- 1Faculty of Agriculture, Shahrood University of Technology, Shahrood, Iran
- 2Department of Plant Breeding, Faculty of Crop Sciences, Sari Agricultural Sciences and Natural Resources University (SANRU), Sari, Iran
- 3Minab Higher Education Center, University of Hormozgan, Bandar Abbas, Iran
- 4Institute of Biological Sciences, University of Talca, Talca, Chile
The TIFY gene family, a key plant-specific transcription factor (TF) family, is involved in diverse biological processes including plant defense and growth regulation. Despite TIFY proteins being reported in some plant species, a genome-wide comparative and comprehensive analysis of TIFY genes in plant species can reveal more details. In the current study, the members of the TIFY gene family were significantly increased by the identification of 18 and six new members using maize and tomato reference genomes, respectively. Thus, a genome-wide comparative analysis of the TIFY gene family between 48 tomato (Solanum lycopersicum, a dicot plant) genes and 26 maize (Zea mays, a monocot plant) genes was performed in terms of sequence structure, phylogenetics, expression, regulatory systems, and protein interaction. The identified TIFYs were clustered into four subfamilies, namely, TIFY-S, JAZ, ZML, and PPD. The PPD subfamily was only detected in tomato. Within the context of the biological process, TIFY family genes in both studied plant species are predicted to be involved in various important processes, such as reproduction, metabolic processes, responses to stresses, and cell signaling. The Ka/Ks ratios of the duplicated paralogous gene pairs indicate that all of the duplicated pairs in the TIFY gene family of tomato have been influenced by an intense purifying selection, whereas in the maize genome, there are three duplicated blocks containing Ka/Ks > 1, which are implicated in evolution with positive selection. The amino acid residues present in the active site pocket of TIFY proteins partially differ in each subfamily, although the Mg or Ca ions exist heterogeneously in the centers of the active sites of all the predicted TIFY protein models. Based on the expression profiles of TIFY genes in both plant species, JAZ subfamily proteins are more associated with the response to abiotic and biotic stresses than other subfamilies. In conclusion, globally scrutinizing and comparing the maize and tomato TIFY genes showed that TIFY genes play a critical role in cell reproduction, plant growth, and responses to stress conditions, and the conserved regulatory mechanisms may control their expression.
Introduction
Transcription factors (TFs) are main regulatory proteins in whole living cells that bind to DNA flanking target genes. An interaction occurs among transcriptional regulators, consisting of chromatin-modifying or remodeling proteins, activating or repressing transcription. In plants, TFs play a significant role in regulating gene expression and plant responses to environmental conditions. TIFY is a particular gene family that is annotated as TFs by its function, also formerly named ZIM (zinc-finger protein expressed in inflorescence meristem; Nishii et al., 2000; Aparicio-Fabre et al., 2013), and is involved in diverse biological processes including plant defense and growth regulation (Xia et al., 2017; Liu et al., 2020). Members of the TIFY gene family contain a common TIFY domain with the conserved motif TIF[F/Y]XG (Vanholme et al., 2007; Xia et al., 2017). TIFY genes are divided into four subfamilies: TIFY subfamily, jasmonate ZIM (JAZ), ZIM-like (ZML), and PEAPOD (PPD) (Bai et al., 2011). The members of the TIFY subfamily possess only the TIFY domain, whereas the JAZ subfamily, in addition to the TIFY domain, has a Jas motif (SLX2FX2KRX2RX5PY) near the C-terminus (Staswick, 2008). ZIM (zinc-finger expressed in inflorescence meristem), and ZML proteins to gether belong to the ZML subfamily, which contains a CCT domain (CONSTANS, CO-like, and TOC1) and a C2C2-GATA zinc-finger domain (Saha et al., 2016). Proteins of the PPD subfamily have a PPD domain in the N-terminals and a changed Jas motif, which replaces the conserved proline–tyrosine in their C-terminals (Chung et al., 2009).
The JAZ proteins suppress TFs such as MYC2, which plays a role in promoting jasmonic acid (JA)–responsive gene transcription in plant cells with low levels of JA. The NINJA/TPS (novel interactor for JAZ/TOPLESS) corepressor complex engages as a molecular mechanism in the suppression of the downstream genes (Pauwels et al., 2010). The JAZ protein plays an important role in the JA signaling pathway (Garrido-Bigotes et al., 2019). The level of JA in a plant cell that is growing naturally is low. The level of JA-isoleucine (JA-Ile) expression is usually enhanced once a plant is subjected to harsh conditions or in development. JAZ proteins link to coronatine-insensitive 1 (COI1) via Skp1/Cullin1/F-box protein COI1 (SCFcoi1) complex-mediated ubiquitination and adjust ubiquitin-26S proteasome destruction. JAZ protein synthesis is induced by JA to prevent TFs’ activity (Katsir et al., 2008; Sun et al., 2017; Garrido-Bigotes et al., 2019). The COI1, MYC2, and JAZ performance in JA signaling resembles the main components of the auxin signaling pathway (Katsir et al., 2008). Furthermore, some TIFY gene family members are involved in modulating the signaling pathways of hormones such as JA and abscisic acid (ABA) (Zhang et al., 2012; Sirhindi et al., 2016). JAZ directly adjusts plant flower initiation, morphology, tanshinone biosynthesis, salvianolic acids, and cotton fiber initiation (Boter et al., 2015; Hu et al., 2016; Pei et al., 2018; Yu et al., 2018). To date, TIFY genes have been recognized in various species, for instance, 27 TIFY genes in maize (Bai et al., 2011), 18 in Arabidopsis (Chung and Howe, 2009), 49 in wheat (Ebel et al., 2018), and 20 in rice (Chung and Howe, 2009). Some functional studies of the TIFY gene family have been performed. AT4G14720 (PPD2) and AT4G14713 (PPD1) are engaged in the coordination of leaf growth (White, 2006). AT4G24470 (ZIM) adjusts hypocotyl and petiole elongation through mediating cell elongation (Shikata et al., 2004), whereas AT1G51600 (ZML2) plays a transcriptional repressor role in the lignin biosynthesis of transgenic maize (Vélez-Bermúdez et al., 2015).
TIFY genes play a remarkable role in leaf growth synchronization and phosphorus-starvation adaptation in Arabidopsis and common bean (White, 2006; Aparicio-Fabre et al., 2013White, 2006). In rice, OsTIFY11b (OsJAZ10) and OsTIFY3 (OsJAZ1) are regulators governing grain size and spikelet development (Hakata et al., 2012; Cai et al., 2014). TIFY proteins are broadly involved in the plant response to abiotic and biotic stresses such as Pseudomonas syringae DC3000 (jaz10 mutants) in Arabidopsis (Demianski et al., 2012) and bacterial blight resistance (OsJAZ8) in rice (Taniguchi et al., 2014). TaJAZ1 overexpression leads to increased bread wheat resistance in biotic stress (Jing et al., 2019); GhJAZ2 overexpression leads to increased sensitivity in transgenic cotton under salt stress (Sun et al., 2017). AtTIFY10a, 10b, and GsTIFY10a (as their wild soybean homologs) and OsJAZ8 (in rice) overexpression play significant adjusting roles in the responses of the plant under alkaline and salt stresses, respectively (Zhu et al., 2014; Peethambaran et al., 2018). All of these results indicate that the TIFY gene family has multiple regulatory roles in cell signaling and regulating plant responses to stresses and so might be precious resources for stress-responsive genes.
Several studies have examined the function of this gene family’s members, but many structural and regulatory aspects of this gene family remain unknown. Tomato and maize are among the most valuable plants, being important in the human food supply. Previously, 30 TIFY genes in maize (Zhang L. et al., 2015) and 20 genes in tomato (Chini et al., 2017) were identified; the studies were mostly accomplished based on gene expression and phylogeny studies. However, in the current study, the number of TIFY genes was elevated using the updated reference genomes of maize and tomato, which comprised 18 and six new members of the TIFY gene family in maize and tomato, respectively. Hence, a genome-wide comparison analysis of the TIFY gene family between tomato (Solanum lycopersicum) as a dicot and maize (Zea mays) as a monocot was performed based on sequence structure, expression, regulatory systems, and protein interaction. Overall, the reported results increase our knowledge of the evolutionary and regulatory mechanisms of the TIFYs and lay the basis for revealing the mechanism of regulatory and future functional analyses related to molecular mechanisms of TIFY genes.
Materials and Methods
Identification of TIFY Gene Family Members
Reference genomes of Z. mays and S. lycopersicum were obtained from the Ensembl platform (Bolser et al., 2017) for the detection of TIFY family members. The hidden Markov model search was fulfilled through the TIFY domain (PF06200) in the query box of the HMMER 3.0 program (E < × 10––5), and the retrieved amino acid sequences were assessed using the SMART1 (Schultz et al., 2000) and Pfam2 (Finn et al., 2010) databases for the identification of the particular TIFY domain. Genomic sequences and the corresponding cDNA of the predicted proteins were also determined using the Phytozome v13.1 database3 (Goodstein et al., 2012), and the locations of TIFY genes on chromosomes were determined using the gene ID in Ensembl Plants (Bolser et al., 2017). The ProtParam program on the ExPASy server4 (Gasteiger et al., 2003) was used to specify the physicochemical characteristics of the TIFY family proteins, such as theoretical isoelectric point (pI) and molecular weight (MW). The CELLO2GO (Yu et al., 2014) and Gene Ontology (GO) (Yu et al., 2006) programs were used at an E < 0.001 to determine GO annotation of TIFY genes and for proteins subcellular localization.
Chromosomal Mapping, Gene Duplications, and Estimation of Ka/Ks Ratio in the Duplicated Pairs
The retrieved TIFY genes were mapped onto the maize and tomato chromosomes according to their predicted positions through Ensembl using MapChart software (Voorrips, 2002). The duplication events among the genes were identified using the alignment of the TIFY coding DNA sequences via ClustalW5 (Larkin et al., 2007). The matrix with the aligned CDS sequences was predicted by BioEdit software (v. 7.2.5) (Hall, 1999). Genes were considered to be duplicated when there was more than 85% identity at their nucleotide sequences (Zheng et al., 2010), which were manually marked on the chromosomal location. The sequence duplications among species were then determined via the Plant Genome Duplication Database using the MCScan v0.8 program (Wang et al., 2012).
The pressure of selection on the duplicated pairs and dividing of homologous TIFY genes were computed by calculating the synonymous (Ks) and nonsynonymous (Ka) exchange rate per site among the gene pairs using the DnaSP v6 software (Rozas et al., 2017). The time of dividing and duplication was appraised by a synonymous mutation rate of λ substitutions per synonymous site per year as T = (Ks/2λ (λ = 6.5 × 10–9)) × 10–6 (Yang et al., 2008). The syntenic relationships of TIFY genes among the orthologous pairs of maize–rice and tomato–Arabidopsis at both gene and chromosome levels were visualized using the Circos software (Krzywinski et al., 2009).
Phylogenetic Analysis, Motif Recognition, and Promoter cis-Elements
Phylogenetic trees of the evolutionary relationships between TIFY protein sequences from maize, rice, barley (Hordeum vulgare), tomato, soybean (Glycine max), and Arabidopsis (downloaded from the Ensembl platform) were constructed using the ClustalW method and maximum likelihood (ML) algorithm with 1,000 bootstrap replications, implemented in the MEGA 6.0 software (Tamura et al., 2013). Finally, the phylogenetic tree was drawn using an interactive tree of life (Letunic and Bork, 2019). The MEME software6 (Bailey et al., 2009) was used to identify the conserved motifs in amino acid sequences of TIFYs based on the following setting: motifs number: 15; minimum width: 6; maximum width: 50.
Promoter Analysis and Protein–Protein Interaction Assay
cis-Elements within the promoter region of TIFY genes were identified using the 2,000-bp upstream region of the ATG start codon in each putative TIFY gene on the PlantCARE server http://bioinformatics.psb.ugent.be/webtools/plantcare/html/7 (Lescot et al., 2002). The STRING v11 program http://stringdb.org8 (Szklarczyk et al., 2019) was used to determine key TIFY genes in the studied plant species, considering GO annotations, and to infer protein-protein interaction networks.
Three-Dimensional Protein Modeling and Molecular Docking of the Protein Pocket Sites
The three-dimensional (3D) protein structures associated with some candidate TIFY proteins of the identified subfamilies were built via iterative template-based fragment assembly simulations in I-TASSER (Yang et al., 2015). The best models were purified using the 3D-refine program (Bhattacharya et al., 2016). Then, the predicted structures were validated using a Ramachandran plot via measuring the backbone dihedral phi (ϕ) and psi (Ψ) angles using the RAMPAGE program (Lovell et al., 2003). For the prediction of the protein pockets and cavities, the refined structures of TIFY proteins were analyzed using P2Rank in the PrankWeb software (Jendele et al., 2019) and the CASTp tool (Tian et al., 2018). Finally, the results were visualized in PyMOL (DeLano, 2002).
In silico Expression Analysis of TIFY Genes Through RNA-Seq Data
For the assessment of maize and tomato TIFY gene transcription, available RNA-seq data were used to retrieve data in response to stimuli in multiple tissues. The fragments per kilobase of exon per million fragments mapped (FPKM) expression values in maize various tissues, in addition to under stimuli exposure, were determined in the maize B73 v4 genome through a gene ID (Zm00001) search in qTeller in MaizeGDB (Portwood et al., 2019) using the previously published reports for multiple tissues (accession IDs PRJNA171684 and SRP010680) (Stelpflug et al., 2016) and under stresses with accession numbers GSE71046 (Forestan et al., 2016) and PRJNA244661 (Waters et al., 2017). For the expression values of TIFY genes in tomato, the RNA-seq transcriptome data related to various tissues in the tomato cultivar Heinz and tomato leaves treated with different bacteria and pathogen-associated molecular patterns were extracted from the tomato functional genomics database9 (Fei et al., 2010). The extracted magnitudes were then log2 transformed and used to generate the heatmaps and Venn diagrams via the TBtools package (Chen et al., 2020). Clustering of heatmaps was performed using complete data and the Euclidean distance method.
Results
Identification of TIFY Genes and GO Annotation
Based on the HMMER result, 48 and 26 nonredundant putative TIFY proteins were identified in the Z. mays and S. lycopersicum genomes, respectively (Supplementary Table 1). According to the protein-specific domain, the recognized TIFY proteins were classified into four subfamilies: JAZ, TIFY-S (also named TIFY), PPD, and ZML; however, the PPD subfamily was not identified in the maize genome (Supplementary Table 1). The putative TIFY proteins in maize ranged from 60 (in Zm00001d024455) to 539 (in Zm00001d005726) amino acids in length, with the MWs ranging from 6.65 to 57.69 kDa in these proteins, respectively. The theoretical pI of the maize TIFY proteins ranged from 4.37 (in Zm00001d048264) to 10.88 (in Zm00001d032009). The identified TIFY proteins in tomato also ranged from 61 (Solyc01g011175) to 427 (Solyc06g065650), with the MW ranging from 6.95 to 44.85 kDa in these proteins, respectively, and the pI values varied from 4.99 (Solyc01g009730) to 10.34 (Solyc01g097060). Most of the identified TIFY proteins in both of the candidate species revealed an alkaline nature (∼73% in maize and ∼69% in tomato; Supplementary Table 1). The results of subcellular localization revealed that the majority of maize TIFY proteins are localized in the nucleus, extracellularly, intracellularly, and organelles (Figure 1), whereas most tomato proteins showed potential to be located in the nucleus, cytoplasm, and mitochondria (Figure 1).
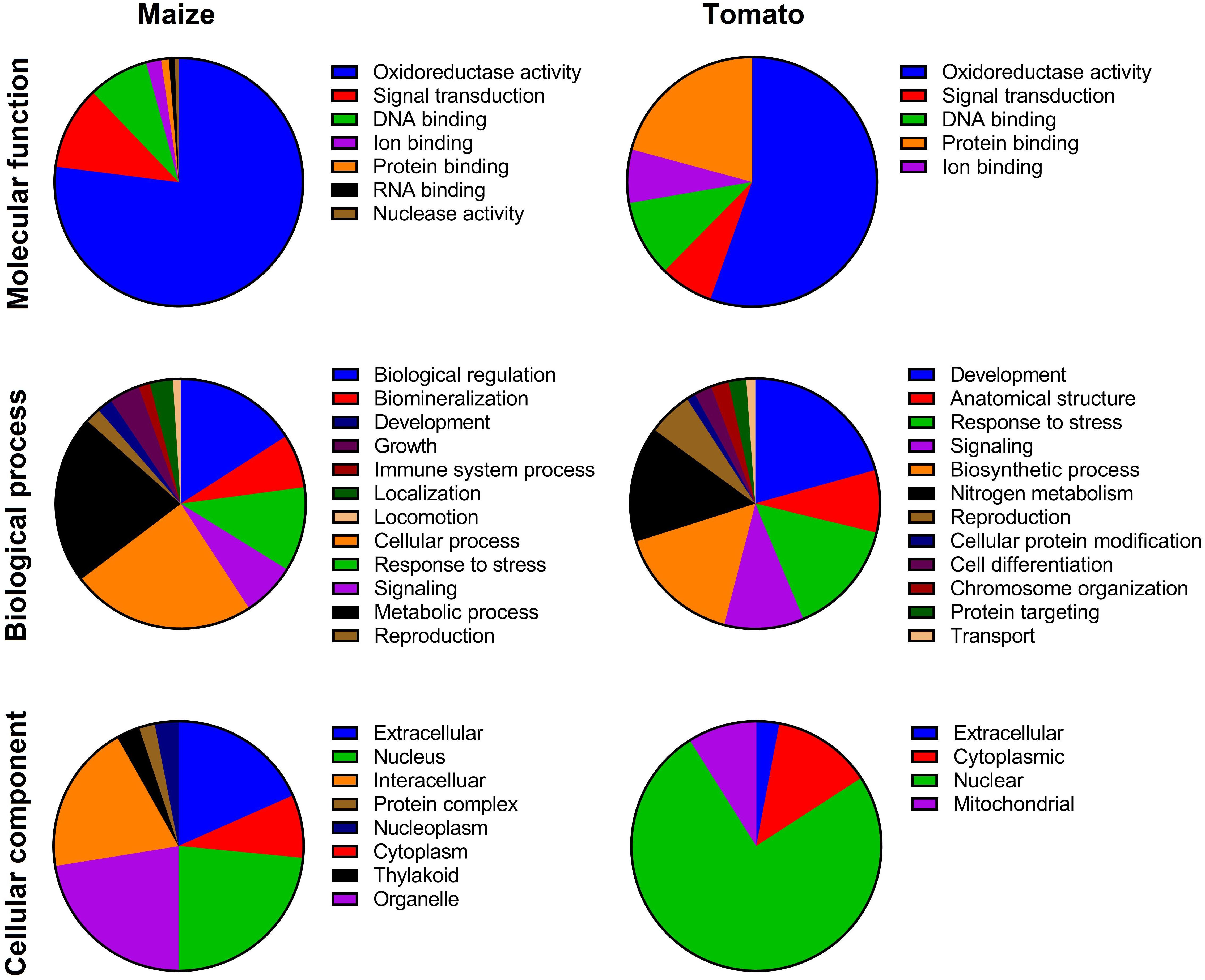
Figure 1. Gene Ontology (GO) of TIFY family members based on molecular function, biological process, and cellular component in maize and tomato. The GO terms were assigned based on a protein sequences search in the CELLO2GO program.
The evaluation of the biological processes mediated by TIFYs evidenced that most of the proteins are probably implicated in growth and developmental processes and response to stimuli in both monocot and dicot plant species (Figure 1). Among the TIFY family proteins, ∼2 and 18% of TIFY proteins showed potential involvement in the regulation of development in maize and tomato, respectively (Figure 1). The roles of TIFYs in reproduction (∼2% in maize and 5% in tomato) and cellular component metabolism (11% in maize and 21% in tomato) were determined through the GO assay. Most of the recognized TIFYs in both plant species were predicted to be involved in the response to adverse conditions. For instance, 8 and 11% of TIFYs in maize and 10 and 14% of these genes in tomato were assumed to be modulating genes during signaling and stress responses, respectively. In the context of molecular functions, most of the TIFY proteins showed potential involvement in oxidoreductase activity (79% in maize and 56% in tomato) and DNA-binding activity (8% in maize and 10% in tomato) (Figure 1). The potential involvement of some TIFY proteins in ion binding (2% in maize and 6% in tomato) was also expected, in addition to their molecular functions in the cell.
Chromosomal Distribution and Gene Duplications
Uneven distribution of the TIFY genes was predicted on 10 maize chromosomes with 11, eight, and eight genes on chromosomes 1, 2, and, 5, respectively (Figure 2A). Chromosomes 3, 8, and 10 in maize also accounted for only two, one, and two TIFY genes, respectively. In the tomato genome, TIFY genes were distributed on 10 of 12 chromosomes, with a high density on chromosomes 1 and 8, which contained nine and four genes, respectively (Figure 2B). Sixteen and three duplicated gene pairs were identified in the TIFY family in maize and tomato, which clustered into five and two groups, respectively (Supplementary Table 2). The segmental duplication events were found to be higher in the maize genome than tomato’s. Among the duplicated clades in maize, groups A, C, and D showed significant duplication events with five, four, and four gene pairs, respectively (Figure 2A and Supplementary Table 2). The duplicated gene pairs in tomato were localized only at chromosomes 1 and 8 (Figure 2B and Supplementary Table 2), whereas the duplicated pairs in maize were found to be distributed on all chromosomes with the highest range of duplication on chromosomes 1 and 2. The intraspecies synteny analysis revealed that all of the duplicated blocks in tomato are collinear, such as Solyc01g009730 and Solyc01g009740. The Ka/Ks ratios of the duplicated paralogous gene pairs cover a domain from 0.491 to 1.830 in maize and 0.570 to 0.775 in tomato, whereas in the maize genome, there are three duplicated blocks containing Ka/Ks > 1 (Supplementary Table 2).
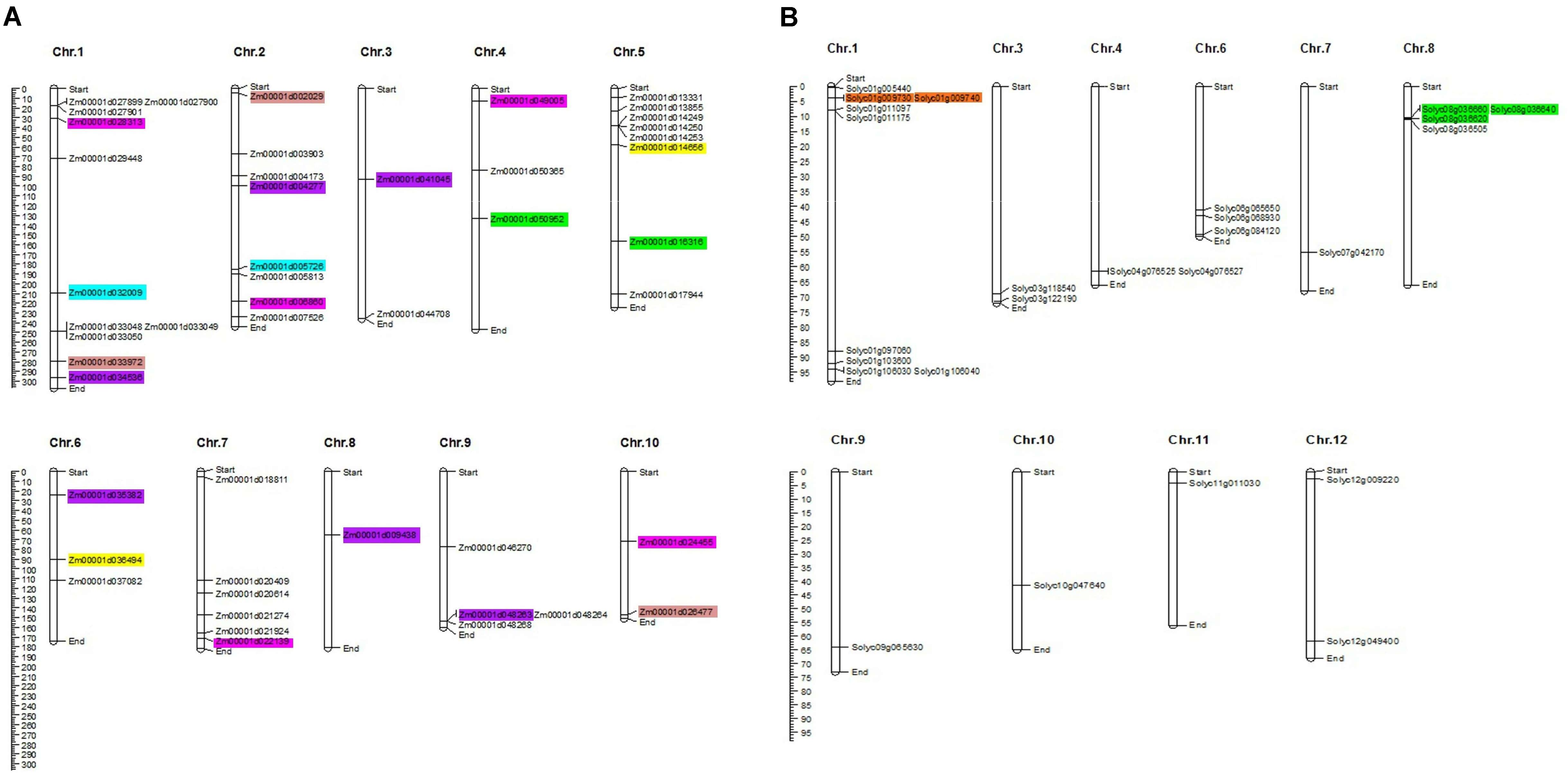
Figure 2. Chromosomal distribution of TIFY genes predicted from the Zea mays (A), and Solanum lycopersicum (B) genomes. The graphical genetic maps were created via the MapChart software, and the duplicated gene pairs are highlighted in the same color.
Phylogenetic Relationships and Conserved Motifs
A total of 169 TIFY protein sequences from three monocot plant species, namely, maize, rice, and barley, along with three dicot species, including tomato, Arabidopsis, and soybean, were employed to assay the phylogenetic relationships that clustered all TIFYs into the seven different groups (Figure 3). Based on the conserved protein motifs, three of 15 motifs (motifs 1, 2, and 3) represent the specific TIFY protein domains in all candidate species (Figure 4). The presence of only motif 2 demonstrates the important functional part of the TIFY-S class; motif 2 with motif 1 represents the JAZ clade, which includes more proteins; and the presence of motifs 1, 2, and 3 demonstrates the ZML group of the TIFY family (Figures 3, 4). In addition, some conserved motifs were observed on the outside of the protein domain. The TIFY proteins belonging to the same phylogenetic class also have some conserved motifs beyond the specific domain region. For instance, motif 7 is shared by the members of the JAZ subfamily. Hence, the motif architectures are approximately conserved in each TIFY-S subfamily, which refer to the conserved and specific functions of the proteins in these clusters. Overall, the members of the JAZ subfamily show high diversity, suggesting relative evolutionary conservation in the cellular function of these proteins from various plant species. High diversity between TIFY genes was observed, indicating that the TIFY gene family originated before the divergence of monocots and dicots.
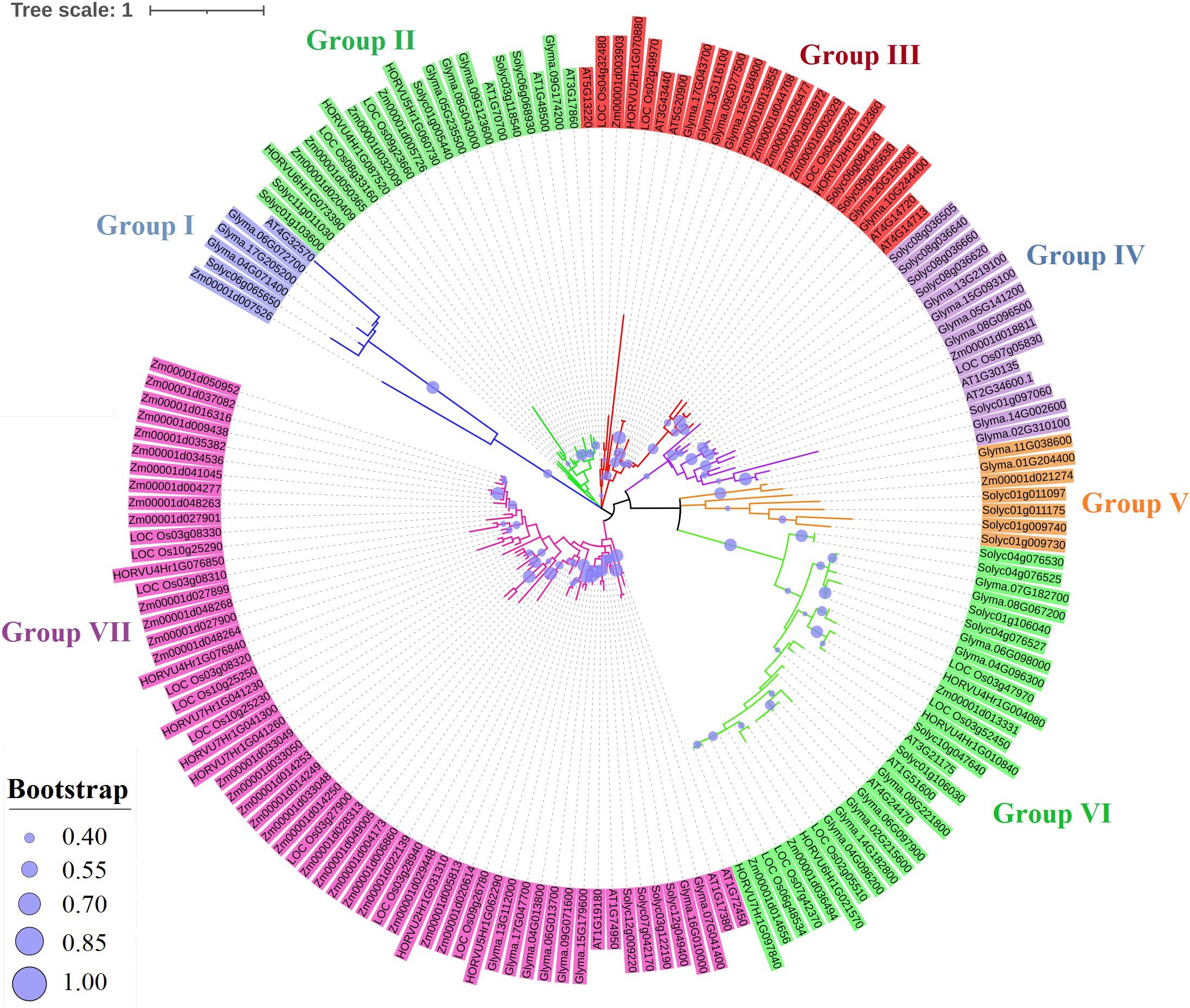
Figure 3. Phylogenetic analysis of TIFY proteins from monocots (maize, barley, and rice) and dicots (tomato, Arabidopsis, and soybean) based on the maximum likelihood (ML) method.
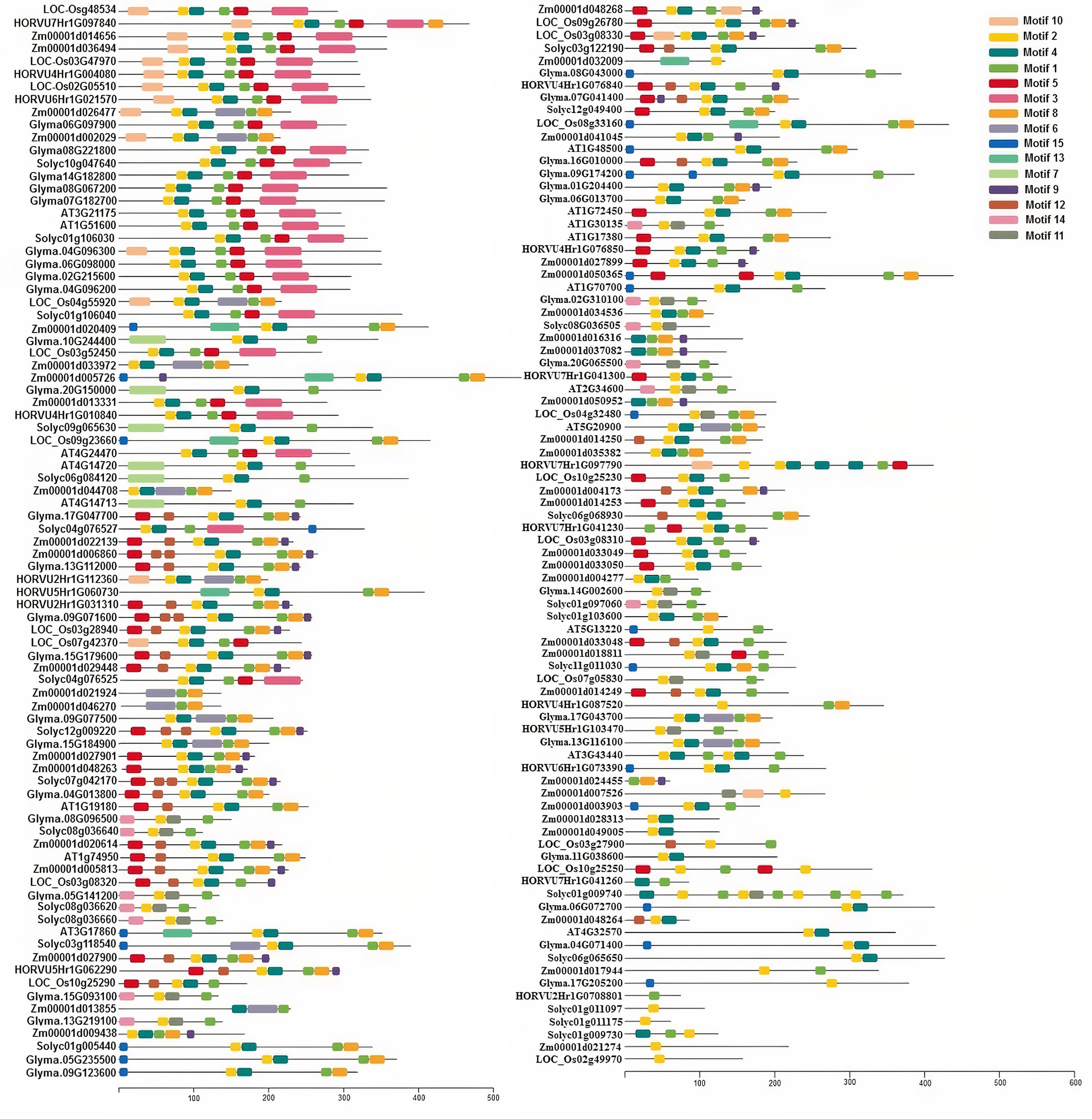
Figure 4. Conserved motif distribution in TIFY proteins from monocots (maize, barley, and rice) and dicots (tomato, Arabidopsis, and soybean). A total of 15 conserved protein motifs were predicted via the MEME program with a length of 6 to 50 amino acid residues. The proteins in the same clade have a similar motif pattern in their structure.
Identification of Blocks Duplicated Between Species With Estimation of the Ka/Ks Ratios
The association of positive Darwinian selection in duplication and divergence, an important parameter for studying the effects of positive selection engagement in gene divergence (Yang et al., 2008), was calculated for the duplicated TIFY genes in maize in comparison with the monocot model plant (Oryza sativa) (Figure 5A) and tomato compared with the dicot model plant (Arabidopsis thaliana) (Figure 5B) as their closest orthologous genes (Supplementary Table 3). As a result, 11 duplicated and 10 triplicated blocks in maize compared with rice species, and three duplications and one triplication in tomato in comparison with Arabidopsis, were identified; the average Ka/Ks ratio for the diverged blocks was estimated to be 0.450 and 0.427 (<1) in maize and tomato, respectively. The duplication event for the TIFY genes was estimated to have occurred approximately 32–137 MYA between maize and rice and 109 to 150 MYA between tomato and Arabidopsis (Supplementary Table 3). Among the closest orthologous TIFY in the grass species, the relatively higher rate of synonymous substitution between maize and rice suggests their earlier divergence about 63 MYA compared to that between tomato and Arabidopsis genes (around 124 MYA). Therefore, the duplication and divergence events among the TIFY genes from monocot species can be considered as a significant aspect in the evolution of this gene family.
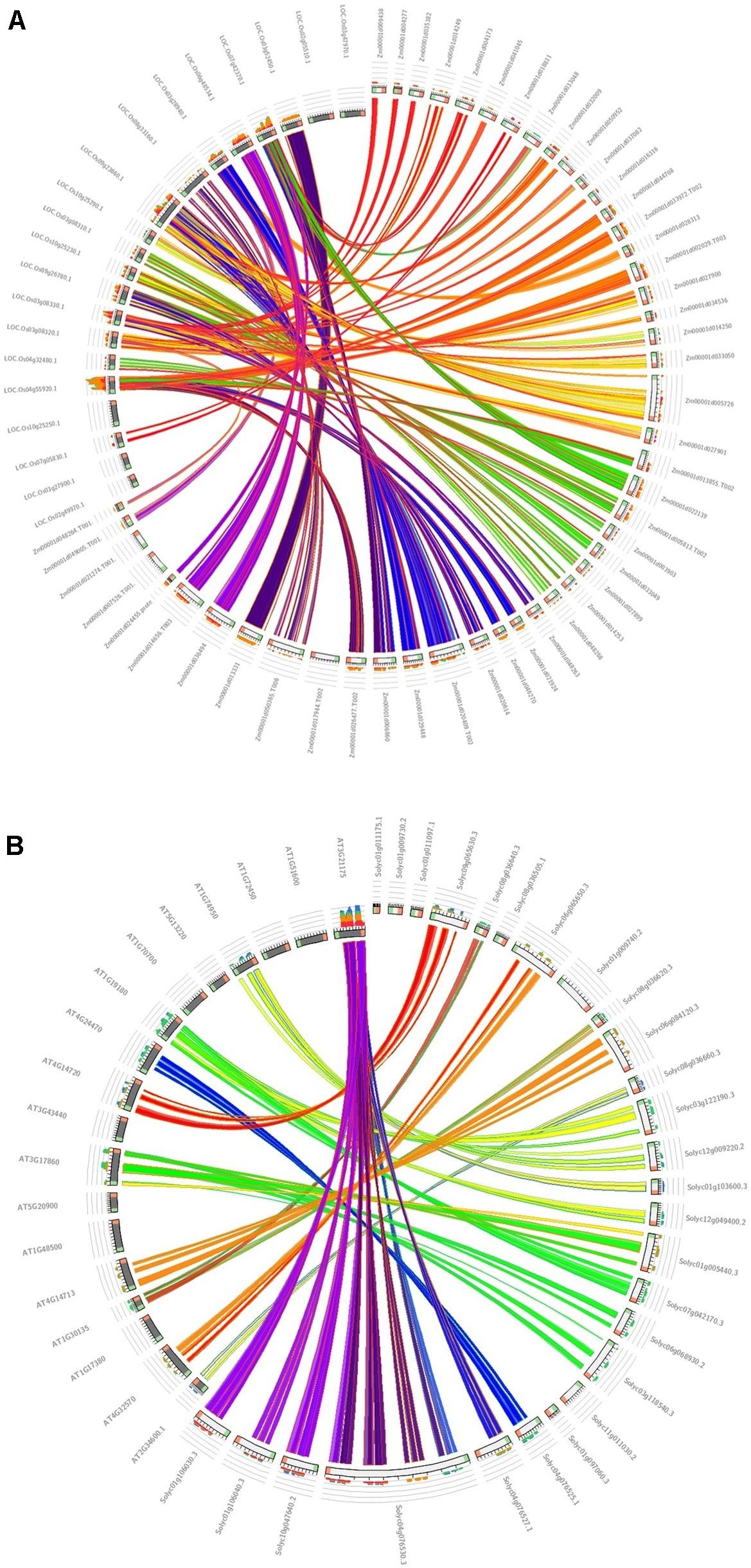
Figure 5. Synteny analysis of TIFY genes. The syntenic blocks of maize TIFY genes are compared with the monocot model plant (O. sativa) (A), and the syntenic blocks of tomato are compared with the dicot model plant (A. thaliana) (B).
Promoter cis-Elements and Protein–Protein Interaction Network
In the present study, several kinds of cis-elements that deal with various phytohormones, abiotic stimulus conditions, and regulation of development were identified in the promoter of TIFY genes (Figure 6). The ABA responsiveness (ABRE), ethylene responsiveness (ERE), and methyl jasmonate responsiveness (MeJA) factors were observed as highly occurring hormone-responding cis-elements approximately in the TIFY genes promoter. The light-responsive G-Box and Box 4, wounding stress-responsive WUN-motif, and stress-responsive MYB elements were detected as the other regulatory cis-elements frequently distinguished in the TIFY genes promoter area, suggesting the important roles of this gene family in stress management in monocot and dicot crops. Moreover, observation of the MBS element, the MYB protein binding region engaged in drought stress coping, and regulation of the flavonoid biosynthetic genes, in some TIFY genes, such as Zm00001d041045, Zm00001d022139, Zm00001d003903, and Zm00001d013331 in maize and Solyc01g103600, Solyc07g042170, Solyc04g076527, and Solyc01g106040 in tomato, confirmed the important role of these genes in anthocyanin/flavonoid production and stimulus coping. The TC-rich repeats (regulating defensive reactions), low-temperature responsive element, TCA element (salicylic acid–responsive), TGA element (auxin-responsive), and W-Box (WRKY TF-binding region, important for responses to abiotic stimuli) were detected as the important abiotic/hormone stress–responsive elements significantly detected in most TIFY genes. Another important finding was the discovery of multiple regulatory cis-elements related to phytohormones and environmental stimuli in the majority of TIFY genes, revealing the role of these genes in plant growth and dealing with stress conditions. In general, the results showed that the distribution of regulatory elements in the promoter region of TIFY genes is similar in both studied species, and the observation of different regulatory elements indicated that this gene family is involved in different cellular pathways.
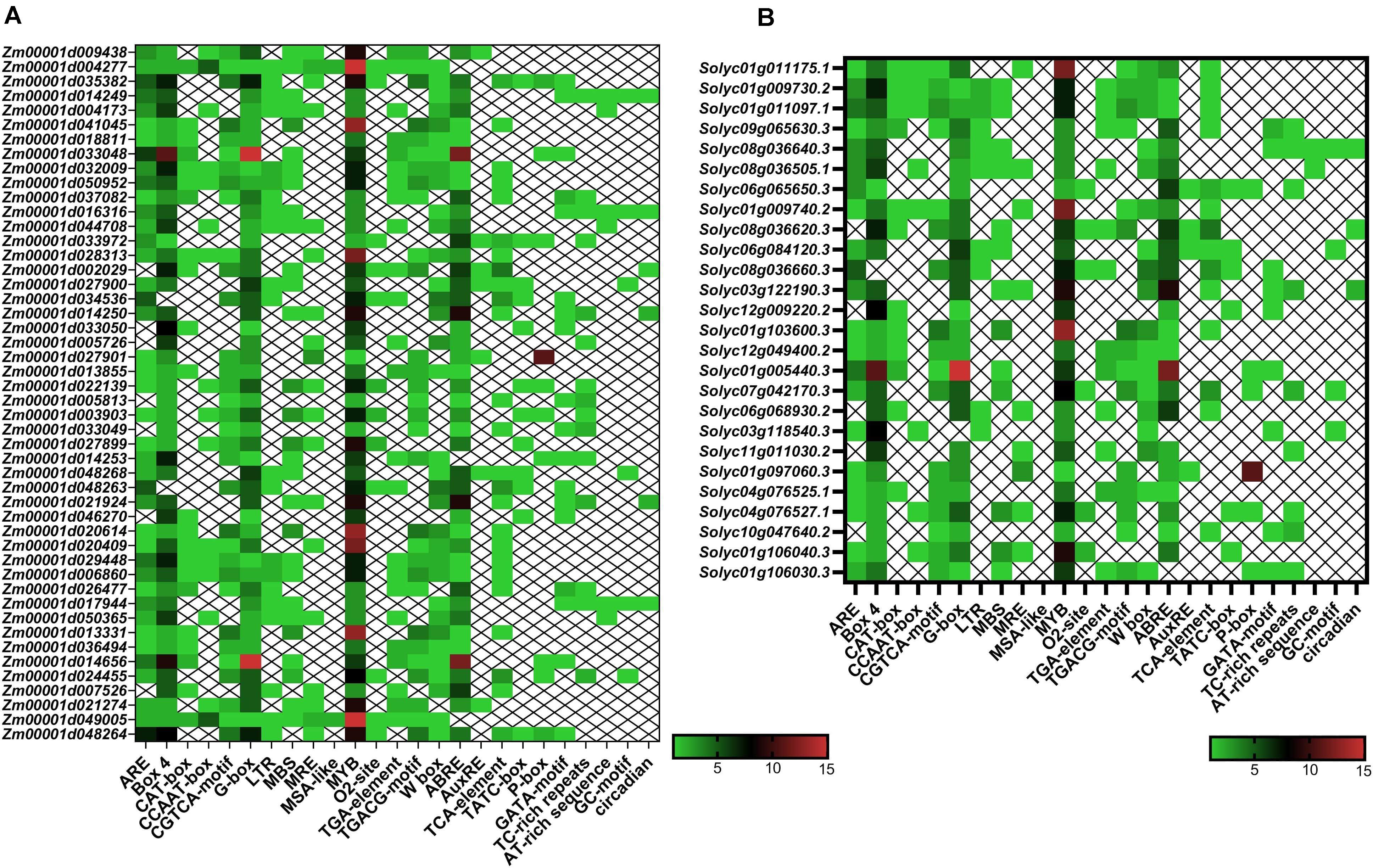
Figure 6. Heatmaps of cis-regulatory elements distribution in the promoter region of TIFY genes of maize (A) and tomato (B). The upstream region (2,000 bp) of the ATG start codon in each putative TIFY gene promoter region was analyzed using the PlantCARE server (Lescot et al., 2002).
The interactome data related to TIFY genes in maize identified two subnetworks, which showed that TIFY proteins interact with the genes engaged in protein dimerization activity, RNA binding, hydrolase activity, and damaged DNA binding (Figure 7A and Supplementary Table 4). The GRMZM2G455115 and GRMZM2G118697 proteins, which are cleavage and polyadenylation specificity factors, were reported to interact with TIFY proteins to regulate posttranscriptional gene silencing by RNA. The DNA ligases GRMZM2G427067 and GRMZM2G137968, which are single-stranded helicases, also revealed a highly confident interaction with TIFYs in maize that contributes to DNA ligation involved in DNA repair, DNA replication, and cellular response to DNA damage. Furthermore, the protein–protein interaction network in tomato showed the interactions between TIFY proteins and TF MYCs and salt-responsive proteins (Figure 7B and Supplementary Table 5). The BIG SEEDS protein BS1, salt-responsive protein SRG, TF MYC, and Pto-responsive gene Prg1 showed a significant contribution with TIFY genes in the regulation of the defense response, plant hormone signal transduction, and multicellular organism development through a hormone-mediated pathway. The interaction of TIFY proteins with the critical coronatine-insensitive Coi1 and allene oxide synthase regulates ubiquitin-dependent protein catabolic and fatty acid biosynthetic processes, which are essential for protein function and plant reproduction and viability. Therefore, our results reveal that TIFY proteins significantly collaborate with the proteins from various metabolisms, which can regulate plant responses to external stimuli and growth.
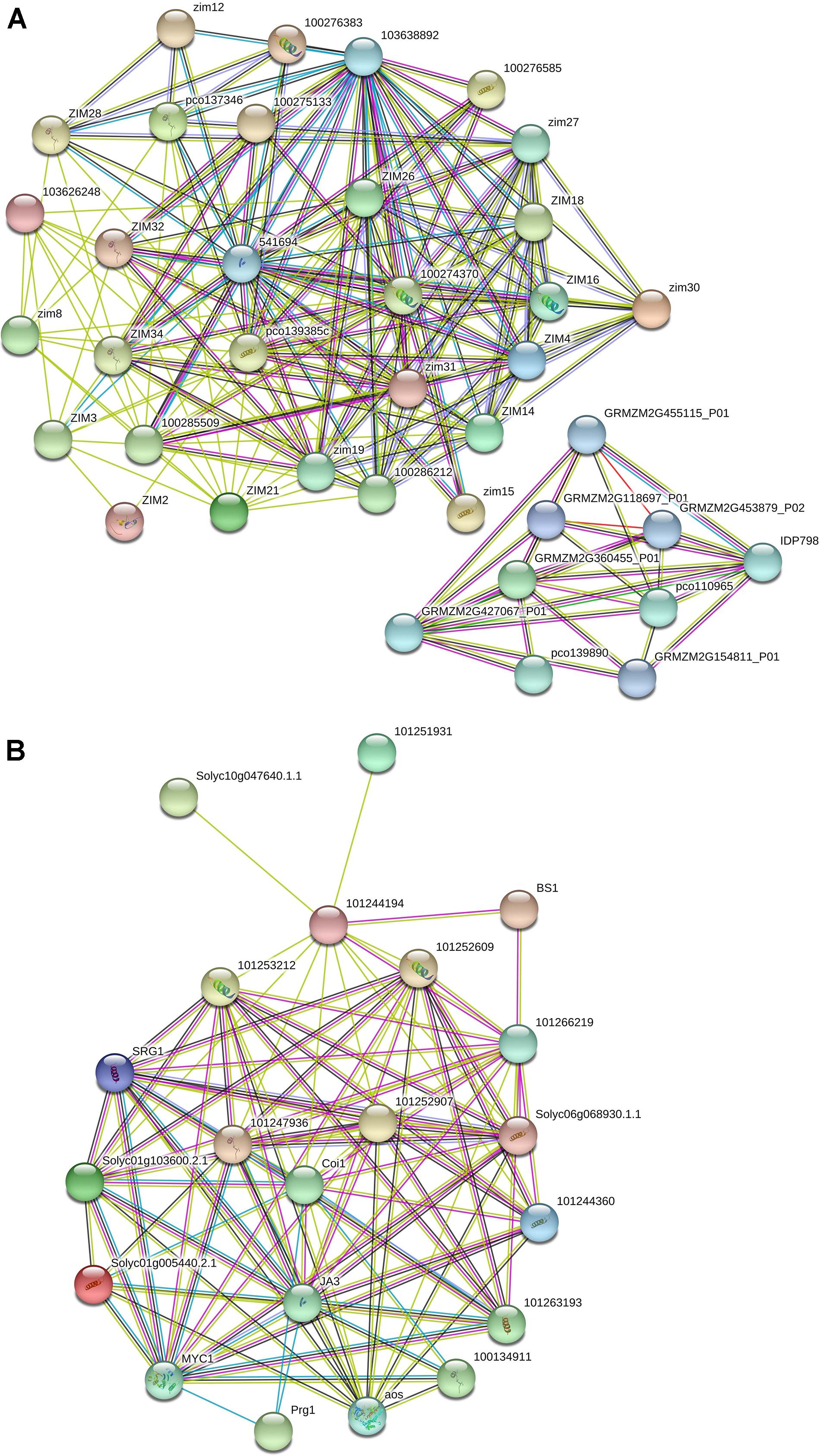
Figure 7. Interaction network of TIFY genes in maize (A) and tomato (B) predicted using the String database.
Homology Modeling of TIFY Proteins and Docking Assay of the Pocket Sites
The 3D structures revealed the presence of the conserved TIFY domain in all of the studied TIFY proteins, which showed a typical 3D frame comprising antiparallel β-sheets followed by parallel α-helixes (Supplementary Figure 1). Topographic features of TIFY proteins were evaluated through the P2Rank program, and major pockets are shown in multiple-colored regions in Figure 8. As a result, different pockets were predicted as the binding region/active sites in the candidate proteins from JAZ, TIFY-S, ZML, and PPD clusters. The amino acid residues present in the pocket sites of TIFY proteins partially differ in each subfamily, although Mg or Ca ions were heterogeneously observed in the center of the active sites of all of the predicted TIFY protein models. In the JAZ subfamily of maize, PRO, ALA, ASN, HIS, ARG, GLY, ASP, and THR were predicted as the important binding residues, whereas SER, THR, PRO, GLU, VAL, LYS, LEU, and TYR were identified as the key residues. Investigation of the predicted pocket sites of TIFY proteins also showed that LEU, CYS, and SER in the TIFY-S of maize and VAL, LYS, GLU, ARG, THR, SER, and LEU in the TIFY-S of tomato have high potential as active binding sites. ARG, THR, LEU, GLN, LYE, and VAL in ZML of maize and SER in ZML of tomato were predicted as the important binding residues. In the PPD subfamily, ARG and GLU showed high potential as key binding residues (Figure 8). Based on our results, the important amino acids found in the pocket sites of all of the candidate TIFY proteins demonstrate the importance of these residues in the positioning on the DNA molecule and, finally, the cellular function performance during various developmental and defensive processes.
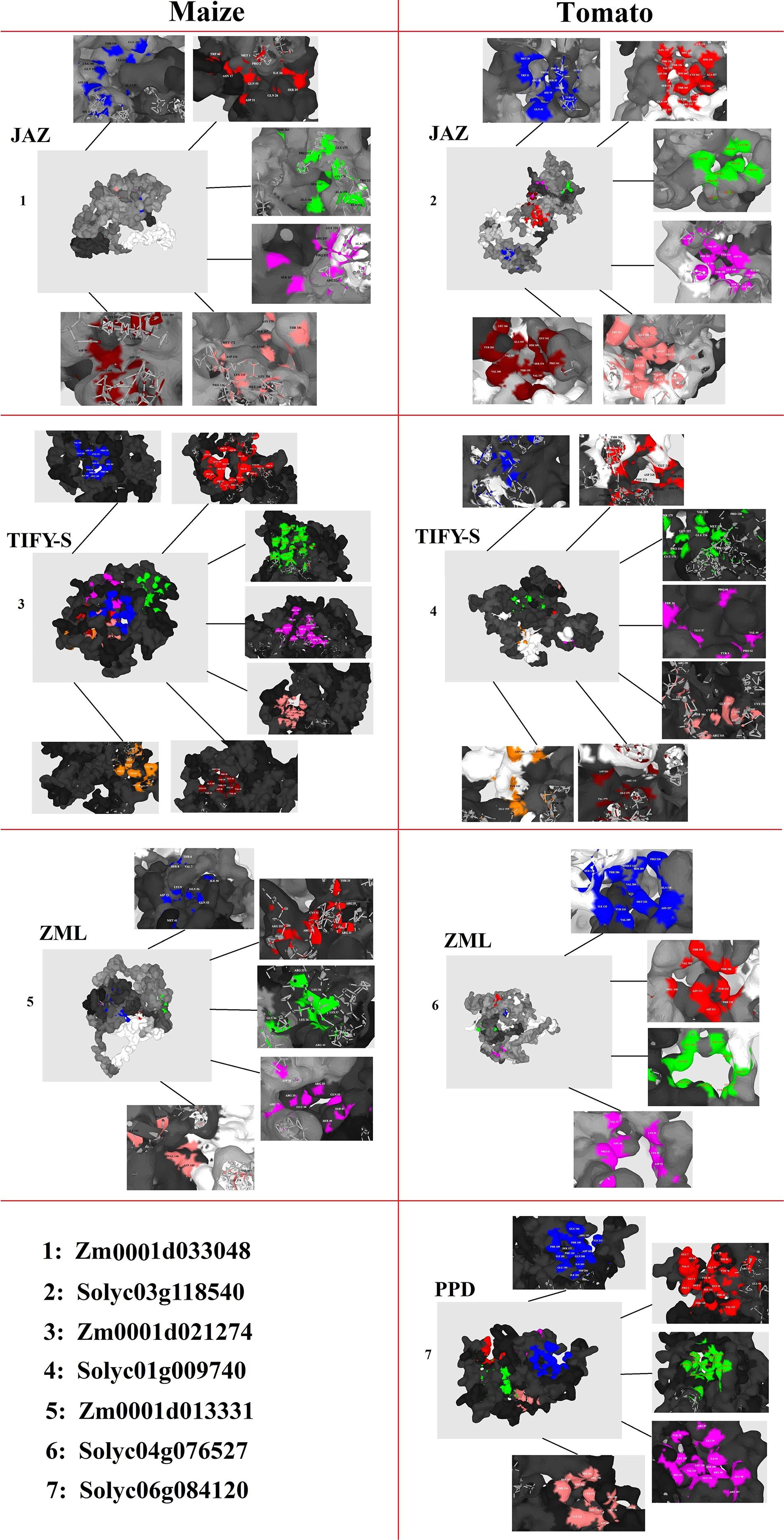
Figure 8. Docking analysis of pocket sites of each subfamily of TIFY proteins including JAZ, TIFY-S, ZML, and PPD.
Expression Assay of TIFYs in Multiple Tissues and Under Abiotic/Biotic Stimuli via RNA-Seq Data
The expression patterns of TIFY genes were investigated under normal growth in multiple tissues in addition to during stress conditions using RNA-seq data sets in maize and tomato. The FPKM values from various parts of roots, leaves, internodes, and seeds in maize were used for identifying genes differentially expressed in these tissues. The results showed a tissue-specific expression pattern of six, one, one, and one TIFY genes in leaf, root, internode, and seed tissues, respectively (Figure 9A). The transcription level of TIFYs could be divided into some major expression groups that contained genes preferentially expressed in all or one of the tissues. In addition to nine genes that were not expressed across the tissues (such as Zm00001d041045, Zm00001d016316, and Zm00001d004277), 11 TIFY genes (e.g., Zm00001d036494, Zm00001d002029, and Zm00001d020409) displayed significant transcription rates in all of the maize tissues, suggesting control of a broad set of genes at the transcriptional level. According to the RNA-seq data related to stress conditions in maize, Zm00001d022139, Zm00001d002029, Zm00001d026477, and Zm00001d048264 were recognized as the TIFYs expressing under all stimulus situations, revealing their important potential in the stress resistance of maize plants (Figure 9A). A total of eight TIFYs, for example, Zm00001d050952, Zm00001d035382, and Zm00001d033048, were not expressed under stress. There were two, one, and one TIFY genes specifically expressed under UV, fungal, and cold stresses, respectively. Nine TIFY genes were also expressed under salt, drought, heat, and cold stresses. Regarding the results, most of the genes with a tendency to express in response to stimuli were from the JAZ and ZML subfamilies, which may reveal the important roles of these genes in dealing with these stimuli.
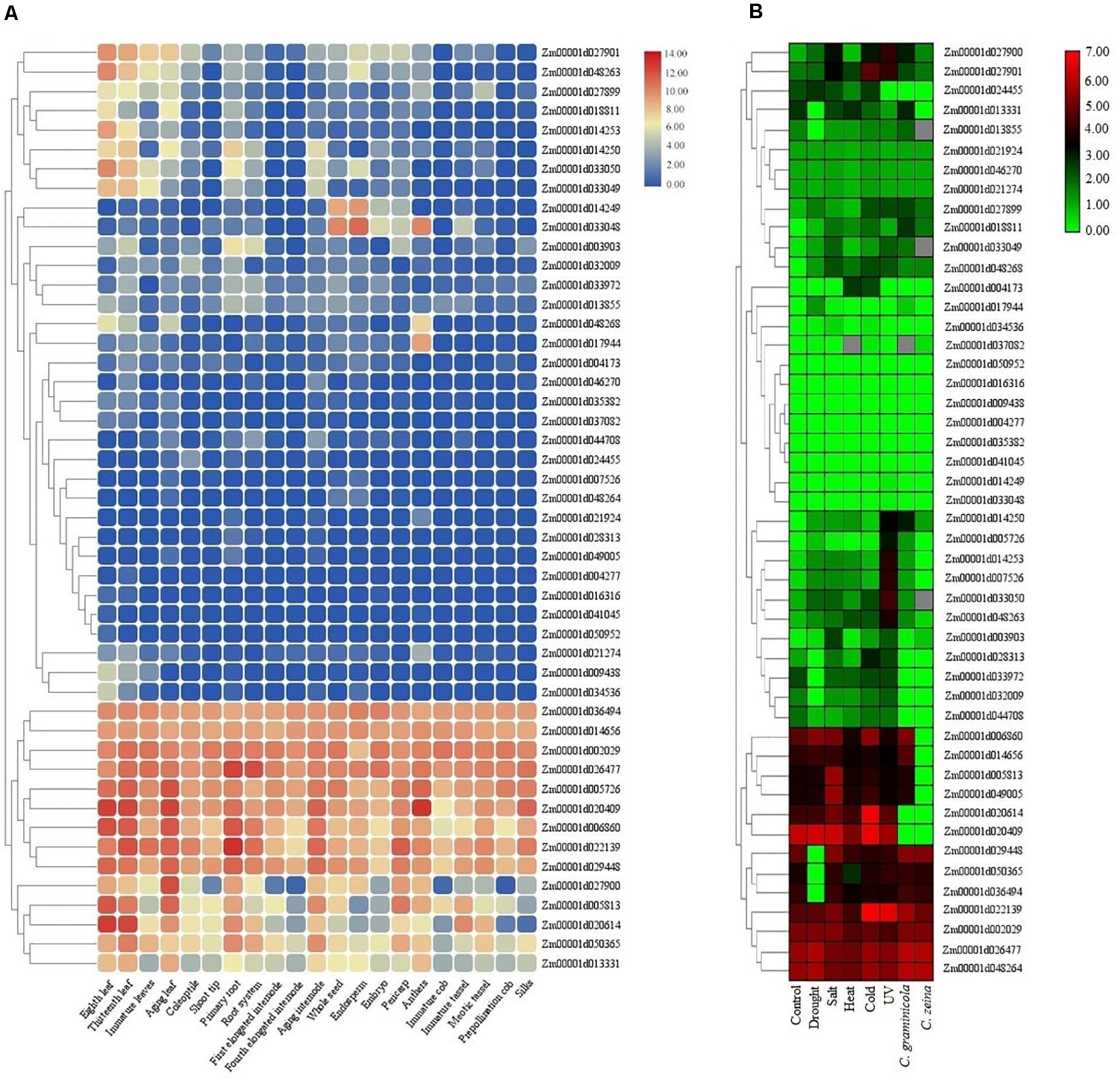
Figure 9. Expression heatmaps of TIFY genes of maize in different tissues (A) and in response to abiotic and biotic stresses (B). The heatmaps were generated based on the log-2–transformed RNA-seq fragments per kilobase of exon per million fragments mapped (FPKM) magnitudes in the maize genome B73 v4.
The RNA-seq experiments were also employed to further verify the expression of the identified TIFY genes in various tissues and under stress in tomato. There were four and one tissue-specific TIFY genes in tomato flower and root tissues, respectively (Figure 10A). A total of 11 TIFY genes, such as Solyc01g005440, Solyc07g042170, and Solyc01g106030, were significantly expressed in all of the candidate tissues in tomato, whereas seven genes, such as Solyc01g011175, Solyc04g076527, and Solyc01g009730, did not reveal any remarkable transcription level in the tomato tissues. The RNA-seq data under stimulus conditions in tomato revealed one TIFY gene responsible for resistance against Agrobacterium tumefaciens; the genes Solyc08g036640 and Solyc08g036620 also demonstrated remarkable expression under exposure to this bacterium (Figure 10B). Nine TIFY genes of tomato were not induced by stress circumstances, whereas 14 genes, such as Solyc01g005440, Solyc11g011030, and Solyc06g065650, demonstrated significant expression in response to all the stimuli (Figure 10B). Solyc08g036660 from the JAZ clade was found to be significantly up-regulated in coping with A. tumefaciens and flagellin 22, suggesting the potential of this gene in dealing with stress. Stress coping in the JAZ protein-encoding genes was significantly greater in comparison with the other subfamilies.
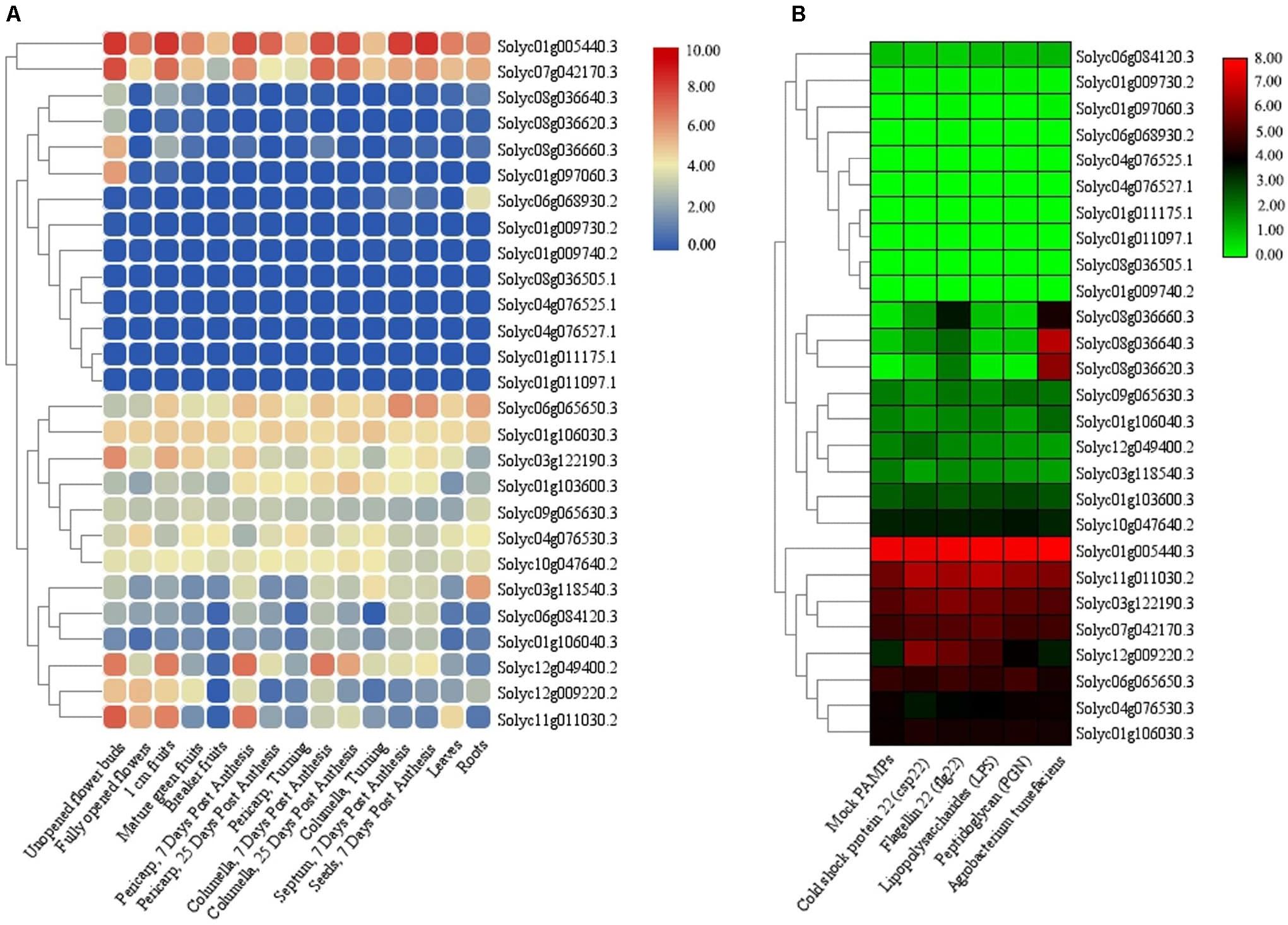
Figure 10. Expression heatmaps of TIFY genes of tomato in different tissues (A) and in response to abiotic and biotic stresses (B). The heatmaps were generated based on the log-2–transformed RNA-seq FPKM magnitudes in the tomato genome.
Discussion
TIFY family proteins, a key TF plant family, have been characterized in different plant species (Vanholme et al., 2007; Aparicio-Fabre et al., 2013; Wang et al., 2020). For instance, 19 members of the TIFY gene family in common bean (Aparicio-Fabre et al., 2013), 49 genes in Triticum aestivum (Ebel et al., 2018), 36 genes in Brassica oleracea (Liu et al., 2020), 36 members in Brassica rapa (Saha et al., 2016), 25 genes in poplar (Wang et al., 2020), 20 genes in rice (Ye et al., 2009), Brachypodium distachyon (Zhang Z. et al., 2015), 50 members in Gossypium hirsutum (Zhao et al., 2016), 34 genes in Glycine soja (Zhu et al., 2013), 30 genes in apple (Li et al., 2015), 21 genes in pear (Ma et al., 2018), and 15 genes in watermelon (Yang et al., 2019) have been identified. Furthermore, in previous studies, 30 TIFY genes in maize (Zhang L. et al., 2015) and 20 genes in tomato (Chini et al., 2017) were recognized. In the current study, 48 and 26 nonredundant putative TIFY genes were identified in the genome of maize (Z. mays) and tomato (S. lycopersicum), respectively. Genome size and ploidy level may affect the number of members of a gene family. For instance, bread wheat, a hexaploid species, and G. hirsutum, a tetraploid plant, contain the most TIFY genes. The identified TIFY proteins are diverse based on their physiochemical properties and domain distribution in maize and tomato. Subcellular localization analysis revealed that the TIFY members of tomato are more located in the nucleus than the TIFY members of maize. Several TIFY members were predicted to be located in organelles, such as chloroplasts and mitochondria (Figure 1). These differences in cellular components indicate that the TIFY gene family developed in an extensive regulatory system in plant cells to control various processes (Bai et al., 2011; Cai et al., 2020). Previous studies indicated that the members of the TIFY gene family are involved in various mechanisms of plant responses to stress and the regulation of plant growth and development (Hakata et al., 2012; Zhou et al., 2015; Sun et al., 2017; Liu et al., 2020; Wang et al., 2020). In the context of biological processes, the TIFY family genes in both candidate plant species are involved in various important processes, such as reproduction, metabolic processes, responses to stresses, and cell signaling (Figure 1). In the context of molecular functions, most identified TIFY family proteins have oxidoreductase activity and DNA-binding TF activity.
High diversity between TIFY genes was observed, indicating that the TIFY family genes originated before the divergence of monocots and dicots. Interestingly, the PPD subfamily was only detected in dicots, supporting the hypothesis that PPD genes are absent in monocots (Ye et al., 2009; Bai et al., 2011). In Arabidopsis, PPD proteins are involved in the regulation of the cell cycle and cell growth (White, 2006). Some other genes in monocots probably compensate for the molecular functions of PPD genes (Bai et al., 2011). In the current study, some conserved motifs were detected from outside of the DNA-binding domain regions that may affect the functioning and cellular localization (Heidari et al., 2020; Rezaee et al., 2020). The duplicated gene pair showed different expression levels in response to stress, indicating that duplicated TIFY genes probably undergo substantial changes in their regulatory mechanisms and/or sequences to assume novel functions (Faraji et al., 2020).
Gene duplication, an evolutionary event for different species, has a significant role in the enlargement of plant TF families (Freeling, 2009; Wang et al., 2013). In the current study, the Ka/Ks ratios of the duplicated paralogous gene pairs showed that all of the duplicated pairs in the TIFY gene family of tomato have been influenced by an intense purifying selection, which could have led to conserved functions or pseudogenization (Juretic et al., 2005), whereas in the maize genome, there were three duplicated blocks containing Ka/Ks > 1, indicating accelerated evolution with positive selection (Faraji et al., 2020). According to intraspecies synteny analysis, all of the duplicated blocks in tomato were collinear, suggesting that these duplication events may have been derived because of the chromosome segmental or large-scale duplication/triplication events (Wang et al., 2018).
Different factors, including interior cavities and protein surface pockets, can affect enzyme activity and DNA–protein interactions (Stank et al., 2016). Prediction of the potential binding sites of proteins can be useful in determining the interaction of proteins and how they are activated (Ahmadizadeh et al., 2020; Faraji et al., 2020). The amino acid residues present in the pocket sites of TIFY proteins partially differ in each subfamily, although the Mg or Ca ions heterogeneously exist in the center of the active sites of all of the predicted TIFY protein models. The SER, GLY, HIS, PRO, GLU, TYR, and ARG amino acids were identified as the important binding residues in the predicted pocket sites of all types of TIFY proteins (Figure 8), which illustrates the potential roles of these proteins in coping with stimuli, in addition to growth and development adjustment, in plant species (Faraji et al., 2020). Proteins with high contents of GLY and PRO residuals play important roles in plants in response to abiotic and biotic stresses (Mousavi and Hotta, 2005). The SER and THR amino acids were predicted as key binding sites in the JAZ protein of tomato. SER, LEU, VAL, and PRO play significantly roles in adjusting the various functions in response to stress (Galili and Höfgen, 2002; Beauregard and Hefford, 2006). The presence of the CYS, VAL, and LYS residues as activating binding sites in almost all TIFY proteins revealed that these proteins may also be involved in sulfur metabolism (Yang et al., 2020). Our results revealed the key binding sites in the protein sequences of each subfamily of TIFY family proteins, which can be used to evaluate the exact function of these proteins. Our findings also indicated that the protein surface pockets in the TIFY family proteins are different in the studied monocot and dicot species and that these differences can affect their associated molecular pathways.
Adverse conditions, such as biotic and abiotic stresses, as limiting factors, affect plant performance. Previous studies revealed that members of the TIFY family, as specific plant TFs, play critical roles in regulating plant responses to adverse environmental conditions (Ebel et al., 2018; Chao et al., 2019; Cai et al., 2020). For instance, overexpression of apple JAZ2 could significantly improve the tolerance to P. syringae pv. tomato DC3000 in Arabidopsis (An et al., 2017). Ebel et al. (2018) found that TIFY genes in durum wheat are involved in the response to different stresses, and TIFY proteins may increase germination under salinity treatment. Regarding the results, most of the genes with a tendency to express in response to adverse conditions were from the JAZ subfamily, which may reveal the important roles of this subfamily in dealing with stimuli (Figures 9, 10). Cai et al. (2020) recently stated that the JAZ genes of tuber mustard are induced by pathogen Plasmodiophora brassicae and salt stress. JAZ9 in rice can interact with the bHLH062 TF to control salt tolerance via affecting the ion transporter genes (Wu et al., 2015). Similarly, in wheat, five JAZ genes were induced under salt stress (Wang et al., 2017). In G. hirsutum, 14 JAZ genes were induced in response to salinity treatment (Sun et al., 2017). The overexpression of maize JAZ14 in Arabidopsis could increase seedling tolerance to hormone treatments, with ABA and JA, and polyethylene glycol stress (Zhou et al., 2015). Based on the expression profile of TIFY genes in both candidate plant species, maize and tomato, we think that JAZ subfamily genes are induced to a greater extent in the response to adverse conditions than other subfamilies. It appears that most genes belonging to this common family in both candidate species have similar expression patterns, indicating that the conserved regulatory mechanisms may control their expression.
Various stimuli responses are controlled by the genes’ transcriptional adjustment, which can be modulated by cis-elements present in the promoter area (Ahmadizadeh and Heidari, 2014; Heidari et al., 2019). In the present study, several kinds of cis-regulatory elements related to cell signaling, the response to biotic and abiotic stresses, and hormone signaling were distinguished in the promoter region of TIFY genes. The presence of light-responsive elements, especially G-Box, indicates that light signals can significantly adjust the TIFY genes’ transcription, which eventually regulates the genes engaged in defensive lines such as flavonoid biosynthesis pathways (Biłas et al., 2016). The Box 4, ABRE, and MYB elements were frequently distinguished in the TIFY genes’ promoter area (Figure 6), suggesting the important roles of this gene family in dealing with stress in monocot and dicot crops. Protein–protein interactions can significantly modulate various cellular functions, such as the replication and transcriptional adjustment of DNA, growth and development, signaling processes, and the coordination of multiple metabolic systems (Fukao, 2012). According to protein–protein interactions, TIFY members in maize can interact with genes involved in DNA replication and the cellular response to DNA damage, whereas TIFY family members in tomato showed significant relationships with TF MYCs and salt-responsive proteins (Figure 7). The activities of the JAZ proteins are associated with jasmonate responses, which suppress the jasmonate signals by interacting with the TFs including MYC2 and MYC3, which control the expression of downstream genes (Yan et al., 2007; Melotto et al., 2008; Bai et al., 2011). According to the results of protein–protein interactions and promoter analysis, we think that TIFY genes play critical roles in cell reproduction, plant growth, and dealing with stress conditions.
Conclusion
In the current study, the TIFY gene family was compared between tomato (as a dicot) and maize (as a monocot) based on sequence, structure, evolutionary, expression, interaction network, and cis-regulatory elements. We identified 48 and 26 nonredundant putative TIFY genes in the genome of maize and tomato, respectively. The identified TIFYs were classified into four subfamilies (JAZ, TIFY-S, PPD, and ZML); PPD subfamily proteins were only detected in dicots. Our results revealed that all of the duplicated pairs in the TIFY gene family of tomato have been influenced by intense purifying selection. The amino acid residues present in the pocket sites of TIFY proteins partially differ in each subfamily, indicating that these proteins have different activities based on their ligand-binding sites. Based on the expression profile of TIFY genes, we found that JAZ subfamily proteins are more involved in the response to stress than other subfamilies. Key cis-regulatory elements were observed in the promoter site of TIFY genes, indicating that the TIFY gene family, a group of plant-specific TFs, is induced by various stimulus. Our findings demonstrate that the TIFY gene family plays important roles in regulating growth and development processes and inducing cell signaling in response to stress. Therefore, the results of this study can be used in future research related to the functional genomics of TIFY genes.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://plants.ensembl.org.
Author Contributions
PH and SF: conceptualization. SF: methodology. PH, SF, and MA: software and writing—original draft preparation. PH, MA, SA, and FM-P: writing—review and editing. FM-P: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
FM-P thanks the Chilean National Fund for Scientific and Technological Development (FONDECYT), Grant No. 1201973.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.657970/full#supplementary-material
Footnotes
- ^ http://smart.embl-heidelberg.de/webcite
- ^ http://pfam.sanger.ac.uk
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ https://web.expasy.org/protparam/
- ^ https://www.genome.jp/tools-bin/clustalw
- ^ http://meme-suite.org/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://stringdb.org
- ^ http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi
References
Ahmadizadeh, M., and Heidari, P. (2014). Bioinformatics study of transcription factors involved in cold stress. Biharean. Biol. 8, 83–86.
Ahmadizadeh, M., Rezaee, S., and Heidari, P. (2020). Genome-wide characterization and expression analysis of fatty acid desaturase gene family in Camelina sativa. Gene Rep. 21:100894. doi: 10.1016/j.genrep.2020.100894
An, X.-H., Hao, Y.-J., Li, E.-M., Xu, K., and Cheng, C.-G. (2017). Functional identification of apple MdJAZ2 in Arabidopsis with reduced JA-sensitivity and increased stress tolerance. Plant Cell Rep. 36, 255–265. doi: 10.1007/s00299-016-2077-9
Aparicio-Fabre, R., Guillén, G., Loredo, M., Arellano, J., Valdés-López, O., Ramírez, M., et al. (2013). Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol. 13:26. doi: 10.1186/1471-2229-13-26
Bai, Y., Meng, Y., Huang, D., Qi, Y., and Chen, M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98, 128–136. doi: 10.1016/j.ygeno.2011.05.002
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208.
Beauregard, M., and Hefford, M. A. (2006). Enhancement of essential amino acid contents in crops by genetic engineering and protein design. Plant Biotechnol. J. 4, 561–574.
Bhattacharya, D., Nowotny, J., Cao, R., and Cheng, J. (2016). 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res. 44, W406–W409.
Biłas, R., Szafran, K., Hnatuszko-Konka, K., and Kononowicz, A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ. Cult. 127, 269–287. doi: 10.1007/s11240-016-1057-7
Bolser, D. M., Staines, D. M., Perry, E., and Kersey, P. J. (2017). “Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomic data,” in Plant Genomics Databases, ed. A. D. J. van Dijk (New York, NY: Springer), 1–31. doi: 10.1007/978-1-4939-6658-5_1
Boter, M., Golz, J. F., Giménez-Ibañez, S., Fernandez-Barbero, G., Franco-Zorrilla, J. M., and Solano, R. (2015). FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27, 3160–3174. doi: 10.1105/tpc.15.00220
Cai, Q., Yuan, Z., Chen, M., Yin, C., Luo, Z., Zhao, X., et al. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 1–13.
Cai, Z., Chen, Y., Liao, J., and Wang, D. (2020). Genome-wide identification and expression analysis of jasmonate ZIM domain gene family in tuber mustard (Brassica juncea var. tumida). PLoS One 15:e0234738. doi: 10.1371/journal.pone.0234738
Chao, J., Zhao, Y., Jin, J., Wu, S., Deng, X., Chen, Y., et al. (2019). Genome-wide identification and characterization of the JAZ gene family in rubber tree (Hevea brasiliensis). Front. Genet. 10:372. doi: 10.3389/fgene.2019.00372
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools-an integrative toolkit developed for interactive analyses of big biological data. bioRxiv[Preprint] doi: 10.1016/j.molp.2020.06.009 bioRxiv: 289660,
Chini, A., Ben-Romdhane, W., Hassairi, A., and Aboul-Soud, M. A. M. (2017). Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS One 12:e0177381. doi: 10.1371/journal.pone.0177381
Chung, H. S., and Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145. doi: 10.1105/tpc.108.064097
Chung, H. S., Niu, Y., Browse, J., and Howe, G. A. (2009). Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70, 1547–1559. doi: 10.1016/j.phytochem.2009.08.022
DeLano, W. L. (2002). Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92.
Demianski, A. J., Chung, K. M., and Kunkel, B. N. (2012). Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 13, 46–57. doi: 10.1111/j.1364-3703.2011.00727.x
Ebel, C., BenFeki, A., Hanin, M., Solano, R., and Chini, A. (2018). Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum durum Td TIFY11a in salt stress tolerance. PLoS One 13:e0200566. doi: 10.1371/journal.pone.0200566
Faraji, S., Filiz, E., Kazemitabar, S. K., Vannozzi, A., Palumbo, F., Barcaccia, G., et al. (2020). The AP2/ERF gene family in Triticum durum: genome-wide identification and expression analysis under drought and salinity stresses. Genes (Basel) 11:1464. doi: 10.3390/genes11121464
Fei, Z., Joung, J.-G., Tang, X., Zheng, Y., Huang, M., Lee, J. M., et al. (2010). Tomato functional genomics database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 39, D1156–D1163.
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222.
Forestan, C., Cigliano, R. A., Farinati, S., Lunardon, A., Sanseverino, W., and Varotto, S. (2016). Stress-induced and epigenetic-mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Sci. Rep. 6, 1–20.
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Galili, G., and Höfgen, R. (2002). Metabolic engineering of amino acids and storage proteins in plants. Metab. Eng. 4, 3–11. doi: 10.1006/mben.2001.0203
Garrido-Bigotes, A., Valenzuela-Riffo, F., and Figueroa, C. R. (2019). Evolutionary analysis of JAZ proteins in plants: an approach in search of the ancestral sequence. Int. J. Mol. Sci. 20:5060. doi: 10.3390/ijms20205060
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186.
Hakata, M., Kuroda, M., Ohsumi, A., Hirose, T., Nakamura, H., Muramatsu, M., et al. (2012). Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 76, 2129–2134. doi: 10.1271/bbb.120545
Hall, T. A. (1999). “BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT,” in Proceedings of the Nucleic Acids Symposium Series, c1979-c2000, Vol. 41, (London: Information Retrieval Ltd), 95–98.
Heidari, P., Ahmadizadeh, M., Izanlo, F., and Nussbaumer, T. (2019). In silico study of the CESA and CSL gene family in Arabidopsis thaliana and Oryza sativa: focus on post-translation modifications. Plant Gene 19:100189. doi: 10.1016/j.plgene.2019.100189
Heidari, P., Mazloomi, F., Nussbaumer, T., and Barcaccia, G. (2020). Insights into the SAM synthetase gene family and its roles in tomato seedlings under abiotic stresses and hormone treatments. Plants 9:586. doi: 10.3390/plants9050586
Hu, H., He, X., Tu, L., Zhu, L., Zhu, S., Ge, Z., et al. (2016). Gh JAZ 2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor Gh MYB 25-like. Plant J. 88, 921–935. doi: 10.1111/tpj.13273
Jendele, L., Krivak, R., Skoda, P., Novotny, M., and Hoksza, D. (2019). PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 47, W345–W349.
Jing, Y., Liu, J., Liu, P., Ming, D., and Sun, J. (2019). Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 9, 1–15.
Juretic, N., Hoen, D. R., Huynh, M. L., Harrison, P. M., and Bureau, T. E. (2005). The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15, 1292–1297. doi: 10.1101/gr.4064205
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., and Howe, G. A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci.U.S.A. 105, 7100–7105. doi: 10.1073/pnas.0802332105
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, X., Yin, X., Wang, H., Li, J., Guo, C., Gao, H., et al. (2015). Genome-wide identification and analysis of the apple (Malus × domestica Borkh.) TIFY gene family. Tree Genet. Genomes 11:808.
Liu, X., Zhao, C., Yang, L., Zhang, Y., Wang, Y., Fang, Z., et al. (2020). Genome-wide identification, expression profile of the TIFY gene family in Brassica oleracea var. capitata, and their divergent response to various pathogen infections and phytohormone treatments. Genes 11:127.
Lovell, S. C., Davis, I. W., Arendall, W. B. III, De Bakker, P. I. W., Word, J. M., Prisant, M. G., et al. (2003). Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct. Funct. Bioinformatics 50, 437–450.
Ma, Y., Shu, S., Bai, S., Tao, R., Qian, M., and Teng, Y. (2018). Genome-wide survey and analysis of the TIFY gene family and its potential role in anthocyanin synthesis in Chinese sand pear (Pyrus pyrifolia). Tree Genet. genomes 14:25.
Melotto, M., Mecey, C., Niu, Y., Chung, H. S., Katsir, L., Yao, J., et al. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988. doi: 10.1111/j.1365-313x.2008.03566.x
Mousavi, A., and Hotta, Y. (2005). Glycine-rich proteins. Appl. Biochem. Biotechnol. 120, 169–174. doi: 10.1385/abab:120:3:169
Nishii, A., Takemura, M., Fujita, H., Shikata, M., Yokota, A., and Kohchi, T. (2000). Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 64, 1402–1409. doi: 10.1271/bbb.64.1402
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Pérez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Peethambaran, P. K., Glenz, R., Höninger, S., Islam, S. M. S., Hummel, S., Harter, K., et al. (2018). Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 18:311. doi: 10.1186/s12870-018-1521-0
Pei, T., Ma, P., Ding, K., Liu, S., Jia, Y., Ru, M., et al. (2018). SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 69, 1663–1678. doi: 10.1093/jxb/erx484
Portwood, J. L., Woodhouse, M. R., Cannon, E. K., Gardiner, J. M., Harper, L. C., Schaeffer, M. L., et al. (2019). MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res. 47, D1146–D1154.
Rezaee, S., Ahmadizadeh, M., and Heidari, P. (2020). Genome-wide characterization, expression profiling, and post-transcriptional study of GASA gene family. Gene Rep. 20:100795. doi: 10.1016/j.genrep.2020.100795
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Saha, G., Park, J.-I., Kayum, M., and Nou, I.-S. (2016). A genome-wide analysis reveals stress and hormone responsive patterns of TIFY family genes in Brassica rapa. Front. Plant Sci. 7:936. doi: 0.3389/fpls.2016.00936
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., and Bork, P. (2000). SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. doi: 10.1093/nar/28.1.231
Shikata, M., Matsuda, Y., Ando, K., Nishii, A., Takemura, M., Yokota, A., et al. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55, 631–639. doi: 10.1093/jxb/erh078
Sirhindi, G., Sharma, P., Arya, P., Goel, P., Kumar, G., Acharya, V., et al. (2016). Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea (Cajanus cajan (L.) Millsp.) under copper stress. J. Plant Biochem. Biotechnol. 25, 301–310. doi: 10.1007/s13562-015-0342-6
Stank, A., Kokh, D. B., Fuller, J. C., and Wade, R. C. (2016). Protein binding pocket dynamics. Acc. Chem. Res. 49, 809–815.
Staswick, P. E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13, 66–71. doi: 10.1016/j.tplants.2007.11.011
Stelpflug, S. C., Sekhon, R. S., Vaillancourt, B., Hirsch, C. N., Buell, C. R., de Leon, N., et al. (2016). An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9, 1–16.
Sun, H., Chen, L., Li, J., Hu, M., Ullah, A., He, X., et al. (2017). The JASMONATE ZIM-domain gene family mediates JA signaling and stress response in cotton. Plant Cell Physiol. 58, 2139–2154. doi: 10.1093/pcp/pcx148
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Taniguchi, S., Hosokawa-Shinonaga, Y., Tamaoki, D., Yamada, S., Akimitsu, K., and Gomi, K. (2014). Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant. Cell Environ. 37, 451–461. doi: 10.1111/pce.12169
Tian, W., Chen, C., Lei, X., Zhao, J., and Liang, J. (2018). CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367.
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The TIFY family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Vélez-Bermúdez, I.-C., Salazar-Henao, J. E., Fornalé, S., López-Vidriero, I., Franco-Zorrilla, J.-M., Grotewold, E., et al. (2015). A MYB/ZML complex regulates wound-induced lignin genes in maize. Plant Cell 27, 3245–3259. doi: 10.1105/tpc.15.00545
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, H., Leng, X., Xu, X., and Li, C. (2020). Comprehensive analysis of the TIFY gene family and its expression profiles under phytohormone treatment and abiotic stresses in roots of Populus trichocarpa. Forests 11:315. doi: 10.3390/f11030315
Wang, N., Xiang, Y., Fang, L., Wang, Y., Xin, H., and Li, S. (2013). Patterns of gene duplication and their contribution to expansion of gene families in grapevine. Plant Mol. Biol. Rep. 31, 852–861. doi: 10.1007/s11105-013-0556-5
Wang, P., Su, L., Gao, H., Jiang, X., Wu, X., Li, Y., et al. (2018). Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 9:64. doi: 10.3389/fpls.2018.00064
Wang, Y., Qiao, L., Bai, J., Wang, P., Duan, W., Yuan, S., et al. (2017). Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genomics 18:152. doi: 10.1186/s12864-017-3582-0
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Waters, A. J., Makarevitch, I., Noshay, J., Burghardt, L. T., Hirsch, C. N., Hirsch, C. D., et al. (2017). Natural variation for gene expression responses to abiotic stress in maize. Plant J. 89, 706–717. doi: 10.1111/tpj.13414
White, D. W. R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci.U.S.A. 103, 13238–13243. doi: 10.1073/pnas.0604349103
Wu, H., Ye, H., Yao, R., Zhang, T., and Xiong, L. (2015). OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 232, 1–12. doi: 10.1016/j.plantsci.2014.12.010
Xia, W., Yu, H., Cao, P., Luo, J., and Wang, N. (2017). Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Front. Plant Sci. 8:493. doi: 10.3389/fpls.2017.00493
Yan, Y., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Yang, Q., Zhao, D., and Liu, Q. (2020). Connections between amino acid metabolisms in plants: lysine as an example. Front. Plant Sci. 11:928. doi: 10.3389/fpls.2020.00928
Yang, S., Zhang, X., Yue, J.-X., Tian, D., and Chen, J.-Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280, 187–198. doi: 10.1007/s00438-008-0355-0
Yang, Y., Ahammed, G. J., Wan, C., Liu, H., Chen, R., and Zhou, Y. (2019). Comprehensive analysis of TIFY transcription factors and their expression profiles under jasmonic acid and abiotic stresses in watermelon. Int. J. Genomics 2019:6813086.
Ye, H., Du, H., Tang, N., Li, X., and Xiong, L. (2009). Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71, 291–305. doi: 10.1007/s11103-009-9524-8
Yu, C., Chen, Y., Lu, C., and Hwang, J. (2006). Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinformatics 64, 643–651.
Yu, C.-S., Cheng, C.-W., Su, W.-C., Chang, K.-C., Huang, S.-W., Hwang, J.-K., et al. (2014). CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One 9:e99368. doi: 10.1371/journal.pone.0099368
Yu, X., Chen, G., Tang, B., Zhang, J., Zhou, S., and Hu, Z. (2018). The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 267, 65–73. doi: 10.1016/j.plantsci.2017.11.008
Zhang, L., You, J., and Chan, Z. (2015). Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 128, 995–1005. doi: 10.1007/s10265-015-0755-2
Zhang, Y., Gao, M., Singer, S. D., Fei, Z., Wang, H., and Wang, X. (2012). Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One 7:e44465. doi: 10.1371/journal.pone.0044465
Zhang, Z., Li, X., Yu, R., Han, M., and Wu, Z. (2015). Isolation, structural analysis, and expression characteristics of the maize TIFY gene family. Mol. Genet. Genomics 290, 1849–1858. doi: 10.1007/s00438-015-1042-6
Zhao, G., Song, Y., Wang, C., Butt, H. I., Wang, Q., Zhang, C., et al. (2016). Genome-wide identification and functional analysis of the TIFY gene family in response to drought in cotton. Mol. Genet. Genomics 291, 2173–2187. doi: 10.1007/s00438-016-1248-2
Zheng, L., Ying, Y., Wang, L., Wang, F., Whelan, J., and Shou, H. (2010). Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 10:166. doi: 10.1186/1471-2229-10-166
Zhou, X., Yan, S., Sun, C., Li, S., Li, J., Xu, M., et al. (2015). A maize jasmonate Zim-domain protein, ZmJAZ14, associates with the JA, ABA, and GA signaling pathways in transgenic Arabidopsis. PLoS One 10:e0121824. doi: 10.1371/journal.pone.0121824
Zhu, D., Bai, X., Luo, X., Chen, Q., Cai, H., Ji, W., et al. (2013). Identification of wild soybean (Glycine soja) TIFY family genes and their expression profiling analysis under bicarbonate stress. Plant Cell Rep. 32, 263–272. doi: 10.1007/s00299-012-1360-7
Keywords: JAZ genes, ZIM subfamily, expression profile, in silico study, phylogenetic analysis
Citation: Heidari P, Faraji S, Ahmadizadeh M, Ahmar S and Mora-Poblete F (2021) New Insights Into Structure and Function of TIFY Genes in Zea mays and Solanum lycopersicum: A Genome-Wide Comprehensive Analysis. Front. Genet. 12:657970. doi: 10.3389/fgene.2021.657970
Received: 24 January 2021; Accepted: 22 March 2021;
Published: 12 May 2021.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Monther Sadder, The University of Jordan, JordanArif Hasan Khan Robin, Bangladesh Agricultural University, Bangladesh
Min Gao, University of Maryland, Baltimore, United States
Copyright © 2021 Heidari, Faraji, Ahmadizadeh, Ahmar and Mora-Poblete. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Parviz Heidari, heidarip@shahroodut.ac.ir; Freddy Mora-Poblete, morapoblete@gmail.com
 Parviz Heidari
Parviz Heidari