
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 03 February 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.812449
 Cun Rui1
Cun Rui1 Xiugui Chen2
Xiugui Chen2 Nan Xu2
Nan Xu2 Jing Wang2
Jing Wang2 Hong Zhang1
Hong Zhang1 Shengmei Li1
Shengmei Li1 Hui Huang2
Hui Huang2 Yapeng Fan2
Yapeng Fan2 Yuexin Zhang2
Yuexin Zhang2 Xuke Lu2
Xuke Lu2 Delong Wang2
Delong Wang2 Wenwei Gao1*
Wenwei Gao1* Wuwei Ye1,2*
Wuwei Ye1,2*Plant 3-ketoacyl-CoA synthase (KCS) gene family catalyzed a β ketoacyl-CoA synthase, which was the rate-limiting enzyme for the synthesis of very long chain fatty acids (VLCFAs). Gossypium barbadense was well-known not only for high-quality fiber, which was perceived as a cultivated species of Gossypium. In this study, a total of 131 KCS genes were identified in four cotton species, there were 38, 44, 26, 23 KCS genes in the G. barbadense, the G. hirsutum, the G. arboreum and G. raimondii, respectively. The gene structure and expression pattern were analyzed. GBKCS genes were divided into six subgroups, the chromosome distribution of members of the family were mapped. The prediction of cis-acting elements of the GBKCS gene promoters suggested that the GBKCS genes may be involved in hormone signaling, defense and the stress response. Collinearity analysis on the KCS genes of the four cotton species were formulated. Tandem duplication played an indispensable role in the evolution of the KCS gene family. Specific expression analysis of 20 GBKCS genes indicated that GBKCS gene were widely expressed in the first 25 days of fiber development. Among them, GBKCS3, GBKCS8, GBKCS20, GBKCS34 were expressed at a high level in the initial long-term level of the G. barbadense fiber. This study established a foundation to further understanding of the evolution of KCS genes and analyze the function of GBKCS genes.
As the basic components of lipids, fatty acids played an important role in plant growth and development and in resisting various stresses. Fatty acids can be classified according to the length of the hydrocarbon chain: short-chain fatty acids with a chain length of four to seven carbons; medium- and long chain fatty acids with a chain length of 8–18 carbons, and very long chain fatty acids (VLCFAs) with a chain length of 20 carbons and above. The biosynthesis of VLCFAs was divided into two stages and was completed in two different parts (plastids and endoplasmic reticulum) of the cell (Haslam and Kunst, 2013). The enzyme that controlled the extension of the fatty acid carbon chain was a complex that includes a variety of enzymes, including 3-ketoacyl-CoA synthase (KCS), β-hydroxyacyl-CoA dehydrogenase (HCD), β-ketoacyl-CoA reductase (KCR), trans-2,3-enoyl-CoA reductase (ECR), these four enzymes participate in the carbon chain elongation reaction and worked together to complete the synthesis of very long chain fatty acids (Kunst and Samuels, 2003; Bach and Faure, 2010). Long chain and very long chain fatty acids had a wide range of physiological functions. They participated in the formation of seed oils, biomembrane lipids and the synthesis of biologically active macromolecular sphingolipids (De Bigault Du Granrut and Cacas, 2016). In Arabidopsis, the hindered synthesis of VLCFAs could cause the apical meristem area of the stem to expand, and the structure of the fused rosette leaves, lateral roots, and branches changes (Faure et al., 1998; Bach et al., 2008; Beaudoin et al., 2009). VLCFAs can act as intercellular signals, inhibit cytokinin synthesis, regulate the proliferation of Arabidopsis stem apex cells (Nobusawa et al., 2013), and can also inhibit the totipotency of Arabidopsis pericylindrical cells and inhibit their formation of callus (Shang et al., 2016).
VLCFAs regulated fiber cell differentiation possibly via a lipid-activated transcriptional mechanism and may play an important role in fiber-cell initiation (Wang et al., 2015). Cotton fiber was a single-celled trichome differentiated from the epidermis of the ovule. It was a good model for studying cell elongation (Pang et al., 2010; Qin and Zhu, 2011). In cotton, in the ovule culture medium, exogenous application of saturated VLCFA can significantly promote the elongation of upland cotton fiber cells. During the development of cotton fiber, several KCS genes were highly upregulated, very long chain fatty acids (C20:0 to C30:0) significantly promoted cotton fiber cell elongation (Qin et al., 2007a; Qin et al., 2007b). Studies on the preferential expression of encoding fatty acid desaturases genes in fiber and exogenous application of linolenic acid (C18:3), soybean L-α-PI (PI: phosphatidylinositol), and phosphatidylinositol monophosphate (PIP) containing PIP 34:3 to promote fiber elongation had shown that fatty acids were involved in fiber elongation (Liu G.-J. et al., 2015).
KCS gene was responsible for the synthesis of wax component precursors and was the key rate-limiting enzyme for the extension of VLCFAs (Fan et al., 2018). Whole genome identification helps to understand and study gene function. 28, 16, 58, and 30 KCS genes were identified in apple (Lian et al., 2020), Citrus sinensis (Yang et al., 2021), Brassica napus (Xue et al., 2020), and Arachis hypogaea (Huai et al., 2020), respectively. There were 21 gene members in the KCS family in Arabidopsis (Joubès et al., 2008). Two domains: an ACP_syn_III_C domain and the Type III polyketide synthase-like protein domain had been proven necessary for KCS (Sagar et al., 2015; Guo et al., 2016; Huai et al., 2020). Researchers divided the KCS protein into four groups: FAE1, KCS1, FHD and CER6 based on the amino acid sequence homology (Costaglioli et al., 2005). The first identified gene in the KCS family was KCS18/FAE1, which could catalyze the elongation of C20 and C22 fatty acids in seeds (Rossak et al., 2001). In Arabidopsis, the gene KCS isolated from Lunaria annua caused a 30-fold increase in nervonic acid (cis-tetracos-15-enoic acid) proportions in seed oils (Guo et al., 2009). Arabidopsis and Brassica carinata were transformed with the Cardamine KCS, the C24:1 content of transgenic plants was increased 15-fold (Taylor et al., 2009). Two KCS genes WSL1 and ONI1 in rice were essential genes for leaf epidermal wax biosynthesis and the normal development of shoots (Lee and Suh, 2015).
Gossypium barbadense was pioneer species in the saline and alkaline land, was well-known for high-quality fiber. The completion of the splicing of the genomes of the four cotton species provides a basis for the comprehensive identification of the KCS genes of the four cotton species (Wang K. et al., 2012; Li et al., 2014; Hu et al., 2019). In our research, the KCS gene was comprehensively identified, and the evolution, chromosome location, and collinearity of the KCS gene were analyzed. It also focused on the analysis of the gene structure and promoter of GBKCS gene, and explored the expression characteristics of GBKCS gene during fiber development. This study established a foundation to further study of the mechanism of action of this family of genes and the biological function of KCS genes in the development of G. barbadense fibers.
The complete genome sequence data of four cotton species, Gossypium barbadense (ZJU), Gossypium hirsutum, (ZJU), Gossypium arboreum (CRI), and Gossypium raimondii (JGI), (Zhu et al., 2017) were obtained from Cotton FGD (Cotton Functional Genomics Database) (https://cottonfgd.org/). The amino acid sequences of KCSs from Arabidopsis thaliana were acquired from TAIR 10 (http://www.arabidopsis.org). The methods (hmmsearch and blastp) were used to identify the KCS proteins in cotton. HMMER (hmmer 3.2.1) was run based the HMM files in the local. Blastp was implemented with e-values of 1e-5 by BLAST (blast-2.12.0+). The amino acid sequences of the 3-Oxoacyl-[acyl-carrier protein (ACP)] synthase III C domain (Pfam: PF08541) and the Type III polyketide synthase-like protein domain (Pfam, PF08392) were used as queries.
To study the evolutionary relationship between KCS genes of different cotton species, the protein sequence was aligned by using the ClustalW program of MEGA7.0 software. The rootless phylogenetic tree was also constructed by MEGA7.0, and the parameters were set as follows: neighboring statistics, bootstrap replication 1,000 to validating the phylogenetic tree, and intraspecific phylogenetic relationships of KCS genes were all constructed by the same method (Tamura et al., 2013). G. barbadense reference genome gff3 downloaded from Cotton FGD file and gene ID file were used to map the chromosome positions of family members through TBtools software, the position of KCSs in the chromosomes of G. hirsutum, G. arboreum and G. raimondii was mapped by the same method.
MEME (http://meme-suite.org/) website was used to identify the conserved sequence of the protein. The maximum motif parameter of the gene is 15, the other of the parameters remain unchanged, and the domain files of the GBKCS family genes were obtained accordingly. The protein domain was analyzed through the TBtools software.
The 2000 bp DNA sequences upstream of GBKCSs gene were obtained from Cotton FGD (Cao et al., 2019; Malik et al., 2020). The cis-regulatory elements in the promoter region of the gene GBKCSs were predicted on the Plant CARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), choosing the hormone (IAA: auxin- AuxRR-core, ABA: abscisic acid- ABRE, SA: salicylic acid, MeJA: methyl jasmonate- CGTCA-motif, TGACG-motif), transcription factor binding, development, circadian control, defense and stress responsiveness, meristem expression related cis-acting elements to analysis. We used TBtools software settng parameters (upstream bases: 2000, check retain only upstream) to check the sequence. The GBKCSs under different stress (salt, 100 mM NaCl, alkaline, 50 mM Na2CO3, treated for 12 h) were selected. RNA-Seq data (PRJNA734700) was downloaded from National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/. nih. gov/). FPKM (fragments per kilobase of exon model per million mapped fragments) were used to analyze the expression level. Following, the summary graph which included cis-acting elements, evolutionary relationships and heatmaps was drawn by using TBtools software.
KCSs gene locus were obtained from the gff3 file. The gff files of the genome were merged as needed. The protein sequence of all the genes were aligned, then the gene pair file of every two cotton species were obtained by using MCScanX. The collinearity analysis results were demonstrated from TBtools by combining the chromosome length files.
The ovule and fiber of cotton (G. barbadense cv Jiza 67) plants was used as experimental material. The flowers were tagged on the day of blooming [0 day post anthesis (DPA)] for collecting cotton fibre samples. Ovules (0 DPA) were carefully peeled off from developing cotton bolls. Fibre samples (5 DPA, 10 DPA, 15 DPA, 20 DPA and 25 DPA) were separated from developing cotton seeds. Ovules and fibre from different plants were tidied and immediately stored in a refrigerator at −80°C for further use.
The total RNA of the ovules and fibre were extracted separately according to the RNAprep Pure Plant Plus Kit (Polysaccharides andPolyphenolics-rich) of TIANGEN instruction. The cDNA of each sample was prepared by TransScript@ All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) instructions. The primer sequences are listed in the Supplementary Table S1. The equipment used for qRT-PCR was Applied Biosystems@ 7,500 Fast, and the fluorescence quantification kit was TransStart Top Green qPCR SuperMix. The ΔΔCt method (Livak and Schmittgen, 2001) was used to calculate the processing results which contained three biological replicates.
In preliminary analysis, 131 KCS genes were identified as KCS family members from four cotton species (G. barbadense, G. hirsutum, G. arboreum, and G. raimondii) (Supplementary Table S2). A total of 38 KCS genes were identified in G. barbadense. The encoded protein ranges from 411 (GBKCS4) to 580 (GBKCS30) amino acids, with pI varying from 8.281 (GBKCS36) to 9.732 (GBKCS4) and MWs varying from 46.015 (GBKCS4) kDa to 65.427 kDa (GBKCS30) (Supplementary Table S3). Through the similarity of the number of genes in two tetraploid and the sum of diploid, we found the KCS gene number of two diploid cotton species was higher than Arabidopsis (Joubès et al., 2008). The result indicated that the KCS family in cotton has undergone enlargement during the evolution. They were named GaCXE1-26, GrCXE1-23, GBKCS1-38, GHCXE1-44, according to their position on the chromosome in four different cotton variants.
To clarify the evolutionary and relationship between G. barbadense (GB), G. hirsutum (GH), G. arboreum (Ga), G. raimondii (Gr), Arabidopsis (At), a total of 152 protein sequences were used for sequence alignment. The evolutionary relationship between each sequence was constructed and the rootless phylogenetic tree was generated by using MEGA7 neighbor-joining method (Figure 1). The KCS proteins were divided according to the number and structural characteristics of the domains in Arabidopsis (Joubès et al., 2008). All the 152 protein sequences were parted into eight subclasses: α, β, γ, δ, ε, ζ, η and θ. The results showed that, in addition to β and η, the KCS proteins of the four cotton species were distributed in each group, and the number of KCS proteins in α, ζ, δ, θ subclasses was much more than that of the other two groups.
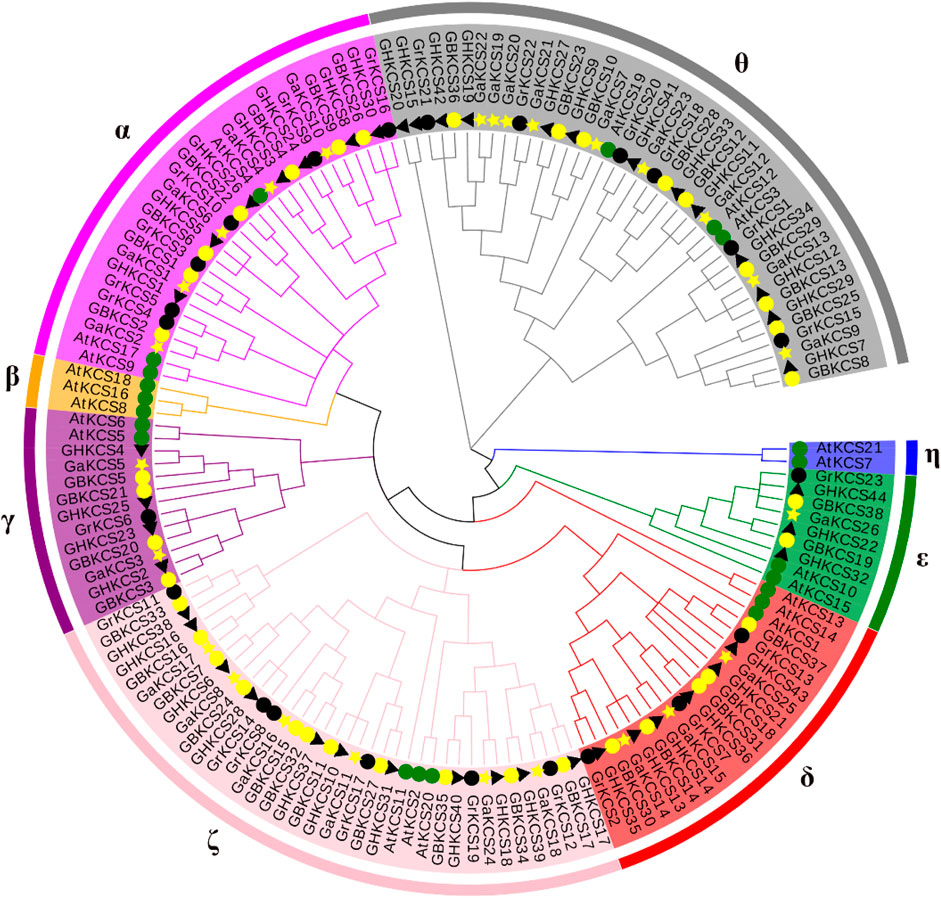
FIGURE 1. Phylogenetic analysis of KCS protein from Gossypium and Arabidopsis. (The rootless phylogenetic tree was constructed by MEGA7.0).
To explore the conservative structure of KCS gene family, the amino acid sequence encoded by the G. barbadense KCS gene was analyzed through the online website MEME. These motifs performed different functions and were distributed in the sequence of each subgroup (Figure 2). Compared with other subgroups, all members (GBKCS19, GBKCS38) of the ε subgroup did not contain motif 10, and consisted of three adjacent exons. In the θ subgroup, three members (GBKCS23, GBKCS10, GBKCS36) lacked motif 15, we also noticed that the GBKCS23, GBKCS10, GBKCS36, GBKCS12, and GBKCS28 of the subgroup did not contain motif 13. In the γ subgroup, the sequences of GBKCS3 and GBKCS21, GBKCS20 and GBKCS5 were highly similar and consisted of the same motifs respectively. GBKCS4, a member of the α group, lacked motif 15, and GBKCS1 had two exons. The similar motif arrangements in the same subgroup implicited that the protein structure was conserved in a specific subgroup, the gene had a certain degree of conservation, and the functions of most conserved motifs remain to be elucidated.
The gff3 file of the genome and the gene ID information were used to understand the specific distribution of genes on chromosomes more intuitively. The KCS genes of four cottons were located on the chromosomes of the corresponding cotton species (Figure 3). The KCS genes of G. arboreum were located on 12 other chromosomes except chromosome 11 (Figure 4B), among them, the number of genes on chromosome 10 was the largest, with a total of six genes (GBKCS19-24). In G. raimondii, D03, D07, D12 of this cotton species had no KCS family members (Figure 4D). The number of genes on the chromosomes of the two subgroups was the same in two tetraploid, and the genes are located in the same D1t and D2t chromosomes. The distribution of the KCS gene on the A subgenome chromosomes of G. barbadense (AD2) and G. hirsutum (AD1) was quite different (Figures 4A,C), that may because the addition or loss of genes through duplication events (entire genome duplication, tandem duplication, fragment duplication).
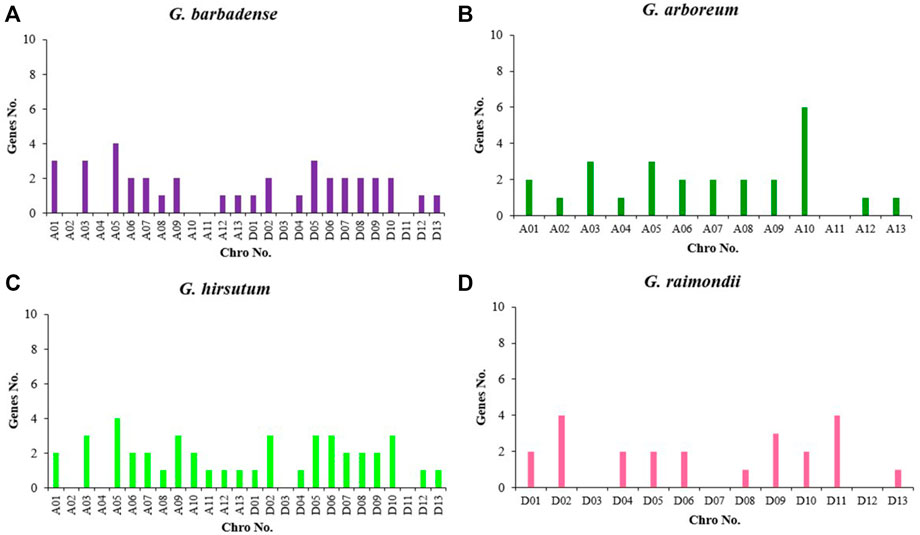
FIGURE 4. Distribution of the number of genes in the KCS genes family of four cotton species. (A) G. barbadense; (B) G. arboreum; (C) G. hirsutum; (D) G. raimondii.
Cis-acting elements occur in the promoter region upstream of the gene, which may bind to transcription factors for regulating gene transcription and responding to different environmental factors. The response element of GBKCSs were identified by using the evolutionary relationship and the 2 kb region file upstream of the start codon. These cis-acting elements were involved in the hormone response, stress induction, and the defense reaction, implying that KCS protein played important roles in hormone response and stress induction in plants. Most of the promoters from GBKCS gene family members contain drought, light, low-temperature, ABA, GA, MeJA, MYB response elements (Figure 5B) (Supplementary Table S5). We can explore and understand the mechanism of gene response to different plant hormones and abiotic stress through promoter analysis. To further investigate the responsive mechanism of GBKCS gene family members against abiotic stresses, RNA-Seq data was used to determine the differences in gene expression of family members under salt-alkaline stress (Figure 5C). Under the salt-alkali stress, the expression level of all members has changed to a certain degree. Except for GBKCS10 and GBKCS23, the expression of other genes in the θ subgroup (Figure 5A) showed a downward trend after salt-alkali stress. Two genes in the ε subgroup (GBKCS19, GBKCS38) showed high expression levels. Interestingly, gene expression changed under different stresses, even though in the same subgroup, the functions of gene members were also diverse. These results indicated that GBKCS members were involved in the regulation of abiotic stress, the response expression of different members was different, and the expression of the same gene under different stress was also different.
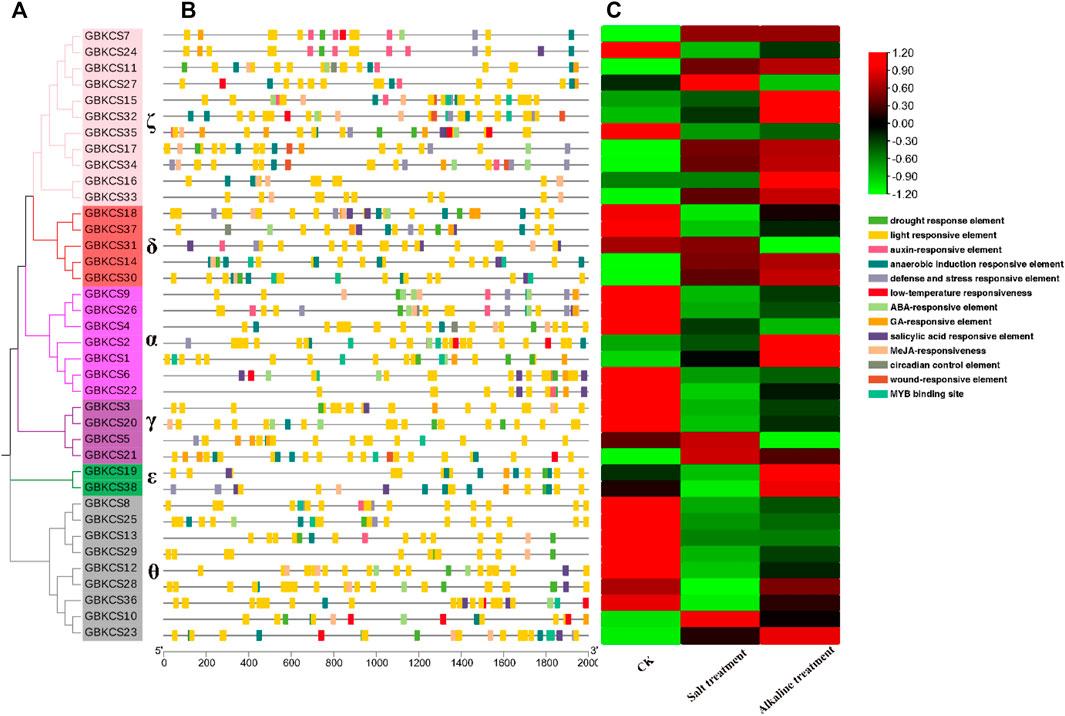
FIGURE 5. Analysis of promoters and differentially expressed genes of GBKCSs family. (A) Phylogenetic tree of GBKCSs; (B) Cis-elements in promoters of GBKCS genes; (C) Differentially expressed genes of GBKCS genes under salt and alkaline stress. (The control sample was designated as CK. The Na2CO3 processed sample designated as Alkaline, and the NaCl processed sample designated as Salt).
Whole genome duplication, segmental duplication and tandem duplication were three processes of gene families arises evolution (Xu et al., 2012). To describe the positional relationship of the homology of genes, the amplification mechanism and the sequence of arrangement, we performed a collinearity analysis on the KCS genes of the four cotton species (Figure 6). MCScanX (Wang Y. et al., 2012) was used to find homologous gene pairs, a total of 680 gene pairs were identified. 2 (GaKCS19- GaKCS20, GaKCS20- GaKCS21) and 1 (GrKCS4- GrKCS5) pair of tandem repeats were identified in Ga-Ga and Gr-Gr, respectively. The two Ga-Ga gene pairs were in the same branch of the evolutionary tree, and the Gr-Gr tandem repeat gene pair was a homologous gene pair. There was a pair of tandem repeats in GB-GB, while no pair was identified in GH-GH. Tandem duplication played an indispensable role in the evolution of the KCS gene family. 15, 7, 66, 59 gene pairs were identified in Ga-Ga, Gr-Gr, GH-GH, GB-GB, respectively. The Gr and Gr collinearity gene pairs were the least among the groups, with seven pairs, meanwhile GB and GH was the most, with 172 pairs. From the perspective of the number of gene pairs, this was consistent with the number of KCS genes in diploid and tetraploid genes.
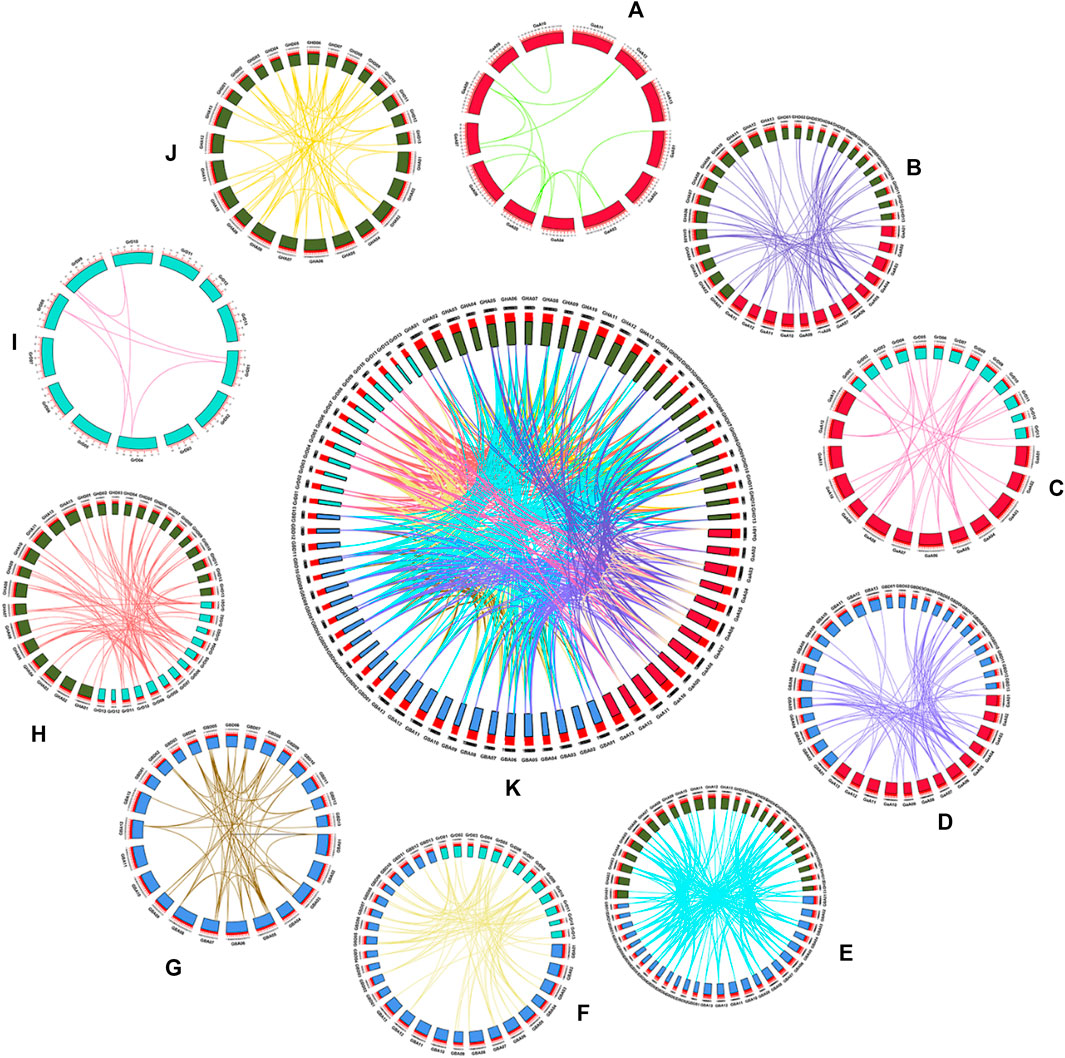
FIGURE 6. The collinearity of KCS genes within and among the four cotton species genomes (Gossypium hirsutum, Gossypium barbadense, Gossypium arboreum and Gossypium raimondii). Chromosomal lines represented by various colors indicate the syntenic regions around the KCS genes. (A) Ga-Ga, (B) Ga-GH, (C) Ga-Gr, (D) Ga-GB, (E) GB-GH, (F) GB-Gr, (G) GB-GB, (H) Gr-GH, (I) Gr-Gr, (J) GH-GH, (K) Summary of 10 combinations of collinearity.
Gene duplication may contribute to the expansion of gene families. The selection pressure of gene duplication gene pairs in the evolution process can be studied by calculating non-synonymous (Ka) to synonymous (Ks) substitution rate. Ka/Ks > 1 and Ka/Ks < 1 were considered positive selection and purification selection, respectively (Hurst, 2002). The homologous gene pairs Ka and Ks and Ka/Ks of four cotton species in 10 combinations (Ga-Ga, Ga-GH, Ga-Gr, Ga-GB, GB-GH, GB-Gr, GB-GB, Gr-GH, Gr-Gr, GH-GH) were measured (Figure 7). In our results, there were four gene pairs with Ka/Ks > 1, and the ratio of 567 gene pairs was less than one (Supplementary Table S4), which meant that the KCS gene family in the four cotton species underwent strong purification selection and remained highly conserved.
Research on KCS wax synthesis genes was mainly focused on oil crops. From the previous evolutionary relationship, motif and promoter element analysis of our research, we found that genes in the same subgroup have the same or similar structure and conservative motifs. In order to explore the potential role of GBKCS gene in the process of fiber development, ovules or fibers of different stages were prepared. According to the evolutionary relationship, a total of 20 genes were selected from each group (Figure 9A). The expression levels of selected genes at different stages of fiber development were finally determined (Figure 8). These genes were α (GBKCS2, GBKCS4, GBKCS9, GBKCS26), γ (GBKCS3, GBKCS20), δ (GBKCS18, GBKCS31, GBKCS37), ε (GBKCS19), ζ (GBKCS11, GBKCS16, GBKCS27, GBKCS33, GBKCS34), θ (GBKCS8, GBKCS12, GBKCS13, GBKCS25, GBKCS29) members of subgroups. In order to show more intuitively the expression patterns of these 20 GBKCS genes at different stages of fiber development, we made a heat map (Figure 9B). The representative genes of the α subgroup showed high expression levels on the 10–20 days, while the expression levels of GBKCS2, GBKCS13 and GBKCS4 were higher at the early stage of fiber development (5 days), and we found that the expression level of GBKCS4 during 10–25 days was lower than that of the ovule. The expression levels of GBKCS3 and GBKCS20 genes from the same subgroup at 10, 15, and 20 days were significantly higher than that of the ovule. The expression levels of genes GBKCS18, GBKCS31 and GBKCS37 were different in different periods. The gene GBKCS16 of the ζ subgroup was only expressed at a high level on the 10 days. The members of the θ group GBKCS8, GBKCS12 and GBKCS25 all expressed high levels on the 10 days.
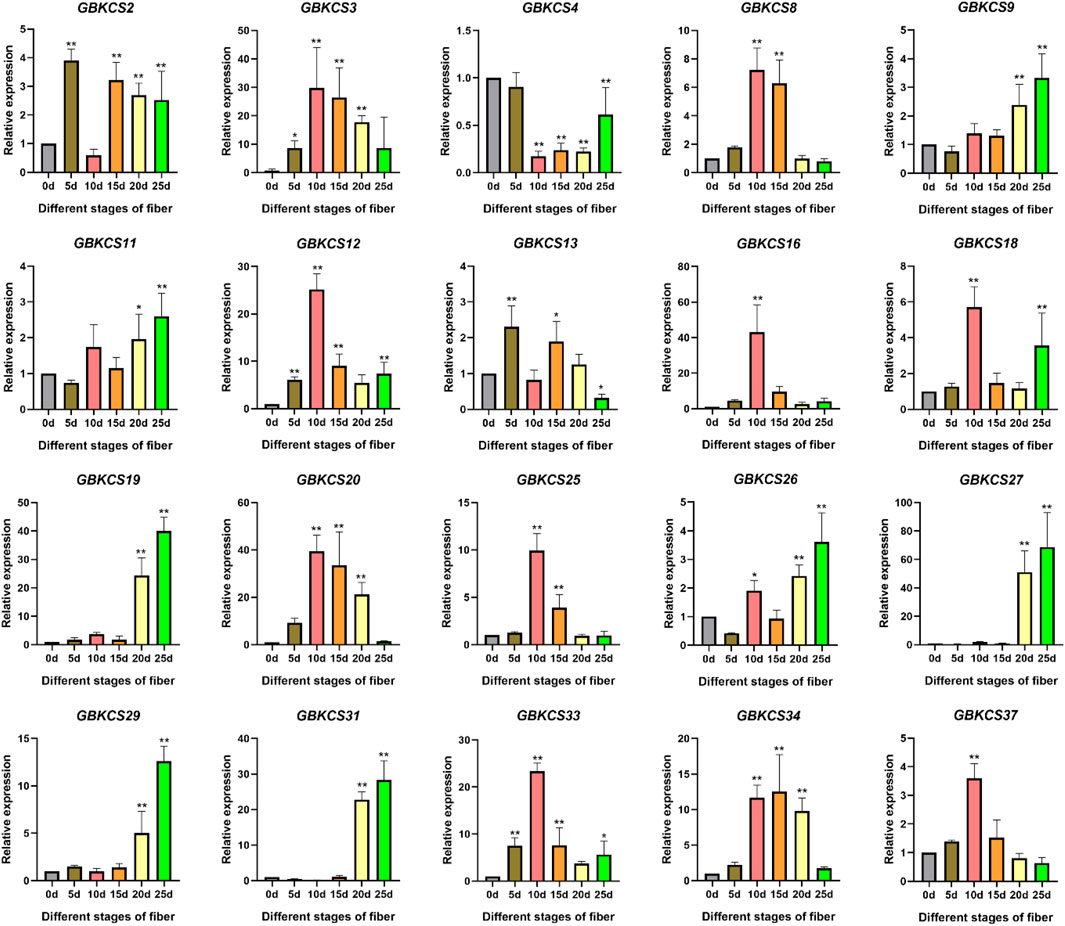
FIGURE 8. Analysis of differentially expressed KCS gene family members in G. barbadense (*p < 0.05, **p < 0.01).
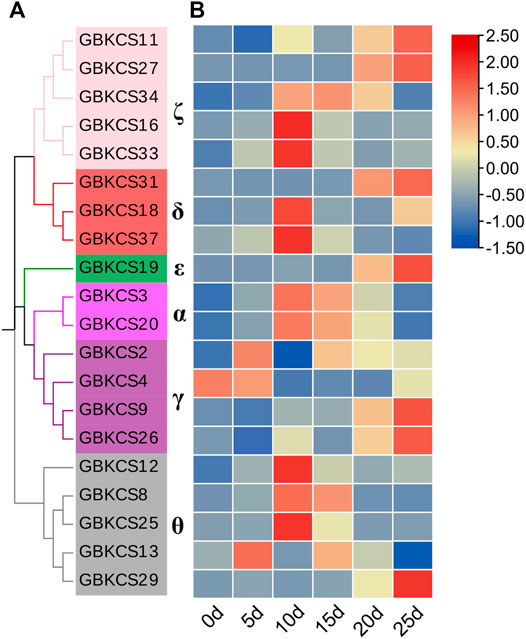
FIGURE 9. Analysis of the expression patterns of 20 genes in the GBKCS gene family. (A) The evolutionary relationship of 20 GBKCS genes. (B) Expression model of 20 GBKCS genes at different stages of fiber development. Each column represented the expression of different genes in the same sample, and each row represented the expression of the same gene in different samples. The heat map was drawn with TBtools software.
For functional analysis of GBKCS proteins, we combined the promoter element and the heat map analysis of saline-alkali stress (Figures 5B,C), and finally GBKCS34 that was expressed at a high level after stress was selected. The homologous gene of GBKCS34 in Arabidopsis was obtained according to blastp. STRING database (https://string-db.org/) was used to construct an interaction network to analyze GBKCS protein function (Figure 10). Overview of GBKCS proteins, more than 90% of the protein were involved in the pathway module of fatty acid biosynthesis, elongation, endoplasmic reticulum (M00415). The proteins were enriched in fatty acid elongation (ko00062) and biosynthesis of secondary metabolites (ko01110) metabolic pathways. According to GBKCS34 promoter analysis, the sequence contained light, low-temperature, GA, ABA, auxin, MeJA responsive element. Take this, the function of GBKCS protein can be further deduced. The function of GBKCS protein was speculated to be based on the study of AtKCSs. The Arabidopsis thaliana ECERIFERUM1 (CER1) protein was an essential element of wax alkane synthesis, GBKCS34 interacted with protein 3-ketoacyl-CoA reductase (KCR1, KCR2), CER1, CER3, CER4, CER10. Studies have reported that AtCER1 and AtCER3 were overexpressed in yeast, and it was found that AtCER1 and AtCER3 interacted to synthesize VLCFAs into alkanes (Bernard et al., 2012). The analysis of the KCS protein promoter elements and protein network interactions would help us understand the protein’s mechanism of action.
KCS gene exists in almost all plant species (Guo et al., 2016). The relationship between individual or group genetic differences and gene redundancy were clarified through research on gene families (Liu H.-J. et al., 2015). Our results showed that 26, 23, 38, 44 KCS genes had been identified in four cotton species (G. arboreum, G. raimondii, G. barbadense, G. hirsutum) (Supplementary Table S2). The number of genes in each species explained the origin of cotton tetraploid to a certain extent. Genes were often acquired and lost in the process of evolution. This increase and deletion of genes leaded to differences in the number of KCS genes in cotton. Based on the evolutionary relationship of the KCS family in Arabidopsis (Joubès et al., 2008), we combined the amino acid sequence to divide the KCS genes in the four cotton species into eight subgroups (α, β, γ, δ, ε, ζ, η and θ) (Figure 1). The analysis results of motif and gene structure (Figure 2) showed that most of GBKCS members were relatively conservative with one exon and no intron structure.
Replication events were one of the main drivers of evolutionary diversification of genomes and genetic systems (Gu et al., 2003). Non-functionalization, sub-functionalization and new functionalization were the three evolutionary directions of proteins (Yang et al., 2017). The total number of Brassica napus KCS genes was less than the sum of Brassica rapa and Brassica oleracea (Xue et al., 2020), and the members of the Brassica napus FAD gene family were also accompanied by a decrease in the number of genes after polyploidization (Xue et al., 2018; Xu et al., 2019). Environmental conditions and artificial selection affected the number of gene family members. The doubling of the gene sequence in cotton would occur with the occurrence of repetitive events, and some redundant genes would be selectively lost or recombined (Li et al., 2015). In our research results, consistent with Brassica napus was that the KCS gene in G. barbadense, G. hirsutum cotton was less than the sum of G. arboreum, and G. raimondii. The function of normal KCS genes may be affected by non-synonymous mutations, and even affected the normal growth of plants. Studies have reported that most of the Citrinae KCS genes were synonymous mutations (Yang et al., 2021). However, WSL1, a member of the rice KCS gene family, which was involved in leaf wax accumulation, the total amount of epidermal wax in its mutant leaves and leaf sheaths was significantly reduced, and VLCFA precursors of C20-C24 decreased in both (Yu et al., 2008). In tomato slcer6 mutants, SlCER6 loss of function provokes a decrease of n- and iso-alkanes with chain lengths of C27 or greater and of anteiso-alkanes with chain lengths of C28 or greater in flower cuticular waxes. This mutation ultimately resulted in impaired fertility and changes in flower shape (Smirnova et al., 2013). Allotetraploid genomes had lower selection pressure than diploid genomes (Li et al., 2015). Existing studies had revealed that purification selection was the main evolutionary force acting on the FAE1 gene (Sun et al., 2013). Consistent with this, our results showed that the Ka/Ks of more than 90% of the KCS gene pairs in the four cotton species were far below one. Therefore, it can be inferred that KCS had undergone purification selection during the evolution of cotton.
VLCFAs were the components of plant cuticle wax layer and cell plasma membrane, which played an important role in the survival and development of plants (Bernard and Joubès, 2013; Xue et al., 2017). KCS genes determined whether fatty acids will be extended and the number and types of VLCFAs produced (Lechelt-Kunze et al., 2003; Dongxin et al., 2015; Ozseyhan et al., 2018). The transcription of CsKCS6 was changed in response to drought stress, salt stress and abscisic acid (ABA) treatment. CsKCS6 was essential in the synthesis of fatty acid precursors involved in wax synthesis, and it also enhanced the tolerance of transgenic Arabidopsis to drought and salt stress (Guo et al., 2020). In our results, some KCS genes of G. barbadense responded to salt-alkaline stress (Figure 5C). Genes such as GBKCS34, GBKCS2, GBKCS1, and GBKCS13 were significantly up-regulated, indicating that the members of the KCS gene family of G. barbadense play a role in saline-alkali abiotic stress.
To have a more comprehensive understanding of the expression patterns of KCS family genes, GBKCS genes were selected to be classified according to the subgroups of evolutionary relationships. The 20 genes we selected from each subgroup were widely expressed in the 0–25 days of fiber development (Figure 8). GBKCS34 was a homologous gene of AtKCS20, all of which belonged to the ζ subgroup. AtKCS20, AtKCS2, AtKCS5 were found to be enzymatically active with endogenous yeast fatty acid substrates and to some extent with externally supplied unsaturated substrates, involved in the synthesis of saturated and monounsaturated fatty acids 20–30 carbon atoms in length (Trenkamp et al., 2004; Paul et al., 2006). The abundance of regulatory elements in the promoter region may be the result of selective pressure (Xiao et al., 2016). Analysis of promoter action elements suggested that pathways such as low temperature induction, light, and plant hormones may regulate the transcription of GBKCS34 by combining these action elements, and then played a regulatory role in these pathways. In addition, the expression of GBKCS34 in 10 DPA, 15 DPA, and 20 DPA was significantly higher than the other three periods, which also indicated that this gene was involved in the development of fiber elongation.
The KCS family genes of the G. barbadense, G. hirsutum, G. arboreum, G. raimondii genomes were analyzed. A total of 131 KCS genes were identified in four cotton species through screening in silico. We analyzed the developmental and evolutionary relationship of the four KCS genes, chromosome location, and collinearity. The gene structure, motif, promoter elements, and expression patterns of G. barbadense KCS were analyzed. The family had undergone purification selection during evolution. Some GBKCS genes responded to salt-alkali stress and were also widely expressed at different stages of fiber development. Furthermore, the high expression of GBKCS3, GBKCS8, GBKCS20 and GBKCS34 in a specific period implied the potential function of these genes in the fiber elongation period. This work had enriched our knowledge of the KCS gene of G. barbadense and laid the foundation for further exploring the mechanism of the KCS family gene in the development of G. barbadense fiber.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
WY designed the project, CR conceived and designed the experiments, writing original draft. XC, NX, and JW provided Methodology, Software. HZ, SL, and YF were responsible for writing-review and editing. HH, YZ, XL, and DW provided the Methodology. WG and WY were mainly responsible for overseeing and revising the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was supported by China Agriculture Research System of MOF and MARA (CARS-15-02) and National natural science foundation of China (general) (31771885).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.812449/full#supplementary-material
Supplementary Table S1 | Primer sequence of GBKCS.
Supplementary Table S2 | KCS members of the four cotton species.
Supplementary Table S3 | Physical properties of GBKCS proteins.
Supplementary Table S4 | Prediction of duplicated gene pairs involved in different combinations from four Gossypium species.
Supplementary Table S5 | Promoter sequence and classification information.
Bach, L., and Faure, J.-D. (2010). Role of Very-Long-Chain Fatty Acids in Plant Development, when Chain Length Does Matter. Comptes rendus biologies 333, 361–370. doi:10.1016/j.crvi.2010.01.014
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., et al. (2008). The Very-Long-Chain Hydroxy Fatty Acyl-CoA Dehydratase PASTICCINO2 Is Essential and Limiting for Plant Development. Proc. Natl. Acad. Sci. 105, 14727–14731. doi:10.1073/pnas.0805089105
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional Characterization of the Arabidopsis β-Ketoacyl-Coenzyme A Reductase Candidates of the Fatty Acid Elongase. Plant Physiol. 150, 1174–1191. doi:10.1104/pp.109.137497
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J.-D., et al. (2012). Reconstitution of Plant Alkane Biosynthesis in Yeast Demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 Are Core Components of a Very-Long-Chain Alkane Synthesis Complex. Plant Cell 24, 3106–3118. doi:10.1105/tpc.112.099796
Bernard, A., and Joubès, J. (2013). Arabidopsis Cuticular Waxes: Advances in Synthesis, export and Regulation. Prog. lipid Res. 52, 110–129. doi:10.1016/j.plipres.2012.10.002
Cao, X., Duan, W., Wei, C., Chen, K., Grierson, D., and Zhang, B. (2019). Genome-Wide Identification and Functional Analysis of Carboxylesterase and Methylesterase Gene Families in Peach (Prunus Persica L. Batsch). Front. Plant Sci. 10, 1511. doi:10.3389/fpls.2019.01511
Costaglioli, P., Joubès, J., Garcia, C., Stef, M., Arveiler, B., Lessire, R., et al. (2005). Profiling Candidate Genes Involved in Wax Biosynthesis in Arabidopsis thaliana by Microarray Analysis. Biochim. Biophys. Acta (Bba) - Mol. Cell Biol. Lipids 1734, 247–258. doi:10.1016/j.bbalip.2005.04.002
De Bigault Du Granrut, A., and Cacas, J.-L. (2016). How Very-Long-Chain Fatty Acids Could Signal Stressful Conditions in Plants? Front. Plant Sci. 7, 1490. doi:10.3389/fpls.2016.01490
Fan, Y., Yuan, C., Jin, Y., Hu, G.-R., and Li, F.-L. (2018). Characterization of 3-Ketoacyl-coA Synthase in a Nervonic Acid Producing Oleaginous Microalgae Mychonastes Afer. Algal Res. 31, 225–231. doi:10.1016/j.algal.2018.02.017
Faure, J. D., Vittorioso, P., Santoni, V., Fraisier, V., Prinsen, E., Barlier, I., et al. (1998). The PASTICCINO Genes of Arabidopsis thaliana Are Involved in the Control of Cell Division and Differentiation. Development 125, 909–918. doi:10.1007/s00429005013410.1242/dev.125.5.909
Gu, Z., Steinmetz, L. M., Gu, X., Scharfe, C., Davis, R. W., and Li, W.-H. (2003). Role of Duplicate Genes in Genetic Robustness against Null Mutations. Nature 421, 63–66. doi:10.1038/nature01198
Guo, H.-S., Zhang, Y.-M., Sun, X.-Q., Li, M.-M., Hang, Y.-Y., and Xue, J.-Y. (2016). Evolution of the KCS Gene Family in Plants: the History of Gene Duplication, Sub/neofunctionalization and Redundancy. Mol. Genet. Genomics 291, 739–752. doi:10.1007/s00438-015-1142-3
Guo, W., Wu, Q., Yang, L., Hu, W., Liu, D., and Liu, Y. (2020). Ectopic Expression of CsKCS6 from Navel orange Promotes the Production of Very-Long-Chain Fatty Acids (VLCFAs) and Increases the Abiotic Stress Tolerance of Arabidopsis thaliana. Front. Plant Sci. 11, 564656. doi:10.3389/fpls.2020.564656
Guo, Y., Mietkiewska, E., Francis, T., Katavic, V., Brost, J. M., Giblin, M., et al. (2009). Increase in Nervonic Acid Content in Transformed Yeast and Transgenic Plants by Introduction of a Lunaria Annua L. 3-Ketoacyl-CoA Synthase (KCS) Gene. Plant Mol. Biol. 69, 565–575. doi:10.1007/s11103-008-9439-9
Haslam, T. M., and Kunst, L. (2013). Extending the story of Very-Long-Chain Fatty Acid Elongation. Plant Sci. 210, 93–107. doi:10.1016/j.plantsci.2013.05.008
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium Hirsutum Genomes Provide Insights into the Origin and Evolution of Allotetraploid Cotton. Nat. Genet. 51, 739–748. doi:10.1038/s41588-019-0371-5
Huai, D., Xue, X., Li, Y., Wang, P., Li, J., Yan, L., et al. (2020). Genome-Wide Identification of Peanut KCS Genes Reveals that AhKCS1 and AhKCS28 Are Involved in Regulating VLCFA Contents in Seeds. Front. Plant Sci. 11, 406. doi:10.3389/fpls.2020.00406
Huai, D., Zhang, Y., Zhang, C., Cahoon, E. B., and Zhou, Y. (2015). Combinatorial Effects of Fatty Acid Elongase Enzymes on Nervonic Acid Production in Camelina Sativa. PLOS ONE 10, e0131755. doi:10.1371/journal.pone.0131755
Hurst, L. D. (2002). The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. 18, 486–487. doi:10.1016/S0168-9525(02)02722-1
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA Elongase Gene Family in Arabidopsis thaliana: Phylogenetic Analysis, 3D Modelling and Expression Profiling. Plant Mol. Biol. 67, 547–566. doi:10.1007/s11103-008-9339-z
Kunst, L., and Samuels, A. L. (2003). Biosynthesis and Secretion of Plant Cuticular Wax. Prog. lipid Res. 42, 51–80. doi:10.1016/s0163-7827(02)00045-0
Lechelt-Kunze, C., Meissner, R. C., Drewes, M., and Tietjen, K. (2003). Flufenacet Herbicide Treatment Phenocopies the Fiddlehead Mutant in Arabidopsis thaliana. Pest Manag. Sci. 59, 847–856. doi:10.1002/ps.714
Lee, S. B., and Suh, M. C. (2015). Advances in the Understanding of Cuticular Waxes in Arabidopsis thaliana and Crop Species. Plant Cell Rep 34, 557–572. doi:10.1007/s00299-015-1772-2
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome Sequence of Cultivated Upland Cotton (Gossypium Hirsutum TM-1) Provides Insights into Genome Evolution. Nat. Biotechnol. 33, 524–530. doi:10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome Sequence of the Cultivated Cotton Gossypium Arboreum. Nat. Genet. 46, 567–572. doi:10.1038/ng.2987
Lian, X.-Y., Wang, X., Gao, H.-N., Jiang, H., Mao, K., You, C.-X., et al. (2020). Genome Wide Analysis and Functional Identification of MdKCS Genes in Apple. Plant Physiol. Biochem. 151, 299–312. doi:10.1016/j.plaphy.2020.03.034
Liu, G.-J., Xiao, G.-H., Liu, N.-J., Liu, D., Chen, P.-S., Qin, Y.-M., et al. (2015a). Targeted Lipidomics Studies Reveal that Linolenic Acid Promotes Cotton Fiber Elongation by Activating Phosphatidylinositol and Phosphatidylinositol Monophosphate Biosynthesis. Mol. Plant 8, 911–921. doi:10.1016/j.molp.2015.02.010
Liu, H.-J., Tang, Z.-X., Han, X.-M., Yang, Z.-L., Zhang, F.-M., Yang, H.-L., et al. (2015b). Divergence in Enzymatic Activities in the Soybean GST Supergene Family Provides New Insight into the Evolutionary Dynamics of Whole-Genome Duplicates. Mol. Biol. Evol. 32, 2844–2859. doi:10.1093/molbev/msv156
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. methods 25, 402–408. doi:10.1006/meth.2001.1262
Malik, W. A., Wang, X., Wang, X., Shu, N., Cui, R., Chen, X., et al. (2020). Genome-wide Expression Analysis Suggests Glutaredoxin Genes Response to Various Stresses in Cotton. Int. J. Biol. macromolecules 153, 470–491. doi:10.1016/j.ijbiomac.2020.03.021
Nobusawa, T., Okushima, Y., Nagata, N., Kojima, M., Sakakibara, H., and Umeda, M. (2013). Synthesis of Very-Long-Chain Fatty Acids in the Epidermis Controls Plant Organ Growth by Restricting Cell Proliferation. Plos Biol. 11, e1001531. doi:10.1371/journal.pbio.1001531
Ozseyhan, M. E., Kang, J., Mu, X., and Lu, C. (2018). Mutagenesis of the FAE1 Genes Significantly Changes Fatty Acid Composition in Seeds of Camelina Sativa. Plant Physiol. Biochem. 123, 1–7. doi:10.1016/j.plaphy.2017.11.021
Pang, C.-Y., Wang, H., Pang, Y., Xu, C., Jiao, Y., Qin, Y.-M., et al. (2010). Comparative Proteomics Indicates that Biosynthesis of Pectic Precursors Is Important for Cotton Fiber and Arabidopsis Root Hair Elongation. Mol. Cell Proteomics 9, 2019–2033. doi:10.1074/mcp.M110.000349
Paul, S., Gable, K., Beaudoin, F., Cahoon, E., Jaworski, J., Napier, J. A., et al. (2006). Members of the Arabidopsis FAE1-like 3-Ketoacyl-CoA Synthase Gene Family Substitute for the Elop Proteins of Saccharomyces cerevisiae. J. Biol. Chem. 281, 9018–9029. doi:10.1074/jbc.M507723200
Qin, Y.-M., Hu, C.-Y., Pang, Y., Kastaniotis, A. J., Hiltunen, J. K., and Zhu, Y.-X. (2007a). Saturated Very-Long-Chain Fatty Acids Promote Cotton Fiber and Arabidopsis Cell Elongation by Activating Ethylene Biosynthesis. The Plant Cell 19, 3692–3704. doi:10.1105/tpc.107.054437
Qin, Y.-M., Pujol, F. M., Hu, C.-Y., Feng, J.-X., Kastaniotis, A. J., Hiltunen, J. K., et al. (2007b). Genetic and Biochemical Studies in Yeast Reveal that the Cotton Fibre-specific GhCER6 Gene Functions in Fatty Acid Elongation. J. Exp. Bot. 58, 473–481. doi:10.1093/jxb/erl218
Qin, Y.-M., and Zhu, Y.-X. (2011). How Cotton Fibers Elongate: a Tale of Linear Cell-Growth Mode. Curr. Opin. Plant Biol. 14, 106–111. doi:10.1016/j.pbi.2010.09.010
Rossak, M., Smith, M., and Kunst, L. (2001). Expression of the FAE1 Gene and FAE1 Promoter Activity in Developing Seeds of Arabidopsis thaliana. Plant Mol. Biol. 46, 717–725. doi:10.1023/A:1011603923889
Sagar, M., Pandey, N., Qamar, N., Singh, B., and Shukla, A. (2015). Domain Analysis of 3 Keto Acyl-CoA Synthase for Structural Variations in Vitis vinifera and Oryza Brachyantha Using Comparative Modelling. Interdiscip. Sci. Comput. Life Sci. 7, 7–20. doi:10.1007/s12539-013-0017-8
Shang, B., Xu, C., Zhang, X., Cao, H., Xin, W., and Hu, Y. (2016). Very-long-chain Fatty Acids Restrict Regeneration Capacity by Confining Pericycle Competence for Callus Formation in Arabidopsis. Proc. Natl. Acad. Sci. USA 113, 5101–5106. doi:10.1073/pnas.1522466113
Smirnova, A., Leide, J., and Riederer, M. (2012). Deficiency in a Very-Long-Chain Fatty Acid β-Ketoacyl-Coenzyme A Synthase of Tomato Impairs Microgametogenesis and Causes Floral Organ Fusion. Plant Physiol. 161, 196–209. doi:10.1104/pp.112.206656
Sun, X., Pang, H., Li, M., Peng, B., Guo, H., Yan, Q., et al. (2013). Evolutionary Pattern of the FAE1 Gene in Brassicaceae and its Correlation with the Erucic Acid Trait. PLoS One 8, e83535. doi:10.1371/journal.pone.0083535
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. 30, 2725–2729. doi:10.1093/molbev/mst197
Taylor, D. C., Francis, T., Guo, Y., Brost, J. M., Katavic, V., Mietkiewska, E., et al. (2009). Molecular Cloning and Characterization of aKCSgene fromCardamine Graecaand its Heterologous Expression inBrassicaoilseeds to Engineer High Nervonic Acid Oils for Potential Medical and Industrial Use. Plant Biotechnol. J. 7, 925–938. doi:10.1111/j.1467-7652.2009.00454.x
Trenkamp, S., Martin, W., and Tietjen, K. (2004). Specific and Differential Inhibition of Very-Long-Chain Fatty Acid Elongases from Arabidopsis thaliana by Different Herbicides. Proc. Natl. Acad. Sci. 101, 11903–11908. doi:10.1073/pnas.0404600101
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012a). The Draft Genome of a Diploid Cotton Gossypium Raimondii. Nat. Genet. 44, 1098–1103. doi:10.1038/ng.2371
Wang, X.-C., Li, Q., Jin, X., Xiao, G.-H., Liu, G.-J., Liu, N.-J., et al. (2015). Quantitative Proteomics and Transcriptomics Reveal Key Metabolic Processes Associated with Cotton Fiber Initiation. J. Proteomics 114, 16–27. doi:10.1016/j.jprot.2014.10.022
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012b). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40, e49. doi:10.1093/nar/gkr1293
Xiao, G.-H., Wang, K., Huang, G., and Zhu, Y.-X. (2016). Genome-scale Analysis of the Cotton KCS Gene Family Revealed a Binary Mode of Action for Gibberellin A Regulated Fiber Growth. J. Integr. Plant Biol. 58, 577–589. doi:10.1111/jipb.12429
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. 109, 1187–1192. doi:10.1073/pnas.1109047109
Xu, L., Zeng, W., Li, J., Liu, H., Yan, G., Si, P., et al. (2019). Characteristics of Membrane-Bound Fatty Acid Desaturase (FAD) Genes in Brassica Napus L. And Their Expressions under Different Cadmium and Salinity Stresses. Environ. Exp. Bot. 162, 144–156. doi:10.1016/j.envexpbot.2019.02.016
Xue, D., Zhang, X., Lu, X., Chen, G., and Chen, Z.-H. (2017). Molecular and Evolutionary Mechanisms of Cuticular Wax for Plant Drought Tolerance. Front. Plant Sci. 8, 621. doi:10.3389/fpls.2017.00621
Xue, Y., Chen, B., Wang, R., Win, A. N., Li, J., and Chai, Y. (2018). Genome-wide Survey and Characterization of Fatty Acid Desaturase Gene Family in Brassica Napus and its Parental Species. Appl. Biochem. Biotechnol. 184, 582–598. doi:10.1007/s12010-017-2563-8
Xue, Y., Jiang, J., Yang, X., Jiang, H., Du, Y., Liu, X., et al. (2020). Genome-wide Mining and Comparative Analysis of Fatty Acid Elongase Gene Family in Brassica Napus and its Progenitors. Gene 747, 144674. doi:10.1016/j.gene.2020.144674
Yang, H., Mei, W., Wan, H., Xu, R., and Cheng, Y. (2021). Comprehensive Analysis of KCS Gene Family in Citrinae Reveals the Involvement of CsKCS2 and CsKCS11 in Fruit Cuticular Wax Synthesis at Ripening. Plant Sci. 310, 110972. doi:10.1016/j.plantsci.2021.110972
Yang, Z., Gong, Q., Qin, W., Yang, Z., Cheng, Y., Lu, L., et al. (2017). Genome-wide Analysis of WOX Genes in upland Cotton and Their Expression Pattern under Different Stresses. BMC Plant Biol. 17, 1–17. doi:10.1186/s12870-017-1065-8
Yu, D., Ranathunge, K., Huang, H., Pei, Z., Franke, R., Schreiber, L., et al. (2008). Wax Crystal-Sparse Leaf1 Encodes a β-ketoacyl CoA Synthase Involved in Biosynthesis of Cuticular Waxes on rice Leaf. Planta 228, 675–685. doi:10.1007/s00425-008-0770-9
Keywords: Gossypium barbadense, 3-ketoacyl-CoA synthase, long chain fatty acids, phylogenetic analysis, expression analysis
Citation: Rui C, Chen X, Xu N, Wang J, Zhang H, Li S, Huang H, Fan Y, Zhang Y, Lu X, Wang D, Gao W and Ye W (2022) Identification and Structure Analysis of KCS Family Genes Suggest Their Reponding to Regulate Fiber Development in Long-Staple Cotton Under Salt-Alkaline Stress. Front. Genet. 13:812449. doi: 10.3389/fgene.2022.812449
Received: 10 November 2021; Accepted: 12 January 2022;
Published: 03 February 2022.
Edited by:
Subhojit Datta, Indian Council of Agricultural Research, IndiaReviewed by:
Dawei Xue, Hangzhou Normal University, ChinaCopyright © 2022 Rui, Chen, Xu, Wang, Zhang, Li, Huang, Fan, Zhang, Lu, Wang, Gao and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wuwei Ye, eWV3MTU4QDE2My5jb20=; Wenwei Gao, Z3d3MDkxMUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.