- College of Life Sciences, Shanxi Normal University, Taiyuan, China
TGA is one of the members of TGACG sequence-specific binding protein family, which plays a crucial role in the regulated course of hormone synthesis as a stress-responsive transcription factor (TF). Little is known, however, about its implication in response to bacterial wilt disease in potato (Solanum tuberosum) caused by Ralstonia solanacearum. Here, we performed an in silico identification and analysis of the members of the TGA family based on the whole genome data of potato. In total, 42 StTGAs were predicted to be distributed on four chromosomes in potato genome. Phylogenetic analysis showed that the proteins of StTGAs could be divided into six sub-families. We found that many of these genes have more than one exon according to the conserved motif and gene structure analysis. The heat map inferred that StTGAs are generally expressed in different tissues which are at different stages of development. Genomic collinear analysis showed that there are homologous relationships among potato, tomato, pepper, Arabidopsis, and tobacco TGA genes. Cis-element in silico analysis predicted that there may be many cis-acting elements related to abiotic and biotic stress upstream of StTGA promoter including plant hormone response elements. A representative member StTGA39 was selected to investigate the potential function of the StTGA genes for further analysis. Quantitative real-time polymerase chain reaction (qRT-PCR) assays indicated that the expression of the StTGAs was significantly induced by R. solanacearum infection and upregulated by exogenous salicylic acid (SA), abscisic acid (ABA), gibberellin 3 (GA3), and methyl jasmonate (MeJA). The results of yeast one-hybrid (Y1H) assay showed that StTGA39 regulates S. tuberosum BRI1-associated receptor kinase 1 (StBAK1) expression. Thus, our study provides a theoretical basis for further research of the molecular mechanism of the StTGA gene of potato tolerance to bacterial wilt.
Introduction
Plants face a variety of challenges and stresses during growth and development. Specific transcription factors (TF) in plants can specifically regulate the expression of plant genes to enhance the ability of plants to adapt to stress environment. In plants, the gene regulatory network which TFs participate in plays a crucial role in the stress response pathway (Li B. et al., 2019). The members of the basic Region-Leucine Zipper (bZIP) gene family are crucial regulators, which play an important role in development, stress adaptation, and hormone synthesis of plants (Azeem et al., 2020; Kumar et al., 2021). The TGACG sequence-specific binding protein family (TGA) TFs belong to a subfamily of bZIP and play an important role in abiotic stress responses during the period of plant growth (Ke et al., 2022). The TGA TFs include large regulatory regions, which may affect DNA-binding by allosteric or electrostatic interaction as for the bZIP (Salladini et al., 2020).
Some TGA members play crucial roles in stress mitigation. Evidence revealed that the role of TGA class II is important during the response of Arabidopsis to control reactive oxygen species (ROS) levels (Ariel et al., 2021). In addition, there was evidence that Cr6+ can cause the binding of TGA3 to the L-cysteine desulfhydrase (LCD) promoter in plants, thus increasing the expression of LCD (Fang et al., 2017). Furthermore, glutaredoxins (GRXs) may play a crucial role in the formation of floral organs with TGA (Rouhier et al., 2015). ROXYs (GRX,CC-type glutaredoxin, named ROXYs in Arabidopsis thaliana) and CC-type GRXs from rice and corn interact with the TGA family to control developmental processes (Gutsche et al., 2017; Uhrig et al., 2017).
An important role of TGAs during plant development has been covered in a variety of plants. It can be seen that the TGA gene family participates in many signaling pathways in plants and plays a crucial role during plant growth. In soybean, the TGA plays a crucial role in response to nitrogen availability (Ullah et al., 2019). Biosynthesis of salicylic acid (SA) is indirectly modulated by TGA1 and TGA4 TF (Budimir, et al., 2020). Nonexpresser of pathogenesis-related genes (NPR1) recruits TGA TFs in the presence of SA and facilitates gene expression to establish plant immunity (Chen et al., 2019). Multiple TGA members have been shown to play a key role in plant immunity. Oryza sativa TGA2 (OsTGA2) can directly regulate defense-related genes (Moon et al., 2018). The key points in the regulation of sunflower resistance by TGA may be involved in the resistance of sunflower to Verticillium dahliae (Guo et al., 2017). In addition, ROXY can be used as TGA-dependent promoter to control the negative regulators of detoxification genes in A. thaliana (Huang et al., 2016). Brassinoidsteroid (BR) induces apoplastic ROS, which activates the TGA2 factor and triggers the metabolism of pesticide residues in tomato (Hou et al., 2019). Taken together, studies of TGA TFs and the TGA family as a whole demonstrate that TGA proteins play important roles in regulating multiple biological processes in plants.
Although there are many studies on TGA, no study has been reported in potato (Solanum tuberosum), which is an important food crop and widely cultivated and consumed all over the world (Gebhardt, 2016). There are many kinds of major diseases affecting potato production worldwide, and bacterial wilt caused by Ralstonia solanacearum is the second one in importance (Barchenger et al., 2022). R. solanacearum is a soil-borne, devastating plant pathogen and uses type III effectors to inhibit the plant immune system (Salanoubat et al., 2002; Qi et al., 2022). This pathogen has a remarkably wide host range and global distribution and is involved in the invasion of plant root to the vessel of xylem and may eventually lead to plant death (Gutsche and Zachgo, 2016; Ferreira et al., 2017).
In the current study, a comprehensive investigation of TGA TFs in potato was conducted, and the analysis of the member distribution, evolutionary model, gene structure, and expression patterns was performed. In addition, we found a representative member S. tuberosum TGA39 (StTGA39) that could be induced by abiotic/biotic stress and has a critical role in enhanced bacterial wilt tolerance in potato during R. solanacearum infection. This will lay a foundation for further research on the function of StTGA genes.
Materials and Methods
Identification and Classification of StTGA Gene Family
StTGA gene identification in the potato genome was performed using BLAST and hidden Markov models (HMM) search methods (Wang et al., 2018). We downloaded the potato genome from the Phytozome database. Briefly, the HMM seed file of delay of germination 1 (DOG1) domain (accession number: PF14144), which belongs to TGA, was downloaded from the Pfam database (http://pfam.xfam.org/). A round of HMM scan was performed for all the obtained hits against the Pfam database. We used the HMMER program to search all putative TGA protein sequences and extracted the corresponding sequence IDs, with expectation value (E-value) set to 1.0. The sequences IDs were submitted to the Spud DB (http://solanaceae.plantbiology.msu.edu/). The results from the two methods were compared, and common sequence IDs were selected. In total, 42 members of the StTGA gene family in potato were predicted and named StTGA01–StTGA42. The information about the renaming StTGA genes in potato is provided in Supplementary Additional File S1. The physicochemical parameters, such as molecular weight and isoelectric points (pIs), and subcellular localization of each StTGA protein were predicted via ExPASy (http://web.expasy.org/protparam/) and PSORT (https://wolfpsort.hgc.jp/) online tools (Liang et al., 2017).
Phylogenetic Analysis, Conserved Motif Analysis, In Silico Chromosome Mapping, and Gene Structure Analysis of StTGA
We constructed the phylogenetic tree by MEGA 7.0 via the neighbor-joining (NJ) method, using 1000 bootstrap iterations and default parameters (Gao et al., 2018). The potato gff3. annotation file was parsed to extract the genome locations of the identified TGA genes. According to this information, we visualized the in silico predicted chromosomal distribution of StTGA genes by TBtools software (Chen et al., 2020). The in silico exon–intron distribution of StTGA genes was predicted through the Gene Structure Display Server (GSDS) website (http://gsds.gao-lab.org/index.php). Conserved motifs of StTGA genes were predicted by the online service Multiple Em for Motif Elicitation (MEME) (Fan et al., 2021a). The information of in silico chromosome location of StTGA was retrieved from potato genome data downloaded from the phytozome database (Chen et al., 2020). The secondary and 3D structure prediction of StTGA proteins was performed using SOPMA and I-TASSER web servers, and the data were captured using Discovery Studio 4.5 (Geourjon and Deléage, 1995; Ambrish et al., 2010). The upstream 2000 bp region of the initiation codon ATG of each StTGA gene was used to search the promoter region in the downloaded sequence and predicted potential cis-acting elements of each binding site on PlantCARE website (Lescot et al., 2002; Zhu et al., 2020).
Collinearity Analysis of StTGAs
Pair-wise all-against-all BLAST was performed for potato, tomato, tobacco, Arabidopsis, and pepper protein sequences. The obtained results and the gff3. annotation file were then determined by the Multiple Collinearity Scan toolkit (MCScanX) for determination of the gene duplication type (Wang et al., 2012; Fan et al., 2021b). Microsynteny relationships between potato and the other four species were analyzed to show the gene homology relationship (Gutsche and Zachgo, 2016), and visualized by TBtools (Chen et al., 2020).
RNA-Seq Analysis of StTGAs
Based on the digital expression RNA-Seq data retrieved by previous methods (Cao et al., 2021), the expression level of StTGAs in different tissues and developmental stages under abiotic/biotic stress was indicated using Fragments Per Kilobase of transcript sequence per Millions base pairs (FPKM), which were retrieved through the website Spud DB (http://solanaceae.plantbiology.msu.edu/) (Cao et al., 2021). The heatmaps of StTGA expression and hierarchical clustering analysis were conducted using the TBtools software.
Furthermore, the co-expression pattern of StTGA genes was analyzed based on the Pearson correlation coefficient (PCC) and graphically presented using the Cytoscape package. The PCC and mutual rank (MR) were calculated according to previous reports (Da et al., 2019). Forty-two potato StTGA differently expressed genes and 24 related genes were analyzed by co-expression network analysis (Renamed in Supplementary Additional File S2). The interaction network of differentially expressed TFs was built by STRING: functional protein association networks (https://string-db.org/) and determined by Cytoscape (Kohl et al., 2011; He et al., 2022). When the PCC was greater than 0.8, it was assumed that they were co-expressed.
Plant Material and Growth Conditions
Potato plants (ZHONG 3,2 n = 48, tetraploid cultivation) used in this study were provided by the Chinese Academy of Agricultural Sciences (CAAS). The pots which were used to culture these plants were 10 cm in diameter and 15 cm in height, and they contained 450 ml peat and vermiculite (3:1, volume ratio). In the process of culture, S. tuberosum (ZHONG 3) wild-type plants were grown under long-day conditions (16 h, 26°C, day/8 h, 18°C, night) on the abovementioned mixed soil. The light intensity was about 3000 lx, and the relative humidity was 75% (Gao et al., 2010; Yu et al., 2021a).
Bacterial Strains and Inoculum
R. solanacearum belongs to PO41 strain (seed type II, race 3 biotype 2) (He et al., 1983), which was cultured in a CPG medium, and then the solution was diluted to 108 cfu/ml (Optical Density, OD600 = 0.2). Potatoes were inoculated by the root injury irrigation method when they grew to the stage of two-week-old potato plants with 7–8 leaves (Yu et al., 2021b). The control group was inoculated with the same amount of water. At 12, 24, 36, 48, 60, 72, 84, 96, 108, and 120 hpi after inoculation, we sampled the leaves of the experimental group and control group. All samples were frozen in liquid nitrogen immediately and then stored at −80°C (Kong et al., 2016). Three replicates were performed for both R. solanacearum and mock inoculation.
Hormone Treatment
When potatoes grew to the stage of two weeks with 7–8 leaves, the leaves were sprayed with 100 μM abscisic acid (ABA), 350 μM, gibberellin 3 (GA3), 50 μM SA, and 50 μM methyl jasmonate (MeJA) (Yu et al., 2021a). At 1, 2, 3, 4, and 5 dpi, we sampled the leaves of each hormone treatment. All samples were frozen in liquid nitrogen immediately and then stored at −80°C until these samples were used to extract RNA (Fan et al., 2021a). Each treatment was repeated three times.
Verification of RNA-Seq Data
The RNA-seq data of the StTGA39 gene were verified by qRT-PCR according to the methods of Schmittgen and Livak, 2008. The leaf samples for RNA extraction were previously stored at −80°C, and the total RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. The mRNA was re-transcribed into cDNA using the Prime Script cDNA Synthesis Kit (TransGen, Beijing, China). Then, cDNA was used as a template for qRT-PCR, which was performed on the Quant Studio 3 qRT-PCR System (Thermo Fisher Scientific, Shanghai, China) (Kong et al., 2016). The following primers were used in the process of qRT-PCR with three replicates for each gene as described previously: StTGA-FP: TCCAGCACATCCAACACC; StTGA-RP: TTCACCAAGATTTCCCAC. ACTIN of potato served as the internal control (GenBank Accession: X55747), and the following primers were used in the process of qRT-PCR with three replicates for each gene as described previously: Actin-F: TATAACGAGCTTCGTGTTGCAC; Actin-R: ACTGGCATACAGCGAAAGAACA.
Staining Treatment and Quantifying Levels of Malondialdehyde
After the potato seedlings were inoculated with R. solanacearum or water for 72 hpi, the leaves were collected for observing the infection of R. solanacearum. The leaves were left in trypan blue solution for 24 h (Yu et al., 2021b). Levels of H2O2 were detected using the DAB staining method by the DAB Substrate Kit (Solarbio, Beijing, China). Leaves were soaked in the solution to stain for 3–10 min under dark conditions (Christensen et al., 1997; Song et al., 2021). The leaves were sampled from inoculated plants by R. solanacearum at 12, 24, 36, 48, 60, 72, 84, 96, 108, and 120 hpi for quantifying levels of MDA. The content of MDA of these leaves was tested by using an MDA Assay Kit (Abbkine, Wuhan, China) according to the previous method (Islamoglu et al., 2018). OD at 532 and 600 nm was determined by a microplate reader.
Y1H Assays
In order to analyze whether there is a TGACG cis-element in the upstream promoter sequence of the gene homologous to S. tuberosum Brassinosteroid insensitive 1 (BRI1)-associated receptor kinase 1 (StBAK1), we analyzed the relationship between StTGA1 and StBAKl by the Y1H assay Kit (Clontech, State of California, United States) in this study. Briefly, the sequence of StBAK1 with 1512 bp was amplified from S. tuberosum cDNA by gene-specific primers and digested by EcoRI (CAATTC) and MluI (ACGCGT) so that the oligonucleotide sequences containing TGA cis-elements and other corresponding promoter sequences were cloned into the pHIS2 to generate the reporter vector. All of the constructs were co-transformed into yeast strain Y187 according to the manufacturer’s instructions (Liu et al., 2016). The full-length open-reading frame (ORF) of StTGA (855bp) was fused to the GAL4 activation domain in the vector pGADT7 digested with EcoRI (CAATTC) and BamHI (GGATCC) to get a fusion protein TGA-pGADT7 (effector vector) (Huang et al., 2010). The others were carried out following the manufacturer’s instructions.
Statistical Analysis
Standard values and standard error of experimental data were calculated using Microsoft Excel 2016 and n = 3 for independent experiments according to a t-test. Analyses of the significance of differences were conducted via the data processing System. The PCC is calculated by the SPSS online services tool (https://spssau.com/indexs.html). PCR efficiency was estimated from the data obtained from the exponential phase of each individual amplification plot. The expression level of each gene of interest (GOI) is presented as 2−∆∆Ct; where ∆∆Ct = ∆CtGOI-∆CtContorl; ∆CtGOI = CtGOI-CtActin; ∆CtContorl = CtControl-CtActin.
Results
Genome-Wide Identification of StTGA Genes
In total, 42 StTGA genes were identified in S. tuberosum, and they were named StTGA01–StTGA42. This study revealed that the largest number of amino acids is 488 in the StTGA gene family, and the relative molecular weight of these genes is between 17,290.85 and 54,371.83 Da. In addition, we predicted the pIs of StTGA protein. The results showed that the pI of StTGA17 is 4.96, which is the lowest in the StTGA protein, while the pI of StTGA01 is 9.1, which is the highest. Furthermore, we speculated the StTGA may be distributed in many parts of the cell and most of StTGA proteins are located in cytoplasm and nucleus (Supplementary Table S1).
In Silico Chromosomal Location, Phylogenetic Relationship, Exon–Intron Structure, and Conserved Motif Analysis
Using TBtools, we drew the location map of StTGA and found they are distributed on four chromosomes of potato (Figure 1A). Chromosome St12 had 29 StTGAs, chromosome St08 contains six, St01 with four, and St09 contain three StTGAs. In order to study the StTGA family, we constructed the phylogenetic tree by MEGA 7 via the NJ method, which is shown in Figure 1B. The StTGA family was divided into six groups (designated classes I to VI) with 9, 7, 3, 12, 4, and 7 StTGAs. Then, the exon–intron structure of StTGAs was further analyzed. The number of exons of StTGA genes was between 1 and 11, of which six genes contained only one exon, and 42.86% of genes had five exons or less Figure 1D. Furthermore, 10 high conserved motifs were predicted in Figure 1C and Figure 1E. Compared to other classes, motifs 10 were exclusively found in classes IV, motifs 9 in classes I and II, and motifs 8 in classes IV and VI, separately Figure 1C.
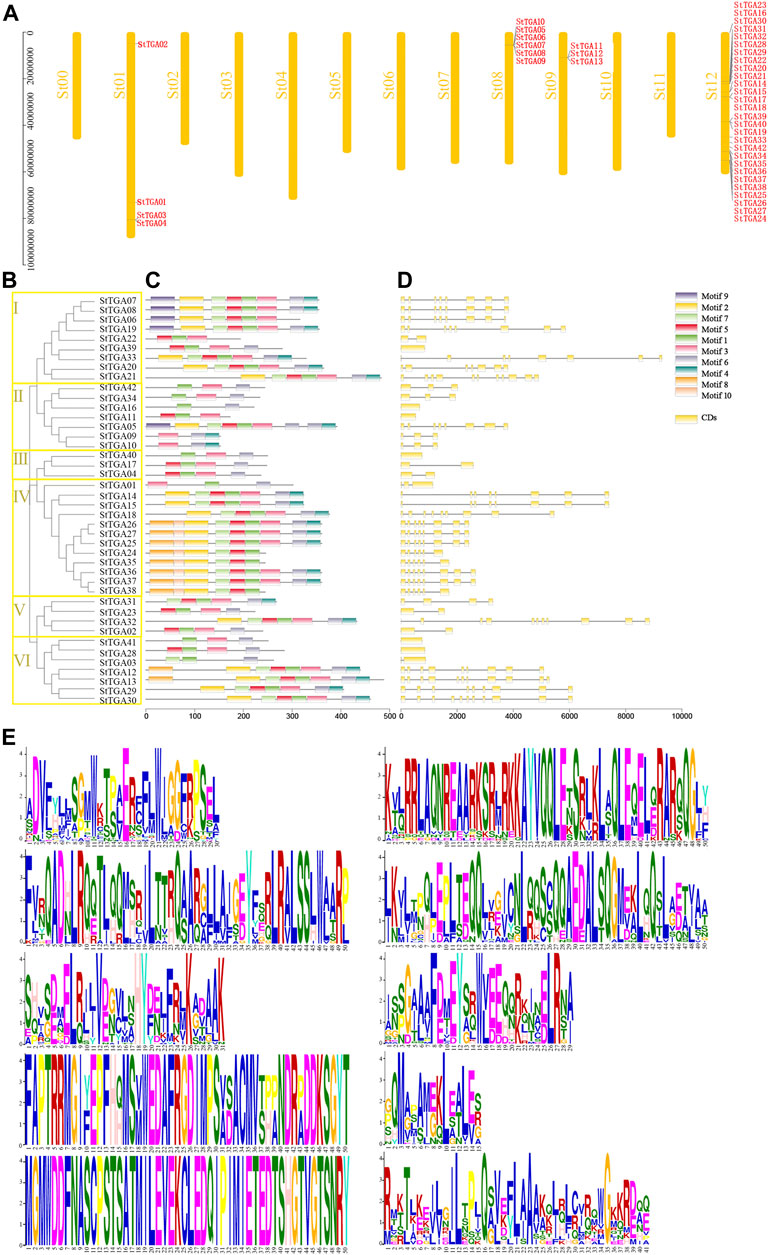
FIGURE 1. Chromosomal location, phylogenetic relationship, exon–intron structure, and conserved motif analysis of StTGA protein. (A) Chromosomal locations of StTGA genes. The gene names of StTGA are shown in red to the right of the chromosome. (B) Phylogenetic tree of potato StTGA based on its amino acid sequence. Branch lines with different colors indicated different subgroups. The proteins on the tree can be divided into six distinct subfamilies, which are indicated by different colored backgrounds. (C) Conserved motif of StTGA proteins analyzed by online program MEME server. Different colored boxes indicated different motifs. (D) Gene structure of StTGA. The exon–intron structure of StTGA genes visualized by online tool GSDS 2.0, yellow boxes indicated exons, and black lines indicated introns. (E) Amino acid composition of 10 conserved motifs.
Cis-Acting Element Prediction Analysis of StTGAs
In order to study the transcriptional regulation, the 2000 bp promoter region upstream of the StTGA genes was extensively analyzed by PlantCARE. The cis-elements responding to abiotic/biotic stresses and hormone treatments with the prediction score being greater than or equal to 5 were considered for further analysis (Figure 2). The results showed that the numbers and distribution patterns of the cis-elements also greatly varied among the promoters. This means that the StTGA genes might have different regulatory mechanisms in expression, implying their functional divergence in responses to abiotic/biotic stress and phytohormone treatments.
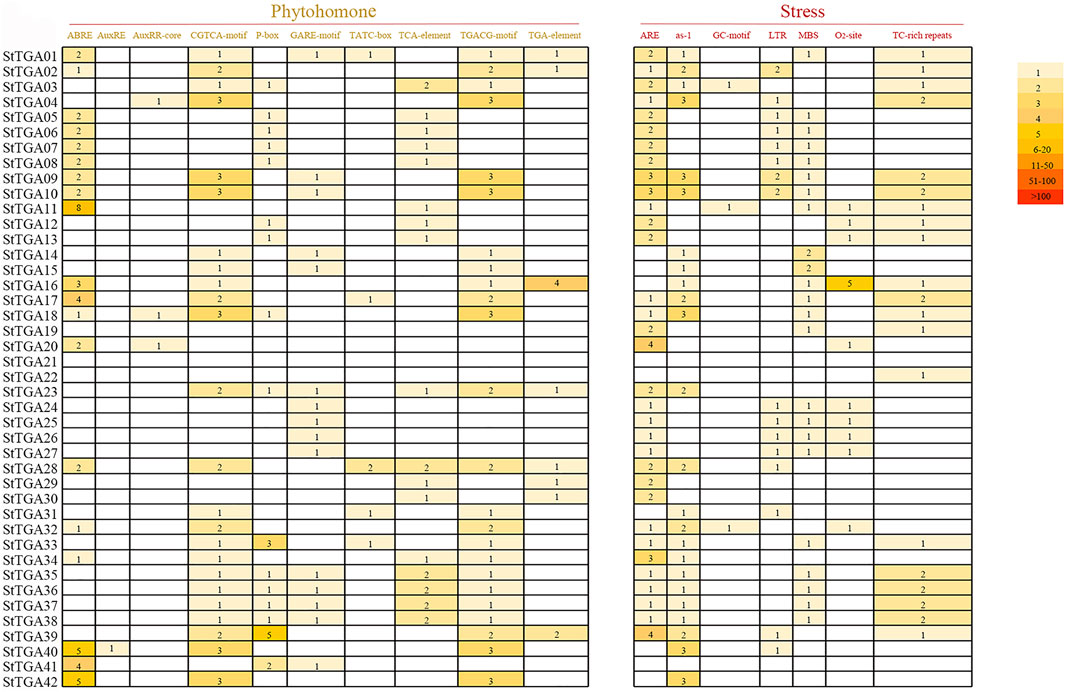
FIGURE 2. Cis-acting elements analysis in the promoter regions of StTGA. The promoter cis-elements were analyzed by PlantCARE, and the number of ciselements was searched in the 2,000 bp region upstream of the translation initiation site of the StTGA gene. Cis-elements were divided into two different types: phytohomone and stress. Expressed by different colors according to their number. The darker the color, the higher the frequency of appearance, and the number indicates the number of cis-elements.
Collinear Analysis of StTGAs
To study the evolutionary relationship of StTGA, we selected S. tuberosum as the core and identified collinearities between tomato, tobacco, Arabidopsis, pepper, and potato TGA genes. There are 46 pairs of one-to-one microsynteny relations (Figure 3). Some of the genes could not be assigned to any of the chromosomes. In addition, 21 orthologous gene pairs were detected between potato and tomato, 20 orthologous genes were found between potato and pepper, 10 orthologous gene pairs were identified between potato and Arabidopsis, and two orthologous gene pairs were detected between potato and tobacco. The collinear relationship among these five species is shown in Supplementary Additional File S3.
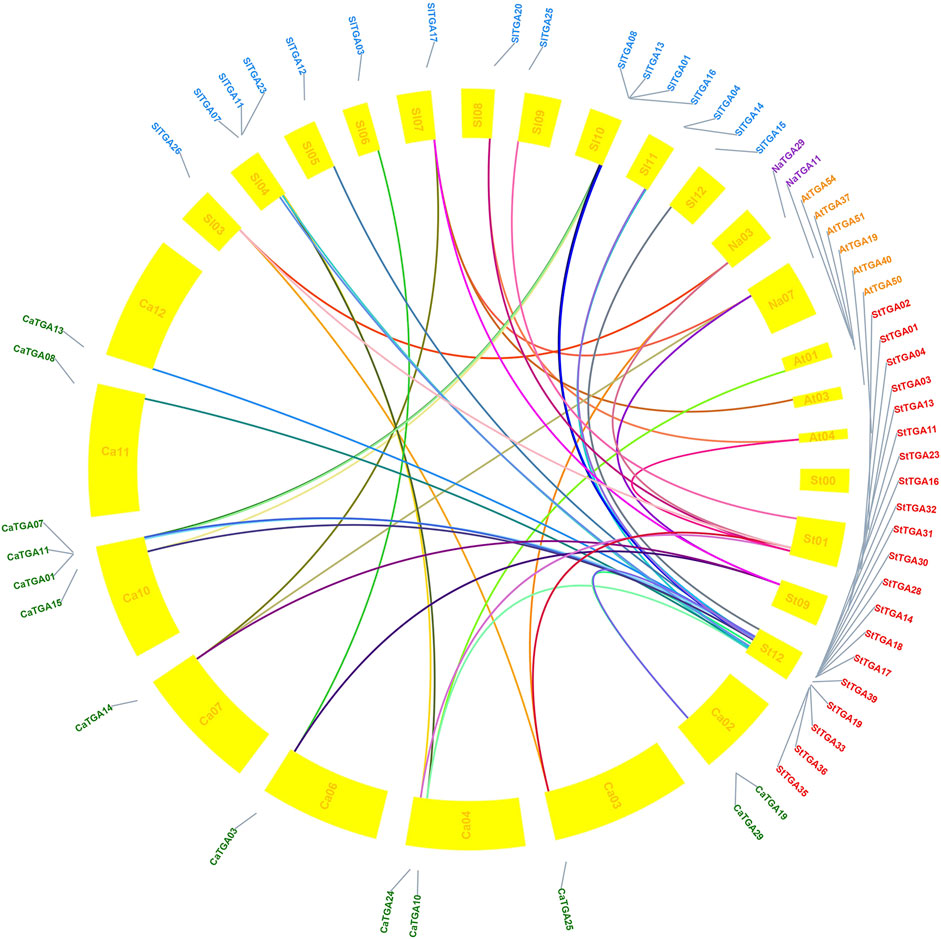
FIGURE 3. Comparative orthologous relationships of TGA from five species. TBtools was used to analyze the gene homology relationship between potato, tomato, pepper, Arabidopsis, and tobacco TGA gene families and visualize the gene homology relationship. TGA genes connecting five species genome are shown in colored links.
Expression Profile of StTGAs
We have retrieved the expression data of StTGA via the RNA-Seq Expression Browser and the heat map of gene FPKM value constructed using the TBtools software. We found that 11 of the StTGAs (StTGA 01, StTGA 02, StTGA03, StTGA 04, StTGA 11, StTGA 16, StTGA 23, StTGA 28, StTGA 34, StTGA 41, and StTGA42) were not expressed in almost all organs. On the contrary, StTGA05, StTGA15, StTGA25, StTGA32, and StTGA36 played roles in different developmental stages and tissues (Figure 4). For verification of RNA-seq data, the potato seedlings were treated with the exogenous hormones ABA, GA3, SA, and MeJA, and the results showed the inducible upregulated expression of the StTGA39 gene (Figure 5). By comparison, the effect of SA was relatively strong, while that of the MeJA, ABA, and GA3 was light for inducing the upregulation.
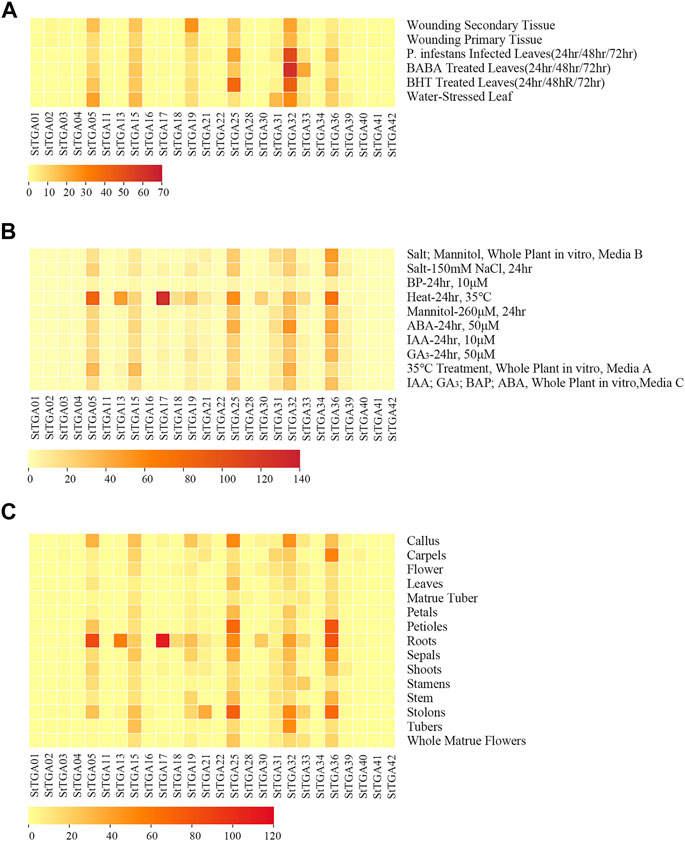
FIGURE 4. Heat map of StTGA genes under different stresses and hormone treatments in potato. The heat map displayed the StTGA expression patterns of treatment in leaves, whole plant, or in different tissues and organs. (A) In leaves, distance values range from 0.00 to 70.00, (B) in the whole plant, distance values range from 0.00 to 140.00, (C) in different tissues or organs indicated to the right of the heat map, distance values range from 0.00 to 120.00. The gene names are indicated at the bottom. Distance value range is depicted by the gradient of colors ranging from light yellow (lowest distance value indicating high similarity between genomes) to red (highest distance value indicating low similarity between genomes). Stress treatment in leaves is indicated to the right of the heat map.
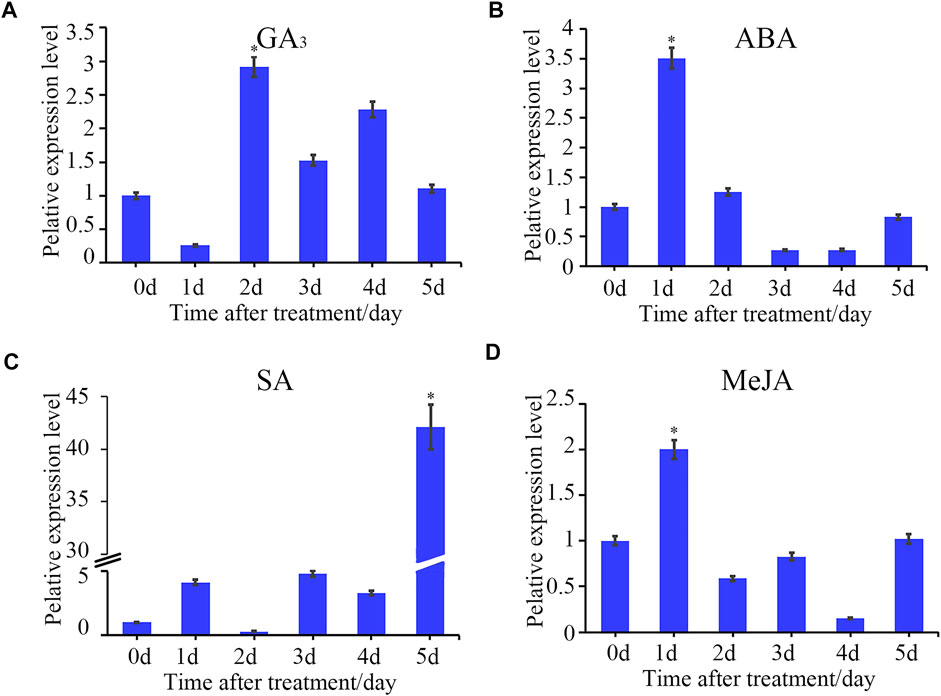
FIGURE 5. Time-course gene expression pattern of StTGA39 induced by exogenous phytohormones. The relative expression level of StTGA39 at different time points in potato inoculated with plant hormones (A) GA3, (B) ABA, (C) SA and (D) MeJA was analyzed by qRT-PCR. mRNA levels were normalized to actin (Student’s t-test, n = 3 independent experiments, data shown are mean ± standard deviation).
Co-Expression Network Construction
To determine the relationship of StTGA proteins and their interactors further, we performed a gene co-expression network analysis based on the STRING database. Twenty-four other potato proteins, such as 60S ribosomal protein L18 (RPLs), DNA-binding proteins (DBPs), and Glutaredoxin (GRXs), were identified Figure 6, which exhibited a co-expression relationship with StTGA (Figure 6A). The co-expression network comprised 36 pairs, and each pair of nodes was directly or indirectly connected to each component (Figure 6A). StTGA and StTGA members, StTGA and other potato proteins presented co-expression relationships, albeit with different weight values (Figure 6B). In particular, StTGA01 (StTGA28), StTGA02 (StTGA40), StTGA05 (StTGA13), etc., presented co-expression relationships (Figure 6A).
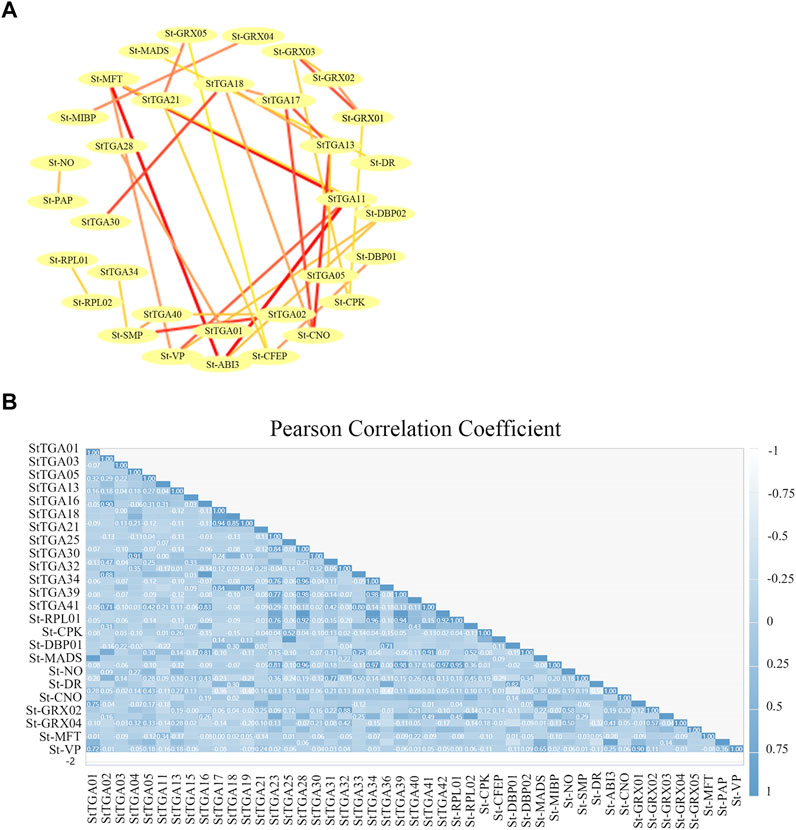
FIGURE 6. Construction of the StTGA co-expression network. (A) Co-expression network analyses are performed to characterize the 36 differentially expressed StTGA. In the network, nodes represented RNAs, while lines represented co-expression and prediction relationship. (B) PCC of all genes in each gene show in this figure. Pearson correlation matrix between all StTGAs. (positive correlations = dark blue with value, negative correlations = light blue, non-significant correlations = white).
Prediction of StTGA Structure
The secondary structure of StTGA39 was predicted by SOPMA. It was found that the a-helix of StTGA protein consists of 188 amino acids, accounting for 67.14%; the random coil of StTGA protein consists of 78 amino acids, accounting for 27.86%; the extended strand of StTGA protein consists of nine amino acids, accounting for 3.21%; and the β-turn of StTGA protein consists of five amino acids, accounting for 1.79% (Figure 7A). A three-dimensional structure of StTGA39 was built with the I-TASSER web server. Five possible models of StTGA protein were predicted on the 3D structure, according to the website modeling with Discovery Studio 4.5 Client. The conservative domains of these models were basically the same (Figure 7B).
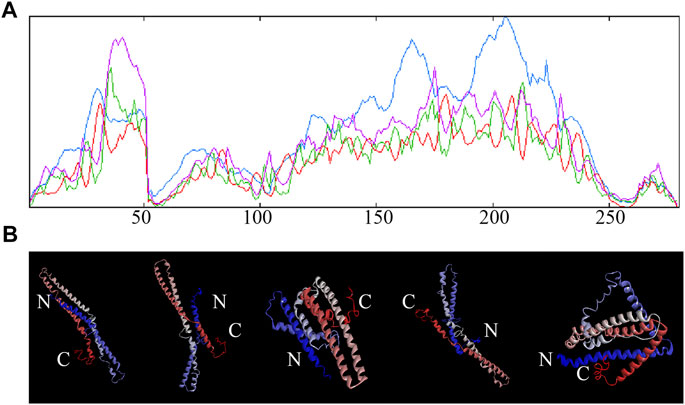
FIGURE 7. Secondary and tertiary structure of potato StTGA39 protein. (A) Secondary structure of StTGA39 was predicted by SOPMA in potato. Blue lines represent alpha-helix, purple lines represent random coil, red lines represent extended strand, and green lines represent β-turn. (B) Three-dimensional structure of StTGA39 was built with the I-TASSER web server.
Effect of R. solanacearum Infection on Potato
In order to confirm whether StTGA39 was induced by R. solanacearum, the induced expression of StTGA39 mRNA in potato seedlings inoculated with R. solanacearum was compared and analyzed by the qRT-PCR method (Figure 8A). We found that the expression level of StTGA39 was low when plants were inoculated with water. In comparison with the control group, the expression increased at 60 hpi and reached the highest level at 72 hpi after inoculating with the highly pathogenic strain PO41. That is to say, the expression of the StTGA39 gene was up-regulated when stressed by R. solanacearum and may play an important role in potato resistance to bacterial wilt. Similar results have been confirmed by other family members of StTGA (Supplementary Additional File S4).
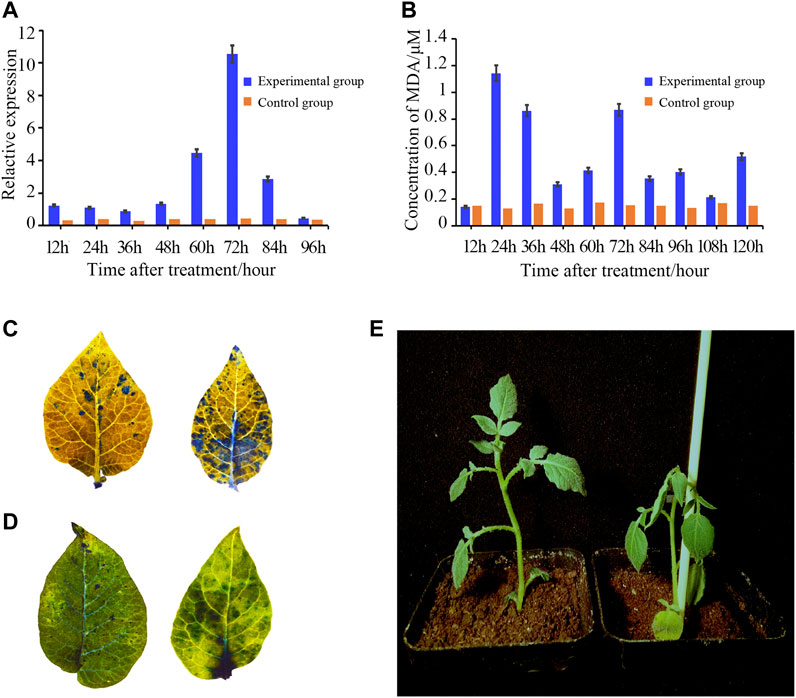
FIGURE 8. Upregulated expression pattern of StTGA39 and concentration of MDA during R. solanacearum infection on potato. (A) Expression pattern of StTGA39 was induced by R. solanacearum. (B) Levels of MDA in potato plants were assessed at 12, 24, 36, 48, 60, 72, 84, 96, 108, and 120 hpi inoculated with R. solanacearum. (C) Phenotype of two-week-old potato leaves after 72 hpi with flg22 treatment was compared with WT. (D) Cell death in potato was detected by histochemical staining with trypan blue staining. (E) Photograph showed the phenotype of the WT and was taken 3 days after R. solanacearum infection. mRNA levels were normalized to actin (Student’s t-test, n = 3 independent experiments, data shown are mean ± standard deviation).
In order to determine the defense response, various histochemical analyses were carried out on potato leaves (Figure 8). Under the stress of R. solanacearum solution of 108 cfu/L, the effect of MDA content in potato seedling leaves is shown in Figure 8B. It could be seen from Figure 8B that with the prolongation of infection time of R. solanacearum, the change of MDA content in potato seedling leaves showed a trend, which was upregulated once at 24 h and then decreased at intervals of 24 h. Callose strengthens the plant cell walls and prevents cells from being invaded by pathogens. Aniline staining analysis and trypan blue analysis showed that the induction of callose deposition and the HR-like cell death was significantly increased in the treated potato leaves (Figure 8C and Figure 8D).
Y1H Assay
BAK1 is a well-established receptor for defense-related genes in plants. To identify if StTGA1 could interact directly with StBAK1, Y1H analysis was used (Figure 9). The yeast expression vector TGA-pGADT7 and BAK-pHis2 were constructed. The results of Y1H showed that all the Y1H gold strains co-transformed with pHIS2 vectors and clones grew on SD (-Leu, -Trp) medium, indicating that the co-transformation was successful. The growth was different at different concentrations of 3-AT-deficient SD (-His, -Leu, -Trp) medium. On SD (-His, -Leu, -Trp) medium, the growth inhibition of the pGADT7/BAK-pHis2 self-activated group deepened with the increase of 3-AT concentration. The growth condition of the TGA-pGADT7/BAK-pHis2 group was slightly better than that of the control group, although the experimental group could not grow on SD (-His, -Leu, -Trp; 90 mM) (Figure 9). The results showed that StTGA1 might bind to StBAK1.
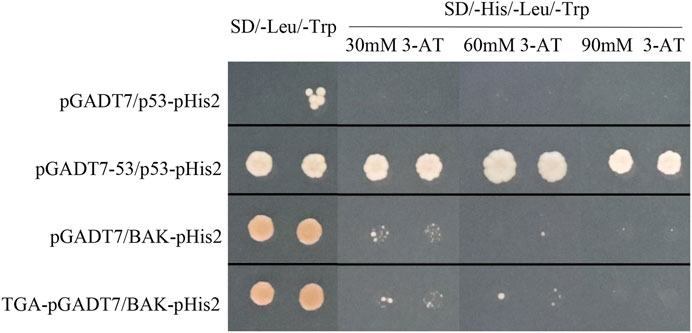
FIGURE 9. Y1H showing the association of StTGA39 with the promoter of the StBAK1-20 gene. Y1H assay for determination of StTGA39-StABAK1-20 interaction. Growth of yeast cells transformed with the effector vector and the reporter vector on SD (-His, -Leu, -Trp) supplemented different concentrations of 3-AT. The positive control is pGADT7-53/p53-pHis2 vector and negative control is pGADT7/p53-pHis2 vector.
Discussion
TGA TFs have been reported to function in various biological processes in plants (Jiang et al., 2021; Martins et al., 2022) and members of the TGA TFs play crucial roles in response to microbial pathogens in plants, such as Arabidopsis, soybean, rice, and tobacco (Pontier et al., 2002; Van et al., 2011; Moon et al., 2018; Ullah et al., 2019). There is a lack of reports on the potential link between TGA TFs and resistance to bacterial wilt caused by R. solanacearum in potato. In the present study, 42 StTGA genes were identified based upon the entire genome sequences of the potato, and the total number of TGA in potato was slightly expanded compared to that in tomato, tobacco, and pepper identified in this study but lower than that in Arabidopsis (Supplementary Additional File S1), which was different from the earlier reports (Ullah et al., 2019). This was mainly due to the continuous updates in the database and the different approaches used by authors. In addition, the phylogenetic analysis and collinearity was carried out. The results indicated that the StTGA family is conserved in the evolutionary history with the TGA TFs orthologs of other plants. This result corresponds well with previous observations in many plant species (Moon et al., 2018; Ullah et al., 2019; Jiang et al., 2021).
In order to investigate the expression patterns in response to different abiotic and biotic stresses, 10 members were selected for further study. The expression heat map demonstrated that the TGA TFs may have played significant and complex roles in potato (Figure 4), which was reflected in at least two aspects. First, the RNA-seq data-based heat map showed that the expression of most of the StTGAs was significantly different under abiotic/biotic stresses, which was consistent with previous studies (Jiang et al., 2021). Second, the same gene showed different or even opposite expression patterns under different stresses or in different tissues. For instance, the StTGA17 gene was upregulated in leaves under heat stress but downregulated under salt stress (Figure 4B). These results illustrate that these genes are widely involved in the response to various stresses and participate in a complex cross-regulatory network and signaling pathways (Duan et al., 2022).
To uncover the possible signal-associated functions of StTGAs in response to hormone stress, we conducted qRT-PCR to analyze their relative expressions under SA, MeJA, ABA, and GA3 treatments (Figure 5). This induction may be related to the upstream cis-element elements in the promoters of the genes. According to the analysis of cis-acting elements (Figure 2) and the results of qRT-PCR (Figure 5), we speculated StTGA could be induced by multiple phytohormones and might play a crucial role in multiple hormone signal pathways. Consistent with our results, some cis-elements play an important role in responses to ABA (Baker et al., 1994; Busk and Pagés, 1998; Li J. et al., 2019). There is an interaction between NPR1 and TGA TFs, which eventually leads to activation of SA-dependent response (Rahman et al., 2012). Studies have shown that a R. solanacearum effector targets TGA TFs to subvert SA signaling (Qi et al., 2022). Furthermore, the JA signal pathway is related to the function of Tripterygium wilfordii hook. F. TGA1 (TwTGA1) (Han et al., 2020). All of these research studies further support our idea that TGA may play a key role in the regulation of these hormone signal pathways.
An important goal in this study was to speculate the function of the StTGA gene in the response mechanism of potato to R. solanacearum. In potato, the invasion of R. solanacearum could cause up-regulation of StTGA gene expression according to Figure 8A. The result showed that StTGA was involved in resisting the invasion of R. solanacearum. Although the detailed mechanism needs further study, to prove this view, our study showed some other data related to resistance. Plants could resolve pathogens such as bacteria by causing ROS burst (Li et al., 2016; Ferreira et al., 2017). Evidence revealed that TGA class II plays an important role in the tolerance response to control ROS levels in Arabidopsis (Ariel et al., 2021). In addition, ROS induces gene expression and stress response and has regulatory roles in a wide range of important plant biological processes (Xia et al., 2010). Consistent with this possibility, R. solanacearum infection can result in enhanced production of ROS and lead to ROS-related oxidative damage (Ferreira et al., 2017). The results of DAB staining confirmed there is a mechanism in S. tuberosum to resist this adverse effect (Figure 8D). Furthermore, MDA can indicate the degree of membrane per-oxidation and further support the results of DAB staining (Figure 8B). The abovementioned views further support StTGA TFs are crucial regulatory factors in potato resistance to bacterial wilt, and our further data on identification of the target gene of this TF seem to support this view.
TGA TFs recognize the TGACG-motif (TGA-binding site) within the promoters of their target genes (Hou et al., 2019). To detect whether StTGA39 binds to the TGACG-motif present in the StBAK1-20 promoter (Supplementary Additional File S5) in yeast, the Y1H assays were performed, and the result suggested that StTGA39 specifically binds to the TGACG-motif in the promoter of the BAK1 gene which plays an essential role in regulated plant immunity (Beg et al., 2013). The results showed that the StBAK1-20 gene involved in the PAMP-triggered immunity (PTI) signal pathway (Beg et al., 2013) and interacted with StTGA in our materials. The abovementioned results indicated that the StTGA can function by combining with the promoter region of StBAK1-20 (Supplementary Additional File S6). To our knowledge, there are few reports on the TGA-promoted transcription of BAKs despite recent reports that TGAs directly activate respiratory burst oxidase homolog D (RBOHD) and pathogenesis-related protein 1 (PR1) expression in Arabidopsis (Qi et al., 2022; Shimizu et al., 2022).
Conclusion
To sum up, we conducted a genome-wide analysis of the StTGA gene family in potato and primarily explored the role of them. A total of 42 StTGA genes were identified from the potato genome. The structure diversity, chromosomal distribution, and evolutionary history of StTGA were comprehensively analyzed. These results extended the understanding on the abundance and diversity of StTGA genes in this important crop, which may serve as a fundamental resource for the molecular breeding of potato. However, the detailed molecular mechanism needs to be further studied. Taken together, this study lays the foundation for further investigation of StTGA in potato.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials; further inquiries can be directed to the corresponding author.
Author Contributions
TT planned and designed the research, analyzed the data, and wrote the manuscript. TT and YS studied gene expression by qRT-PCR. TT, LC, and GL studied the effect of MDA content in potato seedling leaves. GG supervised the research. RY, DY, YS, HW, XL, and GG revised the manuscript. All authors read and approved the final manuscript.
Funding
This study is supported by the National Natural Science Foundation of China (31771858).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (31771858). We thank all of the others in our laboratory for providing useful discussions and technical assistance and all the laboratories whose data were used in our analyses. We are very grateful to the editor and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.894844/full#supplementary-material
References
Azeem, F., Tahir, H., Ijaz, U., and Shaheen, T. (2020). A Genome-wide Comparative Analysis of bZIP Transcription Factors in G. Arboreum and G. Raimondii (Diploid Ancestors of Present-Day Cotton). Physiol. Mol. Biol. Plants 26 (3), 433–444. doi:10.1007/s12298-020-00771-9
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5?-region of Arabidopsis thaliana Cor15a Has Cis-Acting Elements that Confer Cold-, Drought- and ABA-Regulated Gene Expression. Plant Mol. Biol. 24, 701–713. doi:10.1007/BF00029852
Barchenger, D. W., Hsu, Y.-m., Ou, J.-y., Lin, Y.-p., Lin, Y.-c., Balendres, M. A. O., et al. (2022). Whole Genome Resequencing and Complementation Tests Reveal Candidate Loci Contributing to Bacterial Wilt (Ralstonia sp.) Resistance in Tomato. Sci. Rep. 12 (1), 8374. doi:10.1038/s41598-022-12326-x
Budimir, J., Treffon, K., Nair, A., Thurow, C., and Gatz, C. (2020). Redox-active Cysteines in TGACG-BINDING FACTOR 1 (TGA1) Do Not Play a Role in Salicylic Acid- or Pathogen-Induced Expression of TGA1-Regulated Target Genes in Arabidopsis thaliana. BioRxiv 230 (6), 2420–2432. doi:10.1101/2020.01.30.926758
Busk, P. K., and Pagès, M. (1998). Regulation of Abscisic Acid-Induced Transcription. Plant Mol. Biol. 37, 425–435. doi:10.1023/A:1006058700720
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide Analysis of MYB Gene Family in Chinese Bayberry (Morella Rubra) and Identification of Members Regulating Flavonoid Biosynthesis. Front. Plant Sci. 12, 691384. doi:10.3389/fpls.2021.691384
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, J., Mohan, R., Mohan, R., Zhang, Y., Li, M., Chen, H., et al. (2019). NPR1 Promotes its Own and Target Gene Expression in Plant Defense by Recruiting CDK8. Plant Physiol. 181, 289–304. doi:10.1104/pp.19.00124
Da, L., Liu, Y., Yang, J., Tian, T., She, J., Ma, X., et al. (2019). AppleMDO: A Multi-Dimensional Omics Database for Apple Co-expression Networks and Chromatin States. Front. Plant Sci. 10, 1333. doi:10.3389/fpls.2019.01333
Duan, L., Mo, Z., Fan, Y., Li, K., Yang, M., Li, D., et al. (2022). Genome-wide Identification and Expression Analysis of the bZIP Transcription Factor Family Genes in Response to Abiotic Stress in Nicotiana Tabacum L. BMC Genomics 23 (1), 318. doi:10.1186/s12864-022-08547-z
Fan, Y., Yan, J., Lai, D., Yang, H., Xue, G., He, A., et al. (2021a). Genome-wide Identification, Expression Analysis, and Functional Study of the GRAS Transcription Factor Family and its Response to Abiotic Stress in Sorghum [Sorghum Bicolor (L.) Moench]. BMC Genomics 22, 509. doi:10.1186/s12864-021-07848-z
Fan, Y., Yang, H., Lai, D., He, A., Xue, G., Feng, L., et al. (2021b). Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family and its Response to Abiotic Stress in Sorghum [Sorghum Bicolor (L.) Moench]. BMC Genomics 22, 415. doi:10.1186/s12864-021-07652-9
Fang, H., Liu, Z., Long, Y., Liang, Y., Jin, Z., Zhang, L., et al. (2017). The Ca2+/calmodulin2-Binding Transcription Factor TGA3 elevatesLCDexpression and H2S Production to Bolster Cr6+tolerance in Arabidopsis. Plant J. 91 (6), 1038–1050. doi:10.1111/tpj.13627
Ferreira, V., Pianzzola, M. J., Vilaró, F. L., Galván, G. A., Tondo, M. L., Rodriguez, M. V., et al. (2017). Interspecific Potato Breeding Lines Display Differential Colonization Patterns and Induced Defense Responses after Ralstonia Solanacearum Infection. Front. Plant Sci. 8, 1424. doi:10.3389/fpls.2017.01424
Gao, G., Jin, L. P., Xie, K. Y., and Qu, D. Y. (2009). The Potato StLTPa7 Gene Displays a Complex Ca 2+ ‐associated Pattern of Expression during the Early Stage of Potato- Ralstonia Solanacearum Interaction. Mol. Plant Pathol. 10, 15–27. doi:10.1111/j.1364-3703.2008.00508.x
Gao, H., Wang, Z., Li, S., Hou, M., Zhou, Y., Zhao, Y., et al. (2018). Genome-wide Survey of Potato MADS-Box Genes Reveals that StMADS1 and StMADS13 Are Putative Downstream Targets of Tuberigen StSP6A. BMC Genomics 19 (1), 726. doi:10.1186/s12864-018-5113-z
Gebhardt, C. (2016). The Historical Role of Species from the Solanaceae Plant Family in Genetic Research. Theor. Appl. Genet. 129, 2281–2294. doi:10.1007/s00122-016-2804-1
Geourjon, C., and Deléage, G. (1995). SOPMA: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Bioinformatics 11 (6), 681–684. doi:10.1093/bioinformatics/11.6.681
Guo, S., Zuo, Y., Zhang, Y., Wu, C., Su, W., Jin, W., et al. (2017). Large-scale Transcriptome Comparison of Sunflower Genes Responsive to Verticillium dahliae. BMC Genomics 18 (1), 42. doi:10.1186/s12864-016-3386-7
Gutsche, N., Holtmannspötter, M., Maß, L., O'Donoghue, M., Busch, A., Lauri, A., et al. (2017). Conserved Redox-dependent DNA Binding of ROXY Glutaredoxins with TGA Transcription Factors. Plant Direct 1 (6), e00030. doi:10.1002/pld3.30
Gutsche, N., and Zachgo, S. (2016). The N-Terminus of the Floral Arabidopsis TGA Transcription Factor PERIANTHIA Mediates Redox-Sensitive DNA-Binding. Plos One 11 (4), e0153810. doi:10.1371/journal.pone.0153810
Han, J., Liu, H.-t., Wang, S.-c., Wang, C.-r., and Miao, G.-p. (2020). A Class I TGA Transcription Factor from Tripterygium Wilfordii Hook.F. Modulates the Biosynthesis of Secondary Metabolites in Both Native and Heterologous Hosts. Plant Sci. 290, 110293. doi:10.1016/j.plantsci.2019.110293
He, L. Y., Sequeira, L., and Kelman, A. (1983). Characteristics of Strains of Pseudomonas Solanacearumfrom China. Plant Dis. 67 (12), 1357–1361. doi:10.1094/PD-67-1357
He, Z. Q., Li, Y. H., Feng, G. H., Yuan, X. W., Lu, Z. B., Dai, M., et al. (2022). Pharmacological Perturbation of Mechanical Contractility Enables Robust Transdifferentiation of Human Fibroblasts into Neurons. Adv. Sci. 9, 2104682. doi:10.1002/advs.202104682
Herrera-Vásquez, A., Fonseca, A., Ugalde, J. M., Lamig, L., Seguel, A., Moyano, T. C., et al. (2021). TGA Class II Transcription Factors Are Essential to Restrict Oxidative Stress in Response to UV-B Stress in Arabidopsis. J. Exp. Bot. 72 (5), 1891–1905. doi:10.1093/jxb/eraa534
Hou, J., Sun, Q., Li, J., Ahammed, G. J., Yu, J., Fang, H., et al. (2019). Glutaredoxin S25 and its Interacting TGACG Motif-Binding Factor TGA2 Mediate Brassinosteroid-Induced Chlorothalonil Metabolism in Tomato Plants. Environ. Pollut. 255 (2), 113256. doi:10.1016/j.envpol.2019.113256
Huang, L.-J., Li, N., Thurow, C., Wirtz, M., Hell, R., and Gatz, C. (2016). Ectopically Expressed Glutaredoxin ROXY19 Negatively Regulates the Detoxification Pathway in Arabidopsis thaliana. BMC Plant Biol. 16, 200. doi:10.1186/s12870-016-0886-1
Huang, X.-S., Liu, J.-H., and Chen, X.-J. (2010). Overexpression of PtrABF Gene, a bZIP Transcription Factor Isolated from Poncirus Trifoliata, Enhances Dehydration and Drought Tolerance in Tobacco via Scavenging ROS and Modulating Expression of Stress-Responsive Genes. BMC Plant Biol. 10 (1), 230. doi:10.1186/1471-2229-10-230
Islamoglu, H., Cao, R., Teskey, G., Gyurjian, K., Lucar, S., Fraix, M., et al. (2018). Effects of readiSorb L-GSH in Altering Granulomatous Responses against mycobacterium Tuberculosis Infection. Jcm 7, 40. doi:10.3390/jcm7030040
Jiang, H., Gu, S., Li, K., and Gai, J. (2021). Two TGA Transcription Factor Members from Hyper-Susceptible Soybean Exhibiting Significant Basal Resistance to Soybean Mosaic Virus. Ijms 22 (21), 11329. doi:10.3390/ijms222111329
Ke, D., He, Y., Fan, L., Niu, R., Cheng, L., Wang, L., et al. (2022). The Soybean TGA Transcription Factor GmTGA13 Plays Important Roles in the Response to Salinity Stress. Plant Biol. J. 24 (2), 313–322. doi:10.1111/plb.13360
Kim, B. H., Kim, S. Y., and Nam, K. H. (2013). Assessing the Diverse Functions of BAK1 and its Homologs in Arabidopsis, beyond BR Signaling and PTI Responses. Mol. Cells 35 (1), 7–16. doi:10.1007/s10059-013-2255-3
Kohl, M., Wiese, S., and Warscheid, B. (2011). Cytoscape: Software for Visualization and Analysis of Biological Networks. Methods Mol. Biol. 696, 291–303. doi:10.1007/978-1-60761-987-1_18
Kong, C.-Y., Luo, Y.-p., Duan, T.-T., Xue, Z., Gao, X.-D., Zhao, X., et al. (2016). Potato Remorin Gene StREMa4 Cloning and its Spatiotemporal Expression Pattern under Ralstonia Solanacearum and Plant Hormones Treatment. Phytoparasitica 44 (4), 575–584. doi:10.1007/s12600-016-0536-z
Kumar, P., Kumar, P., Sharma, D., Verma, S. K., Halterman, D., and Kumar, A. (2021). Genome-wide Identification and Expression Profiling of Basic Leucine Zipper Transcription Factors Following Abiotic Stresses in Potato (Solanum tuberosum L.). Plos One 16 (3), e0247864. doi:10.1371/journal.pone.0247864
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, Y. V. D., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Li, B., Liu, Y., Cui, X.-Y., Fu, J.-D., Zhou, Y.-B., Zheng, W.-J., et al. (2019). Genome-wide Characterization and Expression Analysis of Soybean TGA Transcription Factors Identified a Novel TGA Gene Involved in Drought and Salt Tolerance. Front. Plant Sci. 10, 549. doi:10.3389/fpls.2019.00549
Li, J., Han, G., Sun, C., and Sui, N. (2019). Research Advances of MYB Transcription Factors in Plant Stress Resistance and Breeding. Plant Signal. Behav. 14 (8), 1613131. doi:10.1080/15592324.2019.1613131
Li, K., Du, Y., and Miao, Y. (2016). Future Challenges in Understanding ROS in Plant Responses to Abiotic Stress. Sci. China Life Sci. 59 (12), 1343–1344. doi:10.1007/s11427-016-0362-7
Liang, Y., Wan, N., Cheng, Z., Mo, Y., Liu, B., Liu, H., et al. (2017). Whole-genome Identification and Expression Pattern of the Vicinal Oxygen Chelate Family in Rapeseed (Brassica Napus L.). Front. Plant Sci. 8, 745. doi:10.3389/fpls.2017.00745
Liu, C., Long, J., Zhu, K., Liu, L., Yang, W., Zhang, H., et al. (2016). Characterization of a Citrus R2R3-MYB Transcription Factor that Regulates the Flavonol and Hydroxycinnamic Acid Biosynthesis. Sci. Rep. 6, 25352. doi:10.1038/srep25352
Martins, D. C., Rubiales, D., and VazPatto, M. C. (2022). Association Mapping of Lathyrus Sativus Disease Response to Uromyces Pisi Reveals Novel Loci Underlying Partial Resistance. Front. Plant Sci. 13 (13), 842545. doi:10.3389/fpls.2022.842545
Moon, S.-J., Park, H. J., Kim, T.-H., Kang, J.-W., Lee, J.-Y., Cho, J.-H., et al. (2018). OsTGA2 Confers Disease Resistance to Rice against Leaf Blight by Regulating Expression Levels of Disease Related Genes via Interaction with NH1. Plos One 13 (11), e0206910. doi:10.1371/journal.pone.0206910
Pontier, D., Privat, I., Trifa, Y., Zhou, J.-M., Klessig, D. F., and Lam, E. (2002). Differential Regulation of TGA Transcription Factors by Post-transcriptional Control. Plant J. 32, 641–653. doi:10.1046/j.1365-313X.2002.01461.x
Qi, P., Huang, M., Hu, X., Zhang, Y., Wang, Y., Li, P., et al. (2022). A Ralstonia Solanacearum Effector Targets TGA Transcription Factors to Subvert Salicylic Acid Signaling. Plant Cell 34, 1666–1683. doi:10.1093/plcell/koac015
Rahman, T. A. E., Oirdi, M. E., Gonzalez-Lamothe, R., and Bouarab, K. (2012). Necrotrophic Pathogens Use the Salicylic Acid Signaling Pathway to Promote Disease Development in Tomato. Mpmi 25 (12), 1584–1593. doi:10.1094/MPMI-07-12-0187-R
Rouhier, N., Cerveau, D., Couturier, J., Reichheld, J.-P., and Rey, P. (2015). Involvement of Thiol-Based Mechanisms in Plant Development. Biochimica Biophysica Acta (BBA) - General Subj. 1850 (8), 1479–1496. doi:10.1016/j.bbagen.2015.01.023
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 5 (4), 725–738. doi:10.1038/nprot.2010.5
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome Sequence of the Plant Pathogen Ralstonia Solanacearum. Nature 415, 497–502. doi:10.1038/415497a
Salladini, E., Jørgensen, M. L. M., Theisen, F. F., and Skriver, K. (2020). Intrinsic Disorder in Plant Transcription Factor Systems: Functional Implications. Ijms 21 (24), 9755. doi:10.3390/ijms21249755
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 3 (6), 1101–1108. doi:10.1038/nprot.2008.73
Shimizu, K., Suzuki, H., Uemura, T., Nozawa, A., Desaki, Y., Hoshino, R., et al. (2022). Immune Gene Activation by NPR and TGA Transcriptional Regulators in the Model Monocot Brachypodium Distachyon. Plant J. 110 (2), 470–481. doi:10.1111/tpj.15681
Song, Y., Zhai, Y., Li, L., Yang, Z., Ge, X., Yang, Z., et al. (2021). BIN2 Negatively Regulates Plant Defence against Verticillium dahliae in Arabidopsis and Cotton. Plant Biotechnol. J. 19, 2097–2112. doi:10.1111/pbi.13640
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular Localization of H2O2 in Plants. H2O2 Accumulation in Papillae and Hypersensitive Response during the Barley-Powdery Mildew Interaction. Plant J. 11 (6), 1187–1194. doi:10.1046/j.1365-313X.1997.11061187.x
Uhrig, J. F., Huang, L.-J., Barghahn, S., Willmer, M., Thurow, C., and Gatz, C. (2017). CC-type Glutaredoxins Recruit the Transcriptional Co-repressor TOPLESS to TGA-dependent Target Promoters in Arabidopsis thaliana. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1860 (2), 218–226. doi:10.1016/j.bbagrm.2016.11.001
Ullah, I., Magdy, M., Wang, L., Liu, M., and Li, X. (2019). Genome-wide Identification and Evolutionary Analysis of TGA Transcription Factors in Soybean. Sci. Rep. 9 (1), 11186. doi:10.1038/s41598-019-47316-z
van Verk, M. C., Neeleman, L., Bol, J. F., and Linthorst, H. J. M. (2011). Tobacco Transcription Factor NtWRKY12 Interacts with TGA2.2 In Vitro and In Vivo. Front. Plant Sci. 2 (32), 1–10. doi:10.3389/fpls.2011.00032
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40, e49. doi:10.1093/nar/gkr1293
Wang, Y., Zhang, N., Li, T., Yang, J., Zhu, X., Fang, C., et al. (2019). Genome-wide Identification and Expression Analysis of StTCP Transcription Factors of Potato (Solanum tuberosum L.). Comput. Biol. Chem. 78, 53–63. doi:10.1016/j.compbiolchem.2018.11.009
Xia, X.-J., Chen, Z., and Yu, J.-Q. (2010). ROS Mediate Brassinosteroids-Induced Plant Stress Responses. Plant Signal. Behav. 5 (5), 532–534. doi:10.4161/psb.10989
Yu, R. M., Chang, Y. N., Song, Y. J., Tian, T., Wang, H. J., and Gao, G. (2021a). Genome-Wide Identification and Expression Analysis of the Zinc Transporter Protein ZIP Family in Potato (Solanum tuberosum). Intl J. Agric. Biol. 24, 1533–1542. doi:10.17957/IJAB/15.1592
Yu, R.-M., Suo, Y.-Y., Yang, R., Chang, Y.-N., Tian, T., Song, Y.-J., et al. (2021b). StMBF1c Positively Regulates Disease Resistance to Ralstonia Solanacearum via It's Primary and Secondary Upregulation Combining Expression of StTPS5 and Resistance Marker Genes in Potato. Plant Sci. 307, 110877. doi:10.1016/j.plantsci.2021.110877
Zhu, C., Zhang, S., Zhou, C., Chen, L., Fu, H., Li, X., et al. (2020). Genome-wide Investigation and Transcriptional Analysis of Cytosine-5 DNA Methyltransferase and DNA Demethylase Gene Families in Tea Plant (Camellia Sinensis) under Abiotic Stress and Withering Processing. Peer J. 8, e8432. doi:10.7717/peerj.8432
Keywords: potato, StTGA, qRT-PCR, MDA, Y1H
Citation: Tian T, Yu R, Suo Y, Cheng L, Li G, Yao D, Song Y, Wang H, Li X and Gao G (2022) A Genome-Wide Analysis of StTGA Genes Reveals the Critical Role in Enhanced Bacterial Wilt Tolerance in Potato During Ralstonia solanacearum Infection. Front. Genet. 13:894844. doi: 10.3389/fgene.2022.894844
Received: 12 March 2022; Accepted: 06 June 2022;
Published: 26 July 2022.
Edited by:
Dwijesh Chandra Mishra, Indian Council of Agricultural Research, IndiaReviewed by:
Yordan Muhovski, Walloon Agricultural Research Centre, BelgiumPaola Gaiero, Universidad de la República, Uruguay
Copyright © 2022 Tian, Yu, Suo, Cheng, Li, Yao, Song, Wang, Li and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Gao, Z2dzeG51QDEyNi5jb20=
 Tian Tian
Tian Tian Ruimin Yu
Ruimin Yu Xinyu Li
Xinyu Li