
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 14 March 2025
Sec. Cardiovascular Imaging
Volume 12 - 2025 | https://doi.org/10.3389/fcvm.2025.1543816
 Juhwan Lee1
Juhwan Lee1 Tao Hu1
Tao Hu1 Michelle C. Williams2
Michelle C. Williams2 Ammar Hoori1
Ammar Hoori1 Hao Wu1
Hao Wu1 Justin N. Kim1
Justin N. Kim1 David E. Newby2
David E. Newby2 Robert Gilkeson3,4
Robert Gilkeson3,4 Sanjay Rajagopalan3
Sanjay Rajagopalan3 David L. Wilson1,4*
David L. Wilson1,4*
Introduction: Non-contrast CT calcium scoring (CTCS) exams have been widely used to assess coronary artery disease. However, their clinical applications in predicting coronary arterial remodeling remain unknown. This study aimed to develop a novel machine learning model to predict positive remodeling (PR) from CTCS scans and evaluate its clinical value in predicting major adverse cardiovascular events (MACE).
Methods: We analyzed data from 1,324 patients who underwent both CTCS and CT angiography. PR was defined as an outer vessel diameter at least 10% greater than the average diameter of the segments immediately proximal and distal to the plaque. We utilized a total of 246 features, including 23 clinical features, 12 Agatston score-derived features, and 211 epicardial fat-omics features to predict PR. Feature selection was performed using Elastic Net logistic regression, and the selected features were used to train a CatBoost machine learning model. Classification performance was evaluated using 1,000 repetitions of five-fold cross-validation and survival analyses, comparing actual and predicted PR in the context of predicting MACE.
Results: PR was identified in 429 patients (32.4%). Using Elastic Net, we identified the top 13 features, including four clinical features, three Agatston score-derived features, and six fat-omics features. Our method demonstrated excellent classification performance for predicting PR, achieving a sensitivity of 80.3 ± 1.7%, a specificity of 89.7 ± 1.7%, and accuracy of 81.9 ± 2.5%. The Agatston-score-derived and fat-omics features provided additional benefits, improving classification performance. Furthermore, our model effectively predicted MACE, with a hazard ratio (HR) of 4.5 [95% confidence interval (CI): 3.2–6.4; C-index: 0.578; p < 0.00001] in the training set and an HR of 3.2 (95% CI: 2.5–4.0; C-index: 0.647; p < 0.00001) in the external validation set.
Conclusion: We developed an innovative machine learning model to predict coronary arterial remodeling from epicardial fat and calcification features from low-cost/no-cost screening CTCS scans. Our results suggest that vast number of CTCS scans can support more informed clinical decision-making and potentially reduce the need for invasive and costly testing for low-risk patients.
Coronary artery disease (CAD) is the leading cause of morbidity and mortality in industrialized countries globally (1, 2), highlighting the urgent need for advanced diagnostic tools to predict and mitigate its progression at an early stage. Early detection and accurate prediction of CAD are crucial for timely intervention and improving patient outcomes. Various imaging techniques have been employed to visualize plaque in coronary vessels and characterize CAD. Among these techniques, coronary computed tomography angiography (CCTA) is considered the most effective for identifying the presence and distribution of CAD (3).
CCTA can identify high-risk plaque features such as low-attenuation plaque, napkin-ring sign, spotty calcification, and positive remodeling (PR), which are indicators of increased risk for cardiovascular events. PR refers to the outward expansion of a coronary artery in response to atherosclerotic plaque buildup, and it has been shown to be a strong indicator of major adverse cardiovascular events (MACE) in several studies (4–8). However, the use of CCTA is often limited by factors such as cost, variability in assessments, the need for contrast agents, and exposure to ionizing radiation. Additionally, the relationship between PR and the likelihood of adverse events is not fully understood.
Non-contrast CT calcium scoring (CTCS) exam can provide direct evidence of coronary atherosclerosis through the detection of calcifications in the coronary arteries. It is recognized by several guidelines as a preferred tool for risk assessment (9, 10). However, to date, no studies have investigated whether the use of non-contrast CTCS could predict high-risk plaque features. While CTCS is well-established for assessing cardiovascular risk, its potential to predict specific disease conditions is not fully utilized, presenting an opportunity to expand its diagnostic capabilities. We hypothesize that quantitative measurements from CTCS are related to high-risk plaque features. Specifically, we identified features in CTCS images associated with the presence of PR. This analysis aims to uncover significant features from CTCS relevant to arterial remodeling and enable further correlative studies with CCTA to identify obstructive disease based on CTCS findings. In this regard, we have developed novel feature sets using CTCS, such as calcium-omics (11) and fat-omics (12), which have shown strong associations with future MACE outcomes. Given that CTCS offers unique perspective on coronary plaque, there is a strong rationale to relate CTCS findings to cardiovascular risk assessed by CCTA.
In this study, we developed a novel machine learning model to predict the presence of PR from low-cost or no-cost screening non-contrast CTCS scans and evaluated its performance using extensive multi-center datasets. We analyzed various clinical features, Agatston score-derived features, and epicardial fat-omics features (12) to identify the most relevant ones. We then trained multiple machine learning models and compared their performance in predicting PR and MACE.
This research was conducted as a sub-study within the Scottish COmputed Tomography of the HEART (SCOT-HEART) multicenter randomized controlled trial (ClinicalTrials.gov. Unique identifier: NCT01149590). The primary trial received approval from the local ethics committee, and all participants provided written informed consent. This specific sub-study was carried out under a data use agreement between the University of Edinburgh, Edinburgh, UK, and the University Hospitals Cleveland Medical Center, Cleveland, Ohio, USA. The main findings of the primary study have been published previously (13–15). In summary, of the 4,146 participants who attended the cardiology outpatient clinic, 2,073 were assigned to the intervention group. Among them, 1,778 underwent CT scans, and 1,324 of these scans were of sufficient quality for analysis in this sub-study. Further details can be found in other publications (7, 16).
All participants underwent both non-contrast electrocardiogram-gated CTCS and contrast-enhanced electrocardiogram-gated CCTA. The imaging was conducted using either 64- or 320-detector row scanners (Brilliance 64 from Philips Medical Systems, Netherlands; Biograph mCT from Siemens, Germany; Aquilion ONE from Toshiba Medical Systems, Japan). Adjustments for tube current, voltage, and the volume of iodine-based contrast agent were made according to each participant's body mass index (BMI).
The assessment of positive remodeling for each coronary segment was conducted using standardized semi-automated software (Autoplaque, Version 2.5, Cedars-Sinai Medical Center). PR was characterized by an outer vessel diameter that was at least 10% greater than the average diameter of the segments directly proximal and distal to the plaque (remodeling index >1.1) (17). The remodeling index was calculated by semi-automatically extracting coronary artery centerlines and identifying the vessel lumen, wall, and plaque components on multi-planar reformatted images, with manual adjustments as necessary. Subsequently, the vessel diameters were measured to determine the remodeling index. In this study, PR was evaluated in the proximal, mid, and distal regions of each major coronary artery, including the left anterior descending (LAD), left circumflex (LCX), and right coronary artery (RCA). A patient was considered to have PR if at least one PR was present in any of these arteries. The reliability of observer agreement in assessing positive remodeling has generally been reported as fair (18). Figure 1 illustrates an example of paired CTCS and CCTA images showing PR in the left anterior descending artery.
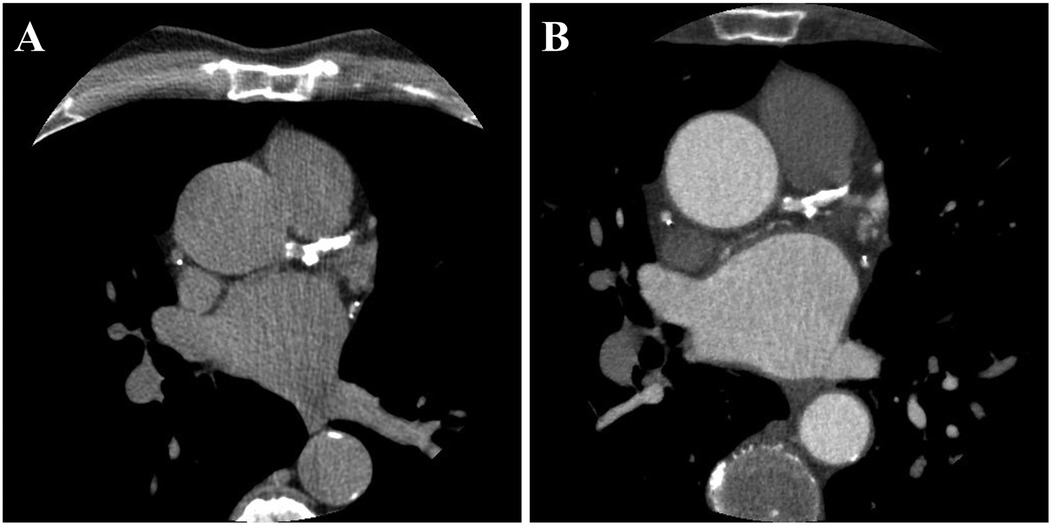
Figure 1. A representative example of paired CTCS (A) and CCTA (B) images showing the presence of PR. PR was observed in all major coronary arteries, including the left anterior descending (LAD), left circumflex (LCX), and right coronary artery (RCA). The total Agatston score was 3,027, distributed as follows: left main, 43; LAD, 1,168; LCX, 633; and RCA, 1,183.
We examined 23 clinical, 12 Agatston score-derived, and 211 fat-omics features to predict the presence of PR.
(1) Clinical features: These included baseline characteristics (e.g., gender, BMI, age, cholesterol, family history, etc.), blood tests (e.g., cholesterol levels), medications (e.g., statin, ACE inhibitor, and Beta blocker), and more. Table 1 lists the clinical features analyzed as predictors of PR.
(2) Agatston score-derived features: These included the Agatston scores for all coronary arteries, the logarithms of the Agatston scores (with 1 added to avoid infinite values), the diffusivity index (19), and high coronary artery calcification (CAC). The diffusivity index was calculated as 1 min the ratio of the Agatston score of the most affected vessel to the total Agatston score, indicating the distribution of coronary artery calcification. A higher diffusivity index signifies a more diffuse distribution, while a lower index indicates that the calcification is concentrated in a single artery. High CAC was defined as positive when the total Agatston score exceeded 1,000. Table 2 lists the Agatston score-derived features.
(3) Fat-omics features: We analyzed 211 handcrafted, pathophysiological-inspired fat-omics features, divided into morphological, intensity, and spatial categories (12). Using our previously developed DeepFat method (20), we segmented the epicardial adipose tissue regions within the pericardium. Morphological features included measurements such as volume, principal axis lengths, and epicardial fat thickness. Intensity features included statistical metrics like the minimum, maximum, and mean Hounsfield units (HU), skewness, and histogram bins. For spatial analysis, we divided the heart area into four equally thick slabs of image slices from top to bottom and four equidistant ribbons from the outer to inner regions. Detailed descriptions of fat-omics features are provided elsewhere (12).
We employed Elastic Net regression (21), which integrates the properties of both Lasso (L1) and Ridge (L2) regularization, for feature selection in our classification models. The mixing parameter (α) was set to 0.5 to equally balance the effects of L1 and L2 regularization. A series of 100 regularization values (λ) was automatically generated on a logarithmic scale, ranging from a maximum value that results in all coefficients being zero to a minimum value. To optimize these hyperparameters and avoid overfitting, we utilized 5-fold cross-validation. During this process, the data was divided into five subsets, with the model iteratively trained on four subsets and validated on the remaining one. This approach ensured the robustness and generalizability of the model to new data.
After identifying the optimal features with Elastic Net regression, we trained a machine learning model using the CatBoost algorithm (22). We optimized the CatBoost model through hyperparameter tuning using a grid search. The hyperparameters tuned included the number of iterations (100), learning rate (0.01), depth (6), L2 leaf regularization (3), random subspace method (0.75), border count (64), loss function (logloss), evaluation metric (F1), bootstrap type (Bernoulli), subsample (0.6), and thread count (1). This grid search enabled us to find the best combination of hyperparameters for our dataset. The hyperparameter search ranges for each model are provided in Supplementary Table S1.
We developed and trained three different models with varying feature combinations: (Model 1) clinical features only, (Model 2) clinical features plus Agatston score-derived features, and (Model 3) clinical features, Agatston score-derived features, and fat-omics features. Each model underwent the same hyperparameter optimization process. All the employed codes, including DeepFat (20), Elastic Net regression for feature selection, and CatBoost machine learning models, are available in our GitHub repository (https://github.com/Thefreak123TH).
To ensure the robustness and reliability of our model, we employed 1,000 iterations of 5-fold cross-validation. This method involved dividing the dataset into five subsets, training the model on four subsets, and validating it on the remaining subset. Repeating this process 1,000 times helped to average out variability and prevent overfitting. The classification performance was evaluated using conventional metrics such as sensitivity, specificity, accuracy, and area under the receiver operating characteristic curve (AUC).
To investigate performance variance, we compared the optimized CatBoost model with three other established methods: Support Vector Machine (SVM) (23), Random Forest (RF) (24), and XGBoost (25). Detailed training information for all machine learning models is provided in Supplementary Table S1. Each model was trained and tested using the same dataset split to ensure a fair comparison. This comprehensive evaluation allowed us to identify the most effective machine learning approach for our classification task.
In addition to conventional metrics, we conducted clinically relevant assessments to evaluate the model's utility in predicting MACE. Following classification, we used Kaplan–Meier survival analysis to estimate survival probabilities and stratify patients based on predicted and actual PRs. Cox proportional hazards models were then fitted using the predicted and actual PRs as covariates to assess their association with MACE. The survival predictions derived from these analyses were compared for consistency and clinical relevance. MACE was defined as a composite outcome of nonfatal myocardial infarction, nonfatal stroke, cardiovascular death, and revascularization. For external validation, we used a subset of the CLARIFY trial (ClinicalTrials.gov. Unique identifier: NCT04075162), comprising 2,316 patients who underwent both CCTA and CTCS.
Continuous variables were presented as mean ± standard deviation, and categorical variables were reported as frequencies. Comparisons between PR and non-PR groups were made using Student's t-test for continuous variables and the Chi-square test for categorical variables. Normality of continuous variables was assessed using the Shapiro–Wilk test. To evaluate the models' performance, we used McNemar's test (for sensitivity and specificity) and the Delong test (for AUC) to compare Model 1 with Model 2 and Model 2 with Model 3. To predict MACE, we utilized Kaplan–Meier survival analysis and Cox proportional hazard modeling, reporting hazard ratios (HR) and 95% confidence intervals (CI). A p-value of less than 0.05 was considered statistically significant. All statistical analyses were conducted using R Studio software (version 2024.04.1, R Foundation for Statistical Computing, Vienna, Austria).
Among the 1,324 patients, PR was identified in 429 patients (32.4%). The PR group had a significantly higher prevalence of males, hypertension, chest pain, hyperlipidemia, and medications (except for calcium blockers) than the non-PR group (p < 0.05). Additionally, age, height, and systolic blood pressure were significantly higher in PR group compared to the non-PR group (p < 0.05). While there was no significant difference in total cholesterol levels, HDL cholesterol was significantly lower in the PR group (p < 0.05). The baseline characteristics of the study population, comparing the PR and non-PR groups, are detailed in Table 1. All the Agatston score-derived features were significantly higher in the PR group compared to the non-PR group (Table 2).
We selected the top 13 features for model 3 using Elastic Net regression, which included four clinical features, three Agatston score-derived features, and six fat-omics features (Figure 2C). Among them, the top three were fat-omics (SR4_Pro_90_70, SR3_Pro_170_150, and SR3_Pro_150_130), highlighting the significant impact of epicardial fat assessment. Notably, all fat-omics features were derived from histograms of HU values (Supplementary Figure S1 and Table S2). Such assessments are related to fat inflammation, thought to be a driver of cardiovascular risk (see Discussion). The best features for Models 1 and 2 were also determined by Elastic Net (Figures 2A,B).
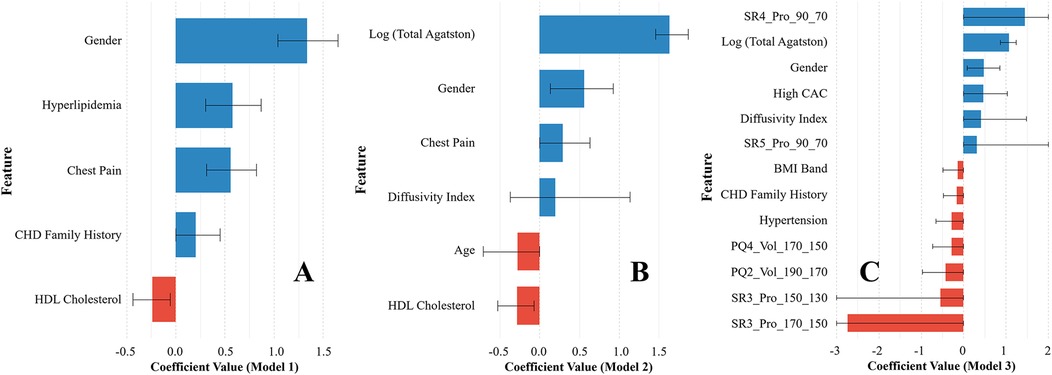
Figure 2. Variable importance plots of the three prediction models [(A) Model 1, (B) Model 2, and (C) Model 3] selected by Elastic Net regression. For Model 3, 13 features were selected and used to train the CatBoost machine learning classification model. The 95% confidence interval (CI) is provided for each feature. The actual CI ranges for SR4_Pro_90_70, SR5_Pro_90_70, SR3_Pro_150_130, and SR3_Pro_170_150 were clipped for better visualization.
Our method (Model 3) demonstrated excellent classification performance for predicting PR, achieving a sensitivity of 80.3 ± 1.7%, a specificity of 89.7 ± 1.7%, an accuracy of 81.9 ± 2.5%, and an AUC of 88.7 ± 1.2% (Table 3). Model 1 had the poorest classification performance, with the lowest values across all metrics. The classification performance improved significantly with the addition of Agatston score-derived features (Model 2) (p < 0.05). Adding fat-omics to Model 2 further improved sensitivity by 10% (from 70.0% to 80.3%) and specificity by 4% (from 85.9% to 89.7%) in Model 3 (p < 0.05). Among the four machine learning methods employed, the CatBoost method exhibited the best classification performance (Table 4). Although SVM showed higher sensitivity with much smaller standard deviation (87.9 ± 0.4%), its specificity was very low (67.9 ± 1.0%) compared to other methods. RF and XGBoost methods demonstrated similar classification results. The classification results of the Models 1–2 obtained using the other machine learning methods are provided in Supplementary Tables S3, S4.
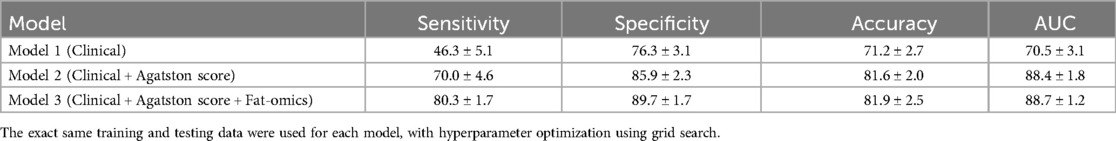
Table 3. Classification results of each model obtained using 1,000 repeated 5-fold cross validation.
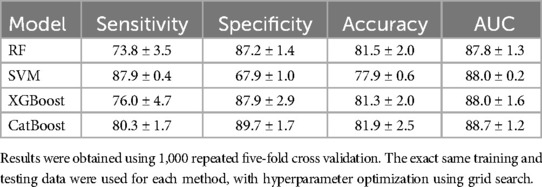
Table 4. Comparison of the proposed method (CatBoost) against other established machine learning approaches, including random forest (RF), support vector machine (SVM), and XGBoost.
Our method demonstrated excellent prediction of MACE. In Kaplan–Meier analyses, both actual and predicted PR significantly differentiated between MACE and no-MACE groups (p < 0.00001) (Figure 3). However, the predicted PR showed a much wider range of 95% CI compared to the actual PR. In Cox proportional hazard analyses, the actual PR had a higher HR (6.5; 95% CI: 4.8–9.0; C-index: 0.722; p < 0.00001) than the predicted PR (4.5; 95% CI: 3.2–6.4; C-index: 0.578; p < 0.00001). When applied to the external validation set (CLARIFY, n = 2,316), our method significantly differentiated between MACE and no-MACE groups (p < 0.00001) (Figure 4), with an HR of 3.2 (95% CI: 2.5–4.0; C-index: 0.647; p < 0.00001).
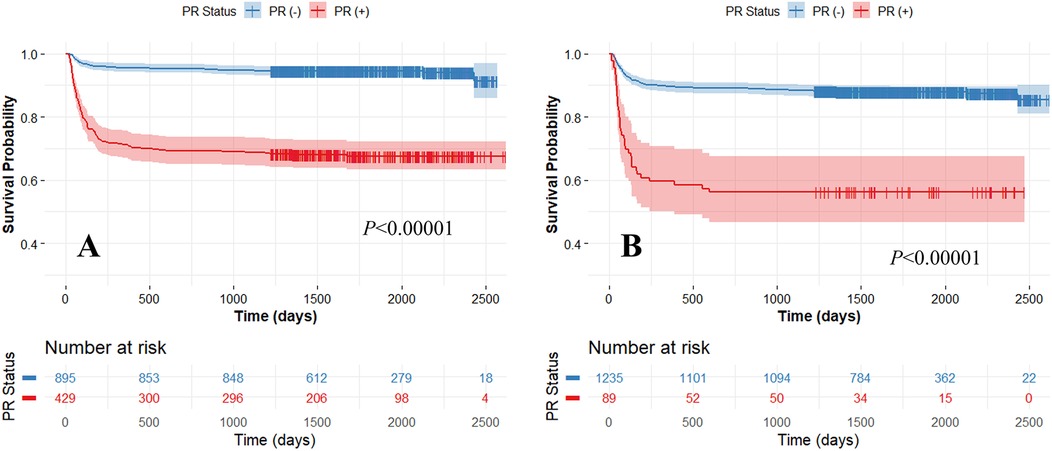
Figure 3. Kaplan-Meier survival curves between actual (A) and predicted (Model 3) (B) positive remodeling (PR) obtained using five-fold cross-validation. High and low-risk groups are split with the median risk. The x-axis represents survival time, and the y-axis represents the survival probability of patients. Both the actual and predicted PRs significantly differentiated between MACE and no-MACE groups (p < 0.00001). The colors are blue (no-MACE) and red (MACE).
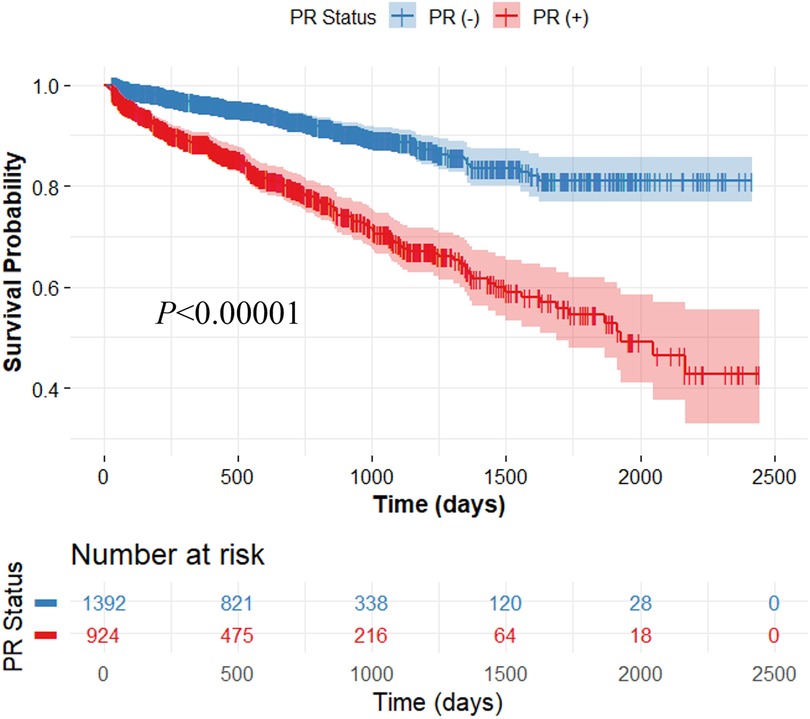
Figure 4. Kaplan-Meier survival curve on the external validation set consisting of 2,316 patients. For survival analysis, the CatBoost classification model (Model 3) was trained using the entire SCOT-HEART data (n = 1,324) and applied to the external validation set. The output of the trained model was used for survival analysis. The x-axis represents survival time, and the y-axis represents the survival probability of patients. The predicted positive remodeling (PR) significantly differentiated between MACE and no-MACE groups (p < 0.00001). The colors are blue (no-MACE) and red (MACE).
We built on our previous studies utilizing CT imaging (11, 12, 20, 26–31) and developed a novel machine learning model to predict the presence of PR from CTCS scans. To the best of our knowledge, this is the first study to examine CTCS imaging features as predictors of the arterial remodeling. The proposed study has several important contributions. (1) We utilized unique feature sets consisting of clinical, Agatston score-derived, and epicardial fat-omics features (12). The most relevant features were then determined using Elastic Net regression. (2) We implemented various state-of-the-art machine learning methods and compared their classification performance with different feature groups. This comprehensive evaluation allowed us to determine the most effective machine learning approach. (3) We further investigated the clinical value of the proposed method for predicting MACE using multi-center datasets (i.e., SCOT-HEART and CLARIFY).
Our fat-omics features had a significant impact on predicting PR, highlighting the importance of epicardial fat assessment. Epicardial fat is a visceral fat deposit located between the heart and the pericardium that can potentially cause local inflammation in the coronary arteries. This inflammation can directly contribute to coronary atherosclerosis and influence arterial remodeling, the process of structural changes in the coronary artery wall. Assessing epicardial fat is crucial because it serves as an active endocrine organ, secreting various adipokines and cytokines that can exacerbate inflammation and promote atherosclerotic changes. Additionally, the presence of epicardial fat has been associated with increased risk of plaque formation, arterial stiffening, and other adverse changes in the coronary arteries (32–34). In our previous study (12), we developed an automated method for the first time to quantitatively assess the epicardial fat deposition (fat-omics) and demonstrated the importance of elevated HU values for predicting MACE. Similarly, in this study, we found that the probabilities of EAT voxels with elevated HU values in the outer layers (e.g., SR4 and SR5) were potentially beneficial for prediction PR. The SR4 and SR5 regions correspond to areas where coronary arteries most likely located, which is why they were identified as strong predictors for PR (SR4_Pro_90_70 and SR5_Pro_90_70). The higher HU range (−90 to −70) is more indicative of lipid-rich necrotic cores, fibrotic-lipid mixed plaques, and inflamed pericoronary adipose tissue, all of which are strongly associated with PR. In contrast, the lower HU range (e.g., −170 to −150) is more reflective of adipose tissue, which may not directly contribute to PR. Inflammation and proteolysis promote outward expansion of the vessel wall, leading to PR. Since these processes are more strongly linked to regions with higher HU values, this explains their predictive importance.
The addition of Agatston score-derived features, particularly the logarithmic total Agatston score and diffusivity index, significantly impacted predicting PR. Higher Agatston scores indicate a greater burden of calcified plaque, reflecting more advanced and extensive coronary artery disease. This is associated with both positive and negative arterial remodeling, where the arterial wall undergoes structural changes in response to plaque accumulation. Positive remodeling can initially preserve the arterial lumen, while negative remodeling can lead to luminal narrowing and increased risk of cardiovascular events. Additionally, a higher diffusivity index indicates more diffuse distribution of calcification across the entire coronary arteries, suggesting that the spread of coronary calcification plays a crucial role in arterial remodeling. The diffuse calcification distribution may exacerbate the remodeling process, making it more challenging to manage and treat coronary artery disease effectively.
The CatBoost model demonstrated superior classification performance compared to other machine learning methods, including RF, SVM, and XGBoost. This enhanced performance can be attributed to several unique features of CatBoost. First, CatBoost is specifically designed to handle categorical features effectively without requiring extensive preprocessing or one-hot encoding, often necessary with other models. This results in better utilization of clinical features, Agatston score-derived features, and fat-omics features. Second, CatBoost employes an ordered boosting technique, which mitigates the prediction shift commonly encountered in traditional gradient boosting algorithms. This leads to more accurate and stable predictions, particularly compared to XGBoost. Additionally, CatBoost efficiently handles missing data and incorporates robust feature importance mechanisms, further enhancing its predictive capabilities. Its ability to manage data with varying distributions and complexities makes it particularly well-suited for medical imaging datasets, where such variability is common. In contrast, SVM showed the highest sensitivity in our experiments but suffered from very poor specificity, likely due to its tendency to overfit on positive instances, resulting in a higher false positive rate. Last, the model's inherent resistance to overfitting, combined with minimal hyperparameter tuning, helped improve classification performance.
Our method demonstrated high reproducibility, validated using extensive multi-center datasets, including those from the SCOT-HEART and CLARIFY trials. This evaluation across multiple centers is crucial as it confirms that the model performs robustly and is generalizable across various patient populations and imaging conditions. Although minor differences were observed in HR and C-index between the training and external validation sets (HR: 4.5, C-index: 0.578 vs. HR: 3.2, C-index: 0.647), our approach effectively predicted MACE (see Figures 3, 4). By employing datasets from different centers for training and testing, we minimized the risk of overfitting to a single-center dataset, enhancing the model's applicability in diverse settings. Our consistent classification performance across multiple independent cohorts suggests its reliability and potential for widespread clinical use in predicting arterial remodeling from CTCS data. Such rigorous validation is essential for establishing confidence in the model's predictions and ensuring its practical effectiveness in real-world clinical scenarios.
This study has a few limitations. First, the relatively small sample size may limit the generalizability of the findings, and results could vary with a larger cohort. Second, despite the full automation of the fat-omics calculations, each CTCS scan assessment required 15–20 min. Future improvements in software automation could streamline this process and enable wider use. Third, integrating additional quantified variables, such as calcium-omics (11), could potentially enhance predictive accuracy.
We developed an innovative machine learning model to predict coronary arterial remodeling from epicardial fat and calcification features from low-cost or no-cost screening non-contrast CTCS scans. The same model also predicted MACE. This suggests that the vast number of CTCS scans available can support more informed clinical decision-making and potentially reduce the need for invasive and expensive tests in low-risk patients.
The original contributions presented in this study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by Institutional Review Board of University Hospitals ClevelandMedical Center, Cleveland, OH, USA. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
JL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. TH: Data curation, Formal analysis, Writing – review & editing. MW: Data curation, Writing – review & editing. AH: Data curation, Formal analysis, Writing – review & editing. HW: Formal analysis, Writing – review & editing. JK: Formal analysis, Writing – review & editing. DN: Data curation, Writing – review & editing. RG: Conceptualization, Writing – review & editing. SR: Conceptualization, Writing – review & editing. DW: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research and/or publication of this article. This project was supported by the National Heart, Lung, and Blood Institute through grants K01HL171795, R01HL167199, and NIH R01HL165218. This research was conducted in space renovated using funds from an NIH construction grant (C06 RR12463) awarded to Case Western Reserve University.
The content of this report is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The grants were obtained via collaboration between Case Western Reserve University and University Hospitals of Cleveland. This work made use of the High-Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. The veracity guarantor, Justin N. Kim, affirms to the best of his knowledge that all aspects of this paper are accurate.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer KP declared a past co-authorship with the author DN to the handling editor.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2025.1543816/full#supplementary-material
1. Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the global burden of disease study 2017. Lancet. (2018) 392:1736–88. doi: 10.1016/S0140-6736(18)32203-7
2. Nichols M, Townsend N, Scarborough P, Rayner M. Cardiovascular disease in Europe 2014: epidemiological update. Eur Heart J. (2014) 35:2950–9. doi: 10.1093/eurheartj/ehu299
3. Gulati M, Levy PD, Mukherjee D, Amsterdam E, Bhatt DL, Birtcher KK, et al. 2021 AHA/ACC/ASE/CHEST/SAEM/SCCT/SCMR guideline for the evaluation and diagnosis of chest pain: a report of the American College of Cardiology/American Heart Association joint committee on clinical practice guidelines. Circulation. (2021) 144:e368–454. doi: 10.1161/CIR.0000000000001029
4. Yang S, Hoshino M, Koo B-K, Yonetsu T, Zhang J, Hwang D, et al. Relationship of plaque features at coronary CT to coronary hemodynamics and cardiovascular events. Radiology. (2022) 305:578–87. doi: 10.1148/radiol.213271
5. Kinoshita D, Suzuki K, Yuki H, Niida T, Fujimoto D, Minami Y, et al. Coronary plaque phenotype associated with positive remodeling. J Cardiovasc Comput Tomogr. (2024) 18:401–7. doi: 10.1016/j.jcct.2024.04.009
6. Nurmohamed NS, Bom MJ, Jukema RA, de Groot RJ, Driessen RS, van Diemen PA, et al. AI-guided Quantitative plaque staging predicts long-term cardiovascular outcomes in patients at risk for atherosclerotic CVD. JACC Cardiovasc Imaging. (2024) 17:269–80. doi: 10.1016/j.jcmg.2023.05.020
7. Williams MC, Moss AJ, Dweck M, Adamson PD, Alam S, Hunter A, et al. Coronary artery plaque characteristics associated with adverse outcomes in the SCOT-HEART study. J Am Coll Cardiol. (2019) 73:291–301. doi: 10.1016/j.jacc.2018.10.066
8. Senoner T, Plank F, Barbieri F, Beyer C, Birkl K, Widmann G, et al. Added value of high-risk plaque criteria by coronary CTA for prediction of long-term outcomes. Atherosclerosis. (2020) 300:26–33. doi: 10.1016/j.atherosclerosis.2020.03.019
9. Orringer CE, Blaha MJ, Blankstein R, Budoff MJ, Goldberg RB, Gill EA, et al. The national lipid association scientific statement on coronary artery calcium scoring to guide preventive strategies for ASCVD risk reduction. J Clin Lipidol. (2021) 15:33–60. doi: 10.1016/j.jacl.2020.12.005
10. Golub IS, Termeie OG, Kristo S, Schroeder LP, Lakshmanan S, Shafter AM, et al. Major global coronary artery calcium guidelines. JACC Cardiovasc Imaging. (2023) 16:98–117. doi: 10.1016/j.jcmg.2022.06.018
11. Hoori A, Al-Kindi S, Hu T, Song Y, Wu H, Lee J, et al. Enhancing cardiovascular risk prediction through AI-enabled calcium-omics. Sci Rep. (2024) 14:11134. doi: 10.1038/s41598-024-60584-8
12. Hu T, Freeze J, Singh P, Kim J, Song Y, Wu H, et al. AI prediction of cardiovascular events using opportunistic epicardial adipose tissue assessments from CT calcium score. ArXiv (2024) arXiv:2401.16190v1.
13. The SCOT-HEART Investigators. Coronary CT angiography and 5-year risk of myocardial infarction. N Engl J Med. (2018) 379:924–33. doi: 10.1056/NEJMoa1805971
14. The SCOT-HEART Investigators. CT coronary angiography in patients with suspected angina due to coronary heart disease (SCOT-HEART): an open-label, parallel-group, multicentre trial. Lancet. (2015) 385:2383–91. doi: 10.1016/S0140-6736(15)60291-4
15. Williams MC, Hunter A, Shah ASV, Assi V, Lewis S, Smith J, et al. Use of coronary computed tomographic angiography to guide management of patients with coronary disease. J Am Coll Cardiol. (2016) 67:1759–68. doi: 10.1016/j.jacc.2016.02.026
16. Williams MC, Kwiecinski J, Doris M, McElhinney P, D’Souza MS, Cadet S, et al. Low-Attenuation noncalcified plaque on coronary computed tomography angiography predicts myocardial infarction. Circulation. (2020) 141:1452–62. doi: 10.1161/CIRCULATIONAHA.119.044720
17. Motoyama S, Sarai M, Harigaya H, Anno H, Inoue K, Hara T, et al. Computed tomographic angiography characteristics of atherosclerotic plaques subsequently resulting in acute coronary syndrome. J Am Coll Cardiol. (2009) 54:49–57. doi: 10.1016/j.jacc.2009.02.068
18. Maroules CD, Hamilton-Craig C, Branch K, Lee J, Cury RC, Maurovich-Horvat P, et al. Coronary artery disease reporting and data system (CAD-RADS TM): inter-observer agreement for assessment categories and modifiers. J Cardiovasc Comput Tomogr. (2018) 12:125–30. doi: 10.1016/j.jcct.2017.11.014
19. Blaha MJ, Budoff MJ, Tota-Maharaj R, Dardari ZA, Wong ND, Kronmal RA, et al. Improving the coronary artery calcium score—addition of regional measures of calcium distribution: multi-ethnic study of atherosclerosis (MESA). JACC Cardiovasc Imaging. (2016) 9:1407–16. doi: 10.1016/j.jcmg.2016.03.001
20. Hoori A, Hu T, Lee J, Al-Kindi S, Rajagopalan S, Wilson DL. Deep learning segmentation and quantification method for assessing epicardial adipose tissue in CT calcium score scans. Sci Rep. (2022) 12:2276. doi: 10.1038/s41598-022-06351-z
21. Zou H, Hastie T. Regularization and variable selection via the elastic net. J Royal Statistical Soc B. (2005) 67:301–20. doi: 10.1111/j.1467-9868.2005.00503.x
22. Prokhorenkova L, Gusev G, Vorobev A, Dorogush AV, Gulin A. Catboost: unbiased boosting with categorical features. Advances in Neural Information Processing Systems. Montréal: Curran Associates, Inc. (2018). p. 6639–49. Available online at: https://proceedings.neurips.cc/paper_files/paper/2018/hash/14491b756b3a51daac41c24863285549-Abstract.html (accessed June 3, 2024).
23. Cortes C, Vapnik V. Support-vector networks. Mach Learn. (1995) 20:273–97. doi: 10.1007/BF00994018
24. Ho TK. Random decision forests. Proceedings of 3rd International Conference on Document Analysis and Recognition. Montreal, Que., Canada: IEEE Comput. Soc. Press (1995). p. 278–82. doi: 10.1109/ICDAR.1995.598994
25. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ‘16. New York, NY, USA: Association for Computing Machinery (2016). p. 785–94. doi: 10.1145/2939672.2939785
26. Song Y, Hoori A, Wu H, Vembar M, Al-Kindi S, Ciancibello L, et al. Improved bias and reproducibility of coronary artery calcification features using deconvolution. JMI. (2023) 10:014002. doi: 10.1117/1.JMI.10.1.014002
27. Hoori A, Freeze J, Singh P, Hu T, Song Y, Wu H, et al. Prediction of major adverse cardiovascular events using comprehensive AI analysis of calcifications and fat depots in CT calcium score images. In: Gimi BS, Krol A, editors. Medical Imaging 2024: Clinical and Biomedical Imaging. San Diego, United States: SPIE (2024). p. 10. doi: 10.1117/12.3003977
28. Hoori A, Hu T, Lee J, Al-kindi S, Rajagopalan S, Wilson DL. An enriched survival study of epicardial adipose tissues risk on major adverse cardiovascular event in CT calcium score images. In: Gimi BS, Krol A, editors. Medical Imaging 2023: Biomedical Applications in Molecular, Structural, and Functional Imaging. San Diego, United States: SPIE (2023). p. 27. doi: 10.1117/12.2653613
29. Wu H, Song Y, Hoori A, Subramaniam A, Lee J, Kim J, et al. Translating non-contrast CT calcium score images to virtual CCTA to aid segmentation of coronary arteries and emyocardium. Medical Imaging 2024: Clinical and Biomedical Imaging. San Diego, CA: SPIE (2024). p. 26–31. doi: 10.1117/12.3006516
30. Hu T, Hoori A, Lee J, Kim J, Song Y, Wu H, et al. AI Predictions of major adverse cardiovascular event using epicardial and paracardial adipose tissue assessments in CT calcium score images. In: Gimi BS, Krol A, editors. Medical Imaging 2024: Clinical and Biomedical Imaging. San Diego, United States: SPIE (2024). p. 11. doi: 10.1117/12.3006789
31. Song Y, Wu H, Lee J, Kim J, Hoori A, Hu T, et al. Pericoronary adipose tissue feature analysis in CT calcium score images with comparison to coronary CTA. (2024) doi: 10.48550/arXiv.2401.15554.
32. Nerlekar N, Brown AJ, Muthalaly RG, Talman A, Hettige T, Cameron JD, et al. Association of epicardial adipose tissue and high-risk plaque characteristics: a systematic review and meta-analysis. J Am Heart Assoc. (2017) 6:e006379. doi: 10.1161/JAHA.117.006379
33. Goeller M, Achenbach S, Marwan M, Doris MK, Cadet S, Commandeur F, et al. Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr. (2018) 12:67–73. doi: 10.1016/j.jcct.2017.11.007
Keywords: high risk plaque features, positive remodeling, computed tomography calcium scoring, fat-omics, machine learning, classification, major adverse cardiovascular events
Citation: Lee J, Hu T, Williams MC, Hoori A, Wu H, Kim JN, Newby DE, Gilkeson R, Rajagopalan S and Wilson DL (2025) Detection of arterial remodeling using epicardial adipose tissue assessment from CT calcium scoring scan. Front. Cardiovasc. Med. 12:1543816. doi: 10.3389/fcvm.2025.1543816
Received: 11 December 2024; Accepted: 3 March 2025;
Published: 14 March 2025.
Edited by:
Francesco Sturla, IRCCS San Donato Polyclinic, ItalyReviewed by:
Alexandru Florea, University Hospital RWTH Aachen, GermanyCopyright: © 2025 Lee, Hu, Williams, Hoori, Wu, Kim, Newby, Gilkeson, Rajagopalan and Wilson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David L. Wilson, ZGx3QGNhc2UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.