
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet. , 31 October 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1009938
This article is part of the Research Topic Individual and Population Specific Variation in Cancer Susceptibility, Prevalence, and Disease Presentation View all 8 articles
 Chuncheng Yi1,2
Chuncheng Yi1,2 Tiandong Li1,2
Tiandong Li1,2 Yajing Shen1,2
Yajing Shen1,2 Peng Wang1,2
Peng Wang1,2 Liping Dai2,3
Liping Dai2,3 Jianxiang Shi2,3
Jianxiang Shi2,3 Keyan Wang2,3
Keyan Wang2,3 Changqing Sun1,4*
Changqing Sun1,4* Hua Ye1,2*
Hua Ye1,2*Background: Reduced DNA repair capacity in nucleotide excision repair (NER) pathways owing to genetic variant may influence cancer susceptibility. According to published studies, variants of NER genes associations with colorectal cancer (CRC) risk were inconclusive. Thus, this meta-analysis aimed to explore the possible association. A trial sequence analysis (TSA) analysis was performed to control the risk of false positive or false negative.
Methods: PubMed, Web of Science, Embase, Cochrane Library, China National Knowledge Network (CNKI), Wanfang Database and Scientific and Technical Journal Database (VIP) were searched to identify relative studies until April 2022. The association was assessed by odds ratio (OR) in Allele, homozygous, heterozygous, dominant, recessive, and over-dominant models. In addition, Begg’s and Egger’s tests, sensitivity analysis, subgroup analysis and TSA analysis were performed.
Results: A total of 29 studies were eventually included in the meta-analysis, including 12,153 CRC patients and 14,168 controls. It showed that excision and repair cross complementary group 1 (ERCC1) rs11615 CC genotype decreased the risk of CRC, compared with TT genotype (CC vs. TT: OR = 0.816, 95% CI = 0.673–0.990, p = 0.039). For ERCC1 rs3212986, the significant impact was detected on increased the risk of CRC in the allele (OR = 1.267, 95% CI = 1.027–1.562, p = 0.027), homozygous (OR = 1.805, 95% CI = 1.276–2.553, p = 0.001), dominant (OR = 1.214, 95% CI = 1.012–1.455, p = 0.037) and recessive (OR = 1.714, 95% CI = 1.225–2.399, p = 0.002) models, especially in the Asian population. The results revealed the association of ERCC2 rs1799793 A allele with a higher risk of CRC (A vs. G: OR = 1.163, 95% CI = 1.021–1.325, p = 0.023). It also showed that ERCC5 rs17655 increased CRC risk in the allele (OR = 1.104, 95% CI = 1.039–1.173, p = 0.001), homozygous (OR = 1.164, 95% CI = 1.018–1.329, p = 0.026), heterozygous (OR = 1.271, 95% CI = 1.018–1.329, p < 0.001), dominant (OR = 1.241, 95% CI = 1.135–1.358, p < 0.001) and over-dominant (OR = 0.828, 95% CI = 0.762–0.900, p < 0.001) models, especially among Asians.
Conclusion: This meta-analysis based on current evidence suggests that the significant association was observed between ERCC1 rs11615, ERCC1 rs3212986, ERCC2 rs1799793, and ERCC5 rs17655 and CRC susceptibility. However, given the limited sample size and the influence of genetic background, studies of a larger scale and well-designed are required to confirm the results.
Globally, colorectal cancer (CRC) has the third highest incidence and second highest mortality rate, with a 5-year relative survival rate of 65% (Sung et al., 2021). By 2030, 2.2 million new cases and 1.1 million cancer deaths will occur, up 60% from today (Arnold et al., 2017). The mechanisms of CRC are multifactorial and multistage, influenced by hereditary, environmental factors, and their interaction (Khan et al., 2017). Smoking, excessive drinking and a diet heavy in red meat are all identified as potential risk factors of CRC (Baroudi and Benammar-Elgaaied, 2016). Besides, approximately 20%–30% of CRC patients have a family history of the disease, which demonstrates that genetic factors have been considered to be necessary in the development of CRC (Butterworth et al., 2006; Rasool et al., 2014).
DNA repair systems play a crucial role in preventing mutations or repairing DNA damage caused by environmental and genetic carcinogens (Sancar et al., 2004; Chatterjee and Walker, 2017). Nucleotide excision repair (NER) pathway is the first and most important step in any DNA repair process. It is a general mechanism for removing helical twisted DNA damage and structure from the genome (Marteijn et al., 2014). Its major types of damage repair involves radiation, chemotherapeutic agents and/or mutagens, such as cyclobutane pyrimidine dimer (CPD) and pyrimidine 6-4 (64 PP) caused by Ultraviolet B (UVB) radiation (Gillet and Schärer, 2006). There are two subpaths in NER. One is transcription-coupled NER(TC-NER) and the other is global genome NER(GG-NER). They are selective about which DNA strands are transcribed in the expressed genes. Many genes have been identified in the NER pathway, such as Xeroderma pigmentosum group C (XPC), XPA, excision and repair cross complementary group 1 (ERCC1), ERCC2/XPD, ERCC4/XPF, ERCC5/XPG (Hanawalt, 2002; Al-Shaheri et al., 2020).
Mutations in NER genes may result in a variety of phenotypes, range from an predisposition to cancer, to neurodevelopmental abnormalities defects associated with premature ageing (Marteijn et al., 2014). For example, XPC patients with defects in GG-NER have a 1,000-fold increased susceptibility to sun-induced skin cancer, as well as an increased risk of various internal tumors (DiGiovanna and Kraemer, 2012). Currently, the relationship between variants of NER gene and CRC risk has been reported in different populations. Some reports have shown that variants in NER genes lead to considerable differences in DNA repair ability between individuals, thus affecting their susceptibility to CRC (Dziki et al., 2017). Many studies have demonstrated the association between ERCC2 rs13181 polymorphism and CRC among Asians (Yeh et al., 2007; Gil et al., 2012; Kabzinski et al., 2015), while others indicated no association (Jelonek et al., 2010; Wang et al., 2010). Paszkowska-Szczur et al. (2015) reported the association between single nucleotide polymorphisms (SNPs) of seven NER genes and CRC risk in the Polish population, which confirmed that variants in XPC rs2228000, ERCC2 rs1799793 and rs238406 might be associated with CRC risk. ERCC5 rs17655 variant related to increased CRC susceptibility in some case-control studies among Polish and Chinese populations (Gil et al., 2012; Su et al., 2019) but not others (Steck et al., 2014; Kabzinski et al., 2015). Due to the inconsistent and incomprehensive results of previous studies, this paper aims to systematically evaluate the association between SNPs in NER genes and the risk of CRC.
This meta-analysis adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement (Page et al., 2021).
The terms used in search strategy were “colorectal OR colonic OR rectal OR colon OR rectum,” “cancer OR carcinoma OR neoplasms OR tumor,” “Single Nucleotide Polymorphism OR Polymorphism OR SNP OR variant OR variation,” and “nucleotide excision repair OR NER OR DNA repair OR excision repair.” In addition, studies published as of 6 April 2022 were extracted from PubMed, Cochrane Library, Web of Science, Embase, China National Knowledge Network (CNKI), Wanfang Database, and Scientific and Technical Journal Database (VIP) to meet the search strategy, regardless of language restrictions. Detailed strategies are shown in Supplementary Information.
Inclusion criteria for the study were as follows: 1) It was a case-control study; 2) The difference in SNP frequency between CRC patients and healthy controls was compared; 3) The studied SNPs were NER gene SNPs; 4) Cancer risk is the outcome; 5) Cases were serum samples from CRC patients who did not receive chemotherapy; 6) The control group included healthy individuals with different age, sex, country, and tumor stage and patients with non-malignant disease.
Two authors (Chuncheng Yi and Tiandong Li) independently extracted the literature. Data extracted from individual papers included: author, publication year, country, ethnicity, sample size, control type, gender composition, age at diagnosis, and details of target SNPs, including genotyping method, genotype frequency, and source from Hardy-Weinberg equilibrium (HWE).
The quality of the studies that met the inclusion criteria was assessed. Two reviewers assessed the methodological quality of each study against the Newcastle-Ottawa scale (NOS) for case-control study (Stang, 2010). Scores greater than six are considered high quality, and the total score was nine.
A 95% confidence interval (95% CI) and odds ratio (OR) were utilized to determine the association between SNPs in NER gene and risk of CRC, and OR was analyzed using the Z test. Six different genetic models were used to test the association, including allelic, homozygous, heterozygous, dominant, recessive, and over-dominant models. Heterogeneity was assessed by Q test and I2 tests. I2 < 25% was considered to be of low heterogeneity, 25%–50% to be of moderate heterogeneity and 50% was considered a high level of heterogeneity. p < 0.05 or I2 > 50% was considered significant for heterogeneity, and a Dersimonian-Laird random effect model was applied if heterogeneity was not detected; otherwise, a Mantel-Haenszel fixed effect model was applied. Subgroup analyses were performed based on ethnicity and source of control to explore potential heterogeneity. A sensitivity analysis was performed to assess the stability of the results by excluding each study individually. The funnel plot evaluated whether publication bias was present (both Begg’s and Egger’s tests). For all statistical analyses, Stata version 14.0 software was used (StataCorp LP, College Station, TX, United States).
To control the risk of false positive or false negative, a trial sequential analysis (TSA) method was performed. If the curve exceeds the traditional boundary but does not cross the TSA boundary, the prompt may make a false positive error; If the curve intersects TSA boundary, the meta-analysis results are stable, even if required information size (RIS), which is the target value needed to make a reliable statistical inference, is not reached; If the curve does not intersect with both the traditional and TSA bounds, no positive or negative conclusion can be drawn; If the curve intersects an invalid line, the hint is meaningless. We performed TSA with a type I error risk of 5%, a power of 90% and a relative risk reduction of 15%, using TSA version 0.9.5.10 software (Copenhagen Trial Unit, Copenhagen, Denmark; http://www.ctu.dk/tsa).
The flow chart of this study is shown in Figure 1. In total, 6,617 relevant articles were initially retrieved in this study including two articles obtained from cited references. Among them, 3,615 duplicated articles, 2,148 articles with the irrelevant subject after reading titles and abstracts, 536 articles with SNPs other than NER gene, 67 review or meta-analysis articles and 32 not case-control studies were excluded. After evaluating the remaining 217 articles, 12 studies with unavailable data or data do not meet the HWE, and 178 studies reporting SNP in less than three studies. Therefore, a total of 29 studies (Moreno et al., 2006; Skjelbred et al., 2006; Stern et al., 2006; Yeh et al., 2007; Pardini et al., 2008; Sliwinski et al., 2009; Wang et al., 2009; Engin et al., 2010; Jelonek et al., 2010; Wang et al., 2010; Canbay et al., 2011; Gil et al., 2012; Liu et al., 2012; Du et al., 2014; Hou et al., 2014; Ni et al., 2014; Steck et al., 2014; Gómez-Díaz et al., 2015; Kabzinski et al., 2015; Paszkowska-Szczur et al., 2015; Sun et al., 2015; Yang et al., 2015; Yueh et al., 2017; Zhang et al., 2017; Zhang et al., 2018; Jin et al., 2019; Su et al., 2019; Balkan et al., 2020; Li et al., 2020) were included with 12,153 CRC patients and 14,168 controls. The characteristics of the studies included in the meta-analysis were showed in Table 1. The genotype distributions of eight SNPs in the five genes in this study agreed with the Hardy-Weinberg equilibrium (p > 0.05). The result of quality assessment is shown in Supplementary Table S1 and all included studies were of high quality. Table 2 shows the genotype frequency distributions of included studies.
Upon analysis of these eight NER-SNPs, we found that four of them were significantly associated with CRC risk. Detailed results are shown in Supplementary Table S2.
This meta-analysis showed that ERCC1 rs11615 homozygous CC genotype, but not the heterozygous CT genotype, decreased the risk of CRC, compared with the wild-type TT genotype (CC vs. TT: OR = 0.816, 95% CI = 0.673–0.990, p = 0.039; CT vs. TT: OR = 0.864, 95% CI = 0.711–1.050, p = 0.141) (Figure 2), however, this association was not shown in subgroup analysis of ethnicity and source of control. For ERCC1 rs3212986, the significant impact was detected on increased the risk of CRC under allele, homozygous, dominant and recessive models (A vs. C: OR = 1.267, 95% CI = 1.027–1.562, p = 0.027; AA vs. CC: OR = 1.805, 95% CI = 1.276–2.553, p = 0.001; AA+AC vs. CC: OR = 1.214, 95% CI = 1.012–1.455, p = 0.037; AA vs. AC+CC: OR = 1.714, 95% CI = 1.225–2.399, p = 0.002) (Figure 3). The stratified analysis by ethnicity and source of control revealed a significantly higher CRC risk was found for the Asian population and the population-based control subgroup (Supplementary Figure S1).
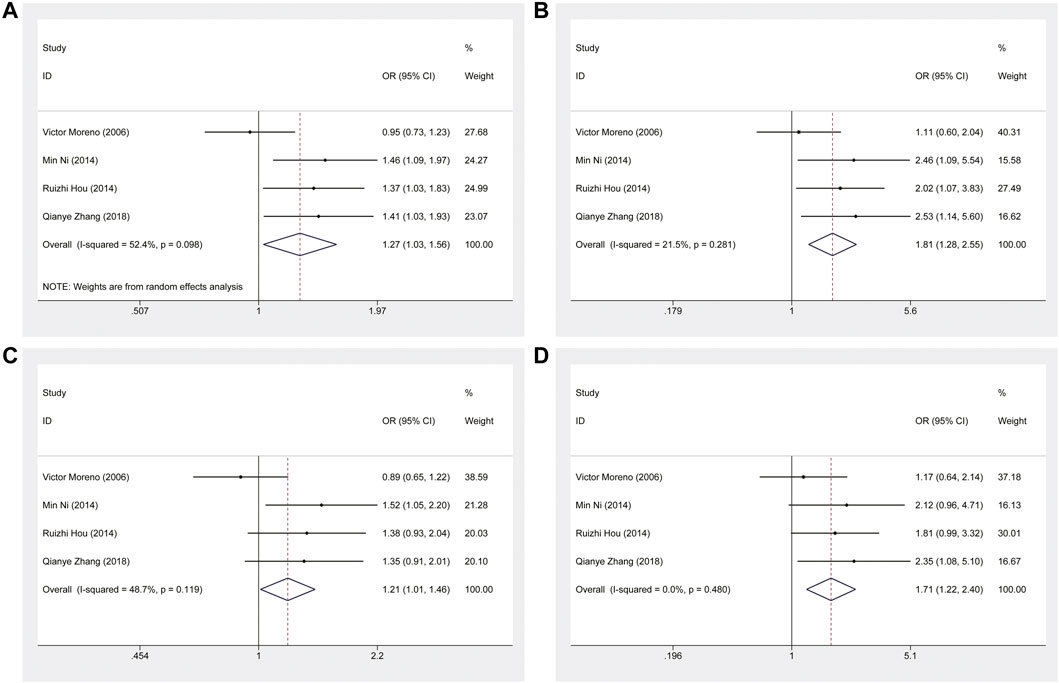
FIGURE 3. Forest plot related to ERCC1 rs3212986 and risk of CRC. (A) Allele model (B) Homozygous model (C) Dominant model (D) Recessive model.
For ERCC2 rs1799793, our meta-analysis showed that compared with G allele, A allele was associated with an increased risk of CRC (A vs. G: OR = 1.163, 95% CI = 1.021–1.325, p = 0.023) (Figure 4). And the association was also shown in the Caucasian population (Supplementary Figure S2). About the risk of CRC and ERCC2 rs13181, there was no significant evidence of an association between them when all eligible studies were included under all genetic models. However, the stratified analysis by ethnicity revealed that ERCC2 rs13181 CC/CA genotype was associated with a significantly increased risk of CRC, compared with AA genotype among the Asian population. (CC+CA vs. AA: OR = 1.205, 95% CI = 1.032–1.408, p = 0.018) (Supplementary Figure S3).
The results demonstrated that significant associations between ERCC5 rs17655 and CRC risk were noted under five genetic models. The correlation results under allele model was OR = 1.104(C vs. G: 95% CI = 1.039–1.173, p = 0.001), homozygous model was OR = 1.164(CC vs. GG: 95% CI = 1.018–1.329, p = 0.026), heterozygous model was OR = 1.271(CG vs. GG: 95% CI = 1.018–1.329, p < 0.001), dominant model was OR = 1.241(CC+CG vs. GG: 95% CI = 1.135–1.358, p < 0.001) and over-dominant model was OR = 0.828(CC+GG vs. CG: 95% CI = 0.762–0.900, p < 0.001) (Figure 5). Moreover, these significant associations exist in the Asian population under five genetic models and in the population-based control and hospital-based control subgroup under four models except the homozygous model (Supplementary Figure S4).
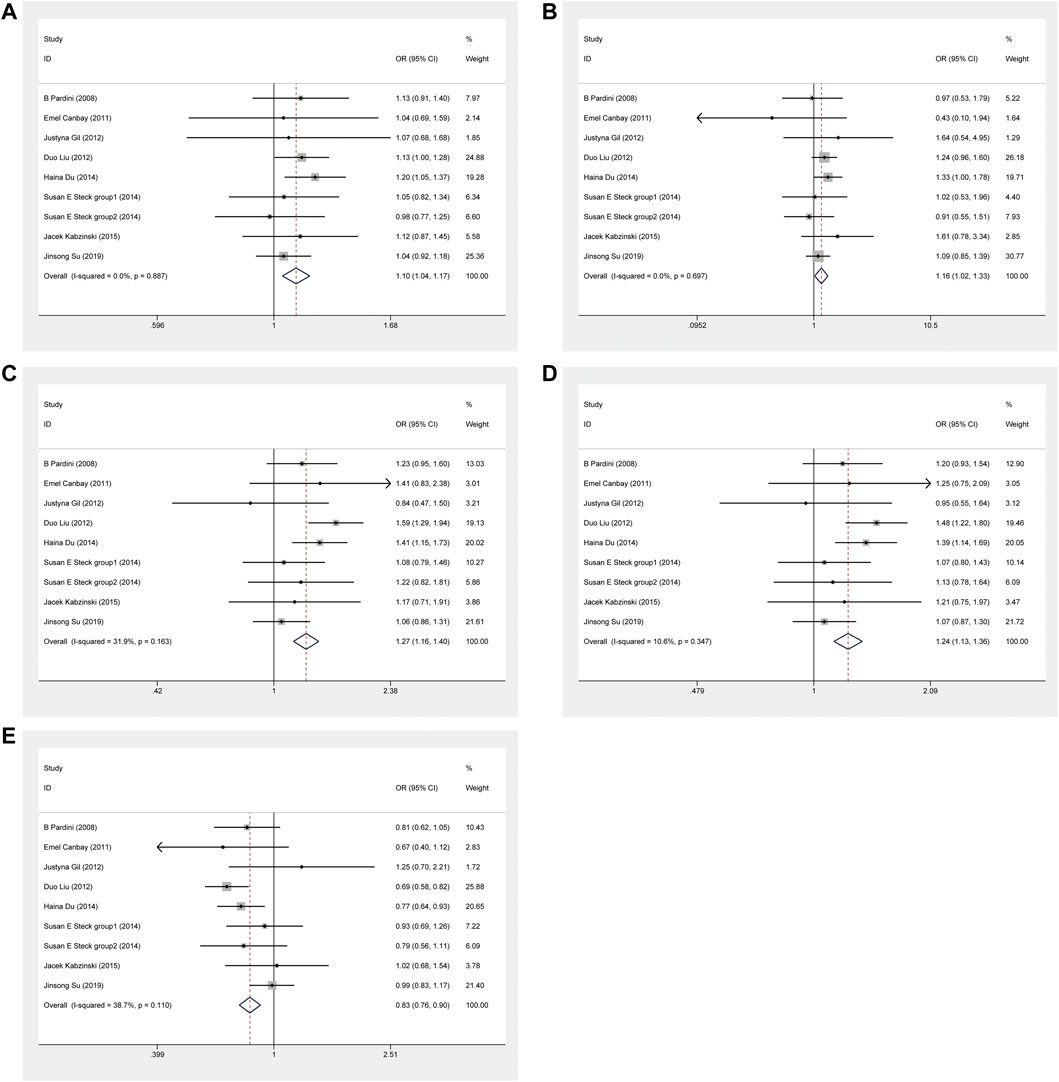
FIGURE 5. Forest plot related to ERCC5 rs17655 and risk of CRC. (A) Allele model (B) Homozygous model (C) Heterozygous model (D) Dominant model (E) Over-dominant model.
No significant association was observed between ERCC4 rs1800067 variant and CRC susceptibility in the six genetic models, nor in the stratified analysis. For XPC rs2228001 and rs2228000 variants, although no significant associations exist in the overall meta-analysis, ethnic subgroup analysis showed that significant correlations were noted in reducing or increasing the risk of CRC in Asian, Caucasian and African populations under different models. The detailed results are shown in Supplementary Figures S5, S6.
Under the allelic model, ERCC1 rs3212986 had high heterogeneity (I2 = 52.4%, p = 0.098). We then assessed the source of heterogeneity by ethnicity and control source. The results showed that heterogeneity among the Asian and population-based control subgroups decreased. Then, sensitivity analyses were performed to assess the stability of the results by deleting each study in sequence. The results showed that none of the studies significantly changed the combined OR, indicating that the combined OR of this variant was robust. Heterogeneity was low for the remaining meaningful SNPs, and the sensitivity analysis results were robust. Supplementary Figures S7–S10 for details.
Begg’s funnel plot and Egger’s regression test were performed to estimate publication bias. The results showed no significant asymmetry of funnel plot shape (Supplementary Figures S7–S10), and Begg’s and Egger’s test also showed that the evaluation of publication bias had no statistical significance.
As shown in Figures 6, 7, although the accumulated information has not reached RIS, the Z curve has crossed the traditional boundary and TSA boundary. The cumulative evidence was sufficient to the association between ERCC1 rs11615 variant and CRC risk under homozygous model, similar to ERCC2 rs1799793. For ERCC1 rs3212986 variant in the four genetic models, the Z curve only crossed the traditional boundary, and the sample size did not reach RIS, indicating that there was a high possibility of false positives in the results. Therefore, more subsequent studies are needed for further verification to make this association valid (Figure 8). For ERCC5 rs17655 variant (Figure 9), its Z curve crossed the traditional boundary and TSA boundary, and the sample size reached RIS, demonstrating that the results were robust.
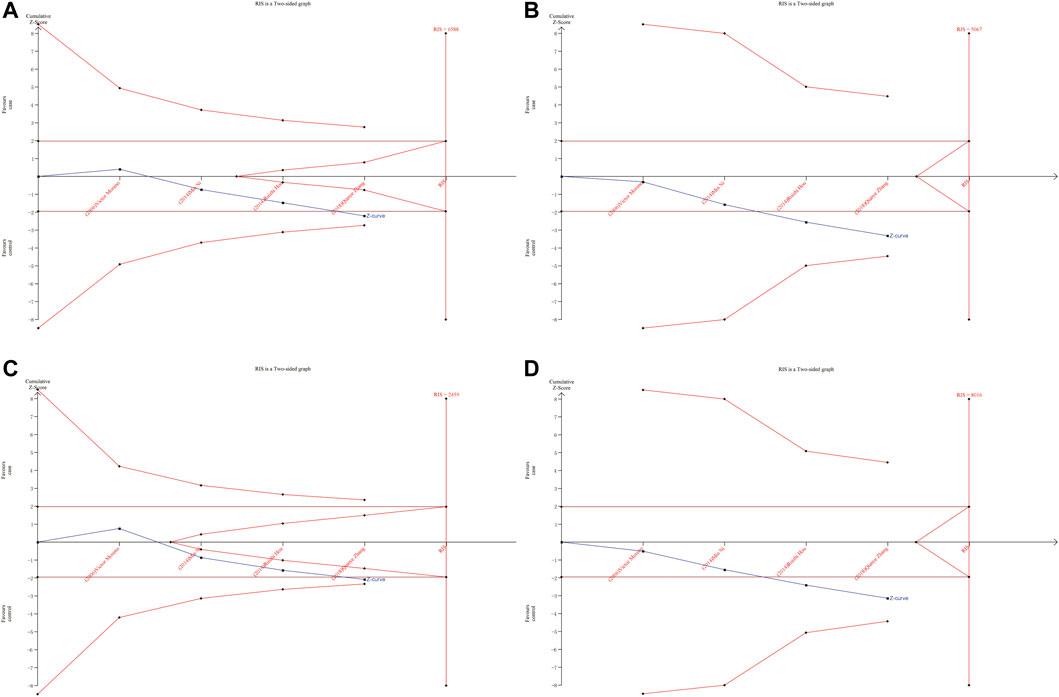
FIGURE 8. TSA for ERCC1 rs3212986 polymorphism and CRC risk. (A) Allele model (B) Homozygous model (C) Dominant model (D) Recessive model.
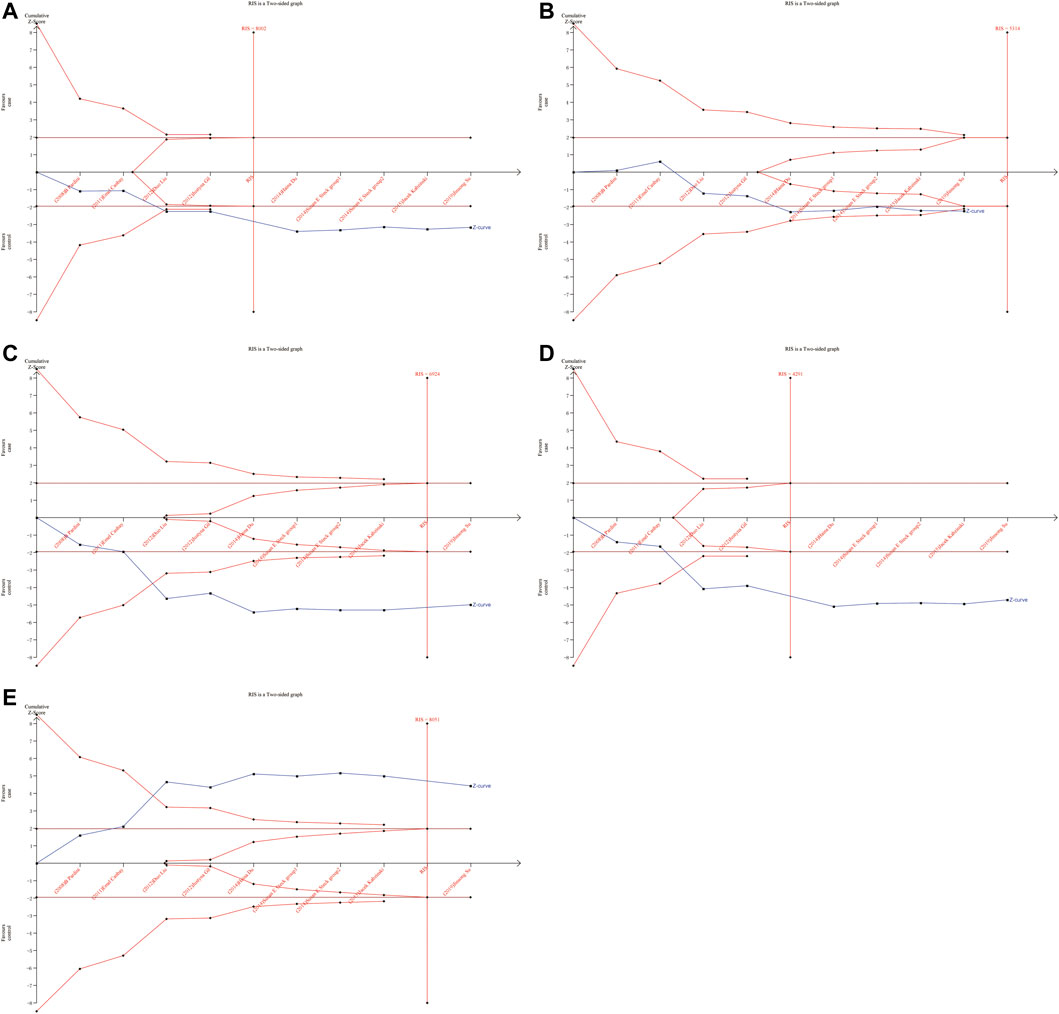
FIGURE 9. TSA for ERCC5 rs17655 polymorphism and CRC risk. (A) Allele model (B) Homozygous model (C) Heterozygous model (D) Dominant model (E) Over-dominant model.
The NER repair pathway is ubiquitous in all living organisms. It eliminates barriers to replication and transcription, as well as structures that may affect DNA stability or integrity (Spivak, 2015). NER repairs a wide range of lesions. Phenotypes of cancer susceptibility observed in GG-NER disorders. The deficiency of TC-NER impairs cell function and induces premature cell death, thus accelerating aging (Marteijn et al., 2014). Therefore, the mechanism of NER pathway and its impact on CRC susceptibility is an important area of research. Previous studies have shown that the expression changes of four NER genes in CRC are significantly different (Gómez-Díaz et al., 2015). In recent years, several epidemiological studies have described associations between the risk of CRC and SNPs in the NER repair pathway (Jelonek et al., 2010; Slyskova et al., 2012), while the results were inconsistent and incomprehensive.
We performed a meta-analysis to comprehensively discover the association between variants in NER gene and CRC susceptibility. After rigorous screening, a total of 29 case-control studies were eligible, including eight SNPs in the NER repair pathway. Our results showed that ERCC1 rs11615, ERCC1 rs3212986, ERCC2 rs1799793, and ERCC5 rs17655 variants were significantly correlated with CRC susceptibility. No significant impact of ERCC2 rs13181 variant, ERCC4 rs1800067 variant, XPC rs2228001 and XPC rs2228000 variants was detected on CRC risk.
ERCC1 enzyme is part of the NER system. ERCC1 protein is essential for maintaining genetic stability. Deletion of ERCC1 protein is associated with cancer susceptibility and has been reported as a biomarker for platinum resistance in colorectal and gastric cancer patients (Kwon et al., 2007). Previous meta-analyses showed that ERCC1 rs11615 variant was not associated with CRC risk (Chen et al., 2017). We added two more published studies to this basis, which showed that ERCC1 rs11615 CC genotype was associated with a reduced occurrence of CRC, but this significance was not shown in further stratified analysis. TSA analysis showed that with the increase in sample size, the overall results tend to be meaningful. Although the cumulative sample size does not reach RIS, the Z curve has intersected with the boundary value, indicating that the current results are highly reliable and the possibility of false positives is not significant. For the ERCC1 rs3212986 variant, our results showed AA genotype was associated with an increased risk of CRC. Further stratified analysis showed that the association persisted in the population-based control and Asian population subgroups. This is consistent with the results of previous studies by Chen et al. (2017). However, TSA analysis showed that Z curve only crossed the traditional boundary, and the total sample size did not reach RIS, indicating that the results had a large possibility of false positive, which requires further verification by follow-up studies to make the association valid.
ERCC2 gene is an essential component of NER, an ATP-dependent 5′-3′ helices, and mutations at different sites of the ERCC2 gene lead to repair and transcriptional defects (Spitz et al., 2001). Many studies have been performed to investigate the association of ERCC2 variants with CRC risk with mixed results, in which the most informative variants are rs1799793 and rs13181 (Lunn et al., 2000). A previous meta-analysis suggested that rs1799793 and rs13181 polymorphisms may not be associated with the development of CRC (Zhang et al., 2011). As for rs13181, our results were consistent with the above findings. However, further subgroup analysis of the association between rs13181 variant and CRC revealed a significantly increased risk in the Asian population under the dominant model. Moreover, A allele was associated with an increased risk of CRC in rs1799793, which persisted in the Caucasian population. This inconsistency may be due to the relatively small sample size of each study and the possible weak effect of polymorphism on CRC risk. It suggests that future studies can further explore the significance of these two variants in different populations.
ERCC5 gene is a structure-specific endonuclease and 5′-3′ exonuclease (Scherly et al., 1993) that are required to participate in two NER subpathways and play a key role in carcinogenesis. ERCC5 deficiency results in defective DNA repair and dysregulation of gene transcription (Koeppel et al., 2004). A previous study systematically estimated the association between ERCC5 rs17655 variant and overall cancer risk showed that ERCC5 rs17655 variant was not associated with cancer risk susceptibility (Zhu et al., 2012). In our meta-analysis, six more studies on CRC were included, and the results showed that rs17655 was significantly associated with increased risk of CRC in all five genetic models Furthermore, this association persisted in the Asian population. Although the detailed mechanism by which the SNPs in ERCC5 are associated with CRC susceptibility is unclear, our findings could provide new insights into the genetic factors underlying cancer susceptibility and carcinogenesis.
In addition, XPC is the main damage sensor in GG-NER. It is involved in DNA damage recognition and DNA repair activation of NER mechanism, including UV-induced damage, in-chain crosslinking, and photoproducts (Christmann et al., 2003). Several significant SNPs have been identified in XPC loci, including rs2228000 and rs2228001. A meta-analysis by Wang et al. (2015) showed no evidence of a significant association between XPC variant and CRC risk, persisted in subgroup analyses of ethnicity and study design, which is consistent with our findings. Our subgroup analysis showed risk or protective significance in different populations, possibly due to relatively small sample size and the influence of different genetic backgrounds. Further studies of homogenous CRC patients and well-matched controls are necessary.
In this meta-analysis, to ensure comprehensive inclusion of the literature, keywords and their synonyms were included in the search. If the word appeared in the title or abstract or even the full text, it was initially included. Then, according to the inclusion criteria, a rigorous screening process was performed. The initial collection of 6,617 articles included many articles whose research topic was not related to CRC, NER, or SNP, as well as some for which the original study data were not available through the original text or by contacting the authors directly. In addition, some SNP studies of NER related genes with only one or two reports were also excluded in order to reduce bias. Finally, A total of 29 case-control studies were eligible. Moreover, to reduce the risk of drawing false positive or false negative conclusion, we performed TSA in this meta-analysis. TSA tests the confidence of the determined results by combining estimates of the size of the information with adjustment thresholds for statistical significance of the cumulative meta-analysis.
There are certain advantages that should be acknowledged. First, we included high-quality case-control studies with genotypes that met the Hardy-Weinberg balance. In addition, there was no significant publication bias in all comparisons, and the sensitivity analysis showed robustness. Nevertheless, there are some limitations to our study. Firstly, the present meta-analysis could not include several relevant studies owing to the lack of raw data, improper publication formats or publication limitations. Secondly, the study was based on unadjusted effect estimates and 95% CI, and lacked information such as age and gender, which may lead to confounding bias. Finally, our meta-analysis did not include environmental factors such as smoking, alcohol consumption or viral infections. It did not examine the effects of interactions between genes or genes and the environment.
In conclusion, our meta-analysis of studies in different populations indicated that ERCC1 rs11615, ERCC1 rs3212986, ERCC2 rs1799793, and ERCC5 rs17655 were significantly associated with CRC risk. Conversely, the results showed no significant association with CRC risk of four SNPs, including ERCC2 rs13181, ERCC4 rs1800067, and two SNPs of XPC (rs2228001 and rs2228000). In future research, the following aspects should be paid attention to. Firstly, there may be a need for further research on different SNP topics in the NER gene, involving broader population regions and sample sizes. Secondly, gene-environment interactions and gene-gene interactions should also be considered in subsequent studies. Finally, functional studies are required to elucidate the underlying mechanisms of NER genetic variants in tumorigenesis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
CY, HY were responsible for study conception and design; CY, TL were responsible for acquisition and analysis of data; CY drafted the first manuscript; YS, PW, LD, JS, KW performed the interpretation; HY, CS helped to revise the manuscript. All authors read and approved the final manuscript.
This study is supported by Zhengzhou Major Project for Collaborative Innovation (18XTZX12007), the Major Project of Science and Technology in Henan Province (161100311400), the key research program of higher education in Henan province (No. 22A330003), the Project for International Training of High-level talents in Henan Province and the Foundation for fostering Characteristics Discipline in 2022.
Thanks to TL for the acquisition and analysis of data and to HY and CS for the revision of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1009938/full#supplementary-material
Al-Shaheri, F. N., Al-Shami, K. M., Gamal, E. H., Mahasneh, A. A., and Ayoub, N. M. (2020). Association of DNA repair gene polymorphisms with colorectal cancer risk and treatment outcomes. Exp. Mol. Pathol. 113, 104364. doi:10.1016/j.yexmp.2019.104364
Arnold, M., Sierra, M. S., Laversanne, M., Soerjomataram, I., Jemal, A., and Bray, F. (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66 (4), 683–691. doi:10.1136/gutjnl-2015-310912
Balkan, E., Bilici, M., Gundogdu, B., Aksungur, N., Kara, A., Yasar, E., et al. (2020). ERCC2 Lys751Gln rs13181 and XRCC2 Arg188His rs3218536 gene polymorphisms contribute to subsceptibility of colon, gastric, HCC, lung and prostate cancer. J. B.U.ON 25 (1), 574–581.
Baroudi, O., and Benammar-Elgaaied, A. (2016). Involvement of genetic factors and lifestyle on the occurrence of colorectal and gastric cancer. Crit. Rev. Oncol. Hematol. 107, 72–81. doi:10.1016/j.critrevonc.2016.08.014
Butterworth, A. S., Higgins Jp Fau - Pharoah, P., and Pharoah, P. (2006). Relative and absolute risk of colorectal cancer for individuals with a family history: A meta-analysis. Eur. J. Cancer 42 (2), 216–227. doi:10.1016/j.ejca.2005.09.023
Canbay, E., Cakmakoglu, B., Zeybek, U., Sozen, S., Cacina, C., Gulluoglu, M., et al. (2011). Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr. Med. Res. Opin. 27 (7), 1295–1302. doi:10.1185/03007995.2011.573544
Chatterjee, N., and Walker, G. C. (2017). Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58 (5), 235–263. doi:10.1002/em.22087
Chen, J., Sun, N., Hu, G., Chen, X., Jiang, J., Wu, H., et al. (2017). Association of ERCC1 polymorphisms with the risk of colorectal cancer: A meta-analysis. Crit. Rev. Eukaryot. Gene Expr. 27 (3), 267–275. doi:10.1615/CritRevEukaryotGeneExpr.2017019713
Christmann, M., Tomicic, M. T., Roos, W. P., and Kaina, B. (2003). Mechanisms of human DNA repair: An update. Toxicology 193 (1-2), 3–34. doi:10.1016/s0300-483x(03)00287-7
DiGiovanna, J. J., and Kraemer, K. H. (2012). Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 132 (32), 785–796. doi:10.1038/jid.2011.426
Du, H., Zhang, X., Du, M., Guo, N., Chen, Z., Shu, Y., et al. (2014). Association study between XPG Asp1104His polymorphism and colorectal cancer risk in a Chinese population. Sci. Rep. 4, 6700. doi:10.1038/srep06700
Dziki, L., Dziki, A., Mik, M., Majsterek, I. A.-O., and Kabzinski, J. (2017). Modulation of colorectal cancer risk by polymorphisms in 51Gln/his, 64Ile/val, and 148Asp/glu of APEX gene; 23Gly/ala of XPA gene; and 689Ser/arg of ERCC4 gene. Gastroenterol. Res. Pract. 2017, 3840243. doi:10.1155/2017/3840243
Engin, A. B., Karahalil, B., Engin, A., and Karakaya, A. E. (2010). Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet. Test. Mol. Biomarkers 14 (4), 559–564. doi:10.1089/gtmb.2009.0195
Gil, J., Ramsey, D., Stembalska, A., Karpinski, P., Pesz, K. A., Laczmanska, I., et al. (2012). The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual's susceptibility to sporadic colorectal cancer. Mol. Biol. Rep. 39 (1), 527–534. doi:10.1007/s11033-011-0767-5
Gillet, L. C., and Schärer, O. D. (2006). Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106 (2), 253–276. doi:10.1021/cr040483f
Gómez-Díaz, B., Gutiérrez-Angulo, M., Valle-Solis, A. E., Linares-González, L. M., and González-Guzmán, R. (2015). Analysis of ERCC1 and ERCC2 gene variants in osteosarcoma, colorectal and breast cancer. Oncol. Lett. 9 (4), 1657–1661. doi:10.3892/ol.2015.2894
Hanawalt, P. C. (2002). Subpathways of nucleotide excision repair and their regulation. Oncogene 21 (58), 8949–8956. doi:10.1038/sj.onc.1206096
Hou, R., Liu, Y., Feng, Y., Sun, L., Shu, Z., Zhao, J., et al. (2014). Association of single nucleotide polymorphisms of ERCC1 and XPF with colorectal cancer risk and interaction with tobacco use. Gene 548 (1), 1–5. doi:10.1016/j.gene.2014.05.025
Jelonek, K., Gdowicz-Klosok, A., Pietrowska, M., Borkowska, M., Korfanty, J., Rzeszowska-Wolny, J., et al. (2010). Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J. Appl. Genet. 51 (3), 343–352. doi:10.1007/BF03208865
Jin, D., Zhang, M., and Hua, H. (2019). Impact of polymorphisms in DNA repair genes XPD, hOGG1 and XRCC4 on colorectal cancer risk in a Chinese Han Population. Biosci. Rep. 39 (1), BSR20181074. doi:10.1042/BSR20181074
Kabzinski, J., Przybylowska, K., Dziki, L., Dziki, A., and Majsterek, I. (2015). An association of selected ERCC2 and ERCC5 genes polymorphisms, the level of oxidative DNA damage and its repair efficiency with a risk of colorectal cancer in Polish population. Cancer Biomark. 15 (4), 413–423. doi:10.3233/CBM-150488
Khan, A. A., Khan, Z., Malik, A., Kalam, M. A., Cash, P., Ashraf, M. T., et al. (2017). Colorectal cancer-inflammatory bowel disease nexus and felony of Escherichia coli. Life Sci. 180, 60–67. doi:10.1016/j.lfs.2017.05.016
Koeppel, F., Poindessous, V., Lazar, V., Raymond, E., Sarasin, A., and Larsen, A. K. (2004). Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clin. Cancer Res. 10 (16), 5604–5613. doi:10.1158/1078-0432.CCR-04-0442
Kwon, H. C., Roh, M. S., Oh, S. Y., Kim, S. H., Kim, M. C., Kim, J. S., et al. (2007). Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann. Oncol. 18 (3), 504–509. doi:10.1093/annonc/mdl430
Li, Y. K., Xu, Q., Sun, L. P., Gong, Y. H., Jing, J. J., Xing, C. Z., et al. (2020). Nucleotide excision repair pathway gene polymorphisms are associated with risk and prognosis of colorectal cancer. World J. Gastroenterol. 26 (3), 307–323. doi:10.3748/wjg.v26.i3.307
Liu, D., Wu, H. Z., Zhang, Y. N., Kang, H., Sun, M. J., Wang, E. H., et al. (2012). DNA repair genes XPC, XPG polymorphisms: Relation to the risk of colorectal carcinoma and therapeutic outcome with oxaliplatin-based adjuvant chemotherapy. Mol. Carcinog. 51 (1), E83–E93. doi:10.1002/mc.21862
Lunn, R. M., Helzlsouer, K. J., Parshad, R., Umbach, D. M., Harris, E. L., Sanford, K. K., et al. (2000). XPD polymorphisms: Effects on DNA repair proficiency. Carcinogenesis 21 (4), 551–555. doi:10.1093/carcin/21.4.551
Marteijn, J. A., Lans, H., Vermeulen, W., and Hoeijmakers, J. H. (2014). Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell. Biol. 15 (7), 465–481. doi:10.1038/nrm3822
Moreno, V., Gemignani, F., Landi, S., Gioia-Patricola, L., Chabrier, A., Blanco, I., et al. (2006). Polymorphisms in genes of nucleotide and base excision repair: Risk and prognosis of colorectal cancer. Clin. Cancer Res. 12 (71), 2101–2108. doi:10.1158/1078-0432.CCR-05-1363
Ni, M., Zhang, W. Z., Qiu, J. R., Liu, F., Li, M., Zhang, Y. J., et al. (2014). Association of ERCC1 and ERCC2 polymorphisms with colorectal cancer risk in a Chinese population. Sci. Rep. 4, 4112. doi:10.1038/srep04112
Page, M., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 372, n71. doi:10.1136/bmj.n71
Pardini, B., Naccarati, A., Novotny, J., Smerhovsky, Z., Vodickova, L., Polakova, V., et al. (2008). DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat. Res. 638 (1-2), 146–153. doi:10.1016/j.mrfmmm.2007.09.008
Paszkowska-Szczur, K., Scott, R. J., Górski, B., Cybulski, C., Kurzawski, G., Dymerska, D., et al. (2015). Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the Polish population. Mol. Biol. Rep. 42 (3), 755–764. doi:10.1007/s11033-014-3824-z
Rasool, S., Rasool V Fau - Naqvi, T., Naqvi T Fau - Ganai, B. A., Ganai Ba Fau - Shah, B. A., and Shah, B. A. (2014). Genetic unraveling of colorectal cancer. Tumour Biol. 35 (6), 5067–5082. doi:10.1007/s13277-014-1713-7
Sancar, A., Lindsey-Boltz La Fau - Unsal-Kaçmaz, K., Unsal-Kaçmaz K Fau - Linn, S., and Linn, S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85. doi:10.1146/annurev.biochem.73.011303.073723
Scherly, D., Nouspikel, T., Corlet, J., Ucla, C., Bairoch, A., and Clarkson, S. G. (1993). Complementation of the DNA repair defect in xeroderma pigmentosum group G cells by a human cDNA related to yeast RAD2. Nature 363 (6425), 182–185. doi:10.1038/363182a0
Skjelbred, C. F., Saebø, M., Wallin, H., Nexø, B. A., Hagen, P. C., Lothe, I. M., et al. (2006). Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: A case control study. BMC Cancer 6, 67. doi:10.1186/1471-2407-6-67
Sliwinski, T., Krupa, R., Wisniewska-Jarosinska, M., Pawlowska, E., Lech, J., Chojnacki, J., et al. (2009). Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population. Tohoku J. Exp. Med. 218 (3), 185–191. doi:10.1620/tjem.218.185
Slyskova, J., Korenkova, V., Collins, A. R., Prochazka, P., Vodickova, L., Svec, J., et al. (2012). Functional, genetic, and epigenetic aspects of base and nucleotide excision repair in colorectal carcinomas. Clin. Cancer Res. 18 (21), 5878–5887. doi:10.1158/1078-0432.CCR-12-1380
Spitz, M. R., Wu, X., Wang, Y., Wang, L. E., Shete, S., Amos, C. I., et al. (2001). Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 61 (4), 1354–1357.
Spivak, G. (2015). Nucleotide excision repair in humans. DNA Repair (Amst) 36, 13–18. doi:10.1016/j.dnarep.2015.09.003
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25 (9), 603–605. doi:10.1007/s10654-010-9491-z
Steck, S. E., Butler, L. M., Keku, T., Antwi, S., Galanko, J., Sandler, R. S., et al. (2014). Nucleotide excision repair gene polymorphisms, meat intake and colon cancer risk. Mutat. Res. 762, 24–31. doi:10.1016/j.mrfmmm.2014.02.004
Stern, M. C., Siegmund, K. D., Conti, D. V., Corral, R., and Haile, R. W. (2006). XRCC1, XRCC3, and XPD polymorphisms as modifiers of the effect of smoking and alcohol on colorectal adenoma risk. Cancer Epidemiol. Biomarkers Prev. 15 (12), 2384–2390. doi:10.1158/1055-9965.EPI-06-0381
Su, J., Zhu, Y., Dai, B., Yuan, W., and Song, J. (2019). XPG Asp1104His polymorphism increases colorectal cancer risk especially in Asians. Am. J. Transl. Res. 11 (2), 1020–1029.
Sun, K., Gong, A., and Liang, P. (2015). Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour Biol. 36 (3), 1549–1559. doi:10.1007/s13277-014-2721-3
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Wang, G., Sun, H., Liu, Z., Huang, R., Chen, Y., Tang, Q., et al. (2015). Lack of associations between XPC polymorphisms and colorectal cancer: A meta-analysis. J. BUON 20 (3), 770–774.
Wang, J., Zhao, Y., Jiang, J., Gajalakshmi, V., Kuriki, K., Nakamura, S., et al. (2010). Polymorphisms in DNA repair genes XRCC1, XRCC3 and XPD, and colorectal cancer risk: A case-control study in an Indian population. J. Cancer Res. Clin. Oncol. 136 (10), 1517–1525. doi:10.1007/s00432-010-0809-8
Wang, L., Lou, Y., Fang, C. Q., and Yuan, Z. W. (2009). Association of single nucleotide polymorphism in XPD with susceptibility of colon cancer in Han population from northeast region of China. Chin. J. CANCER Prev. Treat. 16 (17), 1284–1286.
Yang, H., Li, G., and Li, W. F. (2015). Association between ERCC1 and XPF polymorphisms and risk of colorectal cancer. Genet. Mol. Res. 14 (1), 700–705. doi:10.4238/2015.January.30.13
Yeh, C. C., Sung, F. C., Tang, R. P., Chang-Chieh, C. R., and Hsieh, L. L. (2007). Association between polymorphisms of biotransformation and DNA-repair genes and risk of colorectal cancer in Taiwan. J. Biomed. Sci. 14 (2), 183–193. doi:10.1007/s11373-006-9139-x
Yueh, T. C., Chou, A. K., Gong, C. L., Fu, C. K., Pei, J. S., Wu, M. H., et al. (2017). The contribution of excision repair cross-complementing group 1 genotypes to colorectal cancer susceptibility in taiwan. Anticancer Res. 37 (5), 2307–2313. doi:10.21873/anticanres.11568
Zhang, Q., Zheng, X., Li, X., Sun, D., Xue, P., Zhang, G., et al. (2018). The polymorphisms of miRNA-binding site in MLH3 and ERCC1 were linked to the risk of colorectal cancer in a case-control study. Cancer Med. 7 (14), 1264–1274. doi:10.1002/cam4.1319
Zhang, S., Chen, Z., and Du, W. (2017). Genetic polymorphisms of DNA repair genes XPD, XPC, and XRCC4 in relation to colorectal cancer susceptibility. Chin. J. Clin. Oncol. 44 (8), 365–370. doi:10.3969/j.issn.1000-8179.2017.08.385
Zhang, Y., Ding, D., Wang, X., Zhu, Z., Huang, M., and He, X. (2011). Lack of association between XPD Lys751Gln and Asp312Asn polymorphisms and colorectal cancer risk: A meta-analysis of case-control studies. Int. J. Colorectal Dis. 26 (10), 1257–1264. doi:10.1007/s00384-011-1222-8
Keywords: colorectal cancer, risk, single nucleotide polymorphisms, meta-analysis, nucleotide excision repair gene
Citation: Yi C, Li T, Shen Y, Wang P, Dai L, Shi J, Wang K, Sun C and Ye H (2022) Polymorphisms of nucleotide excision repair genes associated with colorectal cancer risk: Meta-analysis and trial sequential analysis. Front. Genet. 13:1009938. doi: 10.3389/fgene.2022.1009938
Received: 02 August 2022; Accepted: 10 October 2022;
Published: 31 October 2022.
Edited by:
Ranajit Das, Yenepoya University, IndiaReviewed by:
Somsubhra Nath, Presidency University, IndiaCopyright © 2022 Yi, Li, Shen, Wang, Dai, Shi, Wang, Sun and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Ye, eWVodWFAenp1LmVkdS5jbg==; Changqing Sun, c3VuY3FAenp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.