
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 11 June 2019
Sec. Livestock Genomics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00529
This article is part of the Research TopicGenetic Dissection of Important Traits in Aquaculture: Genome-scale Tools Development, Trait Localization and Regulatory Mechanism ExplorationView all 38 articles
 Aglaya García-Angulo1
Aglaya García-Angulo1 Manuel A. Merlo1
Manuel A. Merlo1 María E. Rodríguez1
María E. Rodríguez1 Silvia Portela-Bens1
Silvia Portela-Bens1 Thomas Liehr2
Thomas Liehr2 Laureana Rebordinos1*
Laureana Rebordinos1*Global aquaculture production continues to increase rapidly. One of the most important species of marine fish currently cultivated in Southern Europe is Solea senegalensis, reaching more than 300 Tn in 2017. In the present work, 14 Bacterial Artificial Chromosome (BAC) clones containing candidate genes involved in the immune system (b2m, il10, tlr3, tap1, tnfα, tlr8, trim25, lysg, irf5, hmgb2, calr, trim16, and mx), were examined and compared with other species using multicolor Fluorescence in situ Hybridization (mFISH), massive sequencing and bioinformatic analysis to determine the genomic surroundings and syntenic chromosomal conservation of the genomic region contained in each BAC clone. The mFISH showed that the groups of genes hmgb2-trim25-irf5-b2m; tlr3-lysg; tnfα-tap1, and il10-mx-trim16 were co-localized on the same chromosomes. Synteny results suggested that the studied BACs are placed in a smaller number of chromosomes in S. senegalensis that in other species. Phylogenetic analyses suggested that the evolutionary rate of immune system genes studied is similar among the taxa studied, given that the clustering obtained was in accordance with the accepted phylogenetic relationships among these species. This study contributes to a better understanding of the structure and function of the immune system of the Senegalese sole, which is essential for the development of new technologies and products to improve fish health and productivity.
Global aquaculture production continues to increase rapidly, yet only a small proportion of the aquatic animals and plants being produced are obtained from managed breeding and improvement programs (MacKenzie and Jentoft, 2016). However, the accelerated growth of aquaculture has resulted in adverse effects to the natural environment and to human health. This concern is illustrated by the widespread and, in some cases, unrestricted use of prophylactic antibiotics in this industry, with the objective of preventing bacterial infections resulting from sanitary shortcomings in fish rearing. This practice has resulted in the emergence of antibiotic-resistant bacteria in aquaculture environments, the increase of antibiotic resistance in fish pathogens, the transfer of these resistance determinants to bacteria affecting land animals and to human pathogens, and alterations of the bacterial microbiome in both sediments and water column (Han et al., 2017). All of these are serious and undesirable outcomes.
A viable alternative for avoiding chemicals and preventing economic impact is the administration of immunostimulants, prebiotics and probiotics, which act to reinforce the innate immune system of the farmed fish (Nayak, 2010). A selection program is also an important tool for optimizing the immune capacity of the stocks, developing new technologies and products to improve productivity and to overcome the misuse of antibiotics. Thus, it is essential to understand in depth the structure and function of the fish immune system. However, few studies deal with the immune system in commercially important fish species.
Solea senegalensis is a flatfish species belonging to the Pleuronectiformes order, which comprises about 570 species. It is distributed along the northwestern coast of Africa, as far north as the southwestern coast of the Iberian Peninsula, including the Mediterranean Sea (Díaz-Ferguson et al., 2007). Commercial production of S. senegalensis started in the early 1980’s and this species is considered a promising candidate for the diversification of aquaculture (Chairi et al., 2010; Padrós et al., 2011). In the last 10 years, the production of S. senegalensis in Spain has increased from 32 to 747.15 Tn, which illustrates the rapid growth of interest in production of the species (FishStat, FAO, 2019).
Several studies have already been carried out to improve the production of the Senegalese sole. The high mortality rates at different phases of production and the high incidence of diseases, particularly pasteurellosis and flexibacteriosis, have been critical in recognizing the need advocating for better production methods for the sole (Morais et al., 2014). Comprehensive study of the genes involved in disease resistance should greatly facilitate the solution of these problems.
The elaboration of an integrated genetic map would provide complete information about the localization and structure of genes of interest. This information could be used for comparative genomics purposes, and would constitute the scientific basis for developing improvement programs. In the case of S. senegalensis, the mapping of its genome has been carried out in recent years, using markers such as the minor and major ribosomal genes and other repetitive sequences were first localized using FISH techniques (Cross et al., 2006; Manchado et al., 2006). The elaboration of a BAC library in S. senegalensis has allowed researchers to localize single copy genes (Ponce et al., 2011), and to integrate the cytogenetic map with the physical map obtained by BAC sequencing (García-Cegarra et al., 2013; Merlo et al., 2017; Portela-Bens et al., 2017). In addition, linkage maps were also created in S. senegalensis (Molina-Luzón et al., 2014) and in the closely related species Solea solea (Diopere et al., 2014). A preliminary draft genome for a S. senegalensis female has been published and consisted of 34176 scaffolds with a N50 of 85 kb. Furthermore, this draft genome contained 209 out of the 274 ultra-conserved core eukaryotic genes, with a completeness of 84.3% and an average number of orthologues of 1.31, considering the number of eukaryotic genes discovered into the scaffolds (Manchado et al., 2016).
In S. senegalensis, the gene expression of various genes related to the immune system has been examined, including hepcidin, lysozyme g-type and the TNF gene family (Salas-Leiton et al., 2010, 2012; Núñez-Díaz et al., 2016). An exhaustive expression analysis of genes relevant for the immune system was also undertaken in the closely related species S. solea (Ferraresso et al., 2016). However, knowledge of the gene structure, genomic characterization and localization of immune-related genes is limited. Studies of this kind have been carried out only with the g-type lysozyme (Ponce et al., 2011), myxovirus resistance protein 1, immunoglobulin superfamily member 9b, and semaphorin 7a (García-Cegarra et al., 2013).
In this work, the localization and the genomic organization of 14 BAC clones containing immune-related genes was assessed. Seven out of the 14 BAC clones contain well-known immune-related genes, such as the g-type lysozyme (lysg), myxovirus resistance 1 (mx1), toll-like receptors 3 and 8 (tlr3 and tlr8), beta-2-microglobulin (b2m), interferon regulatory factor 5 (irf5) and tumor necrosis factor α (tnfα). Another four BAC clones were chosen for their relationship with the immune system found in the bibliography, such as antigen peptide transporter 1 (tap1) (Pinto et al., 2011) interleukin-10 (il10) (Zou et al., 2003), and two BAC clones with calreticulin (calr) (Wang et al., 2018). The remaining three were anonymous BAC clones that, in the sequencing and annotation process, were found to contain immune-related genes, such as tripartite motif-containing proteins 16 and 25 (trim16 and trim25) and high mobility group protein B2 (hmgb2). The genes studied belong to both the innate and acquired immune system. The objective of this study was to carry out an analysis of micro-synteny, comparative mapping and phylogenetics between S. senegalensis and relevant aquaculture species. This will allow deepening the knowledge about the structure of the genome and evolutionary trends of the immune system within flatfish species. The results would facilitate future work related to quantitative trait loci (QTL) and gene expression.
A 4D-PCR methodology (Asakawa et al., 1997) was carried out to find and isolate clones bearing targeted gene sequences from a BAC library previously constructed in S. senegalensis (García-Cegarra et al., 2013). Fourteen BAC clones containing immune-related genes were isolated (Table 1). The thirteen candidate genes used to isolate BACs were lysozyme g type (lysg), calreticulin (calr), myxovirus resistance 1 (mx1), toll-like receptors 3, and 8 (tlr3, and tlr8), interferon regulatory factor 5 (irf5), beta-2 microglobulin (b2m), antigen peptide transporter 1 (tap1), interleukin-10 (il10), tumor necrosis factor α (tnf α), tripartite motif-containing protein 25 (trim25), high mobility group protein B2 (hmgb2) and tripartite motif-containing protein 16 (trim16). The PCR conditions were the same as those described in García-Cegarra et al. (2013). BACs are named after the name of the harboring candidate gene.
Positive BAC clones were isolated using the Large Construct Kit (Qiagen, Hilden, Germany), and sent for sequencing using the Illumina sequencing platform (Illumina, San Diego, CA, United States). The sequences were generated on the Miseq equipment, with a configuration of 300 cycles of paired end reads (Lifesequencing S.L., Valencia). The reads were de novo assembled using SPAdes software version 3.11.1. The functional and structural annotation of the gene sequences identified in each BAC clone was carried out in a semi-automated process. Proteins and expressed sequence tags (ESTs) from S. senegalensis and related species were compared. The homologous sequences obtained were used to obtain the best predictions for gene annotation. Finally, all the information available was used to create plausible models and, when possible, functional information was added. Using the Apollo genome editor (Lewis et al., 2002), Signal map software (Roche Applied Science, Penzberg, Germany), and Geneious R11 (Kearse et al., 2012), the results were individually completed and adjusted in the final editing process of the annotation. All BAC clones have been deposited in the GenBank database under the accession numbers AC278047 to AC278120. The structure of some of the genes was compared with those of seven other representative fish species, i.e., Danio rerio (zebrafish), Oreochromis niloticus (tilapia), Gasterosteus aculeatus (stickleback), Seriola dumerili (greater amberjack), Seriola lalandi dorsalis (yellowtail amberjack), Scophthalmus maximus (turbot), and Cynoglossus semilaevis (tongue sole).
Cross-species genome comparisons were carried out at two levels. At the first level, a micro-synteny study was performed using the ENSEMBL database and the NCBI platform. The order of the contigs within each BAC clone of S. senegalensis was estimated using the information provided by these programs. The seven species used for this comparison were the same as those listed above. All BAC clones were analyzed with the exception of the mx BAC clone, where only one gene was found, and the trim16 BAC clone. In the schematic figures each gene is represented by a different color; white color indicates a gene that is different from that found in the Senegalese sole.
At the second level of comparison, a synteny analysis was performed using the CIRCOS software (Krzywinski et al., 2009). The five species used in this analysis are C. semilaevis, S. maximus, O. niloticus, G. aculeatus and D. rerio. The flatfish Paralichthys olivaceus has not been included because the genome assembly is not at chromosome level. These sequences are available in the ENSEMBL database and the NCBI platform. In the case of S. dumerilii and S. lalandi dorsalis, the synteny analysis could not be done because the complete genomes were not available in these databases. In the figures the locations of the genes that make up the BAC clones of S. senegalensis were compared with the location they presented in the other species studied, so that the relationship between the chromosomes of both species appears in the figures. Each BAC clone was represented by a different color so that co-localizations and reorganizations of genes could be better observed.
Chromosome preparations were obtained according to García-Angulo et al. (2018). To prepare FISH probes, BAC clones were grown on LB containing chloramphenicol, at 37°C, overnight. BAC-DNA was extracted using the BACMAXTM DNA purification kit (Epicenter Biotechnologies, Madison, United States), following the manufacturer’s instructions. Insert presence was evaluated by digestion with EcoRI and agarose gel electrophoresis (0.8%). Probes were amplified by DOP-PCR and then labeled by a conventional PCR using four different fluorochromes, i.e., Texas Red (Life Technologies, Carlsbad, California, United States), Spectrum Orange, Fluorescein isothiocyanate (FITC) (Abbott Molecular/ENZO, Illinois, United States), and diethylaminocoumarin (DEAC) (Vysis, Downers Grove, United States), using the protocol described in Liehr (2009).
Chromosome preparations were pre-treated with pepsin solution at 37°C and fixed with paraformaldehyde solution. Finally, preparations were dehydrated with ethanol in a concentration series of 70%, 90%, and 100%, and air-dried. Hybridization and post-hybridization treatment was according to Portela-Bens et al. (2017).
Slides were visualized with a fluorescence microscope (Olympus BX51 and/or Zeiss Axioplan using software of MetaSystems, Altlussheim, Germany) equipped with a digital CCD camera (Olympus DP70) to capture the images.
Before concatenation, the sections with the highest homogeneity in each gene were taken and the substitution saturation degree was also examined in each gene using saturation plots with transitions (s) and transversions (v) implemented in the DAMBE6 software (Xia, 2017). The distance model used was GTR. Saturation is inferred when the index of substitution saturation (ISS) is either larger or not significantly smaller than the critical value (ISS.C). Finally, for phylogenetic analysis were chosen those genes representatives of different immune pathways and present in a wide number of species, in addition to do not present significative substitution saturation. Under these requirements, up to five immune-system genes (tlr3, tlr8, nlrc3, calr, ikbke) were concatenated to perform the phylogenetic analysis. Thirty-four species were included to generate the phylogenetic tree; twenty-two were fish species, including the target species S. senegalensis; ten mammal species, one reptile species and, additionally, Latimeria chalumnae was included as an outgroup to root the tree. The sequences were aligned using the MAFFT program (Katoh and Toh, 2008) following an iterative method of 100 iterations. The final alignment consisted in a total of 7806 positions, in which 2354 were for tlr3, 1725 for tlr8, 1772 for nlrc3, 1284 for calr and 671 for ikbke. The PhyML 3.0 program (Guindon et al., 2010) was used to determine the best-fit phylogenetic model and then to run the model. The resulting best-fit model was the Generalized Time-Reversible (GTR) model (Tavaré, 1986), considering the proportion of invariable sites (+I) and gamma distribution (+G). The statistic used for model selection was the Akaike information criterion (AIC), the value of which was 255435.93458, and the -LnL was -127384.510441. Branch support was tested by the fast likelihood-based method using aLRT SH-like (Anisimova et al., 2011). Finally, the tree was edited in the MEGA7 program (Kumar et al., 2016).
Of the 14 BAC clones analyzed, nine BAC clones were sequenced with a total of 80 genes annotated, and the other five BAC clones had been sequenced previously (Ponce et al., 2011; García-Cegarra et al., 2013; Merlo et al., 2017; García-Angulo et al., 2018). In total, 109 genes were annotated and 24 of the 109 genes (22.01%) were found to be related to the immune system (Table 1).
Using mFISH, the 14 BAC clones were localized on six different chromosome pairs. (Figures 1, 2). BAC clones hmgb2, b2m, irf5, and trim25 co-localized in a metacentric chromosome pair. The BAC clones tlr3 and lysg co-localized in one acrocentric chromosome pair. The BAC clones tap 1 and tnfα co-localized in a second acrocentric chromosome pair; and, lastly, the BAC clones il10, mx and trim16 co-localized in a third acrocentric chromosome pair. Conversely, the BAC clone calr showed a signal on the largest metacentric chromosome, different from that in which the genes hmgb2, b2m, irf5, and trim25 were co-located. The BAC clone tlr8 showed a signal in two different chromosomes pairs, one stronger signal in a submetacentric pair and the other weaker signal in an acrocentric chromosome pair. Only the most intense signal was considered.
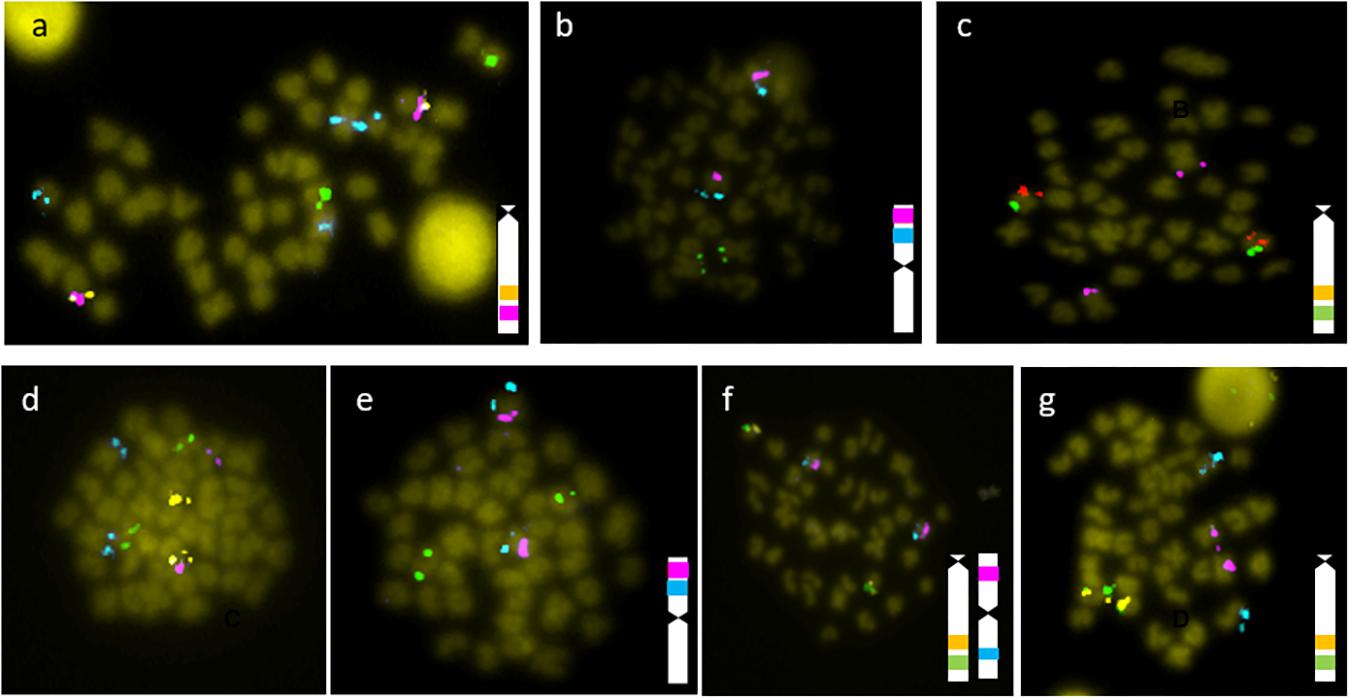
Figure 1. Results of mFISH of the BACs isolated with the following candidate genes: (a) tlr8 (blue), lysg (green), tap1 (pink), tnfα (orange); (b) irf5 (blue), calr (green), b2m (red); (c) trim 16 (green), mx (orange), calr (pink); (d) irf5 (blue), tlr3 (green); tnfα (orange), mx (pink); (e) hmgb2 (blue), lysg (green), b2m (pink); (f) trim25 (blue), tnfα (green), tap1 (orange), b2m (pink); (g) il10 (blue), tnfα (green), tap1 (orange), b2m (pink). In those cases where two or more probes co-localize in one chromosome, a diagrammatic representation is included.
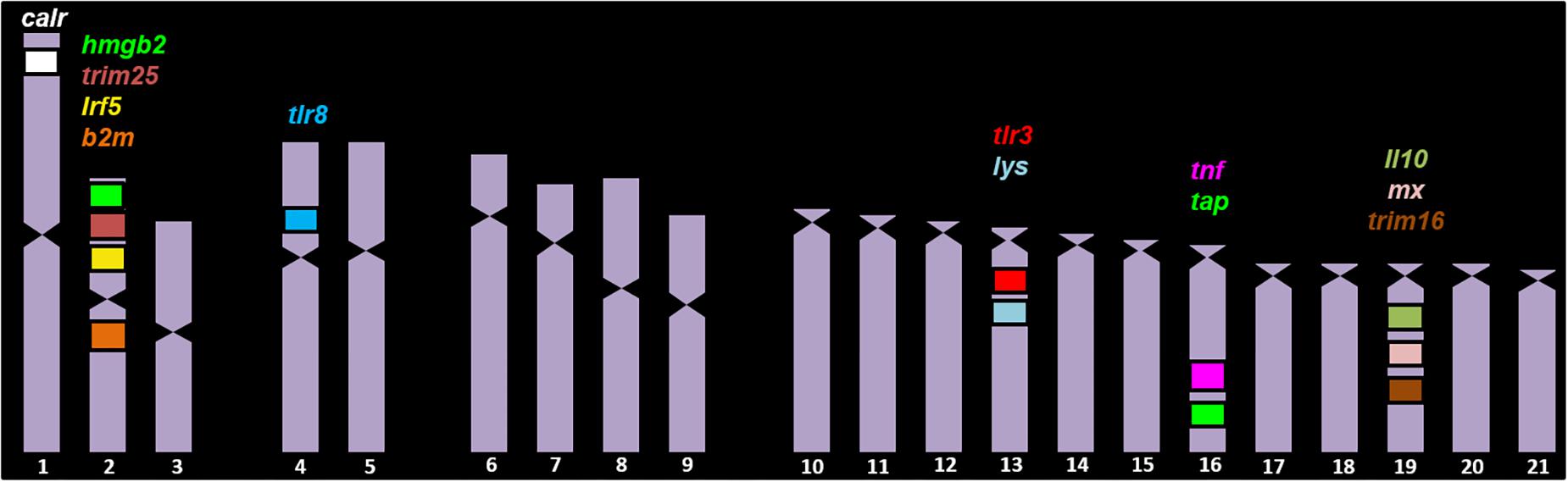
Figure 2. BAC clones analyzed in the chromosomes of S. senegalensis. On chromosome 1 the BAC clone calr was localized; on chromosome 2 there were four BAC clones, with the genes b2ml, b2m, irak3, trim25, hmgb2, and irf5. On chromosomes 4 and 12 the BAC clone 30J4 was localized, with the genes tlr8, tlr7, and nlrc3. The BAC clones 4N21 and 29D4, with the lysg and tlr3 candidate genes, respectively, were found on chromosome 13. On chromosome 16, the BAC clones 53D20 and 71N11 were localized, with the genes tap, hla-drb1, tnfα and pycard. Lastly on chromosome 19 the BAC clones 42P4, 12K6 and 31C1 were localized, with the genes il10, il19, mx, trim16, ikbke.
The genes annotated in Senegalese sole were found in eleven chromosomes in C. semilaevis, S. maximus and O. niloticus, in nineteen chromosomes in D. rerio, and in eleven and five scaffolds in G. aculeatus (Figures 3, 4 and Supplementary Figures S1–S3). The closest species was C. semilaevis: 96.36% of the genes of S. senegalensis were found in seven chromosomes of C. semilaevis (Figure 3). Moreover, D. rerio was the species with the largest number of rearrangements (Supplementary Figure S1).
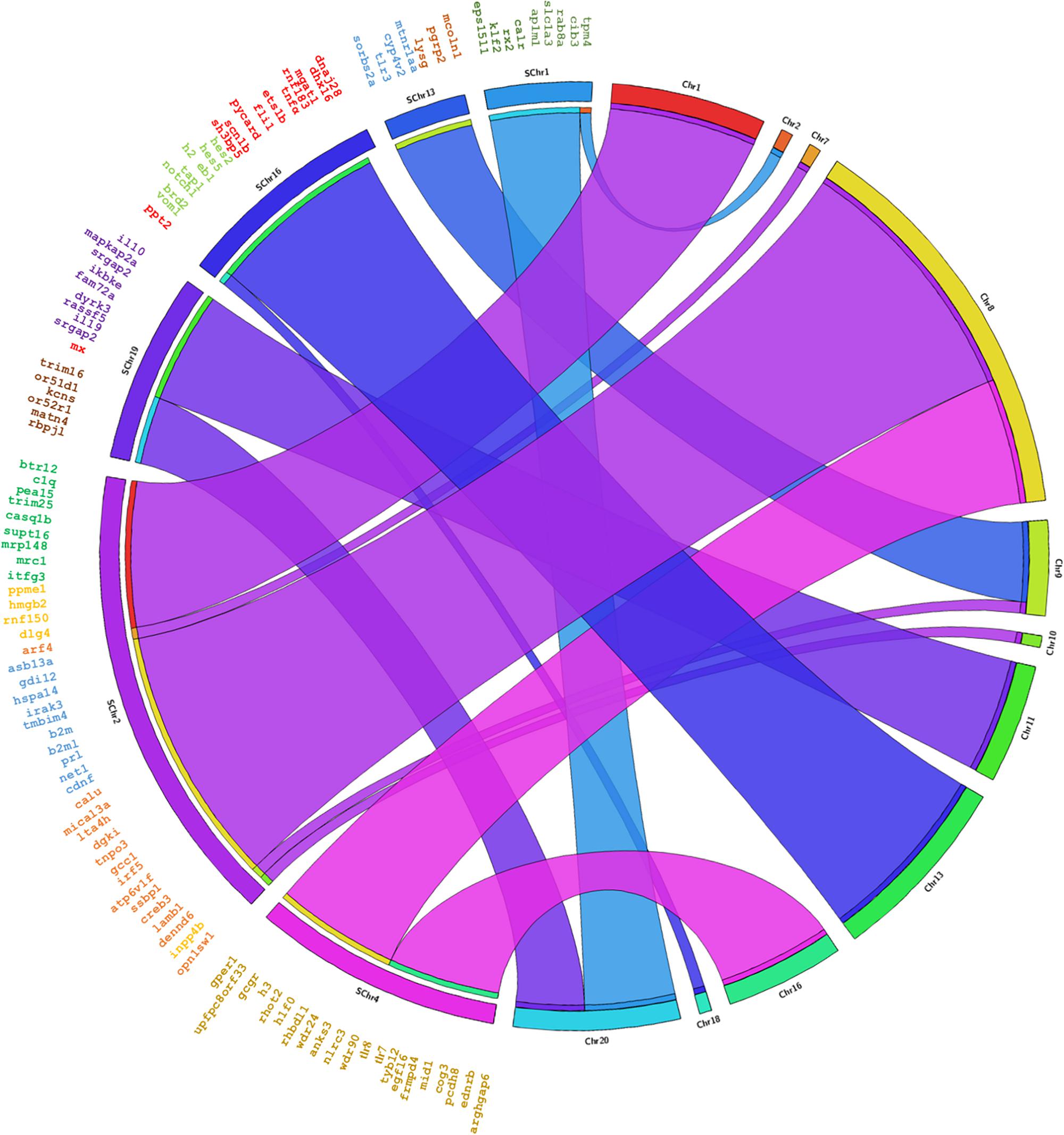
Figure 3. Circos analysis in the species C. semilaevis. On the left side the distribution of the BAC clones of Senegalese sole distributed in chromosomes can be observed. Each BAC clone is represented in a different color. The genes found by annotation are indicated within each BAC clone and their corresponding localization in the C. semilaevis chromosomes is denoted by crossing lines. The BAC clones analyzed are given in Table 1.
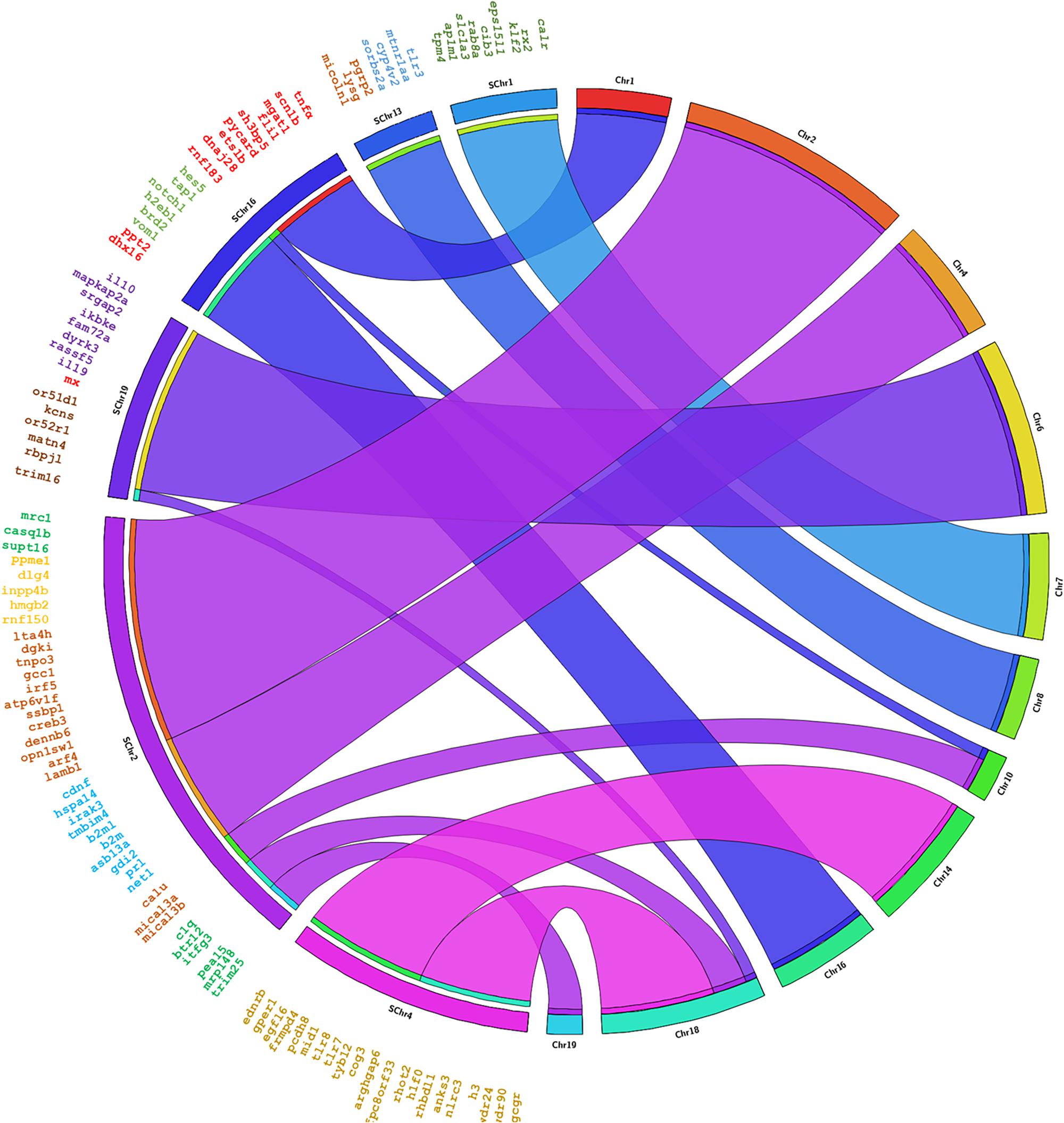
Figure 4. Circos analysis in the species S. maximus. On the left side the distribution of the BAC clones of Senegalese sole distributed in chromosomes can be observed. Each BAC clone is represented in a different color. The genes found by annotation are indicated within each BAC clone and their corresponding localization in the S. maximus chromosomes are denoted by crossing lines. The BAC clones analyzed are given in Table 1.
Considering the comparison with C. semilaevis, large clusters of genes were conserved with S. senegalensis. Hence, chromosome 2 of Senegalese sole seems to correspond mainly with chromosomes 1 and 8 of C. semilaevis; chromosome 4 with chromosomes 8 and 16; and chromosome 19 with chromosomes 11 and 20 (Figure 3). S. maximus also presented large conserved regions with respect to S. senegalensis; chromosomes 1, 13 and 19 of Senegalese sole seem to correspond to chromosomes 7, 8 and 6 of S. maximus, respectively (Figure 4). The comparison with O. niloticus, G. aculeatus and D. rerio showed more gene rearrangements (Supplementary Figures S1–S3).
Several gene co-localizations observed in S. senegalensis also appeared in other species, as the cases of il10-mx-trim16 in S. maximus and O. niloticus, tlr7-tlr8 in all species, tlr3-lysg in C. semilaevis, S. maximus and G. aculeatus, tnfα-tap1 in C. semilaevis, O. niloticus and G. aculeatus, trim25-hmgb2 in C. semilaevis and G. aculeatus, b2m-irf5 in C. semilaevis and D. rerio, b2ml-irf5 in O. niloticus, or hmgb2-irf5 in S. maximus.
The micro-synteny analysis showed that many candidate genes have conserved genomic surroundings, and that, among the genomic regions analyzed, C. semilaevis is the species with greatest homology. The region surrounding genes il10, tlr3, tlr8, nlrc3, and calr were highly conserved in all species (Figures 5a,b,k,e,i). The micro-synteny of BAC clone b2m showed that gene b2m presented one paralog gene (b2ml) in all the species analyzed apart from D. rerio. Curiously, in O. niloticus the paralog gene was present but not the candidate gene (Figure 5f). The most gene rearrangements within the same genomic structure were observed in the micro-synteny of BAC clone hmgb2 (Figure 5c).
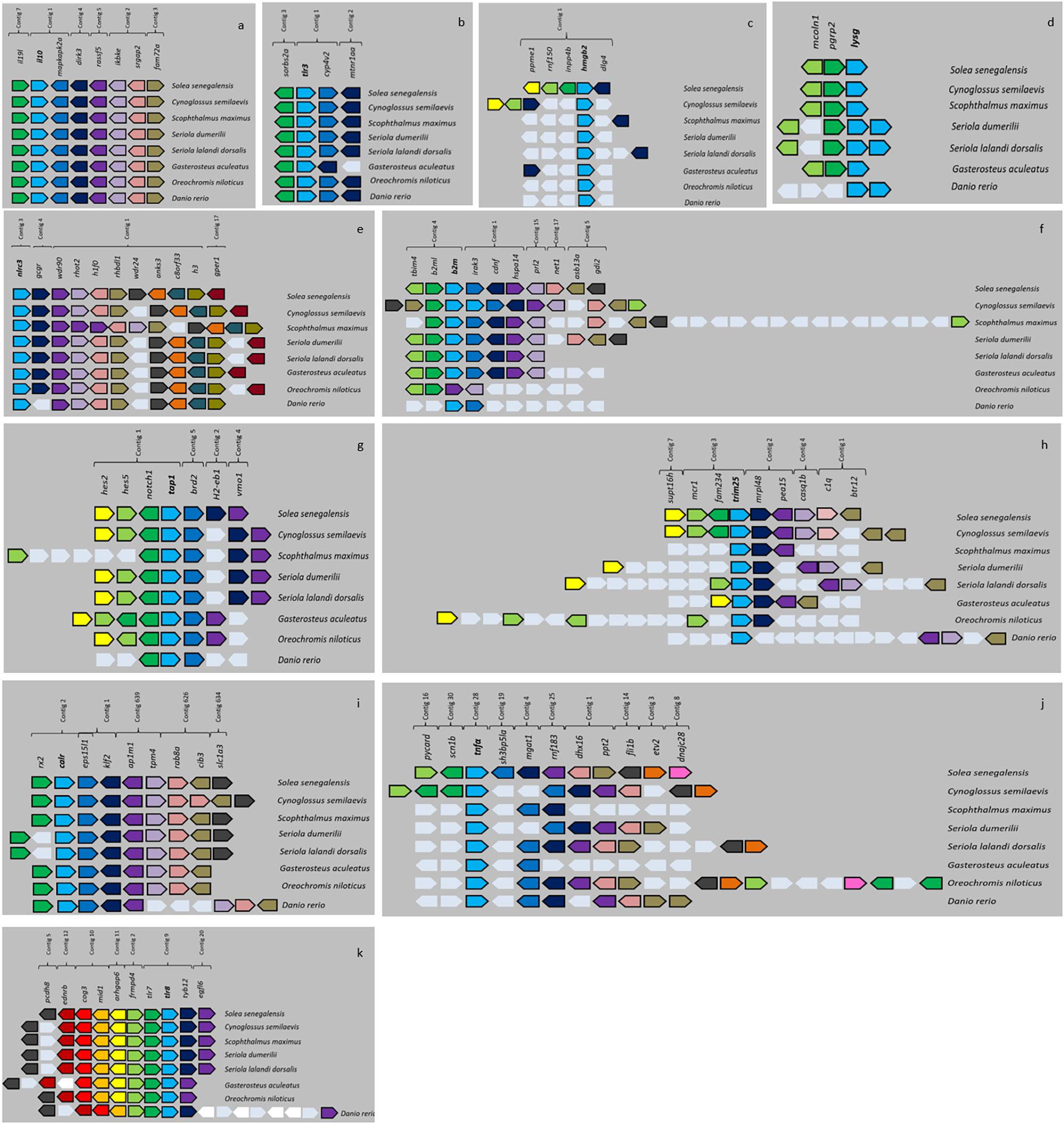
Figure 5. Micro-synteny of 11 BAC clones. Genes of the species C. semilaevis, S. maximus, S. lalandi dorsalis, S. dumerili, G. aculeatus, O. niloticus, and D. rerio are represented in different colors. White color indicates non-synteny genes. (a) BAC clone il10; (b) BAC clone tlr3; (c) BAC clone hmgb2; (d) BAC clone lysg; (e) BAC clone nlrc3; (f) BAC clone b2m; (g) BAC clone tap1; (h) BAC clone trim25; (i) BAC clone calr; (j) BAC clone tnf alfa; (k) BAC clone tlr8.
In some of the BAC clones analyzed, other genes whose function is involved in the immune system were also found. In the BAC clone il10, the genes il19 and ikbke appeared in the same region (Figure 5a). In the BAC clone tap1, the gene hla-drb1 was found (Figure 5g). In the BAC clone tnfα, the gene pycard was observed (Figure 5j). In the BAC clone tlr8, the genes tlr7 and nlrc3 appeared (Figures 5e,k). In the BAC clone lysg, the gene pgrp2 was found (Figure 5d). In the BAC clone b2m, the genes b2m, b2ml and irak3 appeared (Figure 5f). Lastly, in the BAC clone trim25, genes c1q and btr12 were found (Figure 5h). These genomic architectures were not found in all species: only the group of genes il10-il19-ikbke (Figure 5a) and the tandem tlr8-tlr7 were preserved among all the species analyzed (Figure 5k).
To carry out the phylogenetic analysis, up to five candidate genes were concatenated (Supplementary Data Sheet S1). The resulting alignment had 7806 positions, where 5808 and 4975 were variable and parsimony-informative, respectively. The nucleotide frequencies were similar: f(A) = 0.26617; f(C) = 0.27001; f(G) = 0.24585; f(T) = 0.21796; and GTR relative rate parameters were A < – > C 1.30911; A < – > G 3.60083; A < – > T 1.25710; C < – > G 1.05237; C < – > T 4.73877; G < – > T 1.00000, with the proportion of invariable sites at 0.191. Results of substitution saturation tests (Xia et al., 2003) for each gene and for the concatenated alignment did not show any significant saturation, since ISS indices were lower than ISS.C values in all cases (Supplementary Data Sheet S2). The phylogenetic tree showed a good resolution and robust branch support. The phylogeny clearly separated two main clusters: ray-finned fishes and tetrapods (Figure 6). The cluster of tetrapods was divided into mammals and reptiles, and the cluster of ray-finned fishes was divided into Holostei and Teleostei. Within the group of teleosts, S. senegalensis appeared included in a subgroup together with the species C. semilaevis, P. olivaceus, S. maximus, Lates calcarifer and S. dumerili. All these species belong to the Carangaria group.
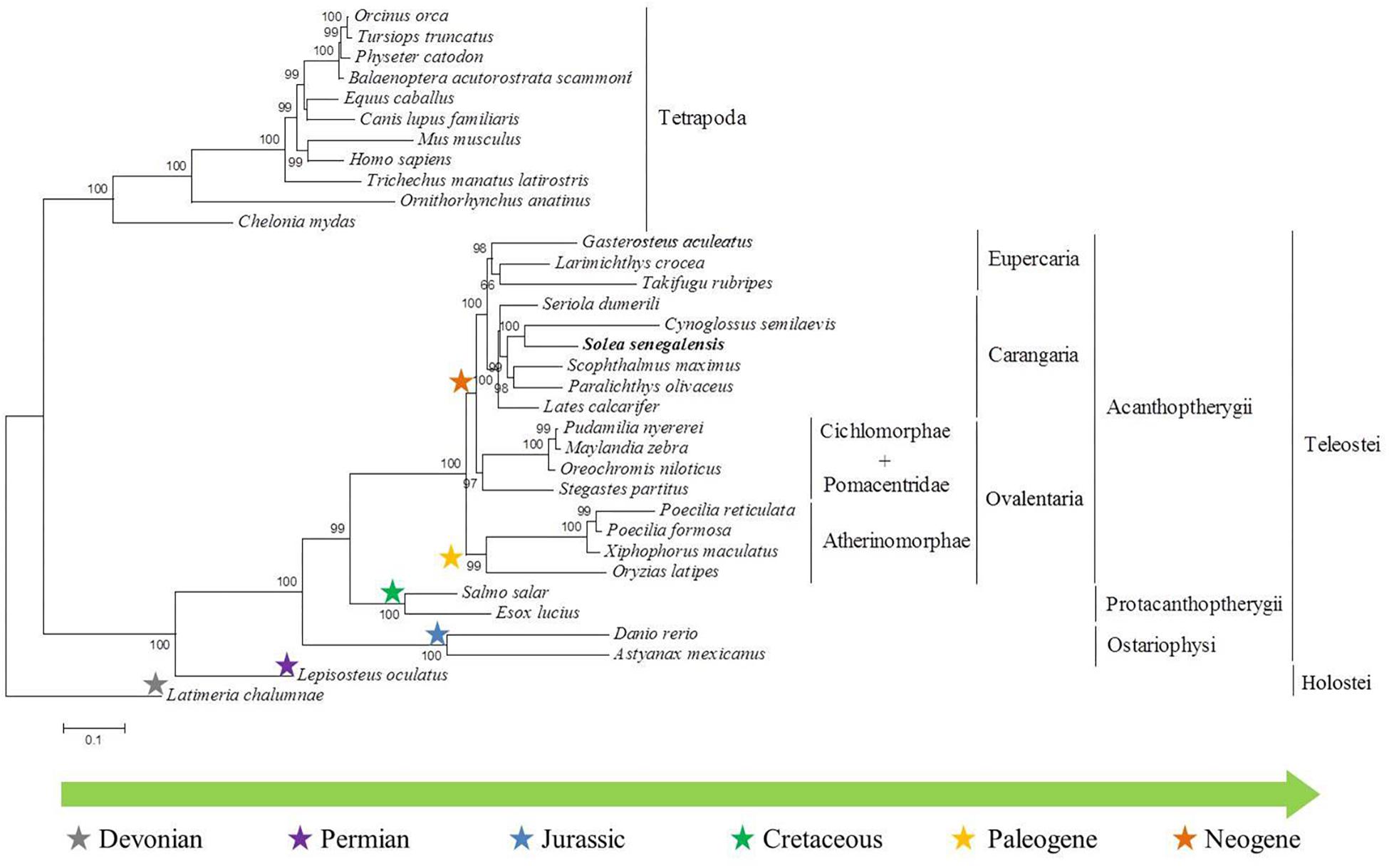
Figure 6. Phylogenetic tree constructed from five immune-system genes (tlr3, tlr8, nlrc3, calr, ikbke) of thirty-four different species.
From the BAC library of S. senegalensis 16 genes related to the immune system have been obtained. These genes, together with those already annotated, make a total of 24 relevant genes (Table 1). The use of BAC libraries has proven to be helpful for characterizing the genome in different species, including the fish species Larimichthys crocea (Ao et al., 2015), S. maximus (Taboada et al., 2014), Ictalurus punctatus (Xu et al., 2007), Oncorhynchus mykiss (Palti et al., 2009), and bivalve species (Zhang et al., 2011; Cross et al., 2018).
Seventeen of 24 of the annotated genes related to the immune system are part of the NF-κB signaling pathway or JAK-STAT signaling pathway, two routes for the immune response. Up to twelve genes have been annotated for the first pathway, including the genes tlr, nlrc3, lysg, pgrp2, il, tnfα, irak3, ikbke, and pycard.
Toll-like receptors (tlr) are a class of pattern recognition receptors (PRR) and their function is to recognize microbial pathogens. In fish species, the tlr genes exhibit distinctive features and large diversity; these differences are probably derived from the diverse evolutionary history of this group and the distinctive environments that they occupy (Palti, 2011). This feature makes this kind of genes good candidates for immunity-response improvement of the aquaculture stocks. The tlr8 gene was found to be composed by only one exon in S. senegalensis, as also occurs in C. semilaevis and G. aculeatus. However, in S. maximus, S. dumerili and D. rerio, tlr8 is composed by two exons and one intron, which could indicate an intron losing process in different lineages during the teleost species evolution. Another class of PRR has been annotated within the same BAC clone as that of genes tlr7 and tlr8, the gene nlrc3. Whereas TLR proteins are extracellular PRRs that recognize extracellular PAMPs, NLR proteins are intracellular PRRs that recognize intracellular PAMPs (Sha et al., 2009). Two additional PRRs were annotated in other BAC clone, the genes lysg and pgrp2. Lysozyme is a conserved molecule in teleosts (Sun et al., 2006) and is an important enzyme of the innate immunity response to bacterial infection. Many factors, such as stress and infection, sexual maturity, nutrition, toxic substances, and others, have been studied in relation to the activity levels of lysozyme in fish (Saurabh and Sahoo, 2008). The PGRP2 protein, as lysozyme, also recognizes and hydrolyzes the peptidoglycan layer (Choi et al., 2019).
The remaining genes of the NF-κB signaling pathway do not participate as PRRs. Instead, the function is downstream of the pathway. The genes il10 and il19 encode for two members of the IL-10 cytokines family, which includes IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26 (Lutfalla et al., 2003). The IL-10 cytokine plays an important role as an anti-inflammatory agent in the innate and adaptive immune system, and IL-19 is a pro-inflammatory cytokine of the innate immune system (Hofmann et al., 2012). The il19 gene of S. senegalensis showed a conserved structure across different teleost species, since it is structured in 5 exons and 4 introns; the exception is G. aculeatus, which shows an additional exon. tnfα, as well as il10 and il19, is another cytokine that is secreted by activated immune-related cells upon induction by various pathogens - parasitic, bacterial and viral (Salazar-Mather and Hokeness, 2006). The irak3 gene encodes for an inteleukin-1 associated-receptor kinase, which, in the orange-spotted grouper (Epinephelus coioides) has been proved to induce the NF-κB activation through TLR signaling (Li et al., 2018). It has been reported in humans that ikbke and pycard genes act as positive and negative regulators, respectively, of the NF-κB activation. In addition, ikbke also participates in the JAK-STAT signaling pathway (Sarkar et al., 2006; Liu et al., 2019).
Aside from ikbke, another five annotated genes participate in the JAK-STAT signaling pathway for the immunity response. Interferons (IFN) are another type of cytokine that are involved in key aspects of the host defense mechanisms (Reboul et al., 1999). Interferon regulatory factors (IRF) were originally identified as transcription factors participating in the regulation of interferon expression (Mamane et al., 1999). It is known that, in C. semilaevis, the irf5 gene may play a role in the immune defense, primarily against intracellular pathogens (Zhang et al., 2015). The irf5 gene structure was conserved across representative species of the Pleuronectiformes order and in O. niloticus and G. aculeatus, composed by 8 exons and 7 introns. Non-etheless, the two representative species of the Seriola genus showed a derived structure composed by 6 exons and 5 introns, but more analyses needs to be done in order to ascertain if this structure is plesiomorphic within the Carangidae family. D. rerio also showed a derived structure composed by 9 exons and 8 introns, which could be representative of such ancient lineage of teleosts. Trim16, trim25 and btr12 genes present orthologs in mammals, but btr genes come from the trim39 gene of mammals. Both trim16 and trim25 are considered the genes from which the so-called fintrims, specific to teleosts, diverged (van der Aa et al., 2009). These three genes belong to the class IV subgroup of TRIM proteins, which are involved in antiviral immunity of the IFN signaling cascade (Boudinot et al., 2011). Two different types of Mx were observed in the European seabass (Dicentrarchus labrax), and both showed antiviral activity, but with different intensity and spatial and temporal patterns (Novel et al., 2013).
Calreticulin is a calcium-binding protein with an important role in the assembly and expression of Major Histocompatibility Complex (MHC) class I molecules (Raghavan et al., 2013). Two calreticulin types that may function as a PRR have been described in the flatfish C. semilaevis, thus demonstrating the antiviral and antibacterial activity of that molecule (Wang et al., 2018).
Regarding the MHC molecules, four additional genes have been annotated. MHC molecules are members of the immunoglobulin superfamily that present pathogen peptides of infected cells and thus initiate the generation of adaptive immunity to pathogens (Zhu et al., 2013). The b2m gene codifies for the beta subunit of the MHC class I and different paralog copies have been observed in several fish species (Kondo et al., 2010; Sun et al., 2015). Two types of b2m genes have been found in S. senegalensis and adjacent to each other. This can be observed in genome databases of fish species, in which the two adjacent copies are annotated as b2m and b2ml. Both genes presented different gene structures in S. senegalensis, since in b2m two exons and one intron were identified, and in b2ml 3 exons and 2 introns. The structure of b2ml was conserved across the other representative teleost species, but not the structure of the b2m gene, which is composed by 3 exons and 2 exons in those species. An expression analysis would be able to conclude definitively if this gene is undergoing a pseudogenization process. The tap1 gene plays an essential role in the antigen presentation MHC class I pathway, transporting peptides from cytosol to the lumen of the endoplasmic reticulum (ER), where the peptides are loaded to MHC class I (Pinto et al., 2011). In the same BAC clone where the tap1 gene is located, another MHC-related gene, hla-drb1 was found, which encodes for the beta subunit of the MHC class II. In primates five different families of MHC class II genes, including the DR family, have been described (Satta et al., 1996).
The c1q gene belongs to the C1 component of the complement system pathway, and binds immunoglobulins attached to pathogen surfaces; it subsequently activates a complement cascade that culminates in the elimination of the infectious agents (Chen et al., 2018).
The hmgb2 gene belongs to a gene family that encodes non-histone chromosomal proteins, and the HMGB2 protein has been demonstrated to display an antibacterial activity in fish due to its ability to bind pathogen DNA (Wang et al., 2019). The hmgb2 gene has 4 exons and 3 introns in S. senegalensis, a structure that is also observed in the other representative teleost species.
The 24 annotated genes related to the immune system are distributed in 28.571% of the chromosome complement of S. senegalensis. This was the lowest value in comparison with those of C. semilaevis (33.333%), O. niloticus (40.909%), S. maximus (45.455%), G. aculeatus (47.619%), and D. rerio (52%). These data clearly show a grouping tendency of immune system genes in the two Soleoidei species, and in S. senegalensis in particular. Such grouping could be a consequence of the reduction and compaction trend observed in the Pleuronectiformes evolution (Cerdá and Manchado, 2013). Moreover, more than 50% of the genes studied fell into two chromosome pairs, which could reflect a certain degree of chromosome specialization in order to facilitate the immune response of the organism. Altogether, non-random proximity patterns could be formed in a way that provides functional advantages in the genomic architecture (Cremer and Cremer, 2010). A study carried out in 2008 indicated that the selection of groups of genes of the immune system could have been an important factor that affected the reorganization of the vertebrate genome (Makino and McLysaght, 2008). However, further analysis including new immune-related genes will be required to prove this hypothesis.
The hybridizations agree with the results obtained for the BAC clones tlr8, calr, lysg and mx that had already been located (Ponce et al., 2011; García-Cegarra et al., 2013; Merlo et al., 2017; García-Angulo et al., 2018). In our study, candidate genes tlr3 and tlr8 were found in different chromosomes. This result has also been observed in all the species studied and in other Pleuronectiformes species such as P. olivaceus (Hwang et al., 2011). The tlr7 and tlr8 locus is highly conserved in vertebrates and these genes are located together in the chromosomes of mammals, birds and fishes (Rauta et al., 2014).
The gene nlrc3 was located within the BAC clone tlr8, but, in all the species analyzed, the gene nlrc3 was found in a chromosome different from the genes tlr7 and tlr8. However, in Tetraodontiformes species (Tetraodon nigroviridis and Takifugu rubripes) the genes tlr7 and nlrc3 were in the same chromosome, which may be due to the compact structure of the genome in the Tetraodontiformes species, since these species have genomes that are among the most compact known in vertebrates (Jaillon et al., 2004). As mentioned before, this same trend could have taken place in the Pleuronectiformes evolution.
As can be deduced from the fish genome databases, the gene tlr3 is linked with genes lysg and pgrp2, and this linkage is conserved throughout many bony fishes, thus representing a genomic cluster that evolves together. As discussed before, the three genes share similar functions, so the linkage could represent an advantage for a more effective immune response. It has been postulated that a conserved group of genes could indicate a functional cluster (Overbeek et al., 1999). The calreticulin BAC clone appears as a region strongly conserved in all the species studied at the level of macro-synteny and micro-synteny, so it could represent another functional cluster. This same situation has been observed with the genes belonging to BAC clone il10, which show a highly conserved synteny in all the species analyzed, including the il19 gene and, in some species, the mx gene. The linkage between il10 and il19 has been described in other fish species (Lutfalla et al., 2003). Another example of conserved linkage among fish species is the tap1-tnfα genes. In S. senegalensis the irf5 gene co-localized with the genes b2m, trim25, and hmgb2 in one metacentric chromosome. Interestingly, this result was not observed in any other of the species studied, although a tendency to co-localize two-to-two was observed.
The results also show that large parts of the genomic regions tend to be conserved in the species most closely related to S. senegalensis, such as the Pleuronectiformes C. semilaevis and S. maximus, with C. semilaevis being the species that presents the most homology between the genomic regions analyzed. Regardless of the genetic distance between the species, a large conserved region of genes present in chromosome 2 of S. senegalensis was found in only one chromosome in the rest of the species analyzed. An exception was observed in stickleback; this is because part of the stickleback genome is assembled at the scaffold level.
The results of micro-synteny revealed a paralog of b2m in most of the species analyzed. It is clear that gene duplication and subsequent diversification have played a major role in the evolution of diversity in molecules of the MHC (Andersson, 1996). In studies carried out in bivalve species such as Crassostrea gigas, the expansion of several gene families related to defense pathways, including protein folding, oxidation and anti-oxidation, apoptosis and immune responses, has been observed (Wang et al., 2012).
The phylogeny results clearly separated two main clusters: ray-finned fishes and tetrapods. The coelacanth was used to root the tree and, as expected, the genetic sequence analyzed is closer to the tetrapods than to the actinopterygian fish. The innate immune system is phylogenetically older than the adaptive system, and it is found, in some form, in all multicellular organisms, whereas the adaptive system is found in all vertebrates except jawless animals (Zarkadis et al., 2001). The concatenated gene sequences used in the phylogenetic analysis belong to both the innate and adaptive immune systems, so it was not possible to set an invertebrate as an outgroup species. However, several studies indicate that the coelacanth provides the ideal outgroup sequence against which tetrapod genomes can be measured (Noonan et al., 2004).
The cluster of tetrapods is divided into mammals and reptiles; the cluster of ray-finned fishes is divided into holostei and teleostei. Ray-finned fishes (Actinopterygii) diverged from the lineage of lobe-finned fishes (Sarcopterygii) about 450 million years ago (Christoffels et al., 2004). The actinopterygians, in turn, are divided into two main groups: holostei, the only representative of which in this tree is Lepisosteus oculatus; and teleostei in which group the other fish analyzed are included. L. oculatus appears Paleozoic era and the first fossil record dates from the Late Permian period. The genome of L. oculatus has a very low evolution rate and it was sequenced to help connect teleost biomedicine to human biology because its lineage represents the unduplicated sister group of teleosts (Anderson et al., 2012).
As these results show, within the clade of the teleosts, the phylogenetic tree is divided into three main groups. In the first group (superorder Ostariophysi), Astyanax mexicanus and D. rerio belong to the cohort Otomorpha and are separated from the cohort Euteleosteomorpha (the rest of those with radiated fins). In the second group (superorder Protacanthoptherygii), Esox lucius belongs to the order of Esociformes; Salmo salar belongs to the order of Salmoniformes. The third group (superorder Acanthoptherygii) is composed of several subgroups: Ovalentaria, Carangaria, and Eupercaria groups.
Similar clustering, with some exceptions, is observed in a phylogeny based on concatenated sequences related to sex determination and reproduction (Portela-Bens et al., 2017). However, other phylogenies made with one immune system-related sequence show unexpected fish-species groupings, like c1q (Zeng et al., 2015), and hmgb2 (Wang et al., 2019). It was established that, for phylogenetics, the concatenation approach yields more accurate trees, even when the different concatenated sequences evolve with different substitution patterns (Gadagkar et al., 2005). The result presented here represents a novel phylogeny based on the concatenation of several immune-related genes of fishes. Moreover, the fish immune system has contributed significantly to a better understanding of the evolutionary history of the immune system (Rauta et al., 2012).
Based on the results obtained, it can be concluded that the immune system genes studied tend to be grouped together in the genome of S. senegalensis, as the 24 immune-related genes annotated were located in only six chromosome pairs. The second conclusion is that S. senegalensis, and the Soleoidei suborder in general, show a higher degree of grouping in the immune-related genes, which could represent an evolutionary advantage. In addition, it seems that large parts of these genomic regions tend to be conserved in the species most closely related to S. senegalensis, particularly the Pleuronectiformes C. semilaevis and S. maximus; and that the rate of variability of the immune system genes studied is not high.
The experimental procedures are ac-cording to the recommendation of the University of Cádiz (Spain) for the use of laboratory animals and the Guidelines of the European Union Council (86/609/EU).
AG-A and TL carried out the multiple FISH. AG-A drafted the manuscript. MR and SP-B isolated the BACs and carried out the bioinformatic analysis. MM constructed the phylogenetic tree and helped with the discussion and drafting. LR conceived and coordinated the study, participated in its design, discussed the results and corrected the manuscript. All authors read and approved the manuscript.
This study has been supported by the Spanish Ministerio de Ciencia e Innovacin MICINN-FEDER (AGL2014-51860-C2-1-P and RTI2018-096847-B-C21) and the Instituto de Investigaciones Marinas (INMAR-UCA). AG-A received a fellowship from the UCA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge their gratitude to the Ensemble team and the Genomicus Browser curators for providing a browser and database that greatly facilitated the use and analyses of the BACs used in this work. We also thank Dr. Manuel Manchado of the IFAPA (El Toruño, Cádiz, Spain) for sharing the BAC library, Mr. Emilio García for technical support with the cultures and chromosomal preparations and Dr. Alberto Arias and Mr. Royston Snart for a critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00529/full#supplementary-material
FIGURE S1 | Circos analysis in the species D. rerio. On the left side the distribution of the BAC clones of Senegalese sole distributed in chromosomes can be observed. Each BAC clone is represented in a different color. The genes found by annotation are indicated within each BAC clone and their corresponding localization in the D. rerio chromosomes are denoted by crossing lines. The BAC clones analyzed are given in Table 1.
FIGURE S2 | Circos analysis in the species O. niloticus. On the left side the distribution of the BAC clones of Senegalese sole distributed in chromosomes can be observed. Each BAC clone is represented in a different color. The genes found by annotation are indicated within each BAC clone and their corresponding localization in the O. niloticus chromosomes are denoted by crossing lines. The clones analyzed are given in Table 1.
FIGURE S3 | Circos analysis in the species G. aculeatus. On the left side the distribution of the BAC clones of Senegalese sole distributed in chromosomes can be observed. Each BAC clone is represented in a different color. The genes found by annotation are indicated within each BAC clone and their corresponding localization in the G. aculeatus chromosomes are denoted by crossing lines. The BAC clones analyzed are given in Table 1.
DATA SHEET S1 | Sequences used for the construction of the phylogenetic tree.
DATA SHEET S2 | Saturation indices and plots with transitions (s) and transversions (v) for each gene and in the concatenated sequence.
Anderson, J. L., Rodríguez Marí, A., Braasch, I., Amores, A., Hohenlohe, P., Batzel, P., et al. (2012). Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by rad mapping and population genomics. PLoS One 7:e40701. doi: 10.1371/journal.pone.0040701
Andersson, L. (1996). “Major histocompatibility complex evolution,” in The Major Histocompatibility Complex Region of Domestic Animal Species, eds L. B. Sckook and S. J. Lamont (Boca Raton: CRC Press, Inc.), 1–16.
Anisimova, M., Gil, M., Dufayard, J.-F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi: 10.1093/sysbio/syr041
Ao, J., Mu, Y., Xiang, L.-X., Fan, D., Feng, M., Zhang, S., et al. (2015). Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 11:e1005118. doi: 10.1371/journal.pgen.1005118
Asakawa, S., Abe, I., Kudoh, Y., Kishi, N., Wang, Y., Kubota, R., et al. (1997). Human BAC library: construction and rapid screening. Gene 191, 69–79. doi: 10.1016/S0378-1119(97)00044-49
Boudinot, P., van der Aa, L. M., Jouneau, L., Du Pasquier, L., Pontarotti, P., Briolat, V., et al. (2011). Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of Zebrafish and Pufferfish. PLoS One 6:e22022. doi: 10.1371/journal.pone.0022022
Cerdá, J., and Manchado, M. (2013). Advances in genomics for flatfish aquaculture. Genes Nutr. 8, 5–17. doi: 10.1007/s12263-012-0312-318
Chairi, H., Ferná Ndez-Diaz, C., Navas, J. I., Manchado, M., Rebordinos, L., and Blasco, J. (2010). In vivo genotoxicity and stress defences in three flatfish species exposed to CuSO 4. Ecotoxicol. Environ. Saf. 73, 1279–1285. doi: 10.1016/j.ecoenv.2010.07.028
Chen, M., Ding, M., Li, Y., Zhong, X., Liu, S., Guo, Z., et al. (2018). The complement component 1 q (C1q) in Nile tilapia (Oreochromis niloticus): functional characterization in host defense against bacterial infection and effect on cytokine response in macrophages. Dev. Comp. Immunol. 87, 98–108. doi: 10.1016/j.dci.2018.05.023
Choi, K.-M., Joo, M.-S., Cho, D. H., Bae, J.-S., Jung, J.-M., Hwang, J. Y., et al. (2019). Characterization of gene expression profiles and functional analysis of peptidoglycan recognition protein 2 from rock bream (Oplegnathus fasciatus). Fish Shellfish Immunol. 84, 1068–1074. doi: 10.1016/j.fsi.2018.11.025
Christoffels, A., Koh, E. G. L., Chia, J., Brenner, S., Aparicio, S., and Venkatesh, B. (2004). Fugu Genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21, 1146–1151. doi: 10.1093/molbev/msh114
Cremer, T., and Cremer, M. (2010). Chromosome territories. Cold Spring Harb. Perspect. Biol. 2:a003889. doi: 10.1101/cshperspect.a003889
Cross, I., Portela-Bens, S., García-Angulo, A., Merlo, M. A., Rodríguez, M. E., Liehr, T., et al. (2018). A preliminary integrated genetic map distinguishes every chromosome pair and locates essential genes related to abiotic adaptation of Crassostrea angulata/gigas. BMC Genet. 19:104. doi: 10.1186/s12863-018-0689-5
Cross, I., Rebordinos, L., and Díaz, E. (2006). Species identification of Crassostrea and Ostrea oysters by polymerase chain reaction amplification of the 5S rRNA gene. J. AOAC Int. 89, 144–148.
Díaz-Ferguson, E., Cross, I., Del Barrios, M. M., and Rebordinos, L. (2007). Genetic relationships among populations of the senegalese sole Solea senegalensis in the Southwestern Iberian Peninsula detected by mitochondrial DNA–restriction fragment length polymorphisms. Trans. Am. Fish. Soc. 136, 484–491. doi: 10.1577/T06-030.1
Diopere, E., Maes, G. E., Komen, H., Volckaert, F. A. M., and Groenen, M. A. M. (2014). A Genetic linkage map of sole (Solea solea): a tool for evolutionary and comparative analyses of exploited (Flat)fishes. PLoS One 9:e115040. doi: 10.1371/journal.pone.0115040
FAO (2019). Fisheries and Aquaculture Statistics. Global Aqualculture Production 1950–2019. Rome: FishStatJ, FAO Fisheries and Aquaculture Department. Available at: http://www.fao.org/fishery/statistics/software/FishStatJ/en
Ferraresso, S., Bonaldo, A., Parma, L., Buonocore, F., Scapigliati, G., Gatta, P. P., et al. (2016). Ontogenetic onset of immune-relevant genes in the common sole (Solea solea). Fish Shellfish Immunol. 57, 278–292. doi: 10.1016/j.fsi.2016.08.044
Gadagkar, S. R., Rosenberg, M. S., and Kumar, S. (2005). Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J. Exp. Zool. Part B Mol. Dev. Evol. 304, 64–74. doi: 10.1002/jez.b.21026
García-Angulo, A., Merlo, M. A., Portela-Bens, S., Rodríguez, M. E., García, E., Al-Rikabi, A., et al. (2018). Evidence for a robertsonian fusion in Solea senegalensis (Kaup, 1858) revealed by zoo-FISH and comparative genome analysis. BMC Genomics 19:818. doi: 10.1186/s12864-018-5216-6
García-Cegarra, A., Merlo, M. A., Ponce, M., Portela-Bens, S., Cross, I., Manchado, M., et al. (2013). A preliminary genetic map in Solea senegalensis (Pleuronectiformes, Soleidae) using BAC-FISH and next-generation sequencing. Cytogenet. Genome Res. 141, 227–240. doi: 10.1159/000355001
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New Algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Han, Y., Wang, J., Zhao, Z., Chen, J., Lu, H., and Liu, G. (2017). Fishmeal application induces antibiotic resistance gene propagation in mariculture sediment. Environ. Sci. Technol. 51, 10850–10860. doi: 10.1021/acs.est.7b02875
Hofmann, S. R., Rösen-Wolff, A., Tsokos, G. C., and Hedrich, C. M. (2012). Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin. Immunol. 143, 116–127. doi: 10.1016/j.clim.2012.02.005
Hwang, S. D., Fuji, K., Takano, T., Sakamoto, T., Kondo, H., Hirono, I., et al. (2011). Linkage mapping of toll-like receptors (TLRs) in Japanese flounder. Paralichthys Olivaceus. Mar. Biotechnol. 13, 1086–1091. doi: 10.1007/s10126-011-9371-x
Jaillon, O., Aury, J.-M., Brunet, F., Petit, J.-L., Stange-Thomann, N., Mauceli, E., et al. (2004). Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957. doi: 10.1038/nature03025
Katoh, K., and Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. doi: 10.1093/bib/bbn013
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kondo, H., Darawiroj, D., Gung, Y.-T.-A., Yasuike, M., Hirono, I., and Aoki, T. (2010). Identification of two distinct types of beta-2 microglobulin in marine fish. Pagrus major and Seriola quinqueradiata. Vet. Immunol. Immunopathol. 134, 284–288. doi: 10.1016/j.vetimm.2009.09.005
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lewis, S. E., Searle, S. M. J., Harris, N., Gibson, M., Lyer, V., Richter, J., et al. (2002). Apollo: a sequence annotation editor. Genome Biol. 3:research0082.1–82.14. doi: 10.1186/gb-2002-3-12-research0082
Li, Y.-W., Han, R., Wang, J.-L., Yang, M., Dan, X.-M., and Li, A.-X. (2018). Molecular identification and functional characterization of IRAK-3 from a teleost fish, the orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 81, 383–389. doi: 10.1016/j.fsi.2018.07.029
Liehr, T. (ed.) (2009). Fluorescence in Situ Hybridization—Application Guide. Berlin: Springuer-Verlag.
Liu, T., Gao, X., and Xin, Y. (2019). Identification of an IKBKE inhibitor with antitumor activity in cancer cells overexpressing IKBKE. Cytokine 116, 78–87. doi: 10.1016/j.cyto.2019.01.005
Lutfalla, G., Crollius, H. R., Stange-Thomann, N., Jaillon, O., Mogensen, K., and Monneron, D. (2003). Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 4:29. doi: 10.1186/1471-2164-4-29
MacKenzie, S., and Jentoft, S. (eds) (2016). Genomics in Aquaculture, 1st Edn. Amsterdam: Elsevier Inc.
Makino, T., and McLysaght, A. (2008). Interacting gene clusters and the evolution of the vertebrate immune system. Mol. Biol. Evol. 25, 1855–1862. doi: 10.1093/molbev/msn137
Mamane, Y., Heylbroeck, C., Génin, P., Algarté, M., Servant, M. J., LePage, C., et al. (1999). Interferon regulatory factors: the next generation. Gene 237, 1–14. doi: 10.1016/S0378-1119(99)00262-260
Manchado, M., Planas, J. V., Cousin, X., Rebordinos, L., and Claros, M. G. (2016). “Current status in other finfish species: Description of current genomic resources for the gilthead seabream (Sparus aurata) and soles (Solea senegalensis and Solea solea),” in Genomics in Aquaculture, eds S. Mackenzie and S. Jentoft (Amsterdam: Elsevier), 195–221.
Manchado, M., Zuasti, E., Cross, I., Merlo, A., Infante, C., and Rebordinos, L. (2006). Molecular characterization and chromosomal mapping of the 5S rRNA gene in Solea senegalensis: a new linkage to the U1, U2, and U5 small nuclear RNA genes. Genome 49, 79–86. doi: 10.1139/g05-068
Merlo, M. A., Iziga, R., Portela-Bens, S., Cross, I., Kosyakova, N., Liehr, T., et al. (2017). Analysis of the histone cluster in Senegalese sole (Solea senegalensis): evidence for a divergent evolution of two canonical histone clusters. Genome 60, 441–453. doi: 10.1139/gen-2016-2143
Molina-Luzón, M. J., López, J. R., Robles, F., Navajas-Pérez, R., Ruiz-Rejón, C., De la Herrán, R., et al. (2014). Chromosomal manipulation in senegalese sole (Solea senegalensis Kaup, 1858): induction of triploidy and gynogenesis. J. Appl. Genet. 56, 77–84. doi: 10.1007/s13353-014-0233-x
Morais, S., Mendes, A. C., Castanheira, M. F., Coutinho, J., Bandarra, N., Dias, J., et al. (2014). New formulated diets for Solea senegalensis broodstock: effects of parental nutrition on biosynthesis of long-chain polyunsaturated fatty acids and performance of early larval stages and juvenile fish. Aquaculture 432, 374–382. doi: 10.1016/j.aquaculture.2014.04.033
Nayak, S. K. (2010). Probiotics and immunity: a fish perspective. Fish Shellfish Immunol. 29, 2–14. doi: 10.1016/j.fsi.2010.02.017
Noonan, J. P., Grimwood, J., Danke, J., Schmutz, J., Dickson, M., Amemiya, C. T., et al. (2004). Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 14, 2397–2405. doi: 10.1101/gr.2972804
Novel, P., Fernández-Trujillo, M. A., Gallardo-Gálvez, J. B., Cano, I., Manchado, M., Buonocore, F., et al. (2013). Two Mx genes identified in European sea bass (Dicentrarchus labrax) respond differently to VNNV infection. Vet. Immunol. Immunopathol. 153, 240–248. doi: 10.1016/j.vetimm.2013.03.003
Núñez-Díaz, J. A., Fumanal, M., Mancera, J. M., Moriñigo, M. A., and Balebona, M. C. (2016). Two routes of infection with Photobacterium damselae subsp. piscicida are effective in the modulation of the transcription of immune related genes in Solea senegalensis. Vet. Immunol. Immunopathol. 179, 8–17. doi: 10.1016/j.vetimm.2016.07.009
Overbeek, R., Fonstein, M., D’Souza, M., Pusch, G. D., and Maltsev, N. (1999). The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U.S.A. 96, 2896–2901. doi: 10.1073/pnas.96.6.2896
Padrós, F., Villalta, M., Gisbert, E., and Estévez, A. (2011). Morphological and histological study of larval development of the Senegal sole Solea senegalensis: an integrative study. J. Fish Biol. 79, 3–32. doi: 10.1111/j.1095-8649.2011.02942.x
Palti, Y. (2011). Toll-like receptors in bony fish: from genomics to function. Dev. Comp. Immunol. 35, 1263–1272. doi: 10.1016/j.dci.2011.03.006
Palti, Y., Luo, M.-C., Hu, Y., Genet, C., You, F. M., Vallejo, R. L., et al. (2009). A first generation BAC-based physical map of the rainbow trout genome. BMC Genomics 10:462. doi: 10.1186/1471-2164-10-462
Pinto, R. D., Pereira, P. J. B., and dos Santos, N. M. S. (2011). Transporters associated with antigen processing (TAP) in sea bass (Dicentrarchus labrax, L.): molecular cloning and characterization of TAP1 and TAP2. Dev. Comp. Immunol. 35, 1173–1181. doi: 10.1016/j.dci.2011.03.024
Ponce, M., Salas-Leiton, E., Garcia-Cegarra, A., Boglino, A., Coste, O., Infante, C., et al. (2011). Genomic characterization, phylogeny and gene regulation of g-type lysozyme in sole (Solea senegalensis). Fish Shellfish Immunol. 31, 925–937. doi: 10.1016/j.fsi.2011.08.010
Portela-Bens, S., Merlo, M. A., Rodríguez, M. E., Cross, I., Manchado, M., Kosyakova, N., et al. (2017). Integrated gene mapping and synteny studies give insights into the evolution of a sex proto-chromosome in Solea senegalensis. Chromosoma 126, 261–277. doi: 10.1007/s00412-016-0589-582
Raghavan, M., Wijeyesakere, S. J., Peters, L. R., and Del Cid, N. (2013). Calreticulin in the immune system: ins and outs. Trends Immunol. 34, 13–21. doi: 10.1016/j.it.2012.08.002
Rauta, P. R., Nayak, B., and Das, S. (2012). Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol. Lett. 148, 23–33. doi: 10.1016/j.imlet.2012.08.003
Rauta, P. R., Samanta, M., Dash, H. R., Nayak, B., and Das, S. (2014). Toll-like receptors (TLRs) in aquatic animals: signaling pathways, expressions and immune responses. Immunol. Lett. 158, 14–24. doi: 10.1016/j.imlet.2013.11.013
Reboul, J., Gardiner, K., Monneron, D., Uzé, G., and Lutfalla, G. (1999). Comparative genomic analysis of the interferon/interleukin-10 receptor gene cluster. Genome Res. 9, 242–250.
Salas-Leiton, E., Anguis, V., Martín-Antonio, B., Crespo, D., Planas, J. V., Infante, C., et al. (2010). Effects of stocking density and feed ration on growth and gene expression in the Senegalese sole (Solea senegalensis): potential effects on the immune response. Fish Shellfish Immunol. 28, 296–302. doi: 10.1016/j.fsi.2009.11.006
Salas-Leiton, E., Coste, O., Asensio, E., Infante, C., Cañavate, J. P., and Manchado, M. (2012). Dexamethasone modulates expression of genes involved in the innate immune system, growth and stress and increases susceptibility to bacterial disease in Senegalese sole (Solea senegalensis Kaup, 1858). Fish Shellfish Immunol. 32, 769–778. doi: 10.1016/j.fsi.2012.01.030
Salazar-Mather, T. P., and Hokeness, K. L. (2006). “Cytokine and chemokine networks: pathways to antiviral defense,” in Current Topics in Microbiology and Immunology, ed. T. E. Lane (Berlin: Springer-Verlag), 18.
Sarkar, A., Duncan, M., Hart, J., Hertlein, E., Guttridge, D., and Wewers, M. (2006). ASC directs NF-κB activation by regulating receptor interacting protein-2 (RIP2) caspase-1 interactions. J. Immunol. 176, 4979–4986.
Satta, Y., Mayer, W. E., and Klein, J. (1996). Evolutionary relationship ofHLA-DRB genes inferred from intron sequences. J. Mol. Evol. 42, 648–657. doi: 10.1007/BF02338798
Saurabh, S., and Sahoo, P. K. (2008). Lysozyme: an important defence molecule of fish innate immune system. Aquac. Res. 39, 223–239. doi: 10.1111/j.1365-2109.2007.01883.x
Sha, Z., Abernathy, J. W., Wang, S., Li, P., Kucuktas, H., Liu, H., et al. (2009). NOD-like subfamily of the nucleotide-binding domain and leucine-rich repeat containing family receptors and their expression in channel catfish. Dev. Comp. Immunol. 33, 991–999. doi: 10.1016/j.dci.2009.04.004
Sun, B. J., Wang, G. L., Xie, H. X., Gao, Q., and Nie, P. (2006). Gene structure of goose-type lysozyme in the mandarin fish Siniperca chuatsi with analysis on the lytic activity of its recombinant in Escherichia coli. Aquaculture 252, 106–113. doi: 10.1016/j.aquaculture.2005.07.046
Sun, Y. N., Su, X. R., Xu, T. J., and Li, T. W. (2015). Genomic characterization and sequence diversity of the β2-microglobulin gene in the miiuy croaker. miichthys miiuy. Genet. Mol. Res. 14, 10249–10257. doi: 10.4238/2015.August.28.9
Taboada, X., Pansonato-Alves, J. C., Foresti, F., Martínez, P., Viñas, A., Pardo, B. G., et al. (2014). Consolidation of the genetic and cytogenetic maps of turbot (Scophthalmus maximus) using FISH with BAC clones. Chromosoma 123, 281–291. doi: 10.1007/s00412-014-0452-452
Tavaré, S. (1986). Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17, 57–86.
van der Aa, L. M., Levraud, J.-P., Yahmi, M., Lauret, E., Briolat, V., Herbomel, P., et al. (2009). A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 7:7. doi: 10.1186/1741-7007-7-7
Wang, G., Jiang, Z., He, S., and Zhang, M. (2018). Two novel calreticulin-related molecules with microbial binding and phagocytosis enhancing capacity in the half-smooth tongue sole, Cynoglossus semilaevis. Fish Shellfish Immunol. 72, 174–180. doi: 10.1016/j.fsi.2017.10.057
Wang, G., Wang, J., Yue, B., Du, X., Du, H., Zhang, M., et al. (2019). High mobility group box 2 of black rockfish Sebastes schlegelii: gene cloning, immunoregulatory properties and antibacterial effect. Fish Shellfish Immunol. 84, 719–725. doi: 10.1016/j.fsi.2018.10.066
Wang, J., He, Y., Li, L., Yang, M., Wang, Q., Zhu, Y., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Xia, X. (2017). DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. J. Hered. 108, 431–437. doi: 10.1093/jhered/esx033
Xia, X., Xie, Z., Salemi, M., Chen, L., and Wang, Y. (2003). An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26, 1–7. doi: 10.1016/S1055-7903(02)00326-323
Xu, P., Wang, S., Liu, L., Thorsen, J., Kucuktas, H., and Liu, Z. (2007). A BAC-based physical map of the channel catfish genome. Genomics 90, 380–388. doi: 10.1016/j.ygeno.2007.05.008
Zarkadis, I. K., Mastellos, D., and Lambris, J. D. (2001). Phylogenetic aspects of the complement system. Dev. Comp. Immunol. 25, 745–762. doi: 10.1016/S0145-305X(01)00034-39
Zeng, Y., Xiang, J., Lu, Y., Chen, Y., Wang, T., Gong, G., et al. (2015). sghC1q, a novel C1q family member from half-smooth tongue sole (Cynoglossus semilaevis): identification, expression and analysis of antibacterial and antiviral activities. Dev. Comp. Immunol. 48, 151–163. doi: 10.1016/j.dci.2014.09.010
Zhang, J., Li, Y., and Hu, Y. (2015). Molecular characterization and expression analysis of eleven interferon regulatory factors in half-smooth tongue sole, Cynoglossus semilaevis. Fish Shellfish Immunol. 44, 272–282. doi: 10.1016/j.fsi.2015.02.033
Zhang, X., Zhao, C., Huang, C., Duan, H., Huan, P., Liu, C., et al. (2011). A BAC-based physical map of zhikong scallop (Chlamys farreri jones et preston). PLoS One 6:e27612. doi: 10.1371/journal.pone.0027612
Zhu, L.-Y., Nie, L., Zhu, G., Xiang, L.-X., and Shao, J.-Z. (2013). Advances in research of fish immune-relevant genes: a comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 39, 39–62. doi: 10.1016/j.dci.2012.04.001
Keywords: Solea senegalensis, bacterial artificial chromosome, immune system, aquaculture, syntenic conservation
Citation: García-Angulo A, Merlo MA, Rodríguez ME, Portela-Bens S, Liehr T and Rebordinos L (2019) Genome and Phylogenetic Analysis of Genes Involved in the Immune System of Solea senegalensis – Potential Applications in Aquaculture. Front. Genet. 10:529. doi: 10.3389/fgene.2019.00529
Received: 09 January 2019; Accepted: 14 May 2019;
Published: 11 June 2019.
Edited by:
Paulino Martínez, University of Santiago de Compostela, SpainReviewed by:
Filippo Biscarini, Italian National Research Council (CNR), ItalyCopyright © 2019 García-Angulo, Merlo, Rodríguez, Portela-Bens, Liehr and Rebordinos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laureana Rebordinos, bGF1cmVhbmEucmVib3JkaW5vc0B1Y2EuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.