- 1College of Life Sciences, Qingdao Agricultural University, Qingdao, China
- 2College of Agronomy, Qingdao Agricultural University, Qingdao, China
DIRIGENT (DIR) genes are key players in environmental stress responses that have been identified in many vascular plant species. However, few studies have examined the VrDIR genes in mungbean. In this study, we characterized 37 VrDIR genes in mungbean using a genome-wide identification method. VrDIRs were distributed on seven of the 11 mungbean chromosomes, and chromosome three contained the most VrDIR genes, with seven members. Thirty-two of the 37 VrDIRs contained a typical DIR gene structure, with one exon; the conserved DIR domain (i.e., Pfam domain) occupied most of the protein in 33 of the 37 VrDIRs. The gene structures of VrDIR genes were analyzed, and a total of 19 distinct motifs were detected. VrDIR genes were classified into five groups based on their phylogenetic relationships, and 13 duplicated gene pairs were identified. In addition, a total of 92 cis-acting elements were detected in all 37 VrDIR promoter regions, and VrDIR genes contained different numbers and types of cis-acting elements. As a result, VrDIR genes showed distinct expression patterns in different tissues and in response to salt and drought stress.
Introduction
Mungbean is an important legume crop that is mainly grown in Asian countries, including India, Thailand, and China (Tomooka et al., 2002). Mungbean is thought to have been domesticated in India and then spread to other countries (Fuller, 2007). Mungbean provides humans with several benefits. First, the roots of mungbean can fix atmospheric nitrogen and thus improve soil fertility and texture (Graham and Vance, 2003). Second, mungbean seeds contain high amounts of protein and nutrients; for this reason, they are widely consumed (Keatinge et al., 2011). The nutritional value of mungbean increases during seed germination because of the degradation of proteins, vitamins, and minerals (El-Adawy et al., 2003). Thus, mungbean sprouts are a favored vegetable in many countries (Tomooka et al., 2002). With the rapid growth of the human population, the demand for mungbean has increased. However, the production of mungbean is affected by many environmental factors, such as salt and drought stress. The lack of knowledge of functional genes in mungbean limits our ability to enhance the resistance of mungbean to adverse environments. Generally, the study of the functions of genes in mungbean provides useful information for improving mungbean plants.
Many genes have been identified to be involved in abiotic and biotic stress, such as DIRIGENT (DIR) genes (Ralph et al., 2007; Guo et al., 2012; Paniagua et al., 2017; Khan et al., 2018; Li et al., 2019; Vaahtera et al., 2019; Han et al., 2020; Hu et al., 2020; Yu et al., 2020; Zhu et al., 2020; Liu et al., 2021). The first DIR reported stipulates stereoselective coupling of two coniferyl alcohol to produce (+) -pinoresinol, provided a one electron oxidase or oxidant is present. It was first identified in Forsythia intermedia (Davin et al., 1997); and then was later reported in Arabidopsis (Kim et al., 2002; Kim et al., 2012), Schizandra chinensis (Kim et al., 2012), Pisum sativum (Seneviratne et al., 2015), and Linum usitatissimum (Dalisay et al., 2015). DIR genes typically contain an exon without an intron, and the conserved DIR domain occupies most of the DIR protein (Corbin et al., 2018). DIR genes are found in almost all vascular plants, including ferns, gymnosperms, and angiosperms (Davin and Lewis, 2000; Ralph et al., 2007; Wu et al., 2009; Li et al., 2014). DIR genes have been studied at the whole genome level in many plant species, including Arabidopsis (25 AtDIRs), rice (54 OsDIRs), pepper (24 CaDIRs), Medicago (45 MtDIRs), Brassica rapa (29 BrDIRs), and soybean (54 GmDIRs) (Ralph et al., 2007; Guo et al., 2012; Thamil Arasan et al., 2013; Khan et al., 2018; Song and Peng, 2019; Ma et al., 2021). DIR genes can be classified into seven groups, designated as DIR-a to DIR-g, based on their evolutionary relationships (Ralph et al., 2006).
The functions of many DIR proteins have been identified. Members of DIR-a subfamily participate in the formation of pinoresinol (Ralph et al., 2006; Corbin et al., 2018). Several DIR-b/d subfamily members are involved in aromatic diterpenoid biosynthesis (Liu et al., 2008; Effenberger et al., 2015) and pterocarpan biosynthesis (Uchida et al., 2017; Meng et al., 2020), and the DIR-e subfamily is thought to participate in Casparian band lignin formation (Hosmani et al., 2013). In addition, many DIR genes have been shown to be involved in biotic stress in plants. For example, the expression of GmDIR22 is induced by Phytophthora sojae infection, and the overexpression of GmDIR22 enhances the resistance of susceptible soybean cultivar ‘Dongnong 50’ to P. sojae by increasing total lignan accumulation (Li et al., 2017). The expression of many DIR genes in spruce (Picea spp) was induced in response to insect attacks (Ralph et al., 2007). Many DIR genes also participate in abiotic stress responses. For example, the expression of ScDIR in sugarcane increases in response to H2O2, PEG, or NaCl stress (Guo et al., 2012). BhDIR1 transcripts accumulate in response to changes in water and temperature stress (Wu et al., 2009). Loss of function of CaDIR7 reduced root activity after salt stress, and the induction of stress-related genes was suppressed in CaDIR7-silenced plants (Khan et al., 2018). Several BrDIR members showed altered expression levels in response to water, ABA, and cold stress, and the expression of many BrDIR genes is correlated with increased lignification under water stress (Arasan et al., 2013).
The study of mungbean VrDIR genes could provide important information for the molecular breeding of mungbean plants. The release of the mungbean draft genome sequence provides essential information for the analysis of VrDIR genes (Kang et al., 2014). In this study, we characterized mungbean VrDIR genes using genome-wide identification and investigated their phylogenetic relationships, gene structures, conserved motifs, gene duplications, cis-acting elements in promoters, and expression profiles in different tissues in response to salt and drought stress. Our study provides key insight into the function of mungbean VrDIR genes in the regulation of abiotic stress.
Materials and Methods
Plant Materials and Growth Conditions
The sequenced mungbean variety VC 1973A was used in all experiments in this study (Kang et al., 2014). Eight tissues, including flowers, pods, leaves, seeds, nodule roots, stems, roots, and shoot apices, were collected from mungbean plants grown in the field as described by Shi et al. (Liu et al., 2021; Shi et al., 2021). Mungbean seedlings were grown in a growth chamber for stress treatment. The growth conditions were as follows: 10 h, 28°C, light/14 h, 25°C dark cycles (light source: white fluorescent lights, ∼100 μmol m−2 s−1), and the humidity was maintained at approximately 30%. Seven-day-old mungbean plants were used for stress treatment. For salt stress treatment, plants were watered with 150 mmol NaCl, and tissues were sampled 9 days after treatment (Zhang et al., 2019; Zhu et al., 2019). For drought stress treatment, 7 day-old plants were watered, and then the plants were grown without irrigation; the control plants were watered every 5 days. After 12 days drought and normal condition treatments, the soil moisture content was measured using the method of weighing the soil before and after drying treatment. The soil moisture content under drought stress was reduced to around 17.29% from 49.86% under normal condition (Supplementary Figure S3). Tissues were collected 12 days after treatment. The shoots and roots were sampled separately and then stored at –80°C for RNA isolation. Three biological replicates were collected for each sample.
Identification of Mungbean VrDIR Genes
The amino acid sequences of 25 Arabidopsis and 54 soybean DIR proteins were used as blast queries against the National Center for Biotechnology Information (NCBI) database to search for mungbean VrDIR candidate genes. All the output genes were analyzed using HMMER to confirm the conserved PF03018 domain (DIR domain) (Simon et al., 2018), and candidate genes containing conserved DIR domains were designated as VrDIR members. The gene ID, genomic length, and amino acid number were obtained from NCBI and mungbean database (https://legumeinfo.org/organism/Vigna/radiata) (http://plantgenomics.snu.ac.kr/mediawiki-1.21.3/index.php/Main_Page). ProtParam software (https://web.expasy.org/protparam/) was used to analyze the molecular weight (MW) of proteins and the theoretical iso-electric point (pI). The chromosome position of each VrDIR gene was obtained from NCBI and visualized using MapChart software (Voorrips, 2002).
Phylogenetic Analysis
The amino acid sequences of DIR proteins from mungbean and other species reported by Corbin et al. (Corbin et al., 2018), including DIR members from Arabidopsis thaliana, Forsythia x intermedia, Gossypium barbadense, Glycyrrhiza echinata, Gossypium hirsutum, Hordeum vulgare, Oryza sativa, Podophyllum peltatum, Picea sitchensis, Sorghum bicolor, Schizandra chinensis, Saccharum hybrid cultivar, Sesamum indicum, Triticum aestivum, Tamarix androssowii, Tsuga heterophylla, Thuja plicata, Linum usitatissimum, Zea mays, Glycine max, Vigna unguiculata, Pisum sativum and Phaseolus vulgaris were aligned using ClustalW2 (Oliver et al., 2005). The phylogenetic tree was constructed using the alignment results of DIR proteins from these species in MEGA 7.0 using the neighbor-joining method with default parameters (Kumar et al., 2016). The classification of mungbean VrDIR genes was carried out as described by Corbin et al. (Corbin et al., 2018).
Gene Duplication Analysis
Gene duplication of VrDIR genes was analyzed using OrthoMCL and Circos software following the methods of Jin et al. (Krzywinski et al., 2009; Fischer et al., 2011; Jin et al., 2019; Jin et al., 2020). Amino acid sequences with a similarity greater than 80% were designated as duplicated gene pairs.
Exon-Intron Organization and Conserved Motif Analyses
The full lengths of each VrDIR genomic sequence and coding sequence were aligned using the Gene Structure Display Server program to analyze the exon-intron organization (Hu et al., 2015), and the UTRs, exons, and introns were shown in different colored boxes. The MEME tool was used to analyze VrDIR-conserved motifs with default parameters (Bailey et al., 2009), and the different motifs were displayed using different colored boxes.
Cis-Acting Element Analysis of VrDIR Promoter Regions
The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the cis-acting elements in VrDIR promoter regions (Lescot et al., 2002). The promoter regions were analyzed using sequences 2 kb upstream of the initiation codon ATG. The cis-acting elements were classified into different groups based on their potential functions.
RNA Isolation, cDNA Synthesis, and Gene Expression Analysis of VrDIR Genes
For RNA isolation and cDNA synthesis, the tissues were prepared as previously described (Li et al., 2021). Gene expression levels were analyzed using quantitative real-time PCR (qRT-PCR) as described by Ma et al. (Ma et al., 2021). All samples were analyzed using three biological replicates. The gene expression levels were normalized to a mungbean Actin gene (Li et al., 2018). All primers used in this study are listed in Supplementary Table S1.
Results
Identification of Mungbean VrDIR Genes
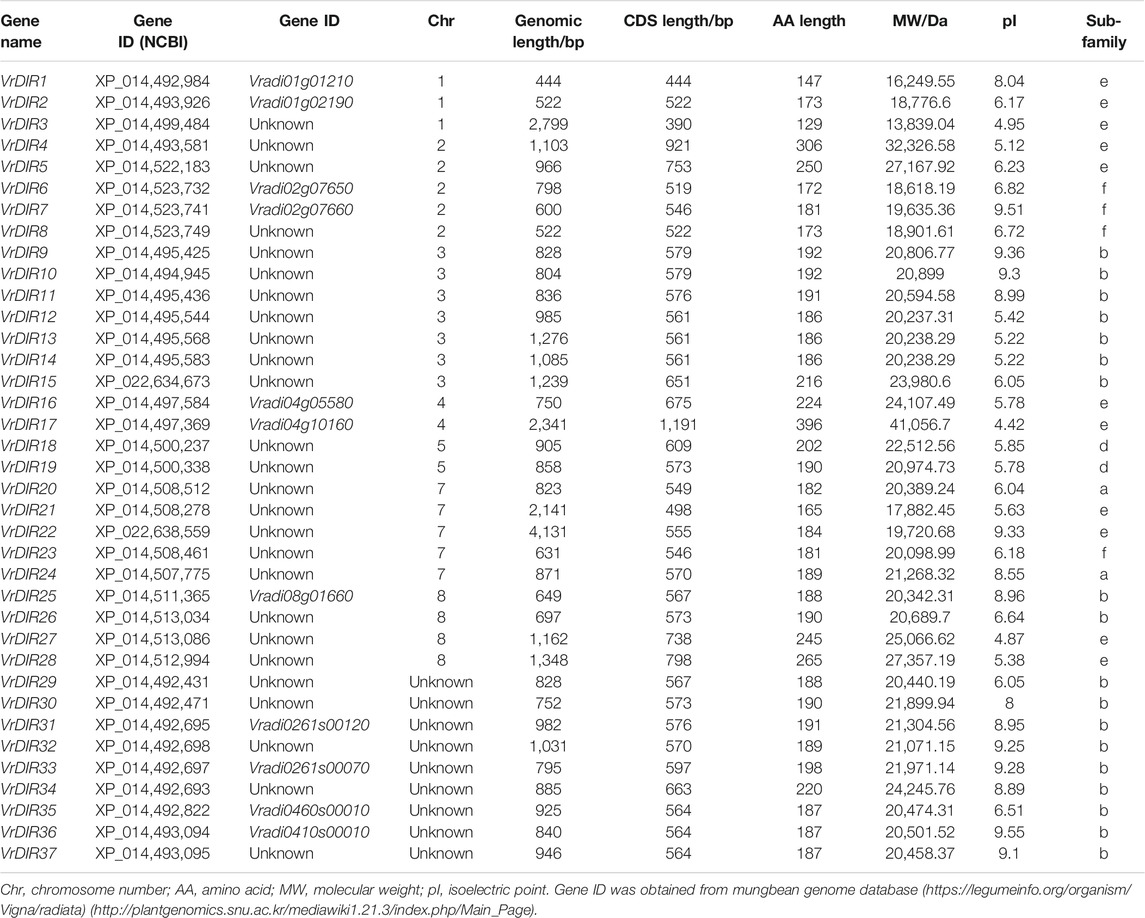
To search for mungbean VrDIR genes, we used the amino acid sequences of 25 Arabidopsis and 54 soybean DIRs as blast queries against the NCBI database. All of the output genes were analyzed using HMMER to confirm the conserved DIR domains (Simon et al., 2018). A total of 37 VrDIR genes were found in mungbean using genome-wide identification (Table 1). The genomic length of VrDIRs ranged from 444 (XP_014,492,984) to 4,131 bp (XP_022,638,559), and twenty-six VrDIR genes had a genomic length less than 1,000 bp. The CDS length ranged from 390 (XP_014,499,484) to 1,191 bp (XP_014,497,369), and the amino acid number varied from 129 to 396. Moreover, 28 out of the 37 VrDIR proteins had less than 200 amino acids, indicating that most VrDIRs are small proteins. The molecular weight of VrDIR proteins apparently ranged from 13,839.04 (XP_014,499,484) to 41,056.7 Da (XP_014,497,369), and the theoretical pI ranged from 4.42 (XP_014,497,369) to 9.55 (XP_014,493,094). In addition, 15 VrDIR proteins were predicted to be alkaline proteins (pI > 7.0), and 22 were predicted to be acidic proteins (pI < 7.0) (Table 1).
The distribution of VrDIRs was uneven among the 11 mungbean chromosomes. Seven of the 11 chromosomes contained VrDIR genes, with the exception of chromosomes 6, 9, 10, and 11 (Table 1, Supplementary Figure S1). VrDIR genes were renamed from VrDIR1 to VrDIR28 based on their chromosome locations, and the other nine VrDIR genes were randomly designated from VrDIR29 to VrDIR37. Among these chromosomes, chromosome three contained the most VrDIR genes, with seven members, followed by chromosomes 2 and 7, with five VrDIRs on each. Several VrDIR genes were located close to each other on the same chromosomes, such as VrDIR4 and VrDIR5.
Phylogenetic Relationships Among VrDIR Genes
We constructed a phylogenetic tree of DIR genes from mungbean, soybean, common bean, cowpea, Arabidopsis, and other species examined by Corbin et al. to investigate the evolutionary relationships between VrDIR genes and DIRs in other species (Corbin et al., 2018). VrDIR genes were classified into five subclades based on their phylogenetic relationships (Figure 1). DIR-a, DIR-b, DIR-d, DIR-e, and DIR-f contained 2 (VrDIR20, VrDIR24), 18 (VrDIR9, VrDIR10, VrDIR11, VrDIR12, VrDIR13, VrDIR14, VrDIR15, VrDIR25, VrDIR26, VrDIR29, VrDIR30, VrDIR31, VrDIR32, VrDIR33, VrDIR34, VrDIR35, VrDIR36, VrDIR37), 2 (VrDIR18, VrDIR19), 11 (VrDIR1, VrDIR2, VrDIR3, VrDIR4, VrDIR5, VrDIR16, VrDIR17, VrDIR21, VrDIR22, VrDIR27, VrDIR28), and 4 (VrDIR6, VrDIR7, VrDIR8, VrDIR23) members, respectively (Figure 1). The homologous genes might have similar functions in different species, and we obtained some information for VrDIRs from the well-studied DIRs in other species based on their evolutionary relationships. VrDIR30 and VrDIR31 were closely related to AtDIR19, the expression level of which changed in response to heat stress (Paniagua et al., 2017), indicating that these two DIR genes might be involved in heat stress responses in mungbean. Moreover, AtDIR9, which is involved in the salt stress response, was classified into the same subgroup as VrDIR17, suggesting that VrDIR17 is potentially involved in the salt stress response (Figure 1). We also constructed a phylogenetic tree using only VrDIR genes and found that VrDIR genes in each group were clustered together in the phylogenetic tree (Figure 2).
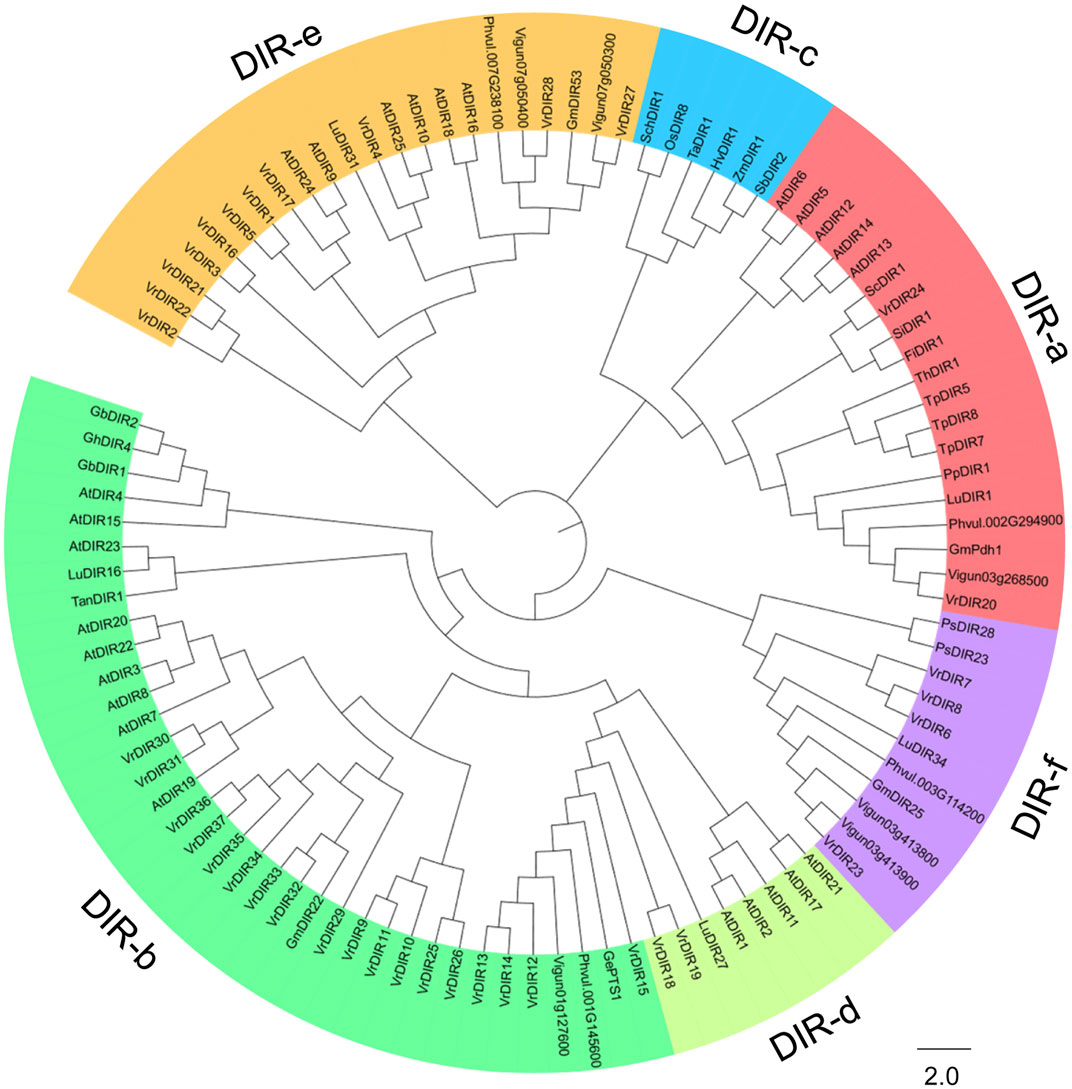
FIGURE 1. Analysis of the evolutionary relationships of DIR proteins from mungbean and other species. The amino acid sequences of DIR proteins from mungbean, Arabidopsis, and other species reported by Corbin et al. (2018) were used to conduct the phylogenetic analysis in MEGA 7.0 using the neighbor-joining method. DIR proteins were classified into six groups based on their phylogenetic relationships, DIR-a to DIR-f, which are indicated by different colors in the phylogenetic tree.
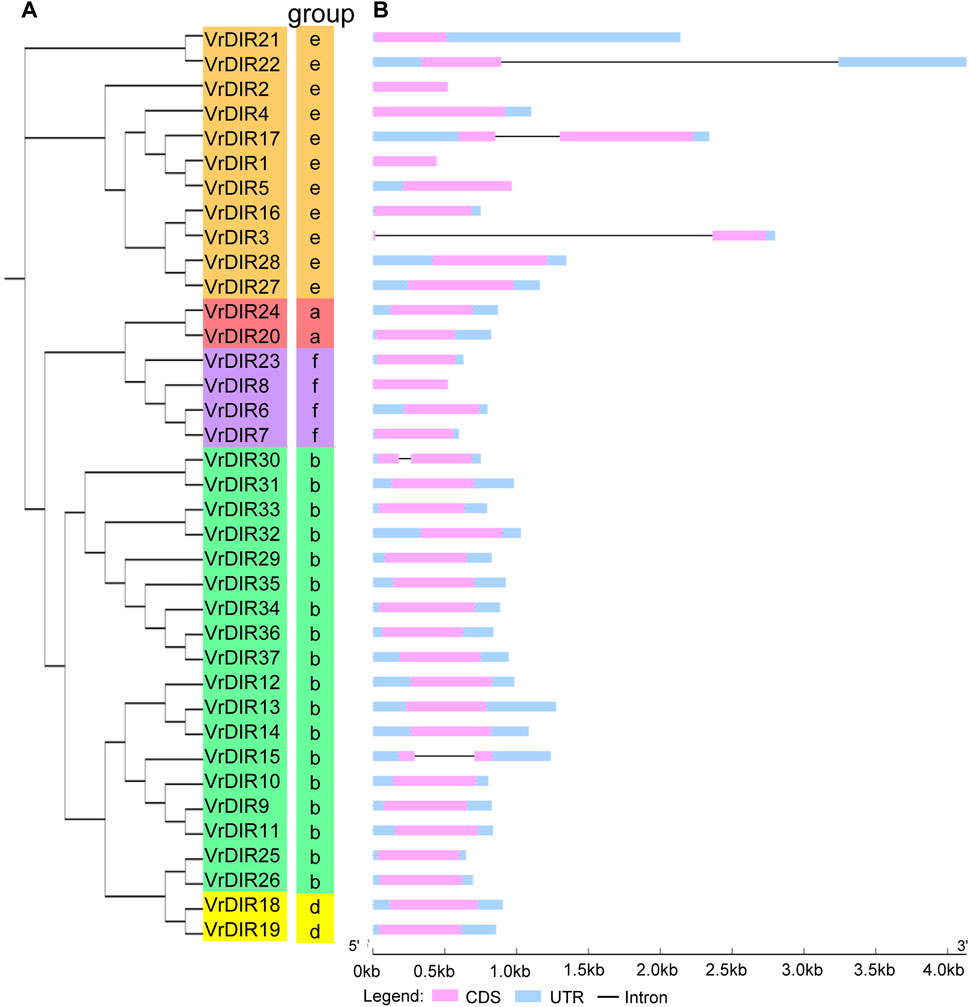
FIGURE 2. Phylogenetic and gene structure analyses of VrDIR genes. (A) Phylogenetic analysis of VrDIR proteins. The phylogenetic tree was constructed in MEGA 7.0 using the neighbor-joining method. (B) Exon-intron organization of VrDIR genes. The light blue, pink, and black boxes indicate UTRs, exons, and introns, respectively. The genomic lengths of VrDIR genes are indicated.
Gene Structure and Conserved Motif Analyses of VrDIR Genes
To investigate the exon-intron organization of VrDIR genes, we used the Gene Structure Display Server program to analyze VrDIR genomic and coding sequences. Thirty-two out of the 37 VrDIRs had a classical DIR gene structure, one exon without introns; the exceptions were VrDIR3, VrDIR15, VrDIR17, VrDIR22, and VrDIR30, which each had one intron (Figure 2). Moreover, most of the VrDIR genes contained UTRs, with the exception of VrDIR1, VrDIR2, and VrDIR8, which had only one exon (Figure 2). The conserved DIR domain occupied the majority of the protein in most VrDIRs, with the exception of DIR-e subfamily members VrDIR4, VrDIR17, VrDIR27, and VrDIR28 (Figure 3). Next, we analyzed the conserved motifs of VrDIR proteins using MEME tools; a total of 19 distinct motifs were detected in all 37 VrDIR proteins (Figure 3, Supplementary Figure S2). Motif one was present in all VrDIR proteins, which might indicate the conserved DIR domain (Figure 3). The differences in the motifs reflect the diversity of VrDIR proteins. For example, motifs three and nine occurred in the DIR-b and DIR-d groups, and motif 19 was only present in two DIR-f genes, which indicates that VrDIR proteins are functionally diverse (Figure 3).
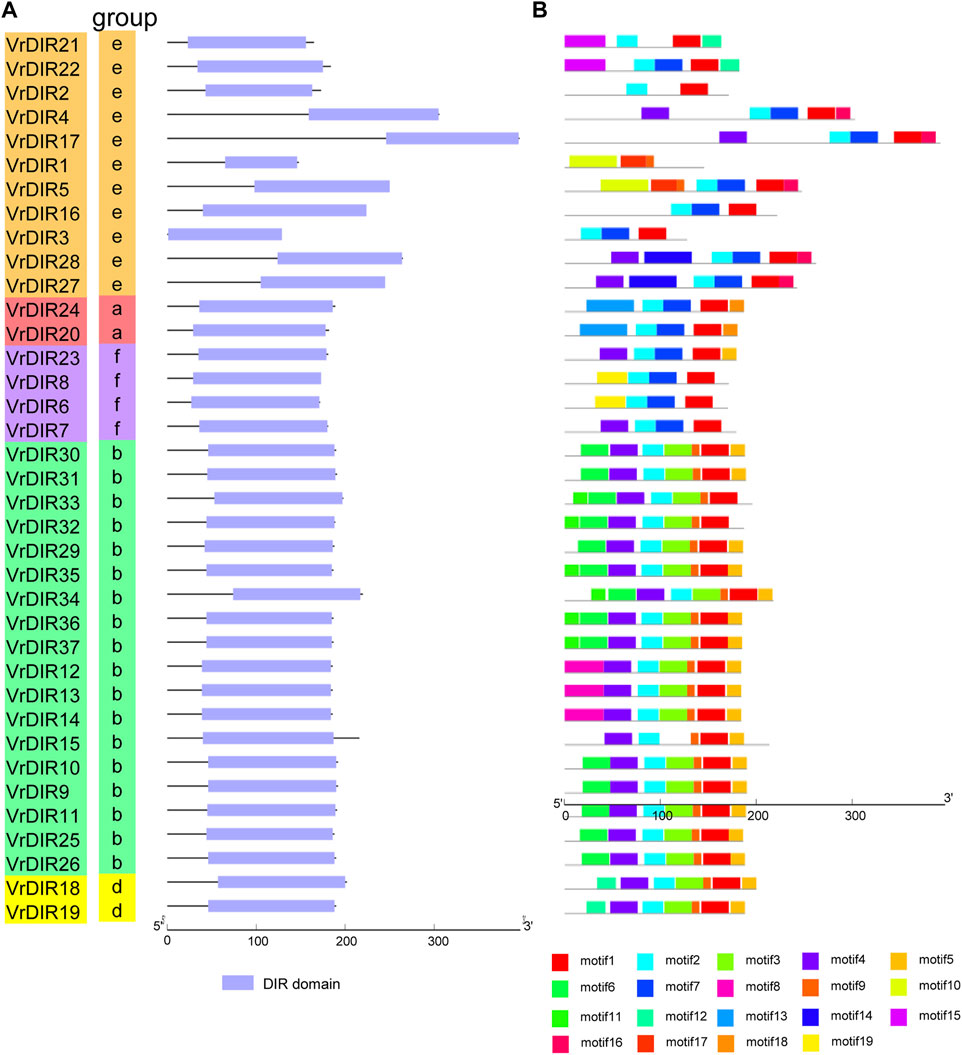
FIGURE 3. Conserved domain and motif analyses in VrDIR proteins. (A) The positions and lengths of the conserved DIR domains in VrDIR proteins. The light blue boxes indicate the conserved DIR domains. (B) The conserved motifs in VrDIR proteins. The 19 motifs are indicated using different colored boxes. The length of each DIR protein is indicated.
Duplication Analysis of VrDIRs
Duplicated gene pairs are produced during whole-genome duplication in plants (Li et al., 2019; Ma et al., 2021). Duplication events were analyzed among VrDIR members. A total of 13 duplicated gene pairs were observed among 37 VrDIR genes, which is consistent with the high similarity observed in mungbean DIR genes. Eleven duplicated gene pairs are shown in Figure 4, including VrDIR1/VrDIR5, VrDIR3/VrDIR16, VrDIR9/VrDIR10, VrDIR9/VrDIR11, VrDIR10/VrDIR11, VrDIR12/VrDIR13, VrDIR12/VrDIR14, VrDIR13/VrDIR14, VrDIR18/VrDIR19, VrDIR25/VrDIR26, and VrDIR27/VrDIR28; the duplicated gene pairs VrDIR36/VrDIR37 and VrDIR32/VrDIR33 were discarded because of a lack of chromosome information. The duplicated genes were clustered into a clade in the phylogenetic tree (Figure 2). Chromosome three contained the most duplicated genes, including VrDIR9, VrDIR10, VrDIR11, VrDIR12, VrDIR13, and VrDIR14; chromosomes 1, 2, 4, 5, and eight contained 2, 1, 1, 2, and four duplicated genes, respectively (Figure 4). VrDIR10/VrDIR11, VrDIR12/VrDIR13, VrDIR13/VrDIR14, VrDIR18/VrDIR19, and VrDIR27/VrDIR28 were derived from tandem duplication events, whereas VrDIR1/VrDIR5 and VrDIR3/VrDIR16 represented interchromosomal duplicated gene pairs (Figure 4).
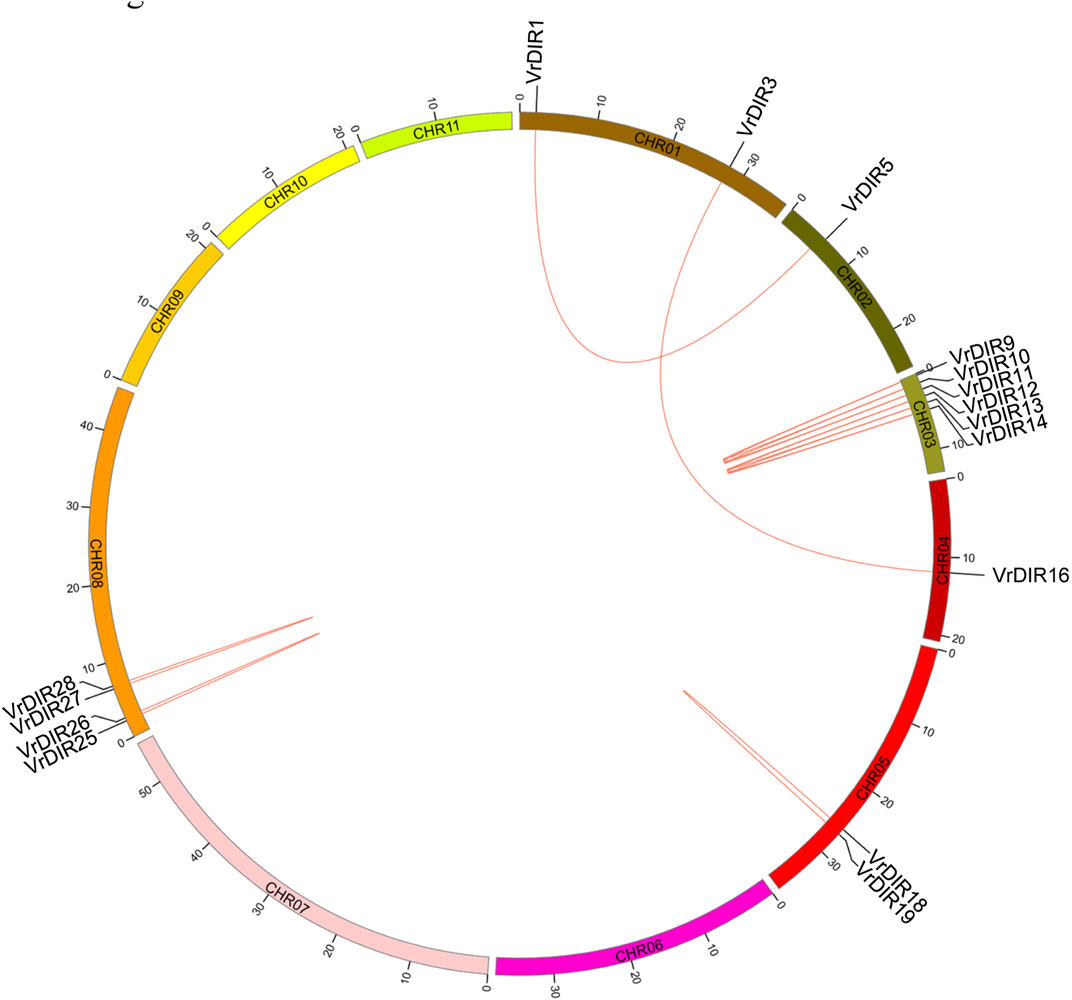
FIGURE 4. Duplication analysis of VrDIR genes. The positions of VrDIR genes in each chromosome are presented, and the duplicated gene pairs are connected using red lines. The length of each chromosome is indicated, and CHR indicates chromosome.
Cis-Acting Element Analysis of VrDIR Promoters
Cis-acting elements mediate various responses by regulating promoter activities; we predicted cis-acting elements in VrDIR promoter regions using sequences 2 kb upstream of each initiation codon. A total of 92 cis-acting elements were found in all 37 VrDIR promoter regions, and 57 cis-acting elements had predicted functions (Supplementary Table S2), which were classified into six different groups according to their potential functions (Figure 5) (Ma et al., 2021). Light-responsive elements were the most abundant in 34 VrDIR genes; in the promoter regions of VrDIR4, VrDIR6, and VrDIR14, hormone-responsive elements were the most abundant, indicating that these three VrDIRs are involved in hormone responses (Figure 5). Moreover, the DIR-b, DIR-d, and DIR-e subfamilies contained all six groups of cis-acting elements, and DIR-a and DIR-f subfamilies contained only five groups of cis-acting elements, with the exception of ‘site-binding related elements’ (Figure 5), which suggests that the expression of these VrDIR genes in response to stress varies. In addition, the cis-acting elements in duplication events differed in some gene pairs, indicating that they might have different functions (Figure 5). For example, the duplicated genes VrDIR13 and VrDIR14 have different numbers of hormone-responsive elements, light-responsive elements, promoter-related elements, and site-binding-related elements, suggesting that these two genes might show different responses to stress (Figure 5, Supplementary Table S2).
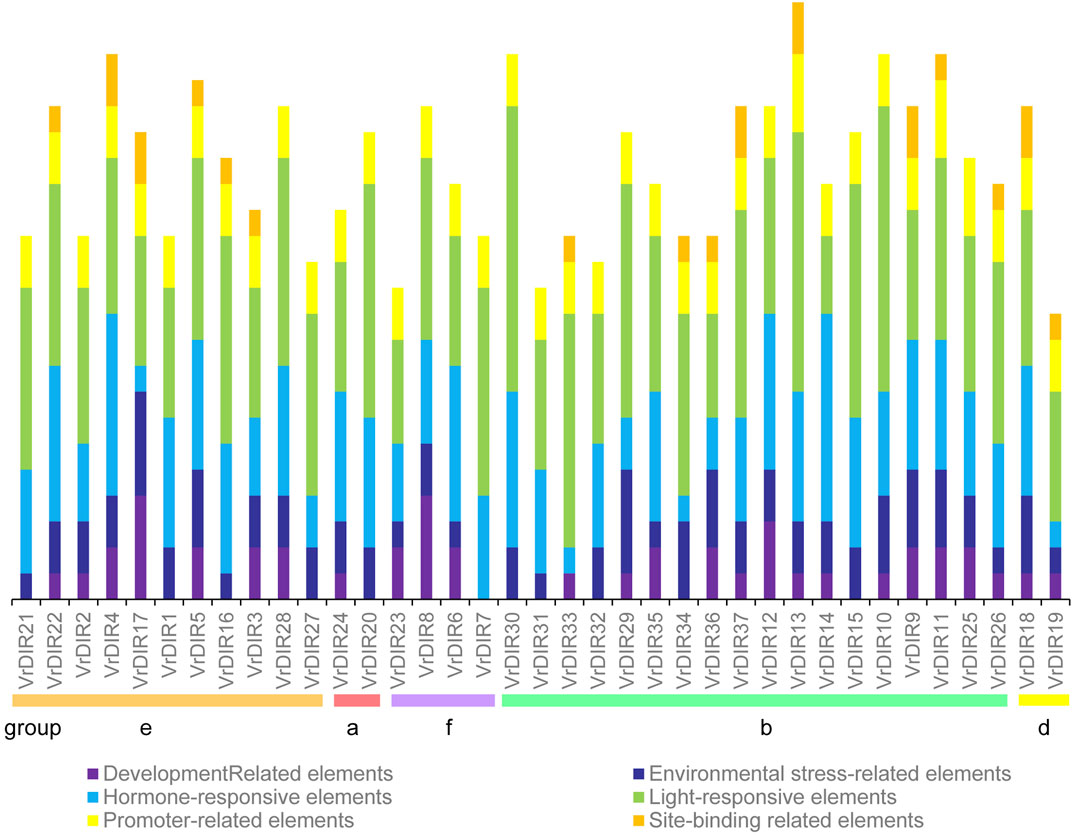
FIGURE 5. Cis-acting element analysis in VrDIR promoter regions. The cis-acting elements were classified into six groups based on their predicted functions as described by Jin et al. (2019). Different cis-acting elements were presented using different colored boxes, and the Y-axis indicates the number of cis-acting elements in VrDIR promoters.
Analysis of VrDIR Expression in Different Tissues
We analyzed the expression levels of VrDIRs in eight different tissues: flowers, pods, leaves, seeds, nodule roots, stems, roots, and shoot apices (Figure 6). The expression levels of VrDIRs varied among tissues. For example, the expression of VrDIR1 and VrDIR3 was low in all tested tissues, indicating that these two genes might have weak functions in these tissues. In contrast, the expression of VrDIR24 and VrDIR31 was high in all tissues (Figure 6), indicating that VrDIR24 and VrDIR31 have important functions in these tissues. In addition, duplicated genes might retain some common functions as well as evolve new functions. Thus, several duplicated genes showed similar expression levels in some tissues, and expression patterns differed in other tissues (Figure 6). For example, VrDIR9 and VrDIR11 showed similar expression patterns in flowers, leaves, stems, and shoot apices, but different expression levels in pods, seeds, nodule roots, and roots, indicating that VrDIR9 and VrDIR11 might have similar functions in some tissues but different roles in other tissues.
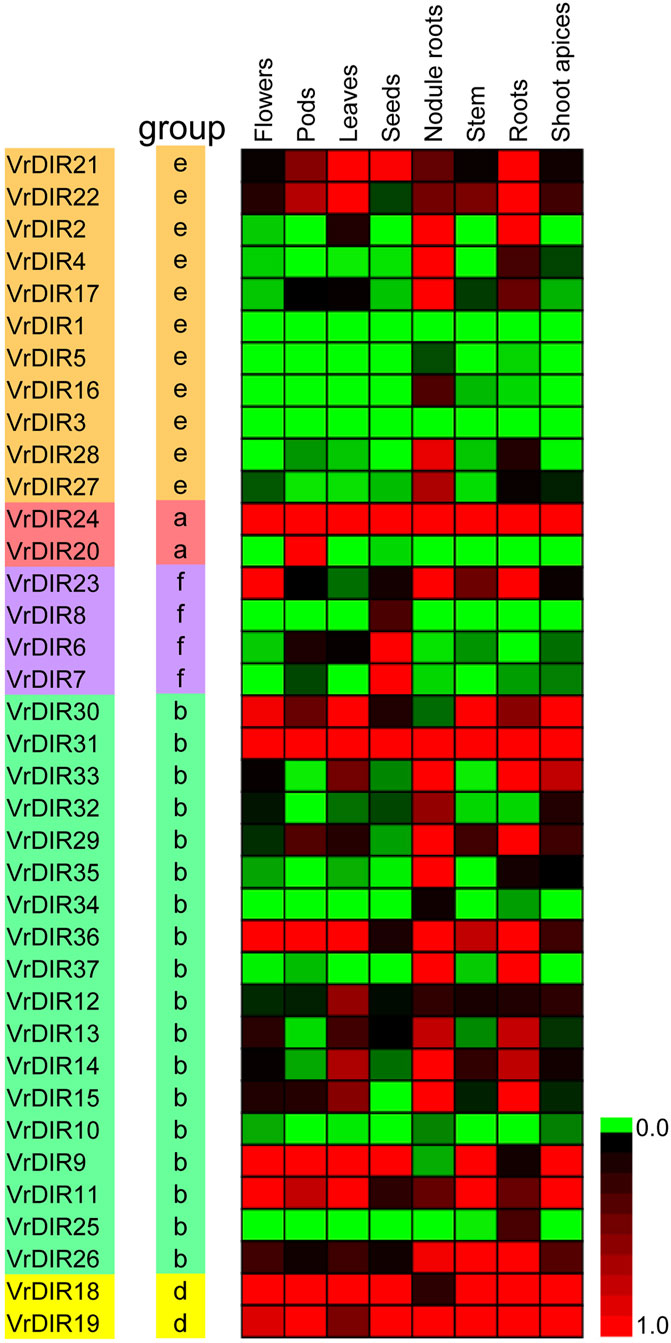
FIGURE 6. Relative expression levels of VrDIR genes in different tissues. Eight tissues, including flowers, pods, leaves, seeds, nodule roots, stems, roots, and shoot apices, were used for analysis. The expression level of VrDIR37 in nodule roots was set as one, and the others were adjusted accordingly. The gene expression results were visualized using a heatmap generated with Multiple Experiment Viewer 4.9.0 (Saeed et al., 2003). The expression levels from 0 to one are indicated by different colors.
Expression of VrDIR Genes in Response to Salt and Drought Stress
Next, we analyzed the expression of VrDIR genes in mungbean shoots and roots under salt and drought stress. The fresh weights of mungbean plants were significantly lower under salt or drought treatment than that under normal conditions which indicated that the growth of mungbean plants was inhibited after salt or drought stress treatment (Supplementary Figure S3). VrDIR genes varied in their responses under different stress conditions (Figures 7, 8). Most of the VrDIR genes showed altered expression levels under both drought and salt stress conditions in shoots and roots, with the exception of VrDIR4, whose expression was not affected by salt or drought stress (Figures 7 and 8). For example, the expression level of VrDIR34 decreased in both shoots and roots under drought stress and increased in roots and decreased in shoots under salt stress (Figures 7 and 8). Some VrDIR genes only exhibited responses to either drought or salt stress. For example, the expression of VrDIR19 decreased in both shoots and roots under drought conditions; however, the expression of VrDIR19 was not affected by salt stress (Figures 7 and 8). Moreover, the expression of VrDIR25 in different tissues did not vary under drought stress but increased and decreased in shoots and roots under salt stress, respectively (Figures 7 and 8), indicating that this VrDIR gene might have different functions in response to drought and salt stress.
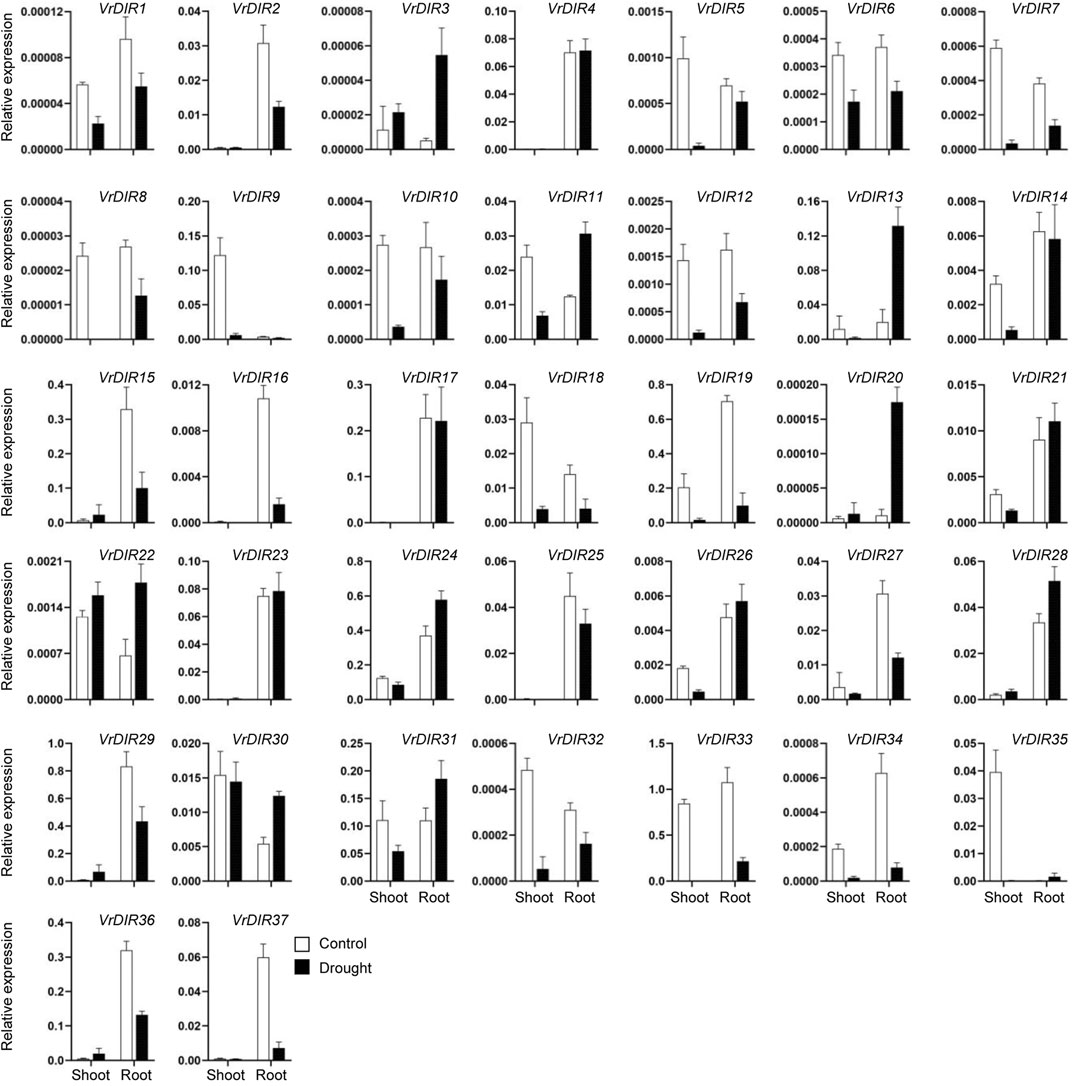
FIGURE 7. Expression levels of VrDIR genes in response to drought stress. The expression of VrDIR genes in shoots and roots grown under normal and drought conditions was analyzed using qRT-PCR. Each sample was analyzed using three biological replicates and normalized to an Actin-expressing gene in mungbean.
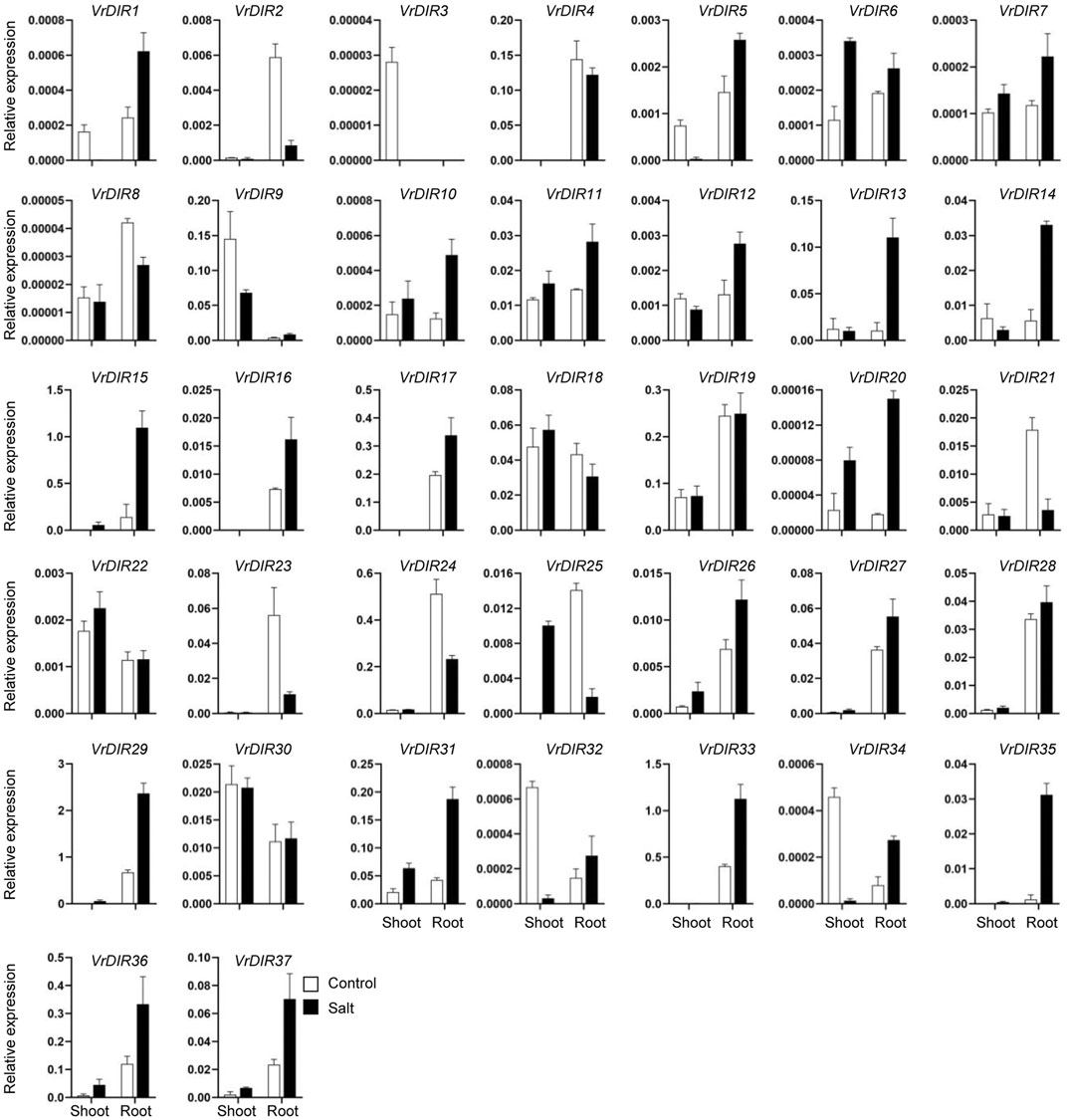
FIGURE 8. Expression levels of VrDIR genes in response to salt stress. The expression of VrDIR genes in shoots and roots grown under normal and salt conditions was analyzed using qRT-PCR. Each sample was analyzed using three biological replicates and normalized to an Actin-expressing gene in mungbean.
Discussion
In previous decades, several studies have characterized DIR genes, and this work has greatly increased our understanding of the responses of plants to stress in many species. Many DIR gene expression studies correlated their involvement in biotic and abiotic stress. In this study, we characterized 37 DIR genes from mungbean and analyzed their expression patterns in response to drought or salt stress.
The typical structure of a DIR gene contains one exon and no introns (Corbin et al., 2018). In mungbean, 32 VrDIR genes contained this typical gene structure, and five VrDIR genes, including VrDIR3, VrDIR15, VrDIR17, VrDIR22, and VrDIR30 (all of which belong to groups DIR-b and DIR-e), contained introns. In contrast, all DIR-a, DIR-d, and DIR-f members contained the typical gene structure (Figure 2), indicating that the gene structure of VrDIR genes is conserved in these three subfamilies. In soybean, eight DIR genes contain introns, seven of which belong to groups DIR-b and DIR-e (Ma et al., 2021). The conserved DIR domain occupies the majority of typical DIR proteins (Corbin et al., 2018), and the DIR domain in several mungbean DIRs, including VrDIR4, VrDIR17, VrDIR27, and VrDIR28, made up less than half of the protein (Figure 3), all of which belonged to group DIR-e. These results indicated that the gene structure differed in DIR-b and DIR-e members; other group members show high conservation in their sequences. In addition, some motifs, such as motif 1, were observed in all VrDIR proteins, indicating the conservation of VrDIR genes. However, some motifs are only present in specific subfamilies, such as motif three and motif 9, which reflects the high functional diversity of VrDIR proteins in different subfamilies (Figure 3).
The legume plants were considered to be evolved from the same origin million years ago, and the genomes of different species have been modified in various ways over evolutionary time (Schmutz et al., 2010; Young et al., 2010; Kang et al., 2014). Mungbean and Medicago underwent one round of whole-genome duplication. Mungbean contained 37 DIRs, and Medicago plants contained 45 DIRs (Table 1) (Young et al., 2010; Song and Peng, 2019), indicating that some DIR genes might have been lost in mungbean during their evolutionary history. Soybean has undergone two rounds of whole-genome duplication and had 54 GmDIR genes, which is less than twice the number in mungbean or Medicago (Schmutz et al., 2010; Ma et al., 2021). These results suggest that the numbers of DIR genes have changed extensively during the evolutionary history of legumes. Moreover, DIRs were grouped into seven groups (DIR-a to DIR-g) in flax (Linum usitatissimum L.); they were only grouped into five groups in mungbean and did not have a DIR-c member, which only occurs in monocot species, nor a DIR-g member, which only occurs in flax (Corbin et al., 2018), indicating that these two groups were lost during mungbean evolution. Mungbean contained two DIR-d group members, VrDIR18 and VrDIR19 (Figure 1). However, soybean has no DIR-d members, indicating that the types of DIR genes changed during the evolution of legumes (Ma et al., 2021). In addition, the VrDIR genes in some subfamilies underwent several duplication events during evolution. For example, all of the duplicated gene pairs were present in the DIR-b, DIR-d, and DIR-e subfamilies, whereas no duplication events were observed in the DIR-a and DIR-f subfamilies (Figure 4), which reflects the evolutionary diversity of these subfamilies. Chromosome three contained the most duplicated genes, and most of the VrDIR genes located on chromosome three had duplicated genes, with the exception of VrDIR15 (Figure 4), indicating that chromosome three might contain the original genes of many duplicated VrDIR genes (Li et al., 2019). VrDIRs in the same subfamily showed distinct expression patterns in different tissues, which suggests that these VrDIRs are functionally diverse. For example, the DIR-a subfamily members VrDIR20 and VrDIR24 are thought to be involved in pinoresinol formation (Ralph et al., 2006; Corbin et al., 2018). VrDIR24 showed high expression levels in all tested tissues, whereas VrDIR20 was only highly expressed in pods. The different VrDIR subfamilies contained different types of cis-acting elements. For example, DIR-b, DIR-d, and DIR-e subfamilies contained all six groups of cis-acting elements, whereas the DIR-a and DIR-f subfamilies contained only five groups, which might be responsible for the different expression patterns of VrDIR genes in different tissues (Figures 5, 6). Although VrDIR1 and VrDIR3 promoters contained many cis-acting elements, they showed extremely low expression levels in all tested tissues (Figure 6). The expression levels of genes are affected by many factors aside from cis-acting elements, such as temporal and spatial factors. VrDIR1 and VrDIR3 might be expressed at other time points in response to the environment. Duplicated genes might have the same origin and similar functions. Thus, VrDIR9 and VrDIR11 might retain similar functions in flowers, leaves, stems, and shoot apices and evolve new functions in pods, seeds, nodule roots, and roots based on their expression patterns (Figure 6). Moreover, mungbean had 13 VrDIR duplicated gene pairs, nearly half of that in soybean, which contained 24 GmDIR duplicated gene pairs (Ma et al., 2021), indicating that the evolution of duplicated gene pairs might be conserved in these two legumes. In Arabidopsis, 16 of the 25 AtDIR genes are highly expressed in the roots (Paniagua et al., 2017); in contrast, only a portion of VrDIR genes was highly expressed in mungbean roots (Figure 6), indicating that the functions of many DIR genes in Arabidopsis and mungbean have diverged. Moreover, homologous genes might have similar functions in different species. The expression of AtDIR9 changed under salt stress, and the expression of its close homologs VrDIR1, VrDIR5, and VrDIR17 increased under salt stress in roots; the expression of VrDIR1 and VrDIR5 in the shoots decreased. The expression of AtDIR5 is altered under salt and drought stress (Paniagua et al., 2017), and the expression of its close homologs VrDIR20 and VrDIR24 was altered in the shoots and roots (Figures 7, 8). The expression of AtDIR5 is altered in response to methyl jasmonate, wounding, and oxidative stress (Paniagua et al., 2017), suggesting that VrDIR20 and VrDIR24 might be involved in the regulation of these various types of stress in mungbean.
In sum, we identified and characterized 37 VrDIR genes in mungbean and characterized the phylogenetic relationships, exon-intron organization, conserved motifs, duplication events, cis-acting elements, and expression patterns in different tissues. We also analyzed the expression patterns of VrDIR genes in response to drought and salt stress. Our study provides basic information for future studies of VrDIR genes and their role in stress responses.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
HoZ conceived and designed the research. WX, TL and HuZ conducted the experiments and analyzed the data. HoZ wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was funded by the National Science Foundation of Shandong Province (grant ZR2020QC120), the Opening Foundation of Shandong Dry-land Farming Technology Key Laboratory (grant 2219006), and the Qingdao Agricultural University Scientific Research Foundation (grant 6631119010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Suk-Ha Lee at Seoul National University, Seoul, Korea, for supplying mungbean VC 1973A seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.658148/full#supplementary-material
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Corbin, C., Drouet, S., Markulin, L., Auguin, D., Lainé, É., Davin, L. B., et al. (2018). A Genome-wide Analysis of the Flax (Linum usitatissimum L.) Dirigent Protein Family: from Gene Identification and Evolution to Differential Regulation. Plant Mol. Biol. 97 (1-2), 73–101. doi:10.1007/s11103-018-0725-x
Dalisay, D. S., Kim, K. W., Lee, C., Yang, H., Rübel, O., Bowen, B. P., et al. (2015). Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 78 (6), 1231–1242. doi:10.1021/acs.jnatprod.5b00023
Davin, L. B., and Lewis, N. G. (2000). Dirigent Proteins and Dirigent Sites Explain the Mystery of Specificity of Radical Precursor Coupling in Lignan and Lignin Biosynthesis. Plant Physiol. 123 (2), 453–462. doi:10.1104/pp.123.2.453
Davin, L. B., Wang, H.-B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S., et al. (1997). Stereoselective Bimolecular Phenoxy Radical Coupling by an Auxiliary (Dirigent) Protein without an Active center. Science 275 (5298), 362–367. doi:10.1126/science.275.5298.362
Effenberger, I., Zhang, B., Li, L., Wang, Q., Liu, Y., Klaiber, I., et al. (2015). Dirigent Proteins from Cotton ( Gossypium sp.) for the Atropselective Synthesis of Gossypol. Angew. Chem. Int. Ed. 54 (49), 14660–14663. doi:10.1002/anie.201507543
El-Adawy, T. A., Rahma, E. H., El-Bedawey, A. A., and El-Beltagy, A. E. (2003). Nutritional Potential and Functional Properties of Germinated Mung Bean, Pea and Lentil Seeds. Plant Foods Hum. Nutr. 58 (3), 1–13. doi:10.1023/B:QUAL.0000040339.48521.75
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). Using OrthoMCL to Assign Proteins to OrthoMCL-DB Groups or to Cluster Proteomes into New Ortholog Groups. Curr. Protoc. Bioinformatics, 1–19. doi:10.1002/0471250953.bi0612s35
Fuller, D. Q. (2007). Contrasting Patterns in Crop Domestication and Domestication Rates: Recent Archaeobotanical Insights from the Old World. Ann. Bot. 100 (5), 903–924. doi:10.1093/aob/mcm048
Graham, P. H., and Vance, C. P. (2003). Legumes: Importance and Constraints to Greater Use. Plant Physiol. 131 (3), 872–877. doi:10.1104/pp.017004
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 Zinc finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 11, 115. doi:10.3389/fpls.2020.00115
Hosmani, P. S., Kamiya, T., Danku, J., Naseer, S., Geldner, N., Guerinot, M. L., et al. (2013). Dirigent Domain-Containing Protein Is Part of the Machinery Required for Formation of the Lignin-Based Casparian Strip in the Root. Proc. Natl. Acad. Sci. 110 (35), 14498–14503. doi:10.1073/pnas.1308412110
Hu, B., Deng, F., Chen, G., Chen, X., Gao, W., Long, L., et al. (2020). Evolution of Abscisic Acid Signaling for Stress Responses to Toxic Metals and Metalloids. Front. Plant Sci. 11, 909. doi:10.3389/fpls.2020.00909
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an Upgraded Gene Feature Visualization Server. Bioinformatics 31 (8), 1296–1297. doi:10.1093/bioinformatics/btu817
Jin, H., Tang, X., Xing, M., Zhu, H., Sui, J., Cai, C., et al. (2019). Molecular and Transcriptional Characterization of Phosphatidyl Ethanolamine-Binding Proteins in Wild Peanuts Arachis Duranensis and Arachis Ipaensis. BMC Plant Biol. 19 (1), 484. doi:10.1186/s12870-019-2113-3
Jin, H., Xing, M., Cai, C., and Li, S. (2020). B-box Proteins in Arachis Duranensis: Genome-wide Characterization and Expression Profiles Analysis. Agronomy 10, 23. doi:10.3390/agronomy10010023
Jin-long, G., Li-ping, X., Jing-ping, F., Ya-chun, S., Hua-ying, F., You-xiong, Q., et al. (2012). A Novel Dirigent Protein Gene with Highly Stem-specific Expression from Sugarcane, Response to Drought, Salt and Oxidative Stresses. Plant Cel Rep 31 (10), 1801–1812. doi:10.1007/s00299-012-1293-1
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B.-K., et al. (2014). Genome Sequence of Mungbean and Insights into Evolution within Vigna Species. Nat. Commun. 5, 5443. doi:10.1038/ncomms6443
Keatinge, J. D. H., Easdown, W. J., Yang, R. Y., Chadha, M. L., and Shanmugasundaram, S. (2011). Overcoming Chronic Malnutrition in a Future Warming World: the Key Importance of Mungbean and Vegetable Soybean. Euphytica 180 (1), 129–141. doi:10.1007/s10681-011-0401-6
Khan, A., Li, R.-J., Sun, J.-T., Ma, F., Zhang, H.-X., Jin, J.-H., et al. (2018). Genome-wide Analysis of Dirigent Gene Family in Pepper (Capsicum Annuum L.) and Characterization of CaDIR7 in Biotic and Abiotic Stresses. Sci. Rep. 8 (1), 5500. doi:10.1038/s41598-018-23761-0
Kim, K.-W., Moinuddin, S. G. A., Atwell, K. M., Costa, M. A., Davin, L. B., and Lewis, N. G. (2012). Opposite Stereoselectivities of Dirigent Proteins in Arabidopsis and Schizandra Species. J. Biol. Chem. 287 (41), 33957–33972. doi:10.1074/jbc.M112.387423
Kim, M. K., Jeon, J.-H., Davin, L. B., and Lewis, N. G. (2002). Monolignol Radical-Radical Coupling Networks in Western Red Cedar and Arabidopsis and Their Evolutionary Implications. Phytochemistry 61 (3), 311–322. doi:10.1016/s0031-9422(02)00261-3
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 19 (9), 1639–1645. doi:10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Li, N., Zhao, M., Liu, T., Dong, L., Cheng, Q., Wu, J., et al. (2017). A Novel Soybean Dirigent Gene GmDIR22 Contributes to Promotion of Lignan Biosynthesis and Enhances Resistance to Phytophthora Sojae. Front. Plant Sci. 8, 1185. doi:10.3389/fpls.2017.01185
Li, Q., Chen, J., Xiao, Y., Di, P., Zhang, L., and Chen, W. (2014). The Dirigent Multigene Family in Isatis Indigotica: Gene Discovery and Differential Transcript Abundance. BMC Genomics 15 (1), 388. doi:10.1186/1471-2164-15-388
Li, S., Ding, Y., Zhang, D., Wang, X., Tang, X., Dai, D., et al. (2018). Parallel Domestication with a Broad Mutational Spectrum of Determinate Stem Growth Habit in Leguminous Crops. Plant J. 96 (4), 761–771. doi:10.1111/tpj.14066
Li, S., Wang, R., Jin, H., Ding, Y., and Cai, C. (2019). Molecular Characterization and Expression Profile Analysis of Heat Shock Transcription Factors in Mungbean. Front. Genet. 9, 736. doi:10.3389/fgene.2018.00736
Li, S., Wang, X., Xu, W., Liu, T., Cai, C., Chen, L., et al. (2021). Unidirectional Movement of Small RNAs from Shoots to Roots in Interspecific Heterografts. Nat. Plants 7 (1), 50–59. doi:10.1038/s41477-020-00829-2
Liu, C., Zhang, Q., Zhu, H., Cai, C., and Li, S. (2021). Characterization of Mungbean CONSTANS-LIKE Genes and Functional Analysis of CONSTANS-LIKE 2 in the Regulation of Flowering Time in Arabidopsis. Front. Plant Sci. 12, 608603. doi:10.3389/fpls.2021.608603
Liu, J., Stipanovic, R. D., Bell, A. A., Puckhaber, L. S., Magill, C. W., Sun, W., et al. (2008). Stereoselective Coupling of Hemigossypol to Form (+)-gossypol in Moco Cotton Is Mediated by a Dirigent proteinComprehensive Multiomics Analysis Reveals Key Roles of NACs in Plant Growth and Development and its Environmental Adaption Mechanism by Regulating Metabolite Pathways. Phytochemistry 69 (18), 3038–3042. doi:10.1016/j.ygeno.2020.08.038
Ma, X., Xu, W., Liu, T., Chen, R., Zhu, H., Zhang, H., et al. (2021). Functional Characterization of Soybean (Glycine max) DIRIGENT Genes Reveals an Important Role of GmDIR27 in the Regulation of Pod Dehiscence. Genomics 113, 979–990. doi:10.1016/j.ygeno.2020.10.033
Meng, Q., Moinuddin, S. G. A., Kim, S.-J., Bedgar, D. L., Costa, M. A., Thomas, D. G., et al. (2020). Pterocarpan Synthase (PTS) Structures Suggest a Common Quinone Methide-Stabilizing Function in Dirigent Proteins and Proteins with Dirigent-like Domains. J. Biol. Chem. 295 (33), 11584–11601. doi:10.1074/jbc.RA120.012444
Oliver, T., Schmidt, B., Nathan, D., Clemens, R., and Maskell, D. (2005). Using Reconfigurable Hardware to Accelerate Multiple Sequence Alignment with clustalW. Bioinformatics 21 (16), 3431–3432. doi:10.1093/bioinformatics/bti508
Paniagua, C., Bilkova, A., Jackson, P., Dabravolski, S., Riber, W., Didi, V., et al. (2017). Dirigent Proteins in Plants: Modulating Cell wall Metabolism during Abiotic and Biotic Stress Exposure. J. Exp. Bot. 68 (13), 3287–3301. doi:10.1093/jxb/erx141
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER Web Server: 2018 Update. Nucleic Acids Res. 46 (W1), W200–W204. doi:10.1093/nar/gky448
Ralph, S. G., Jancsik, S., and Bohlmann, J. (2007). Dirigent Proteins in conifer Defense II: Extended Gene Discovery, Phylogeny, and Constitutive and Stress-Induced Gene Expression in spruce (Picea spp.). Phytochemistry 68 (14), 1975–1991. doi:10.1016/j.phytochem.2007.04.042
Ralph, S., Park, J.-Y., Bohlmann, J., and Mansfield, S. D. (2006). Dirigent Proteins in conifer Defense: Gene Discovery, Phylogeny, and Differential Wound- and Insect-Induced Expression of a Family of DIR and DIR-like Genes in spruce (Picea spp.). Plant Mol. Biol. 60 (1), 21–40. doi:10.1007/s11103-005-2226-y
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a Free, Open-Source System for Microarray Data Management and Analysis. Biotechniques 34 (2), 374–378. doi:10.2144/03342mt01
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome Sequence of the Palaeopolyploid Soybean. Nature 463 (7278), 178–183. doi:10.1038/nature08670
Seneviratne, H. K., Dalisay, D. S., Kim, K.-W., Moinuddin, S. G. A., Yang, H., Hartshorn, C. M., et al. (2015). Non-host Disease Resistance Response in Pea (Pisum Sativum) Pods: Biochemical Function of DRR206 and Phytoalexin Pathway Localization. Phytochemistry 113, 140–148. doi:10.1016/j.phytochem.2014.10.013
Shi, R., Xu, W., Liu, T., Cai, C., and Li, S. (2021). VrLELP Controls Flowering Time under Short-Day Conditions in Arabidopsis. J. Plant Res. 134 (1), 141–149. doi:10.1007/s10265-020-01235-7
Song, M., and Peng, X. (2019). Genome-wide Identification and Characterization of DIR Genes in Medicago Truncatula. Biochem. Genet. 57 (4), 487–506. doi:10.1007/s10528-019-09903-7
Thamil Arasan, S. K., Park, J.-I., Ahmed, N. U., Jung, H.-J., Hur, Y., Kang, K.-K., et al. (2013). Characterization and Expression Analysis of Dirigent Family Genes Related to Stresses in Brassica. Plant Physiol. Biochem. 67, 144–153. doi:10.1016/j.plaphy.2013.02.030
Tomooka, N., Vaughan, D. A., Moss, H., and Maxted, N. (2002). The Asian Vigna: Genus Vigna Subgenus Ceratotropis Genetic Resources. Dordrecht: Kluwer Academic Publishers, 270.
Uchida, K., Akashi, T., and Aoki, T. (2017). The Missing Link in Leguminous Pterocarpan Biosynthesis Is a Dirigent Domain-Containing Protein with Isoflavanol Dehydratase Activity. Plant Cel Physiol 58 (2), 398–408. doi:10.1093/pcp/pcw213
Vaahtera, L., Schulz, J., and Hamann, T. (2019). Cell wall Integrity Maintenance during Plant Development and Interaction with the Environment. Nat. Plants 5, 924–932. doi:10.1038/s41477-019-0502-0
Voorrips, R. E. (2002). MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 93 (1), 77–78. doi:10.1093/jhered/93.1.77
Wu, R., Wang, L., Yan, Z., Qi, D., and Deng, D. (2009). Cloning and Expression Analysis of a Dirigent Protein Gene from the Resurrection Plant Boea Hygrometrica. Prog. Nat. Sci. 19, 347–352. doi:10.1016/j.pnsc.2008.07.010
Young, N. D., Debellé, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2010). The Medicago Genome Provides Insight into the Evolution of Rhizobial Symbioses. Nature 480 (7378), 520–524. doi:10.1038/nature10625
Yu, Y., Qian, Y., Jiang, M., Xu, J., Yang, J., Zhang, T., et al. (2020). Regulation Mechanisms of Plant Basic Leucine Zippers to Various Abiotic Stresses. Front. Plant Sci. 11, 1258. doi:10.3389/fpls.2020.01258
Zhang, H., Gao, X., Zhi, Y., Li, X., Zhang, Q., Niu, J., et al. (2019). A Non‐tandem CCCH ‐type Zinc‐finger Protein, IbC3H18, Functions as a Nuclear Transcriptional Activator and Enhances Abiotic Stress Tolerance in Sweet Potato. New Phytol. 223, 1918–1936. doi:10.1111/nph.15925
Zhu, D., Hou, L., Xiao, P., Guo, Y., Deyholos, M. K., and Liu, X. (2019). VvWRKY30, a Grape WRKY Transcription Factor, Plays a Positive Regulatory Role under Salinity Stress. Plant Sci. 280, 132–142. doi:10.1016/j.plantsci.2018.03.018
Keywords: mungbean, VrDIR, gene expression, salt stress, drought stress
Citation: Xu W, Liu T, Zhang H and Zhu H (2021) Mungbean DIRIGENT Gene Subfamilies and Their Expression Profiles Under Salt and Drought Stresses. Front. Genet. 12:658148. doi: 10.3389/fgene.2021.658148
Received: 25 January 2021; Accepted: 10 September 2021;
Published: 22 September 2021.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Karthikeyan Adhimoolam, Jeju National University, South KoreaNorman George Lewis, Washington State University, United States
Copyright © 2021 Xu, Liu, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Zhu, emh1aG9uZ0BxYXUuZWR1LmNu
 Wenying Xu1
Wenying Xu1 Tong Liu
Tong Liu Hong Zhu
Hong Zhu