
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 14 November 2019
Sec. Evolutionary Developmental Biology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00437
This article is part of the Research Topic From Meristems to Floral Diversity: Developmental Options and Constraints View all 11 articles
 Ze Hong Lee1
Ze Hong Lee1 Yoshitaka Tatsumi1
Yoshitaka Tatsumi1 Yasunori Ichihashi2,3
Yasunori Ichihashi2,3 Takamasa Suzuki4
Takamasa Suzuki4 Arisa Shibata5
Arisa Shibata5 Ken Shirasu5
Ken Shirasu5 Nobutoshi Yamaguchi1,2
Nobutoshi Yamaguchi1,2 Toshiro Ito1*
Toshiro Ito1*The appropriate timing of the termination of floral meristem activity (FM determinacy) determines the number of floral organs. In Arabidopsis, two transcription factors, CRABS CRAW (CRC) and SUPERMAN (SUP), play key roles in FM determinacy. CRC belongs to the YABBY transcription factor family, whose members contain a zinc finger and a helix-loop-helix domain. The crc mutation causes the formation of unfused carpels and leads to an increase in carpel number in sensitized backgrounds. The SUP gene encodes a C2H2-type zinc-finger protein, and sup mutants produce extra carpels and stamens. However, the genetic interaction between CRC and SUP is not fully understood. Here, we show that these two transcription factors regulate multiple common downstream genes during stamen development. The crc sup double mutant had significantly more stamens and carpels than the parental lines and an enlarged floral meristem. Transcriptome data have implicated several cytokinin- and auxin-related genes as well as stress- and metabolic-related genes to function downstream of CRC and SUP during stamen development. The regulation of common downstream genes of CRC and SUP might contribute to the initiation of an appropriate number of stamens and to subsequent growth and development.
Plant development is dependent on the persistent activity of pluripotent meristematic cells that are responsible for organ formation (Laux et al., 1996). In angiosperms, flower development initiates from the floral stem-cell pool that is located in the center of the floral meristem (FM). The FMs produce an appropriate population of cells to initiate a fixed number of floral organs before FM activity terminates (FM determinacy) (Lenhard et al., 2011). In Arabidopsis, the FM is maintained by activity of the WUSCHEL (WUS) homeodomain protein and CLAVATA (CLV) ligand–receptor system (Laux et al., 1996; Brand et al., 2000; Schoof et al., 2000). The WUS–CLV negative feedback loop controls the balance between stem-cell renewal and organ formation and determines the size and number of floral organs, which are sepals, petals, stamens, and carpels (Schoof et al., 2000; Sun et al., 2009). Loss-of-function mutants of any of the CLV genes results in enlarged FMs and consequently leads to an increase in floral organ number (Clark et al., 1993, 1995; Kayes and Clark, 1998). Conversely, wus mutants have fewer floral organs. Thus, the WUS–CLV negative feedback loop is essential to control FM activity.
The homeotic gene AGAMOUS (AG) determines when FM activity ceases and limits the size of the FM (Yanofsky et al., 1990; Lohmann et al., 2001; Lenhard et al., 2011). In loss-of-function ag mutants, the FM overproliferates, resulting in an increase in floral organ number or in a “flower within flower” phenotype, due to the failure of FM termination (Yanofsky et al., 1990). The AG protein binds to the WUS promoter and represses WUS expression via the deposition of repressive histone marks (Liu et al., 2011; Guo et al., 2018). During FM termination, AG represses WUS expression by directly inducing two key downstream targets, KNUCKLES (KNU) and CRABS CLAW (CRC) (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Payne et al., 2004; Gomez-Mena et al., 2005; Lee et al., 2005; Sun et al., 2014; Bollier et al., 2018). Previous studies have demonstrated that the crc knu double mutant shows prolonged WUS expression and produces flowers with reiterated floral organs, such as stamens and carpels (Breuil-Broyer et al., 2016; Yamaguchi et al., 2017). The KNU gene encodes a C2H2 zinc-finger protein with a conserved transcriptional repressor motif (Payne et al., 2004; Sun et al., 2009, 2014, 2019; Bollier et al., 2018). The KNU transcriptional repressor interacts with Polycomb group proteins and deposits repressive histone marks at the WUS locus (Sun et al., 2019). Alternatively, CRC belongs to the YABBY family of transcription factors, members of which contain a zinc finger and a helix-loop-helix domain (Bowman and Smyth, 1999; Baum et al., 2001; Lee et al., 2005). We previously identified two CRC direct and auxin-related targets, which are responsible for FM termination (Yamaguchi et al., 2017, 2018): TORNADO2 (TRN2) encodes a transmembrane protein of the tetraspanin family and controls auxin homeostasis (Cnops et al., 2006; Chiu et al., 2007) and YUCCA4 (YUC4) is one of the 11 YUCCA genes that encode flavin monooxygenases involved in tryptophan-dependent auxin biosynthesis (Cheng et al., 2006, 2007). The downregulation of TRN2 and upregulation of YUC4 by CRC terminates FM proliferation and triggers floral organ formation (Yamaguchi et al., 2017, 2018).
Other factors have been reported to be involved in FM termination (Uemura et al., 2017): SUPERMAN (SUP) encodes a C2H2 zinc-finger protein, which has been proposed to act as a boundary gene that specifies the separation between floral whorls 3 and 4 (Bowman et al., 1992; Sakai et al., 1995). The loss-of-function sup mutation leads to the formation of supernumerary stamens at the expense of carpel tissues (Sakai et al., 1995, 2000; Prunet et al., 2017). Ectopic SUP expression causes altered auxin and cytokinin phenotypes (Nibau et al., 2010). SUP interacts with CURLY LEAF, one of the Polycomb group (PcG) protein components (Xu et al., 2018). Then, the SUP-PcG complex represses two auxin biosynthesis genes, YUCCA1 (YUC1) and YUC4, to limit stamen number through deposition of H3K27me3 (Zhao et al., 2001; Cheng et al., 2006, 2007; Sassi and Vernoux, 2013; Xu et al., 2018).
Organ formation is controlled by a complex network of interactions between plant hormones. One simple model of hormone interaction is via antagonism. In antagonistic interactions, the components of one plant hormone signaling pathway interact with those of another hormone. For example, auxin inhibits cytokinin signaling. One cytokinin signaling component, ARABIDOPISIS HISTIDINE PHOSPHOTRANSFER PROTEIN6 (AHP6), is functionally important in auxin–cytokinin crosstalk (Mähönen et al., 2006; Moreira et al., 2013; Besnard et al., 2014a,b). Auxin signaling downstream of AHP6 inhibits cytokinin signaling to regulate the positioning of floral primordia (Moreira et al., 2013; Besnard et al., 2014a,b). Although it is generally reported that auxin and cytokinin function antagonistically, recent studies have revealed that they also act synergistically during gynoecium formation (Wolters and Jurgens, 2009; El-Showk et al., 2013; Schaller et al., 2015; Müller et al., 2017; Reyes-Olalde et al., 2017a,b). Although these findings have demonstrated that the interplay between auxin and cytokinin via AHP6 is important for floral primordium positioning and gynoecium formation, the role of AHP6 in FM determinacy and floral organ initiation remains unclear.
Subsequent floral organ growth and development might also be affected by CRC and/or SUP, potentially via the transcriptional regulation of downstream genes. These might include, for example, the auxin-responsive gene AUX/IAA19, which regulates stamen elongation (Tashiro et al., 2009; Ghelli et al., 2018), and REPRODUCTIVE MERISTEM (REM) genes, which are potentially involved in the early stages of flower development and are often transcriptionally regulated by well-known key floral regulators (Mantegazza et al., 2014). Despite the importance of many genes involved in the regulation of floral organ growth and development, little is known concerning their regulation.
Here, we report the genetic interaction between SUP and CRC. Molecular genetic analysis revealed that SUP and CRC cooperatively fine-tune hormone signaling and various stress or metabolic events to regulate stamen formation during flower development.
The Arabidopsis thaliana mutants crc-1 and sup-5 used in this study were in the Landsberg erecta (Ler) ecotype background. The crc-1, and sup-5 lines were described previously (Gaiser et al., 1995; Bowman and Smyth, 1999; Jacobsen et al., 2000; Sakai et al., 2000; Yamaguchi et al., 2017, 2018). The double mutants were generated by genetic crossing and genotyped by PCR in subsequent generations. Seeds were sown on soil and stratified at 4°C for 3–7 days. Plants were grown at 22°C under 24 h of continuous light. Plants to be directly compared were grown side-by-side to minimize environmental differences within the growth chamber.
The first 5–10 flowers at developmental stage 13 (according to Smyth et al., 1990) were harvested for phenotyping open flowers (Figures 1, 2). To measure flower size, flowers of Ler, crc-1, sup-5, and crc-1 sup-5 were removed with forceps and fixed onto agar, and photos were taken from above. Flower size was measured using Image J (http://imagej.nih.gov/ij/) software. Thirty flowers (five flowers each from six individual plants) from each genotype were measured. The number of floral organs (sepals, petals, stamen, and carpels) in the wild type (Ler), crc-1, sup-5, and crc-1 sup-5 of stage 13 flowers was counted under a dissecting microscope. Forty flowers (five each from eight individual plants) were counted for each genotype. To test for statistical significance, one-way ANOVA was followed by the post-hoc Tukey HSD test.
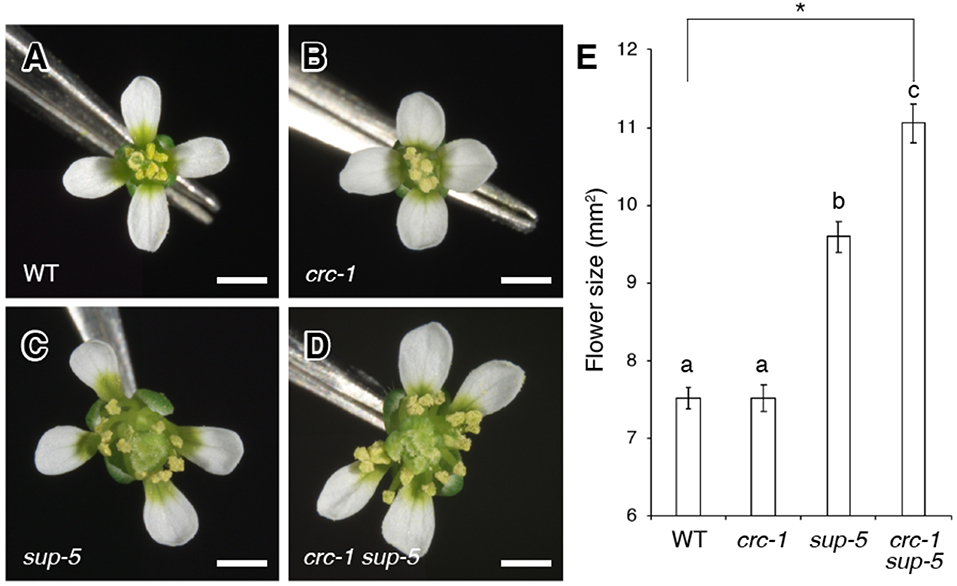
Figure 1. Comparison of flower size among the wild type, crc-1, sup-5, and crc-1 sup-5 at floral stage 13. (A–D) Top view of flowers. (A) The wild type (WT), (B) crc-1, (C) sup-5, and (D) crc-1 sup-5. Scale bars represent 500 μm. (E) Quantification of flower size. Mean ± SEM are shown. The asterisk indicates significant difference based on one-way ANOVA. The same letters indicate non-significant differences, whereas different letters indicate significant differences based on the post-hoc Tukey HSD test (p < 0.01).
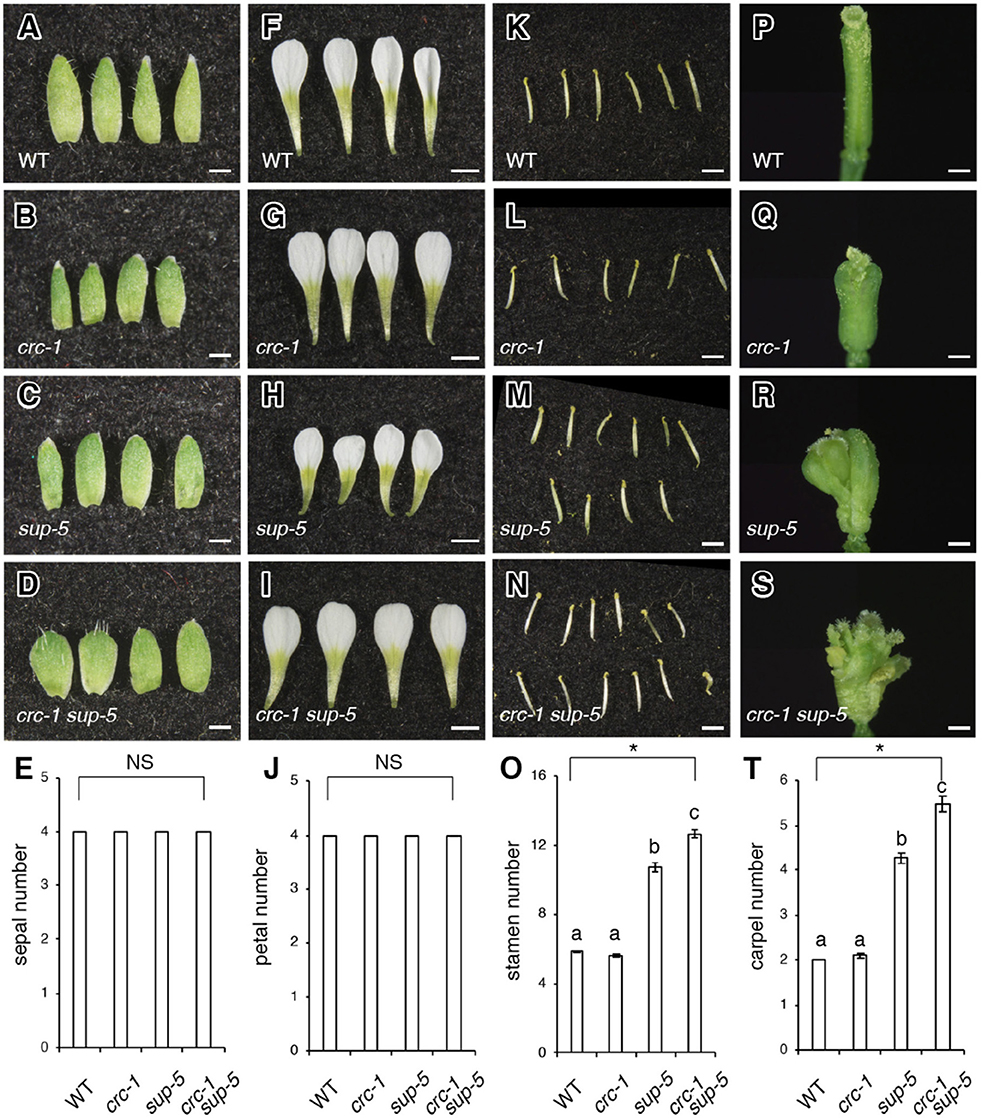
Figure 2. Comparison of floral organ number among the wild type, crc-1, sup-5, and sup-5 crc-1 at floral stage 13. (A–D) Side view of sepals. (A) The wild type (WT), (B) crc-1, (C) sup-5, and (D) crc-1 sup-5. Scale bars represent 500 μm. (E) Quantification of sepal number. (F–I) Side view of petals. (F) The wild type, (G) crc-1, (H) sup-5, and (I) crc-1 sup-5. Scale bars represent 1 mm. (J) Quantification of petal number. (K–N) Side view of stamens. (K) Wild type, (L) crc-1, (M) sup-5, and (N) crc-1 sup-5. Scale bars represent 1 mm. (O) Quantification of stamen number. (P–S) Side view of carpels. (P) The wild type, (Q) crc-1, (R) sup-5, and (S) crc-1 sup-5. Scale bars represent 500 μm. (T) Quantification of carpel number. Mean ± SEM are shown. Scale bars represent 500 μm. Asterisks indicate significant differences based on one-way ANOVA. The same letters indicate non-significant differences, whereas different letters indicate significant differences based on the post-hoc Tukey HSD test (p < 0.01).
To measure the size of the FM, inflorescences 1–3 cm tall were harvested immediately after bolting. Inflorescences were fixed with FAA overnight. The resulting inflorescences were dehydrated in an ethanol series (50, 60, 70, 80, 90, 95, and 100%; not <20 min each). The fixed samples were then removed from 100% ethanol and placed in Technovit 7100 resin (Heraeus) before overnight incubation for polymerization. Eight, 10-μm thick sections were prepared using a RM2255 microtome (Leica Microsystems) for each genotype or floral developmental stage. Significance was tested using the Student's t-test and one-way ANOVA followed by the post-hoc Tukey HSD test.
For RNA extraction, floral buds up to floral stage 8 from inflorescences 1–3 cm tall were harvested. Five biological replicates were harvested from wild-type (Ler), crc-1, sup-5, and crc-1 sup-5 backgrounds. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), and genomic DNA was removed using an RNase-Free DNase Set (Qiagen). Library preparation and sequencing were performed as described previously (Uemura et al., 2017; Ichihashi et al., 2018). The created libraries were sequenced by next-generation sequencing (Illumina), and the produced bcl files were then converted into fastq files by bcl2fastq (Illumina). Mapping of sequences to the Arabidopsis TAIR10 genome was performed using Bowtie with the following options (“—all—best—strata—trim5 8”). The number of reads for each reference was then counted, and the false discovery rate (FDR), log concentration (Conc) and log fold-change (FC) were obtained using the edge R package (Robinson et al., 2010). To determine DEGs, FDR with adjusted p < 0.05 was used. The data were deposited into the DNA Data Bank of Japan (DRA008874).
To identify common differentially expressed genes (DEGs), online software (bioinformatics.psb.ugent.be/webtools/Venn/) was used to calculate the overlap between DEGs lists in all mutant backgrounds. Furthermore, Gene Ontology (GO) term analysis was performed using online software agriGO v2.0 (systemsbiology.cau.edu.cn/agriGOvs/) (Tian et al., 2017), followed by REVIGO (Reduced + Visualize Gene Ontology; revigo.irb.hr/) (Supek et al., 2011) to reduce the redundant GO terms.
For RNA extraction, floral buds up to floral stage 8 from inflorescences 1- to 3-cm tall were harvested. Plants from mutant backgrounds (crc-1, sup-5, and crc-1 sup-5) and the controls [wild-type (Ler)] were grown side-by-side. Approximately 100 mg of floral bud tissue was prepared and frozen immediately after trimming, without fixation. Tissues were kept at −80°C until use (<5 months). RNA extraction was performed using the RNeasy Plant Mini Kit (Qiagen). Genomic DNA was then removed using an RNase-Free DNase Set (Qiagen) to minimize contamination by genomic DNA. The RNA concentration was determined with an IMPLEN NanoPhotometer P-Class spectrophotometer. Synthesis of cDNA was performed with a PrimeScript first-strand cDNA Synthesis Kit (Takara) using <5 μg total RNA, 50 μM oligo dT primer and 200 U PrimeScript RTase with RNase Inhibitor, at 42°C for 30 min. The resulting cDNA was quantified by a LightCycler 480 (Roche) using FastSmart Essential DNA Green Master Mix (Roche) and Cq values were obtained. The expression levels of AHP6 (AT1G80100), IAA19 (AT3G15540), REM25 (AT5G09780), and TPPI (AT5G10100) were quantified; EIF4A-1 (AT3G13920) was used for the normalization of signals. Five biological replicates were performed and similar results were obtained. The primers used in this study were as follows: AHP6-FW, 5′-CAGCTGGAGCAGCAGAGAAT-3′; AHP6-RV, 5′-TTTCGCTTCGGTAGCTTATAACACA-3′; IAA19-FW, 5′-GATCTAGCCTTTGCTCTTGATAAGC-3′; IAA19-RV, 5′-ATGACTCTAGAAACATCCCCCAAG-3′; REM25-FW, 5′-CTTGGGAGACCACGAGTTTCTTA-3′; REM25-RV, 5′-TTTTGACACGACTAGAAGAAGCGAA-3′; TPPI-FW, 5′-TACAG GTTCGGTCGGTATTAAAGAA-3′; TPPI-RV, 5′-TTGTTAGTGTTCCCAAATCCAAGTG-3′; EIF4-FW, 5′-ACCAGGCGTAAGGTTGATTG-3′; EIF4-RV, 5′-GGTCCATGTCTCCGTGAGTT-3′.
To analyze the genetic interaction between SUP and CRC, we generated a crc-1 sup-5 double mutant and compared the size of flowers between the wild type, crc-1, sup-5, and crc-1 sup-5 (Figures 1A–E). A wild-type flower consists of four sepals, four petals, six stamens, and two carpels (Figure 1A). The mean size of wild-type flowers was 7.5 ± 0.1 mm2 (Figure 1E). Similar to the wild type, the crc-1 single mutant had a fixed number of four types of floral organs (Figure 1B). The size of crc-1 flowers was 7.5 ± 0.2 mm2 (Figure 1E), which was not significantly different from wild-type and crc-1 flowers (p > 0.01) (Figure 1E). As reported previously (Uemura et al., 2017), sup-5 plants produce significantly larger flowers (9.6 ± 0.2 mm2) than the wild type (p < 0.01) (Figures 1C,E). An increase in the size of sup-5 mutant flowers was accompanied by the presence of extra whorls of stamens, due to sustained floral stem-cell activity (Figure 1C) (Xu et al., 2018). In crc-1 sup-5 double mutants, a large number of stamens and carpels arose from whorls 3 and 4 (Figure 1C). The mean size of crc-1 sup-5 flowers was 11.1 ± 0.2 mm2 (Figures 1D,E) and was thus significantly larger than that of the wild type or either single mutant (p < 0.01) (Figure 1E).
To investigate the genetic interaction between CRC and SUP further, the number of organs was counted in wild-type, crc-1, sup-5, and crc-1 sup-5 double mutant flowers (Figures 2A–T). Wild-type flowers had four sepals, four petals, six stamens, and two carpels (Figures 2A,E,F,J,K,O,P,T). Similarly, the crc mutant produced four sepals, four petals, and six stamens (Figures 2B,E,G,J,L,O,Q,T). Although crc-1 mutants produced three or even occasionally four carpels, the mean carpel number for crc-1 was 2.1 ± 0.0 (Figure 2T), which did not differ significantly from the wild type. Indeed, there was no significant difference in the number of all four floral organs between the wild type and crc-1 mutants (p > 0.01) (Figures 2E,J,O,T). No significant difference was observed in the number of sepals and petals between wild-type and sup-5 flowers (p > 0.01) (Figures 2A,C,E,F,H,J). However, the mean numbers of stamens and carpels in sup-5 were 10.7 ± 0.3 and 4.3 ± 0.1, respectively (Figures 2K,M,O,P,R,T), significantly higher than those of the wild type (p < 0.01) (Figures 2O,T). Similar to the wild type or the parental lines, crc-1 sup-5 double mutant flowers also produced four sepals and four petals (p > 0.01) (Figures 2A–J) but produced significantly more stamens and carpels than the wild type or either single mutant (p < 0.01) (Figures 2K–T). Thus, the combination of crc with sup enhanced the sup phenotype.
Next, we aimed to determine whether the changes in the size of flowers and/or the number of floral organs correlated with changes in FM height and width. Therefore, we quantified the height (from the groove between sepal primordia and the FM to the top of the floral meristem) and the width (between the two grooves along the lateral axis) of the FM by sectioning (Figures 3A–O). In wild-type plants, mean FM height and width at stage 3 were 12.0 ± 0.5 and 58.0 ± 1.1 μm, respectively (Figures 3A,E), and were 29.2 ± 0.9 and 75.2 ± 2.0 μm, respectively, at stage 5 (Figures 3A,E). In crc-1 mutants, no significant difference in FM height and width at stage 3 or stage 5 was observed compared to wild-type plants (p > 0.01) (Figures 3A,B,E–G,J). Similarly, no significant difference in FM height and width was observed between wild-type and sup-5 FMs at stage 3 (p > 0.01). Although FM height in the wild type and sup-5 mutants did not differ significantly (p > 0.01) at stage 5, there was a significant difference in FM width (p < 0.01) (Figures 3F,J). In crc-1 sup-5, FM height and width were similar to in sup-5 by stage 5 (Figures 3C–E,H–J). A significant difference in FM width was observed between sup-5 and crc-1 sup-5 FMs at stage 6, but no difference in height (p < 0.01, Student's t-test). Therefore, combining the crc mutation with sup enhanced the sup mutant FM width phenotype.
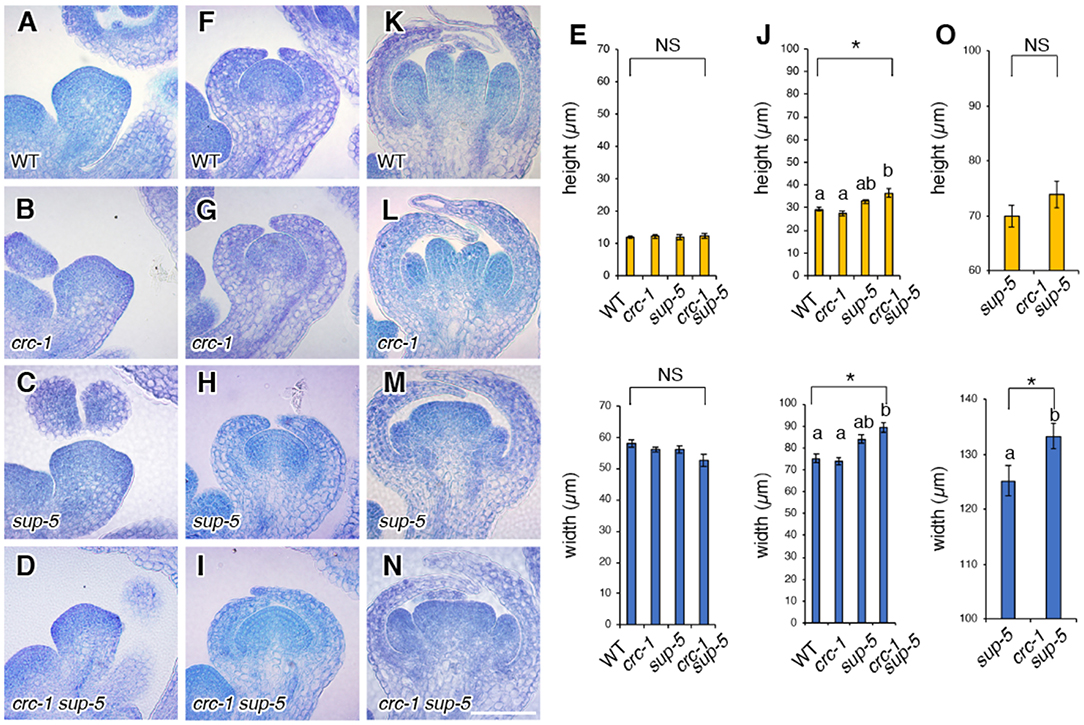
Figure 3. Comparison of floral meristem height and width among the wild type, crc-1, sup-5, and sup-5 crc-1 at different floral stages. (A–D) Vertical cross sections of stage 3 floral meristems. (A) The wild type, (B) crc-1, (C) sup-5, and (D) crc-1 sup-5. (E) Quantification of floral meristem height and width at stage 3. (F–J) Vertical cross sections of floral meristems at stage 5. (F) The wild type, (G) crc-1, (H) sup-5, and (I) crc-1 sup-5. (J) Quantification of floral meristem height and width at stage 5. (K–O) Vertical cross sections of floral meristems at stage 6. (K) The wild type, (L) crc-1, (M) sup-5, and (N) crc-1 sup-5. (O) Quantification of floral meristem height and width at stage 6. Mean ± SEM are shown. Scale bars represent 50 μm. For multiple comparisons, asterisks indicate significant differences based on one-way ANOVA. The same letters indicate non-significant differences, whereas different letters indicate significant differences based on the post-hoc Tukey HSD test (p < 0.01). For single comparisons, the p-value was determined using the Student's t-test (*p < 0.01).
RNA-seq was performed to examine the genetic interaction at molecular levels in crc, sup, and crc sup (DRA008874). Approximately 10 M reads were sequenced per sample, which were then mapped onto the Arabidopsis TAIR 10 genome and differentially expressed genes (DEGs) were identified. In crc-1 mutants, 263 genes were differentially expressed compared to the wild type (FDR < 0.05) (Figure 4A; Tables S1, S2). Similarly, 65 and 281 genes in total were differentially expressed in sup and crc sup mutants, respectively (FDR < 0.05) (Figures 4B,C; Tables S1, S2).
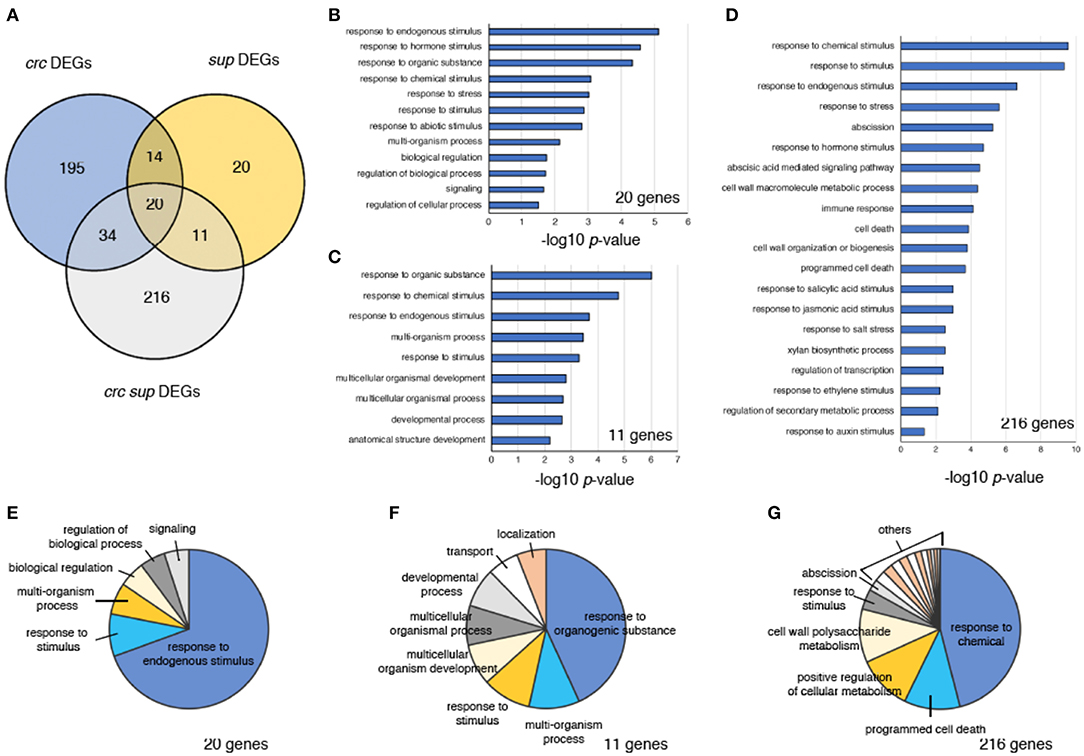
Figure 4. Characterization of DEGs in crc sup. (A) Venn diagram showing the number of DEGs in crc, sup, and crc sup mutants. (B–D) GO term enrichment analysis of categories containing 20 genes (B), 11 genes (C), and 216 genes (D). Selected terms determined by their –log10-adjusted p-values are shown. (E–G) Pie charts showing summarized GO terms by REVIGO in categories of 20 genes (E), 11 genes (F), and 216 genes (G) within the GO term category “biological process”.
To identify genes involved in the enhancement of the sup phenotype in the crc sup double mutant, we focused on three different categories of DEGs, containing either 20 genes, 11 genes, or 216 genes (Figures 4B–G). The “20 genes” category contained the DEGs common to all crc-1, sup-1, and crc-1 sup-5 backgrounds (Figure 4A). This category contained genes that were differentially expressed in single mutants and whose expression was further altered in the double mutant. To examine the probable functions of these 20 genes, GO term enrichment analysis was performed using agriGO v2.0 online software (Tian et al., 2017). Stimulus-related GO terms such as “response to hormone stimulus,” “response to hormone stimulus,” and “response to stimulus” were identified (Figure 4B; Table S3). A further reduction of redundant GO terms by REVIGO categorized ~70% of the GO terms as “response to endogenous stimulus” (Figure 4E; Table S4). The “11 genes” category contained the DEGs shared by sup-5 and crc-1 sup-5 mutants. This category consisted of the downstream genes of SUP, whose expression was affected by combination with the crc mutation. In addition to stimulus-related terms, this category of GO terms also contained development-related GO terms, such as “developmental process” and “anatomical structure development” (Figures 4C,F; Tables S3, S4). The “216 genes” category contained genes that were differentially expressed only in crc-1 sup-5. The GO terms in this category included “response to chemical stimulus,” “response to stimulus,” and “response to hormone stimulus,” which were also present in the “20 genes” and “11 genes” categories (Figures 4B–D; Tables S3, S4). In addition, unique GO terms such as “abscission,” “programmed cell death,” “immune response,” and “cell wall organization or biogenesis” were present (Figure 4D; Tables S3, S4).
To identify the genes involved in supernumerary stamen initiation in crc-1 sup-5 double mutants, we computationally identified genes involved in stamen development using a published transcriptome dataset (Alves-Ferreira et al., 2007; GSE8864). Previous studies have determined the global expression profile of Arabidopsis stamen development using ap3, spl/nzz, and ms1 mutants. Among 247 DEGs identified in Figure 4, 47 genes were predicted to be expressed at early, intermediate, and late stages of stamen development (Figure 5A; Table S5). Out of 47 identified genes, 37 were differentially expressed in ap3 (early stage) (Figure 5A). This is consistent with the functions of CRC or SUP, since both genes are highly expressed from early stages of flower development. Furthermore, an additional 21 and 8 identified genes were differentially expressed in nzz/spl (intermediate stage) and ms1 (late stage), respectively (Figure 5A).
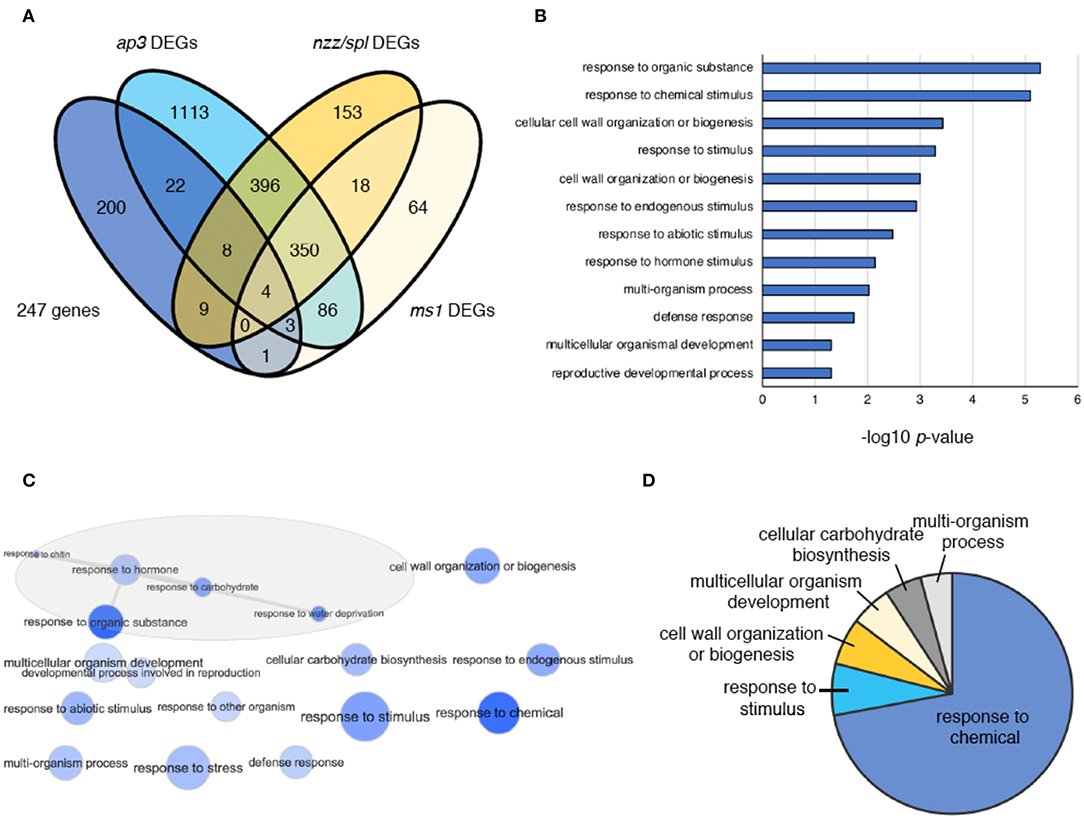
Figure 5. DEGs in crc sup, ap3, nzz/spl, and ms1 flowers. (A) Venn diagram showing 247 DEGs in ap3, nzz/spl, and ms1. (B) GO term enrichment analysis of 47 genes coregulated by CRC and SUP and related to stamen development. Selected terms determined by their –log10-adjusted p-values are shown. (C) The interactive graph view of 47 genes generated by REVIGO. (D) Pie charts showing summarized GO terms by REVIGO for 47 genes within the GO-term category “biological process”.
To understand the function of the 47 genes involved in stamen development, GO term analysis was performed (Figure 5B). The enriched terms included “response to organic substance,” “response to chemical stimulus,” “cell wall organization or biogenesis,” “response to stimulus,” “response to hormone,” “multicellular organismal development,” and “reproductive developmental process” (Figure 5B; Table S6). The interactive graph view of 47 genes generated by REVIGO identified a cluster of GO terms that contain five different terms: “response to organic substance,” “response to hormone,” “response to chitin,” “response to carbohydrate,” and “response to water deprivation” (Figure 5C). After removing redundant GO terms, ~70% of GO terms were involved in “response to chemical” (Figure 5D; Table S7).
Based on RNA-seq, SUP was expressed at normal levels in crc and, similarly, CRC expression was unaffected in sup (FDR < 0.05). Thus, it is unlikely that either transcription factor transcriptionally regulates the expression of the other (Figure S1). All of the 47 DEGs in crc-1 sup-5 that overlapped with the list of DEGs from the stamen transcriptome dataset were categorized into two different clusters by K-means clustering (Figure 6A). Cluster 1 contained 28 genes, which were highly expressed in all mutant backgrounds, whereas cluster 2 contained genes that were downregulated in all mutant backgrounds (Figures 6A,B). Compared to the subtle difference in differential gene expression observed in either crc-1 or sup-5 single mutants, the expression levels of 47 genes were greatly affected in crc-1 sup-5 double mutants.
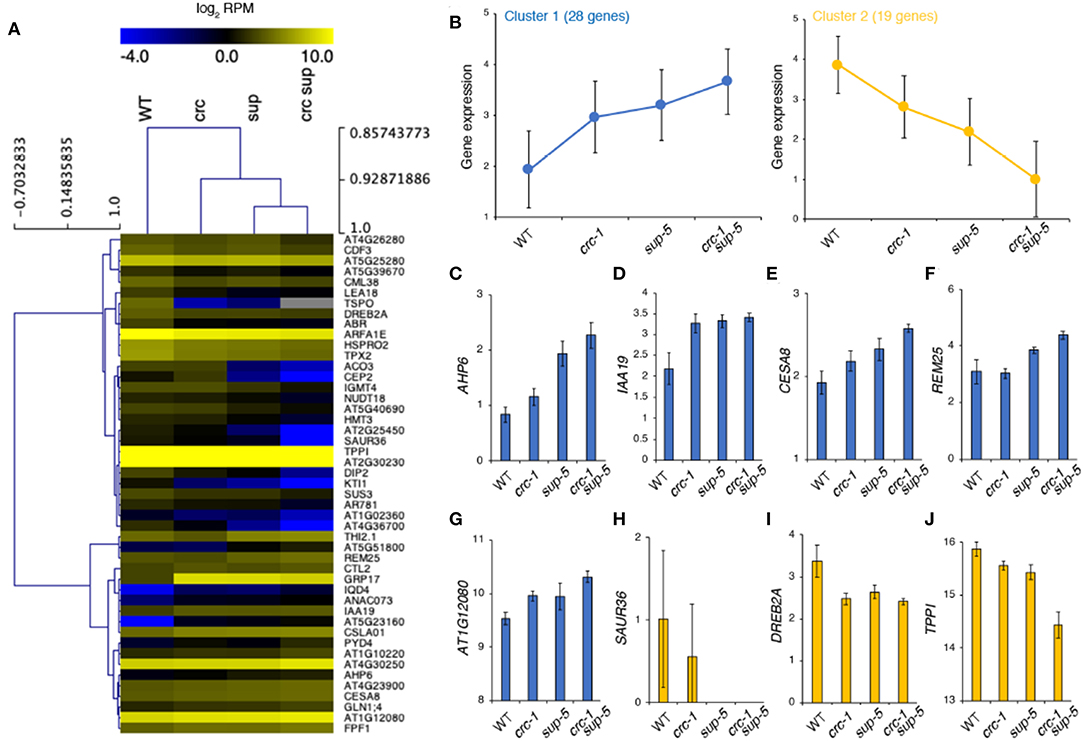
Figure 6. Clustering of high-confidence downstream targets regulated by CRC and SUP. (A) Clustering of 47 genes during stamen formation. (B) Gene expression from two different clusters: cluster 1 (left) and cluster 2 (right). (C–J) Expression level of genes according to RNA-seq. AHP6 (C), IAA19 (D), CESA8 (E), REM25 (F), AT1G12080 (G), SAUR36 (H), DREB2A (I), and TPPI (J). Mean ± SEM (log2 of reads per million mapped reads [RPM]) are shown.
The upregulated genes identified in crc-1 sup-5 might be direct targets of SUP because SUP is a transcriptional repressor (Figures 6C–J). These genes might also be directly regulated by CRC, since CRC can act as a bifunctional transcription factor (Yamaguchi et al., 2017, 2018; Gross et al., 2018). Consistent with the GO term analysis, hormone-related genes such as AHP6 and IAA19 were highly expressed in the single mutants (Figures 6C,D) (Nakamura et al., 2003; Tatematsu et al., 2004; Mähönen et al., 2006; Besnard et al., 2014a,b). These two genes were also more highly expressed in crc sup than in either single mutant. The expression levels of CELLULOSE SYNTHASE8 (CESA8), REPRODUCTIVE MERISTEM25 (REM25), and AT1G12080 (Turner and Somerville, 1997; Chen et al., 2005; Mantegazza et al., 2014) were also higher in crc-1 sup-5 (Figures 6E–G). Among the potential downstream genes, the roles of CESA8 and REM25 have been well studied during cell wall organization and reproductive development, respectively.
By contrast, some of the genes related to hormonal regulation, such as SMALL AUXIN UPREGULATED36 (SAUR36) and DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) (Sakuma et al., 2006; Kim et al., 2011; Hou et al., 2013; Stamm and Kumar, 2013), were downregulated (Figures 6H–J). One sugar metabolism regulator, TREHALOSE-6-PHOSPHATE PHOSPHATASE I (TPPI) (Schluepmann et al., 2004), has also been identified as the downregulated target in the mutant backgrounds. The RNA-seq data were confirmed by RT-qPCR (Figure S2).
Floral meristem determinacy and meristematic cell differentiation are two critical steps in flower development (Sablowski, 2015; Bommert and Whipple, 2017; Xu et al., 2019). Disruption in either or both of these processes caused by the misexpression of FM regulators eventually results in abnormal floral phenotypes (Lohmann et al., 2001; Ma, 2005). Both SUP and CRC are highly conserved genes in Angiosperms and function as FM activity and identity genes in distinct spatio-temporal manners (Sun and Ito, 2015). Compared to sup mutants, crc mutants do not have altered floral organ number, despite the occasional presence of more than two carpels (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). However, our data show that combination of the crc mutation with sup-5 resulted in the formation of sterile flowers with significantly more stamens and carpels. The crc mutation enhanced FM indeterminacy in sup-5 mutants, leading to an increase in FM size. Because the increased number of stamens in sup mutants is at the expense of carpel tissues due to the expansion of the APETALA3 gene expression domain into the fourth whorl, an increase in FM size, leading to the formation of more carpels, might increase the rate of floral organ identity conversion, coupled with hormonal changes (Prunet et al., 2017). The sterility of crc sup double mutant flowers was probably due to a failure in establishing carpel polarity caused by crc. Polarity changes further enhance the carpel defect phenotypes in sup single mutants, which produce apically open carpels with abnormal ovule integument development (Gaiser et al., 1995; Baker et al., 1997; Eshed et al., 1999; Breuil-Broyer et al., 2016).
Compared to the Arabidopsis CRC gene, its orthologs in other species play broader and prominent roles during plant growth and development (Nagasawa et al., 2003; Yamaguchi et al., 2004; Li et al., 2011). No clear defects in FM determinacy and no homeotic defects are observed in crc single mutants (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Baum et al., 2001; Nagasawa et al., 2003). By contrast, mutation of CRC orthologs in other plant species causes multiple phenotypic defects during the vegetative and reproductive stages. For example, one well-characterized CRC ortholog is DROOPING LEAF (DL) in rice (Oryza sativa). Molecular studies have identified the gene functions that specify carpel identity and regulate midrib leaf formation in rice (Yamaguchi et al., 2004). To date, no crc alleles that show floral homeotic or leaf defects as strong as those in dl mutants have been identified in Arabidopsis. When combined with mutants of other key regulators of floral meristem activity or development, these higher-order crc multiple mutants show synergistic effects and produce flowers with extra floral whorls or floral organs with abnormal phenotypes (Eshed et al., 2001; Prunet et al., 2008; Zuniga-Mayo et al., 2012). In sup crc double mutants, aberrant FM indeterminacy and floral organ identity were observed. Although CRC is only expressed in the abaxial region of carpels prior to FM termination, the crc mutation enhances not only FM determinacy, but also the initiation of floral organs such as stamens or carpels. Thus, it is conceivable that phytohormones are involved in the genetic interaction between CRC and SUP because phytohormones exert multiple roles in a non-cell-autonomous manner. Recent studies have identified roles for CRC and SUP in hormone homeostasis (Yamaguchi et al., 2017, 2018; Xu et al., 2018). Hormonal regulation by CRC might also explain why its orthologs have various functions in different plant species.
Based on RNA-seq, SUP was expressed at normal levels in crc and similarly, CRC expression was unaffected in sup (FDR < 0.05). Thus, it is unlikely that either transcription factor transcriptionally regulates the expression of the other (Figure S1).
Cytokinins maintain meristem activity by controlling cell division (Riou-Khamlichi et al., 1999; Werner et al., 2001; Yang et al., 2002; Zhang et al., 2013). Consistent with this finding, we identified AHP6 as a common downstream gene of CRC and SUP. It has been reported that supernumerary stamen primordia in sup are formed at stage 7 (Prunet et al., 2017). Considering that CRC is expressed from floral stage 6 onwards (Bowman and Smyth, 1999; Lee et al., 2005), the effect of the crc mutation in the sup background and AHP6 misexpression was observed after stage 6. Stage-specific gene expression and binding tests using a synchronized system, together with the expression of multiple marker genes, will provide greater insight into how stamen number is defined by CRC and SUP via AHP6.
IAA19, which plays a key role in controlling stamen elongation (Tashiro et al., 2009; Ghelli et al., 2018), was also differentially expressed in crc-1 sup-5 plants. This suggests that SUP might be important not only for early stamen formation, but also for subsequent stamen growth. The crc mutation affects the expression of IAA19, which is expressed in stamens, even though CRC is not expressed in these organs. It is unclear how this regulation occurs, but might involve non-cell-autonomous effects.
The REM25 and TPPI genes were also differentially expressed in crc-1 sup-5 (Mantegazza et al., 2014). Based on previous in situ hybridization data, REM25 is highly expressed in stamen and carpel primordia at floral stage 6. Since SUP, CRC, and REM25 have overlapping expression domains (Sakai et al., 1995; Bowman and Smyth, 1999; Lee et al., 2005), SUP and CRC might be upstream regulators of REM25. Genetic redundancy and the physical linkage of REM loci hamper functional studies of REM family genes (Mantegazza et al., 2014); therefore, the generation of mutants via CRISPR/Cas9 might contribute to understanding the function of genes within this family. TPPI is required for the appropriate establishment of organ boundaries (Lor, 2014), which is consistent with the regulation of organ boundary genes, such as CUC2 (Xu et al., 2018), by SUP. However, the exact function of TPPI during flower development is largely unknown and it is relevant to study the molecular function of TPPI during stamen/carpel boundary specification.
The datasets generated for this study can be found in the DRA008874.
ZL, YT, NY, and TI conceived and designed the experiments. ZL and YT performed the phenotypic experiments. ZL extracted RNA. YI, AS, and KS generated RNA-seq libraries. TS performed sequencing. NY conducted transcriptome analyses and wrote the paper.
This work was supported by a grant from the Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology (JPMJPR15QA), JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (No. 18H04782), JSPS KAKENHI Grant-in-Aid for Scientific Research B (No. 18H02465), Grant-in-Aid for challenging Exploratory Research (No. 19K22431), the NAIST foundation and the LOTTE Foundation to NY, Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology (JPMJPR15Q2) and Cabinet Office, Government of Japan, Cross-ministerial Strategic Innovation Promotion Program (SIP), Technologies for Smart Bio-industry and Agriculture to YI, JSPS KAKENHI Grant-in-Aid for Scientific Research C (No. 15H05955) to TS, JSPS KAKENHI Grant-in-Aid for Scientific Research S (No. 15H05959, 17H06172) to KS, JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (No. 17H05843, 18H04839), JSPS KAKENHI Grant-in-Aid for Scientific Research A (No. 15H02405), Grant-in-Aid for challenging Exploratory Research (No. 18K19342) to TI.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Akie Takahashi, Hiroko Egashira, Kyoko Sunuma, Mayumi Nara, Mikiko Higashiura, Taeko Kawakami, and Yuka Kadoya, for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00437/full#supplementary-material
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386.
Alves-Ferreira, M., Wellmer, F., Banhara, A., Kumar, V., Riechmann, J. L., and Meyerowitz, M. M. (2007). Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145, 747–762. doi: 10.1104/pp.107.104422
Baker, S. C., Robinson-Beers, K., Villanueva, J. M., Gaiser, J. C., and Gasser, C. S. (1997). Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145, 1109–1124.
Baum, S. F., Eshed, Y., and Bowman, J. L. (2001). The Arabidopsis nectary is an ABC-independent floral structure. Development 128, 4657–4667. Available online at: https://dev.biologists.org/content/128/22/4657.long
Besnard, F., Refahi, Y., Morin, V., Marteaux, B., Brunoud, G., Chambrier, P., et al. (2014a). Cytokinin signaling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421. doi: 10.1038/nature12791
Besnard, F., Rozier, F., and Vernoux, T. (2014b). The AHP6 cytokinin signaling inhibitor mediates an auxin-cytokinin crosstalk that regulates the timing of organ initiation at the shoot apical meristem. Plant Signal. Behav. 9:e28788. doi: 10.4161/psb.28788
Bollier, N., Sicard, A., Leblond, J., Latrasse, D., Gonzalez, N., Gévaudant, F., et al. (2018). At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83–100. doi: 10.1105/tpc.17.00653
Bommert, P., and Whipple, C. (2017). Grass inflorescence architecture and meristem determinacy. Semin. Cell Dev. Biol. 79, 37–47. doi: 10.1016/j.semcdb.2017.10.004
Bowman, J. L., Sakai, H., Jack, T., Weigel, D., Mayer, U., and Meyerowitz, E. M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–515.
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396.
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Breuil-Broyer, S., Trehin, C., Morel, P., Boltz, V., Sun, B., Chambrier, P., et al. (2016). Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male–female boundary, flower meristem termination and carpel compartmentalization. Ann. Bot. 117, 905–923. doi: 10.1093/aob/mcw023
Chen, Z. Z., Hong, X. H., Zhang, H. R., Wang, Y. Q., Li, X., Zhu, J. K., et al. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43, 273–283. doi: 10.1111/j.1365-313X.2005.02452.x
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. doi: 10.1105/tpc.107.053009
Chiu, W. H., Chandler, J., Cnops, G., Van Lijsebettens, M., and Werr, W. (2007). Mutations in the TORNADO2 gene affect cellular decisions in the peripheral zone of the shoot apical meristem of Arabidopsis thaliana. Plant Mol. Biol. 63, 731–744. doi: 10.1007/s11103-006-9105-z
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067.
Cnops, G., Neyt, P., Raes, J., Petrarulo, M., Nelissen, H., Malenica, N., et al. (2006). The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell 18, 852–866. doi: 10.1105/tpc.105.040568
El-Showk, S., Ruonala, R., and Helariutta, Y. (2013). Crossing paths: Cytokinin signalling and crosstalk. Development 140, 1373–1383. doi: 10.1242/dev.086371
Eshed, Y., Baum, S. F., and Bowman, J. L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. doi: 10.1016/S0092-8674(00)81651-7
Eshed, Y., Baum, S. F., Perea, J. V., and Bowman, J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. doi: 10.1016/S0960-9822(01)00392-X
Gaiser, J. C., Robinson-Been, K., and Gasser, C. S. (1995). The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7, 333–345. doi: 10.2307/3869855
Ghelli, R., Brunetti, P., Napoli, N., De Paolis, A., Cecchetti, V., Tsuge, T., et al. (2018). A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell 30, 620–637. doi: 10.1105/tpc.17.00840
Gomez-Mena, C., de Folter, S., Costa, M. M., Angenent, G. C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132, 429–438. doi: 10.1242/dev.01600
Gross, T., Broholm, S., and Becker, A. (2018). CRABS CLAW acts as a bifunctional transcription factor in flower development. Front. Plant Sci. 9:835. doi: 10.3389/fpls.2018.00835
Guo, L., Cao, X., Liu, Y., Li, J., Li, Y., Zhang, K., et al. (2018). A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 94, 1083–1097. doi: 10.1111/tpj.13921
Hou, K., Wu, W., and Gan, S. S. (2013). SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 161, 1002–1009. doi: 10.1104/pp.112.212787
Ichihashi, Y., Fukushima, A., Shibata, A., and Shirasu, K. (2018). High impact gene discovery: simple strand-specific mRNA library construction and differential regulatory analysis based on gene co-expression network, in Plant Transcription Factors. Methods in Molecular Biology, Vol. 1830, ed N.. Yamaguchi, 163–189 (New York, NY: Humana Press).
Jacobsen, S. E., Sakai, H., Finnegan, E. J., Cao, X., and Meyerowitz, E. M. (2000). Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr. Biol. 10, 179–186. doi: 10.1016/S0960-9822(00)00324-9
Kayes, J. M., and Clark, S. E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851.
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Laux, T., Mayer, K. F. X., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Lee, J. Y., Baum, S. F., Alvarez, J., Patel, A., Chitwood, D. H., and Bowman, J. L. (2005). Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17, 25–36. doi: 10.1105/tpc.104.026666
Lenhard, M., Bohnert, A., Jürgens, G., and Laux, T. (2011). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. doi: 10.1016/S0092-8674(01)00390-7
Li, H. F., Liang, W. Q., Yin, C. S., Zhu, L., and Zhang, D. B. (2011). Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 156, 263–274. doi: 10.1104/pp.111.172080
Liu, X., Kim, Y. J., Müller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23, 3654–3670. doi: 10.1105/tpc.111.091538
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Lor, J. L. (2014). Function of TPPI and TPPJ in Arabidopsis boundary regions (Unpublished masters dissertation). University of California, Riverside, USA.
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Mähönen, A. P., Bishopp, A., Higuchi, M., Nieminen, K. M., Kinoshita, K., Tormakangas, K., et al. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311, 94–98. doi: 10.1126/science.1118875
Mantegazza, O., Gregis, V., Mendes, M. A., Morandini, P., Alves-Ferreira, M., Patreze, C. M., et al. (2014). Analysis of the Arabidopsis REM gene family predicts functions during flower development. Ann. Bot. 114, 1507–1515. doi: 10.1093/aob/mcu124
Moreira, S., Bishopp, A., Carvalho, H., and Campilho, A. (2013). AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PLoS ONE 8:e56370. doi: 10.1371/journal.pone.0056370
Müller, C. J., Larsson, E., Spichal, L., and Sundberg, E. (2017). Cytokinin-auxin crosstalk in the gynoecial promodium ensures correct domain patterning. Plant Physiol. 175, 1144–1157. doi: 10.1104/pp.17.00805
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H. Y., Sakai, H., et al. (2003). SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Nakamura, A., Higuchi, K., Goda, H., Fujiwara, M. T., Sawa, S., Koshiba, T., et al. (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133, 1–11. doi: 10.1104/pp.103.030031
Nibau, C., Stilio, V. S. D., Wu, H. M., and Cheung, A. Y. (2010). Arabidopsis and tobacco SUPERMAN regulate hormone signaling and mediate cell proliferation and differentiation. J. Exp. Bot. 62, 949–961. doi: 10.1093/jxb/erq325
Payne, T., Johnson, S. D., and Koltunow, A. M. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131, 3737–3749. doi: 10.1242/dev.01216
Prunet, N., Morel, P., Thierry, A. M., Eshed, Y., Bowman, J. L., Negrutiu, I., et al. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20, 901–919. doi: 10.1105/tpc.107.053306
Prunet, N., Yang, W., Das, P., Meyerowitz, E. M., and Jack, T. P. (2017). SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl. Acad. Sci. U.S.A. 114, 7166–7171. doi: 10.1073/pnas.1705977114
Reyes-Olalde, J. I., Zuniga-Mayo, V. M., Marsch-Martinez, N., and de Folter, S. (2017a). Synergistic relationship between auxin and cytokinin in the ovary and the participation of the transcription factor SPATULA. Plant Signal. Behav. 12:e1376158. doi: 10.1080/15592324.2017.1376158
Reyes-Olalde, J. I., Zúñiga-Mayo, V. M., Serwatowska, J., Chavez Montes, R. A., Lozano-Sotomayor, P., Herrera-Ubaldo, H., et al. (2017b). The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 13:e1006726. doi: 10.1371/journal.pgen.1006726
Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J. A. H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. doi: 10.1126/science.283.5407.1541
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sablowski, R. (2015). Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 66, 1065–1073. doi: 10.1093/jxb/eru514
Sakai, H., Krizek, B. A., Jacobsen, S. E., and Meyerowitz, E. M. (2000). Regulation of SUP expression identifies multiple regulators involved in Arabidopsis floral meristem development. Plant Cell 12, 1607–1618. doi: 10.1105/tpc.12.9.1607
Sakai, H., Medrano, L. J., and Meyerowitz, E. M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203. doi: 10.1038/378199a0
Sakuma, Y., Maruyama, K, Qin, F, Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Sassi, M., and Vernoux, T. (2013). Auxin and self-organization at the shoot apical meristem. J. Exp. Bot. 64, 2579–2592. doi: 10.1093/jxb/ert101
Schaller, G. E., Bishopp, A., and Kieber, J. J. (2015). The Yin-Yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63. doi: 10.1105/tpc.114.133595
Schluepmann, H., van Dijken, A., Aghdasi, M., Wobbes, B., Paul, M., and Smeekens, S. (2004). Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 135, 879–890. doi: 10.1104/pp.104.039503
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F. X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Smyth, D. R., Bowman, J. L., and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. doi: 10.1105/tpc.2.8.755
Stamm, P., and Kumar, P. P. (2013). Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 32, 759–769. doi: 10.1007/s00299-013-1406-5
Sun, B., and Ito, T. (2015). Regulation of floral stem cell termination in Arabidopsis. Front. Plant Sci. 6:17. doi: 10.3389/fpls.2015.00017
Sun, B., Looi, L. S., Guo, S., He, Z., Gan, E. S., Huang, J., et al. (2014). Timing mechanism dependent on cell division is invoked by polycomb eviction in plant stem cells. Science 343:1248559. doi: 10.1126/science.1248559
Sun, B., Xu, Y., Ng, K. H., and Ito, T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23, 1791–1804. doi: 10.1101/gad.1800409
Sun, B., Zhou, Y., Cai, J., Shang, E., Yamaguchi, N., Xiao, J., et al. (2019). Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell 31, 1488–1505. doi: 10.1105/tpc.18.00450
Supek, F., Bosnjak, M., Skunca, N., and Smuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800. doi: 10.1371/journal.pone.0021800
Tashiro, S., Tian, C. E., Watahiki, M. K., and Yamamoto, K. T. (2009). Changes in growth kinetics of stamen filaments cause inefficient pollination in massugu2, an auxin insensitive, dominant mutant of Arabidopsis thaliana. Physiol. Plant 137, 175–187. doi: 10.1111/j.1399-3054.2009.01271.x
Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M. K., Harper, R. M., et al. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393. doi: 10.1105/tpc.018630
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Turner, S. R., and Somerville, C. R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in secondary cell wall. Plant Cell 9, 689–701. doi: 10.1105/tpc.9.5.689
Uemura, A., Yamaguchi, N., Xu, Y., Wee, W. Y., Ichihashi, Y., Suzuki, T., et al. (2017). Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis. Plant Reprod. 31, 89–105. doi: 10.1007/s00497-017-0315-0
Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. U.S.A. 98, 10487–10492. doi: 10.1073/pnas.171304098
Wolters, H., and Jurgens, G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305–317. doi: 10.1038/nrg2558
Xu, Y., Prunet, N., Gan, E. S., Wang, Y., Stewart, D., Wellmer, F., et al. (2018). SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37:e97499. doi: 10.15252/embj.201797499
Xu, Y., Yamaguchi, N., Gan, E. S., and Ito, T. (2019). When to stop: an update on molecular mechanisms of floral meristem termination. J. Exp. Bot. 70, 1711–1718. doi: 10.1093/jxb/erz048
Yamaguchi, N., Huang, J., Tatsumi, Y., Abe, M., Sugano, S. S., Kojima, M., et al. (2018). Chromatin-mediated feed-forward regulation of YUCCA4 expression by AGAMOUS and CRABS CLAW directs gynoecium formation. Nat. Commun. 9:5290. doi: 10.1038/s41467-018-07763-0
Yamaguchi, N., Huang, J., Xu, Y., Tanoi, K., and Ito, T. (2017). Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 8:1125. doi: 10.1038/s41467-017-01252-6
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H. Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16, 500–509. doi: 10.1105/tpc.018044
Yang, J., Zhang, J., Huang, Z., Wang, Z., Zhu, Q., and Liu, L. (2002). Correlation of cytokinin levels in the endosperms and roots with cell number and cell division activity during endosperm development in rice. Ann. Bot. 90, 369–377. doi: 10.1093/aob/mcf198
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Zhang, W., Swarup, R., Bennett, M., Schaller, G. E., and Kieber, J. J. (2013). Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 23, 1979–1989. doi: 10.1016/j.cub.2013.08.008
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Keywords: Arabidopsis, CRABS CLAW, cytokinin, floral meristem, flower, SUPERMAN
Citation: Lee ZH, Tatsumi Y, Ichihashi Y, Suzuki T, Shibata A, Shirasu K, Yamaguchi N and Ito T (2019) CRABS CLAW and SUPERMAN Coordinate Hormone-, Stress-, and Metabolic-Related Gene Expression During Arabidopsis Stamen Development. Front. Ecol. Evol. 7:437. doi: 10.3389/fevo.2019.00437
Received: 28 August 2019; Accepted: 25 October 2019;
Published: 14 November 2019.
Edited by:
Annette Becker, University of Giessen, GermanyReviewed by:
Nayelli Marsch-Martinez, Center for Research and Advanced Studies (CINVESTAV), MexicoCopyright © 2019 Lee, Tatsumi, Ichihashi, Suzuki, Shibata, Shirasu, Yamaguchi and Ito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toshiro Ito, aXRvdEBicy5uYWlzdC5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.