- 1Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, China
- 2Taizhou Hospital of Chinese Medicine, Taizhou, China
- 3Laboratory of Molecular Biology, Central Laboratory, Jiangsu Province Hospital of Chinese Medicine, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, China
Background: Many studies have assessed the potential link between interleukin-6 polymorphisms and susceptibility to allergic diseases. However, the results are still conflicting. Therefore, a comprehensive meta-analysis can not only resolve differences but also provide clues for future projects.
Methods: A systematic electronic search was conducted on the databases of Web of Science, PubMed, and Cochrane Library to retrieve all published studies. Revman and Stata software were used for statistical analysis.
Results: This meta-analysis included 11 studies. The results revealed that there was a statistically significant association between IL-6 rs1800795 polymorphism and the risk of asthma and allergic rhinitis in the general population. Subgroup analyses demonstrated that rs1800795 affected allergic diseases risk in different populations.
Conclusion: Our findings suggested that IL-6 rs1800795 was associated with allergic diseases susceptibility among Asians and Caucasians in opposite trends, and it might influence the risk of asthma and allergic rhinitis. None of the IL-6 polymorphisms were shared risk variants of allergic diseases.
Introduction
Allergic diseases, including allergic rhinitis, allergic asthma, and atopic dermatitis, often coexist in the same individual and are considered multifactorial inflammatory diseases (Blumenthal, 2012; Pinart et al., 2014; Ferreira et al., 2017). Allergic diseases have become major health problems worldwide, resulting in significant economic and social burdens (Asher et al., 2006; Nutten, 2015). The mechanism and pathological characteristics of allergic diseases are similar. Genes, environment factors, and the mutual effects among them can be the underlying mechanism of the progress and expression of these diseases (Blumenthal 2012; Galli and Tsai 2012; Ferreira et al., 2017). It shows that initial manifestations of atopic dermatitis often occur early in life and frequently ahead of other allergic diseases like asthma or allergic rhinitis (Nutten 2015), partially due to a shared genetic origin (Ferreira et al., 2017). In order to identify shared risk variants, the nature of individual genes recognized as susceptible factors for allergic diseases have been summarized, and the list of these genetic factors may expand significantly with the latest emergence of genome-wide association approaches (Vercelli 2008; Cheng et al., 2014; Ferreira et al., 2017).
Interleukin-6 (IL-6) is an effective pro-inflammatory cytokine, which can be released in response to immune attacks or tissue damages and stimulate a wide range of innate and adaptive immune responses (Tanaka et al., 2014). Overexpression of IL-6 can cause chronic inflammatory disorders and potentially fatal hyperinflammation (Spencer et al., 2019), as seen in advanced coronavirus disease 2019 (COVID-19) (Mehta et al., 2020). More and more evidence shows that IL-6 can modulate the differentiation and activation of T cells and induce the production of Th2 cytokines, which is an important signal for coordinating chronic inflammation and adaptive immunity (Rincon and Irvin 2012). Furthermore, IL-6 and transforming growth factor-β (TGF-β) together induce the production of Th17 cells while inhibiting the differentiation of Treg cells (Tanaka et al., 2014). Previous studies have found that IL-6 is related to the occurrence of allergic diseases such as asthma and atopic dermatitis (Gentile et al., 2001; Zhang et al., 2014), and it also has been shown that IL-6 can increase nasal secretion in patients with allergic rhinitis (Gentile et al., 2001).
Human IL-6 is composed of 212 amino acids, including a 28-amino-acid signal peptide, and its gene has been located on chromosome 7p21 (Kämäräinen et al., 2008); glycosylation accounts for the size of 21–26 kDa of natural IL-6 (Tanaka et al., 2014). Recent studies demonstrated the association between IL-6 gene polymorphisms and allergic diseases risk. There are many verified single-nucleotide polymorphisms (SNPs) in the IL6 gene in the dbSNP database, but only three SNPs, including −174 G/C (rs1800795), −572 G/C (rs1800796), and −597 G/A (rs1800797) restriction sites have been extensively studied (Kämäräinen et al., 2008; Cash et al., 2010). Many studies aimed to evaluate the relationship between the IL-6 polymorphisms and allergic diseases risk in different populations (Reich et al., 2003; Settin et al., 2008; Trajkov et al., 2008; Xiaomin et al., 2008; Mahdaviani et al., 2009; Daneshmandi et al., 2012; Gharagozlou et al., 2013; Kosugi et al., 2013; Nasiri et al., 2014; Zhao et al., 2016; Babusikova et al., 2017). Nevertheless, the outcomes were still elusive. Hence, the purpose of the present research lies at generalizing the existing proof to clarify the exact association between IL-6 gene polymorphisms and the risk of allergic diseases.
Previously, two meta-analyses examined the association between IL-6 rs1800795 polymorphism and asthma risk (Li et al., 2015; Zhu et al., 2020). However, these two articles did not study the relationship between other allergic diseases with IL6 gene polymorphisms and did not concern the two other polymorphisms of the IL-6 (rs1800796 and rs1800797), although the associations between these SNPs and asthma have been reported. Thus, we performed this meta-analysis based on the accumulating evidence to estimate the link between IL-6 gene polymorphisms and allergic diseases risk (including allergic rhinitis, allergic asthma, and atopic dermatitis). Besides these, we carried out isolated meta-analyses for each allergic disease to check the impact of IL-6 SNPs on each disease and also to identify whether IL-6 SNPs were shared risk variants of allergic diseases. Therefore, our meta-analysis was the first study on genetic associations of IL-6 rs1800795, rs1800796, and rs1800797 polymorphisms with allergic diseases.
Materials and Methods
Our research was carried out as per the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guideline (Liberati et al., 2009).
Literature Review and Inclusion Criteria
We used the keywords below to search for suitable studies from PubMed, Web of Science, and Cochrane Library: “IL-6,” “Interleukin-6,” “polymorphism,” “variant,” “variation,” “mutation,” “SNP,” “asthma,” “atopic dermatitis,” and “allergic rhinitis.” In February 2021, we conducted the initial literature search, and the recent renewal was completed in June 2021. Additionally, our team went through the bibliographies of the acquired studies to obtain more underlying related articles.
The selection standards of this study were as follows: 1) case–control study was designed to explore the association between IL-6 SNPs and allergic diseases risk; 2) published research data can be adopted to speculate an odds ratio (OR) and 95% confidence interval (CI); and 3) full text was provided. If any of the standards below apply, the research will be excluded: 1) incomplete or inaccessible study data; 2) summaries or meta-analyses; and 3) research that was not based on human beings. In case that identical studies were accepted by several periodicals, we would merely include the latest complete studies for analysis.
Data Extraction and Quality assessment
The entire necessary data were obtained from articles according to the normalized list of the information below: first author's name, publishing time, country, ethnicity, disease type, diagnosis basis, sample scale, genotype approach, and genotypical calculation for all polymorphic results. The p-value of Hardy–Weinberg equilibrium (HWE) was computed as well according to a formula. Our team adopted the Newcastle–Ottawa Scale (NOS) criteria to assess the quality of each study sequence (Zeng et al., 2015). According to the scores, studies were identified as inferior (0–3), ordinary (4–6), and superior (7–9) studies. Information obtainment and quality evaluation were carried out by two different researchers. Any disagreement between the two researchers was discussed till there was an agreement.
Statistical Analysis
The entire statistic assays were analyzed via use of Revman 5.4 (The Nordic Cochrane Centre, Denmark) and Stata 15.1 (StataCorp LP, America). We used ORs and 95% CIs to assess the tightness of the connection between IL-6 polymorphisms and allergic diseases risks. If p ≤ 0.05, it would be deemed as important on statistics. For all SNPs, the dominant comparison, recessive comparison, allele comparison, homozygote comparison, and heterozygote comparison were used to speculate the impacts. In detail, defined comparisons are as follows: for −174 G/C (rs1800795) and −572 G/C (rs1800796): dominant comparison (CC+GC vs. GG), recessive comparison (CC vs. CG+GG), allele comparison (C vs. G), heterozygote comparison (GC vs. GG), and homozygote comparison (CC vs. GG). For −597 G/A (rs1800797): dominant comparison (AA+GA vs. GG), recessive comparison (AA vs. AG+GG), allele comparison (A vs. G), heterozygote comparison (GA vs. GG), and homozygote comparison (AA vs. GG).
The heterogeneity between studies was assessed through use of the I2 statistics. If I2 > 50%, it meant that there was heterogeneity; the random-effects model was required to calculate the combination of OR. Otherwise, the fixed-effects model was employed (Higgins and Thompson 2002). Simultaneously, a χ2 test was adopted to test if the genotypical distribution of included studies accorded with the HWE, and p > 0.05 showed that it coincided. Moreover, the subgroup study was carried out as per ethnicity, age, and disease type. The steadiness of synthesized outcomes was assessed via sensitive analysis, and potential publication biases were assessed via funnel plots and Egger's test (Egger et al., 1997).
Results
Characteristics of Included Studies
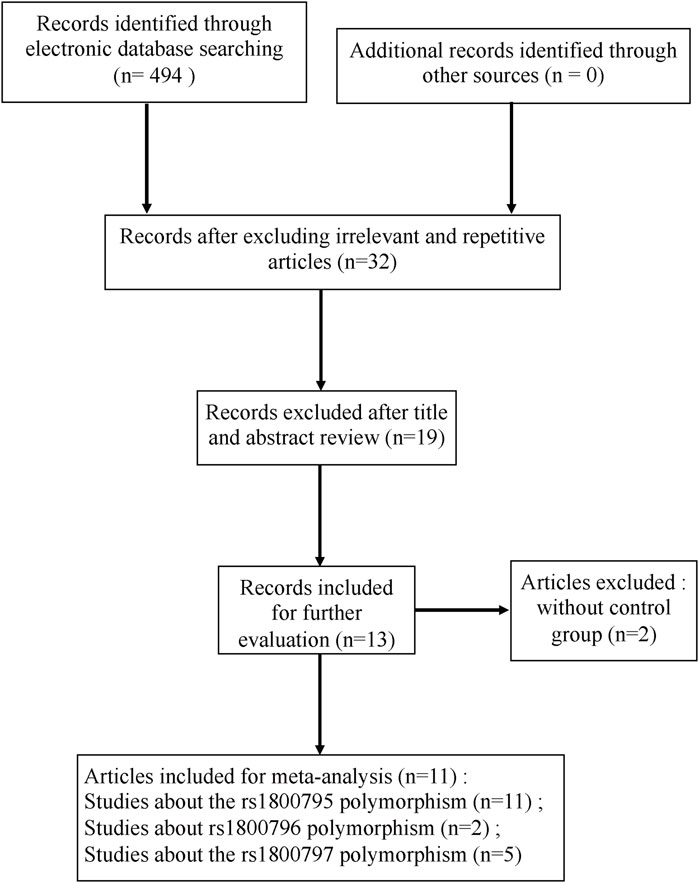
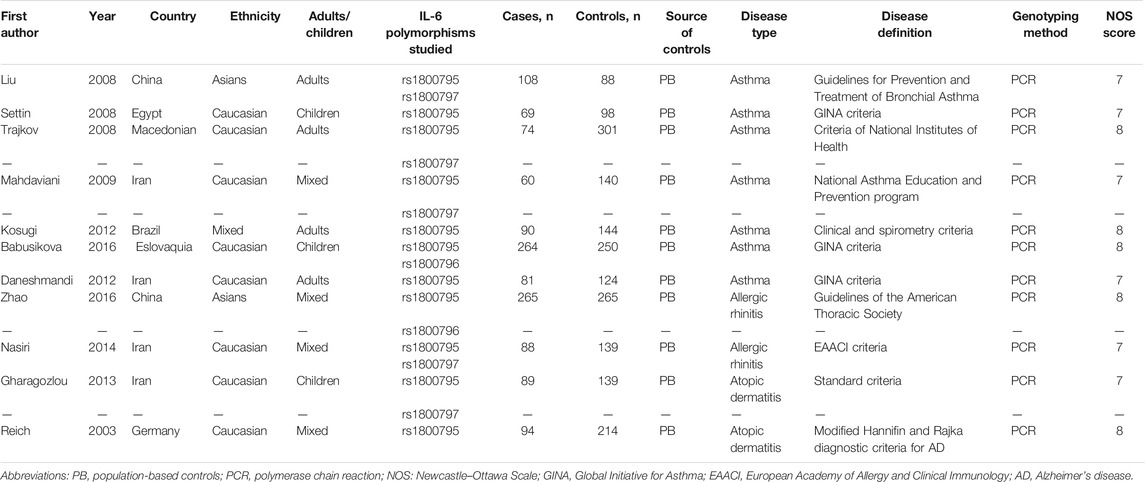
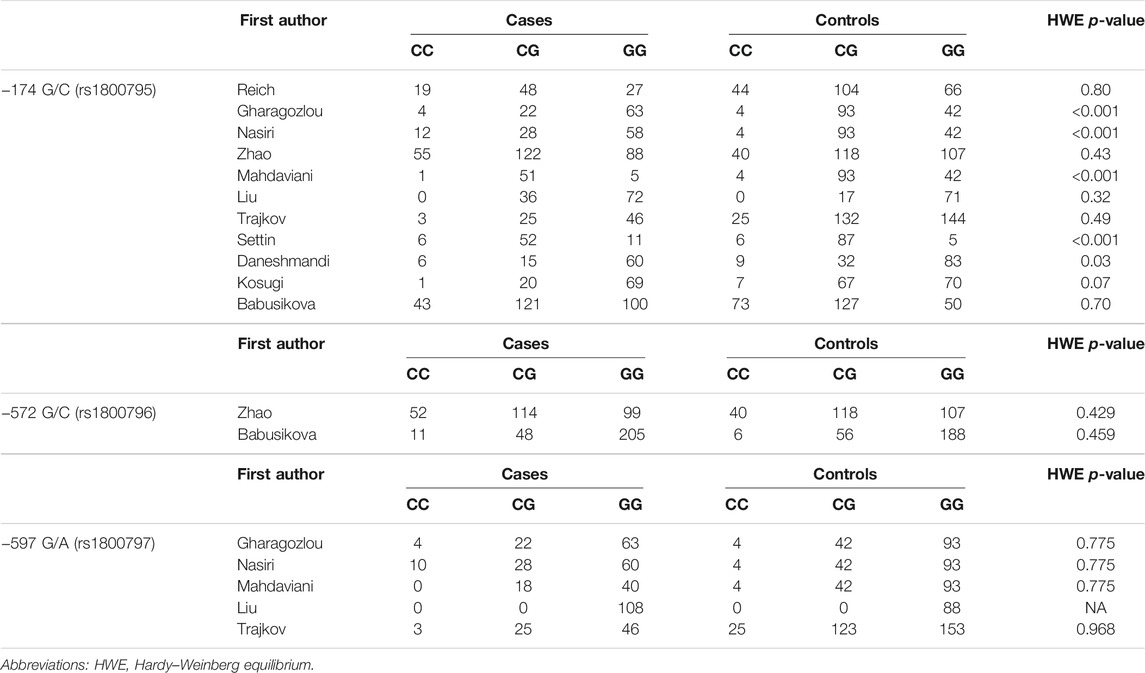
The search and screening process of this study is depicted in Figure 1 (Liberati et al., 2009). A total of 494 studies were selected, 462 studies were removed after excluding irrelevant and repetitive articles, then 32 articles were assessed for eligibility, and after reading headlines and abstracts, 19 articles were eliminated. Two others were excluded for lacking controls. Finally, a total of 11 eligible studies were included in this meta-analysis. The NOS score of the included studies was 7–8 points, which proved that the methodological quality of all included studies was generally good. In the included studies, PCR was used as a genotyping method. Tables 1 and 2 summarize the main characteristics of the included studies and the frequency of genotype distribution.
Meta-Analysis of the Association Between −174 G/C rs1800795 Polymorphism and Overall Allergic Diseases Risk
Eleven articles included 1,282 patients with allergic disease, and 1,902 controls were analyzed to find the relationship between rs1800795 polymorphism and overall allergic diseases risk. Our results did not show significant association of rs1800795 polymorphism and overall allergic diseases risk in the general population (Table 3). However, subgroup analyses based on ethnicities demonstrated that rs1800795 polymorphism was associated with the decreased risk of overall allergic diseases in Caucasian populations under dominant comparison, allele comparison, and heterozygote comparison (dominant comparison: OR = 0.56, 95% CI =0.32, 0.96, p = 0.04; allele comparison: OR = 0.72, 95% CI = 0.54, 0.97, p = 0.03; heterozygote comparison: OR = 0.54, 95% CI =0.29, 0.98, p = 0.04), while in Asian populations, rs1800795 polymorphism was associated with an increased risk of overall allergic diseases in dominant comparison, allele comparison, and homozygote comparison (dominant comparison: OR = 1.53, 95% CI =1.05, 2.23, p = 0.03; allele comparison: OR = 1.39, 95% CI =1.06, 1.83, p = 0.02; homozygote comparison: OR = 1.67, 95% CI =1.02, 2.74, p = 0.04) (Table 3 and Figure 2).
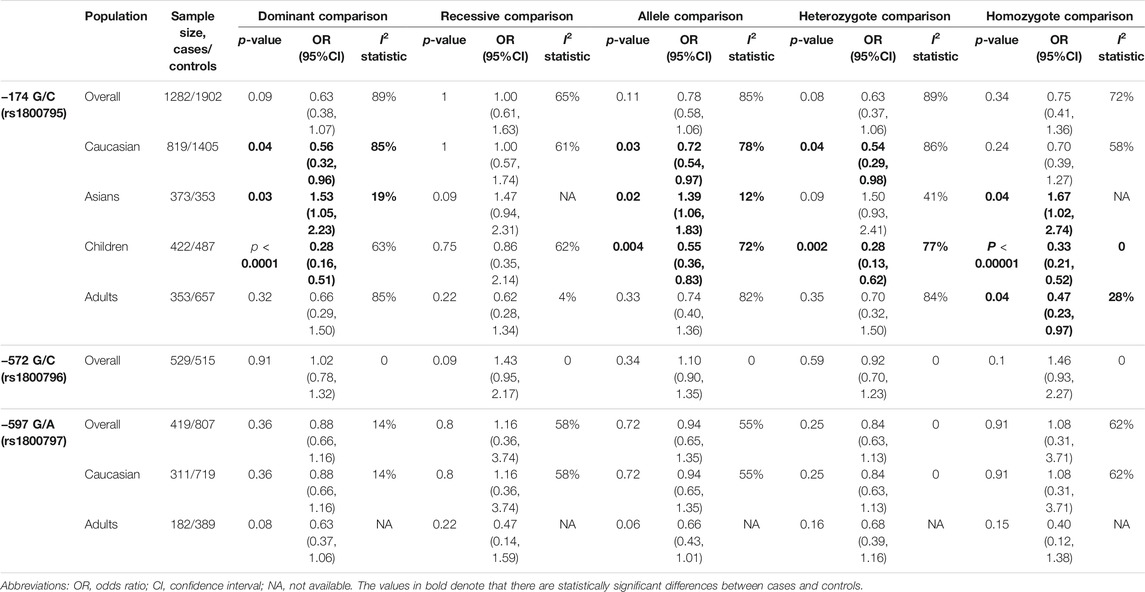
TABLE 3. Results of pooled ORs in the meta-analysis of the association between IL-6 gene polymorphisms and overall allergic disease.
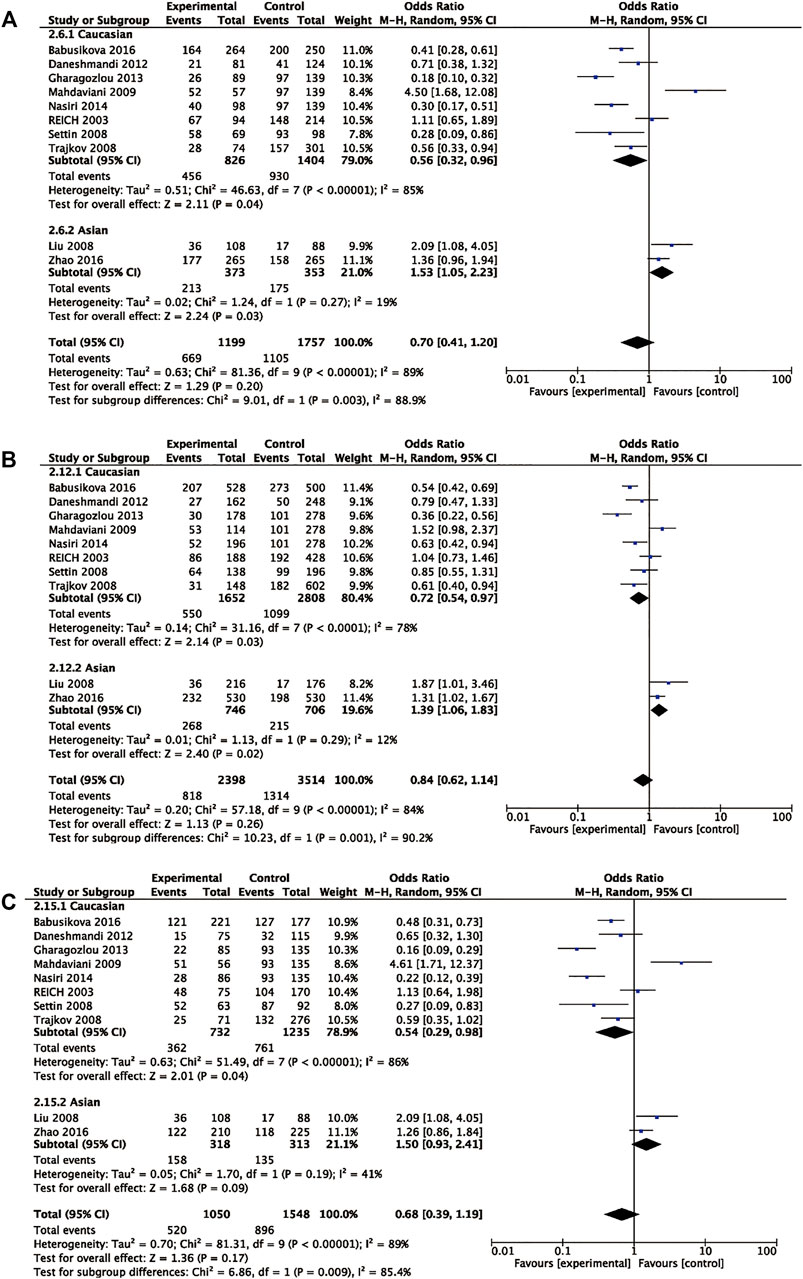
FIGURE 2. Pooled OR and 95% CI of individual studies and pooled data for the association between IL6 rs1800795 and allergic diseases risk in different ethnicity subgroups for (A) dominant comparison, (B) allele comparison, and (C) heterozygote comparison.
Subgroup analyses on the basis of age demonstrated that IL6 rs1800795 variant was associated with a decreased risk of childhood allergic diseases in dominant comparison, allele comparison, heterozygote comparison, and homozygote comparison (dominant comparison: OR = 0.28, 95% CI = 0.16, 0.51, p < 0.0001, allele comparison: OR = 0.55, 95% CI = 0.36, 0.83, p = 0.004, heterozygote comparison: OR = 0.28, 95% CI = 0.13, 0.62, p = 0.002, homozygote comparison: OR = 0.33, 95% CI = 0.21, 0.52, p < 0.00001). The decreased risk of allergic diseases was related to rs1800795 polymorphism across homozygote comparison (homozygote comparison: OR = 0.47, 95% CI = 0.23, 0.97, p = 0.04) (Table 3 and Figure 3).
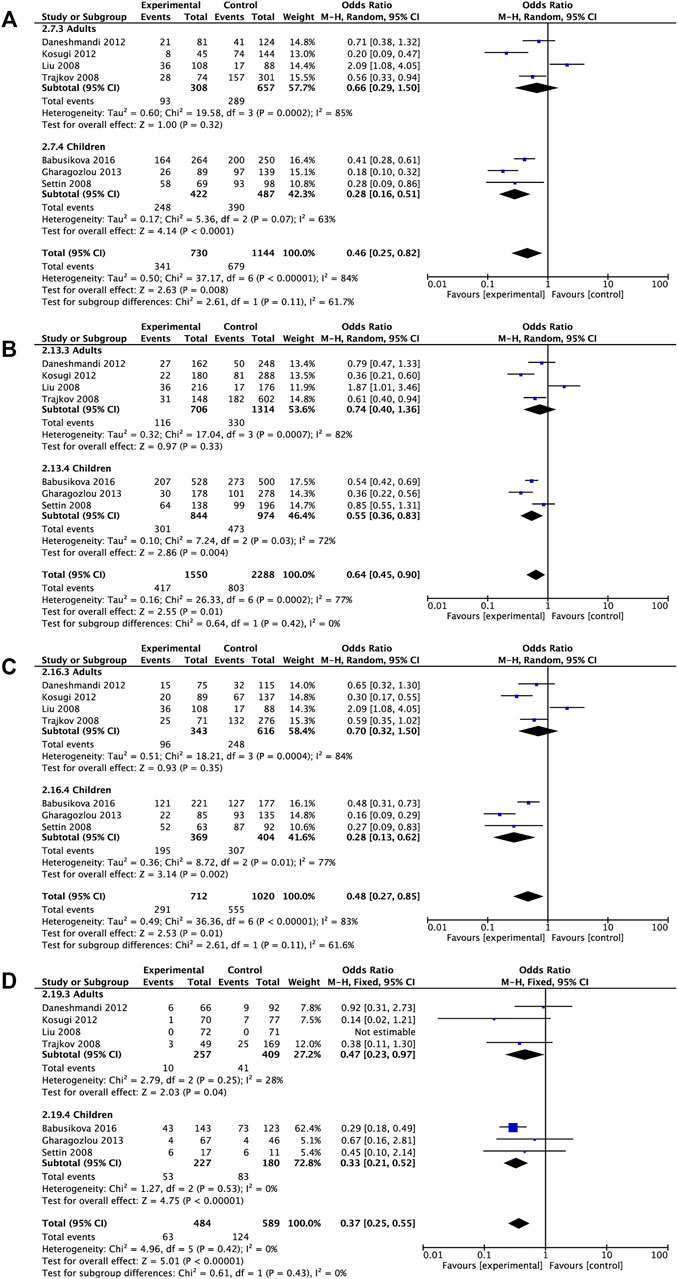
FIGURE 3. Pooled OR and 95% CI of individual studies and pooled data for the association between IL6 rs1800795 polymorphism and overall allergic diseases risk in different age subgroups for (A) dominant comparison, (B) allele comparison, (C) heterozygote comparison, and (D) homozygote comparison.
Meta-Analysis of the Association Between −572 G/C rs1800796 Polymorphism and Overall Allergic Diseases Risk
Two articles including 529 patients and 515 controls were analyzed to investigate the association between rs1800796 polymorphism and overall allergic diseases risk. Of the two articles, one was about asthma, reporting 264 patients and 250 controls, and the other was about allergic rhinitis, including 265 patients and 265 controls. Since one study was on Caucasian children and the other was on mixed-age Asians, subgroup analysis based on age and ethnicity was not feasible. The pooled analyses indicated that under any comparison, there was no significant association between the rs1800796 polymorphism and the risk of allergic diseases (Table 3). This negative result may be due to insufficient research, so more research was needed to corroborate our conclusion.
Meta-Analysis of the Association Between nt565 G/A rs1800797 Polymorphism and Overall Allergic Diseases Risk
Five articles including 419 patients and 807 controls were analyzed to find the association between rs1800797 polymorphism and allergic diseases risk. Among the five articles, three were about asthma, one about allergic rhinitis, and one about atopic dermatitis. In any of our comparison analyses, no significant association was found between the rs1800797 polymorphism and the risk of overall allergic diseases (Table 3). Further stratification analysis by ethnicity and age revealed that no significantly decreased or increased risks of overall allergic diseases were found in any genetic model (Table 3). This result may still be caused by the lack of research, which needs more research to confirm.
Meta-Analysis of the Association Between IL-6 Polymorphisms and the Risk of Each Allergic Disease
Considering the different pathogenic and epigenetic potential factors of the three allergic diseases, the relationship between IL-6 gene polymorphisms and the risk of each disease may be different. Therefore, it is necessary to study each form of allergy separately and classify them separately to test the effectiveness of the study in each category.
Seven studies involving 746 asthmatic patients and 1,145 controls were analyzed to determine the association of rs1800795 and asthma risk. The results revealed that rs1800795 polymorphism was significantly associated with the risk of asthma under the recessive comparison and homozygote comparison (recessive comparison: OR = 0.55, 95% CI = 0.39, 0.77, p = 0.0006, homozygote comparison: OR = 0.37, 95% CI = 0.25, 0.55, p< 0.00001) (Table 4 and Figure 4).
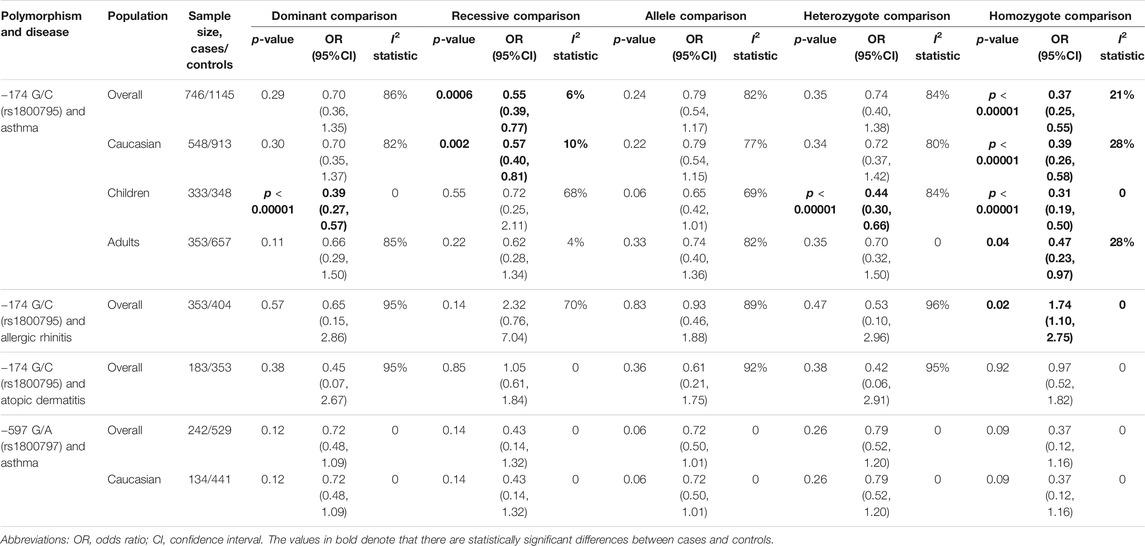
TABLE 4. Results of pooled ORs in the meta-analysis of the association between IL-6 gene polymorphisms and each allergic disease.
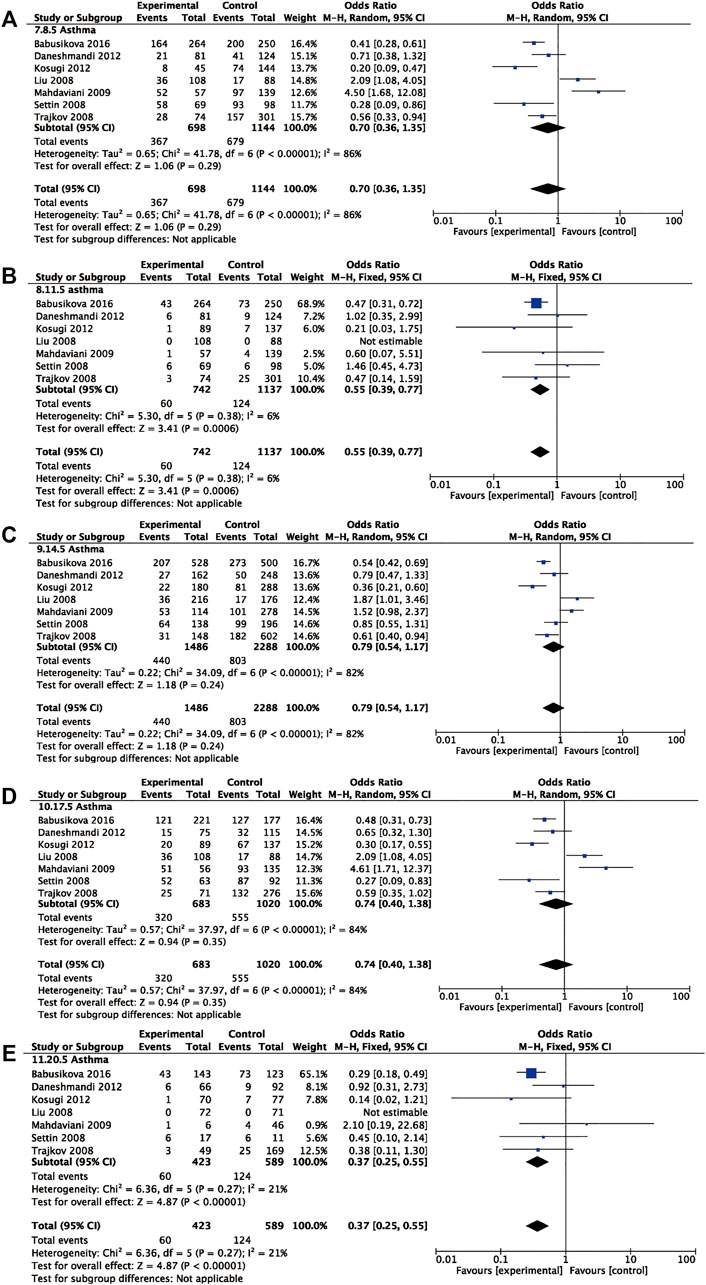
FIGURE 4. Pooled OR and 95% CI of individual studies and pooled data for the association between IL-6 rs1800795 polymorphism and asthma risk for (A) dominant comparison, (B) recessive comparison, (C) allele comparison, (D) heterozygote comparison, and (E) homozygote comparison.
Further subgroup analyses were used for ethnicity and age. The rs1800795 polymorphism decreased the risk of asthma in Caucasians across the recessive comparison and homozygote comparison (recessive comparison: OR = 0.57, 95% CI = 0.40, 0.81, p = 0.002, homozygote comparison: OR = 0.39, 95% CI = 0.26, 0.58, p < 0.00001) and was associated with a decreased risk of asthma in children in the dominant comparison, heterozygote comparison, and homozygote comparison (dominant comparison: OR = 0.39, 95% CI = 0.27, 0.57, p < 0.0001; heterozygote comparison: OR = 0.44, 95% CI = 0.30, 0.66, p < 0.0001, homozygote comparison: OR = 0.31, 95% CI = 0.19, 0.50, p < 0.00001). The same trend was found in the adults under the homozygote comparison (homozygote comparison: OR = 0.47, 95% CI = 0.23, 0.97, p = 0.04) (Table 4).
Two case–control studies including 353 patients with allergic rhinitis and 404 controls were utilized to analyze the relationship between IL-6 rs1800795 and allergic rhinitis. The result showed that IL-6 rs1800795 polymorphism increased the risk for allergic rhinitis in homozygote comparison (homozygote comparison = 1.74, 95% CI = 1.10, 2.75, p = 0.02).
Two case–control studies, including 183 patients and 353 controls, were utilized to analyze the association between rs1800795 and atopic dermatitis. The results showed that there was no significant association between IL-6 rs1800795 polymorphism and atopic dermatitis risk (Table 4).
Three case–control studies, including 242 asthmatic patients and 529 controls, were utilized to analyze the association between rs1800797 and asthma. The results showed no association between the polymorphism and risk of asthma in any of the models (Table 4).
Publication Bias
Egger's test and Funnel plots were performed to measure the publication bias. The funnel plots' shapes of rs1800795 did not show any evidence of obvious asymmetry. Meanwhile, the results of Egger's tests also showed no evidence of publication biases. The associative outcomes of rs1800796 and 1800797 were less than 10 studies, so they were precluded from publication bias testing (Supplementary Figures S8 and S9) .
Sensitivity Analysis
We performed sensitivity analysis by deleting one eligible study at a time (Supplementary Figures S4–S7). The significance of the pooled ORs was not affected by any single study in each comparison for rs1800796, rs1800797, while for rs1800795, the combined ORs of some comparisons were changed significantly after removing Mahdaviani's (Mahdaviani et al., 2009), Babusikova's (Babusikova et al., 2017), and Liu's (Xiaomin et al., 2008) studies in the general, adult, and Caucasian populations. We also excluded the studies with a deviation of HWE in the controls, and some results changed, which indicated that the results of these comparisons were controversial and needed more studies to verify in the future (Supplementary Tables S2 and S3).
Discussion
Human genome research emphasizes the role of common genetic variations in the etiology and pathogenesis of allergic diseases (Asher et al., 2006; Nutten 2015). The IL-6 gene has three promoter variants, rs1800795, rs1800796, and rs1800797, which have been proved to affect the transcription and secretion of IL-6 and are suspected to be risk factors for allergic diseases (Fishman et al., 1998; Kämäräinen et al., 2008). Considering the inconsistent results of published studies, we included all eligible publications for the meta-analysis to assess the link between IL-6 polymorphisms and the risk of allergic diseases.
Excluding the results with poor stability, our findings indicated no evidence of the association of IL-6 rs1800795, rs1800796, or rs1800797 polymorphism with overall allergic diseases risk in general study populations. However, in the ethnicity-based subgroup analysis, the presence of rs1800795 SNP in Caucasians reduced the risk of allergic diseases but elevated the risk of allergic diseases in Asians. The trends of association in Asians and Caucasians were contrary to each other, indicating that there might be large differences in the genotype distribution of rs1800795 polymorphism among different ethnicities. These may be due to geographical differences, distinct dietary habits, ethnic diversity, the influence of ethnicity on IL-6 gene expression, and serum IL-6 levels. Stratified by age, IL-6 rs1800795 was negatively related to susceptibilities of overall allergic diseases in both adults and children.
When we studied the association between IL-6 gene polymorphisms and each allergic disease separately, we found that the rs1800795 polymorphism was associated with a lower risk of asthma but was associated with increased susceptibility to allergic rhinitis. The age-based and ethnicity-based subgroup analyses in asthma indicated that IL-6 rs1800795 was a protective factor against asthma in Caucasians—children and adults.
We included allergic rhinitis, atopic dermatitis, and asthma in this study, hoping to explore whether IL-6 polymorphisms were the common risk variants of allergic diseases. According to our results, the rs1800795 polymorphism had the opposite effect on the susceptibility to asthma and allergic rhinitis and had no effect on the risk of atopic dermatitis. IL-6 rs1800796 and rs1800797 polymorphisms were not related to the risk of allergic diseases. All of these results indicated that IL-6 polymorphisms might not be the shared risk variants of allergic diseases. This conclusion may not be accurate enough due to the lack of studies, since there were only two studies on allergic rhinitis and two on atopic dermatitis.
It is known that the pathogenesis of allergic diseases involves the complex interaction of genetic and environmental factors, and ethnicities seem to have a significant impact on the prevalence of allergic diseases (Kim et al., 2020). It is reported that compared to the Asians, asthma risk was significantly greater for Black/African American, American Indian and Alaska Native (AIAN), White, and multiracial respondents (Hall et al., 2020). A study from the Netherlands found that Caucasians were less likely to have allergic rhinitis than most other ethnicities (Hoffmans et al., 2018). The above prompted us that the morbidity of asthma and allergic rhinitis showed opposite trends in Caucasians and Asians, which was consistent with our result that the association between IL-6 rs1800795 polymorphism and the risk of allergic disease showed opposite trends in different ethnicities. We speculate that the IL-6 rs1800795 polymorphism may be the same as factors such as residence, lifestyle, and eating habits (Hoffmans et al., 2018; Hall et al., 2020), which is the reason for the different susceptibilities of different ethnic groups to asthma and allergic rhinitis. However, more studies are still needed to prove this.
IL-6 rs1800795 polymorphism was reported to be close to the sites of many transcription factors, including NF-κB and NF-IL6. Guanine-containing oligonucleotides at position −174 have more DNA protein interactions than cytosine-containing oligonucleotides (Ray et al., 1990; Terry et al., 2000). Therefore, they have a higher affinity for transcription factors, resulting in increased IL-6 gene expression (Fishman et al., 1998). IL-6 has emerged as an important regulator of effector CD4 T cell fate, promoting IL-4 production during Th2 differentiation, inhibiting Th1 differentiation, and, together with TGF-β, promoting Th17 cell differentiation (Zhu et al., 2010; Rincon and Irvin 2012). Based on these, we hypothesized that rs1800795 polymorphism could affect the differentiation of Th1 and Th17 cells by affecting the expression of IL-6, thereby affecting the risk of allergic diseases.
It is reported that rs1800796 does not contain any effective homology with any identified transcription factor binding site; this may be why rs1800796 was not associated with the risk of allergic diseases. The IL-6 rs1800797 polymorphism is another mutation in the IL-6 gene promoter region. Studies have shown that it can regulate the effect of rs1800795 on transcriptional activity (Terry et al., 2000; Rivera-Chavez et al., 2003), but our study did not reflect the influence of rs1800797 on the risk of allergic diseases.
As for evaluation of heterogeneities, for IL-6 rs1800796 and rs1800797 polymorphisms, we observed that heterogeneities were slight. However, moderate to severe heterogeneities were observed among eligible studies in certain comparisons for rs1800795. In the further subgroup analysis, we found that the heterogeneity of rs1800795 was decreased in Asians, Caucasians, children, and adults, which indicated that distributions of rs1800795 polymorphism varies greatly from population to population. Moreover, differences in ethnicity and age could affect genetic association between IL6 rs1800795 polymorphism and allergic diseases. Therefore, the genetic associations between rs1800795 polymorphism and allergic diseases may be ethnic-specific and age-specific.
There are several points to consider in our findings. Firstly, in the sensitivity analysis, we found that the removal of Liu's (Li et al., 2015) study would have a significant effect on the results in the overall population. Considering that the study of Liu was in line with HWE, it might be that the difference in ethnicity caused this poor stability. The significantly reduced heterogeneity and sensitivity in the ethnicity-based subgroup analysis confirmed this. So the genetic associations between polymorphisms in IL-6 and allergic diseases risk may be ethnically specific, and maybe we should not try to generalize the combined results of our positive findings to a wider population. Secondly, the pathogenesis of allergic diseases is very complex. Therefore, our team highly suggests that other studies should carry out monomer-type studies and explore underlying gene–gene mutual effects to further reveal the impacts of genes on allergic diseases susceptibility (Steen 2012; Nishi et al., 2016). Thirdly, we aimed to investigate more allergic diseases such as eczema or urticaria or allergic conjunctivitis at the beginning. However, we did not find studies on the relationship between these diseases and IL-6 gene polymorphisms. Moreover, there is also a lack of research on allergic rhinitis and atopic dermatitis. Maybe scholars should pay more attention to relative allergic diseases in the future.
Previous relevant meta-analyses were reported by Zhu et al. (2020) and Li et al. (2015), which were partly consistent with our results in the association between IL-6 rs1800795 and asthma risk. In addition, our study has been expanded in the following aspects. Through the inclusion of studies on three kinds of allergic diseases and three IL-6 polymorphisms, we explored whether the IL-6 SNPs were shared risk variants of these allergic diseases. Moreover, we conducted subgroup analyses on ethnicity, age, and disease type. Hence, our findings seem to be more convincing and abundant.
Our research also had some unavoidable defects. First of all, only 11 studies were analyzed, and the specimen scale was very limited, resulting in insufficient outcome dependability. Second, environmental factors might impact the association between IL-6 polymorphisms and allergic diseases as well. Unfortunately, the majority of selected studies merely highlight the association, so it is hard to analyze the gene–environment mutual effects. Third, IL-6 polymorphisms may be associated with the severity and susceptibility of these diseases, but we did not perform a meta-analysis on this, owing to the lack of relevant data in the included studies. Fourth, the sample size of the current study is still insufficient, especially in the subgroup analysis. Therefore, large and well-designed studies are needed to validate our findings. Fifth, we excluded the gray literature from the analysis because its quality is difficult to evaluate. Considering the above mentioned limitations, our findings should be interpreted with caution.
Conclusion
This meta-analysis demonstrated that rs1800795 polymorphism of IL-6 gene might be a risk factor for allergic rhinitis. In addition, IL-6 rs1800795 polymorphism was associated with allergic diseases susceptibility among Asians. However, the opposite trend appeared in Caucasians. IL-6 rs1800795 polymorphism was a protective factor against asthma, while it was associated with increased susceptibility to allergic rhinitis. Neither rs1800795, rs1800796, nor rs1800797 was a shared risk variant of overall allergic diseases. However, further well-designed studies with a bigger specimen scale are needed to verify the discoveries in this work. Additionally, more studies are required as well to reveal the possible molecular mechanisms of the discoveries in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YY and JX conceived and designed the study. YY, LT, and SW conducted the literature review. YY, BW, XS, and ZX analyzed the data. YY, LL, and SS drafted the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81774267 and No. 82004265).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.822091/full#supplementary-material
Abbreviations
CI, confidence interval; IL, interleukin; NOS, Newcastle–Ottawa Scale; OR, odds ratio; PRISMA, Preferred Reporting Items for Systematic reviews and Meta-Analyses; SNP, single-nucleotide polymorphism; TGF-β, transforming growth factor-β.
References
Asher, M. I., Montefort, S., Björkstén, B., Lai, C. K., Strachan, D. P., Weiland, S. K., et al. (2006). Worldwide Time Trends in the Prevalence of Symptoms of Asthma, Allergic Rhinoconjunctivitis, and Eczema in Childhood: ISAAC Phases One and Three Repeat Multicountry Cross-Sectional Surveys. The Lancet 368, 733–743. doi:10.1016/s0140-6736(06)69283-0
Babusikova, E., Jurecekova, J., Jesenak, M., and Evinova, A. (2017). Association of Gene Polymorphisms in Interleukin 6 in Infantile Bronchial Asthma. Archivos de Bronconeumología (English Edition) 53, 381–386. doi:10.1016/j.arbr.2016.11.006
Blumenthal, M. N. (2012). Genetic, Epigenetic, and Environmental Factors in Asthma and Allergy. Ann. Allergy Asthma Immunol. 108, 69–73. doi:10.1016/j.anai.2011.12.003
Cash, H., Relle, M., Menke, J., Brochhausen, C., Jones, S. A., Topley, N., et al. (2010). Interleukin 6 (IL-6) Deficiency Delays Lupus Nephritis in MRL-FaslprMice: The IL-6 Pathway as a New Therapeutic Target in Treatment of Autoimmune Kidney Disease in Systemic Lupus Erythematosus. J. Rheumatol. 37, 60–70. doi:10.3899/jrheum.090194
Cheng, D., Hao, Y., Zhou, W., and Ma, Y. (2014). The Relationship between Interleukin-18 Polymorphisms and Allergic Disease: a Meta-Analysis. Biomed. Res. Int. 2014, 290687. doi:10.1155/2014/290687
Daneshmandi, S., Pourfathollah, A. A., Pourpak, Z., Heidarnazhad, H., and Kalvanagh, P. A. (2012). Cytokine Gene Polymorphism and Asthma Susceptibility, Progress and Control Level. Mol. Biol. Rep. 39, 1845–1853. doi:10.1007/s11033-011-0927-7
Egger, M., Smith, G. D., Schneider, M., and Minder, C. (1997). Bias in Meta-Analysis Detected by a Simple, Graphical Test. Bmj 315, 629–634. doi:10.1136/bmj.315.7109.629
Ferreira, M. A., Vonk, J., Vonk, J. M., Baurecht, H., Marenholz, I., Tian, C., et al. (2017). Shared Genetic Origin of Asthma, hay Fever and Eczema Elucidates Allergic Disease Biology. Nat. Genet. 49, 1752–1757. doi:10.1038/ng.3985
Fishman, D., Faulds, G., Jeffery, R., Mohamed-Ali, V., Yudkin, J. S., Humphries, S., et al. (1998). The Effect of Novel Polymorphisms in the Interleukin-6 (IL-6) Gene on IL-6 Transcription and Plasma IL-6 Levels, and an Association with Systemic-Onset Juvenile Chronic Arthritis. J. Clin. Invest. 102, 1369–1376. doi:10.1172/jci2629
Galli, S. J., and Tsai, M. (2012). IgE and Mast Cells in Allergic Disease. Nat. Med. 18, 693–704. doi:10.1038/nm.2755
Gentile, D. A., Yokitis, J., Angelini, B. L., Doyle, W. J., and Skoner, D. P. (2001). Effect of Intranasal challenge with Interleukin-6 on Upper Airway Symptomatology and Physiology in Allergic and Nonallergic Patients. Ann. Allergy Asthma Immunol. 86, 531–536. doi:10.1016/s1081-1206(10)62901-8
Gharagozlou, M., Farhadi, E., Khaledi, M., Behniafard, N., Sotoudeh, S., Salari, R., et al. (2013). Association between the Interleukin 6 Genotype at Position -174 and Atopic Dermatitis. J. Investig. Allergol. Clin. Immunol. 23, 89–93.
Hall, T., Rooks, R., and Kaufman, C. (2020). Intersections of Adverse Childhood Experiences, Race and Ethnicity and Asthma Outcomes: Findings from the Behavioral Risk Factor Surveillance System. Int. J. Environ. Res. Public Health 17 (21), 8236. doi:10.3390/ijerph17218236
Higgins, J. P. T., and Thompson, S. G. (2002). Quantifying Heterogeneity in a Meta-Analysis. Statist. Med. 21, 1539–1558. doi:10.1002/sim.1186
Hoffmans, R., Wagemakers, A., van Drunen, C., Hellings, P., and Fokkens, W. (2018). Acute and Chronic Rhinosinusitis and Allergic Rhinitis in Relation to Comorbidity, Ethnicity and Environment. PloS one 13, e0192330. doi:10.1371/journal.pone.0192330
Kämäräinen, O.-P., Solovieva, S., Vehmas, T., Luoma, K., Riihimäki, H., Ala-Kokko, L., et al. (2008). Common Interleukin-6 Promoter Variants Associate with the More Severe Forms of Distal Interphalangeal Osteoarthritis. Arthritis Res. Ther. 10, R21. doi:10.1186/ar2374
Kim, J. T., Kim, H. S., Chun, Y. H., Yoon, J.-s., and Kim, H. H. (2020). Effect of Multi-Ethnicity and Ancestry on Prevalence of Allergic Disease. J. Microbiol. Immunol. Infect. 53, 640–646. doi:10.1016/j.jmii.2018.10.004
Kosugi, E. M., de Camargo-Kosugi, C. M., Hirai, E. R., Mendes-Neto, J. A., Gregorio, L. C., Guerreiro-da-Silva, I. D. C., et al. (2013). Interleukin-6 -174 G/C Promoter Gene Polymorphism in Nasal Polyposis and Asthma. Rhin 51, 70–76. doi:10.4193/rhin12.166
Li, F., Xie, X., Li, S., Ke, R., Zhu, B., Yang, L., et al. (2015). Interleukin-6 Gene -174G/C Polymorphism and Bronchial Asthma Risk: A Meta-Analysis. Int. J. Clin. Exp. Med. 8, 12601–12608.
Liberati, A., Altman, D. G., Tetzlaff, J., Mulrow, C., Gøtzsche, P. C., Ioannidis, J. P. A., et al. (2009). The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies that Evaluate Health Care Interventions: Explanation and Elaboration. J. Clin. Epidemiol. 62, e1–e34. doi:10.1016/j.jclinepi.2009.06.006
Mahdaviani, S. A., Rezaei, N., Moradi, B., Dorkhosh, S., Amirzargar, A. A., and Movahedi, M. (2009). Proinflammatory Cytokine Gene Polymorphisms Among Iranian Patients with Asthma. J. Clin. Immunol. 29, 57–62. doi:10.1007/s10875-008-9232-1
Mehta, P., McAuley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., and Manson, J. J. (2020). COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. The Lancet 395, 1033–1034. doi:10.1016/s0140-6736(20)30628-0
Nasiri, R., Movahedi, M., Amirzargar, A. A., Hirbod-Mobarakeh, A., Farhadi, E., Ansaripour, B., et al. (2014). Association of Interleukin 6 Single Nucleotide Polymorphisms with Allergic Rhinitis. Int. J. Pediatr. Otorhinolaryngol. 78, 1426–1429. doi:10.1016/j.ijporl.2014.04.035
Nishi, A., Milner, D. A., Giovannucci, E. L., Nishihara, R., Tan, A. S., Kawachi, I., et al. (2016). Integration of Molecular Pathology, Epidemiology and Social Science for Global Precision Medicine. Expert Rev. Mol. Diagn. 16, 11–23. doi:10.1586/14737159.2016.1115346
Nutten, S. (2015). Atopic Dermatitis: Global Epidemiology and Risk Factors. Ann. Nutr. Metab. 66, 8–16. doi:10.1159/000370220
Pinart, M., Benet, M., Annesi-Maesano, I., von Berg, A., Berdel, D., Carlsen, K. C. L., et al. (2014). Comorbidity of Eczema, Rhinitis, and Asthma in IgE-Sensitised and Non-IgE-sensitised Children in MeDALL: A Population-Based Cohort Study. Lancet Respir. Med. 2, 131–140. doi:10.1016/s2213-2600(13)70277-7
Ray, A., LaForge, K. S., and Sehgal, P. B. (1990). On the Mechanism for Efficient Repression of the Interleukin-6 Promoter by Glucocorticoids: Enhancer, TATA Box, and RNA Start Site (Inr Motif) Occlusion. Mol. Cel Biol 10, 5736–5746. doi:10.1128/mcb.10.11.5736-5746.1990
Reich, K., Westphal, G., König, I. R., Mössner, R., Schupp, P., Gutgesell, C., et al. (2003). Cytokine Gene Polymorphisms in Atopic Dermatitis. Br. J. Dermatol. 148, 1237–1241. doi:10.1046/j.1365-2133.2003.05307.x
Rincon, M., and Irvin, C. G. (2012). Role of IL-6 in Asthma and Other Inflammatory Pulmonary Diseases. Int. J. Biol. Sci. 8, 1281–1290. doi:10.7150/ijbs.4874
Rivera-Chavez, F. A., Peters-Hybki, D. L., Barber, R. C., and O'Keefe, G. E. (2003). Interleukin-6 Promoter Haplotypes and Interleukin-6 Cytokine Responses. Shock 20, 218–223. doi:10.1097/00024382-200309000-00004
Settin, A., Zedan, M., Farag, M., Ezz El Regal, M., and Osman, E. (2008). Gene Polymorphisms of IL-6−174 G/C and IL-1Ra VNTR in Asthmatic Children. Indian J. Pediatr. 75, 1019–1023. doi:10.1007/s12098-008-0161-z
Spencer, S., Köstel Bal, S., Egner, W., Lango Allen, H., Raza, S. I., Ma, C. A., et al. (2019). Loss of the Interleukin-6 Receptor Causes Immunodeficiency, Atopy, and Abnormal Inflammatory Responses. J. Exp. Med. 216, 1986–1998. doi:10.1084/jem.20190344
Tanaka, T., Narazaki, M., and Kishimoto, T. (2014). IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harbor Perspect. Biol. 6, a016295. doi:10.1101/cshperspect.a016295
Terry, C. F., Loukaci, V., and Green, F. R. (2000). Cooperative Influence of Genetic Polymorphisms on Interleukin 6 Transcriptional Regulation. J. Biol. Chem. 275, 18138–18144. doi:10.1074/jbc.m000379200
Trajkov, D., Mirkovska-Stojkovikj, J., Arsov, T., Petlichkovski, A., Strezova, A., Efinska-Mladenovska, O., et al. (2008). Association of Cytokine Gene Polymorphisms with Bronchial Asthma in Macedonians. Iran J. Allergy Asthma Immunol. 7, 143–156.
Van Steen, K. (2012). Travelling the World of Gene-Gene Interactions. Brief. Bioinform. 13, 1–19. doi:10.1093/bib/bbr012
Vercelli, D. (2008). Discovering Susceptibility Genes for Asthma and Allergy. Nat. Rev. Immunol. 8, 169–182. doi:10.1038/nri2257
Xiaomin, L., Fenglin, C., Jianmin, H., Yuzhi, S., Binsheng, G., and Yingmei, Z. (2008). Correlation between Genetic Polymorphism of Cytokine Genes, Plasma Protein Levels and Bronchial Asthma in the Han People in Northern China. J. Asthma 45, 583–589. doi:10.1080/02770900802032925
Zeng, X., Zhang, Y., Kwong, J. S. W., Zhang, C., Li, S., Sun, F., et al. (2015). The Methodological Quality Assessment Tools for Preclinical and Clinical Studies, Systematic Review and Meta-Analysis, and Clinical Practice Guideline: A Systematic Review. J. Evidence-Based Med. 8, 2–10. doi:10.1111/jebm.12141
Zhang, Z., Xiao, C., Gibson, A. M., Bass, S. A., and Khurana Hershey, G. K. (2014). EGFR Signaling Blunts Allergen-Induced IL-6 Production and Th17 Responses in the Skin and Attenuates Development and Relapse of Atopic Dermatitis. J. Immunol. 192, 859–866. doi:10.4049/jimmunol.1301062
Zhao, N., Liu, H. J., Sun, Y. Y., and Li, Y. Z. (2016). Role of Interleukin-6 Polymorphisms in the Development of Allergic Rhinitis. Genet. Mol. Res. 15. doi:10.4238/gmr.15016987
Zhu, J., Yamane, H., and Paul, W. E. (2010). Differentiation of Effector CD4 T Cell Populations. Annu. Rev. Immunol. 28, 445–489. doi:10.1146/annurev-immunol-030409-101212
Keywords: IL-6, allergic diseases, meta-analysis, polymorphism, allergic rhinitis, allergic asthma, atopic dermatitis
Citation: Yang Y, Xiao J, Tang L, Wang B, Sun X, Xu Z, Liu L and Shi S (2022) Effects of IL-6 Polymorphisms on Individual Susceptibility to Allergic Diseases: A Systematic Review and Meta-Analysis. Front. Genet. 13:822091. doi: 10.3389/fgene.2022.822091
Received: 25 November 2021; Accepted: 18 January 2022;
Published: 16 March 2022.
Edited by:
Loredana Bury, University of Perugia, ItalyReviewed by:
Mirko Spiroski, Spiroski Scientific Foundation, North MacedoniaAhmad Setteen, Mansoura University, Egypt
Copyright © 2022 Yang, Xiao, Tang, Wang, Sun, Xu, Liu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suofang Shi, anNzc2YyMDA2QDEyNi5jb20=; Li Liu, anNzenl5ZnpAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Ying Yang
Ying Yang Jingxin Xiao2†
Jingxin Xiao2† Lingling Tang
Lingling Tang Zhongchi Xu
Zhongchi Xu