- 1School of Materials Science and Engineering, Shanghai Institute of Technology, Shanghai, China
- 2Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai, China
- 3Department of Reproductive Medicine, Shandong First Medical University, Affiliated Jinan Central Hospital, Jinan, Shandong, China
- 4Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University, Shanghai, China
- 5Department of Radiation Oncology, Fudan University Shanghai Cancer Center, Fudan University, Shanghai, China
- 6Department of Traditional Chinese Medicine, Jinshan Hospital, Fudan University, Shanghai, China
- 7Centre for Artificial Intelligence Driven Drug Discovery, Faculty of Applied Sciences, Macao Polytechnic University, Macao, China
- 8Digital Diagnosis and Treatment Innovation Center for Cancer, Institute of Translational Medicine, Shanghai Jiao Tong University, Shanghai, China
- 9Ninth People’s Hospital, Shanghai Key Laboratory of Stomatology and Shanghai Research Institute of Stomatology, National Clinical Research Center of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 10Institute of Clinical Science, Zhongshan Hospital, Fudan University, Shanghai, China
The success rate of drug development today remains low, with long development cycles and high costs, especially in areas such as oncology, neurology, immunology, and infectious diseases. Single-cell omics, encompassing transcriptomics, genomics, epigenomics, proteomics, and metabolomics enable the analysis of gene expression profiles and cellular heterogeneity from the perspective of individual cells, offering a high-resolution view of their functional diversity. These technologies can help reveal disease mechanisms, drug target identification and validation, selection of preclinical models and candidate drugs, and clinical decision-making based on disease response to drugs, all at the single-cell level. The development of deep learning technology has provided a powerful tool for research in drug discovery based on single-cell techniques, which has evolved with the advent of large-scale public databases to predict drug responses and targets. In addition, traditional Chinese medicine (TCMs) research has also entered the era of single-cell technology. Single-cell omics technologies offer an alternative way in deciphering the mechanisms of TCMs in disease treatment, revealing drug targets, screening new drugs, and designing combinations of TCMs. This review aims to explore the application of single-cell omics technologies in drug screening and development comprehensively, highlighting how they accelerate the drug development process and facilitate personalized medicine by precisely identifying therapeutic targets, predicting drug responsiveness, deciphering mechanisms of action. It is also concluded that drug development process and therapeutic efficacy of drugs can be improved by combining single-cell omics and artificial intelligence techniques.
1 Introduction
Drug discovery is a complex and time-consuming process that often involves multiple stages, including target identification, primary screening, drug optimization, and clinical trials (Van de Sande et al., 2023; Vemula et al., 2023). Each stage faces significant challenges: target identification requires an in-depth understanding of the molecular mechanism of the disease; the preliminary screening stage requires efficient compound screening technology; and the drug optimization stage requires balancing the efficacy and safety of the drug. In addition, clinical trials need to rigorously verify the effectiveness and safety of drugs. The entire process is not only time-consuming, but also requires a large amount of capital investment and has a high failure rate (Shaker et al., 2021; Sarkar et al., 2023).
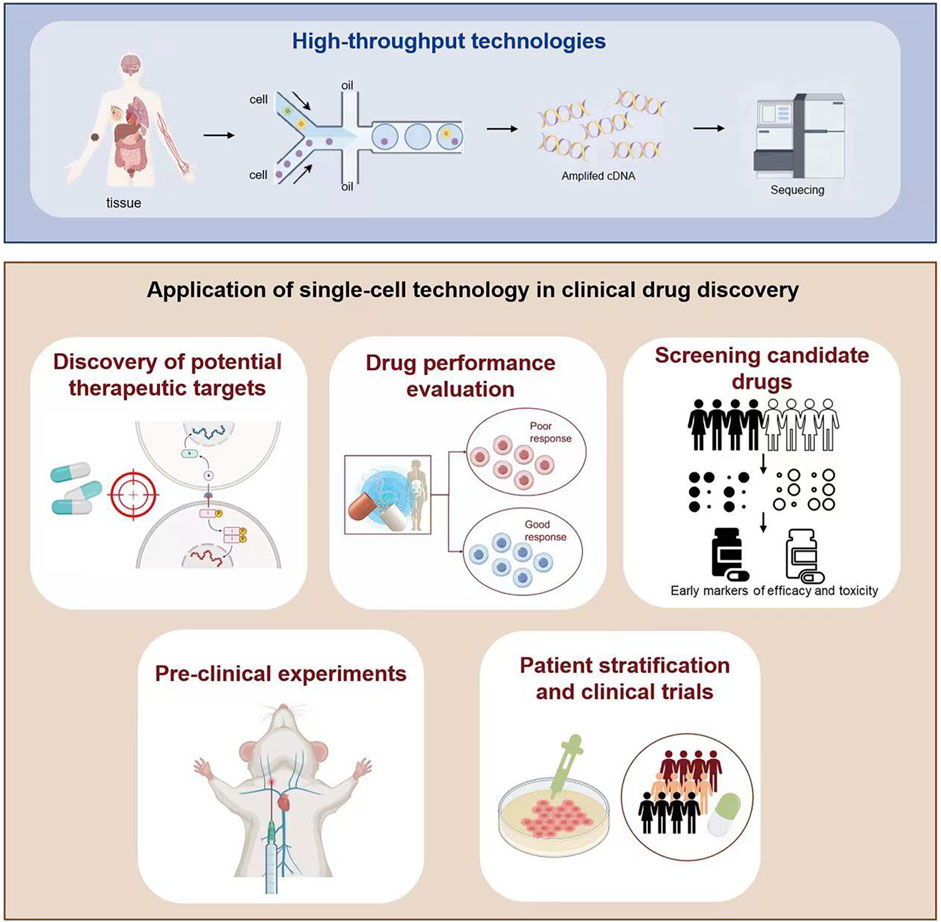
Since 2009 (Tang et al., 2009), single-cell sequencing technology has experienced rapid development and is widely used in clinical medical research and drug discovery (Van de Sande et al., 2023). Compared with bulk sequencing, single-cell sequencing technology has significant advantages. Single-cell technology can comprehensively describe the biological characteristics and status of individual cells and their interactions with the tissue environment, such as single-cell genome, single-cell transcriptome, single-cell epigenetic group, etc. Other single-cell technologies, such as single-cell proteomics, T-cell and B-cell receptor sequencing, and single-cell metabolomics, further expand the scope of single-cell research (Figure 1A).
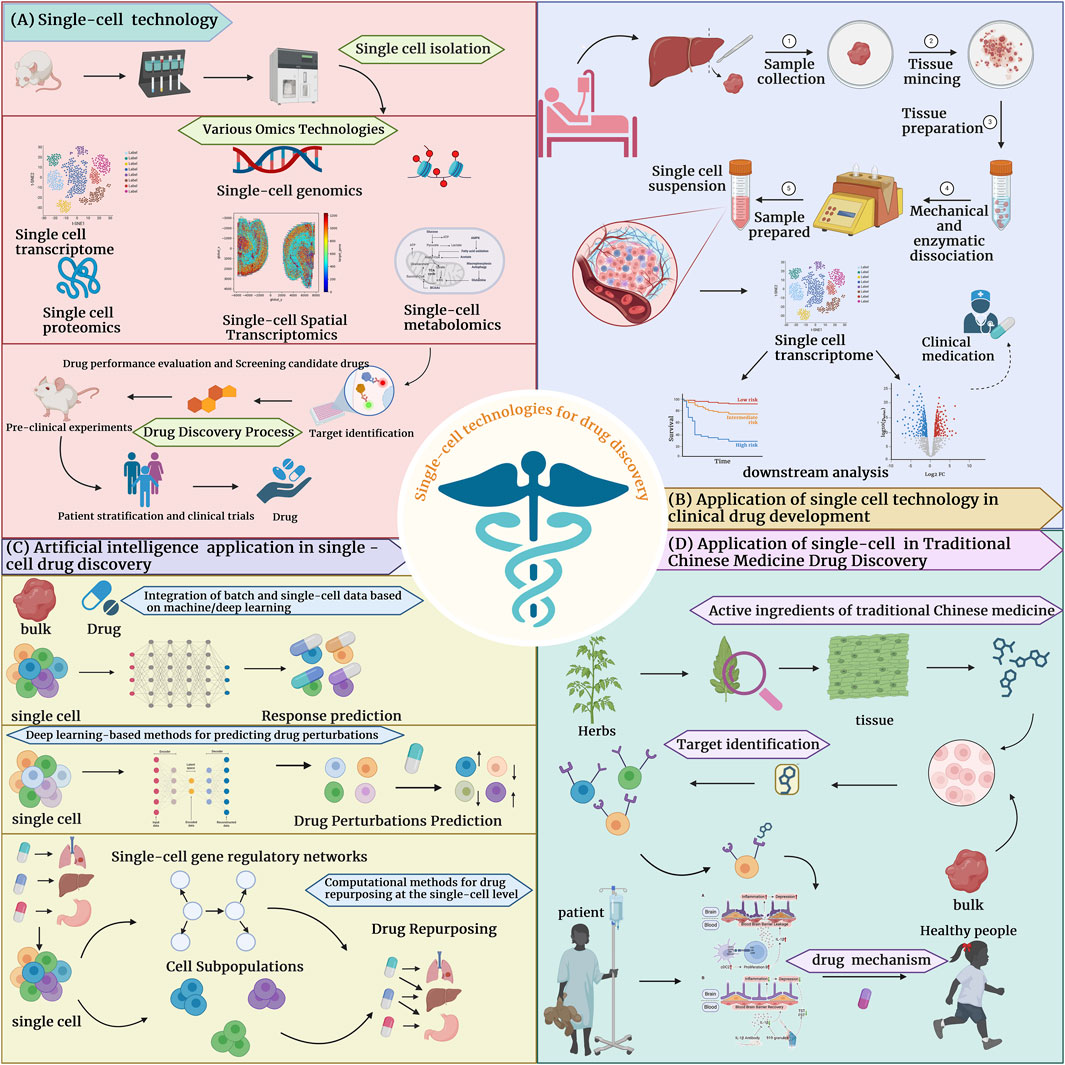
Figure 1. Application of single-cell technology in drug development (A) The process of drug development by combining single-cell technology with one or more omics technologies. (B) The application of single-cell technology in clinical drug development in tumors and non-tumors. (C) Single-cell technology combined with computational methods for drug discovery, including 1. Methods based on deep, machine learning to integrate batch and SC data, 2. Deep learning-based drug perturbation prediction methods and drug repurposing computational methods to predict drug response. (D) Single-cell technology combined with traditional Chinese medicine for drug development, explained in three categories: research on active ingredients of traditional Chinese medicine, targets of traditional Chinese medicine, and elucidation of the pharmacological mechanisms of traditional Chinese medicine.
The application of single-cell technology in drug discovery has significant advantages (Nassar et al., 2021; Saviano et al., 2020). It helps identify and validate disease-related targets by providing precise cell-specific data, improving the efficiency and success rate of drug development (Figure 1B; Figure 3). Single-cell technology is widely used in the treatment of tumors (Zheng L. et al., 2021; Wagner et al., 2019), neurological diseases (Yang et al., 2024; Liu et al., 2020), immune-mediated diseases (Zhang et al., 2019; Nehar-Belaid et al., 2020), and infectious diseases (Wang C. et al., 2022; Wang Y. et al., 2022), providing new research perspectives and drug development possibilities. Through sing-cell technology, researchers can reveal the cellular heterogeneity of diseases (Liang et al., 2022; Ma et al., 2021) and identify key therapeutic targets to develop new treatments. This technology has made significant progress in screening and validating drug candidates, identifying a variety of potential drug candidates and providing directions for future drug development. Finally, single-cell technologies show great promise in analyzing disease phenotypes, enable large-scale disease modeling and drug testing, and help to optimize therapeutic interventions in clinical trials (Wu et al., 2022; Eggenhuizen et al., 2024).
In past research, it is common to combine computational methods with high throughput omics data to predict drug effectiveness and disease progression (Partin et al., 2023; Maeser et al., 2021). However, these methods often lack understanding of cellular heterogeneity and molecular mechanisms at the single-cell level, limiting their effectiveness in identifying specific cell types in response to drugs. In recent years, there have been more research on algorithms for applying deep learning and machine learning to single-cell data (Figure 1C; Figure 4). In the field of drug discovery, the applications of deep learning or machine learning are mainly divided into drug perturbation prediction, integrate batch and drug response prediction and drug repurposing. In terms of predicting drug perturbations at the single-cell level, significant progress has been made using deep learning frameworks, including variational autoencoder (VAE) (Lotfollahi et al., 2019), transformer (Theodoris et al., 2023), etc., to simulate cellular responses to various perturbations. Integrating batch and single-cell data is critical to improve the accuracy of drug response predictions. It is a common practice to use deep transfer learning (DTL) to transfer drug response information from batch data (such as cell lines) to single-cell data (Zheng et al., 2023; Tang Z. et al., 2023; Chen J. et al., 2022). Single-cell technology has also revolutionized drug repurposing strategies. Utilizing the differential genetic information of disease-related cell subpopulations and more detailed gene regulatory network information provided by single-cell technology, computational methods (He et al., 2023; Hsieh et al., 2023; Oubounyt et al., 2023) can more accurately screen candidate drugs, improve development efficiency, and provide new treatment strategies for diseases.
Before the single-cell era, the drug development of TCMs mainly relied on traditional bulk data analysis, and the effects and mechanisms of action of TCMs components were obtained by studying the entire tissue or sample (Yang et al., 2021; Shang et al., 2023). However, traditional method has many limitations, and it is difficult to screen the active ingredients of TCMs, and it is difficult to reveal the differences in the effects of TCMs between different cell types and complex biological processes. Combining single-cell technology, researchers have revealed the role of TCMs in drug development from the perspective of its active ingredients, targets, and mechanisms of action. (Figure 1D; Figure 5).
2 Single-cell technology
In the drug discovery process, the application of single-cell technologies has brought significant advantages to improving the effectiveness and safety of drugs (Van de Sande et al., 2023). These technologies can provide unprecedented cellular resolution, revealing cell subpopulations and molecular mechanisms that are difficult to discover with traditional methods (Mitra-Kaushik et al., 2021). Using single-cell technologies, researchers can more accurately assess the effects of drugs in different cell types and their potential side effects, thereby optimizing drug design, improving therapeutic effects, and reducing adverse reactions (Aissa et al., 2021). In the following parts of this section, we will introduce single-cell technologies including single-cell genomics, single-cell transcriptomics, and single-cell epigenomics (Figure 2).
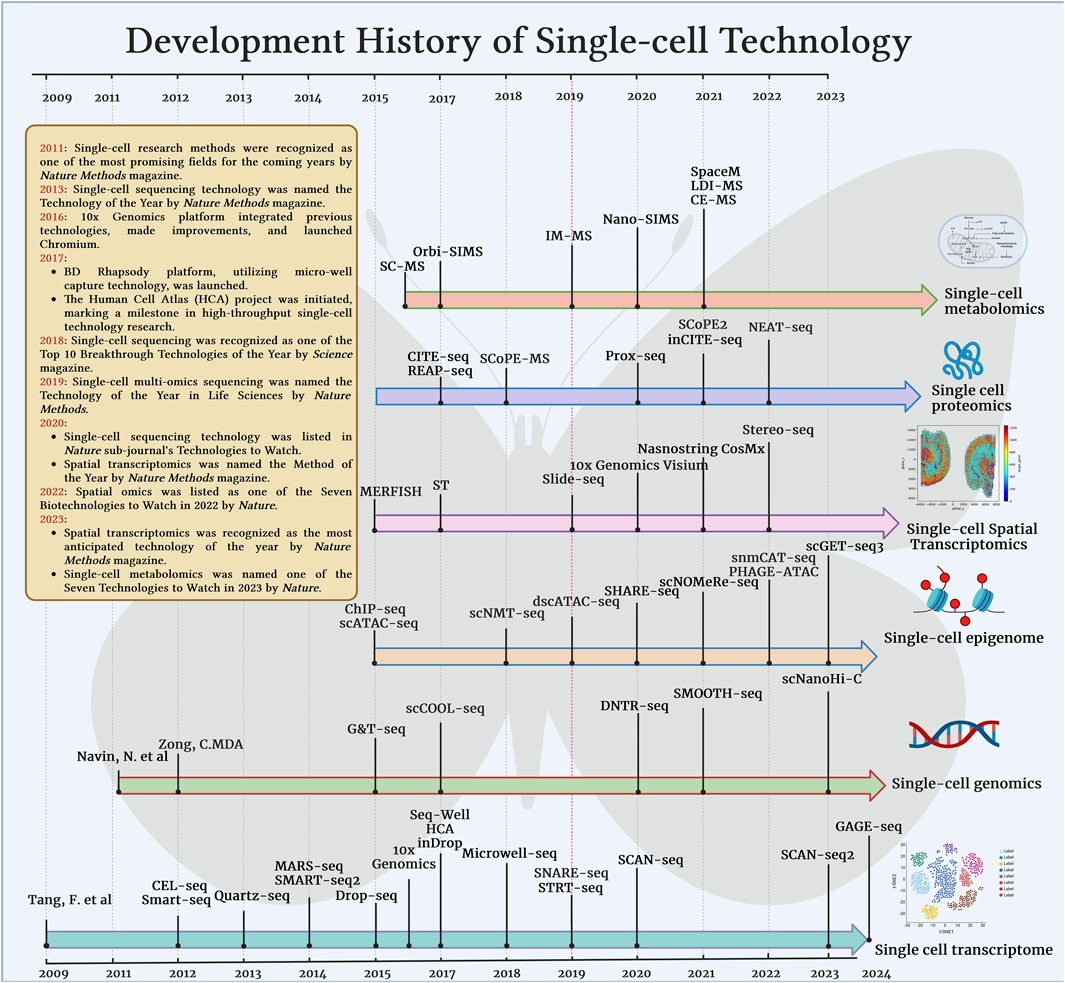
Figure 2. Development and major milestones of single-cell technology in various omics technologies such as transcriptome, genome, epigenetic group, proteome, spatial transcriptome, metabolome.
2.1 Single-cell DNA sequencing technology
Single-cell DNA sequencing technology is an advanced technology that can perform genome analysis at the single cell level. By sequencing the genome of a single cell, it reveals cell-to-cell heterogeneity and genome variation. The core steps of this technology include single-cell isolation, genome amplification, and high-throughput sequencing. Navin, N. et al. first sequenced the genome of a single cell in 2011 (Navin et al., 2011). With the development of second-generation and third-generation sequencing technologies, single-cell whole genome sequencing such as SMOOTH-seq (Fan et al., 2021), Digital-WGS (Ruan et al., 2020), Refresh-seq (Wang et al., 2024a), etc., have been used in drug discovery.
2.2 Single-cell RNA sequencing technology
The evolution of single-cell RNA sequencing technology (scRNA-seq) began with the work (Tang et al., 2009) in 2009 and was first applied to small-scale single-cell analysis in 2011, followed by rapid development and commercialization. SMART-seq2 (Picelli et al., 2014) technology has the advantage of improving sequencing coverage and sensitivity for full-length transcript sequencing. In recent years, the emergence of technologies such as Drop-seq (Macosko et al., 2015), Seq-Well (Gierahn et al., 2017), and Microwell-seq (Han et al., 2018)has further improved the throughput and accuracy of single-cell sequencing. With the continuous development of these technologies, scRNA-seq has become a key tool for studying cellular heterogeneity and complex biological systems, promoting the rapid development of biomedical research and precision medicine.
2.3 Single-cell epigenome sequencing
Single-cell epigenome sequencing is an advanced technology that studies epigenetic modifications at the single-cell level. It can reveal epigenetic changes in different cell types and states and gain in-depth understanding of the mechanisms that regulate gene expression. Epigenetic modifications include histone modifications, chromatin accessibility, and DNA methylation, which play a key role in regulating gene expression and cell function. The earliest technologies such as Hi-C (Nagano et al., 2013), ATAC-seq (Buenrostro et al., 2015a; Buenrostro et al., 2015b), and ChIP-seq (Rotem et al., 2015), with the development of single-cell technology, can reveal the genomic structure, mutations, and variations of single cells.
2.4 Single-cell spatial transcriptome technology
Spatial transcriptome technology is a cutting-edge technology for studying gene expression and its spatial distribution at the level of individual cells, aiming to reveal the spatial heterogeneity of cells in tissues and their functional characteristics. This technology combines scRNA-seq and spatial information capture to achieve detailed analysis of complex tissue structures by preserving the position and gene expression data of each cell on the tissue section. Generally speaking, it can be divided into two categories. The first category is image-based technology, such as in situ hybridization and in situ sequencing, which includes MERFISH (Zhang et al., 2021), seqFISH (Shah et al., 2018) and Nanostring CosMx. The second category is capture and sequencing-based technology, represented by 10x Visium, slide-seq (Rodriques et al., 2019) and Stereo-seq (Chen A. et al., 2022). In recent years, some technologies have reached the level of spatial transcriptome sequencing with subcellular resolution, such as 10x Xenium, nanostring CosMx SMI.
2.5 Other single-cell technologies
Other single-cell technologies include single-cell proteomics, T cell receptor (TCR) and B cell receptor (BCR) sequencing, and single-cell metabolomics sequencing technology. Single-cell proteomics (Li L. et al., 2018) combines single-cell separation, protein marker detection and high-sensitivity mass spectrometry analysis to provide protein expression profiles and interaction networks for each cell, and is applied in cancer (Li L. et al., 2018; Mund et al., 2022), immunology (Bassez et al., 2021), neuroscience (Goto-Silva and Junqueira, 2021). TCR and BCR sequencing technologies analyze the variable regions of T cell and B cell receptors through high-throughput sequencing, revealing the diversity and antigen specificity of immune cells, which is of great significance for understanding the role of the immune system in infection (Wang P. et al., 2021), autoimmune diseases (Zheng F. et al., 2021) and cancer immunotherapy (He et al., 2022). Single-cell metabolomics (Shaojie et al., 2023) uses high-sensitivity mass spectrometry technology to detect and analyze metabolites at the single-cell level, revealing metabolic heterogeneity and metabolic networks between cells, and promoting a deeper understanding of cell function and dynamic changes. In addition, single-cell multi-omics technologies have been developed, such as scNanoCOOL-seq (Lin et al., 2023), CITE-seq (Stoeckius et al., 2017), etc. These technologies also have a catalytic effect on drug discovery.
3 Application of single-cell technology in clinical drug discovery
Drug discovery is a complex, multi-stage process that takes years from target identification to drug launch (Figure 3), and single-cell technology plays an important role in drug development in both oncology and non-oncology applications (Zeng Q. et al., 2023; Huang et al., 2023). We primarily emphasized the significant advantages of single-cell omics technology in discovering drug targets and investigating drug resistance mechanisms by analyzing tissue heterogeneities.
3.1 Tumors
In clinical research, drug discovery is a comprehensive task, particularly for tumors such as lung cancer (Ruiz-Cordero and Devine, 2020; Meijer et al., 2022), liver cancer (Li and Wang, 2016) and glioblastoma (El Atat et al., 2023). Unlike common diseases, tumor drug discovery faces challenges related to tumor heterogeneity and resistance (Dagogo-Jack and Shaw, 2018; Vasan et al., 2019), which traditional methods are unable to overcome. However, the application of single-cell omics is addressing these obstacles.
3.1.1 Discovery of potential therapeutic targets under tumor heterogeneity and tumor microenvironment
Specific biomarkers and new drug targets for effective targeted therapy or immunotherapy are the key to the treatment of diseases and drug development. In the early stage of drug development, single cell sequencing technology can find effective biomarkers and potential drug targets more accurately from the gene or protein level by in-depth study of tumor heterogeneity and tumor microenvironment.
By integrating scRNA-seq and bulk RNA sequencing, Zhang et al. (Zhang L. et al., 2022) studied the differences in gene amplification, cell composition and expression module across different types of lung cancer such as lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). They identified 10 sub-cluster markers such as AQP5 and KPNA2, which could potentially serve as drug targets for early diagnosis and treatment of lung cancer. Additionally, using scRNA-seq and spatial transcriptome RNA sequencing data, Nicotinamide N-Methyltransferase (NNMT) and Hypoxia inducible lipid droplet-associated (HILPDA) have been identified as promising therapeutic targets in clear cell renal cell carcinoma (ccRCC), emphasizing their association with hypoxia and metabolic pathways (Zhang et al., 2024). In gastric cancer (GC) research, by generating cell maps of 10 patients with GC, IL-17+ tumor-associated stromal cells (TASCs) were found to potentially promote tumor progression through IL-17, IL-22 and IL-26 signaling pathway, highlighting the potential of targeting IL-17+ cells and related signaling pathways as drug targets and therapeutic strategies for GC (Sun et al., 2022). In the study of uveal melanoma (UM), CD8+ T cells were found to predominantly express the checkpoint marker LAG3, suggesting LAG3 as a potential candidate target for immune checkpoint blocking in high-risk UM patients (Durante et al., 2020). Similarly, Zhu et al. (Zhu et al., 2023a) developed inflammation-related prognostic signals (IRPS) using scRNA-seq data in prostate cancer, identifying six genes significantly associated with tumor immunity, therapeutic response and drug screening. Among these, PTGIR exhibited strong affinity for multiple anti-tumor drugs, potentially serving as a biomarker for candidate drugs.
3.1.2 Drug resistance research
It is important to evaluate the drug sensitivity, effectiveness and drug resistance before clinical trials. Studies have shown that tumor heterogeneity is an important reason for drug resistance of tumor cells. Drug-resistant cells will gradually replace drug-sensitive cells with the progress of chemotherapy. Single-cell sequencing technology can clarify the mechanism of drug action at the single cell level and can effectively evaluate the dynamic changes between different cell molecules in tumor microenvironment under the condition of drug treatment.
Previous studies have shown the drug resistance may attribute to both cancer cells and microenvironment cells. Treatment-resistant glioblastoma patients exhibited an increased presence of high-stemness tumor cells and a reduced proportion of microglia (Wu et al., 2023). Similarly, the cell cycle activated melanoma tumor cells demonstrated high level of treatment resistance (Randic et al., 2023). The characteristics of mesenchymal stem cells were found to be related to the effectiveness of anti-tumor treatment in GC (Shen et al., 2023). The heterogeneity of tumor cells was found as the cause of tyrosine kinase inhibitor treatment resistance in tumor patients with somatic mutations of receptor tyrosine kinase (Aissa et al., 2021). Malignant B cell subsets are enriched in drug-resistant multiple myeloma (MM), and exhausted CD8+ T cells and unique CD16+ myeloid cells are related to the progress of drug-resistant MM (Wang HN. et al., 2021). Similarly, type Ⅲ malignant B cells also play an important role in the drug resistance mechanism of Mantle cell lymphoma (MCL). Some drug target genes, such as BTK, FCGR2A, FCGR2B and FCGR3A, are highly expressed in patients with MCL, which indicates that the activity of B cells is enhanced during and after drug resistance (Wang et al., 2020).
3.1.3 Screening anti-tumor drugs
Screening drugs plays a vital role in drug development, but it takes a lot of work and time. Single cell sequencing technology can reflect the mechanism of drug action at the cellular and molecular level, provide the most original data for screening candidate drugs, which is helpful to the discovery of candidate drugs and promote the development of new drugs.
In prior studies, researchers identified specific cellular subgroups or disease-associated differentially expressed genes by analyzing scRNA-seq data under various disease conditions. This analysis facilitated drug screening and the discovery of candidate drugs. For instance, single-cell transcriptome sequencing was used to analyze the cellular composition and gene expression in diabetic kidney disease (DKD), leading to the identification of candidate drugs relevant to DKD. Researchers evaluated the efficacy of the top three candidate drugs through cellular and animal experiments (Li et al., 2024). In addition, by integrating scRNA-seq and scATAC-seq technologies, we analyzed the changes in gene expression and chromatin state of prostate cancer cell lines before and after enzalutamide (ENZ) treatment, identified seven key gene signatures, and nominated combination drug candidates through drug-gene network analysis to enhance early treatment response or overcome ENZ resistance (Fan et al., 2023). Furthermore, scRNA-seq revealed that 177Lu-FAP6-DOTA is a reliable candidate for treating solid tumors, demonstrating high affinity for FAP-expressing cells, significantly increased tumor doses, and effective tumor growth suppression with minimal toxicity (Lindeman et al., 2023).
3.1.4 Pre-clinical models with single-cell
At present, a key way to develop anti-tumor drugs is to use preclinical animal models, and many important discoveries are achieved through preclinical animal models, utilizing single-cell technology.
For example, using scRNA-seq, researchers examined CD45+ tumor infiltrating lymphocytes in mouse model of CRC treated with AB680, a selective inhibitor of CD73 extracellular enzyme, to block tumorigenic ATP adenosine signals as a candidate immunotherapy (Kim et al., 2021). Similarly, in DKD, scRNA-seq revealed diverse responses among renal cell types to various treatment regimens in mouse models. These findings suggest the potential of SGLT2i inhibitors to regulate proximal tubule splicing, providing insights into their therapeutic mechanisms (Wu et al., 2022).
Although preclinical animal models have played a crucial role in drug research and development, their variability in immune responses and interspecies differences often limit their ability to accurately recapitulate responses seen in human tumor patients, posing ongoing challenges. Therefore, linking human and animal models is essential. Single-cell technology allows for the mapping of cellular landscapes across different animal models, enabling comparisons with patient-derived maps to identify models that mimic similar tumor microenvironment characteristics for use in preclinical studies of specific drugs under development. For instance, analysis of myeloid cells in the lung tissues from non-small cell lung cancer patients and mouse models using scRNA-seq revealed conservation of monocyte and dendritic cell subtypes across species but heterogeneity among macrophage subsets between humans and mice (Han et al., 2022). Similarly, systematic evaluation of single-cell transcription regulation maps in cynomolgus monkeys and other non-human primate models compared to humans highlights differences in cell composition, organ heterogeneity, and spatiotemporal gene expression. These insights provide foundational understanding for assessing drug responses and disease mechanisms relevant to human health (Qu et al., 2022).
3.1.5 Patient stratification and clinical trials for precision medicine
In clinical trials, analyzing patients’ genetic backgrounds at the single-cell level help researchers accurately identify patient subgroups suitable for specific drugs, guiding precise clinical medication and advancing.
For instance, using scRNA-seq to analyze breast cancer cell lines, researchers generated a single-cell map of breast cancer (Gambardella et al., 2022). Deconvolution algorithms were then employed to determine cell composition, enabling patient stratification based on cell lines. By integrating in vitro drug screening results from cell lines and single-cell data, this approach aids in optimizing existing treatments. In another study, the heterogeneity of triple-negative breast cancer (TNBC) tumors was comprehensively revealed, highlighting key clinical correlations and influencing factors (Ge et al., 2024). This research proposed a new strategy of combining LINE-1 inhibition with immunotherapy for TNBC, offering potential biomarkers for predicting patient treatment responses. For non-small cell lung cancer (NSCLC), single-cell datasets were integrated to create a high-resolution view of 44 major cell types/states within the tumor microenvironment (TME) (Salcher et al., 2022). This detailed single-cell composition allowed for precise tumor classification and patient stratification into four immune phenotypes: immune-deserted, myeloid, B cell, and T cell subtypes. These insights are significant for enhancing the efficacy of tumor immunotherapy in NSCLC. Additionally, the analysis of plasma cells from patients with advanced refractory multiple myeloma (aRRMM) undergoing treatment with selinexor combined with dexamethasone or bortezomib combined with dexamethasone used scRNA-seq maps in conjunction with clinical trials (Cohen et al., 2021). This method stratified patients based on their resistance to anti-MM treatment, defining new resistance biomarkers, potentially supporting personalized treatment decisions and uncovering new therapeutic targets.
3.2 The role of single cell techniques in drug development for non-cancerous diseases
Single-cell technology has extensive applications in the treatment of non-cancerous diseases, such as neurological disorders (e.g., Alzheimer’s disease (AD), Parkinson’s disease (PD), multiple sclerosis (MS)), immune-mediated diseases (e.g., rheumatoid arthritis (RA), systemic lupus erythematosus (SLE)), and infectious diseases (e.g., COVID-19, HIV/AIDS). This technology provides new perspectives for studying the mechanisms underlying these diseases and opens up new possibilities for drug development (Roostaei et al., 2017; Hammond et al., 2019; Mathys et al., 2019; Mathys et al., 2017). It is particularly important in target identification, target validation, preclinical research, drug screening, biomarker discovery, and understanding patient stratification and treatment responses.
3.2.1 Discovery of potential therapeutic targets
3.2.1.1 Neurological disorders
The emergence of scRNA-seq has enabled researchers to explore the impact of AD on cellular heterogeneity. In AD research, preventing dysfunction of astrocytic organelles is considered a crucial direction in the therapeutic strategies for AD (Galea et al., 2022). Dang et al. utilized scRNA-seq to reveal that emotional and cognitive impairments in AD are associated with ferroptosis in astrocytes. They identified FTH1 and SAT1 as critical regulators influencing ferroptosis in astrocytes. This study provides insights into the pathophysiological processes at the cellular level following AD and underscores potential therapeutic targets for treating the disease (Dang et al., 2022). At single-cell resolution, the characterization of in vitro human dopamine neurons reveals composition and cell-type specific responses to genetic and cytotoxic stress (Fernandes et al., 2020). By combining human brain snRNAseq data with tau mouse model data, researchers studied neurons and glial cells in neurodegenerative PD (Min et al., 2023). They identified therapeutic targets with potential and validated these targets in vivo using a tau fly model. In addition, research on multiple sclerosis (MS) also demonstrates the power of single-cell analysis. Single-cell analysis of immune cells in the cerebrospinal fluid during relapses of multiple sclerosis patients suggests that targeting heterogeneous cell populations could potentially serve as therapeutic targets for MS (Ostkamp et al., 2022).
3.2.1.2 Infectious diseases
scRNA-seq has established a high-resolution map of blood antigen-presenting cells (APCs) in COVID-19 pneumonia patients, revealing multi-process defects in antiviral immunity, specifically in certain subsets of dendritic cells (DCs). These findings lay the groundwork for novel therapies aimed at restoring defective APC functions in COVID-19 patients. Many studies have already developed numerous adjuvants targeting DC subsets and have also considered DCs in the development of prophylactic vaccines (Bryant et al., 2019; Saxena and Bhardwaj, 2017). In addition, the application of single-cell technology in HIV research has also brought new hope for drug discovery. Additionally, research utilizing single-cell techniques characterized mature monocyte subsets from uninfected individuals, HIV-infected individuals, and individuals with HIV who remain uninfected despite exposure, without ART. This study identified therapeutic targets for blocking HIV monocyte entry into tissues (Leon-Rivera et al., 2020).
3.2.2 Drug performance evaluation
3.2.2.1 Neurological disorders
In the study of drug performance evaluation, single-cell RNA sequencing (scRNA-seq) technology has provided important insights into AD. By generating more representative human microglia models (MDMi) and utilizing scRNA-seq to analyze their expression profiles, we can gain valuable insights into the AD. For example, examining cytokine responses post-treatment with anti-inflammatory drugs (dasatinib and spironolactone) can provide crucial therapeutic information (Cuni-Lopez et al., 2024).
3.2.2.2 Immune-mediated diseases
Similarly, the application of single-cell technology in immune-mediated diseases has also shown its great potential. Hedman et al. analyzed and compared the effects of methotrexate (MTX) treatment in RA patients and healthy controls to study the contribution of immune cells to RA therapy (Hedman et al., 2023). Rheumatoid arthritis synovial single-cell spatial analysis also identified specific macrophage cellular states associated with treatment response (Julien et al., 2023). In vitro experiments suggest that non-steroidal anti-inflammatory drugs (NSAIDs) effectively block TNFα-induced synovial macrophage responses (Kuo et al., 2019), and different subpopulations of synovial macrophages regulate inflammation and resolution in RA (104). The research used single-cell transcriptomics to profile synovial tissue macrophages (STM) and revealed STM subpopulations (MerTKpos STMs) in remission, associated with an increased risk of disease flare after treatment cessation (Alivernini et al., 2020). Therapeutic modulation of MerTKpos STM subpopulations could therefore be a potential treatment strategy for RA.
3.2.2.3 Infectious diseases
Moreover, the immune cell landscape of COVID-19 also revealed how neutrophils are dynamically regulated or programmed in COVID-19 patients receiving dexamethasone treatment, enabling more effective responses to viral infection (Chen Y. et al., 2023).
3.2.3 Screening candidate drugs
3.2.3.1 Neurological disorders
In recent years, numerous studies have made significant progress in screening and validating candidate drugs for the treatment of various diseases. Yin et al. found that BRI2’s ability to regulate APP and TREM2 processing, along with its cell-autonomous roles in neurons and microglia, makes it a promising candidate for the development of therapeutic drugs for AD and AD-related dementias (Yin and D’Adamio, 2023). The further application of single-cell technology has also achieved important results in the study of multiple sclerosis (MS). Analysis of AD-specific gene-related signaling pathways from astrocytes isolated from the entorhinal cortex of AD patients revealed that anti-rheumatic drugs can reverse gene characteristics associated with AD in astrocytes (Pushparaj et al., 2021). Dimethyl fumarate (DMF) is commonly used to suppress relapses in MS patients. Studies have shown that it can reduce a T helper population expressing GM-CSF, highlighting its relevance as a therapeutic target (Galli et al., 2019).
3.2.3.2 Infectious diseases
Transcriptomic analysis of monocytes from survivors and deceased COVID-19 patients identified key host response pathways overactivated in non-survivors. Based on this, tacrolimus, zotarolimus, and nintedanib were identified as three potent candidate drugs for the treatment of critically ill patients upon hospital admission (Singh et al., 2022). High-throughput screening of FDA-approved drugs revealed multiple COVID-19 entry inhibitors, including imatinib, meclofenamic acid, and quinacrine dihydrochloride. These drugs significantly inhibited COVID-19 infection at physiologically relevant levels (Han et al., 2021). By applying single-cell techniques to these studies, scientists were not only able to identify and validate potential drug candidates for the treatment of COVID-19, but also made similar advances in HIV research. A study employing scRNA-seq data predicted drugs that could reverse the altered monocyte-derived signatures. Doxycycline and sunitinib are considered promising candidate drugs for further research and development (Knoll et al., 2023). Collectively, these studies demonstrate the potential of different candidate drugs in treating various diseases, providing valuable insights and directions for future drug development.
3.2.4 Pre-clinical experiments
3.2.4.1 Neurological disorders
In recent years, there has been substantial attention directed towards understanding the roles of neuroinflammation and immune regulation in a range of neurodegenerative and autoimmune diseases. A study simulated the effects of pharmacological compounds on human microglial subtypes across various in vitro and in vivo model systems. It found that the topoisomerase I inhibitor camptothecin reduced a subtype of microglia characterized by high CD74/MHC expression. This subtype may play a role in the early stages of AD specifically during the initial accumulation of amyloid-beta pathology (Haage et al., 2024). Recently, another method was reported for in vitro modeling of human microglial subtypes (Dolan et al., 2023). This involved exposing iPSC-derived microglia (iMG) to selected central nervous system (CNS) substrates, followed by associating the induced microglial subtypes with human microglial subtypes derived from snRNAseq datasets (Gazestani et al., 2023).
Research on early relapsing-remitting MS patients’ blood and CSF-derived cell transcriptome changes, along with animal models, revealed that IL-11/IL-11R signaling transduction in monocytes is a therapeutic target for RRMS (Seyedsadr et al., 2023). Identification of the overall changes in microglia or astrocytes associated with the development of brain tissue lesions in MS provides therapeutic targets to limit lesion progression and demyelination. Through the Mertk-KO mouse model, it has been demonstrated that Mertk, a gene highly expressed by microglial cells, can alter the risk of MS and is necessary for effective remyelination (Shen et al., 2021). Using an MS mouse model with doxycycline-inducible CXCL1 expression from astrocytes, it was revealed that neutrophils may contribute to demyelination through multiple pathways in white matter, offering new therapeutic targets for improving demyelination (Skinner et al., 2022).
3.2.4.2 Immune-mediated diseases
Furthermore, utilizing human and murine in vitro models, it has been determined that inhibiting macrophage glycolysis can ameliorate autoantibody-induced inflammation. Targeting macrophage metabolism may thus represent an effective therapeutic strategy for lupus nephritis (Jing et al., 2020). Research has also shown that Sm-specific Tregs (Sm-Tregs) effectively suppress ex vivo Sm-specific pro-inflammatory responses and inhibit disease progression in a humanized mouse model of lupus nephritis (Eggenhuizen et al., 2024). Sm-Tregs represent a promising therapeutic approach for SLE.
3.2.5 Patient stratification and clinical trials
3.2.5.1 Neurological disorders
Single-cell analysis can aid in identifying disease subgroups targeted for treatment. A study in treating SLE (Der et al., 2017) confirmed a significant reduction in IFN scores in SLE patients responding to treatment. In SLE, the IFN signature can categorize patients into two groups: SLE1, characterized by low expression levels of IFN-induced genes, and SLE2, characterized by high expression levels of IFN-induced genes (Ronnblom and Leonard, 2019). In SLE2 patients, a cytotoxic CD4 T cell subset has been identified (Trzupek et al., 2021). A recent attempt to reduce heterogeneity in brain organoids involves generating midbrain-like organoids from primitive neural stem cells (NSCs), mimicking phenotypes associated with PD. Disease phenotypes responded to disease-specific therapies, supporting the potential of this method in large-scale disease modeling and drug testing.
3.2.5.2 Immune-mediated diseases
ScRNA-seq analysis of cultured rheumatoid arthritis fibroblast-like synoviocytes (RA-FLS) in a simulated RA inflammatory milieu ± ABT-317 characterizes the molecular effects of the selective JAK1 inhibitor (ABT-317). Jak inhibition is effective in downregulating several proinflammatory pathways induced by conditioned media stimulation in FLS cultures (Son et al., 2023).
3.2.5.3 Infectious diseases
In infectious diseases, single-cell technology also shows important potential of patient stratification. Su et al. conducted a single-cell multi-omics analysis of peripheral blood mononuclear cells (PBMCs), identifying a sharp transition in disease state between mild and moderate COVID-19, suggesting that moderate COVID-19 may provide the most effective environment for therapeutic intervention (Su et al., 2020). Single-cell technology analysis of the immune landscape in the bronchioalveolar lavage of humans infected with COVID-19 revealed the critical role of macrophage-driven innate immunity in resolving COVID-19 infection (Singh et al., 2022). At the same time, significant progress has been made in its application in HIV research. Bradley et al. utilized scRNA-seq to analyze cellular gene expression in HIV latency primary cell models. The data indicate that the expression of HIV proviruses in the latent reservoir is influenced by host cell transcriptional programs, modulation of which therapeutically can reverse or reinforce HIV latency (Bradley et al., 2018).
4 Application of single-cell technology combined with deep learning and machine learning in drug development and drug repurposing
In the realm of drug development, the integration of single-cell technologies with computational methods and drug repurposing methods is revolutionizing the field (Theodoris et al., 2023; Zheng et al., 2023). Single-cell technologies provide high-resolution data at the cellular level, enabling researchers to gain a deep understanding of cellular heterogeneity and molecular mechanisms. By incorporating computational methods, researchers can leverage sophisticated algorithms and deep learning models to extract valuable insights from vast amounts of single-cell data, thereby accelerating the identification of drug targets and the screening of potential drug candidates (Lotfollahi et al., 2019; Zhu et al., 2023b; Wang et al., 2024b). Additionally, single-cell technologies play a pivotal role in drug repurposing by analyzing how different cell types respond to existing drugs, uncovering new therapeutic potentials and indications. This multidisciplinary approach not only enhances the efficiency and success rate of new drug development but also paves the way for personalized medicine and precision therapies (Zhu et al., 2023b) (Figure 4).

Figure 4. Single-cell technology combined with computational methods to predict drug response (A) Batch and SC data integration based on deep learning (B) Batch and SC data integration based on deep learning (C) Drug perturbation prediction method based on deep learning (D) Computational method for drug reuse at the cellular level.
4.1 Computational methods for drug discovery at the single-cell level
Computational methods based on various machine learning (ML) and deep learning (DL) frameworks have shown great potential in drug discovery (Jarada et al., 2020; Maeser et al., 2024; Adam et al., 2020; Wu et al., 2020). These methods utilize high-throughput screening (HTS) data and molecular profiles of cancer cell lines (CCL) to infer drug sensitivity (Adam et al., 2020). Recent studies using scRNA-seq have emphasized that subpopulations within heterogeneous tumors are closely related to drug resistance and disease progression (Dagogo-Jack and Shaw, 2018). Therefore, there is a need for drug discovery targeted at specific cell types. In this context, computational methods for inferring drug activity at the single-cell level have demonstrated significant potential for application.
The transcriptomic profiles of CCLs are based on bulk sequencing, hence they cannot be directly applied to scRNA-seq data for effective prediction of drug responses by cell type. Therefore, current algorithm development strategies focus on deriving knowledge from bulk CCL data and exploiting and discovering new insights within scRNA-seq data. To achieve this goal, some algorithms focus on integrating bulk and single-cell data. Moreover, some methods predict drug perturbations in single-cell transcriptomes, which can also support drug development targeted at specific cell types.
4.1.1 Deep learning-based methods for integrating bulk and single-cell data
SCAD (24), SpaRx (Tang Z. et al., 2023), and scDEAL (26) are methods utilize deep transfer learning (DTL) to integrate bulk and single-cell data and predict drug responses in cell types within single-cell or spatial transcriptomics (ST). The core step of these deep learning methods is transferring the drug response predictor trained on the source domain (e.g., cell lines) to the target domain (e.g., single cells). The methods differ primarily in their implementation of domain transfer.
The SCAD model employs adversarial discriminative domain adaptation, featuring a feature extractor and a domain discriminator. The domain discriminator challenges the feature extractor to develop domain-invariant features, enhancing the transferability of the drug response label classifier from the source to the target domain. This approach improves drug sensitivity prediction across different cellular states and identifies potential combination therapy strategies through the IntegratedGradients method. SpaRx employs a graph-based domain adaptation model, using a mutual nearest neighbors method for cell line similarity in the source domain and nearest neighbors for spatial cell graphs in the target domain. It features a multi-head graph transformer as the feature extractor and introduces dynamic adversarial adaptation learning, enhancing the prediction of drug responses on ST data. scDEAL adopts a different strategy for transfer learning. It uses two feature extractors to extract low-dimensional feature representations from the source domain and the target domain and predicts drug responses at the source domain feature level using a drug response predictor, which is a Multilayer Perceptron (MLP). Subsequently, it minimizes the maximum mean discrepancy (MMD) loss between gene features from the two extractors to achieve knowledge transfer from the source to the target domain, ultimately using the drug response predictor to predict drug responses at the single-cell level.
It is important to note that these deep learning approaches come with limitations. The optimal model structure can vary significantly across different drugs and datasets, requiring substantial computational resources which may impede their practical use. Furthermore, each method necessitates fine-tuning of neural network parameters specific to each drug and dataset, limiting the ability to simultaneously evaluate multiple drugs or perform large-scale drug screenings.
4.1.2 Machine learning-based integration of bulk and single-cell data
The scIDUC (Zhang et al., 2023) method combines drug-gene signatures with machine learning techniques to infer drug responses at the cellular level. Before integrating bulk and single-cell data, drug response-related genes (DRGs) are first identified from the bulk data. Then, canonical correlation analysis is used to integrate these datasets, identifying common gene expression patterns between the two and adjusting for existing differences. Non-negative matrix factorization can also be applied to achieve dataset integration. Subsequently, a linear regression model is constructed based on these integrated data. scIDUC features simple parameter adjustments, avoiding the complex computations involved in the deep learning methods mentioned previously. However, traditional machine learning methods often rely on a predefined set of key genes, which might not always capture the complex biological variability necessary to predict drug sensitivity accurately.
4.1.3 Deep learning-based methods for predicting drug perturbations
scGEN (22) and Geneformer (Theodoris et al., 2023) are two deep learning-based methods for predicting single-cell perturbations, but they are based on distinct theoretical frameworks. scGEN is a deep learning model utilizing variational autoencoders to predict single-cell responses to perturbations like drugs or infections. It works by encoding gene expression data into latent vectors, calculating perturbation vectors from differences between conditions, and then using these for predictions. Geneformer, based on transformer architecture, pre-trains on millions of single-cell transcriptomes, using masked gene prediction to model cell states under various conditions, including disease. In summary, these two methods have achieved deep learning-based predictions of single-cell perturbations, demonstrating powerful data processing and analytical capabilities through their adaptability and large-scale learning. However, they face challenges in computational resource demands and interpretability, thus practical application may require consideration of their complexity and dependence on high-quality data.
4.2 Computational methods for drug repurposing at the single-cell level
Drug Repurposing (DRP) (Roessler et al., 2021; Issa et al., 2021; Parvathaneni et al., 2019) is a strategy for finding new uses for existing drugs in different diseases. This approach not only reduces the cost and time of drug development, but also provides new hope for treating complex diseases. The application of single-cell technology in drug repurposing brings new breakthroughs. First, single-cell technology is able to reveal in detail the intercellular heterogeneity in diseases, as well as the differentially expressed genes (DEGs) between disease and normal in specific cellular subpopulations. Screening of drugs on cellular subpopulations makes the drug repurposing process more precise and effective. Second, the gene regulatory network constructed from single-cell data, combined with drug target information, can more accurately predict and infer the effectiveness of drugs, and the acquired effective alternative drugs can be verified by further in vitro experiments (Figure 4).
4.2.1 Cellular subcluster and differential gene based drug reutilization methods
ASGARD (27) is a single-cell drug reutilization calculation method that recommends drugs by comprehensively analyzing all cell clusters in a patient’s body and defining a drug score. ASGARD clusters and annotates the cells to get cell subgroups, DEGs are obtained for normal versus diseased cells. These genes are potential drug targets, and further used to screen existing drugs that can reverse these differentially expressed genes. ASGARD calculates drug scores and assess drug efficacy across multiple cell clusters. In a COVID-19 study, ASGARD analyzed scRNA-seq data from critically ill patients to predict and validate drugs such as rasinamide and enalapril, which helped to reduce mortality in critically ill COVID-19 patients.
scDrug (Hsieh et al., 2023) is an effective computational method in the repurposing of drugs for the treatment of hepatocellular carcinoma (HCC). After tumor cells are segmented by scDrug to identify tumor subclusters, survival analysis is performed on the differentially expressed genes in these subclusters and predicted the responses to different drugs. In a hepatocellular carcinoma research, scDrug identified tumor subpopulations with malignant characteristics and predicted drug candidates with high sensitivity to these tumor subpopulations in the PRISM database. Finally, based on the LINCS L1000 data (Musa et al., 2019), scDrug suggested combination therapy strategies (including dasatinib, torin, etc.) that could effectively inhibit the growth of the tumor subpopulations.
4.2.2 Single-cell gene regulatory network-based approach to drug repurposing
SCANet is an approach that infers differential key regulatory networks and mechanisms in scRNA-seq data to hypothesize drug repurposing candidates (Oubounyt et al., 2023). SCANet uses weighted gene co-expression network analysis (WGCNA) to identify gene modules, construct transcription factor gene regulatory networks, and perform cis-regulatory analysis to ensure accuracy. To achieve drug repurposing, SCANet constructs heterogeneous networks by accessing multiple databases to identify potential drug targets and candidate drugs. scDrugPrio is a computational framework for constructing inflammatory disease network models using single-cell RNA sequencing data for drug prioritization and repurposing (Schafer et al., 2024). scDrugPrio provides final drug rankings by calculating intracellular and extracellular centrality of drug-differentially expressed gene interaction networks and synthesizing centrality scores for all cell types.
5 The role of single-cell technology combined with TCMs in drug development
5.1 Research on active ingredients of TCMs
The active ingredients of TCMs are bioactive molecules with therapeutic effects. These components, including saponins (Sun et al., 2009; Rao and Sung, 1995), alkaloids (Li et al., 2023), flavonoids (Panche et al., 2016), etc., have biological function such as anti-inflammatory, anti-cancer or immunomodulatory effects. The application of single-cell technology on TCMs elucidates the synthesis mechanism of active ingredients in plants and its potential for high-throughput screening of active ingredients.
Single-cell technology has revealed the biosynthetic pathway of monoterpene indole alkaloids (MIA) in Catharanthus roseus (Li et al., 2023). It detected the specific expression patterns of MIA pathway genes in Catharanthus roseus leaves and roots and determined the clustered distribution of these genes on chromosomes. The results provide a basis for the biosynthesis and drug development of anti-cancer components in Catharanthus roseus.
Screening of active ingredients in TCMs is another important task. In recent years, single-cell technology has been used to screen and measure TCMs, such as artemisinin (Chen J. et al., 2023), quercetin (Zhao et al., 2023) and the natural substances frankincense and myrrh (Liu et al., 2023). single-cell technology allows a more in-depth analysis of the effects of their active ingredients at cellular levels. Meanwhile, along with the development of drug response testing technologies, single-cell omics have great potential for large-scale screening of active ingredients in TCMs.
5.2 Targets of action of TCMs
The identification of the targets of action of TCMs is extremely critical to unravelling their therapeutic mechanisms and improving their efficacy and safety. Single-cell studies can help resolve the heterogeneity of different cell types in tissues, trace the signaling pathways affected by TCM components, and discover new biomarkers and therapeutic targets.
A study on the treatment of non-small cell lung cancer with Jin-Fu-An Decoction (JFAD) showed (Tang Y. et al., 2023) that JFAD can promote the transformation of anti-inflammatory M2 macrophages to pro-inflammatory M1 macrophages, enhance the anti-tumor immune response in the tumor microenvironment, through downregulating the β-catenin signaling pathway. Network pharmacology analysis further showed that the targets of JFAD involved 53 compounds and 263 target proteins, among which β-catenin was identified as a key target.
Celastrol, a pentacyclic triterpenoid compound derived from the roots of the celastrol plant, has shown potential therapeutic effects in animal models for a variety of inflammatory diseases such as rheumatoid arthritis. scRNA-seq revealed that celastrol inhibits B-cell migration and humoral immune responses by disrupting the COMMD3/8 complex, thereby halting the progression of rheumatoid arthritis (Shirai et al., 2023). This discovery highlights the COMMD3/8 complex as a potential drug target for the treatment of autoimmune diseases.
Using single-cell multi-omics technology, researchers identified the HMOX1 gene as a key target for the TCMs Sini decoction (SND) in the treatment of sepsis (Gu et al., 2024). SND contains 116 active ingredients, among which quercetin and kaempferol act on the HMOX1 gene through related pathways and play a key role in the prognosis of sepsis treatment. In addition, SND may treat sepsis by regulating the interaction between immune cells through multiple mechanisms. These results provide new targets for the precision treatment of sepsis and help develop drugs for severe infections, demonstrating the potential of combining TCMs with modern multi-omics methods.
5.3 Elucidation of pharmacological mechanisms of TCMs
An important issue in the development of TCM is the elucidation of their pharmacological mechanisms. In the past decades, tissue sequencing technology and protein structure analysis have been applied to research TCM mechanisms, such as artemisinin (Meshnick, 2002; Nkereuwem et al., 2013) periwinkle alkaloids (Gigant et al., 2005) and quercetin (Li et al., 2016). In recent years, single-cell technology is leading the elucidation of TCMs mechanisms of action from a new perspective and a cellular resolution level.
Acute promyelocytic leukemia (APL) is one of the most aggressive types of leukemia. Compound Andrographis Natural Ganoderma Lucidum Tablets is an effective oral arsenic agent for the treatment of APL. Using single-cell technology to analyze the mouse APL model, it was found that the combination of arsenic trioxide, tanshinone IIA, and indocyanine (ATI) had the strongest therapeutic effect (Zhang X. et al., 2022). ATI improved normal hematopoiesis in leukemic mice by regulating the expression of related genes in bone marrow mesenchymal stromal cells (BMSCs). This study clarifies the potential mechanism of ATI regulating BMSCs from the perspective of the overall hematopoietic microenvironment and broadens the understanding of ATI compatibility in BMSCs.
Tanshinone is a commonly used drug for the treatment of myocardial infarction and has antioxidant, anti-inflammatory and inhibitory effects on myocardial fibrosis (Li Y. et al., 2018; Xu and Liu, 2013; Sun and Tang, 2014). Recent studies have identified macrophage subpopulations with potential therapeutic targets through scRNA-seq of tanshinone IIA-treated mouse hearts. Tanshinone IIA significantly reduced the proportion of certain key macrophage subpopulations, suggesting the possibility of attenuating myocardial infarction by modulating the activity of these cells.
Cycloastragenol (CAG) is an active ingredient extracted from astragalus, which has the effects of inhibiting tumor cell proliferation, inducing apoptosis, inhibiting invasion and metastasis, and anti-inflammatory effects (Li et al., 2017; Wang et al., 2019). Single-cell technology showed that CAG enhanced the killing ability of CD8+T cells by inhibiting histone B (CTSB)-mediated MHC-I degradation (Deng et al., 2022) and could also improve immune cell function. These findings provide new potential targets for the development of immune cell targeted therapeutic measures.
5.4 Prospects for the future of TCMs
TCMs treatment emphasizes the consideration of individual differences and holistic treatment. Single-cell technology can reveal the response of TCMs at the cellular level, enabling researchers to understand the specific response of various types of cells to the components of TCMs, thus promoting the realization of personalized treatment plans (Figure 5). In addition, single-cell technology can also monitor the effects of TCM components on single cells or specific cell types, which can help to predict and assess the potential toxicity and side effects, thus improving the safety of the treatment. Finally, single-cell technology provides a new method for screening and verifying the active ingredients and their mechanisms of action in TCM, accelerating the discovery and development of new drugs. Taken together, single-cell technology shows a broad application prospect in the treatment of diseases in TCM, which is expected to greatly promote the modernization and internationalization of TCM (Jia-yun et al., 2021; Yang et al., 2022).
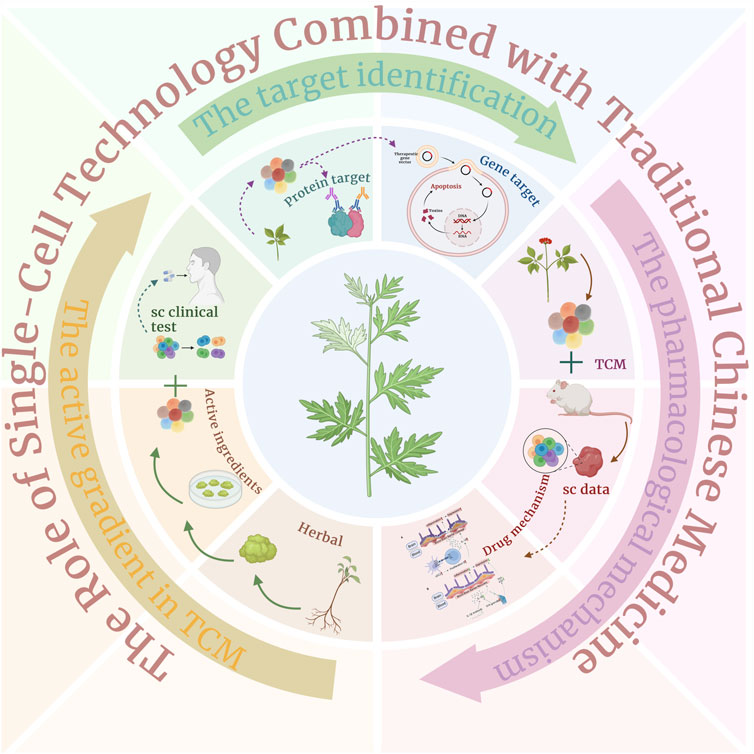
Figure 5. Single-cell technology combined with traditional Chinese medicine for drug development, detailed introduction of three processes: active ingredients of traditional Chinese medicine, prediction of traditional Chinese medicine targets, and research on the mechanism of traditional Chinese medicine.
6 Discussion
Single-cell technology has brought significant advantages to drug development. It can reveal cell subpopulations and molecular mechanisms that are difficult to discover using traditional methods. Considering each step of drug discovery, it can optimize drug design, improve therapeutic effects and reduce side effects. Single-cell technology helps to identify specific disease-causing cells and molecular targets in clinical drug development, thereby developing precise targeted therapies. Furthermore, combined with advanced machine learning and deep learning methods, utilizing valuable information from large amounts of single-cell data can accelerate the identification of drug targets, screening of drug candidates, and drug repurposing processes. In the field of TCMs, single-cell technology is revolutionizing people’s understanding, from the screening of active ingredients to the identification of drug targets and the pharmacological mechanisms of TCMs.
One of the development trends of single-cell technology is the continuous improvement of resolution (Zeng H. et al., 2023; Garrido-Trigo et al., 2023). With the advancement of sequencing technology and data processing capabilities, we are now able to capture changes in gene expression in single cells with higher precision. For example, the latest scRNA-seq technology can analyze gene expression at the subcellular level and reveal the complex dynamic processes inside cells. This high-resolution data helps us to identify the specific responses of cells under specific pathological conditions, thereby discovering potential therapeutic targets (Gottschlich et al., 2023; Lambo et al., 2023). Secondly, the development of multi-omics analysis has greatly expanded our understanding of cell functions and interactions. Single-cell multi-omics technology combines multiple omics data such as genomics, transcriptomics, epigenomics and proteomics, allowing us to comprehensively analyze the information of single cells at different levels. For example, by simultaneously analyzing gene expression and epigenetic modifications of single cells, we can better understand gene regulatory mechanisms and their role in diseases (Mathys et al., 2023; Vandereyken et al., 2023; Gao et al., 2023). This integrated data not only reveals the complexity of cell function, but also provides clues for new therapeutic strategies. Single-cell spatial transcriptome technology is another important development trend. This technology combines scRNA-seq and spatial information capture technology to analyze gene expression patterns in three-dimensional space. This means that we can perform gene expression analysis at cellular resolution while preserving the spatial structure of the tissue. The latest spatial transcriptomic technologies, such as 10x Genomics’ Visium, slide-seq, and Stereo-seq, have been able to achieve subcellular resolution. The development of these technologies has enabled us to construct a three-dimensional gene expression panorama of complex organs, such as the brain (Shi et al., 2023) and embryo (Bergmann et al., 2022; Lu et al., 2023). This panorama reveals differences in gene expression in different regions within the organ, providing us with a new perspective to precision medicine and drug development.
Single-cell technology, as a high-resolution biological analysis tool, has made significant strides in cancer research in recent years, particularly in elucidating tumor heterogeneity and treatment resistance (Zhang et al., 2024; Wu et al., 2023; Randic et al., 2023; Shen et al., 2023). However, its application in chronic disease therapies is relatively nascent yet promising. Single-cell studies in chronic disease fields primarily focus on understanding disease mechanisms, identifying novel therapeutic targets, and optimizing personalized treatment strategies. For instance, neurological disorders (Roostaei et al., 2017; Hammond et al., 2019; Mathys et al., 2019; Mathys et al., 2017), immune-mediated diseases (Zhang et al., 2019; Cheng et al., 2021; Cheung et al., 2019), and infectious diseases (Zhao et al., 2021; Wilk et al., 2020; Xu et al., 2020) all stand to benefit from the application of single-cell technology. Despite the considerable success of single-cell technology in cancer research, it faces challenges and research gaps in chronic disease fields. Chronic disease samples are typically more complex and heterogeneous, necessitating higher standards of technical standardization and cost-effective solutions. Compared to cancer, chronic diseases exhibit greater cell types and state diversity, thus complicating data interpretation and pattern recognition. With further advancements and expanded applications, single-cell technology is expected to play a critical role in future drug development for chronic diseases.
In the field of TCMs, drug development research combined with single-cell technology has significant advantages. Single-cell technology can provide high-resolution data, reveal the specific mechanism of action of TCMs at the cellular level, and help to identify and verify the therapeutic targets of active ingredients of TCMs (Gu et al., 2024; Zhang X. et al., 2022). However, although single-cell technology has shown great potential in TCMs research, its application is still in its early stages. More research is needed to verify the effectiveness of single-cell technology in revealing the mechanisms and therapeutic effects of active ingredients in TCMs, and to ensure its reliability and feasibility in practical applications. In future studies, the combined use of TCMs with chemotherapy drugs and targeted drugs has great potential.
At last, the combination of single-cell technology with computational methods and drug repurposing methods is revolutionizing this field. However, how to effectively integrate and interpret rich data, especially when combining single-cell data with deep learning and machine learning models, the quality of data and the optimization of algorithms still need further research. In terms of drug perturbation prediction, the two deep learning methods introduced in this article (Lotfollahi et al., 2019; Theodoris et al., 2023) require a lot of computing resources and high-quality data to achieve optimal performance. The same problem also exists in the integration of batch single-cell data and drug repurposing methods. Secondly, the complexity of the computational model and the training process require rich expertise, which may limit its widespread application. In addition, disease progression and drug response are dynamic processes (Barrett et al., 2022; Tang et al., 2021), so it is necessary to consider time series single-cell data, and how to integrate these dynamic changes into the model remains a challenge. Finally, although single-cell technology can reveal the heterogeneity between cells, the mechanisms of complex biological systems are extremely complex, and a single computational model may not be able to fully capture them, thus affecting the prediction accuracy of drug repurposing.
Author contributions
AZ: Writing–original draft. JZ: Writing–original draft. YX: Writing–original draft. LG: Writing–original draft. FD: Writing–original draft. YL: Writing–original draft. PG: Writing–original draft. HT: Writing–review and editing. LT: Writing–review and editing. XZ: Writing–review and editing, Writing–original draft. JH: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82170045 to JH); the Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZLCX20212301to JH); the Macao Polytechnic University Internal Research Grant (RP/FCSD-02/2022).
Acknowledgments
We have used the GPT, version GPT4-O sourced from OpenAI to polish our manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adam, G., Rampasek, L., Safikhani, Z., Smirnov, P., Haibe-Kains, B., and Goldenberg, A. (2020). Machine learning approaches to drug response prediction: challenges and recent progress. NPJ Precis. Oncol. 4, 19. doi:10.1038/s41698-020-0122-1
Aissa, A. F., Islam, A., Ariss, M. M., Go, C. C., Rader, A. E., Conrardy, R. D., et al. (2021). Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 12 (1), 1628. doi:10.1038/s41467-021-21884-z
Alivernini, S., MacDonald, L., Elmesmari, A., Finlay, S., Tolusso, B., Gigante, M. R., et al. (2020). Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat. Med. 26 (8), 1295–1306. doi:10.1038/s41591-020-0939-8
Barrett, J. S., Nicholas, T., Azer, K., and Corrigan, B. W. (2022). Role of disease progression models in drug development. Pharm. Res. 39 (8), 1803–1815. doi:10.1007/s11095-022-03257-3
Bassez, A., Vos, H., Van Dyck, L., Floris, G., Arijs, I., Desmedt, C., et al. (2021). A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27 (5), 820–832. doi:10.1038/s41591-021-01323-8
Bergmann, S., Penfold, C. A., Slatery, E., Siriwardena, D., Drummer, C., Clark, S., et al. (2022). Spatial profiling of early primate gastrulation in utero. Nature 609 (7925), 136–143. doi:10.1038/s41586-022-04953-1
Bradley, T. F. G., Haynes, B. F., Margolis, D. M., and Browne, E. P. (2018). Single-cell analysis of quiescent hiv infection reveals host transcriptional profiles that regulate proviral latency. Cell Rep. 25 (1), 107–117.e3. doi:10.1016/j.celrep.2018.09.020
Bryant, C. E. S. S., Kong, B., Papadimitrious, M. S., Fromm, P. D., Hart, D. N. J., Bryant, C. E., et al. (2019). Dendritic cells as cancer therapeutics. Seminars cell and Dev. Biol. 86, 77–88. doi:10.1016/j.semcdb.2018.02.015
Buenrostro, J. D., Wu, B., Chang, H. Y., and Greenleaf, W. J. (2015a). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109 (21), 9 1–21. doi:10.1002/0471142727.mb2129s109
Buenrostro, J. D., Wu, B., Litzenburger, U. M., Ruff, D., Gonzales, M. L., Snyder, M. P., et al. (2015b). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523 (7561), 486–490. doi:10.1038/nature14590
Chen, A., Liao, S., Cheng, M., Ma, K., Wu, L., Lai, Y., et al. (2022b). Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185 (10), 1777–1792.e21. doi:10.1016/j.cell.2022.04.003
Chen, J., Gao, P., Xiao, W., Cheng, G., Krishna, S., Wang, J., et al. (2023b). Multi-omics dissection of stage-specific artemisinin tolerance mechanisms in Kelch13-mutant Plasmodium falciparum. Drug Resist Updat 70, 100978. doi:10.1016/j.drup.2023.100978
Chen, J., Wang, X., Ma, A., Wang, Q. E., Liu, B., Li, L., et al. (2022a). Deep transfer learning of cancer drug responses by integrating bulk and single-cell RNA-seq data. Nat. Commun. 13 (1), 6494. doi:10.1038/s41467-022-34277-7
Chen, Y., Li, X., Liu, S., Ao, W., Lin, J., Li, Z., et al. (2023a). An atlas of immune cell transcriptomes in human immunodeficiency virus-infected immunological non-responders identified marker genes that control viral replication. Chin. Med. J. Engl. 136 (22), 2694–2705. doi:10.1097/CM9.0000000000002918
Cheng, L., Wang, Y., Wu, R., Ding, T., Xue, H., Gao, C., et al. (2021). New insights from single-cell sequencing data: synovial fibroblasts and synovial macrophages in rheumatoid arthritis. Front. Immunol. 12, 709178. doi:10.3389/fimmu.2021.709178
Cheung, P., Khatri, P., Utz, P. J., and Kuo, A. J. (2019). Single-cell technologies - studying rheumatic diseases one cell at a time. Nat. Rev. Rheumatol. 15 (6), 340–354. doi:10.1038/s41584-019-0220-z
Cohen, Y. C., Zada, M., Wang, S. Y., Bentur, O. S., Chubar, E., Cohen, A., et al. (2021). Single cell RNA sequencing in patients enrolled in a selinexor clinical trial reveals overexpression of alternative nuclear export pathways associated with resistance to selinexor in refractory multiple myeloma. Blood 138, 2725. doi:10.1182/blood-2021-149701
Cuni-Lopez, C., Stewart, R., Oikari, L. E., Nguyen, T. H., Roberts, T. L., Sun, Y., et al. (2024). Advanced patient-specific microglia cell models for pre-clinical studies in Alzheimer's disease. J. Neuroinflammation 21 (1), 50. doi:10.1186/s12974-024-03037-3
Dagogo-Jack, I., and Shaw, A. T. (2018). Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15 (2), 81–94. doi:10.1038/nrclinonc.2017.166
Dang, Y., He, Q., Yang, S., Sun, H., Liu, Y., Li, W., et al. (2022). FTH1- and SAT1-induced astrocytic ferroptosis is involved in Alzheimer's disease: evidence from single-cell transcriptomic analysis. Pharm. (Basel) 15 (10), 1177. doi:10.3390/ph15101177
Deng, G., Zhou, L., Wang, B., Sun, X., Zhang, Q., Chen, H., et al. (2022). Targeting cathepsin B by cycloastragenol enhances antitumor immunity of CD8 T cells via inhibiting MHC-I degradation. J. Immunother. Cancer 10 (10), e004874. doi:10.1136/jitc-2022-004874
De Lima, J., Boutet, M-A., Bortolotti, O., Chépeaux, L-A., Glasson, Y., Dumé, A-S., et al. (2023). Spatial mapping of rheumatoid arthritis synovial niches reveals specific macrophage networks associated with response to therapy. bioRxiv Prepr. Serv. Biol. doi:10.1101/2023.10.20.563040
Der, E., Ranabothu, S., Suryawanshi, H., Akat, K. M., Clancy, R., Morozov, P., et al. (2017). Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2 (9), e93009. doi:10.1172/jci.insight.93009
Dolan, M. J., Therrien, M., Jereb, S., Kamath, T., Gazestani, V., Atkeson, T., et al. (2023). Exposure of iPSC-derived human microglia to brain substrates enables the generation and manipulation of diverse transcriptional states in vitro. Nat. Immunol. 24 (8), 1382–1390. doi:10.1038/s41590-023-01558-2
Durante, M. A., Rodriguez, D. A., Kurtenbach, S., Kuznetsov, J. N., Sanchez, M. I., Decatur, C. L., et al. (2020). Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 11 (1), 496. doi:10.1038/s41467-019-14256-1
Edikpo, N., Ghasi, S., Elias, A., and Oguanobi, N. (2013). Artemisinin and biomolecules: the continuing search for mechanism of action. Mol. Cell Pharmacol. 5 (2), 75–89. doi:10.4255/mcpharmacol.13.09
Eggenhuizen, P. J., Cheong, R. M. Y., Lo, C., Chang, J., Ng, B. H., Ting, Y. T., et al. (2024). Smith-specific regulatory T cells halt the progression of lupus nephritis. Nat. Commun. 15 (1), 899. doi:10.1038/s41467-024-45056-x
El Atat, O., Naser, R., Abdelkhalek, M., Habib, R. A., and El Sibai, M. (2023). Molecular targeted therapy: a new avenue in glioblastoma treatment. Oncol. Lett. 25 (2), 46. doi:10.3892/ol.2022.13632
Fan, H., Li, J., Manuel, A. M., and Zhao, Z. (2023). Enzalutamide-induced signatures revealed by epigenetic plasticity using single-cell multi-omics sequencing in prostate cancer. Mol. Ther. Nucleic Acids 31, 648–661. doi:10.1016/j.omtn.2023.02.022
Fan, X., Yang, C., Li, W., Bai, X., Zhou, X., Xie, H., et al. (2021). SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 22 (1), 195. doi:10.1186/s13059-021-02406-y
Fernandes, H. J. R., Patikas, N., Foskolou, S., Field, S. F., Park, J. E., Byrne, M. L., et al. (2020). Single-cell transcriptomics of Parkinson's disease human in vitro models reveals dopamine neuron-specific stress responses. Cell Rep. 33 (2), 108263. doi:10.1016/j.celrep.2020.108263
Galea, E., Weinstock, L. D., Larramona-Arcas, R., Pybus, A. F., Giménez-Llort, L., Escartin, C., et al. (2022). Multi-transcriptomic analysis points to early organelle dysfunction in human astrocytes in Alzheimer's disease. Neurobiol. Dis. 166, 105655. doi:10.1016/j.nbd.2022.105655
Galli, E., Hartmann, F. J., Schreiner, B., Ingelfinger, F., Arvaniti, E., Diebold, M., et al. (2019). GM-CSF and CXCR4 define a T helper cell signature in multiple sclerosis. Nat. Med. 25 (8), 1290–1300. doi:10.1038/s41591-019-0521-4
Gambardella, G., Viscido, G., Tumaini, B., Isacchi, A., Bosotti, R., and di Bernardo, D. (2022). A single-cell analysis of breast cancer cell lines to study tumour heterogeneity and drug response. Nat. Commun. 13 (1), 1714. doi:10.1038/s41467-022-29358-6
Gao, Y., Chi, Y., Chen, Y., Wang, W., Li, H., Zheng, W., et al. (2023). Multi-omics analysis of human mesenchymal stem cells shows cell aging that alters immunomodulatory activity through the downregulation of PD-L1. Nat. Commun. 14 (1), 4373. doi:10.1038/s41467-023-39958-5
Garrido-Trigo, A., Corraliza, A. M., Veny, M., Dotti, I., Melon-Ardanaz, E., Rill, A., et al. (2023). Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease. Nat. Commun. 14 (1), 4506. doi:10.1038/s41467-023-40156-6
Gazestani, V., Kamath, T., Nadaf, N. M., Burris, S. J., Rooney, B., Junkkari, A., et al. (2023). Early Alzheimer's disease pathology in human cortex is associated with a transient phase of distinct cell states. bioRxiv. doi:10.1101/2023.06.03.543569
Ge, L. P., Jin, X., Ma, D., Wang, Z. Y., Liu, C. L., Zhou, C. Z., et al. (2024). ZNF689 deficiency promotes intratumor heterogeneity and immunotherapy resistance in triple-negative breast cancer. Cell Res. 34 (1), 58–75. doi:10.1038/s41422-023-00909-w
Gierahn, T. M., Wadsworth, M. H., Hughes, T. K., Bryson, B. D., Butler, A., Satija, R., et al. (2017). Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14 (4), 395–398. doi:10.1038/nmeth.4179
Gigant, B., Wang, C., Ravelli, R. B., Roussi, F., Steinmetz, M. O., Curmi, P. A., et al. (2005). Structural basis for the regulation of tubulin by vinblastine. Nature 435 (7041), 519–522. doi:10.1038/nature03566
Goto-Silva, L., and Junqueira, M. (2021). Single-cell proteomics: a treasure trove in neurobiology. Biochim. Biophys. Acta Proteins Proteom 1869 (7), 140658. doi:10.1016/j.bbapap.2021.140658
Gottschlich, A., Thomas, M., Grunmeier, R., Lesch, S., Rohrbacher, L., Igl, V., et al. (2023). Single-cell transcriptomic atlas-guided development of CAR-T cells for the treatment of acute myeloid leukemia. Nat. Biotechnol. 41 (11), 1618–1632. doi:10.1038/s41587-023-01684-0
Gu, Y., Li, Z., Li, H., Yi, X., Liu, X., Zhang, Y., et al. (2024). Exploring the efficacious constituents and underlying mechanisms of sini decoction for sepsis treatment through network pharmacology and multi-omics. Phytomedicine 123, 155212. doi:10.1016/j.phymed.2023.155212
Haage, V., Tuddenham, J. F., Comandante-Lou, N., Bautista, A., Monzel, A., Chiu, R., et al. (2024). A pharmacological toolkit for human microglia identifies Topoisomerase I inhibitors as immunomodulators for Alzheimer's disease. New York, NY: bioRxiv.
Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., et al. (2019). Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50 (1), 253–271. doi:10.1016/j.immuni.2018.11.004
Han, X., Wang, R., Zhou, Y., Fei, L., Sun, H., Lai, S., et al. (2018). Mapping the mouse cell atlas by microwell-seq. Cell 173 (5), 1307. doi:10.1016/j.cell.2018.05.012
Han, Y., Duan, X., Yang, L., Nilsson-Payant, B. E., Wang, P., Duan, F., et al. (2021). Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589 (7841), 270–275. doi:10.1038/s41586-020-2901-9
Han, Y., Yang, L., Lacko, L. A., and Chen, S. (2022). Human organoid models to study SARS-CoV-2 infection. Nat. Methods 19 (4), 418–428. doi:10.1038/s41592-022-01453-y
He, B. X. Y., Liang, H., Huang, Q., Du, Y., Li, Y., Garmire, D., et al. (2023). ASGARD is A Single-cell guided pipeline to aid repurposing of drugs. Nat. Commun. 14 (1), 993. doi:10.1038/s41467-023-36637-3
He, J., Xiong, X., Yang, H., Li, D., Liu, X., Li, S., et al. (2022). Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response. Cell Res. 32 (6), 530–542. doi:10.1038/s41422-022-00627-9
Hedman, A. K., Winter, E., Yoosuf, N., Benita, Y., Berg, L., Brynedal, B., et al. (2023). Peripheral blood cellular dynamics of rheumatoid arthritis treatment informs about efficacy of response to disease modifying drugs. Sci. Rep. 13 (1), 10058. doi:10.1038/s41598-023-36999-0
Hsieh, C. Y., Wen, J. H., Lin, S. M., Tseng, T. Y., Huang, J. H., Huang, H. C., et al. (2023). scDrug: from single-cell RNA-seq to drug response prediction. Comput. Struct. Biotechnol. J. 21, 150–157. doi:10.1016/j.csbj.2022.11.055
Huang, D., Ma, N., Li, X., Gou, Y., Duan, Y., Liu, B., et al. (2023). Advances in single-cell RNA sequencing and its applications in cancer research. J. Hematol. Oncol. 16 (1), 98. doi:10.1186/s13045-023-01494-6
Issa, N. T., Stathias, V., Schurer, S., and Dakshanamurthy, S. (2021). Machine and deep learning approaches for cancer drug repurposing. Semin. Cancer Biol. 68, 132–142. doi:10.1016/j.semcancer.2019.12.011
Jarada, T. N., Rokne, J. G., and Alhajj, R. (2020). A review of computational drug repositioning: strategies, approaches, opportunities, challenges, and directions. J. Cheminform 12 (1), 46. doi:10.1186/s13321-020-00450-7
Jia-yun, CHEN, Cheng-chao, X. U., and Ji-gang, WANG (2021). A new research paradigm in modernization of traditional Chinese medicine: single cell pharmacology. Acta Pharm. Sin. 56 (12), 3300–3312. doi:10.16438/j.0513-4870.2021-1010
Jing, C., Castro-Dopico, T., Richoz, N., Tuong, Z. K., Ferdinand, J. R., Lok, L. S. C., et al. (2020). Macrophage metabolic reprogramming presents a therapeutic target in lupus nephritis. Proc. Natl. Acad. Sci. U. S. A. 117 (26), 15160–15171. doi:10.1073/pnas.2000943117
Kim, M., Min, Y. K., Jang, J., Park, H., Lee, S., and Lee, C. H. (2021). Single-cell RNA sequencing reveals distinct cellular factors for response to immunotherapy targeting CD73 and PD-1 in colorectal cancer. J. Immunother. Cancer 9 (7), e002503. doi:10.1136/jitc-2021-002503
Knoll, R., Bonaguro, L., Dos Santos, J. C., Warnat-Herresthal, S., Jacobs-Cleophas, M. C. P., Blumel, E., et al. (2023). Identification of drug candidates targeting monocyte reprogramming in people living with HIV. Front. Immunol. 14, 1275136. doi:10.3389/fimmu.2023.1275136
Kuo, D., Ding, J., Cohn, I. S., Zhang, F., Wei, K., Rao, D. A., et al. (2019). HBEGF(+) macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci. Transl. Med. 11 (491), eaau8587. doi:10.1126/scitranslmed.aau8587
Lambo, S., Trinh, D. L., Ries, R. E., Jin, D., Setiadi, A., Ng, M., et al. (2023). A longitudinal single-cell atlas of treatment response in pediatric AML. Cancer Cell 41 (12), 2117–2135.e12. doi:10.1016/j.ccell.2023.10.008
Leon-Rivera, R., Morsey, B., Niu, M., Fox, H. S., and Berman, J. W. (2020). Interactions of monocytes, HIV, and ART identified by an innovative scRNAseq pipeline: pathways to reservoirs and HIV-associated comorbidities. mBio 11 (4). doi:10.1128/mBio.01037-20
Li, C., Wood, J. C., Vu, A. H., Hamilton, J. P., Rodriguez Lopez, C. E., Payne, R. M. E., et al. (2023). Single-cell multi-omics in the medicinal plant Catharanthus roseus. Nat. Chem. Biol. 19 (8), 1031–1041. doi:10.1038/s41589-023-01327-0
Li, J., Yang, Y., and Jiang, L. (2017). Cycloastragenol inhibits the proliferation of colorectal cancer cells by regulating telomerase and p53. J. Cancer Res. Clin. Oncol. 143 (1), 11–19.
Li, L., and Wang, H. (2016). Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 379 (2), 191–197. doi:10.1016/j.canlet.2015.07.018
Li, L., Yan, S., Lin, B., Shi, Q., and Lu, Y. (2018a). Single-cell proteomics for cancer immunotherapy. Adv. Cancer Res. 139, 185–207. doi:10.1016/bs.acr.2018.04.006
Li, Y., Gao, S., Guo, Z., Chen, Z., Wei, Y., Li, Y., et al. (2024). Screening of potential drugs for the treatment of diabetic kidney disease using single-cell transcriptome sequencing and connectivity map data. Biochem. Biophysical Res. Commun. 725, 150263. doi:10.1016/j.bbrc.2024.150263
Li, Y., Shen, X., Tang, X., and Cheng, X. (2018b). Antioxidant and cardioprotective effects of tanshinone IIA in myocardial ischemia and reperfusion injury in rats. J. Ethnopharmacol. 224, 217–226.
Li, Y., Yao, J., Han, C., Yang, J., Chaudhry, M. T., Wang, S., et al. (2016). Quercetin, inflammation and immunity. Nutrients 8 (3), 167. doi:10.3390/nu8030167
Liang, Y., Tan, Y., Guan, B., Guo, B., Xia, M., Li, J., et al. (2022). Single-cell atlases link macrophages and CD8(+) T-cell subpopulations to disease progression and immunotherapy response in urothelial carcinoma. Theranostics 12 (18), 7745–7759. doi:10.7150/thno.77281
Lin, J., Xue, X., Wang, Y., Zhou, Y., Wu, J., Xie, H., et al. (2023). scNanoCOOL-seq: a long-read single-cell sequencing method for multi-omics profiling within individual cells. Cell Res. 33 (11), 879–882. doi:10.1038/s41422-023-00873-5
Lindeman, S. D., Mukkamala, R., Horner, A., Tudi, P., Booth, O. C., Huff, R., et al. (2023). Fibroblast activation protein-targeted radioligand therapy for treatment of solid tumors. J. Nucl. Med. 64 (5), 759–766. doi:10.2967/jnumed.122.264494
Liu, T., Bai, M., Liu, M., Li, T., Liao, Y., Zhao, C., et al. (2023). Novel synergistic mechanism of 11-keto-β-boswellic acid and Z-Guggulsterone on ischemic stroke revealed by single-cell transcriptomics. Pharmacol. Res. 193, 106803. doi:10.1016/j.phrs.2023.106803
Liu, W., Venugopal, S., Majid, S., Ahn, I. S., Diamante, G., Hong, J., et al. (2020). Single-cell RNA-seq analysis of the brainstem of mutant SOD1 mice reveals perturbed cell types and pathways of amyotrophic lateral sclerosis. Neurobiol. Dis. 141, 104877. doi:10.1016/j.nbd.2020.104877
Lotfollahi, M., Wolf, F. A., and Theis, F. J. (2019). scGen predicts single-cell perturbation responses. Nat. Methods 16 (8), 715–721. doi:10.1038/s41592-019-0494-8
Lu, T., Ang, C. E., and Zhuang, X. (2023). Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 186 (10), 2275–2279. doi:10.1016/j.cell.2023.04.006
Ma, L., Wang, L., Khatib, S. A., Chang, C. W., Heinrich, S., Dominguez, D. A., et al. (2021). Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 75 (6), 1397–1408. doi:10.1016/j.jhep.2021.06.028
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161 (5), 1202–1214. doi:10.1016/j.cell.2015.05.002
Maeser, D., Gruener, R. F., and Huang, R. S. (2021). oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform 22 (6), bbab260. doi:10.1093/bib/bbab260
Maeser, D., Zhang, W., Huang, Y., and Huang, R. S. (2024). A review of computational methods for predicting cancer drug response at the single-cell level through integration with bulk RNAseq data. Curr. Opin. Struct. Biol. 84, 102745. doi:10.1016/j.sbi.2023.102745
Mathys, H., Adaikkan, C., Gao, F., Young, J. Z., Manet, E., Hemberg, M., et al. (2017). Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21 (2), 366–380. doi:10.1016/j.celrep.2017.09.039
Mathys, H., Davila-Velderrain, J., Peng, Z., Gao, F., Mohammadi, S., Young, J. Z., et al. (2019). Single-cell transcriptomic analysis of Alzheimer's disease. Nature 570 (7761), 332–337. doi:10.1038/s41586-019-1195-2
Mathys, H., Peng, Z., Boix, C. A., Victor, M. B., Leary, N., Babu, S., et al. (2023). Single-cell atlas reveals correlates of high cognitive function, dementia, and resilience to Alzheimer's disease pathology. Cell 186 (20), 4365–4385.e27. doi:10.1016/j.cell.2023.08.039
Meijer, J. J., Leonetti, A., Airo, G., Tiseo, M., Rolfo, C., Giovannetti, E., et al. (2022). Small cell lung cancer: novel treatments beyond immunotherapy. Semin. Cancer Biol. 86 (Pt 2), 376–385. doi:10.1016/j.semcancer.2022.05.004
Meshnick, S. R. (2002). Artemisinin: mechanisms of action, resistance and toxicity. Int. J. Parasitol. 32 (13), 1655–1660. doi:10.1016/s0020-7519(02)00194-7
Min, Y., Wang, X., Is, O., Patel, T. A., Gao, J., Reddy, J. S., et al. (2023). Cross species systems biology discovers glial DDR2, STOM, and KANK2 as therapeutic targets in progressive supranuclear palsy. Nat. Commun. 14 (1), 6801. doi:10.1038/s41467-023-42626-3
Mitra-Kaushik, S., Mehta-Damani, A., Stewart, J. J., Green, C., Litwin, V., and Gonneau, C. (2021). The evolution of single-cell analysis and utility in drug development. AAPS J. 23 (5), 98. doi:10.1208/s12248-021-00633-6
Mund, A., Coscia, F., Kriston, A., Hollandi, R., Kovacs, F., Brunner, A. D., et al. (2022). Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 40 (8), 1231–1240. doi:10.1038/s41587-022-01302-5
Musa, A., Tripathi, S., Dehmer, M., and Emmert-Streib, F. (2019). L1000 viewer: a search engine and web interface for the LINCS data repository. Front. Genet. 10, 557. doi:10.3389/fgene.2019.00557
Nagano, T., Lubling, Y., Stevens, T. J., Schoenfelder, S., Yaffe, E., Dean, W., et al. (2013). Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502 (7469), 59–64. doi:10.1038/nature12593
Nassar, S. F., Raddassi, K., and Wu, T. (2021). Single-cell multiomics analysis for drug discovery. Metabolites 11 (11), 729. doi:10.3390/metabo11110729
Navin, N., Kendall, J., Troge, J., Andrews, P., Rodgers, L., McIndoo, J., et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472 (7341), 90–94. doi:10.1038/nature09807
Nehar-Belaid, D., Hong, S., Marches, R., Chen, G., Bolisetty, M., Baisch, J., et al. (2020). Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat. Immunol. 21 (9), 1094–1106. doi:10.1038/s41590-020-0743-0
Ostkamp, P., Deffner, M., Schulte-Mecklenbeck, A., Wunsch, C., Lu, I. N., Wu, G. F., et al. (2022). A single-cell analysis framework allows for characterization of CSF leukocytes and their tissue of origin in multiple sclerosis. Sci. Transl. Med. 14 (673), eadc9778. doi:10.1126/scitranslmed.adc9778
Oubounyt, M., Adlung, L., Patroni, F., Wenke, N. K., Maier, A., Hartung, M., et al. (2023). Inference of differential key regulatory networks and mechanistic drug repurposing candidates from scRNA-seq data with SCANet. Bioinformatics 39 (11). doi:10.1093/bioinformatics/btad644
Panche, A. N., Diwan, A. D., and Chandra, S. R. (2016). Flavonoids: an overview. J. Nutr. Sci. 5, e47. doi:10.1017/jns.2016.41
Partin, A., Brettin, T. S., Zhu, Y., Narykov, O., Clyde, A., Overbeek, J., et al. (2023). Deep learning methods for drug response prediction in cancer: predominant and emerging trends. Front. Med. (Lausanne) 10, 1086097. doi:10.3389/fmed.2023.1086097
Parvathaneni, V., Kulkarni, N. S., Muth, A., and Gupta, V. (2019). Drug repurposing: a promising tool to accelerate the drug discovery process. Drug Discov. Today 24 (10), 2076–2085. doi:10.1016/j.drudis.2019.06.014
Picelli, S., Faridani, O. R., Bjorklund, A. K., Winberg, G., Sagasser, S., and Sandberg, R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9 (1), 171–181. doi:10.1038/nprot.2014.006
Pushparaj, P. N., Kalamegam, G., Wali Sait, K. H., and Rasool, M. (2021). Decoding the role of astrocytes in the entorhinal cortex in Alzheimer's disease using high-dimensional single-nucleus RNA sequencing data and next-generation knowledge discovery methodologies: focus on drugs and natural product remedies for dementia. Front. Pharmacol. 12, 720170. doi:10.3389/fphar.2021.720170
Qu, J., Yang, F., Zhu, T., Wang, Y., Fang, W., Ding, Y., et al. (2022). A reference single-cell regulomic and transcriptomic map of cynomolgus monkeys. Nat. Commun. 13 (1), 4069. doi:10.1038/s41467-022-31770-x
Randic, T., Magni, S., Philippidou, D., Margue, C., Grzyb, K., Preis, J. R., et al. (2023). Single-cell transcriptomics of NRAS-mutated melanoma transitioning to drug resistance reveals P2RX7 as an indicator of early drug response. Cell Rep. 42 (7), 112696. doi:10.1016/j.celrep.2023.112696
Rao, A. V. S. M. K., and Sung, M. K. (1995). Saponins as anticarcinogens. J. Nutr. 125, 717S-724S–24. doi:10.1093/jn/125.3_Suppl.717S
Rodriques, S. G., Stickels, R. R., Goeva, A., Martin, C. A., Murray, E., Vanderburg, C. R., et al. (2019). Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363 (6434), 1463–1467. doi:10.1126/science.aaw1219
Roessler, H. I., Knoers, N., van Haelst, M. M., and van Haaften, G. (2021). Drug repurposing for rare diseases. Trends Pharmacol. Sci. 42 (4), 255–267. doi:10.1016/j.tips.2021.01.003
Ronnblom, L., and Leonard, D. (2019). Interferon pathway in SLE: one key to unlocking the mystery of the disease. Lupus Sci. Med. 6 (1), e000270. doi:10.1136/lupus-2018-000270
Roostaei, T., Nazeri, A., Felsky, D., De Jager, P. L., Schneider, J. A., Pollock, B. G., et al. (2017). Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer's disease. Mol. Psychiatry 22 (2), 287–295. doi:10.1038/mp.2016.35
Rotem, A., Ram, O., Shoresh, N., Sperling, R. A., Goren, A., Weitz, D. A., et al. (2015). Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33 (11), 1165–1172. doi:10.1038/nbt.3383
Ruan, Q., Ruan, W., Lin, X., Wang, Y., Zou, F., Zhou, L., et al. (2020). Digital-WGS: automated, highly efficient whole-genome sequencing of single cells by digital microfluidics. Sci. Adv. 6 (50), eabd6454. doi:10.1126/sciadv.abd6454
Ruiz-Cordero, R., and Devine, W. P. (2020). Targeted therapy and checkpoint immunotherapy in lung cancer. Surg. Pathol. Clin. 13 (1), 17–33. doi:10.1016/j.path.2019.11.002
Salcher, S., Sturm, G., Horvath, L., Untergasser, G., Kuempers, C., Fotakis, G., et al. (2022). High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell 40 (12), 1503–1520.e8. doi:10.1016/j.ccell.2022.10.008
Sarkar, C., Das, B., Rawat, V. S., Wahlang, J. B., Nongpiur, A., Tiewsoh, I., et al. (2023). Artificial intelligence and machine learning technology driven modern drug discovery and development. Int. J. Mol. Sci. 24 (3), 2026. doi:10.3390/ijms24032026
Saviano, A., Henderson, N. C., and Baumert, T. F. (2020). Single-cell genomics and spatial transcriptomics: discovery of novel cell states and cellular interactions in liver physiology and disease biology. J. Hepatol. 73 (5), 1219–1230. doi:10.1016/j.jhep.2020.06.004
Saxena, M., and Bhardwaj, N. (2017). Turbocharging vaccines: emerging adjuvants for dendritic cell based therapeutic cancer vaccines. Curr. Opin. Immunol. 47, 35–43. doi:10.1016/j.coi.2017.06.003
Schafer, S., Smelik, M., Sysoev, O., Zhao, Y., Eklund, D., Lilja, S., et al. (2024). scDrugPrio: a framework for the analysis of single-cell transcriptomics to address multiple problems in precision medicine in immune-mediated inflammatory diseases. Genome Med. 16 (1), 42. doi:10.1186/s13073-024-01314-7
Seyedsadr, M., Wang, Y., Elzoheiry, M., Shree Gopal, S., Jang, S., Duran, G., et al. (2023). IL-11 induces NLRP3 inflammasome activation in monocytes and inflammatory cell migration to the central nervous system. Proc. Natl. Acad. Sci. U. S. A. 120 (26), e2221007120. doi:10.1073/pnas.2221007120
Shah, S., Takei, Y., Zhou, W., Lubeck, E., Yun, J., Eng, C. L., et al. (2018). Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell. 174 (2), 363–376. doi:10.1016/j.cell.2018.05.035
Shaker, B., Ahmad, S., Lee, J., Jung, C., and Na, D. (2021). In silico methods and tools for drug discovery. Comput. Biol. Med. 137, 104851. doi:10.1016/j.compbiomed.2021.104851
Shang, L., Wang, Y., Li, J., Zhou, F., Xiao, K., Liu, Y., et al. (2023). Mechanism of Sijunzi Decoction in the treatment of colorectal cancer based on network pharmacology and experimental validation. J. Ethnopharmacol. 302 (Pt A), 115876. doi:10.1016/j.jep.2022.115876
Shaojie, Q. D. M., Zhang, X., Zhang, Yi, and Bai, Yu (2023). Methods developments of mass spectrometry based single cell metabolomics. TrAC Trends Anal. Chem. 164, 117086. doi:10.1016/j.trac.2023.117086
Shen, K., Chen, B., and Gao, W. (2023). Integrated single-cell RNA sequencing analysis reveals a mesenchymal stem cell-associated signature for estimating prognosis and drug sensitivity in gastric cancer. J. cancer Res. Clin. Oncol. 149 (13), 11829–11847. doi:10.1007/s00432-023-05058-6
Shen, K., Reichelt, M., Kyauk, R. V., Ngu, H., Shen, Y. A., Foreman, O., et al. (2021). Multiple sclerosis risk gene Mertk is required for microglial activation and subsequent remyelination. Cell Rep. 34 (10), 108835. doi:10.1016/j.celrep.2021.108835
Shi, H., He, Y., Zhou, Y., Huang, J., Maher, K., Wang, B., et al. (2023). Spatial atlas of the mouse central nervous system at molecular resolution. Nature 622 (7983), 552–561. doi:10.1038/s41586-023-06569-5
Shirai, T., Nakai, A., Ando, E., Fujimoto, J., Leach, S., Arimori, T., et al. (2023). Celastrol suppresses humoral immune responses and autoimmunity by targeting the COMMD3/8 complex. Sci. Immunol. 8 (81), eadc9324. doi:10.1126/sciimmunol.adc9324
Singh, D. K., Aladyeva, E., Das, S., Singh, B., Esaulova, E., Swain, A., et al. (2022). Myeloid cell interferon responses correlate with clearance of SARS-CoV-2. Nat. Commun. 13 (1), 679. doi:10.1038/s41467-022-28315-7
Skinner, D. D., Syage, A. R., Olivarria, G. M., Stone, C., Hoglin, B., and Lane, T. E. (2022). Sustained infiltration of neutrophils into the CNS results in increased demyelination in a viral-induced model of multiple sclerosis. Front. Immunol. 13, 931388. doi:10.3389/fimmu.2022.931388
Son, Y., Korenfeld, D., Harvey, B., Wang, J., Suarez-Fueyo, A., Chang, D., et al. (2023). Pos1025 combination of bulk and single cell rnaseq analyses to reveal mechanisms of abt-317 (jak inhibitor) on synovial fibroblasts. United Kingdom: BMJ Publishing Group Ltd.
Stoeckius, M., Hafemeister, C., Stephenson, W., Houck-Loomis, B., Chattopadhyay, P. K., Swerdlow, H., et al. (2017). Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14 (9), 865–868. doi:10.1038/nmeth.4380
Su, Y., Chen, D., Yuan, D., Lausted, C., Choi, J., Dai, C. L., et al. (2020). Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 183 (6), 1479–1495.e20. doi:10.1016/j.cell.2020.10.037
Sun, H. X., Xie, Y., and Ye, Y. P. (2009). Advances in saponin-based adjuvants. Vaccine 27 (12), 1787–1796. doi:10.1016/j.vaccine.2009.01.091
Sun, K., Xu, R., Ma, F., Yang, N., Li, Y., Sun, X., et al. (2022). scRNA-seq of gastric tumor shows complex intercellular interaction with an alternative T cell exhaustion trajectory. Nat. Commun. 13 (1), 4943. doi:10.1038/s41467-022-32627-z
Sun, Y., and Tang, Z. (2014). Tanshinone IIA inhibits cardiac remodeling induced by acute myocardial infarction through TGF-β1/Smad3 pathway inhibition. J. Pharmacol. Sci. 126 (4), 350–358.
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6 (5), 377–382. doi:10.1038/nmeth.1315
Tang, Y., Sun, Z., Wu, S., Zhang, C., Zhang, Y., and Cao, Y. (2023b). Jin-Fu-An decoction manipulation of macrophage polarization via β-catenin (CTNNB1) synergizes with cisplatin in lung cancer. Biomed. Pharmacother. 168, 115828. doi:10.1016/j.biopha.2023.115828
Tang, Z., Liu, X., Li, Z., Zhang, T., Yang, B., Su, J., et al. (2023a). SpaRx: elucidate single-cell spatial heterogeneity of drug responses for personalized treatment. Brief. Bioinform 24 (6), bbad338. doi:10.1093/bib/bbad338
Tang, Z., Yu, Y., Ng, K., Sow, D., Hu, J., and Mei, J. (2021). Disease network delineates the disease progression profile of cardiovascular diseases. J. Biomed. Inf. 115, 103686. doi:10.1016/j.jbi.2021.103686
Theodoris, C. V., Xiao, L., Chopra, A., Chaffin, M. D., Al Sayed, Z. R., Hill, M. C., et al. (2023). Transfer learning enables predictions in network biology. Nature 618 (7965), 616–624. doi:10.1038/s41586-023-06139-9
Trzupek, D., Lee, M., Hamey, F., Wicker, L. S., Todd, J. A., and Ferreira, R. C. (2021). Single-cell multi-omics analysis reveals IFN-driven alterations in T lymphocytes and natural killer cells in systemic lupus erythematosus. Wellcome Open Res. 6, 149. doi:10.12688/wellcomeopenres.16883.2
Vandereyken, K., Sifrim, A., Thienpont, B., and Voet, T. (2023). Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24 (8), 494–515. doi:10.1038/s41576-023-00580-2
Van de Sande, B., Lee, J. S., Mutasa-Gottgens, E., Naughton, B., Bacon, W., Manning, J., et al. (2023). Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 22 (6), 496–520. doi:10.1038/s41573-023-00688-4
Vasan, N., Baselga, J., and Hyman, D. M. (2019). A view on drug resistance in cancer. Nature 575 (7782), 299–309. doi:10.1038/s41586-019-1730-1
Vemula, D., Jayasurya, P., Sushmitha, V., Kumar, Y. N., and Bhandari, V. (2023). CADD, AI and ML in drug discovery: a comprehensive review. Eur. J. Pharm. Sci. 181, 106324. doi:10.1016/j.ejps.2022.106324
Wagner, J., Rapsomaniki, M. A., Chevrier, S., Anzeneder, T., Langwieder, C., Dykgers, A., et al. (2019). A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177 (5), 1330–1345. doi:10.1016/j.cell.2019.03.005
Wang, C., Huyan, T., Zhou, X., Zhang, X., Duan, S., Gao, S., et al. (2022a). Development of single-cell transcriptomics and its application in COVID-19. Viruses 14 (10), 2271. doi:10.3390/v14102271
Wang, H. N., Yang, J., Xie, D. H., Liang, Z., Wang, Y., Fu, R. Y., et al. (2021b). Single-cell RNA sequencing infers the role of malignant cells in drug-resistant multiple myeloma. Clin. Transl. Med. 11 (12), e653. doi:10.1002/ctm2.653
Wang, L., Mo, S., Li, X., He, Y., and Yang, J. (2020). Single-cell RNA-seq reveals the immune escape and drug resistance mechanisms of mantle cell lymphoma. Cancer Biol. Med. 17 (3), 726–739. doi:10.20892/j.issn.2095-3941.2020.0073
Wang, P., Jin, X., Zhou, W., Luo, M., Xu, Z., Xu, C., et al. (2021a). Comprehensive analysis of TCR repertoire in COVID-19 using single cell sequencing. Genomics 113 (2), 456–462. doi:10.1016/j.ygeno.2020.12.036
Wang, Y., Chen, X., Tang, N., Guo, M., and Ai, D. (2024b). Boosting clear cell renal carcinoma-specific drug discovery using a deep learning algorithm and single-cell analysis. Int. J. Mol. Sci. 25 (7), 4134. doi:10.3390/ijms25074134
Wang, Y., Chen, Y., Gao, J., Xie, H., Guo, Y., Yang, J., et al. (2024a). Mapping crossover events of mouse meiotic recombination by restriction fragment ligation-based Refresh-seq. Cell Discov. 10 (1), 26. doi:10.1038/s41421-023-00638-9
Wang, Y., Han, X., and Jiang, C. (2019). Cycloastragenol inhibits metastasis and invasion in human colorectal cancer cells via the inhibition of the Wnt/β-catenin pathway. Oncol. Rep. 41 (1), 274–282.
Wang, Y., Wang, X., Luu, L. D. W., Li, J., Cui, X., Yao, H., et al. (2022b). Single-cell transcriptomic atlas reveals distinct immunological responses between COVID-19 vaccine and natural SARS-CoV-2 infection. J. Med. Virol. 94 (11), 5304–5324. doi:10.1002/jmv.28012
Wilk, A. J., Rustagi, A., Zhao, N. Q., Roque, J., Martinez-Colon, G. J., McKechnie, J. L., et al. (2020). A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26 (7), 1070–1076. doi:10.1038/s41591-020-0944-y
Wu, H., Gonzalez Villalobos, R., Yao, X., Reilly, D., Chen, T., Rankin, M., et al. (2022). Mapping the single-cell transcriptomic response of murine diabetic kidney disease to therapies. Cell Metab. 34 (7), 1064–1078.e6. doi:10.1016/j.cmet.2022.05.010
Wu, H., Guo, C., Wang, C., Xu, J., Zheng, S., Duan, J., et al. (2023). Single-cell RNA sequencing reveals tumor heterogeneity, microenvironment, and drug-resistance mechanisms of recurrent glioblastoma. Cancer Sci. 114 (6), 2609–2621. doi:10.1111/cas.15773
Wu, Z., Lawrence, P. J., Ma, A., Zhu, J., Xu, D., and Ma, Q. (2020). Single-cell techniques and deep learning in predicting drug response. Trends Pharmacol. Sci. 41 (12), 1050–1065. doi:10.1016/j.tips.2020.10.004
Xu, G., Qi, F., Li, H., Yang, Q., Wang, H., Wang, X., et al. (2020). The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 6, 73. doi:10.1038/s41421-020-00225-2
Xu, S., and Liu, P. (2013). Tanshinone II-A: new perspectives for old remedies. Expert Opin. Ther. Pat. 23 (2), 149–153. doi:10.1517/13543776.2013.743995
Yang, B., Hu, S., Jiang, Y., Xu, L., Shu, S., and Zhang, H. (2024). Advancements in single-cell RNA sequencing research for neurological diseases. Mol. Neurobiol. doi:10.1007/s12035-024-04126-3
Yang, P. H., Jin, L. J., Liao, J., Shao, X., Cheng, J. Y., Li, L., et al. (2022). Modern research on Chinese medicine based on single-cell omics: technologies and strategies. Zhongguo Zhong Yao Za Zhi 47 (15), 3977–3985. doi:10.19540/j.cnki.cjcmm.20220601.702
Yang, Z., Zhang, Q., Yu, L., Zhu, J., Cao, Y., and Gao, X. (2021). The signaling pathways and targets of traditional Chinese medicine and natural medicine in triple-negative breast cancer. J. Ethnopharmacol. 264, 113249. doi:10.1016/j.jep.2020.113249
Yin, T., and D’Adamio, L. (2023). BRI2-mediated regulation of TREM2 processing in microglia and its potential implications for Alzheimer's disease and related dementias. bioRxiv. doi:10.1101/2023.06.14.544924
Zeng, H., Huang, J., Ren, J., Wang, C. K., Tang, Z., Zhou, H., et al. (2023b). Spatially resolved single-cell translatomics at molecular resolution. Science 380 (6652), eadd3067. doi:10.1126/science.add3067
Zeng, Q., Mousa, M., Nadukkandy, A. S., Franssens, L., Alnaqbi, H., Alshamsi, F. Y., et al. (2023a). Understanding tumour endothelial cell heterogeneity and function from single-cell omics. Nat. Rev. Cancer 23 (8), 544–564. doi:10.1038/s41568-023-00591-5
Zhang, F., Wei, K., Slowikowski, K., Fonseka, C. Y., Rao, D. A., Kelly, S., et al. (2019). Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20 (7), 928–942. doi:10.1038/s41590-019-0378-1
Zhang, L., Zhang, Y., Wang, C., Yang, Y., Ni, Y., Wang, Z., et al. (2022a). Integrated single-cell RNA sequencing analysis reveals distinct cellular and transcriptional modules associated with survival in lung cancer. Signal Transduct. Target Ther. 7 (1), 9. doi:10.1038/s41392-021-00824-9
Zhang, M., Eichhorn, S. W., Zingg, B., Yao, Z., Cotter, K., Zeng, H., et al. (2021). Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598 (7879), 137–143. doi:10.1038/s41586-021-03705-x
Zhang, W., Maeser, D., Lee, A., Huang, Y., Gruener, R. F., Abdelbar, I. G., et al. (2023). Inferring therapeutic vulnerability within tumors through integration of pan-cancer cell line and single-cell transcriptomic profiles. bioRxiv. doi:10.1101/2023.10.29.564598
Zhang, X., Chen, H., Huang, X., Xu, H., Li, Y., Yuan, H., et al. (2022b). Single-cell transcriptomics profiling the compatibility mechanism of As(2)O(3)-indigo naturalis formula based on bone marrow stroma cells. Biomed. Pharmacother. 151, 113182. doi:10.1016/j.biopha.2022.113182
Zhang, Y., Huang, X., Yu, M., Zhang, M., Zhao, L., Yan, Y., et al. (2024). The integrate profiling of single-cell and spatial transcriptome RNA-seq reveals tumor heterogeneity, therapeutic targets, and prognostic subtypes in ccRCC. Cancer Gene Ther. 31, 917–932. doi:10.1038/s41417-024-00755-x
Zhao, Q., Zheng, Y., Zhao, D., Zhao, L., Geng, L., Ma, S., et al. (2023). Single-cell profiling reveals a potent role of quercetin in promoting hair regeneration. Protein Cell 14 (6), 398–415. doi:10.1093/procel/pwac062
Zhao, X. N., You, Y., Cui, X. M., Gao, H. X., Wang, G. L., Zhang, S. B., et al. (2021). Single-cell immune profiling reveals distinct immune response in asymptomatic COVID-19 patients. Signal Transduct. Target Ther. 6 (1), 342. doi:10.1038/s41392-021-00753-7
Zheng, F., Xu, H., Zhang, C., Hong, X., Liu, D., Tang, D., et al. (2021b). Immune cell and TCR/BCR repertoire profiling in systemic lupus erythematosus patients by single-cell sequencing. Aging (Albany NY) 13 (21), 24432–24448. doi:10.18632/aging.203695
Zheng, L., Qin, S., Si, W., Wang, A., Xing, B., Gao, R., et al. (2021a). Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374 (6574), abe6474. doi:10.1126/science.abe6474
Zheng, Z., Chen, J., Chen, X., Huang, L., Xie, W., Lin, Q., et al. (2023). Enabling single-cell drug response annotations from bulk RNA-seq using SCAD. Adv. Sci. (Weinh) 10 (11), e2204113. doi:10.1002/advs.202204113
Zhu, W., Huang, J., Wu, J., Wu, C., Ye, F., Li, X., et al. (2023a). Inflammation-related signature for prognostic prediction, tumor immune, genomic heterogeneity, and drug choices in prostate cancer: integrated analysis of bulk and single-cell RNA-sequencing. Heliyon 9 (11), e21174. doi:10.1016/j.heliyon.2023.e21174
Keywords: single cell technology, drug discovery, deep learning, machine learning, traditional Chinese medicine
Citation: Zhang A, Zou J, Xi Y, Gao L, Deng F, Liu Y, Gao P, Tong HHY, Tan L, Zou X and Hao J (2024) Single-cell technology for drug discovery and development. Front. Drug Discov. 4:1459962. doi: 10.3389/fddsv.2024.1459962
Received: 05 July 2024; Accepted: 04 October 2024;
Published: 18 October 2024.
Edited by:
Linheng Li, Stowers Institute for Medical Research, United StatesReviewed by:
Hongyuan Zhang, The University of Manchester, United KingdomSoma Samanta, University of Michigan, United States
Copyright © 2024 Zhang, Zou, Xi, Gao, Deng, Liu, Gao, Tong, Tan, Zou and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Hao, amhhb0BmdWRhbi5lZHUuY24=; Xin Zou, YWxidW14aW5AcXEuY29t; Lianjiang Tan, dGFubGlhbmppYW5nQDEyNi5jb20=
†These authors have contributed equally to this work
‡Present address: Fulan Deng, Centre for Artificial Intelligence Driven Drug Discovery, Faculty of Applied Sciences, Macao Polytechnic University, Macao, China
 Anzhuo Zhang1†
Anzhuo Zhang1† Henry H. Y. Tong
Henry H. Y. Tong Xin Zou
Xin Zou Jie Hao
Jie Hao