- 1Department of Mathematics, Faculty of Mathematics and Natural Sciences, University of Bergen, Bergen, Norway
- 2Computational Biology Unit, University of Bergen, Bergen, Norway
- 3School of Physics and Astronomy, College of Engineering and Physical Science, University of Birmingham, Birmingham, United Kingdom
- 4Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, United Kingdom
- 5Centre for Computational Biology, University of Birmingham, Birmingham, United Kingdom
Bioinformatics is a highly interdisciplinary subject, with substantial and growing influence in health, environmental science and society, and is utilised by scientists from many diverse academic backgrounds. Education in bioinformatics therefore necessitates effective development of skills in interdisciplinary collaboration, communication, ethics, and critical analysis of research, in addition to practical and technical skills. Insights from bioinformatics training can additionally inform developing education in the tightly aligned and emerging disciplines of data science and artificial intelligence. Here, we describe the design, implementation, and review of a module in a UK MSc-level bioinformatics programme attempting to address these goals for diverse student cohorts. Reflecting the philosophy of the field and programme, the module content was designed either as “diversity-addressing”—working toward a common foundation of knowledge—or “diversity-exploiting”—where different student viewpoints and skills were harnessed to facilitate student research projects “greater than the sum of their parts.” For a universal introduction to technical concepts, we combined a mixed lecture/immediate computational practical approach, facilitated by virtual machines, creating an efficient technical learning environment praised in student feedback for building confidence among cohorts with diverse backgrounds. Interdisciplinary group research projects where diverse students worked on real research questions were supervised in tandem with interactive contact time covering transferable skills in collaboration and communication in diverse teams, research presentation, and ethics. Multi-faceted feedback and assessment provided a constructive alignment with real peer-reviewed bioinformatics research. We believe that the inclusion of these transferable, interdisciplinary, and critical concepts in a bioinformatics course can help produce rounded, experienced graduates, ready for the real world and with many future options in science and society. In addition, we hope to provide some ideas and resources to facilitate such inclusion.
Introduction
Bioinformatics is “one of the fastest-growing interdisciplinary sciences of [...] the early 21st century” (Ai et al., 2012). The topic is defined in various different ways, but a well-known description is: “applying informatics techniques (derived from disciplines such as applied maths, computer science, and statistics) to understand and organize the information associated with [biological] molecules” (Luscombe et al., 2001). A central theme throughout such definitions and descriptions is the interdisciplinary nature of the topic. Bioinformatics projects, like projects in broader data science fields, often involve collaborations between researchers and stakeholders from diverse scientific and societal backgrounds, necessitating training in cross-disciplinary working and a set of communication and co-operation skills in addition to technical competence.
As a modern and highly applied academic field, bioinformatics teaching and practice is also tightly connected with broader trends in biomedical and environmental science and society. The unthinking use of technology in the era of big data, data science, and artificial intelligence can lead to a plethora of scientific, societal, and ethical problems (Ioannidis, 2005). In particular, the growing presence of bioinformatics both in health technology and environmental science means that errors in analysis and critical interpretation can have profound consequences for human health and environmental policy, respectively, (Goodman and Cava, 2008; Fulekar, 2009). New and developing issues associated with the acquisition, storage, and analysis of biological samples and personal data also necessitates an introduction to the ethics of bioinformatics research (Goodman and Cava, 2008; Taneri, 2011; Amer, 2017). Responsible bioinformatics requires not just a knowledge of tools but a broader critical mindset for continual evaluation of the approaches that one takes (Pevzner and Shamir, 2009; Goodman and Dekhtyar, 2014). Furthermore, as a comparatively new field, the alignment between taught bioinformatics skills with the changes in perception of, and structure in, the graduate workforce has yet to equilibrate (Ranganathan, 2005; Hersh, 2008; Tomlinson, 2008; Mulder et al., 2018; Attwood et al., 2019). Several regions have explicitly invested broadly in developing bioinformatics skills to address this gap in the research and industrial sectors (Koch and Fuellen, 2008; Tastan Bishop et al., 2014). Importantly, bioinformatics should not be the privilege of the selected few “bioinformaticians,” but be embraced by scientists across the life sciences; thus enabling them to better use their data, or at least understand what data science can do for them. In aligning educational approaches to societal needs, a pervasive question is whether bioinformatics education involves (and should aspire to be) “educating scientists” or “training technicians” (Hack and Kendall, 2005b; Attwood et al., 2019). Clearly, bioinformatics education can involve a diverse set of challenges, from the spread of desired outcomes and corresponding curriculum contents, through the genuine risks associated with misunderstanding, to the diversity of student cohorts.
Due to the comparative youth of the discipline, pedagogy in the field is young, necessarily technology-coupled and research-focused, and debated (Pevzner and Shamir, 2009; Magana et al., 2014). As the discipline evolved from disconnected individual approaches to its own field, the formalisation of bioinformatics teaching followed Altman (Altman, 1998) who called for a set of topics for the “ideal” bioinformatics graduate program. Even this early set of topics suggested pedagogical challenges, including a disparate range of subjects and having to cater for students from a range of backgrounds and with diverse skill sets, from biologists to computer scientists. Since then, the field and its associated pedagogical literature has developed quickly. Curriculum proposals have expanded and diversified, remaining debated (Koch and Fuellen, 2008; Pevzner and Shamir, 2009; Magana et al., 2014; Tastan Bishop et al., 2014) but increasingly informed by surveys of perceived knowledge and skill gaps in academia and industry (Mulder et al., 2018; Attwood et al., 2019). The “shrinking half-life of knowledge” (Gonzalez, 2004) is noticeable every day in the discipline. A striking example of this during the design of the course was in a module lead discussion about recommended reading for students, where the difficulty of constructing a reading list was noted, given that specific methodological details in books run the risk of becoming outdated over the timescale of publication. Indeed, Magana et al. (2014) proposed that these rapid technical advances should be quickly integrated into education and training, to ensure that students emerge “ready to take jobs and conduct research in these emerging fields.”
This evolution of the discipline from the ad hoc application of disconnected approaches to its own recognised discipline has been mirrored in the evolution of bioinformatics teaching in higher education institutions. As Furge et al. (2009) discuss, and Magana et al. (2014) survey, bioinformatics teaching at an institution typically follows one or both of two paradigms: (i) decentralised bioinformatics content in courses contained primarily within other topics; and (ii) centralised courses with a largely bioinformatics focus. The fast development of the field, and its rapid inclusion into many different biological topics, raises questions about how best to include bioinformatics into existing teaching (Cummings and Temple, 2010) and whether vertical or horizontal integration paradigms should be followed (Furge et al., 2009). Debate on good practice in bioinformatics teaching continues to spark new initiatives and proposed “best practices” (Cattley and Arthur, 2007; Schneider et al., 2011; Welch et al., 2014). Of particular note given the aforementioned issues in bioinformatics applications was a striking absence of ethics content in bioinformatics curricula (Taneri, 2011).
In tandem, bioinformatics as a general discipline (and the related and emerging field of data science) aligns tightly with the connectivist paradigm proposed by (Siemens, 2005). Problems in bioinformatics are often best solved through international, technology-assisted connectivism: the use of community messageboard resources; distributed and open-access software; and the global distribution of source data itself. Correspondingly and almost tautologically, computer-based learning is generally deemed essential in bioinformatics education (Cummings and Temple, 2010). Siemens (2005) highlights a role for “diverse teams of varying viewpoints [as] a critical structure for completely exploring ideas,” aligning with team and active learning and making a strength of diversity in the classroom.
Finally, bioinformatics not only provides excellent opportunities for research-led teaching (St Clair and Visick, 2010; Brown, 2016), but indeed is hard to imagine being taught in any other way (Goodman and Dekhtyar, 2014). Challenge-based learning, including “student-centred approaches such as inquiry learning and collaborative learning” was identified as the most common mode of bioinformatics education in a survey of approaches (Magana et al., 2014). Engagement with open research questions can demonstrate valuable cutting-edge concepts to students as well as serving to motivate and justify the reason for learning about these topics and exposing them to a realistic representation of the field beyond education.
These ideas can be synthesised and distilled to identify a set of positive principles for bioinformatics education (and broader data science education), although the weighting of these principles varies across studies. These general positive principles emphasise problem-based learning and self-learning (Ranganathan, 2005; St Clair and Visick, 2010) and often adopt a jointly cognitivist and constructivist strategy with (i) an intensive introduction to the range of necessary concepts and (ii) subsequent focus on a subset of skills motivated by a specific problem (St Clair and Visick, 2010; Goodman and Dekhtyar, 2014), following a constructive alignment approach (Biggs, 1996). The goal of the project outlined in this report was to realise this paradigm while supporting student development of ethics, critical analysis, and transferable skills, thereby furnishing graduates with technical and transferable skills that increase their diversity of options in the workplace and in society (Mulder et al., 2018; Attwood et al., 2019). We hope here to provide insights, references, and resources that may be of value in the design of other bioinformatics and broader courses aiming for this goal in the future.
Interdisciplinary and Transferable Skills in a Bioinformatics Course: Curriculum Design and Review
The module described in this article is titled “Interdisciplinary Group Project in Bioinformatics,” and is a 20-credit part of a recently-designed UK MSc Bioinformatics course (UK FHEQ Level 7). This course takes graduates from a range of topics (computer science through biology), with a 2:1 entry requirement. At the time of writing, cohort sizes were in the range of 10–30 students with an even gender balance (48% female), and a geographical spread of 62% from the UK, 27% from other European countries, and others from the USA, Middle Eastern, and Asian regions (the latter proportion has increased since the time of writing). The other modules are Essentials of Mathematics and Statistics (20 credits —where core mathematical and statistical concepts are introduced, and computational approaches are developed further); Genomics & Next Generation Sequencing (20 credits); Data Analytics & Statistical Machine Learning (20 credits); Metabolomics and advanced (omics) technologies (20 credits); Computational Biology for Complex Systems (20 credits); and an Individual Research Project (60 credits). The details of specific bioinformatic tools are presented in these other modules.
The interdisciplinary module was designed around the paradigm of constructive alignment (Biggs, 1996), with the essential aspects of the above section which were desired to be captured:
• Progression through a taxonomy of learning (Krathwohl, 2002), from knowledge of tools to their application and critical evaluation;
• Introduction to a research environment;
• Group work facilitating peer-assisted, active learning (McKinney, 2010) and necessitating the development of co-operation and communication across a range of backgrounds (Pevzner and Shamir, 2009);
• Development of skills that are valuable outside the specific academic field of bioinformatics.
We considered the delivery of a genuine bioinformatic research project by an interdisciplinary group of students as the ideal “desired result” that would provide evidence of these targets (Wiggins et al., 2005; Magana et al., 2014). To this end, a format was decided where students work in groups of 2–4 to address an open-ended research question under the supervision of an academic at the institution. This reflects two principles for the promotion of learning from Merrill (2002) highlighted for bioinformatics by Cummings and Temple (2010): learners are “engaged in solving real-world problems” and “new knowledge is applied by the learners.” Taught, discussion, and practical exercise content is presented in tandem, involving introductory technical skills and transferable concepts including how to prepare scientific publications, posters, and presentations, as well as on the ethics requirements of research. The formal module outcomes are given in Table 1 and linked to the general QAA principles for an FHEQ level 7 course and for biosciences qualifications in Appendix A (Hack and Kendall, 2005a).
In the design of this module, the ideas from the introduction above have naturally crystallised into a multi-faceted strategy. There is a role for a limited cognitivist paradigm in those situations where a correct answer does exist (for example, choosing an appropriate statistical test or a particular piece of software) which can be used to build a consistent knowledge platform across a diverse class. We can then naturally build upon this knowledge foundation to progress chronologically through Bloom's taxonomy of learning (Bloom et al., 1956; Krathwohl, 2002). Through introductory technical lectures, students acquire knowledge (specific tools, statistical tests, and so on, building a consistent platform across the diverse student backgrounds in the class), and comprehension (how these specific processes relate to each other and to scientific questions). Motivated by an open research question (Cummings and Temple, 2010; Brown, 2016), and working in diverse teams (Siemens, 2005), the students will develop, apply, synthesise, and critique their own bioinformatics approaches (Krathwohl, 2002) through a jointly social constructivist and connectivist strategy (Kim, 2001; Siemens, 2005). They apply these technical concepts in parallel with the interdisciplinary skills in collaboration, communication, and ethics that are developed through the module as they design, implement, critique, and present a real group research project (St Clair and Visick, 2010). The social constructivism inherent in the group project structure (Kim, 2001) is harmonious with the targets of developing communication and teamwork skills, and with learning via “belonging to something” (Hodkinson et al., 2004) via shared problem-solving (Collin and Valleala, 2005).
The students must demonstrate extensive and critical knowledge of ideas in the field in order to design their strategy to address the research question, which is, by construction, an original application of this knowledge. In addition, they must creatively deal with typical issues that arise from a real research project such as absence of anticipated data, computational-logistical issues and teams shrinking as students left the course.
The module is geared toward goals of developing self-direction and acquisition of transferable skills including ethics and communication, supported by peers and supervisors while developing an individual trajectory. The module is reviewed every year with module leads, student representatives, and an external examiner. In harmony with positive feedback from students and the external examiner, the outcomes of this review so far have been to keep the learning outcomes the same while redesigning some timetabling aspects to give students more time to independently absorb ideas (Appendix B.II). Future monitoring of the evolving literature in the field and utilisation of instructor and peer experience on the role of the subject in society allows ongoing reflection on this design.
Interdisciplinary and Transferable Skills in a Bioinformatics Course: Module Content
A consistent challenge in bioinformatics teaching is the diversity of student cohorts and the corresponding technical backgrounds. This module aimed to address this challenge with a dual approach. First, a “diversity-addressing,” non-examinable set of foundation lectures and exercises ensuring that important core concepts from both biology and computing, as well as important transferable and interdisciplinary skills, were met and engaged with by all students. Second, a “diversity-exploiting” strategy where the strengths of students' diverse backgrounds were brought together to be greater than the sum of their parts.
We will outline the component parts of the module here, with further details of and reflection on specific content and references in Appendix C, and some links to online resources in Appendix D.
Introductory Philosophy: Information and Life
The first “diversity-addressing” foundational content required non-biologists to meet core biological concepts and biologists to meet key informational concepts. This was addressed by first providing a set of basic biology lectures with an “information and life” flavour, and next providing an entry-level set of coupled lectures and exercises on core technologies. The learning objectives for this section were simply for students to be exposed to an information-centric picture of biology, and to the core types of information in molecular biology and our ways of measuring them. The topics and discussion points covered are described in Appendix C.
Teaching Computational Core Concepts With Virtual Machines
The first technical aspect of this module is an initial intensive introduction to various central software approaches in bioinformatics. The learning objectives for this section were a basic familiarity with command-line tools for operating systems usage, data curation, and organisation; the fundamentals of programming in Python including loops, conditionals, and functions; fundamentals and data structures in R; and a primer on version control with Git. Based on previous experience and positive feedback from students, and following the paradigm of active learning (McKinney, 2010), we employed intrinsically mixed lecture delivery and computational practical classes. Here, students are introduced to a set of computational ideas (for example, the structure of a file system, or a particular set of commands) and then immediately perform a task involving those ideas on their laptops. The idea is to couple the theory with an immediate practical application, thus reinforcing the concept in student's minds (Yazedjian and Kolkhorst, 2007) and giving them first-hand constructive experience of a genuine application. Here, both an instructor and peers are on hand to help with troubleshooting, breaking down barriers of anonymity (Michaelsen et al., 2002) and facilitating peer-assisted learning (Johnston, 1997).
An issue in previous experiences with this mode of working was inconsistency: what worked on one student's computer might not work on another; some may use Macs and others Windows machines; and so on. As Dahlö et al. (2015) point out, “if a student tries to run a command from the instructions and it results in an error message, it can be hard for a beginner to figure out what went wrong... every problem they encounter with the operating system steals focus from the subject being taught.” This is perhaps not the only issue; it can be highly demoralising for students who, faced with an unfamiliar interface or application, fail to get something working for technical reasons. This issue is amplified by the diverse student backgrounds in a bioinformatics class with a range of computational abilities and experience. This was anecdotally illustrated to one of us (IGJ) in a separate, undergraduate computational biology practical session: students faced with between-computer inconsistency were both frustrated and disillusioned about the “rigour” of the topic (and of the instructors).
To pre-empt this issue, we use a “virtual machine” (VM) environment. VMs are simulations of a complete computer system that can be constructed exactly as the instructors require and are accessed remotely by the students using their laptops (Figure 1). We therefore ensure that each student sees the exact same computational platform, and prior to the class ensure that all commands and approaches involved will function as expected. The instructor's own behaviour on the same VM environment is projected to the class. Taught content in this environment included an introduction to the command line, programming in Python and R, and version control using Git (details in Appendix C).
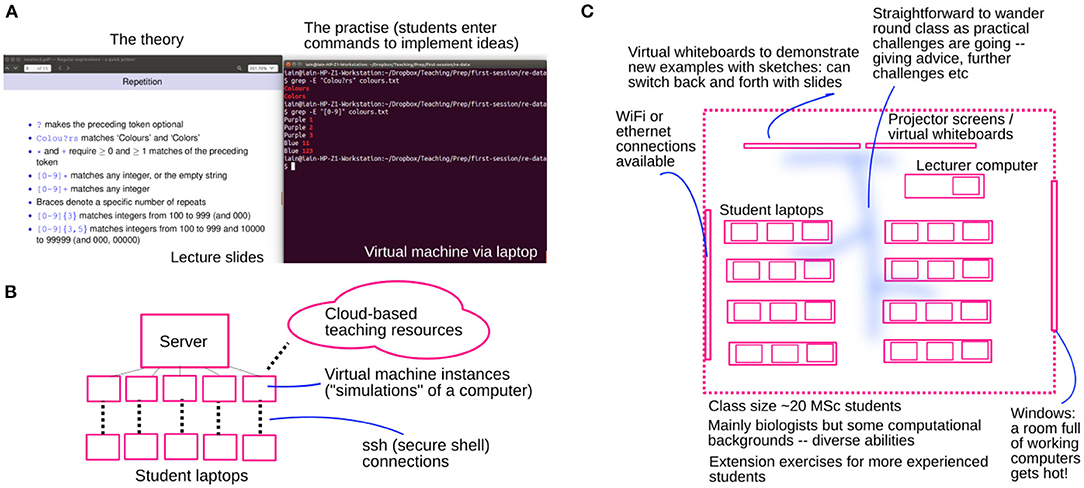
Figure 1. Physical and computational structure of the virtual machine learning space. The specific VM environment was part of a wider infrastructure in total comprising 48 nodes with between 20 and 40 cores and 128+GB RAM each. The VMs presented to the students had 2 virtual CPUs and 14GB RAM. Secure Shell (SSH) connections (via Putty on Windows or the terminal on Mac/Linux machines) with graphics forwarding enabled was used to connect to these VMs, which presented students with a Bash command line interface through which Bash commands, Python code, R code, and other software could be run. (A) Students's eye view. (B) Virtual teaching space. (C) Physical teaching space.
Experience with the joint student-instructor exploration of topics with the VM teaching environment was very positive. The infrastructure needed to access VMs took some time to install, but the students were informed of this before the session, more staff were present to help, and a light-hearted environment prevailed through this short technical phase. Subsequently, content delivery was smoother and completely consistent between teacher and student (and between students), increasing accessibility across the diverse student cohort. Students readily helped each other out with specific commands in the uniform environment; individual and group exercises progressed very smoothly. The environment was explicitly praised in student feedback, with positive reception of “activities in the [programming] sessions to reinforce our learning” as “a good way to understand what I have learnt.” Particularly encouraging was praise for the approach which “really helped build confidence” and “meant students from a non-computing background could follow along without feeling too lost” (Appendix B.I). Throughout, the instructor was present in the classroom to observe working dynamics; we found generally that students naturally allocated work evenly between partners and readily cooperated, and regularly mixed group structures to ensure no particular imbalances were “frozen” throughout the sessions. Few issues occurred when students later migrated to work on their own local machines, likely because of the consistency of the development environments and interfaces with which they were presented; some indeed chose to continue with the VM environment for future work.
Interestingly, while VMs are used in computer science education (Lathan et al., 2002), there seems to be limited literature examining their use in bioinformatics teaching (Afgan et al., 2015; Dahlö et al., 2015). The VM environment has several potentially powerful strengths and extensions, one of which is its natural provision of data for learning analytics. Commands that students enter in the VM are stored and can be retrieved: these data can be used to explore, for example, which commands students experience the most trouble with, and the amount of time students spend on different exercises, so that attention can be focused on more problematic areas.
Interdisciplinary and Transferable Skills in Bioinformatics
The learning outcomes in Table 1 require several skills that are naturally “transferable” in nature (although take some specific forms in the field of bioinformatics). These include (i) the ability to work effectively in an interdisciplinary team; (ii) to present the results of the project in written and oral form; and (iii) to demonstrate critical awareness of the implications of research in biology-related fields, including an understanding of ethics.
To address these goals, we held an ongoing series of seminars and exercises on the topics of: (i) structuring a scientific investigation, logistics of bioinformatic investigation, and communication in interdisciplinary groups; (ii) scientific writing and delivering research; and (iii) scientific methods and philosophies, bioethics, and ethics in bioinformatics. These topics (detailed below) were presented in a series of 2-h joint lectures and exercise classes, as with the technical content delivered previously.
Structuring and Logistics of Investigation, and Scientific Communication
The introduction to structuring an investigation was intended to give students a grounding in some recommended steps (and a possible ordering of those steps) for a successful research project, prior to their embarking on the group project itself. These steps were presented as “phases”: specifically, introductory, exploratory, development, science, analysis, delivery. Flexibility in this ordering—including loops—was explicitly supported. We particularly focused on introductory (literature searching, discussion, crystallisation of research questions) and exploratory (here, data curation and exploration) phases as important for new researchers to take time over. A real project from one of us (IGJ) was presented to the students to illustrate the many backs-and-forth between these phases that research can take, and to reinforce that backtracking, abandoning research paths, and diverging from the original plan are natural dynamics in research (subset shown in Figure 2A). General principles—realistic milestones, codifying and updating goals, keeping colleagues and PIs up to date—were also highlighted.
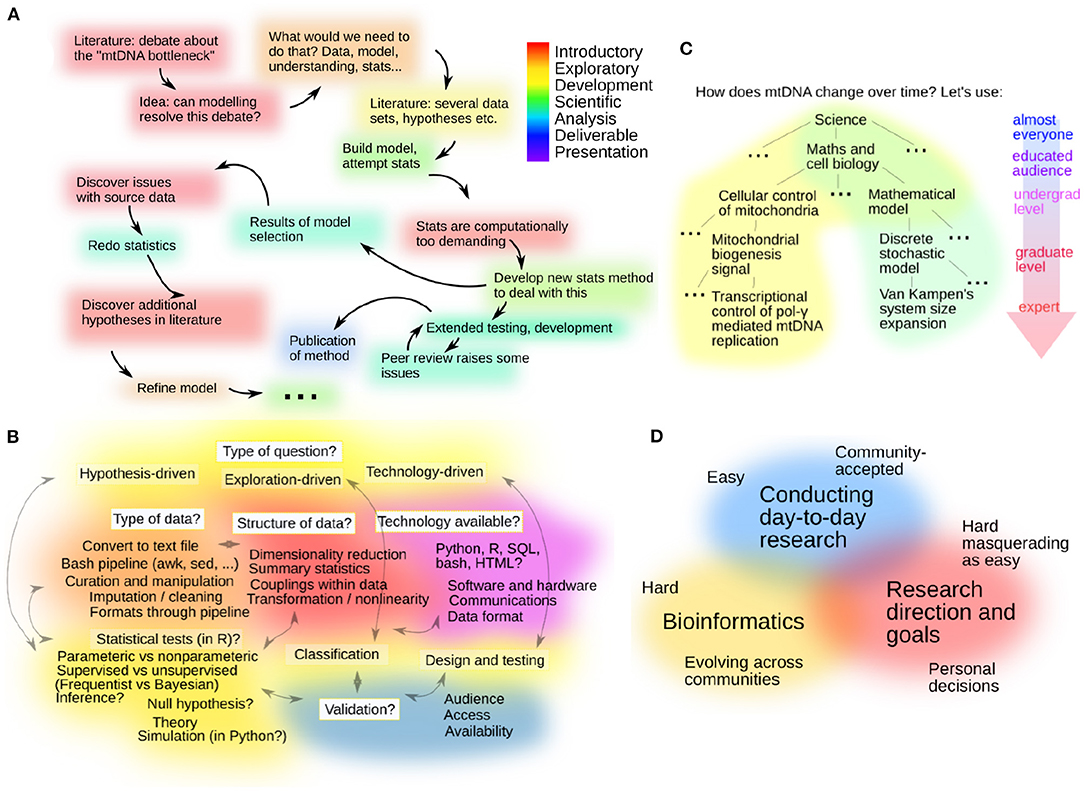
Figure 2. Illustrative concepts in interdisciplinary and transferable topics. (A) The illustrative evolution of a real research project, showing branching and backstepping between ideas, emphasising to students the nonlinearity of research and the imperfect state of knowledge at different points. Colours broadly refer to the different labelled phases of investigation. (B) Putative “decision bush” for logistics of a bioinformatic investigation, illustrating some different types of research projects, the (coarse-grained, and sometimes overlapping) general approaches that can be used in different cases, and some questions that can be used to choose a direction for a particular project. (C) A “phylogeny of jargon” for interdisciplinary communication in bioinformatics groups, with the “comfort zones” of two different researchers and their intersection for a specific subset of topics. (D) A fuzzy Venn diagram of ethical perspectives between general “day-to-day” science (where principles of core good practice are generally accepted), specific bioinformatics practice (where principles of good practice are evolving and new issues are arising), and individual researcher priorities (where personal ethics require consideration).
In the subsequent “logistics” discussion, we focused on “what is the question?”, “what data is available?”, and “what tools and technologies can we use?”. In particular we highlighted that a bioinformatics question can be scientific (“does gene expression change in disease?”), exploratory (“what happens after exercise?”), or technological (“can we build a tool to interrogate and visualise this dataset?”). Students were encouraged to contribute thoughts on choosing bioinformatics pipelines as well as statistical and visualisation approaches appropriate for the combination of question and data (hypothesis-driven vs. exploratory, categorical/continuous/high-dimensional data, and so on). In particular, the philosophical difference between hypothesis testing and exploratory, hypothesis-free research (as is often the case in bioinformatics, where data-driven exploration is used as a process for hypothesis generation) was highlighted. A “decision bush” (Figure 2B) was suggested as an amalgamation of these overlapping routes. We also focused on the message that results should be robust with respect to choices made in the analysis pipeline (e.g., particular parameters, thresholds, etc., should give the same qualitative results).
Bridging these topics in research design and the students' actual group projects, we next discussed communication in and to interdisciplinary groups. Students were encouraged through lectures and exercises to consider a “phylogeny” of technical language, starting with very basic “ancestral” terms like “life” and “science” and progressing, as a participant's specialisation increases, to more technical terms “gene,” “stochastic model,” and so on (Figure 2C). Students were encouraged to identify the ‘common ancestor' terms for a particular topic with a given imaginary conversational partner (a schoolchild, a physicist, an experimental collaborator) and build terminology from there. We also discussed “unspoken” conventions in different disciplines that may be graphical (for example, the box plots and significance stars that are ubiquitous in biology but rare in physics), verbal (different abbreviations), quantitative (different interpretations of statistical significance, p < 0.05 in biology vs. roughly p < 3 × 10−7 in physics—the 5σ threshold for “discovery.”) Differences in scientific philosophy (heavily hypothesis-focused in biology, exploratory model-based in physics, data-driven exploration in bioinformatics) and career priorities (publication strategy and emphasis of bibliometrics including impact factor, authorship conventions, and expected grant incomes were also mentioned as potential sources of tension in interdisciplinary groups. These discussions drew on the instructor's experience of working in different academic fields and different departments (specifically physics, maths, and biology departments).
To develop skills in research communication, the students took part in an interactive scientific writing exercise hosted by institutional experts, with followup class discussion on the basics of scientific publication, posters, and talks. Students were asked to name journals that they had come across; this growing list was augmented with some of the instructor's suggestions in both interdisciplinary science and the bioinformatics field. Bibliometrics, audiences, publication costs, and other issues in choosing a journal were discussed. Mensh and Kording's “Ten Simple Rules for Structuring Papers” in PLoS Computational Biology (Mensh and Kording, 2017) were introduced, leading to ideas for structuring striking research posters, and general principles for giving a good scientific talk following Alon's brief but highly targeted paper (Alon, 2009). Issues in scientific publication, including preprints, open access, double-blind review, and gender bias (Borsuk et al., 2009) were also briefly presented.
Scientific Methods and Philosophies, and Ethics in Bioinformatics
Linking with the previous mentions of different philosophies of science in different disciplines, a brief overview of general scientific philosophy was discussed with students. This included a historical perspective illustrated through evolving theories of astronomy, a discussion of deductive, inductive, and abductive reasoning, and Kuhn's structure of scientific revolutions (Kuhn, 2012). The discussion then turned to why these ideas are important for bioinformatics. As a comparatively new field, bioinformatics can be viewed as a (gentle) revolution, where new paradigms of interdisciplinarity, computational focus, and hypothesis-free investigation are reshaping traditional modes of biological investigation (Hack and Kendall, 2005a). The risk of overemphasis on buzzwords and newer, less interpretable ways of working (as with any exciting new discipline) was discussed, with some focus on “mistakes from early Genome-Wide Association Studies (GWAS)” (Lambert and Black, 2012) and highlighting the need for critical analysis of pipelines and published results. It was striking how many students considered the appearance of work in a peer-reviewed journal as a signature of “gospel truth;” the importance of critically assessing published methodologies and interpretations was emphasised through several case studies. These included the dramatic observed differences between lab animals and their wild relatives (Abolins et al., 2017); the fact that mice respond differently to male and female human researchers (Sorge et al., 2014); the well-known critique of statistical approaches in biomedical sciences suggesting that “most published research is false” (Ioannidis, 2005); and the reproducibility crisis across scientific disciplines (Collaboration et al., 2015) including drug discovery for cancer, where only 11% of pre-clinical research results were found to be reproducible (Begley and Ellis, 2012), and Phase II success rates had fallen to 18% in 2011 (Prinz et al., 2011).
This coverage of issues in scientific publishing and practice led naturally to a discussion of ethics in bioinformatics research, framed initially via the broad categories in Figure 2D. The students were asked to construct a “code of practice”—a set of tenets that they considered to describe ethically sound research. These points were then compared with the instructor's set of ideas. Frequently there was substantial overlap, with students demonstrating an awareness of plagiarism, forgery, and transparency. Topics highlighted by the instructor that were less commonly noted by students included personal aggression and discrimination in research interactions, distinguishing evidence from speculation, and appropriate assignment of credit via citations or other referencing. The subsequent taught content is detailed in Appendix C, and included the role and funding of research in society, “doing good,” acquisition and use of data in bioinformatics, international and cross-cultural aspects of bioinformatics data and results, and benefit sharing.
Previous studies have highlighted a surprising absence of ethical topics in bioinformatics courses (Taneri, 2011), and given the comparative youth of the field overall, and the ongoing debate about curriculum content, it is perhaps not surprising that a consensus set of central topics has yet to be decided (Amer, 2017). We propose the above topics – general bioethics; the role of research in society; the risk of biological misclassifications; the global sampling and benefits of bioinformatic research; and the use of personal data, not as a fixed set of recommended discussion points but as an illustrative sub-curriculum to get students thinking, discussing, and critiquing some relevant and important issues in the developing field.
Interdisciplinary Group Projects
During the summer prior to the course start in September, academic staff in the natural, sports, and medical sciences divisions were contacted with a soliciting email offering the opportunity to propose bioinformatics projects. The requested response was a detailed abstract for a project outlining the research question and data to be used, as well as some key references for interested students. The suggested format was weekly supervisory meeting with the project's principal investigator (PI) from October to March. Projects were vetted by the module lead for bioinformatics alignment (though no projects to date have been proposed that were unsuitable) and compiled into a booklet. Students were presented with this booklet and asked to rank their top five choices by order of preference. Students were then allocated projects based on their disciplinary and geographical background, their propensity for active participation in class discussion and Q&A, and their preferred rankings (see below).
The goal in student assignment was to expose students to a diversity of backgrounds and personalities within a group. Diversity of backgrounds was typically easy to achieve; groupings mixed biologists and non-biologists where possible, and biologists of different sub-disciplines where not. UK/non-UK and European/non-European undergraduate locations were also mixed.
Grouping personalities was more challenging. The instructor at this stage in the course had only had limited (though intensive) exposure to the students. At such early stages of the programme we deemed it desirable to have “some but not too much” difference between the members of a group. Some diversity had many advantages: requiring the students to put into practice the module content on working in interdisciplinary groups; reflecting an accurate picture of a real bioinformatics collaboration (Biggs, 1996; Wiggins et al., 2005; Cummings and Temple, 2010); working with different priorities, personalities, and team roles (Belbin, 2012; Salazar et al., 2012); and requiring compromise between different parties' interests and goals. However, we strove to avoid situations where, for example, one domineering personality was paired with several less bombastic partners. While such situations certainly exist in the broader world, we believed that this would prevent all students getting the broadest set of experiences from the group project.
Following existing approaches to explore and develop bioinformatics skills with projects (St Clair and Visick, 2010), the idea here was to expose students to open-ended research questions that were not pre-choreographed, with the associated requirements of project development, management, critical analysis, and overcoming unforeseen hurdles. Projects run in this module included: learning pathways of disease progression for precision medicine; predicting responses to endurance-exercise training; control and function of beta cells; constructing an “atlas of the phosphatasome;” using ecological and evolutionary dynamics to forecast responses to global change; detour of proteins in cancer; “metabolism across space and time” and its interpretation; horizontal gene transfer in bacteria; and “moving toward predictive evolution.” These ranged from more classical bioinformatics (RNA-seq data, enrichment analysis, gene ontologies, pathways) to more unusual goals such as constructing a web resource for a class of proteins (the phosphatasome atlas above). This latter project was particularly interesting: the small group consisted of one student from a computer science background and one from a biology background, leading to mutual reinforcement and cross-fertilisation of ideas, and leading to an output deliverable (a fully functional database and rich graphical web interface populated with biological instances) rather greater than the sum of its parts. This project also drew particularly positive peer responses, with other students explicitly noting the benefits of the joint expertise of the researchers.
These projects took place while students were also attending taught content on different topics (modules above). This led to time management issues for some groups, particularly at times where these other topics were being assessed. Two projects involved, to different degrees, an anticipated issue: heterogeneity in workload (or perceived workload) within the group. Some students raised concerns that their colleagues were not sufficiently contributing to the project. The concern was less often around how much research could be achieved and more often about how group marking would reward these colleagues. As discussed below, ongoing review of the assessment structure for this module will seek to address this.
Interdisciplinary and Transferable Skills in a Bioinformatics Course: Assessment Design and Review
Several targets were borne in mind when designing and reviewing the assessment structure and its alignment with our goals and learning outcomes. Vitally, and given that “assessment is the curriculum as far as students are concerned” (Ramsden, 2003), we wished to “support worthwhile learning” (Gibbs and Simpson, 2005). The learning outcomes for this module (Table 1), as discussed above, emphasise teamwork, communication, and transferable skills in addition to the technical completion of a research project. Based on our perceptions of the wider role of bioinformatics and its associated skill set in society, we wanted this worthwhile learning to reflect what graduates may require in the workplace as well as in academic settings. We wished to ensure that teamwork was rewarded while also allowing individual flair, and to adequately reward different strengths in a diverse cohort. This approach is inspired by the constructive alignment paradigm (Biggs, 1996) of “testing bioinformatics by doing bioinformatics”—and “doing bioinformatics” necessarily involves a combination of transferable and technical skills (Pevzner and Shamir, 2009; Goodman and Dekhtyar, 2014).
Two particularly pertinent points from Gibbs and Simpson (2005) naturally crystallise a reflection on assessment, with our priorities and responses outlined in Figure 3. We designed this strategy both according to constructive alignment principles and to reflect employment—delivering a project as a team, with a mode of individual assessment, and with skills in research and communication (both to peers and lay audiences) jointly tested. Regarding feedback as an integral part of the learning process (Cramp, 2011), this strategy helps guide the students' work and develops their experience of a research environment (Evans, 2013). Following the “feed-forward” and “feed-up” strategy (Hounsell et al., 2008), the feedback on their oral presentation and write-ups also prepares them for similar assessment of an individual project later in the course, and is envisaged to help them refine their communication skills more generally for work in academia and broader sectors.
Campbell and Nehm (2013) draw attention to the lack of evidence of reliable, validated assessment in a survey of bioinformatics courses. We ensured that independent assessment of each of these tasks was performed by two experienced and non-collaborating academics, and overseen by an external examiner; we attempted to follow the peer-review model as closely as possible for the write-up assessment while also grading according to defined marking criteria which were visible to the students throughout the project. The two examining academics were pedagogically-trained bioinformatics experts from within the institution with whom the course outline, philosophy, and objectives had previous been discussed. In particular, the model of an objective peer review process was highlighted. We encouraged examiners to note particular specific points for improvement in oral work, and to detail technical issues with the research itself, to ensure that assessment points could be translated into actionable tasks for improvement. Examining all presentations required several days of work; for larger cohorts, the required resource will be higher, and more examiners and/or more dedicated time will be required. Further validation of the assessment structure here will be an important part of future course development. Reflections on the performance of students in this assessment structure are given in Appendix E.
We have received positive feedback both from students and our external examiner on the multi-faceted nature of these assessment principles, suggesting that the this core assessment structure can be retained into the future. However, following the concerns of unequal contributions above, there clearly exists a tension between the pros of representing a genuine group work experience (which always has the risk of unequal contributions) and the cons of perceived unfairness in the resulting assessment. We are exploring suggestions from colleagues and our external examiner about making the structure more robust to situations like student dropouts or “defectors” in group work.
Conclusions and Reflection Points
In the design and maintenance of a bioinformatics course, both general and discipline-specific pedagogical principles can be leveraged. General principles include the rational design of classes, modules, and environments to follow a principled taxonomy of learning (Krathwohl, 2002), assessment structures that “support worthwhile learning” (Gibbs and Simpson, 2005), and consideration of humanist aspects of teaching style (Rogers et al., 2013)—latter perhaps being particularly important in a largely connectivist topic (Siemens, 2005) where a social constructivist paradigm can develop and reinforce collaborative skills and make deliverables greater than the sum of their parts (Kim, 2001). Discipline-specific debate on pedagogical ideas and technology in bioinformatics is somewhat more recent but provides valuable overall “best practice” ideas (Saravanan and Shanmughavel, 2007; Pevzner and Shamir, 2009; Gnimpieba et al., 2013). Ongoing monitoring of the societal priorities for technical and scientific skills in bioinformatics, by region and discipline (Hersh, 2008; Koch and Fuellen, 2008; Tastan Bishop et al., 2014), is also valuable in the design and continuing review of learning outcomes that will benefit students not just in the field of bioinformatics but as members of and contributors to broader society.
Siemens (2005) challenges us: “what adjustments need to made with learning theories when technology performs many of the cognitive operations previously performed by learners?”. Particularly in the connectivist world of bioinformatics, it can potentially be easy to lose a human connection with a class. Indeed, if all knowledge is available via online sources like MOOCs (Ding et al., 2014), what sort of teacher-student relationship is appropriate or needed? Answers to this question are fluid and evolving. Our own experience through this course suggested that the presence of the instructor in the classroom leads to more rapid and tailored responses to technical questions; improving access across a diverse student group; more opportunities to start and catalyse discussions in social learning (Hodkinson and Hodkinson, 2004); a valuable humanist connection with the class (Rogers et al., 2013) when discussing transferable, teamwork, and communication skills, and particularly ethical topics; and the benefits of personal experience in the field. This picture is supported in part by student responses which praised instructor “helpfulness” and particularly accessibility (Appendix B). However, development of bioinformatics teaching must necessitate continual reflection on this question.
Student feedback (Appendix B) has been positive about the aspects of the module discussed above, with some particularly positive notes about increasing accessibility to, and confidence in, a diverse cohort (some examples of comments received were: “really helped build confidence,” “meant students from a non-computing background could follow along without feeling too lost.”) The merit of “Learning by doing” was also explicitly praised. Positive responses for teaching delivery included finding “lecturers to be very helpful and willing to give breaks every 45 min with a chance to ask questions—very helpful,” “Very enthusiastic and knowledgeable,” “Engaging at all times,” and the informal but pleasing “Mr Johnston was very useful and with good vibes.” The intensive nature of information delivery is reflected in “The speed of speaker is a little bit quickly [sic] for me to follow,” suggesting continuing work on this trade-off between enthusiasm and clarity is desirable. These themes begin to answer the earlier question about the merit of the teacher-student relationship in a connectivist topic (Siemens, 2005). Going forward, we are reviewing structural suggestions that students proposed to facilitate smoother learning and will continue to assess and reflect as the course goes on.
Encouragingly, the inclusion of interdisciplinary concepts in bioinformatics teaching was overwhelmingly positively received by students. Responses to “Was the content of the lectures and practicals...” involved 13 “highly relevant” (the highest response), 1 “quite useful” (second highest response), 1 split between these two responses, and none for “limited relevance” or “useless.” Teaching style was also received positively, in addition to the qualitative points above, with 9 “excellent” and 6 “good” responses and no lower scores. We believe that this positive feedback and the successes of the student collaborations via group project work supports our goal of creating and supporting an environment where students learn the value of interdisciplinary collaboration and acquire skills and experience to implement this way of work. Students have gone on to a diverse range of later employment, mirroring the broad need for bioinformaticians and data scientists in a broad range of fields including a range of “big” established as well as small to medium sized enterprises in pharmaceuticals, the food and chemical industry, health, and more. Course alumni have gone on to work in the NHS (reflecting an ongoing transformation into the area of genomics and big data), genetics labs and commercial biotechnology companies, within the UK and internationally. Several alumni are working in UK and international forensics, where new genetic testing and data analysis is increasingly demanding bioinformatics skills. More alumni are now in PhD programmes where bioinformatics, interdisciplinary research skills, and experience with real-world research are in very high demand—destinations include several Russell Grou publishing.
Going forward, we intend to provide increasing numbers of projects with external partners, including industries and institutions. While this expansion will come with increased challenges of monitoring projects, it will progress even further toward the constructive alignment principle of teaching bioinformatics by doing (real-world) bioinformatics in interdisciplinary settings (Biggs, 1996; Pevzner and Shamir, 2009; Goodman and Dekhtyar, 2014). Connections and experience with industry and other institutions will also provide useful links for potential student destinations after the course (Mulder et al., 2018; Attwood et al., 2019). Future axes of expansion of the course include geography (expanding to international campuses), content (including reviewing integration with an overlapping MSc Health Data Science) and format (including expansion into distance learning and apprenticeship models). We also plan to continually review the Python and R training aspects of this initial module with respect to ongoing debate on introductory coding teaching. Further parallel course design will development and implement an equivalent module to that outlined here in these more diverse settings.
To conclude, we hope that this report has served to argue the value of including interdisciplinary and transferable concepts in bioinformatics education, and indeed that the field can provide excellent opportunities to develop research-led, group learning (Goodman and Dekhtyar, 2014; Magana et al., 2014) and expansive discussion on important ethical issues in bioinformatics (Taneri, 2011). The diversity of bioinformatics student cohorts can pose challenges (which we can combat through “diversity-addressing” approaches) but also present opportunities for constructive learning and group research (which we can harness using “diversity-exploiting” approaches) that prepares students for a wide range of future activities in society.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
This study has received full ethical approval from the Science, Technology, Engineering, and Mathematics Ethical Review Committee at the University of Birmingham, application number ERN_20-0228.
Author Contributions
All authors contributed to curriculum design. J-BC and IJ designed assessment. IJ drafted the manuscript. All authors edited and wrote the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
IJ is grateful to the Educational Development team at the University of Birmingham for their support in preparing this report. The authors thank the module leads for supporting the development and delivery of this course. The current details of the course can be found on the webpage here https://www.birmingham.ac.uk/postgraduate/courses/taught/med/bioinformatics.aspx.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2022.826951/full#supplementary-material
References
Abolins, S., King, E. C., Lazarou, L., Weldon, L., Hughes, L., Drescher, P., et al. (2017). The comparative immunology of wild and laboratory mice, mus musculus domesticus. Nat. Commun. 8:14811. doi: 10.1038/ncomms14811
Afgan, E., Sloggett, C., Goonasekera, N., Makunin, I., Benson, D., Crowe, M., et al. (2015). Genomics virtual laboratory: a practical bioinformatics workbench for the cloud. PLoS ONE 10:e0140829. doi: 10.1371/journal.pone.0140829
Ai, Y.-C., Firth, N., and Jermiin, L. (2012). Teaching bioinformatics: a student-centred and problem based approach. Int. J. Innov. Sci. Math. Educ. (formerly CAL-Laborate Int.) 10.
Altman, R. B. (1998). A curriculum for bioinformatics: the time is ripe. Bioinformatics (Oxford, England) 14, 549–550. doi: 10.1093/bioinformatics/14.7.549
Amer, S. (2017). “Ethical concerns regarding the use of bioinformatics and computational genomics,” in Proceedings of the International Conference on e-Learning, e-Business, Enterprise Information Systems, and e-Government (EEE) (Athens: The Steering Committee of The World Congress in Computer Science, Computer), 58–62.
Archer, J. C. (2010). State of the science in health professional education: effective feedback. Med. Educ. 44, 101–108. doi: 10.1111/j.1365-2923.2009.03546.x
Attwood, T. K., Blackford, S., Brazas, M. D., Davies, A., and Schneider, M. V. (2019). A global perspective on evolving bioinformatics and data science training needs. Briefings Bioinf. 20, 398–404. doi: 10.1093/bib/bbx100
Begley, C. G., and Ellis, L. M. (2012). Drug development: raise standards for preclinical cancer research. Nature 483, 531. doi: 10.1038/483531a
Biggs, J. (1996). Enhancing teaching through constructive alignment. High. Educ. 32, 347–364. doi: 10.1007/BF00138871
Bloom, B. S. (1956). Taxonomy of Educational Objectives. Vol. 1: Cognitive Domain. (New York, NY: McKay), 20–24.
Borsuk, R. M., Aarssen, L. W., Budden, A. E., Koricheva, J., Leimu, R., Tregenza, T., et al. (2009). To name or not to name: the effect of changing author gender on peer review. BioScience 59, 985–989. doi: 10.1525/bio.2009.59.11.10
Brown, J. A. (2016). Evaluating the effectiveness of a practical inquiry-based learning bioinformatics module on undergraduate student engagement and applied skills. Biochem. Mol. Biol. Educ. 44, 304–313. doi: 10.1002/bmb.20954
Campbell, C. E., and Nehm, R. H. (2013). A critical analysis of assessment quality in genomics and bioinformatics education research. CBE Life Sci. Educ. 12, 530–541. doi: 10.1187/cbe.12-06-0073
Cattley, S., and Arthur, J. W. (2007). Biomanager: the use of a bioinformatics web application as a teaching tool in undergraduate bioinformatics training. Briefings Bioinf. 8, 457–465. doi: 10.1093/bib/bbm039
Collaboration, O. S. (2015). Estimating the reproducibility of psychological science. Science 349:aac4716. doi: 10.1126/science.aac4716
Collin, K., and Valleala, U. M. (2005). Interaction among employees: how does learning take place in the social communities of the workplace and how might such learning be supervised? J. Educ. Work 18, 401–420. doi: 10.1080/13639080500327873
Cramp, A. (2011). Developing first-year engagement with written feedback. Active Learni. High. Educ. 12, 113–124. doi: 10.1177/1469787411402484
Cummings, M. P., and Temple, G. G. (2010). Broader incorporation of bioinformatics in education: opportunities and challenges. Briefings Bioinf. 11, 537–543. doi: 10.1093/bib/bbq058
Dahlö, M., Haziza, F., Kallio, A., Korpelainen, E., Bongcam-Rudloff, E., and Spjuth, O. (2015). Biolmg. org: a catalog of virtual machine images for the life sciences. Bioinf. Biol. Insights 9:BBI–S28636. doi: 10.4137/BBI.S28636
Ding, Y., Wang, M., He, Y., Ye, A. Y., Yang, X., Liu, F., et al. (2014). “ioinformatics: introduction and methods,” a bilingual massive open online course (mooc) as a new example for global bioinformatics education. PLoS Comput. Biol. 10:e1003955. doi: 10.1371/journal.pcbi.1003955
Evans, C. (2013). Making sense of assessment feedback in higher education. Rev. Educ. Res. 83, 70–120. doi: 10.3102/0034654312474350
Fulekar, M. (2009). Bioinformatics: Applications in Life and Environmental Sciences. Heidelberg: Springer Science & Business Media.
Furge, L. L., Stevens-Truss, R., Moore, D. B., and Langeland, J. A. (2009). Vertical and horizontal integration of bioinformatics education: a modular, interdisciplinary approach. Biochem. Mol. Biol. Educ. 37, 26–36. doi: 10.1002/bmb.20249
Gibbs, G., and Simpson, C. (2005). Conditions under which assessment supports students' learning. Learn. Teach. High. Educ. 1:3–31.
Gnimpieba, E. Z., Jennewein, D., Fuhrman, L., and Lushbough, C. M. (2013). Bioinformatics knowledge transmission (training, learning, and teaching): overview and flexible comparison of computer based training approaches. arXiv preprint arXiv:1310.8383.
Gonzalez, C. (2004). The role of blended learning in the world of technology. Retrieved December 10:2004.
Goodman, A. L., and Dekhtyar, A. (2014). Teaching bioinformatics in concert. PLoS Comput. Biol. 10:e1003896. doi: 10.1371/journal.pcbi.1003896
Goodman, K. W., and Cava, A. (2008). Bioethics, business ethics, and science: bioinformatics and the future of healthcare. Cambridge Quart. Healthcare Ethics 17, 361–372. doi: 10.1017/S096318010808050X
Hack, C., and Kendall, G. (2005a). Bioinformatics: current practice and future challenges for life science education. Biochem. Mol. Biol. Educ. 33, 82–85. doi: 10.1002/bmb.2005.494033022424
Hack, C., and Kendall, G. (2005b). Bioinformatics education in the uk: are we educating scientists or training technicians? Biosci. Bull. 15:10.
Hersh, W. (2008). Health and biomedical informatics: opportunities and challenges for a twenty-first century profession and its education. Yearbook Med. Inf. 17, 157–164. doi: 10.1007/978-0-387-78703-9_2
Hodkinson, H., and Hodkinson, P. (2004). Rethinking the concept of community of practice in relation to schoolteachers' workplace learning. Int. J. Train. Develop. 8, 21–31. doi: 10.1111/j.1360-3736.2004.00193.x
Hodkinson, P., Biesta, G., and James, D. (2004). “Towards a cultural theory of college-based learning,” in Manchester Annual Conference of the British Educational Research Association. Available online at: http://education.exeter.ac.uk/tlc/docs/publications/Conf/BERA%2004/ALL_CONF_BERA04_TOWARDSCULTTHEORY_09.04.htm
Hounsell, D., McCune, V., Hounsell, J., and Litjens, J. (2008). The quality of guidance and feedback to students. High. Educ. Res. Develop. 27, 55–67.
Ioannidis, J. P. (2005). Why most published research findings are false. PLoS Med. 2:e124. doi: 10.1371/journal.pmed.0020124
Johnston, S. (1997). Using peer support: implications for student and lecturer. Aspects Educ. Train. Technol. 29, 69–76.
Koch, I., and Fuellen, G. (2008). A review of bioinformatics education in germany. Briefings Bioinf. 9, 232–242. doi: 10.1093/bib/bbn006
Krathwohl, D. R. (2002). A revision of bloom's taxonomy: an overview. Theory Into Pract. 41, 212–218. doi: 10.1207/s15430421tip4104_2
Kuhn, T. S. (2012). The Structure of Scientific Revolutions: 50th Anniversary Edition. University of Chicago Press.
Lambert, C. G., and Black, L. J. (2012). Learning from our gwas mistakes: from experimental design to scientific method. Biostatistics 13, 195–203. doi: 10.1093/biostatistics/kxr055
Lathan, C. E., Tracey, M. R., Sebrechts, M. M., Clawson, D. M., and Higgins, G. A. (2002). Using virtual environments as training simulators: Measuring transfer. Handbook of Virtual Environments: Design, Implementation, and Applications, New Jersey, NJ: Lawrence Erlbaum Mahwah, 403–414.
Luscombe, N. M., Greenbaum, D., and Gerstein, M. (2001). What is bioinformatics? a proposed definition and overview of the field. Methods Inf. Med. 40, 346–358. doi: 10.1055/s-0038-1634431
Magana, A. J., Taleyarkhan, M., Alvarado, D. R., Kane, M., Springer, J., and Clase, K. (2014). A survey of scholarly literature describing the field of bioinformatics education and bioinformatics educational research. CBE Life Sci. Educ. 13, 607–623. doi: 10.1093/her/13.4.623
McKinney, K. (2010). Active Learning. Illinois State University. Center for Teaching, Learning & Technology. Available online at: https://www.phy.ilstu.edu/pte/311content/activelearning/activlearn1.html
Mensh, B., and Kording, K. (2017). Ten simple rules for structuring papers. PLoS Comput. Biol. 13:e1005619.
Merrill, M. D. (2002). First principles of instruction. Educ. Technol. Res. Develop. 50, 43–59. doi: 10.1007/BF02505024
Michaelsen, L. K., Stanley, C., and Porter, M. (2002). Team learning in large classes. Engaging Large Classes Strategies and Techniques for College Faculty, 27:67.
Mulder, N., Schwartz, R., Brazas, M. D., Brooksbank, C., Gaeta, B., Morgan, S. L., et al. (2018). The development and application of bioinformatics core competencies to improve bioinformatics training and education. PLoS Comput. Biol. 14:e1005772. doi: 10.1371/journal.pcbi.1005772
Pevzner, P., and Shamir, R. (2009). Computing has changed biology–biology education must catch up. Science 325, 541–542. doi: 10.1126/science.1173876
Prinz, F., Schlange, T., and Asadullah, K. (2011). Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Disc. 10:712. doi: 10.1038/nrd3439-c1
Ranganathan, S. (2005). Bioinformatics education–perspectives and challenges. PLoS Comput. Biol. 1:e52. doi: 10.1371/journal.pcbi.0010052
Rogers, C. R., Lyon, H. C., and Tausch, R. (2013). On Becoming an Effective Teacher: Person-Centered Teaching, Psychology, Philosophy, and Dialogues With Carl R. Rogers and Harold Lyon. London, UK: Routledge.
Salazar, M. R., Lant, T. K., Fiore, S. M., and Salas, E. (2012). Facilitating innovation in diverse science teams through integrative capacity. Small Group Res. 43, 527–558. doi: 10.1177/1046496412453622
Säljö, R. (1982). Learning and Understanding: A Study of Differences in Constructing Meaning From a Text, Vol. 41. Gothenburg: Humanities Pr.
Saravanan, V., and Shanmughavel, P. (2007). E-learning as a new tool in bioinformatics teaching. Bioinformation 2:83. doi: 10.6026/97320630002083
Schneider, M. V., Walter, P., Blatter, M.-C., Watson, J., Brazas, M. D., Rother, K., et al. (2011). Bioinformatics training network (btn): a community resource for bioinformatics trainers. Briefings Bioinf. 13, 383–389. doi: 10.1093/bib/bbr064
Sorge, R. E., Martin, L. J., Isbester, K. A., Sotocinal, S. G., Rosen, S., Tuttle, A. H., et al. (2014). Olfactory exposure to males, including men, causes stress and related analgesia in rodents. Nat. Methods 11:629. doi: 10.1038/nmeth.2935
St Clair, C., and Visick, J. (2010). Exploring Bioinformatics:A Project-based Approach. Burlington, MA: Jones and Bartlett Publishers. USA.
Taneri, B. (2011). Is there room for ethics within bioinformatics education? J. Comput. Biol. 18, 907–916. doi: 10.1089/cmb.2010.0187
Tastan Bishop, Ö., Adebiyi, E. F., Alzohairy, A. M., Everett, D., Ghedira, K., Ghouila, A., et al. (2014). Bioinformatics education–perspectives and challenges out of africa. Briefings Bioinf. 16, 355–364. doi: 10.1093/bib/bbu022
Tomlinson, M. (2008). The degree is not enough : students' perceptions of the role of higher education credentials for graduate work and employability. Brit. J. Sociol. Educ. 29, 49–61. doi: 10.1080/01425690701737457
Welch, L., Lewitter, F., Schwartz, R., Brooksbank, C., Radivojac, P., Gaeta, B., et al. (2014). Bioinformatics curriculum guidelines: toward a definition of core competencies. PLoS Comput. Biol. 10:e1003496. doi: 10.1371/journal.pcbi.1003496
Keywords: bioinformatics, computational biology, MSc, interdisciplinary/multi-disciplinary, transferability, curriculum, assessment
Citation: Johnston IG, Slater M and Cazier J-B (2022) Interdisciplinary and Transferable Concepts in Bioinformatics Education: Observations and Approaches From a UK MSc Course. Front. Educ. 7:826951. doi: 10.3389/feduc.2022.826951
Received: 01 December 2021; Accepted: 24 January 2022;
Published: 24 February 2022.
Edited by:
Jorge Rodríguez-Becerra, Universidad Metropolitana de Ciencias de la Educacin, ChileReviewed by:
Carlos C. Goller, North Carolina State University, United StatesNicolas Palopoli, National University of Quilmes, Argentina
John R. Jungck, University of Delaware, United States
Copyright © 2022 Johnston, Slater and Cazier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iain G. Johnston, iain.johnston@uib.no
 Iain G. Johnston
Iain G. Johnston
