AvS (Table 3).
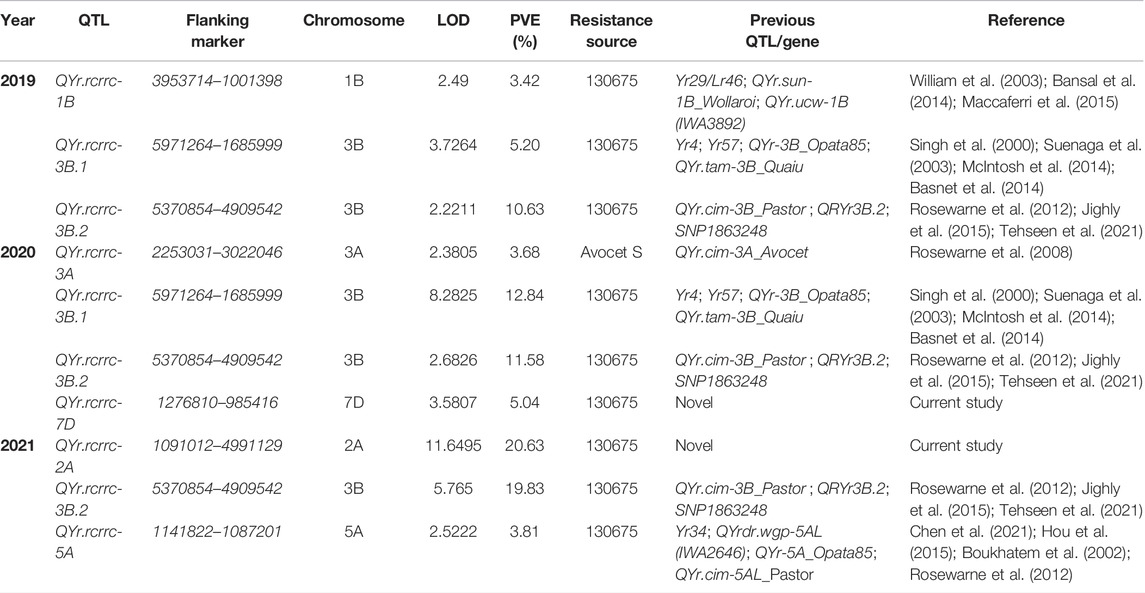
TABLE 3. Quantitative trait loci (QTLs) associated with adult plant stripe rust resistance in DH population in different environments.
QTL Region Physical Positions and Candidate Gene Prediction
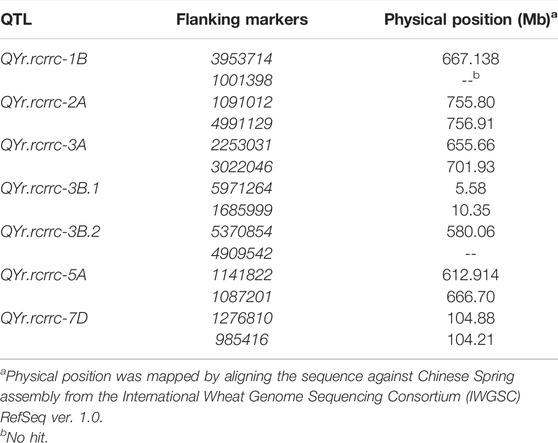
The alignment of significant QTL markers with reference genome confirmed their physical positions according to chromosome assignments (Table 4). The largest physical distance of 53.7 Mb spanned between the flanking markers of QYr.rcrrc-5A. The QYr.rcrrc-5A also spanned a large interval on the genetic map compared with other QTLs (Figure 2). The physical distances between QYr.rcrrc-2A, QYr.rcrrc-3B.1, and QYr.rcrrc-7D were 1.1, 4.7, and 0.6 Mb, respectively. The expressed genes between the flanking markers of the QTL were identified using the BLASTn searches from the flanking markers sequences (Table 5). The high confidence genes which were previously reported to be associated with disease resistance were selected as candidate genes.
Discussion
Stripe rust is a devastating disease of wheat and could result in 100% yield loss under high disease pressure (Manickavelu et al., 2016). Historically, it was considered a disease in wet and cool climates but with the emergence of new races adapted to high temperatures the disease has sporadically spread to areas that were once considered unsuitable for its growth and disease development (Hovmøller et al., 2016; Ali et al., 2017). The most effective strategy to manage the continuous appearance of new stripe rust races is genetic resistance and the development of lines harboring both minor and major genes (Chen et al., 2014). The plant breeders tend to stack multiple different traits in elite backgrounds; therefore, breeding for one trait is not always simple and even in the presence of highly resistant germplasm breeders do not necessarily always utilize it due to the undesirable linkage drag associated with resistance locus. Hence, to circumvent the potential linkage drag, the breeders focus on identifying and mapping resistance genes from elite breeding lines with accumulated favorable morphological and agronomic traits. The current study used an advanced breeding line with a distinct resistance response to stripe rust from the IWWIP breeding program.
Overall, 10 QTLs in seven genomic regions across the three environments (years) were detected in the current study. The phenotypic variance explained by the QTL ranged from 3.4 to 20.6% confirming their significant effects in reducing stripe rust severity. The significant QTLs detected in the study were compared with the previously published stripe rust known genes and QTL based on their chromosome location, physical position, pedigree, linked markers, and rust resistance.
The QYr.rcrrc-1B detected in the current study on chromosome 1B overlapped the several previously reported Yr QTL and an APR gene Yr29 (William et al., 2003; Bansal et al., 2014; Maccaferri et al., 2015). The pleiotropic locus Yr29/Lr46/Sr58 on chromosome 1B has been widely used in breeding programs around the world, including CIMMYT wheat germplasm (Gebrewahid et al., 2020). The Yr29/Lr46/Sr58 locus is associated with a wide-spectrum resistance level explaining 2.9–74.5% of the phenotypic variation in different bi-parental mapping populations and under different environmental trials (Zhang et al., 2019). The stripe rust Yr29 is a slow rusting adult plant resistance gene and its effect decreases with the increase in the inoculum load. This could be the plausible reason why QYr.rcrrc-1B was detected only in the 2019 crop year. QYr.rcrrc-1B is most likely the Yr29/Lr46/Sr58 complex.
The QYr.rcrrc-2A detected in the current study does not correspond to any of the previously identified Yr QTL and/or genes. A seedling resistant gene Yr1 is also located on the long arm of chromosome 2A; however, according to the physical position of Yr1, the gene and the QYr.rcrrc-2A are 15.4 Mb apart at the distal end. QYr.wpg-2A.6 (IWA966) a minor effect APR QTL was also reported on chromosome 2A and is the closest QTL to QYr.rcrrc-2A and the two are 6.79 Mb apart (Naruoka et al., 2015). Since QYr.rcrrc-2A is a major QTL and does not overlap with any of the previously reported Yr QTL/gene; furthermore, the closest gene to QYr.rcrrc-2A is Yr1 which is a seedling resistant gene and is ineffective against the Warrior (PstS7) race used in the study. Therefore, based on the physical locations of the close-by QTL and resistance pattern of the nearby genes, the QTL QYr.rcrrc-2A is considered novel.
QYr.rcrrc-3A was found significant to the stripe rust resistance in the field. The locus overlapped a previously reported Yr QTL QYr.cim-3A_Avocet (Rosewarne et al., 2008). The QYr.cim-3A_Avocet was found in a RIL population derived from a cross between AvS and Pastor, the two QTLs expressed similar total phenotypic variation and the source of resistance in both the populations was cultivar AvS. Therefore, based on the physical overlapping positions, a similar effect of QTL, and sources of resistance in both studies, it was concluded that both QYr.rcrrc-3A and QYr.cim-3A_Avocet represent the same genomic region.
Two stable QTLs such as QYr.rcrrc-3B.1 and QYr.rcrrc-3B.2 were detected on the short and long arms of chromosome 3B, respectively. QYr.rcrrc-3B.1 lies in the same genomic region as the several Yr QTLs and genes reported earlier on the short arm of chromosome 3B (Singh et al., 2000; Suenaga et al., 2003; Basnet et al., 2014; McIntosh et al., 2014). The seedling resistance gene Yr4 is avirulent on one of the pathotypes that is PstS7 used in the study for artificial inoculation. Based on the physical position and virulence/avirulence pattern it is likely that QYr.rcrrc-3B.1 represent is linked to Yr4; however, further studies are required to confirm the relationship as some studies have reported a different APR locus in the absence of Yr4 (Buerstmayr et al., 2014). QYr.rcrrc-3B.2, the second stable QTL detected in all three field experiments was found on the long arm of chromosome 3B and overlapped the previously reported QYr.cim-3B_Pastor, QRYr3B.2, and SNP1863248 (Rosewarne et al., 2012; Jighly et al., 2015; Tehseen et al., 2021). The three previously reported QTLs conferred APR; therefore, it is likely that QYr.rcrrc-3B.2 is linked to these QTLs.
A minor effect of QTL QYr.rcrrc-5A was detected on the long arm of chromosome 5A and overlapped the same genomic region previously reported to be linked with several APR and high-temperature adult plant (HTAP) Yr QTL (Boukhatem et al., 2002; Rosewarne et al., 2012; Hou et al., 2015). An APR gene with a moderate level of resistance is also located in the same genomic location (Chen et al., 2021). Since QYr.rcrrc-5A is a minor effect on QTL and Yr34 also shows moderate resistance it is likely that QYr.rcrrc-5A is linked with Yr34; however, further genetic analysis is required to confirm the relationship as no source of resistance with Yr34 was used in the differential set for race typing of the pathotypes used in the current study.
Two Yr resistant genes and a seedling resistance marker are previously reported on chromosome 7D (Maccaferri et al., 2015; Bulli et al., 2016; Tehseen et al., 2021). However, the locus QYr.rcrrc-7D detected in the current study is outside the genomic regions of the two genes and the QTL. The approximate distance between QYr.rcrrc-7D and the gene Yr33 and the seedling resistant locus QYr.7D_seedling is 33Mb and 17 Mb, respectively. Thus, based on the physical distances QYr.rcrrc-7D is a novel QTL region.
Regarding the predicted proteins in the current study, the candidate genes include NB-ARC domain proteins, which are involved in pathogen recognition and subsequent activation of plants’ defense mechanisms (Van Ooijen 1999; Van Ooijen et al., 2008; Steele et al., 2019); protein kinase domain proteins, which modify other proteins and are vital in several signaling and regulatory pathways in addition to apoptosis and cell division (Brueggeman et al., 2008); and leucine-rich repeats (LRR), which play a vital role in plants’ defense mechanism and are typically annotated to resistance genes (Jones and Jones 1997; Yuan et al., 2018), F-box domain proteins; they are involved in plant vegetative and reproductive growth and development. These proteins are reported to regulate cell death and defense when the pathogen is recognized in the tobacco and tomato plant (van den Burg et al., 2008), and NAC domain proteins which are involved in several processes, including the formation of secondary walls, senescence, and abiotic and biotic stresses (Puranik et al., 2012; Ng et al., 2018; Yuan et al., 2019). All candidate genes have been previously reported to play role in the plant’s defense mechanism; therefore, it is highly likely that they could be one of the candidate genes for stripe rust resistance. However, these putative candidate proteins should be used with caution as they are not the only proteins found within the confidence intervals of the linked markers but are the ones that have been reported to be involved in plant defense and disease and/or stress resistance mechanisms.
Marker-assisted breeding (MAB) is a valuable tool and is being utilized in many breeding programs around the world for different kinds of crops. MAB allows successful introgression of biotic and abiotic stress-resistant genes in high-yielding susceptible backgrounds (Ren et al., 2012). Therefore, detection of significant and tightly linked markers is desirable, which can be converted into breeder-friendly markers to be utilized in the breeding programs through MAB. In this study, we identified seven QTLs associated with APR to stripe rust across environments including QYr.rcrrc-1B, QYr.rcrrc-2A, QYr.rcrrc-3A, QYr.rcrrc-3B.1, QYr.rcrrc-3B.2, QYr.rcrrc-5A, and QYr.rcrrc-7D, and they were closely linked to SNP markers 3953714, 1091012, 2253031, 5971264, 5370854, 1141822, and 1276810, respectively. With new extensive research and cloning of APR genes, the overall function of the APR genes is better understood. However, the durability of any APR gene or the combination of APR genes is still a mystery and is based on prediction and time (Lowe et al., 2011). Nevertheless, the QTL reported in the current study particularly QYr.rcrrc-2A and QYr.rcrrc-7D were new QTL for APR to stripe rust. They should enhance the genetic basis of resistance to stripe rust, and their closely linked markers can be converted into breeder-friendly markers and utilized in MAB and stacking of multiple APR genes in common wheat backgrounds for durable resistance to stripe rust.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Conceptualization: KN; methodology: KN and MMT; software: MMT; validation: KN, FT, and MT; formal analysis: MMT; investigation: KN, MMT, EK, ON, and IO; resources: BA and HR; data curation: KN, MMT, and EK; writing—original draft preparation: MMT; writing—review and editing: KN and FT; project administration: KN, FT, and MT; and funding acquisition: KN and FT. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) with the project number 117O049 and the CGIAR Research Program on Wheat (WHEAT) administrated by the International Maize and Wheat Improvement Center (CIMMYT) and the Durable Rust Resistance in Wheat (DGGW) project administrated by the Cornell University and funded by the Bill & Melinda Gates Foundation and the United Kingdom Department for International Development, Grant no. OPP1133199.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Umara Sahar Rana (University of Agriculture Faisalabad) for her valuable advice and support during the genetic data analysis in this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.900558/full#supplementary-material
Reference
Ali, S., Gladieux, P., Leconte, M., Gautier, A., Justesen, A. F., Hovmøller, M. S., et al. (2014). Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen Puccinia Striiformis f.Sp. Tritici. Plos Pathog. 10, e1003903. doi:10.1371/journal.ppat.1003903
Ali, S., Rodriguez-Algaba, J., Thach, T., Sørensen, C. K., Hansen, J. G., Lassen, P., et al. (2017). Yellow Rust Epidemics Worldwide Were Caused by Pathogen Races from Divergent Genetic Lineages. Front. Plant Sci. 8, 1057. doi:10.3389/fpls.2017.01057
Appels, R., Eversole, K., Appels, R., Eversole, K., Feuillet, C., Keller, B., et al. (2018). Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 361. doi:10.1126/science.aar7191
Bansal, U. K., Kazi, A. G., Singh, B., Hare, R. A., and Bariana, H. S. (2014). Mapping of Durable Stripe Rust Resistance in a Durum Wheat Cultivar Wollaroi. Mol. Breed. 33, 51–59. doi:10.1007/s11032-013-9933-x
Basnet, B. R., Singh, R. P., Ibrahim, A. M. H., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C., et al. (2014). Characterization of Yr54 and Other Genes Associated with Adult Plant Resistance to Yellow Rust and Leaf Rust in Common Wheat Quaiu 3. Mol. Breed. 33, 385–399. doi:10.1007/s11032-013-9957-2
Boukhatem, N., Baret, P. V., Mingeot, D., and Jacquemin, J. M. (2002). Quantitative Trait Loci for Resistance against Yellow Rust in Two Wheat-Derived Recombinant Inbred Line Populations. Theor. Appl. Genet. 104, 111–118. doi:10.1007/s001220200013
Boyd, L. A. (2005). Can Robigus Defeat an Old Enemy? - Yellow Rust of Wheat. J. Agric. Sci. 143, 233–243. doi:10.1017/S0021859605005095
Brar, G. S., Fuentes-Dávila, G., He, X., Sansaloni, C. P., Singh, R. P., and Singh, P. K. (2018). Genetic Mapping of Resistance in Hexaploid Wheat for a Quarantine Disease: Karnal Bunt. Front. Plant Sci. 9, 1497. doi:10.3389/fpls.2018.01497
Brueggeman, R., Druka, A., Nirmala, J., Cavileer, T., Drader, T., Rostoks, N., et al. (2008). The Stem Rust Resistance Gene Rpg5 Encodes a Protein with Nucleotide-Binding-Site, Leucine-Rich, and Protein Kinase Domains. Proc. Natl. Acad. Sci. U.S.A. 105, 14970–14975. doi:10.1073/pnas.0807270105
Buerstmayr, M., Matiasch, L., Mascher, F., Vida, G., Ittu, M., Robert, O., et al. (2014). Mapping of Quantitative Adult Plant Field Resistance to Leaf Rust and Stripe Rust in Two European winter Wheat Populations Reveals Co-location of Three QTL Conferring Resistance to Both Rust Pathogens. Theor. Appl. Genet. 127, 2011–2028. doi:10.1007/s00122-014-2357-0
Bulli, P., Zhang, J., Chao, S., Chen, X., and Pumphrey, M. (2016). Genetic Architecture of Resistance to Stripe Rust in a Global Winter Wheat Germplasm Collection. G3: Genes, Genomes, Genet. 6, 2237–2253. doi:10.1534/g3.116.028407
Burdon, J. J., Barrett, L. G., Rebetzke, G., and Thrall, P. H. (2014). Guiding Deployment of Resistance in Cereals Using Evolutionary Principles. Evol. Appl. 7, 609–624. doi:10.1111/eva.12175
Chen, S., Hegarty, J., Shen, T., Hua, L., Li, H., Luo, J., et al. (2021). Stripe Rust Resistance Gene Yr34 (Synonym Yr48) Is Located within a Distal Translocation of Triticum Monococcum Chromosome 5AmL into Common Wheat. Theor. Appl. Genet. 134, 2197–2211. doi:10.1007/s00122-021-03816-z
Chen, W., Wellings, C., Chen, X., Kang, Z., and Liu, T. (2014). Wheat Stripe (Yellow) Rust Caused byPuccinia Striiformisf. sp.Tritici. Mol. Plant Pathol. 15, 433–446. doi:10.1111/mpp.12116
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., et al. (2017). An Improved Assembly and Annotation of the Allohexaploid Wheat Genome Identifies Complete Families of Agronomic Genes and Provides Genomic Evidence for Chromosomal Translocations. Genome Res. 27, 885–896. doi:10.1101/gr.217117.116
Cobo, N., Pflüger, L., Chen, X., and Dubcovsky, J. (2018). Mapping QTL for Resistance to New Virulent Races of Wheat Stripe Rust from Two Argentinean Wheat Cultivars. Crop Sci. 58, 2470–2483. doi:10.2135/cropsci2018.04.0286
de Vallavieille-Pope, C., Ali, S., Leconte, M., Enjalbert, J., Delos, M., and Rouzet, J. (2012). Virulence Dynamics and Regional Structuring of Puccinia Striiformis F. Sp. Tritici in France between 1984 and 2009. Plant Dis. 96, 131–140. doi:10.1094/pdis-02-11-0078
Doyle, J. J., and Doyle, J. L. (1987). A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull 19, 11–15.
Ellis, J. G., Lagudah, E. S., Spielmeyer, W., and Dodds, P. N. (2014). The Past, Present and Future of Breeding Rust Resistant Wheat. Front. Plant Sci. 5, 641. doi:10.3389/fpls.2014.00641
Gebrewahid, T. W., Zhang, P., Zhou, Y., Yan, X., Xia, X., He, Z., et al. (2020). QTL Mapping of Adult Plant Resistance to Stripe Rust and Leaf Rust in a Fuyu 3/Zhengzhou 5389 Wheat Population. Crop J. 8, 655–665. doi:10.1016/j.cj.2019.09.013
Godoy, J. G., Rynearson, S., Chen, X., and Pumphrey, M. (2018). Genome-Wide Association Mapping of Loci for Resistance to Stripe Rust in North American Elite Spring Wheat Germplasm. Phytopathology 108, 234–245. doi:10.1094/PHYTO-06-17-0195-R
Hou, L., Chen, X., Wang, M., See, D. R., Chao, S., Bulli, P., et al. (2015). Mapping a Large Number of QTL for Durable Resistance to Stripe Rust in winter Wheat Druchamp Using SSR and SNP Markers. PLoS One 10, e0126794. doi:10.1371/journal.pone.0126794
Hovmøller, M. S., Sørensen, C. K., Walter, S., and Justesen, A. F. (2011). Diversity of Puccinia Striiformis on Cereals and Grasses. Annu. Rev. phytopathology 49, 197. doi:10.1146/annurev-phyto-072910-095230
Hovmøller, M. S., Walter, S., Bayles, R. A., Hubbard, A., Flath, K., Sommerfeldt, N., et al. (2016). Replacement of the European Wheat Yellow Rust Population by New Races from the centre of Diversity in the Near-Himalayan Region. Plant Pathol. 65, 402–411. doi:10.1111/ppa.12433
Hulbert, S., and Pumphrey, M. (2014). A Time for More Booms and Fewer Busts? Unraveling Cereal-Rust Interactions. MPMI 27, 207–214. doi:10.1094/MPMI-09-13-0295-FI
Jighly, A., Oyiga, B. C., Makdis, F., Nazari, K., Youssef, O., Tadesse, W., et al. (2015). Genome-wide DArT and SNP Scan for QTL Associated with Resistance to Stripe Rust (Puccinia Striiformis F. Sp. Tritici) in Elite ICARDA Wheat (Triticum aestivum L.) Germplasm. Theor. Appl. Genet. 128, 1277–1295. doi:10.1007/s00122-015-2504-2
Jones, D. A., and Jones, J. D. G. (1997). The Role of Leucine-Rich Repeat Proteins in Plant Defences. Adv. Bot. Res. 24, 89–167. doi:10.1016/s0065-2296(08)60072-5
Li, H., Vikram, P., Singh, R. P., Kilian, A., Carling, J., Song, J., et al. (2015). A High Density GBS Map of Bread Wheat and its Application for Dissecting Complex Disease Resistance Traits. BMC genomics 16, 1–15. doi:10.1186/s12864-015-1424-5
Liu, S., Huang, S., Zeng, Q., Wang, X., Yu, R., Wang, Q., et al. (2021). Refined Mapping of Stripe Rust Resistance Gene YrP10090 within a Desirable Haplotype for Wheat Improvement on Chromosome 6A. Theor. Appl. Genet. 134, 2005–2021. doi:10.1007/s00122-021-03801-6
Lowe, I., Cantu, D., and Dubcovsky, J. (2011). Durable Resistance to the Wheat Rusts: Integrating Systems Biology and Traditional Phenotype-Based Research Methods to Guide the Deployment of Resistance Genes. Euphytica 179, 69–79. doi:10.1007/s10681-010-0311-z
Maccaferri, M., Zhang, J., Bulli, P., Abate, Z., Chao, S., Cantu, D., et al. (2015). A Genome-wide Association Study of Resistance to Stripe Rust (Puccinia Striiformis F. Sp. Tritici) in a Worldwide Collection of Hexaploid spring Wheat (Triticum aestivum L.). G3 (Bethesda) 5, 449–465. doi:10.1534/g3.114.014563
Manickavelu, A., Joukhadar, R., Jighly, A., Lan, C., Huerta-Espino, J., Stanikzai, A. S., et al. (2016). Genome Wide Association Mapping of Stripe Rust Resistance in Afghan Wheat Landraces. Plant Sci. 252, 222–229. doi:10.1016/j.plantsci.2016.07.018
McIntosh, R. A., Dubcovsky, J., Rogers, J. W., Morris, C. F., Appels, R., and Xia, X. C. (2014). Catalogue of Gene Symbols for Wheat: 2013-14 Supplement. Annu. wheat Newsl. 58, 4. doi:10.1016/j.orbis.2013.11.004
Muleta, K. T., Bulli, P., Rynearson, S., Chen, X., and Pumphrey, M. (2017). Loci Associated with Resistance to Stripe Rust (Puccinia Striiformis F. Sp. Tritici) in a Core Collection of spring Wheat (Triticum aestivum). PLoS ONE 12, e0179087. doi:10.1371/journal.pone.0179087
Naruoka, Y., Garland-Campbell, K. A., and Carter, A. H. (2015). Genome-wide Association Mapping for Stripe Rust (Puccinia Striiformis F. Sp. Tritici) in US Pacific Northwest winter Wheat (Triticum aestivum L.). Theor. Appl. Genet. 128, 1083–1101. doi:10.1007/s00122-015-2492-2
Ng, D., Abeysinghe, J., and Kamali, M. (2018). Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int. J. Mol. Sci. 19, 3737. doi:10.3390/ijms19123737
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC Proteins: Regulation and Role in Stress Tolerance. Trends Plant Science 17, 369–381. doi:10.1016/j.tplants.2012.02.004
Rajaram, S. (2015). “Challenges in Wheat Research and Development,” in The International Dimension of the American Society of Agronomy: Past and Future (John Wiley & Sons), 14, 39–47. doi:10.2134/2010.internationaldimension.c6
Ren, Y., Li, Z., He, Z., Wu, L., Bai, B., Lan, C., et al. (2012). QTL Mapping of Adult-Plant Resistances to Stripe Rust and Leaf Rust in Chinese Wheat Cultivar Bainong 64. Theor. Appl. Genet. 125, 1253–1262. doi:10.1007/s00122-012-1910-y
Rimbert, H., Darrier, B., Navarro, J., Kitt, J., Choulet, F., Leveugle, M., et al. (2018). High Throughput SNP Discovery and Genotyping in Hexaploid Wheat. PLOS ONE 13, e0186329. doi:10.1371/journal.pone.0186329
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., Herrera-Foessel, S. A., Forrest, K. L., Hayden, M. J., et al. (2012). Analysis of Leaf and Stripe Rust Severities Reveals Pathotype Changes and Multiple Minor QTLs Associated with Resistance in an Avocet × Pastor Wheat Population. Theor. Appl. Genet. 124, 1283–1294. doi:10.1007/s00122-012-1786-x
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., and Rebetzke, G. J. (2008). Quantitative Trait Loci for Slow-Rusting Resistance in Wheat to Leaf Rust and Stripe Rust Identified with Multi-Environment Analysis. Theor. Appl. Genet. 116, 1027–1034. doi:10.1007/s00122-008-0736-0
Saari, E. E., and Wilcoxson, R. D. (1974). Plant Disease Situation of High-Yielding dwarf Wheats in Asia and Africa. Annu. Rev. phytopathology 12, 49–62. doi:10.1146/annurev.py.12.090174.000405
Sadasivaiah, R. S., Orshinsky, B. R., and Kozub, G. C. (1999). Production of Wheat Haploids Using Anther Culture and Wheat X maize Hybridization Techniques. Cereal Res. Commun. 27, 33–40. doi:10.1007/bf03543916
Singh, R. P., Huerta-Espino, J., and Rajaram, S. (2000). Achieving Near-Immunity to Leaf and Stripe Rusts in Wheat by Combining Slow Rusting Resistance Genes. Acta phytopathologica et entomologica hungarica 35, 133
Steele, J. F. C., Hughes, R. K., and Banfield, M. J. (2019). Structural and Biochemical Studies of an NB-ARC Domain from a Plant NLR Immune Receptor. PloS one 14, e0221226. doi:10.1371/journal.pone.0221226
Steele, K. A., Humphreys, E., Wellings, C. R., and Dickinson, M. J. (2001). Support for a Stepwise Mutation Model for Pathogen Evolution in Australasian Puccinia Striiformis f.Sp. Tritici by Use of Molecular Markers. Plant Pathol. 50, 174–180. doi:10.1046/j.1365-3059.2001.00558.x
Stubbs, R. W., Prescott, J. M., Saari, E. E., and Dubin, H. J. (1986). Cereal Disease Methodology Manual. México: Centro Internacional de Mejoramento de Maiz y Trigo (CIMMYT), 1–46.
Suenaga, K., Singh, R. P., Huerta-Espino, J., and William, H. M. (2003). Microsatellite Markers for Genes Lr34/Yr18 and Other Quantitative Trait Loci for Leaf Rust and Stripe Rust Resistance in Bread Wheat. Phytopathology 93, 881–890. doi:10.1094/phyto.2003.93.7.881
Tadesse, W., Ogbonnaya, F. C., Jighly, A., Nazari, K., Rajaram, S., and Baum, M. (2014). Association Mapping of Resistance to Yellow Rust in Winter Wheat Cultivars and Elite Genotypes. Crop Sci. 54, 607–616. doi:10.2135/cropsci2013.05.0289
Tehseen, M. M., Kehel, Z., Sansaloni, C. P., Lopes, M. d. S., Amri, A., Kurtulus, E., et al. (2021). Comparison of Genomic Prediction Methods for Yellow, Stem, and Leaf Rust Resistance in Wheat Landraces from Afghanistan. Plants 10, 558. doi:10.3390/plants10030558
Tehseen, M. M., Tonk, F. A., Tosun, M., Amri, A., Sansaloni, C. P., Kurtulus, E., et al. (2021). Genome-Wide Association Study of Resistance to PstS2 and Warrior Races of Puccinia Striiformis F. Sp. Tritici (Stripe Rust) in Bread Wheat Landraces. Plant. Genome 14, e20066. doi:10.1002/tpg2.20066
van den Burg, H. A., Tsitsigiannis, D. I., Rowland, O., Lo, J., Rallapalli, G., MacLean, D., et al. (2008). The F-Box Protein ACRE189/ACIF1 Regulates Cell Death and Defense Responses Activated during Pathogen Recognition in Tobacco and Tomato. The Plant Cell 20, 697–719. doi:10.1105/tpc.107.056978
Van Ooijen, G., Mayr, G., Kasiem, M. M. A., Albrecht, M., Cornelissen, B. J. C., and Takken, F. L. W. (2008). Structure-function Analysis of the NB-ARC Domain of Plant Disease Resistance Proteins. J. Exp. Bot. 59, 1383–1397. doi:10.1093/jxb/ern045
Van Ooijen, J. W. (1999). LOD Significance Thresholds for QTL Analysis in Experimental Populations of Diploid Species. Heredity 83, 613–624. doi:10.1038/sj.hdy.6886230
Wang, M., Wang, S., Liang, Z., Shi, W., Gao, C., and Xia, G. (2018). From Genetic Stock to Genome Editing: Gene Exploitation in Wheat. Trends Biotechnology 36, 160–172. doi:10.1016/j.tibtech.2017.10.002
William, M., Singh, R. P., Huerta-Espino, J., Islas, S. O., and Hoisington, D. (2003). Molecular Marker Mapping of Leaf Rust Resistance Gene Lr46 and its Association with Stripe Rust Resistance Gene Yr29 in Wheat. Phytopathology 93, 153–159. doi:10.1094/phyto.2003.93.2.153
Wu, J., Yu, R., Wang, H., Zhou, C. e., Huang, S., Jiao, H., et al. (2021). A Large‐scale Genomic Association Analysis Identifies the Candidate Causal Genes Conferring Stripe Rust Resistance under Multiple Field Environments. Plant Biotechnol. J. 19, 177–191. doi:10.1111/pbi.13452
Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., et al. (2002). A Draft Sequence of the rice Genome (Oryza Sativa L. Ssp. Indica). science 296, 79–92. doi:10.1126/science.1068037
Yuan, N., Yuan, S., Li, Z., Zhou, M., Wu, P., Hu, Q., et al. (2018). Stress Induced Factor 2, a Leucine-Rich Repeat Kinase Regulates Basal Plant Pathogen Defense. Plant Physiol. 176, 3062–3080. doi:10.1104/pp.17.01266
Yuan, X., Wang, H., Cai, J., Li, D., and Song, F. (2019). NAC Transcription Factors in Plant Immunity. Phytopathology Res. 1, 1–13. doi:10.1186/s42483-018-0008-0
Zhang, P., Li, X., Gebrewahid, T.-W., Liu, H., Xia, X., He, Z., et al. (2019). QTL Mapping of Adult-Plant Resistance to Leaf and Stripe Rust in Wheat Cross Sw 8588/thatcher Using the Wheat 55k Snp Array. Plant Dis. 103, 3041–3049. doi:10.1094/PDIS-02-19-0380-RE
Keywords: QTL mapping, yellow rust, adult plant resistance, doubled haploid (DH), wheat
Citation: Tehseen MM, Tonk FA, Tosun M, Randhawa HS, Kurtulus E, Ozseven I, Akin B, Nur Zulfuagaoglu O and Nazari K (2022) QTL Mapping of Adult Plant Resistance to Stripe Rust in a Doubled Haploid Wheat Population. Front. Genet. 13:900558. doi: 10.3389/fgene.2022.900558
Received: 20 March 2022; Accepted: 13 April 2022;
Published: 11 May 2022.
Edited by:
Muhammad Abdul Rehman Rashid, Government College University, Faisalabad, PakistanReviewed by:
Qiang Li, Northwest A and F University, ChinaAsif Saeed, University of Agriculture, Faisalabad, Pakistan
Kaixiang Chao, Yuxi Normal University, China
Muhammad Abu Bakar Zia, Niğde Ömer Halisdemir University, Turkey
Muhammad Azhar Nadeem, Sivas University of Science and Technology, Turkey
Copyright © 2022 Tehseen, Tonk, Tosun, Randhawa, Kurtulus, Ozseven, Akin, Nur Zulfuagaoglu and Nazari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Massub Tehseen, bWFzc3ViLnRlaHNlZW5AZ21haWwuY29t; Kumarse Nazari, ay5uYXphcmlAY2dpYXIub3Jn
 Muhammad Massub Tehseen
Muhammad Massub Tehseen Fatma Aykut Tonk1
Fatma Aykut Tonk1 Ezgi Kurtulus
Ezgi Kurtulus Kumarse Nazari
Kumarse Nazari