- Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome, Italy
The problem of transforming Real World Data into Real World Evidence is becoming increasingly important in the frameworks of Digital Health and Personalized Medicine, especially with the availability of modern algorithms of Artificial Intelligence high computing power, and large storage facilities.Even where Real World Data are well maintained in a hospital data warehouse and are made available for research purposes, many aspects need to be addressed to build an effective architecture enabling researchers to extract knowledge from data.We describe the first year of activity at Gemelli Generator RWD, the challenges we faced and the solutions we put in place to build a Real World Data laboratory at the service of patients and health researchers. Three classes of services are available today: retrospective analysis of existing patient data for descriptive and clustering purposes; automation of knowledge extraction, ranging from text mining, patient selection for trials, to generation of new research hypotheses; and finally the creation of Decision Support Systems, with the integration of data from the hospital data warehouse, apps, and Internet of Things.
1 Introduction–Real World Evidence and Digital Health
The huge availability of data from different generic and special purpose information technology (IT) systems in today’s healthcare process is profoundly impacting knowledge management for medical specialists, by providing new insight and understanding in all diagnostic and prognostic domains. This will progressively help reshaping the care process to design personalized therapies and improve quality of care.
By leveraging the continuous flow of data produced during routine clinical practice–what is now generically defined as the generation of Real World Evidence - researchers and medical staff have at reach new and better ways to take decisions. A large amount of information from several patients’ clinical histories can be readily available, providing the base to identify disease and care patterns, linking biomarkers to outcomes, and creating reference frameworks where a physician can relate the health status of a patient with other patients’ histories, use this newly-acquired knowledge to design fit-for-purpose therapies and improve quality of care through more personalized treatments. Apart from the conventional source of data, great attention has been indeed focused on the collection of data from real life situations, producing a prominent change in the process of reviewing and interpreting data (Lewis et al., 2017 et al.).
The emergence of a new era in personalized and translational medicine has been brought about by the verification of new data sets through the accessibility of omics data, ranging from health records, lab reports, bio-images to more innovative data harvesting approaches (Cesario et al., 2021a).
Shifting the perspective towards the data side, we observe that the definition and common understanding about Real World Data (RWD) in healthcare has been so far quite controversial (Hiramatsu et al., 2021).
This was recently further analyzed by the FDA, who extensively defined health RWD as the data acquired from medical records, surveys, mobile applications, registries and administrative or insurance related databases (https://www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence). Even the optimal use and exploitation of RWD, especially in the context of the European union, is a closely discussed topic (Gill et al., 2016).
As a result, Evidence based Medicine which uses randomized Clinical trials to answer a given research question could potentially be supplemented by the new RWD based translational approach.
In truth, despite the similarities, these two methods differ from each other with regards to the quality of data, mechanisms of data collection as well as data interpretation and actionability. The incorporation of RWD data management AI based models is necessary and will therefore serve as an improved and more ideal option in supporting decision making in personalized, translational and multi-omics predictive systems. This approach will enhance the collection and processing of substantial amounts of data which were previously inaccessible, thereby giving clinicians the ability to decipher the relationship between different, uncorrelated variables to improve patient care and boost their decisional capacity (Abernethy et al., 2010).
The need for Clinical data scientists will soon become compelling for the investigation of novel AI-based research applications, and the analysis of medical data. We are in the middle of the digital transformation in healthcare, a huge, challenging transition. The creation of integrated frameworks–combination of technology, process, skills and organization - transforming data and information into actionable knowledge is not unique of healthcare: It is at the core of digital transformation for any industry and knowledge domain.
Yet, healthcare is probably one of the most challenging domains for digital transformation, due to general and specific sources of complexity, which can be summarized as follows:
• Data integration and transformation: healthcare systems constantly generate data of quite different nature, with significant technical challenges (implementing efficient extraction and transformation process); quality related issues (ensuring data have a well understood meaning, and introducing effective validation process); and the ambition to render this information effective and understandable for the end user (with proper integration, visualization and fruition methods)
• Skill and organization: to ensure that technical solutions are directed to solve the right problems, and that these are used by non-IT experts (the medical staff) in the most effective way, transformation initiatives must leverage the right mix of skills with a user-oriented, incremental approach
• Ethical and privacy challenges: protection of patient’s privacy and strong, unquestionable ethical criteria, with constant focus of benefit for patients and care, must be continuously validated and tracked, leveraging independent bodies and experts, with strict adherence to regulatory norms and robust practices engrained in daily execution from all actors
We will share our experience in designing and implementing an approach trying to address all these aspects in a coordinated way. The result is the creation of an execution framework where a large hospital, with high volumes of care and extensive pathologies treatment, can leverage digital transformation, analytics and AI to progress both in clinical research and practice.
This project is being executed at Fondazione Policlinico Universitario Agostino Gemelli, one of the largest Italian hospitals with care and clinical research missions, recently ranked as 45th clinical center at European level. Under the name of GENERATOR REAL WORLD DATA Facility, this initiative is supporting many research groups, and provides data analysis and modelling expertise, structured methods for data governance and processing, project design and implementation, management of ethical and privacy aspects.
This work is organized as follows:
• Firstly, we analyze some key challenges and how GENERATOR RWD is addressing them:
• Data integration and management, definition of replicable approach to organize disease-specific knowledge domain and research projects.
• Focus on privacy-preserving methods and ethical aspects.
Skills and organization development, in terms of education, project organization and role mapping.
• Then we show how analysis and models are developed starting from data organized for the specific study/pathology domain; we go through some use cases to describe the execution framework and assets exploited: AI and Machine Learning; data visualization and decision support tools; digital platforms for patient journey support.
• Subsequently, we provide an overview of the value creation patterns which are actively pursued through GENERATOR (such as research projects; transnational consortia initiatives; industrial co-development) and how effective data governance methods can be implemented to be consistent with privacy goals in open collaboration scenarios.
• Finally, we share our perspective on future project steps and areas to be addressed and strengthened, given that the project is evolving day by day.
2 Part 1: Key Challenges and Enabling Factors
2.1 Data Integration and Data Management
In a modern hospital, digital information comes from vastly different data sources, often tailored to the needs of the specific procedure originating them. Typical examples are:
• Structured or semi-structured data sources with administrative information (admission, discharge, transfers between wards, billing codes ).
• Structured or semi-structured data sources (laboratory exams results, vital parameters measurements ).
• Unstructured data sources (clinical notes and observations collected during the hospitalization and at the time of discharge).
• Imaging data typically stored on RIS-PACS (Radiology Information System/Picture Archiving and Communication System) in Digital Imaging and Communications in Medicine (DICOM) format.
Data-storing and the data-exchange processes involve a deep understanding of communication standards such as Clinical Document Architecture (CDA), Health Level Seven (HL7), and Fast Healthcare Interoperability Resources (FHIRE) to name the most relevant. For these reasons, a data integration layer between the data sources and the Research and Developement (R&D) personnel is particularly important to extract valuable information and create added value associated to the mere data collection and data exchange processes.
This is compounded with the need to implement and integrate innovative approaches to provide clinicians and researchers with Real World information to gain a more complete picture and understanding of the patient by capturing their unique characteristics (Ahmed et al., 2020; Cesario et al., 2021b). To fully implement and benefit from this innovative approach, integration of mixed and heterogeneous data from several domains and contexts should be addressed, including personal factors, such as education, lifestyle, physical functions, environmental and social elements, and individual preferences (Cesario et al., 2021c). Thus, in this perspective, electronic health records (EHR) need to be augmented by assorted data sources, among which tools like questionnaires, wearable devices and mobile applications, which collect Patient Reported Outcomes (PROs), and Patient Reported Experiences (PREs), innovative and promising RWD sources. Even if the technological progress of the last decades gave a significant boost to this approach, the collection, integration and analysis of Real World Data (RWD) from varied sources in an effective way is still one of the biggest challenges that Personalized Medicine and, specifically, Gemelli Generator is addressing.
This novel approach is a great step forward in patient management, especially those with overly complex situations; multimorbidity, polypharmacotherapy, frailty and chronic disease (Cesario et al., 2021c).
Information science has defined a paradigm to describe and manage this complexity: Big Data.
2.2 The 4 Vs of Big Data
The concept of Big Data is based on four fundamental metrics, namely Volume, Velocity, Variety, Veracity.
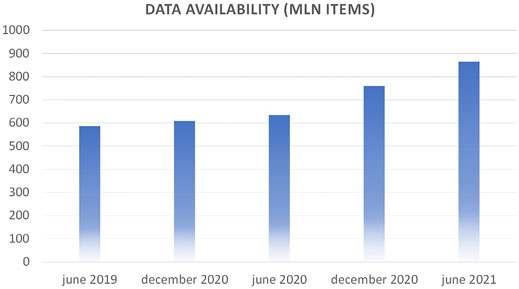
Volume is defined as the raw amount of data items that are present in the Data Warehouse (DWH) at a given time. At the time of writing, Gemelli Generator can count on over 800 million atomic data items related to over 15 years of patient history, lab tests, physician visits at Policlinico Gemelli. Figure 1 shows the progression in the last years.
Velocity is related to the amount of new data items that are made available in a given time interval and to the frequency of updates of the models learnt from data. Gemelli Generator data storage and processing take advantage of a dedicated cluster, with high performance, automated backup procedures and high availability.
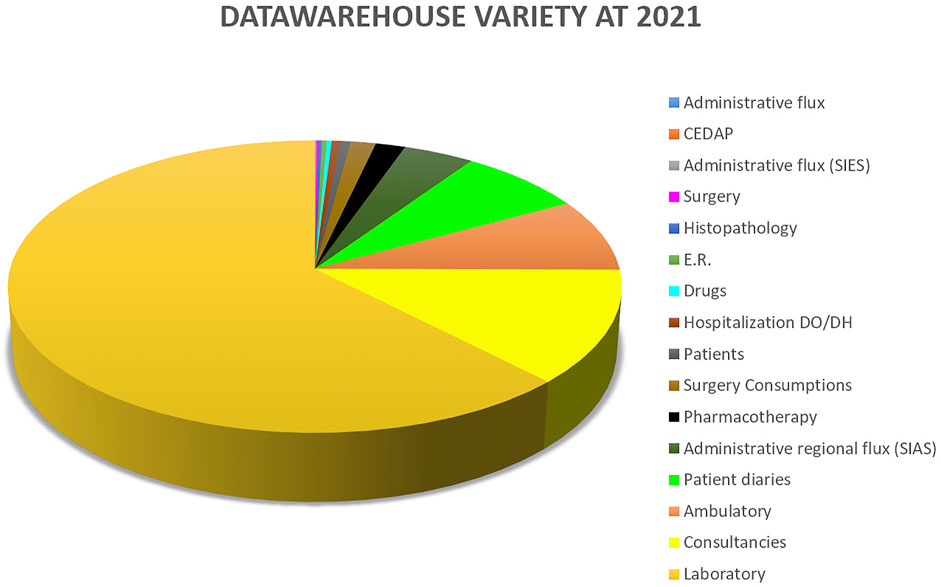
Variety measures the diversity of data types that are seen in the data warehouse. Figure 2 shows the distribution of data sources available to Gemelli Generator.
Integration with data from other sources, like the world of -omics (e.g.: Radiomics: information from automated analysis of diagnostic images such as CT-Scans, Magnetic Resonance Imaging (MRI’s), Positron Emission Tomography (PT-scans), radiograms) enhances this parameter and gives medical professionals the opportunity to design elaborate, complex studies relying on many different aspects of the patient.
Veracity concerns the quality of the data which is of paramount importance in any study. Two key issues must be addressed. First, semantic consistency must be preserved: when we choose a variable to be included in a study, we want to be sure that the meaning we are assuming for that covariate is the same with which it was collected. This aspect will be explored in one of the following sections.
Another, not less important, issue arises when some data items are simply wrong. Automated services, called Bots, are in place to intercept patently wrong values, based on the acceptable domain of the variables: nobody is 4 m tall, or lives 200 years; there is no stage 7 in cancer classification, and so on. Finally, missing values are easy to spot, and can be treated with well-known data science techniques, like imputations, where appropriate.
2.3 Real World Data: Implementing Rapid Learning
Initially proposed in 2007 in Health Affairs (Etheredge 2014) the Rapid Learning approach is based on the availability of a large amount of Real World Data: patient data collected by Hospital IT systems during ordinary day by day clinical practice. While the golden standard for clinical research is the canonical clinical trial, there is clinical data that is routinely recorded into the hospital systems, thus presenting no additional effort to the researcher (Lambin et al., 2013). Use of this kind of data is complementary to the clinical trial approach, allowing for different research models to be exploited (Lambin et al., 2015).
For these reasons, implementation of the Rapid Learning paradigm is one of the strategic goals that Gemelli Generator has established for its first development phase. This goal is being pursued through the implementation of a number of services to clinical research, of which some examples are listed below.
The first example is that of retrospective studies based on large numbers of patients aimed at building predictive models and decision support systems. Availability of integration pathways with heterogeneous data sources allows for the creation of models based, for instance, on clinical data, exams, diagnostic imaging, genomics, real-life data, and more. The considerable number of variables involved in these models asks for consequently adequate numbers of patients made available to the study. It is easy to see how this can only be achieved in a Big Data environment, possibly with the collaboration of many territorially distributed hospitals.
Another option offered by Rapid Learning based on Real World Data is that of evaluating changes in clinical guidelines for specific pathologies. This is based on the idea that, after a change in guidelines comes into effect, objective benefits should be observed in the general population of patients to whom the guidelines are applicable (Figure 3). Availability of a large amount of patient data and adequate analysis tools are crucial to this on-field evaluation step.
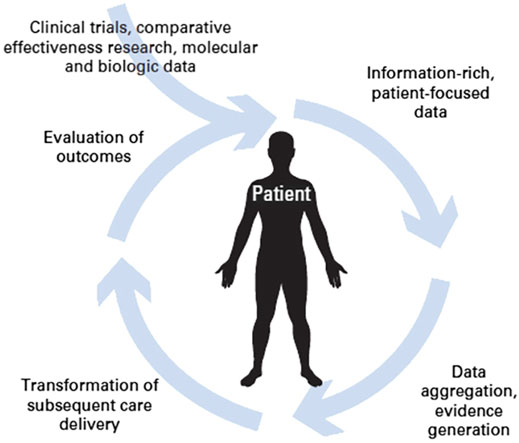
FIGURE 3. The circular path of Rapid Learning (courtesy of P. Lambin et al., 2015).
Generation of new evidence and research hypotheses is another possibility offered by the Rapid Learning approach based on Real World Data. This can be seen as complementary to the evaluation of changes in clinical guidelines. While in that case Real World Data were used to help evaluate the effectiveness of some changes in clinical guidelines, here we see things from the other side: we offer clinical researchers tools for a quick evaluation of a research hypothesis. In this way, intuitions can be tested at a low cost, and if the idea is confirmed, a solid clinical trial can be built around it (Figure 4).
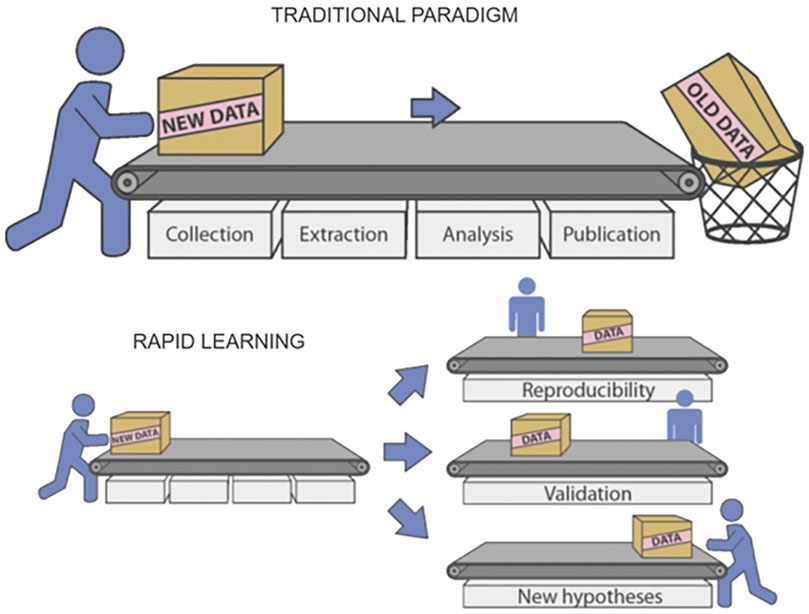
FIGURE 4. Multiple reusability of data and results in Rapid Learning (courtesy of P. Lambin et al., 2015).
For given pathologies, the considerable number of patient data accrued through time in the hospital data warehouse means that many distinct groups of patients are well represented. These groups can be identified by clustering algorithms. Clinicians can study these clusters, understand the differences between them, and finally personalize therapies based on what cluster a given patient belongs to. This can be regarded as an effective approach to personalized medicine: the idea that the choice of a therapy should not be exclusively based on the identification of the pathology, but that individual factors should also enter the equation and lead to the choice of more effective options, when available, for the specific patient.
Availability of a well-organized large amount of clinical data is also key for the selection of patients for clinical trials. Patients to be included in a trial are specified by a set of attributes, which can range from simple ones, like age, to more elaborate, like the results of some given clinical tests. The process of selecting eligible patients from those available is often long and error prone. An automated approach based, when necessary, on the AI-guided interpretation of clinical reports via text mining and Natural Language Processing techniques, makes this process fast and safe, leaving the clinicians only a final check and refinement to be performed on a reduced subset of candidates. It is easy to see the advantages both in terms of quick availability of the results and cost effectiveness of the approach.
Finally, it is important to remember that in a RWD environment like Gemelli Generator, new data are made available by the hospital DWH on a daily basis. This means that new, updated versions of predictive models and decision support systems can be created periodically, giving researchers a dynamic view through time. At the same time, this is an ideal data source for clinical management quality assurance processes, which are cyclical by nature.
Managing and exploiting the huge amount of data made available by the hospital information infrastructure, and implementing the Rapid Learning Paradigm, is the task of the Gemelli Generator infrastructure.
2.4 The Gemelli Generator Infrastructure
The Gemelli GENERATOR infrastructure was designed and implemented, with the main purpose of managing, centralizing and standardizing the variety of data through a data curation process.
The infrastructure is composed of three main parts: the Data Warehouse/data lake layer, the data marts layer and the data analytics layer (Figure 5).
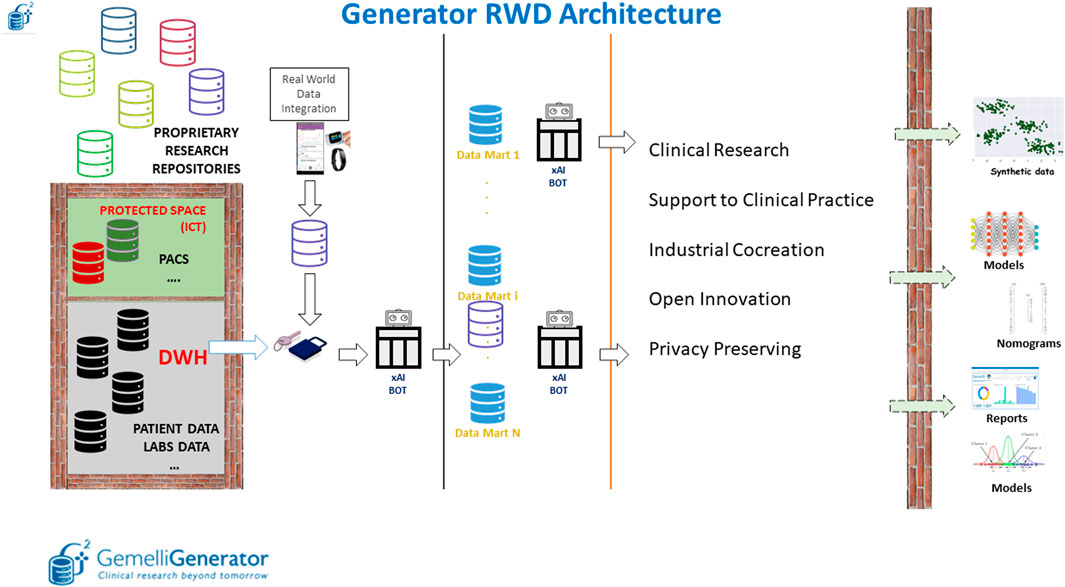
FIGURE 5. Overview of the Generator Real World Data Architecture composed of three main blocks: data lake layer, data integration layer and data analytics layer.
The first layer, within the protected space of our ICT (Information and Communications Technology) department, depicted in Figure 5 as an impassable wall, is characterized by the collection of longitudinal clinical data by producing specific views (such as information on admissions, reports on various clinical areas, information on laboratory analyses, etc.) that are essentially replicas of the archives that are filled daily in the course of clinical practice.
These views and external data sources, which are automatically maintained and updated daily, are made accessible to the Real world data facility in pseudonymized form. Data made available in the data lake, are enriched in the Gemelli Generator infrastructure by external data sources, which may be related to departmental archives (not directly connected to the computerized medical record) and external devices for the collection of Real World data.
This diverse variety of data sources is used as input of the second part of the infrastructure, responsible for data qualification, validation, quality check and creation of thematic data marts.
In our perspective, a data mart is a complete representation of the available data on a specific area of knowledge (examples are COVID-19, breast oncology, chronic lung disease). This type of approach allows the individual researcher to interrogate the specific data mart to carry out several studies without having to invest additional time in data retrieval and curation. Pseudonymized data defined based on a specific ontology, described in more detail in the following section, are queried and integrated through the implementation of specific Extract, Transform and Load (ETL) procedures. The use of such procedures allows the integration of clinical data from different applications through the creation of relational databases. Data quality is checked through the application of ad-hoc Bots.
Finally, the third layer has the main purpose of extracting knowledge and value from the different data marts through the application of machine learning and AI models to develop prognostic and diagnostic predictive tools to be put at the service of the clinician and the patient as clinical decision support systems through user-friendly interfaces.
Our data marts are hosted in SAS databases located on a high availability cluster node. Local SQL Server and SQL Server Lite are also used for specific aims. Computations are performed on dedicated nodes on the cluster, which also features the availability of CUDA processors for parallel machine learning. Software tools include SAS 9.4, SAS VIYA, R, Python 3 with machine learning libraries, C#, Julia 1.6 with machine learning and optimization libraries (mainly Convex.jl). For data visualization tasks, we employ a Python/Django or a Python/Dash infrastructure in order to allow quick plug-and-play dashboard that can be modified on the fly and propagated on a load-balancer for easy scalability. As many of our services are designed as web-services, we are also able to allow clinicians to connect from nearly any device with an internet browser, and as such offer notable flexibility in displaying the results of our projects.
In the following paragraphs we will briefly describe how the second layer interfaces with the first, to obtain an effective standardization of data collection through the definition of automated, high-quality ontologies, using Bots.
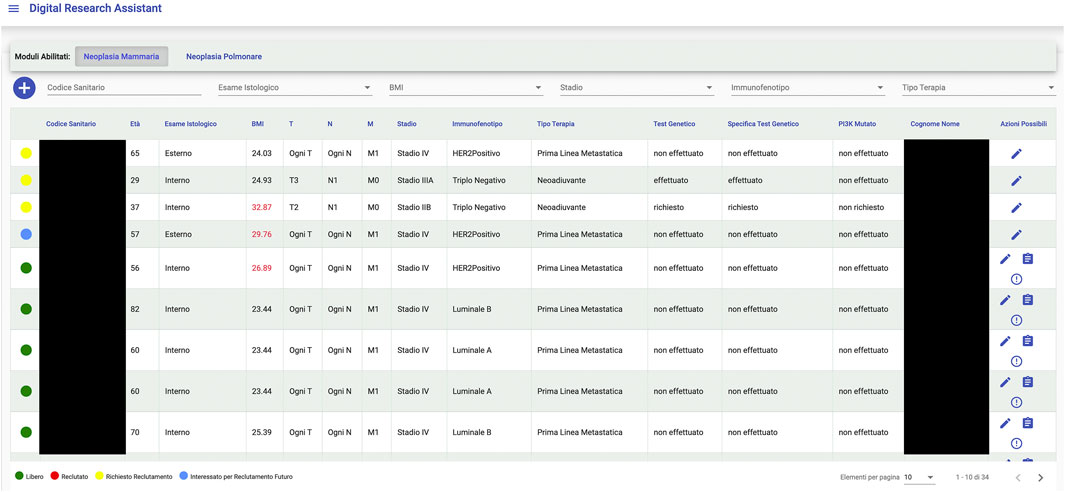
An interesting example of what a Big Data framework like Gemelli Generator can achieve is found in the integration project with a tool, designed and developed by Fondazione Policlinico Universitario Agostino Gemelli (FPUAG) for internal use, called Digital Research Assistant (Figure 6). The tool was conceived to enhance communication among clinicians and achieve optimization of patients’ accrual in clinical trials (Cesario et al., 2021d).
A connection with Generator RWD has been designed, to give clinicians the opportunity to query the Generator RWD data marts and check, for each selected patient, for availability of Real World Data that can add information to what is already present at the Digital Research Assistant (DRA) level and suggest new research hypotheses, based on ready-to-use, quality checked data to the clinical professional.
2.5 Standardized Data Collection
One of the crucial aspects of the second layer of the Generator infrastructure is the definition of a standardized data collection whose main goal is to normalize clinical and Real World data in terms of data structures, interpretation, and coding. This systematization, which makes use of internationally approved codes (such as International Classification disease (ICD), Diagnosis Related Group (DRG) etc.), is designed to overcome the limitations of a complicated technical language. Moreover, this approach effectively overcomes language barriers making international collaborations easier to achieve. For each of the projects using the Generator infrastructure, an Excel template is compiled in which for each of the variables to be included in the study, the clinical area to which it belongs (laboratory analysis, pathological anatomy, radiology, etc.), the name of the field, the definition of each field, the unit of measurement when applicable, data type and validation pattern, the names of the various applications from which to extract the information and, in case of free-text report, the lists of search terms are defined.
The use of a standardized data collection in the creation of large databases can help interoperability among different databases, homogenization of data analysis and facilitate international cooperation. These are typically specific to the area of clinical application (12–16 Lancellotta et al., 2020; Alitto et al., 2017; Chiesa et al., 2020; Marazzi et al., 2021; Tagliaferri et al., 2018) and represent the first phase of any clinical research project using the Generator infrastructure.
Automation of the ETL process: the extraction Bots.
As discussed in the earlier paragraphs, once the ontology has been defined and the ETL procedures for the creation of thematic data marts have been implemented, it is crucial to hook up a whole series of services that guarantee automatic feeding and high data quality. Indeed, to make this process possible and maintainable, it is essential to put in place services that automatically control and securely access the information stored in the data marts called Bots. With the term Bot we define all those automated services that allow to feed in real-time thematic data marts, to convert a textual data source into a structured table, to check the quality and consistency of data and, finally, to apply and update predictive models.
One of the most critical applications of Bots in clinical practice is the process of transforming textual data into structured tables, this workflow linked to the Generator infrastructure is shown in Figure 7. This workflow constitutes the Bot called unstructured data extractor. These tasks are typically used to filter, normalize and clean the report to identify key information to be made structured. In more detail, once the part of the report which we want to analyze has been identified, it is mandatory to divide the text into small parts called tokens. The tokenization process in fact, is the process of breaking text documents apart into those pieces. The second fundamental textual normalization process is stemming, which reduces a word to its root form. The last process of normalizing the text allows the elimination of stop words which are a set of commonly used words in a language. Once the report has been normalized and standardized, the Part of speech tagging is the process of finding and tagging the part of speech of every token in a document. All the above steps are preparatory to the identification of the concepts to be extracted from the reports and to be structured to generate the final table.
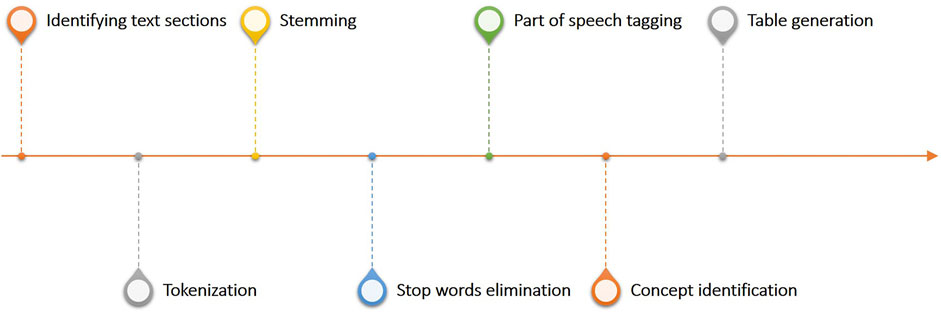
FIGURE 7. Overview of the text analytics workflow linked to the generator infrastructure and used to process and structure free text reports.
Regarding the quality of the data that will make up the various data marts, dedicated Bots called data validators, have been set up according to the type of variable to be checked. In the case of EHR laboratory test data, for example, we can define the acceptable range for each test to identify possible out-of-threshold values that may be considered suspicious and worthy of a dedicated investigation. In the generator infrastructure, in the case of structured clinical data, there are Bots to check for outliers based on acceptability ranges defined with the clinicians and cross-correlation techniques to check the consistency of the variables. In the case of data coming from free text sources, the situation is more complex. For this type of source, we have provided the possibility of using dedicated user interfaces, which allow the clinician to have direct access to each individual report accompanied by the structured information generated by the Bot.
2.7 Data Privacy and Data Protection
The GENERATOR RWD research facility relies on a structured interface and integration layer with the operational IT systems of the hospital, as described in the earlier section. From the personal data protection perspective, this means that a “privacy by design” approach is implemented. As previously noted, ICT hospital unit and GENERATOR RWD have developed pseudonymization procedures that convert patient sensitive information into encrypted data, so that the creation of thematic data marts and studies are always executed using pseudonymized data. If study outcomes are to be linked to clinical process (for care validation and traceability purposes) the medical staff and ICT can define back-tracking processes to verify results on specific patient data, but this is outside of GENERATOR RWD mode of operation. In addition, the upfront pseudonymization procedures are periodically tested and upgraded to strengthen security measures.
The legal framework to define and test that such privacy-by-design approach is effective and compliant with patient data protection is the EU Regulation 2016/679 (General Data Protection Regulation) and corresponding Italian decrees (196/2003 and updated 101/2018). In line with such legal framework, Generator RWD infrastructure has also been validated through a specific impact analysis (DPIA, Data Protection Impact Assessment), jointly performed by the hospital Data Protection Officier (DPO) and ICT organization, to verify technical implementations (hardware and software infrastructure, user management policies, potential security vulnerability) and execution processes (roles and responsibilities, document classification, document management, physical access and policies, … ).
When a new study is designed and the data model/ontology defined, both the GENERATOR RWD project team and the clinical lead put significant focus on fulfilling data minimization principles and data governance. During the feasibility phase the mapping of data sources for input variables is delivered (both in terms of IT systems and originating units). This allows traceability of study contributions and–at study completion–careful validation of the GDPR minimization principle, strictly related to the purpose of processing.
2.8 Establishing a Multidisciplinary Environment
As highlighted in the section on infrastructure, the multidisciplinary aspect is a key part in the development of Gemelli Generator. Data scientists, statistics experts, mathematicians, medical doctors from different fields needed to find a common ground and a shared language. In this perspective, the core structure at Gemelli Generator originates from a research group that grew up within the Oncology PhD path at UCSC (Università Cattolica del Sacro Cuore) side by side with clinicians of Policlinico Gemelli IRCCS. In this way, the senior data scientists at Gemelli Generator, all of which are post-doctoral professionals, have a great aptitude for working on research projects alongside with doctors, and at the same time take advantage of an education path that ranged from research methodologies, to advanced algorithmics, image analysis methods, clinical approaches and medical language.
Conversely, the clinical professionals were able to build a common communication framework with the data scientists, and through it, a way of representing their needs, ideas, intuitions. In this framework, a particular type of medical doctor was defined, denominated “ambassador”, with skills complementary to those of the senior data scientists, and the ability to communicate easily with them. The ambassador is today the first contact point in the interaction between a doctor and the Gemelli Generator facility. From that point on, data scientists work with doctors and become proxies that translate the needs of the clinicians into technical requirements and help in the execution of the research project.
The result we see today is that the doctors need not delve into technicalities, and that clinical concepts are understood and transformed into actions by Gemelli Generator Data Scientists.
3 From Data to Knowledge Discovery: execution framework and use cases.
The execution layer is a part of the Gemelli generator Infrastructure, dedicated to the implementation and application of Statistical Learning and Machine Learning algorithms to data analysis.
Below are some examples of use cases that have already been implemented in the Generator infrastructure. The impact of these preliminary experiences on daily clinical practice and their real value in terms of healthcare management has still to be fully measured and validated in larger test frameworks. Nevertheless, the potentialities expressed in the proves of concept shown below, represent an ideal starting point to set up an advanced AI based environment, with the aim to foster research (section 3.1); achieve better clinical results thanks to efficient clinical decision making support (sections 3.2-.4); ensure adherence to guidelines and good practice (section 3.5); enable innovative biomarkers (section 3.6) and offer overall better services and life experiences to the patients (section 3.7) through the application of robust analysis methodologies (section 3.8) in a robust ethics and privacy preserving structure (section 3.8).
3.1 Patient Selection for Trials
In many branches of medicine, the development of increasingly effective therapeutic strategies is based on clinical trials of new treatments. The introduction in clinical practice of new drugs is subject to the demonstration of efficacy of new molecules in prospective randomized trials. In the absence of therapeutic strategies aimed at healing, as for example is the case in a relevant number of advanced oncological diseases, facilitating the access of patients to clinical trials allows the early use of innovative drugs and at the same time prove their usefulness. In light of these considerations, optimizing the accrual process of patients into clinical trials is an essential goal towards managing the well-being of patients and improving quality of care.
The specific enrollment of patients in clinical trials is supervised by physicians who, based on the patient’s condition and the characteristics of the disease, evaluate the possibility of having the patient access a specific clinical trial currently active in their treatment center. Typically, this process is done manually with a considerable investment of time by the clinical staff who have to select the patient and have to communicate his or her placement within a specific study.
To develop a concrete solution to this problem, we connected a series of Bots to the Generator infrastructure to kickstart a 3-phased protocol based on these three individual components: a pre-processing Bot, a successive data filtration Bot and a selection dashboard.
In the first phase, the pre-configured data-source connections of Generator allowed to quickly access a large number of different databases and collected the contained data that was subsequently pre-processed in order to conform to pre-defined ontologies and guarantee data quality standard.
A second data filtration Bot was then employed to process the now cleaned dataset by creating specific filter-oriented tables based on the inclusion criteria for the specific study, with the intent of tagging any patient not included in the aforementioned criteria. It is worth noting that the filtration Bot incorporates a notable amount of finely tuned text mining procedures, that automate and greatly relieve the burden placed on the healthcare operators tasked with identifying and reading the relevant patient files. Patients are never specifically excluded but only tagged as non-recruitable, in order to allow any end-user clinicians the leeway to make specific clinical decisions based on the circumstances.
The final dashboard then incorporated the outputs of the previous Bots and allowed to interactively view specific information about all suggested patients, and manually exclude or include patients based on the clinical experience of the involved clinicians, greatly reducing the time-requirements needed to create a viable patient-accrual database.
3.2 Avatars: patient clustering as a decision support system.
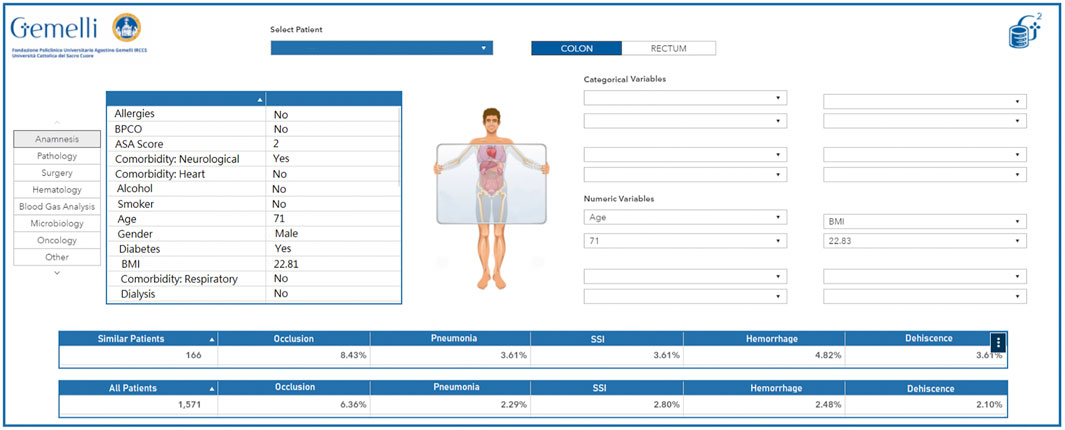
The wealth of information and real world data that are currently collected daily during the treatment pathways followed by patients lend themselves well to be employed in models or representations aimed at generating personalized-healthcare protocols, treatment suggestions and risk analyses. To this point, we aimed to give clinicians a more accurate overview of the characteristics of the patients they see (and saw in the past) in their daily practice, and at the same time allow a more in-depth interaction with the patients by presenting easily readable summaries of treatment options, with their correlated risks, earlier patients with the same characteristics had undergone. Creating digital representations (sometimes called Avatars) of physical patients is seen as a primary need to accomplish this goal. As a proof-of-concept, we started developing a self-updating retrospective patient-clusterization engine for colorectal cancer patients, based on the integration between both structured and unstructured clinical data with available real world data, functioning both as an interactive clinical support system, and as a hypothesis generator through a data exploration and simulation dashboard.
As an initial cohort, all colorectal cancer patients treated in our hospital from 2015 to 2019 (a total of 2,641 patients) were selected for this study. To extract data from our hospital’s operational data-warehouses and production databases, we exploited our Generator exchange infrastructure within our IT-based Real World Data support environment. We then integrated specific algorithms into the data extraction pipeline to sanitize, clean and restructure the information flow, while applying text mining and natural language processing technologies to the unstructured texts. The results of this preprocessing were then refactored through a specifically designed ontology to reveal any duplicate info. This methodology results in the creation of Data Marts which are continuously and automatically updated with new data.
The developed algorithm and its underlying infrastructure classify any newly identified patients based on the available retrospective data, with the possibility of overriding or manually modifying specific characteristics based on the inputs by the clinician using the interface. The resulting information presented through the dynamic interface allows to thoroughly explore any retrospective patient present in the database, to infer the best course of action based on historical data and the experience of the clinician. Additionally, the included interface also allows a more generalized exploration workflow, that may even be decoupled from any specific reference patient, to act as a hypothesis generator for the user, by clustering information based on custom criteria and as such allowing to generate an exploratory analysis of the available wealth of information.
In its simplest form, this tool can be seen as a virtual physician: a colleague with high availability, a perfect memory, and the ability to quickly and accurately retrieve “similar patients” and present their statistics in a graphical interface. Of course, the definition of “similar patient” rests on a suitable study conducted by specialized medical doctors and data scientists together and is implemented technically via the adoption of a correct clustering technology. An example of avatar representation is available in Figure 8. This dashboard allows to quickly visualize the current characteristics of the selected patient as they were extracted by the Generator infrastructure. Additionally, the clinician can compare the current patient (or a dynamic selection of his characteristics) to the full pool of past patients and visualize specific desired outcomes (in this case, a selection of perioperative adverse effects) in relation to the entire pool of patients or a smaller pool of patients deemed analogous to the current patient.
3.3 Decision Support Systems
The development of measures that allow treating physicians to deliver tailored treatment, leads the transformation from a population-based treatment toward a personalized medicine concept with an essential role of Decision Support Systems (DSS): tools that supply clinicians with deeper information, based on elaboration of patient data, to help them have a precise, structured view of every single patient. Prediction tools such as nomograms, either actionable online or offline, have the potential to improve patient outcomes through enhancing the consistency and quality of clinical decision-making, facilitating equitable and cost-effective distribution of finite resources and encouraging behaviour change, thus having a significant impact on care. Our vision is to innovate healthcare (HC) delivery for patients by providing an individualized treatment and care pathway through a clinical decision-support system that will match AI with patient-centred metrics. We see a future in which end-users, including patients, care givers and HC providers, will benefit from a novel and multidisciplinary approach of optimal care delivery for patient.
3.4 Guardian Bots
One of the crucial aspects for the validation and application in clinical practice of predictive models is to identify a fast pipeline in order to accelerate the adoption of these tools by clinicians. The design and implementation of automated Bots that automatically extract, feed and update the data that make-up the underlying data mart is one of the challenges we faced.
As an example, we have developed an automated service to alert clinicians if the probability of an adverse event for a patient is increased over a given threshold, based on continuously updated patient data. In our applications, this service is a predictive model linked to the computerized medical record to provide clinicians with the most up-to-date result possible.
We call this model Guardian Bot.-We deployed a specific use case to test its benefits and challenges in the healthcare process in the area of infectious diseases. An increase of healthcare-associated infections caused by multidrug-resistant organisms (MDRO) is currently observed. One of the main causes of the emergence of a MDRO infection is an overuse of antibiotics. Therefore, saving useless antibiotic treatment is currently a priority from a public health point of view. The evaluation of the risk of having a bloodstream infection will allow both activating faster treatment decisions (when the risk is significantly high) or to save useless resources in terms of diagnostic tests and treatments, also limiting the potential for side effects (when the risk is significantly low).
The aim of our Proof-of-concept called Bacterium was to develop a predictive system for an early detection of Blood Stream Infection (BSI) in hospitalized patients by analysis clinical, laboratory and microbiological variables. By using the Gemelli Generator Real World Data infrastructure previously described, a total number of 8,500 consultancies of 7,500 hospitalized patients from 2016 to 2018 were included in the study. An intensive activity of text mining procedures has been used in the project by analyzing more than 40 million text reports. Outcome of the study is the implementation of prediction models used by clinicians to support their decision in terms of type and duration of antibiotic treatment.
The final deliverable of the Proof-of-Concept (PoC) was the integration of the model in a web-based page accessible directly from the electronical medical record (Figure 9) in order to maximize the use and the transferability in daily practice of the system developed. The implementation of the dashboard allows the clinician to use the model directly at the patient’s bedside without the need to manually enter variables identified as possible predictors of bloodstream infection. By simply entering the patient’s health code, the clinician will see on the screen the probability of blood stream infection risk and the corresponding risk level (red light elevated risk, orange medium risk and green light low risk). Using the interface, the clinician can consult the value of each of the variables considered in the model and can change the value of some of them if a more up-to-date examination value is available.
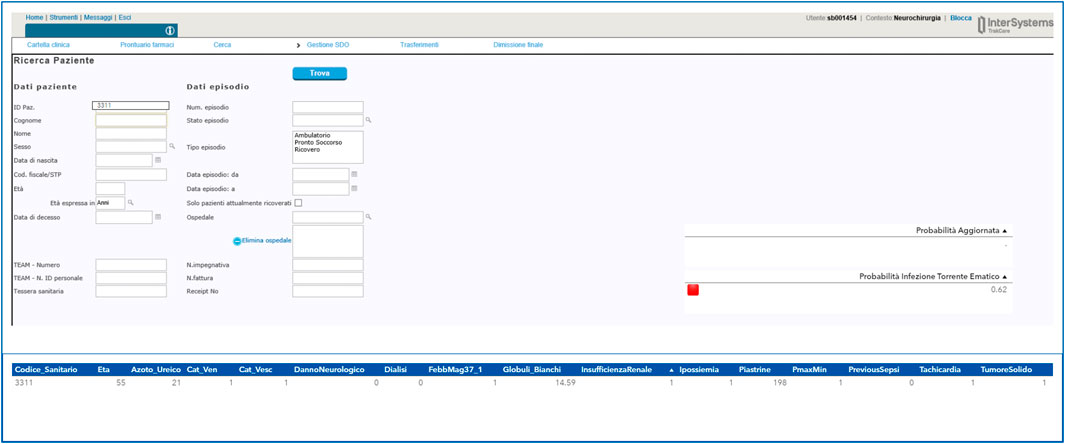
FIGURE 9. Example of a user interface developed for the Guardian Bot linked to the computerized medical record.
3.5 Conformance Checking
In a hospital setting a great amount of process-oriented data are stored. These data typically arise from inpatient admission, transfer between wards and discharge, but they also include therapies and treatment administration, imaging or laboratory exams requested and executed. The technique that is typically used to analyse this data is called process mining. It is a data-driven process analysis technique. It consists of analysing in detail the business processes as they are carried out in everyday reality, with the aim of mapping them, discovering their strengths, weaknesses, and deviations from standard processes. For process mining to work on these data, each event must be marked with a timestamp so that the sequence of events can be represented in a chronological process. One of the most valuable analysis to be performed on these kinds of data is conformance checking analysis. Conformance checking on processes is a family of process mining techniques (Van der Aalst 2016) that compare a process model with an event log of the same process (Van der Aalst 2011).
It is used to check if the actual execution of a process, as recorded in the event log, conforms to the model and vice versa. The interpretation of non-conformance depends on the purpose of the model. Discrepancies between model and log indicate that the model needs to be improved to capture reality better. Recently a growing interest in these techniques has gained ground in the healthcare domain with collaborative workshops and initiatives such as in (Gatta et al., 2017; Gatta et al., 2020). One of the most interesting applications in healthcare is to compare the real clinical pathway of a patient with a process model to check how much a specific healthcare provider is adhering to a clinical guideline in order to assess the quality of care and improve daily practice accordingly (Lenkowicz et al., 2018). As research group we developed a software tool that implements the process mining analysis pipeline, which can be found in (Gatta et al., 2017).
3.6 Radiomics
The use and role of medical imaging technologies has greatly expanded during the last decade from that of a mainly diagnostic, qualitative tool, to acquiring a central role in the context of individualized medicine, with a quantitative value. Several studies have been developed to analyze and quantify different imaging features (e.g. descriptors of intensity distribution, spatial relationships between the various intensity levels, texture heterogeneity patterns, shape descriptors etc.) and the relationships of the tumor to surrounding tissues, to identify relationships with treatment outcomes or gene expressions (Lambin et al., 2012; Kumar et al., 2012). In this scenario, radiomics is emerging as an innovative technique of high throughput extraction of quantitative features from standard radiological images, creating high dimensional dataset followed by data mining approaches for improved decision support (Parekh and Jacobs, 2016; Zwanenburg et al., 2020; Gatta et al., 2018). We hypothesize that radiomics features, being a robust quantification of imaging phenotypes, will potentially add layer in early and accurate diagnosis, prognostication and treatment stratification, with promising first evidence such as in (Akinci D Antonoli et al., 2020, Nero et al., 2020). As research group we developed a software tool that implements the radiomics analysis pipeline (Figure 10), which can be found in (Gatta et al., 2017).
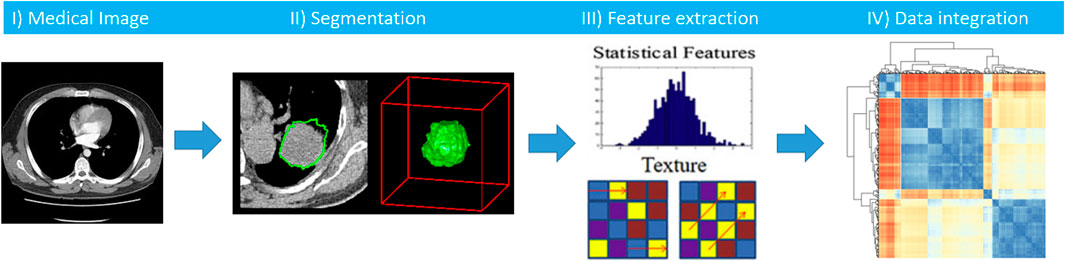
FIGURE 10. Workflow of radiomics analysis includes segmentation of a medical image, features extraction, and integration of radiomics features with other clinical biomarkers.
3.7 Patient Service
Models of patient management and treatment delivery are rapidly changing. The increasing adoption of the integrative Predictive, Preventive, Personalized and Participatory (P4) Medicine foresees a significant change in the role of patients, who become main actors in their own care pathway (Hood and Flores 2012; Flores et al., 2013).
This approach is naturally leading to an innovative assistance model and completely new services, among which digital tools that enable a much closer interaction between healthcare professionals and patients/caregivers have a key role. In this context, the use of advanced remote assistance systems able to collect PROMs (Patients Reported Outcome Measures) and PREMs (Patient Reported Experience Measurements) represents an innovative and valuable opportunity.
Gemelli Generator is indeed developing with the healthcare professionals several digital tools for a new remote collaborative model to continuously monitor the patients in their daily life and early capture the evolution of their disease, in addition to establish proximity and relief.
RWD from several domains like symptoms, Quality of Life, psychological and disease status evaluations, functional tests and treatment adherence are collected by dedicated mobile Apps.
These digital systems are often enriched by integrating the use of wearable devices for the continuous measurement of objective parameters and vitals (Figure 11).
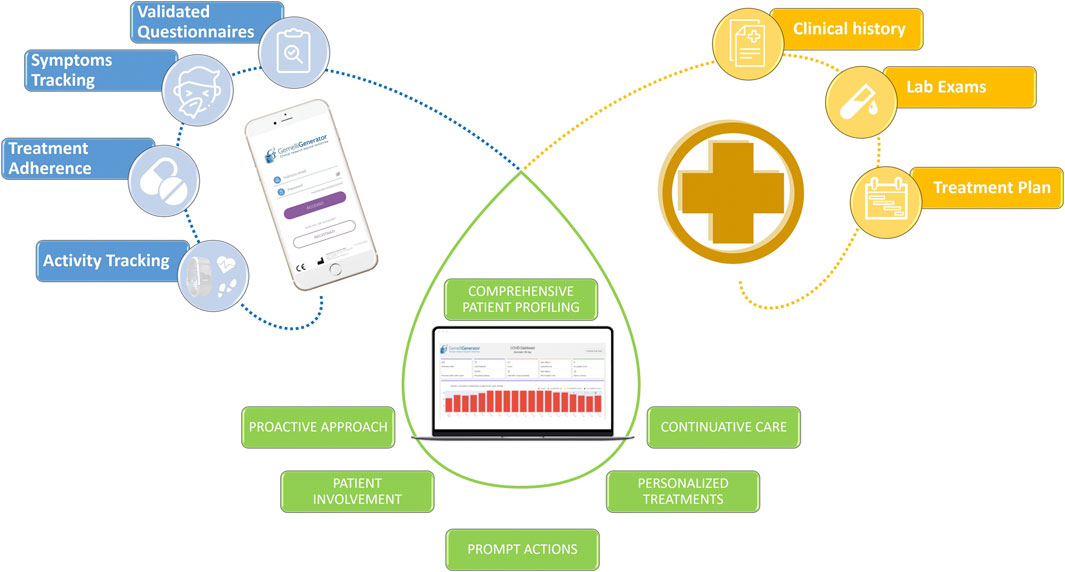
FIGURE 11. Example of the integration of heterogeneous data with the aim of offering innovative and more efficient patient services through the use of digital tools.
Critical situations are immediately identified and all RWD are integrated with hospital clinical data in an e-Clinical platform by means of AI algorithms and translated in useful insights for healthcare professionals for an effective individual patient management.
3.8 Traceability of Methods and Results
Any scientific result must be, by definition, reproducible. This is true both when research results are published in a peer-reviewed paper, and when they form the deliverable of a project developed with an industrial partner. In any case, we need to be able to prove that our results come from a reproducible methodology applied to authentic data (National Academies of Sciences Engineering, and Medicine, 2019). Also, in the scenario where a data-driven method is exploited within clinical practices (e.g., an AI algorithm that supports decision from the clinical staff) this concept of traceability is the foundation for an audit process that may also lead to certify the method as a medical device. Traceability process is a matter evolving in terms of requisites, methods and tools. In general terms, which must be addressed at multiple levels: data must be tracked up to the single patient and clinical event; algorithms–and parameters - used in the research process must be perfectly identified; algorithms implementation and software tools must be identified and accessible, in combination with scripts and workflow used in the study. Lastly, when the study is executed to fulfill a contractual obligation with an external party, some form of mapping between such contract terms and the step-by-step view of the workflow must be secured.
Ideaey, when running the data through the algorithm chain we should obtain the same results as in the published research, with a small caveat for stochastic algorithms (like stochastic gradients and Monte Carlo sampling methods), for which a “close approximation” compromise is accepted for mathematical reasons involving the random elements introduced by the algorithm.
Fulfilling this multi-layered demand for traceability and reproducibility t can be harder than it appears. In Generator research protocols, we are pursuing different approaches, also depending on the context of the specific study, with the aim to define standardized, yet adaptive approaches that can be replicated across studies.
The easiest solution for data is to take a snapshot of the research dataset, duly anonymized, and archive it in an ad-hoc long-term backup solution. We still need means to remap the records in the backup dataset back to the physical patient. Given its importance, this traceability process is directly linked to the second layer of the Generator infrastructure but, there is a constraint: as already noted, patient data in Generator data marts are pseudonymized with a unique ID. But this ID is remappable, at the ICT Datawarehouse level, to a unique patient. In this way, patient privacy is enforced because the remapping happens at a level higher than Gemelli Generator, which in this way keeps its patient-agnostic character intact.
Coming to algorithms and methods (Alston and Rick 2020), one might argue that the research paper itself or a professionally written documentation are already sufficient to ensure that the same computation pipeline can be reproduced later. While this is theoretically true, things can be harder when you want to run the data through the pipeline and hopefully obtain the same results. Data scientists know from direct experience that as time passes new versions of operating systems, packages, libraries, are made available, sometimes involving a change in function invocation signature, different algorithm implementations, or even unrecoverable conflicts with other components. Sometimes, software products are no longer maintained and at the worst possible moment you discover that they cannot be installed on the current version of a given operating system. In brief: there are several things that can go wrong, especially when years have passed since when the study was completed.
A possible solution to this issue, which we are testing within the Generator infrastructure is containerization. A Docker container, including both the research environment and the pseudonymized data is stored in a secure backup area, ready to be mounted and run to repeat the exact study pipeline.
Finally, a certification both of content and of date can be applied to the container, using technologies like the blockchain, making it impossible to alter the container and configuring in this way a real asset which, in the case of an industrial contract, can be shown as proof of fulfillment of a contract obligation. This is one of the aspects that we will explore in the near future as a possible solution to this issue, also in the context of multiparty efforts and research consortia.
This solution for data is probably the best way to obtain safe, privacy-safe reproducibility while keeping impact low and minimizing risks.
3.9 Generalizability, Explainability and Ethical issues as challenges for AI in Medical Research.
AI offers powerful tools to researchers to extract knowledge from vast amounts of data. This is especially true when data are available in massive quantities and the number of independent variables is high: a scenario in which Gemelli Generator is often involved. One of the most notable examples of the wide applicability of AI is given by Deep Learning, the way we call Neural Networks with more than one hidden layer (Boldrini et al., 2019a).
The peculiar ability of deep neural networks to build models from data was certified by theorems proved between 1989 and 1999 (Cybenko 1989), but see also the nice visual demonstration at the website http://neuralnetworksanddeeplearning.com/chap4.html, showing mathematically that any multivariate function can be simulated with arbitrary precision by a deep neural network of adequate complexity, provided that an adequate amount of data is available. This is one of the reasons for the impressing development of this technique, especially after computing power, fast storage, and data science libraries began to be available at reasonable costs. Similar considerations can be made about other AI techniques, such as Random Forests, an ensemble method of classification and regression consisting in the construction, based on training data, of several decision trees, that are then asked to vote on new, never seen before instances that need to be classified automatically at a later time. Both these methods are powerful and widely used, but they share a common problem: that of explainability.
It has been recognized during the second decade of this century (https://www.darpa.mil/program/explainable-artificial-intelligence) that, although complex models, typical of AI and machine learning, like deep neural networks and random forests, are more expressive and can capture higher levels of complexity, there is a price to pay: they tend to function as black boxes (Abernethy et al., 2010). Let’s consider a linear regression problem. the model we obtain is directly explainable: the weights in the expression tell us the importance and effect of each covariate in the model, and the theory of hypothesis testing gives us a lot of information on how much we should trust the model based uniquely on the data used to fit it. Quite similar is the situation with logistic regression, talking about classifiers. Those models are known as transparent.
But with machine learning models, it’s a different story. Deep neural networks, applied both to regression and classification problems, are all but transparent. There is no direct way to associate the coefficients, for example, to some definite effect in the model. The same goes for random forests, our other example of classifier. These are called opaque models.
Matt Durek (https://www.darpa.mil/program/explainable-artificial-intelligence) expresses all the doubts about the interpretation of AI models behaviour with six questions, that any user can ask when seeing AI at work (see the supplemental material for further information). These questions can be considered the starting point of the discipline called eXplainable Artificial Intelligence (XAI) (Guidotti et al., 2018).
In our experience, this is indeed a critical issue. Traditionally, clinical research relies on the well-established apparatus of statistics and epidemiology. As shown in our basic examples about linear and logistic regressions, those disciplines offer transparent methods and solid tools for testing and validating their results. With AI, this is not readily available. A paradigm change is happening, and we do not want to give up on the opportunities that new Machine Learning (ML) technologies offer; at the same time, we are looking for ways to understand better the meaning of AI methods (Amparore et al., 2021). A new emerging discipline called Algorethics (Benanti 2019; Benanti 2020) studies the problem of using opaque automated methods to take important decisions with a big ethical impact. Clinical research is one of the fields in which this problem is felt strongly: a doctor has the right to know on what basis an AI driven Decision Support System is producing its output: any unjustified choice is unacceptable in a clinical setting.
For these reasons, our choice at Gemelli Generator is to use opaque methods only when their performance is higher than that of an equivalent transparent method, or when an equivalent transparent method is not available. Nevertheless, there are specific fields of ML for clinical research in which this is often the case, as in Radiomics applications. As an example of how we can understand the functioning of an opaque algorithm, heat maps can be generated to show what areas of a medical image contributed most to a given classification, giving an insight to medical doctors to better understand and possibly formulate new clinical hypotheses. Figure 12 shows the use of this method to visualize the areas of a fluorescein angiography of the fundus oculi that were most relevant for the output of a deep learning algorithm trained on images to predict retinal detachment in prematurely born infants (Lepore et al., 2020).
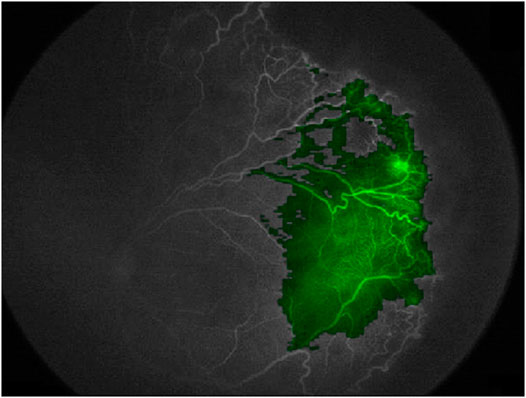
FIGURE 12. Using a heatmap to visualize areas of an image that contributed to the output of a deep learning model (Fluoresceine Angiography Imaging of Fundus Oculi from (Lepore et al., 2020))
When opaque methods are used, cross validation techniques are mandatory to prevent overfitting, the typical behaviour of neural networks to fit “too well” learning data, only to fail dangerously on unseen data.
At the end of the learning phase, an external validation on unseen data is performed to test the ability of the model to work correctly on new data (generalizability).
4 From Knowledge to Value
4.1 Research-Industry Cooperation in Digital Medicine
The examples of digital healthcare cooperation with industrial partners are growing fast, since both large clinical centers and enterprises (e.g., in Pharma or Electro-medical industries) are now fully aware of the potential from data-driven approaches for applied research.
From the process/methodology standpoint, the most relevant challenges for this type of project are related to privacy and ethical aspects. Therefore, it is key that within a framework like Generator, particular care is directed to the following topics:
Clear definition of study aims, in-depth description and evaluation of patient benefits, and focus on how the social community and different stakeholders will see these benefits translated into quality of care, personalized care, etc.
Methods and algorithms must be detached from individual patient information and detailed clinical history. Technical measures for data privacy and data protection must follow an extremely high standard. Research workflow must be auditable and repeatable.
Leveraging the role of the Open Innovation Unit (OIU) as prime interface to external cooperation, GENERATOR RWD has already started some key industry-oriented research projects. The OIU is a branch of Policlinico Gemelli Scientific Directorate (IRCCS). Their staff is mainly composed of clinical specialists and business development experts, who work as a shared service to the rest of the organization, and especially in support of the Hospital Departments to facilitate relationships with medical/pharmaceutical industry and the expanded innovation ecosystem (Universities, Research Centers, Service Providers). They help transforming innovation ideas and pilot programs into structured programs and support the clinical staff throughout the full cycle from early creation to deployment. In doing so, they support program/project design, budgeting and contractual phases. The high degree of innovation intrinsic of these partnerships require novel approaches to the project architecture and contract setup, very much relying on concepts like co-creation/co-development, as well as agile project management approaches (such as design thinking; minimum viable products; agile and scrum methods). Therefore, these projects are developed starting from the ultimate beneficiary perspective (the patient and his/her caregiver), the gaps and pain points that are impeding better care and outcomes; and a series of data/evidence driven initiatives that allow to exploit the partnership knowledge base and expertise to create disease prevention algorithms, methods to simulate prognostic responses, and new integrated care model based on extensive adoption of digital platforms.
As per the general approach for data privacy/protection within GENERATOR, none of these projects give access to patient data. Retrospective analysis delivers secondary data analysis which can provide aggregated evidence which are useful for incidence/severity scoring; learning algorithms are translated in synthetic data/methods which are not attached to individual patient data; the adoption of new therapies supported by digital methods is always experimented in the context of trials that engage candidate patients through informed consent and continuous, transparent engagement.
Moreover, the last layer of the Generator infrastructure provides a whole series of technologies and tools such as distributed learning and the sandbox (which will be explored in more detail in the following paragraphs with two use cases) that allow corporate or academic partners to work on data without having direct access to it. Furthermore, the GENERATOR team has put in place dedicated technical frameworks providing additional data protection in the context of external cooperation and multi-centric initiatives.
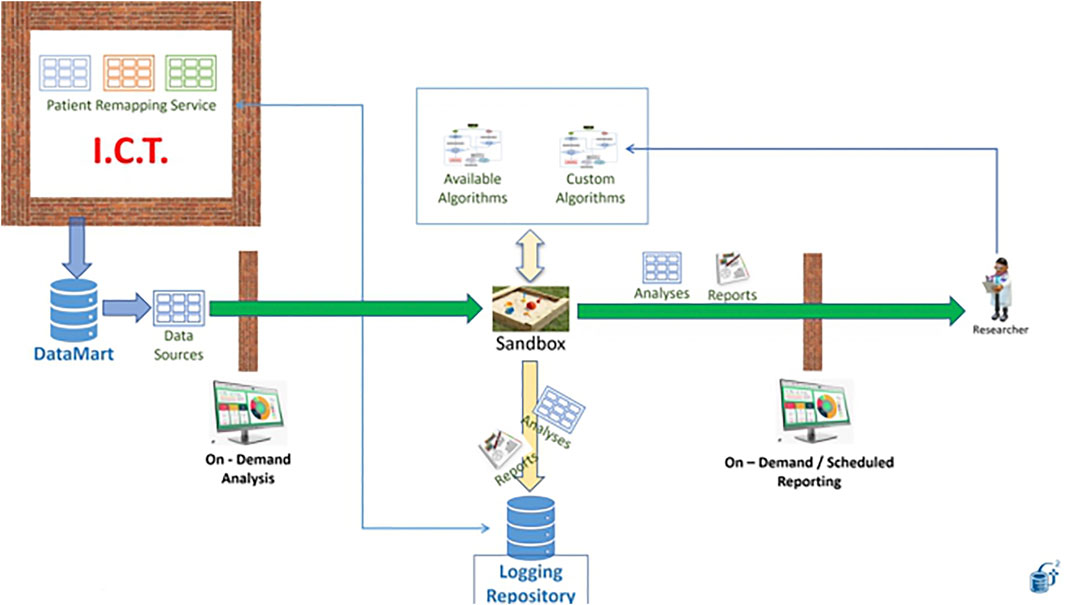
4.2 Industrial Research on Patient Data With Elevated Privacy Requirements: The Sandbox
In our first use case, an external party wants to learn a model from our data. They have the workflow and need the data. Instead of giving them the data, we ask them to containerize the workflow and put it at work in the Generator Sandbox. In Figure 13, the needed dataset is made available to the workflow thanks to the link with an ad-hoc Datamart (left). The desired workflow can be designed, developed and tested outside Generator using synthetic data produced through the application of perturbative methods to real patient data to mimic the same structure and distributions that the workflow will work on once installed in the sandbox (see for example (Goncalves et al., 2020). A list of typical algorithms is already made available by the Sandbox (top of Figure 13).
Results are produced in the form of reports that are made available to the external party on demand or at scheduled times (right).
Generator data scientists reserve the right to inspect the output of the workflow, which is certified for datetime and content in a dedicated certified repository (bottom). the possibility of making this content blockchain certified is currently under study.
The sandbox is a neutral space in which Generator data scientists need not access the algorithms, while third parties do not see data. Availability of a certified repository of all past runs of the workflow and of the results obtained is key to the establishment of a constructive relation between Generator and external partners, with the result that knowledge extraction becomes possible without endangering intellectual property, data ownership and patient rights.
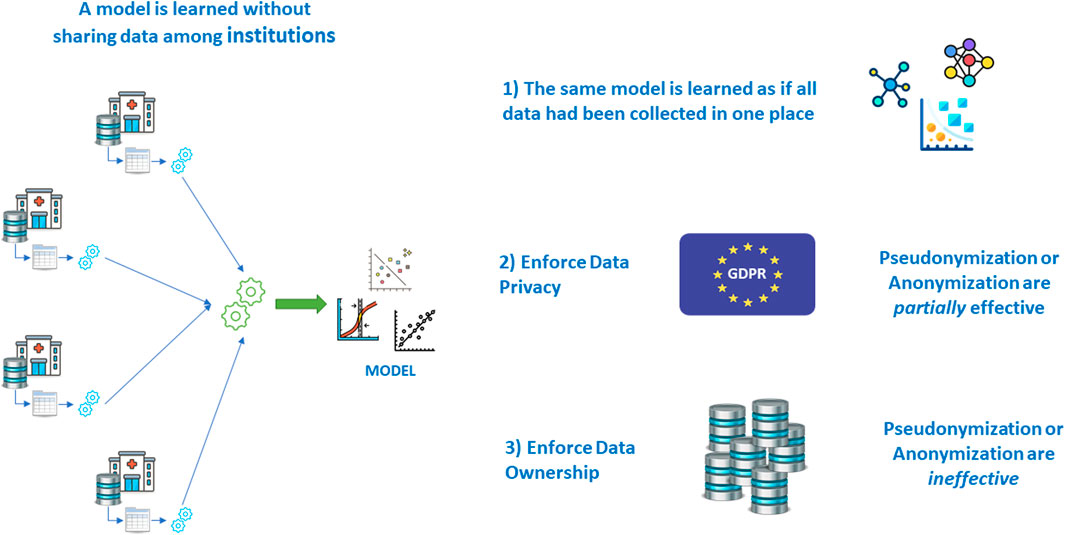
4.3 Multicentric Research Without Sharing Patient Data: Federated Learning
It is a known fact that the most robust models are learned and validated on large volumes of data. This is even more important when the number of variables grows, and when machine learning models are used, that do not include significance tests like those of inferential statistics and hypothesis testing.
For this reason, multi-centric studies are regarded as a solution: learning and validating the same model on patients’ data from different institutions allows for an even greater volume and variability, once the semantic alignment has been achieved via the adoption of a suitable terminological system to harmonize the real meaning of covariates coming from various places.
But then another problem arises: that of centralizing the data in one place to perform the learning and validation effort.
As seen in the earlier paragraphs, letting real patient data out of the hospital walls poses a series of problems; if all other issues are overcome, legislation makes it hard in some countries, even impossible in others. Fortunately, there are mathematical techniques that allow learning a model without sharing data, achieving the same result that would have been obtained with data sharing (Deist et al., 2020) (Figure 14).
For learning models based on the gradient methods family (also including Newton Raphson method, and proximal gradient for non–differentiable terms as in the norm-1 regularize in the LASSO (Least Absolute Shrinkage and Selection Operator)), associativity is exploited (Lu et al., 2015). Stochastic gradient and mini batch methods are used for Neural Networks and Deep Learning (Boldrini et al., 2019b), averaging methods for Bayesian Networks (Sieswerda et al., 2020), while ensemble methods, like Random Forests, are naturally distributable. More advanced methods can be leveraged, like the ADMM (Alternate Directions Method of Multipliers) (Boyd et al., 2011; Damiani et al., 2015), making vertically partitioned datasets and checkerboard partitioned data tractable (Figure 15).
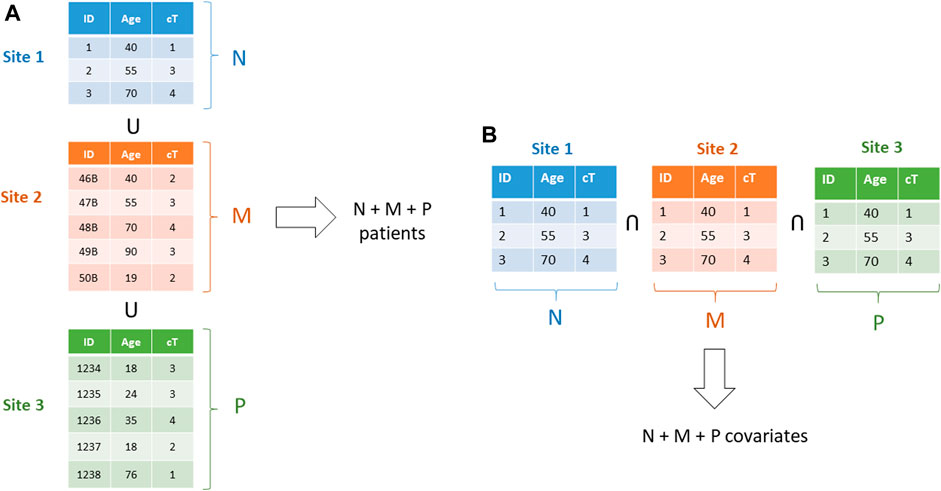
FIGURE 15. (A)–A horizontally partitioned dataset: each one of the three datasets includes full patients. b)–a vertically partitioned dataset: each institution includes a subset of the covariates for all patients.
Preliminary analysis (Damiani et al., 2018) and Validation (Shi et al., 2019) are also fundamental parts of the workflow, and their distributed versions are available for different algorithms.
Data Scientists working at Gemelli Generator have gained experience on Distributed Learning during their PhD years and are able to be part in distributed efforts in international groups as partners, leaders, and developers of ad-hoc solutions for new cases. A few DL efforts are currently ongoing with partners across the world.
4.4 Gemelli Generator and International Cooperation
The architecture and project framework implemented at Generator aims also at enabling innovative projects in the context of multi-country advanced research initiatives such as European-funded programs such as Horizon Europe and similar ones.
There are recurrent challenges during design and implementation phases of such multi-party initiatives, that sometime becomes showstoppers for these important collaborative efforts.
Integration of skills is critical in the design phase, to have a clear end-to-end view (medical expertise combined with technology and process view) of overall feasibility, incremental delivery of results and clear identification of attention points and complexity that consortia must address.
Moreover, a clear definition of the logical components of the project is key for the success of such initiatives in a large consortium. The availability of standard blocks for execution in terms of technical steps; the definition of data model, data and model flows, and the enforcement of security and privacy practices that can be adopted from all Consortium members in a standardize fashion is a major enabler and dependency.
We have experienced that the introduction of Generator architecture and process blueprint can be an accelerator, given that some of the assets needed in design and implementation are already available as generic artefacts, and can accelerate the learning curve virtually for all project partners.
5 Discussion
The Gemelli Generator Real World Data experience stems from two well established assets: availability of a huge amount of historical patient data, well maintained, constantly updated, and made easily accessible through the ICT structure of Policlinico Gemelli; and a group of data scientists and doctors that built together, during 7 years of tight collaboration, a multidisciplinary approach to knowledge extraction from Real World Data in clinical research.
In designing and developing the Gemelli Generator Infrastructure, a few challenges had to be faced, and some founding principles were chosen since the beginning: the Big Data approach for data management; the Rapid Learning paradigm to extract value from data in a multidisciplinary, evolving, personalized medicine-oriented framework; a flexible standardization of clinical data via the use of formal and non-formal ontologies; a high level of automation in the data transfer; a “privacy by design” approach, as dictated by the GDPR, also in view of the protection of data ownership and intellectual property.
From the beginning, a few data marts were established and, correspondingly, clinical studies of different kinds were conducted, to serve as proof of concept for clinical researchers that were interested in working with real world data. At the same time, a parallel line of work was created to deal with four Horizon 2020 projects that the facility is currently involved in. A few industrial collaborations were also started, which gave the facility the chance of testing the feasibility and appropriateness of the structural choices, with a special emphasis on patient privacy protection and data ownership.
During this first year of activity, the approach was confirmed to be solid, and the interest in using research services offered by Gemelli Generator RWD has been indeed quite high, both from internal investigators, the Gemelli Hospital clinical researchers, and from external research subjects of different nature: academic, industrial, and institutional. The wide range of aspects touched by the projects proposed by our partners has confirmed the importance of a multidisciplinary, personalized approach, which in turn results in several time-consuming activities to be performed. This is the main take-away message of these 12 months, which leads the facility to invest in the automation of some data management tasks. We provided a broad overview of the most relevant Generator projects (published during this first year of activity) in the supplemental materials under. Additionally, all of the four Horizon 2020 projects in which Generator is involved aim at producing measurable, health improvement-related KPIs, as such we intend to be able to demonstrate the tangible clinical benefits of our infrastructure thanks to these projects. The timeframe of those project is between two to 4 years.
For these reasons, our wish for the short–medium term is to enable some data management functions to be fully or at least partially automated, starting with the Sandbox and Federated Learning infrastructures, which will be operative in September 2021, while three other development lines have been launched for the creation of an AI based data catalog to rationalize and optimize data selection and data mart updating, for streamlining the process of data value creation, and for building a certifiable research reproducibility framework.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AD, CM, JL, NC, LB, LT, AM and AL contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript. AD, SP, AC, PS, VV supervised and revised the work. All authors provided critical feedback and helped shape the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcomp.2021.768266/full#supplementary-material.
References
Abernethy, A. P., Etheredge, L. M., Ganz, P. A., Wallace, P., German, R. R., Neti, C., et al. (2010). Rapid-learning System for Cancer Care. Jco 28, 4268–4274. doi:10.1200/JCO.2010.28.5478
Ahmed, Z., Mohamed, K., Zeeshan, S., and Dong, X. (2020). Artificial Intelligence with Multi-Functional Machine Learning Platform Development for Better Healthcare and Precision Medicine. Database 2020, baaa010. doi:10.1093/database/baaa010
Akinci D'Antonoli, T., Farchione, A., Lenkowicz, J., Chiappetta, M., Cicchetti, G., Martino, A., et al. (2020). CT Radiomics Signature of Tumor and Peritumoral Lung Parenchyma to Predict Nonsmall Cell Lung Cancer Postsurgical Recurrence Risk. Acad. Radiol. 27, 497–507. doi:10.1016/j.acra.2019.05.019
Alitto, A., Gatta, R., Vanneste, B., Vallati, M., Meldolesi, E., Damiani, A., et al. (2017). PRODIGE: PRediction Models in prOstate Cancer for Personalized meDIcine challenGE. Future Oncol. 13, 2171–2181. doi:10.2217/fon-2017-0142
Alston, J. M., and Rick, J. A. (2020). A Beginner's Guide to Conducting Reproducible Research. Bull. Ecol. Soc. Am. 102. doi:10.1002/bes2.1801
Amparore, E., Perotti, A., and Bajardi, P. (2021). To Trust or Not to Trust an Explanation: Using LEAF to Evaluate Local Linear XAI Methods. Peerj Comput. Sci. 16 (7), e479. doi:10.7717/peerj-cs.479
Benanti, P. (2020). Digital age. Teoria del cambio d’epoca. Persona, famiglia e società. Milano, Italy: San Paolo Edizioni Press.
Benanti, P. (2019). le macchine sapienti. Intelligenze artificiali e decisioni umane. Milano, Italy: Marietti Press.
Boldrini, L., Bibault, J. E., Masciocchi, C., Shen, Y., and Bittner, M. I. (2019b). Deep Learning: A Review for the Radiation Oncologist. Front. Oncol. 9. doi:10.3389/fonc.2019.00977
Boldrini, L., Bibault, J. E., Masciocchi, C., Shen, Y., and Bittner, M. I. (2019a2019). Deep Learning: a Review for the Radiation Oncologist. Front. Oncol. 9, 977. doi:10.3389/fonc.2019.00977
Boyd, S., Parikh, N., Chu, E., Peleato, B., and Eckstein, J. (2011). Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers. Foundations Trends Machine Learn. 3, 1–122. doi:10.1561/2200000016
Cesario, A., D’Oria, M., and Scambia, G. (2021a). La Medicina Personalizzata tra Ricerca e Cura. Milano: FrancoAngeli s.r.l.Press.
Cesario, A., D’Oria, M., Bove, F., Privitera, G., Boškoski, I., Pedicino, D., et al. (2021c). Personalized Clinical Phenotyping through Systems Medicine and Artificial Intelligence. Jpm 11, 265. doi:10.3390/jpm11040265
Cesario, A., D’Oria, M., Calvani, R., Picca, A., Pietragalla, A., Lorusso, D., et al. (2021b). The Role of Artificial Intelligence in Managing Multimorbidity and Cancer. Jpm 11, 314. doi:10.3390/jpm11040314
Cesario, A., Simone, I., Paris, I., Boldrini, L., Orlandi, A., Franceschini, G., et al. (2021d). Development of a Digital Research Assistant for the Management of Patients' Enrollment in Oncology Clinical Trials within a Research Hospital. Jpm 11, 244. doi:10.3390/jpm11040244
Chiesa, S., Tolu, B., Longo, S., Nardiello, B., Capocchiano, N. D., Rea, F., et al. (2020). A New Standardized Data Collection System for Brain Stereotactic External Radiotherapy: the PRE.M.I.S.E Project. Future Sci. OA 6, FSO596. doi:10.2144/fsoa-2020-0015
Cybenko, G. (19891989). Approximation by Superpositions of a Sigmoidal Function. Math. Control. Signal. Syst. 2, 303–314. doi:10.1007/BF02551274
Damiani, A., Vallati, M., Gatta, R., Dinapoli, N., Jochems, A., Deist, T., et al. (2015). Distributed Learning to Protect Privacy in Multi-Centric Clinical Studies. Artificial Intelligence in Medicine. Lecture Notes in Computer Science. Italy, Pavia. 17/06/15-20/06/15, 9105. Springer, Cham. doi:10.1007/978-3-319-19551-3_8
Damiani, A., Masciocchi, C., Boldrini, L., Gatta, R., Dinapoli, N., Lenkowicz, J., et al. (2018). Preliminary Data Analysis in Healthcare Multicentric Data Mining: a Privacy-Preserving Distributed Approach. J. E-Learning Knowledge Soc. 14 (1). doi:10.20368/1971-8829/1454
Deist, T. M., Dankers, F. J. W. M., Ojha, P., Scott Marshall, M., Janssen, T., Faivre-Finn, C., et al. (2020). Distributed Learning on 20 000+ Lung Cancer Patients - the Personal Health Train. Radiother. Oncol. 144, 189–200. doi:10.1016/j.radonc.2019.11.019
Etheredge, L. M. (2014). Rapid Learning: a Breakthrough Agenda. Health Aff. 33, 1155–1162. doi:10.1377/hlthaff.2014.0043
Flores, M., Glusman, G., Brogaard, K., Price, N., and Hood, L. (2013). P4 Medicine: How Systems Medicine Will Transform the Healthcare Sector and Society. Personalized Med. 10, 565–576. doi:10.2217/pme.13.5
Gatta, R., Vallati, M., Dinapoli, N., Masciocchi, C., Lenkowicz, J., Cusumano, D., et al. (2018). Towards a Modular Decision Support System for Radiomics: A Case Study on Rectal Cancer. Artif. Intelligence Med. 96. doi:10.1016/j.artmed.2018.09.003
Gatta, R., Lenkowicz, J., Vallati, M., Rojas, E., Damiani, A., Sacchi, L., et al. (2017). Innovative R Library for Performing Process Mining in MedicinepMineR: An. Proceedings of the Conference on Artificial Intelligence in Medicine. Springer. doi:10.1007/978-3-319-59758-4_42
Gatta, R., Vallati, M., Fernandez-Llatas, C., Martinez-Millana, A., Orini, S., Sacchi, L., et al. (2020). What Role Can Process Mining Play in Recurrent Clinical Guidelines Issues? A Position Paper. Ijerph 17, 6616. doi:10.3390/ijerph17186616
Gill, J. L., Avouac, B., Duncombe, R., Hutton, J., Jahnz-Rozyk, K., Schramm, W., et al. (2016). The Use of Real World Evidence in the European Context: An Analysis of Key Expert Opinion. London, United Kingdom: London School of Economics. doi:10.21953/LSE.68442
Goncalves, A., Ray, P., Soper, B., Stevens, J., Coyle, L., and Sales, A. P. (2020). Generation and Evaluation of Synthetic Patient Data. BMC Med. Res. Methodol. 20, 108. doi:10.1186/s12874-020-00977-1
Guidotti, R., Monreale, A., Ruggieri, S., Turini, F., Giannotti, F., and Pedreschi, D. (2018). A Survey of Methods for Explaining Black Box Models. ACM Comput. Surv. 51, 5. doi:10.1145/3236009
Hiramatsu, K., Barrett, A., and Miyata, Y. (2021). Current Status, Challenges, and Future Perspectives of Real-World Data and Real-World Evidence in Japan. Drugs - Real World Outcomes. doi:10.1007/s40801-021-00266-3
Hood, L., and Flores, M. (2012). A Personal View on Systems Medicine and the Emergence of Proactive P4 Medicine: Predictive, Preventive, Personalized and Participatory. New Biotechnol. 29, 613–624. doi:10.1016/j.nbt.2012.03.004
Kumar, V., Gu, Y., Basu, S., Berglund, A., Eschrich, S. A., Schabath, M. B., et al. (2012) Radiomics: the Process and the Challenges Magn Reson Imaging. 30, 1234–1248. doi:10.1016/j.mri.2012.06.010
Lambin, P., Rios-Velazquez, E., Leijenaar, R., Carvalho, S., van Stiphout, R. G., Granton, P., et al. (2012). Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 48, 441–446. doi:10.1016/j.ejca.2011.11.036
Lambin, P., Roelofs, E., Reymen, B., Velazquez, E. R., Buijsen, J., Zegers, C. M. L., et al. (2013). 'Rapid Learning Health Care in Oncology' - an Approach towards Decision Support Systems Enabling Customised Radiotherapy'. Radiother. Oncol. 109, 159–164. doi:10.1016/j.radonc.2013.07.007
Lambin, P., Zindler, J., Vanneste, B., van de Voorde, L., Jacobs, M., Eekers, D., et al. (2015). Modern Clinical Research: How Rapid Learning Health Care and Cohort Multiple Randomised Clinical Trials Complement Traditional Evidence Based Medicine. Acta Oncologica 54, 1289–1300. doi:10.3109/0284186X.2015.1062136
Lancellotta, V., Guinot, J. L., Fionda, B., Rembielak, A., Di Stefani, A., Gentileschi, S., et al. (2020). SKIN-COBRA (Consortium for Brachytherapy Data Analysis) Ontology: The First Step towards Interdisciplinary Standardized Data Collection for Personalized Oncology in Skin Cancer. jcb 12, 105–110. doi:10.5114/jcb.2020.94579
Lenkowicz, J., Gatta, R., Masciocchi, C., Casà, C., Cellini, F., Damiani, A., et al. (2018). Assessing the Conformity to Clinical Guidelines in Oncology: An Example for the Multidisciplinary Management of Locally Advanced Colorectal Cancer Treatment. Management Decis. 56, 2172–2186. doi:10.1108/MD-09-2017-0906
Lepore, D., Ji, M., Pagliara, M., Lenkowicz, J., Capocchiano, N. D., Tagliaferri, L., et al. (2020). Convolutional Neural Network Based on Fluorescein Angiography Images for Retinopathy of Prematurity Management. Translational Vis. Sci. Technology 7. doi:10.1167/tvst.9.2.37
Lewis, J. R. R., Kerridge, I., and Lipworth, W. (2017). Use of Real-World Data for the Research, Development, and Evaluation of Oncology Precision Medicines. JCO Precision Oncol. 1, 1–11. doi:10.1200/PO.17.00157
Lu, C. L., Wang, S., Ji, Z., Wu, Y., Xiong, L., Jiang, X., et al. (2015). WebDISCO: a Web Service for Distributed Cox Model Learning without Patient-Level Data Sharing. J. Am. Med. Inform. Assoc. 22, 1212–1219. doi:10.1093/jamia/ocv083
Marazzi, F., Tagliaferri, L., Masiello, V., Moschella, F., Colloca, G. F., Corvari, B., et al. (2021). GENERATOR Breast DataMart-The Novel Breast Cancer Data Discovery System for Research and Monitoring: Preliminary Results and Future Perspectives. Jpm 11, 65. doi:10.3390/jpm11020065
National Academies of Sciences Engineering, and Medicine, (2019). Reproducibility and Replicability in Science. Washington, DC: The National Academies Press. doi:10.17226/25303
Nero, C., Ciccarone, F., Boldrini, L., Lenkowicz, J., Paris, I., Capoluongo, E. D., et al. (2020). Germline BRCA 1-2 Status Prediction through Ovarian Ultrasound Images Radiogenomics: a Hypothesis Generating Study (PROBE Study). Sci. Rep. 10. doi:10.1038/s41598-020-73505-2
Parekh, V., and Jacobs, M. A. (2016). Radiomics: a New Application from Established Techniques. Expert Rev. Precis Med. Drug Dev. 1, 207–226. doi:10.1080/23808993.2016.1164013
Shi, Z., Foley, K., Mey, J., Spezi, E., Whybra, P., Crosby, T., et al. (2019). External Validation of Radiation-Induced Dyspnea Models on Esophageal Cancer Radiotherapy Patients. Front. Oncol. 9, 1411. doi:10.3389/fonc.2019.01411
Sieswerda, M. S., Bermejo, I., Geleijnse, G., Aarts, M., Lemmens, V., Ruysscher, D., et al. (2020). Predicting Lung Cancer Survival Using Probabilistic Reclassification of TNM Editions with a Bayesian Network. JCO Clin. Cancer Inform. 4, 436–443. doi:10.1200/CCI.19.00136
Tagliaferri, L., Gobitti, C., Colloca, G. F., Boldrini, L., Farina, E., Furlan, C., et al. (2018). A New Standardized Data Collection System for Interdisciplinary Thyroid Cancer Management: Thyroid COBRA. Eur. J. Intern. Med. 53, 73–78. doi:10.1016/j.ejim.2018.02.012
Van der Aalst, W. (2011). Process Mining: Discovery, Conformance and Enhancement of Business Processes. Berlin: Springer Press.
Keywords: big data and analytics, real world data, healthcare infrastructure, experience and current status, personalized medical care, artificial intelligence, real world data architecture
Citation: Damiani A, Masciocchi C, Lenkowicz J, Capocchiano ND, Boldrini L, Tagliaferri L, Cesario A, Sergi P, Marchetti A, Luraschi A, Patarnello S and Valentini V (2021) Building an Artificial Intelligence Laboratory Based on Real World Data: The Experience of Gemelli Generator. Front. Comput. Sci. 3:768266. doi: 10.3389/fcomp.2021.768266
Received: 31 August 2021; Accepted: 12 November 2021;
Published: 07 December 2021.
Edited by:
Stefan Sebastian Busnatu, Carol Davila University of Medicine and Pharmacy, RomaniaReviewed by:
Alexandru Burlacu, Grigore T. Popa University of Medicine and Pharmacy, RomaniaIon-Gheorghe Petrovai, Babeș-Bolyai University, Romania
Copyright © 2021 Damiani, Masciocchi, Lenkowicz, Capocchiano, Boldrini, Tagliaferri, Cesario, Sergi, Marchetti, Luraschi, Patarnello and Valentini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. D. Capocchiano, bmlrb2xhZGluby5jYXBvY2NoaWFub0Bwb2xpY2xpbmljb2dlbWVsbGkuaXQ=
 A. Damiani
A. Damiani C. Masciocchi
C. Masciocchi J. Lenkowicz
J. Lenkowicz N. D. Capocchiano
N. D. Capocchiano L. Boldrini
L. Boldrini L. Tagliaferri
L. Tagliaferri A. Luraschi
A. Luraschi V. Valentini
V. Valentini