- 1School of Electronic and Information Engineering, Anhui Architecture University, Hefei, Anhui, China
- 2Department of Medical Technology, Anhui Medical College, Hefei, China
- 3Department of Cardiology, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 4Anhui Public Health Clinical Center, Hefei, China
- 5School of Advanced Manufacturing Engineering, Hefei University, Hefei, China
- 6Anhui Provincial Key Laboratory of Environmental Pollution Control and Resource Reuse, Hefei, Anhui, China
- 7Anhui International Joint Research Center for Ancient Architecture Intellisensing and MultiDimensional Modeling, Hefei, China
Background: Multi-wavelength transmittance spectroscopy, in combination with the artificial neural network, has been a novel tool used to identify and classify microorganisms in recent years.
Methods: In our work, the transmittance spectra in the region from 200 to 900 nm for four bacterial species of interest, Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), Klebsiella pneumoniae (K.pneumoniae), and Salmonella typhimurium (S. typhi), were recorded using an ultraviolet–visible spectrophotometer. Considering too much redundant data on the full-wave band spectra, the characteristic wavelength variables were selected using the competitive adaptive reweighting sampling (CARS) algorithm. Spectra of the initial training set of these targeted microorganisms were used to create identification models representing the spectral variability of each species using four kinds of neural networks, namely, backpropagation (BP), radial basis function network (RBF), generalized regression neural network (GRNN), and probabilistic neural network (PNN).
Results: The blinded isolate spectra of targeted species were identified using the four identification models given above. Compared to fullband modeling, after using CARS to screen the wavelength variables, four identification models are established for the 35 preferred characteristic wavelengths, and the prediction performance of the four models is notably improved. Among them, the CARS–PNN model is the best, and the identification rates of all targeted bacteria were achieved with 100% accuracy; the calculation time is just approximately 0.04 s.
Discussion: The use of CARS can effectively remove useless information from the spectra, reduce model complexity, and enhance model prediction performance. Multi-wavelength transmission spectroscopy, combined with the CARS–PNN method, can provide a new method for the rapid detection of bacteria in water and could be readily extended for bacterial microbiological detection in blood and food.
1 Introduction
Pathogens are important sources causing human and animal diseases. A large number of outbreaks due to bacterial infections could occur via water contaminated by bacteria (Rajapaksha et al., 2019; Nnachi et al., 2022). Pathogenic Escherichia coli, Campylobacter jejuni, Shigella, and Salmonella typhi may cause polar gastroenteritis, dysentery, and typhoid, respectively, in human and animals. Furthermore, E. coli O157:H7 and Shigella have been proven as the causes of waterborne disease outbreaks worldwide, including the E. coli contamination of drinking water in Canada, which led to 2300 symptomatic residents and seven deaths (Meinhardt, 2006). In August 2020, contaminated tap water in Shouxian county, Anhui province, infected 493 people with Shigella bacteria, who successively suffered from fever, vomiting, abdominal pain, and diarrhea (Gao, 2021).
The rapid and accurate detection of bacteria in water is of great importance in controlling and preventing waterborne diseases and outbreaks. The techniques that are currently being used for bacteria detection include isolated cultures (Sieuwerts et al., 2008; Azzam et al., 2014), immunofluorescence assay (IFA) (Kuhn et al., 2010), polymerase chain reaction (PCR) (Zhang et al., 2011; Gensberger et al., 2015; Menu et al., 2018), and enzyme-linked immunosorbent assay (ELISA) (Välimaa et al., 2015; Sakamoto et al., 2018). The results from these methods are relatively accurate. However, they require complex sample pretreatment, are time-consuming, require expensive biological reagents, and cannot achieve real-time and on-site detection. Therefore, a simple, rapid, and accurate method for the detection of waterborne bacteria is greatly needed.
Multi-wavelength transmission spectroscopy (MWTS) is a new non-invasive testing spectroscopy technology. It is generated by the direct interaction of light with the cell structure and chemical composition and can be used for the characterization of the suspended matter in water. At present, multi-wavelength transmission spectroscopy combined with stoichiometry has a certain degree of research capability on the classification and identification of microorganisms. For example, multi-wavelength transmission spectroscopy combined with the multivariate statistical analysis can identify four bacterial microorganisms of clinical significance with high accuracy (Smith et al., 2012). Ultraviolet–visible (UV–Vis) spectra combined with a support vector machine have been used to identify five different bacterial species, and the accuracy rate reached 100% (Feng et al., 2021). Artificial neural networks (ANNs) are computer programs that mimic the way the human brain thinks, aiming at classifying or predicting things by imitating the way the human brain processes information. It has the characteristics of self-learning, self-organization, self-adaptation, and fault tolerance and is especially suitable for the processing of unknown and uncertain non-linear problems. As the most important branch of artificial intelligence, ANNs have many applications in classification and regression, such as image recognition (Elish, 2019; Zhang et al., 2023) and model building (Ho et al., 2019; Liu et al., 2019).
The objectives of this study were to investigate the feasibility of using MWTS to detect bacteria in water, including Escherichia coli, Staphylococcus aureus, Klebsiella pneumonia, and Salmonella typhimurium, and to differentiate among bacterial species using artificial neural networks (that is, backpropagation [BP], radial basis function [RBF] network, generalized regression neural network [GRNN], and probabilistic neural network [PNN]).
2 Materials and methods
2.1 Preparation of bacterial samples
All strains of E. coli (China Center of Industrial Culture Collection (CICC) #10389), Staphylococcus aureus (CICC #21648), K. pneumonia (CICC #21106), Salmonella typhi (CICC #21913), and E. coli (DH5α) were obtained from the China Center of Industrial Culture Collection. The bacterial suspension was made by the process that involved the activation of bacteria; culturing at 37°C in the beef extract peptone medium (pH 7.0), containing 0.3% beef extract, 1% peptone, 0.5% NaCl, and 2% agar; expanding propagation in the solution culture; centrifugation using a centrifuge; and washing in sterilized deionized water.
2.2 Spectroscopy measurements
For each bacterial suspension, a series of bacterial test samples of different concentrations were obtained by diluting them with deionized water for determining the multi-wavelength transmission spectroscopy measurement. The multi-wavelength transmission spectra of five bacterial suspensions were recorded using a UV/visible spectrophotometer (UV-2550 Shimadzu, Kyoto, Japan) in the wavelength range of 200–900 nm. The instrument-recorded spectra represent the averages of three replicate measurements with 1-nm sampling intervals and medium scan speed. To eliminate the effect of inhomogeneity in the suspending medium, sterilized deionized water was used as the reference solution. All measurements were conducted using a 1-cm path length and 3.5-mL volume quartz cuvettes at room temperature.
2.3 Spectral pretreatment
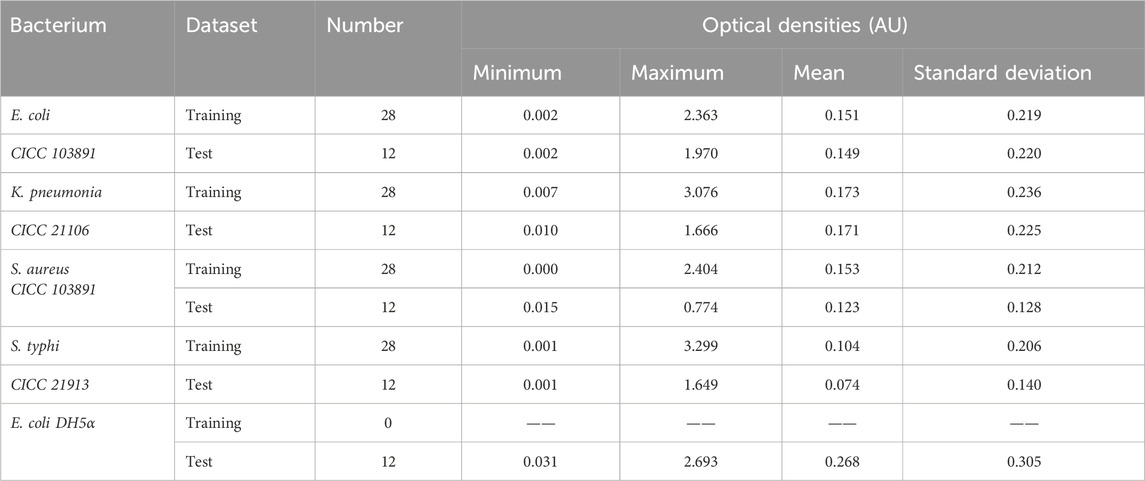
To eliminate bacterial concentration effects, the transmission spectra were normalized with the average optical density between 200 and 900 nm. With sample set partitioning based on the joint x–y distance (SPXY), 40 spectra of each bacterial suspension were performed by selecting 28 spectra as the training set; the remaining spectra were assigned to the test set to ensure the selected optimal spectra characterizing each bacterial species. In order to verify the ability of the model to discriminate between the species within the genus of bacteria, 12 spectra of Escherichia coli (DH5α) were used as the test set. The spectral statistics for the training and test sets of five bacterial species are reported in Table 1.
As indicated in Table 1, the minimum and maximum optical densities divided into the test set were within the range of the values given in the training set, indicating that the partition of sample sets was reasonable using the SPXY algorithm, and the established discrimination model could also produce good results.
2.4 Bacterial identification models
The generation of an accurate identification model for each microorganism must be based on a set of spectra that represents as many spectral variations as possible, which can be exhibited by each microorganism. The optical density at each wavelength point in the multi-wavelength transmission spectrum of bacterial suspensions is a function of the number, shape, size, internal structure, and the chemical composition of bacteria (Hu et al., 2017). Therefore, the bacterial identification model is characterized by the measured wavelength, and the optical density at the wavelength is the characteristic value, that is, the sample set is (λi and τ(λi)).
ANNs are constructed as complex networks of interconnected artificial neurons, which are used to estimate or approximate the functions that can depend on a large number of inputs and are usually unknown (Slabbinck et al., 2009; Wang et al., 2010; Gargouri et al., 2012). The output signal of each artificial neuron is given by the following:
where
where x is the input value, which is determined by continuously adjusting the weights and biases of the network; the error between the network output and the expected output is minimized.
In the RBF neural network, the activation function is expressed by the Gaussian function (also known as the radial basis function):
where x is the input value, c is the center of the Gaussian function, and sigma is the standard deviation of the Gaussian function.
In the GRNN, the activation function is also expressed by the Gaussian function:
where x represents the input value and sigma is the standard deviation.
In the PNN, the activation function is given by the probability density function:
where x is the input value, mu is the mean of the Gaussian function, and sigma is the standard deviation of the Gaussian function. By adjusting mu and sigma, the shape and width of the probability density function can be controlled, improving the fitting ability and smoothness of the neural network.
3 Results and discussion
3.1 Multi-wavelength transmission spectra for bacteria
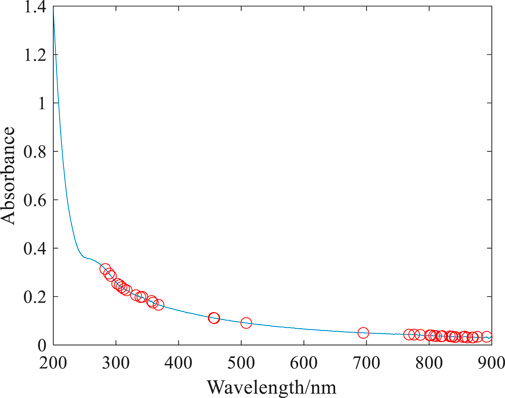
The normalized optical density spectra of E. coli, K. pneumonia, S. aureus, and S. typhi in the region from 200 to 900 nm are shown in Figure 1. A qualitative comparison of the spectra of the four bacteria reveals the discriminating power among them using the spectroscopy approach. It can be seen that the spectral patterns of K. pneumonia, E. coli, and S. typhi have similar features with their peak at approximately 260 nm, which is likely due to the similarities in shape, aggregation state, and chemical composition of these microorganisms (Garcia-Rubio et al., 2004; Alupoaei and García-Rubio, 2005). However, when their optical densities at some wavelength points are distinct, it is due to the differences in the size and content of the chemical composition (Adams et al., 1989). With the differences of the first three microorganisms, the spectra of S. aureus have no apparent absorption peak at 260 nm; this is because S. aureus cells are spherical and show different spectral features (Mattley et al., 2000). Clearly, the spectra of the four microorganisms are almost overlapped in the region from 450 to 600 nm; however, there are differences in the optical density in the wavelength range (250–450 nm and 600–900 nm). These spectral fingerprints are able to distinguish among different bacterial species with the help of artificial neural networks.
3.2 Discrimination model construction and validation based on the full-spectrum band
The optical density spectra in the full range of the wavelength (200–900 nm) were selected as the analysis data, and the four bacterial species in the experiment were classified into species numbers: E. coli as species 1, K. pneumonia as species 2, S. aureus as species 3, and S. typhi as species 4. The bacteria identification model was established using the corresponding training set (28 spectra were selected from the full set of spectra available for each species, with a total of 112 training sets).
ANNs are usually composed of three neuron layers: one input layer, one hidden layer, and one output neuron layer. The number of input and output neurons correspond to the optical density for each wavelength (n = 701) and the number of bacterial species present in the dataset. After multiple-experiment comparative analyses, the parameters of the BP neural network are set as follows: the number of the neural units is 6, and the epoch limits, the target error, and the learning rate are set to 1000, 1e-6, and 0.01, respectively. The parameters controlled the threshold size in the neural network of the RBF, GRNN, and PNN, which are set to 0.003, 0.003, and 0.0013, respectively.
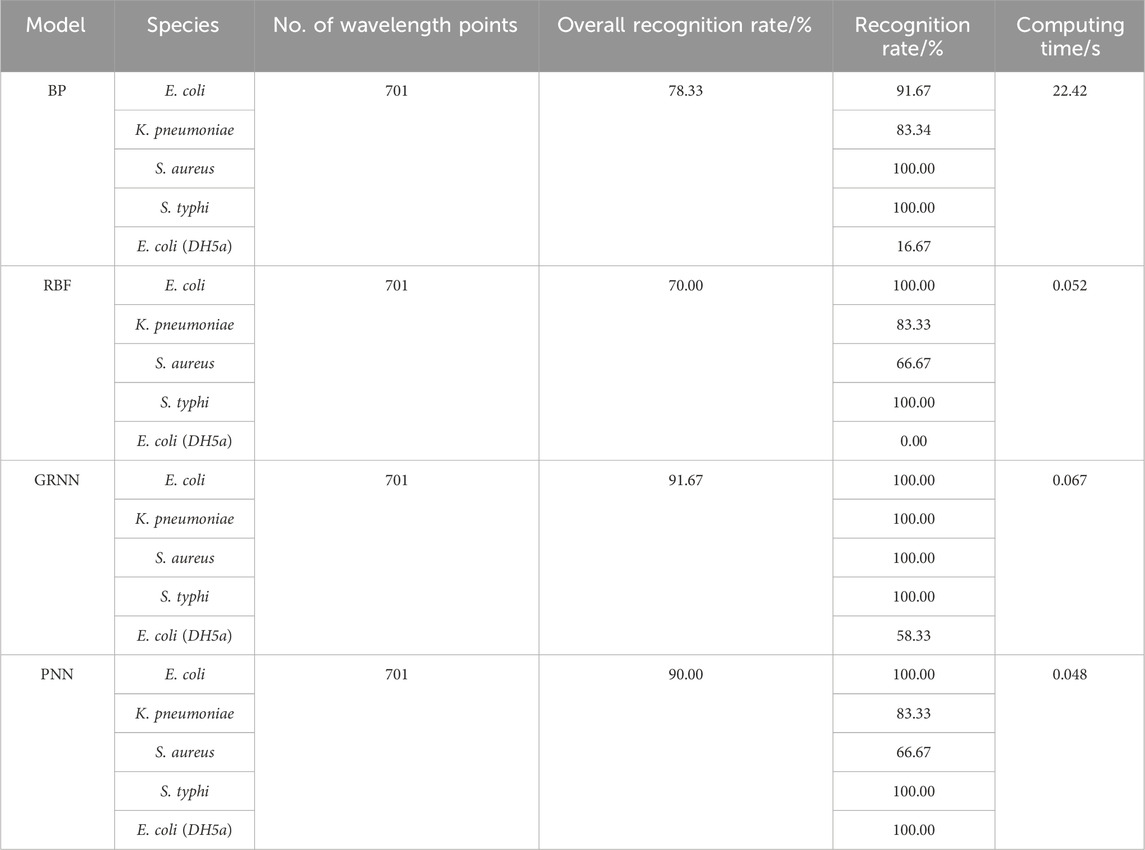
In order to verify the accuracy and reliability of bacterial discrimination models, 60 spectra in the test set were analyzed. A performance breakdown for the individual classes is displayed in the confusion matrix (Figure 2). It can be seen that the overall identification rates of targeted bacterial species for four neural networks are 78.33%, 70.00%, 91.67%, and 90.00%, respectively.
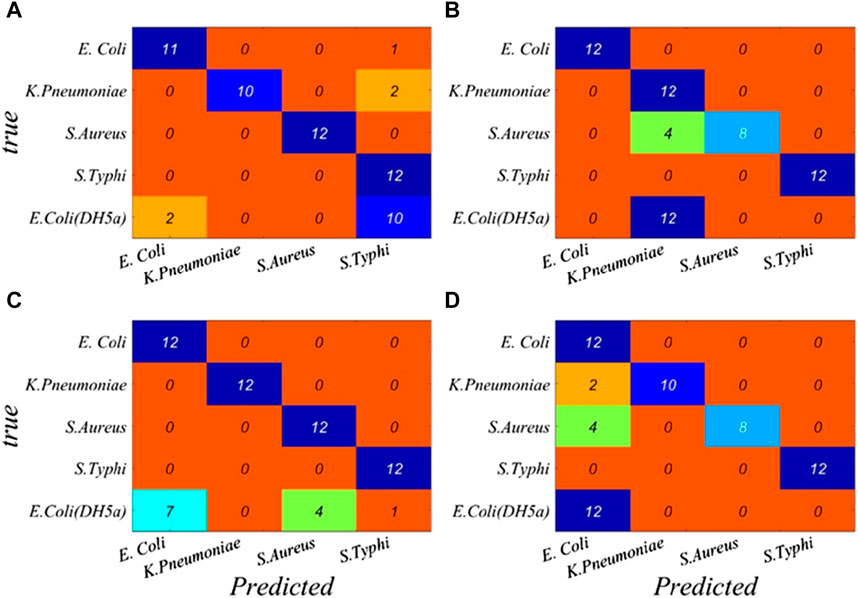
FIGURE 2. Performance breakdown for individual classes based on the full band is displayed in the confusion matrix. (A) BP, (B) RBF, (C) GRNN, and (D) PNN.
The classification results of four neural networks based on the full band are presented in Table 2. The classification rates of the BP neural network for S. typhi and S. aureus both achieve 100.00%, whereas the classification rates of E. coli (CICC 103891), K. pneumonia, and E. coli (DH5α) are just 91.67%, 83.34%, and 16.67%, respectively. The classification rates of the RBF neural network for E. coli (CICC 103891) and S. typhi achieve 100.00%, whereas the classification rates of K. pneumonia, S. aureus, and E. coli (DH5α) are just 83.33%, 66.67%, and 0.00%, respectively. For the GRNN, E. coli (CICC 103891), K. pneumonia, S. aureus, and S. typhi achieve correct classification rates of 100.00%; however, the classification rate of E. coli (DH5α) is 58.33%. For the PNN, E. coli (CICC 103891), S. typhi, and E. coli (DH5α) achieve correct classification rates of 100.00%, whereas the classification rates of K. pneumonia and S. aureus are just 83.33% and 66.67%, respectively.
It can be seen that the performance of four identification models needs to be improved. Since there are as many as 701 wavelength variables in the full spectrum, some variables contain irrelevant information, which is unfavorable for classification modeling and leads to a reduced prediction accuracy. Therefore, reducing the complexity of the model and extracting the wavelength variables related to the sample information can get a better classification model.
3.3 Discrimination model construction and validation based on the characteristic wavelengths
3.3.1 Feature wavelength selection
In order to improve the bacterial recognition rate and reduce the required time for ANNs to process data, we used the competitive adaptive reweighting sampling (CARS) method. It is also used to extract the wavelengths with notable differences in the optical density values of the target microorganisms from the wavelength variables of the full spectrum, which were used as the new training sets for the four neural networks.
The main steps of the CARS algorithm (Li et al., 2019; Yuan et al., 2020; Yang et al., 2023) are as follows: first, the Monte Carlo sampling (MCS) method was used to sample n times, and 80% of the samples were randomly selected from the sample set as the correction set each time to establish the PLS model; the wavelength variables with less absolute weight of the regression coefficients were removed, and the number of variables removed was determined using the exponentially decreasing function (EDF). Then, the wavelength variables were screened using the adaptive reweighted sampling (ARS) technique; finally, the PLS model was established by each newly generated variable subset, and the variables with the smallest root-mean-squared error of the cross-validation value (RMSECV) comprised the optimal variable subset.
In the experiment, the parameters of the CARS algorithm are set as follows: the number of times Monte Carlo sampling was carried out was 40 times, the number of folds in half for cross-validation was 10, and the selected best variable criteria was 0. The screening procedure for wavelength variables is shown in Figure 3. The abscissa stands for the sampling number and the ordinate stands for the number of wavelength variables, RMSECV, and the regression coefficient of the wavelength variable.
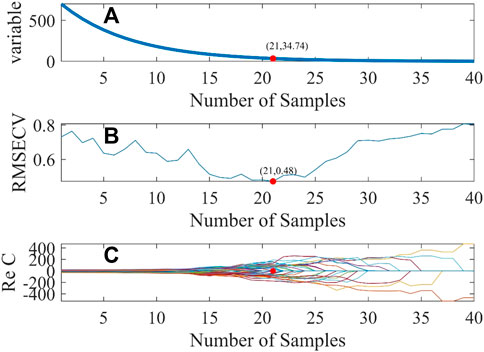
FIGURE 3. Diagram of the CARS variable selection process. Changes in variable, root mean square error of Cross-Validation, regression coefficients with the number of sampling times (A) the number of wavelength variables (B) root mean square error of cross-validation (C) regression coefficients.
As indicated in Figure 3, with the increase in the sampling times, the number of wavelength variables gradually decreases until the optimal number of sampling times is selected. The smaller the RMSECV, the better the number of sampling times is. When RMSECV is 0.48, the corresponding optimal sampling number is 21, and then, 35 characteristic wavelength variables can be screened from 701 wavelength variables. These characteristic wavelengths are mainly concentrated in the wavelength range of 250–400 nm and 750–900 nm, as shown by the red hollow dot in Figure 4.
3.3.2 Evaluation and analysis of improved identification models
A performance breakdown for individual classes based on the characteristic wavelengths is displayed in the confusion matrix (Figure 5). It can be seen that the overall identification rates of the targeted bacterial species for CARS–BP, CARS–RBF, CARS–GRNN, and CARS–PNN models are 93.33%, 90.00%, 91.67%, and 100.00%, respectively, which demonstrates that the transmission spectra of microorganisms combined with the CARS–PNN model can be utilized for microorganism identification, eliminating the need for culturing or specialized reagents.
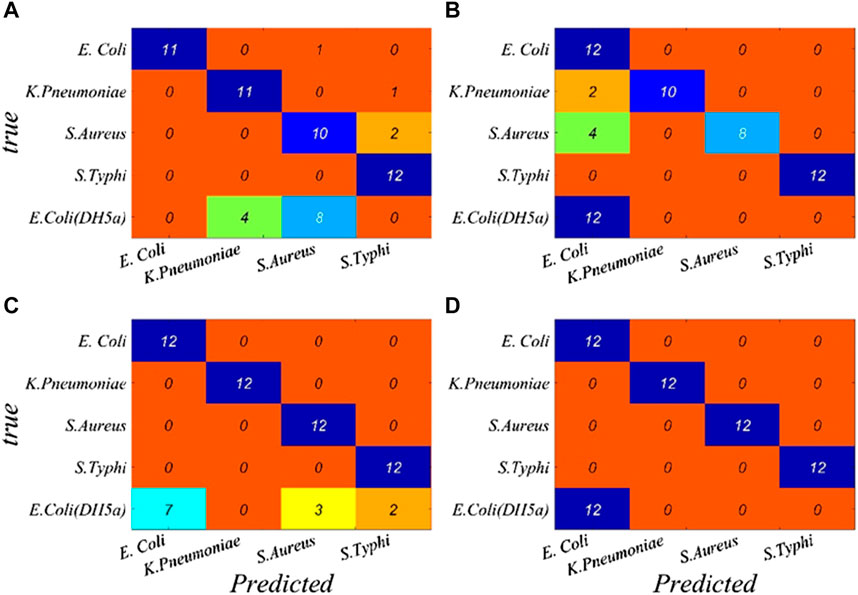
FIGURE 5. Performance breakdown for individual classes based on the characteristic wavelengths is displayed in the confusion matrix. (A) BP, (B) RBF, (C) GRNN, and (D) PNN.
The results are shown in Table 3; after wavelength variable screening using CARS, the recognition performance of the GRNN model remains unchanged, but the required computing time is reduced. The recognition performance of the BP, RBF, and PNN models are, respectively, improved, and the computation time is also reduced. This is because full-wavelength spectra often contain redundant information, which is unfavorable for classification modeling. It can be seen that selecting feature wavelength variables can not only reduce the model complexity but also enhance the prediction accuracy.
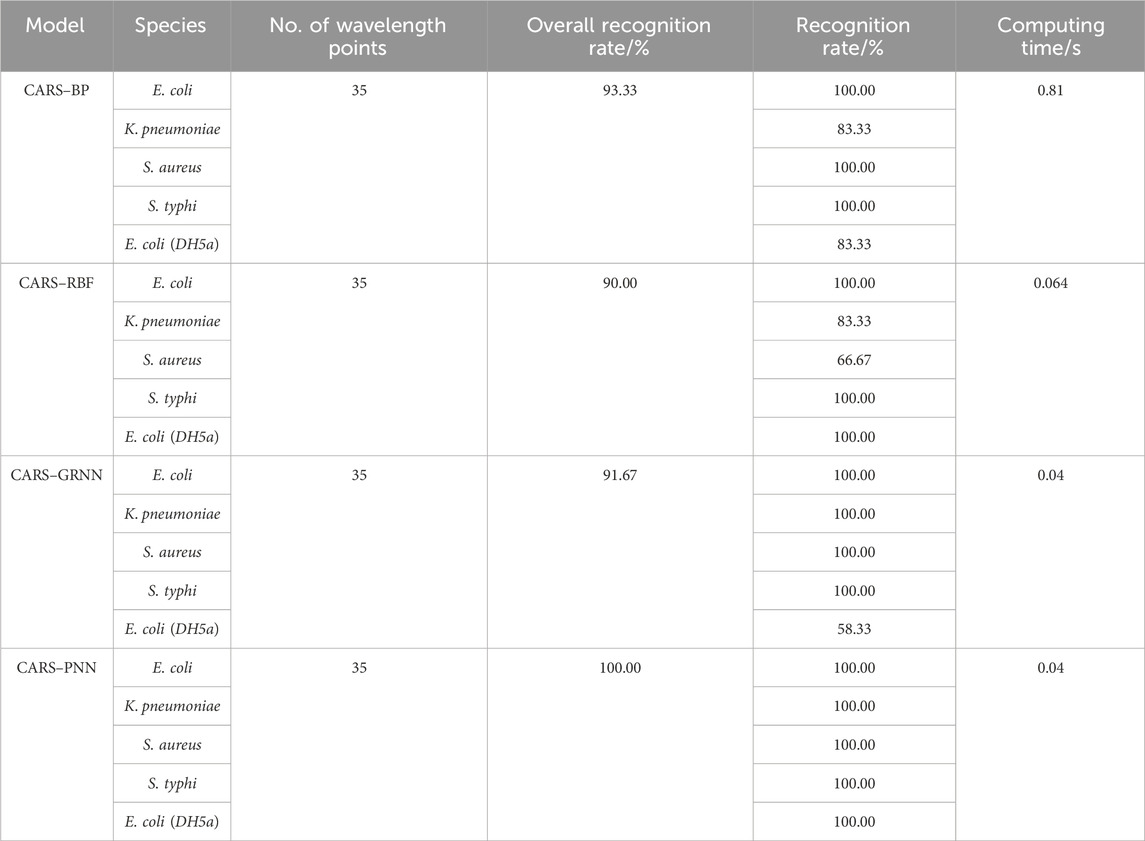
TABLE 3. Identification rate and calculation time of four neural networks based on the characteristic wavelengths.
In terms of the recognition accuracy and calculation time, the CARS–PNN model is the best. The identification accuracy of the CARS–PNN method for E. coli (CICC 10389), K. pneumonia, S. aureus, and S. typhi reaches 100.00%, and the calculation time is just 0.04 s. It demonstrates that the bacterial identification model based on the CARS–PNN has high accuracy. The recognition rate of E. coli (DH5α) is also 100.00%, which proves that the recognition model shows the certain stability of inter-genus recognition and potential to recognize bacteria of the same genus outside the training set.
5 Conclusion
In this paper, common bacterial microorganisms in water are chosen as the research object; four identification models of BP, RBF, GRNN, and PNN are established, respectively; and the wavelength variables are selected optimally using the CARS method; the identification performance of the four models for target bacteria is compared. The results show that compared with the full-band model, the four neural network models based on the characteristic wavelength can improve the performance of bacterial recognition; the CARS–PNN model is the best, the identification accuracy of the five tested bacteria is 100.00%, and the computing time is only 0.04 s. This study demonstrates that multi-wavelength transmission spectroscopy combined with the CARS–PNN model can effectively identify bacterial microorganisms, providing a simple, accurate, and reagent-free new method for the rapid identification of bacteria in water.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding authors.
Author contributions
YH: writing–original draft and writing–review and editing. YuZ: visualization and writing–review and editing. DH: writing–review and editing. NZ: data curation, formal analysis, and writing–review and editing. LX: resources and writing–review and editing. WL: methodology and writing–review and editing. JX: investigation and writing–original draft. YiZ: formal analysis and writing–original draft. PY: project administration and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Nos 62105002 and 51978003), the Key Research Project of Natural Science in Anhui Province (Nos KJ 2020A0471 and 2022AH050249), the Key Research and Development Projects of Anhui Province (No. 2022a05020043), and the College Students Innovation and Entrepreneurship Training Program (Nos S202310878131 and S202210878108).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, R. L., Knowler, J. T., and Leader, D. P. (1989). The biochemistry of nucleic acids. 10th edition. New York: McGraw Hill.
Alupoaei, C. E., and García-Rubio, L. H. (2005). An interpretation model for the UV-VIS spectra of microorganisms. Chem. Eng. Comm. 192, 198–218. doi:10.1080/00986440590473326
Azzam, R., Strohschön, R., Baier, K., Lu, L., Wiethoff, K., Bercht, A. L., et al. (2014). “Water quality and socio-ecological vulnerability regarding urban development in selected case studies of megacity Guangzhou, China,” in Megacities (Guangzhou, China: Our Global Urban Future), 33–58.
Elish, M. O. (2019). Comparison of different types of ANNs for identification of vulnerable web components. Springer, 1042–1055.
Feng, C., Zhao, N., Yin, G., Gan, T., and Chen, X. (2021). Study on multi-wavelength transmission spectral feature extraction combined with support vector machine for bacteria identification. Spectrosc. Spectr. Analysis 41 (9), 2940–2944. doi:10.3964/j.issn.1000-0593(2021)09-2940-05
Gao, Y. (2021). Comparative analysis of real-time fluorescent quantitative PCR and conventional bacterial identification for detection of enteropathogenic bacteria. Guide China Med. 19 (14), 126–127. doi:10.15912/j.cnki.gocm.2021.14.058
Garcia-Rubio, L. H., Alupoaei, C. E., Olivares, J. A., Stark, P., Garcia-Lopez, A., Stephans, C., et al. (2004). “A new spectroscopy method for in situ rapid detection and classification of micro-organisms,” in Proceedings volume 5585, chemical and biological point sensors for homeland defense II (Philadelphia, PA, United States: SPIE), 88–97.
Gargouri, N., Masmoudi, A. D., Masmoudi, D. S., and Abid, R. (2012). A new GLLD operator for mass detection in digital mammograms. J. Biomed. Imaging 2012, 4. doi:10.1155/2012/765649
Gensberger, E. T., Gössl, E.-M., Antonielli, L., Sessitsch, A., and Kostić, T. (2015). Effect of different heterotrophic plate count methods on the estimation of the composition of the culturable microbial community. PeerJ 3, e862. doi:10.7717/peerj.862
Ho, C.-S., Jean, N., Hogan, C. A., Blackmon, L., Jeffrey, S. S., Holodniy, M., et al. (2019). Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 10, 4927. doi:10.1038/s41467-019-12898-9
Hu, Y., Zhao, N., Gan, T., Duan, J., Yu, H. J., Meng, D., et al. (2017). Analytic method on characteristic parameters of bacteria in water by multiwavelength transmission spectroscopy. J. Spectrosc. 2017, 1–7. doi:10.1155/2017/4039048
Kuhn, W., Armstrong, D., Atteberry, S., Dewbrey, E., Smith, D., and Hooper, N. (2010). Usefulness of the Paralens™ fluorescent microscope adaptor for the identification of mycobacteria in both field and laboratory settings. open Microbiol. J. 4, 30–33. doi:10.2174/1874285801004010030
Li, G., Gao, X., Xiao, n., and Xiao, Y. (2019). Estimation soil organic matter contents with hyperspectra based on sCARS and RF algorithms. Chin. J. Luminescence 40, 1030–1039. doi:10.3788/fgxb20194008.1030
Liu, Z., Wu, D., Liu, Y., Han, Z., Lun, L., Gao, J., et al. (2019). Accuracy analyses and model comparison of machine learning adopted in building energy consumption prediction. Energy Explor. Exploitation 37, 1426–1451. doi:10.1177/0144598718822400
Mattley, Y., Leparc, G., Potter, R., and García-Rubio, L. (2000). Light scattering and absorption model for the quantitative interpretation of human blood platelet spectral data. Photochem. Photobiol. 71, 610–619. doi:10.1562/0031-8655(2000)0710610lsaamf2.0.co2
Meinhardt, P. L. (2006). Recognizing waterborne disease and the health effects of water contamination: a review of the challenges facing the medical community in the United States. J. Water Health 4, 27–34. doi:10.2166/wh.2006.0041
Menu, E., Mary, C., Toga, I., Raoult, D., Ranque, S., and Bittar, F. (2018). Evaluation of two DNA extraction methods for the PCR-based detection of eukaryotic enteric pathogens in fecal samples. BMC Res. notes 11, 206–6. doi:10.1186/s13104-018-3300-2
Nnachi, R. C., Sui, N., Ke, B., Luo, Z., Bhalla, N., He, D., et al. (2022). Biosensors for rapid detection of bacterial pathogens in water, food and environment. Environ. Int. 166, 107357. doi:10.1016/j.envint.2022.107357
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., and Chapman, J. (2019). A review of methods for the detection of pathogenic microorganisms. Analyst 144, 396–411. doi:10.1039/c8an01488d
Sakamoto, S., Putalun, W., Vimolmangkang, S., Phoolcharoen, W., Shoyama, Y., Tanaka, H., et al. (2018). Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 72, 32–42. doi:10.1007/s11418-017-1144-z
Sieuwerts, S., De Bok, F. A., Mols, E., De Vos, W. M., and van Hylckama Vlieg, J. (2008). A simple and fast method for determining colony forming units. Lett. Appl. Microbiol. 47, 275–278. doi:10.1111/j.1472-765x.2008.02417.x
Slabbinck, B., De Baets, B., Dawyndt, P., and De Vos, P. (2009). Towards large-scale FAME-based bacterial species identification using machine learning techniques. Syst. Appl. Microbiol. 32, 163–176. doi:10.1016/j.syapm.2009.01.003
Smith, J. M., Huffman, D. E., Acosta, D., Serebrennikova, Y., García-Rubio, L., and Leparc, G. F. (2012). Reagent-free bacterial identification using multivariate analysis of transmission spectra. J. Biomed. Opt. 17, 1070021–1107002. doi:10.1117/1.jbo.17.10.107002
Välimaa, A.-L., Tilsala-Timisjärvi, A., and Virtanen, E. (2015). Rapid detection and identification methods for Listeria monocytogenes in the food chain–a review. Food control. 55, 103–114. doi:10.1016/j.foodcont.2015.02.037
Wang, W., Peng, Y., and Zhang, X. (2010). Study on modeling method of total viable count of fresh pork meat based on hyperspectral imaging system. Spectrosc. Spectr. Analysis 30, 411–415. doi:10.3964/j.issn.1000-0593(2010)02-0411-05
Yang, J., Ma, X., Guan, H., Yang, C., Zhang, Y., Li, G., et al. (2023). A recognition method of corn varieties based on spectral technology and deep learning model. Infrared Phys. Technol. 128, 104533. doi:10.1016/j.infrared.2022.104533
Yuan, L.-m., Mao, F., Huang, G., Chen, X., Wu, D., Li, S., et al. (2020). Models fused with successive CARS-PLS for measurement of the soluble solids content of Chinese bayberry by vis-NIRS technology. Postharvest Biol. Technol. 169, 111308. doi:10.1016/j.postharvbio.2020.111308
Zhang, A., Yan, Z., Liu, L., and Liu, Y. (2011). Distribution of four kinds of typical enteric pathogens in different surface waters. J. Earth Environ. 2, 272–277.
Keywords: multi-wavelength transmission spectroscopy, ANNs, bacteria, CARS, species identification
Citation: Hu Y, Zhu Y, Hu D, Zhou N, Xiu L, Li W, Xie J, Zhang Y and Yan P (2024) Rapid identification of bacteria in water by multi-wavelength transmittance spectroscopy and the artificial neural network. Front. Environ. Sci. 11:1274605. doi: 10.3389/fenvs.2023.1274605
Received: 09 August 2023; Accepted: 22 December 2023;
Published: 12 January 2024.
Edited by:
Björn Vinnerås, Swedish University of Agricultural Sciences, SwedenReviewed by:
Jingjing Liu, Northeast Electric Power University, ChinaWei gao Zhao, Tianjin University, China
Copyright © 2024 Hu, Zhu, Hu, Zhou, Xiu, Li, Xie, Zhang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dun Hu, aHVkdW5AbWFpbC51c3RjLmVkdS5jbg==
†These authors share first authorship
 Yuxia Hu
Yuxia Hu Yunhao Zhu
Yunhao Zhu Dun Hu
Dun Hu Na Zhou3,4
Na Zhou3,4 Yiming Zhang
Yiming Zhang