- 1Lankenau Institute for Medical Research, Wynnewood, PA, United States
- 2Department of Basic and Translational Sciences, University of Pennsylvania School of Dental Medicine, Philadelphia, PA, United States
- 3Materials Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 4Department of Bioengineering and Materials Science and Engineering, UC Berkeley, Berkeley, CA, United States
Bone injuries and fractures reliably heal through a process of regeneration with restoration to original structure and function when the gap between adjacent sides of a fracture site is small. However, when there is significant volumetric loss of bone, bone regeneration usually does not occur. In the present studies, we explore a particular case of volumetric bone loss in a mouse model of human periodontal disease (PD) in which alveolar bone surrounding teeth is permanently lost and not replaced. This model employs the placement of a ligature around the upper second molar for 10 days leading to inflammation and bone breakdown and closely replicates the bacterially induced inflammatory etiology of human PD to induce bone degeneration. After ligature removal, mice are treated with a timed-release formulation of a small molecule inhibitor of prolylhydroxylases (PHDi; 1,4-DPCA) previously shown to induce epimorphic regeneration of soft tissue in non-regenerating mice. This PHDi induces high expression of HIF-1α and is able to shift the metabolic state from OXPHOS to aerobic glycolysis, an energetic state used by stem cells and embryonic tissue. This regenerative response was completely blocked by siHIF1a. In these studies, we show that timed-release 1,4-DPCA rapidly and completely restores PD-affected bone and soft tissue with normal anatomic fidelity and with increased stem cell markers due to site-specific stem cell migration and/or de-differentiation of local tissue, periodontal ligament (PDL) cell proliferation, and increased vascularization. In-vitro studies using gingival tissue show that 1,4-DPCA indeed induces de-differentiation and the expression of stem cell markers but does not exclude the role of migrating stem cells. Evidence of metabolic reprogramming is seen by the expression of not only HIF-1α, its gene targets, and resultant de-differentiation markers but also the metabolic genes Glut-1, Gapdh, Pdk1, Pgk1 and Ldh-a in the periodontal tissue.
Introduction
Humans, like virtually all mammals, heal tissue and organ injuries by the process of scarring with limited restoration of normal anatomical integrity and functionality. This is in contrast to species such as newts, salamanders and other vertebrates which heal perfectly through the process of regeneration. In these examples, bone and soft tissues are replaced creating indistinguishable replicas of lost or damaged tissues (1–3). There are several routes to mammalian regeneration being actively considered for regenerative therapies, usually involving the use of stem cells with or without bioscaffolds (4–9). In a dental context, pulp stem cells as an autologous graft and the use of compounds such as high molecular weight hyaluronic acid can affect periodontal bone healing (10–12). However, the possibility of a systemically acting drug which alone could induce regeneration in multiple tissues would be an intriguing therapy.
The path to identifying such a drug began with the serendipitous observation that the MRL mouse strain, long employed in autoimmunity studies, possessed an unusual capacity for tissue regeneration. Through-and-through ear pinna wounds used as lifelong mouse identifiers healed by fully closing without scarring within 30 days. All tissue types found in the ear including epidermis, dermis, blood vessels, nerve, glands, cartilage and hair follicles were restored (13, 14). Furthermore, multiple studies showed that this regenerative phenotype extended to MRL cornea, tendon, cartilage, muscle, fat, and other tissues (15–18). However, bone injuries were largely unexplored.
Insights into the biological basis of the regenerative phenotype of the MRL mouse came from the fact that the adult MRL employs a strongly aerobic glycolytic metabolism in the basal state also seen in embryos and stem cell niches. This metabolic state was enhanced during regenerative wound healing but not normal wound repair (16, 19, 20). One well-known molecule that regulates aerobic glycolysis is hypoxia-inducible factor (HIF-1α; 21, 22), which was highly upregulated in the MRL upon initiation of injury (23). Blocking HIF-1α using siHIF1a completely blocked the regenerative response (23).
The term “metabolic reprogramming” has been generally used to describe the metabolic state of tumor cells relative to normal cells (24–27). However, it has also been used to describe changes in metabolism necessary for proliferating cells in general. Here, glycolysis, with increased lactic acidosis, glucose consumption, and amino acid and nucleic acid synthesis, is preferred over oxidative phosphorylation. By transiently increasing the level of HIF-1α, a major positive regulator of aerobic glycolysis, through blocking prolyl hydroxylases (PHDs) that negatively control HIF-1α levels, the PHD inhibitor (PHDi), 1,4-DPCA, was employed as a regeneration activator.
In the current set of studies, we explored the possibility that 1,4-DPCA (28) could also have a positive effect on the regeneration of bone and soft tissue of the jaw in mice. The use of 1,4-DPCA delivered in a hydrogel or coupled to PEG was previously shown by our laboratories to lead to a regenerative healing response resulting in closure of ear holes and replacement of soft tissue in mice (16, 23, 29) indistinguishable from that seen in MRL mice. This outcome was also seen in a second soft tissue target, enhanced DPCA-induced liver regeneration (30). We employed a mouse model of periodontal disease induced by placing a ligature around the upper second molar for 10 days, leading to an oral bacterial accumulation and bone degeneration, followed by removal of that ligature (31–33). 1,4-DPCA coupled to a PEG gel was then administered systemically followed by a second dose of drug administered 8 days later. After 20 days post-ligature removal and administration of drug, we saw full replacement of alveolar bone and gingival soft tissue.
Using micro-CT analysis of bone damage and regeneration combined with immunohistochemistry and gene expression, we explored the DPCA effect (23, 29) for over 220 days in longitudinal studies to determine if full regeneration was achieved and maintained. In addition to the rapid recovery of alveolar bone architecture and gingival soft tissue there was significant regeneration of the periodontal ligament (PDL) which attaches the tooth to bone and in the pulp of the teeth. Here, we found increased cell proliferation, HIF-1α levels, bone-PDL interactions, vascularization, and stem cell markers. These included 1) scleraxis, a transcription factor expressed in tendon progenitor populations, mature tendon and PDL fibroblasts (34–37), and 2) neurofilament, a structural protein of mature nerve fibers, seen in the pulp and PDL (38). The known pulp markers of progenitor cell populations shown to be mesenchymal stem cells (MSC), alpha smooth muscle actin (aSMA) and CD44 (39–42), were also increased after drug treatment. Furthermore, the possibility of tissue de-differentiation followed by re-differentiation into mature tissue, a hallmark of classic epimorphic regeneration with the expression of Oct3/4, Nanog, Pax7, Sox2, and CD34, was supported by these results and had previously been noted in ear pinna regeneration (1–3, 23, 29).
In the broadest sense, bone is the biological “bioscaffold” which structurally supports the soft tissues of the body. The ability to regenerate bone through the upregulation of HIF-1α by a systemically acting drug extends the range of possibilities of regenerative therapies.
Results
Ligature-induced bone degeneration
Micro-CT scanning was performed on C57BL/6 (B6) female mice 2 days prior to placement of ligature (day minus 2) around the upper (maxillary) left 2nd molar (Figure 1Aa,b, ref. 31). Mice were re-scanned on day 5 (with ligature present) and again on day 10 (immediately after ligature removal). Image analysis showed significant bone degeneration within 5 days continuing to day 10 (Figure 1Ba–c). The ligature was well tolerated with no obvious ill effects such as reduced chow ingestion. Graphed values of bone loss are seen in Figure 1C.
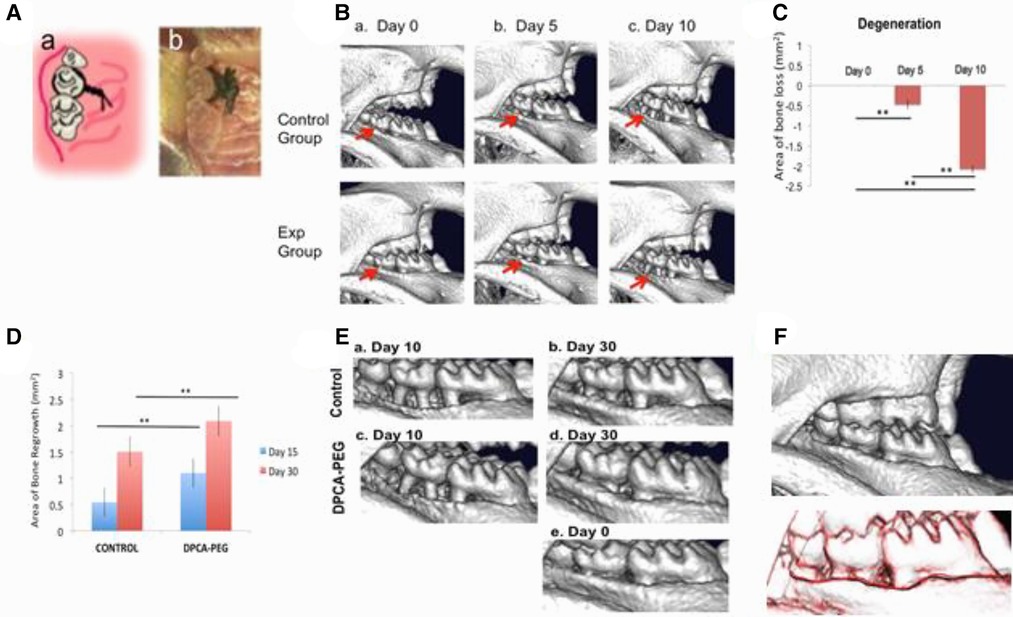
Figure 1. Degeneration of the mouse jawbone in the presence of ligature followed by regeneration of the jawbone post-ligature and post-drug. (Aab) The ligature-induced periodontitis model. 5–0 silk suture was passed through the interdentium between the maxillary first molar, the second molar and the third molar using Dumont forceps. Suture was tied firmly using a triple-knot and excess suture was cut using spring scissors as seen in the cartoon (a) and photomicrograph (b). Taken from Ref. (31). (Ba–c) Micro-CT scans of jaws from mice during the 10 day ligature period for days 0, 5, and 10. The control group image is representative of the group (n = 3) that will not receive drug (control, upper row) and the experimental group image is representative of the group (n = 3) that will receive drug (experimental, lower row). The red arrows show the maxillary left second molars with bone degeneration extending to the adjacent first and third molars. For visual clarity, the images are inverted 180 degrees (now: mandible top, maxilla bottom). (C) The area of degeneration was determined for all animals tested and shown for day 5 and day 10 after ligature placement. The y-axis is: area of bone loss (mm2); (n = 7); error bars represent standard errors; for days 0–5, p = 0.00289; for days 0–10, p = 7.33084 × 10−11; and for days 5–10, p = 3.60818 × 10−7. (**) Represents p < 0.01. In (D), a graph of area of bone growth is seen for mice post–ligature but not given drug (no drug control) vs. mice given drug (DPCA-PEG). Mice were injected with DPCA-PEG subcutaneously on days 0 and 8 after ligature placement and removal. MicroCT scans were obtained on day 15 of the experiment (day 5 post-ligature) and on day 30 (day 20 post-ligature). Here, a statistical analysis of the area of bone growth (mm2) is seen. Significant differences are found between the no drug control mice (n = 10) and DPCA-PEG-treated mice (p = 0.00253) on day 15 (blue bars) (n = 12). The same is true on day 30, where DPCA-PEG-treated mice showed highly significant differences from non-drug-treated controls (p = 0.00612) (red bars). Area analysis was performed as described in the Materials and Methods. The y-axis = area of bone regrowth (mm2); the error bars represent standard errors; and p values are represented as (*) = p < 0.05; (**) = p < 0.01. Mouse jaws analyzed were n = 10 for ligature, no drug; n = 12 for ligature/plus drug. In (E) micro-CT data shows a representative mouse maxilla which had ligature removed on day 10, scanned on day 10 and then re-scanned on day 30 (Ea,b) as compared to a representative mouse maxilla receiving ligature and DPCA-PEG drug seen on day 10 and day 30 (Ec,d). The level of regrowth in the drug-treated group shows an almost, if not complete, return to what is seen before the start of the experiment (da0) (Ed,e). In (F), there was no change in bone histology approximately 6 months later. Mice injected with DPCA-PEG drug were kept for additional observation as they aged. Over six months after the day 30 scan, mice were rescanned (upper panel) and then compared to the day 30 scan by overlaying the two scans on da 30 and da 220 (lower panel). The black line is the da 30 scan and the red line is the da 220 scan. Though shown as two lines, they are exactly overlapping. This result is representative of three mice.
Changes in bone regrowth in mice given DPCA-containing drug
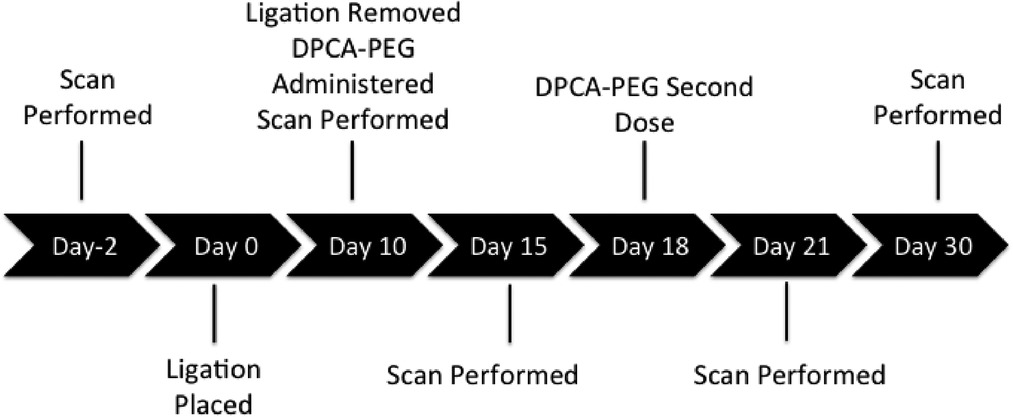
At day 10, ligature was removed, and mice were separated into two groups: 1) the control group treated no further and 2) a group injected twice (days 10 and 18) with DPCA-PEG (29). Mice were then micro-CT-imaged on days 15, 21, and 30 (Supplementary Figure S1).
Three separate experiments were carried out, and micro-CT results quantified. The key measure of alveolar bone loss and subsequent bone regeneration is derived by calculating changes in bone area assessed from lateral images using the cementum-enamel junction (CEJ) and crest of the buccal alveolar bone as anatomic landmarks (Figures 1D,E and Supplementary Figure S1) to determine changes in each jaw. Statistical analyses were performed, and statistically significant differences in bone area of treated and control groups were seen. These differences demonstrate the efficacy of DPCA-PEG in reversing induced periodontal disease in this mouse model. In Supplementary Figure S1, jaws are also shown digitally overlaid and aligned on top of one another to show changes in bone growth. In Figure 1D, treatments and degree of increase/decrease in area in mm2 is seen with highly significant changes after drug treatment.
Thus, DPCA-PEG induced rapid and significant bone regrowth. Comparing jaw from the day ligature is removed where extreme jawbone degeneration is seen by day 10 (Figure 1Ea,c), one can see dramatic bone re-growth (Figures 1D,Ed, day 20 after ligature removal). Regrowth of alveolar bone in mice given DPCA-PEG is nearly, if not completely, recovered compared to images taken before ligature and drug treatment, day 0 (Figure 1Ee). Not only does bone length and apparent bulk volume return to normal within 30 days, but morphology of regenerated bone is indistinguishable from normal alveolar bone. Thus, the thickened boney alveolar ridge adjacent to the crowns is fully restored in drug-treated mice.
It should also be noted that control bone itself does grow back, albeit to a significantly less extent, consistent with earlier observations (43, 44). This is not surprising since it has been long established that many rodent species continually wear down tooth crowns with subsequent regrowth of tooth roots and surrounding alveolar bone (see Discussion).
Finally, three ligature plus DPCA-PEG-injected mice were examined for any long-term effects or reversal of healed injury. After approximately 6 months, bone morphology was still stable. As seen in Figure 1F, no reversal of healing nor adverse effects of drug on the health of the mice were seen.
Soft tissue changes post-ligature and post-drug
Bone loss in PD is preceded by bacterially induced inflammatory changes in soft tissue (gingiva) characterized often by swelling and bleeding on probing. As bone loss continues, the inflammatory state is intensified with breakdown of attachment fibers between supporting alveolar bone and roots of the teeth. The combination of direct bone loss and breakdown of periodontal (PDL) fibers leads to tooth mobility and eventual tooth loss. It was thus important to observe whether drug-induced regenerative therapy restored not only bone but also soft tissue integrity.
In Figures 2Aa–c,Da, normal tissue histology around the tooth is shown with rete pegs in the epidermis, un-inflamed dermis, rich cellular pulp, and a PDL surrounding the full tooth root.
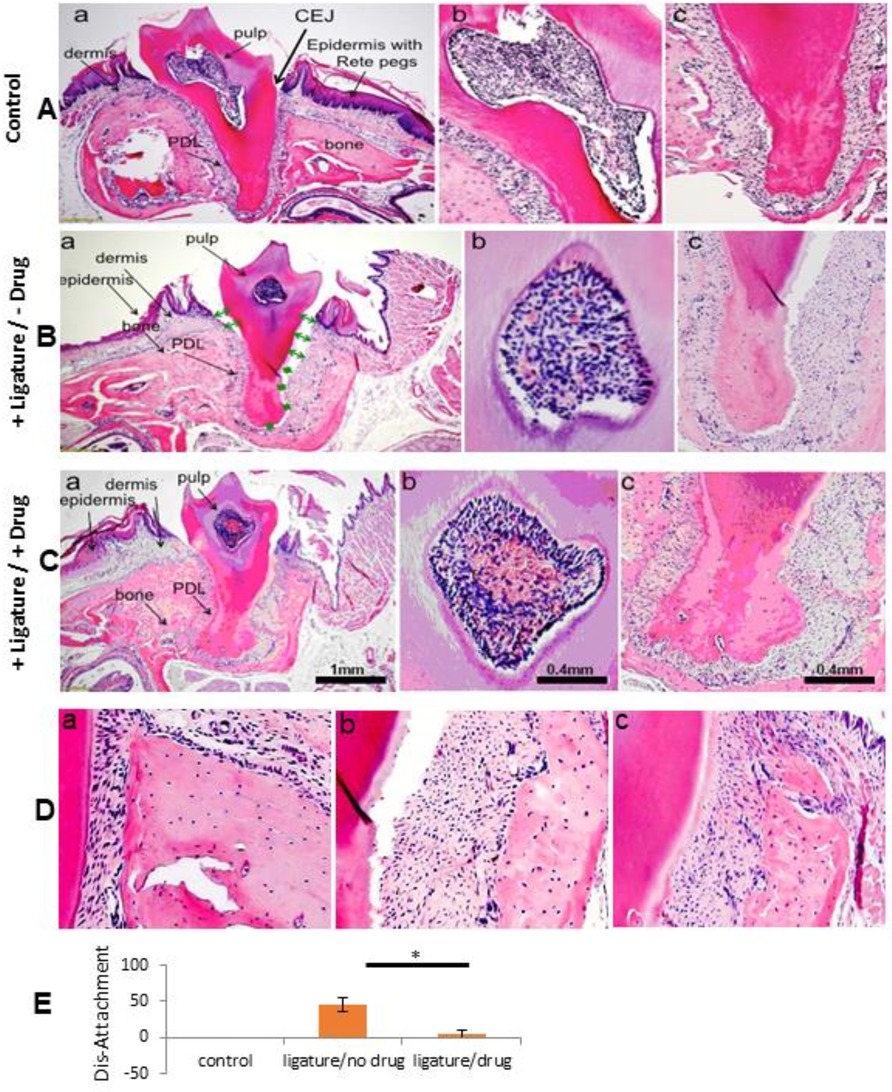
Figure 2. Histological analysis of normal and 20 days post-ligature +/- drug-treated jaw tissue. H&E-stained lateral jaw sections of a maxillary second molar are seen. The scale bar = 1mm for (A-Ca). The scale bar = 0.4 mm for all other photomicrographs, marked and unmarked. In row A, a normal mouse maxilla is seen (Aa-c). In row B, a maxilla from a +ligature/-drug-treated mouse is seen (Ba-c), and in row C, a maxilla from a +ligature/+DPCA-PEG-treated mouse (Ca-c) is seen on experimental day 30 (day 20 post ligature) and all samples are representative of 3 different mice for each condition. (a's) tooth and surrounding tissue; (b's) pulp; (c's) PDL. (D) A high magnification of bone from a normal maxilla (Da), from a maxilla of a +ligature/-drug-treated mouse (Db), and from a maxilla of a +ligature/+DPCA-PEG-treated mouse (Dc). In the normal control mouse (Aa) is seen a thick gingival dermis and epidermis with Rete pegs, a rich periodontal ligament attachment extending from root tip to cemento-enamel junction (CEJ), with periodontal ligament fibers and fibroblasts attached from tooth to normal bone (Da), along with pulp chamber and its rich cellular composition. The pathology induced from a silk ligature around the root adjacent to the crown is seen in (Aab). (B,C). In the +ligature-treated/-drug-treated mouse (Bac), a space between PDL and tooth extending to both sides of the tooth apex (green arrows, stars) is seen. Breakdown of dermis and epidermis surrounding the tooth crown with obvious dis-attachment of PDL to root surface results in a deep invagination extending > 60% of the length of the CEJ to tooth APEX. PDL is totally eliminated on the right of (Bac) and partially obliterated on the left side. Epidermis and dermis are tattered and edematous. Bone shows porous changes (Db) and clinically this tooth would be highly mobile within the tooth socket and correspond in humans to advanced PD involvement within an indication for extraction. The jaw in (Ca) is from a +ligature/+drug treated (2 doses DPCA-PEG, da0, da8 post-ligature) mouse. Here, PDL is attached to bone (Dc) and tooth, dermis is richer than without drug-treatment. Pulp shows higher levels of blood cells and vessels. A higher magnification of pulp shows differences with different treatments (Ab,Bb,Cb). After drug treatment, the pulp is richer with higher levels of red cells and angiogenesis compared to both normal and ligature alone-treated mice (pink/red).
In Figure 2Ba–c, tissue from ligature-only-treated mice is seen on day 30. There is ablation of PDL root-bone attachment down to the root apex (green arrows). Furthermore, there is increased porosity of the surrounding alveolar bone (Figure 2Db). The human clinical correlate might be extreme PD with tooth extraction indicated. Compared to some alveolar bone recovery seen by micro-CT (Figure 1Ea,b), soft tissue recovery is poor by day 30 (Figures 2Ba,c,E).
Figure 2Ca–c shows tissue after ligature plus drug (day 30). Cell-rich reattached PDL fully surrounding the root (Figures 2Cc,E), alveolar bone with less porosity (Figure 2Dc), and a highly vascularized pulp (Figure 2Cb) is found. PDL fibers have returned (Figures 2Cc,Dc).
Vascular changes in pulp and increases in stem cell markers post drug
Twenty days after drug initiation (day 30), there was an almost 8-fold increase in cellularity and vascularity in B6 pulp after drug vs. no drug control (Figures 2Cb, 3A). Neurofilament IHC, used to measure nerve in highly innervated pulp, showed increases in B6 mouse jaws after ligature plus drug (Figure 3E). Increases in early stem cell markers such as OCT3/4 and PAX7 (Figures 3C,D) and the mesenchymal stem cell marker aSMA (Figure 3F) were also seen. Two markers, SOX2 and CD34, showed no differences with or without drug.
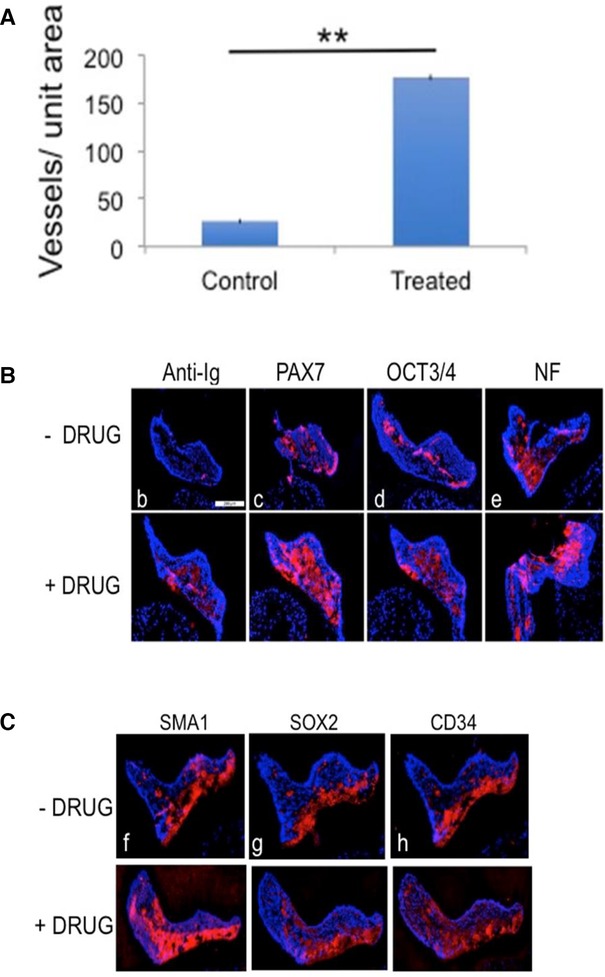
Figure 3. Stem cell marker expression in pulp after ligature and drug treatment. In (A), a graph of the level of angiogenesis in the pulp on day 30 (Figures 2Bb–Cb) is presented. The level of red in the pulp was determined by determining the number of pixels of red (IHC staining) using Photoshop as compared to the number of pulp area pixels (blue, DAPI) and is shown as a graph of the level of red in (A). The y-axis is the vessel area/pulp area in the control and drug-treated mice; n = 4; the error bars are standard errors; the p = 0.0027127 and (**) represents p < 0.01. Stem cell or de-differentiation marker expression for PAX7, OCT3/4, aSMA (SMA1), SOX2, and CD34 was determined by IHC (red) in the pulp and is seen in (B–H). The anti-Ig control is lower in drug-untreated than treated samples. The scale bar = 200 um for all photomicrographs. The pulp was also examined for expression levels of neurofilament (NF), an intermediate nerve fiber filament and marker of innervation (E). Quantitative staining using Photoshop CS6 determined the # of red pixels over the total number of pixels in the pulp giving % positive staining; (n = 4) and percentage of red pixels to total pixels determined. For (B–H), the no drug control vs. plus drug experimental is seen for (B) (0.7% vs. 6%); (C) (14% vs. 38%); (D) (9% vs. 21%); (E) (17.5% vs. 27%); (F) (19% vs. 38%); (G) (12.5% vs. 13%); and (H) (18% vs. 18%). Two markers, SOX2 and CD34, did not show a difference with and without drug on day 30.
Changes in PDL markers
A closer examination of B6 PDL showed that after ligature plus drug, the number of PDL fibroblasts increased two-fold compared to ligature alone. This was carried out by examining longitudinal cross-sections (Figure 4A) and transverse cross-sections (Supplementary Figure S2), both giving the same result.
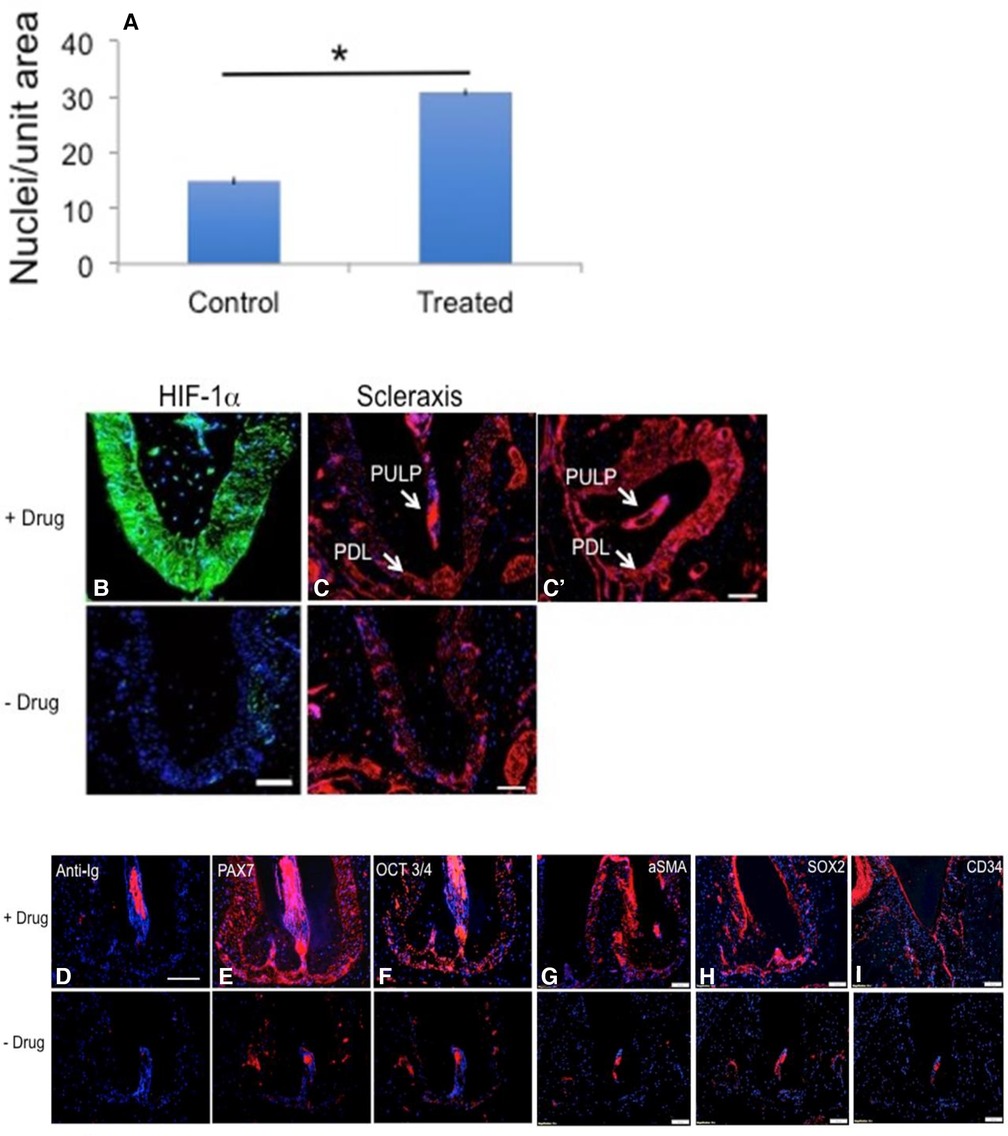
Figure 4. Stem cell marker expression in PDL after ligature and drug treatment. PDL analysis was carried out in B6-treated mice. In (A), in B6 mice, the number of PDL fibroblast nuclei were counted from Figures 2Bc,Cc and are two-fold greater in the drug-treated B6 mice. The Y-axis is the number of nuclei/unit area; error bars are standard errors; n = 4; *p < 0.01. In (B,C,C'), longitudinal jaw cross-sections from ligated-plus-drug (upper panels) vs. ligated only (lower panels) respectively were stained with antibody to HIF-1α (green) and scleraxis (red) with PDL staining levels higher in sections from drug-treated vs. no drug mice (for scleraxis, 11% vs. 4% red pixels; as described in Figure 3) (C). The scale bar = 100 um. In (E–I), antibodies to the stem cell or de-differentiation markers PAX7, OCT3/4, aSMA, SOX2, and CD34 stained more highly in the drug-treated PDL (upper panels) compared to the non-drug-treated PDL (lower panels). Anti-Ig controls showed no staining in the PDL except in the root canal (D). The scale bar = 100 um for all photomicrographs except Fig 4d where the scale bar = 200 um.
Given that 1,4-DPCA blocks PHDs and leads to HIF-1α stabilization, we noted high HIF-1α expression levels in PDL (Figure 4B). High expression levels of scleraxis, a marker of activated PDL (Figures 4C,C′), were observed post ligature plus drug in the pulp chamber and canal. In previous studies (23, 29), after DPCA-PEG treatment of ear pinna wounds and a regenerative response, we noted expression of de-differentiation and immature cell markers. Here, we saw in both PDL and pulp including the apical pulp canal, all of which contain stem cell progenitors, an increased immunostaining for stem cell markers PAX7 and OCT3/4 (Figures 4E,F) and MSC markers aSMA, SOX2, and CD34 (Figures 4G–I). Interestingly, SOX2 and CD34 showed no changes in the pulp (Figures 3G,H).
The cellular location of stem cell markers (cytoplasm vs. nucleus) could not be determined using tissue sections. Therefore, in vitro analysis of stem cell marker OCT ¾ location in cell culture was examined pre- and post-addition of 1,4-DPCA (Figure 5). Using gingival cells in the absence of DPCA (Figures 5A,A′), we found OCT3/4 expression in the cytoplasm but not nucleus. After treatment with DPCA (100 uM) in vitro, all cells showed both nuclear and cytoplasmic localization of OCT3/4 (Figures 5B,B′). The amount of OCT3/4 staining increased after DPCA treatment.
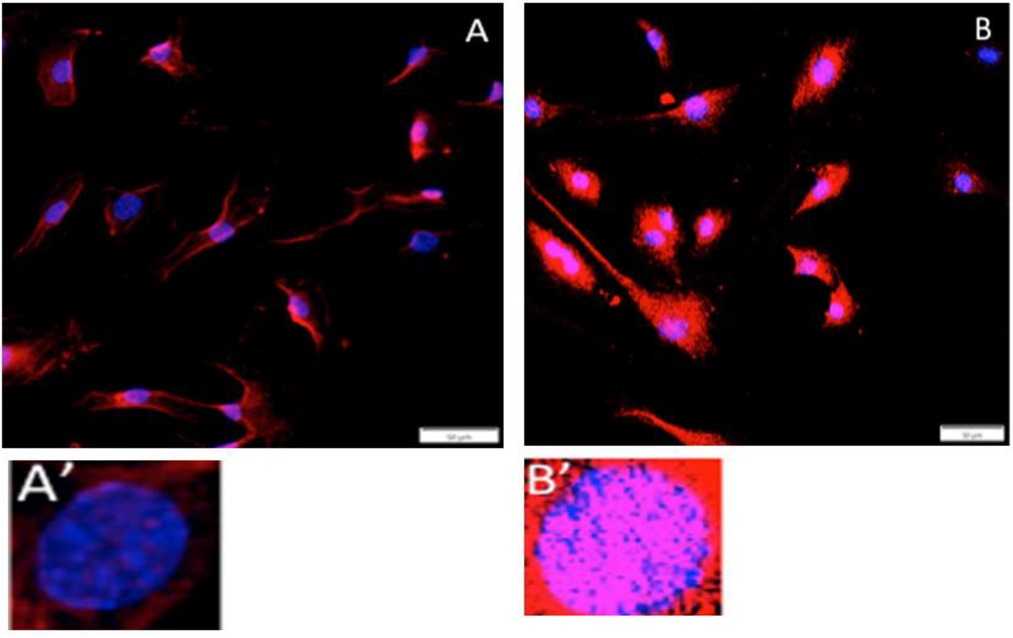
Figure 5. In vitro nuclear expression of stem cell or de-differentiation markers. Gingival fibroblast-like cells from normal B6 female mice were grown on coverslips, treated without (A,A') or with 1,4-DPCA (100 uM) (B,B') overnight in culture, and stained with antibody to OCT3/4 (red) and the nuclei stained with DAPI (blue). OCT3/4 staining is seen in the cytoplasm in DPCA-untreated cells and seen increased in the cytoplasm but now also found in the nucleus in DPCA-treated cells. The scale bars = 50um.
Metabolic reprogramming
Treatment of mice with DPCA-PEG also led to gene expression changes in jaw tissue in molecules related to the aerobic glycolysis metabolic state (Figure 6). Gapdh, Glut1, Pdk1, Pgk1, and Ldha showed increased mRNA expression levels in ligature + plus drug-treated mouse jaw tissue (da 30) obtained from isolated paraffin-embedded tissue sections as seen in Figure 2 containing bone, tooth, gingiva, and PDL. All genes showed the same expression patterns with high levels after ligature plus drug but low expression levels with ligature or drug alone.
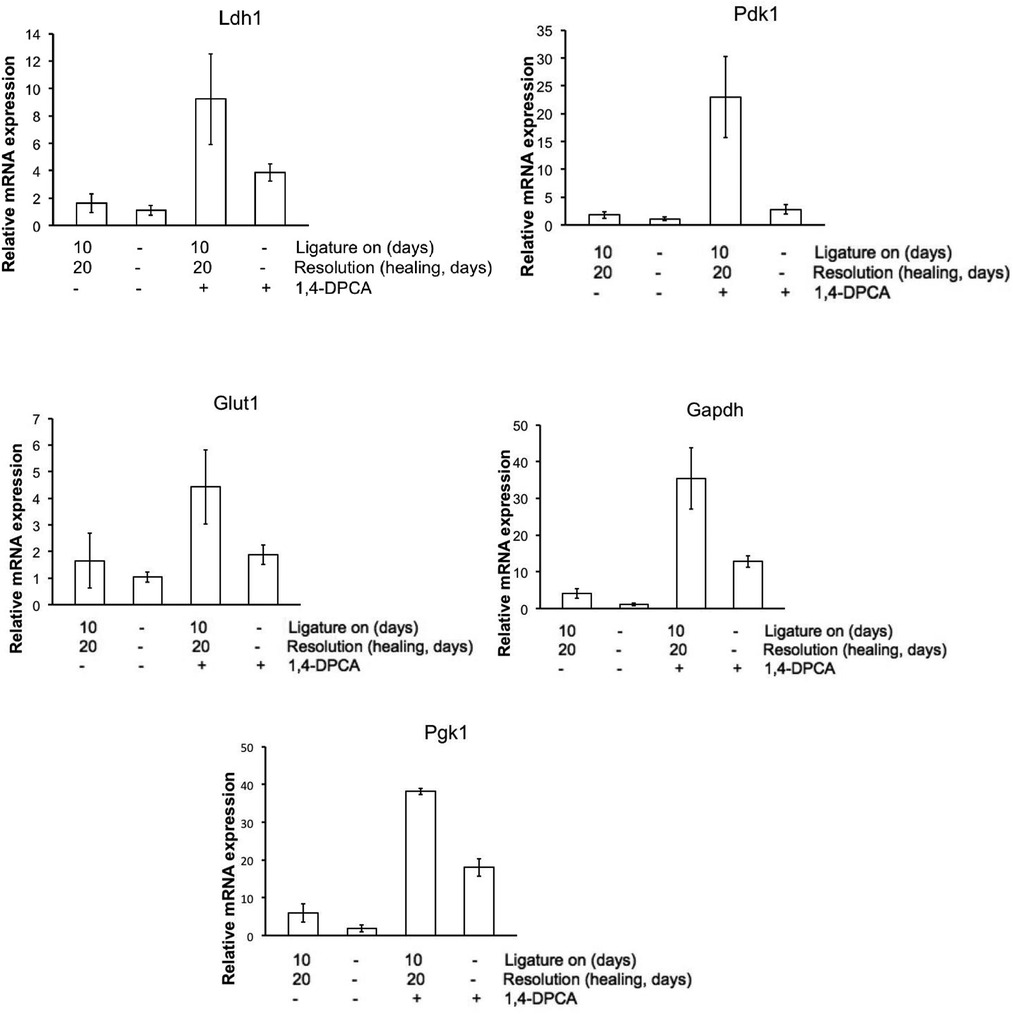
Figure 6. Metabolic gene expression in jaw tissue. mRNA was extracted from 2 sets of jaw tissue as seen in Figure 2. QPCR was carried out and results show that for all metabolic genes associated with aerobic glycolysis, high expression levels were seen in tissue from mice treated with both 1,4-DPCA and with ligature placed round the second molar. These results were done in triplicate (n = 3). The p-value differences between ligature with DPCA (3rd bar) and without DPCA (1st bar) were for LDHa, p = 0.0439*; Pdk1, p = 0.0095**; Glut1, p = 0.0922 !; Gapdh, p = 0.0103*; and Pgk1, p = 0.0001**. These values were (a) highly suggestive (!), (b) significant (*), and (c) highly significant (**).
Discussion
Induced periodontal disease as a model for bone and soft tissue regeneration
Experimental bone injuries in vivo generally require surgical incisions through overlaying skin, fascia and muscle. Periodontal disease, however, arises from a bacterial overgrowth of resident intra-oral micro-organisms external to soft tissues (the gums) of the oral cavity with subsequent host inflammatory responses leading to massive lesions of underlying alveolar bone. An induced periodontal disease model is therefore attractive from the perspectives of both basic bone regeneration biology and as a translational stepping stone in the treatment of a common human disease.
With declining incidence of caries, periodontal disease (PD) has emerged as the most common cause of tooth loss affecting 30%–60% of the adult population, presenting a major clinical challenge (45, 46) with the loss of supporting alveolar bone around the roots of teeth as well as the destruction of adjacent soft tissues (47) and can involve a single tooth or the entire dentition. PD etiology is due to a number of identified periodontopathic organisms found in the gingival crevice (31–33, 47) combined with host susceptibility factors such as diabetes (48, 49).
The challenge in advanced PD, where bone loss may exceed 50%–80% of the length of the tooth root, is to halt further progression and achieve a functionally and esthetically acceptable outcome. Thus, a primary goal is to restore lost alveolar bone and soft tissues to the original state without loss of teeth (50, 51). It is for this reason that we have explored PHDi-based regenerative drug-induced bone and soft tissue regeneration in a PD model.
The role of HIF-1α and changes in vascularization
The biphasic high expression of HIF-1α and its target genes was shown to be a critical molecular tissue regeneration pathway as determined from previous soft tissue studies (16, 23, 29). These target genes include Pdk, Ldha, Pkm2, Gapdh, and Pgk1, which are all glycolytic enzymes (Figure 6) and lead to metabolic remodeling or reprogramming (21, 22, 24–27). These molecules required two signals for elevated expression, drug + injury (ligature). HIF-1α was identified as the molecule responsible for the broad regenerative ability of the MRL mouse from multiple directions. First, in genetic mapping studies of MRL mice, a reduced level of the gene RNF7, part of the ubiquitin ligase complex, necessary for proteolysis and lowered protein expression levels of HIF-1α (52), proved to be a candidate gene associated with regenerative healing (53). Furthermore, MRL mice were found to be metabolically more embryonic using aerobic glycolysis regulated by HIF-1α target genes (19, 20). Finally, siHIF1a completely suppressed ear hole closure both in the MRL and non-regenerative mice treated with 1,4-DPCA-hydrogel (23).
One of the many intriguing aspects of HIF-1α elevation is the de-differentiation of mature cell populations to an immature state determined by changes in molecular markers such as Nanog and Oct ¾ regulated by HIF-1α and aerobic glycolysis (54–61). This state characterizes cells in the amphibian regenerative blastema and is thought to be key to the regenerative response (62). Also, HIF-1α is responsible for an enhanced vascularization response, producing molecules such as VEGF and HMOX1. It is clear in the tooth pulp that vascular tissue is significantly increased 15–20 days after DPCA-PEG administration (Figures 2Cb, 3A). This change in vascularity could lead to an increased number of stem cells migrating into the tooth as noted by stem cell marker increase (Figures 3B–F). On the other hand, DPCA-PEG could lead to de-differentiation with increased stem cell markers and growth as seen in the ear accompanying regenerative ear hole closure (23, 29).
Induction of stem cell markers
Many molecular and cellular markers of regeneration observed in spontaneously regenerating species such as the newt and axolotl (1–3) and in the MRL mouse (19) were indistinguishable from those seen in DPCA-PEG-treated mice (23, 29, 16). Our previous studies largely focused on soft tissue, specifically in the ear pinna and included regrowth of hair follicles and cartilage (23, 29). In the PD model studied here, not only soft tissue but also bone is affected by the drug. Multiple early impacts (5 days post-ligature) using a different form of the drug, DPCA-hydrogel, included increased Treg FOXP3 + populations and lowered inflammatory cytokines (63). In the current study using DPCA-PEG, between 20 days and as long as 220 days post-ligature, the PDL, which is key to supporting the teeth and securing them to the bone, showed complete re-attachment to the teeth. Increased numbers of PDL fibroblasts in the drug group are also seen. PDL also showed increased levels of scleraxis, a transcription factor considered to be a PDL marker (34–37) associated with osteocytes and cementoblasts (36).
Another dental soft tissue target, the pulp, is considered to be a source of stem cells and contains dentinoblasts which re-line the inside of the tooth dentin. We noted after drug that the pulp vasculature as well as stem cell markers were increased. As mentioned above, whether this is due to a de-differentiation process in the pulp or to increased numbers of stem cells in the over-abundant vasculature is not clear. We also noted an increase of neurofilament (38), a nerve marker, which is present in normal pulp and increased after drug.
Stem cell populations have been identified that are associated with teeth in the pulp chamber, at the base of growing roots, and in the PDL (40). Progenitor cell populations have been identified previously in periodontal tissue and express mesenchymal stem cell markers such as STRO-1, CD146, CD44, and αSMA (30–33, 64–66). These progenitors exhibit many stem-cell-like features, including small size, responsiveness to stimulating factors, slow cycle time and the ability to generate multiple mesenchymal lineages (67–69). In addition, neural crest-derived cells have been identified in the periodontal ligament and the pulp chamber using markers such as Slug, AP2 alpha, HNK-1, p75NTR and Nestin (70–74).
Alpha SMA has been found in stem cells and regenerating tissue and in blood vessel pericytes and myoepithelial cells involving force-generating function. During mandible development, aSMA was found in the dental follicle, in the periostin-positive area along with RUNX2 positive cells, and localizes to the alveolar bone region suggesting involvement in bone formation (75).
We tested multiple stem cell markers such as OCT3/4, PAX7, SOX2, CD34 and aSMA in the jaw. These de-differentiation markers were found in pulp and PDL and increased after DPCA-PEG. Again, these markers could be due to the dedifferentiation of mature cells (1–3, 23, 30, 54–61) or could be markers of stem cell populations in the pulp and PDL as discussed above (76). Cross talk between the pulp and periodontal ligament should not be ruled out since pulp and PDL are anatomically connected via the apical root vasculature.
Although stem cell markers were increased in PDL with DPCA- PEG, it was difficult to see cellular sub-structure and intra-cellular location of these stem cell markers. We grew gingival-derived fibroblast-like cells to analyze the staining of OCT ¾ with and without addition of DPCA. Here, we found that cells before DPCA showed staining in the cytoplasm, but after DPCA, staining was seen in the nucleus where Oct ¾ acts as a transcription factor and is key to self-renewal of undifferentiated embryonic stem cells and is a specific marker for dedifferentiation (51).
Taken together, our present work has shown the ability of systemic DPCA-PEG administration to regenerate severely degraded alveolar bone and soft tissue with remarkable anatomic fidelity within 20 days post drug.
It should be noted that in humans, regenerative recovery of bone is seen but is usually limited to bone fractures which are aligned with a gap of less than 1 mm (77, 78). On the other hand, it is also known that both the maxillary and mandibular incisor teeth of rodents kept in animal colonies on soft chow often elongate (33, 34) and we are seeing a background growth effect. In studies reported here, however, the regenerative contribution of the drug is clearly distinguishable from the background.
In conclusion, this study extends our previous work on drug-mediated stabilization of HIF-1α to achieve soft tissue regeneration in mice (16, 23, 29, 59) in new directions. First, systemic DPCA-PEG rapidly reverses severe bone loss in an anatomically complex structure, alveolar bone of the maxilla, leading to a nearly perfect replica of healthy bone and associated soft tissue such as PDL with long-term maintenance. This induced bone loss occurs in an experimental system which emulates the bacterial etiology of the human disease, periodontitis.
Materials and methods
Study design
We used the inbred mouse strain, C57BL/6 female, to study the effect of a small-molecule inhibitor of PHDs on the in vivo expression of HIF-1α and the impact on quantitative regenerative maxillary bone growth (63) and soft tissue regeneration including PDL and pulp. A 5–0 silk ligature model was used to induce periodontitis in the mice, and DPCA–PEG was tested and subcutaneously implanted in the back of the neck at multiple time points (Figure 7). End points of the study were previously determined to be a minimal of 30 days and up to 220 days after injury and included key indices of tissue regeneration such as bone regrowth as measured by micro-CT scanning, soft tissue regrowth after H&E histological analysis including epidermis, gingiva, PDL attachment and cell number, and markers of regeneration determined by immunohistochemistry of jaw tissue for stem cell markers, and HIF-1α, neurofilament, and scleraxis and RT-PCR analysis of gingiva. These parameters involved physical measurements of growth and gene expression at the RNA and protein levels. Tissue was coded and different laboratory personnel were involved in doing ligatures, scanning, tissue preparation, and data analysis.
Animals
C57BL/6 female mice, 9 weeks of age, were obtained from Taconic Laboratories. The experiment was done with 2 groups of 3–4 mice based on the previous work of Hajishengallis et al. (31, 63) and was repeated twice for a total of 10 mice in each group.
Food and water were provided ad libitum. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of LIMR and were performed in compliance with institutional, state, and federal policies.
Tissue culture
Primary fibroblast-like cell lines from gingival tissue were established from B6 female mice by plating in dispase and then collagenase and then grown in DMEM-10% FBS supplemented with 2 mM L-glutamine and 100 IU/ml penicillin streptomycin and maintained at 37 °C, 5% CO2, and 21% O2. The cells were washed and only adherent fibroblasts maintained. Cells were split 1 : 5 as needed to maintain exponential growth and avoid contact inhibition. Passage numbers were documented and cells from early passages (<P20) frozen in liquid nitrogen and used in the described experiments.
For immunohistochemical staining, primary gingival fibroblasts were grown on coverslips in DMEM with 10% FBS at 37 °C in a humidified 5% CO2 incubator. Before staining, cells were incubated with DPCA overnight. The coverslips were rinsed with 1× PBS, and the cells were fixed in cold methanol (−20 °C) for 10 min, rinsed with 1× PBS, treated with 0.1% Triton-X100, and then incubated with the appropriate primary and secondary antibodies (Supplementary Table S1). Photomicrographs were produced using the fluorescent microscope (Olympus AX70) and a DP74 camera with CellSens Standard software for image analysis.
Drug application
DPCA-PEG was synthesized as described (29) and was injected subcutaneously on day 0 and day 8 using 25 ul of gel #10/injection or 50 ug of DPCA. The time course of injections and scans performed during longitudinal experiments can be seen in Figure 7.
Preparation and immunohistochemistry of jaw tissue
Tissue from upper jaws was fixed in Prefer fixative (the active ingredient is glyoxal) (Anatech) for 5 days and then washed in H20. Jaw tissue was then decalcified using UltraPure 0.5 M EDTA, pH 8.0, for 5 weeks with changes in EDTA solution twice a week. Tissue was put in ETOH and then embedded in paraffin and cut to 5 μm thick sections. Before staining, slides were dewaxed in xylene and rehydrated. Tissue sections were then treated with 3% H2O2 and nonspecific binding was blocked with 4% BSA (A7906; Sigma) for 1 h. Primary antibodies and matched secondary antibodies used for IHC are shown in Supplementary Table S1. Photomicrographs were produced using the fluorescent microscope (Olympus AX70) with an Olympus DP74 color camera using CellSens imaging software.
For histological stains, tissue sections were treated as above and stained with hematoxylin (Leica Microsystems, #3801562) and eosin (Leica Microsystems, #3801602). The slides were washed, rehydrated, cleared with xylene and coverslipped with Permount mounting media (Fisher, SP15-500). Staining was visualized using an Olympus (AX70) microscope in bright field for H&E and fluorescence.
Induction of periodontal disease
For ligature placement, mice were anesthetized with ketamine/xylazine, a 5–0 silk ligature was placed around the upper left second molar of the maxilla according to an established procedure (31) generating a dental plaque-retentive milieu that reliably and quantitatively produces a periodontal bone lesion. Importantly, this ligature procedure induces not only bacterial overgrowth but also selective expansion of periodontopathic microorganisms, mimicking quantitative and qualitative (dysbiotic) microbiome alterations, the same etiology as that in human disease (32, 33). Upon placement of the ligature, the 5–0 silk ligation accumulates dental plaque and oral bacteria creating a local inflammatory state in the surrounding gums and bone of the tooth. Ketamine/xylazine was used to remove the ligature and mice were re-scanned. Mice were subsequently scanned for further analysis. During this time, mice were kept on a normal diet of mouse chow and were monitored daily for any signs of physical discomfort in accordance with the Lankenau Institute for Medical Research (LIMR) Animal Care Policies and Procedures Manual. At each time point, animals were imaged and analyzed with the area analysis described below. On four occasions the ligature model was not completed due to the 5–0 silk ligation falling off of the tooth. These mice were excluded from the study and the remaining mice were randomly distributed before any DPCA-PEG injections.
Ketamine/xylazine mixture
Stock solutions of ketamine (100 mg/ml, Hanna) and xylazine (20 mg/ml, Hanna) are prepared using PBS as a diluent, at a ratio of 3:1:16, and is vortexed and used immediately.
Micro-CT scanning
For scanning, mice were anesthetized using isoflurane (Henry Schein) 2%–4% concentration in 100% O2 for 3 min in an anesthesia chamber. Upon establishing anesthesia, mice were placed into the microCT FX (PerkinElmer; ref 80) tray with the isoflurane nose cone. 3D images were collected and rendered with a voltage of 90 kV, a CT current of 160 μA and a live current of 80 μA for 17 s for a total dose of 11 milliGY (79, 80). This constitutes the pre-intervention baseline.
Buccal images were analyzed unlike original experiments done by measuring the palatal side without micro-CT (31, 63). They were normalized to the Micro-CT HA D4.5 phantom from QRM (Quality Assurance in Radiology and Medicine GmbH).
Micro-CT analysis
An analytical method to quantitate longitudinal changes in bone morphology was designed and implemented. 3D renderings were obtained using Quantum software (Perkin Elmer) and subsequently converted to 2D images, which were then superimposed in Photoshop (CC 2019) for quantitation of bone morphological changes.
RNA isolation and quantitative qPCR
Total RNA isolation from paraffin-embedded jaw samples was performed using RNeasy DSP FFPE Kit (Qiagen), according to the manufacturer's protocol. cDNA was synthesized from 500 ng of total RNA using Invitrogen SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific) according to the manufacturer's instructions. qPCR was performed with Applied Biosystems SYBR Green PCR Master Mix (Thermo Fisher Scientific). In brief, a 10 μl mixture was used containing 5 μl SYBR Green PCR Master Mix, 0.5 μl forward and reverse primer, 2.5 μl sterile water, and 2 μl of complementary DNA template. The real-time PCR was performed using Applied Biosystems QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer's instructions. Gene-specific primers (Supplementary Table S2) for detection and quantification of murine genes investigated in this study were purchased from Integrated DNA Technologies. All data were normalized to 18S rRNA and quantitative measurements were obtained using the ΔΔCt method.
Statistical analysis
For multiple-group comparisons, data were analyzed by one-way ANOVA followed by Tukey's multiple comparison test. A two-tailed Student's t-test was used for two-group comparisons. P values <0.05 were considered to be statistically significant and values P <0.01 were considered highly significant. All statistical analyses were performed using RStudio (version 1.1.463) with the stats and stats4 packages (versions 3.5.1).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding author/s
Ethics statement
The animal study was reviewed and approved by IACUC from the Lankenau Institute for Medical Research.
Author contributions
EZ, TK and EHK designed, performed experiments, analyzed, interpreted, and graphed data, and prepared the manuscript. GH provided expert advice and experimental design, together with PBM interpreted the data, and critically reviewed the manuscript. PBM and JC provided drug development and experimental data and constructs; YZ and KB carried out all Swiss Webster studies, AA carried out all micro-CT scanning and QPCR analysis; SB provided animal care and tissue preparation. KB carried out histology and immunohistochemistry studies. DS provided clinical dental expertise and manuscript preparation. All authors contributed to the article and approved the submitted version.
Acknowledgments
This work was supported by the grant DE021104 from NIDCR (NIH) and the grant PR180789: W81XWH-19-1-0467 from DOD. EZ and SB were supported by the Stanley and Fiona Druckenmiller Family Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2022.992722/full#supplementary-material.
Abbreviations
B6, C57BL/6 mouse; CEJ, cemento-enamel junction; DPCA, 1,4-dihydrophenonthrolin-4-one-3-carboxylic acid; Gapdh, glyceraldehyde-3-phosphate dehydrogenase: Glut-1, glucose transporter type 1; HIF-1α; hypoxia-inducible factor-1alpha; Ldh-a, lactate dehydrogenase a; MRL, Murphy Roths large mouse; Pdk1, pyruvate dehydrogenase kinase; PD, periodontal disease; PDL, periodontal ligament; Pgk1, phosphoglycerate kinase 1; PHDi, prolyl hydroxylase inhibitor.
References
1. Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. (2005) 310:1919–23. doi: 10.1126/science.1115200
2. Stocum DL, Cameron JA. Looking proximally and distally: 100 years of limb regeneration and beyond. Dev Dyn. (2011) 240:943–68. doi: 10.1002/dvdy.22553
3. Sousounis K, Qi F, Yadav MC, Millán JL, Toyama F, Chiba C, et al. A robust transcriptional program in newts undergoing multiple events of lens regeneration throughout their lifespan. eLife. (2015) 2:4. doi: 10.7554/eLife.09594
4. D’Aquino R, De Rosa A, Lanza V, Tirino V, Laino L, Graziano A, et al. Human mandible bone defect repair by the grafting of dental pulp stem/progenitor cells and collagen sponge biocomplexes. Eur Cell Mater. (2009) 18:75–83. doi: 10.22203/eCM.v018a07
5. La Noce M, Paino F, Spina A, Naddeo P, Montella R, Desiderio V, et al. Dental pulp stem cells: state of the art and suggestions for a true translation of research into therapy. J Dent. (2014) 42:761–8. doi: 10.1016/j.jdent.2014.02.018
6. Shi X, Mao J, Liu Y. Pulp stem cells derived from human permanent and deciduous teeth: biological characteristics and therapeutic applications. Stem Cells. (2020) 9:445–64. doi: 10.1002/sctm.19-0398
7. Weissman IL. Stem cell: units of development, units of regeneration, and units of evolution. Cell. (2000) 100:157–68. doi: 10.1016/S0092-8674(00)81692-X
8. Blanpain C, Fuchs E. Plasticity of epithelial stem cells in tissue regeneration. Science. (2014) 344:1242281. doi: 10.1126/science.1242281
9. Mao AS, Mooney DJ. Regenerative medicine: current therapies and future directions. PNAS. (2015) 112:14452–9. doi: 10.1073/pnas.1508520112
10. Dahiya P, Kamal R. Hyaluronic acid: a boon in periodontal therapy. N Am J Med Sci. (2013) 5:309–15. doi: 10.4103/1947-2714.112473
11. Casale M, Moffa A, Vella P, Sabatino L, Capuano F, Salvinelli B, et al. Hyaluronic acid: perspectives in dentistry. A systematic review. Int J Immunopath Pharmacol. (2016) 29:572–82. doi: 10.1177/0394632016652906
12. Derwich M, Mitus-Kenig M, Pawlowska E. Mechanisms of action and efficacy of hylauronic acid, corticosteroids, and platelet-rich plasma in the treatment of TMJ osteoarthritis – A systemic review. Int J Mol Sci. (2021) 22:7405. doi: 10.3390/ijms22147405
13. Clark LD, Clark RK, Heber-Katz E. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol. (1998) 88:35–4. doi: 10.1006/clin.1998.4519
14. McBrearty BA, Desquenne-Clark L, Zhang X-M, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound healing trait. Proc Natl Acad Sci USA. (1998) 95:11792–7. doi: 10.1073/pnas.95.20.11792
15. Edwards RG. From embryonic stem cells to blastema and MRL mice. Reprod BioMed Online. (2008) 16:425–61. doi: 10.1016/S1472-6483(10)60605-0
16. Heber-Katz E. Oxygen, metabolism, and regeneration – lessons from mice. Trends Mol Med. (2017) 23:1024–36. doi: 10.1016/j.molmed.2017.08.008
17. Heydemann A. The super super-healing MRL mouse strain. Front Biol. (2012) 7:522–53. doi: 10.1007/s11515-012-1192-4
18. Heber-Katz E, Messersmith P. Drug delivery and epimorphic salamander-type regeneration. Adv Drug Deliv Rev. (2018) 129:254–61. doi: 10.1016/j.addr.2018.02.006
19. Naviaux RK, Le TP, Bedelbaeva K, Leferovich J, Gourevitch D, Sachadyn P, et al. Retained features of embryonic metabolism in the adult MRL mouse. Mol Genet Metab. (2009) 96:133–44. doi: 10.1016/j.ymgme.2008.11.164
20. Heber-Katz E, Naviaux R. The MRL mouse: A model of regeneration and cancer. In: Berger NA, editors. Murine models, energy balance, and cancer. Springer (2015). p. 47–64.
21. Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci USA. (1993) 90:4304–8. doi: 10.1073/pnas.90.9.4304
22. Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-Dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci. (2019) 20:238. doi: 10.3390/ijms20020238
23. Zhang Y, Bedelbaeva K, Strehin I, Gourevitch D, Messersmith PB, Heber-Katz E. Drug-induced regeneration in adult mice. Sci Transl Med. (2015) 7:290. doi: 10.1126/scitranslmed.3010228
24. Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res. (2015) 34:111. doi: 10.1186/s13046-015-0221-y
25. Jacquet P, Stephanou A. Metabolic reprogramming, questioning, and implications for cancer. Biology (Basel). (2021) 10:129. doi: 10.3390/biology10020129
26. Kim J. Regulation of immune cell functions by metabolic reprogramming. J Immunol Res. (2018) 2018:8605471:1–12. doi: 10.1155/2018/8605471
27. Murphy M, O’Neill LAJ. How should we talk about metabolism? Nat Immunol. (2020) 21:713–5. doi: 10.1038/s41590-020-0691-8
28. Franklin TJ, Morris WP, Edwards PN, Large MS, Stephenson R. Inhibition of prolyl 4-hydroxylase in vitro and in vivo by members of a novel series of phenanthrolinones. Biochem J. (2001) 353(Pt 2):333–8. doi: 10.1042/bj3530333
29. Cheng J, Amin D, Latona J, Heber-Katz E, Messersmith PB. Supramolecular polymer hydrogels for drug-induced tissue regeneration. ACS Nano. (2019) 13:5493–501. doi: 10.1021/acsnano.9b00281
30. Latona J, Shah A, Cheng J, Messersmith P, Heber-Katz E. Enhanced liver regeneration after partial hepatectomy in mice treated with a prolyl hydroxylase inhibitor. Am J Transplant. (2017) 17(Suppl 3).
31. Abe T, Hajishengallis G. Optimization of the ligature-induced periodontitis model in mice. J Immunol Methods. (2013) 394:49–54. doi: 10.1016/j.jim.2013.05.002
32. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. (2015) 15:30–44. doi: 10.1038/nri3785
33. Graves DT, Fine D, Teng YT, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol. (2008) 35:89. doi: 10.1111/j.1600-051X.2007.01172.x
34. Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. (2007) 236:1677–82. doi: 10.1002/dvdy.21179
35. Inoue M, Ebisawa K, Itaya T, Sugito T, Yamawaki-Ogata A, Sumita Y, et al. Effect of GDF-5 and BMP-2 on the expression of tendon/ ligamentogenesis-related markers in human PDL-derived cells. Oral Dis. (2012) 18:206–12. doi: 10.1111/j.1601-0825.2011.01871.x
36. Rogulic H, Mathews BG, Yang W, Cvija H, Mina M, Kalajzic I. In vivo identification of periodontal progenitor cells. J Dent Res. (2013) 92:709–15. doi: 10.1177/0022034513493434
37. Takimoto A, Kawatsu M, Yoshimoto Y, Kawamoto T, Seiryu M, Takano-Yamamoto T. Scleraxis and osterix antagonistically regulate tensile force-responsive remodeling of the periodontal ligament and alveolar bone. Development. (2015) 142:787–96. doi: 10.1242/dev.116228
38. Sato O, Maeda T, Kobayashi S, Iwanaga T, Fujita T, Takahashi Y. Innervation of periodontal ligament and dental pulp in the rat incisor: an immuno-histochemical investigation of neurofilament protein and glia-specific S-100 protein. Cell Tissue Res. (1987) 251:13–21. doi: 10.1007/BF00215442
39. Tatullo M, Marrelli M, Kevin M, Shakesheff KM, White LJ. Dental pulp stem cells: function, isolation and applications in regenerative medicine. J Tissue Eng Regen Med. (2015) 9:1205–627. doi: 10.1002/term.1899
40. Wilson KR, Kang I-H, Baliga U, Xiong Y, Chatterjee S, Moore E, et al. Hematopoietic stem cells as a novel source of dental tissue cells. Sci Rep. (2018) 8:8026. doi: 10.1038/s41598-018-26258-y
41. Leonardi R, Lanteri E, Stivala F, Travali S. Immunolocalization of CD44 adhesion molecules in human periradicular lesions. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2000) 89:480–5. doi: 10.1016/S1079-2104(00)70129-6
42. Leonardi R, Loreto C, Caltabiano R, Caltabiano C. Immunolocalization of CD44s in human teeth. Acta Histochem. (2006) 108:425–9. doi: 10.1016/j.acthis.2006.06.006
43. Merzel J, Salmon CR. Growth and the modeling/remodeling of the alveolar bone of the rat incisor. Anat Rec. (2008) 291:827–34. doi: 10.1002/ar.20703
44. Dontas IA, Tsolakis AI, Khaldi L, Patra E, Lyritis GP. Malocclusion in aging wistar rats. JAALAS. (2010) 49:22–6.20122311
45. Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. (2012) 91:914–23. doi: 10.1177/0022034512457373
46. Chapple IL. Time to take periodontitis seriously. Br Med J. (2014) 348:g2645. doi: 10.1136/bmj.g2645
47. Beikler T, Flemmig TF. Oral biofilm-associated diseases: trends and implications for quality of life, systemic health and expenditures. Periodontol 2000. (2011) 55:87–103. doi: 10.1111/j.1600-0757.2010.00360.x
48. Casanova L, Hughes F, Preshaw P. Diabetes and periodontal disease. BDJ Team. (2015) 1:15007. doi: 10.1038/bdjteam.2015.7
49. Mealey BL. Periodontal disease and diabetes: a two-way street. JADA. (2006) 147:526–31. doi: 10.14219/jada.archive.2006.0404
50. Foster BL, Somerman MJ. Regenerating the periodontium: is there a magic formula? Orthod Craniofac Res. (2005) 8:285–91. doi: 10.1111/j.1601-6343.2005.00351.x
51. Murakami S. Periodontal tissue regeneration by signaling molecule(s): what role does basic fibroblast growth factor (FGF-2) have in periodontal therapy? Periodontol 2000. (2011) 56:188–208. doi: 10.1111/j.1600-0757.2010.00365.x
52. Tan M, Gu Q, He H, Parmarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1α ubiquination and degradation. Oncogene. (2008) 27:1404–11. doi: 10.1038/sj.onc.1210780
53. Cheverud JM, Lawson HA, Bouckaert K, Kossenkov A, Showe L, Cort L, et al. Fine-mapping quantitative trait loci affecting murine external ear tissue regeneration in the LG/J by SM/J advanced intercross line. Heredity. (2014) 112:508–18. doi: 10.1038/hdy.2013.133
54. Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. (2003) 113:631–42. doi: 10.1016/S0092-8674(03)00393-3
55. Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. (2000) 24:372–6. doi: 10.1038/74199
56. Dunwoodie SL. The role of hypoxia in development of the mammalian embryo. Dev Cell. (2009) 17:755–73. doi: 10.1016/j.devcel.2009.11.008
57. Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. (2010) 7:150–61. doi: 10.1016/j.stem.2010.07.007
58. Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, et al. HIF Induces human embryonic stem cell markers in cancer cells. Cancer Res. (2011) 71:4640–52. doi: 10.1158/0008-5472.CAN-10-3320
59. Ehnes DD, Hussein AM, Ware CB, Mathieu J, Ruohola-Baker H. Combinatorial metabolism drives the naïve to primed pluripotent chromatin landscape. Exp Cell Res. (2020) 389(2):111913. doi: 10.1016/j.yexcr.2020.111913
60. Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. (2013) 140:2535–47. doi: 10.1242/dev.091777
61. Axelson H, Fredlund E, Ovenberger M, Landberg G, Pahlman S. Hypoxia-induced dedifferentiation of tumor cells – a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin Cell Dev Biol. (2005) 16:554–63. doi: 10.1016/j.semcdb.2005.03.007
62. Call MK, Grogg MW, Tsonis PA. An eye on regeneration. Anat Rec B New Anat. (2005) 287:42–8. doi: 10.1002/ar.b.20084
63. Nagai K, Ideguchi H, Kajikawa T, Xiaofei Li X, Chavakis T, Cheng J, et al. An injectable hydrogel-formulated inhibitor of prolyl-4-hydroxylase promotes T regulatory cell recruitment and enhances alveolar bone regeneration during resolution of experimental periodontitis. FASEB J. (2020):1–15 34:13726–40. doi: 10.1096/fj.202001248R
64. Kemoun P, Laurencin-Dalicieux S, Rue J, Vaysse F, Romeas A, Arzate H, et al. Localization of STRO-1, BMP-2/-3/-7, BMP receptors and phosphorylated smad-1 during the formation of mouse periodontium. Tissue Cell. (2007) 39:257–66. doi: 10.1016/j.tice.2007.06.001
65. Chen SC, Marino V, Gronthos S, Bartold PM. Location of putative stem cells in human periodontal ligament. J Periodontal Res. (2006) 41:547–53. doi: 10.1111/j.1600-0765.2006.00904.x
66. Xu J, Wang W, Kapila Y, Lotz J, Kapila S. Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev. (2009) 18:487–96. doi: 10.1089/scd.2008.0113
67. San Miguel SM, Fatahi MR, Li H, Igwe JC, Aguila HL, Kalajzic I. Defining a visual marker of osteoprogenitor cells within the periodontium. J Periodontal Res. (2010) 45:60–70. doi: 10.1111/j.1600-0765.2009.01201.x
68. Mrozik K, Gronthos S, Shi S, Bartold PM. A method to isolate, purify, and characterize human periodontal ligament stem cells. Methods Mol Biol. (2010) 666:269–84. doi: 10.1007/978-1-60761-820-1_17
69. McCulloch CA. Progenitor cell populations in the periodontal ligament of mice. Anat Rec. (1985) 211:258–62. doi: 10.1002/ar.1092110305
70. Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. (2004) 364:149–55. doi: 10.1016/S0140-6736(04)16627-0
71. Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod Craniofac Res. (2005) 8:191–9. doi: 10.1111/j.1601-6343.2005.00331.x
72. Kaku M, Komatsu Y, Mochida Y, Yamauchi M, Mishina Y, Ko CC. Identification and characterization of neural crest-derived cells in adult periodontal ligament of mice. Arch Oral Biol. (2012) 57:1668–75. doi: 10.1016/j.archoralbio.2012.04.022
73. Volponi AA, Pang Y, Sharpe PT. Stem cell-based biological tooth repair and regeneration. Trends Cell Biol. (2010) 20:715–22. doi: 10.1016/j.tcb.2010.09.012
74. Cohen F-M, Sun H-H, Lu H, Yu Q. Stem cell-delivery therapeutics for periodontal tissue regeneration. Biomaterials. (2012) 33:6320–44. doi: 10.1016/j.biomaterials.2012.05.048
75. Hosoya A, Nakamura H, Ninomiya T, Yoshiba K, Yoshiba N, Nakaya H, et al. Immunohistochemical localization of a-smooth muscle actin during rat molar tooth development. J Histochem Cytochem. (2006) 54:1371–8. doi: 10.1369/jhc.6A6980.2006
76. Shetty H, Kakade A, Shetty S, Neelakantan NS, Desai R, Beri K. Immuno histochemical characterization of stem cell and differentiation markers of the dental pulp of human natal teeth. Future Sci OA. (2018) 4:FS0342. doi: 10.4155/fsoa-2018-0062
77. Einhorn TA, Gerstenfeld L. Fracture healing: mechanisms and interventions. Nat Rev Rheumatol. (2015) 11:45–54. doi: 10.1038/nrrheum.2014.164
78. Galatz LM, Gerstenfeld LC, Heber-Katz E, Sa R. Tendon regeneration and scar formation: the concept of scarless healing. J Orthopedic Surg. (2015) 33:823–31. doi: 10.1002/jor.22853
79. Elmer Perkin. (2013). Quantum FX: Low Dose Computed Tomography [Product Note]. Available at: https://www.perkinelmer.com/labsolutions/resources/docs/BRO_010486_01_PRD_QuantumFX.pdf
Keywords: alveolar bone, GAPDH, GLUT-1, HIF-1α, ldh-a: micro-CT, mice, oct3/4, periodontal disease, periodontal ligament, PGK1, PDK1, PHDI, pulp, 1, 4-DPCA
Citation: Zebrowitz E, Aslanukov A, Kajikawa T, Bedelbaeva K, Bollinger S, Zhang Y, Sarfatti D, Cheng J, Messersmith PB, Hajishengallis G and Heber-Katz E (2022) Prolyl-hydroxylase inhibitor-induced regeneration of alveolar bone and soft tissue in a mouse model of periodontitis through metabolic reprogramming. Front. Dent. Med 3:992722. doi: 10.3389/fdmed.2022.992722
Received: 12 July 2022; Accepted: 20 October 2022;
Published: 25 November 2022.
Edited by:
Bjorn Olsen, Harvard Medical School, United StatesReviewed by:
Takehito Ouchi, Tokyo Dental College, JapanZhipeng Fan, Capital Medical University, China
© 2022 Zebrowitz, Aslanukov, Kajikawa, Bedelbaeva, Bollinger, Zhang, Sarfatti, Cheng, Messersmith, Hajishengallis and Heber-Katz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ellen Heber-Katz, heberkatz@icloud.com
†Present Addresses: Elan Zebrowitz, New York Medical College, Valhalla, NY, United States
Sam Bollinger, Cancer Biology Graduate Group, Stanford, CA, United States
Yong Zhang, Analytical Research & Development, Merck & Co, Kenilworth, NJ, Uniited States
Jing Cheng, Alcon Laboratories, Duluth, GA, United States
‡These authors share first authorship
Specialty Section: This article was submitted to Regenerative Dentistry, a section of the journal Frontiers in Dental Medicine
 Elan Zebrowitz
Elan Zebrowitz