- 1School of Pharmacy, Hangzhou Normal University, Hangzhou, China
- 2Key Laboratory of Elemene Class Anti-Cancer Chinese Medicines, Engineering Laboratory of Development and Application of Traditional Chinese Medicines, Collaborative Innovation Center of Traditional Chinese Medicines of Zhejiang Province, Hangzhou Normal University, Hangzhou, China
MYB superfamily is one of the most abundant families in plants, and plays critical role in plant growth, development, metabolism regulation, and stress response. Curcuma wenyujin is the main source plant of three traditional Chinese medicines, which are widely used in clinical treatment due to its diverse pharmacological activities. In present study, 88 CwMYBs were identified and analyzed in C. wenyujin, including 43 MYB-related genes, 42 R2R3-MYB genes, two 3R-MYB genes, and one 4R-MYB gene. Forty-three MYB-related proteins were classified into several types based on conserved domains and specific motifs, including CCA1-like type, R-R type, Myb-CC type, GARP-like type, and TBR-like type. The analysis of motifs in MYB DBD and no-MYB regions revealed the relevance of protein structure and function. Comparative phylogeny analysis divided 42 R2R3-MYB proteins into 19 subgroups and provided a reference for understanding the functions of some CwMYBs based on orthologs of previously characterized MYBs. Expression profile analysis of CwMYB genes revealed the differentially expressed genes responding to various abiotic stresses. Four candidate MYB genes were identified by combining the results of phylogeny analysis and expression analysis. CwMYB10, CwMYB18, CwMYB39, and CwMYB41 were significantly induced by cold, NaCl, and MeJA stress treatments. CwMYB18 and CwMYB41 were proved as regulators with activity of transcriptional activation, whereas CwMYB39 and CwMYB10 were not. They may participate in the response to abiotic stresses through different mechanisms in C. wenyujin. This study was the first step toward understanding the CwMYB family and the response to abiotic stresses in C. wenyujin.
Introduction
Curcuma wenyujin Y.H. Chen et C. Ling is a member of the Zingiberaceae family and mainly cultivated in Wenzhou City, Zhejiang Province, P. R. China. Because of their various pharmacological activities, the dried rhizomes and root tubers of C. wenyujin have been used as traditional Chinese medicine (TCM) for over 1,000 years (Zhai et al., 2019; Li et al., 2021). Drought, high salt, extreme temperatures, and other stresses all impact the yield and quality of C. wenyujin. The v-myb avian myeloblastosis viral oncogene homolog (MYB) superfamily is one of most significant transcription factor (TF) families in plants, which regulates plant growth, development, and a variety of physiological/biochemical processes (Cao et al., 2020; Wang et al., 2021; Xiao et al., 2021).
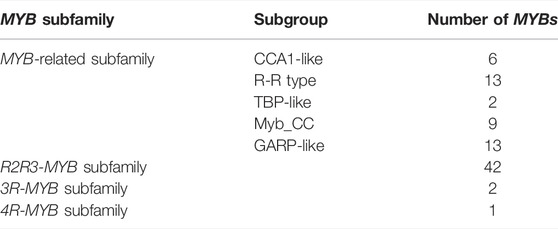
There is a conserved DNA binding domain (DBD) in MYB proteins, made up of 1–4 imperfect amino acid sequence repeats i.e. “R”. There are approximately 52 amino acids in each “R”, forming a very similar folding architecture with three well-defined α-helixes. The helix-turn-helix hydrophobic core was formed by the second and third helices of each “R” with three regularly spaced tryptophans (W) or other hydrophobic residues. MYB superfamily is divided into four subfamilies based on the number of “R” in MYB DBD: MYB-related subfamily gene with a single or partial “R”, R2R3-MYB subfamily gene with “R2” and “R3”, 3R-MYB subfamily gene with “R1”, “R2”, and “R3”, as well as 4R-MYB subfamily gene with four “R1/R2” (Dubos et al., 2010).
The R2R3-MYB subfamilies from Arabidopsis thaliana and Oryza sativa, the model plants for dicotyledons and monocotyledons respectively, have been well identified and characterized (Dubos et al., 2010; Katiyar et al., 2012). Many R2R3-MYBs were reported to regulate the primary and secondary metabolism, cell fate and identity, developmental processes, and response to biotic, and abiotic stresses (Li X. et al., 2019; Nguyen et al., 2019; Cao et al., 2020; Xiao et al., 2021). For example, Arabidopsis MYB30 acts as a critical negative regulator promoting PIF4 and PIF5 protein accumulation in the light to regulate photomorphogenic development (Yan et al., 2020). AtMYB111 plays a role as a positive regulator in salt stress response, depending on its regulation on flavonoid synthesis by activating the transcription of chalcone synthase (CHS), flavanone carboxylase (F3H), and flavonol synthase 1 (FLS1) (Li B. et al., 2019). In rice, MORE FLORET 1, an MYB transcription factor, interacts with TOPLESS-RELATED PROTEINs (TPRs) proteins to regulate the organ identity and spikelet determinacy by repressing the expression of downstream target genes (Ren et al., 2020). A rice R2R3-MYB (OsC1) transcriptional regulator helps to ameliorate oxidative stress in plants by increasing anthocyanin accumulation (Upadhyaya et al., 2021). Besides, more and more R2R3-MYB genes were identified and characterized in other plants. In Rosa multiflora, RmMYB108 is positively involved in response to cold, freeze, high salt, and drought stresses. RmMYB108 overexpressed in Arabidopsis improves the cold tolerance by reducing plant damage and promoting plant growth (Dong et al., 2021). CmMYB15 regulates the biosynthesis of lignin to enhance the resistance of chrysanthemum to aphids (An et al., 2019). PlMYB108 from Paeonia lactiflora overexpressed in tobacco plants increases the flavonoid accumulation, antioxidant enzyme activities, and photosynthesis to confer drought tolerance (Wu et al., 2021).
MYB-related subfamily can be divided into several types based on the different structure of “R” in MYB BDB: CCA1-like type, R-R type, TRF-like type, TBP-like type, I-box like type, CPC-like type, GARP-like type, and so on (Du et al., 2013). CCA1-like MYB in MYB DBD contains a conserved “R” with the motif SHAQK (y/f)F. Rice CIRCADIAN CLOCK ASSOCIATED1 (OsCCA1) positively regulates expression of TEOSINTE BRANCHED1 (OsTB1), DWARF14 (D14), and IDEAL PLANT ARCHITECTURE1 (IPA1) to repress tiller-bud outgrowth (Wang et al., 2020). Moreover, CCA1-like MYB protein GmABAS1 enhances abscisic acid (ABA) sensitivity by acting as a transcriptional repressor of the target gene ABI in ABA signal pathway (Ku et al., 2020). TBP-like MYBs contain a conserved motif LKDKW(R/K) (N/T) in “R”. Most of the known TBP-like genes encode telomere-binding proteins (Du et al., 2013). The Myb-CC type MYBs have an “R” with the SHAQK (y/f)F motif in MYB BDB and the Myb_CC_LHEQLE domain (pfam14379) in no-MYB region (Gu et al., 2022). ZmMYB-CC10 enhances tolerance to drought stress by directly activating ZmAPX4 expression, thereby reducing H2O2 content (Zhang et al., 2022). SlPHL1 is a newly identified MYB-CC TF from tomato, which participates in Pi-starvation signaling by directly upregulating the PSI genes to elevate root hair growth, promote APase activity, and favor Pi uptake (Zhang et al., 2021). MYB DBD of GARP-like protein contains a conserved “R” with the motif SHLQK/MxR. There are two viewpoints about GARP-like proteins: 1) those that belong to MYB-related subfamily due to a similar motif but have distant relationships; and 2) those that do not belong to MYB-related subfamily due to the only “W” residue in conserved MYB DBD (Du et al., 2013; Safi et al., 2017). Arabidopsis overexpressing GhGLK1 (a GARP-like gene) showed more adaptability to drought and cold treatments with the less leaf damage and lower ion permeability (Liu et al., 2021). Interestingly, R-R type MYB-related proteins were the exception, with two far apart “R” located in the N-terminal and middle of the sequence, respectively. The second “R” in middle of the sequence contains a SHAQK (y/f)F motif similar to that in CCA1-like MYBs. In Arabidopsis, AtDIV2, encoding an R-R type MYB TF, plays negative role in salt stress and is required for ABA signaling (Fang et al., 2018). In addition, 3R-MYB and 4R-MYB subfamilies are the smaller classes that often contain a few members in most plants.
Because of the roles of MYB genes in plant growth, development, metabolism regulation, and stress response, it is of great significance to study MYB family in C. wenyujin and its expression patterns in different tissues and under different stresses. With transcriptome sequence data of C. wenyujin, MYB family was systematically identified and analyzed including gene classification, evolutionary relationship, conserved domain, and motif composition for the first time. The spatial and temporal expression profiles were also analyzed, as well as the differential expression profiles of MYB genes in response to methyl jasmonate (MeJA), cold, and salt stresses. The findings will provide a comprehensive understanding of MYB family, and lay the theoretical groundwork for investigating MYBs’ potential roles in C. wenyujin.
Materials and Methods
Plant Materials and Stress Treatment
C. wenyujin was used as the plant material in this study. The germplasm was obtained from Wenzhou City, Zhejiang Province, P. R. China (14 m altitude, 27°47′N, and 120°37′E). The original plants and their macroscopic characteristics were authenticated by Professor Zengxi Guo, who works at the Institute of Food and Drug Control in Zhejiang, China. Seedlings were cultured in Murashige and Skoog (MS) solid medium in an incubator (12 h light/12 h dark cycle at 22°C) for 1 month to trefoil stage to perform the expression analysis of MYB genes with abiotic stresses. Seedlings were cultured with 200 mM NaCl solution as salt stress treatment for 24 h. For MeJA treatment, seedlings were cultured in solution containing 250 μM MeJA for 24 h. For cold stress treatment, seedlings were cultured in an incubator at 4°C for 24 h. All samples were collected at the time points (0, 1, 3, 6, 12, and 24 h), frozen in liquid nitrogen, and then stored at -80 °C for subsequent RNA extraction.
Identification of MYBs in C. wenyujin
The Hidden Markov Model (HMM) profile of MYB DBD (Accession No. PF00249) was downloaded from the Pfam database (http://pfam.xfam.org) and used as a query to identify CwMYBs in the transcriptome data of C. wenyujin, utilizing TB tools v1.09’ simple HMMER search with E-value ≤ 1e−5 (Chen et al., 2020; Khaksar and Sirikantaramas, 2021). The transcriptome data of C. wenyujin (Accession No. CRA000632) was available in the Genome Sequence Archive (GSA) database (https://ngdc.cncb.ac.cn/gsa/). The redundant sequences were removed utilizing the R program with a cutoff value of identity percentage > = 80%. MYB DBD of identified protein sequences was confirmed using online software SMART version 9 (http://smart.embl-heidelberg.de) and sequence search tool Pfam 35.0 (http://pfam.xfam.org/search/sequence) with default parameters. The identified MYB proteins were divided into four subfamilies based on the number of “R” in MYB DBD. ClustalX 2.1 software was used to perform multiple sequence alignment of conserved MYB DBD (Larkin et al., 2007). The sequence logos of conserved MYB DBD were created with the online software Weblogo 2.8.2 (http://weblogo.berkeley.edu/logo.cgi). Molecular weight (Mw) and theoretical isoelectric point (pI) of MYBs were predicted using the online software “Compute pI/Mw tool” (https://web.expasy.org/compute_pi/). The subcellular location information was predicted utilizing the online software WOLF PSORT (https://wolfpsort.hgc.jp/).
Phylogenetic Analysis and Conserved Motif Analysis
Multiple amino acids sequence alignments of CwMYBs were performed using the software ClustalW 2.1 with the default parameters. The phylogenetic tree was generated by the neighbor-joining (NJ) method with 1,000 bootstrap replicates in MEGA 7.0 (Kumar et al., 2016). The online tool MEME 5.4.1 program (http://meme-suite.org/index.html) was used to detect conserved motifs of CwMYBs with the following parameters: zero or one per sequence, motif width ranges of 6–60 amino acids, and 20 as the maximum number of motifs. The default values were used for the remaining options. Only motifs with an E-value of < 1e−2 were kept for further analysis. The map was accomplished with TB tools v1.09 (Chen et al., 2020).
Expression Profiles Analysis of MYB Genes
The RNA-seq data (https://www.ncbi.nlm.nih.gov/sra/, CRA003702) of fresh leaf and tuber samples from mature plants was used to analyze the spatial and temporal expression profiles of MYBs in C. wenyujin (Chen et al., 2021). The RNA-seq data (https://www.ncbi.nlm.nih.gov/sra/, CRA006461) was used to analyze the expression profiles of MYBs in C. wenyujin leaves with MeJA treatment. Transcripts per million reads (TPM) were obtained from RNA-seq data. Heatmaps were generated from log2 based TPM values using TB tools v1.09’ heatmap illustrator (Chen et al., 2020). The differentially expressed genes (DEGs) were screened with the following parameters: TPM >0, |log2Fold chang| >1, and p-value < 0.05.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from different samples with an RNAprep Pure Extraction Kit (DP441, TIANGEN, Beijing, China). First-strand cDNA was synthesized with PrimeScript™RT reagent Kit with gDNA Eraser (Takara, China). The CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, CA, United States) was used to perform quantitative real-time polymerase chain reaction (qRT-PCR) using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The program parameters were as follows: 30 s at 95°C, 45-cycles of 10 s at 95°C, 30 s at 58°C, and 30 s at 72°C, and then 65–95°C for melting curve detection. Expression data was analyzed with the comparative 2−ΔΔCt method (Livak and Schmittgen, 2001). Statistical analysis was performed using SPSS v17.0 software (IBM, New York, US). Statistical significance was set at *p < 0.05 and **p < 0.01. 18S rRNA was used as the internal control. The primers in this assay were listed in Supplementary Table S1.
Yeast One-Hybrid Experiment
The Clontech Matchmaker™ Yeast One-Hybrid system (TBUSA, CA, United States), a GAL4-based yeast one-hybrid system, was employed to examine the transactivation activity. The complete open reading frames (ORF) and various truncated ORFs of MYBs were amplified by PCR (Supplementary Table S1). These fragments were then inserted into the pGBKT7 vector to construct corresponding recombinant plasmids. The plasmid pGBKT7 was used as a negative control. All of these plasmids were transformed into the yeast strain AH109, respectively. The transformation and screening were carried out following the users’ manual (Clontech, United States).
Results
Identification and Classification of CwMYBs in C. wenyujin
All the nucleotides and amino acid sequences of 88 CwMYBs identified from C. wenyujin, were listed in Supplementary Tables S2–S4. The MYB proteins were classified into four subfamilies based on the number of “R” in conserved MYB-DBD (Table 1). Forty-two CwMYBs containing typically R2 (-W-x19-W-x19-W-) and R3 (-F/I/L/M-x18-W-x18-W-) in conserved MYB-DBD were classified into the typical R2R3-MYB subfamily and named CwMYB1-42 (Figure 1). Two CwMYBs containing three “R” and one CwMYB containing four “R” in conserved MYB-DBD were classified into 3R-MYB subfamily and 4R-MYB subfamily respectively, referred as CwMYB3R1-2 and CwMYB4R1. A total of forty-three CwMYBs were classified into MYB-related subfamily with a single or partial “R” in MYB DBD, and named CwMYBR1-43. MYB-related subfamily was further divided into different groups due to the variable of “R” in MYB DBD. CwMYBR1-6, which contained the motif SHAQK (y/f)F in the “R” of the conserved MYB DBD, belonged to the CCA1-like group (Figure 2A). Two “R” were located in the N-terminus and middle of the sequences in CwMYBR7-19 respectively, separated by a long distance. The third tryptophan residue (W) in the first “R” was replaced by tyrosine residue (Y) (Figure 2B). The second “R” contained the motif SHAQK (y/f)F (Figure 2C). They are classified into R-R-type group in MYB-related subfamily. CwMYBR20 and CwMYBR21 were members of TBP-like group with a motif LKDKW(R/K) (N/T) in “R” of MYB DBD (Figure 2D). Based on a motif SHLQK/MxR in MYB DBD and the other domain Myb_CC_LHEQLE (pfam14379) in the no-MYB region, CwMYBR22-30 were identified as Myb-CC type MYB-related proteins (Figures 2E,F). Because of the variant MYB domain with motif SHLQK/MxR, CwMYBR31-43 were classified into GAPR-like group (Figure 2G). The predicted MYB proteins’ amino acid sequences ranged from 172 (CwMYB1) to 1,054 (CwMYB3R1), with the molecular weights ranged from 19.39 kDa (CwMYB1) to 115.44 kDa (CwMYB3R1). The theoretical isoelectric point (pI) ranged from 5.01 (CwMYB3R1) to 9.92 (CwMYBR12). The computed parameters of identified CwMYBs including Mw, pI, and subcellular localization were listed in Supplementary Table S5.
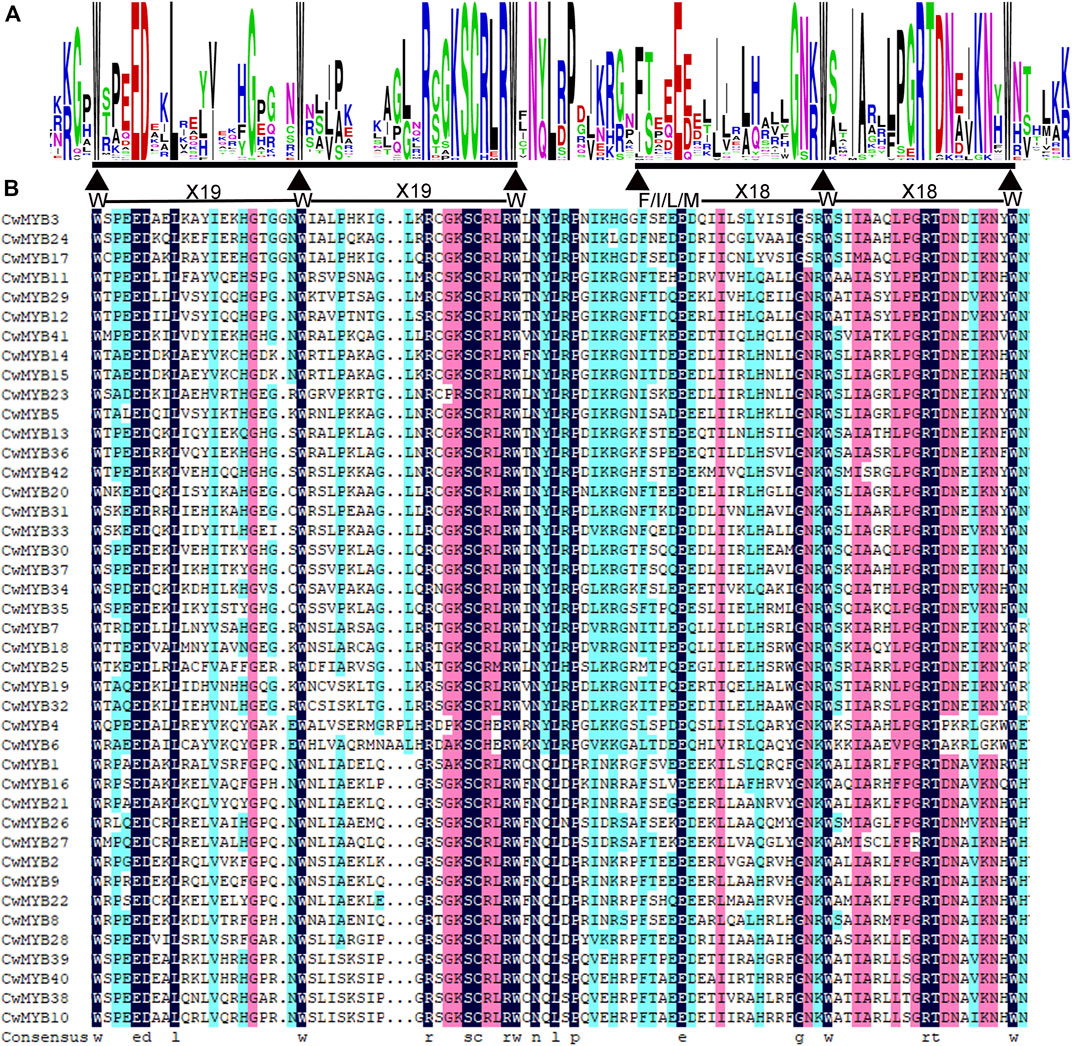
FIGURE 1. Conserved domains in R2R3 type CwMYB1-42 from C. wenyujin. (A) The sequence logos of conserved domains R2 and R3 repeat. Letter height indicates the sequence’s content and conservation at the corresponding position. Triangles in the MYB domain represent conserved amino acid residues or motifs. Weblogo 3 online software was used to create the image. (B) The conserved domain R2 (-W-X19-W-X19-W-) and R3 (-F/I/L/M-X18-W-X18-W-) repeats. The amino acid residues with a black background indicate 100% identity in the CwMYB1-42 protein sequences.
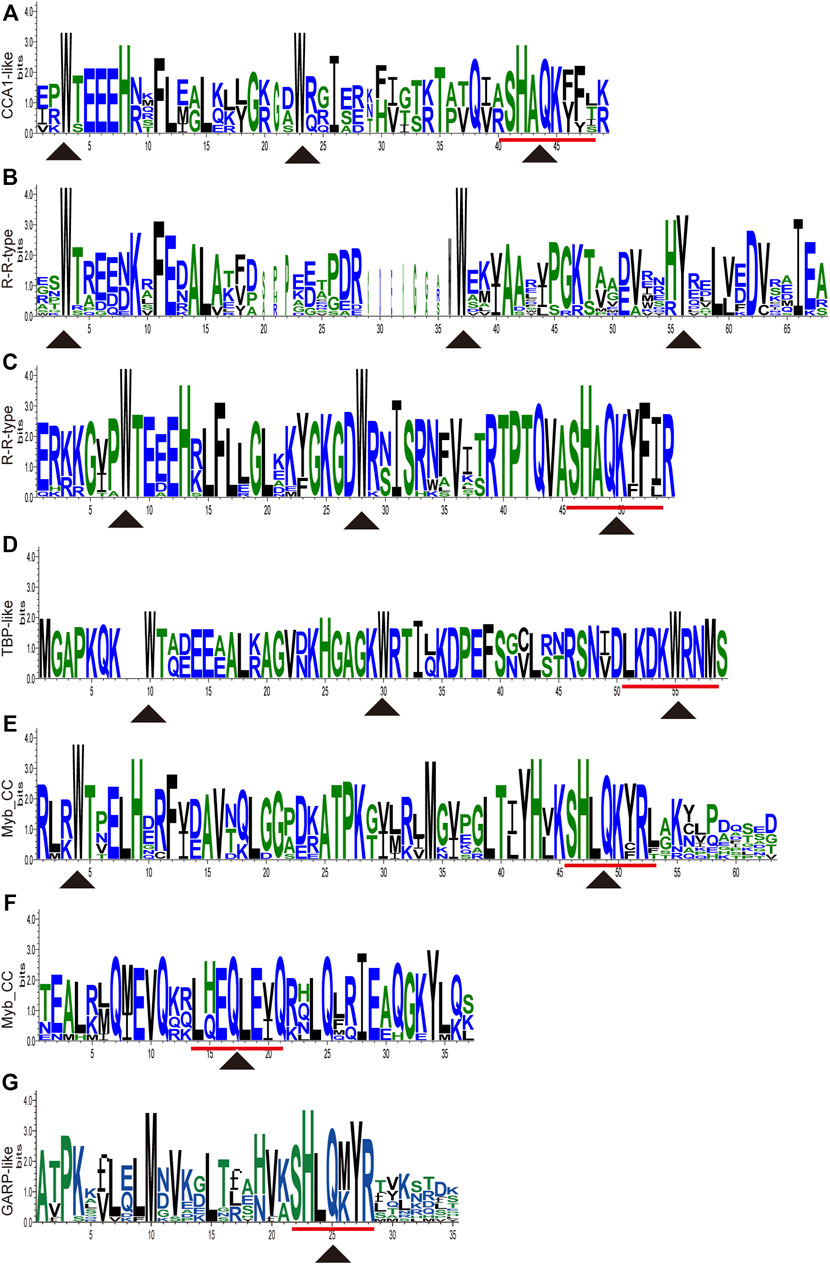
FIGURE 2. Sequence logos of conserved domains in MYB-related proteins from C. wenyujin. (A) CCA1-like type MYB-related proteins. (B,C) R-R type MYB-related proteins. (D) TBP-like type MYB-related proteins. (E,F) Myb_CC type MYB-related proteins. (G) GARP-like type MYB-related proteins. Y axis indicates the sequence content and its conservation at the corresponding position. Triangles indicate conserved amino acid residues or motifs in the MYB domain. The picture was drawn by TB tools software.
Phylogenetic and Motif Composition Analysis of MYB Proteins in C. wenyujin
In order to investigate the evolutionary relationship of CwMYB proteins in C. wenyujin, phylogenetic tree was constructed with the software MEGA 7.0 (Supplementary Figure S1). R2R3-MYBs, R-related MYBs, and 4R-MYBs were divided into different groups, whereas 3R-MYBs were divided into R2R3-MYBs group, indicating a closer evolutionary relationship. This result supports the hypothesis: 3R-MYBs evolve from R2R3-MYBs by acquiring an “R1”, or R2R3-MYBs evolve from 3R-MYBs by losing an “R1” (Jiang et al., 2004). Meanwhile, the phylogenetic relationship along with motif composition of R2R3-MYBs and R-related MYBs were also analyzed. The results revealed that the motifs distribution of MYB proteins supported the evolutionary relationship. Motif logos were illustrated in Supplementary Figures S2, S3. For R2R3-MYB proteins analysis (Figure 3), all MYB DBD contained motif 1 and motif 2, as well as motif 8 or motif 3 excepting CwMYB4/6/25. Non-MYB regions, outside of the MYB DBD, were highly variable and disordered, but similar in the same evolutionary cluster. Most CwMYBs contained one motif near MYB DBD such as motif 10/16/5. Besides, some CwMYBs had motifs located in the extreme C-terminal region such as CwMYB2/9/21/22/16/126/27/14/15, or throughout their non-MYB regions such as CwMYB10/38/39/40. It is worth noting that CwMYB10/38/39/40 contained a shorter EAR (LxLxL) motif in motif 9, which implies that this MYB may work as a repressor in gene transcription (Kagale and Rozwadowski, 2011).
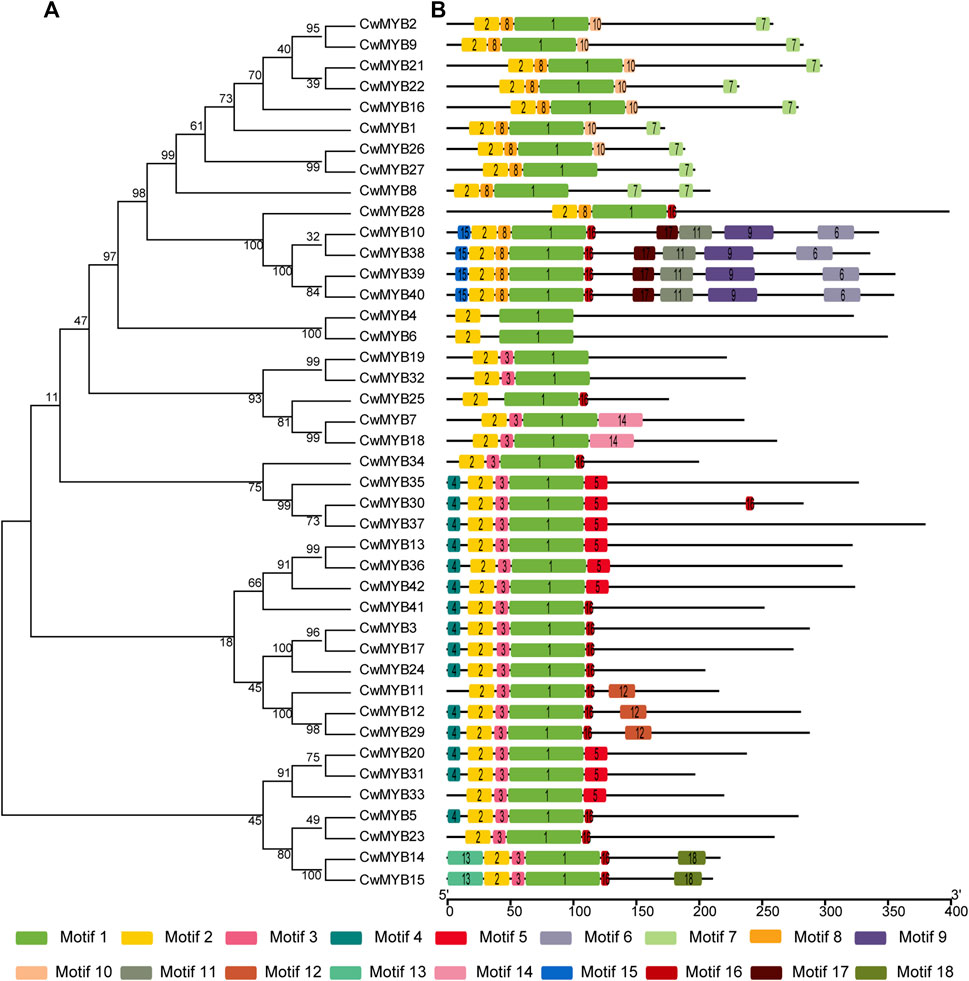
FIGURE 3. Phylogenetic tree and motif compositions of R2R3 type CwMYB1-42 proteins in C. wenyujin. (A) A phylogenetic tree of CwMYB1-42 was constructed using MEGA 7 software coupled with the Neighbor-Joining method with a bootstrap of 1,000 replicates. (B) Schematic diagrams of motif compositions. Colored boxes indicate different motifs. The picture was drawn by TB tools software.
MYB DBD domains of MYB-related proteins were less conservative than R2R3-MYBs, with an irregular distribution not always located in the N-terminal region (Figure 4). For example, CCA1-like MYB DBDs contained motifs 1/11 (CwMYBR1/4/6) or motifs 6/1 (CwMYBR2/3/5). The R-R-type MYB DBDs had motifs 10/4 (CwMYBR14/18/16/10/8/9) or motifs 3/4 (CwMYBR7/11/15/12/1317/19) in the first “R” structure, and motifs 6/1 in the second “R” structure, respectively. The MYB DBDs of Myb-CC type and GAPR-like MYBs were composed of motifs 3/2, while TBP-like MYB DBDs contained motif 13. Non-MYB regions outside of the MYB DBDs were also highly variable and disordered, same as R2R3-MYB proteins.
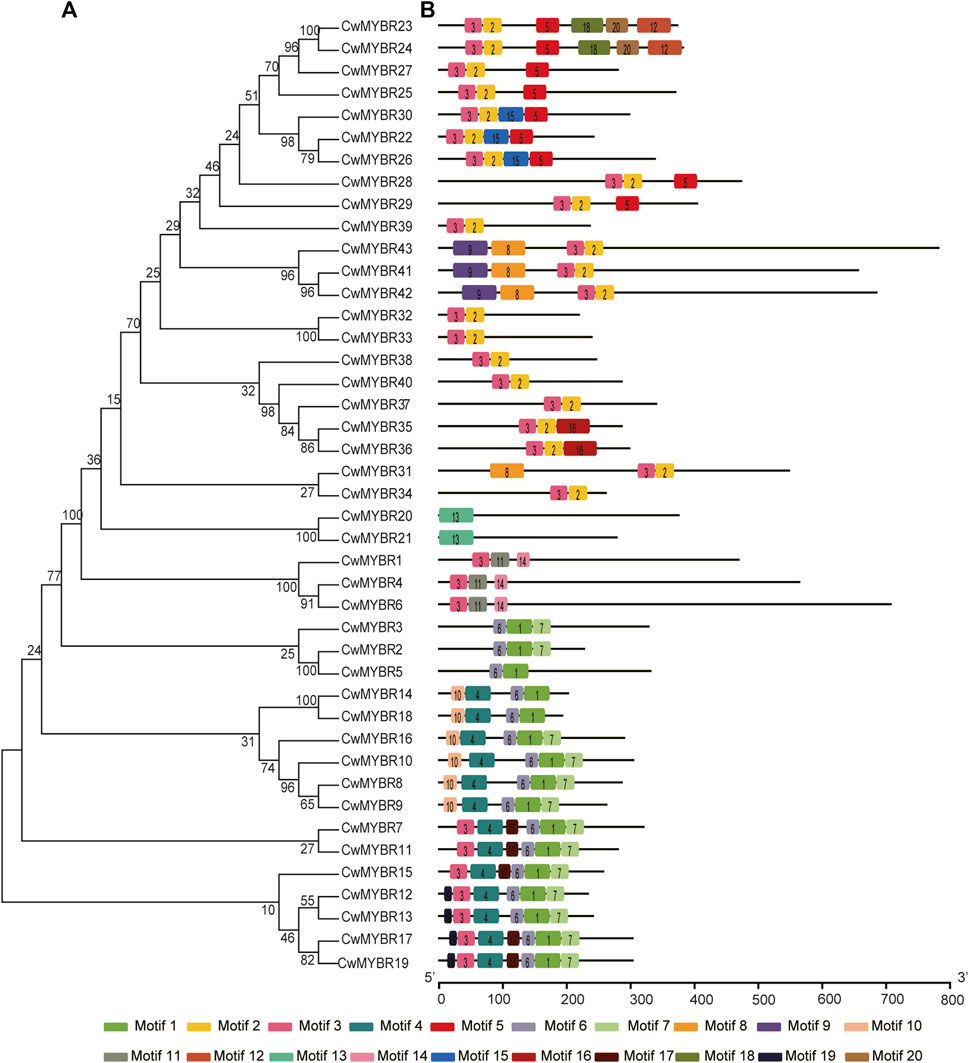
FIGURE 4. Phylogenetic tree and motif compositions of MYB-related proteins in C. wenyujin. (A) A phylogenetic tree of 43 MYB-related proteins was constructed by using MEGA 7 software coupled with Neighbor-Joining method with a bootstrap of 1,000 replicates. (B) Schematic diagrams of motif compositions. Colored boxes indicate different motifs. The picture was drawn by TB tools software.
Phylogeny of R2R3-MYBs Subfamily in Different Plants
To understand the evolving relationship, the phylogenetic tree was constructed using R2R3-MYB proteins from C. wenyujin, A. thaliana, and O. sativa (Figure 5; Supplementary Table S6). Based on the conserved MYB DBD at the N-terminus and no-MYB regions at the C-terminus, AtMYBs had been classified into 23 subgroups (Dubos et al., 2010). According to the AtMYBs classification in Arabidopsis, thirty-six R2R3-MYBs of C. wenyujin were divided into 13 groups: S1, S2, S4, S5, S10, S13, S14, S16, S20, S21, S22, S23, and S24. Homologous genes in different plants are generally assumed to perform similar biological functions (Cai et al., 2012). CwMYB7/18 in subgroup 20, CwMYB10/38/39/40 in subgroup 22, and CwMYB41 in subgroup 2 could all play a role in abiotic stress responses. The evolutionary relationship of CwMYBs with AtMYBs and OsMYBs provided more valuable references for the functional prediction of CwMYBs in C. wenyujin. In addition, six R2R3-CwMYBs with AtMYBs and OsMYBs were distributed into four groups named C1, C3, C4, and C5, in which no MYBs have been characterized. It is worth noting that CwMYB14, CwMYB15, and CwMYB23 were classified into C2 which did not contain MYBs from Arabidopsis and rice, indicating the divergence of evolution in C. wenyujin, Arabidopsis, and rice.
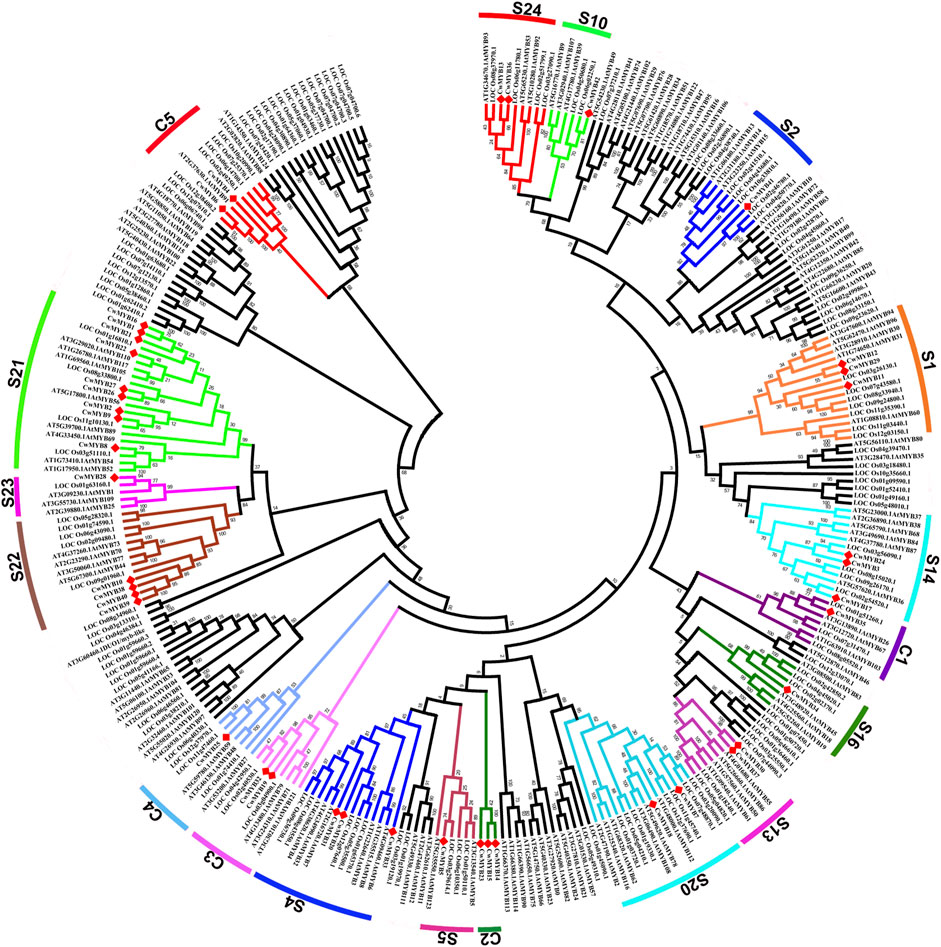
FIGURE 5. Phylogenetic tree of R2R3-MYBs in C. wenyujin, Arabidopsis thaliana, and Oryza sativa. The sequences contain 42 CwR2R3-MYBs in C. wenyujin, 126 AtR2R3-MYBs in Arabidopsis, and 130 OsR2R3-MYBs in rice. Forty-two full length CwMYB1-42 were divided into 13 subgroups (S) and 5 classes (C1-C5). The picture was generated using MEGA 7 software coupled with Neighbor-Joining method with a bootstrap of 1,000 replicates.
Expression Profiles of CwMYBs in Different Tissues
To investigate the expression patterns of MYB family genes in tuber and leaf, a heatmap was constructed based on TPM values from reported RNA-seq data. Finally, the expression patterns of 26 R2R3-MYB genes and 38 MYB-related genes were generated (Figure 6A; Supplementary Figure S4; Supplementary Table S7). The results revealed that the expression level of R2R3-MYBs in group I was higher in the leaf than that in the tuber. In contrast, R2R3-MYBs expression was lower in the leaf than that in the tuber in group II. MYB-related genes in group IV were expressed more in the tuber than in the leaf, whereas MYB-related genes in group VI were expressed less in the tuber than in the leaf. Differential expression analysis with strict standards revealed that 9 R2R3-MYB genes and 19 MYB-related genes were expressed in the tuber and leaf with significant difference (Supplementary Table S7).
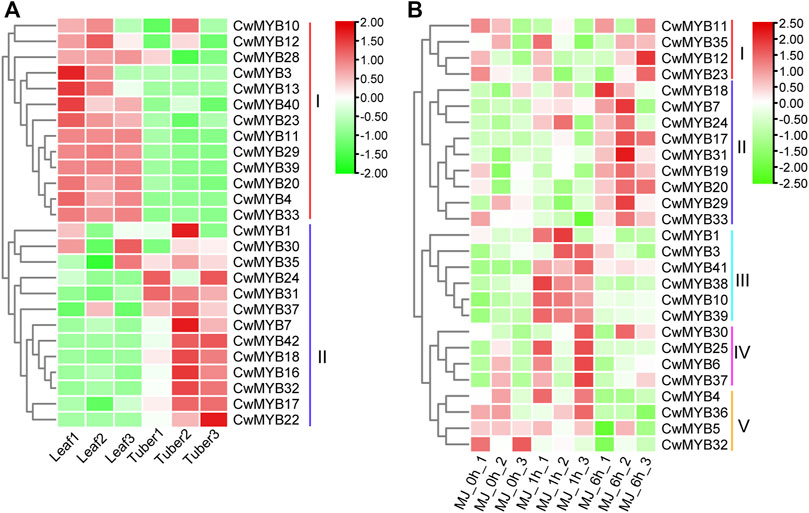
FIGURE 6. Expression profiles analysis of R2R3-CwMYB genes (A) R2R3-CwMYB gene expression patterns in tuber and leaf of C. wenyujin. (B) R2R3-CwMYB gene expression patterns in C. wenyujin with MeJA induction. Heatmap was created by TB tool using the transcript per million (TPM) values. The red and green cells represent the highest and lowest expression levels, respectively.
Expression Profiles of CwMYB Genes With MeJA Treatment
To explore the expression patterns of MYB family genes with MeJA treatment, a heatmap was constructed based on TPM values from reported RNA-seq data. Finally, the expression patterns of 27 R2R3-CwMYB genes and 28 MYB-related genes were generated. The expression analysis results of R2R3-CwMYB genes revealed that the expression profiles in groups II, III, and IV were up-regulated (Figure 6B). R2R3-MYB expression levels were upregulated 1 h after MeJA induction in groups III and IV, while 6 h after MeJA induction in group II. Instead, with 1 and 6 h of MeJA treatment, the expression levels of R2R3-MYBs were down-regulated in groups I and V respectively. In the expression analysis of MYB-related genes (Supplementary Figure S5), the expression profiles in groups I and II were upregulated after 6 and 1 h of MeJA induction, respectively. Concurrently, the expression profiles of MYB genes were downregulated in groups III and IV. Differential expression analysis with strict standards revealed that 5 R2R3-MYB genes and 8 MYB-related genes were involved in the response to MeJA treatment (Supplementary Table S8).
The Expression Levels and Transcriptional Activation Analysis of Four Candidate MYBs in C. wenyujin
The MYB protein is well known for its role in plant response to various abiotic stresses. In this study, candidate MYB genes CwMYB10/18/39/41 were screened based on the results of phylogeny and expression analysis. The expression levels of four candidate CwMYBs were determined using qRT-PCR with cold, NaCl and MeJA treatments. According to the findings, expression levels of all four MYB genes were significantly increased by cold, NaCl, and MeJA treatments (Figure 7). As a result, these MYB proteins may act as positive regulators in C. wenyujin in response to various abiotic stresses.
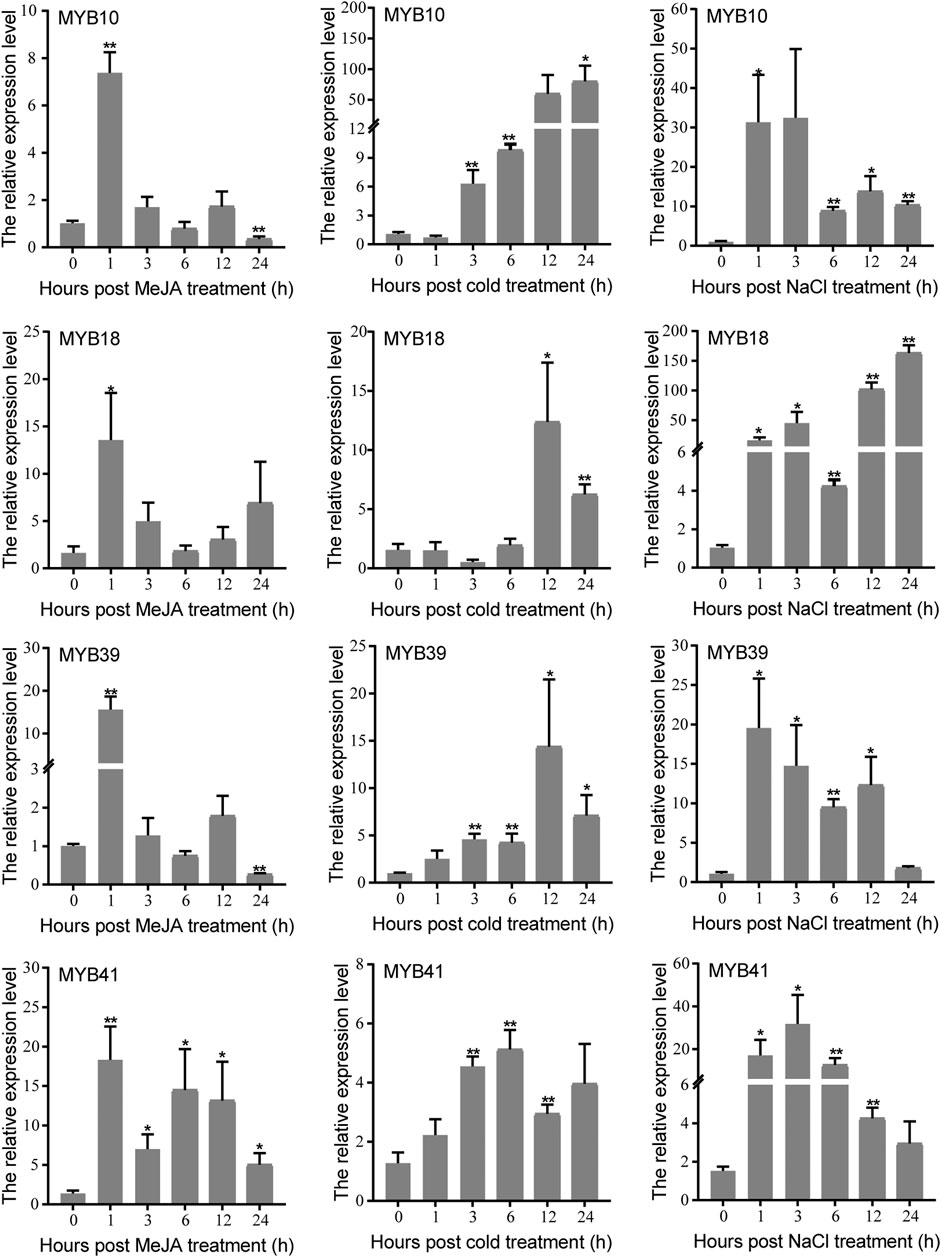
FIGURE 7. Expression levels of four candidate R2R3-type CwMYBs under cold, NaCl, and MeJA treatments. Three biological replicates were performed. Vertical bars refer to ±SE (n = 3). Asterisks indicate significant differences (*p < 0.05; **p < 0.01).
To investigate the transactivation activity of MYB TFs, the yeast one-hybrid experiment was performed. The complete and various truncated ORFs of MYB were cloned into the pGBKT7 plasmid to obtain recombinant plasmids GAL4BD-CwMYB. Then recombinant plasmids were transformed into the yeast strain AH109 respectively, to examine the transactivation ability (Figure 8). The recombinant strains GAL4BD-CwMYB18 and GAL4BD-CwMYB41 grew well and turned blue on SD-Trp/His/Ade medium with X-α-galactoside (X-α-gal), while recombinant strains GAL4BD-CwMYB10 and GAL4BD-CwMYB39 cannot develop normally. The recombinant strains harbouring truncated CwMYB10Δ361 (only with the C-terminus of CwMYB10) grew well and turned blue on SD-Trp/His/Ade medium with X-α-gal. The recombinant strains harbouring various truncated CwMYB39 remained unable to grow on SD-Trp/His/Ade medium with X-α-gal. The results confirmed the activity of transcriptional activation for complete CwMYB18, CwMYB41 and truncated CwMYB10Δ361, but not for complete CwMYB10, CwMYB39 and truncated CwMYB39.
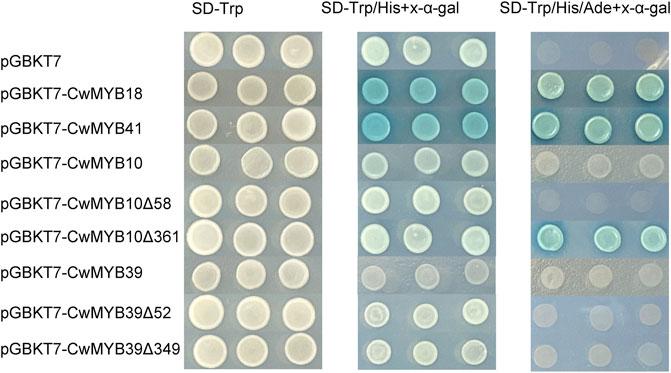
FIGURE 8. Analysis of the transactivation activity of four candidates R2R3-type CwMYBs in yeast. Recombinant plasmids were transformed into yeast strain AH109, and the transformant strains were screened by SD/−Trp, SD/−Trp/−His + X-a-gal, and SD/−Trp/−His/−Ade + Xa-gal media.
Discussion
Plants are frequently subjected to harsher environments. After long-term evolution, they have evolved highly efficient mechanisms to adapt to stress conditions. Plants TFs function as key regulators of gene expression networks, controlling various developmental and physiological processes in plants (Meraj et al., 2020; Manna et al., 2021; Romani and Moreno, 2021). MYB family is one of the largest transcription factor families in plants. The large size of MYB family reflects their importance and functional diversity in regulating plant physiological and biochemical processes. Previous studies depicted that MYBs are involved in plant development, cell shape and petal morphogenesis, cellular proliferation and differentiation, trichome development, phenylpropanoid metabolism, hormone responses, biotic and abiotic stress responses, etc (Baillo et al., 2019; Cao et al., 2020; Wang et al., 2021; Xiao et al., 2021).
Nevertheless, MYBs in C. wenyujin are entirely unknown to date. Studies on the identification and characterization of MYBs would provide insights into the understanding of CwMYB family and their response to abiotic stresses in C. wenyujin. Based on RNA-seq data, 88 CwMYBs were identified from C. wenyujin for the first time, including 43 MYB-related genes, 42 R2R3-MYB genes, two 3R-MYB genes, and one 4R-MYB genes (Table 1 and Supplementary Tables S2–S5). The identification of CwMYB superfamily members may be limited due to the lackness of C. wenyujin genome data. The genome sequencing of C. wenyujin in the future will be a significant step forward in comprehensively understanding of the CwMYB superfamily.
The domains and motifs of transcription factors are often associated with transcriptional activity, protein-protein interactions, and DNA binding (Millard et al., 2019). R2R3-MYB proteins featured a highly conserved MYB DBD at the N-terminus containing “R2” and “R3” structures. The first tryptophan (W) of “R3” was frequently replaced by phenylalanine (F), isoleucine (I), leucine (L), or methionine (M). However, the three tryptophans (W) of “R2” were highly conserved (Ambawat et al., 2013) (Figure 1). The MYB DBD of MYB-related protein was variable and randomly distributed not always located in the N-terminal region. The third tryptophan (W) of “R” structure is often replaced by specific motif, such as SHAQK (y/f)F, LKDKW(R/K) (N/T), SHLQK/MxR, or Myb_CC_LHEQLE, which is the criterion for MYB subfamily grouping (Du et al., 2013) (Figure 2). Different MYB DBDs have variations in the DNA-binding preferences, but it’s highly similar for DNA-binding specificity of MYBs with close evolutionary relationships. It suggests that other properties of proteins contribute to functional differentiation (Millard et al., 2019). Non-MYB regions are essential for explaining the highly functional diversity because of the more significant sequence diversity than MYB DBDs. Motif composition analysis of CwMYB proteins revealed differences of motifs in the non-MYB regions, indicating the functional diversity (Figures 3, 4). As we all know, protein function is closely related to its structure. Unfortunately, most of these conserved motifs are orphan and have not been linked to specific molecular functions yet (Millard et al., 2019).
The R2R3-MYB subfamily is the largest one in MYB superfamily, and the corresponding function research is also comprehensive and clear in various plants (Ambawat et al., 2013). Many studies suggest that the homologous MYB TFs in different species may play similar roles. The functional research of MYB proteins on model plants such as Arabidopsis and rice, is relatively clear. In Arabidopsis, 126 R2R3-MYBs have been classified into 23 subgroups based on the no-MYB regions of C-terminal. The members in the same subgroup play a similar regulatory role in plant growth and development processes. In subgroup 2, AtMYB14 and AtMYB15 were reported to regulate cold tolerance in Arabidopsis by affecting expression of CBF genes (Agarwal et al., 2006; Chen et al., 2013). In subgroup 20, AtMYB62, AtMYB112, AtMYB2, and AtMYB108 are involved in response to drought, salinity, high light, and Pi-starvation stresses through hormone dependent or independent mechanisms in Arabidopsis (Devaiah et al., 2009; Cui et al., 2013; Lotkowska et al., 2015; Jia et al., 2020). In subgroup 22, AtMYB44 participates in various hormone mediated responses to biotic and abiotic stress in Arabidopsis (Nguyen and Cheong, 2018; Nguyen et al., 2019). AtMYB73 is a negative regulator of salt overly sensitive (SOS) gene. Loss of AtMYB73 causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis (Kim et al., 2013). According to phylogenetic relationships of CwMYBs and AtMYBs (Figure 5), it was speculated that the homologous genes CwMYB41 in subgroup 2, CwMYB10 and CwMYB39 in subgroup 22, as well as CwMYB18 in subgroup 20 may play roles in abiotic and biotic stress responses in C. wenyujin. Besides, the significantly increased expression level of CwMYB10, CwMYB39, and CwMYB41 under MeJA stress further indicated that they may play roles as transcriptional regulators (Figure 6). A subsequent experiment revealed that CwMYB10, CwMYB18, CwMYB39, and CwMYB41 were significantly induced with different expression patterns under cold, NaCl, and MeJA treatments (Figure 7). The activity of transcriptional activation was confirmed for complete CwMYB18 and CwMYB41, while not for complete CwMYB39 and CwMYB10. Therefore, CwMYB18 and CwMYB41 proteins participated in transcriptional regulation as transcriptional activators in C. wenyujin. Furthermore, truncated CwMYB10Δ361 lacking the N-terminus exhibited transcriptional activation activity, whereas all kinds of truncated CwMYB39 did not (Figure 8). Motif analysis revealed that CwMYB39 and CwMYB10 proteins contained an EAR motif (LxLxL), which implies that this MYB may work as a repressor in gene transcription (Kagale and Rozwadowski, 2011). CwMYB39 and CwMYB10 belonging to subgroup 22, have a close evolutionary relationship with AtMYB44 and AtMYB73 that have been thoroughly studied. Complete AtMYB44 and EAR (LxLxL) motif-mutated Atmyb44 have no activity of transcriptional self-activating. AtMYB44 functions as a repressor of gene transcription in stress responses. For example, AtMYB44 interacts with TOPLESS-RELATED (TPR) corepressors to repress negative regulators PP2Cs, thus playing a positive regulator in the ABA-mediated stress responses (Nguyen and Cheong, 2018). AtMYB44 represses the transcription of drought response gene AtLEA4-5 under normal conditions, but is eliminated in response to osmotic stress (Nguyen et al., 2019). Not coincidentally, AtMyb73 acts as a negative regulator in response to salt stress by repressing SOS induction in Arabidopsis (Kim et al., 2013). Therefore, the regulation of CwMYB10 and CwMYB39 in response to abiotic stresses as the transcription repressors may rely on their modification or interaction with other cofactors.
In conclusion, the first transcriptome-wide identification and analysis of CwMYBs in C. wenyujin will serve as a foundation for future functional characterization. Following the successful construction of the C. wenyujin genetic transformation system, the future research will focus on the functional characterization of candidate regulators CwMYB10, CwMYB18, CwMYB39, and CwMYB41.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
QW designed the research, participated in all experiments and analysis, and wrote the manuscript. XY and TX participated in its design and helped to draft the manuscript. RC, TH, and SZ participated in partial data analysis. XW and YL participated in plants culture and samples collection. YL and KL performed gene expression and transcriptional activity experiments. All authors read and approved the manuscript.
Funding
The work was supported by the Natural Science Foundation of Zhejiang province (LQ19C020002), the National Natural Science Foundation of China (81903742), and the start-up research program from HZNU (2018QDL054).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thank Dr. Zheng (Wenzhou, Zhejiang province, P.R. China) for providing Curcuma wenyujin Y.H. Chen et C. Ling materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.894928/full#supplementary-material
References
Agarwal, M., Hao, Y., Kapoor, A., Dong, C.-H., Fujii, H., Zheng, X., et al. (2006). A R2R3 Type MYB Transcription Factor Is Involved in the Cold Regulation of CBF Genes and in Acquired Freezing Tolerance. J. Biol. Chem. 281 (49), 37636–37645. doi:10.1074/jbc.M605895200
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). MYB Transcription Factor Genes as Regulators for Plant Responses: an Overview. Physiol. Mol. Biol. Plants 19 (3), 307–321. doi:10.1007/s12298-013-0179-1
An, C., Sheng, L., Du, X., Wang, Y., Zhang, Y., Song, A., et al. (2019). Overexpression of CmMYB15 Provides chrysanthemum Resistance to Aphids by Regulating the Biosynthesis of Lignin. Hortic. Res. 6, 84. doi:10.1038/s41438-019-0166-y
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 10 (10), 771. doi:10.3390/genes10100771
Cai, H., Tian, S., and Dong, H. (2012). Large Scale In Silico Identification of MYB Family Genes from Wheat Expressed Sequence Tags. Mol. Biotechnol. 52 (2), 184–192. doi:10.1007/s12033-011-9486-3
Cao, Y., Li, K., Li, Y., Zhao, X., and Wang, L. (2020). MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 9 (3), 61. doi:10.3390/biology9030061
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, R., Wei, Q., Liu, Y., Wei, X., Chen, X., Yin, X., et al. (2021). Transcriptome Sequencing and Functional Characterization of New Sesquiterpene Synthases from Curcuma Wenyujin. Arch. Biochem. Biophys. 709, 108986. doi:10.1016/j.abb.2021.108986
Chen, Y., Chen, Z., Kang, J., Kang, D., Gu, H., and Qin, G. (2013). AtMYB14 Regulates Cold Tolerance in Arabidopsis. Plant Mol. Biol. Rep. 31 (1), 87–97. doi:10.1007/s11105-012-0481-z
Cui, F., Brosché, M., Sipari, N., Tang, S., and Overmyer, K. (2013). Regulation of ABA Dependent Wound Induced Spreading Cell Death by MYB 108. New Phytol. 200 (3), 634–640. doi:10.1111/nph.12456
Devaiah, B. N., Madhuvanthi, R., Karthikeyan, A. S., and Raghothama, K. G. (2009). Phosphate Starvation Responses and Gibberellic Acid Biosynthesis Are Regulated by the MYB62 Transcription Factor in Arabidopsis. Mol. Plant 2 (1), 43–58. doi:10.1093/mp/ssn081
Dong, J., Cao, L., Zhang, X., Zhang, W., Yang, T., Zhang, J., et al. (2021). An R2R3-MYB Transcription Factor RmMYB108 Responds to Chilling Stress of Rosa Multiflora and Conferred Cold Tolerance of Arabidopsis. Front. Plant Sci. 12, 696919. doi:10.3389/fpls.2021.696919
Du, H., Wang, Y.-B., Xie, Y., Liang, Z., Jiang, S.-J., Zhang, S.-S., et al. (2013). Genome-wide Identification and Evolutionary and Expression Analyses of MYB-Related Genes in Land Plants. DNA Res. 20 (5), 437–448. doi:10.1093/dnares/dst021
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi:10.1016/j.tplants.2010.06.005
Fang, Q., Wang, Q., Mao, H., Xu, J., Wang, Y., Hu, H., et al. (2018). AtDIV2, an R-R-type MYB Transcription Factor of Arabidopsis, Negatively Regulates Salt Stress by Modulating ABA Signaling. Plant Cel Rep 37 (11), 1499–1511. doi:10.1007/s00299-018-2321-6
Gu, B.-J., Tong, Y.-K., Wang, Y.-Y., Zhang, M.-L., Ma, G.-J., Wu, X.-Q., et al. (2022). Genome-wide Evolution and Expression Analysis of the MYB-CC Gene Family in Brassica Spp. PeerJ 10, e12882. doi:10.7717/peerj.12882
Jia, T., Zhang, K., Li, F., Huang, Y., Fan, M., and Huang, T. (2020). The AtMYB2 Inhibits the Formation of Axillary Meristem in Arabidopsis by Repressing RAX1 Gene under Environmental Stresses. Plant Cel Rep 39 (12), 1755–1765. doi:10.1007/s00299-020-02602-3
Jiang, C., Gu, J., Chopra, S., Gu, X., and Peterson, T. (2004). Ordered Origin of the Typical Two- and Three-Repeat Myb Genes. Gene 326, 13–22. doi:10.1016/j.gene.2003.09.049
Kagale, S., and Rozwadowski, K. (2011). EAR Motif-Mediated Transcriptional Repression in Plants. Epigenetics 6 (2), 141–146. doi:10.4161/epi.6.2.13627
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide Classification and Expression Analysis of MYB Transcription Factor Families in rice and Arabidopsis. BMC Genomics 13, 544. doi:10.1186/1471-2164-13-544
Khaksar, G., and Sirikantaramas, S. (2021). Transcriptome-wide Identification and Expression Profiling of the ERF Gene Family Suggest Roles as Transcriptional Activators and Repressors of Fruit Ripening in Durian. PLoS One 16 (8), e0252367. doi:10.1371/journal.pone.0252367
Kim, J. H., Nguyen, N. H., Jeong, C. Y., Nguyen, N. T., Hong, S.-W., and Lee, H. (2013). Loss of the R2R3 MYB, AtMyb73, Causes Hyper-Induction of the SOS1 and SOS3 Genes in Response to High Salinity in Arabidopsis. J. Plant Physiol. 170 (16), 1461–1465. doi:10.1016/j.jplph.2013.05.011
Ku, Y.-S., Ni, M., Muñoz, N. B., Xiao, Z., Lo, A. W.-Y., Chen, P., et al. (2020). ABAS1 from Soybean Is a 1R-Subtype MYB Transcriptional Repressor that Enhances ABA Sensitivity. J. Exp. Bot. 71 (10), 2970–2981. doi:10.1093/jxb/eraa081
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23 (21), 2947–2948. doi:10.1093/bioinformatics/btm404
Li, B., Fan, R., Guo, S., Wang, P., Zhu, X., Fan, Y., et al. (2019). The Arabidopsis MYB Transcription Factor, MYB111 Modulates Salt Responses by Regulating Flavonoid Biosynthesis. Environ. Exp. Bot. 166, 103807. doi:10.1016/j.envexpbot.2019.103807
Li, X., Guo, C., Ahmad, S., Wang, Q., Yu, J., Liu, C., et al. (2019). Systematic Analysis of MYB Family Genes in Potato and Their Multiple Roles in Development and Stress Responses. Biomolecules 9 (8), 317. doi:10.3390/biom9080317
Li, Y., Wu, Y., Li, Y., and Guo, F. (2021). Review of the traditional uses, phytochemistry, and pharmacology of Curcuma wenyujin Y. H. Chen et C. Ling. J. Ethnopharmacology 269, 113689. doi:10.1016/j.jep.2020.113689
Liu, J., Mehari, T. G., Xu, Y., Umer, M. J., Hou, Y., Wang, Y., et al. (2021). GhGLK1 a Key Candidate Gene from GARP Family Enhances Cold and Drought Stress Tolerance in Cotton. Front. Plant Sci. 12, 759312. doi:10.3389/fpls.2021.759312
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lotkowska, M. E., Tohge, T., Fernie, A. R., Xue, G.-P., Balazadeh, S., and Mueller-Roeber, B. (2015). The Arabidopsis Transcription Factor MYB112 Promotes Anthocyanin Formation during Salinity and under High Light Stress. Plant Physiol. 169 (3), 1862–1880. doi:10.1104/pp.15.00605
Manna, M., Thakur, T., Chirom, O., Mandlik, R., Deshmukh, R., and Salvi, P. (2021). Transcription Factors as Key Molecular Target to Strengthen the Drought Stress Tolerance in Plants. Physiologia Plantarum 172 (2), 847–868. doi:10.1111/ppl.13268
Meraj, T. A., Fu, J., Raza, M. A., Zhu, C., Shen, Q., Xu, D., et al. (2020). Transcriptional Factors Regulate Plant Stress Responses through Mediating Secondary Metabolism. Genes 11 (4), 346. doi:10.3390/genes11040346
Millard, P. S., Kragelund, B. B., and Burow, M. (2019). R2R3 MYB Transcription Factors - Functions outside the DNA-Binding Domain. Trends Plant Sci. 24 (10), 934–946. doi:10.1016/j.tplants.2019.07.003
Nguyen, N. H., and Cheong, J.-J. (2018). AtMYB44 Interacts with TOPLESS-RELATED Corepressors to Suppress Protein Phosphatase 2C Gene Transcription. Biochem. Biophysical Res. Commun. 507 (1-4), 437–442. doi:10.1016/j.bbrc.2018.11.057
Nguyen, N. H., Nguyen, C. T. T., Jung, C., and Cheong, J.-J. (2019). AtMYB44 Suppresses Transcription of the Late Embryogenesis Abundant Protein Gene AtLEA4-5. Biochem. Biophysical Res. Commun. 511 (4), 931–934. doi:10.1016/j.bbrc.2019.03.006
Ren, D., Rao, Y., Yu, H., Xu, Q., Cui, Y., Xia, S., et al. (2020). MORE FLORET1 Encodes a MYB Transcription Factor that Regulates Spikelet Development in Rice. Plant Physiol. 184 (1), 251–265. doi:10.1104/pp.20.00658
Romani, F., and Moreno, J. E. (2021). Molecular Mechanisms Involved in Functional Macroevolution of Plant Transcription Factors. New Phytol. 230 (4), 1345–1353. doi:10.1111/nph.17161
Safi, A., Medici, A., Szponarski, W., Ruffel, S., Lacombe, B., and Krouk, G. (2017). The World According to GARP Transcription Factors. Curr. Opin. Plant Biol. 39, 159–167. doi:10.1016/j.pbi.2017.07.006
Upadhyaya, G., Das, A., and Ray, S. (2021). A rice R2R3‐MYB ( OsC1 ) Transcriptional Regulator Improves Oxidative Stress Tolerance by Modulating Anthocyanin Biosynthesis. Physiologia Plantarum 173 (4), 2334–2349. doi:10.1111/ppl.13583
Wang, F., Han, T., Song, Q., Ye, W., Song, X., Chu, J., et al. (2020). The rice Circadian Clock Regulates Tiller Growth and Panicle Development through Strigolactone Signaling and Sugar Sensing. Plant Cell 32 (10), 3124–3138. doi:10.1105/tpc.20.00289
Wang, X., Niu, Y., and Zheng, Y. (2021). Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Ijms 22 (11), 6125. doi:10.3390/ijms22116125
Wu, Y., Li, T., Cheng, Z., Zhao, D., and Tao, J. (2021). R2R3-MYB Transcription Factor PlMYB108 Confers Drought Tolerance in Herbaceous Peony (Paeonia Lactiflora Pall.). Ijms 22 (21), 11884. doi:10.3390/ijms222111884
Xiao, R., Zhang, C., Guo, X., Li, H., and Lu, H. (2021). MYB Transcription Factors and its Regulation in Secondary Cell wall Formation and Lignin Biosynthesis during Xylem Development. Ijms 22 (7), 3560. doi:10.3390/ijms22073560
Yan, Y., Li, C., Dong, X., Li, H., Zhang, D., Zhou, Y., et al. (2020). MYB30 Is a Key Negative Regulator of Arabidopsis Photomorphogenic Development that Promotes PIF4 and PIF5 Protein Accumulation in the Light. Plant Cell 32 (7), 2196–2215. doi:10.1105/tpc.19.00645
Zhai, B., Zhang, N., Han, X., Li, Q., Zhang, M., Chen, X., et al. (2019). Molecular Targets of β-elemene, a Herbal Extract Used in Traditional Chinese Medicine, and its Potential Role in Cancer Therapy: A Review. Biomed. Pharmacother. 114, 108812. doi:10.1016/j.biopha.2019.108812
Zhang, G., Li, G., Xiang, Y., and Zhang, A. (2022). The Transcription Factor ZmMYB-CC10 Improves Drought Tolerance by Activating ZmAPX4 Expression in maize. Biochem. Biophysical Res. Commun. 604, 1–7. doi:10.1016/j.bbrc.2022.02.051
Keywords: MYB family, phylogeny analysis, expression pattern, transcriptional activation, C. wenyujin
Citation: Wei Q, Liu Y, Lan K, Wei X, Hu T, Chen R, Zhao S, Yin X and Xie T (2022) Identification and Analysis of MYB Gene Family for Discovering Potential Regulators Responding to Abiotic Stresses in Curcuma wenyujin. Front. Genet. 13:894928. doi: 10.3389/fgene.2022.894928
Received: 12 March 2022; Accepted: 08 April 2022;
Published: 25 April 2022.
Edited by:
Muhammad Abdullah, Shanghai Jiao Tong University, ChinaCopyright © 2022 Wei, Liu, Lan, Wei, Hu, Chen, Zhao, Yin and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiuhui Wei, d3FoXzI2OEAxNjMuY29t; Xiaopu Yin, eWlueHBAaHpudS5lZHUuY24=; Tian Xie, eGJzQGh6bnUuZWR1LmNu
 Qiuhui Wei
Qiuhui Wei Yuyang Liu1,2
Yuyang Liu1,2 Tianyuan Hu
Tianyuan Hu Rong Chen
Rong Chen Xiaopu Yin
Xiaopu Yin Tian Xie
Tian Xie