- 1Center for Dark Energy Biosphere Investigations, University of Southern California, Los Angeles, CA, United States
- 2Division of Natural Sciences, Maryville College, Maryville, TN, United States
- 3School of Marine and Atmospheric Sciences, Stony Brook University, Stony Brook, NY, United States
- 4Horn Point Laboratory, University of Maryland Center for Environmental Science, Cambridge, MD, United States
- 5School of Life Sciences, Arizona State University, Tempe, AZ, United States
- 6Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA, United States
- 7MIT-WHOI Joint Program in Oceanography/Applied Ocean Science and Engineering, Cambridge, Woods Hole, MA, United States
- 8School of Biological Sciences, University of East Anglia, Norwich, United Kingdom
- 9Thermophile Research Unit, Te Aka Mātuatua - School of Science, University of Waikato, Hamilton, New Zealand
- 10Environmental Science, Policy and Management Department, University of California, Berkeley, CA, United States
- 11Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA, United States
- 12Biology, Chemistry and Environmental Studies Department, Center for Environmental Research and Coastal Oceans Monitoring (CERCOM), Molloy College, Rockville Centre, NY, United States
- 13Interface Geochemistry, GFZ German Research Centre for Geosciences, Helmholtz Centre Potsdam, Potsdam, Germany
- 14Plant and Microbial Biology Department, University of California, Berkeley, CA, United States
- 15Department of Biological Sciences - Marine and Environmental Biology, University of Southern California, Los Angeles, CA, United States
Lockdowns and “stay-at-home” orders, starting in March 2020, shuttered bench and field dependent research across the world as a consequence of the global COVID-19 pandemic. The pandemic continues to have an impact on research progress and career development, especially for graduate students and early career researchers, as strict social distance limitations stifle ongoing research and impede in-person educational programs. The goal of the Bioinformatics Virtual Coordination Network (BVCN) was to reduce some of these impacts by helping research biologists learn new skills and initiate computational projects as alternative ways to carry out their research. The BVCN was founded in April 2020, at the peak of initial shutdowns, by an international group of early-career microbiology researchers with expertise in bioinformatics and computational biology. The BVCN instructors identified several foundational bioinformatic topics and organized hands-on tutorials through cloud-based platforms that had minimal hardware requirements (in order to maximize accessibility) such as RStudio Cloud and MyBinder. The major topics included the Unix terminal interface, R and Python programming languages, amplicon analysis, metagenomics, functional protein annotation, transcriptome analysis, network science, and population genetics and comparative genomics. The BVCN was structured as an open-access resource with a central hub providing access to all lesson content and hands-on tutorials (https://biovcnet.github.io/). As laboratories reopened and participants returned to previous commitments, the BVCN evolved: while the platform continues to enable “a la carte” lessons for learning computational skills, new and ongoing collaborative projects were initiated among instructors and participants, including a virtual, open-access bioinformatics conference in June 2021. In this manuscript we discuss the history, successes, and challenges of the BVCN initiative, highlighting how the lessons learned and strategies implemented may be applicable to the development and planning of future courses, workshops, and training programs.
Introduction
As governments across the world began issuing various lockdowns, “stay-at-home” orders, and quarantines to curtail the spread of the coronavirus, academic labs that were considered nonessential were shuttered. It was unclear how long these closures would last, but initial estimates assumed at least 6–8 weeks. However, social distancing restrictions and other mitigation protocols ultimately minimized laboratory activities in many parts of the world for far longer. We developed the Bioinformatics Virtual Coordination Network (BVCN) due to the concerns raised on social media in mid-March 2020 about the impact that lab closures would have on the progress of graduate students and early-career researchers. This network is a collective of international early career microbiology researchers; with experience and expertise in bioinformatics and computational biology. Our initial mission was to provide an outlet for bench and field researchers to learn computational methods to pursue alternative research during laboratory closures or analyze data generated after returning to normal research activities. At the start of a global pandemic, accomplishing this goal required a flexible approach that could quickly ramp up to meet the needs of an international audience.
The BVCN joins a growing number of bioinformatics training resources (e.g., Wibberg et al., 2019; https://datacarpentry.org/) that have been prescribed as a necessity to teach life science researchers the skills required for large-scale data analysis (Attwood et al., 2017; Barone et al., 2017; Batut et al., 2018; Williams et al., 2019). The BVCN has a microbiology centric approach, with many of the core concepts and skills applicable to wider life science data analysis (Welch et al., 2014; Wilson Sayres et al., 2018). In the early phases of developing the BVCN, content distribution was envisioned as live events, combining microlectures on specific topics with hands-on demonstrations that could function as standalone lessons or part of an extended series on a broad topic. We developed a platform that built on the successes of previous short-form training courses and workshops (DIBSI: http://ivory.idyll.org/dibsi/toc.html, ECOGEO: doi. org/10.17504/protocols.io.fjjbkkn, STAMPS: https://mblstamps.github.io/, etc.) and then adapted these resources to accommodate the needs of a group of instructors and learners spread throughout the world. What evolved was a decentralized platform that has persisted beyond the end of lesson development, which coincided with the relaxing of lockdown orders and a return to in-person laboratory research.
We provide details on the critical aspects of what made the BVCN a success, along with the challenges we encountered and reflections on how to mitigate those challenges. The BVCN leveraged the varied expertise of its instructors to use multiple online tools and platforms to design material and learning environments tailored to a specific topic. As one of the successes of the BVCN model, this provided experience to learners on how to use computational resources that mirrored those required for genuine research. These lessons are freely available and mutable through a public GitHub repository (https://biovcnet.github.io/). The BVCN made early commitments to establishing an inclusive environment with explicit goals to lower the entry barriers for researchers to participate in computational biology. Now that formal lesson developments have ended, we have a library that allows interested learners to pursue self-led, “point of need” instruction of microbial bioinformatics and computational biology when suitable for their particular research needs and questions.
Implementation of the Education Program
In March 2020, the BVCN lead, Dr. Benjamin Tully, put out an announcement on Twitter to gauge interest in teaching and learning bioinformatics during the pandemic. Within a few days, the announcement had gathered interest from more than 50 computational biology educators and several hundred participants. Following an introductory virtual meeting among interested instructors that discussed possible avenues for distributing lessons and the breadth of topics to include, we created a BVCN Slack workspace (Teckchandani, 2018; https://slack.com/). As instructors, we self-assigned into topics and chose one person to act as a coordinator for each topic. Based on the breadth of bioinformatics research disciplines represented by the instructors, we chose to include lessons addressing Unix, R programming, amplicons, metagenomics, functional annotation, transcriptomics, network analysis, population genetics and comparative genomics, and Python programming (Table 1). With an aim to promote reproducible science and good open data practice, we also created the Reproducibility Challenge (see below) to encourage learners to reproduce bioinformatic results from published research. We organized communication amongst the instructors through Slack and Zoom (https://zoom.us/), with collaborative editing on Google Docs and Sheets within a shared Google Drive (https://www.google.com/drive/), accessible to all instructors. We selected GitHub (https://github.com/) as a central repository for collaboratively creating and editing tutorial material and YouTube (https://youtube.com/) for sharing recorded microlectures. Instructors leading each topic had the flexibility and independence to plan lessons in the formats most relevant to the strengths and weaknesses of the specific topic and/or lesson. While topics functioned semi-independently, weekly meetings between all instructors and coordination with the Project Lead and the Coordinating Committee helped foster the sharing of resources and creative approaches.
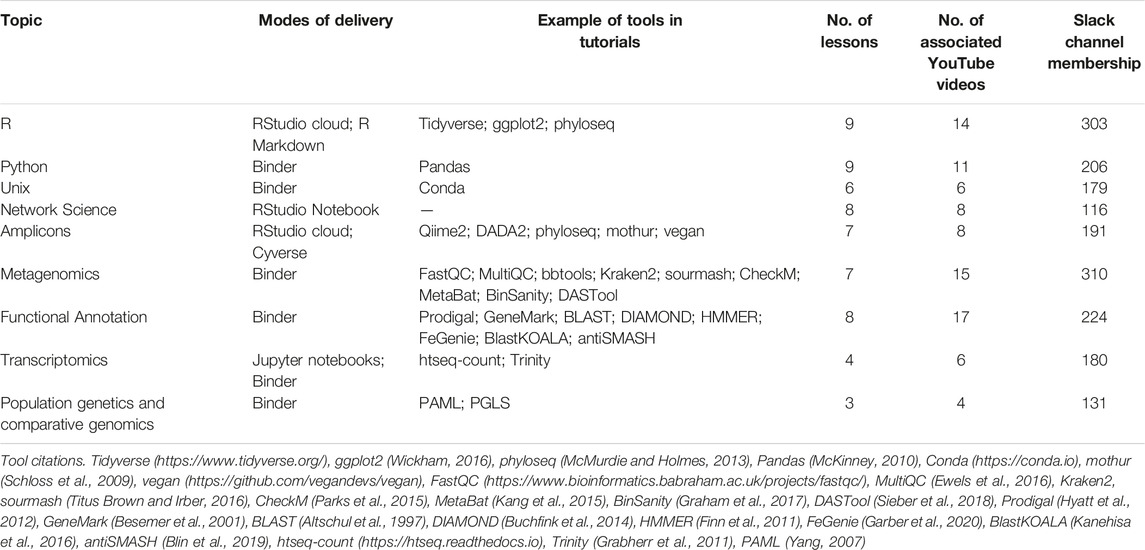
TABLE 1. List of BVCN topics with how material was delivered, and examples of core concepts reviewed (https://github.com/biovcnet/biovcnet.github.io/wiki).
We started creating lessons in April continuing through August 2020, and developed lessons for each topic in various formats, with a majority scheduled and planned as synchronous learning events, either as a lesson, short-format tutorial, or live Q and A discussion. At its peak, all topics produced about one lesson per week. We recorded all of our lessons and made these available on YouTube (https://www.youtube.com/channel/UC5qVqcvUPfgPQWOhBaR_Low), along with datasets, code, and any other necessary materials. When applicable, short-format tutorials were executed in shareable computing environments (see below: platforms and tools), with the explicit goal of enabling learners to complete tutorial material without needing to modify their local computing environments, while also providing a genuine experience equivalent to what would be required for the participants’ own research. All of BVCN was implemented with a forward-looking approach; our materials are available indefinitely and can be accessed and modified by others. The BVCN Slack workspace, organized in channels based on topic, continues to be a community resource where users can post broad or specific questions regarding their research and have conversations between learners and instructors.
Platforms and Tools Incorporated Into the Lesson Plans
Each topic of the BVCN has common features, as well as unique teaching platforms and systems determined by the set of instructors for that topic. We decided in the early planning meetings that every course should contain a live component, when possible, for learners interested in a synchronous approach to the lessons, and all lectures and tutorials should be recorded for asynchronous viewing at the convenience of the learners. Components needed to follow along asynchronously, such as interactive code and links to video lessons, were collated by lesson and organized through the wiki page (https://github.com/biovcnet/biovcnet.github.io/wiki). Lecture components tended to be conceptual in nature, providing essential background to the tools that we used in the tutorials. Tutorials provided an example of an applied approach using standard toolsets for accomplishing a specific computational task. For example, Lesson 4 of the metagenomics topic includes a lecture, recorded live, on the principles of read mapping and some of the main tools and algorithms used in this practice. We followed this lecture with two short-format tutorials for the read mapping tools Bowtie2 (Langmead and Salzberg, 2012) and BBmap (Bushnell et al., 2017). We delivered synchronous content as online meetings on Zoom. Meetings varied between formal microlectures and less formal Q and A sessions. Synchronous meeting planning, posting of asynchronous content, discussions about lesson content, and direct responses to learner questions were all hosted through Slack.
Each topic had a different set of instructors and within these topics the instructors chose the specific delivery platforms for the interactive course content. Two main strategies emerged (Table 1). Most topics packaged content using interactive Binder (https://mybinder.org/) environments which included installed dependencies, tools, and data, which gave learners access to an online Unix terminal interface that could be used to perform small-scale tasks on real data. The major advantage of using Binder environments is the ease with which tutorials can be launched regardless of a student’s local computing resources and avoiding any software incompatibility, as Binder operates in the cloud using a web browser. However, this flexibility comes with the tradeoff that Binder environments are limited in the size of the data that can be stored and the processing power that can be applied. Other topics, like the R programming and amplicon channels, used the RStudio Cloud platform which offers similar functionality to Binder but is optimized to reflect an R coding environment. Budgetary restrictions put an upper limit on the amount of content we could release using the RStudio Cloud (https://rstudio.cloud/) platform (e.g., capacity for number of participants, storage, RStudio Projects, etc.). Other topics (e.g., network science) encouraged students to download lesson content from the BVCN GitHub and perform local analysis, which had the advantage of no explicit limits on the number of lessons, but required learners to possess some proficiency in setting up local computing environments before they could interact with the content.
Establishment of Diversity and Equity Statement
As a community-led project, we aspire to make the BVCN a safe and open workspace where bioinformatics can be taught and learnt by a diverse audience. Our founding document, the BVCN Code of Conduct (https://biovcnet.github.io/code-of-conduct/), describes our commitment to diversity and inclusion. Building off of the codes of conduct of multiple other open-source communities, the document’s strength is that it details how the BVCN will provide accessible content, what the community expects of its members, and provides detailed examples as to what the community defines as acceptable and unacceptable behaviors. This code of conduct drove how we worked within the BVCN, enhancing the virtual learning experience of a wider global audience. We actively worked towards decreasing entry barriers into bioinformatics, including knowledge access, support, and representation. We see this as especially important for reaching and providing access to an international community of early career researchers. For many, the access to resources, such as textbooks, workshops, training courses, research experience, and computational infrastructure, can prevent interested scientists from pursuing bioinformatics.
Our primary avenue for promoting the BVCN was through professional networks on Twitter and word of mouth. Access to the growing archive of lessons and short-format tutorials, along with the Slack workspace detailing active lesson deployment, helped to facilitate access to formal support, information exchange, and collaborations. This approach aimed to lower the entry barriers between awareness (i.e., promotion) and the use of BVCN content as a resource (i.e., ease of access to archived and ongoing support), which is identified as a key guiding principle for bioinformatics education (Williams et al., 2010). The high degree of connectivity between instructors and learners laid a strong foundation for establishing mentee-mentor relationships. The combined experiences of our instructors, who represent diversity in gender, ethnicity, career stage, and academic background, provided opportunities for learners to observe representation within the field. Gender-balance at the instructor level (∼59% of instructors identify as women) offered the potential for same-gender mentoring, which has been shown to increase belonging, self-efficacy, and retention in STEM and engineering fields (Dennehy and Dasgupta, 2017).
Despite these efforts, we have identified areas of current and future improvement within our approach to diversity and inclusion. One aspect that we feel could have a significant impact on learner engagement would be to increase efforts to develop formal mentor-mentee and/or peer-to-peer partnerships with a targeted effort to enhance the recruitment of historically excluded groups. In order to have the most impact, and truly expand on the BVCN’s mission of increasing representation of historically excluded groups in bioinformatics, we need to continue to work to enhance the visibility and accessibility of our resources and support network. We are committed to reducing entry barriers through continuous improvements in participant recruitment and retention through purposeful global outreach (see below: ongoing collaborations) to raise awareness about the free resources and support supplied through the BVCN.
Evolution of Approaches as Topics Emerge
As instructors, learners, and topics harmonized over our various platforms, custom lesson plans were crafted. One common challenge learners expressed was how to select the “correct” tool or pipeline for an analysis and then incorporate it into their own research. Many topics provided opinions about “best practices” and highlighted multiple approaches that allowed learners to pick and choose how they approached complex topics. We regularly emphasized that there is often more than one way to approach a problem without sacrificing the quality of the results. To demonstrate this more thoroughly, we addressed the question “Which tool(s) should one use to perform amplicon analysis” by developing lessons that showcased the most commonly used environments for the analysis of 16 S/18 S rRNA gene or intergenic spacer region (ITS) amplicons. This series of lessons kicked off with a live Q and A session where several instructors discussed the methods by which we conduct amplicon sequence analysis (https://youtu.be/egkCswqQMWM). The goal of this “fireside chat” was to demonstrate that our individual bioinformatic practices vary and shed light on current trends within the interest. We then designed parallel lessons using an identical initial dataset. These data were processed using the tutorials from Happy Belly Bioinformatics (Lee, 2019), which implements the DADA2 pipeline (Callahan et al., 2016) in R (R Core Team. R, 2020) and the QIIME2 pipeline (Bolyen et al., 2019) on the cloud-computing infrastructure platform CyVerse (Merchant et al., 2016).
Discussions highlighting variations in approaches typically occur at conferences or in more casual settings in the workplace; without these interactions, learners working through their first datasets may feel isolated. Conversations about alternative approaches, which occurred during recorded lessons, on Slack, or during scheduled office hours, were a critical part in building a community of learning within the BVCN. Even after lesson development ended and the BVCN shifted to an asynchronous learning format, the Slack workspace continues to serve as a valuable resource for learners who may have recently completed the coursework and are pursuing follow-up questions regarding how to apply tools to their own datasets.
Asynchronous Audience and Ongoing Membership
As might be expected for an initiative started in response to a global event that caused the forced shutdown of laboratory research, the highest level of audience engagement occurred in the initial phases of government-ordered shutdowns. Using the metric of YouTube channel views, it is clear that the BVCN continues to reach an audience interested in accessing our archive of video and written tutorial materials. We collected viewership data from YouTube (accessed April 29, 2021) for the BVCN channel which hosts 95 videos (lectures and tutorials) and has 562 subscribers. The videos had a total of 16,325 cumulative views and over 1,700 cumulative watch hours. BVCN content was developed and deployed almost entirely between April 6 and August 4, 2020, approximately during the height of laboratory lockdowns. Monthly viewings peaked at 2,774 views during May 2020, shortly after we started, and dropped through August 2020. Since the formal end of BVCN lesson development, we have sustained approximately 1,000 views per month (Figure 1A). By cumulative views, metagenomics, R, functional annotation, amplicons, and Unix were the top five viewed subject playlists (Figure 1B).
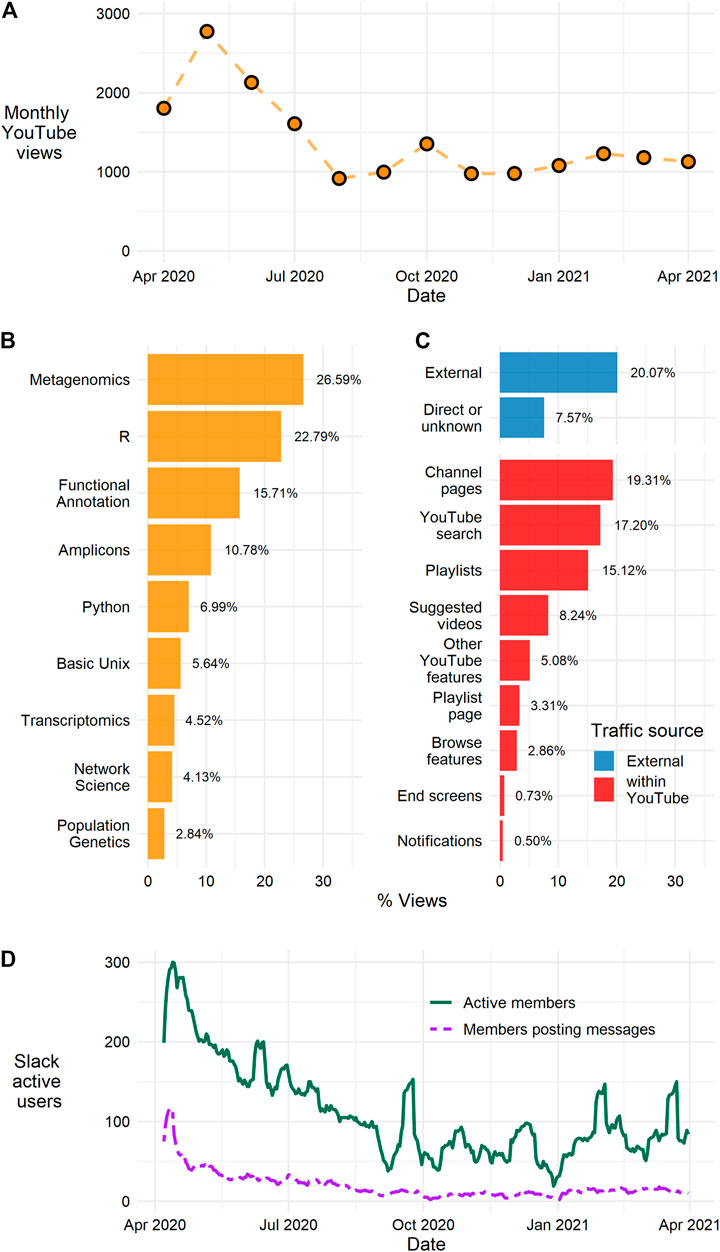
FIGURE 1. Overview of the number of views of BVCN YouTube videos and Slack membership. (A) Monthly viewing figures for the BVCN YouTube channel; the total accumulated views for the channel is 16,325 views (accessed 29 April 2021), views per month peaked in May 2020 at 2774, and viewing numbers have fallen but stabilized at an average of over 1,000 views per month from August 2020 to present. (B) The percentage of total views for the BVCN channel split by subject playlists; the Metagenomics and R playlists are the first and second most viewed playlists accounting for almost half of the total views for the BVCN channel. (C) The percentage of total views for the BVCN channel split by “traffic source” top bars (blue) indicate views came from sources external to YouTube, bottom bars (red) indicate views that originate within the YouTube website. The top four bars (External, Direct or unknown, Channel pages, YouTube search) are views that originate from “searches with intent” viewers actively searching for BVCN videos these account for 64% of total views. (D) The seven-day rolling average for Slack user activity; active members are those who used the Slack workspace in the last seven days (solid green line), and the number of users that have posted messages to the workspace (dashed purple line). Slack use peaked in April 2020 and stabilized at between 50 and 100 active users per month from September 2020.
A four-week window was selected from 15 February to March 14, 2021 to assess what type of users were accessing the video content. During this period, 488 unique viewers and 48 returning viewers visited the channel. YouTube provides information on where “traffic sources” originate from, quantifying how viewers reached videos on the BVCN channel (e.g., external links, visiting the channel page, YouTube search, etc.). About a fifth of traffic to YouTube originated from external links, likely learners directed from the BVCN wiki or social media posts. Roughly three-quarters of total views originated from within YouTube (Figure 1C), but around 64% of total traffic (internal and external to YouTube) came from “searches with intent” (i.e., users searching or accessing BVCN content directly; Figure 1C).
The BVCN Slack workspace remains active and provides a central location for 630 members of the community to exchange ideas, share resources, and troubleshoot problems (Figure 1D). Activity within Slack mirrors that of the views tracked by YouTube. Weekly active members spiked in the first month following the start of lockdowns, but steadily declined through September 2020. Since that time, the number of weekly members has stabilized to between 50 and 100 participants who are engaging with the workspace either through reading public posts, participating in private channels, or direct messaging. Data collected by Slack from a recent 30-days window (April 5 to May 3, 2020) indicates that most of this participation (47%) is based on users accessing messages in public channels and not governed strictly by direct messages (37%) or private channels (16%).
Reproducibility Challenge
In collaboration with Drs. Harriet Alexander and Maria Pachiadaki, instructors for a graduate-level course in bioinformatics at the Woods Hole Oceanographic Institution (WHOI), we developed a research task for students to enhance their hands-on bioinformatics skills. Building off of the existing knowledge base of the WHOI course, students would select a bioinformatics analysis in a publication and attempt to recreate the outputs as presented by the authors. The goal was to encourage learners to consider what would be required to create reproducible research and was accomplished by providing intermediate learners a platform to perform analysis on an existing dataset with the goal of reproducing published results. Additionally, we sought to foster growth within the learner community and provide learners with a network for peer-to-peer mentorship while working on a formalized collaborative project. We created an accompanying tutorial website (https://alexanderlabwhoi.github.io/BVCNReproducibility/intro.html) explaining some of the fundamental stages of a bioinformatics project, such as data curation/management, tool installation, and reproducible coding platforms, and provided two example datasets and exemplar code which learners could use to reproduce published research results. Learners and instructors were matched with other interested partners through an online form and learners were provided an example timeline to complete the research task. Ultimately, few learners participated in the project based on the provided timeline. While one project was successful and resulted in establishing a network of researchers working on a collaborative project, many projects suffered from scheduling challenges and time constraints. As with many facets of the global pandemic, it appeared that participants became overwhelmed with responsibilities and were more comfortable with using the materials for asynchronous learning.
Discussion
Simultaneous Building of Learning Resources and a Community
The impromptu, grassroots nature of the BVCN came with a number of challenges, which were compounded by the COVID-19 pandemic. We attempted to simultaneously develop a novel learning resource while building a community–two tasks with an immense amount of complexity even under normal conditions. To this end, we saw several areas for which the BVCN has accomplished the goals laid out in early planning phases:
1) Provide a platform that encourages the dissemination of knowledge from experienced bioinformaticians and computational biologists to those new in the field. This pedagogical approach leveraged our collective research experiences to help guide learners with hesitancy about using bioinformatic techniques towards a place where they felt comfortable exploring these concepts. We aggregated a number of introductory resources (e.g., the Happy Belly Bioinformatics Unix tutorial (Lee, 2019)) and paired them with lessons that advanced in complexity, allowing learners to see the progression from introduction to practical use.
2) Implementation of a community educational resource that has previously been limited in accessibility due to numerous constraints. While there are initiatives of learning these types of skills, many of them occur through short course formats (Attwood et al., 2017) that may have restrictions related to attendance, cost, or location. We took advantage of the limitations imposed by the pandemic by providing a resource that learners can access at their own pace, with sole limitations being access to an internet connection and personal computer, while still interacting with the instructors and their colleagues. The pedagogy of the lessons is also designed with an active learning approach in mind. Conceptual lectures are paired with tutorial content which allow learners to actively explore bioinformatic tools and techniques which tend to have better learning outcomes (Markant et al., 2016). Broadly, our goal with the BVCN was to assist in bridging life science researchers without experience in bioinformatics/computational biology towards an understanding of the tasks involved in this type of research. The persistent nature of the BVCN platform can assist in achieving this goal for years to come.
3) Establishing an international community, oriented around microbiology, bioinformatics, and computational biology. In many instances, forming national and international collaborations amongst early career researchers occur at meetings and conferences. As with many things, the pandemic has prevented these events, which may ultimately have an impact on these types of relationships in the long term. With the BVCN, we have built an international community, predominantly of early career researchers, that has the capacity to persist beyond the months of active lesson development and act as a meeting place for bench/wet-lab and/or field scientists to interface with bioinformaticians and computational biologists. Further, the instructors and learners of the BVCN have initiated multiple projects, activities, and collaborations that would not have occurred had it not been for the efforts of the platform (see below: Emergent Collaborative Projects).
Continued Challenges to Building an Online Learning Initiative
Some of the challenges we experienced appear to be specific to the pandemic and reflected in other similar experiences, but there are others which we could have effectively addressed during the initiation of the BVCN had the Project Lead, Coordinating Committee, and instructors been aware of such issues arising in other online initiatives. Future bioinformatics training initiatives may benefit from considering issues that we encountered highlighted below:
1) Rapidly declining attendance/interest in organized live sessions for topics after the first several weeks. As supported by viewership on YouTube and active member data on Slack, many learners shifted away from active participation and towards an asynchronous learning approach. While videos from all sections continue to accrue views, participation in the interactive Zoom sections tapered off after the initial pandemic lockdowns (June/July 2020). While additional live sessions mirroring standard “office hours” were established, these sessions also saw a rapid decline in attendance. Successes like the merged amplicon and R lesson (described above) resulted from active feedback from learners, but the decrease in instructor-learner interactions had impacts on our ability to develop customized BVCN lessons. Some small groups of learners and instructors have continued to meet through 2021 (e.g., weekly network science office hour). Without a direct incentive for attending lessons consistently, learners adjusted to a mode of knowledge sharing suitable for their personal research needs. This was likely driven by the ability of BVCN lessons to provide “point-of-need” training on specific concepts and tools, more suitable for researchers as they actively perform analyses. As noted before, many existing bioinformatics workshops have solved this by selecting a limited cohort of students to be fully immersed in the learning experience but inherently restrict the number of participants.
2) No system or time to iteratively improve lessons or strategies. The speed at which the BVCN was deployed was in direct response to the emotions and fears spun out of the early phases of the pandemic. As such, we prioritized the consistent production of new lessons. The trade-offs to this approach were clear. While it provided resources for learners immediately, it meant that our limited time was focused on producing additional content, not necessarily on returning to improve, refine, or iterate previous content. An alternative could have been to form our instructor base for the BVCN and spend several months producing and refining lesson content prior to release, though the downside would have been the trajectory of the pandemic itself, where most BVCN instructors and learners saw their availability and priorities shift back towards their teaching and/or research commitments, as conditions in the pandemic stabilized (see below: collaborations with KBase).
3) Difficulty assessing the degree to which lessons, stylistic choices, use of computational infrastructure, etc. assisted learners in achieving their goals. This challenge likely is a result of the initial infrastructure we established. Efforts were made from the onset to make all elements open access and reduce the “cost” of participation within the BVCN, which we saw as sufficient to encourage ongoing participation. However, without formal checkpoints or feedback routes, it was difficult to assess the needs of learners. During the peak of activity, it would have been useful if we had implemented post-lesson assessments that could have tracked sentiments and/or encouraged learners to engage in the future direction of lessons.
Emergent Collaborative Projects
One of our continuing successes from the BVCN has been the number of collaborative projects that have been initiated as a direct result of the interaction between instructors and learners. One example of such a collaboration emerged from the network science topic. In that group, discussions between instructors and learners led to the conclusion that the current “best practice” for network analysis did not address many common statistical considerations of time-series analysis. One learner, in particular, with support from two of our instructors, took the lead on developing a novel approach that is robust to common statistical artifacts and that works with unevenly spaced time-series data. Another collaborative project initiated between the instructors from WHOI and the University of Southern California has seen the development of multiple complementary approaches for the large-scale analysis of eukaryotic metagenome-assembled genomes. Additionally, the relationships created between instructors has led to additional opportunities for cross-pollination between fields. For example, an instructor-led seminar series for the Center for Dark Energy Biosphere Investigations tapped BVCN instructors as presenters for a seminar about novel and upcoming bioinformatic approaches.
One of the current ongoing projects is to continue the educational legacy of the BVCN. As the development of new content in the BVCN channels slowed, we looked to new and alternative ways our community could facilitate bioinformatics learning more broadly. We identified a gap in bioinformatics education: while tools and some workflows may have good documentation, a holistic overview of entire bioinformatics projects, including the decisions as to which tools to use and how to combine them, was missing. To address this education gap, we envisioned a virtual conference focused on open-science methods, where featured speakers outlined their bioinformatics pipelines and provided open data and code alongside their talks. In this format, the speakers would be asked to put special emphasis on the many difficult decisions and trade-offs that go into shaping and executing a project. In addition to filling a gap in the bioinformatics education infrastructure, we identified during the early exploratory stage of conference planning that a virtual conference offered opportunities to make the event more accessible to a wide audience in ways a traditional conference never could. As part of these continuing efforts, the BVCN will welcome over 200 attendees from around the world to “A BVCN Training Conference: Holistic Bioinformatic Approaches used in Microbiome Research” in June 2021 supported by the Code for Science and Society Event Fund (https://eventfund.codeforscience.org/).
The BVCN represents one of many educational initiatives started during the COVID-19 pandemic. Early in the development of the BVCN, we collaborated with the educational directive led by Drs. Ellen Dow and Elisha Wood-Charlson at the Department of Energy Systems Biology knowledgebase (KBase). Multiple conversations were held to determine how the BVCN and KBase could support each other’s educational initiatives. The BVCN concentrated on short-format tutorials that utilized command line or coding examples, while KBase built educational material that used their cloud-based, graphical user interface (GUIs). BVCN content was directed at early careers researchers and the KBase educational directive, supported by tenured and tenure-track faculty, developed lesson plans for undergraduate microbiology majors (Dow et al., 2021). These lessons form a complement to those created by the BVCN and reflect the strength of formal support through a funded sponsor and long-term planning for lesson development and maintenance.
Concluding Remarks
The popularity of the BVCN makes it clear that there is a need for the bioinformatic training of life scientists, including microbiologists, at all career stages. With the BVCN, we have laid the groundwork for filling one of the most persistent gaps in bioinformatic training, moving beyond introductory content towards intermediate levels of expertise. We provided a grassroots response to the needs of the community at a time of heightened global uncertainty, and we hope that future educational initiatives, either direct descendants of the BVCN or inspired by, may be able to satisfy the continued need for these resources and take the lessons and approaches applied in this initiative to achieve further success.
Data Availability Statement
The datasets presented in this study can be found in online repositories. Data on YouTube and Slack usage, along with code used to produce figures, can be found at https://github.com/biovcnet/BVCN_stats
Author Contributions
BT, JB, AC, JC, AG, SH, AK, PL, AM, ES, DS, ES, CT, LV-A, and JL wrote and edited the article. BT founded the Bioinformatics Virtual Coordination Network. The BVCN Instructor Consortium consists of Michael D. Lee, Harriet Alexander, R. Eric Collins, Maria Pachiadaki, Adelaide Rhodes, Wayne Decatur, who along with the authors of the article, contributed lessons and/or expertise during the Bioinformatics Virtual Coordination Network.
Funding
BT was supported by the Center for Dark Energy Biosphere Investigations (OCE-0939654). The BVCN Training Conference was funded in full by a grant from Code for Science and Society, made possible by grant number GBMF8449 from the Gordon and Betty Moore Foundation. AM was supported by Smart Ideas award (UOWX1602) from the New Zealand Ministry of Business, Innovation and Employment and the Rutherford Foundation Royal Society Te Aparangi Postdoctoral Fellowship (20-UOW-006). ETS. was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, Genomic Science Program, Scientific Focus Area award SCW1632 to Jennifer Pett-Ridge and award DE-SC0014079 to Mary K. Firestone. CT acknowledges financial support from the German Helmholtz Recruiting Initiative (award number: I-044–16-01) and from the European Research Council Synergy Grant (“Deep Purple” grant # 856416) awarded to Liane G Benning. AK was supported by the U.S. Department of Energy Computational Science Graduate Fellowship (DE-SC0020347). DS was supported by the Netherlands Organisation for Scientific Research, Rubicon award 019.153LW.039 and the US Department of Energy, Office of Science, Office of Biological and Environmental Research under award number DE-SC0016469 to Victoria J. Orphan. JL was supported by a postdoctoral fellowship in marine microbial ecology from Simons Foundation Award 653212. The Center for Dark Energy Biosphere Investigations (OCE-0939564) supported the participation of SH through a C-DEBI Postdoctoral Fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a New Generation of Protein Database Search Programs. Nucleic Acids Res. 25, 3389–3402. doi:10.1093/nar/25.17.3389
Attwood, T. K., Blackford, S., Brazas, M. D., Davies, A., and Schneider, M. V. (2017). A Global Perspective on Evolving Bioinformatics and Data Science Training Needs. Brief. Bioinform. 20, 398–404. doi:10.1093/bib/bbx100
Barone, L., Williams, J., and Micklos, D. (2017). Unmet Needs for Analyzing Biological Big Data: A Survey of 704 NSF Principal Investigators. Plos Comput. Biol. 13, e1005755. doi:10.1371/journal.pcbi.1005755
Batut, B., Hiltemann, S., Bagnacani, A., Baker, D., Bhardwaj, V., Blank, C., et al. (2018). Community-Driven Data Analysis Training for Biology. Cell Syst 6, 752–e1. doi:10.1016/j.cels.2018.05.012
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a Self-Training Method for Prediction of Gene Starts in Microbial Genomes. Implications for Finding Sequence Motifs in Regulatory Regions. Nucleic Acids Res. 29, 2607–2618. doi:10.1093/nar/29.12.2607
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 47, W81–W87. doi:10.1093/nar/gkz310
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37, 852–857. doi:10.1038/s41587-019-0209-9
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 12, 59–60. doi:10.1038/nmeth.3176
Bushnell, B., Rood, J., and Singer, E. (2017). BBMerge - Accurate Paired Shotgun Read Merging via Overlap. PLoS ONE 12, e0185056. doi:10.1371/journal.pone.0185056
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 13, 581–583. doi:10.1038/nmeth.3869
Dennehy, T. C., and Dasgupta, N. (2017). Female Peer Mentors Early in College Increase Women's Positive Academic Experiences and Retention in Engineering. Proc. Natl. Acad. Sci. U S A. 114, 5964–5969. doi:10.1073/pnas.1613117114
Dow, E. G., Wood-Charlson, E. M., Biller, S. J., Paustian, T., Schirmer, C. S., Sheik, W., et al. (2021). Bioinformatic teaching resources - for educators, by educators - using KBase, a free, user-friendly, open source platform. Front. Educ. doi:10.3389/feduc.2021.711535
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 32, 3047–3048. doi:10.1093/bioinformatics/btw354
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Garber, A. I., Nealson, K. H., Okamoto, A., McAllister, S. M., Chan, C. S., Barco, R. A., et al. (2020). FeGenie: A Comprehensive Tool for the Identification of Iron Genes and Iron Gene Neighborhoods in Genome and Metagenome Assemblies. Front. Microbiol. 11, 37. doi:10.3389/fmicb.2020.00037
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 29, 644–652. doi:10.1038/nbt.1883
Graham, E. D., Heidelberg, J. F., Tully, B. J., and Tully, B. J. (2017). BinSanity: Unsupervised Clustering of Environmental Microbial Assemblies Using Coverage and Affinity Propagation. PeerJ 5, e3035–19. doi:10.7717/peerj.3035
Hyatt, D., LoCascio, P. F., Hauser, L. J., and Uberbacher, E. C. (2012). Gene and Translation Initiation Site Prediction in Metagenomic Sequences. Bioinformatics 28, 2223–2230. doi:10.1093/bioinformatics/bts429
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 428, 726–731. doi:10.1016/j.jmb.2015.11.006
Kang, D. D., Froula, J., Egan, R., and Wang, Z. (2015). MetaBAT, an Efficient Tool for Accurately Reconstructing Single Genomes from Complex Microbial Communities. PeerJ 3, e1165–15. doi:10.7717/peerj.1165
Langmead, B., and Salzberg, S. L. (2012). Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
Lee, M. (2019). Happy Belly Bioinformatics: an Open-Source Resource Dedicated to Helping Biologists Utilize Bioinformatics. Jose 2, 53. doi:10.21105/jose.00053
Markant, D. B., Ruggeri, A., Gureckis, T. M., and Xu, F. (2016). Enhanced Memory as a Common Effect of Active Learning. Mind, Brain Educ. 10, 142–152. doi:10.1111/mbe.12117
McKinney, W. (2010). Data Structures for Statistical Computing in Python. Proc. 9th Python Sci. Conf., 56–61. doi:10.25080/Majora-92bf1922-00a
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 8, e61217. doi:10.1371/journal.pone.0061217
Merchant, N., Lyons, E., Goff, S., Vaughn, M., Ware, D., Micklos, D., et al. (2016). The iPlant Collaborative: Cyberinfrastructure for Enabling Data to Discovery for the Life Sciences. Plos Biol. 14, e1002342. doi:10.1371/journal.pbio.1002342
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 25, 1043–1055. doi:10.1101/gr.186072.114
R Core Team. R (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing Mothur: Open-Source, Platform-independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75, 7537–7541. doi:10.1128/AEM.01541-09
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of Genomes from Metagenomes via a Dereplication, Aggregation and Scoring Strategy. Nat. Microbiol. 3, 836–843. doi:10.1038/s41564-018-0171-1
Teckchandani, A. (2018). Slack: A Unified Communications Platform to Improve Team CollaborationSlack: A Unified Communications Platform to Improve Team Collaboration, Available at https://slack.Com/. Amle 17, 226–228. doi:10.5465/amle.2018.0061
Titus Brown, C., and Irber, L. (2016). Sourmash: a Library for MinHash Sketching of DNA. JOSS 1, 27–31. doi:10.21105/joss.00027
Welch, L., Lewitter, F., Schwartz, R., Brooksbank, C., Radivojac, P., Gaeta, B., et al. (2014). Bioinformatics Curriculum Guidelines: toward a Definition of Core Competencies. Plos Comput. Biol. 10, e1003496. doi:10.1371/journal.pcbi.1003496
Wibberg, D., Batut, B., Belmann, P., Blom, J., Glöckner, F. O., Grüning, B., et al. (2019). The de.NBI/ELIXIR-DE training platform - Bioinformatics training in Germany and across Europe within ELIXIR. F1000Res 8, 1877. doi:10.12688/f1000research.20244.1
Williams, J. J., Drew, J. C., Galindo-Gonzalez, S., Robic, S., Dinsdale, E., Morgan, W. R., et al. (2019). Barriers to Integration of Bioinformatics into Undergraduate Life Sciences Education: A National Study of US Life Sciences Faculty Uncover Significant Barriers to Integrating Bioinformatics into Undergraduate Instruction. PLoS ONE 14, e0224288. doi:10.1371/journal.pone.0224288
Williams, J. M., Mangan, M. E., Perreault-Micale, C., Lathe, S., Sirohi, N., and Lathe, W. C. (2010). OpenHelix: Bioinformatics Education outside of a Different Box. Brief Bioinform 11, 598–609. doi:10.1093/bib/bbq026
Wilson Sayres, M. A., Hauser, C., Sierk, M., Robic, S., Rosenwald, A. G., Smith, T. M., et al. (2018). Bioinformatics Core Competencies for Undergraduate Life Sciences Education. PLoS ONE 13, e0196878. doi:10.1371/journal.pone.0196878
Keywords: bioinformatics, educational initiative, computational biology, microbiology, open source, free educational resource
Citation: Tully BJ, Buongiorno J, Cohen AB, Cram JA, Garber AI, Hu SK, Krinos AI, Leftwich PT, Marshall AJ, Sieradzki ET, Speth DR, Suter EA, Trivedi CB, Valentin-Alvarado LE and Weissman J (2021) The Bioinformatics Virtual Coordination Network: An Open-Source and Interactive Learning Environment. Front. Educ. 6:711618. doi: 10.3389/feduc.2021.711618
Received: 18 May 2021; Accepted: 17 September 2021;
Published: 14 October 2021.
Edited by:
Hugo Verli, Federal University of Rio Grande do Sul, BrazilReviewed by:
Aristóteles Góes-Neto, Federal University of Minas Gerais, BrazilMiguel Rocha, University of Minho, Portugal
Copyright © 2021 Tully, Buongiorno, Cohen, Cram, Garber, Hu, Krinos, Leftwich, Marshall, Sieradzki, Speth, Suter, Trivedi, Valentin-Alvarado and Weissman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin J. Tully, dHVsbHkuYmpAZ21haWwuY29t
 Benjamin J. Tully
Benjamin J. Tully Joy Buongiorno
Joy Buongiorno Ashley B. Cohen
Ashley B. Cohen Jacob A. Cram
Jacob A. Cram Arkadiy I. Garber5
Arkadiy I. Garber5 Sarah K. Hu
Sarah K. Hu Arianna I. Krinos
Arianna I. Krinos Philip T. Leftwich
Philip T. Leftwich Alexis J. Marshall
Alexis J. Marshall Elizabeth A Suter
Elizabeth A Suter Christopher B. Trivedi
Christopher B. Trivedi Luis E. Valentin-Alvarado
Luis E. Valentin-Alvarado JL Weissman
JL Weissman