
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Drug Saf. Regul., 10 November 2022
Sec. Advanced Methods in Pharmacovigilance and Pharmacoepidemiology
Volume 2 - 2022 | https://doi.org/10.3389/fdsfr.2022.1021068
This article is part of the Research TopicComputational Methods and Systems to Support Decision Making in PharmacovigilanceView all 7 articles
Understanding and mitigating medication errors is critical for ensuring patient safety and improving patient care. Correctly identifying medication errors in the United States Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) reports can be difficult because of the complexities of medication error concepts. We took a user-centered design approach to support the medication error categorization workflow process with artificial intelligence (AI). We developed machine learning models to categorize medication error terms. The average F1-score, precision, recall, and area under the precision recall curve for 18 Medical Dictionary for Regulatory Activities (MedDRA) Lower Level Term (LLT) relating to medication errors were 0.88, 0.92. 0.85, and 0.83 respectively. We developed a framework to help evaluate opportunities for artificial intelligence integration in the medication error categorization workflow. The framework has four attributes: technical deployment, process rigidity, AI assistance, and frequency. We used the framework to compare two AI integration opportunities and concluded that the quality assurance (QA) opportunity to be a more feasible initial option for AI integration. We then extended these insights into the development and user testing of a prototype application. The user testing identified the highlighting and commenting capabilities of the application to be more useful and sliders and similar report suggestions to be less useful. This suggested that different AI interactions with human highlighting should be explored. While the medication error quality assurance prototype application was developed for supporting the review of direct FAERS reports, this approach can be extended to assist in the workflow for all FAERS reports.
The United States Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) receives more than 100,000 reports each year associated with a suspected medication error. A medication error is generally defined as any preventable event that may cause or lead to inappropriate medication use or patient harm while the medication is in the control of the health care professional, patient, or consumer (National Coordinating Council for Medication Error Prevention and Reporting, 2022); medication errors are one of the leading causes of avoidable adverse events in global healthcare systems, with costs estimated at $42 billion annually (The World Health Organization, 2017). Accurate coding of the medication error information in the reports is critical for the FDA to identify and mitigate the errors to minimize the risk of adverse events. However, coding the information in the reports can be challenging because of the complexities of medication error concepts (e.g., individual definitions of a medication error, understanding of root causes and contributing factors related to errors, or incomplete or overlapping coding terminologies). These challenges can result in inconsistent and incorrect coding of the medication error information, which necessitates exploring artificial intelligence techniques to improve coding practices.
It is important to first understand the workflow for medication error categorization at a high level to provide context for AI system integration opportunities. Medication error reports are voluntarily submitted by healthcare providers and consumers to FAERS either directly through the MedWatch program (referred to as “direct reports”) using various channels (e.g., online reporting, postal mail, email, fax) (United States. Food and Drug Administration, 2022) or through drug manufacturers who submit the reports electronically using standardized regulatory reporting forms. Workflow for medication error categorization of direct reports primarily consists of three steps: Intake and triage, Case processing, and Data analytics and publishing, Figure 1. These processes are supported by the transactional platform in FAERS. Intake and triage: First, direct reports are reviewed by a pharmacist to make sure the report has the necessary elements (e.g., identifiable reporter, event, and drug product) for a valid report to be entered into FAERS (reports not involving a drug product such as a dietary supplement or device are forwarded to the appropriate FDA center to be entered in their respective reporting systems). Case Processing: The reports are then sent to case processing where the report information (approximately 50 different data fields) is entered into the FAERS system, and the drug product name(s) and manufacturer name is validated. The reports are then routed to clinicians who read the free-text case narrative and use the Medical Dictionary for Regulatory Activities (MedDRA) to manually select the appropriate medication error, adverse event, and product quality Lower Level Terms (LLT). Data Analytics and Publishing: After the case processing is complete, the reports are available to safety analysts who rely on the codes to screen and retrieve reports for safety signal detection and evaluation to determine if regulatory action is needed to mitigate a medication error. The coded reports are also used by the public in a variety of ways to support the analysis, research, and identification issues or trends that may impact public safety. In addition, the reports are manually reviewed by the FDA for coding, quality assurance, and to provide feedback to the coders to help with training and learning.
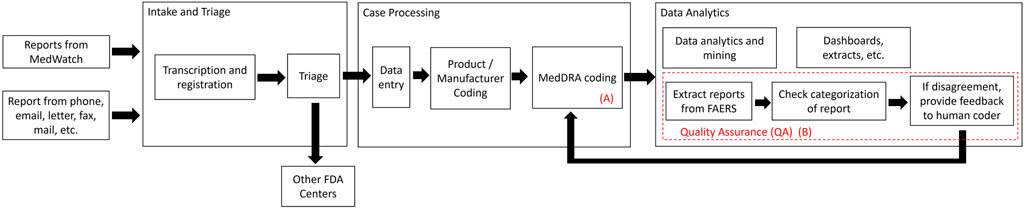
Figure 1. Summary of medication error categorization workflow for direct FAERS reports. Two opportunities for AI integration are identified as (A) human MedDRA coding of FAERS reports and (B) quality assurance review process.
There have been several calls for using natural language processing (NLP), machine learning (ML), and artificial intelligence (AI) approaches to help identify and categorize adverse events and medication errors. Much work has been done on the development of algorithms and techniques to categorize and identify adverse drug events in FAERS reports (Botsis et al., 2014; Combi et al., 2018; Eskildsen et al., 2020). A recent review of 14 publications provided examples where NLP supported the identification of adverse drug reactions (Pilipiec et al., 2022). In addition to FAERS reports adverse drug events were able to be extracted from drug labels and Vaccine Adverse Event Reporting System (VAERS) reports using NLP and rule-based techniques (Botsis et al., 2013; Ly et al., 2018; Bayer et al., 2021; Du et al., 2021). Vaccine Adverse Event Text Miner (VaeTM) is an example of a text mining system developed to extract safety concepts from VAERS reports (Botsis et al., 2011; Botsis et al., 2012; Baer et al., 2016). More recent works have explored the contributions of NLP to support adverse event reporting workflows and operational insights through the use of decision support systems and information visualization applications (Botsis et al., 2016; Spiker et al., 2020). These recent works aims to tackle the important challenges of translating technical advances into operations to support workflow (Ball and Dal Pan, 2022).
Complexities in real-world workflows often contain important information external to reports that impact both the modeling process and application of such models (Kreimeyer et al., 2021). In fact, challenges with AI initiatives and integration of AI into workflow had been discussed as early as the 2000s (Myers and Berry, 1999). More recently, discussions have focused on strategies and frameworks to help software and application developers and stakeholders evaluate AI workflow integration. A four step approach has been proposed for understanding the technologies that are involved, creating a portfolio of projects, piloting projects, and scaling up (Davenport and Ronanki, 2018). Additionally, Translational Evaluation of Healthcare AI (TEHAI) has been developed as a framework to evaluate the operationalization of clinical AI models (Reddy et al., 2021). TEHAI has a foundation in translational research and contains three main components: capability, utility, and adoption. TEHAI can support starting steps in a user-centered design process by providing a framework to help identify user needs and the specific context of an application’s use. Although TEHAI is a very comprehensive framework, it is lengthy and resource intensive to use in its entirety. In this work, we created a high-level framework to specifically help decision makers and stakeholders evaluate early prototyping opportunities for integrating AI models into a ‘human-in-the-loop’ workflow. We leverage TEHAI and other software development frameworks with a socio-technical lens to infer the value, benefit over cost, associated with integrating AI models into a real-world workflow (Norman and Draper, 1986; Sittig and Singh, 2010; Reddy et al., 2021).
In this paper, we leverage the vast knowledge of adverse drug reaction ML and NLP work to address the challenges of categorizing medication errors. The contributions of this paper are three-fold. First, we develop ML models to categorize MedDRA LLTs relating to medication errors in FAERS free-text narratives. Second, we present a framework to help evaluate AI workflow integration opportunities and challenges in the medication error workflow. Third, we present and evaluate an AI prototype design to support the medication error quality assurance process.
We used FAERS direct reports received between 1/1/2017 and 10/29/2021 that were coded with at least one MedDRA medication error LLT for the modeling. For the initial development, we focused on the modeling of medication error LLTs that occurred in at least 1% of reports.
We extracted free-text between templated language from the free-text case narratives. Templated language is introduced to narratives when reporters use a form or templates when completing their report. When these reports are submitted to FAERS, the free-text responses as well as the templated language (i.e., headers) in the structured forms or templates are concatenated together into a string. We accounted for different templates and structured languages amongst all form types. Reports will have different amounts, variations, and spacing of templated language depending on how the report was completed. We took a dynamic programming approach to extract text between templated language. We first created a list of possible templated language. We then indexed the beginning and end location in the string of all occurring templated language with more than five words. We lastly ordered the index and extracted the text between sequential templated language. Conditions are included to check and include narratives with both templated language and narratives that do not start with templated language. The extracted free-text is lowercased and stemmed. Stemming is the process of reducing derived or inflected words to a base form (Friedman and Johnson, 2006). For example, “fly,” “flying,” and “flies” will be stemmed as “fli.” Numbers, punctuations, and extra white space are removed. We then generated term frequency-inverse document frequency (TF-IDF) feature vectors from the cleaned free-text. TF-IDF is a statistical measure that evaluates how relevant a word is to a document in a collection of documents (Ramos, 2003). The TF-IDF score of a word in a document is calculated by multiplying two metrics: the number of times a word appeared in a document and the inverse document frequency of the word across a set of documents. TF-IDF is a popular method to translate text to numerical features and is used in the subsequent model development (Ramos, 2003).
Each LLT (label) model was developed using an 80/20 split of the data: 80% of the data was used for training and validation and 20% reserved for testing. Models were developed using eXtreme Gradient Boosting (XGBoost). We choose XGBoost because it had better performance during initial sample testing when compared to logistic regression and random forest, two other popular machine learning algorithms. XGBoost is a decision tree-based boosting ensemble machine learning algorithm (Chen and Guestrin, 2016). In a boosting algorithm, many weak learners, which are simple classification models that perform only slightly better than random chance, are trained to correctly classify the observations that were incorrectly classified in the previous rounds of training. XGBoost uses shallow trees as weak learners (Chen and Guestrin, 2016). Each model was trained using 5-fold cross validation and evaluated on test precision, recall, F1-score, and area under precision-recall curve.
We leveraged TEHAI and AI integration frameworks to infer value components, defined as technical deployment, process rigidity, AI assistance, and frequency to identify opportunities to prototype an AI system into a medication error categorization workflow.
The time and resources required to develop and deploy an AI system are similar to software product development considerations in real-world settings (Myers and Berry, 1999; Davenport and Ronanki, 2018). However, AI system product design is different from regular software product design, largely in the maintenance of data, models, and user feedback (Davenport and Ronanki, 2018). In general, integrating an AI system into existing software systems would be a higher technical deployment cost compared to deploying a stand-alone AI system.
We define process rigidity as the level of variability of how end-users accomplish tasks, a reflection on understanding workflow in social technical systems (Myers and Berry, 1999). Challenges with integrating AI projects with existing workflows and processes is a common problem faced by AI initiatives (Davenport and Ronanki, 2018). Established workflows where end-users all follow the same process and use the same tools could show high process rigidity. On the other hand, workflows where end-users can have more autonomy would be low process rigidity.
The assistance of an AI system in an existing workflow can be realized in time and resource savings or other measures of support (Reddy et al., 2021). This attribute is viewed in consideration of current workflows and the additional benefit or support an AI system can provide. In the exploratory evaluation process, assistance can be reflective of both measured and perceived benefits by an end-user.
We define frequency of a deployed AI system as the number of end-users and regularity of use (Reddy et al., 2021). An AI system that is used by many end-users daily would have high frequency. An AI system used by one or two individuals monthly would have low frequency. We conducted semi-structured interviews with stakeholders involved in the medication error categorization workflow to evaluate and identify opportunities for AI system integration using the AI workflow integration framework.
We used guerilla usability testing, sometimes referred to as hallway usability testing, for initial iterative design process development of the prototype AI system to support the medication error categorization workflow. Guerilla usability testing is used to gather immediate feedback on interactions or the flow of an application or website when needed. The benefits of this form of testing are best realized during the initial low-fidelity design to influence further iterations (Nielsen and Guerrilla, 1994). Guerilla testing was used among four participants who were experienced in reviewing FAERS reports. This testing involved the utilization of a scenario followed by a series of short tasks to guide participants through a workflow comprised of placeholder text and did not contain medication error report data. Participants were asked to use the ‘think aloud’ method to provide feedback on the prototype and open discussion related to the interaction that the user was experiencing. These responses were then analyzed and used to change existing interactions and modify future prototype designs to prepare for formal usability testing with participants of the target users.
After the iterative design development of a functional prototype, 1-h long formal usability testing sessions were conducted with participants involved in the medication error categorization workflow. These sessions were comprised of a scenario and tasks to walk the participants through the prototype. Like the guerilla testing stage, participants were asked to “think aloud” and pause between tasks to provide feedback during the session. The placeholder text was replaced with synthetic report information to provide a more realistic experience for the participants. During testing, participants’ responses were recorded through digital notetaking and were then synthesized to influence future tool updates with a focus on design and workflow. After the completion of the session tasks, each participant was asked to complete the System Usability Scale (SUS). The SUS is a post-test questionnaire that is commonly used to perceive the usability of an entire system and can suggest problematic parts of a design (Bangor et al., 2008).
The 11,524 free-text case narratives were used in the development of 18 medication error LLT models, summarized in Table 1. The most common LLTs were “product storage error,” “incorrect dose administered,” and “inappropriate schedule of drug administration” 6.9%, 5.9%, and 5.3% respectively. The average F1-score was 0.88 (0.89 median, 0.06 standard deviation) with “wrong technique in product usage process” and “wrong injection technique” having the highest F1-score, 0.99 and 0.98 respectively. The average precision was 0.92 (0.93 median, 0.05 standard deviation). The average recall was 0.85 (0.85 median, 0.08 standard deviation). The average area under the precision-recall curve was 0.83 (0.88 median, 0.18 standard deviation). “Wrong technique in product usage process” and “wrong injection technique” consistently had the highest performance across metrics. On the other hand, “drug prescribing error” and “drug dose prescribing error” tended to have the worst performance across metrics.
Semi-structured interviews were conducted with six stakeholders involved in the medication error workflow integration process. Using the AI workflow integration framework, two primary opportunities for the integration of AI into the current workflow were considered. The first opportunity is during the initial human coding of FAERS reports, Figure 1A. The second opportunity is during the quality assurance review process, Figure 1B. These two options were identified by the stakeholders as being able to directly benefit from the medication error categorization models. We review both options using the developed framework, Table 2.
During the semi-structured interviews, it was determined that a direct integration of an AI system into the human coder system would be significant (Technical deployment: High) and that the coding process between the intake and coders was well established (Process rigidity: High). After data entry, the reports were routed to the coders. This was where the free-text case narrative was read by human clinicians and the appropriate medication error, adverse event, or product quality issue was manually coded at the MedDRA LLT level. The current process occurs frequently although the number of reports submitted to FAERs can greatly vary week to week (Frequency: High). Coders can refer to MedDRA supporting documentation for guidance with coding medication errors (Medical Dictionary for Regulatory Activity, 2022). An AI system to support in this coding process could be beneficial to ensure consistency in selecting appropriate LLTs and reduce the need to refer to additional documentation, however human coders will still need to input and manually review other information about the report (Assistance: Medium).
The quality assurance (QA) process is highlighted with the dashed red box in Figure 1. Stakeholders interviewed described this as a very labor intense process with varying frequency. As a result, an AI system to support the QA process would have high assistance (Assistance: High) though the frequency of use would be lower (Frequency: Medium-Low). In addition, the current manual QA process largely relies on structured data elements to first filter reports in the FAERS applications (Process rigidity: Low). Instead of spot checking for QA, the AI system could function as a surveillance system to push disagreements to users for review. Lastly, through stakeholder discussions, it was determined the cost associated with the technical deployment and overcoming process rigidity is much lower for QA. We concluded the QA opportunity to be a more feasible opportunity and incremental step for AI integration (Davenport and Ronanki, 2018).
We present the design and functionality of the Medication Error QA prototype application, in Figure 2. The prototype will integrate into the QA workflow by providing a stand-alone application to help quality assurance reviewers and safety analysts more quickly inspect, comment, and alert coders to reports that may need recoding. The intention is that this application can serve as a stand-alone platform to both extract reports from the FAERS database and provide feedback to coders thereby replacing the need to use different applications to accomplish the same task. The features of the prototype discussed below are: report overview, flagging slider and similarity slider, highlighting and comment on reports, and feedback to coders.
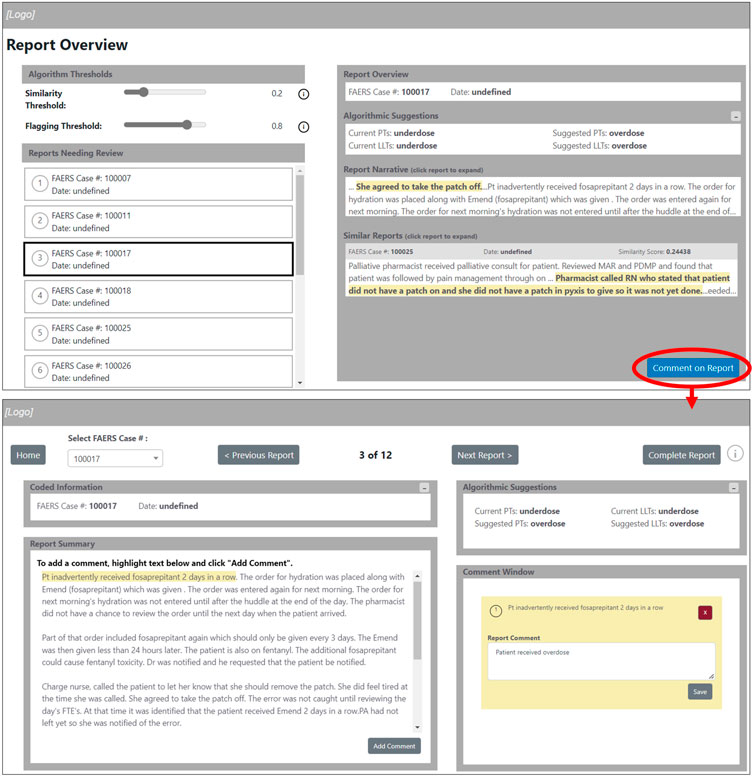
Figure 2. Medication error quality assurance prototype application screens. The arrow indicates the transition between the “Report Overview” screen and the report annotation screen. In the example figure, the human coded the report using the “underdose” LLT while the AI model suggests the “overdose” LLT.
The “Report Overview” screen will give the user the ability to review each report prior to annotating as well as reviewing any information that was previously coded, Figure 2. The reports will be separated by “Reports Needing Review” and “Completed Reports” to indicate which reports still need to be completed by the user. By selecting a report, the user can review the summary of the report and previously coded information, as well as review coding suggestions from the algorithm for similar reports. As the reports are annotated, they are shown as completed using a check mark on the ‘Report Overview’ screen and are available for review under completed reports. This will provide the user the ability to review the completed reports for reference or if any further changes need to be made. This feature can be used as a final review step and will provide the ability to correct possible coding errors.
The flagging slider and similarity slider available on the “Report Overview” screen, Figure 2, will allow the user to interact with the AI models. The flagging slider will change the threshold at which a report is flagged, or identified, as having inconsistent or incorrect MedDRA LLT codes. A flagged report would need additional human review. A higher flagging threshold will result in fewer reports being flagged (more specific and higher precision) while a lower flagging threshold will result in more reports being flagged (higher recall) as inconsistent or incorrect LLT codes. The similarity slider will impact the amount of similar reports that are presented to users in the ‘Similar Reports’ window. A high similarity threshold will result in few similar reports and a low similarity threshold will result in more similar reports. The highlighting of phrases in the reports is intended to help users identify meaningful phrases as prioritized by the algorithm.
This application provides the user the ability to highlight and comment on specific phrases in a case narrative. The user can comment on a highlighted section of the report which will be saved in the “Comment Window.” These comments can be edited or deleted. The “Coded Information” window will provide an easy reference for the user to understand further context surrounding the report. The “Algorithm Suggestions” window will provide suggested codes to apply to the report based on information from similar reports as well as the context of the current report being reviewed.
Once the user completes reviewing and commenting on the reports, the option of sending or exporting the annotated reports will become available on the “Report Overview” screen. Although this feature was not implemented at the time of user testing, the intention of this feature is to construct an e-mail containing the completed annotated reports that can then be reviewed by designated users. This will accomplish the goal of sharing annotated information from an expert to coders.
Individual usability testing sessions were conducted virtually with six participants. The average years of experience with coding or reviewing postmarketing reports amongst the participants was 7.1 years (6.5 median). The participants included one regulatory scientist, one informatic pharmacist, two managers, and two directors involved in the medication error categorization workflow. Several usability themes emerged from the feedback during the hour-long usability testing sessions.
The feature of highlighting and commenting on the text within the report summary was easy and useful for many of the participants. Participants could follow the on-screen instructions and stated that this feature was useful in identifying specific regions of the report that would justify a different code related to directly submitted reports. [P2] and [P6] remarked that the interaction and multi-colored notes were easy to keep their comments organized and reminded them of other software that they were familiar with. In addition, participants made several recommendations for improvement. [P3] and [P5] wanted to have the ability to make comments without highlighting text and commenting on the codes that were documented. Maintaining the order and color consistencies of the comments was also important to several participants [P1, P3, P4, and P6].
All participants noted the need for additional context within both the report overview and report comment screens. All participants stated that the report summary alone did not contain enough information for proper analysis and feedback. Providing more context about the report would reduce the ambiguity of the individual report summary. [P1], [P3], and [P6] requested further information regarding the suspect drug product labeling and the full FAERS report in an PDF format when available. With this information, the context surrounding the initial codes could be further understood while justifying any changes. In addition, more information related to the number of reports that were flagged as miscoded was also requested by all participants. [P1] remarked that having a count of reports “needing review” and “completed reports” would help “stay on track with the number of reports to review.” Clearer color coding of the completed reports was also suggested by [P4] and [P6] and the gray boxes and gray font would need to be changed to reflect principles of accessibility within the design.
No participants used the threshold sliders while reviewing the report narrative. [P1], [P3], and [P3] believed its use to be unclear and that the terminology used was not similar to that used by the coders of the report. [P1] mentioned that a legend or a way to find more information about the use of the sliders could help with any confusion, however it was also stated that he or she still would not use them within their review workflow.
All participants were not certain of the purpose of the similar reports that were available to review under the report summary. [P3] and [P6] believed them to be interesting, though noted that they would not be useful within their workflow for report review. The highlighted text within the similar report window was not considered helpful to many of the participants, as it was unclear how the highlighted text was associated with the report summary. [P2] and [P3] recommended a different way to observe the similarities among the reports by highlighting specific words or phrases within the similar reports that were closely related to the coder’s terminology. This would allow the reviewer the ability to understand similar reports that were related to the case narrative. However, the similar reports would not have been used within several of the participants’ workflows [P1, P2, P4, and P5].
The SUS is considered a valid and reliable questionnaire and was given to the participants after the usability test. All six participants completed the SUS, where each question was given an adjective rating that was associated with a numerical score of 1–5, Figure 3. The overall SUS score after testing was 50, which fell within the marginal acceptability range for use and shows a need for further design iteration to increase user experience and usability (Bangor et al., 2009).
The feedback of the stand-alone prototype application has been positive. One of the more useful features was the highlighting and commenting capabilities. These features helped participants focus their attention and communicate their intentions in the feedback loop. It is interesting to note that this feature parallels the motivation behind the AI highlighting of text in similar reports. However, participants generally did not find the similar reports to be useful and did not use the sensitivity and specificity sliders. This suggests that different AI interactions with human highlighting should be explored, such as AI evaluation of human highlighted text.
While the medication error prototype application is designed to support the medication error categorization workflow for direct reports, this application and associated models can be expanded to support the review and analysis for all FAERS reports, including those submitted by manufacturers. There is considerable focus on using ML to support the pharmacovigilance lifecycle and understanding how an integrated AI system can help with the ingestion and data integrity of data is paramount to both realizing practical impact and operational insights (Bate and Luo, 2022). The majority of FAERS reports are electronically submitted to the FDA from manufacturers and consistent analysis and review of these reports can help ensure data and coding integrity both between and within manufacturers. As consumer and healthcare behaviors change, proactively monitoring all FAERS reports will be crucial in helping safety, healthcare, and regulatory personnel identify and address new and emerging medication errors and patient safety concerns.
We recognize that the AI workflow integration framework generalizes many of the variables and interactions between variables involved in the deployment of an AI system. We also recognize that this framework does not include considerations like AI ethics and privacy highlighted by other frameworks (Reddy et al., 2021). However, for AI system to be useful and accepted by users, evaluating the AI alone is not sufficient. It is critical to also consider how the AI system was implemented (Li et al., 2020). The AI workflow integration framework provided a ‘light-weight’ method to help facilitate discussion amongst decision makers in the early prototype phase. This framework is not to replace formal usability testing, product development cycles, and implementation scientific methods but rather provide a stepping-stone in the process. Lastly, we recognize the limitations with usability testing with synthetic data and will include real-world data in subsequent usability sessions.
We used a user-center design approach to integrate AI in the medication error categorization workflow. As AI models improve, technologies advance, and workflows change, there will be new and different opportunities for human machine integration. The AI workflow integration framework was helpful in the initial decision making for prototyping and prioritizing a stand-alone application over a more integrated option. This framework builds upon and can complement existing product development and usability frameworks for stakeholders when exploring options early in the integration cycle.
The data analyzed in this study is subject to the following licenses/restrictions: FAERS free-text case narratives are used to generate the AI models. FAERS free-text case narratives are not publically available. Requests to access these datasets should be directed to https://www.fda.gov/regulatory-information/freedom-information/how-make-foia-request.
The studies involving human participants were reviewed and approved by the MedStar Health Research Institute. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
AF has substantial contributions to the conception and design of the work, acquisition, analysis and interpretation of data. AF has been involved in the drafting, revising, and final approval of the work. CB has substantial contributions to the design of the work, acquisition, analysis and interpretation of data. CB has been involved in the drafting, revising, and final approval of the work. VV has substantial contributions to the design of the work, acquisition, and analysis of data. VV has been involved in the drafting, revising, and final approval of the work. SD has substantial contributions to the design of the work, analysis and interpretation of data. SD has been involved in the drafting, revising, and final approval of the work. DK has substantial contributions to the design of the work, analysis and interpretation of data. DK has been involved in the drafting, revising, and final approval of the work. JW has substantial contributions to the design of the work, acquisition, analysis and interpretation of data. JW has been involved in the drafting, revising, and final approval of the work.
This work was funded by the US Food and Drug Administration (Contract Number: 75F40121C00089).
We want to acknowledge the tremendous support from the MedStar Health team members, Raj Ratwani, Zach Hettinger, and Christopher Washington on this project. In addition, we want to thank the support, guidance, and insights from the US Food and Drug Administration team.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baer, B., Nguyen, M., Woo, E. J., Winiecki, S., Scott, J., Martin, D., et al. (2016). Can natural language processing improve the efficiency of vaccine adverse event report review? Methods Inf. Med. 55 (02), 144–150. doi:10.3414/ME14-01-0066
Ball, R., and Dal Pan, G. (2022). Artificial intelligence” for pharmacovigilance: Ready for prime time? Drug Saf. 45 (5), 429–438. doi:10.1007/s40264-022-01157-4
Bangor, A., Kortum, P., and Miller, J. (2008). An empirical evaluation of the system usability scale. Int. J. Hum. Comput. Interact. 24 (6), 574–594. doi:10.1080/10447310802205776
Bangor, A., Kortum, P., and Miller, J. (2009). Determining what individual SUS scores mean: Adding an adjective rating scale. J. usability Stud. 4 (3), 114–123.
Bate, A., and Luo, Y. (2022)., 45. Springer, 403–405.Artificial intelligence and machine learning for safe medicinesDrug Saf.
Bayer, S., Clark, C., Dang, O., Aberdeen, J., Brajovic, S., Swank, K., et al. (2021). ADE eval: An evaluation of text processing systems for adverse event extraction from drug labels for pharmacovigilance. Drug Saf. 44 (1), 83–94. doi:10.1007/s40264-020-00996-3
Botsis, T., Buttolph, T., Nguyen, M. D., Winiecki, S., Woo, E. J., and Ball, R. (2012). Vaccine adverse event text mining system for extracting features from vaccine safety reports. J. Am. Med. Inf. Assoc. 19 (6), 1011–1018. doi:10.1136/amiajnl-2012-000881
Botsis, T., Jankosky, C., Arya, D., Kreimeyer, K., Foster, M., Pandey, A., et al. (2016). Decision support environment for medical product safety surveillance. J. Biomed. Inf. 64, 354–362. doi:10.1016/j.jbi.2016.07.023
Botsis, T., Nguyen, M. D., Woo, E. J., Markatou, M., and Ball, R. (2011). Text mining for the vaccine adverse event reporting system: Medical text classification using informative feature selection. J. Am. Med. Inf. Assoc. 18, 631–638. [Internet]. [cited 2014 Sep 15];18(5):631–8. Available from: aHR0cDovL3d3dy5wdWJtZWRjZW50cmFsLm5paC5nb3YvYXJ0aWNsZXJlbmRlci5mY2dpP2FydGlkJiN4MDAwM2Q7MzE2ODMwMCYjeDAwMDI2O3Rvb2wmI3gwMDAzZDtwbWNlbnRyZXomI3gwMDAyNjtyZW5kZXJ0eXBlJiN4MDAwM2Q7YWJzdHJhY3Q=. doi:10.1136/amiajnl-2010-000022
Botsis, T., Scott, J., Goud, R., Toman, P., Sutherland, A., and Ball, R. (2014). “Novel algorithms for improved pattern recognition using the US FDA Adverse Event Network Analyzer. Stud. Health. Technol. Inform. 205, 1178–1182.
Botsis, T., Woo, E. J., and Ball, R. (2013). Application of information retrieval approaches to case classification in the vaccine adverse event reporting system. Drug Saf. 36 (7), 573–582. doi:10.1007/s40264-013-0064-4
Chen, T., and Guestrin, C. (2016). “Xgboost: A scalable tree boosting system,” in Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, 785–794.
Combi, C., Zorzi, M., Pozzani, G., Moretti, U., and Arzenton, E. (2018). From narrative descriptions to MedDRA: Automagically encoding adverse drug reactions. J. Biomed. Inf. 84, 184–199. doi:10.1016/j.jbi.2018.07.001
Davenport, T. H., and Ronanki, R. (2018). Artificial intelligence for the real world. Harv Bus. Rev. 96 (1), 108–116.
Du, J., Xiang, Y., Sankaranarayanapillai, M., Zhang, M., Wang, J., Si, Y., et al. (2021). Extracting postmarketing adverse events from safety reports in the vaccine adverse event reporting system (VAERS) using deep learning. J. Am. Med. Inf. Assoc. 28 (7), 1393–1400. doi:10.1093/jamia/ocab014
Eskildsen, N. K., Eriksson, R., Christensen, S. B., Aghassipour, T. S., Bygsø, M. J., Brunak, S., et al. (2020). Implementation and comparison of two text mining methods with a standard pharmacovigilance method for signal detection of medication errors. BMC Med. Inf. Decis. Mak. 20, 94–11. doi:10.1186/s12911-020-1097-0
Friedman, C., and Johnson, S. B. (2006). natural language and text processing in biomedicine, 312–343.
Kreimeyer, K., Dang, O., Spiker, J., Muñoz, M. A., Rosner, G., Ball, R., et al. (2021). Feature engineering and machine learning for causality assessment in pharmacovigilance: Lessons learned from application to the FDA Adverse Event Reporting System. Comput. Biol. Med. 135, 104517. doi:10.1016/j.compbiomed.2021.104517
Li, R. C., Asch, S. M., and Shah, N. H. (2020). Developing a delivery science for artificial intelligence in healthcare. NPJ Digit. Med. 3 (1), 107–113. doi:10.1038/s41746-020-00318-y
Ly, T., Pamer, C., Dang, O., Brajovic, S., Haider, S., Botsis, T., et al. (2018). Evaluation of natural language processing (NLP) systems to annotate drug product labeling with MedDRA terminology. J. Biomed. Inf. 83, 73–86. doi:10.1016/j.jbi.2018.05.019
Medical Dictionary for Regulatory Activity (2022). International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human UsePoints to consider documents and MedDRA best practices document. Available at: https://www.meddra.org/how-to-use/support-documentation/english.
Myers, K. L., and Berry, P. M. (1999). “At the boundary of workflow and AI,” in Proc AAAI 1999 workshop on agent-based systems in the business context.
National Coordinating Council for Medication Error Prevention and Reporting (2022). What is a medication error?. Available at: https://www.nccmerp.org/about-medication-errors
Nielsen, J., and Guerrilla, H. C. I. (1994). Using discount usability engineering to penetrate the intimidation barrier. Cost-justifying usability, 245–272.
Pilipiec, P., Liwicki, M., and Bota, A. (2022). Using machine learning for pharmacovigilance: A systematic review. Pharmaceutics 14 (2), 266. doi:10.3390/pharmaceutics14020266
Ramos, J. (2003). “Using TF-IDF to determine word relevance in document queries,” in Proceedings of the first instructional conference on machine learning. Available from: aHR0cDovL2NpdGVzZWVyeC5pc3QucHN1LmVkdS92aWV3ZG9jL2Rvd25sb2FkP2RvaSYjeDAwMDNkOzEwLjEuMS4xMjE=.
Reddy, S., Rogers, W., Makinen, V-P., Coiera, E., Brown, P., Wenzel, M., et al. (2021). Evaluation framework to guide implementation of AI systems into healthcare settings. BMJ Health Care Inf. 28 (1), e100444. doi:10.1136/bmjhci-2021-100444
Sittig, D. F., and Singh, H. (2010). A new socio-technical model for studying health information technology in complex adaptive healthcare systems. Qual. Saf. Health Care 19 (3), i68–i74. doi:10.1136/qshc.2010.042085
Spiker, J., Kreimeyer, K., Dang, O., Boxwell, D., Chan, V., Cheng, C., et al. (2020). Information visualization platform for postmarket surveillance decision support. Drug Saf. 43 (9), 905–915. doi:10.1007/s40264-020-00945-0
The World Health Organization (2017). Medication Without Harm: WHO Global Patient Safety Challenge. World Heal. Organ., 4654.
U.S. Food and Drug Administration (2022). MedWatch: The FDA safety information and adverse event reporting program. [Internet]. [cited 2022 Aug 10]. Available from: aHR0cHM6Ly93d3cuZmRhLmdvdi9zYWZldHkvbWVkd2F0Y2gtZmRhLXNhZmV0eS1pbmZvcm1hdGlvbi1hbmQtYWR2ZXJzZS1ldmVudC1yZXBvcnRpbmctcHJvZ3JhbQ==.
Keywords: natural language processing (computer science), machine learning, ML, human-in-the-loop (HITL), quality assurance, pharmacovigilance (MeSH), medication error, usability and user experience
Citation: Fong A, Bonk C, Vasilchenko V, De S, Kovich D and Wyeth J (2022) Exploring opportunities for AI supported medication error categorization: A brief report in human machine collaboration. Front. Drug Saf. Regul. 2:1021068. doi: 10.3389/fdsfr.2022.1021068
Received: 17 August 2022; Accepted: 26 October 2022;
Published: 10 November 2022.
Edited by:
Taxiarchis Botsis, Johns Hopkins Medicine, United StatesReviewed by:
Pantelis Natsiavas, Institute of Applied Biosciences, GreeceCopyright © 2022 Fong, Bonk, Vasilchenko, De, Kovich and Wyeth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Allan Fong, YWxsYW4uZm9uZ0BtZWRzdGFyLm5ldA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.