- Department of Biological Sciences, University of North Carolina at Charlotte, Charlotte, NC, United States
Echinoderm larvae have served as a fundamental system for understanding development and life history evolution over much of the last century. In the last few decades, our understanding of echinoderm larvae has expanded to the microbiota that they associate with. These symbionts and the communities that they form in relation to echinoderm larval host are the focus of this review. Our synthesis of the literature suggests three primary themes. First, larval echinoderms associate with “subcuticle bacteria” that appear to colonize select tissue types. Second, the bacterial communities associated with larval echinoderms exhibit compositional shifts that are correlated with several fundamental properties of larval biology (e.g., development and morphological plasticity) and ecology (e.g., feeding environment). Third, echinoderm larvae exhibit specific responses to pathogenic bacteria that may aid in maintaining the symbiont community and avoid dysbiosis. To our knowledge, no studies have focused on whether climate-related stressors impact the composition of these symbiont communities or how changes in bacteria may modulate response by larvae to these environmental stressors. Lastly, we conclude by outlining techniques that need to be established in echinoderm larvae to transition from correlations between larvae and their associated microbiota to the function of these symbionts.
Echinoderms And Their Larvae
The phylum Echinodermata is characterized by their pentaradial symmetry and global distribution in marine ecosystems. This group first appeared in the Cambrian and is composed of ~7,000 extant species (Appeltans et al., 2012) that are grouped into five classes: Crinoidea (feather stars and sea lilies), Holothuroidea (sea cucumbers), Asteroidea (sea stars), Ophiuroidea (brittle stars), and Echinoidea (sea urchins). Echinoderm life cycles are primarily bi-phasic, where the adults reside on the sea floor and the embryonic stages are suspended in the water column (Thorson, 1950; Mileikovsky, 1971; Young and Chia, 1987). This phylum is, perhaps, most recognized for its remarkable diversity of larval forms that have fascinated biologists for more than a century (Levin and Bridges, 1995).
Echinoderm development, in general, follows either a lecithotrophic (non-feeding) or planktotrophic (feeding) trajectory (Thorson, 1950; Mileikovsky, 1971; Strathmann, 1985). Lecithotrophs develop from relatively large and energy-rich eggs (~300 μm to 1 mm in diameter, or where s ≥ 1; Vance, 1973; McEdward and Miner, 2006) with sufficient maternal investment to complete development and undergo metamorphosis. The developmental period for lecithotrophs typically lasts a few days and, due to a shorter pelagic larval duration, can result in marginal dispersal between populations (Thorson, 1950; Mileikovsky, 1971; Strathmann, 1985). Planktotrophs, on the other hand, develop from more energy-poor eggs (~100 to ~300 μm in diameter, or where s < 1; Vance, 1973; McEdward and Miner, 2006) with sufficient maternal input to complete embryogenesis and develop into larvae with initial feeding structures. The remaining energetic supply required for larval development and metamorphosis is acquired from exogenous resources (e.g., phytoplankton, detritus, and other particles) that are concentrated by a cilia-lined feeding apparatus (Strathmann, 1987; Feehan et al., 2018). These particulates are often dilute, leading to a pelagic larval duration lasting weeks to months and, in some cases, years (Thorson, 1950; Mileikovsky, 1971; Strathmann, 1985; Olson and Olson, 1989).
Nearly four decades ago, Rivkin et al. (1986) recognized that the planktotrophic larvae of the asteroid Porania antarctica selectively interacts with the environmental microbiota through bacterivory. It has since been observed that additional planktotrophic echinoderms exhibit bacterivory and that this feeding mode is hypothesized to be important, but not essential, to the metabolic requirements of the larva (Pearse et al., 1991; Douillet, 1993; Ayukai, 1994; Moal et al., 1996; Gosselin and Qian, 1997). Based on maximum clearance rates and particle abundance, it is estimated that echinoderm larvae interact with ~20 million bacteria each day by feeding alone (Carrier et al., 2018a). It is, however, unknown which bacterial (or other microbial) groups that echinoderm larvae may target and whether these microbes are selected strictly for bacterivory or as a symbiont that may be acquired by horizontal transmission.
Just prior to recognizing that echinoderm larvae were bacterivorous, Cameron and Holland (1983) observed that bacteria were living inside the tissues of healthy larvae. In this review we synthesize the properties of these bacteria and how they relate to the biology and ecology of the echinoderm larval host. In the first section, we provide an overview of our understanding of echinoderm larvae and their bacterial symbionts through the lens of microscopy and next-generation sequencing. In the second section, we summarize how larval-associated microbiota may be relevant in coping with anthropogenic stressors and outline the techniques needed to transition toward understanding the function of these symbionts.
Bacterial Symbionts Of Echinoderm Larvae
Over the last few decades our understanding of echinoderm larvae and their microbes has gone through two primary phases. The first phase uses microscopy and focuses on “subcuticle bacteria” (or, due to their location within larval tissues, could be also be characterized as endosymbionts but this has not been explicitly tested). The second phase has developed in recent years and uses next-generation sequencing and other molecular tools to characterize larval-associated bacterial communities. This phase may be further divided into two focal points: the dynamics of these bacterial communities under different ecological conditions and the immune responses of the larval host when faced with pathogenic bacteria.
Subcuticle Bacteria
While determining how to preserve the thin cuticle overlying the epidermis for transmission and scanning electron microscopy, Holland and Nealson (1978) observed that adults for each of the five echinoderm classes had a high abundance of what they called ‘subcuticle bacteria.' Holland and Nealson (1978) did not test whether embryonic or larval stages also contained subcuticle bacteria. They did, however, speculate on the nature of transmission for echinoderms, stating that because the eggs are “unattached to any follicle cells, and no bacteria have ever been observed on or in echinoderm eggs” that “it is probable that each new generation of such echinoderms acquires its subcuticular bacteria from the surrounding sea water.” Moreover, Holland and Nealson (1978) suggested that, if acquired during the embryonic or larval stages, the echinoderm must select for these symbiotic bacteria from a diverse community of environmental microbiota.
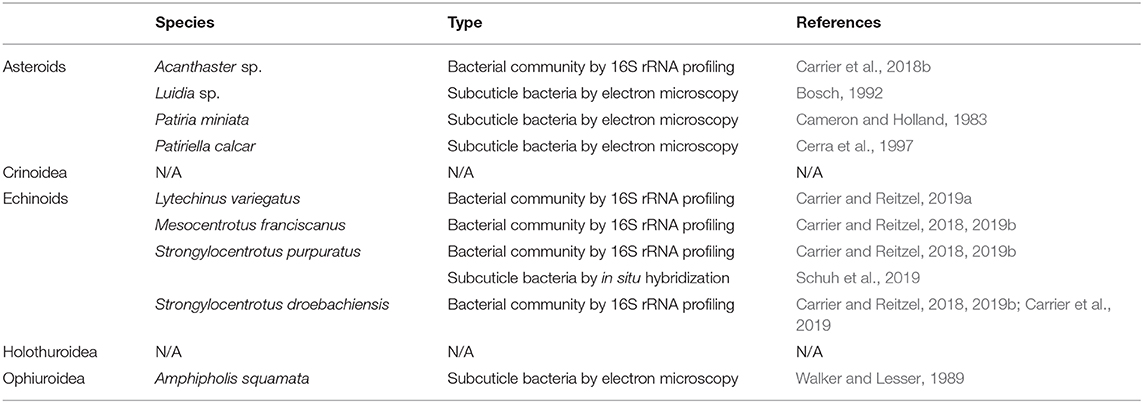
Shortly thereafter and on multiple occasions since these original observations, studies have identified subcuticle bacteria in the developmental stages of echinoderms. These symbionts, thus far, have been found in three asteroids (Cameron and Holland, 1983; Bosch, 1992; Cerra et al., 1997), one ophiuroid (Walker and Lesser, 1989), and one echinoid (Heyland et al., 2018; Schuh et al., 2019) (Figure 1; Table 1). In these five species, subcuticle bacteria have been observed within the mouth and gut lumen, out-pockets of the extracellular matrix that surrounds the larval body, embedded in the inner layer of the secondary cuticle of the rudiment epidermis, and are engulfed and, in some cases, digested by epidermal cells (Cameron and Holland, 1983; Walker and Lesser, 1989; Bosch, 1992; Cerra et al., 1997; Heyland et al., 2018; Schuh et al., 2019).
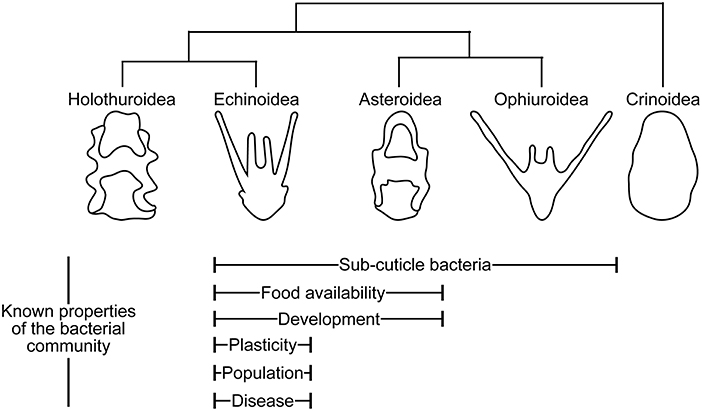
Figure 1. Echinoderm larvae and their symbionts. Properties of echinoderm larval biology and ecology with known correlated shifts in the associated bacterial communities.
The function of these subcuticle bacteria remains essentially unknown. Two cases, however, may suggest that these symbionts interact with and functionally benefit the larval host. First, the brittle star Amphipholis squamata has a “vestigial” pluteus (i.e., greatly reduced larval arms and lacking a ciliated mouth) that is brooded within their central plate. Using transmission and scanning electron microscopy, Walker and Lesser (1989) found that a rod-shaped Octadecabacter was abundant and actively dividing within the tissues of this “vestigial” pluteus (Morrow et al., 2018). This Octadecabacter, which strictly associates with these larvae and is not found in the environment, can uptake dissolved free amino acids that are then incorporated into bacterial proteins and increase the total amino acid uptake for A. squamata (Walker and Lesser, 1989; Lesser and Blakemore, 1990; Lesser and Walker, 1992). Second, clonal larvae of the sea star Luidia collected from the Gulf Stream had one to three rod-shaped morphotypes of subcuticle bacteria (Bosch, 1992). For Luidia as well as Acanthaster, some symbionts were located in the gut and auto-fluoresced (Bosch, 1992; Galac et al., 2016; Carrier et al., 2018b), suggesting the potential ability to be phototrophic. Collectively, these examples suggest, but do not show explicitly, that bacterial symbionts may have metabolic functions that could potentially benefit the larval host.
Bacterial Communities
The subcuticle bacteria discussed above are a portion of the collection of bacteria associated with echinoderm larvae. Using next-generation sequencing, larval-associated bacterial communities have been reported for six species of echinoderm larvae: two asteroids (Galac et al., 2016; Carrier et al., 2018b) and four echinoids (Carrier and Reitzel, 2018, 2019a,b; Carrier et al., 2019) (Figure 1; Table 1). In general, these bacterial communities are composed of a couple hundred bacterial species (i.e., Operational Taxonomic Units or OTUs, as defined by ≥99% similarity of the phylogenetically-conserved 16S rRNA gene) (Galac et al., 2016; Carrier and Reitzel, 2018, 2019b; Carrier et al., 2018b, 2019). The predominant bacterial families encompassing these diverse communities are the α- and γ-Proteobacteria (Proteobacteria) and the Flavobacteriaceae (Bacteroidetes) (Galac et al., 2016; Carrier et al., 2018b, 2019; Carrier and Reitzel, 2019a,b).
Like many other studies of animal and plant microbes, the bacterial communities associated with echinoderm larvae are species-specific and taxonomically distinct from the environmental microbiota (Galac et al., 2016; Carrier and Reitzel, 2018; Carrier et al., 2018b), suggesting that these communities are, at least in part, selected by the host. This host-specificity, however, appears to be lost when larvae are cultured under traditional laboratory settings for rearing the developmental stages of marine invertebrates (i.e., fine-filtered or artificial saltwater) (Schuh et al., 2019). Specifically, Strongylocentrotus purpuratus larvae cultured under traditional laboratory settings associate with bacterial communities that are less diverse in total taxa and the phylogenetic breadth of those taxa, and retain ~40% of the OTUs harbored by “wild-type” counterparts (Schuh et al., 2019). This implies that studying larval-associated bacterial communities is most accurately performed at near-natural conditions, such as by filtering ambient seawater to 5-μm to remove most debris and planktonic predators while retaining the environmental microbiota (Carrier and Reitzel, 2018; Hodin et al., 2019).
Larval-associated bacterial communities are variable in community membership and composition but exhibit non-random shifts that correlate with multiple components of larval biology and ecology. These communities, for example, are established on unfertilized eggs but not the sperm of sea urchins (Carrier and Reitzel, 2019b; Schuh et al., 2019). When cultured using coarsely (5-μm) filtered seawater, echinoderm larvae exhibit a development-based succession in symbiont composition and, using fluorescent in situ hybridization, these bacteria localize in the mouth and gut lumen (Carrier and Reitzel, 2019b; Schuh et al., 2019). Following fertilization, the diversity of these communities increases by ~20% during the early embryonic stages and decreased by nearly ~85% following hatching and through metamorphosis (Carrier and Reitzel, 2019b). From egg to hatching, the early embryonic stages appear to converge taxonomically with the environmental microbiota but then exhibit a host-mediated selection by diverging from this community following the onset of feeding (Carrier and Reitzel, 2019b). In cases where embryonic development includes asexual reproduction (e.g., cloning), the larval clones deviate little from the parent larva by maintaining a high proportion of particular bacteria, including phototropic species (Galac et al., 2016; Carrier et al., 2018b).
The six species of echinoderm larvae with profiled bacterial communities are planktotrophs, and by definition, are required to feed. Five of these species (echinoids: Strongylocentrotus purpuratus, S. droebachiensis, Mesocentrotus franciscanus, and Lytechinus variegatus; asteroid: Acanthaster sp.) have been differentially fed to test whether community composition varies with food quantity. In each case, bacterial communities are diet-specific with well-fed larvae distinguished from diet-restricted treatments (Carrier and Reitzel, 2018, 2019a; Carrier et al., 2018b). When diet-restriction is prolonged, larval sea urchins compensate by elongating their feeding arms to increase water filtration capacity (Hart and Strathmann, 1994; Miner, 2004; McAlister and Miner, 2018). This change in morphology is correlated with a shift in the composition of the bacterial symbionts, such that larval urchins associate with phenotype-specific microbiota (Carrier and Reitzel, 2018, 2019a; Carrier et al., 2018b).
Establishment of a phenotype-specific bacterial community for larvae of the sea urchin L. variegatus follows a four-stage succession (Carrier and Reitzel, 2019a). First, larvae across degrees of food availability associated with bacterial communities similar in both composition and structure. Second, different food environments (i.e., algal concentrations) induced diet-specific bacterial communities in both membership and composition. Third, the bacterial communities of diet-restricted larvae associated with similar bacterial communities that are also distinct from that of well-fed larvae, with the latter coinciding with a reduction in community diversity. Lastly, composition and structure are maintained from the prior successional stage and now correlate with the short- and long-arm phenotypes (Carrier and Reitzel, 2019a). This suggests that changes in the larval-associated bacterial community shifts prior to the expression of the environmentally elicited morphological phenotypes and that microbial communities may respond to environmental variation more quickly than morphological changes.
Recent research also suggests that microbial communities differ not only between species but also between populations (e.g., ascidians: Dishaw et al., 2014; seaweed: Marzinelli et al., 2015; fish: Llewellyn et al., 2016; sponges: Marino et al., 2017). Differential feeding of larvae of the echinoid S. droebachiensis from three populations in different ocean basins showed parallel responses that resulted in diet-specific bacterial communities (Carrier et al., 2019). Despite each population associating with a diet-specific bacterial community, variation in membership and community composition correlated more strongly with geographic location (Carrier et al., 2019). Moreover, when comparing the taxonomic membership between populations, 20–30% of bacterial taxa were specific to a single location while ~10% were shared between all three locations (Carrier et al., 2019). Collectively, these data suggest that larvae for a given species associates with a population-specific bacterial community. It is, however, worth noting that this comparison was not performed using common garden culturing, and that it would be expected that these communities are more taxonomically similar but still population-specific, when cultured using identical seawater.
Pathogenic Bacteria
Biological responses to foreign “particles” by echinoderm larvae were first recognized in the late nineteenth century (Metchnikoff, 1891; Tauber, 2003). More recently, echinoderm larvae have been used as a comparative system to define the cellular and molecular mechanisms of immunity when combating pathogenic bacteria. Due to the availability of a genome (Sodergren et al., 2006), the majority of this work has used the echinoid Strongylocentrotus purpuratus. The immune response by S. purpuratus to pathogenic bacteria was recently reviewed by Buckley and Rast (2017) and Heyland et al. (2018). We refer the reader to these in-depth reviews for the molecular underpinnings of larval immunity, as this section will focus on the ecological components.
From the amoebic disease (Paramoeba invadens) of S. droebachiensis in Nova Scotia (Scheibling and Stephenson, 1984; Feehan et al., 2013) to the major epizootic that decimated Diadema antillarum throughout the Caribbean (Lessios et al., 1984; Lessios, 2016) or, more recently, the densovirus-associated wasting disease of asteroids (Hewson et al., 2014; Harvell et al., 2019), the ecological impacts of disease on echinoderms have been well-documented over the last few decades (Feehan and Scheibling, 2014). Less of this work, however, has focused on embryonic and larval echinoderms. Echinoderm larvae potentially interact with ~20 million bacteria each day by feeding alone (Carrier et al., 2018a), and a portion of these bacteria may be consumed through bacterivory, be symbionts acquired by horizontal transmission, or pathogenic bacteria that require an immune response.
When faced with variation in food quantity, larval echinoids exhibit a trade-off in the expression of immune and metabolic genes (Carrier et al., 2015, 2018a), such that well-fed larvae upregulate metabolism and suppress immunity. High food availability is also suitable environmental conditions for bacteria to express pathogenic characteristics. If faced with a pathogen in these feeding conditions, food-induced suppression of the immune system may result in a suboptimal physiological response, and larvae may be less able to regulate the associated microbiota. Echinoid larvae may then be at risk of pathogen-induced disease or exhibiting dysbiosis, both of which are hypothesized to be precursors to larval mortality (Carrier et al., 2018a).
When faced with pathogenic bacteria, echinoid larvae exhibit immune responses, for example, by expressing genes in the interleukin 17 (IL17) complex (Ho et al., 2016; Buckley and Rast, 2017; Buckley et al., 2017). IL17s are known to both serve as a primary barrier to foreign bodies and to regulate the composition of larval-associated microbiota (Buckley and Rast, 2017). For S. purpuratus, exposure to the pathogen V. diazotrophicus coincides with successive expression of gut epithelial-specific IL17 subtypes to prevent progression of V. diazotrophicus and maintain the gut microbiota (Buckley and Rast, 2017; Buckley et al., 2017). When S. purpuratus larvae are made nearly germ-free, they become more susceptible to Vibrio-induced infections and mortality than counterparts with their native bacteria (Schuh et al., 2019). Pathogens that elicit a response appear to be lineage-specific, as not all Vibrio species or strains induce the expression of the IL17s (Buckley and Rast, 2017; Buckley et al., 2017). Such responses by the larval host may contribute to maintaining homeostatic symbioses (e.g., Mortzfeld and Bosch, 2017), but the functional underpinnings for larval echinoderms have yet to be determined (but see Ho et al., 2016; Buckley and Rast, 2017; Buckley et al., 2017).
Larval Echinoderms In a Changing Environment
Our understanding of echinoderm larvae and their relationship with microbial symbionts has been studied at ambient conditions. Marine invertebrates and their life history stages are, however, encountering a suite of anthropogenic stressors (Byrne et al., 2018) that may disrupt homeostatic symbioses (e.g., Rosenberg et al., 2007). To our knowledge, no studies have focused on how climate-related stressors (e.g., temperature and pH) affect the associated microbiota of echinoderm larvae. Similar to the cnidarian planula (Mortzfeld et al., 2015) and sponge amphiblastula (Webster et al., 2011), we hypothesize that an acclimation response to abiotic or biotic stressors would include shifts in symbiont composition. This, in particular, provides larvae with an opportunity to acquire bacterial symbionts with genes that are novel to the larval hologenome and that may aid in ameliorating physiological stress. Whether at ambient conditions or facing climate-related stressors, the function of larval-associated symbionts as well as if the host benefits from these partnerships remains uncertain.
Determining if and how microbial symbionts contribute to the larval holobiont requires a transition from 16S rRNA profiling to functional studies (Williams and Carrier, 2018). The functional potential and expression profiles of microbial symbionts may be assessed using metagenomics (e.g., Slaby et al., 2017) and metatranscriptomics (e.g., Moitinho-Silva et al., 2014). The impact that particular taxa or the symbiont community have on the larval host may then be assessed by generating microbe-free larvae through an antibiotic treatment or gnotobiotic chambers (Rawls et al., 2004; Bates et al., 2006; Smith et al., 2007; Gloeckner et al., 2013; Leigh et al., 2016; Schuh et al., 2019) and adding back single or a mix of culturable taxa (e.g., Domin et al., 2018). Such techniques can be coupled with established visualization approaches (electron microscopy: Cerra et al., 1997; fluorescent in situ hybridization: Schuh et al., 2019) to define the spatial distribution of these symbiont and which tissues they colonize. Lastly, viruses, archaea, and fungi are also functionally important members of host-associated microbial communities (e.g., Webster and Thomas, 2016). Similar molecular and sequencing approaches can and should be used to characterize if and how they interact with the larval host as well as other microbial groups within the microbiome.
Conclusion
Our understanding of symbioses between larval echinoderms and microbes has primarily developed in the last few years; yet, in this time we suggest that three primary themes have materialized. First, larval echinoderms associate with subcuticle bacteria that appear to colonize select tissues. Second, the bacterial communities associated with larval echinoderms exhibit compositional shifts that are correlated with several fundamental properties of larval biology and ecology. Third, the echinoderm larval host exhibits strict responses to pathogenic microbiota that may aid in maintaining the symbiont community to avoid dysbiosis.
As echinoderms larvae continue to serve as a fundamental system for understanding development and life history evolution (Love and Strathmann, 2018), this diversity in form and function may act as a strong foundation to understand how and to what extent bacteria and other microbes influence the many dimensions of larval biology (Hammer et al., 2019). In particular, these diverse developmental approaches enable the fields of animal-microbe symbiosis and larval biology to test for unique functional links between host and symbiont during, for example, morphological plasticity and life history transitions. And in a time where it is becoming clear that the bacterial flora cannot be ignored, we must remember that echinoderm larvae have always and will continue to live and evolve in a sea of microbes (Walne, 1956; Zilber-Rosenberg and Rosenberg, 2008; McFall-Ngai et al., 2013; Bordenstein and Theis, 2015).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. Specifically, TC drafted and edited the manuscript and AR edited the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
A special thanks to the editors of Symbiosis in a Changing Environment for the invitation and the reviewers for providing critical feedback to an earlier version of this manuscript. TC was supported by an NSF Graduate Research Fellowship and AR was supported by Human Frontier Science Program Award RGY0079/2016.
References
Appeltans, W., Ahyong, S. T., Anderson, G., Angel, M. V., Artois, T., Bailly, N., et al. (2012). The magnitude of global marine species diversity. Curr. Biol. 22, 2189–2202. doi: 10.1016/j.cub.2012.09.036
Ayukai, T. (1994). Ingestion of ultraplankton by the planktonic larvae of the crown-of-thorns starfish, Acanthaster planci. Biol. Bull. 186, 90–100. doi: 10.2307/1542039
Bates, J. M., Mittge, E., Kuhlman, J., Baden, K. N., Cheesman, S. E., and Guillemin, K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 297, 374–386. doi: 10.1016/j.ydbio.2006.05.006
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Bosch, I. (1992). Symbiosis between bacteria and oceanic clonal sea star larvae in the western North Atlantic Ocean. Mar. Biol. 114, 495–502. doi: 10.1007/BF00350041
Buckley, K. M., Ho, E. C. H., Hibino, T., Schrankel, C. S., Schuh, N. W., Wang, G. Z., et al. (2017). IL17 factors are early regulators in the gut epithelium during inflammatory response to Vibrio in the sea urchin larva. Elife 6:e23481. doi: 10.7554/eLife.23481.025
Buckley, K. M., and Rast, J. P. (2017). An organismal model for gene expression networks in the gut-associated immune response. Front. Immunol. 8:1297. doi: 10.3389/fimmu.2017.01297
Byrne, M., Ross, P. M., Dworjanyn, S. A., and Parker, L. (2018). “Larval ecology in the face of changing climate—impacts of ocean warming and ocean acidification,” in Evolutionary Ecology of Marine Invertebrate Larvae, eds T. J. Carrier, A. M. Reitzel, and A. Heyland (Oxford: Oxford University Press), 251–272. doi: 10.1093/oso/9780198786962.003.0017
Cameron, R. A., and Holland, N. D. (1983). Electron microscopy of extracellular materials during the development of a seastar, Patiria miniata (Echinoderm: Asteroidea). Cell Tissue Res. 243, 193–200. doi: 10.1007/BF00217412
Carrier, T. J., Dupont, S., and Reitzel, A. M. (2019). Geographic location and food availability offer differing levels of influence on the bacterial communities associated with larval sea urchins. FEMS Microbiol. Ecol. 95:fiz103. doi: 10.1093/femsec/fiz103
Carrier, T. J., King, B. L., and Coffman, J. A. (2015). Gene expression changes associated with the developmental plasticity of sea urchin larvae in response to food availability. Biol. Bull. 228, 171–180. doi: 10.1086/BBLv228n3p171
Carrier, T. J., Macrander, J., and Reitzel, A. M. (2018a). A microbial perspective on the life-history evolution of marine invertebrate larvae: if, where, and when to feed. Mar. Ecol. 39:e12490. doi: 10.1111/maec.12490
Carrier, T. J., and Reitzel, A. M. (2018). Convergent shifts in host-associated microbial communities across environmentally elicited phenotypes. Nat. Commun. 9:952. doi: 10.1038/s41467-018-03383-w
Carrier, T. J., and Reitzel, A. M. (2019a). Shift in bacterial taxa precedes morphological plasticity in a larval echinoid. Mar. Biol. 166:164. doi: 10.1101/640953
Carrier, T. J., and Reitzel, A. M. (2019b). Bacterial community dynamics during embryonic and larval development of three confamilial echinoids. Mar. Ecol. Prog. Ser. 611, 179–188. doi: 10.3354/meps12872
Carrier, T. J., Wolfe, K., Lopez, K., Gall, M., Janies, D. A., Byrne, M., et al. (2018b). Diet-induced shifts in the crown-of-thorns (Acanthaster sp.) larval microbiome. Mar. Biol. 165:157. doi: 10.1007/s00227-018-3416-x
Cerra, A., Byrne, M., and Hoegh-Guldberg, O. (1997). Developments of the hyaline layer around the planktonic embryos and larvae of the asteroid Patiriella calcar and the presence of associated bacteria. Invertebr. Reprod. Dev. 31, 337–343. doi: 10.1080/07924259.1997.9672594
Dishaw, L. J., Flores-Torres, J., Lax, S., Gemayel, K., Leigh, B., Melillo, D., et al. (2014). The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS ONE 9:e93386. doi: 10.1371/journal.pone.0093386
Domin, H., Zurita-Gutiérrez, Y. H., Scotti, M., Buttlar, J., Hentschel, U., and Fraune, S. (2018). Predicted bacterial interactions affect in vivo microbial colonization dynamics in Nematostella. Front. Microbiol. 9:728. doi: 10.3389/fmicb.2018.00728
Douillet, P. (1993). Bacterivory in Pacific oyster Crassostrea gigas larvae. Mar. Ecol. Prog. Ser. 98, 123–134. doi: 10.3354/meps098123
Feehan, C. J., Grauman-Boss, B. C., Strathmann, R. R., Dethier, M. N., and Duggins, D. O. (2018). Kelp detritus provides high-quality food for sea urchin larvae. Limnol. Oceanogr. 63, 5299–5306. doi: 10.1002/lno.10740
Feehan, C. J., Johnson-Mackinnon, J., Scheibling, R. E., Lauzon-Guay, J.-S., and Simpson, A. B. G. (2013). Validating the identity of Paramoeba invadens, the causative agent of recurrent mass mortality of sea urchins in Nova Scotia, Canada. Dis. Aquat. Org. 103, 209–227. doi: 10.3354/dao02577
Feehan, C. J., and Scheibling, R. E. (2014). Effects of sea urchin disease on coastal marine ecosystems. Mar. Biol. 161, 1467–1485. doi: 10.1007/s00227-014-2452-4
Galac, M. R., Bosch, I., and Janies, D. A. (2016). Bacterial communities of oceanic sea star (Asteroidea: Echinodermata) larvae. Mar. Biol. 163:162. doi: 10.1007/s00227-016-2938-3
Gloeckner, V., Lindquist, N., Schmitt, S., and Hentschel, U. (2013). Ectyoplasia ferox, an experimentally tractable model for vertical microbial transmission in marine sponges. Microb. Ecol. 65, 462–474. doi: 10.1007/s00248-012-0142-7
Gosselin, L. A., and Qian, P.-Y. (1997). Can bacterivory alone sustain larval development in the polychaete Hydroides elegans and the barnacle Balanus amphitrite? Mar. Ecol. Prog. Ser. 161, 93–101. doi: 10.3354/meps161093
Hammer, T. J., Sanders, J. G., and Fierer, N. (2019). Not all animals need a microbiome. FEMS Microbiol. Lett. 336:fnz117. doi: 10.1093/femsle/fnz117
Hart, M. W., and Strathmann, R. R. (1994). Functional consequences of phenotypic plasticity in echinoid larvae. Biol. Bull. 186, 291–299. doi: 10.2307/1542275
Harvell, C. D., Montecino-Latorre, D., Caldwell, J. M., Burt, J. M., Bosley, K., Keller, A., et al. (2019). Disease epidemic and a marine heat wave are associated with the continental-scale collapse of a pivotal predator (Pycnopodia helianthoides). Sci. Adv. 5:eaau7042. doi: 10.1126/sciadv.aau7042
Hewson, I., Button, J. B., Gudenkauf, B. M., Miner, B., Newton, A. L., Gaydos, J. K., et al. (2014). Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. U.S.A. 111, 17278–17283. doi: 10.1073/pnas.1416625111
Heyland, A., Schuh, N. W., and Rast, J. P. (2018). “Sea urchin larvae as a model for postembryonic development,” in Marine Organisms as Model Systems in Biology and Medicine, eds M. Kloc and J. Z. Kubiak (Cham: Springer), 137–161. doi: 10.1007/978-3-319-92486-1_8
Ho, E., Buckley, K. M., Schrankel, C. S., Schuh, N. W., Hibino, T., Solek, C. M., et al. (2016). Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol. Cell Biol. 94, 861–874. doi: 10.1038/icb.2016.51
Hodin, J., Heyland, A., Mercier, A., Pernet, B., Cohen, D. L., Hamel, J.-F., et al. (2019). Culturing echinoderm larvae through metamorphosis. Methods Cell Biol. 150, 125–169. doi: 10.1016/bs.mcb.2018.11.004
Holland, N. D., and Nealson, K. H. (1978). The fine structure of the echinoderm cuticle and subcuticular bacteria of echinoderms. Acta Zool. 59, 169–185. doi: 10.1111/j.1463-6395.1978.tb01032.x
Leigh, B. A., Liberti, A., and Dishaw, L. J. (2016). Generation of germ-free Ciona intestinalis for studies of gut-microbe interactions. Front. Microbiol. 7:2092. doi: 10.3389/fmicb.2016.02092
Lesser, M. P., and Blakemore, R. P. (1990). Description of a novel symbiotic bacterium from the brittle star, Amphipholis squamata. Appl. Environ. Microbiol. 56, 2436–2440. doi: 10.1128/AEM.56.8.2436-2440.1990
Lesser, M. P., and Walker, C. W. (1992). Comparative study of the uptake of dissolved amino acids in sympatic brittlestars with and without endosymbiotic bacteria. Comp. Biochem. Physiol. 101, 217–223. doi: 10.1016/0305-0491(92)90182-Q
Lessios, H. A. (2016). The great Diadema antillarum die-off: 30 years later. Ann. Rev. Mar. Sci. 8, 267–283. doi: 10.1146/annurev-marine-122414-033857
Lessios, H. A., Robertson, D. R., and Cubit, J. D. (1984). Spread of Diadema mass mortality through the Caribbean. Science 226, 335–337. doi: 10.1126/science.226.4672.335
Levin, L. A., and Bridges, T. S. (1995). “Pattern and diversity in reproduction and development,” in Ecology of Marine Invertebrate Larvae, ed L. R. Mcedward (Boca Raton, FL: CRC Press), 1–48.
Llewellyn, M. S., Mcginnity, P., Dionne, M., Letourneau, J., Thonier, F., Carvalho, G., et al. (2016). The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J. 10, 1280–1284. doi: 10.1038/ismej.2015.189
Love, A. C., and Strathmann, R. R. (2018). “Marine invertebrate larvae: model life histories for development, ecology, and evolution,” in Evolutionary Ecology of Marine Invertebrate Larvae, eds T. J. Carrier, A. M. Reitzel, and A. Heyland (Oxford: Oxford University Press), 306–321. doi: 10.1093/oso/9780198786962.003.0021
Marino, C. M., Pawlik, J. R., LóPez-Legentil, S., and Erwin, P. M. (2017). Latitudinal variation in the microbiome of the sponge Ircinia campana correlates with host haplotype but not anti-predatory chemical defense. Mar. Ecol. Prog. Ser. 565, 53–66. doi: 10.3354/meps12015
Marzinelli, E. M., Campbell, A. H., Valdes, E. Z., Vergés, A., Nielsen, S., Wernberg, T., et al. (2015). Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17, 4078–4088. doi: 10.1111/1462-2920.12972
McAlister, J. S., and Miner, B. G. (2018). “Phenotypic plasticity of feeding structures in marine invertebrate larvae,” in Evolutionary Ecology of Marine Invertebrate Larvae, eds T. J. Carrier, A. M. Reitzel, and A. Heyland (Oxford: Oxford University Press), 103–123. doi: 10.1093/oso/9780198786962.003.0008
McEdward, L. R., and Miner, B. (2006). Estimation and interpretation of egg provisioning in marine invertebrates. Integr. Comp. Biol. 46, 224–232. doi: 10.1093/icb/icj026
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Loso, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Metchnikoff, E. (1891). Lectures on the Comparative Pathology of Inflammation: Delivered at the Pasteur Institute in 1891. London: Kegan Paul, Trench, Trubner and Company.
Mileikovsky, S. A. (1971). Types of larval development in marine bottom invertebrates, their distribution and ecological significance: a re-evalution. Mar. Biol. 10, 193–213. doi: 10.1007/BF00352809
Miner, B. G. (2004). Evolution of feeding structure plasticity in marine invertebrate larvae: a possible trade-off between arm length and stomach size. J. Exp. Mar. Biol. Ecol. 315, 117–125. doi: 10.1016/j.jembe.2004.09.011
Moal, J., Samain, J. F., Corre, S., Nicolas, J. L., and Glynn, A. (1996). Bacterial nutrition of great scallop larvae. Aquac. Int. 4, 215–223. doi: 10.1007/BF00117383
Moitinho-Silva, L., Seridi, L., Ryu, T., Voolstra, C. R., Ravasi, T., and Hentschel, U. (2014). Revealing microbial functional activities in the Red Sea sponge Stylissa carteri by metatranscriptomics. Environ. Microbiol. 16, 3683–3698. doi: 10.1111/1462-2920.12533
Morrow, K. M., Tedford, A. R., Pankey, M. S., and Lesser, M. P. (2018). A member of the Roseobacter clade, Octadecabacter sp., is the dominant symbiont in the brittle star Amphipholis squamata. FEMS Microbiol. Ecol. 94:fiy030. doi: 10.1093/femsec/fiy030
Mortzfeld, B. M., and Bosch, T. C. G. (2017). Eco-Aging: stem cells and microbes are controlled by aging antagonist FoxO. Curr. Opin. Microbiol. 38, 181–187. doi: 10.1016/j.mib.2017.06.009
Mortzfeld, B. M., Urbanski, S., Reitzel, A. M., Kunzel, S., Technau, U., and Fraune, S. (2015). Response of bacterial colonization in Nematostella vectensis to development, environment and biogeography. Environ. Microbiol. 18, 1764–1781. doi: 10.1111/1462-2920.12926
Olson, R. R., and Olson, M. H. (1989). Food limitation of planktotrophic marine invertebrate larvae: does it control recruitment success? Annu. Rev. Ecol. Syst. 20, 225–247. doi: 10.1146/annurev.es.20.110189.001301
Pearse, J. S., Bosch, I., Pearse, V. B., and Basch, L. V. (1991). Bacterivory by bipinnarias: in the Antarctic but not in California. Am. Zool. 31:6A. doi: 10.1093/icb/31.1.65
Rawls, J. F., Samuel, B. S., and Gordon, J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 101, 4596–4601. doi: 10.1073/pnas.0400706101
Rivkin, R. B., Bosch, I., Pearse, J. S., and Lessard, E. J. (1986). Bacterivory: a novel feeding mode for asteroid larvae. Science 233, 1311–1314. doi: 10.1126/science.233.4770.1311
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Scheibling, R. E., and Stephenson, R. L. (1984). Mass mortality of Strongylocentrotus droebachiensis (Echinodermata: Echinoidea) off Nova Scotia, Canada. Mar. Biol. 78, 153–164. doi: 10.1007/BF00394695
Schuh, N. W., Carrier, T. J., Schrankel, C. S., Reitzel, A. M., Heyland, A., and Rast, J. P. (2019). Bacterial exposure mediates developmental plasticity and resistance to lethal Vibrio lentus infection in purple sea urchin (Strongylocentrotus purpuratus) larvae. Front. Immunol. 10:3014. doi: 10.3389/fimmu.2019.03014
Slaby, B. M., Hackl, T., Horn, H., Bayer, K., and Hentschel, U. (2017). Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 11, 2465–2478. doi: 10.1038/ismej.2017.101
Smith, K., Mccoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. doi: 10.1016/j.smim.2006.10.002
Sodergren, E., Weinstock, G. M., Davidson, E. H., Cameron, R. A., Gibbs, R. A., Weinstock, G. M., et al. (2006). The genome of the sea urchin Strongylocentrotus purpuratus. Science 314, 941–952. doi: 10.1126/science.1133609
Strathmann, R. R. (1985). Feeding and nonfeeding larval development and life-history evolution in marine invertebrates. Annu. Rev. Ecol. Syst. 16, 339–361. doi: 10.1146/annurev.es.16.110185.002011
Strathmann, R. R. (1987). “Larval feeding,” in Reproduction of Marine Invertebrates: General Aspects, Seeking Unity in Diversity, eds A. C. Giese, J. S. Pearse, and V. B. Pearse (Palo Alto, CA: Blackwell Scientific Publications), 465–550.
Tauber, A. L. (2003). Metchnikoff and the phagocytosis theory. Nat. Rev. Mol. Cell Biol. 4, 897–901. doi: 10.1038/nrm1244
Thorson, G. (1950). Reproductive and larval ecology of marine bottom invertebrates. Biol. Rev. 25, 1–45. doi: 10.1111/j.1469-185X.1950.tb00585.x
Vance, R. R. (1973). On reproductive strategies in marine benthic invertebrates. Am. Nat. 107, 339–352. doi: 10.1086/282838
Walker, C. W., and Lesser, M. P. (1989). Nutrition and development of brooded embryos in the brittlestar Amphipholis squamata: do endosymbiotic bacteria play a role? Mar. Biol. 103, 519–530. doi: 10.1007/BF00399584
Walne, P. R. (1956). Bacteria in experiments on rearing oyster larvae. Nature 178:91. doi: 10.1038/178091a0
Webster, N. S., Botte, E. S., Soo, R. M., and Whalan, S. (2011). The larval sponge holobiont exhibits high thermal tolerance. Environ. Microbiol. Rep. 3, 756–762. doi: 10.1111/j.1758-2229.2011.00296.x
Webster, N. S., and Thomas, T. (2016). The sponge hologenome. MBio 7:e00135-16. doi: 10.1128/mBio.00135-16
Williams, E. A., and Carrier, T. J. (2018). “An-omics perspective on marine invertebrate larvae,” in Evolutionary Ecology of Marine Invertebrate Larvae, eds T. J. Carrier, A. M. Reitzel, and A. Heyland (Oxford: Oxford University Press), 288–304. doi: 10.1093/oso/9780198786962.003.0019
Young, C. M., and Chia, F.-S. (1987). “Abundance and distribution of pelagic larvae as influenced by predation, behavior, and hydrographic factors,” in Reproduction of Marine Invertebrates IX. General Aspects: Seeking Unity in Diveristy, eds A. C. Giese, J. S. Pearse, and V. B. Pearse (Palo Alto, CA: Blackwell Scienfic Publications), 385–463.
Keywords: microbiome, host-microbe, endosymbiont, marine invertebrate, development, life history, plasticity, evolutionary ecology
Citation: Carrier TJ and Reitzel AM (2020) Symbiotic Life of Echinoderm Larvae. Front. Ecol. Evol. 7:509. doi: 10.3389/fevo.2019.00509
Received: 26 August 2019; Accepted: 16 December 2019;
Published: 22 January 2020.
Edited by:
Anne Duplouy, Lund University, SwedenReviewed by:
Lucia Pita, GEOMAR Helmholtz Center for Ocean Research Kiel, GermanyClaudia Pogoreutz, Universität Konstanz, Germany
Copyright © 2020 Carrier and Reitzel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tyler J. Carrier, dGNhcnJpZTFAdW5jYy5lZHU=
 Tyler J. Carrier
Tyler J. Carrier Adam M. Reitzel
Adam M. Reitzel