- 1Department of Computer Science, Colorado State University, Fort Collins, CO, United States
- 2Department of Mathematics, Colorado State University, Fort Collins, CO, United States
Artificial neural networks (ANNs) are sensitive to perturbations and adversarial attacks. One hypothesized solution to adversarial robustness is to align manifolds in the embedded space of neural networks with biologically grounded manifolds. Recent state-of-the-art works that emphasize learning robust neural representations, rather than optimizing for a specific target task like classification, support the idea that researchers should investigate this hypothesis. While works have shown that fine-tuning ANNs to coincide with biological vision does increase robustness to both perturbations and adversarial attacks, these works have relied on proprietary datasets—the lack of publicly available biological benchmarks makes it difficult to evaluate the efficacy of these claims. Here, we deliver a curated dataset consisting of biological representations of images taken from two commonly used computer vision datasets, ImageNet and COCO, that can be easily integrated into model training and evaluation. Specifically, we take a large functional magnetic resonance imaging (fMRI) dataset (BOLD5000), preprocess it into representational dissimilarity matrices (RDMs), and establish an infrastructure that anyone can use to train models with biologically grounded representations. Using this infrastructure, we investigate the representations of several popular neural networks and find that as networks have been optimized for tasks, their correspondence with biological fidelity has decreased. Additionally, we use a previously unexplored graph-based technique, Fiedler partitioning, to showcase the viability of the biological data, and the potential to extend these analyses by extending RDMs into Laplacian matrices. Overall, our findings demonstrate the potential of utilizing our new biological benchmark to effectively enhance the robustness of models.
1 Introduction
Over the last decade, the landscape of state-of-the-art neural networks has shifted at a near continuous rate. But, within the last few years, the discourse around how to achieve the state-of-the-art has shifted from an emphasis on architecture to an emphasis on robust learned representations. In this work, we note that what constitutes a “good” representation to optimize for is still a matter of debate—we posit that one promising representation to strive for is the one employed by the biological brain. Our contributions include the curation of a new dataset to facilitate measuring the similarity of neural network representations with biological representations, an investigation of the biological fidelity of several state-of-the-art models' representations, and the establishment of a new evaluative benchmark to facilitate further research into aligning artificial and biological neural representations.
Investigations into how similar a trained neural network is to a biological brain have been unfolding since the early days of the neural network boom (Yamins et al., 2013, 2014; Hong et al., 2016; Kheradpisheh et al., 2016; Yamins and DiCarlo, 2016). Prior work has shown that representations closer to the biological brain are more robust to adversarial attacks (Li et al., 2019), are adaptable to new tasks in a zero-shot context (Schrimpf et al., 2018; Blanchard et al., 2019), and have gains in task performance that emerge quicker than when learning representations without biological similarity (Blanchard, 2019; Blanchard et al., 2019). Given this pedigree, one would be remiss not to wonder why research into these comparisons is so rare. Unfortunately, most biological datasets are proprietary or too small (Chang et al., 2019) and without this resource, neither researchers nor practitioners can further investigate this phenomenon. Further, state-of-the-art models have traditionally been assessed by their accuracy on key datasets while evaluations of how well-embedded representations generalize to new tasks is a relatively recent phenomenon (Radford et al., 2021).
Additionally, many of these works simply focus on post-hoc evaluations. There are relatively few works investigating how to optimize networks to achieve biological representations (Elsken et al., 2018; Hsu et al., 2018; Liu et al., 2018; Pham et al., 2018; Bashivan et al., 2019). Even recent efforts to learn strong representations focus on unsupervised methods that allow massive amounts of data to be used for training, with the hope that stronger representations will emerge (Radford et al., 2021). We hypothesize that a large biological dataset would facilitate a deeper investigation into the viability of biological representations for artificial neural networks. Of particular interest to this community is the potential for deeper investigations into how to optimize for biologically grounded manifolds—current methodologies utilize representational similarity analysis (RSA) (Kriegeskorte et al., 2008), but recent work has suggested methods to adopt and expand these methods (Jamil et al., 2023) by redefining the core data structures of RSA, representational dissimilarity matrices (RDMs), into weighted graphs.
Following the methodology pioneered by Jamil et al. (2023), we demonstrate that network representations drift further away from biological representations when networks are optimized for task performance. This is in agreement with Kumar et al. (2022), who found an inverse-U relationship exists between ImageNet classification accuracy of a network and its perceptual similarity score (Zhang et al., 2018). We posit that these mirror critiques of prominent research groups like Google's DeepMind, where Goh et al. (2021) identified the viability of an adversarial typographic attack where simply writing the incorrect word on an object sufficed for causing misclassifications. In a blog post discussing the attack, Goh et al. (2021) suggested,
“this attack exploits the way image classification tasks are constructed. While images may contain several items, only one target label is considered true, and thus the network must learn to detect the most ‘salient' item in the frame. The adversarial patch exploits this feature by producing inputs much more salient than objects in the real world. Thus, when attacking object detection or image segmentation models, we expect a targeted toaster patch to be classified as a toaster, and not to affect other portions of the image.”
It is true that assessing state-of-the-art models has always been important for both practitioners adapting those models to their own tasks and researchers seeking to understand and push toward better models (Kingma and Welling, 2013); however, the advent of works like CLIP, from Radford et al. (2021), have ushered in a new era driven by evaluating neural networks on how adaptable their learned representations are to new tasks in a zero-shot context. This work provides the tools for researchers to take this idea further providing biologically viable target representations that can be factored into the optimization of networks. As illustrated in Figure 1, our contributions include:
• The presentation and re-release of the BOLD5000 dataset (Chang et al., 2019), which has been fully processed to facilitate evaluating and optimizing neural networks on biologically grounded representations of data. Our efforts culminate in one of the largest biological datasets for vision ever released, which will facilitate widespread investigations into optimal representations.
• The application of a previously unexplored graph-based technique, the Fiedler algorithm, to this preprocessed dataset, demonstrating its versatility as an evaluation metric.
• The introduction of a framework which allows researchers to better fine-tune, evaluate, and select models for robustness. Ultimately, the products of this work will facilitate future research into how robust representations manifest, and methods for optimizing networks to achieve trustworthy and adversarially robust results.
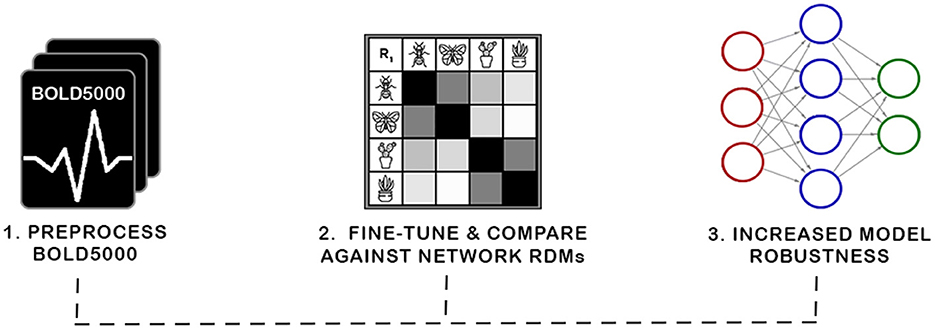
Figure 1. In this work, we present a new biologically grounded representation for evaluation and optimization of neural representations. Prior work has shown such representations correspond with robustness to adversarial attacks and task generalization. The curation of this new benchmark required preprocessing the BOLD5000 data into representational dissimilarity matrices (RDMs) and establishing a framework for investigating biological representations. We then investigate the viability of our discovered representation with a novel application of Fiedler partitioning on the data to demonstrate the potential of the biological representation for adversarial robustness.
2 Related work
Here, we detail prior works that investigate biological representation benchmarks. In particular, we focus on methods that investigate “neuro-similarity,” i.e., the similarity of an artificial neural network's (ANN's) learned representation to a benchmark of the biological brain. First, we examine metrics of neuro-similarity, then, efforts to increase neuro-similarity, and conclude with an investigation of works that link biologically consistent ANNs and robustness.
2.1 Metrics of neuro-similarity
Representational similarity analysis (RSA) is a popular tool measuring neuro-similarity where similarity metrics are derived from representational dissimilarity matrices (RDMs) (Kriegeskorte et al., 2008). ANN activations and neural data can be abstracted into RDMs for a set of stimuli. If two RDMs are created using the same stimuli set, they can be directly compared to one another by measuring the similarity of the consistency across that stimuli set. Two established metrics that capitalize on RSA for measuring the neuro-similarity of ANNs are human-model similarity (HMS) (Blanchard et al., 2019) and Brain-Score (Schrimpf et al., 2018).
HMS (Blanchard et al., 2019) evaluates the neuro-similarity between fMRI data and ANNs as the Spearman correlation between the averaged fMRI RDM and an ANN's RDM. The metric was validated on self-supervised predictive coding networks—a form of ANN composed of convolutional long short-term memory (LSTM) units designed to mimic predictive coding employed by biological visual systems. They found that models with higher HMS exhibited higher performance on next-frame prediction (the self-supervised task the networks were trained on) and were more robust to other tasks that networks were not trained for, such as object matching. They also found that HMS could be accurately measured early in the training process, and they proposed that it could be utilized for “early stopping”, i.e., training could be abandoned before the weights fully converged.
Similar to HMS, Brain-Score (Schrimpf et al., 2018) is a composite neural and behavioral benchmark set, which uses multiple evaluation metrics to score and rank ANNs according to how brain-like their visual object-recognition mechanisms are. To accomplish this, the internal representations of ANNs trained on ImageNet were compared for similarity against neural recordings taken from the V4 and IT cortical areas of macaque monkeys. From this, Dense-Net169, COREnet-S, and ResNet-101 were found to be the most brain-like, though Brain-Score was unable to reveal why.
HMS is the most similar to our methodology because we too use publicly available fMRI data, but a major limitation of HMS is that it only utilizes 92 stimuli, making it unsuitable to train with since networks quickly overfit to the small sample. These metrics are a great starting point for measuring neural similarity—however, to improve model robustness, more specific metrics need to be created. To effectively achieve this, datasets similar to this one must have as little noise in them as possible, something we address with BOLD5000.
In addition to RSA, there has also been research into psychophysical comparison metrics between ANNs and the human vision system. Jacob et al. (2021) found that ANNs trained for object recognition tasks were susceptible to some of the same visual illusions as the human visual system, such as mirror confusion, while other effects were absent. Gomez-Villa et al. (2020) found that ANNs trained for low-level visual tasks such as denoising and deblurring demonstrate human-like contrast similarity, but noted that deeper, more flexible network architectures did not demonstrate the same similarity. Human-like contrast similarity was also found in a variety of ANN architectures trained for different tasks (Li et al., 2022; Akbarinia et al., 2023).
2.2 Increasing neuro-similarity
Multiple methods have been investigated to affect an increase in the neuro-similarity of ANNs. One approach is the tailoring of image training datasets to achieve a distribution of input stimuli that more closely matches what may be experienced in nature (Aliko et al., 2020; Mehrer et al., 2021; Roads and Love, 2021; Allen et al., 2022). This approach is based on observations that training datasets designed for machine vision applications are crafted for domain specific applications, or otherwise contain internal biases in their distribution of subject matter that do not match what is in nature (Smith and Slone, 2017). A specific example of such a bias is the fact that ImageNet (Deng et al., 2009), one of the most widely used image classification datasets in the field, contains 120 categories of dog breeds, but lacks any categories for humans. By creating datasets with more natural image distributions, researchers have been able to significantly improve the neuro-similarity of the DNNs trained on these datasets.
While this approach does improve neuro-similarity in the trained models and demonstrates the potential of DNNs to achieve higher levels of neuro-similarity, it may not always be feasible or desirable to augment every dataset with a great enough volume of images, or images of the correct type, to achieve a distribution that matches the natural world. For example, domain specific datasets, such as for medical imaging research, don't have a complementary input set in nature to draw from. Datasets for machine vision research are also growing in size constantly and it may not be cost-effective or efficient to increase their size to a point where a natural distribution is achieved. However, these domain-specific models can potentially still benefit from greater neuro-similarity.
One architectural approach to increasing neuro-similarity is divisive normalization, which seeks to replicate how neighboring neurons normalize their activations non-linearly (Miller et al., 2021; Veerabadran et al., 2021; Hernández-Cámara et al., 2023).
It has been demonstrated that DNN models with greater neuro-similarity perform better at some tasks than models with lesser neuro-similarity. One exciting example of this, and the inspiration for this paper was work done by Li et al. (2019), who improved the robustness of a deep convolutional neural network (DCNN) to image noise via fine-tuning with an additional loss function that favored greater neuro-similarity. These experiments were conducted using a dataset derived from two-photon excitation microscopy (2PEF) of mice brains—they released the code to enable the fine-tuning but did not release the data itself. The fine-tuning was enabled via RDM comparisons—however, unlike Brain-Score and HMS, they approximated complete RDMs during training by only creating an RDM for a subset of stimuli. Constructing an entire RDM during training is computationally expensive because activations for each of the stimuli must be collected and compared.
2.3 Linking neuro-similarity to robustness
Despite initial findings that improving neuro-similarity could increase robustness (Li et al., 2019), none of the known evaluation metrics explicitly measure this improvement. We think this is an area where some could be created. We propose that robustness should be measured via psychophysics (RichardWebster et al., 2018, 2019). This evaluation focuses on evaluating robustness across a range of different noise levels. It also focuses on explainable and trustworthy evaluations of networks—by exploring a multitude of different noise types, the evaluation reveals specific weaknesses that networks are susceptible to, e.g., in the domain of face recognition, RichardWebster et al. (2018) found that FaceNet was surprisingly susceptible to brown noise, while other methods were not.
3 Materials and methods
3.1 BOLD5000
BOLD5000, one of the largest, publicly available fMRI datasets, was created to address three areas of neural dataset design: create a dataset of sufficient size to enable fine-tuning a deep neural network (DNN), have a greater diversity of images and image categories than is normally present in a neural study, and provide an overlap between the stimulus images used in the fMRI trials and the training image datasets of DNNs to allow for a more direct comparison of DNN and human brain activations (Chang et al., 2019).
Similar to most other human fMRI brain scan datasets, BOLD5000 is composed of stimuli images pulled from existing machine vision image datasets (Geirhos et al., 2018; Allen et al., 2022). In total, it consists of fMRI brain scans from four participants (CSI1-4) who were presented with 4,916 real-world images from three commonly used computer vision datasets: 1,916 from ImageNet (Deng et al., 2009), 2,000 from Common Objects in Context (COCO) (Lin et al., 2014), and 1,000 custom images of scenes from categories inspired by Scene UNderstanding (SUN) (Xiao et al., 2010). Collectively, these datasets span a wide variety of categories and consist of images of real-world indoor and outdoor scenes and objects either centered in or interacting with complex real-world scenes.
All selected images were resized, cropped to 375 × 375, and adjusted for even luminance. For each input dataset, exemplar images were hand-selected by the BOLD5000 authors on a per-category basis. Subjects then engaged in 15 functional MRI sessions, where all images were presented on a single trial basis, except for a subset of 113, for which unique neural representation data was collected.
During the original BOLD5000 study, one participant (CSI4) did not complete the entire experiment. As a result, CSI4 is typically discarded from studies using the BOLD5000 (Sexton and Love, 2022). However, because there are already only three complete participants to begin with, and because the majority of the stimuli images are only presented once, this study incorporates CSI4's partial data into a mean subject using the RDMs calculated as part of the RSA analysis (Section 3.4).
A second release of the BOLD5000 dataset occurred in 2021 (Chang et al., 2021). The major difference with the second release was the re-processing of the beta values for the fMRI sessions using the GLMSingle toolbox (Prince et al., 2022). The goal of the second release was to increase the reliability of the beta value estimates.
3.2 Preprocessing
All betas were provided in NIfTI format, divided by subject and session. The image coordinate transforms provided within the file header did not correspond to the transforms used for brainmasks, ROI masks, and T1w anatomical images from the original BOLD5000 release. This transform information is required for several other processing steps, including the re-application of the functional region of interest (ROI) masks from the original release of the BOLD5000 and application of the two new ROI atlases, vcAtlas (Rosenke et al., 2018) and visfAtlas (Rosenke et al., 2021), to the four participant brains. We solved this issue by intuiting that the provided NIfTI files were derived from the same fMRIPrep (Esteban et al., 2019) derivatives as the original BOLD5000, thus allowing us to utilize the same alignments and brainmasks. The affine transforms from the original BOLD5000 brainmasks were applied to the GLMSingle beta files and the results were visually checked against both the original brainmasks and the T1w anatomical scans of the participants to confirm good alignment. The generation of a global brainmask intersection was also required for each of the four subjects across all sessions. RSA analysis calculates distance metrics for each pair of input stimuli and therefore requires that the input vectors for each of the stimuli have the same number of dimensions (in the case of fMRI, dimension is a voxel). The BOLD5000 is somewhat unique in that it is largely made up of single presentations of each stimulus, and the order of the stimuli is randomized across multiple sessions for each participant. This poses a challenge because even very minor positional changes between sessions can lead to the introduction of invalid voxel values, especially around the very edge or pial surface of the brain.
The fMRIPrep pipeline uses a number of advanced tools to correct for any changes (Esteban et al., 2019), however, it was found that the participant brain masks provided in the original BOLD5000 release still resulted in invalid voxels being included for some trials. To address this issue, a global mask was calculated for each participant using the intersection of the valid voxels for each input across all sessions. These global participant brain masks were applied to every ROI to ensure that no invalid voxel data entered into the RSA calculations.
3.3 FreeSurfer
FreeSurfer is an incredibly powerful suite of tools originally developed with the goal of reconstructing cortical surface models from T1w anatomical scans (Fischl, 2012). A further goal of this original development in reconstructing the cortical surface is finding alignments between subject brains based on cortical folding patterns. It is this alignment functionality that makes the FreeSurfer a vital component of the fMRIPrep (Esteban et al., 2019) pipeline used in the original BOLD5000 release (Chang et al., 2019).
As follow-on researchers, we leverage these FreeSurfer derivatives to extract additional information from the dataset. We use FreeSurfer to parcellate a reconstructed cortical surface based on its folding patterns using specially crafted atlases. We used this functionality to identify and extract additional areas relevant to vision based on structural connectivity or functional response to images using vcAtlas and visfAtlas respectively. Our analysis is concerned with comparing the BOLD activations of voxels in volumetric space. Thus, several steps were required to convert these surface atlases into volumetric ROI masks.
First, the labels from the atlases were resampled from the standard fsaverage surface to each of the subjects' cortical surfaces. This is accomplished using the mri_surf2surf command. With the labels for each atlas and ROI now resampled onto the subjects' cortical surfaces, the labels were used to define a volumetric ROI as the volume of gray matter that makes up the cortex beneath the cortical surface label. This is accomplished with the mri_label2vol command with projection fraction set to include 100% of the volume between the pial and white matter surfaces. The output of this function is a series of volumetric ROI masks in NIfTI format, similar to the ROI masks from the original BOLD5000. All ROI masks generated using FreeSurfer also had the global mask for each participant applied to them to ensure that only valid voxels would be extracted for a given ROI.
3.4 RSA
After preprocessing and utilizing FreeSurfer to identify ROIs, we create RDMs from the neural data. We construct RDMs in accordance with established methodology (Kriegeskorte et al., 2008; Blanchard et al., 2019). Here, we briefly summarize the process:
RDM construction. Given a single feature f and a single stimulus s, v = f(s), where v is the value of feature f in response to s. Likewise, the vector
can represent the feature values of a collection of n features, f1, f2, ..., fn, in response to s. If one expands the representation of s to a set of m stimuli S = s1, s2, ..., sm, the natural extension of is the set of feature value collections , in which si∈S is paired with for each i = 1, 2, ..., m. The last step prior to constructing an RDM is to define the dissimilarity score between any two and . We use the symmetric function
where is the mean of the features in . An RDM R may then be constructed from S, V, and ψ as:
3.4.1 Biological similarity metric
The methodology for comparing a network to a biologically constructed RDM is simple: After constructing an RDM R1 for the network following the procedure outlined in Section 3.4 using the same stimuli set S, one can compute the similarity to the biological RDM R2 with the function
where is the flattened RDM and ρ corresponds with a similarity metric, e.g., Pearson's correlation. Note, many works suggest estimating the RDM during training by only considering a subset of the stimuli (Li et al., 2019).
3.4.2 rsatoolbox package
rsatoolbox is a Python package for RSA (Nili et al., 2014). Originally developed for Matlab, rsatoolbox is under active development and can be used for the generation and comparison of RDMs, the creation and evaluation of multiple types of models with various statistical tools, and visualization tools. All fMRI RDMs, RDM comparisons, and models were performed with rsatoolbox (Initial RDM generation for the ANNs were generated using functionality built into the Net2Brain tool as detailed in Section 3.7.).
3.5 ImageNet in BOLD5000
The use of images from the ImageNet dataset in the BOLD5000 presents a unique opportunity because the ImageNet Large Scale Visual Recognition Challenge (ILSVC) benchmark remains the standard benchmark and training dataset for image classification models such as those included in this paper. Prior to the BOLD5000 data, representations of neurological data tended to be collected for simple stripped-back stimuli such as a clearly cut-out image against a gray background. While these simple stimuli enabled research comparing biological representations to artificial representations (e.g., Blanchard et al., 2019), they had limited additional uses. For example, these stimuli were too simple and too few for fine-tuning networks to exhibit biologically consistent embeddings. The use of complex images like those within the ImageNet dataset may be non-ideal for traditional fMRI research, but they enable a wealth of experiments examining artificial neural networks (ANNs).
ImageNet classes are based on the WordNet synset hierarchy. In theory, this synset hierarchy can be used to establish the relationships between image classes. However, there are known deficiencies in the WordNet structure and most researchers resort to creating custom “supercategories” that combine multiple synsets. The original BOLD5000 paper used four supercategories for t-SNE analysis: Objects, Food, Living Inanimate, and Living Animate (Chang et al., 2019). For this work, five new supercategories were chosen that attempt to create more logical pairings for comparison. Animate subjects were divided into “vertebrate” and “invertebrate” supercategories, inanimate classes were divided into “artifact” (man-made) and “natural object”, and a final “place” category was included to align with the emphasis on scenes in the BOLD5000. Table 1 summarizes the supercategories created for this project and the hypernyms used to define each supercategory. Each of the ImageNet synset labels were sorted into a supercategory by matching the synset's hypernyms to one of the supercategory hypernyms.
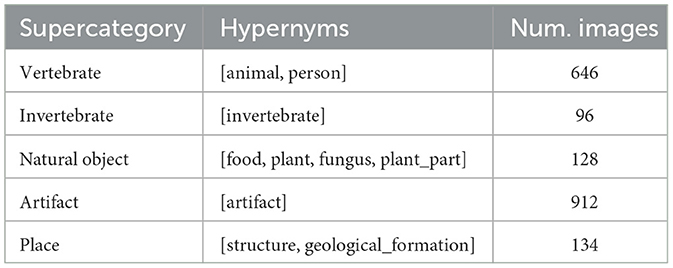
Table 1. Five supercategories were created by combining the synset labels from the ImageNet stimuli.
3.6 Categorical model analysis
While the end goal of our RSA analysis is to compare the biological data from the BOLD5000 fMRI trials to ANNs, RSA also allows us to leverage other types of dissimilarity models such as the supercategories within ImageNet as described in Section 3.5. First, categorical RDMs are generated for each supercategory as illustrated in Figure 2. These consist of an RDM where all images of the same category are assigned the minimum distance/dissimilarity for a given metric and all images from other categories are assigned the maximum distance/dissimilarity for a given metric.
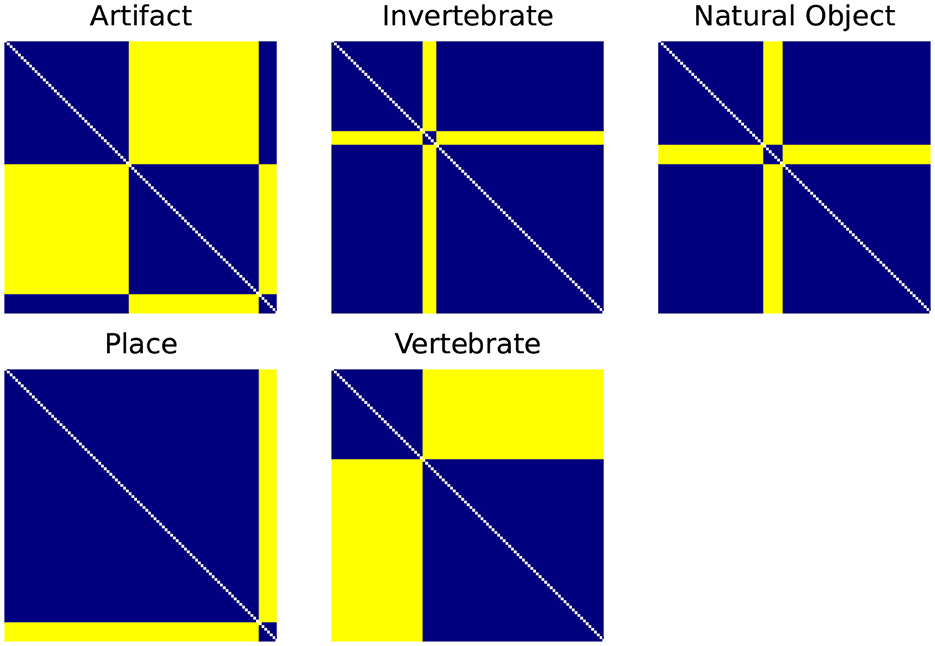
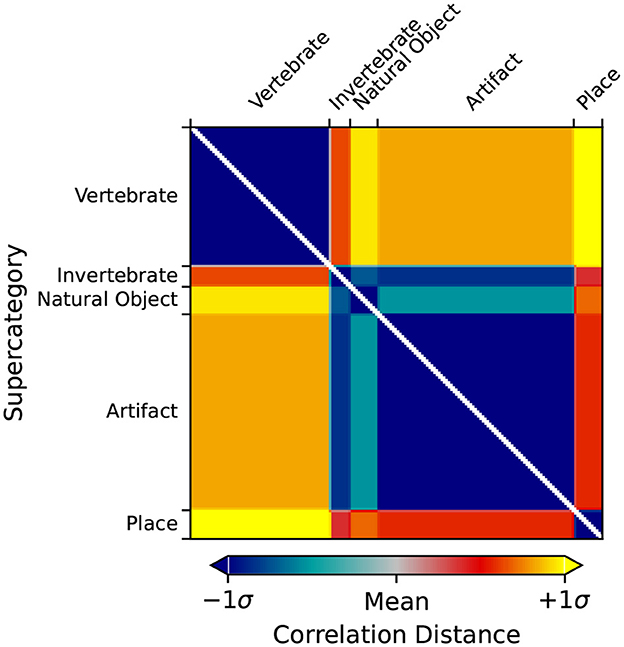
Figure 2. Categorical RDMS for each of the ImageNet supercategories. Categorical RDMs consist of an RDM where all images of the same category are assigned the minimum distance/dissimilarity for a given metric (i.e., for the 1-r distance metric, 0) and all images from other categories are assigned the maximum distance/dissimilarity for a given metric (i.e., 1).
Using rsatoolbox's weighted model functionality, the individual category RDMs are linearly fit to the Mean Subject RDMs for the vcAtlas ROIs. The model weights were then used to predict the final categorical model shown in Figure 3. This categorical model is a representation of the relative similarities of each of the supercategories as perceived by the human brain. Categorical models such as this can act as a reference point for later RSA analysis because it relies on additional structural information that is embedded into the ImageNet image labels.
3.7 Net2Brain
Here, we link our preprocessed data and subsequent evaluations to Net2Brain, a toolbox for researching the internal geometric representations of artificial deep neural networks, particularly convolution neural networks, using RSA. One of the strengths of Net2Brain is the very extensive set of over 600 models that it is preconfigured to pull down, extract activations from, and calculate RDMs for. Net2Brain is able to pull models not only from the official PyTorch model zoo, but also from timm, the Pytorch Image Models library created by Ross Wightman. All of the aforementioned 600+ models available to Net2Brain come pre-trained and are fully ready for activation extraction. All of the stimuli from the BOLD5000 are made available to Net2Brain and once it pulls down the pre-trained model in question, it presents each of the BOLD5000 images to the model as input and performs a forward pass. The model activations from each of the model's convolutional layers are then extracted and stored to disk. Once all of the activations have been extracted, RDMs for each of the convolutional layers are calculated. As of the time of writing, the toolbox enables creating RDMs using Pearson's correlation, and there are plans to add various other distance metrics.
3.7.1 Model selection
Of the over 600 models available, four were chosen based on a couple of criteria. First, due to the limitations in the architecture of both Net2Brain and rsatoolbox, the calculation of RDMs required substantial amounts of memory given the number of unique stimuli in the BOLD5000. There was, therefore a relative size limit to the number of output activations in a model given the memory limits of available hardware. The second criterion was to achieve a representative sampling of ANN model architectures that are designed for image classification tasks and pre-trained on the ImageNet dataset over time. The four models chosen were: AlexNet (Krizhevsky et al., 2012), the progenitor of all subsequent deep convolutional neural networks, ResNet50 (He et al., 2016), which introduced skip connections to neural network architectures, MobileNetv2 (Sandler et al., 2018), which was specifically designed to perform well even on restricted hardware such as mobile devices, and finally EfficientNet (Pham et al., 2018), which expands on the same architectural concepts present in MobilNet with efficient network scaling.
3.8 Fiedler vector partitioning
In this section, we detail how we employ Fiedler partitioning, a graph-based technique, on the processed data. Fiedler partitioning aims to partition a graph into two distinct groups by utilizing the Fiedler vector, which corresponds to the second smallest eigenvector of the Laplacian (Fiedler, 1973, 1975).
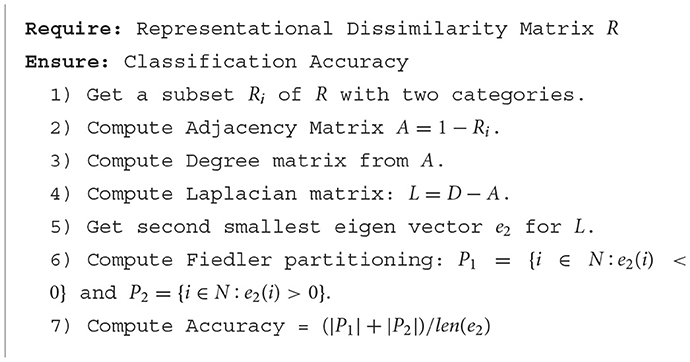
We analyzed individual RDMs for three BOLD5000 participants (CSI1-3), and a mean RDM (averaged subject data) for fMRI data specific to the Left-Hand Fusiform Gyrus 3 (LHFG3). Each RDM is composed of the supercategories described in Table 1. From these supercategories, we first extracted subsets of each class pairing. We then applied Fiedler partitioning to these RDMs and recorded the classification accuracy for each class in a pair. The pseudo code for finding the Fiedler partitioning accuracy for an RDM is detailed in Algorithm 1. Bias corrected and accelerated (BCa) bootstrap intervals were calculated for each binary classification.
4 Results
4.1 Categorical model analysis
Each of the vcAtlas ROIs from the mean subject was compared back against the predicted categorical model to determine which ROI best represents the supercategorical structure of the data. Figure 4 shows the correlation of each of the ROIs to the categorical model. In the case of the BOLD5000 data, the LHFG3 is the best exemplar of the categorical model.
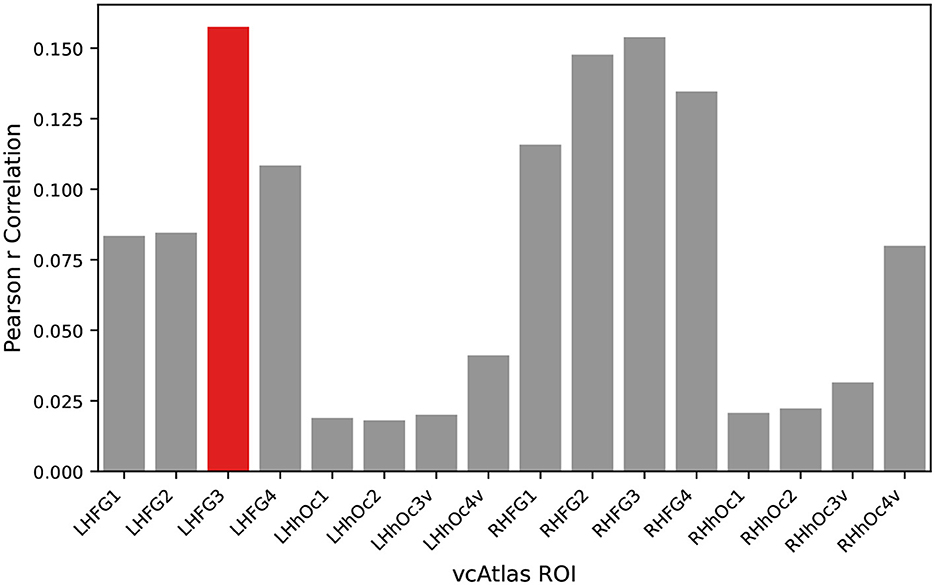
Figure 4. An exemplar ROI is chosen from the available vcAtlas ROIs by comparing its Pearson correlation to the categorical model (Figure 3). The Left-Hand Fusiform Gyrus 3 (LHFG3) (highlighted in red), was found to have the highest correlation with the categorical model.
Figure 5 shows the RDM for LHFG3 from the mean subject with the images sorted by supercategory. Comparing LHFG3 Figure 5 to the categorical model Figure 3, is it clear how the supercategory representations cluster in a similar way. This can be explored further through the comparison of RDM correlations.
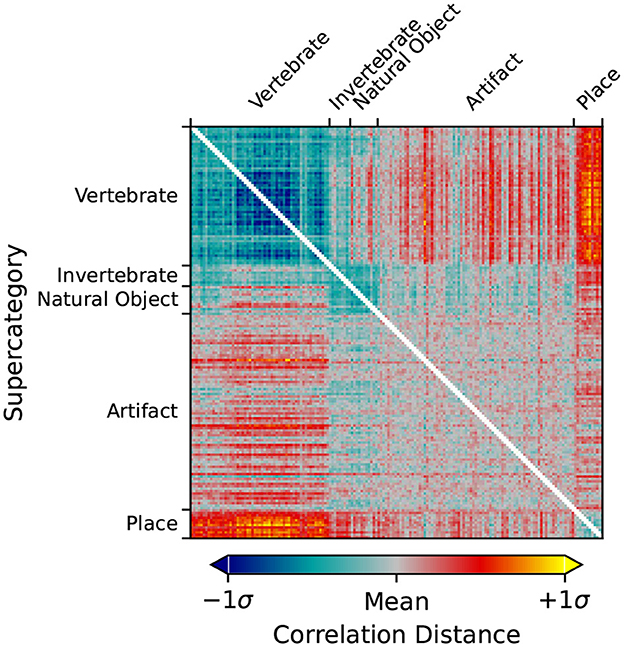
Figure 5. RDM of the Left-Hand Fusiform Gyrus 3 (LHFG3) ROI calculated from the mean subject using the correlation distance metric. Image inputs are sorted by their ImageNet supercategory. Clustering of similar images within supercategories is visible, as is dissimilarity between supercategories.
Figure 6 shows the RDMs of the layer with the highest correlation to the categorical model for each of the four ANNs investigated. When visually comparing the categorical model, Figure 3, the mean subject fMRI response, Figure 5 and the ANN responses, Figure 6, a correspondence between the representation of the supercategories is evident.

Figure 6. RDMs from each of the four ANNs ordered by ImageNet supercategory. Each RDM is taken from the ANN layer with the highest correlation to the categorical model calculated in Section 3.6.
4.1.1 ANN vs. human fMRI RDM comparison
Direct comparison of RDMs can be accomplished through a number of similarity measures. Here, we report Pearson correlation, an established standard for use in RSA (Kriegeskorte et al., 2008). Table 2 presents the Pearson correlation between the categorical model and each of the four ANNs under test. Bootstrap resampling of the input stimuli was performed to assess that all results were statistically significant (p < 0.001). All comparisons and statistical evaluations were performed using the rsatoolbox package (Section 3.4.2).

Table 2. Comparison of mean subject LHFG3 ROI RDM to categorical model and ANN RDMs with bootstraped p-values.
An unexpected result of this analysis is the inverse relationship between model age and its biological similarity. AlexNet (Krizhevsky et al., 2012), the model that kicked off the deep convolutional neural network revolution in machine vision, has the highest biological similarity of the models tested, and EfficientNet (Tan and Le, 2019), the most modern and highest performing classification model, has by far the lowest biological similarity.
4.1.2 Comparing human fMRI ROIs to individual ANN layers
In Figure 7, we break down our evaluation layer by layer in order to provide fine-grained details on which components of the trained network best exhibits biological similarity.
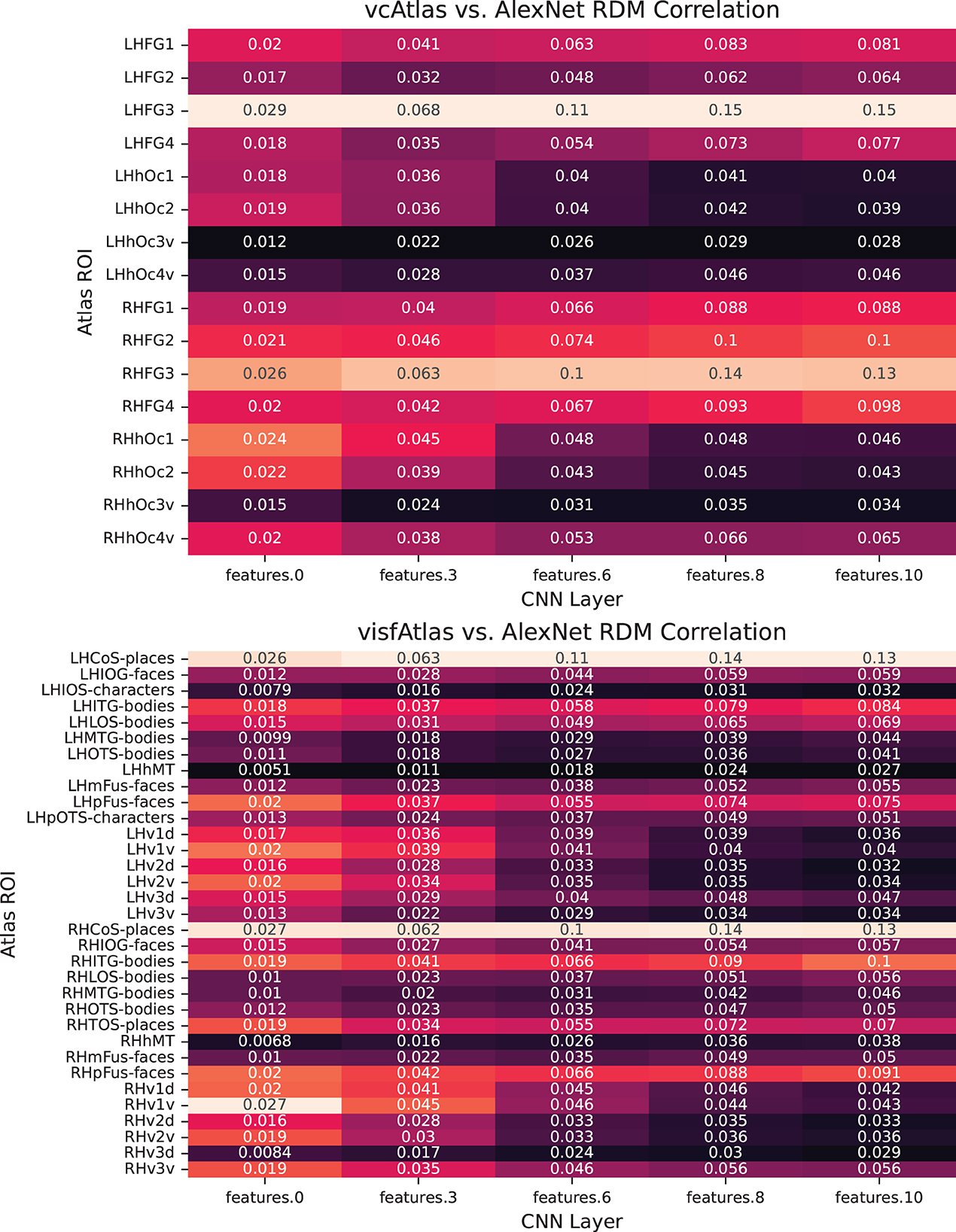
Figure 7. Pairwise analysis of each of the layers of AlexNet to ROIs of the new vcAtlas and visfAtlas cortical atlases. In the vcAtlas comparison it can be seen that while the LHFG3 ROI does dominate the comparison, there is a correlation between the first two layers of AlexNet with the early visual cortex in Oc1 and Oc2.
One of the goals in reprocessing the BOLD5000 dataset using the vcAtlas (Rosenke et al., 2018) and visfAtlas (Rosenke et al., 2021) maps was to enable future research into comparing how various components of an ANN, such as individual convolutional layers, can be compared to specialized structures in biological representations. For example, consider the theoretical concept behind the ventral visual stream in the human brain is that visual information flows from the early visual cortex at the back of the brain forward into the Fusiform Gyrus. Along the way, the visual stimuli is decoded in increasingly higher order representations. Our findings give credence to the observation that deep convolutional neural networks mimic some of what occurs with this process.
The human brain also has a number of very specialized areas for certain tasks such as facial recognition in the Fusiform Face Area (FFA) (Kanwisher et al., 1997), one of the ROIs included in the visfAtlas. The goal is to provide the data so that these specialized areas of the brain can be used to analyze and train equivalently specialized components of ANNs.
4.2 Fiedler partitioning
Figure 8 displays the Fiedler partitioning accuracies for each of the four ANNs from our experiments, and Figure 9 shows the partition accuracy for the biological data. All accuracies illustrate the separability of class pairs—the results indicate that the human subjects consistently achieved higher classification accuracy when discriminating between the vertebrate class and the invertebrate, natural object, and place categories. This shows that the feature embeddings in the LHFG3 are well-clustered for those categories.
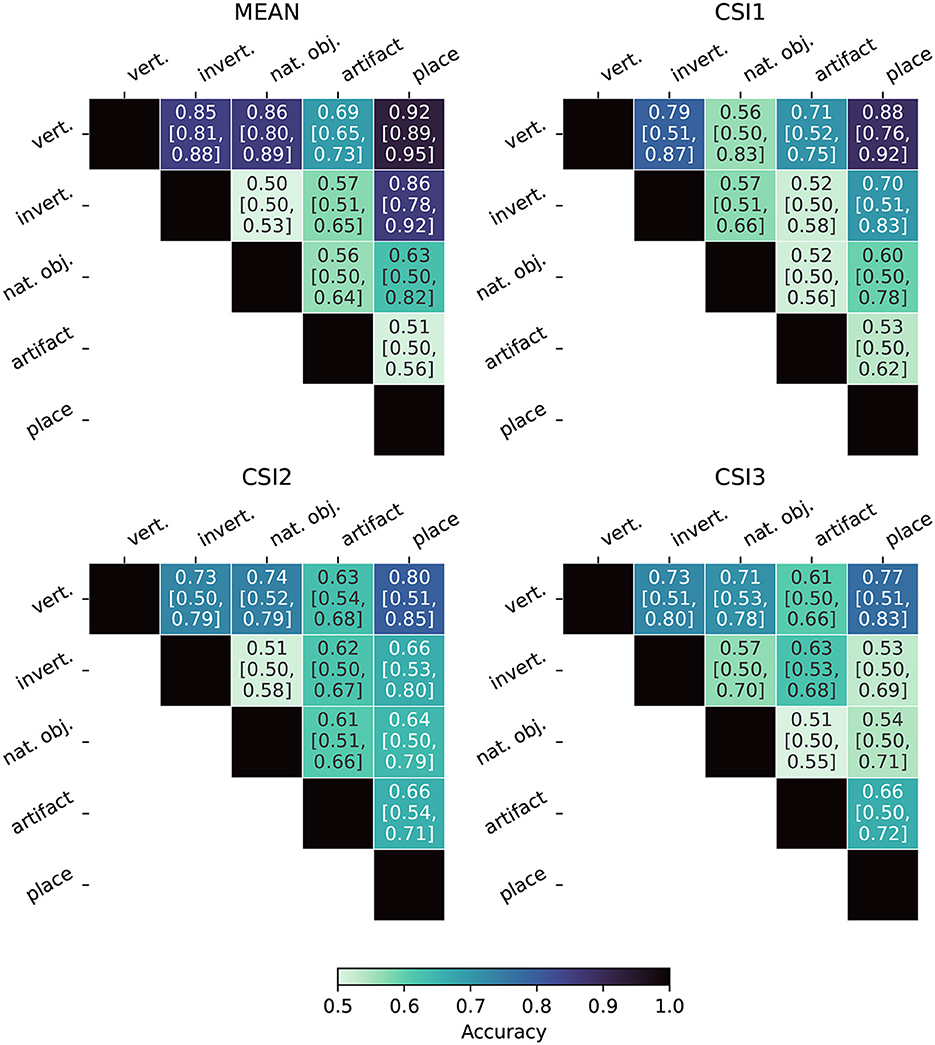
Figure 8. Fiedler partitioning accuracy for ANNs. Partitioning is performed on the RDM of the layer with the highest correlation to the categorical model developed in Section 3.6. Accuracy is reported along with Bonferroni-corrected BCa bootstrap intervals (CIs) (accuracy [lower CI bound, upper CI bound]). AlexNet and ResNet demonstrate significantly higher accuracy and significance across all supercategory comparisons.
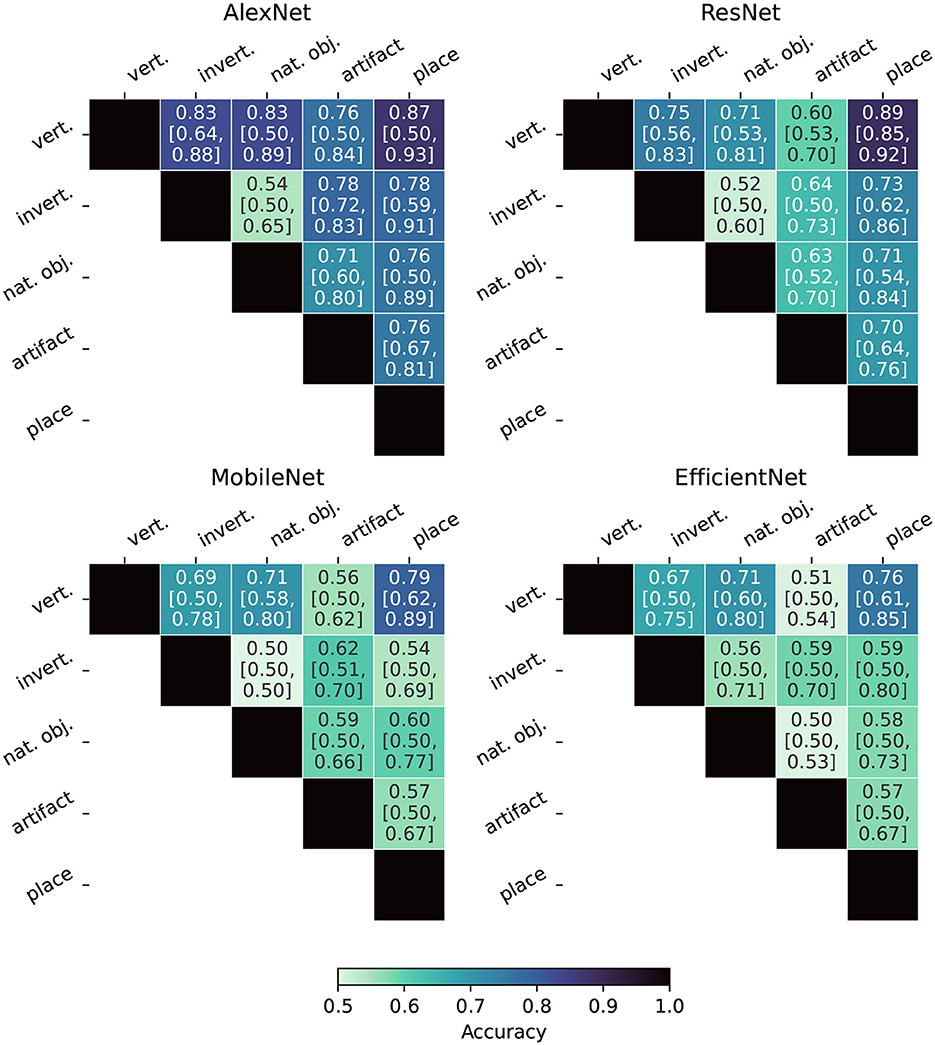
Figure 9. Fiedler partitioning accuracy for fMRI subjects. Partitioning is performed on the RDM of the LHFG3 ROI. Accuracy is reported along with Bonferroni-corrected BCa bootstrap intervals (CIs) (accuracy [lower CI bound, upper CI bound]). The mean subject demonstrates both higher accuracy and statistical significance when compared to each of the three complete participants.
Bonferroni-corrected BCa confidence intervals are also reported in Figures 8, 9. Because Fiedler partitioning is applied against supercategory pairs, the null hypothesis corresponds to an accuracy of 0.50. An interesting phenomenon that can be seen in the results is that even if the Fiedler partitioning achieves high levels of accuracy, we may not be able to reject the null hypothesis due to the lower bootstrap confidence interval being at or near 0.50. This can be seen both in the fMRI subject results and the ANN results. Analysis of the bootstrap sample distribution shows this is sometimes the result of a bimodal distribution of results, where a small cluster of failed partitions will be present at or near 0.5 accuracy, with the rest scoring much higher. This effect is likely due to a combination of label noise in the BOLD5000 dataset and how the Fiedler vector is calculated.
Label noise arises when the two ImageNet images selected for each synset category contains more than just the intended subject, or when other factors, such as the crop of the image that was used for presentation obscure or make the intended subject unclear. An example of this is the fact that many of the stimuli images in the BOLD5000 which fall under the supercategory “artifact” depict people holding the object with a full human face visible in the image. The presence of human faces in images that are meant to depict inanimate objects causes significant unintended brain activation in areas such as the FFA (Kanwisher et al., 1997).
The second factor contributing to this phenomenon is the fact that Fiedler partitioning is not an operation that is performed on individual stimuli, but on the RDM as a whole. The Fiedler vector is calculated as the second smallest eigenvector of the Laplacian of the RDM. Therefore, if a bootstrap sample is composed of a set of images with sufficient label noise, the Fiedler vector will partition the entire RDM orthogonal to the intended supercategories. An example of this was found with the above “artifact” supercategory example. When applied solely to the stimuli images from the “artifact” supercategory in an unsupervised manner, the Fiedler partitioning algorithm spontaneously partitions the images into a group that contains humans in the image and a group that contains just inanimate objects in the frame.
Overall, our findings indicate that Fiedler partitioning effectively identifies supercategory clusters in the RDMs of human fMRI subjects and ANNs. Similar to our findings with the RDM comparisons, a surprising trend emerges with the ANNs. AlexNet, the oldest of the ANNs, produces a far higher Fiedler partitioning accuracy than the newer models. EfficientNet-b0, in particular, does not produce results significantly above noise for most of the supercategory pairings.
5 Discussion and future work
ANNs have long suffered from decreased performance as a result of their sensitivity to random noise and adversarial attacks. Recent works have shown that fine-tuning a network representation to align with a biological standard fortifies networks against both noise and adversarial corruptions of images (Blanchard et al., 2019; Li et al., 2019). However, exploration of these ideas has been limited by the unavailability of public datasets: prior works have relied on private datasets (Li et al., 2019) or datasets with a limited number of stimuli (Blanchard et al., 2019). The BOLD5000 dataset has always been a promising resource for investigating just this, but the data was not packaged for use by researchers without a strong neuroscience background. Here, we eliminate this barrier—our curation and investigations of the BOLD5000 data will now enable the broader community to explore the viability of biological representation in networks.
An important result of our analysis is that recent, more advanced, neural networks such as EfficientNet (Tan and Le, 2019) have lower neuro-similarity to human fMRI responses than the much older and simpler AlexNet, despite also performing much better on the standard ImageNet Large Scale Visual Recognition Challenge (ILSVRC).
The discovery that ANNs are diverging from their biological inspiration is not, in and of itself, surprising and is supported by other recent research (Gomez-Villa et al., 2020; Kumar et al., 2022). It does emphasize the fundamental question of whether or not neuro-similarity is an asset, a hindrance, or simply a non-factor. Are these newer models performing better on an, admittedly artificial, metric because of their neuro-dissimilarity or in spite of it? Humans are not susceptible to the same adversarial attacks that ANNs have been shown to be susceptible, so it's possible this divergence in the geometry of ANN embedding spaces from their human counterparts is opening up new avenues of attack.
To put a finer point on it, are more advanced models achieving higher accuracy by focusing on minutia instead of the complete composition, e.g., the features being extracted from an image of one of ImageNet's many dog breed classes focused on things like fur texture and color as a way to correctly classify the breed, the source of the image classification, or on the fact that the image depicts a four-legged creature with two eyes and other mammalian features? Having a fragmented embedding space that emphasizes minutia is likely to make a model more susceptible to adversarial attacks. To use the example above, a model that has been overfit to the point where it only focuses on fur pattern features to identify something as a dog, could be tricked into misidentifying a common artifact such as a box, by covering it with a fake fur texture or image.
We expect images containing similar features to elicit activations that are closer together within the embedding space while dissimilar activations should exist further apart—Goh et al. (2021) investigated the presence of this phenomenon, finding certain neuron groups in CLIP activated or deactivated in response to similar concepts. Fiedler partitioning of an RDM should be able to exploit this clustering of like embeddings to get us in the ballpark of a reasonable classification by selecting an appropriate class category regardless of whether or not there is a strong correlation between the ANN and the biological benchmark. By demonstrating that this works well for a model like AlexNet, but not for a model like EfficientNet, we imply that these more advanced models are not creating the expected clusters within their embedding space. This leads to the question of how these new ANNs are actually structuring their feature space or whether they are extracting a similar set of features at all. Our work shows that ANNs trained for classification performance are evolving internal embedding space geometries more dissimilar from the human vision system and that these embedding spaces lack a geometry that clusters like image subjects together. We can either conclude that state-of-the-art ANNs are creating a novel way to learn and store image feature representations, or we must conclude that embedding spaces are becoming more disjoint because of the singular push to maximize classification accuracy.
Since learned representations like (Goh et al., 2021) do seem to demonstrate this phenomenon with CLIP embeddings, and since CLIP embeddings match or surpass the performance of the models we evaluate (Radford et al., 2021), it seems likely that the biological ideal does correspond with robustness. However, a full investigation of the viability of the biological benchmark is beyond the scope of this work—and likely beyond the scope of any singular work. Instead, a wealth of future research is needed to tease out the intricacies of what kinds of representations correspond with robustness. The most impactful outcome of this work is the facilitation of these future research projects via a shared, publicly available dataset that allows researchers and practitioners to scrutinize the evidence for a biologically grounded representation, and investigate alternatives.
Finally, the curation of this data also facilitates additional uses of the data: modeling neural processes and creating new biologically consistent architectures. Neural networks are the premier means for modeling neural data. However, it has also been shown that current architectures have largely plateaued (Storrs et al., 2021) and that all networks are equally predictive of the human inferior temporal cortex. This is problematic because these models still fail to predict certain properties of visual processing (Storrs et al., 2021). Our data could facilitate the creation of neural network designs that are biologically grounded. Previously, work has shown that networks deliberately modeled on neural phenomena exhibit higher biological consistency than traditional CNNs (Blanchard, 2019), which corresponds with higher performance. However, even this work would vastly benefit from expanding methods for comparing with biological benchmarks via novel techniques like extending RDMs into Laplacian matrices (Jamil et al., 2023).
6 Conclusion
Here, we establish a new biological benchmark for embedded representations. Our experiments on our benchmark establish the viability of utilizing this data to enhance the robustness of learned representations to inputs like adversarial attacks. Specifically, our experiments with Fiedler partitioning showcase how biologically grounded representations facilitate interwoven separability and clustering of data. As part of this work, we release our curated data and a framework to facilitate further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.5061/dryad.wpzgmsbtr.
Author contributions
WP: Conceptualization, Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. KS: Investigation, Writing – original draft, Visualization. HJ: Investigation, Methodology, Software, Visualization, Writing – original draft. NC: Software, Visualization, Writing – original draft. NB: Conceptualization, Investigation, Project administration, Software, Supervision, Writing – original draft. MK: Supervision, Writing – review & editing. CP: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported by Defense Advanced Research Projects Agency (DARPA) HR00112290074.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akbarinia, A., Morgenstern, Y., and Gegenfurtner, K. R. (2023). Contrast sensitivity function in deep networks. Neural Netw. 164, 228–244. doi: 10.1016/j.neunet.2023.04.032
Aliko, S., Huang, J., Gheorghiu, F., Meliss, S., and Skipper, J. I. (2020). A naturalistic neuroimaging database for understanding the brain using ecological stimuli. Sci. Data 7, 347. doi: 10.1038/s41597-020-00680-2
Allen, E. J., St-Yves, G., Wu, Y., Breedlove, J. L., Prince, J. S., Dowdle, L. T., et al. (2022). A massive 7T fMRI dataset to bridge cognitive neuroscience and artificial intelligence. Nat. Neurosci. 25, 116–126. doi: 10.1038/s41593-021-00962-x
Bashivan, P., Tensen, M., and Dicarlo, J. (2019). “Teacher guided architecture search,” in 2019 IEEE/CVF International Conference on Computer Vision (ICCV) (Seoul), 5319–5328. doi: 10.1109/ICCV.2019.00542
Blanchard, N., Kinnison, J., Richard Webster, B., Bashivan, P., and Scheirer, W. J. (2019). “A neurobiological evaluation metric for neural network model search,” in 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (Long Beach, CA), 5399–5408. doi: 10.1109/CVPR.2019.00555
Blanchard, N. T. (2019). Quantifying internal representation for use in model search (Ph.D. thesis). University of Notre Dame, Notre Dame, IN, United States.
Chang, N., Pyles, J., Prince, J., Tarr, M., and Aminoff, E. (2021). BOLD5000 Release 2.0. Carnegie Mellon University. doi: 10.1184/R1/14456124. Available online at: https://kilthub.cmu.edu/articles/dataset/BOLD5000_Release_2_0/14456124/2
Chang, N., Pyles, J. A., Marcus, A., Gupta, A., Tarr, M. J., and Aminoff, E. M. (2019). BOLD5000, a public fMRI dataset while viewing 5000 visual images. Sci. Data 6, 49. doi: 10.1038/s41597-019-0052-3
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei, L. (2009). “ImageNet: a large-scale hierarchical image database,” in 2009 IEEE Conference on Computer Vision and Pattern Recognition (Miami, FL), 248–255. doi: 10.1109/CVPR.2009.5206848
Elsken, T., Metzen, J. H., and Hutter, F. (2018). Neural architecture search: a survey. J. Mach. Learn. Res. 20, 1997–2017. doi: 10.48550/arXiv.1808.05377
Esteban, O., Markiewicz, C. J., Blair, R. W., Moodie, C. A., Isik, A. I., Erramuzpe, A., et al. (2019). fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116. doi: 10.1038/s41592-018-0235-4
Fiedler, M. (1975). A property of eigenvectors of nonnegative symmetric matrices and its application to graph theory. Czechoslovak Math. J. 25, 619–633.
Geirhos, R., Temme, C. R. M., Rauber, J., Schütt, H. H., Bethge, M., and Wichmann, F. A. (2018). “Generalisation in humans and deep neural networks,” in Advances in Neural Information Processing Systems, eds S. Bengio, H. Wallach, H. Larochelle, K. Grauman, N. Cesa-Bianchi, and R. Garnett (Curran Associates, Inc.).
Goh, G., Cammarata, N., Voss, C., Carter, S., Petrov, M., Schubert, L., et al. (2021). Multimodal neurons in artificial neural networks. Distill. 6, e30. doi: 10.23915/distill.00030
Gomez-Villa, A., Martín, A., Vazquez-Corral, J., Bertalmío, M., and Malo, J. (2020). Color illusions also deceive CNNs for low-level vision tasks: analysis and implications. Vision Res. 176, 156–174. doi: 10.1016/j.visres.2020.07.010
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Las Vegas, NV: IEEE), 770–778.
Hernández-Cámara, P., Vila-Tomás, J., Laparra, V., and Malo, J. (2023). Neural networks with divisive normalization for image segmentation. Pattern Recogn. Lett. 173, 64–71. doi: 10.1016/j.patrec.2023.07.017
Hong, H., Yamins, D. L. K., Majaj, N. J., and DiCarlo, J. J. (2016). Explicit information for category-orthogonal object properties increases along the ventral stream. Nat. Neurosci. 19, 613–622. doi: 10.1038/nn.4247
Hsu, C.-H., Chang, S.-H., Liang, J.-H., Chou, H.-P., Liu, C.-H., Chang, S.-C., et al. (2018). MONAS: multi-objective neural architecture search using reinforcement learning. arXiv [Preprint]. arXiv:1806.10332. doi: 10.48550/arXiv.1806.10332
Jacob, G., Pramod, R. T., Katti, H., and Arun, S. P. (2021). Qualitative similarities and differences in visual object representations between brains and deep networks. Nat. Commun. 12, 1872. doi: 10.1038/s41467-021-22078-3
Jamil, H., Liu, Y., Caglar, T., Cole, C., Blanchard, N., Peterson, C., et al. (2023). “Hamming similarity and graph Laplacians for class partitioning and adversarial image detection,” in 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) (Vancouver, BC), 590–599. doi: 10.1109/CVPRW59228.2023.00066
Kanwisher, N., McDermott, J., and Chun, M. M. (1997). The fusiform face area: a module in human extrastriate cortex specialized for face perception. J. Neurosci. 17, 4302–4311.
Kheradpisheh, S. R., Ghodrati, M., Ganjtabesh, M., and Masquelier, T. (2016). Deep networks can resemble human feed-forward vision in invariant object recognition. Sci. Rep. 6, 32672. doi: 10.1038/srep32672
Kingma, D. P., and Welling, M. (2013). Auto-encoding variational Bayes. arXiv [Preprint]. arXiv:1312.6114. doi: 10.48550/arXiv.1312.6114
Kriegeskorte, N., Mur, M., and Bandettini, P. (2008). Representational similarity analysis - connecting the branches of systems neuroscience. Front. Syst. Neurosci. 2, 4. doi: 10.3389/neuro.06.004.2008
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). “ImageNet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems (Curran Associates, Inc.)
Kumar, M., Houlsby, N., Kalchbrenner, N., and Cubuk, E. D. (2022). Do better ImageNet classifiers assess perceptual similarity better? arXiv [Preprint]. arXiv:2203.04946. doi: 10.48550/arXiv.2203.04946
Li, Q., Gomez-Villa, A., Bertalmío, M., and Malo, J. (2022). Contrast sensitivity functions in autoencoders. J. Vision 22, 8. doi: 10.1167/jov.22.6.8
Li, Z., Brendel, W., Walker, E., Cobos, E., Muhammad, T., Reimer, J., et al. (2019). “Learning from brains how to regularize machines,” in Advances in Neural Information Processing Systems, Vol. 32 (Curran Associates, Inc.).
Lin, T.-Y., Maire, M., Belongie, S., Hays, J., Perona, P., Ramanan, D., et al. (2014). “Microsoft COCO: common objects in context,” in Computer Vision – ECCV 2014, eds D. Fleet, T. Pajdla, B. Schiele, and T. Tuytelaars (Cham: Springer International Publishing), 740–755.
Liu, C., Zoph, B., Neumann, M., Shlens, J., Hua, W., Li, L.-J., et al. (2018). “Progressive neural architecture search,” in Proceedings of the European Conference on Computer Vision (ECCV), 19–34.
Mehrer, J., Spoerer, C. J., Jones, E. C., Kriegeskorte, N., and Kietzmann, T. C. (2021). An ecologically motivated image dataset for deep learning yields better models of human vision. Proc. Natl. Acad. Sci. U.S.A. 118, e2011417118. doi: 10.1073/pnas.2011417118
Miller, M., Chung, S., and Miller, K. D. (2021). “Divisive feature normalization improves image recognition performance in AlexNet,” in International Conference on Learning Representations.
Nili, H., Wingfield, C., Walther, A., Su, L., Marslen-Wilson, W., and Kriegeskorte, N. (2014). A toolbox for representational similarity analysis. PLoS Comput. Biol. 10, e1003553. doi: 10.1371/journal.pcbi.1003553
Pham, H., Guan, M., Zoph, B., Le, Q., and Dean, J. (2018). “Efficient neural architecture search via parameters sharing,” in Proceedings of the 35th International Conference on Machine Learning (PMLR), 4095–4104.
Prince, J. S., Charest, I., Kurzawski, J. W., Pyles, J. A., Tarr, M. J., and Kay, K. N. (2022). GLMsingle: a toolbox for improving single-trial fMRI response estimates. bioRxiv [Preprint]. doi: 10.1101/2022.01.31.478431
Radford, A., Kim, J. W., Hallacy, C., Ramesh, A., Goh, G., Agarwal, S., et al. (2021). “Learning transferable visual models from natural language supervision,” in Proceedings of the 38th International Conference on Machine Learning (PMLR), 8748–8763.
RichardWebster, B., Anthony, S. E., and Scheirer, W. J. (2019). PsyPhy: a psychophysics driven evaluation framework for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 41, 2280–2286. doi: 10.1109/TPAMI.2018.2849989
RichardWebster, B., Kwon, S. Y., Clarizio, C., Anthony, S. E., and Scheirer, W. J. (2018). “Visual psychophysics for making face recognition algorithms more explainable,” in Proceedings of the European Conference on Computer Vision (ECCV), 252–270.
Roads, B. D., and Love, B. C. (2021). “Enriching ImageNet with human similarity judgments and psychological embeddings,” in 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (Nashville, TN: IEEE), 3546–3556.
Rosenke, M., van Hoof, R., van den Hurk, J., Grill-Spector, K., and Goebel, R. (2021). A probabilistic functional atlas of human occipito-temporal visual cortex. Cereb. Cortex 31, 603–619. doi: 10.1093/cercor/bhaa246
Rosenke, M., Weiner, K. S., Barnett, M. A., Zilles, K., Amunts, K., Goebel, R., et al. (2018). A cross-validated cytoarchitectonic atlas of the human ventral visual stream. Neuroimage 170, 257–270. doi: 10.1016/j.neuroimage.2017.02.040
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and Chen, L.-C. (2018). “MobileNetV2: inverted residuals and linear bottlenecks,” in 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (Salt Lake City, UT: IEEE), 4510–4520.
Schrimpf, M., Kubilius, J., Hong, H., Majaj, N. J., Rajalingham, R., Issa, E. B., et al. (2018). Brain-score: which artificial neural network for object recognition is most brain-like? BioRxiv [Preprint]. 407007. doi: 10.1101/407007
Sexton, N. J., and Love, B. C. (2022). Reassessing hierarchical correspondences between brain and deep networks through direct interface. Sci. Adv. 8, eabm2219. doi: 10.1126/sciadv.abm2219
Smith, L. B., and Slone, L. K. (2017). A developmental approach to machine learning? Front. Psychol. 8, 2124. doi: 10.3389/fpsyg.2017.02124
Storrs, K. R., Kietzmann, T. C., Walther, A., Mehrer, J., and Kriegeskorte, N. (2021). Diverse deep neural networks all predict human inferior temporal cortex well, after training and fitting. J. Cogn. Neurosci. 33, 2044–2064. doi: 10.1162/jocn_a_01755
Tan, M., and Le, Q. (2019). “EfficientNet: rethinking model scaling for convolutional neural networks,” in Proceedings of the 36th International Conference on Machine Learning, Proceedings of Machine Learning Research, eds K. Chaudhuri and R. Salakhutdinov (PMLR), 6105–6114.
Veerabadran, V., Raina, R., and de Sa, V. R. (2021). “Bio-inspired learnable divisive normalization for ANNs,” in SVRHM 2021 Workshop@NeurIPS.
Xiao, J., Hays, J., Ehinger, K. A., Oliva, A., and Torralba, A. (2010). “SUN database: large-scale scene recognition from abbey to zoo,” in 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (San Francisco, CA), 3485–3492. doi: 10.1109/CVPR.2010.5539970
Yamins, D. L., Hong, H., Cadieu, C., and DiCarlo, J. J. (2013). “Hierarchical modular optimization of convolutional networks achieves representations similar to macaque IT and human ventral stream,” in Advances in Neural Information Processing Systems, eds C. J. Burges, L. Bottou, M. Welling, Z. Ghahramani, and K. Q. Weinberger (Curran Associates, Inc.).
Yamins, D. L. K., and DiCarlo, J. J. (2016). Using goal-driven deep learning models to understand sensory cortex. Nat. Neurosci. 19, 356–365. doi: 10.1038/nn.4244
Yamins, D. L. K., Hong, H., Cadieu, C. F., Solomon, E. A., Seibert, D., and DiCarlo, J. J. (2014). Performance-optimized hierarchical models predict neural responses in higher visual cortex. Proc. Natl. Acad. Sci. U.S.A. 111, 8619–8624. doi: 10.1073/pnas.1403112111
Keywords: brain-inspired neural networks, computational neuroscience, deep learning, geometric analysis, object recognition, functional MRI, Fiedler partitioning, human visual system
Citation: Pickard W, Sikes K, Jamil H, Chaffee N, Blanchard N, Kirby M and Peterson C (2023) Exploring fMRI RDMs: enhancing model robustness through neurobiological data. Front. Comput. Sci. 5:1275026. doi: 10.3389/fcomp.2023.1275026
Received: 09 August 2023; Accepted: 20 November 2023;
Published: 19 December 2023.
Edited by:
Yunye Gong, SRI International, United StatesReviewed by:
Jesús Malo, University of Valencia, SpainPeter C. Doerschuk, Cornell University, United States
Copyright © 2023 Pickard, Sikes, Jamil, Chaffee, Blanchard, Kirby and Peterson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathaniel Blanchard, bmF0aGFuaWVsLmJsYW5jaGFyZEBjb2xvc3RhdGUuZWR1
 William Pickard
William Pickard Kelsey Sikes1
Kelsey Sikes1 Nathaniel Blanchard
Nathaniel Blanchard Michael Kirby
Michael Kirby Chris Peterson
Chris Peterson