
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 04 February 2025
Sec. Clinical Microbiology
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1531018
This article is part of the Research Topic Advances in the Diagnosis and Management of Infectious Diseases View all 14 articles
Two factors frequently impede accurate bacterial identification using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS): inadequate bacterial abundance in real samples and bacterial combinations. For MALDI-TOF MS analysis and libraries for bacterial identification, time-consuming culture procedures are necessary to achieve sufficient concentration and isolation of a single bacterium. When dealing with hazardous bacteria like Brucella, which are more difficult to handle and cure, this problem becomes even more crucial. To overcome these obstacles, Fe3O4 magnetic nanoparticles (MNPs) linked with Brucella-specific antibodies and MALDI-TOF MS analysis have been used to create a quick and accurate technique for direct bacterial separation and identification in complex samples. This method allows MNPs to immune-selectively collect Brucella cells, which are then deactivated and ready for MALDI-TOF MS analysis by a formic acid/acetonitrile wash. Rabbits were used to manufacture brucella antibodies, which have effectively adsorbed onto the MNPs–protein A. Any particular Brucella bacteria found in the media might be absorbed by this MNPs–protein A–antibody immunoprobe. The concentration of Brucella bacterial cells increases the protein spectrum’s visibility by a factor of 103, making it possible to quickly identify Brucella spp. without first growing them in cultural conditions. This method has been successfully used to achieve a limit of detection (LOD) of 50 CFU/mL in an aqueous medium and genuine sample—milk. The diagnostic time for this harmful bacterium is greatly decreased because the entire procedure from bacterial isolation to species identification is finished in less than 60 min. High sensitivity and specificity are demonstrated by the immunoassay–MS approach, as the spectral pattern it produces matches well-known databases like SPECLUST and Ribopeaks.
The world’s health is being threatened by infectious germs, which have the capacity to start pandemics in the future (Nieto and Salvetti, 2014; Cloeckaert, 2024). Bacteria have historically caused a wide range of infectious diseases that impact people, animals, and plants (Dharmarajan et al., 2022; Tiedje et al., 2022; Glajzner et al., 2024). An estimated 420,000 deaths are attributed to foodborne pathogens alone each year, with the largest burden occurring in areas of poverty (Olivares et al., 2013; Schaumburg et al., 2019; de Oliveira et al., 2024). Therefore, bacterial illnesses must be properly diagnosed and treated in order to protect the public’s health (Zia and Alkheraije, 2023). Brucellosis is also known as “Mediterranean fever”, “undulant fever,” and “Malta fever”. Because of its low infectious dosage and ability to spread through inhaling contaminated aerosols, it is considered an infectious bacterial zoonosis (Qin et al., 2023; Zhang et al., 2024). Rapid and accurate identification of pathogenic microorganisms is necessary to comprehend these diseases (Wang et al., 2022; Kang et al., 2024). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) utilizes a mass spectrum library derived from sufficient microbial proteins, allowing the matching of the obtained mass spectrum of one bacteria with known profiles for species identification (Cheng et al., 2016; Kurli et al., 2018; Rahi and Vaishampayan, 2020; Han et al., 2021; Lorente-Leal et al., 2022; Peng et al., 2022; Tsuchida and Nakayama, 2022; Becker and Lupetti, 2023; Foster and Khaiboullina, 2023).
Notable commercial kits include the developed fast BACpro® II kit (Nittobo Medical Co., Tokyo, Japan) (Wang et al., 2016; Oviaño et al., 2021), the Vitek MS blood culture kit (bioMérieux, Inc.) (Nomura et al., 2020), and the Sepsityper® kit (Bruker Daltonics) (Perše et al., 2022). For bacterial analysis in microbiology labs, including cultivation, inactivation, isolation, and data interpretation, the Food and Drug Administration (FDA) has authorized MALDI-TOF MS (Bauermeister et al., 2022; Li et al., 2022; Haider et al., 2023; Pastrone et al., 2023).
A potential method for quick and precise microbiological identification is the combination of MNPs and MALDI-TOF MS (Kumari et al., 2023). Detection limits as low as 10² CFU/mL are made possible by MNPs (Xiao et al., 2022; Abafogi et al., 2024). This technique improves operating speed and accuracy by streamlining processes by doing away with the requirement to culture samples, particularly for highly virulent pathogens such as Brucella (Ha and Kim, 2022). Furthermore, MNPs’ capacity to functionalize and attach selectively to bacterial cells enhances detection specificity in complicated biological samples, contributing to their high selectivity (Houser et al., 2024). Because of protein A’s high affinity for the Fc region of immunoglobulins (IgGs), antibodies on MNPs can be effectively immobilized, greatly enhancing their ability to ensnare target microorganisms (Xu et al., 2019).
Serological tests are usually preferred because determining the source of brucellosis is crucial due to its current threats and takes a lot of time, often requiring highly qualified specialists (Padilla et al., 2010). Serological techniques, however, frequently encounter obstacles like a lack of standardization, validation issues, and restrictions on precisely identifying the species and strain of Brucella implicated (Yagupsky et al., 2019).
In our last study, we used bioinformatics in conjunction with MALDI-TOF MS proteomics analysis to find biomarkers for brucellosis (Hamidi et al., 2022). Additionally, we used MALDI-TOF MS in conjunction with single-chain variable fragment (scFv) antibody-conjugated MNPs as highly sensitive and selective probes to quickly and precisely detect and identify the fig mosaic virus (Soleimani Mashhadi et al., 2020).
Using Fe₃O₄ MNPs modified with protein A and particular antibodies produced in rabbits, an immunoaffinity probe was created in this study to selectively isolate Brucella bacteria from contaminated samples at low concentration levels. This was followed by identification using the MALDI-TOF MS technique. This proposed technique increased the limit of detection (LOD) of Brucella bacteria by a factor of 1,000 in aqueous medium and milk samples.
In this investigation, the following substances were used: CHEM LAB Co. (Zedelgem, Belgium) provided the sodium carbonate and sodium dodecyl sulfate (SDS). Sigma-Aldrich Co. (Altenburg, USA) provided the magnetic iron oxide nanoparticles (Fe3O₄ MNPs), acrylamide, N,N’-methylene bisacrylamide, sinapinic acid, and α-cyano-4-hydroxycinnamic acid (CHCA).
The following additional reagents were obtained from Merck Co. (Darmstadt, Germany): tetramethylethylenediamine (TEMED), Tris (hydroxymethyl) aminomethane, silver nitrate, tetraethyl orthosilicate (TEOS), N,N-dimethylformamide (DMF), succinic anhydride, disodium phosphate, potassium dihydrogen phosphate, glycine, ammonia solution (28%), acetonitrile (HPLC grade), absolute ethanol, formic acid, and trifluoroacetic acid (TFA).
The supplier of formaldehyde was Ghatranshimi Co. in Tehran, Iran. The supplier of ammonium persulfate (APS) was GE Healthcare Co. (Chicago, USA). We purchased 3-aminopropyltriethoxysilane (APTES), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), and N-hydroxysuccinimide (NHS) from Exir GmbH Co. (Vienna, Austria). Lastly, BIOCHEM Chemopharma Co. (Burgundy, France) was the supplier of potassium chloride and sodium chloride.
Initially, 100 mg of MNPs was subjected to a 10-min sonication. After adding ethanol (EtOH), water, ammonia, and TEOS, the mixture was sonicated for 3 h at room temperature. After that, ethanol was used to wash the MNPs. After adding APTES, water, ammonia, and EtOH, the mixture was sonicated for an hour at room temperature and then washed with ethanol to aminate the MNPs.
After the MNPs were cleaned, succinic anhydride was added, and the reaction was left to continue stirring all night. EDAC and NHS were added to the succinylated MNPs after they had been dissolved in 50 mM phosphate-buffered saline (PBS, pH 6.6) to activate the carboxyl groups (Mahmoud et al., 2005).
Following washing, the suspension was vortexed for 3 h and 2 mL of 500 ppm protein A solution was added (Ramandi et al., 2022). Two milliliters of 50 mM PBS (pH 6.6) was used to wash the MNPs three times for 10 min each time. For the SDS-PAGE analysis, 30 μL of each sample was kept. A Tris-HCl buffer (pH 8) was used to quench the MNPs’ unreacted sites.
The buffer was switched out for 50 mM PBS (pH 7.4) in order to encapsulate the antibody. After adding 2 mL of a 500 ppm Brucella antibody solution to the MNPs–protein A complex, the mixture was vortexed for 3 h. Two milliliters of 50 mM PBS (pH 7.4) was used to wash the MNPs three times for 10 min each time. Once more, 30 μL of every sample was saved for SDS-PAGE examination. For bacterial enrichment, the resultant MNPs–protein A–antibody complex was utilized (Figure 1).
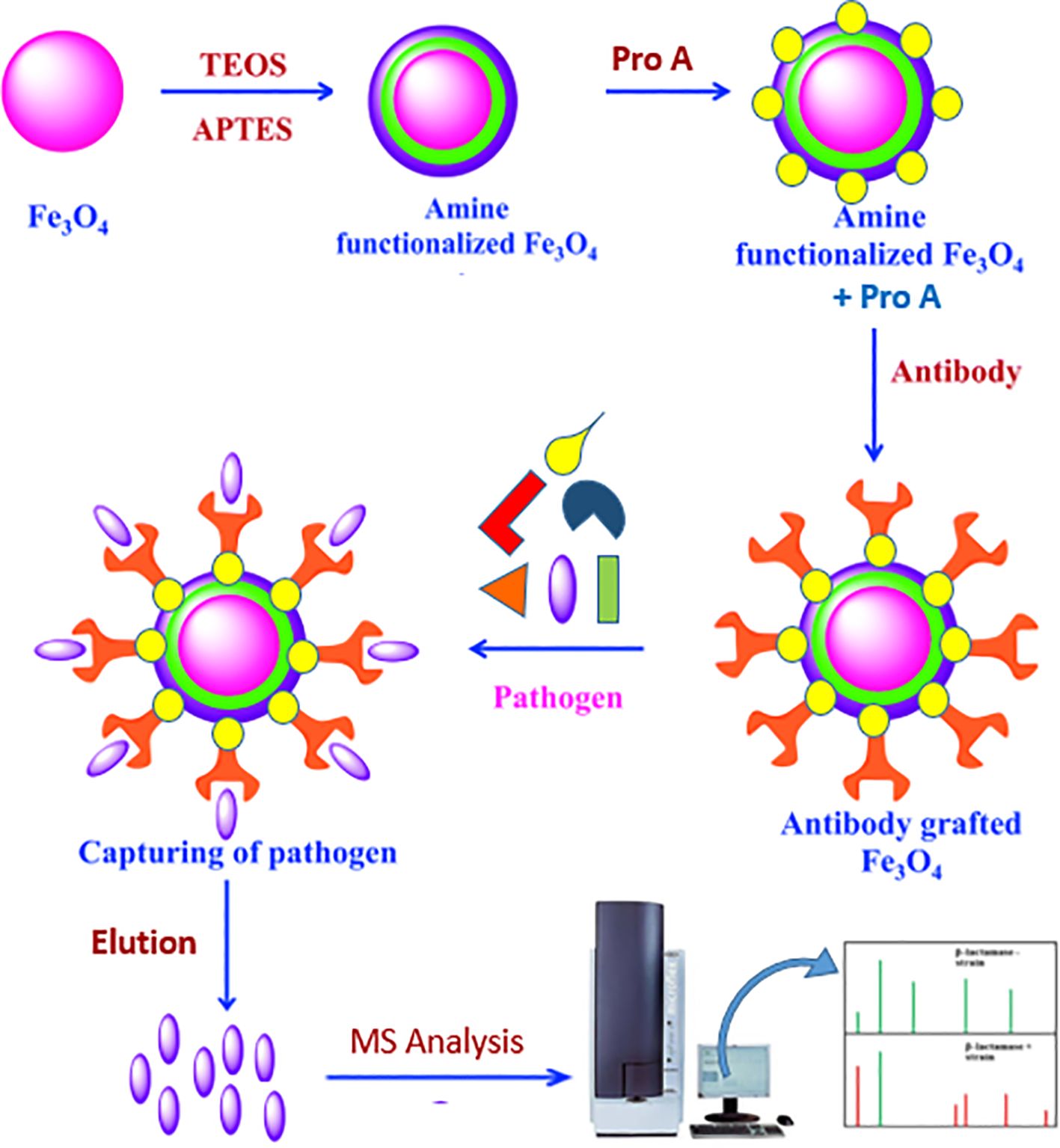
Figure 1. Activation of MNPs and immobilization of protein A and adsorption of antibody on it: TEOS (tetraethyl orthosilicate) and APTES (3-aminopropyltriethoxysilan).
Using the Bio-Rad system (Hercules, CA, USA), one-dimensional SDS-PAGE was performed. A 15% polyacrylamide gel made with Tris-glycine buffer was loaded with protein samples. For the best protein band separation, electrophoresis was carried out at a steady voltage.
To see the separated proteins, a silver nitrate staining procedure was used on the gel following electrophoresis. In short, the gel was sensitized in a sodium thiosulfate solution after being fixed in a methanol, acetic acid, and water solution to maintain the protein bands. After staining the gel with silver nitrate, it was developed with a developer solution based on formaldehyde until protein bands were visible. The gel was submerged in a stop solution to stop the reaction. High-sensitivity protein band detection was made possible by this procedure, guaranteeing that the separated proteins could be seen for further examination.
The reference strain Brucella melitensis 16M was obtained from the Razi Vaccine and Serum Research Institute (Karaj, Iran). The bacteria were cultured on Brucella agar plates (BBL Microbiological Systems, Cockeysville, MD, USA), a selective medium specifically designed to support the growth of Brucella species. The plates were incubated under aerobic conditions at 37°C for 6 days to allow for sufficient bacterial growth.
To ensure optimal bacterial proliferation, the agar plates were prepared fresh and maintained in sterile conditions. Colony morphology was observed daily to confirm the growth characteristics of B. melitensis 16M, and any contamination was ruled out through visual inspection. After the incubation period, bacterial colonies were harvested under aseptic conditions and prepared for downstream applications, such as protein extraction, immunoassay development, or mass spectrometric analysis.
A fresh overnight culture was diluted in PBS to create a bacterial solution with a concentration of 108 CFU/mL to generate antibodies against B. melitensis. Two doses of this suspension were administered to New Zealand White rabbits, with a 3-week gap between the first and booster shots. Two weeks following the booster injection, jugular vein blood samples were taken to extract antibody production. MNPs were employed to extract B. melitensis antibodies from the serum, which were then utilized for the following procedures.
The initial sample was diluted to reach a concentration of 50 billion bacteria after bacterial growth, then subsequent dilutions (50 to 5×108 CFU/mL) were made. Different concentrations of MNPs–protein A–antibody were applied to 5×106 CFU/mL of bacterial suspension to maximize bacterial separation. Bacteria attached to the MNPs–protein A–antibody conjugates were collected after 30 min of incubation at 37°C with constant shaking. They were then rinsed for 10 min with 1,000 μL of PBS (pH 7.4) and then again for 10 min with 1,000 μL of deionized water. After resuspending the particles in 15 μL of 70% formic acid and MS-grade acetonitrile, they were shaken to break down the bacterial cell walls and extract the proteins. The supernatant was spotted on a MALDI plate using the CHCA matrix for analysis.
An Applied Biosystems 4800 MALDI-TOF MS equipped with a Nd laser (200 Hz, AB Sciex, Canada) was used to obtain the mass spectra of bacterial proteins. CHCA (10.0 mg/mL in 2.5% TFA and 50% acetonitrile). The sample solution was added to the MALDI plate after 1 μL of the matrix solution. With a mass range of 2–20 kDa, the analysis was carried out in positive ion linear mode. Each sample received between 600 and 1,000 laser pulses, and the Data Explorer software (version 4.0) was used to create the average mass spectrum.
For bacterial inactivation and extraction, we employed a formic acid–ethanol extraction strategy. A bacterial suspension containing 6 to 10 colonies in 600 μL of water was vortexed for 10 s, followed by the addition of 1,000 μL of absolute ethanol. The mixture was carefully mixed and incubated at 20–25°C for 30 min and then centrifuged at 10,000 rpm for 10 min. The supernatant was removed, and the pellet was resuspended in 10 μL of 70% formic acid. Ten microliters of acetonitrile was added to ensure thorough mixing.
After centrifuging the mixture for 2 min at 11,000 rpm and 20–25°C, the supernatant was gathered and placed in a separate tube. The supernatant was put onto Brucella agar and cultured for 6 days at 37°C in order to evaluate the vitality of the bacteria. The effectiveness of the chemical procedures in deactivating Brucella spp. was validated by the lack of bacterial colonies.
The SDS-PAGE analysis revealed successful immobilization of protein A on MNPs. The presence of protein A was confirmed in the supernatant, wash 1, and wash 2, indicating that protein A was effectively fixed on the MNPs and was ready for antibody binding to capture specific antigens (S1). Additionally, Brucella antibodies are produced in rabbits and purified via MNPs–protein A. The SDS-PAGE gel demonstrating the MNPs–protein A–antibody compared to pure antibodies is illustrated (S2). The non-covalent bond formed between protein A and the Brucella antibody allows for the potential purification of the Brucella antibody through elution from the MNPs–protein A–antibody surface.
A dilution of 5×10³ CFU/mL of Brucella bacteria was incubated with the antibody-coated substrate, and the results were compared to a substrate devoid of antibodies to verify the development of the antibody–antigen bond. The proteins of the Brucella bacteria that were isolated from the antibody-coated substrate produced the anticipated protein spectrum, while no such spectrum was seen from the substrate without antibodies. Additionally, MNPs–protein A–antibody was given to 5×10³ CFU/mL of Escherichia coli to verify its specificity, and no peaks were observed. Considering the necessity to optimize the amount of MNPs–protein A–antibody for isolation, we tested different substrate concentrations (0.5, 1, 2, and 5 mg/mL) using 5×103 CFU/mL of Brucella bacteria (Table 1). Significant variations in protein spectrum intensity across the different substrate amounts led to the conclusion that 2 mg/mL was the ideal substrate concentration.
The antibody-coated substrate was also kept in PBS buffer at 4°C for 3 months to evaluate its shelf life. Effective contact with bacteria was validated by post-storage analysis, which also showed that the produced MNPs–protein A–antibody remained functional for a minimum of 3 months.
To assess the pre-concentration effectiveness of the MNPs–protein A–antibody, both with the optimal quantity of MNPs–protein A–antibody after extraction using MALDI-TOF MS and without trapping the bacteria on the substrate, bacterial proteins were examined at various dilutions. The colony count in the initial suspension was 5×108 CFU/mL. To extract the Brucella bacteria, several aqueous bacterial suspensions were made at dilutions ranging from 5×10⁸ to 50 CFU/mL. A LOD of 5×104 and 50 CFU/mL in the aqueous environment without and with pre-concentration, respectively, was found in the mass spectra derived from these investigations.
To verify the analysis of B. melitensis 16M, the obtained mass spectra were compared to reference spectra, which showed a sizable number of similar peaks. They were generated in an aqueous medium to continue a series of dilutions that interacted with 2 mg/mL of the substrate, ranging from 5×10⁸ to 50 CFU/mL of the original bacterial suspension. After the proteins from the Brucella bacteria were extracted, the data were examined and are shown in Table 2. The average standard deviation (SD) was 12.3, and the linear discriminating range (LDR) was 107, signifying a noteworthy 103-fold increase in the concentration coefficient.
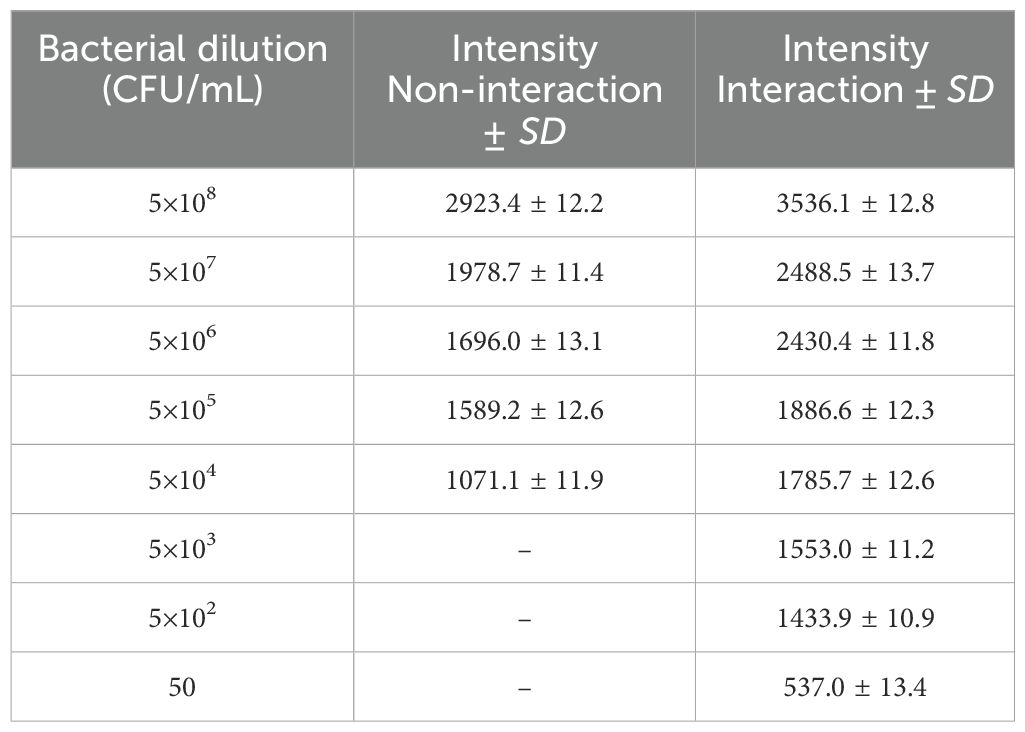
Table 2. Evaluate the pre-concentration efficacy of the MNPs–protein A–antibody, without trapping the bacteria on the substrate and with the optimized amount of MNPs–protein A–antibody following extraction via MALDI-TOF MS.
We referred to the average total viable count in urban samples, which is reported to be between 108 and 105 CFU/mL (Berhe et al., 2020; Mukhopadhyay et al., 2024), in order to further modify this method for biological applications. Therefore, we made bacterial dilutions ranging from 5×104 to 50 CFU/mL and added them to low-fat pasteurized milk as an actual sample analysis. Following the established procedure, we extracted the protein after permitting contact with the substrate. Figure 2 displays the outcome of the 50 CFU/mL dilution analysis.
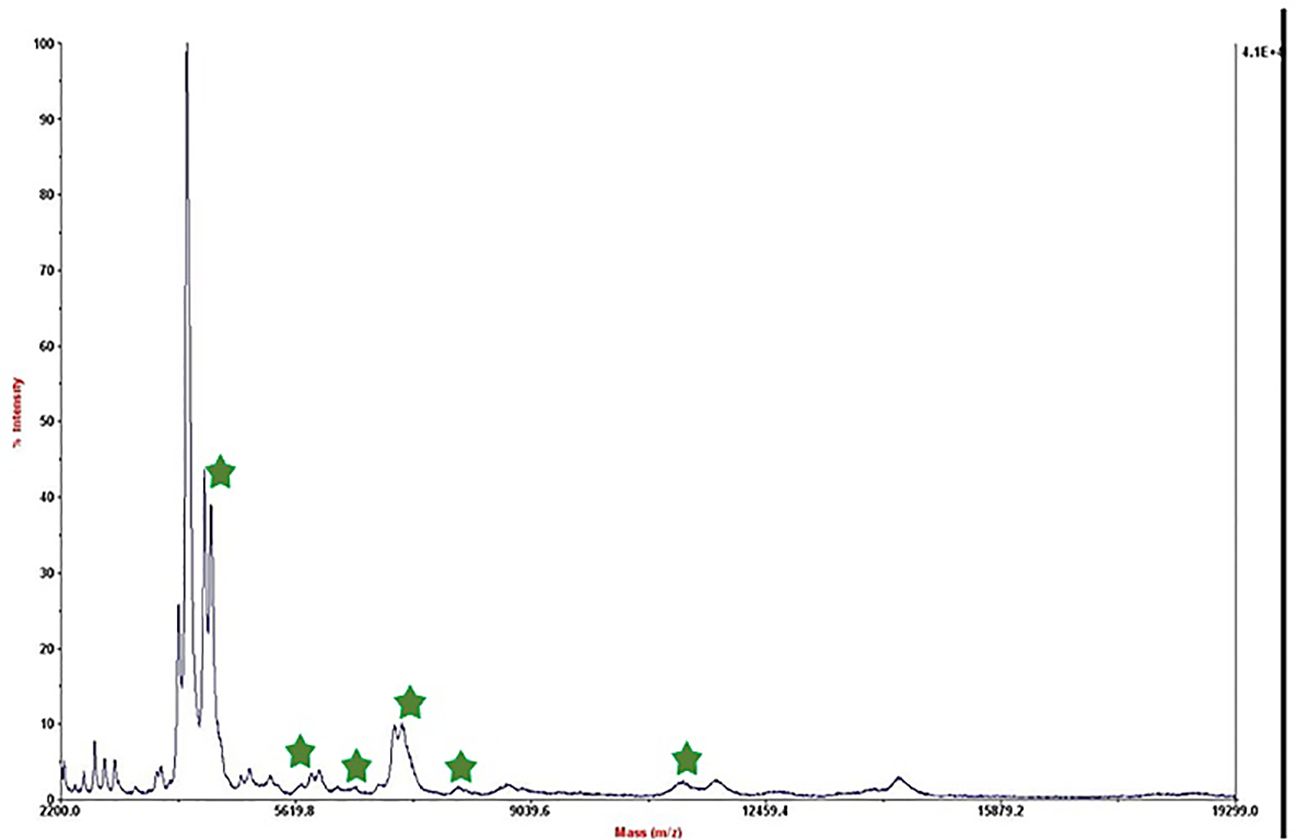
Figure 2. Mass spectrum of B. melitensis in 50 CFU/mL dilution in milk after interaction with MNPs–protein A–antibody.
The mass spectrum displayed significant interference from milk proteins (Šebela, 2022; Cuccato et al., 2022), prompting the exploration of several strategies to enhance the detection of bacterial protein peaks. The approaches employed included centrifugation of contaminated milk, followed by dilution of the sediment in water prior to interaction with the substrate (Punyapornwithaya et al., 2009; Tiedje et al., 2022), washing with a Tween-20-containing buffer, and adding additional washing steps (Barreiro et al., 2012). These methods were applied to bacterial dilutions of 5×104 to 50 CFU/mL, resulting in notable improvements, as shown in Figure 3. Ongoing efforts are focused on optimizing the analysis of smaller bacterial quantities. It means that the LOD of this technique is 50 CFU/mL and we can detect bacteria in milk samples. The significance of this method lies in its potential for rapid and accurate pathogen identification, thereby aiding medical professionals and contributing to public health. This report details our progress to date, with continued work aimed at validating the method for practical applications in clinical settings.
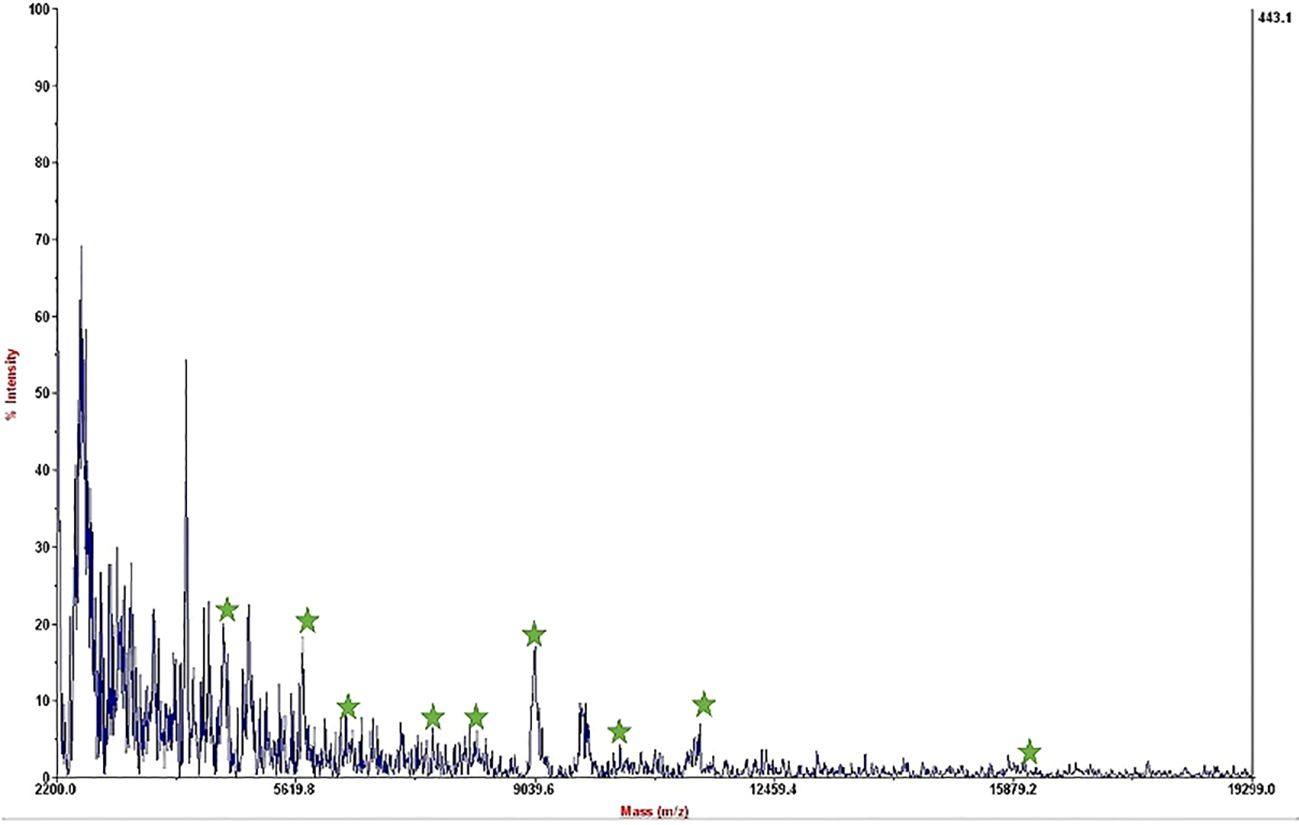
Figure 3. Mass spectrum of B. melitensis in 50 CFU/mL dilution in milk after interaction with MNPs–protein A–antibody and improvement.
We found that 2 mg/mL of MNPs–protein A–antibody is the ideal concentration for efficiently isolating pathogens in complex matrices like milk, where proteins and lipids can make isolation difficult. Our findings concur with those of Xiao et al (Xiao et al., 2022), who emphasized the significance of optimal MNP concentrations for raising the sensitivity of detection. Using MNPs in conjunction with immunoassay methods, we were able to detect B. melitensis in milk and water samples with an LOD of 50 CFU/mL. The application of polydopamine-coated MNPs for automated sepsis detection was recently demonstrated by Zhang et al., who achieved remarkable sensitivity with detection limits as low as 10² CFU/mL across a variety of bacterial species in blood samples (Houser et al., 2024).
Similar to our strategy of using MNPs for bacterial collection from milk, this degree of sensitivity highlights how well MNPs work to capture and concentrate germs from complicated matrices. Furthermore, Chen et al. reported that MNPs grafted with antibodies can greatly increase detection sensitivity (Chen et al., 2022). Our findings that antibody-functionalized MNPs improve the overall sensitivity of bacterial protein spectra by a factor of 10³ detection and expedite the isolation procedure are corroborated by this evidence. When compared to traditional culture methods, our method of using MALDI-TOF MS in conjunction with MNPs provides rapid identification capabilities in less than 60 min, which is a significant reduction in the time needed for bacterial identification. For prompt clinical decision-making and efficient patient care, this quick turnaround is essential.
Additionally, the significant problem of low bacterial abundance in samples is resolved by integrating MNPs with MALDI-TOF MS. Furthermore, new research highlights the adaptability and efficiency of iron oxide-based MNPs in the identification and management of bacteria (Svadlakova et al (Svadlakova et al., 2020)). Improvements in magnetic nanoparticle-based microfluidic systems that allow for the quick and accurate identification of harmful bacteria in a variety of sample types, including food and water, were covered by Han et al (Han et al., 2021). Furthermore, Ha et al. described methods that increase sensitivity and specificity even in complex food matrices (Ha and Kim, 2022) and demonstrated how magnetic nanoparticles improve immunoassays for pathogen detection.
By offering prompt and precise bacterial identification that is essential for efficient patient care and treatment, this quick turnaround has the potential to completely transform clinical microbiology. Although this approach may have a significant impact on clinical microbiology, its wider application will require additional verification of its precision and consistency across a range of bacterial strains and clinical settings.
Adapting this technique to a range of diseases requires high-affinity antibodies that target specific pathogen indicators, such as lipopolysaccharides or cell wall proteins for bacteria and capsid or envelope proteins for viruses. These antibodies ensure the sensitivity and specificity of the immunoaffinity enrichment. The high-throughput, rapid, and cost-effective technique of MALDI-TOF mass spectrometry enhances bacterial identification. However, in settings with limited resources, its high initial costs, maintenance needs, and reliance on skilled personnel may make it less accessible. Continuous sample preparation and efficient processes are necessary for scalability. Despite these challenges, MALDI-TOF offers scalable, cost-effective diagnostics with exceptional mass accuracy and precision.
Although MNPs have several drawbacks when used in therapeutic settings, they provide intriguing applications for bacterial enrichment. It is difficult to achieve specificity across a variety of bacterial populations; thus, current research attempts to improve the identification of mixed pathogens by developing particular substrates based on patient clinical circumstances and protein biomarkers (Sandrin and Demirev, 2018; Yang et al., 2018; Han et al., 2021). The efficiency of magnetic separation is affected by factors such as flux and magnetic field strength, which can be improved by synthesizing smaller MNPs and their quantity optimization for bacterial extraction (Socas-Rodríguez et al., 2020). Furthermore, complex biological matrices often interfere with binding and enrichment processes, requiring preconcentration methods such as centrifugation to clean up samples (Xu et al., 2019). Moreover, implementing MNP-based methods in clinical settings requires compliance with regulatory standards, which presents challenges in standardizing protocols across laboratories (Takallu et al., 2024). It will be essential to highlight these challenges to advance the use of MNPs in routine microbiological diagnostics.
Using this method within the context of an automated diagnostic platform or a field-deployable kit, MS is thus a viable strategy for advancing microbial diagnostics. Future research should involve the development of automated magnetic separation techniques that will improve the method’s efficiency as shown by various studies that have used vancomycin- and allantoin-conjugated MNPs for the rapid concentration of bacteria from the complex samples (Han et al., 2021; Abafogi et al., 2024). In addition, the development of portable diagnostic kits that contain MNPs will help in the identification of pathogens in the affected regions (Wen et al., 2017). The specificity, sensitivity, and selectivity of MNPs can be increased further to capture a wider range of bacterial strains through the use of new conjugation techniques (Gao et al., 2016). In general, these developments can be viewed as having the capacity to enhance microbial diagnostic capabilities in the clinical setting with speed and efficiency.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The experiments were conducted in accordance with the ethical standards of the National Institute of Genetic Engineering and Biotechnology (NIGEB) (IR.NIGEB.EC.1394.8.10).
AS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Writing – original draft. RN: Writing – review & editing, Data curation, Methodology, Resources. AG: Writing – review & editing, Supervision, Validation, Visualization.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors would like to thank Dr. Autosa AliAhmadi, Dr. MohammadAli As’habi, Dr. Hamideh Hamidi, Ms. Negar Saeedi, Dr. Marjan Talebi, Mr. Mohammad-Amin Ensandoost, and Mr. Saeed Nowrozi for their valuable and helpful comments to improve this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1531018/full#supplementary-material
Abafogi, A. T., et al. (2024). Automated sepsis detection with vancomycin-and allantoin-polydopamine magnetic nanoparticles. Sci. Rep. 14, 3693. doi: 10.1038/s41598-024-54236-0
Barreiro, J. R., et al. (2012). Nonculture-based identification of bacteria in milk by protein fingerprinting. Proteomics 12, 2739–2745. doi: 10.1002/pmic.201200053
Bauermeister, A., Mannochio-Russo, H., Costa-Lotufo, L. V., Jarmusch, A. K., Dorrestein, P. C. (2022). Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 20, 143–160. doi: 10.1038/s41579-021-00621-9
Becker, K., Lupetti, A. (2023). MALDI-TOF MS in microbiological diagnostics: future applications beyond identification. Front. Microbiol. 14, 1204452. doi: 10.3389/fmicb.2023.1204452
Berhe, G., Wasihun, A. G., Kassaye, E., Gebreselasie, K. (2020). Milk-borne bacterial health hazards in milk produced for commercial purpose in Tigray, northern Ethiopia. BMC Public Health 20, 1–8. doi: 10.1186/s12889-020-09016-6
Chen, Y.-J., et al. (2022). Microneedle patches integrated with lateral flow cassettes for blood-free chronic kidney disease point-of-care testing during a pandemic. Biosens. Bioelectron. 208, 114234. doi: 10.1016/j.bios.2022.114234
Cheng, K., Chui, H., Domish, L., Hernandez, D., Wang, G. (2016). Recent development of mass spectrometry and proteomics applications in identification and typing of bacteria. PROTEOMICS–Clinical Appl. 10, 346–357. doi: 10.1002/prca.201500086
Cloeckaert, A. (2024). Insights in infectious agents and disease: 2022. Front. Microbiol. 15, 1443636. doi: 10.3389/fmicb.2024.1443636
Cuccato, M., Divari, S., Sacchi, P., Girolami, F., Cannizzo, F. T. (2022). MALDI-TOF mass spectrometry profiling of bovine skim milk for subclinical mastitis detection. Front. Vet. Sci. 9, 1009928. doi: 10.3389/fvets.2022.1009928
de Oliveira, L. M. A., Ribeiro, R. L., Ganda, E. (2024). Foodborne bacterial pathogens under the One Health perspective-antimicrobial resistance, epidemiology, virulence, and zoonotic impact. Front. Cell. Infection Microbiol. 14, 1379188. doi: 10.3389/fcimb.2024.1379188
Dharmarajan, G., et al. (2022). The animal origin of major human infectious diseases: what can past epidemics teach us about preventing the next pandemic? Zoonoses 2, 989. doi: 10.15212/ZOONOSES-2021-0028
Foster, T., Khaiboullina, S. (2023). Community series-innovative approaches in diagnosis of emerging/re-emerging infectious diseases, volume II. Front. Microbiol. 14, 1193841. doi: 10.3389/fmicb.2023.1193841
Gao, X.-L., et al. (2016). Non-selective separation of bacterial cells with magnetic nanoparticles facilitated by varying surface charge. Front. Microbiol. 7, 1891. doi: 10.3389/fmicb.2016.01891
Glajzner, P., Bernat, A., Jasińska-Stroschein, M. (2024). Improving the treatment of bacterial infections caused by multidrug-resistant bacteria through drug repositioning. Front. Pharmacol. 15, 1397602. doi: 10.3389/fphar.2024.1397602
Ha, Y., Kim, I. (2022). Recent developments in innovative magnetic nanoparticles-based immunoassays: from improvement of conventional immunoassays to diagnosis of COVID-19. BioChip J. 16, 351–365. doi: 10.1007/s13206-022-00064-1
Haider, A., Ringer, M., Kotroczó, Z., Mohácsi-Farkas, C., Kocsis, T. (2023). The current level of MALDI-TOF MS applications in the detection of microorganisms: a short review of benefits and limitations. Microbiol. Res. (Pavia). 14, 80–90. doi: 10.3390/microbiolres14010008
Hamidi, H., Bagheri Nejad, R., Es-Haghi, A., Ghassempour, A. (2022). A combination of MALDI-TOF MS proteomics and species-unique biomarkers’ discovery for rapid screening of brucellosis. J. Am. Soc Mass Spectrom. 33, 1530–1540. doi: 10.1021/jasms.2c00110
Han, H., Sohn, B., Choi, J., Jeon, S. (2021). Recent advances in magnetic nanoparticle-based microfluidic devices for the pretreatment of pathogenic bacteria. Biomed. Eng. Lett. 11, 297–307. doi: 10.1007/s13534-021-00202-y
Houser, B. J., et al. (2024). Bacterial binding to polydopamine-coated magnetic nanoparticles. ACS Appl. Mater. Interfaces. doi: 10.1021/acsami.4c11169
Kang, H., et al. (2024). Rapid identification of bloodstream infection pathogens and drug resistance using Raman spectroscopy enhanced by convolutional neural networks. Front. Microbiol. 15, 1428304. doi: 10.3389/fmicb.2024.1428304
Kumari, M., Klodzinska, S. N., Chifiriuc, M. C. (2023). Microbe-nanoparticle interactions: a mechanistic approach. Front. Microbiol. 14, 1273364. doi: 10.3389/fmicb.2023.1273364
Kurli, R., et al. (2018). Cultivable microbial diversity associated with cellular phones. Front. Microbiol. 9, 1229. doi: 10.3389/fmicb.2018.01229
Li, B., et al. (2022). Performance evaluation and clinical validation of optimized nucleotide MALDI-TOF-MS for mycobacterial identification. Front. Cell. Infect. Microbiol. 12, 1079184. doi: 10.3389/fcimb.2022.1079184
Lorente-Leal, V., et al. (2022). MALDI-TOF mass spectrometry as a rapid screening alternative for non-tuberculous mycobacterial species identification in the veterinary laboratory. Front. Vet. Sci. 9, 827702. doi: 10.3389/fvets.2022.827702
Mahmoud, K. A., Long, Y., Schatte, G., Kraatz, H. (2005). Rearrangement of the active ester intermediate during HOBt/EDC amide coupling. Eur. J. Inorg. Chem. 2005, 173–180. doi: 10.1002/ejic.200400504
Mukhopadhyay, M., Malviya, J., Barik, A., Asthana, N. (2024). Assessing the microbial contamination levels in milk samples from rural and urban areas: A focus on raisen and bhopal districts. Macromol. Symp. 413, 2300093. doi: 10.1002/masy.202300093
Nieto, K., Salvetti, A. (2014). AAV vectors vaccines against infectious diseases. Front. Immunol. 5, 5. doi: 10.3389/fimmu.2014.00005
Nomura, F., Tsuchida, S., Murata, S., Satoh, M., Matsushita, K. (2020). Mass spectrometry-based microbiological testing for blood stream infection. Clin. Proteomics 17, 1–11. doi: 10.1186/s12014-020-09278-7
Olivares, J., et al. (2013). The intrinsic resistome of bacterial pathogens. Front. Microbiol. 4, 103. doi: 10.3389/fmicb.2013.00103
Oviaño, M., et al. (2021). Multicenter evaluation of rapid BACpro® II for the accurate identification of microorganisms directly from blood cultures using MALDI-TOF MS. Diagnostics 11, 2251.
Padilla, P. F., Nielsen, K., Ernesto, S. L., Ling, Y. W. (2010). Diagnosis of brucellosis. Open Vet. Sci. J. 4, 46–60. doi: 10.2174/1874318801004010046
Pastrone, L., et al. (2023). Evaluation of two different Preparation protocols for MALDI-TOF MS nontuberculous mycobacteria identification from Liquid and Solid Media. Microorganisms 11, 120. doi: 10.3390/microorganisms11010120
Peng, J., Tang, Y.-W., Xiao, D. (2022). Progress in pathogen identification based on mass spectrometry. Front. Cell. Infection Microbiol. 11, 813133. doi: 10.3389/fcimb.2021.813133
Perše, G., et al. (2022). Sepsityper® kit versus in-house method in rapid identification of bacteria from positive blood cultures by MALDI-TOF mass spectrometry. Life 12, 1744.
Punyapornwithaya, V., Fox, L. K., Gay, G. M., Hancock, D. D., Alldredge, J. R. (2009). The effect of centrifugation and resuspension on the recovery of Mycoplasma species from milk. J. Dairy Sci. 92, 4444–4447. doi: 10.3168/jds.2009-2182
Qin, S., et al. (2023). Case report: A case of brucellosis misdiagnosed as coronavirus disease 2019/influenza in China. Front. Public Heal. 11, 1186800. doi: 10.3389/fpubh.2023.1186800
Rahi, P., Vaishampayan, P. (2020). MALDI-TOF MS application in microbial ecology studies. Front. Microbiol. 10, 2954. doi: 10.3389/fmicb.2019.02954
Ramandi, N. F., et al. (2022). Study of Glutathione S-transferase-P1 in cancer blood plasma after extraction by affinity magnetic nanoparticles and monitoring by MALDI-TOF, IM-Q-TOF and LC-ESI-Q-TOF MS. J. Chromatogr. B 1190, 123091.
Sandrin, T. R., Demirev, P. A. (2018). Characterization of microbial mixtures by mass spectrometry. Mass Spectrom. Rev. 37, 321–349. doi: 10.1002/mas.21534
Schaumburg, F., Carrell, C. S., Henry, C. S. (2019). Rapid bacteria detection at low concentrations using sequential immunomagnetic separation and paper-based isotachophoresis. Anal. Chem. 91, 9623–9630. doi: 10.1021/acs.analchem.9b01002
Šebela, M. (2022). Biomolecular profiling by MALDI-TOF mass spectrometry in food and beverage analyses. Int. J. Mol. Sci. 23, 13631. doi: 10.3390/ijms232113631
Socas-Rodríguez, B., Herrera-Herrera, A. V., Asensio-Ramos, M., Rodríguez-Delgado, M.Á. (2020). Recent applications of magnetic nanoparticles in food analysis. Processes 8, 1140. doi: 10.3390/pr8091140
Soleimani Mashhadi, I., Safarnejad, M. R., Shahmirzaie, M., Aliahmadi, A., Ghassempour, A. (2020). Conjugation of single-chain variable fragment antibody to magnetic nanoparticles and screening of fig mosaic virus by MALDI TOF mass spectrometry. Anal. Chem. 92, 10460–10469. doi: 10.1021/acs.analchem.0c01119
Svadlakova, T., et al. (2020). Proinflammatory effect of carbon-based nanomaterials: In vitro study on stimulation of inflammasome NLRP3 via destabilisation of lysosomes. Nanomaterials 10, 418. doi: 10.3390/nano10030418
Takallu, S., et al. (2024). Nanotechnology improves the detection of bacteria: Recent advances and future perspectives. Heliyon. doi: 10.1016/j.heliyon.2024.e32020
Tiedje, J. M., et al. (2022). Microbes and climate change: a research prospectus for the future. MBio 13, e00800–e00822. doi: 10.1128/mbio.00800-22
Tsuchida, S., Nakayama, T. (2022). MALDI-based mass spectrometry in clinical testing: Focus on bacterial identification. Appl. Sci. 12, 2814. doi: 10.3390/app12062814
Wang, H., et al. (2016). Evaluation of the Bruker Biotyper matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system for identification of clinical and environmental isolates of Burkholderia pseudomallei. Front. Microbiol. 7, 415. doi: 10.3389/fmicb.2016.00415
Wang, Y.-C., Lee, Y.-T., Matsuura, K., Liu, X., Cheng, C.-M. (2022). Detection nanodevices for infectious diseases. Front. Bioeng. Biotechnol. 10, 962746. doi: 10.3389/fbioe.2022.962746
Wen, C.-Y., et al. (2017). Efficient enrichment and analyses of bacteria at ultralow concentration with quick-response magnetic nanospheres. ACS Appl. Mater. Interfaces 9, 9416–9425. doi: 10.1021/acsami.6b16831
Xiao, F., et al. (2022). Rapid enrichment and detection of Staphylococcus aureus in milk using polyethyleneimine functionalized magnetic nanoparticles. Microchem. J. 178, 107388. doi: 10.1016/j.microc.2022.107388
Xu, C., Akakuru, O. U., Zheng, J., Wu, A. (2019). Applications of iron oxide-based magnetic nanoparticles in the diagnosis and treatment of bacterial infections. Front. Bioeng. Biotechnol. 7, 141. doi: 10.3389/fbioe.2019.00141
Yagupsky, P., Morata, P., Colmenero, J. D. (2019). Laboratory diagnosis of human brucellosis. Clin. Microbiol. Rev. 33, 10–1128. doi: 10.1128/CMR.00073-19
Yang, Y., Lin, Y., Qiao, L. (2018). Direct MALDI-TOF MS identification of bacterial mixtures. Anal. Chem. 90, 10400–10408. doi: 10.1021/acs.analchem.8b02258
Zhang, X., Zhang, D., Zhang, X., Zhang, X. (2024). Artificial intelligence applications in the diagnosis and treatment of bacterial infections. Front. Microbiol. 15, 1449844. doi: 10.3389/fmicb.2024.1449844
Keywords: immunoassay-mass spectrometry, Brucella spp., magnetic nanoparticles, milk, MALDI-TOF MS
Citation: Sharif A, Nejad RB and Ghassempour A (2025) Immunoassay–mass spectrometry to identify Brucella melitensis. Front. Cell. Infect. Microbiol. 15:1531018. doi: 10.3389/fcimb.2025.1531018
Received: 19 November 2024; Accepted: 09 January 2025;
Published: 04 February 2025.
Edited by:
Diana Manolescu, Victor Babes University of Medicine and Pharmacy, RomaniaReviewed by:
Mirella Luciani, Experimental Zooprophylactic Institute of Abruzzo and Molise G. Caporale, ItalyCopyright © 2025 Sharif, Nejad and Ghassempour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alireza Ghassempour, YV9naGFzc2VtcG91ckB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.