- 1Key Laboratory of Grassland Resources (IMAU), Key Laboratory of Forage Cultivation, Processing and High Efficient Utilization, College of Grassland, Resource and Environmental Science, Ministry of Education, Ministry of Agriculture, Inner Mongolia Agricultural University, Hohhot, China
- 2Institute of Animal Science, The Chinese Academy of Agricultural Sciences, Beijing, China
Plants of the Agropyron genus are important pasture resources, and they also play important roles in the ecological restoration. Chloroplast genomes are inherited from maternal parents, and they are important for studying species taxonomy and evolution. In this study, we sequenced the complete chloroplast genomes of five typical species of the Agropyron genus (eg., A. cristatum × A. desertorum Fisch. Schult, A. desertorum, A. desertorum Fisch. Schult. cv. Nordan, A. michnoi Roshev, and A. mongolicum Keng) using the Illumina NovaSeq platform. We found that these five chloroplast genomes exhibit a typical quadripartite structure with a conserved genome arrangement and structure. Their chloroplast genomes contain the large single-copy regions (LSC, 79,613 bp-79,634 bp), the small single-copy regions (SSC, 12,760 bp-12,768 bp), and the inverted repeat regions (IR, 43,060 bp-43,090 bp). Each of the five chloroplast genomes contains 129 genes, including 38 tRNA genes, eight rRNA genes, and 83 protein-coding genes. Among them, the genes trnG-GCC, matK, petL, ccsA, and rpl32 showed significant nucleotide diversity in these five species, and they may be used as molecular markers in taxonomic studies. Phylogenetic analysis showed that A. mongolicum is closely related to A. michnoi, while others have a closer genetic relationship with the Triticum genus.
Introduction
The Agropyron genus contains many important wild species, which is a close relative of the Triticum genus. Different species of the Agropyron genus have distinct chromosome ploidies, including diploid, PP, 2n = 2x = 14; tetraploid, PPPP, 2n = 4x = 28; and hexaploid, PPPPPP, 2n = 6x = 42 (Dewey, 1984; Fordlloyd et al., 2011). Most species of the Agropyron genus are excellent sources of forage and habitat for livestock and wildlife. They are also valuable for weed control, habitat use, soil stabilization, and watershed management (Wang, 2011). Species of Agropyron also possess a lot of valuable traits, such as tolerance to drought and cold, resistance to diseases, and high yield (Asay et al., 1985; Dong et al., 1992; Lu et al., 2015). They are high-quality forage for grassland improvement and valuable genetic resources for wheat improvement (Ochoa et al., 2015; Sun et al., 2021).
The Agropyron genus contains several major species, namely, A. cristatum, A. mongolicum, A. desertorum, A. michnoi, and A. cristatum × A. desertorum Fisch. Schult. Among them, A. cristatum is originated from Iran and distributed in arid, semi-arid, and alpine regions (Dewey, 1984). A. mongolicum, a unique species in China, is mainly distributed in Shaanxi, Inner Mongolia, Gansu, Ningxia, Shanxi, and other areas of China (Guo, 1987). Both A. cristatum and A. mongolicum have a diploidy group, but these two species are distinct in general morphology (Dewey, 1984; Dewey,1981). In traditional taxonomy, the species of Agropyron is mainly recognized by the spike morphology. A. mongolicum differs from A. cristatum by its narrow, linear spikes. All other diploid accessions within the Agropyron genus are similar to A. cristatum with broad spikes. These two species could hybridize easily, and the F1 hybrids displayed better characteristics (Dewey, 1984; Hsiao et al., 1986). Among the species of the Agropyron genus distributed in China, A. cristatum and A. mongolicum are genetically differentiated species, while A. desertorum is the intermediate one. A. desertorum, A. mongolicum, and A. michnoi are the offspring species of A. cristatum that share the same basic genome from the counterpart based on SSR analysis (Che et al., 2015). A. cristatum × A. desertorum Fisch. Schult. is a distant hybrid of the induced tetraploid A. cristatum and the natural tetraploid A. desertorum (Dewey and Pendse, 1968; Asay and Johnson, 1990).
Chloroplast is a vital organelle in plants, which plays an important role in photosynthesis. Chloroplast is involved in plant physiology and development, the biosynthesis of amino acids, nucleotides, fatty acids, phytohormones, vitamins, and the assimilation of sulfur and nitrogen (Daniell et al., 2016a). The chloroplast genome contains genes with functions in many processes of photosynthesis and other metabolism. The chloroplast genome possesses highly conserved DNA sequences at a low substitution level (especially in inverted repeat regions). Therefore, the chloroplast genome is ideal for phylogenetic inference at species and higher orders. The chloroplast genome makes a large contribution to the search of phylogeny in the plant family and solves the group evolution relationship from phylogeny (Daniell et al., 2016a). Similarly, it is essential to study the chloroplast genome of the Agropyron genus for a better understanding on its classification, phylogenetic relationship, and variation among species.
In this study, in order to provide comprehensive insights into the evolution of the chloroplast genomes of the Agropyron genus, we sequenced the chloroplast genomes of five Agropyron species. In addition, we conducted comparative analyses on the chloroplast genome of five Agropyron species and eleven other related species. Therefore, our study provides valuable information for the phylogenetic relationship and species identification within the Agropyron genus.
Materials and Methods
Sampling, Chloroplast DNA Extraction, and Sequencing
Agropyron plants were grown at the pasture experimental station of the Inner Mongolia Agricultural University, located in Hohhot, Inner Mongolia Autonomous Region. The experimental station had a typical temperate continental climate with precipitation and heat in the same season. The average annual temperature was 8°C, and the frost-free period was 130–140 d. The soil was sandy loam, with medium fertility and a pH of 7.0–7.5.
Fresh leaves were collected and snap-frozen in liquid nitrogen and then stored at−80°C until DNA extraction. DNA extraction was performed using the modified CTAB method (Doyle, 1987). DNA quality was assessed using a nanodrop spectrophotometer. Genomic DNA samples were mechanically fragmented, and then, fragments were detected by agarose gel electrophoresis. The library for sequencing was amplified by PCR, and sequencing was performed using an Illumina NovaSeq platform with PE150 based on sequencing by synthesis (SBS).
Chloroplast Genome Annotation
Fastp (version 0.20.0, https://github.com/OpenGene/fastp) software was used to filter the raw data by removing the joint sequences and low-quality reads. High-quality clean data were assembled according to chloroplast genome sequences of reference species to obtain chloroplast sequence assembly results (Langmead and Salzberg, 2012). Two methods were used to annotate chloroplast genomes to improve the accuracy. First, the Prodigal v2.6.3 was used for annotation of chloroplast CDS (https://www.github.com/hyattpd/Prodigal), and the hmmer v3.1 b2 (http://www.hmmer.org/) software and aragorn v1.2.38 (http://130.235.244.92/ARAGORN/) were used to predict rRNA and tRNA. Second, based on the gene sequences of available related species in the NCBI, gene sequences of Agropyron were extracted, and then, BLAST v2.6 (https://blast.ncbi.nlm.nih.gov/Blast. cgi) was used to obtain the second round annotation. Then, different genes from the two annotation results were manually checked, the wrong annotations and redundant annotations were removed, and the multi-exon boundary and the final annotation were accomplished. The final step was carried out by Gene Pioneer Biotechnologies (Nanjing, China).
Analysis of the Ka/Ks Ratio
The Ka/Ks ratio is a reflection of selection pressures. Mafft v7.310 (https://mafft.cbrc jp/alignment/software/) was used for the sequence alignment, and the software KaKs Calculator v2.0 (https://sourceforge.net/projects/kakscalculator2/) was used to calculate Ka and Ks values (Zhang et al., 2006).
Analyses of Dispersed Repeats and SSR
Vmatch V2.3.0 (http://www.vmatch.de/) software was used in combination with Perl scripts to identify duplicate sequences. The parameters were set as: minimum length = 30 bp hamming distance = 3, and the identification was in four forms: forward, palindromic, reverse, and complement. Simple sequence repeats (SSRs) are a class of markers containing several nucleotides (usually one–six) as repeats. SSR markers in chloroplast genomes are called cpSSR markers. We used the software MISA v1.0 (MIcroSAtellite identification tool, http://pgrc.ipk-gatersleben.de/misa/misa.html) for cpSSR analysis.
Analysis of Sequence Variation
Mafft software (Auto mode) was used for global alignment of homologous gene sequences in different species. MEGA X (Kumar et al., 2008) was used to calculate the level of SNV (single nucleotide variation) and the average genetic distance of chloroplast genome sequences. DnaSP v6 (Julio) was used to determine the nucleotide diversity with parameter settings as follows: a step length of 200 bp and window length of 800 bp (Rozas et al., 2003).
Comparison of the IR/SC Boundary
SC/IR boundary data were calculated based on the annotated information of the chloroplast genomes of five species of Agropyron, and the simple chloroplast genome structure diagrams of five species were drawn according to the statistical results (Daniell et al., 2016b). The boundary information of the four regions of chloroplast genomes of the Agropyron was compared, and the contraction and expansion of IR regions were observed.
Analysis on the Phylogenetic Relationship
A genome-wide evolutionary tree was constructed. The ring sequence was analyzed with the same starting point, and the full-length sequences were used to carry out the multiple sequence alignment with MAFFT software (v7.427, - auto mode), followed by correction with trim Al (v1.4. Rev15). By using the software RAxML v8.2.10 (https://cme.h-its.org/exelixis/software.html) and choosing the GTRGAMMA model with a rapid bootstrap analysis, a maximum likelihood evolutionary tree was constructed with a bootstrap of 1,000 (Katoh et al., 2005).
Results
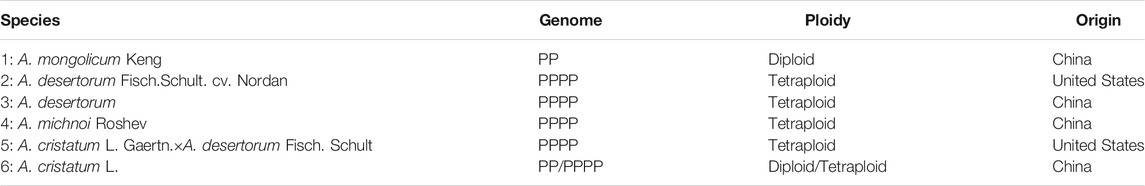
Phenotypic Identification of the Six Agropyron Species
Among all species investigated in this study, A. mongolicum Keng, A. desertorum, A. michnoi, and A. cristatum are widely distributed in China (Table 1). A. cristatum × A. desertorum Fisch. Schult and A. desertorum Fisch. Schult. have been used in the construction of artificial grasslands in northern China. The angles between spikelets and cobs are different among the six species, with the angle gradually increased in an order of A. mongolicum, A. desertorum Fisch. Schult, A. desertorum, A. michnoi, A. cristatum× A. desertorum Fisch. Schult, and A. cristatum (Figure 1). The smallest angle is found in A. mongolicum, whereas the largest angle, which is close to right angle, is found in A. cristatum. The spikelet of A. mongolicum is long, thin, and sparsely arranged. The spikelets of A. michnoi and A. cristatum are short. The spikelet of A. cristatum× A. desertorum Fisch. Schult. is the biggest (Figure 1). Although they showed different phenotypes, the phenological periods of the six Agropyron species are basically the same, and they could be crossed among each other in nature, except for A. mongolicum.

FIGURE 1. Phenotype of the spikelet of six Agropyron species as numbered in Table 1.
Features of the Chloroplast Genomes in Agropyron
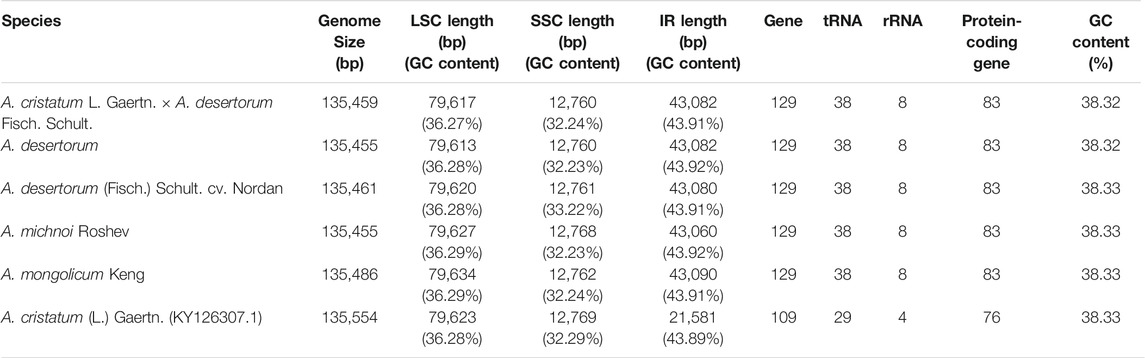
The sizes of the chloroplast genome of A. cristatum× A. desertorum Fisch. Schult, A. desertorum, A. desertorum Fisch. Schult, A. michnoi, and A. mongolicum are 135,459 bp, 135,455 bp, 135,461 bp, 135,455 bp, and 135,486 bp in length, respectively. We also compared them with the available chloroplast genome of A. cristatum (KY126307.1) (Chen et al., 2018), which is 135,554 bp in length. Our results showed that the lengths of these five Agropyron species are similar.
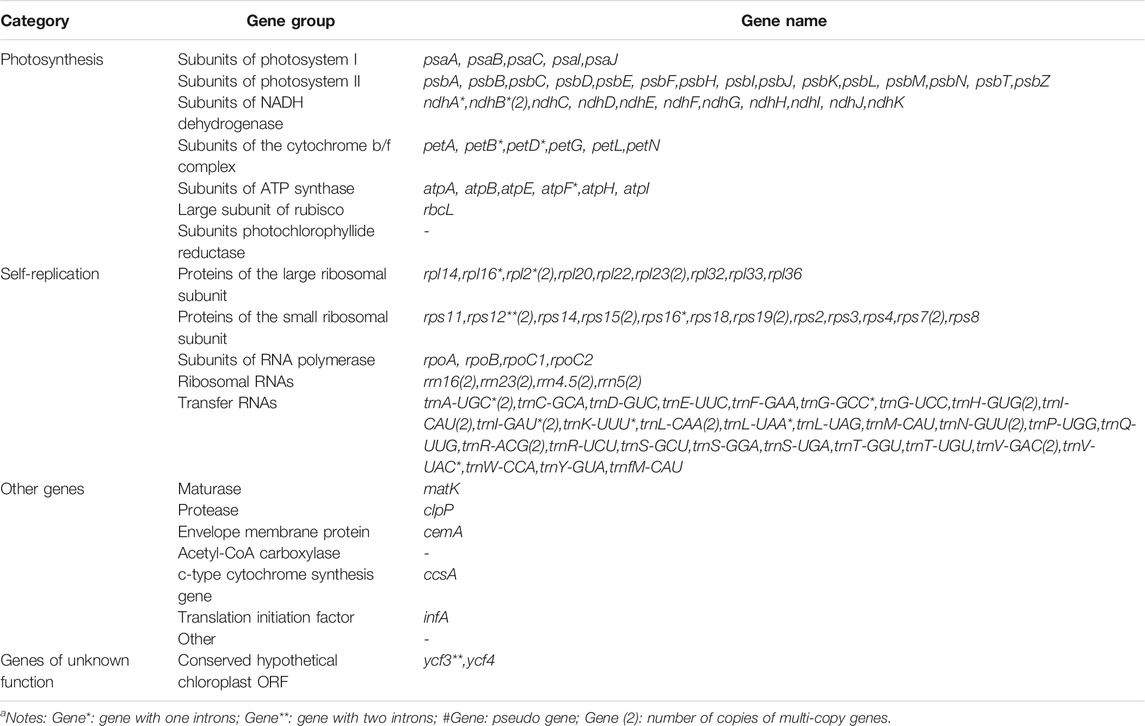
Each of the five genomes contains 129 genes, including 38 tRNA genes, eight rRNA genes, and 83 protein-coding genes. As a comparison, the gene number of A. cristatum chloroplast genome (KY126307.1) is different from other Agropyron species, which is short and contains only 109 genes, including 29 tRNA genes, four rRNA genes, and 76 protein-coding genes (Chen et al., 2018) (Tables 2, 3). The GC contents of A. cristatum × A. desertorum Fisch. Schult, and A. desertorum are exactly the same (38.32%), and the GC contents of the other four Agropyron species are the same (Table 2, Supplementary Tables S1–S5).
The complete chloroplast genomes of the Agropyron species exhibit a typical single-circular molecule with a four-segmented structure (Figure 2), consisting of a large single-copy region (LSC), a small single-copy region (SSC), and the inverted repeat regions a and b (IRa and IRb), which is similar to many other plant species (Pasquale et al., 2015; Qiu et al., 2019). In every Agropyron chloroplast genome, the IR region contains eight rRNA genes, 16 tRNA genes, and 17 protein-coding genes; the SSC region contains one tRNA gene and 10 protein-coding genes, while the LSC region contains 21 tRNA genes and 56 protein-coding genes. Though the GC contents of the complete chloroplast genome are similar in different Agropyron species (38.32–38.33%, Table 2), the GC contents in IR regions (43.89–43.92%) are significantly higher than those in the SSC region (36.27–36.29%) and LSC region (32.23–33.22%), and this pattern is consistent with these five Agropyron genomes (Table 2).
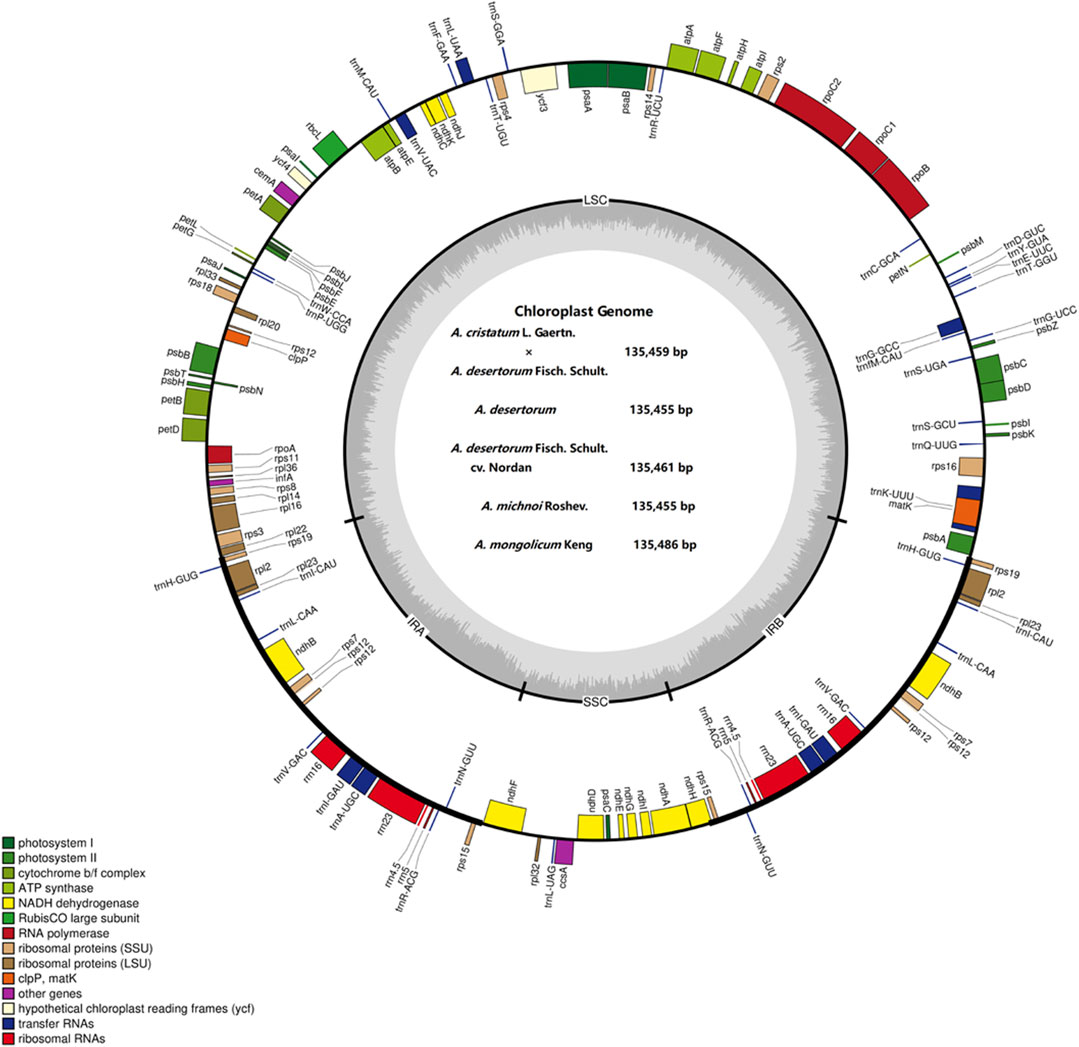
FIGURE 2. Representative map of the Agropyron chloroplast genome. The plus encoding gene is on the outside of the circle, and the minus encoding gene is on the inside of the circle. The inner gray circle represents the GC content.
Analysis of the Ka/Ks Ratio
Nucleotide substitutions may or may not lead to changes in amino acids. If the substitution does not cause an amino acid change, then it is a synonymous mutation, but if the substitution does cause an amino acid change, it is a non-synonymous mutation. Non-synonymous mutations in general are the forces of natural selection. The ratio of the non-synonymous mutation rate (Ka) to synonymous mutation rate (Ks) is used to determine the selection outcomes. If the ratio is greater than 1, it indicates a positive selection. If the ratio is less than 1, it indicates a purifying selection. Therefore, we analyzed the Ka and Ks values for each genes of the chloroplast genome and compared the Ka/Ks ratios among these six species of Agropyron (Supplementary Tables S6–15). It showed that these species do not have obvious nucleotide changes for almost all the genes without available Ka/Ks values (Supplementary Tables S6–15). However, the Ka/Ks values for the ropC2 gene in A. michnoi and the ccsA gene in A. mongolicum were relatively high when compared with other species (Figure 3, Supplementary Tables S6–12), indicating these two genes contained obvious mutations during evolution.
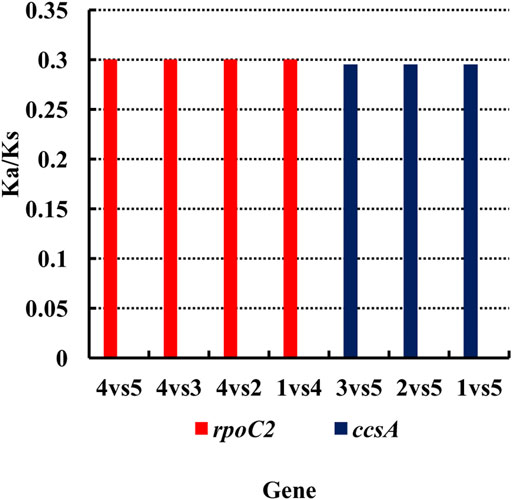
FIGURE 3. Analysis of the Ka/Ks ratio of Agropyron. X-axis: name of the gene. Y-axis: value of Ka/Ks. 1–5: A. cristatum × A. desertorum Fisch. Schult, A. desertorum, A. desertorum Fisch. Schult, A. michnoi, and A. mongolicum. The red bars indicate the rpoC2 gene, and the blue bars indicate the ccsA gene.
Analysis of Dispersed Repeats and SSR
The dispersed repeats of Agropyron are similar except for A. mongolicum and A. cristatum, in which the forward repeats of 30–35 bp are different from that in other Agropyron chloroplast genomes. Different from other species, chloroplast genomes of Agropyron have no reverse or complementary repeats (Figure 4). There are more forward repeats than palindromic repeats in the chloroplast genomes of Agropyron.
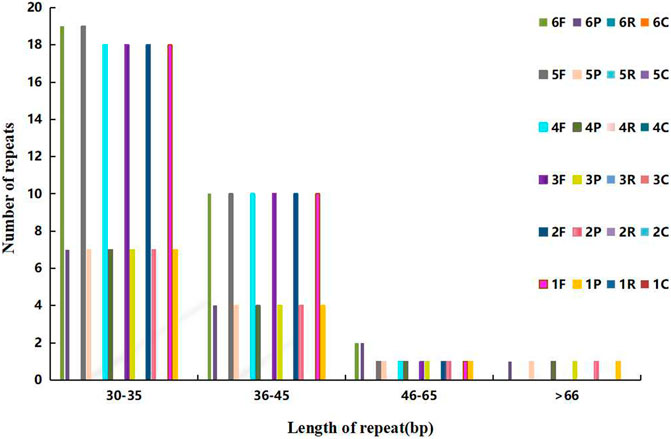
FIGURE 4. Distributions of the dispersed repeat. The x-coordinate is the length of dispersed repeats, and the y-coordinate is the number of dispersed repeats. 1–6: A. cristatum× A. desertorum Fisch. Schult, A. desertorum, A. desertorum Fisch. Schult, A. michnoi, A. mongolicum, and A. cristatum. F stands for forward repeats, P for palindromic repeats, R for reverse repeats, and C for complementary repeats.
Simple sequence repeats (SSRs) consist of six or less than six base repeats and distribute widely in chloroplast genomes, and mono-, di-, tri-, tetra-, and penta-nucleotide repeats are found in five species of Agropyron. In total, 171, 171, 170, 169, 173, and 169 SSRs were found in A. cristatum × A. desertorum Fisch. Schult, A. desertorum, A. desertorum Fisch. Schult, A. michnoi, A. mongolicum, and A. cristatum, respectively (Figure 5). The number of mononucleotide repeats is the largest, similar as observed in other plant species. The mono-nucleotide and tetra-nucleotide SSRs in A. mongolicum are slightly higher than other species. Penta-nucleotide SSRs are only found in A. cristatum × A. desertorum Fisch. Schult. and A. desertorum (Figure 5). Most mono-nucleotide SSRs are polyadenine (poly A) or polythymine (poly T), accounting for about 95%. The result also shows that A. cristatum × A. desertorum Fisch. Schult. is the closest species to A. desertorum from the quantity and length of repeated sequences.
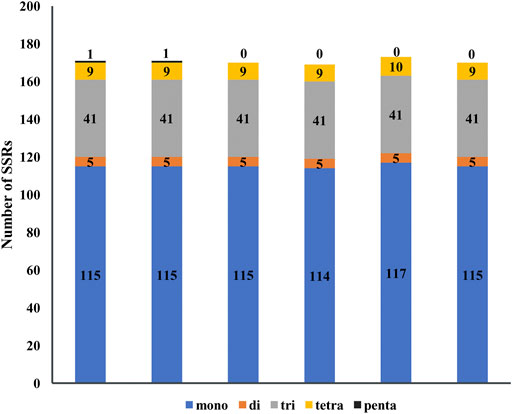
FIGURE 5. Analysis of simple sequence repeats in the chloroplast genome of six Agropyron species. Numbers indicate SSR types detected in each species. X-axis: name of the species. Y-axis: number of SSR.
Analysis on Sequence Variation
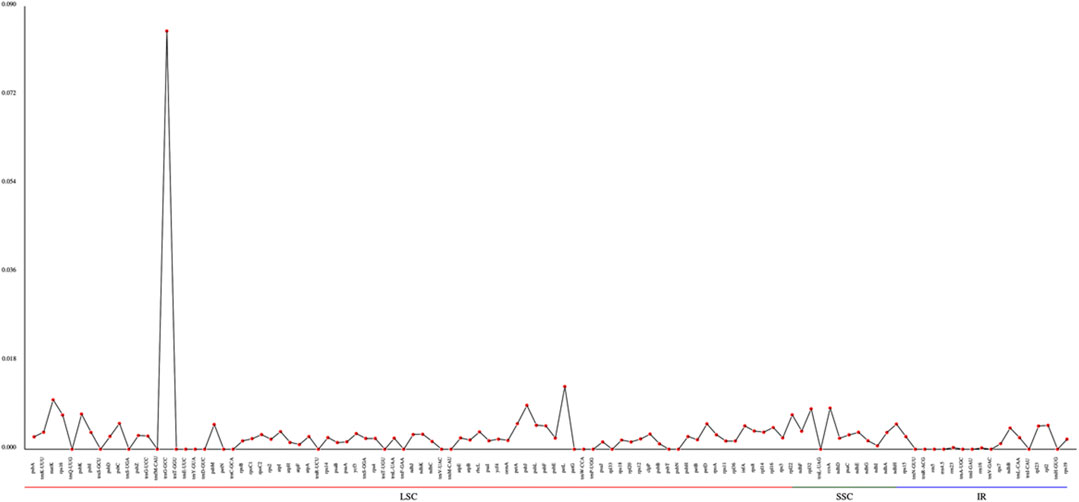
The Pi values (nucleotide diversity) can reveal the variation size of the nucleic acid sequence in different species, and regions with high degree of variations can be used as potential molecular markers for population genetics. Using the Mafft software, we performed global alignment of homologous genes in different species. Our results showed that the trnG-GCC gene in the LSC region has the largest diversity, with the maximum value of 0.0849. The differences of nucleotide diversity in other regions are not obvious. The nucleotide diversity in LSCs and SSCs is substantially higher than that in IRs (Figure 6, Supplementary Table S17).
We compared the chloroplast genome of A. cristatum × A. desertorum Fisch. Schult. with other nine species, using the software CGVIEW (https://stothard.afns.ualberta.ca/cgviewserver). The default parameters were used for the comparative analysis of chloroplast genome structures for proximal species. Our results showed that the structures of the species of Agropyron are similar (Figure 7).
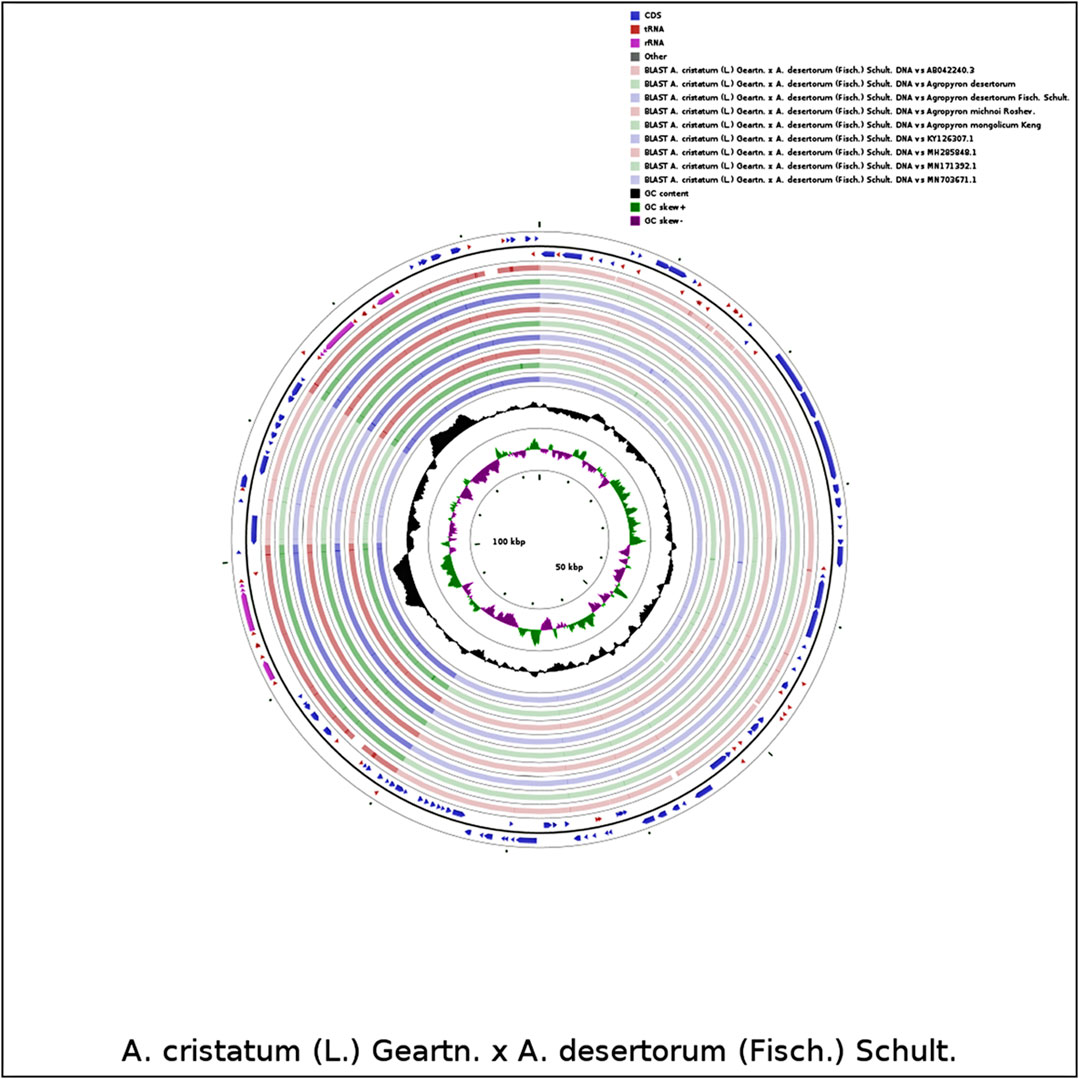
FIGURE 7. Comparative analysis of chloroplast genome structures. The two outermost circles describe the length and direction of genes in the genome; the circles inside represent similar results compared with other reference genomes; the black circles represent the GC content.
Analysis of the IR/SC Boundary
The chloroplast genomic DNA of Agropyron is a two-link molecule, and its size, structure, and gene capacity are highly conserved within four regions (LSC, SSC, IRa, and IRb). We compared the conserved four regions of ten species from the tribe Triticeae, including wheat, barley, and other eight species from the Agropyron genus (Figure 8). In comparison, the chloroplast genome of wheat is slightly smaller than that of Agropyron, whereas the chloroplast genome of barley is larger than that of Agropyron (Figure 8). In both wheat and barley, the LSC region is longer than that of Agropyron. The IR regions of the wheat chloroplast genome are shorter than those of the other nine species. The gene content and arrangement in the 10 species are the same in the IR region, which contain the rps19 and rps15 genes. In Figure 8, the rps19 gene is located in JLB (junction of LSC/IR) at 39 bp to 57 bp, and rps15 has 313 bp to 362 bp extending to JSB (junction of SSC/IRb). The ndhH gene spans the JSA (junction of SSC/IRA) region, and its length also reflects changes in the JSA region. The length distribution in six Agropyron species is the same. The ndhF gene is deviated from the JSB (IRB/SSC) region in the species studied, ranging from 39 bp in six species to 89 bp in A. cristatum (135,489 bp). The rpl22 gene is deviated from the JLB (IRB/LSC) region, ranging from 33 bp to 42 bp.
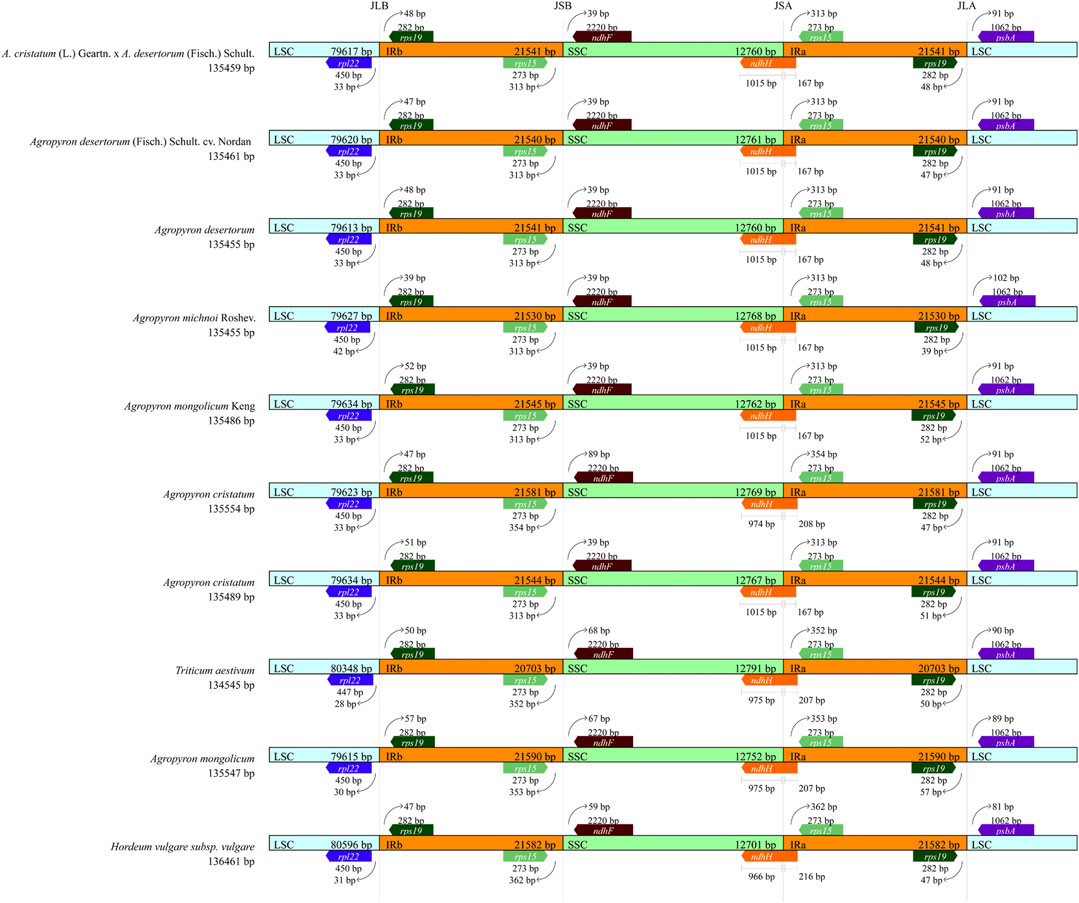
FIGURE 8. Analysis of IR boundary changes of chloroplast genomes. The thin lines represent the connection points of each region, and the map shows information about the genes near the connection points.
Phylogenetic Analysis
We selected 16 species in different genera from Triticeae and generated a phylogenetic tree, including nine species from Agropyron, two from Elymus, two from Triticum, one from Roegneria C. Koch, one from Hordeum Linn, and one from Thinopyrum (Figure 9). It was clear that these species were divided into two clades: the Hordeum genus is clustered in one clade, and all others are clustered in another clade (Figure 9). The species of the Agropyron genus were grouped in one subclade, while species from Elymus, Pseudoroegneria, Thinopyrum, and Triticum are in the other subclade. A. mongolicun Keng is close to A. michnoi, which is consistent with the structure and the SSR of these two species. A. desertorum Fisch. Schult, A. desertorum, and A. cristatum × A. desertorum Fisch. Schult. are in the same group (Figure 9). A. desertorum is more close to A. cristatum × A. desertorum Fisch. Schult, because the latter is a hybrid generated by crossing A. desertorum with A. cristatum.
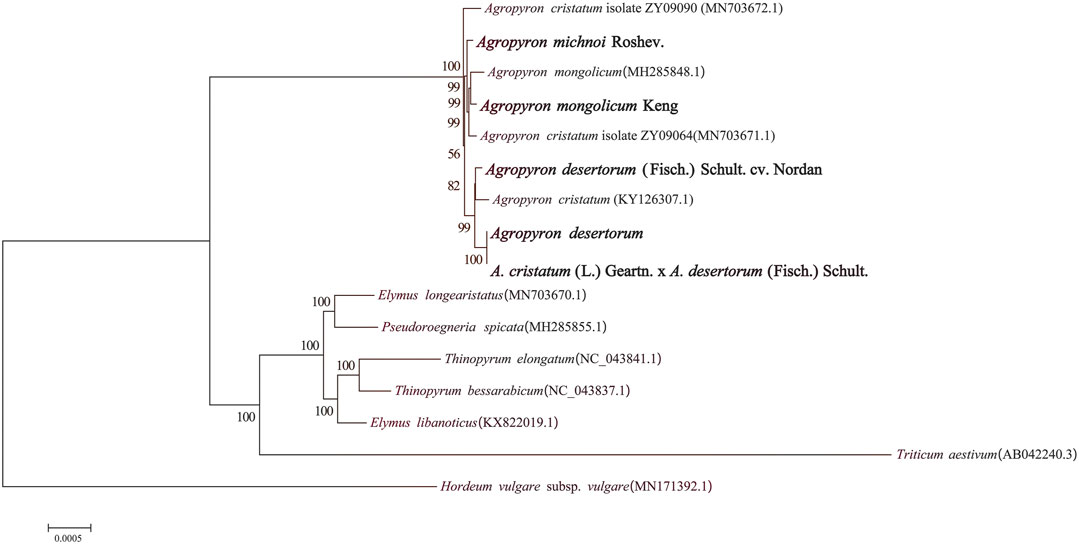
FIGURE 9. Phylogenetic tree of five species of Agropyron and related taxa based on the complete chloroplast genomes.
Discussion
Characteristics of the Chloroplast Genomes of Agropyron Species
The objective of this study is to investigate the relationships among Agropyron species using chloroplast genome sequences. Similar to other plant species, the chloroplast genomes of Agropyron species show a four-segmented structure, consisting of a large single-copy region, a small single-copy region, and inverted repeat regions. Agropyron is closely related to Triticum genus, and its chloroplast genome features are similar to that of Triticum as well (Middleton et al., 2014). The sizes of Agropyron chloroplast genomes are comparable to previously reported complete chloroplast genomes of many angiosperms (Qian et al., 2013). Generally, the chloroplast genome size of angiosperms ranges from 72 to 217 kb. Although A. cristatum is close to A. desertorum Fisch. Schult. in the phylogenetic relationship, their gene numbers are different.
Generally, the common repeat type is the mono-nucleotide repeat, followed by the di-, tri-, and tetra-repeats, but penta-nucleotide repeats are not common. SSRs detected in chloroplast genomes of Agropyron are highly conserved in types and numbers, and most mono-nucleotide repeats in chloroplast genomes of Agropyron comprise base A or T, which is similar as reported in other plant species (Li et al., 2019). The SSRs are ideal genetic markers in plant molecular studies (Khan et al., 2019), and the SSRs are not completely uniform in Agropyron; thus, they can be used in the investigations of genetic diversity and genetic structure of Agropyron.
Comparative Analysis of Chloroplast Genomes
During the evolution, contraction, and expansion of the IR, LSC and SSC regions are the main reason that leads to the size difference in chloroplast genomes (Yao et al., 2015). In this study, the difference of IR and SC regions of chloroplast genomes of Agropyron ranges from 33 bp to 313 bp. The ndhH gene crosses the JSA, which was also observed in other species of the Triticeae genus, such as H. vulgare, B. vulgaris (Chen et al., 2018), and Festuca (Qiu et al., 2019). The ndhH gene is sensitive to high light stress, which may have changed dramatically during evolution of land plants (Peng et al., 2011). The ycf1 gene crosses the JSA, which is also common in other families and genera, such as Fagaceae (Liu et al., 2019), Liliales (Nguyen et al., 2015), Zingiberaceae (Gui et al., 2020), and Anacardiaceae (Wang et al., 2020). As indicated by the Ka/Ks value and nucleotide diversity, changes in the rpoC2 gene in A. michnoi are greater than other species of Agropyron than the other four species and A. cristatum (KY 126307). Similarly, the Ka/Ks ratio of ccsA in A. mongolicum is greater than other species, except A. michnoi. For all the species of the Agropyron genus, the value of Ka/Ks is less than 0.5, indicating they are under purifying selection.
Based on the nucleotide diversity, we discovered the highly variable gene region trnG-GCC, which might be potentially informative molecular markers for species characterization of Agropyron.
Phylogenetic Relationship of Agropyron
Chloroplast genomes have large amounts of genetic information and are easier than nuclear genomes for filtrating the single copy gene (Zhang and Li, 2011). So chloroplast genomes are widely used in plant evolutionary biology. Our analyses revealed that A. michnoi is close to A. mongolicum and that the polymorphic sites are rich in chloroplast genomes of Agropyron, including the mono-nucleotide site, indels, and SSRs (Figure 5). Plants of the Agropyron genus consist of the P genomes (Dewey, 1984). Plants with the P genome can be used to improve agronomic traits such as drought, cold, and disease resistance in wheat and other crops (Dong et al., 1992). Meanwhile, the P genome can also be used for species classification; for example, A. cristatum and A. mongolicum are found to be two species with significant genetic differentiation when the P genomes were analyzed. This point was also supported by the phylogenetic tree with the chloroplast genome that these two species were close but still separated in the tree (Figure 9). It was shown that A. desertorum is most likely derived from A. cristatum and A. mongolicum (Figure 9). Meanwhile, the bred variety A. desertorum Fisch. Schult. is close to A. desertorum, but A. cristatum × A. desertorum Fisch. Schult shows a closer relationship with A. desertorum than with A. desertorum Fisch. Schult., indicating that the chloroplast genome of A. desertorum plays a dominant role during evolution.
Conclusion
We sequenced and assembled chloroplast genomes of five species from the Agropyron genus and compared their sequence at multiple levels. We found that the chloroplast genomes of Agropyron were different in the length, gene structure, and GC content. These chloroplast genomes of Agropyron are comparable with chloroplast genomes of other species within Triticeae and provide more insights into chloroplast genome evolution within Triticeae. From the phylogenetic tree, we can infer that they have a common ancestor. Based on the comparative analysis of chloroplast genomes of Agropyron, the intergenic Ka/Ks ratios are less than 0.5, suggesting that species of Agropyron are under a purifying selection. Therefore, our study lays a foundation for better understanding of the chloroplast genomes of Agropyron in species evolution, genetics, and phylogenetic relationships.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HH, RQ, YL, and XZ assembled sequences and analyzed the data; HH wrote the manuscript; BD and XC collected the plant material; YZ and YP conceived the research and revised the manuscript. YL, XZ, YZ, CG, HH, and RQ have contributed equally to this work. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 32160326) and the Inner Mongolia Autonomous Region Science and Technology Project (No. 2020GG0176).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully appreciate Professor Jiangqi Wen for help with language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.832809/full#supplementary-material
References
Asay, K. H., Dewey, D. Z., Gomm, F. B., Johnson, D. A., and Carlson, J. R. (1985). Registration of 'Hycrest' Crested Wheatgrass. Crop Sci. 25 (2), 368–369. doi:10.2135/cropsci1985.0011183X002500020054x
Asay, K. H., and Johnson, D. A.(1990). Genetic Variances for Forage Yield in Crested Wheatgrass at Six Levels of Irrigation. Crop Sci., 30, 79–82. doi:10.2135/cropsci1990.0011183X003000010018x
Che, Y. Y., Yang, Y., Yang, X., Li, X., and Li, L. (2015). Phylogenetic Relationship and Diversity Among Agropyron Gaertn. Germplasm Using SSRs Markers. Plant Syst. Evol. 301, 163–170. doi:10.1007/s00606-014-1062-4
Chen, N., Sha, L. N., Dong, Z. Z., Tang, C., Wang, Y., and Kang, H. Y. (2018). Complete Structure and Variation of the Chloroplast Genome of Agropyron Cristatum (L.) Gaertn. Gene 640, 86–96. doi:10.1016/j.gene.2017.10.009
Daniell, H., Chan, H. T., Pasoreck, E. K., and Bonini, N. M. (2016a). Vaccination via Chloroplast Genetics: Affordable Protein Drugs for the Prevention and Treatment of Inherited or Infectious Human Diseases. Annu. Rev. Genet., 50, 595–618. doi:10.1146/annurev-genet-120215-035349
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016b). Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 17, 134. doi:10.1186/s13059-016-1004-2
Dewey, D. R., and Pendse, P. C. (1968). Hybrids between Agropyron Desertorum and Induced-Tetraploid Agropyron Cristatum. Crop Sci. 8, 607–611. doi:10.2135/cropsci1968.0011183X000800050030x
Dewey, D. R. (1981). Cytogenetics of Agropyron Ferganense and its Hybrids with Six Species of Agropyron, Elymus, and Sitanion. Am. J. Bot. 68, 2161537–2252197.
Dewey, D. R. (1984). The Genomic System of Classification as a Guide to Intergeneric Hybridization with the Perennial Triticeae. New York, NY: Plenum Press, 209–279. doi:10.1007/978-1-4613-2429-4_9
Dong, Y. S., Zhou, R. H., Xu, S. J., Li, L. H., Cauderon, Y., and Wang, R. R. C. (1992). Desirable Characteristics in Perennial Triticeae Collected in China for Wheat Improvement. Hereditas (Landskrona) 116, 175–178. doi:10.1111/j.1601-5223.1992.tb00224.x
Doyle, J. J. (1987). A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 19, 11–15. Avaliable at http://irc.igd.cornell.edu/Protocols/DoyleProtocol.pdf.doi:10.1016/s0021-9258(18)50745-6
Fordlloyd, B. V., Schmidt, M., Armstrong, S. J., Barazani, O., Engels, J., Hadas, R., et al. (2011). Crop Wild Relatives-Undervalued, Underutilized and under Threat. Bioscience 61, 559–565. doi:10.1525/bio.2011.61.7.10
Gui, L. J., Jiang, S. F., Xie, D. F., Yu, L., Huang, Y., Zhang, Z. J., et al. (2020). Analysis of Complete Chloroplast Genomes of Curcuma and the Contribution to Phylogeny and Adaptive Evolution. Gene 732, 144355. doi:10.1016/j.gene.2020.144355
Hsiao, C., Wang, R., and Dewey, D. R. (1986). Karyotype Analysis and Genome Relationships of 22 Diploid Species in the Tribe Triticeae. Can. J. Genet. Cytol. 28, 109–120. doi:10.1139/g86-015
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 33 (2), 511–51822. doi:10.1093/nar/gki198
Khan, A., Asaf, S., Khan, A. L., Al-Harrasi, A., Al-Sudairy, O., AbdulKareem, N. M., et al. (2019). First Complete Chloroplast Genomics and Comparative Phylogenetic Analysis of Commiphora Gileadensis and C. Foliacea: Myrrh Producing Trees. PLoS ONE 14, e0208511. doi:10.1371/journal.pone.0208511
Kumar, S., Nei, M., Dudley, J., and Tamura, K. (2008). MEGA: a Biologist-Centric Software for Evolutionary Analysis of DNA and Protein Sequences. Brief. Bioinform. 9, 299–306. doi:10.1093/bib/bbn017
Langmead, B., and Salzberg, S. L. (2012). Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/nmeth.1923
Li, D. M., Zhao, C. Y., and Liu, X. F. (2019). Complete Chloroplast Genome Sequences of Kaempferia Galanga and Kaempferia Elegans: Molecular Structures and Comparative Analysis. Molecules 24, 474. doi:10.3390/molecules24030474
Liu, X., Chang, E. M., Liu, J. F., Huang, Y. N., and Jiang, Z. P. (2019). Complete chloroplast genome sequence and phylogenetic analysis of Quercus bawanglingensis Huang, Li et Xing, a vulnerable oak tree in China. Forest 10, 587. doi:10.3390/f10070587
Lu, Y. Q., Wu, X., Yao, M., Zhang, J., Liu, W., and Yang, X. (2015). Genetic Mapping of a Putative Agropyron Cristatum-derived Powdery Mildew Resistance Gene by a Combination of Bulked Segregant Analysis and Single Nucleotide Polymorphism Array. Mol. Breed. 35 (3), 96. doi:10.1007/s11032-015-0292-7
Middleton, C. P., Senerchia, N., Stein, N., Akhunov, E. D., Keller, B., Wicker, T., et al. (2014). Sequencing of Chloroplast Genomes from Wheat, Barley, rye and Their Relatives Provides a Detailed Insight into the Evolution of the Triticeae Tribe. PLoS ONE 9, e85761. doi:10.1371/journal.pone.0085761
Nguyen, P. A., Kim, J. S., and Kim, J. H. (2015). The Complete Chloroplast Genome of Colchicine Plants (Colchicum Autumnale L. And Gloriosa Superba L.) and its Application for Identifying the Genus. Planta 242 (1), 223–237. doi:10.1007/s00425-015-2303-7
Ochoa, V., Madrid, E., Said, M., Rubiales, D., and Cabrera, A. (2015). Molecular and Cytogenetic Characterization of a Common Wheat-Agropyron Cristatum Chromosome Translocation Conferring Resistance to Leaf Rust. Euphytica 201, 89–95. doi:10.1007/s10681-014-1190-5
Pasquale, L. C., Domenico, D. P., Donatella, D., Giovanni, G. V., and Gabriella, S. (2015). Complete Chloroplast Genome of the Multifunctional Crop globe Artichoke and Comparison with Other Asteraceae. PLoS ONE 10, e120589. doi:10.1371/journal.pone.0120589
Peng, L. W., Yamamoto, H., and Shikanai, T. (2011). Structure and Biogenesis of the Chloroplast NAD(P)H Dehydrogenase Complex. Biochim. Biophys. Acta 1807, 945–953. doi:10.1016/j.bbabio.2010.10.015
Qian, J., Song, J. Y., Gao, H. H., Zhu, Y. J., Xu, J. A., Pang, X. H., et al. (2013). The Complete Chloroplast Genome Sequence of the Medicinal Plant Salvia Miltiorrhiza. PLoS ONE 8, e57607. doi:10.1371/journal.pone.0057607
Qiu, Y. J., Hirsch, C. D., Yang, Y., and Watkins, E. (2019). Towards Improved Molecular Identification Tools in fine Fescue (Festuca L., Poaceae) Turfgrasses Nuclear Genome Size, Ploidy, and Chloroplast Genome Sequencing. Front. Genet. 1012, 1223. doi:10.3389/fgene.2019.01223
Rozas, J., Sánchezdelbarrio, J. C., Messeguer, X., and Rozas, R. (2003). Dnasp, Dna Polymorphism Analyses by the Coalescent and Other Methods. Bioinformatics 19, 2496–2497. doi:10.1079/9780851994758.0139
Sun, Y., Lyu, M., Han, H., Zhou, S., Lu, Y., Liu, W., et al. (2021). Identification and fine Mapping of Alien Fragments Associated with Enhanced Grain Weight from Agropyron Cristatum Chromosome 7P in Common Wheat Backgrounds. Theor. Appl. Genet. 134, 3759–3772. doi:10.1007/s00122-021-03927-7
Wang, L., He, N., Li, Y., Fang, Y. M., and Zhang, F. L. (2020). Complete Chloroplast Genome Sequence of Chinese Lacquer Tree (Toxicodendron Vernicifluum, Anacardiaceae) and its Phylogenetic Significance. Biomed. Res. Int. 2020, 9014873. doi:10.1155/2020/9014873
Wang, R. R. C. (2011). Agropyron and Psathyrostachys. Wild Crop Relatives: Genomic Breed. Resour.. 77–108. doi:10.1007/978-3-642-14228-4_2
Yao, X. H., Tang, P., Li, Z. Z., Li, D. W., Liu, Y. F., and Huang, H. W. (2015). The First Complete Chloroplast Genome Sequences in Actinidiaceae: Genome Structure and Comparative Analysis. PLoS ONE 10, e0129347. doi:10.1371/journal.pone.0129347
Zhang, Y. J., and Li, D. Z. (2011). Advances in Phylogenomics Based on Complete Chloroplast Genomes. Plant Divers. 33, 365–375. doi:10.1631/jzus.B1000171
Keywords: Agropyron, chloroplast genome, phylogenetic relationship, comparative analysis, illumina sequencing
Citation: Han H, Qiu R, Liu Y, Zhou X, Gao C, Pang Y and Zhao Y (2022) Analysis of Chloroplast Genomes Provides Insights Into the Evolution of Agropyron. Front. Genet. 13:832809. doi: 10.3389/fgene.2022.832809
Received: 10 December 2021; Accepted: 06 January 2022;
Published: 25 January 2022.
Edited by:
Zefeng Yang, Yangzhou University, ChinaReviewed by:
Hui Song, Qingdao Agricultural University, ChinaNaichong Chen, Oklahoma State University, United States
Copyright © 2022 Han, Qiu, Liu, Zhou, Gao, Pang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhen Pang, cGFuZ3lvbmd6aGVuQGNhYXMuY24=; Yan Zhao, emhhb3lhbm5tZ0BpbWF1LmVkdS5jbg==
 Huijie Han
Huijie Han Rui Qiu1
Rui Qiu1 Yongzhen Pang
Yongzhen Pang Yan Zhao
Yan Zhao