- 1Maternal and Child Research Institute, Shunde Women and Children's Hospital, Guangdong Medical University, Foshan, China
- 2Department of Medicine, Shunde Women and Children's Hospital, Guangdong Medical University, Foshan, China
- 3Department of Pediatrics, Shunde Women and Children's Hospital, Guangdong Medical University, Foshan, China
Background: Iron responsive element binding protein 2 (IREB2) variants may be involved in the pathogenesis of chronic obstructive pulmonary disease (COPD). Recently, many studies have been performed on IREB2 susceptibility variants, including rs2568494, rs2656069, rs10851906, rs12593229, and rs13180, associated with COPD. However, inconsistent findings have been reported. The aim of our research was to determine the association of IREB2 SNPs with COPD.
Methods: A comprehensive meta-analysis was performed to accurately estimate the association between IREB2 variants and COPD among four different genetic models.
Results: This meta-analysis included a total of 4,096 patients and 5,870 controls. Here, we investigated the 5 IREB2 variants to identify COPD risk. Our results indicate that rs2568494 was associated with an increased risk of COPD for the dominant model (AA+GA vs. GG: OR = 1.150, 95% CI: 1.5–1.304, P = 0.029); rs2656069 was associated with a decreased risk of COPD for the recessive model (GG vs. AA+AG: OR = 0.589, 95% CI: 0.440–0.789; P = 0.000), additive model (GG vs. AA: OR =0.641, 95% CI: 0.441–0.931; P = 0.020), and allele model (G vs. A: OR = 0.812, 95% CI: 0.668–0.988; P = 0.037); and rs10851906 was associated with a decreased risk of COPD for the recessive model (GG vs. AA+AG: OR = 0.732, 95% CI: 0.560–0.958; P = 0.023) and additive model (GG vs. AA: OR = 0.777, 95% CI: 0.637–0.947; P = 0.012).
Conclusion: Our findings suggest that the IREB2 rs2568494 minor alleles A may be a genetic factor in susceptibility to COPD. In addition, the minor alleles G of rs2656069 and rs10851906 appear to have a protective effect.
Introduction
Chronic obstructive pulmonary disease (COPD) is a complex human genetic disease characterized by pulmonary dysfunction and incomplete reversibility of airflow obstruction (Rabe et al., 2007). It is predicted to become the fourth leading cause of death by 2030 (Mathers and Loncar, 2006). It is well-known that COPD is the result of interactions between genetic susceptibility and environmental factors. Although smoking is an important environmental factor in COPD, the COPD susceptibility of smokers varies, and the disease can also appear in non-smokers. These results suggest that genetic factors may be among the reasons for individual susceptibility.
The IREB2 gene codes for an iron-binding protein (IRP2), which participates in maintaining iron homeostasis in human cells. A previous study reported that iron in the lungs has been shown to increase with age and that smokers had higher concentrations of iron in the lungs (Ghio et al., 1997). Thus, abnormalities in IREB2 expression or functioning might contribute to iron metabolism disorders. This iron imbalance may lead to oxidative damage. Furthermore, an excess of iron has an influence on local inflammation in the lungs, which may be related to the pathogenesis of COPD. Interestingly, several genome-wide association studies have identified a IREB2 genomic region associated with COPD (DeMeo et al., 2009; Hancock et al., 2010; Pillai et al., 2010). Moreover, the study reported that three IREB2 SNPs were associated with COPD patients, and increased expression levels of IREB2 mRNA have been detected in the lungs of smokers and COPD patients (DeMeo et al., 2009), which showed IREB2 as a COPD susceptibility gene.
Over the past few years, many SNPs (rs2568494, rs2656069, rs10851906, rs12593229, and rs13180) in IREB2 gene studies have different degrees of reproducibility. Numerous studies have investigated the relation to IREB2 SNPs and COPD susceptibility (Chappell et al., 2011; Guo et al., 2011; Cho et al., 2012; Hardin et al., 2012; Zhou et al., 2012; Arja et al., 2014; Ding et al., 2015; Ziolkowska-Suchanek et al., 2015; Korytina et al., 2016; Nedeljkovic et al., 2018). However, the findings in these studies were inconsistent. Conflicting results may be due to the different populations and small sample sizes of each individual study. Therefore, the purpose of this meta-analysis was to collect all eligible studies to determine the association between IREB2 gene polymorphism and COPD susceptibility.
Materials and Methods
Literature Search
The PubMed and Web of Science databases were systematically searched for relevant studies published before July 30, 2020, using the following key words: (1) “IREB2” or “rs2568494” or “polymorphism” and “COPD”; (2) “IREB2” or “rs2656069” or “polymorphism” and “COPD”; (3) “IREB2” or “rs10851906” or “polymorphism” and “COPD”; (4) “IREB2” or “rs12593229” or “polymorphism” and “COPD”; and (5) “IREB2” or “rs13180” or “polymorphism” and “COPD.” The search had no language restrictions. All studies were assessed by reading the title and abstract, and irrelevant studies were excluded. Then, the full texts of the remaining studies were assessed to determine their eligibility.
Inclusion and Exclusion Criteria
The study inclusion criteria were as follows: (1) The study was a cohort or a case-control study; (2) The studies assessed the association between IREB2 SNPs (rs2568494, rs2656069, rs10851906, rs12593229, and rs13180) and COPD; (3) COPD was diagnosed depending on the distinct clinical criteria. The inclusion criteria for stable COPD comprised airflow limitation as indicated by post-bronchodilator FEV1/FVC <70% and FEV1 ≤ 70% of normal predicted values. The severity of airflow limitation was classified according to the GOLD standard (Global Initiative for Chronic Obstructive Lung Disease) as follows: II-moderate, III-severe, and IV-very severe; (4) The control subjects were in Hardy-Weinberg equilibrium (HWE); (5) The selected studies provided SNP genotype amounts or sufficient data for calculation; and (6) The selected studies provided an odds ratio (OR) with a 95% confidence interval (CI) or sufficient data for calculation (He et al., 2015).
The exclusion criteria were as follows: (1) The study was not a cohort or a case-control study; (2) the study lacked particular raw or calculated data for OR with a 95% CI; (3) study on the repeatability (if the study population overlaps, only the study with the largest sample size was selected); and (4) the control subjects were not in a HWE.
Data Extraction
The following data were independently extracted from the included studies and entered into a database to ensure the veracity of the data: first author, country, year of publication, origin, the numbers of COPD cases and controls, subjects' age and gender, BMI, pack years, FEV1 (%), FEV1/FVC, the distribution of genotype and alleles, ORs with 95% CI, or ability to calculate the OR and 95% CI.
Statistical Analysis
HWE was examined by Pearson's chi-squared test (Shen et al., 2015; Zhang et al., 2015). Four genetic models were used in the study: the dominant model, the recessive model, the additive model and the allele model. Genetic heterogeneity was evaluated using the Q-test and I2-test. I2 statistics range from 0 to 100%. Significant heterogeneity was defined as P < 0.01 and I2 > 50% (Li et al., 2016; Liu et al., 2017; Han et al., 2019). ORs with corresponding 95% CIs were calculated using the fixed effects model (Mantel-Haenszel) when no significant heterogeneity was observed; otherwise, a random effects model was used. The Z-test was used to test the significance of the ORs. Additionally, Egger's and Begg's tests were used to assess publication bias. All statistical analyses were performed using STATA v.16.0 software (Stata Corporation, Texas, USA).
Results
Study Inclusion and Characteristics
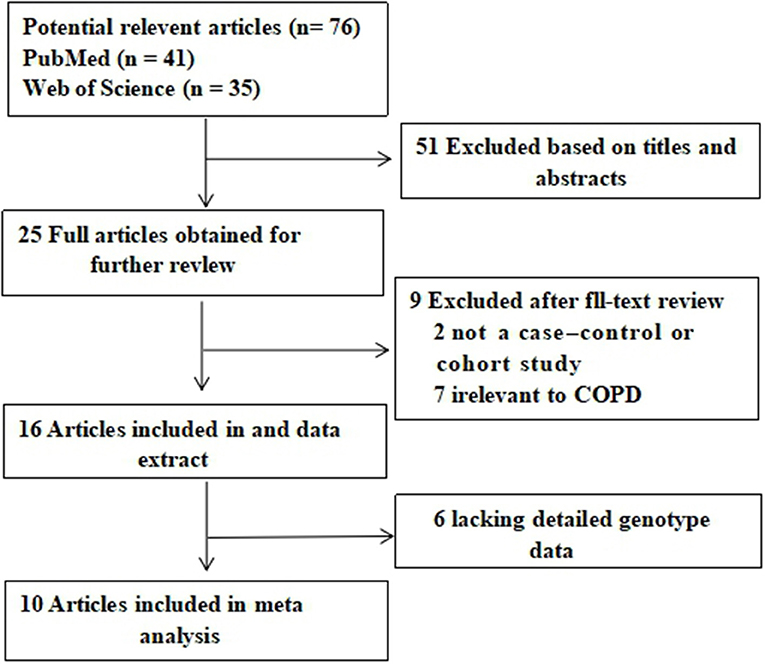
A total of 41 and 35 potential studies were retrieved from the PubMed and Web of Science databases. Using the inclusion and exclusion criteria, we selected 10 articles for meta-analysis. A flow chart of the study selection process is shown in Figure 1. Table 1 shows the major characteristics of each included study. Supplementary Tables 1–5 of the Supplementary Materials show the OR and 95% CI values of the four genotypes of each SNP. The control subjects of each study were in accordance with HWE.
Heterogeneity Analysis
Genetic heterogeneity was evaluated using Cochran's Q statistic and I2 statistic.
rs2568494
Low heterogeneity among studies (Chappell et al., 2011; Guo et al., 2011; Zhou et al., 2012; Arja et al., 2014; Ziolkowska-Suchanek et al., 2015) was detected in the dominant model (AA+GA vs. GG: I2 = 30.6%, P = 0.217), and a moderate degree of heterogeneity was identified among studies under the recessive model (AA vs. GG+GA: I2 = 67.6%, P = 0.015), additive model (AA vs. GG; I2 = 66.2%, P = 0.019), and allele model (A vs. G: I2 = 60.3%, P = 0.039) (Figure 2, Supplementary Figure 1).
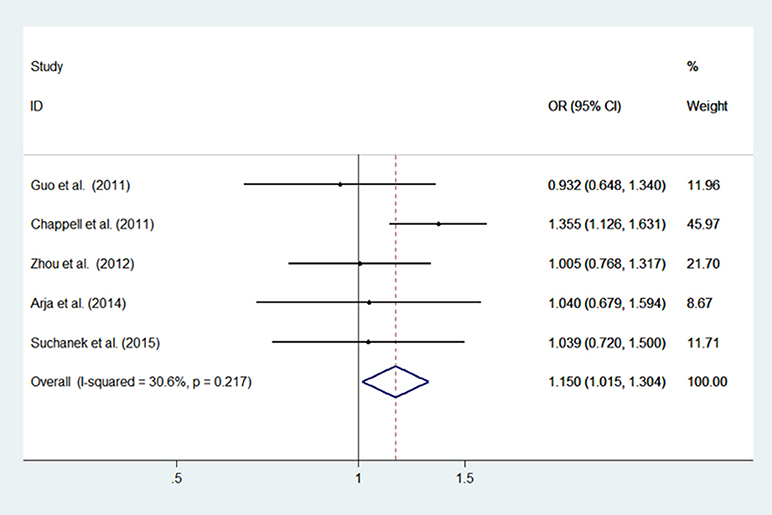
Figure 2. Meta-analysis with a fixed effects model for the association between the IREB2 rs2568494 polymorphism and COPD susceptibility (dominant model, AA+GA vs. GG). OR, odds ratio; CI, confidence interval; I-squared, measure to quantify the degree of heterogeneity in meta-analyses.
rs2656069
Low heterogeneity among studies (Chappell et al., 2011; Zhou et al., 2012; Arja et al., 2014; Ziolkowska-Suchanek et al., 2015) was detected in the recessive model (GG vs. AA+AG: I2 = 20.5%, P = 0.287), and a moderate degree of heterogeneity was found among studies under the dominant model (GG+AG vs. AA: I2 = 69.2%, P = 0.021), additive model (GG vs. AA; I2 = 53.1%, P = 0.094), and allele model (G vs. A: I2 = 66.6%, P = 0.030) (Figure 3, Supplementary Figure 2).
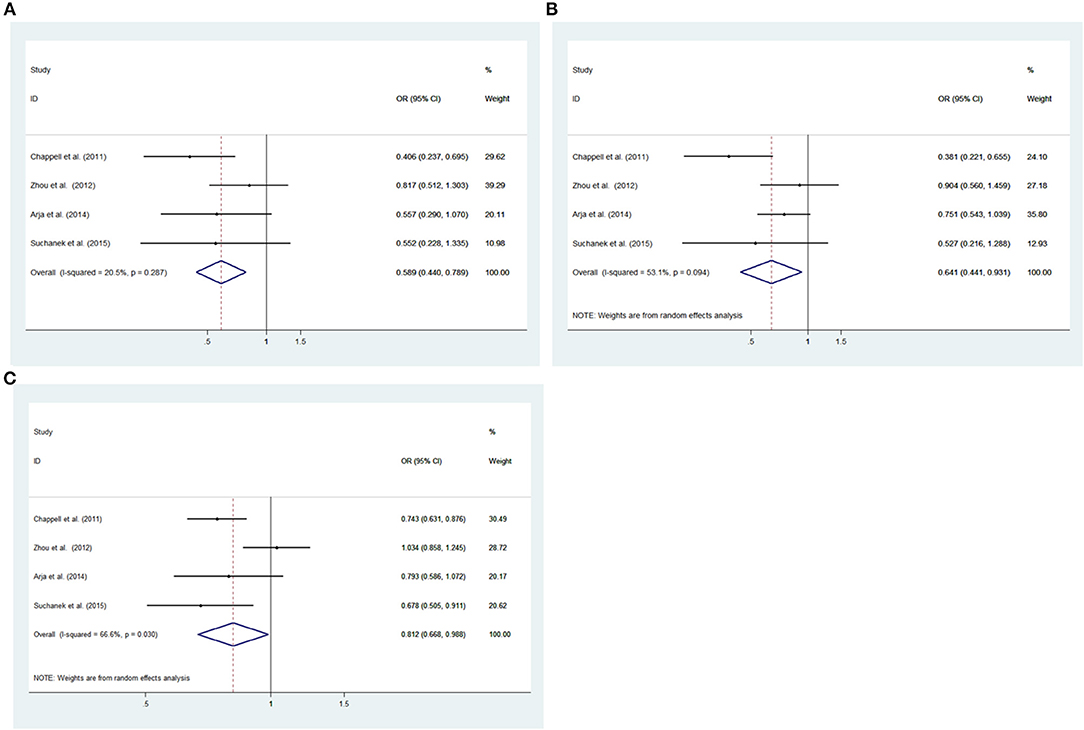
Figure 3. Meta-analysis for the association between the IREB2 rs2656069 polymorphism and COPD susceptibility. (A) Recessive model, GG vs. AA+AG (fixed effects model). (B) Additive model, GG vs. AA (random effects model). (C) Allele model, G vs. A (random effects model). OR, odds ratio; CI, confidence interval; I-squared, measure to quantify the degree of heterogeneity in meta-analyses.
rs10851906
Low heterogeneity among studies (Chappell et al., 2011; Zhou et al., 2012; Arja et al., 2014; Ziolkowska-Suchanek et al., 2015) was detected in the recessive model (GG vs. AA+AG: I2= 0.0%, P = 0.634), additive model (GG vs. AA; I2 = 0.0%, P = 0.883) and allele model (G vs. A: I2 = 32.7%, P = 0.216); there was a moderate degree of heterogeneity among studies under the dominant model (GG+AG vs. AA: I2 = 67.0%, P = 0.028) (Figure 4, Supplementary Figure 3).
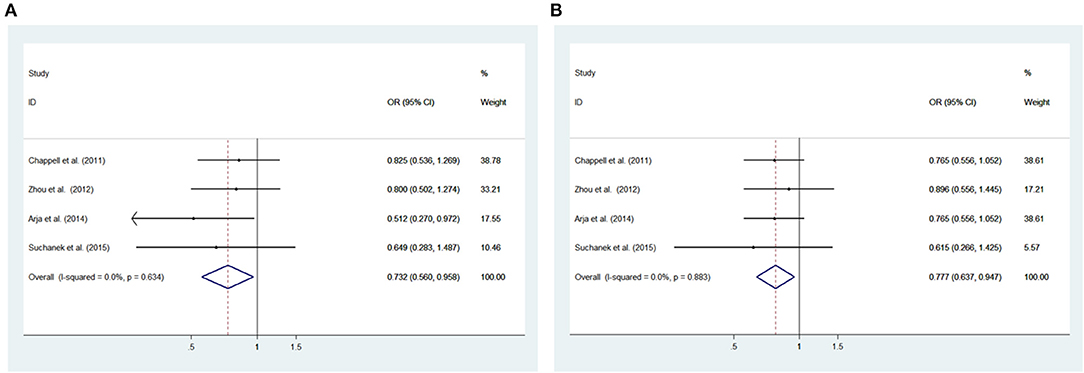
Figure 4. Meta-analysis with a fixed effects model for the association between the IREB2 rs10851906 polymorphism and COPD susceptibility. (A) Recessive model, GG vs. AA + AG. (B) Additive model, GG vs. AA. OR, odds ratio; CI, confidence interval; I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
rs12593229
High heterogeneity among studies (Chappell et al., 2011; Zhou et al., 2012; Arja et al., 2014) was detected in the dominant model (TT+GT vs. GG: I2 = 75.0%, P = 0.018), recessive model (TT vs. GG+GT: I2 = 75.0%, P = 0.018), additive model (TT vs. GG; I2 = 83.7%, P = 0.002), and allele model (T vs. G: I2= 80.5%, P = 0.006) (Supplementary Figure 4).
rs13180
High heterogeneity among studies (Zhou et al., 2012; Ding et al., 2015; Ziolkowska-Suchanek et al., 2015; Korytina et al., 2016) was detected in the dominant model (TT+ CT vs. CC: I2 = 62.5%, P = 0.046), recessive model (TT vs. CC+CT: I2 = 76.4%, P = 0.005), additive model (TT vs. CC: I2 = 77.7%, P = 0.004); a high degree of heterogeneity was also found among studies (Cho et al., 2012; Hardin et al., 2012; Zhou et al., 2012; Ding et al., 2015; Ziolkowska-Suchanek et al., 2015; Korytina et al., 2016; Nedeljkovic et al., 2018) under the allele model (T vs. C: I2 = 84.2%, P = 0.000) (Supplementary Figure 5).
Meta-Analysis Results
rs2568494
A fixed effects model was used to analyze the dominant model; the recessive, additive, and allele models were analyzed with a random effects model. The results showed a significant difference between COPD patients and controls for the dominant model (AA+GA vs. GG: OR = 1.150, 95% CI: 1.5–1.304, P = 0.029). No significant associations were found under the recessive model (AA vs. GG+GA: OR = 0.941, 95% CI: 0.556–1.593; P = 0.822), additive model (AA vs. GG; OR = 1.125, 95% CI: 0.0.746–1.696; P = 0.575), and allele model (A vs. G; OR = 1.049, 95% CI: 0.880–1.251; P = 0.590) between patients and controls (Figure 2, Supplementary Figure 1).
rs2656069
To analyze the recessive model using the fixed effects model; the dominant, additive, and allele models using the random effects model. There had a significant difference between COPD patients and controls for the recessive model (GG vs. AA+AG: OR = 0.589, 95% CI: 0.440–0.789; P = 0.000), additive model (GG vs. AA: OR = 0.641, 95% CI: 0.441–0.931; P = 0.020) and allele model (G vs. A: OR = 0.812, 95% CI: 0.668–0.988; P = 0.037). There was no significant associations under the dominant model (GG+AG vs. AA: OR = 0.884, 95% CI: 0.682–1.145, P =0.351) between patients and controls (Figure 3, Supplementary Figure 2).
rs10851906
The recessive and additive models were analyzed by the fixed effects model; the dominant and allele models were analyzed with a random effects model. The results indicated significant associations between COPD patients and controls for the recessive model (GG vs. AA+AG: OR = 0.732, 95% CI: 0.560–0.958; P = 0.023) and additive model (GG vs. AA: OR = 0.777, 95% CI: 0.637–0.947; P = 0.012). We did not observe significant associations under the dominant model (GG+AG vs. AA: OR = 0.868, 95% CI: 0.618–1.220, P = 0.416) and allele model (G vs. A: OR = 0.903, 95% CI: 0.794–1.026; P = 0.118) between patients and controls (Figure 4, Supplementary Figure 3).
rs12593229
After meta-analysis with random effects. We did not identify significant difference between COPD patients and controls under the dominant model (TT+GT vs. GG: OR = 0.882, 95% CI: 0.628–1.238, P =0.468), recessive model (TT vs. GG+GT: OR = 0.882, 95% CI: 0.605–1.285; P = 0.513), additive model (TT vs. GG: OR = 0.854, 95% CI: 0.551–1.326; P = 0.483) and allele model (T vs. G: OR = 0.891, 95% CI: 0.699–1.134; P = 0.348) between patients and controls (Supplementary Figure 4).
rs13180
we performed a meta-analysis using random effects. There were no significant difference between COPD patients and controls under the dominant model (TT + CT vs. CC: OR = 1.106, 95% CI: 0.821–1.492, P = 0.507), recessive model (TT vs. CC+ CT: OR = 1.133, 95% CI: 0.816–1.572; P = 0.457), additive model (TT vs. CC: OR = 1.170, 95% CI: 0.746–1.836; P = 0.495), and allele model (T vs. C: OR = 0.934, 95% CI: 0.767–1.137; P = 0.526) between patients and controls (Supplementary Figure 5).
Publication Bias
There was no significant publication bias in any of the genetic models according to Begg's and Egger's tests (all P > 0.05, data not shown), and the funnel plot was symmetrical, as the studies did not coagulate into one quadrant of the funnel (Figure 5).
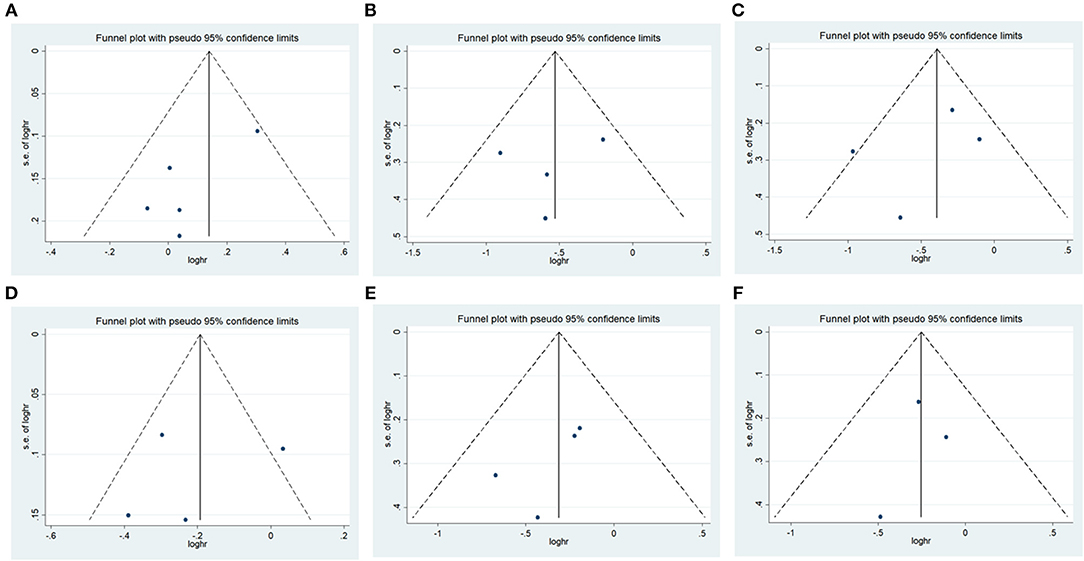
Figure 5. Funnel plot of the odds ratios in the meta-analysis. (A) IREB2 rs2568494 dominant model, AA + GA vs. GG. (B) IREB2 rs2656069 recessive model, GG vs. AA + AG. (C) IREB2 rs2656069 additive model, GG vs. AA. (D) IREB2 rs2656069 allele model, G vs. A. (E) IREB2 rs10851906 recessive model, GG vs. AA + AG. (F) IREB2 rs10851906 additive model, GG vs. AA.
Discussion
Genetic factors play a major role in COPD susceptibility. The region of chromosome 15q25 includes several genes that influence the development of COPD. Among them, IREB2 is particularly attractive for studying the genetic factors of COPD. In this study, 5 SNPs among COPD patients and healthy controls from previous studies were investigated to evaluate the association of IREB2 polymorphism with COPD. The results showed that IREB2 rs2568494 was associated with COPD susceptibility. The minor alleles G by rs2656069 and rs10851906 were protective factors for COPD.
Here, the results showed a significant association of rs2568494 with COPD under the dominant model (AA+GA vs. GG: OR = 1.150, P = 0.029). This result has confirmed previous observations suggesting that IREB2 rs2568494 may play a role in COPD (DeMeo et al., 2009). In addition, two other SNPs (rs2656069 and rs10851906) in IREB2 have been previously reported, and the minor alleles (G) for both of these SNPs were associated with a decreased risk of COPD. Our results also showed a significant difference between COPD patients and controls for the recessive model (GG vs. AA+AG: OR = 0.589, P =0.000) additive model (GG vs. AA: OR = 0.641, P = 0.020), and allele model (G vs. A: OR = 0.812, P = 0.037) in rs2656069. There was a significant difference between COPD patients and controls for the recessive model (GG vs. AA+AG: OR = 0.732, P = 0.023) and additive model (GG vs. AA: OR = 0.777, P = 0.012) in rs10851906. These results showed that GG for both SNPs was associated with a decreased risk of COPD, which was in the same direction as previous reports (Qiu et al., 2011). However, we found a lack of association between IREB2 rs12593229 and rs13180 and COPD susceptibility. Thus, our meta-analysis provides further evidence to support the role of the IREB2 genomic region in COPD pathogenesis, and the role of IREB2 deserves further investigation.
There are still some limitations in our study. On the one hand, due to the limited examination of IREB2 variants in COPD, the sample sizes were comparatively small, which might not provide sufficient statistical power to assess the association between the IREB2 gene and COPD susceptibility; thus, there is an urgent need to conduct research using large samples across the world. On the other hand, owing to the limitation of the original information, we did not perform a smoking status-stratified analysis, which have the impact on association analysis. We known that the genetic factors of COPD may be complex, with the contribution of environmental factors and multiple genes (Seifart and Plagens, 2007). Thus, it is an important prompt for future study to evaluate whether other risk factors together with the IREB2 gene influence COPD susceptibility.
Conclusion
To our knowledge, this is the comprehensive study to assess the role of the IREB2 rs2568494, rs2656069, rs10851906, rs12593229, and rs13180 variants in COPD. The current results show that there was a significant association between the IREB2 gene rs2568494 polymorphism and susceptibility to COPD, suggesting that the presence of minor alleles A may be a genetic factor in susceptibility to COPD. Rs2656069 and rs10851906 were associated with a decreased risk of COPD, and the minor alleles G of both SNPs may appear to have a protective effect.
Author Contributions
QZ, QC, and DZ were responsible for the statistical analysis, study design, and manuscript preparation. RG, DX, and SJ managed the literature searches and analyses. The study was supervised by RC, YW, and GM. All authors contributed to the article and approved the submitted version.
Funding
Support for this work includes funding from the National Natural Science Foundation of China (81670252, 81770034, and 81873649), Guangdong Basic and Applied Basic Foundation (2019A1515011306 and 2020A1515010240), and 2016 Talent Assistance Project of Guangdong (4YF17006G).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.598053/full#supplementary-material
References
Arja, C., Ravuri, R. R., Pulamaghatta, V. N., Surapaneni, K. M., Raya, P., Adimoolam, C., et al. (2014). Genetic determinants of chronic obstructive pulmonary disease in South Indian male smokers. PLoS ONE 9:e89957. doi: 10.1371/journal.pone.0089957
Chappell, S. L., Daly, L., Lotya, J., Alsaegh, A., Guetta-Baranes, T., Roca, J., et al. (2011). The role of IREB2 and transforming growth factor beta-1 genetic variants in COPD: a replication case-control study. BMC Med. Genet. 12:24. doi: 10.1186/1471-2350-12-24
Cho, M. H., Castaldi, P. J., Wan, E. S., Siedlinski, M., Hersh, C. P., Demeo, D. L., et al. (2012). A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet. 21, 947–957. doi: 10.1093/hmg/ddr524
DeMeo, D. L., Mariani, T., Bhattacharya, S., Srisuma, S., Lange, C., Litonjua, A., et al. (2009). Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am. J. Hum. Genet. 85, 493–502. doi: 10.1016/j.ajhg.2009.09.004
Ding, Y., Yang, D., Xun, X., Wang, Z., Sun, P., Xu, D., et al. (2015). Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Hainan population: a case-control study. Int. J. Chron. Obstruct. Pulmon. Dis. 10, 7–13. doi: 10.2147/COPD.S73042
Ghio, A. J., Pritchard, R. J., Dittrich, K. L., and Samet, J. M. (1997). Non-heme (Fe3+) in the lung increases with age in both humans and rats. J. Lab. Clin. Med. 129, 53–61. doi: 10.1016/S0022-2143(97)90161-X
Guo, Y., Lin, H., Gao, K., Xu, H., Deng, X., Zhang, Q., et al. (2011). Genetic analysis of IREB2, FAM13A and XRCC5 variants in Chinese Han patients with chronic obstructive pulmonary disease. Biochem. Biophys. Res. Commun. 415, 284–287. doi: 10.1016/j.bbrc.2011.10.042
Han, Z., Wang, T., Tian, R., Zhou, W., Wang, P., Ren, P., et al. (2019). BIN1 rs744373 variant shows different association with Alzheimer's disease in Caucasian and Asian populations. BMC Bioinformatics 20(Suppl. 25):691. doi: 10.1186/s12859-019-3264-9
Hancock, D. B., Eijgelsheim, M., Wilk, J. B., Gharib, S. A., Loehr, L. R., Marciante, K. D., et al. (2010). Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52. doi: 10.1038/ng.500
Hardin, M., Zielinski, J., Wan, E. S., Hersh, C. P., Castaldi, P. J., and Schwinder, E. (2012). CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am. J. Respir. Cell Mol. Biol. 47, 203–208. doi: 10.1165/rcmb.2012-0011OC
He, D., Ma, L., Feng, R., Zhang, L., Jiang, Y., Zhang, Y., et al. (2015). Analyzing large-scale samples highlights significant association between rs10411210 polymorphism and colorectal cancer. Biomed. Pharmacother. 74, 164–168. doi: 10.1016/j.biopha.2015.08.023
Korytina, G. F., Akhmadishina, L. Z., Viktorova, E. V., Kochetova, O. V., and Viktorova, T. V. (2016). IREB2, CHRNA5, CHRNA3, FAM13A & hedgehog interacting protein genes polymorphisms & risk of chronic obstructive pulmonary disease in Tatar population from Russia. Indian J. Med. Res. 144, 865–876. doi: 10.4103/ijmr.IJMR_1233_14
Li, Y., Song, D., Jiang, Y., Wang, J., Feng, R., Zhang, L., et al. (2016). CR1 rs3818361 polymorphism contributes to alzheimer's disease susceptibility in chinese population. Mol. Neurobiol. 53, 4054–4059. doi: 10.1007/s12035-015-9343-7
Liu, G., Xu, Y., Jiang, Y., Zhang, L., Feng, R., and Jiang, Q. (2017). PICALM rs3851179 variant confers susceptibility to alzheimer's disease in chinese population. Mol. Neurobiol. 54, 3131–3136. doi: 10.1007/s12035-016-9886-2
Mathers, C. D., and Loncar, D. (2006). Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3:e442. doi: 10.1371/journal.pmed.0030442
Nedeljkovic, I., Carnero-Montoro, E., Lahousse, L., van der Plaat, D. A., de Jong, K., Vonk, J. M., et al. (2018). Understanding the role of the chromosome 15q25.1 in COPD through epigenetics and transcriptomics. Eur. J. Hum. Genet. 26, 709–722. doi: 10.1038/s41431-017-0089-8
Pillai, S. G., Kong, X., Edwards, L. D., Cho, M. H., Anderson, W. H., Coxson, H. O., et al. (2010). Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 182, 1498–1505. doi: 10.1164/rccm.201002-0151OC
Qiu, W., Cho, M. H., Riley, J. H., Anderson, W. H., Singh, D., Bakke, P., et al. (2011). Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS ONE 6:e24395. doi: 10.1371/journal.pone.0024395
Rabe, K. F., Hurd, S., Anzueto, A., Barnes, P. J., Buist, S. A., Calverley, P., et al. (2007). Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 176, 532–555. doi: 10.1164/rccm.200703-456SO
Seifart, C., and Plagens, A. (2007). Genetics of chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2, 541–550.
Shen, N., Chen, B., Jiang, Y., Feng, R., Liao, M., Zhang, L., et al. (2015). An updated analysis with 85,939 samples confirms the association between CR1 rs6656401 polymorphism and Alzheimer's disease. Mol. Neurobiol 51, 1017–1023. doi: 10.1007/s12035-014-8761-2
Zhang, S., Zhang, D., Jiang, Y., Wu, L., Shang, H., Liu, J., et al. (2015). CLU rs2279590 polymorphism contributes to Alzheimer's disease susceptibility in Caucasian and Asian populations. J. Neural. Transm. 122, 433–439. doi: 10.1007/s00702-014-1260-9
Zhou, H., Yang, J., Li, D., Xiao, J., Wang, B., Wang, L., et al. (2012). Association of IREB2 and CHRNA3/5 polymorphisms with COPD and COPD-related phenotypes in a Chinese Han population. J. Hum. Genet 57, 738–746. doi: 10.1038/jhg.2012.104
Keywords: chronic obstructive pulmonary disease, IREB2, polymorphism, meta analysis, susceptibility
Citation: Zeng Q, Chen Q, Zou D, Guo R, Xiao D, Jiang S, Chen R, Wang Y and Ma G (2020) Different Associations Between the IREB2 Variants and Chronic Obstructive Pulmonary Disease Susceptibility. Front. Genet. 11:598053. doi: 10.3389/fgene.2020.598053
Received: 23 August 2020; Accepted: 20 October 2020;
Published: 16 November 2020.
Edited by:
Qinghua Jiang, Harbin Institute of Technology, ChinaCopyright © 2020 Zeng, Chen, Zou, Guo, Xiao, Jiang, Chen, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Riling Chen, Y2hlbnJsMzE5QDE2My5jb20=; Yajun Wang, d2FuZ3lhanVueTE5NzdAYWxpeXVuLmNvbQ==; Guoda Ma, c2loYW4xMTA3QDEyNi5jb20=
†These authors have contributed equally to this work
 Qiaoli Zeng
Qiaoli Zeng Qikang Chen1†
Qikang Chen1† Dehua Zou
Dehua Zou Runmin Guo
Runmin Guo Dawei Xiao
Dawei Xiao Shaohu Jiang
Shaohu Jiang Yajun Wang
Yajun Wang Guoda Ma
Guoda Ma