- 1Department of Plant Science, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2Suzhou Polytechnic Institute of Agriculture, Suzhou, China
- 3Jiangbei Grape Research Institute of Shandong Province, Shandong, China
- 4Institute of Grape Science and Engineering, College of Horticulture, Qingdao Agricultural University, Qingdao, China
Plant-specific YABBY (YAB) transcription factors play multiple roles in plant growth and development process. However, no comprehensive study has been performed in grapevines, especially to determine their roles in berry development and abiotic stress response. A total of seven VviYABs allocated to six chromosomal positions in grapevines were identified and classified into five subfamilies based on phylogenetic and structural analysis. Promoter element analysis and tissue-specific transcriptional response of VviYABs suggested that VviYABs might play vital roles in plant growth and development. VviYAB1, 2, 3, and 5 showed significantly higher expression levels in vegetative/green organs than in mature/woody tissues, implying that VviYABs might be involved in the regulatory switch from immature to mature developmental phases. The expression of VviYAB1, 2, 3, and VviFAS were gradually downregulated during berry developmental and ripening, which can be considered as putative molecular biomarkers between vegetative/green and mature/woody samples, and were used to identify key developmental and metabolic processes in grapevines. Furthermore, VviYAB1 expression was not markedly increased by gibberellic acid (GA3) treatment alone, but displayed significant upregulation when GA3 in combination with N-(2-chloro-4-pyridyl)-N′-phenylurea (CPPU) were applied, suggesting an involvement of VviYAB1 in fruit expansion by mediating cytokinin signaling pathway. Additionally, microarray and RNA-seq data suggested that VviYABs showed transcriptional regulation in response to various abiotic and biotic stresses, including salt, drought, Bois Noir, Erysiphe necator, and GLRaV-3 infection. Overall, our results provide a better understanding of the classification and functions of VviYABs during berry development and in response to abiotic and biotic stresses in grapevines.
Introduction
YABBY (YAB) is a small transcription factor gene family, which is specific seed plants (Bowman and Smyth, 1999; Shamimuzzaman and Vodkin, 2013). The YAB genes consist of two conserved domains: an N-terminal zinc-finger motif and a C-terminal YABBY domain (helix–loop–helix motif) (Golz and Hudson, 1999; Siegfried et al., 1999). In Arabidopsis, YAB genes specify abaxial cell fate in lateral organs (Villanueva et al., 1999; Bowman, 2000). However, several reports have shown that YABs in monocots have evolved diverse functions in other developmental processes (Liu et al., 2007; Yang et al., 2016).
The Arabidopsis genome comprises six YAB genes, which are divided into five subfamilies: CRABS CLAW (CRC), FILAMENTOUS FLOWER (FIL)/YAB3, INNER NO OUTER (INO), YAB2, and YAB5 (Bowman, 2000; Eckardt., 2010; Sarojam et al., 2010). Among them, FIL, YAB2, YAB3, and YAB5 function redundantly to control the development of lateral organs (Siegfried et al., 1999; Yamada et al., 2011; Yang et al., 2018). In Arabidopsis, FIL is responsible for the flower formation and development (Chen et al., 1999; Sawa et al., 1999a, Sawa et al., 1999b). Similarly, the TOB1-related YAB genes in rice have conserved functions in flower development(Tanaka et al., 2017). The CRC gene in Arabidopsis promotes carpel polarity and nectary development (Alvarez and Smyth, 1999), and its orthologs in rice and Pisum sativum also regulate the carpel morphogenesis (Yamaguchi et al., 2004; Fourquin et al., 2014). INO plays a crucial role in the regulation of the outer integument growth (Villanueva et al., 1999). In rice, both OsYAB1 and OsYAB4 participate in controlling the gibberellin pathway, and overexpression of the rice OsYAB1 and OsYAB4 leads to a semi-dwarf phenotype and increases in the number of stamens (Dai et al., 2007; Yang et al., 2016).
YAB genes participate in fruit morphogenesis during fruit development. For example, downregulation of the FASCIATED (FAS) gene, encoding a YAB transcription factor, results in an increase of locule number in modern tomato varieties (Cong et al., 2008). In grapevines, VviYAB5 displays different expression patterns between tricarpellate and bicarpellate ovaries, indicating that the expression of VviYAB5 is associated with the formation of tricarpellate fruit (Xing et al., 2016). Furthermore, growing evidences show that YAB genes play important roles in plant abiotic stress responses (Inal et al., 2017; Zhao et al., 2017). For example, the soybean GmYAB10 negatively regulates the drought and salt tolerance (Zhao et al., 2017).
The grapevine is among the most globally cultivated and economically important fruit crops worldwide (Leng et al., 2017; Jiu et al., 2021; Jiu et al., 2022; Xu et al., 2021). A preliminary study in Vitis pseudoreticulata showed that VpYAB1 was involved in leaf development, and ectopic expression of VpYAB1 led to the loss of dorso-ventral polarity in the leaf blade (Xiang et al., 2013). However, comprehensive investigations are needed to evaluate the function of YAB gene family in Vitis vinifera. In the present study, we observed the genomic organization and transcript patterns of the YAB members in grapevines and focused on the roles of VviYAB genes in fruit development and stress response. Our results will facilitate further investigation into the functions of the VviYAB and may have potential improvement for molecular breeding programs in grapevines.
Materials and Methods
Identification and Analysis of VviYABs Family in Grapevines
The hidden Markov model (HMM) profile of the YABBY (PF04690) was downloaded from the Pfam database (http://pfam.xfam.org/) and used to survey the grapevine genome database (CRIBI; http://genomes.cribi.unipd.it/grape/) using HMMER3.1 as in previous reports (Leng et al., 2019). All protein sequences of VviYABs were identified based on the YABBY domain using Simple Modular Architecture Research Tool (SMART) database (http://smart.embl-heidelberg.de/) and National Center for Biotechnology Information-Conserved Domain Database (NCBI-CDD) search (https://www.ncbi.nlm.nih.gov/cdd www.ncbi.nlm.nih. gov/cdd). The physical and chemical parameters as well as subcellular location of VviYABs were determined using ProtParam (http://web.expasy.org/protparam/) and WoLF PSORT (https://www. genscript.com/wolf-psort.html), respectively. These members of the VviYABs family were named according to the rules described by Grimplet et al. (2014).
Sequence Alignments and Phylogenetic Analysis
Sequences of Arabidopsis and tomato YAB proteins were retrieved from The Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org/) and the Solanaceae Genomics Network (https://solgenomics.net/), respectively. Multiple sequence alignments of these YAB proteins were performed using ClustalX. A phylogenetic tree based on the full-length amino acid (aa) sequences was constructed by using MEGA 5.0 with the neighbor-joining (NJ) method. Motif analyses of VviYABs were performed by using MEME tool (http://meme.sdsc.edu/meme/website/intro.html) as in previous reports (Wei et al., 2020).
Chromosomal Locations, Gene Structure, and Gene Duplication
Chromosomal positions of VviYAB genes were obtained from the Grape Genome CRIBI website (http://genomes.cribi.unipd.it/). Exon–intron structures were determined using the Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn) by comparing the cDNA sequences with their corresponding genomic sequences. Tandem duplicated genes were identified as adjacent paralogous on an individual grape chromosome, with no more than one intervening gene (Wei et al., 2020). The syntenic blocks within the grape genome, as well as among grapevines, Arabidopsis, and tomato, were detected using MCScanX software according to our previous research (Leng et al., 2019; Jiu et al., 2020).
Promoter Analysis
The nucleotide sequences of VviYAB genes were obtained from the grapevine genome database (CRIBI; http://genomes.cribi.unipd.it/grape/) in this study. The upstream 1,500-bp region from the translation initiation site for all VviYABs was regarded as the putative promoter sequence (Badawi et al., 2019; Feng et al., 2021). The cis-acting elements of VviYAB genes were predicted using PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Postel et al., 2002).
Expression Patterns of VviYABs in Various Organs and Different Berry Developmental Stages
The microarray expression atlas of VviYAB genes in various organs and developmental stages were acquired from the Gene Expression Omnibus (GEO) datasets (GSE36128; Fasoli et al., 2012). The average expression value of each gene in all tested organs were analyzed and graphically drawn with MeV software (Saeed et al., 2006).
The transcriptional expression patterns of VviYAB genes at 13 different fruit developmental stages were collected from GEO datasets (GSE98923; Fasoli et al., 2018). Furthermore, other transcriptional expression analyses based on RNA-seq data were downloaded from GEO datasets (GSE62744 and GSE62745), which contained 10 grapevine varieties at four berry development stages (Massonnet et al., 2017).
Plant Growth Condition, Hormone, and Stress Treatment
Four-year-old “Fujiminori” grapevines (V. vinifera × V. labrusca) were used as the experimental material, which were maintained in the Qingdao Agricultural University fruit farm, Qingdao, China. Eight grapevine samples, including young leaf, senescent leaf, green stem, woody stem, bud burst (green tip), winter bud, berry post-fruit set, and berry ripening were collected and used for tissue-specific expression. Furthermore, grapevine berries were collected at four developmental stages, including the pea-sized berry stage at 20 days after flowering (DAF, pea_sized), the berries beginning just prior to veraison (pre_veraison), the berry-softening at the end of veraison (end_veraison), and the fully ripe berry stage at harvest (ripe). Three biological replicates were used for different developmental stages of grapevine organs.
For abscisic acid (ABA) and ethylene treatments, 60 DAF of grape berries were immersed in 100 mg/L ABA and 500 mg/L ethephon (ETH, an ethylene-releasing reagent), and the control berries were immersed in deionized water. Berries were sampled at 0 (green), 10 (pre_veraison), 20 (end_veraison), and 30 days (ripening) after treatment. Three biological replicates were used for different harvest dates of ABA and ethylene treatments. All samples were immediately frozen and stored at −80°C until use. For gibberilic acid (GA3) and forchlorfenuron (CPPU) treatment, grapevine RNA-seq datasets were retrieved from published supplemental data sets (Xu et al., 2019), which contained three treatments, i.e., water (CK), 25 mg/L GA3 + 0 mg/L CPPU, and 25 mg/L GA3 + 10 mg/L CPPU (GA3 + CPPU), at full-bloom stage.
Expression analyses in response to abiotic and biotic stresses were downloaded from the NCBI GEO datasets based on microarray data (series matrix accession numbers GSE31594, GSE31677, GSE6404, GSE12842, and GSE31660). To further validate the expression profiles of VviYABs in response to different abiotic stress treatments, grapevine RNA-seq data in response to waterlogging and drought stress were retrieved from NCBI database (SRA accession no. SRP070475 and SRP074162, respectively) (Haider et al., 2017; Zhu et al., 2018). RNA-seq data for expression profiles in response to salt were retrieved from published supplemental datasets (Guan et al., 2018).
qRT-PCR Analysis
Total RNA samples were isolated using the modified cetyltrimethyl ammonium bromide (CTAB) method (Leng et al., 2015a; Leng et al., 2015b; Jiu et al., 2019). Subsequently, 1.0 μg of total RNA was used to synthesize the first-strand cDNA using the Prime Script RT reagent Kit (TaKaRa Biotechnology, Dalian, China), and the resulting cDNA was diluted 10-fold before performing qRT-PCR assay. qRT-PCR reactions (20 μl) contained 10 μl SYBR Green Supermix (Bio-Rad), 0.4 μl 10 μM forward primer, 0.4 μl 10 μM reverse primer, and 2 μl cDNA template. The mixture was placed in an Applied Biosystems (ABI) 7500 FAST Real-Time PCR System, and the amplification was conducted using the program described by (Jiu et al. 2016, Jiu et al., 2021). Each pair of qRT-PCR primers was validated by cloning and sequencing of the product using this pair of primers. Each reaction was performed in triplicate. Actin (AB073011) was used as an internal standard to normalize the expression levels. Relative expression levels were estimated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). The gene-specific primers for the VviYAB genes are listed in Supplementary Table S1.
Subcellular Localization
The full-length cDNA of the VviCRC, VviFAS, and VviYAB3 were amplified using the primers (Supplementary Table S1) and cloned into the binary vector pHB including two cauliflower mosaic virus (CaMV) 35S promoter, a translation enhancer, and a green fluorescence protein (GFP) fluorescent protein tag, respectively, to generate three fusion constructs (35S-VviCRC-GFP, 35S-VviFAS-GFP, and 35S-VviYAB3-GFP). After the three identified sequences, the control vector (pHB) and three fusion constructs were transformed into Agrobacterium tumefaciens GV3101 strains and subsequently agroinfiltrated into the leaves of 3- to 5-week-old Nicotiana benthamiana plants. Localization of fluorescent proteins were observed 3–5 days after infiltration using a confocal laser scanning microscope (Zeiss LSM 780, Germany) according to the manufacturer's instructions.
Statistical Analysis
The experiment was performed using a completely randomized design with three biological replicates. The data were statistically analyzed using the SAS software package (Version 9.2, SAS Institute Inc., Cary, NC, United States). For the qRT-PCR analysis, data are shown as means ± standard deviation (SD) of three biological replicates.
Results
Identification and Analysis of Full-Length VviYABs in Grapevines
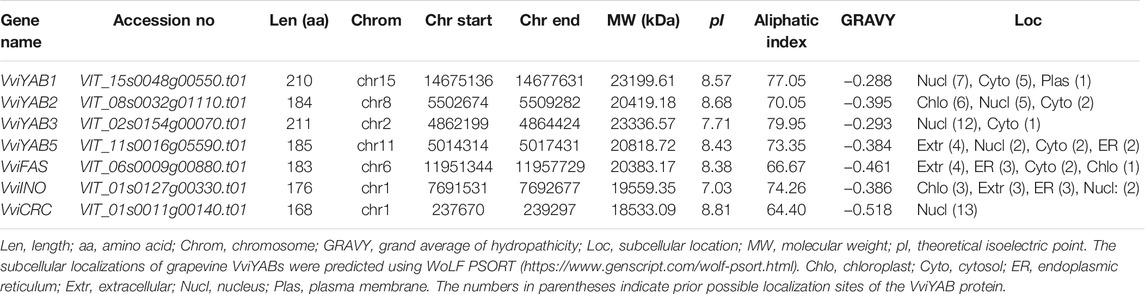
To identify the YABs from the grapevine genome, local BLAST and HMM tools were used as described in previous studies (Leng et al., 2019; Wei et al., 2020). According to the presence of conserved YABBY domain, seven VviYABs were identified in the grapevine and named based on their homology to the Arabidopsis and tomato YAB members (Table 1). All seven VviYABs were unevenly distributed to 6 out of 19 grapevine chromosomal positions (Supplementary Figure S1). Detailed parameters of VviYAB proteins are listed in Table 1. Grapevine VviYAB proteins varied from 168 (VviCRC) to 211 (VviYAB3) aa in length. The grand average of hydropathicity (GRAVY) is less than 0 for all VviYAB proteins, implying that YAB proteins possess a hydrophilic nature (Table 1).
Phylogenetic, Structural, and Synteny Analysis of VviYAB Genes
To understand the evolutionary relationship between YAB proteins, a phylogenetic tree consisting of 30 YAB proteins from four different species was constructed using MEGA (7.0) (Figure 1). As shown in Figure 1, YAB members were further divided into five subgroups (CRC, INO, YAB1/YAB3, YAB2, and YAB5), which is consistent with previous reports in Arabidopsis and tomato (Bowman, 2000; Huang et al., 2013). VviYABs showed a closer relationship to dicotyledons (Arabidopsis and tomato) than monocotyledons (rice). Interestingly, no proteins from rice, the monocotyledon species, was found in the YAB5 subgroup, suggesting that YABs have diversified in the evolution of different species. In addition, a maximum of YAB proteins was observed in the YAB1/YAB3 subgroup, indicating that the YAB1/YAB3 subgroup was primarily attributed to the expansion of the YAB gene family.
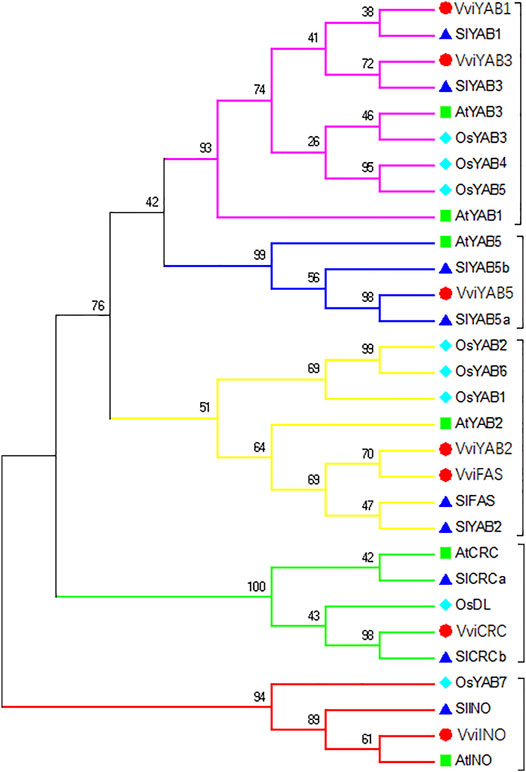
FIGURE 1. Phylogenetic relationships of YAB genes among four plant species. YABs were divided into five subfamilies (YAB1/YAB3, YAB2, YAB5, CRC, and INO) The neighbor-joining method was used, and the bootstrap values were set at 1,000.
All VviYABs contained a C-terminal YABBY domain (Figure 2B and Supplementary Figure S2), which was similar with previously identified YAB proteins. A conserved zinc finger domain was situated near N-terminal (Figure 2B and Supplementary Figure S2). Furthermore, similar protein structures and motif compositions were observed within the groups (Figure 2B and Supplementary Figure S3). For example, motif 3 was present in the C-terminal of YAB2 and YAB1/YAB3 subgroup (Figure 2B). Phylogenetic analysis also showed a close relationship between VviFAS and VviYAB2 and between VviYAB1 and VviYAB3 subgroups (Figure 2A), which was consistent with the similarity in motif composition. Moreover, VviFAS and VviYAB2 as well as VviYAB1 and VviYAB3 showed similar exon structure (Figure 2C), which further supported the analysis of phylogenetic relationship and motif compositions.
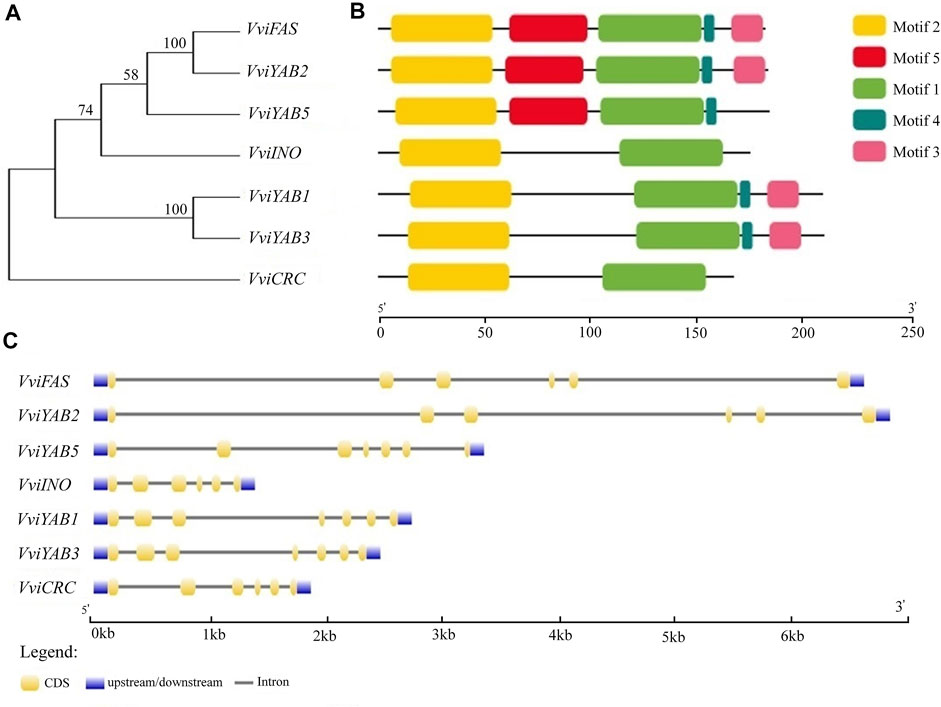
FIGURE 2. Phylogenetic analysis, gene structure, and conserved motifs of YAB family in grapevine. (A) The conserved YABBY domain sequences of VviYAB proteins constructed a neighbor-joining (NJ) phylogenetic tree, and the bootstrap test was performed with 1,000 iterations. (B) Distribution of conserved motifs of VviYAB proteins. Different motifs are shown by different colors numbered 1 to 5. See legend for detailed color. (C) Exon–intron structure of VviYAB genes. Blue indicates untranslated 5′- and 3′- regions, yellow indicates exons; black indicates introns.
In addition, some of YABBY domains among grapevines, tomatoes, and Arabidopsis showed pairwise relationships. A total of 17 pairs of syntenic relations were identified among the three species, including seven grapevine VviYABs, five Arabidopsis AtYABs, and eight tomato SlYABs (Supplementary Figure S4 and Table S2). VviYAB3, AtYAB1, and SlYAB1, belonging to YAB1/YAB5 group, are linked to at least three syntenic events and might indicate a high conservation of group YAB1/YAB3 members. The number of synteny events between grapevines and tomatoes (six synteny events) was greater than that between grapevines and Arabidopsis (three synteny events), indicating a high conservation of YABs between grapevines and tomatoes.
Functional Analysis of Cis-Elements in the Promoter of VviYAB Genes
The evaluation of cis-elements in promoters will provide a better understanding of the gene function and transcriptional regulation. As shown in Supplementary Table S3, two basic cis-elements, TATA-box and CAAT-box, were widely distributed in the promoters of all VviYAB genes. Several growth-related cis-elements including the meristem expression element (CAT-box), the endosperm-specific expression element (GCN4_motif and Skn-1_motif), and the circadian regulator element (circadian) were identified in the promoters of three, five, and six VviYABs, respectively (Figure 3). In hormone-related cis-regulatory elements, the MeJA-responsive element (CGTCA-motif and TGACG-motif), the SA-responsive element (TCA-element), the ABA-responsive element (ABRE), ethylene responsive element (ERE), GA-responsive element (GARE-motif, P-box, and TATC-box), and auxin-responsive element (TGA-element) were identified in the promoter region of VviYABs (Figure 3 and Supplementary Table S3). Some stress-related cis-regulatory elements, which were associated with anaerobic induction (ARE), fungal elicitor (Box-W1), drought (MBS), and stress responsiveness (TC-rich repeats), were found in six, four, three, and five VviYABs, respectively (Figure 3 and Supplementary Table S3).
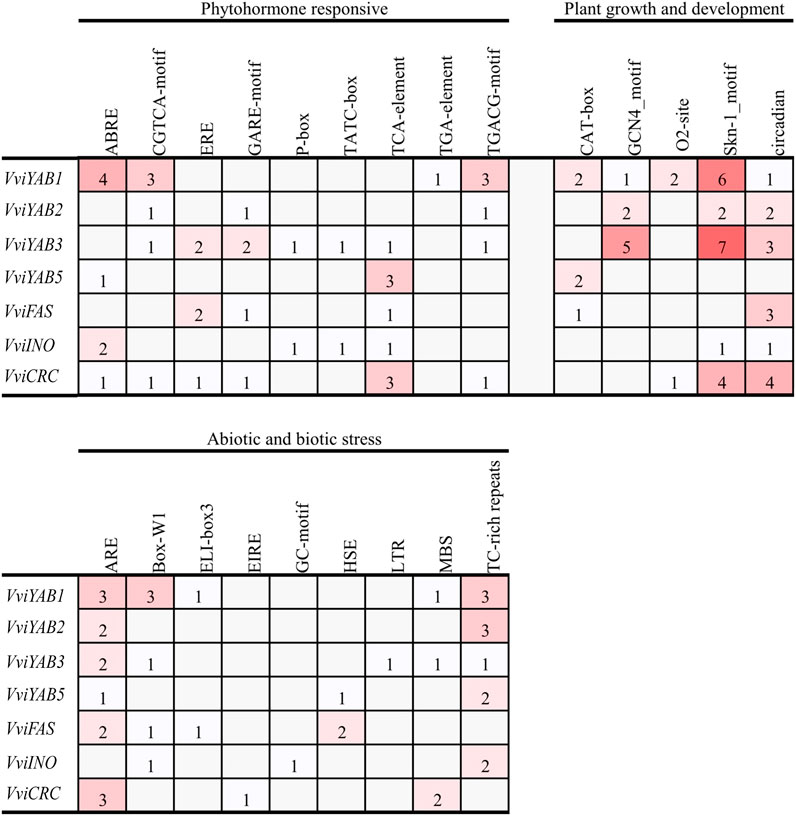
FIGURE 3. The cis-regulatory elements analysis of promoter region of VviYABs. The number of each cis-acting element in the promoter region (1.5 kb upstream of the translation start site) of VviYAB genes. Based on the functional annotation, the cis-acting elements were classified into three major classes: plant growth and development, phytohormone responsive, and abiotic and biotic stresses-related cis-acting elements.
Expression Patterns of VviYAB Genes in Different Tissues
To investigate the functions of the VviYABs, we estimated their expression levels in 42 various organs/tissues, which were obtained from GEO datasets (GSE36128) by previous microarray analysis (Fasoli et al., 2012). Figure 4A showed that the expression of VviCRC was restricted to floral organs, such as well-developed inflorescences and carpels (Figure 4A and Supplementary Table S4), which was in good agreement with previous reports in Arabidopsis (Bowman and Smyth, 1999; Villanueva et al., 1999). VviYAB1 and VviYAB3 from the same subgroup exhibited high expression levels in leaf-derived organs, including buds, and leaves (Figure 4A and Supplementary Table S4), implying an involvement of subgroup YAB1/YAB3 members in leaf development. Additionally, VviFAS and VviYAB2 showed much higher expression levels in young berries, buds, and floral organs. By comparison, VvINO showed lower expression levels in all tested tissues. Remarkably, several VviYABs, including VviYAB1, 2, 3, and 5, were highly expressed in vegetative/green tissues, implying a potential regulatory function during the developmental transition.
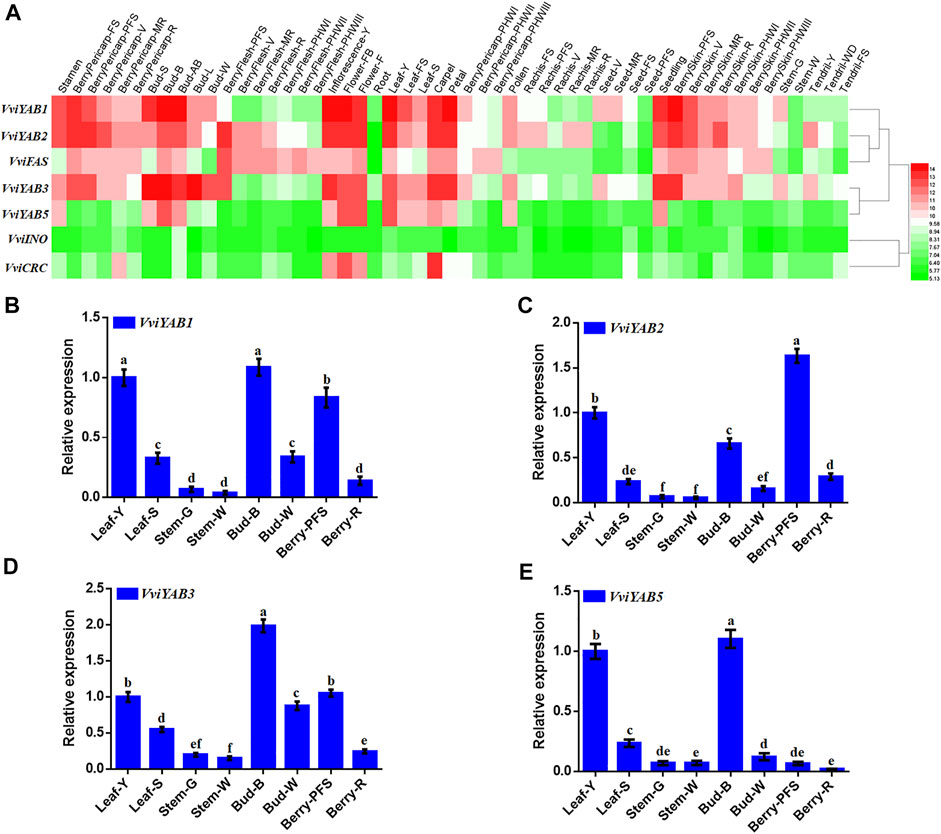
FIGURE 4. Expression profile of VviYABs in various tissues and developmental stages. (A) Expression of VviYABs in the V. vinifera cv “Corvina” atlas. Expression data were normalized based on the mean expression value of each gene in all analyzed tissues. Genes were hierarchically clustered based on average Pearson's distance metric and “average linkage” method. Red and green boxes indicate high and low expression levels, respectively, for each gene. (B–E) qPCR validation of VviYAB gene expression in four vegetative/green and four mature/woody organs, respectively. Transcripts were normalized to the expression of the actin gene. The data are shown as means ± standard deviation (SD) of three replicates. Bud-AB, bud after burst; Bud-B, bud burst; Bud-W, winter bud; Bud-L, latent bud; Bud-S, bud swell; Flower-F, flowering; Flower-FB, flowering begins; FS, fruit set; Inflorescence-Y, young inflorescence with single flowers separated; Inflorescence-WD, well-developed inflorescence; Leaf-FS, mature leaf; Leaf-S, senescing leaf; Leaf-Y, young leaf; MR, mid-ripening; R, ripening; PFS, post fruit set; Stem-G, green stem; Stem-W, woody stem; V, veraison..
To further understand the expression distinction of VviYAB genes between vegetative/green and mature/woody organs by microarray data, the expression patterns of four VviYABs were validated by qRT-PCR in four vegetative/green and four mature/woody organs. As expected, qRT-PCR results were highly consistent with the microarray data and showed significantly different expression between vegetative/green and mature/woody samples (Figures 4B–E). For example, all four VviYABs showed significantly higher expression levels in vegetative/green organs (young leaf, bud burst, and berry post-fruit set) than in mature/woody tissues (senescencing leaf, winter bud, and berry ripening), in the stem (Figures 4B–E). These results implied that VviYABs might be involved in regulatory switch from the immature to the mature developmental phase. Furthermore, four VviYABs (VviYAB1, 2, and 3 and VviFAS) had higher expression in young berries and then showed a gradual decrease from veraison to ripening stage (Figure 4 and Supplementary Table S4), indicating that these genes might be involved in early fruit development and morphogenesis in grapevines.
Expression Patterns of VviYAB Genes During Berry Development and Ripening
To reveal the putative roles of VviYABs during fruit development and ripening, we firstly focused on the expression profiles of VviYAB genes from GEO datasets (GSE98923) (Fasoli et al., 2018), which contained 13 different developmental stages from fruit set to full maturity (Figure 5A and Supplementary Table S5). As shown in Figure 5A, the most obvious correlation was that the expressions of four VviYABs (VviYAB1, 2, and 3 and VviFAS) were negatively related to berry development (from fruit set to full maturity). For example, VviFAS and VviYAB2 showed sharply downregulated expression from immature to the mature berry in grapevines. The expression levels of VviYAB1 and VviYAB3 were very similar to those of VviFAS and VviYAB2, but their expression levels were significantly lower. These results further supported the idea that VviYABs promoted the immature-to-mature transition during grapevine berry development. In addition, VviYAB5, VviCRC, and VviINO were always very low or undetected levels during the whole grapevine berry ripening process, indicating that they might not be involved in the regulatory switch during grapevine berry development.
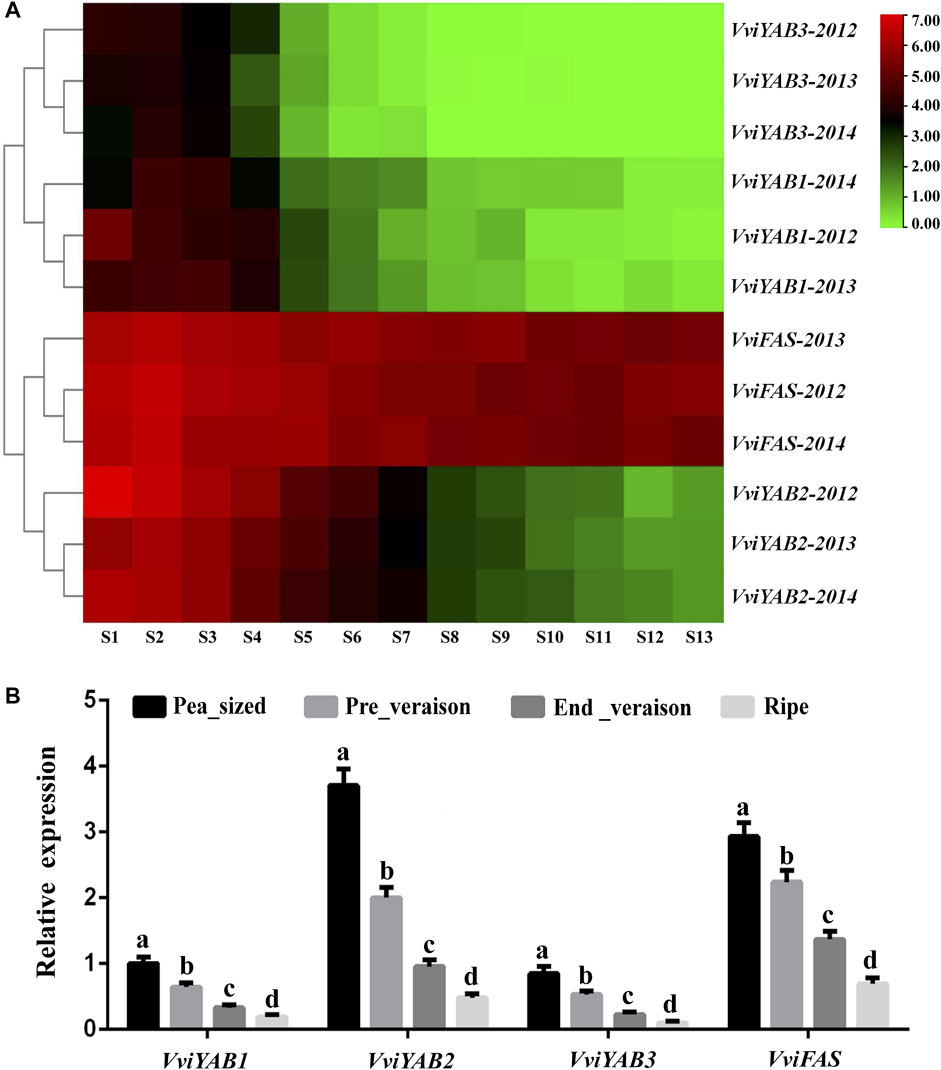
FIGURE 5. Expression profile of four VviYAB genes during developmental and ripening berry in grapevine. (A) Expression of VviYAB genes in the V. vinifera cv “Cabernet Sauvignon” atlas. Berries were collected at 10-day intervals in 2012 and weekly in 2013 and 2014, beginning at fruit set and continuing until harvest (24.5°Brix). S represents stage. Expression data were normalized based on the mean expression value of each gene from fruit set to full maturity. The mean expression values were again normalized using logarithm with the base of 2 using the Heml software. Gene names are displayed to the right of each row. (B) qPCR validation of VviYABs expression in four berry development stages. Transcripts were normalized to the expression of the actin gene. The data are shown as means ± SD of three replicates.
In order to validate regulatory mechanisms involved in the switch between immature and mature berries, qRT-PCR analysis of four VviYABs (VviYAB1, 2, and 3 and VviFAS) was further carried out at four berry development stages, including pea_sized, pre_veraison, end_veraison, and ripe. We found that qRT-PCR results were completely consistent with those of RNA-seq data (Figure 5B). All four VviYABs were highly expressed in immature berries and showed a significant decrease in mature berries, indicating a regulatory switch during berry development and ripening. To obtain more information of VviYABs during berry development transition, the transcription accumulation patterns among 10 different grapevine varieties were obtained from previous transcriptome sequencing data (GSE62744 and GSE62745), which also included four different berry developmental stages (Massonnet et al., 2017). The expression of three VviYABs (VviYAB5, VviCRC, and VviINO) were not detected during the entire grapevine berry ripening process (Supplementary Figure S5), which corresponded with the data from a previous RNA-Seq analysis (Figure 5A). VviYAB1, 2, and 3 and VviFAS remained relatively highly expressed in immature berries (pea-sized berry and pre_veraison stage) and decreased gradually in mature berries (end_veraison and ripe stage) (Supplementary Figure S5). All these results implied that these four VviYABs might play fundamental shift roles from the immature to mature berry developmental processes.
VviYABs in Response to Exogenous ABA, Ethylene, GA3, and CPPU Hormones
To date, the role of YAB proteins in hormonal signaling pathways is scarce in grapevines. To reveal the potential roles of the VviYABs in response to ABA and ethylene treatments, four members (VviYAB1, 2, and 3 and VviFAS) with high expression levels of VviYABs were analyzed by qRT-PCR (Figure 6). Our results demonstrated that ABA and ethylene treatments significantly suppressed the expression levels of VviYAB1 and VviYAB3 from Pre-Verasion to ripening period. VviYAB2 was markedly repressed from pre-verasion to ripening period following the enthylene treatment, but it did not respond markedly to the ABA treatment at pre-veraison and end-veraison periods. However, VviFAS was notably activated by enthylene treatment from pre-verasion to ripening period. In addition, the expression of VviFAS was significantly induced at pre-verasion and ripening periods, while it was inhibited at the end-veraison stage following the ABA treatment (Figure 6). Interestingly, the four VviYABs (VviYAB1, 2, and 3 and VviFAS) did not markedly differ in the presence of enthylene and ABA at the green berry period.
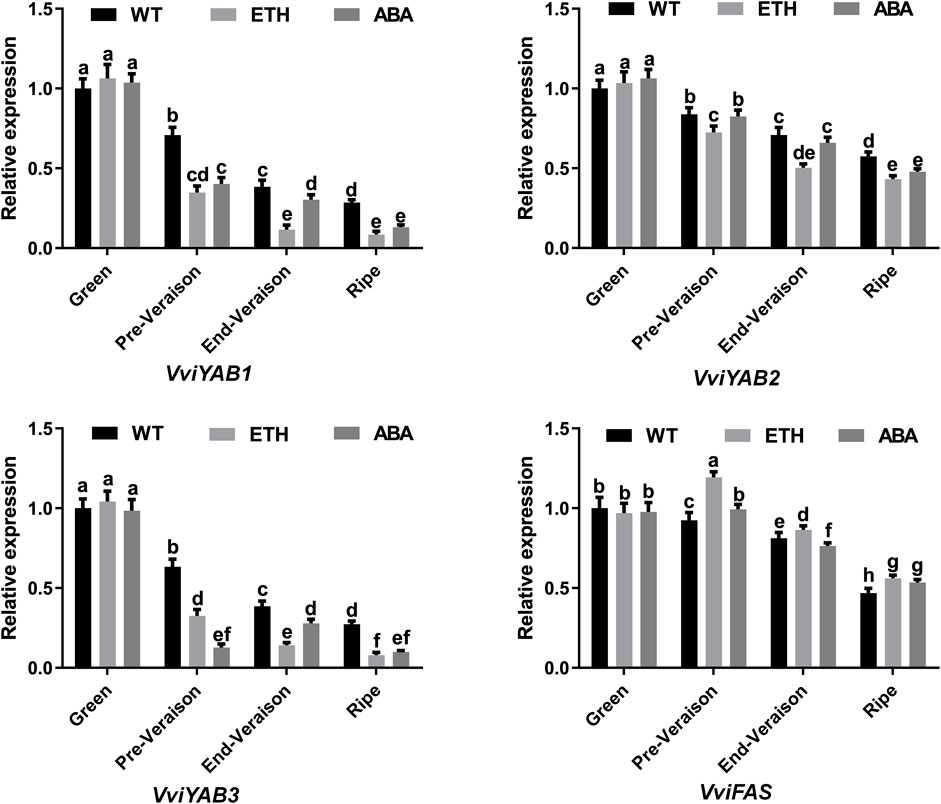
FIGURE 6. Expression profiles of VviYAB genes in response to ABA and ethylene treatments. Transcripts were normalized to the expression of the actin gene. The data are shown as means ± SD of three replicates.
Currently, GA3 and CPPU are usually used conducted to induce seedless fruit set, increase fruit size, and inhibit fruit russet (Xu et al., 2019). Therefore, we used previously published RNA-seq data to investigate in depth the crucial role of the VviYABs in response to GA3 and CPPU (Xu et al., 2019). As shown in Supplementary Table S6, the expression of VviYAB1 was nearly unchanged by GA3 treatment alone. When GA3 in combination with CPPU were applied, VviYAB1 expression was significantly increased suggesting an involvement of VviYAB1 in fruit expansion by mediating cytokinin signaling pathway. Furthermore, VviYAB3 expression was significantly increased by both GA3 and GA3 + CPPU treatments, implying that it may participate in a more complex regulatory network.
VviYABs in Response to Different Abiotic and Biotic Stresses
Expression data of VviYABs against various abiotic stresses, cold, salt, and polyethylene glycol (PEG), were obtained from the NCBI GEO datasets (GSE31594 and GSE31677) (Figure 7 and Supplementary Table S7). VviYAB1 and VviYAB5 showed a sharp downregulation in response to short-term salt and drought treatments. VviFAS was downregulated after 24 h under short-term salt and drought stress conditions. Conversely, VviYAB2 upregulated after 8h under drought stress but was strongly repressed after 24 h under salt stress (Figure 7A and Supplementary Table S7). Interestingly, VviYAB1, VviYAB5, and VviFAS were strongly repressed during the final phase of the long-term stress period (16 days after treatment), whereas VviYAB2 did not respond to either the long-term water or salt stress (Figure 7B and Supplementary Table S7).
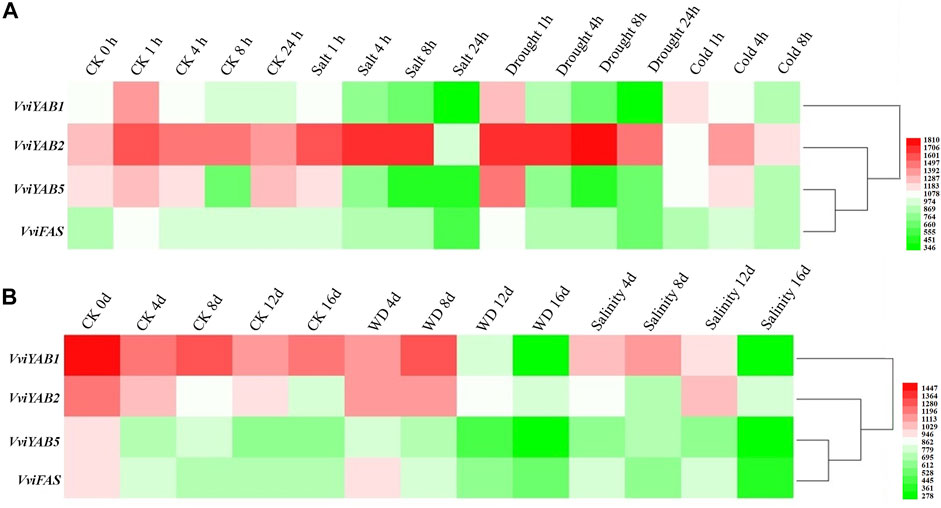
FIGURE 7. Expression patterns of VviYAB genes in response to abiotic stresses. Microarray analysis of VviYAB genes in the V. vinifera cv. Cabernet Sauvignon were downloaded from the NCBI GEO datasets (GSE31594 and GSE31677) and graphically represented with MeV software. (A) V. vinifera cv. Cabernet Sauvignon plants grown in a hydroponic drip system were treated with 120 mM salt, polyethylene glycol (PEG), cold (5 °C), or untreated. Shoots with leaves were collected at 0, 1, 4, and 8 h for all treatments and at 24 h for all treatments except cold (GEO series GSE31594). (B) Potted V. vinifera cv. Cabernet Sauvignon in the greenhouse were exposed to a water-deficit stress (WD) by withholding water or a salt stress by watering plants with a saline solution for 16 days. Non-stressed, normally watered plants served as the control for both treatments. Shoot tips were harvested every 4 days (0, 4, 8, 12, and 16 days) (GEO series GSE31677).
Expression profiles of VviYABs were also compared in response to three host-pathogen interaction experiments, such as Bois Noir phytoplasma (GSE12842), Erysiphe necator (GSE6404), and leaf-roll-associated virus-3 (GLRaV-3) (GSE31660) (Fung et al., 2008; Albertazzi et al., 2009; Vega et al., 2011). The expression aboundance of VviYAB1 was always lower than VviYAB2, 5 and VviFAS after inoculation either in cv. Cabernet Sauvignon (susceptible variety) or in cv. Norton (tolerant variety). The transcriptional abundance of VviYAB5 in response to E. necator infection showed a slightly decrease in cv. Norton and Cabernet Sauvignon (Figures 8A, B and Supplementary Table S8). Under Bois Noir phytoplasma, four VviYABs exhibited significantly decreased expression in the susceptible variety (cv. Chardonnay) than that of tolerant one (cv. Incrocio Manzoni) (Figure 8C and Supplementary Table S9). Finally, four VviYABs showed a slightly downregulated expression after GLRaV-3 infection in veraison stage (Figure 8D and Supplementary Table S10).
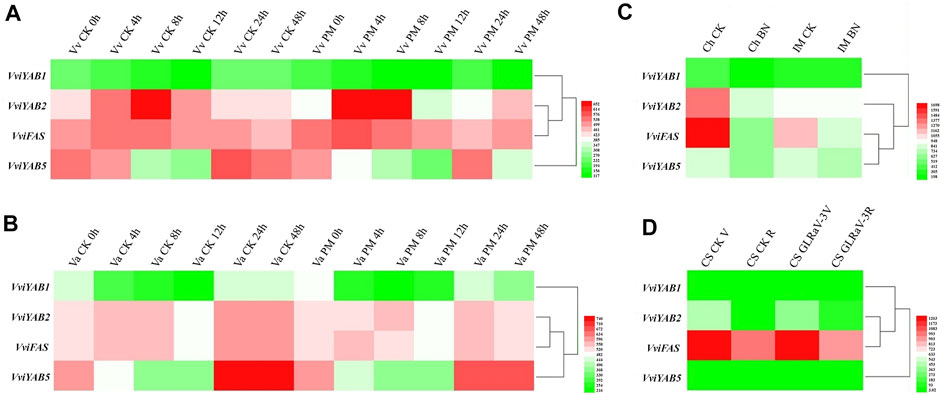
FIGURE 8. Expression patterns of VviYAB genes in response to biotic stresses. (A, B) The susceptible variety V. vinifera cv. Cabernet Sauvignon” and the resistant variety V. aestivalis cv “Norton” plants were grown in an environmental chamber and inoculated with Erysiphe necator conidiospores (PM). Inoculated leaves were harvested at 0, 4, 8, 12, 24, and 48 h after inoculation (GSE6404). (C) Field-grown plants of V. vinifera cv. Chardonnay (Ch) and Incrocio Manzoni (IM) naturally infected with Bois Noir phytoplasma (BN), compared to healthy samples (GSE12842). (D) V. vinifera cv. Cabernet Sauvignon (CS)” was infected with GLRaV-3 during veraison (CS GLRaV-3V) and ripening (CS GLRaV-3R) stages of berry development (GSE31660).
To obtain more information and validate the expression pattern, other RNA-seq data were collected from four previous studies (Haider et al., 2017; Guan et al., 2018; Zhu et al., 2018). Under drought stress, the expression levels of four VviYAB genes (VviYAB1, 2, 3, and 5) were significantly downregulated (Supplementary Figure S6 and Table S11), which is similar with the previously published expression files, indicating that VviYAB1 and 5 were strongly repressed by short- and long-term drought stress periods (Figure 7). In response to waterlogging stress, five VviYABs (VviYAB1, 2, 3, 5, and VviFAS) a decreasing expression trend (Supplementary Figure S6 and Table S11). In addition, VviYAB1, 2, 3 and VviFAS showed different degrees of reduction under salt microarray and RNA-seq data (Supplementary Figure S6 and Table S11). These results suggest that VviYABs play important roles in the response different abiotic and biotic stresses.
Subcellular Localizations
The subcellular localization of proteins is desirable for exploring their biological functions. To investigate the VviYAB functions, their subcellular localizations were authenticated using the fluorescent protein-tagging method. Firstly, the full-length open reading frames (ORFs) lacking the stop codon of three VviYABs were merged to the N-terminal the GFP driven by CaMV 35S promoter, generating fusion proteins 35S-VviCRC-GFP, 35S-VviFAS-GFP, and 35S-VviYAB3-GFP which were agroinfiltrated into leaves of 3- to 5-week-old N. benthamiana plants. The vector containing GFP alone (35S: GFP) was used as a control. Fluorescence microscopy exhibited that the control was uniformly dispersed throughout the cell, whereas 35S-VviCRC-GFP, 35S-VviFAS-GFP, and 35S-VviYAB3-GFP fusion proteins were detected in the nucleus and cytoplasm (Figure 9). The results demonstrate that VviCRC, VviFAS, and VviYAB3 are distributed in the cell nucleus and cytoplasm, suggesting their functional similarity.
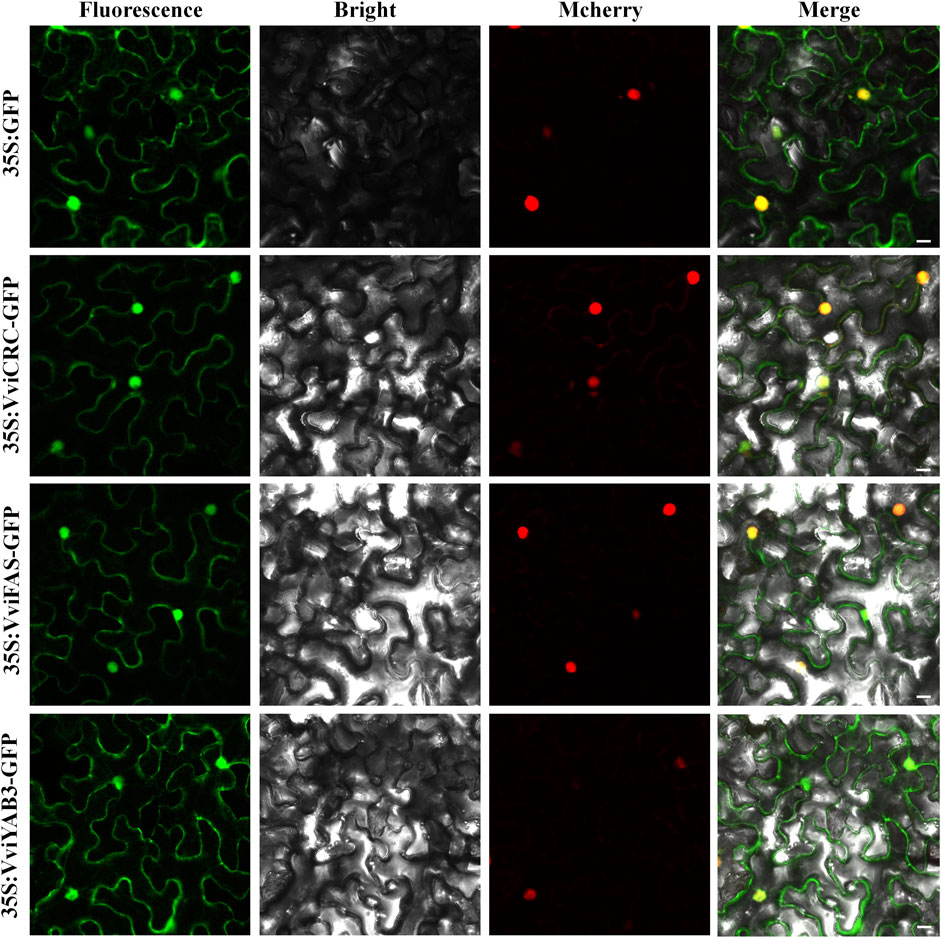
FIGURE 9. The leaves of 3-to 5-week-old Nicotiana benthamiana plants were transiently transformed with control, 35S:VviCRC-GFP, 35S:VviFAS-GFP, and 35S:VviYAB3-GFP. The NLS-mCherry tagged red fluorescent protein was co-expressed. Bar: 25 μm.
Discussion
Evolution and Structure of the VviYABs Family in Grapevines
Plant-specific YAB family plays an important role in abaxial cell development and lateral organ growth (Golz et al., 2004). In addition, YABs have been implicated in plant morphogenesis and stress response. Previous genomic studies have shown that there are 6 YAB members in Arabidopsis, 8 in rice, 9 in tomato (Huang et al., 2013), 12 in pakchoi (Hou et al., 2019), and 17 in soybean (Zhao et al., 2017). A total of seven VviYABs were identified from the grapevine genome, indicating that the grapevine YAB gene family lacked expansion during evolution.
Furthermore, 30 YAB proteins from four plant species, including both dicotyledons and monocotyledons, were divided into five subfamilies according to evolutionary relationships and structural features (Figure 1). As expected, VviYAB proteins showed a closer relationship to dicots than to monocots (Figure 1), suggesting that the YAB family was functionally diversified during the evolution of monocots and dicots. Genes from the same subfamily, such as VviYAB1 and VviYAB3, or VviFAS and VviYAB2, showed similar motif compositions and exon–intron structures (Figure 2), indicating that these gene pairs may have similar functions.
Gene duplication is the primary force that determines the evolutionary mechanisms (Cannon et al., 2004). No segmental and tandem duplication was identified in grapevine YAB family. A total of 17 pairs of syntenic relations were identified among grapevine, tomato, and Arabidopsis. Furthermore, the orthologous genes commonly share a similar structure and biological function. Tomato FAS is strongly associated with fruit shape classification. Grapevine VviFAS was found to share a collinearity relationship with tomato FAS genes, suggesting that they probably possess similar functions.
Potential Roles of VviYAB Genes Involved in the Immature-to-Mature Transition
YAB genes regulate plant growth, development, and morphogenesis, such as floral organ development and fruit morphology (Chen et al., 1999; Monforte et al., 2014). The expression pattern of AtCRC was mostly restricted to carpels and nectaries, while CRC plays a critical role in carpel morphogenesis, floral determinacy, and nectary specification in Arabidopsis (Bowman and Smyth, 1999). Moreover, CRC orthologs were found to be involved in leaf vascular development and carpel identity specification in Poaceae (Fourquin et al., 2014). In grapevines, VviCRC was preferentially highly expressed in well-developed inflorescences and carpels, which was consistent with the expression profile reported in Arabidopsis and Poaceae (Bowman, 2000; Fourquin et al., 2014), signifying that CRC and its orthologs may perform conserved functions during evolution after speciation. The AtINO gene is essential for the growth of the outer integument of the ovule (Villanueva et al., 1999; Simon et al., 2012). Moreover, a previous report showed that VviYABs, including VviINO gene were expressed at relatively high levels during ovule growth of seedless cultivars, indicating that they participate in the development of ovules (Zhang et al., 2019). However, our data demonstrated that VviINO exhibited low expression level in tested tissues of seeded cultivars, which was inconsistent with the findings of Zhang et al. (2019). This may be attributed to the difference in ovule development between seedless and seeded grapevine cultivars.
Previous reports demonstrated that YAB transcription factors regulated vegetative and reproductive development in Arabidopsis (Siegfried et al., 1999). In this study, most VviYAB genes (VviYAB1, 2, 3, and VviFAS) had relatively high expression levels in floral organs, seedling and early-stage fruits, young leaves, and buds, implying that VviYABs have conserved functions and are a broad regulator of grapevines and Arabidopsis. VviYAB5 was precisely highly transcribed during an early stage of burst buds and young leaves, which revealed that VviYAB5 might be involved in the early-stage bud and leaf development. Notably, VviYAB1, 2, 3, and 5 were strongly downregulated during the transition from vegetative/green to mature/woody development by microarray data and qRT-PCR assay, indicating that these VviYABs inhibited the transition to the mature development phase rather than being activated to implement this developmental program. Taken together, the above-mentioned results revealed that VviYABs can be considered as putative positive markers in vegetative/green tissues and negative markers of mature/woody tissues.
Potential Roles of VviYAB Genes in Berry Development and Morphogenesis
The development and maturation of grapevine berry is a complex and dynamic process, which is mainly controlled by a series of transcription factor regulatory networks (Kuhn et al., 2014). Previous studies have shown that FAS and LOCULE NUMBER (LC) control the number of fruit locules, which ultimately influence both fruit shape and size (Rodríguez et al., 2011; Azzi et al., 2015). SlYAB2, the homologous gene of FAS, showed differential gene expression between small- and medium-sized tomato fruit varieties (Bartley and Ishida, 2003). In grapevines, the expression of VviYAB2 was repressed in the fleshless berry mutant, which showed significantly reduced fruit size due to a lack of pericarp development (Fernandez et al., 2006; Fernandez et al., 2007). In the present study, VviYAB2 and VviFAS were strongly expressed in early stages of berry development and then decreased dramatically during berry development and ripening (Figure 6), suggesting that these members were important candidates for studying the diversification of berry morphogenesis. Similar with VviYAB2 and VvFAS, but to a lesser extent, the expression patterns of VviYAB1 and VviYAB3, two members of the YAB1/YAB3 subgroup, were also negatively related to berry shape or size. Similarities of these expression patterns indicated that these VviYABs were likely to have similar functions with SlFAS, which could regulate berry shape or size.
In addition, these four VviYABs (VviYAB1, 2, and 3 and VviFAS) also showed a significant decrease in mature berries and marked the berry developmental transition from immature to mature. These VviYABs can also be considered as putative stage-specificgrapevine berry biomarkers and were used to identify important developmental and metabolic processes in different conditions and organisms.
Potential Roles of VviYAB in Response to Various Phytohormone in Grapevines
Both ethylene and ABA are likely to play important roles, and their interplay may be required to control the berry maturation process (Chervin et al., 2004; Kuhn et al., 2014). Previous studies have shown that both ethylene and ABA could promote berry ripening, and the contents of ethylene and ABA have an increase at the ripening phase (Sun et al., 2010; Wheeler et al., 2019). For example, ethylene biosynthesis related genes are upregulated by ABA treatment (Kuhn et al., 2014). A decrease in berry ABA content was observed following 1-methylcyclopropene (1-MCP) treatment, a specific inhibitor of ethylene receptors (Sun et al., 2010). The role of VviYAB genes in the exogenous application of ABA and ethylene is still poorly understood. Previous studies havedepicted that three BcYAB genes are upregulated by exogenous ABA treatment in Pak-choi (Hou et al., 2019), implying that YABs play a potential role in regulating the ABA signaling pathway. In the current investigation, the expression levels of VviYAB1 and VviYAB3 were decreased with respect to berry ripening phase, indicating their role as negative regulators of grape berry ripening. However, the expression levels of VviYAB1 and VviYAB3 were sharply increased by exogenous GA3 and CPPU treatment. It is well known that GA3 and CPPU are usually applied to increase berry size in vineyards. The markedly increased expression of VviYAB1 and VviYAB3 under the GA3 and CPPU treatments further supported by the possibility that VviYAB genes might play an important role in grape berry expansion. These findings demonstrate that VviYABs are involved in fruit development by multiple mediating hormone signaling pathways.
VviYABs in Response to Abiotic and Biotic Stresses in Grapevines
Transcriptional regulation is a key indicator of plant responses to series of environmental and biotic stresses. Transcription factors bind to specific cis-elements in the promoter region of the target gene, thereby regulating the function of a particular gene. Previous studies have shown that YABs are involved in various abiotic stresses. For example, GmYAB10 were negatively regulated in drought and salinity responses (Zhao et al., 2017). Most of GhYABs in cotton are downregulated under drought and salinity stress, indicating that they may be negative regulators of cotton resistances to drought and salt stresses (Yang et al., 2018). In our study, a series of stress response cis-acting elements, such as ARE, MBS, and TC-rich, frequently occurred in the promoter regions of VviYABs (Supplementary Table S3). These elements are mainly involved in drought, salt, and waterlogging stresses. All VviYAB genes contained at least one of the stress response cis-acting elements, indicating their potential functions in response to abiotic stresses. For example, five VviYABs were downregulated under waterlogging stress, and all of genes contained ARE elements in their promoters. Furthermore, we found that all VviYABs were remarkably downregulated under drought and waterlogging treatments, indicating that these genes might negatively regulate drought and waterlogging responses.
Regarding biotic stresses, VviYABs showed low response in resistant varieties compared to susceptible genotypes, which was consistent with the previous results that overall changes in the global transcriptome were usually lower in the resistant genotypes (Fung et al., 2008; Albertazzi et al., 2009). After E. necator infection, we observed that VviYAB5 expression showed a slightly decrease in cv. Norton (resistant genotype) and Cabernet Sauvignon (susceptible genotype) (Figures 8A, B and Supplementary Table S8). The results suggested that VviYAB5 always responded to E. necator infection in both susceptible and resistant grapevine varieties, although the strong induction expression of the transcript did not occur under E. necator infection.
All VviYAB genes (VviYAB1, 2, and 5 and VviFAS) were more strongly decreased in susceptible variety Chardonnay than in the resistant variety Incrocio Manzoni (Figure 8C and Supplementary Table S9), indicating their role in plant defense against Bois Noir phytoplasma. Additionally, our VviYABs were slightly downregulated after GLRaV-3 infection at the veraison stage (Figure 8D and Supplementary Table S10). The current findings show that VviYABs mediate the grapevine defense response against a series of pathogenic infections. Although the microarray and RNA-seq data demonstrated that the VviYAB genes responded to different stress conditions, further experiments are needed to validate their putative role and the crosstalk between various environmental stresses.
Conclusion
In this study, seven VviYABs were identified, and their evolution relationship and expression patterns were analyzed systematically in grapevines. Numerous cis-acting elements and tissue expression analysis indicated that VviYABs were involved in complex regulatory networks controlling growth, development, and responses to abiotic and biotic stresses. Notably, VviYAB1, 2, 3, and 5 showed significantly higher expression levels in vegetative/green organs than in mature/woody tissues, implying the involvement of regulatory switch mechanisms that might stimulate the transition from the immature to the mature developmental phase. Furthermore, the dramatic downregulation expression of four VviYABs (VviYAB1, 2, and 3 and VviFAS) marked the berry developmental transition from immature to mature program. These findings demonstrated that these VviYAB genes can be considered as putative molecular biomarkers between vegetative/green and mature/woody samples and were used to identify important developmental and metabolic processes in grapevines. In addition, the results suggested an involvement of VviYAB1 in fruit expansion by mediating cytokinin signaling pathway. Microarray and RNA-seq data showed that VviYABs play important roles in response to abiotic and biotic stresses. Our results will pave the way for further studies on the functional conservation and divergence in the VviYAB gene family and provide potential candidates for fruit shape and plant abiotic stress tolerance in the grapevine germplasm.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
Conceptualization, XL and GL; methodology, YZ and PH; qPCR validation and subcellular localizations, SJ, YH and YX; writing—original draft preparation, SJ; writing—review and editing, all authors; supervision, XL and GL. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFD1001405-02), the China Postdoctoral Science Foundation (2019M661866), the Breeding Plan of Shandong Provincial Qingchuang Research Team (2019), the Natural Science Foundation of Jiangsu Province (SBK2020020595), the Natural Science Research Project of Higher Education Institutions of Jiangsu Province (20KJD210001), the science-technology benefiting people project of Qingdao (21-1-4-ny-14-nsh), National Natural Science Foundation of China (NSFC) (Grant No. 32102346), Shanghai Sailing Program (21YF1422100), and Startup Fund for Young Faculty at SJTU (21X010500643).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.762221/full#supplementary-material
References
Albertazzi, G., Milc, J., Caffagni, A., Francia, E., Roncaglia, E., Ferrari, F., et al. (2009). Gene Expression in grapevine Cultivars in Response to Bois Noir Phytoplasma Infection. Plant Sci. 176, 792–804. doi:10.1016/j.plantsci.2009.03.001
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, Two Arabidopsis Genes that Control Carpel Development in Parallel with AGAMOUS. Development 126, 2377–2386. doi:10.1242/dev.126.11.2377
Azzi, L., Deluche, C., Gévaudant, F., Frangne, N., Delmas, F., Hernould, M., et al. (2015). Fruit Growth-Related Genes in Tomato. J. Exp. Bot. 66, 1075–1086. doi:10.1093/jxb/eru527
Badawi, M. A., Agharbaoui, Z., Zayed, M., Li, Q., Byrns, B., Zou, J., et al. (2019). Genome‐Wide Identification and Characterization of the Wheat Remorin ( Ta REM) Family during Cold Acclimation. Plant Genome 12 (2), 180040. doi:10.3835/plantgenome2018.06.0040
Bartley, G. E., and Ishida, B. K. (2003). Developmental Gene Regulation during Tomato Fruit Ripening and In-Vitro Sepal Morphogenesis. BMC Plant Biol. 3, 4. doi:10.1186/1471-2229-3-4
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a Gene that Regulates Carpel and Nectary Development in Arabidopsis, Encodes a Novel Protein with Zinc Finger and Helix-loop-helix Domains. Development 126, 2387–2396. doi:10.1242/dev.126.11.2387
Bowman, J. L. (2000). The YABBY Gene Family and Abaxial Cell Fate. Curr. Opin. Plant Biol. 3, 17–22. doi:10.1016/s1369-5266(99)00035-7
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi:10.1186/1471-2229-4-10
Chen, Q., Atkinson, A., Otsuga, D., Christensen, T., Reynolds, L., and Drews, G. N. (1999). The Arabidopsis FILAMENTOUS FLOWER Gene is Required for Flower Formation. Development 126, 2715–2726. doi:10.1242/dev.126.12.2715
Chervin, C., El-Kereamy, A., Roustan, J. P., Latché, A., Lamon, J., and Bouzayen, M. (2004). Ethylene Seems Required for the berry Development and Ripening in Grape, a Non-climacteric Fruit. Plant Sci. 167, 1301–1305. doi:10.1016/j.plantsci.2004.06.026
Cong, B., Barrero, L. S., and Tanksley, S. D. (2008). Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication. Nat. Genet. 40, 800–804. doi:10.1038/ng.144
Dai, M., Zhao, Y., Ma, Q., Hu, Y., Hedden, P., Zhang, Q., et al. (2007). The Rice YABBY1 Gene is Involved in the Feedback Regulation of Gibberellin Metabolism. Plant Physiol. 144, 121–133. doi:10.1104/pp.107.096586
Eckardt, N. A. (2010). YABBY Genes and the Development and Origin of Seed Plant Leaves. Plant Cell 22, 2103. doi:10.1105/tpc.110.220710
Fasoli, M., Dal Santo, S., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The Grapevine Expression Atlas Reveals a Deep Transcriptome Shift Driving the Entire Plant into a Maturation Program. Plant Cell 24, 3489–3505. doi:10.1105/tpc.112.100230
Fasoli, M., Richter, C. L., Zenoni, S., Bertini, E., Vitulo, N., Dal Santo, S., et al. (2018). Timing and Order of the Molecular Events Marking the Onset of Berry Ripening in grapevine. Plant Physiol. 178, 1187–1206. doi:10.1104/pp.18.00559
Feng, G., Han, J., Yang, Z., Liu, Q., Shuai, Y., Xu, X., et al. (2021). Genome-wide Identification, Phylogenetic Analysis, and Expression Analysis of the SPL Gene Family in Orchardgrass (Dactylis Glomerata L.). Genomics 113, 2413–2425. doi:10.1016/j.ygeno.2021.05.032
Fernandez, L., Romieu, C., Moing, A., Bouquet, A., Maucourt, M., Thomas, M. R., et al. (2006). The Grapevine Fleshless Berry Mutation. A Unique Genotype to Investigate Differences between Fleshy and Nonfleshy Fruit. Plant Physiol. 140, 537–547. doi:10.1104/pp.105.067488
Fernandez, L., Torregrosa, L., Terrier, N., Sreekantan, L., Grimplet, J., Davies, C., et al. (2007). Identification of Genes Associated with Flesh Morphogenesis during grapevine Fruit Development. Plant Mol. Biol. 63, 307–323. doi:10.1007/s11103-006-9090-2
Fourquin, C., Primo, A., Martínez-Fernández, I., Huet-Trujillo, E., and Ferrándiz, C. (2014). The CRC Orthologue from Pisum sativum Shows Conserved Functions in Carpel Morphogenesis and Vascular Development. Ann. Bot. 114, 1535–1544. doi:10.1093/aob/mcu129
Fung, R. W. M., Gonzalo, M., Fekete, C., Kovacs, L. G., He, Y., Marsh, E., et al. (2008). Powdery Mildew Induces Defense-Oriented Reprogramming of the Transcriptome in a Susceptible but Not in a Resistant Grapevine. Plant Physiol. 146, 236–249. doi:10.1104/pp.107.108712
Golz, J. F., and Hudson, A. (1999). Plant Development: YABBYs Claw to the Fore. Curr. Biol. 9, R861–R863. doi:10.1016/s0960-9822(00)80047-0
Golz, J. F., Roccaro, M., Kuzoff, R., and Hudson, A. (2004). GRAMINIFOLIA Promotes Growth and Polarity of Antirrhinum leaves. Development 131, 3661–3670. doi:10.1242/dev.01221
Grimplet, J., Adam-Blondon, A. F., Bert, P. F., Bitz, O., Cantu, D., Davies, C., et al. (2014). The grapevine Gene Nomenclature System. BMC Genomics 15 (1), 1077. doi:10.1186/1471-2164-15-1077
Guan, L., Haider, M., Khan, N., Nasim, M., Jiu, S., Fiaz, M., et al. (2018). Transcriptome Sequence Analysis Elaborates a Complex Defensive Mechanism of grapevine (Vitis vinifera L.) in Response to Salt Stress. Int. J. Mol. Sci. 19 19, 4019. doi:10.3390/ijms19124019
Haider, M. S., Zhang, C., Kurjogi, M. M., Pervaiz, T., Zheng, T., Zhang, C., et al. (2017). Insights into Grapevine Defense Response against Drought as Revealed by Biochemical, Physiological and RNA-Seq Analysis. Sci. Rep. 7, 13134. doi:10.1038/s41598-017-13464-3
Hou, H., Wu, P., Gao, L., Zhang, C., and Hou, X. (2019). Characterization and Expression Profile Analysis of YABBY Family Genes in Pak-Choi (Brassica Rapa ssp. Chinensis) under Abiotic Stresses and Hormone Treatments. Plant Growth Regul. 87, 421–432. doi:10.1007/s10725-019-00475-5
Huang, Z., Van Houten, J., Gonzalez, G., Xiao, H., and van der Knaap, E. (2013). Genome-wide Identification, Phylogeny and Expression Analysis of SUN, OFP and YABBY Gene Family in Tomato. Mol. Genet. Genomics 288, 111–129. doi:10.1007/s00438-013-0733-0
Inal, B., Büyük, İ., Ilhan, E., and Aras, S. (2017). Genome-wide Analysis of Phaseolus vulgaris C2C2-YABBY Transcription Factors under Salt Stress Conditions. 3 Biotech. 7, 302. doi:10.1007/s13205-017-0933-0
Jiu, S., Guan, L., Leng, X., Zhang, K., Haider, M. S., Yu, X., et al. (2021). The Role of VvMYBA2r and VvMYBA2w Alleles of the MYBA2 Locus in the Regulation of Anthocyanin Biosynthesis for Molecular Breeding of Grape (Vitis spp.) Skin Coloration. Plant Biotechnol. J. 19 (6), 1216–1239. doi:10.1111/pbi.13543
Jiu, S., Wang, C., Zheng, T., Liu, Z., Leng, X., Pervaiz, T., et al. (2016). Characterization of VvPAL-like Promoter from Grapevine Using Transgenic Tobacco Plants. Funct. Integr. Genomics 16 (6), 595–617. doi:10.1007/s10142-016-0516-x
Jiu, S., Xu, Y., Wang, J., Haider, M. S., Xu, J., Wang, L., et al. (2022). Molecular Mechanisms Underlying the Action of Strigolactones Involved in Grapevine Root Development by Interacting with Other Phytohormone Signaling. Sci. Hortic. 293, 110709. doi:10.1016/j.scienta.2021.110709
Jiu, S., Xu, Y., Wang, J., Wang, L., Liu, X., Sun, W., et al. (2020). The Cytochrome P450 Monooxygenase Inventory of Grapevine (Vitis Vinifera L.): Genome-wide Identification, Evolutionary Characterization and Expression Analysis. Front. Genet. 11, 44. doi:10.3389/fgene.2020.00044
Jiu, S., Xu, Y., Wang, J., Wang, L., Wang, S., Ma, C., et al. (2019). Genome-wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera. Front. Genet. 10, 1276. doi:10.3389/fgene.2019.01276
Kuhn, N., Guan, L., Dai, Z. W., Wu, B. H., Lauvergeat, V., Gomès, E., et al. (2014). Berry Ripening: Recently Heard through the Grapevine. J. Exp. Bot. 65, 4543–4559. doi:10.1093/jxb/ert395
Leng, X., Jia, H., Sun, X., Shangguan, L., Mu, Q., Wang, B., et al. (2015a). Comparative Transcriptome Analysis of Grapevine in Response to Copper Stress. Sci. Rep. 5, 17749. doi:10.1038/srep17749
Leng, X., Mu, Q., Wang, X., Li, X., Zhu, X., Shangguan, L., et al. (2015b). Transporters, Chaperones, and P-type ATPases Controlling Grapevine Copper Homeostasis. Funct. Integr. Genomics 15, 673–684. doi:10.1007/s10142-015-0444-1
Leng, X., Wang, P., Wang, C., Zhu, X., Li, X., Li, H., et al. (2017). Genome-wide Identification and Characterization of Genes Involved in Carotenoid Metabolic in Three Stages of Grapevine Fruit Development. Sci. Rep. 7, 4216. doi:10.1038/s41598-017-04004-0
Leng, X., Wei, H., Xu, X., Ghuge, S. A., Jia, D., Liu, G., et al. (2019). Genome-wide Identification and Transcript Analysis of TCP Transcription Factors in Grapevine. BMC Genomics 20, 786. doi:10.1186/s12864-019-6159-2
Liu, H. L., Xu, Y. Y., Xu, Z. H., and Chong, K. (2007). A rice YABBY Gene, OsYABBY4, Preferentially Expresses in Developing Vascular Tissue. Dev. Genes Evol. 217, 629–637. doi:10.1007/s00427-007-0173-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Massonnet, M., Fasoli, M., Tornielli, G. B., Altieri, M., Sandri, M., Zuccolotto, P., et al. (2017). Ripening Transcriptomic Program in Red and White Grapevine Varieties Correlates with Berry Skin Anthocyanin Accumulation. Plant Physiol. 174, 2376–2396. doi:10.1104/pp.17.00311
Monforte, A. J., Diaz, A., Caño-Delgado, A., and van der Knaap, E. (2014). The Genetic Basis of Fruit Morphology in Horticultural Crops: Lessons from Tomato and Melon. J. Exp. Bot. 65, 4625–4637. doi:10.1093/jxb/eru017
Postel, D., Vanlemmens, P., Gode, P., Ronco, G., and Villa, P. (2002). Plant CARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Rodríguez, G. R., Muños, S., Anderson, C., Sim, S. C., Michel, A., Causse, M., et al. (2011). Distribution of SUN, OVATE, LC, and FAS in the Tomato Germplasm and the Relationship to Fruit Shape Diversity. Plant Physiol. 156, 275–285. doi:10.1104/pp.110.167577
Saeed, A. I., Bhagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). TM4 Microarray Software Suite. Method Enzymol. 411, 134–193. doi:10.1016/s0076-6879(06)11009-5
Sarojam, R., Sappl, P. G., Goldshmidt, A., Efroni, I., Floyd, S. K., Eshed, Y., et al. (2010). Differentiating Arabidopsis Shoots from Leaves by Combined YABBY Activities. Plant Cell 22, 2113–2130. doi:10.1105/tpc.110.075853
Sawa, S., Ito, T., Shimura, Y., and Okada, K. (1999a). FILAMENTOUS FLOWER Controls the Formation and Development of Arabidopsis Inflorescences and Floral Meristems. Plant Cell 11, 69–86. doi:10.1105/tpc.11.1.69
Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E. H., and Okada, K. (1999b). FILAMENTOUS FLOWER, a Meristem and Organ Identity Gene of Arabidopsis, Encodes a Protein with a Zinc finger and HMG-Related Domains. Genes Dev. 13, 1079–1088. doi:10.1101/gad.13.9.1079
Shamimuzzaman, M., and Vodkin, L. (2013). Genome-wide Identification of Binding Sites for NAC and YABBY Transcription Factors and Co-regulated Genes during Soybean Seedling Development by ChIP-Seq and RNA-Seq. BMC Genomics 14, 477. doi:10.1186/1471-2164-14-477
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., and Bowman, J. L. (1999). Members of the YABBY Gene Family Specify Abaxial Cell Fate in Arabidopsis. Development 126, 4117–4128. doi:10.1242/dev.126.18.4117
Simon, M. K., Williams, L. A., Brady-Passerini, K., Brown, R. H., and Gasser, C. S. (2012). Positive- and Negative-acting Regulatory Elements Contribute to the Tissue-specific Expression of INNER NO OUTER, a YABBY-type Transcription Factor Gene in Arabidopsis. BMC Plant Biol. 12, 214. doi:10.1186/1471-2229-12-214
Sun, L., Zhang, M., Ren, J., Qi, J., Zhang, G., and Leng, P. (2010). Reciprocity between Abscisic Acid and Ethylene at the Onset of Berry Ripening and after Harvest. BMC Plant Biol. 10, 257. doi:10.1186/1471-2229-10-257
Tanaka, W., Toriba, T., and Hirano, H. Y. (2017). Three TOB1-related YABBY Genes Are Required to Maintain Proper Function of the Spikelet and Branch Meristems in Rice. New Phytol. 215, 825–839. doi:10.1111/nph.14617
Vega, A., Gutiérrez, R. A., Peña-Neira, A., Cramer, G. R., and Arce-Johnson, P. (2011). Compatible GLRaV-3 Viral Infections Affect Berry Ripening Decreasing Sugar Accumulation and Anthocyanin Biosynthesis in Vitis vinifera. Plant Mol. Biol. 77, 261–274. doi:10.1007/s11103-011-9807-8
Villanueva, J. M., Broadhvest, J., Hauser, B. A., Meister, R. J., Schneitz, K., and Gasser, C. S. (1999). INNER NO OUTER Regulates Abaxial- Adaxial Patterning in Arabidopsis Ovules. Genes Dev. 13, 3160–3169. doi:10.1101/gad.13.23.3160
Wei, H., Wang, P., Chen, J., Li, C., Wang, Y., Yuan, Y., et al. (2020). Genome-wide Identification and Analysis of B-BOX Gene Family in Grapevine Reveal its Potential Functions in Berry Development. BMC Plant Biol. 20, 72. doi:10.1186/s12870-020-2239-3
Wheeler, S., Loveys, B., Ford, C., and Davies, C. (2019). The Relationship between the Expression of Abscisic Acid Biosynthesis Genes, Accumulation of Abscisic Acid and the Promotion of Vitis vinifera L. Berry Ripening by Abscisic Acid. Aust. J. Grape Wine R. 15, 195–204. doi:10.1111/j.1755-0238.2008.00045.x
Xiang, J., Liu, R. Q., Li, T. M., Han, L. J., Zou, Y., Xu, T. F., et al. (2013). Isolation and Characterization of Two VpYABBY Genes from Wild Chinese Vitis pseudoreticulata. Protoplasma 250, 1315–1325. doi:10.1007/s00709-013-0514-y
Xing, J. Y., Lu, L., Wen, T. J., and Hu, J. F. (2016). The Correlation between VvYABBY5 Expression and the Ontogeny of Tricarpellate Fruit in 'Xiangfei' Grapevine (Vitis vinifera). J. Hortic. Sci. Biotechnol. 91, 308–315. doi:10.1080/14620316.2016.1157006
Xu, Y., Hou, X., Feng, J., Khalil-Ur-Rehman, M., and Tao, J. (2019). Transcriptome Sequencing Analyses Reveals Mechanisms of Eliminated Russet by Applying GA3 and CPPU on ‘Shine Muscat’ Grape. Scientia Horticulturae 250, 94–103. doi:10.1016/j.scienta.2019.02.048
Xu, Y., Wang, J., Wang, R., Wang, L., Zhang, C., Xu, W., et al. (2021). The Role of Strigolactones in the Regulation of Root System Architecture in grapevine (Vitis vinifera L.) in Response to Root-Restriction Cultivation. Int. J. Mol. Sci. 22 (16), 8799. doi:10.3390/ijms22168799
Yamada, T., Yokota, S. y., Hirayama, Y., Imaichi, R., Kato, M., and Gasser, C. S. (2011). Ancestral Expression Patterns and Evolutionary Diversification of YABBY Genes in Angiosperms. Plant J. 67, 26–36. doi:10.1111/j.1365-313x.2011.04570.x
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H.-Y. (2004). The YABBY Gene DROOPING LEAF Regulates Carpel Specification and Midrib Development in Oryza Sativa. Plant Cell 16, 500–509. doi:10.1105/tpc.018044
Yang, C., Ma, Y., and Li, J. (2016). The Rice YABBY4 Gene Regulates Plant Growth and Development through Modulating the Gibberellin Pathway. J. Exp. B. 67, 5545–5556. doi:10.1093/jxb/erw319
Yang, Z., Gong, Q., Wang, L., Jin, Y., Xi, J., Li, Z., et al. (2018). Genome-wide Study of YABBY Genes in Upland Cotton and Their Expression Patterns under Different Stresses. Front. Genet. 9, 33. doi:10.3389/fgene.2018.00033
Zhang, S., Wang, L., Sun, X., v, Y., Yao, J., Nocker, S., et al. (2019). Genome-Wide Analysis of the YABBY Gene Family in Grapevine and Functional Characterization of VvYABBY4. Frontiers in Plant Science 10, 1207. doi:10.3389/fpls.2019.01207
Zhao, S. P., Lu, D., Yu, T. F., Ji, Y. J., Zheng, W. J., Zhang, S. X., et al. (2017). Genome-wide Analysis of the YABBY Family in Soybean and Functional Identification of GmYABBY10 Involvement in High Salt and Drought Stresses. Plant Physiol. Biochem. 119, 132–146. doi:10.1016/j.plaphy.2017.08.026
Keywords: grapevine, genome-wide analysis, VviYABs, berry development, abiotic and biotic stresses
Citation: Jiu S, Zhang Y, Han P, Han Y, Xu Y, Liu G and Leng X (2022) Genome-Wide Identification and Expression Analysis of VviYABs Family Reveal Its Potential Functions in the Developmental Switch and Stresses Response During Grapevine Development. Front. Genet. 12:762221. doi: 10.3389/fgene.2021.762221
Received: 21 August 2021; Accepted: 02 December 2021;
Published: 03 February 2022.
Edited by:
Million Tadege, Oklahoma State University, United StatesReviewed by:
Xiaojiao Han, Chinese Academy of Forestry, ChinaDa-Long Guo, Henan University of Science and Technology, China
Copyright © 2022 Jiu, Zhang, Han, Han, Xu, Liu and Leng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gengsen Liu, Z3NsaXVAcWF1LmVkdS5jbg==; Xiangpeng Leng, bGVuZ3BlbmcyMDA4QDE2My5jb20=
 Songtao Jiu
Songtao Jiu Yanping Zhang2
Yanping Zhang2 Xiangpeng Leng
Xiangpeng Leng