- 1Department of Biosciences, Rice University, Houston, TX, United States
- 2Symphonytech Biologics, Philadelphia, PA, United States
- 3Indian Institute of Science Education and Research, Berhampur, India
Nearly 50% of the human genome is derived from transposable elements (TEs). Though dysregulated transposons are deleterious to humans and can lead to diseases, co-opted transposons play an important role in generating alternative or new DNA sequence combinations to perform novel cellular functions. The appearance of an adaptive immune system in jawed vertebrates, wherein the somatic rearrangement of T and B cells generates a repertoire of antibodies and receptors, is underpinned by Class II TEs. This review follows the evolution of recombination activation genes (RAGs), components of adaptive immunity, from TEs, focusing on the structural and mechanistic similarities between RAG recombinases and DNA transposases. As evolution occurred from a transposon precursor, DNA transposases developed a more targeted and constrained mechanism of mobilization. As DNA repair is integral to transposition and recombination, we note key similarities and differences in the choice of DNA repair pathways following these processes. Understanding the regulation of V(D)J recombination from its evolutionary origins may help future research to specifically target RAG proteins to rectify diseases associated with immune dysregulation.
Introduction
Transposable elements (TEs) are pervasive in all kingdoms of life. There are two major classes based on their mechanism of transposition (Figure 1): Class I TEs, also known as retrotransposons, and Class II TEs, also known as DNA transposons. Class I TEs replicate via a copy-and-paste mechanism through an RNA intermediate (Boeke et al., 1985; Finnegan, 1989; Kazazian, 2004). This class is further divided into two subclasses: LTRs (long terminal repeats) and non-LTRs. LTRs include endogenous retrovirus (ERV) retrotransposons and non-LTRs include both long and short interspersed nuclear elements (LINEs/L1s and SINEs). Class II DNA transposons mobilize through a cut-and-paste mechanism wherein the original sequence is excised from the genome before being inserted at another site. A subclass of DNA transposons, Helitrons, mobilize through a peel-and-paste mechanism with a circular DNA intermediate (Bourque et al., 2018).
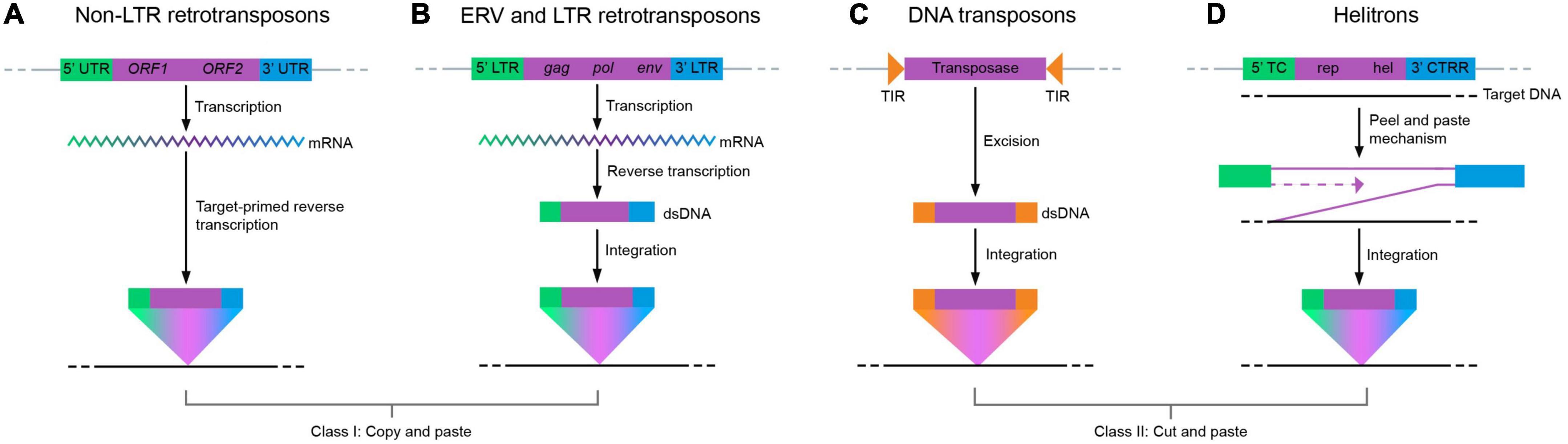
Figure 1. Mechanism of action of the two classes of transposons. Class I TEs mobilize through an RNA intermediate. They can be further divided into two subcategories: non-LTRs (long terminal repeats) and LTRs/ERVs (endogenous retroviruses). (A) A prominent example of a non-LTR retrotransposon is LINE1/L1 (long interspersed nuclear element 1), which uses the process of target-primed reverse transcription carried out by the self-encoded proteins ORF1p and ORF2p. This process involves a nick in the target DNA by an endonuclease followed by reverse transcription and integration, both of which are enabled by the enzymatic activities of ORF2p. (B) ERV and LTR retrotransposons are flanked by LTRs and utilize a double-stranded DNA intermediate (dsDNA) prior to integration into the target site. Class II TEs require excision of genomic DNA with the help of self-encoded transposases. Subsequently, transposases help insert the excised DNA into a target site. (C) DNA transposons excise at the TIR, which is subsequently inserted as dsDNA in the target site. (D) Helitrons transpose by the rolling circle mechanism, involving strand invasion and DNA replication, without excision of the donor transposon DNA.
Functionally, TEs have both negative and positive effects on host cell fitness. In this review, we present the beneficial and detrimental effects of TEs. In particular, we focus on how vertebrate adaptive immunity arose through harnessing genes that arose by TE insertion. These recombination activation genes (RAGs) are considered to be derived from Class II TEs (Huang et al., 2016). This statement is made on the basis that the structural domains and biochemistry of RAG action are strikingly similar to that of Class II TEs (Carmona and Schatz, 2017). In this review, we discuss the contribution of TEs in the evolution of V(D)J recombination, present structural and functional comparisons between transposase and V(D)J recombinase, put forth several unanswered questions, and finally discuss how this knowledge may aid treatment of immune disorders.
Negative and Positive Effects of Transposable Element Activity
Unconstrained TEs have been linked to many human diseases. A common origin for tumorigenic activity is by transposons hopping into essential genes, activating proto-oncogenes, or inactivating tumor suppressor genes. In this way, the new insertion of TEs can impact regulatory regions, altering or disrupting gene function and spatiotemporal dynamics. Another possibility is that ectopic recombination can lead to duplications, deletions, or insertions, all of which cause genomic instability (Economou-Pachnis and Tsichlis, 1985; Meischl and Roos, 1998; Meischl et al., 2000; Claverie-Martin et al., 2005; Burns and Boeke, 2012; Tarallo et al., 2012; Li et al., 2013; Burns, 2017; Guo et al., 2018; Ardeljan et al., 2020; Rodriguez-Martin et al., 2020). Further, while it was thought transposons are active only in the germ line with exceptions in the brain (Muotri et al., 2005; Goodier, 2014), next generation sequencing has demonstrated that retrotransposons can be reactivated in somatic tissues under pathological conditions (Hancks and Kazazian, 2016).
Reports of TE mobilization are not just confined to humans (Bourque et al., 2018). Mus musculus, Danio rerio, Drosophila melanogaster, Escherichia coli, and Arabidopsis thaliana (Huang et al., 2012; Moschetti et al., 2020; Chang et al., 2022), among other widely studied organisms, are known to have TE activity with both detrimental and beneficial functions. However, some TE functions have arisen uniquely for certain species over the course of evolution. Adaptive immunity, a crucial function that arose through TE co-option, is restricted in its species distribution (Mitra et al., 2013; Morales Poole et al., 2017).
In mammals, DNA transposons are generally not active, excepting a few species (Mitra et al., 2013; Gallus et al., 2015). In humans, DNA transposons occupy almost 3% of the genome and are inert and fossilized (Lander et al., 2001). By comparison, retroelements are more active in normal physiology. Out of 500,000 copies of L1, the only autonomously active Class I TE, around 100 are active in humans, and are associated with 124 disease-causing insertions (Hancks and Kazazian, 2012). L1 contains two open reading frames that encode ORF1p and ORF2p, both of which are required for retrotransposition. In addition to its own retrotransposition, ORF2p-encoded reverse transcriptase can also be hijacked by other RNAs in trans (e.g., SINEs-Alu) to complete their retrotransposition activity (Stenz, 2021). These retroelements are controlled by the host at multiple stages of the transcription cycle, through transcriptional repression, RNA degradation, and other mechanisms (Bourque et al., 2018).
Since DNA transposons are not endogenously active in humans, their dysregulated activity is not known to be associated with disease; as retroelements are active, their disease associations are much clearer. Hemophilia was the first human disease shown to be caused by a de novo L1 insertion (Kazazian et al., 1988). Since this initial report, studies have elucidated similar disease-causing roles for retrotransposons in a wide range of disorders. For example, ORF1p has RNA chaperone activities (Martin, 2010), and its expression has been documented in various cancers. In specific, L1 integration into the adenomatous polyposis coli (APC) gene stratifies with colon cancer, and demethylation of a LINE promoter causes constitutive activation of the MET protein in bladder cancer (Ogino et al., 2008; Wolff et al., 2010). Further, L1 elements can evade genomic DNA methylation, enabling them to contribute new L1 insertions (Burns and Boeke, 2012; Hancks and Kazazian, 2016; Burns, 2017; Rodriguez-Martin et al., 2020). Moreover, a wide spectrum of neurodegenerative diseases also involve TEs (Saleh et al., 2019; Tam et al., 2019).
Despite their dangerous effects when unchecked, several TEs have become co-opted into the human genome over time. One explanation for the gradual loss of deleterious activity is that TEs were modified during domestication and lost their ability to autonomously transpose. In fact, several retroviruses have been repurposed into protein-coding genes and small RNAs (Figure 2A), providing important cellular functions. There are many examples of TE co-option and domestication, a few of which we present below. (i) Retrovirus-derived sequences modulate genetic circuits in embryonic development (Gerdes et al., 2016; Chuong, 2018). ERV gag-pol functions as regulators during placental development (Fu et al., 2019; Figure 2B). (ii) ERVs provide transcription regulatory regions for interferon genes. (iii) L1 retroelement mobilization in the brain is thought to diversify neuronal cell populations (Muotri et al., 2005; Coufal et al., 2009; Baillie et al., 2011). (iv) The neuronal gene arc, derived from gag proteins of the Ty3/gypsy retrotransposon, mediates synaptic plasticity (Pastuzyn et al., 2018; Figure 2C). (v) TEs affect the expression of genes at both transcriptional and post-transcriptional levels by providing alternative promoters or small RNAs (Chuong et al., 2016; Petri et al., 2019). (vi) TEs provide cis-regulatory sequences, which alter gene expression by acting as enhancers (Bejerano et al., 2006; Chuong et al., 2013, 2016), providing transcription factor binding sites (Wang et al., 2007; Sundaram et al., 2014; Ito et al., 2017), or acting as insulators (Bourque et al., 2008; Figure 2D). (vii) RAGs, derived from the protoRAG DNA transposon, catalyze V(D)J recombination in the vertebrate immune system (Figure 2E; Hencken et al., 2012). This allows for the generation of a vast repertoire of antigen binding receptors in lymphocytes.
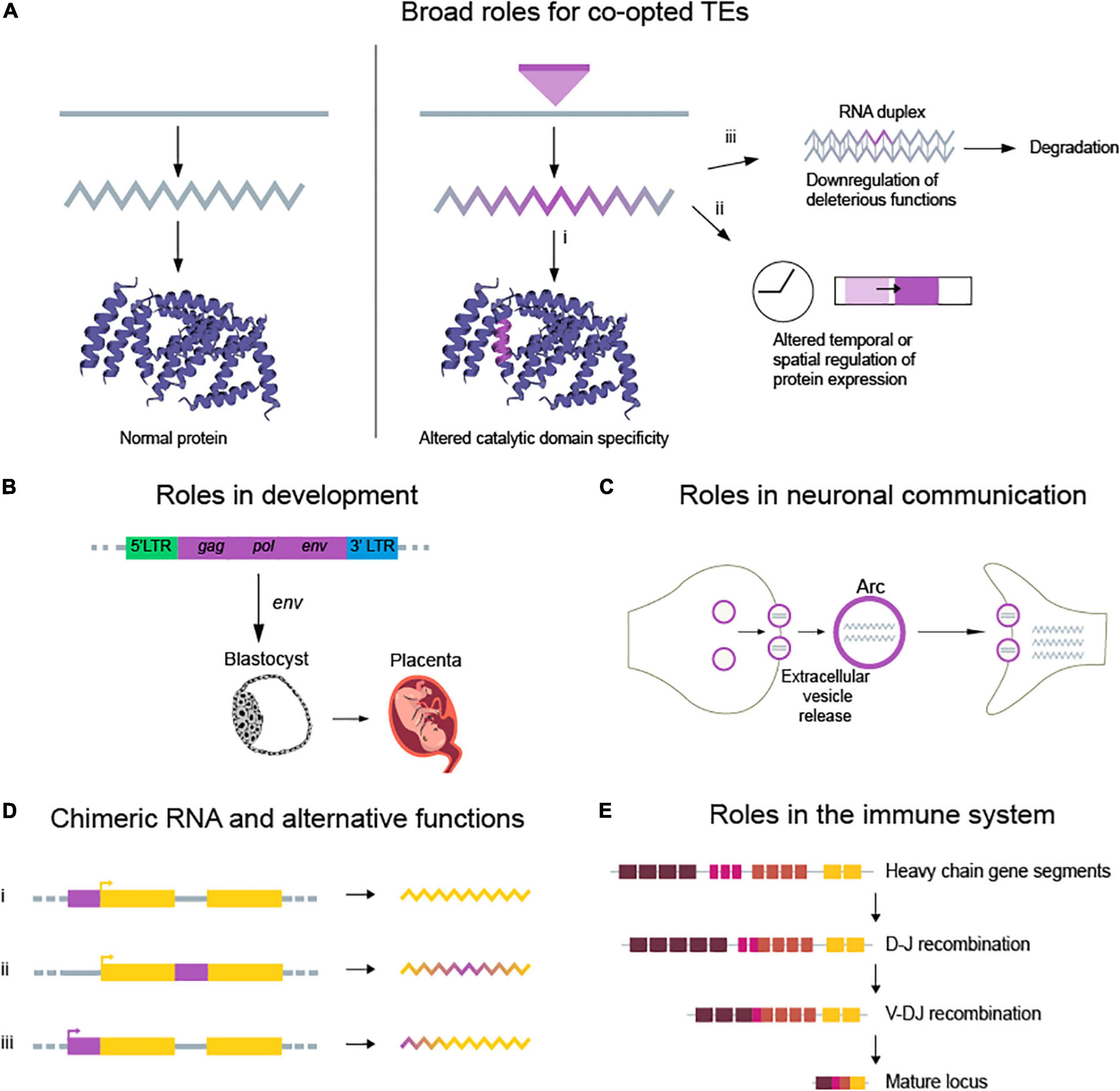
Figure 2. Examples of beneficial TE functions that arose through co-option. (A) Beneficial TE functions can be broadly classified into three categories: introducing new gene functions, regulating existing functions, or downregulating negative functions. (i) New gene functions are typically introduced through co-option of TEs, repurposing natural cut/paste or copy/paste activities, which can result in new beneficial functions. (ii) Altered spatiotemporal gene expression occurs through insertion or modification of cis-regulatory elements, which can introduce binding sites for transcription and regulatory factors. (iii) Downregulation of deleterious genes can be achieved at transcriptional and post-transcriptional levels through small RNA hybridization and degradation of specific targets. (B) Upregulation of ERVs during embryonic development provides cis and trans elements for the expression of genes involved in pluripotency. (C) The Arc protein, derived from the gag protein of retrotransposons, forms capsid-like structures that deliver mRNA during neuronal communication. (D) TEs regulate the expression of neighboring genes in various ways (i) by acting as enhancers, (ii) by modulating splicing junctions, and (iii) by functioning as promoters. (E) RAG1 and RAG2, thought to be derived from TEs, mediate V(D)J recombination. This process generates the diverse repertoire of antigen receptors in immune cells.
DNA Transposons: A Substrate for the Evolution of Vertebrate Adaptive Immunity
The appearance of V(D)J recombination in jawed vertebrates was a pathbreaking step in the evolution of the adaptive immune system (Jones and Gellert, 2004; Cooper and Alder, 2006). Crystallography and electron microscopy studies suggest that the transposase enzyme is the evolutionary progenitor of RAG1 and RAG2 (Liu et al., 2019). Functionally, RAG recombinases initiate V(D)J recombination in developing lymphocytes by generating DNA double stranded breaks (DSBs) at Recombination Signal Sequences (RSSs) (Schatz and Swanson, 2011), which are thought to have evolved from the terminal inverted repeats (TIRs) of Transib transposons (Huang et al., 2016). During V(D)J recombination, the 12 RSS and 23 RSS pair into a synaptic complex with RAG, which catalyzes nick formation at the RSSs and enables DSB formation (van Gent et al., 1996). Further, the endonuclease activity of RAG1/RAG2 utilizes RNase H-mediated trans-esterification chemistry that is shared with transposases. Such structural comparisons between DNA transposons and the V(D)J recombination machinery are summarized in Figure 3. Notably, unlike the cut-and-paste mechanism of DNA transposition, V(D)J recombination circularizes the ends of the excised segments in the signal joint to protect the genome against hazardous insertions.
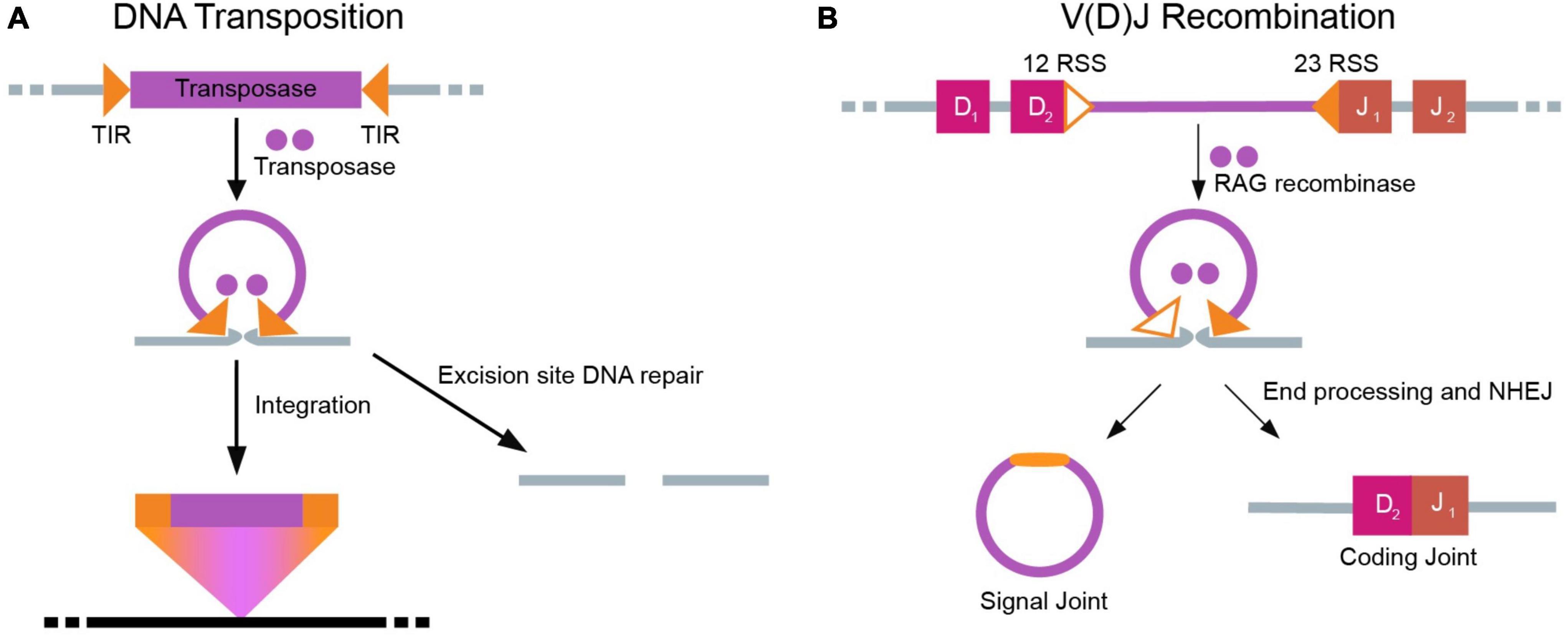
Figure 3. Parallel mechanisms of DNA transposition and V(D)J recombination. (A) Self-encoded transposases excise a DNA transposon, including flanking terminal inverted repeats (TIRs). Subsequently, the transposon inserts into a new DNA sequence. The excision site must now be repaired through host DNA repair machinery. (B) In V(D)J recombination, V, D, and J segments are separated by intervening recombination signal sequences (RSSs) with similarity to transposons TIRs. RAG1/RAG2 recognize the RSS using the 12–23 rule, enabling cleavage of the intervening segment. The two ends of DNA that are separated (D and J ends as shown here) are subsequently joined through end processing and NHEJ to form the signal and coding joint.
Transib and protoRAG from Branchiostoma belcheri (common Lancelet) (Huang et al., 2016) are considered to be the two most promising RAG TE precursor candidates. Based on structural and functional analysis, Transib is an ancient evolutionary ancestor to RAGs, while protoRAG is a proximal ancestor with more features in common with RAG. While Transib and protoRAG both contain TIRs that are similar to RSSs, only protoRAG genes are homologous to both RAG1 and RAG2 and encode proteins with structural and functional similarities to vertebrate RAG.
Regulation of V(D)J Recombination
One of the drawbacks of the generation of a large repertoire of lymphocyte receptors with small energy investment in germline coding capacity is the generation of DNA DSBs. Therefore, to safeguard the genome and ensure that recombination is tightly restricted by cell type and developmental stage, multiple layers of DNA repair and regulation have evolved. In this section, we review important mechanisms that have evolved to regulate the activity of recombinases to preserve genomic integrity in adaptive immune system function.
One key characteristic of RAG function is its highly ordered catalysis. V(D)J recombination of immunoglobulin and T cell receptor (TCR) genes during B and T cell development requires specific recognition of the 12/23 asymmetric RSS by the RAG1 endonuclease. The RAG2 protein assists RAG1 in DNA binding and cleavage (Liu et al., 2019). Since there are often many V, D, and J domains to choose from, permutation of the V(D)J sequences results in diverse B and T cell receptors. These newly rearranged receptors then facilitate negative and positive selection (Klein et al., 2014). The elegant orchestration of these highly regulated processes culminates in a mature immune repertoire.
Complete antigen receptor production involves two sequential gene segment rearrangements, one for each receptor chain locus. The immunoglobulin heavy chain and TCR β-chain loci are the first to be assembled in pre-B-cells and double negative thymocytes, respectively. Assembly occurs serially, with D to J rearrangement followed by V to DJ rearrangement. Following these steps, recombination of the immunoglobulin light chain and TCR α-chain loci is initiated. The highly ordered nature of recombination events creates important checkpoints in lymphocyte development to identify the formation of non-functional receptors. DNA cleavage by RAG recombinases is also regulated by chromatin structure, trans factors, the spatial localization of the antigen receptor loci, and transcription factors. Cis elements, such as promoters and enhancers, precisely direct transcription of V(D)J loci by interacting with transcription factors. Enhancers and promoters are so essential in this process that their deletion causes a complete block in recombination events and a loss of germline transcription. Further, transcriptional control restricts the expression of RAGs to the progenitor stages of B and T cell development, thereby ensuring that V(D)J recombination occurs solely in developing B and T lymphocytes (Gellert, 2002; Krangel, 2003; Schlissel, 2003; Del Blanco et al., 2011).
DNA Transposase Precedes Recombination Activation Gene Recombinase: Structural and Functional Justifications
The dominant hypothesis for RAG evolution explains the origin of adaptive immunity through the domestication and co-option of the protoRAG transposon. This hypothesis gained support from converging lines of evidence, including the in vitro ability of RAGs to mediate efficient transposition of RSS-flanked DNA insertions (Agrawal et al., 1998; Hiom et al., 1998). Structurally, the ProtoRAG DNA transposon contains BbRAG1L (homologous to RAG1) and BbRAG2L (homologous to RAG2). RAG1 contains several key domains, including an N-terminal RING/Zn finger, nonamer binding domain, and a catalytic core (Kim et al., 2015). BbRAG1L retains sequence similarity with the core domain of RAG1 (Carmona and Schatz, 2017). Importantly, the nonamer binding domain of RAG replaced the general DNA-binding domain of BbRAG1L (Zhang et al., 2019), suggesting that as the transposon was co-opted its function was gradually constrained. Vertebrate RAG2 is composed of a core domain (six Kelch beta-stranded motifs that together form a beta-propeller), an acid hinge, and a plant homeodomain (PHD) finger (Carmona and Schatz, 2017). BbRAG2L retains some similarity to the core domain but not the PHD finger of RAG2. The PHD confers the novel ability to read histone post-translational modifications to regulate gene expression (Jain et al., 2020), explaining the increased regulation of vertebrate RAG2. Structural comparisons cannot be complete without a discussion of substrates, and in fact it was the similarity between TIRs flanking DNA transposons and RSSs that prompted Tonegawa et al. to initially propose that RAGs originated by TE domestication (Sakano et al., 1979; Tonegawa, 1983; Fugmann, 2010; Huang et al., 2016). A good visualization of structural comparisons between protoRAG and RAG can be found in Zhang et al. (2019).
In addition to structural parallels, functional parallels exist between the two machineries. A key functional similarity between protoRAG and RAG is their end product (Huang et al., 2016). The cleaved TIR ends that are generated from protoRAG-induced DNA cleavage can undergo transposition or be joined together to form structures resembling the signal joints formed during V(D)J recombination. The non-TIR ends generated by the same mechanism resemble the coding joints formed during V(D)J recombination (Carmona and Schatz, 2017). Analysis of functional conservation lends further support to the notion that the co-option of protoRAG has added restrictions on the typically deleterious effects of unchecked transposition (Etchegaray et al., 2021). In particular, RAG-specific coupled cleavage activity, adherence to the 12/23 rule, and suppressed transposition, all impose restraints on the full extent of RAG action in vertebrate adaptive immunity (Zhang et al., 2019). These descriptions of structural and functional similarities, and the tighter regulation of RAG function, highlight that the evolutionary process of co-option was involved in the vertebrate acquisition of V(D)J recombination.
DNA Repair in V(D)J Recombination and Transposition: How are they Similar?
Since the core functionality of transposition and RAG-mediated recombination depends on the generation of (often double-stranded) chromosomal breaks, DNA repair mechanisms are essential at the excision site (Schlissel et al., 1993; Hedges and Deininger, 2007; Helmink and Sleckman, 2012). Transposon-induced breaks and RAG-induced breaks are different regarding how each is repaired; there are two pathways responsible for the repair of transposon-induced breaks, but only one pathway for the repair of RAG-induced breaks (Izsvak et al., 2004; Munoz-Lopez and Garcia-Perez, 2010). Transposons have two options for DNA repair: (i) non-homologous end joining (NHEJ), which closes DSBs without great specificity, and (ii) homologous recombination (HR), which typically copies information present on the sister chromatid to fill the break (Figure 4A). In case either of these mechanisms is inhibited, the other can usually take its place. In contrast, the RAG machinery relies exclusively on NHEJ to repair DSBs (Lee et al., 2004; Figure 4B). This reliance on NHEJ facilitates the generation of a large assortment of B and T cell receptors. Use of the HR machinery would dampen this diversity. Irrespective of the cause of DSBs, both transposons and RAGs rely on the host’s repair pathways to fix these breaks (Izsvak et al., 2004).
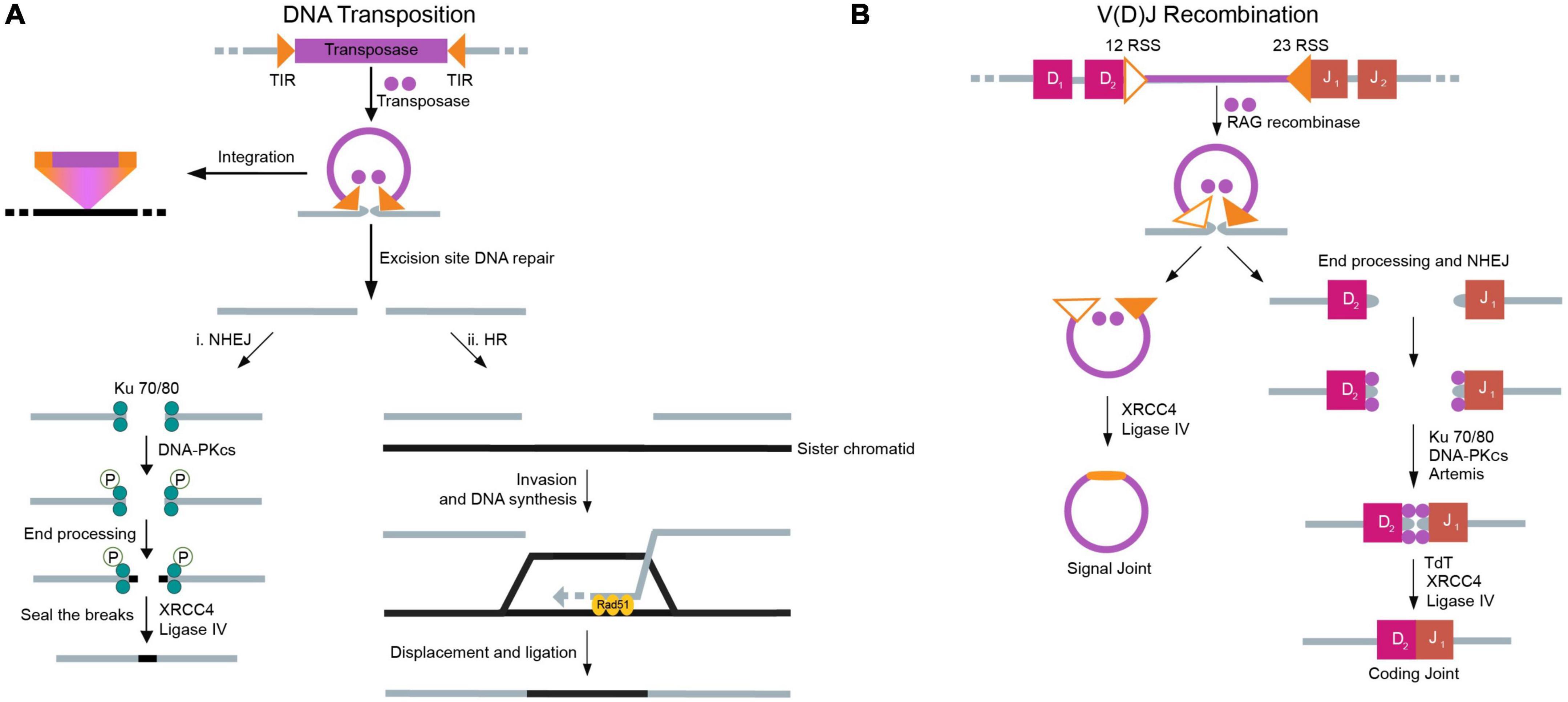
Figure 4. Mechanisms of double-strand break repair for DNA transposition and V(D)J recombination. DNA repair occurs to seal double stranded breaks (DSBs) created in the donor strand due to DNA transposition and V(D)J recombination. (A) In DNA transposition, once the transposon has been excised, the DSB can be repaired using either NHEJ or HR, with preference generally given to NHEJ. (i) In NHEJ, the process starts with the binding of the Ku 70/80 heterodimer to the broken DNA ends. A phosphorylation cascade initiated by DNA PKcs results in the recruitment of various proteins in the NHEJ pathway that are responsible for both the stability of the DNA complex and end processing. Finally, DNA ligase IV and XRCC4 act to seal the breaks in the DNA strand. (ii) In the case of HR, a sister chromatid or homologous chromosome serves as a template to repair the DSB. The process is contingent on the DNA-binding activity of Rad51. After replication based on the template strand, the donor DNA reseals. (B) In V(D)J recombination, the DSB must be repaired using NHEJ. The NHEJ process in V(D)J recombination generally involves the same components as are used in the repair of DSBs caused by transposons. Proteins specific to the V(D)J recombination process are highlighted.
In greater detail, NHEJ typically involves DNA-dependent protein kinase (DNA-PK), DNA ligase IV, and X-ray repair cross-complementing protein 4 (XRCC4) (Drouet et al., 2005). DNA-PK is composed of a catalytic subunit (PKcs) and a Ku heterodimer that binds specifically to free DNA ends (Gottlieb and Jackson, 1993). Once the heterodimer binds to a DSB, it recruits other protein factors crucial for NHEJ. These factors subsequently phosphorylate various proteins in the NHEJ pathway, some of which are involved in the stability of the DNA complex, and others in end processing. DNA end processing occurs by various proteins depending on the nature of the break. In the case of lymphocytes, and therefore relevant to the RAG machinery, terminal deoxynucleotidyl transferase (Tdt) and the Artemis endonuclease add/remove nucleotides from DNA ends (Davis and Chen, 2013). Once end processing is complete, DNA ligase IV and XRCC4 (a scaffold protein) seal the breaks. Specific to the RAG machinery, these proteins operate by joining coding and signal ends; XRCC4 is strictly required for coding end joining, but signal end joining can proceed without it in some cases (Roy et al., 2012). Often, the steps are flexible and depend on the nature of the DSB, although the recruitment of the Ku heterodimer is a crucial first step (Helmink and Sleckman, 2012; Davis and Chen, 2013).
To repair breaks caused by DNA transposons, HR is also an option. HR strongly relies on Rad51 for appropriate functioning. This process involves trimming the DSB to a 3’ overhang, after which Rad51 binds to the DNA, in a process termed pre-synapsis. Pre-synapsis is followed by synapsis, during which junctions form, eventually yielding a D-loop intermediate. From here, pathways diverge as to how the strands are processed. However, all routes use a template (typically a sister chromatid or homologous chromosome) to “refill” the DSB (Li and Heyer, 2008; Shrivastav et al., 2008).
Since transposon-induced DSBs can be repaired using either NHEJ or HR, a decision must be made on which pathway to use. Between different transposons, differences exist regarding how each one prefers to close DSBs. For example, the Sleeping Beauty transposon excision site is repaired using NHEJ factors (including Ku) and ataxia-telangiectasia mutated (ATM) (a protein kinase with roles in DSB signaling). HR is used to close the break only in the absence of both Ku and ATM (Izsvak et al., 2004; Brandsma and Gent, 2012). Another common transposon is Drosophila P-elements, whose excision site strongly prefers HR for DSB repair (Gloor et al., 2000). This process is probably to avoid deleterious and/or non-templated mutations in the germline. NHEJ remains the dominant process by which breaks are repaired in mammalian somatic cells (Ochmann and Ivics, 2021).
Just as a balance must be obtained between TE activity and genomic integrity, a balance must be struck between NHEJ and HR to repair DSBs. There are two reasons why NHEJ might be preferred over HR for non-co-opted TEs: (1) generation of increased diversity through error-prone joining, and (2) an increase in efficiency. Since NHEJ is inherently non-templated, there is potential for greater genetic diversity (Weterings and Chen, 2008), thereby greater functional diversity in the target sequence after insertion. Since transposons are evolutionarily thought of as parasitic, increased TE diversity maximizes their survival fitness. NHEJ also places greater emphasis on speed (Mao et al., 2008), trading quality for rapid sealing of potentially deleterious DSBs.
Since the priority for co-opted TEs (potentially including Drosophila P-elements that function in the fly germ line) is non-hazardous TE function, co-opted TEs have special requirements to preserve DNA. These TEs would only be selected for if the insertions were beneficial and did not damage the host, underscoring the preference for HR over NHEJ in situations of exaptation. It would be interesting to further dissect the possibility of the co-option of P-elements in Drosophila germline.
However, the emergence of adaptive immunity further complicates the discussion (Cui and Meek, 2007). The critical need for an increased diversity of antigen receptors prompted the proto-RAG induced DSBs to choose NHEJ over HR. Mechanistically, one dominant hypothesis for why NHEJ is exclusively used is that the post-cleavage complex (composed of RAG proteins, DNA signal ends, and blunt DNA hairpin coding ends) shepherds the generated signal ends. The RAG proteins disallow signal ends from accessing the HR pathway by holding on to the ends rather than allowing for their early release (Lee et al., 2004). Further, V(D)J recombination is restricted to the G1 phase of the cell cycle, as the RAG2 enzyme that is necessary for the process is destroyed during the G1-S transition (Li et al., 1996). While NHEJ can happen during the G1 phase (Takata et al., 1998), HR cannot occur efficiently during G1 due to the lack of a sister chromatid (Johnson and Jasin, 2000). As such, only NHEJ is observed to repair V(D)J recombination-induced breaks.
Unanswered Questions and Future Directions
In searching for whether and how V(D)J recombination retains structural and functional parallels to TEs, we looked at other processes necessary for adaptive immunity that involve DNA modifications: N/P-nucleotide addition (addition of palindromic sequences and non-genomic nucleotide sequences in the junctions between V, D and J gene segments of the immunoglobulins and T cells), somatic hypermutation, and isotype class switching. For each process, we reviewed the nature of the DNA modification to determine whether it might have originated from TE co-option.
Based on current knowledge, it appears that none of these processes have a proximal TE comparison. N/P-nucleotide addition generates diversity in joining regions between different domains of B-cell and T-cell receptors (Meier and Lewis, 1993; Repasky et al., 2004). This process involves NHEJ DNA repair through the action of Tdt, but no excision and repair steps directly resembling TEs (Repasky et al., 2004; Sandor et al., 2004). For further details on how N/P-nucleotide addition generates diversity see Srivastava and Robins (2012). Somatic hypermutation involves several factors that form a complex responsible for single nucleotide replacement to generate antibody diversity. This process, initiated by the targeting of cytidine deaminase to newly rearranged genes, does not involve excision and repair analogous to V(D)J recombination or TE action (Pilzecker and Jacobs, 2019). Isotype class switching is the process by which IgG, IgE, and IgA antibodies can be produced by mature B-cells. This process requires excision and repair, with the intervening constant domains deleted, which is quite similar to V(D)J recombination (Stavnezer et al., 2008). However, the complex structurally lacks specific recognition sites flanking the constant domains, and endonucleases perform a large portion of the cutting, splicing, and repair activity (Xu et al., 2014). As such, it is conceivable that V(D)J recombination stands on its own, uniquely having TE predecessors in basal organisms that were co-opted in jawed vertebrates over the course of evolution.
In writing this review, we have encountered numerous questions spanning several domains that warrant further study. These selected questions may prompt future studies on TEs and immunological processes:
• TEs are known to have a bias (Cutter et al., 2005; Green et al., 2012). Does the sequence of a TE contain a code for bias? If so, how can it be manipulated?
• As discussed previously, there are several protein components involved in appropriate NHEJ DSB repair. Which of these components are necessary and/or sufficient for DNA repair after transposition and V(D)J recombination? Does manipulation of these components influence the efficiency or functioning of TEs or RAGs?
• Can we repurpose transposase inhibitors as solutions to RAG-related immune dysregulation? More specifically, can we repurpose HIV reverse transcriptase inhibitors as therapeutic solutions to lymphoblastic leukemia and related cancers?
• Experimentally, is it possible to determine the conditions under which TEs will preferentially repair DSBs using HR over NHEJ?
• p53, a trans-repressor of transposons (Tiwari et al., 2017, 2018, 2020), regulates the DNA double strand break response. How might p53 and DNA transposons have interacted during the co-option of RAG recombinase?
• As this review has mentioned, studying how V(D)J recombination has arisen from TE co-option suggests that RAG predecessors have become increasingly regulated in the course of evolution. How can this knowledge of possible RAG origins be applied to human health and disease?
To this final question, our knowledge of TEs may be harnessed for therapeutic potential. For instance, many diseases result from RAG mutations. In cases of immune deficiency, TEs synthetically targeted to the RAG locus might restore RAG recombinase function, thereby ameliorating immune system deficits. Another value in considering these lines of questioning is their contribution to basic science. Generating transgenic organisms has long been time-consuming and fraught with failure. Understanding and manipulating the insertion biases of TEs can generate tools to increase the efficiency of generating transgenic organisms. Collectively, the study of how TEs and immunological processes intersect sheds light on many unexplored areas and offers exciting possibilities for advancing basic science and therapeutics.
Author Contributions
RS, NK, NC, and BT drafted and edited the manuscript. All the authors approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Tarun Krishnan for his input. We would also like to thank Vihang Ghalsasi, Attreyi Bhattacharya, and Shreyas Shridharan for their help with text editing. Finally, we would like to thank Smita Harne and Smritie Sheth for their help in figure design.
References
Agrawal, A., Eastman, Q. M., and Schatz, D. G. (1998). Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751. doi: 10.1038/29457
Ardeljan, D., Wang, X., Oghbaie, M., Taylor, M. S., Husband, D., Deshpande, V., et al. (2020). LINE-1 ORF2p expression is nearly imperceptible in human cancers. Mob. DNA 11:1. doi: 10.1186/s13100-019-0191-2
Baillie, J. K., Barnett, M. W., Upton, K. R., Gerhardt, D. J., Richmond, T. A., De Sapio, F., et al. (2011). Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537. doi: 10.1038/nature10531
Bejerano, G., Lowe, C. B., Ahituv, N., King, B., Siepel, A., Salama, S. R., et al. (2006). A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441, 87–90. doi: 10.1038/nature04696
Boeke, J. D., Garfinkel, D. J., Styles, C. A., and Fink, G. R. (1985). Ty elements transpose through an RNA intermediate. Cell 40, 491–500. doi: 10.1016/0092-8674(85)90197-7
Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., Hammell, M., et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19:199. doi: 10.1186/s13059-018-1577-z
Bourque, G., Leong, B., Vega, V. B., Chen, X., Lee, Y. L., Srinivasan, K. G., et al. (2008). Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762. doi: 10.1101/gr.080663.108
Brandsma, I., and Gent, D. C. (2012). Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr. 3, 9. doi: 10.1186/2041-9414-3-9
Burns, K. H. (2017). Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424. doi: 10.1038/nrc.2017.35
Burns, K. H., and Boeke, J. D. (2012). Human transposon tectonics. Cell 149, 740–752. doi: 10.1016/j.cell.2012.04.019
Carmona, L. M., and Schatz, D. G. (2017). New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J. 284, 1590–1605. doi: 10.1111/febs.13990
Chang, N. C., Rovira, Q., Wells, J. N., Feschotte, C., and Vaquerizas, J. M. (2022). Zebrafish transposable elements show extensive diversification in age, genomic distribution, and developmental expression. Genome Res. doi: 10.1101/gr.275655.121
Chuong, E. B. (2018). The placenta goes viral: Retroviruses control gene expression in pregnancy. PLoS Biol 16:e3000028. doi: 10.1371/journal.pbio.3000028
Chuong, E. B., Elde, N. C., and Feschotte, C. (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. doi: 10.1126/science.aad5497
Chuong, E. B., Rumi, M. A., Soares, M. J., and Baker, J. C. (2013). Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329. doi: 10.1038/ng.2553
Claverie-Martin, F., Flores, C., Anton-Gamero, M., Gonzalez-Acosta, H., and Garcia-Nieto, V. (2005). The Alu insertion in the CLCN5 gene of a patient with Dent’s disease leads to exon 11 skipping. J. Hum. Genet. 50, 370–374. doi: 10.1007/s10038-005-0265-5
Cooper, M. D., and Alder, M. N. (2006). The evolution of adaptive immune systems. Cell 124, 815–822. doi: 10.1016/j.cell.2006.02.001
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Yeo, G. W., Mu, Y., Lovci, M. T., et al. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131. doi: 10.1038/nature08248
Cui, X., and Meek, K. (2007). Linking double-stranded DNA breaks to the recombination activating gene complex directs repair to the nonhomologous end-joining pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 17046–17051. doi: 10.1073/pnas.0610928104
Cutter, A. D., Good, J. M., Pappas, C. T., Saunders, M. A., Starrett, D. M., and Wheeler, T. J. (2005). Transposable element orientation bias in the Drosophila melanogaster genome. J. Mol. Evol. 61, 733–741. doi: 10.1007/s00239-004-0243-0
Davis, A. J., and Chen, D. J. (2013). DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2, 130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02
Del Blanco, B., Garcia, V., Garcia-Mariscal, A., and Hernandez-Munain, C. (2011). Control of V(D)J recombination through transcriptional elongation and changes in locus chromatin structure and nuclear organization. Genet. Res. Int. 2011:970968. doi: 10.4061/2011/970968
Drouet, J., Delteil, C., Lefrancois, J., Concannon, P., Salles, B., and Calsou, P. (2005). DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J. Biol. Chem. 280, 7060–7069. doi: 10.1074/jbc.M410746200
Economou-Pachnis, A., and Tsichlis, P. N. (1985). Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res. 13, 8379–8387. doi: 10.1093/nar/13.23.8379
Etchegaray, E., Naville, M., Volff, J. N., and Haftek-Terreau, Z. (2021). Transposable element-derived sequences in vertebrate development. Mob DNA 12:1. doi: 10.1186/s13100-020-00229-5
Finnegan, D. J. (1989). Eukaryotic transposable elements and genome evolution. Trends Genet. 5, 103–107. doi: 10.1016/0168-9525(89)90039-5
Fu, B., Ma, H., and Liu, D. (2019). Endogenous retroviruses function as gene expression regulatory elements during mammalian pre-implantation embryo development. Int. J. Mol. Sci. 20:790. doi: 10.3390/ijms20030790
Fugmann, S. D. (2010). The origins of the RAG genes – From transposition to V(D)J recombination. Semin. Immunol. 22, 10–16. doi: 10.1016/j.smim.2009.11.004
Gallus, S., Hallstrom, B. M., Kumar, V., Dodt, W. G., Janke, A., Schumann, G. G., et al. (2015). Evolutionary histories of transposable elements in the genome of the largest living marsupial carnivore, the Tasmanian devil. Mol. Biol. Evol. 32, 1268–1283. doi: 10.1093/molbev/msv017
Gaymes, T. J., North, P. S., Brady, N., Hickson, I. D., Mufti, G. J., and Rassool, F. V. (2002). Increased error-prone non homologous DNA end-joining–a proposed mechanism of chromosomal instability in Bloom’s syndrome. Oncogene 21, 2525–2533. doi: 10.1038/sj.onc.1205331
Gellert, M. (2002). V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71, 101–132. doi: 10.1146/annurev.biochem.71.090501.150203
Gerdes, P., Richardson, S. R., Mager, D. L., and Faulkner, G. J. (2016). Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 17:100. doi: 10.1186/s13059-016-0965-5
Gloor, G. B., Moretti, J., Mouyal, J., and Keeler, K. J. (2000). Distinct P-element excision products in somatic and germline cells of Drosophila melanogaster. Genetics 155, 1821–1830. doi: 10.1093/genetics/155.4.1821
Goodier, J. L. (2014). Retrotransposition in tumors and brains. Mob DNA 5:11. doi: 10.1186/1759-8753-5-11
Gottlieb, T. M., and Jackson, S. P. (1993). The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72, 131–142. doi: 10.1016/0092-8674(93)90057-w
Green, B., Bouchier, C., Fairhead, C., Craig, N. L., and Cormack, B. P. (2012). Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3:3. doi: 10.1186/1759-8753-3-3
Guo, C., Jeong, H. H., Hsieh, Y. C., Klein, H. U., Bennett, D. A., De Jager, P. L., et al. (2018). Tau activates transposable elements in Alzheimer’s disease. Cell Rep. 23, 2874–2880. doi: 10.1016/j.celrep.2018.05.004
Hancks, D. C., and Kazazian, H. H. Jr. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203. doi: 10.1016/j.gde.2012.02.006
Hancks, D. C., and Kazazian, H. H. Jr. (2016). Roles for retrotransposon insertions in human disease. Mob. DNA 7:9. doi: 10.1186/s13100-016-0065-9
Hedges, D. J., and Deininger, P. L. (2007). Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat. Res. 616, 46–59. doi: 10.1016/j.mrfmmm.2006.11.021
Helmink, B. A., and Sleckman, B. P. (2012). The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 30, 175–202. doi: 10.1146/annurev-immunol-030409-101320
Hencken, C. G., Li, X., and Craig, N. L. (2012). Functional characterization of an active Rag-like transposase. Nat. Struct. Mol. Biol. 19, 834–836. doi: 10.1038/nsmb.2338
Hiom, K., Melek, M., and Gellert, M. (1998). DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94, 463–470. doi: 10.1016/s0092-8674(00)81587-1
Huang, C. R., Burns, K. H., and Boeke, J. D. (2012). Active transposition in genomes. Annu. Rev. Genet. 46, 651–675. doi: 10.1146/annurev-genet-110711-155616
Huang, S., Tao, X., Yuan, S., Zhang, Y., Li, P., Beilinson, H. A., et al. (2016). Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell 166, 102–114. doi: 10.1016/j.cell.2016.05.032
Ito, J., Sugimoto, R., Nakaoka, H., Yamada, S., Kimura, T., Hayano, T., et al. (2017). Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 13:e1006883. doi: 10.1371/journal.pgen.1006883
Izsvak, Z., Stuwe, E. E., Fiedler, D., Katzer, A., Jeggo, P. A., and Ivics, Z. (2004). Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13, 279–290. doi: 10.1016/s1097-2765(03)00524-0
Jain, K., Fraser, C. S., Marunde, M. R., Parker, M. M., Sagum, C., Burg, J. M., et al. (2020). Characterization of the plant homeodomain (PHD) reader family for their histone tail interactions. Epigenetics Chromatin 13:3. doi: 10.1186/s13072-020-0328-z
Johnson, R. D., and Jasin, M. (2000). Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407. doi: 10.1093/emboj/19.13.3398
Jones, J. M., and Gellert, M. (2004). The taming of a transposon: V(D)J recombination and the immune system. Immunol. Rev. 200, 233–248. doi: 10.1111/j.0105-2896.2004.00168.x
Kazazian, H. H. Jr. (2004). Mobile elements: drivers of genome evolution. Science 303, 1626–1632. doi: 10.1126/science.1089670
Kazazian, H. H. Jr., Wong, C., Youssoufian, H., Scott, A. F., Phillips, D. G., and Antonarakis, S. E. (1988). Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166. doi: 10.1038/332164a0
Kim, M. S., Lapkouski, M., Yang, W., and Gellert, M. (2015). Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature 518, 507–511. doi: 10.1038/nature14174
Klein, L., Kyewski, B., Allen, P. M., and Hogquist, K. A. (2014). Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391. doi: 10.1038/nri3667
Krangel, M. S. (2003). Gene segment selection in V(D)J recombination: accessibility and beyond. Nat. Immunol. 4, 624–630. doi: 10.1038/ni0703-624
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Lee, G. S., Neiditch, M. B., Salus, S. S., and Roth, D. B. (2004). RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117, 171–184. doi: 10.1016/s0092-8674(04)00301-0
Li, W., Prazak, L., Chatterjee, N., Gruninger, S., Krug, L., Theodorou, D., et al. (2013). Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 16, 529–531. doi: 10.1038/nn.3368
Li, X., and Heyer, W. D. (2008). Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113. doi: 10.1038/cr.2008.1
Li, Z., Dordai, D. I., Lee, J., and Desiderio, S. (1996). A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity 5, 575–589. doi: 10.1016/s1074-7613(00)80272-1
Liu, C., Yang, Y., and Schatz, D. G. (2019). Structures of a RAG-like transposase during cut-and-paste transposition. Nature 575, 540–544. doi: 10.1038/s41586-019-1753-7
Mao, Z., Bozzella, M., Seluanov, A., and Gorbunova, V. (2008). Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7, 1765–1771. doi: 10.1016/j.dnarep.2008.06.018
Martin, S. L. (2010). Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol 7, 706–711. doi: 10.4161/rna.7.6.13766
Meier, J. T., and Lewis, S. M. (1993). P nucleotides in V(D)J recombination: a fine-structure analysis. Mol. Cell Biol. 13, 1078–1092. doi: 10.1128/mcb.13.2.1078-1092.1993
Meischl, C., Boer, M., Ahlin, A., and Roos, D. (2000). A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur. J. Hum. Genet. 8, 697–703. doi: 10.1038/sj.ejhg.5200523
Meischl, C., and Roos, D. (1998). The molecular basis of chronic granulomatous disease. Spring. Semin. Immunopathol. 19, 417–434. doi: 10.1007/bf00792600
Mitra, R., Li, X., Kapusta, A., Mayhew, D., Mitra, R. D., Feschotte, C., et al. (2013). Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc. Natl. Acad. Sci. U S A. 110, 234–239. doi: 10.1073/pnas.1217548110
Morales Poole, J. R., Huang, S. F., Xu, A., Bayet, J., and Pontarotti, P. (2017). The RAG transposon is active through the deuterostome evolution and domesticated in jawed vertebrates. Immunogenetics 69, 391–400. doi: 10.1007/s00251-017-0979-5
Moschetti, R., Palazzo, A., Lorusso, P., Viggiano, L., and Marsano, R. M. (2020). “What You Need, Baby, I Got It”: Transposable Elements as Suppliers of Cis-Operating Sequences in Drosophila. Biology (Basel) 9:25 doi: 10.3390/biology9020025
Munoz-Lopez, M., and Garcia-Perez, J. L. (2010). DNA Transposons: Nature and Applications in Genomics. Curr. Genom. 11, 115–128. doi: 10.2174/138920210790886871
Muotri, A. R., Chu, V. T., Marchetto, M. C., Deng, W., Moran, J. V., and Gage, F. H. (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910. doi: 10.1038/nature03663
Ochmann, M. T., and Ivics, Z. (2021). Jumping Ahead with Sleeping Beauty: Mechanistic Insights into Cut-and-Paste Transposition. Viruses 13:76. doi: 10.3390/v13010076
Ogino, S., Nosho, K., Kirkner, G. J., Kawasaki, T., Chan, A. T., Schernhammer, E. S., et al. (2008). A Cohort Study of Tumoral LINE-1 Hypomethylation and Prognosis in Colon Cancer. J. Natl. Cancer. Inst. 100, 1734–1738. doi: 10.1093/jnci/djn359
Pastuzyn, E. D., Day, C. E., Kearns, R. B., Kyrke-Smith, M., Taibi, A. V., McCormick, J., et al. (2018). The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 172, 275–288e18. doi: 10.1016/j.cell.2017.12.024
Petri, R., Brattas, P. L., Sharma, Y., Jonsson, M. E., Pircs, K., Bengzon, J., et al. (2019). LINE-2 transposable elements are a source of functional human microRNAs and target sites. PLoS Genet 15:e1008036. doi: 10.1371/journal.pgen.1008036
Pilzecker, B., and Jacobs, H. (2019). Mutating for Good: DNA Damage Responses During Somatic Hypermutation. Front. Immunol. 10:438. doi: 10.3389/fimmu.2019.00438
Repasky, J. A., Corbett, E., Boboila, C., and Schatz, D. G. (2004). Mutational Analysis of Terminal Deoxynucleotidyltransferase- Mediated N-Nucleotide Addition in V(D)J Recombination. J. Immunol. 172, 5478–5488. doi: 10.4049/jimmunol.172.9.5478
Rodriguez-Martin, B., Alvarez, E. G., Baez-Ortega, A., Zamora, J., Supek, F., Demeulemeester, J., et al. (2020). Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 52, 306–319. doi: 10.1038/s41588-019-0562-0
Roy, S., Andres, S. N., Vergnes, A., Neal, J. A., Xu, Y., Yu, Y., et al. (2012). XRCC4’s interaction with XLF is required for coding (but not signal) end joining. Nucleic Acids Res. 40, 1684–1694. doi: 10.1093/nar/gkr1315
Sakano, H., Huppi, K., Heinrich, G., and Tonegawa, S. (1979). Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288–294. doi: 10.1038/280288a0
Saleh, A., Macia, A., and Muotri, A. R. (2019). Transposable Elements, Inflammation, and Neurological Disease. Front. Neurol. 10:894. doi: 10.3389/fneur.2019.00894
Sandor, Z., Calicchio, M. L., Sargent, R. G., Roth, D. B., and Wilson, J. H. (2004). Distinct requirements for Ku in N nucleotide addition at V(D)J–and non–V(D)J–generated double-strand breaks. Nucleic Acids Res. 32, 1866–1873. doi: 10.1093/nar/gkh502
Schatz, D. G., and Swanson, P. C. (2011). V(D)J Recombination: Mechanisms of Initiation. Annu. Rev. Genet. 45, 167–202. doi: 10.1146/annurev-genet-110410-132552
Schlissel, M., Constantinescu, A., Morrow, T., Baxter, M., and Peng, A. (1993). Double-strand signal sequence breaks in V(D)J recombination are blunt, 5’–phosphorylated, RAG-dependent, and cell cycle regulated. Genes. Dev. 7, 2520–2532. doi: 10.1101/gad.7.12b.2520
Schlissel, M. S. (2003). Regulating antigen-receptor gene assembly. Nat. Rev. Immunol. 3, 890–899. doi: 10.1038/nri1225
Shrivastav, M., De Haro, L. P., and Nickoloff, J. A. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res. 18, 134–147. doi: 10.1038/cr.2007.111
Srivastava, S. K., and Robins, H. S. (2012). Palindromic Nucleotide Analysis in Human T Cell Receptor Rearrangements. PLoS One 7:e52250. doi: 10.1371/journal.pone.0052250
Stavnezer, J., Guikema, J. E., and Schrader, C. E. (2008). Mechanism and Regulation of Class Switch Recombination. Annu. Rev. Immunol. 26, 261–292. doi: 10.1146/annurev.immunol.26.021607.090248
Stenz, L. (2021). The L1-dependant and Pol III transcribed Alu retrotransposon, from its discovery to innate immunity. Mol. Biol. Rep. 48, 2775–2789. doi: 10.1007/s11033-021-06258-4
Sundaram, V., Cheng, Y., Ma, Z., Li, D., Xing, X., Edge, P., et al. (2014). Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976. doi: 10.1101/gr.168872.113
Takata, M., Sasaki, M. S., Sonoda, E., Morrison, C., Hashimoto, M., Utsumi, H., et al. (1998). Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508. doi: 10.1093/emboj/17.18.5497
Tam, O. H., Ostrow, L. W., and Gale Hammell, M. (2019). Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob. DNA 10:32. doi: 10.1186/s13100-019-0176-1
Tarallo, V., Hirano, Y., Gelfand, B. D., Dridi, S., Kerur, N., Kim, Y., et al. (2012). DICER1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88. Cell 149, 847–859. doi: 10.1016/j.cell.2012.03.036
Tiwari, B., Jones, A. E., and Abrams, J. M. (2018). Transposons, p53 and Genome Security. Trends Genet. 34, 846–855. doi: 10.1016/j.tig.2018.08.003
Tiwari, B., Jones, A. E., Caillet, C. J., Das, S., Royer, S. K., and Abrams, J. M. (2020). p53 directly represses human LINE1 transposons. Genes Dev. 34, 1439–1451. doi: 10.1101/gad.343186.120
Tiwari, B., Kurtz, P., Jones, A. E., Wylie, A., Amatruda, J. F., Boggupalli, D. P., et al. (2017). Retrotransposons Mimic Germ Plasm Determinants to Promote Transgenerational Inheritance. Curr. Biol. 27, 3010–3016e3013. doi: 10.1016/j.cub.2017.08.036
Tonegawa, S. (1983). Somatic generation of antibody diversity. Nature 302, 575–581. doi: 10.1038/302575a0
van Gent, D. C., Ramsden, D. A., and Gellert, M. (1996). The RAG1 and RAG2 Proteins Establish the 12/23 Rule in V(D)J Recombination. Cell 85, 107–113. doi: 10.1016/s0092-8674(00)81086-7
Wang, T., Zeng, J., Lowe, C. B., Sellers, R. G., Salama, S. R., Yang, M., et al. (2007). Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad Sci. U.S.A. 104, 18613–18618. doi: 10.1073/pnas.0703637104
Weterings, E., and Chen, D. J. (2008). The endless tale of non-homologous end-joining. Cell Res. 18, 114–124. doi: 10.1038/cr.2008.3
Wolff, E. M., Byun, H. M., Han, H. F., Sharma, S., Nichols, P. W., Siegmund, K. D., et al. (2010). Hypomethylation of a LINE-1 Promoter Activates an Alternate Transcript of the MET Oncogene in Bladders with Cancer. PLoS Genet. 6:e1000917. doi: 10.1371/journal.pgen.1000917
Xu, J., Husain, A., Hu, W., Honjo, T., and Kobayashi, M. (2014). APE1 is dispensable for S-region cleavage but required for its repair in class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 111, 17242–17247. doi: 10.1073/pnas.1420221111
Keywords: transposable elements, V(D)J recombination, adaptive immunity, RAG recombinase, DNA transposons
Citation: Shridharan RV, Kalakuntla N, Chirmule N and Tiwari B (2022) The Happy Hopping of Transposons: The Origins of V(D)J Recombination in Adaptive Immunity. Front. Ecol. Evol. 10:836066. doi: 10.3389/fevo.2022.836066
Received: 15 December 2021; Accepted: 22 March 2022;
Published: 26 May 2022.
Edited by:
Manvendra Singh, Helmholtz Association of German Research Centers (HZ), GermanyReviewed by:
René Massimiliano Marsano, University of Bari Aldo Moro, ItalyElgion Lucio Silva Loreto, Federal University of Santa Maria, Brazil
Copyright © 2022 Shridharan, Kalakuntla, Chirmule and Tiwari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhavana Tiwari, YnRpd2FyaUBpaXNlcmJwci5hYy5pbg==
†These authors have contributed equally to this work
 Rishi V. Shridharan
Rishi V. Shridharan Neha Kalakuntla
Neha Kalakuntla Narendra Chirmule
Narendra Chirmule Bhavana Tiwari
Bhavana Tiwari