
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 09 August 2023
Sec. Clinical Microbiology
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1196084
This article is part of the Research TopicDesign and development of new therapeutics against infectious diseases using computational and experimental approachesView all 13 articles
 Siqi Shen1,2
Siqi Shen1,2 FeiFei Ren1,2
FeiFei Ren1,2 Haiming Qin1,2
Haiming Qin1,2 Ihtisham Bukhari1,2
Ihtisham Bukhari1,2 Jing Yang3
Jing Yang3 Dafang Gao3
Dafang Gao3 Arthur C. Ouwehand4
Arthur C. Ouwehand4 Markus J. Lehtinen4
Markus J. Lehtinen4 Pengyuan Zheng1,2*
Pengyuan Zheng1,2* Yang Mi1,2*
Yang Mi1,2*Purpose: To determine the role of Lactobacillus strains and their combinations in inhibiting the colonization of H. pylori and gastric mucosa inflammation.
Methods: Human gastric adenocarcinoma AGS cells were incubated with H. pylori and six probiotic strains (Lactobacillus acidophilus NCFM, L. acidophilus La-14, Lactiplantibacillus plantarum Lp-115, Lacticaseibacillus paracasei Lpc-37, Lacticaseibacillus rhamnosus Lr-32, and L. rhamnosus GG) and the adhesion ability of H. pylori in different combinations was evaluated by fluorescence microscopy and urease activity assay. Male C57BL/6 mice were randomly divided into five groups (uninfected, H. pylori, H. pylori+NCFM, H. pylori+Lp-115, and H. pylori+NCFM+Lp-115) and treated with two lactobacilli strains (NCFM and Lp-115) for six weeks. H. pylori colonization and tissue inflammation statuses were determined by rapid urease test, Hematoxylin-Eosin (HE) staining, immunohistochemistry, and qRT-PCR and ELISA.
Results: L. acidophilus NCFM, L. acidophilus La-14, L. plantarum Lp-115, L. paracasei Lpc-37, L. rhamnosus Lr-32, and L. rhamnosus GG reduced H. pylori adhesion and inflammation caused by H. pylori infection in AGS cells and mice. Among all probiotics L. acidophilus NCFM and L. plantarum, Lp-115 showed significant effects on the H. pylori eradication and reduction of inflammation in-vitro and in-vivo. Compared with the H. pylori infection group, the mRNA and protein expression levels of IL-8 and TNF-α in the six Lactobacillus intervention groups were significantly reduced. The changes in the urease activity (ureA and ureB) for 1-7h in each group showed that L. acidophilus NCFM, L. acidophilus La-14, L. plantarum Lp-115, and L. rhamnosus GG effectively reduced the colonization of H. pylori. We observed a higher ratio of lymphocyte and plasma cell infiltration into the lamina propria of the gastric mucosa and neutrophil infiltration in H. pylori+NCFM+Lp-115 mice. The infiltration of inflammatory cells in lamina propria of the gastric mucosa was reduced in the H. pylori+NCFM+Lp-115 group. Additionally, the expression of IFN-γ was decreased significantly in the NCFM and Lp-115 treated C57BL/6 mice.
Conclusions: L. acidophilus NCFM and L. plantarum Lp-115 can reduce the adhesion of H. pylori and inhibit the gastric inflammatory response caused by H. pylori infection.
Helicobacter pylori infect over 50% of the population worldwide, and the World Health Organization (WHO) has listed H. pylori as a class I carcinogen since 1994 (Hooi et al., 2017; Shah et al., 2021). H. pylori infection is closely related to the occurrence and development of various gastrointestinal diseases, such as chronic gastritis, peptic ulcer, gastric cancer, and gastric mucosa associated lymphoid tissue lymphoma (Sugano et al., 2015; Liu et al., 2018; Liou et al., 2020; Robinson and Atherton, 2021). Currently, the primary method to eradicate H. pylori is a quadruple therapy based on a proton pump inhibitor, two antibiotics, and a bismuth agent (Fallone et al., 2016; Chey et al., 2017). However, the antibiotic resistance rate of H. pylori has increased, and the side effects of the eradication therapy can be severe (Savoldi et al., 2018). Therefore, searching for novel and efficient H. pylori management options has become an urgent aim (Fallone et al., 2019).
Studies on probiotics and H. pylori have made significant progress recently, thus, increasingly being used in routine clinics (Suez et al., 2019; Sousa et al., 2022). Currently, blends of probiotics are the most widely studied, but little is known about the antagonistic or synergistic effects of the different probiotic strains (Vieira et al., 2013; Ouwehand et al., 2018; Simon et al., 2021). To manage H. pylori infection, the Maastricht VI/Florence Consensus Report mentioned that only some probiotics could effectively reduce gastrointestinal side effects in H. pylori eradication therapy, suggesting strain-specific efficacy (Malfertheiner et al., 2022). However, the European Society of Paediatric Gastroenterology and Hepatology later updated the guidelines. They considered that the existing evidence was insufficient to support the routine use of single or compound probiotic strains in treating H. pylori to reduce adverse reactions and improve the eradication rate (Jones et al., 2017). Therefore, probiotics are mainly used as an adjunct to H. pylori eradication therapy, and only a few reports are available for using probiotics as a single treatment for H. pylori infection, and further investigations are warranted.
The applications of certain probiotics, such as lactobacilli, fecal bacteria, Bifidobacterium spp., Saccharomyces spp., and Bacillus licheniformis, to assist in H. pylori eradication have been incorporated into H. pylori treatment guidelines (Shi et al., 2019). These probiotics attenuate the gastrointestinal adverse effects of H. pylori eradication therapy, but whether they can improve H. pylori eradication rates is controversial (Liu et al., 2018). Meta-analyses on the efficacy of multiple probiotic strains in treating H. pylori have shown the most significant effects with lactobacilli (Lu et al., 2016; McFarland et al., 2016). In related studies of using lactobacilli for treating H. pylori infection, certain lactobacilli such as Lactobacillus acidophilus, Lacticaseibacillus rhamnosus, Lactiplantibacillus plantarum, Lacticaseibacillus paracasei, Limosilactobacillus reuteri and Lactobacillus delbrueckii subsp. bulgaricus can effectively manage H. pylori infection (Zhao et al., 2018; Chen et al., 2019; Yoon et al., 2019; Asgari et al., 2020; Lin et al., 2020; Dargenio et al., 2021) but underlying mechanisms are not well explained. However, it has been speculated that lactobacilli interfere with the adhesion of H. pylori to the mucosa and down-regulate the immune and inflammatory mediators (Keikha and Karbalaei, 2021).
In this study, six lactobacilli strains with good acid, bile salt, and digestive enzyme resistance, combined with good mucosal adhesion were used in screening experiments to identify probiotics that inhibit the adhesion and inflammatory response to H. pylori. We tested the selected probiotics in the H. pylori infected AGS cell line and mouse models. The results of the cell model experiments provided a basis for probiotic strain selection for the eradication of H. pylori in the mouse model.
H. pylori P12 and H. pylori P12-GFP strains were provided by the Max Planck Institute for Infection Biology and H. pylori SS1 (ATCC 43504) (Lee et al., 1997) was provided by the University of Western Australia (UWA), Australia. H. pylori strains were cultured on Columbia agar containing 7% sterile defibrinated sheep blood (Bianzhen, Nanjing, China), 20μg/ml vancomycin (Meilunbio, Dalian, China), 10μg/ml polymyxin (Meilunbio), 10μg/ml amphotericin B (Meilunbio), 10μg/ml trimethoprim (Sigma, St. Louis, USA), then placed in an incubator containing 5% O2 and 10% CO2, cultured at 37°C, subcultured once every three days, and used for the experiment after subculture. The tested lactobacilli were provided by Danisco China (Shanghai, China): Lactobacillus acidophilus NCFM (ATCC 7003969), L. acidophilus La-14 (ATCC SD5212), Lactiplantibacillus plantarum Lp-115 (ATCC SD5209), Lacticaseibacillus paracasei Lpc-37 (ATCC SD5275), Lacticaseibacillus rhamnosus Lr-32 (ATCC SD5217) and L. rhamnosus GG (ATCC 53103). After the Gram Staining Kit (Solarbio, Beijing, China) was used to identify the bacterial morphology, the lactobacilli were cultured anaerobically in MRS broth (Solarbio) at 37 °C 48 hours and then subcultured (Figure S1).
The human gastric adenocarcinoma cell line (AGS) was purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 medium (Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum at 37°C and 5% CO2 in a humidified incubator.
Fifty male C57BL/6 mice, Specific Pathogen Free (SPF), four weeks old, were purchased from Zhejiang Vital River Laboratory Animal Technology Co. Ltd. (Zhejiang, China). All the animals were housed under standard conditions (SPF grade animal room with individually ventilated cages; temperature range from 23°C to 25°C, humidity range from 50% to 60%, 12/12 hours light/dark cycle, food and water were provided ad libitum). The experimental steps and ethics were approved by the ethics committee of the Fifth Affiliated Hospital of Zhengzhou University (KY2022002).
After infection, the culture medium was collected by centrifugation at 12000x rpm for 5 min to remove cell debris and bacteria and collect the supernatant. The concentration of interleukin (IL)-8 and tumor necrosis factor (TNF)-α were detected by human IL-8 ELISA Kit (Elabscience, Wuhan, China) and human TNF-α ELISA Kit (Elabscience, Houston, TX, USA) respectively, by following the guidelines of the manufacturer. All experiments were performed in triplicate.
H. pylori P12 and lactobacilli were cultured overnight in BHI (Thermo Fisher) and MRS broth. After centrifugation at 5000 x rpm for 8 min and 4000 x rpm for 5 min, the supernatant was discarded, and bacteria were harvested and resuspended in 1 ml serum-free RPMI 1640 medium.
Overnight cultured AGS cells were co-incubated with lactobacilli (multiplicity of infection (MOI) = 100) and H. pylori P12 (MOI = 100) for 6h. After the incubation period, the supernatant was harvested for ELISA and cells were harvested for RNA isolation.
AGS cells were cultured in two 12 well plates and incubated with lactobacilli (MOI = 100) and H. pylori P12-GFP (MOI = 100) for 6h. At the end of incubation, one plate of the cells was washed thrice with PBS and photographed by fluorescence and white light. A urease detection reagent was prepared by adding concentrated hydrochloric acid into the PBS (pH=7.4) to adjust the solution to pH=6.8. Urea (Solarbio) and phenol red (Solarbio) were weighed and added to reach concentrations of 110mmol/L and 10mg/L, respectively, and then dissolved by vigorously shaking. To the other 12well plates, 1 ml urease detection reagent was added. After 1-7 hours of reaction, 80μl medium was withdrawn to record the absorbance value at 540nm(Shmuely et al., 2004; Rokka et al., 2008; Tharmalingam et al., 2014; Yang et al., 2020).
AGS cells and mouse gastric mucosal tissue were lysed with RNAiso plus (TaKaRa, Kyoto, Japan), and gastric mucosal tissue needed to be assisted by an ultrasonic crusher. According to the manufacturer’s instructions, cDNA was converted using the ReverTra Ace qPCR RT Kit (TOYOBO, Shanghai, China). qRT-PCR was performed using 2×ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) in a Roche Lightcycler480II system based on the manufacturer’s recommendations. Primer sequences were designed in NCBI Primer-BLAST (Table 1). The relative gene expression was determined using the 2-ΔΔCt method. All experiments were repeated thrice.
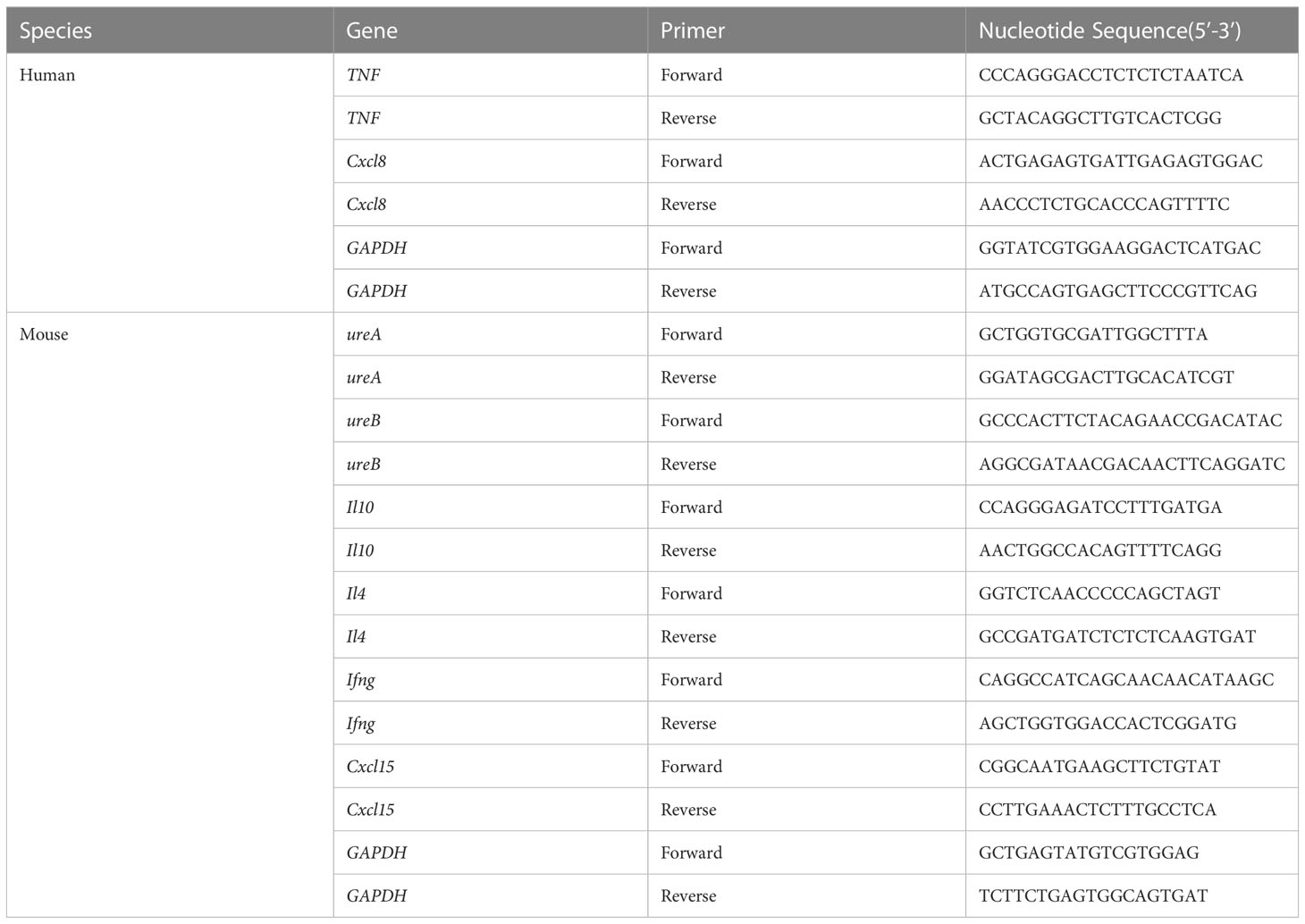
Table 1 Primers for the quantification of inflammatory factors in AGS cells and mouse gastric tissue.
SPF C57BL/6 mice (male 4 weeks old) were obtained from Zhejiang Vital River Laboratory Animal Technology Co., Ltd. (Zhejiang, China) and fed on Laboratory Rodent Diet 5001. The mice were randomly divided into 5 groups (Uninfected, H. pylori SS1, H. pylori SS1+L. acidophilus NCFM, H. pylori SS1+L. plantarum Lp-115, H. pylori SS1+L. acidophilus NCFM and L. plantarum Lp-115) of 10 individuals each after one week of adaptation. The control group was gavaged with PBS, and the experimental groups were gavaged with H. pylori SS1 (1*109 CFU, 0.2ml/piece) only or combined with the corresponding lactobacilli (1*109 CFU, 0.2ml/piece) (1:1) or together (1:1:1) once every other day for 6 weeks (Figure 1A).
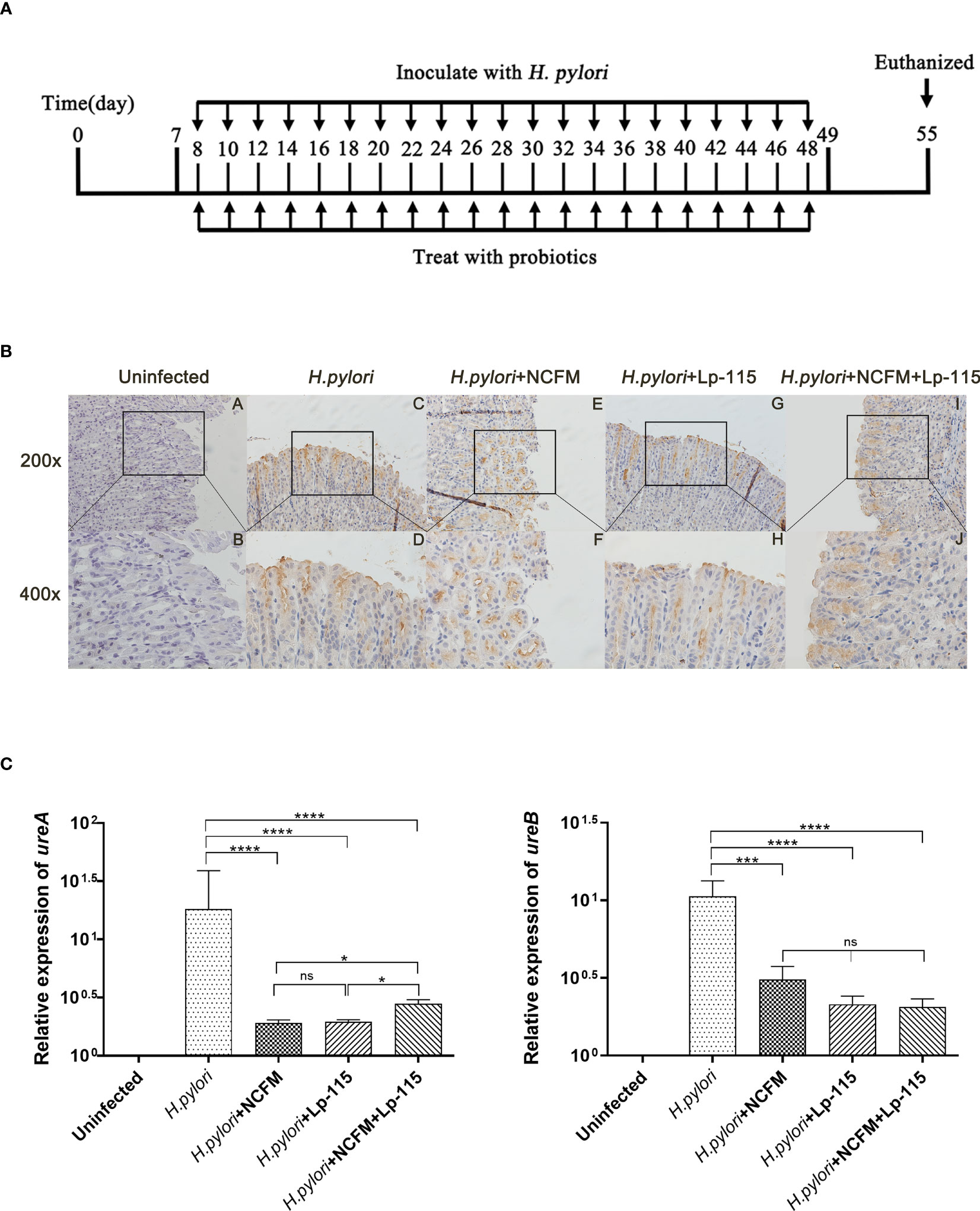
Figure 1 Lactobacillus acidophilus NCFM and Lactiplantibacillus plantarum Lp-115 suppress Helicobacter pylori adhesion in mice. (A) Mice were fed with H. pylori SS1 only or with L. acidophilus NCFM and/or L. plantarum Lp-115 for 6 weeks. (B) The colonization of H. pylori was identified by immune histochemistry. Uninfected group; H. pylori group; H. pylori+ NCFM group; H. pylori + Lp-115 group; H. pylori +NCFM+Lp-115 group. (C) The colonization of H. pylori was identified by the expression of ureA and ureB by qRT-PCR. Mice were coinfected with H. pylori SS1 and L. acidophilus NCFM and/or L. plantarum Lp-115. The mRNA levels of ureA and ureB were determined as described. Each experiment result shows the mean ± standard deviation of three independent experiments. * (P< 0.05); **(P<0.01); ***(P<0.001); ns, not significant.
Mouse gastric mucosa was analyzed for urease activity using the rapid urease Kit (Sanqiang, Fujian, China) as instructed by the manufacturer. The color change was determined, when the solution turns pink or red, the urease test is positive; when it remains yellow, the urease test is negative.
Gastric tissues were fixed with 4% paraformaldehyde, embedded in paraffin, cut into 4mm sections, and stained by HE and immunohistochemical methods. The Chronic gastritis histological grade scale was used to evaluate the samples (Fang et al., 2018). Five histological changes were graded: H. pylori, chronic inflammation, activity, atrophy, and intestinal metaplasia. Each histological change was classified into one of four grades: none, mild, moderate, and severe. According to the new Sydney system, the degree of inflammation and lymphocyte infiltration of gastric tissue after H. pylori infection were evaluated (Kim et al., 2020; Maluf et al., 2020). Immunohistochemistry for H. pylori was performed using a Rabbit anti-H. pylori polyclonal antibody (Cell Marque) (ZSGB, Beijing, China), and the colonies of H. pylori in mouse gastric mucosa were identified.
All statistical analyses were performed using GraphPad Prism 9 software. During the processing of experimental data, the values whose deviation from the average value of the same group of data exceeded three times the standard deviation were considered outliers and eliminated. We performed one-way ANOVA on raw and lg-converted data to compare the multi groups. The Bonferroni and Tukey tests were conducted to calculate the statistical significance among the groups. P-value < 0.05 was considered significant for all statistical analyses.
To compare the effect of the six selected lactobacilli interfering with H. pylori adhesion, AGS cells were co-infected with H. pylori P12-GFP and the six lactobacilli strains (MOI=100) respectively for 6 hours. After infection, cells and GFP-positive H. pylori were measured under fluorescence microscopy. Upon comparative observation under fluoroscopy, the amount of GFP-positive H. pylori in the lactobacilli groups was less than in the H. pylori only infected group. Furthermore, the morphology of AGS cells was relatively normal in the L. acidophilus NCFM and L. plantarum Lp-115 treated cells compared with the other lactobacilli treated cells, indicating less cell stress on the AGS cells compared with other lactobacilli strains (Figure 2A).
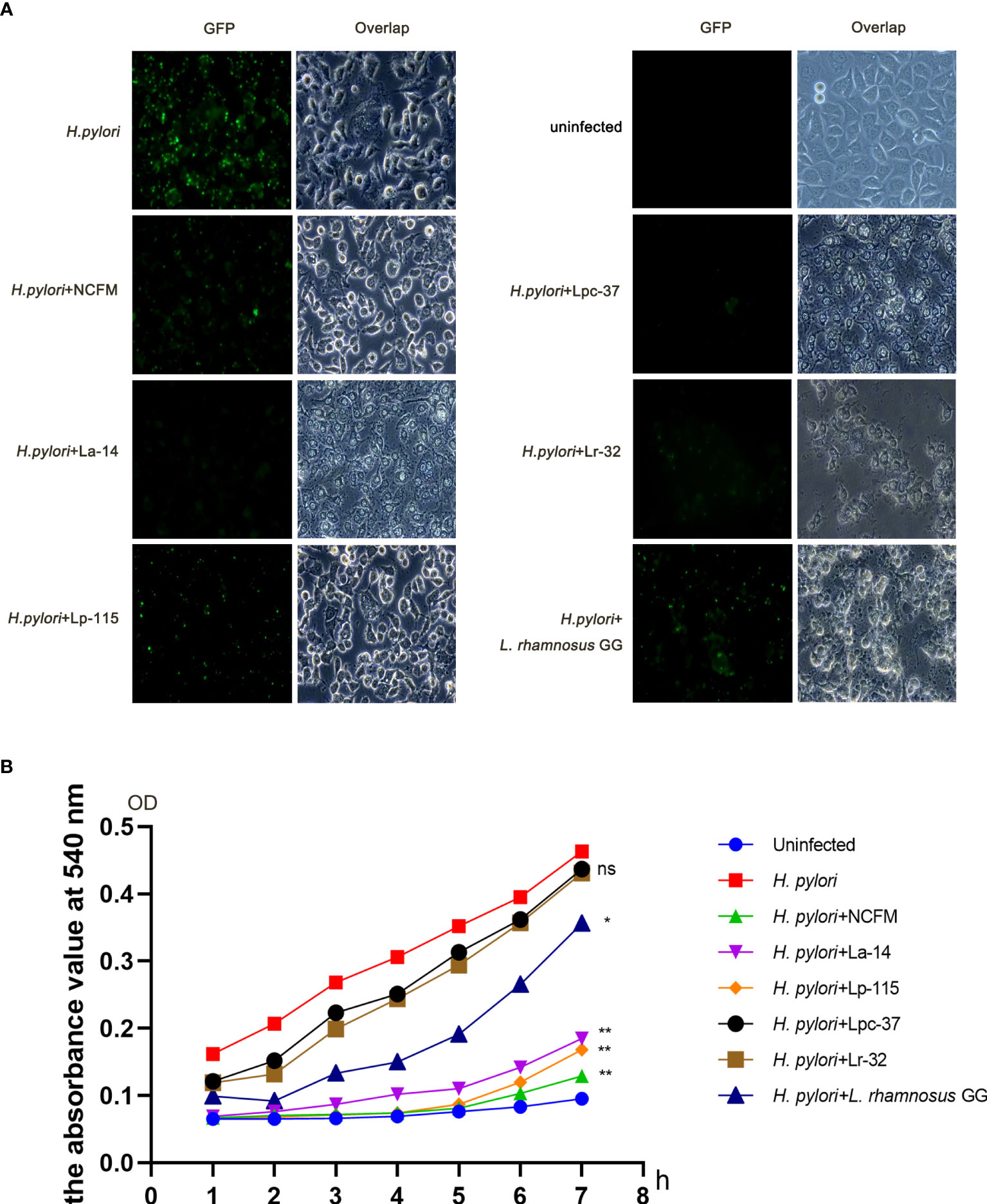
Figure 2 Six lactobacilli strains inhibit the adherence of Helicobacter pylori to AGS cells. (A) Fluorescence microscope images showing the colonization of H. pylori P12-GFP on AGS cells alone or upon intervention with six lactobacilli: Lactobacillus acidophilus NCFM, L. acidophilus La-14, Lactiplantibacillus plantarum Lp-115, Lacticaseibacillus paracasei Lpc-37, Lacticaseibacillus rhamnosus Lr-32 and L. rhamnosus GG). (B) Urease activity assay shows interference with the colonization of H. pylori P12-GFP on AGS cells in 1-7 hours by the six tested probiotic strains. *(P<0.05); **(P<0.01), compared to H. pylori. ns, not significant.
To further quantify the inhibition effect between different lactobacilli, we used a urease activity assay for the co-infection plate and recorded the absorbance at 540nm. The absorbance values of the six intervention groups were decreased to different degrees from 1 to 7 hours compared with that of the H. pylori infected group. For further quantification analysis, the absorbance values at each group’s 7h time point were taken into a bar chart for statistical analysis. The absorbance values of the seventh hour were statistically analyzed. The data suggest that samples of L. acidophilus NCFM, L. acidophilus La-14, L. plantarum Lp-115 and L. rhamnosus GG had significant differences compared with samples of H. pylori (P<0.05), indicating that these four lactobacilli strains can effectively reduce the colonization of H. pylori (Figure 2B).
To compare the inhibitory effect between the six probiotic strains, we performed a co-infection model of H. pylori and the strains (MOI=100) in AGS cell culture for 6 hours. We analyzed the mRNA and protein levels of IL-8 (Cxcl8) and TNF-α (TNF) to determine whether these probiotic strains transcriptionally regulate the inflammatory markers. The expression of these markers is commonly altered upon H. pylori infection. Expression of Cxcl8 and TNF mRNA with all six probiotic strains was significantly reduced compared with the H. pylori infected group (P<0.001). In Cxcl8, Lp-115 showed significant difference than other probiotics except NCFM, while pattern of NCFM was significantly lower than La-14 and Lpc-37 (P<0.01). The comparison of other probiotics including La-14, Lpc-37, Lr32 and GG did not show any significant difference (P>0.01). In TNF, only Lpc-37 was found to have significant difference than other probiotics in the group (P<0.01) (Figure 3A).
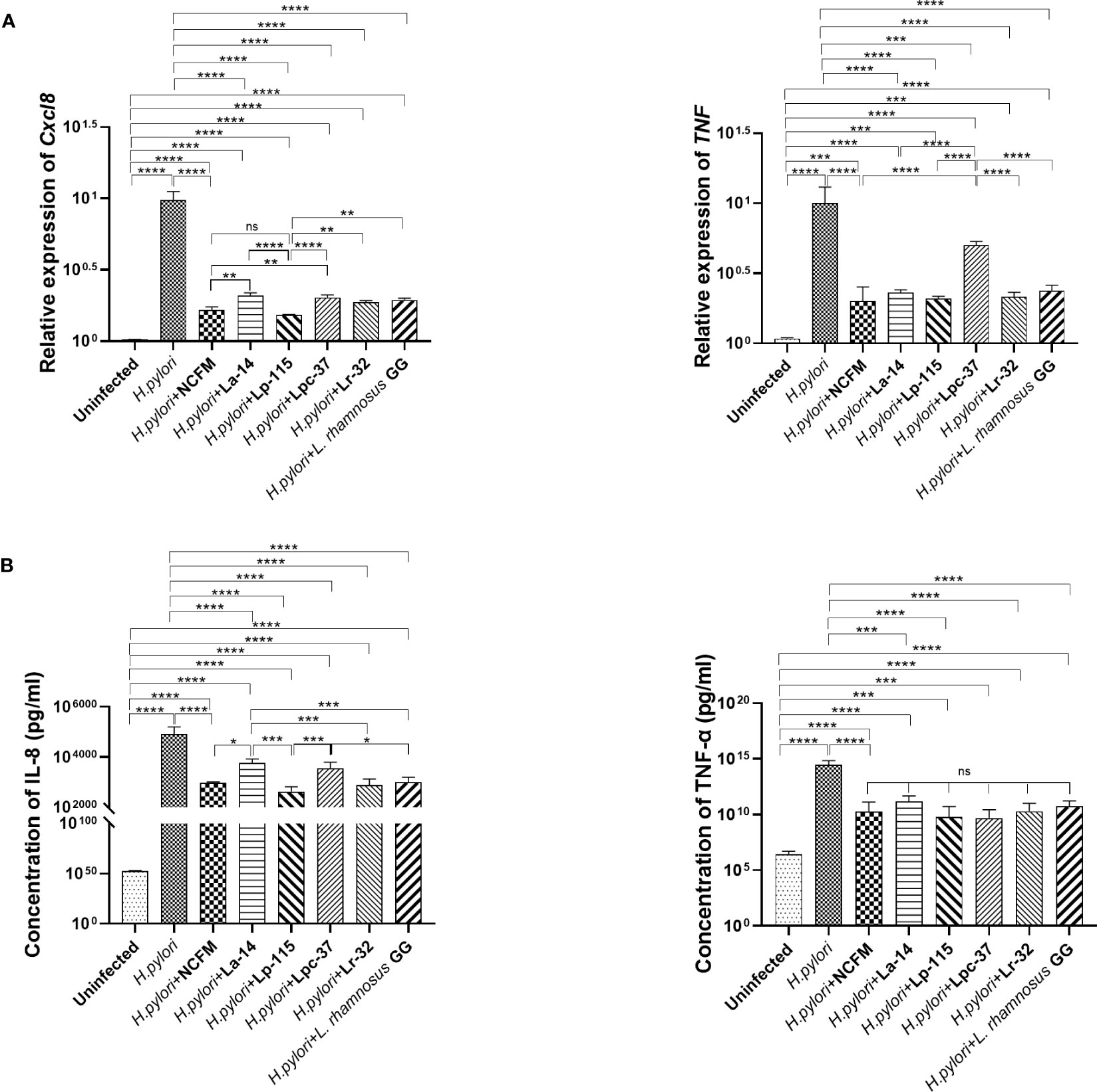
Figure 3 Inhibitory effects of the six probiotic strains on Helicobacter pylori-induced inflammation in AGS cell line. AGS cells were co-infected with the probiotic strains: Lactobacillus acidophilus NCFM, L. acidophilus La-14, Lactiplantibacillus plantarum Lp-115, Lacticaseibacillus paracasei Lpc-37, Lacticaseibacillus rhamnosus Lr-32 and L. rhamnosus GG and H. pylori P12 at a multiplicity of infection (MOI) 100 for 6 hours. (A) The mRNA levels of Cxcl8 and TNF the cells and (B) the protein concentrations of IL-8 and TNF-α in the supernatant. The results of each experiment are shown as mean ± standard deviation of three independent experiments. *(P<0.05); **(P<0.01); ***(P<0.001); ****p<0.0001; ns, not significant.
Consistent with the mRNA results, IL-8 and TNF-α in the cell supernatant were significantly decreased in the probiotic treatments compared with the H. pylori-infected group (P<0.001). In IL-8, NCFM was significantly lower than La-14, while Lp-115 showed significant difference than the La-14 and Lpc-37 (P<0.01). In TNF-a, the inhibitory effect of the six selected probiotic strains did not show significant differences (Figure 3B). Combined with the anti-adhesion results, L. acidophilus NCFM and L. plantarum Lp-115 had a good effect and less cell stress. Therefore, L. acidophilus NCFM and L. plantarum Lp-115 were further selected for validation in the H. pylori infected mouse model.
To further validate whether L. acidophilus NCFM and L. plantarum Lp-115 alone or in combination can counteract H. pylori colonization and attenuate gastric inflammation in vivo, C57BL/6 mice were infected with H. pylori SS1 and co-administered L. acidophilus NCFM and L. plantarum Lp-115 with for 6 weeks (Figure 1A). After co-administration the mice were euthanized, and the gastric tissues were assessed for H. pylori infection by the rapid urease test (Figure S2). The H. pylori adhesion on gastric tissues was then analyzed by immunohistochemistry (IHC) assays (Figure 1B), which showed that L. acidophilus NCFM and/or L. plantarum Lp-115 intervention groups had comparably less H. pylori adhesion than the H. pylori infected group. To further validate the H. pylori colonization in different groups, the mRNA expression of ureA and ureB was tested from gastric tissues of all groups (Figure 1C), which showed that L. acidophilus NCFM and/or L. plantarum Lp-115 intervention groups had less expression of ureA and ureB compared with H. pylori infected group (P<0.001). These results show that L. acidophilus NCFM and L. plantarum Lp-115 alone or combined can reduce H. pylori colonization on gastric mucosa in mice.
To further investigate whether L. acidophilus NCFM and L. plantarum Lp-115 can attenuate H. pylori colonization and gastric inflammation in vivo, gastric tissues from the five study groups were analyzed by Hematoxylin-Eosin staining (HE stain). Compared with the uninfected group, the gastric lamina propria of mice in the H. pylori infected group showed more lymphocyte, plasma cell, and neutrophil infiltration in the active phase. Incidentally, the mice also had local thinning of the mucosal layer, reduction of the glands propria, and thickening of the muscularis mucosae (Figure 4A). Compared with the H. pylori group, the inflammatory cells infiltrating the lamina propria of the gastric mucosa in the H. pylori + NCFM group, H. pylori + Lp-115 group, and H. pylori+NCFM+Lp-115 group were reduced to different degrees, which indicated that L. acidophilus NCFM and L. plantarum Lp-115 could ameliorate the H. pylori induced gastric inflammation (Figure 4A).
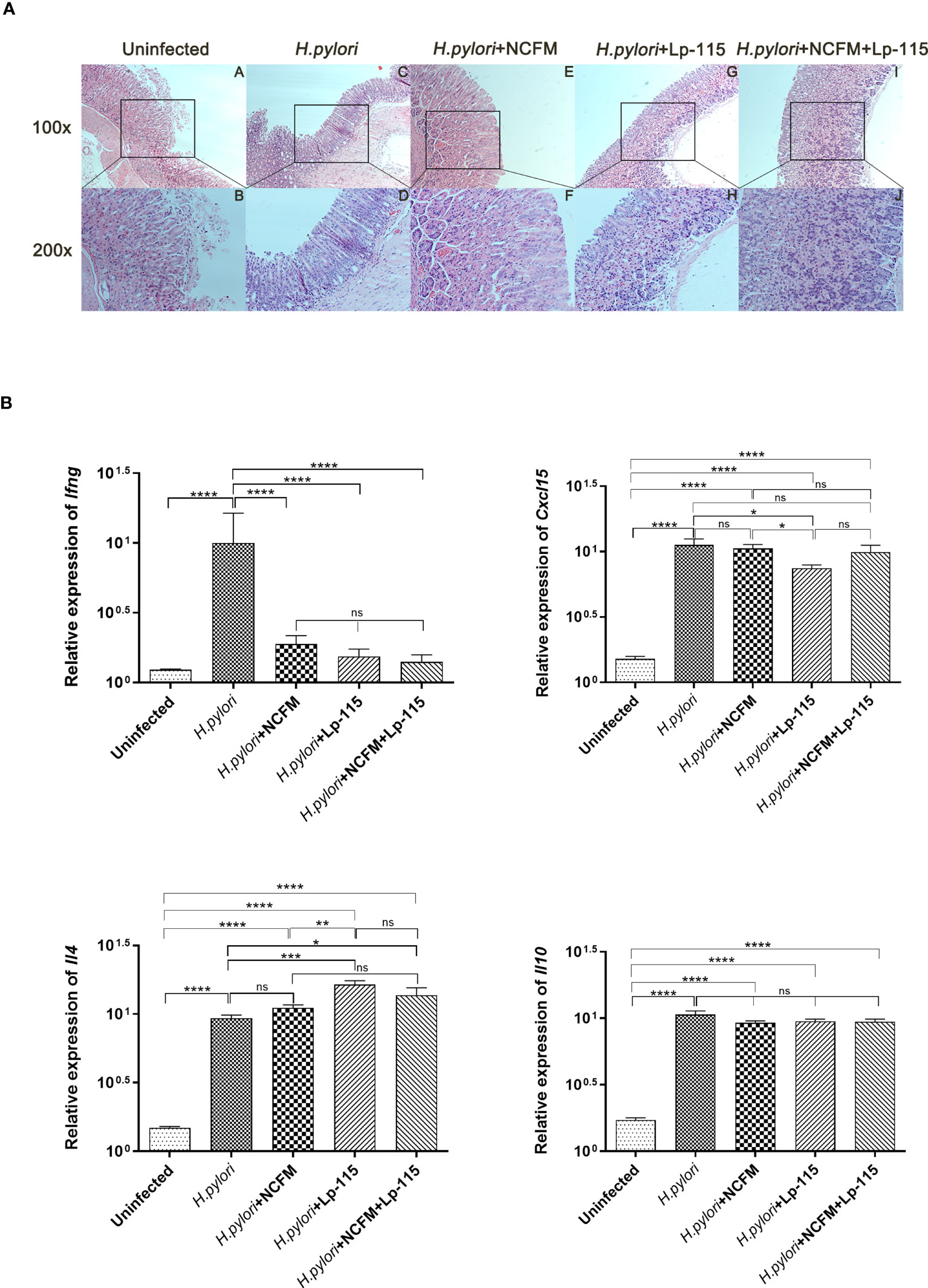
Figure 4 Lactobacillus acidophilus NCFM and Lactiplantibacillus plantarum Lp-115 suppress Helicobacter pylori inflammation in mice. (A) The inflammation of H. pylori was identified by HE. Uninfected group; H. pylori group; H. pylori+ NCFM group; H. pylori + Lp-115 group; H. pylori +NCFM+Lp-115 group. (B) The mRNA expression level of Il4, Cxcl15, Il10, and Ifng in each group. ns(P≥0.05); *(P<0.05); **(P<0.01); ***(P<0.001); ****(P<0.0001); ns, not significant.
To further verify if NCFM and/or Lp-115 can inhibit H. pylori induced Th1 type inflammation, the mRNA expression of Il4 (IL-4), Cxcl15 (CXCL15), Il10 (IL-10), and Ifng (IFN-γ) was measured by qRT-PCR from mouse gastric mucosal tissue. The results showed that NCFM and Lp-115 reduced the expression of Ifng and promoted the expression of Il4 induced by H. pylori in C57BL/6 mice. In Il4 expression, the difference between the Lp-115 and H. pylori groups was the most significant (P<0.001). In Cxcl15, only the Lp-115 group was significantly different from the H. pylori group (P<0.01); therefore, the overall anti-inflammatory effect of Lp-115 was more pronounced than that of NCFM. The expression of Cxcl15 in the group receiving the combination of Lp-115 and NCFM was not significantly different from that in the H. pylori group. The expression of Il4 was lower than that in the Lp-115 group (Figure 4B), indicating that the two LAB strains had no compound effect in improving inflammation caused by H. pylori infection. In conclusion, NCFM and/or Lp-115 can reduce H. pylori induced Th1 type inflammation (Ifng expression) in C57BL/6 mice and tend to transform Th2 type inflammation (Il4 expression).
Currently, quadruple therapy is the standard treatment for H. pylori eradication, but it has drawbacks. There is an increasing incidence of drug resistance and the misuse of antibiotics for H. pylori eradication can cause gastrointestinal disorders, gastrointestinal microbiota dysbiosis and other adverse effects (Hu et al., 2017). Modulation of the gastrointestinal microecology by microbial agents could represent a novel therapy or adjunct therapy for the current quadruple treatment. Furthermore, previous studies have shown that probiotics can modulate immune function, balance normal gastrointestinal microbiota, and reduce the side effects of antibiotics (Lu et al., 2016; Wang et al., 2017; Goderska et al., 2018), but may also inhibit H. pylori adhesion and gastric inflammation, suggesting beneficial effects. Although probiotics have advantages in aiding the eradication of H. pylori infection during conventional therapy, potential risks still exist. For some immunocompromised people, some strains of lactobacilli under certain rare conditions can cause infections (Liong, 2008). Therefore, selecting safe and suitable probiotic strains to support H. pylori eradication and management therapy is necessary.
Previous studies have shown that selected strains of lactobacilli can inhibit adherence of H. pylori to the gastric mucosa (Song et al., 2019; Zuo et al., 2019). In our study, we screened six probiotic strains and found that L. acidophilus NCFM and/or L. plantarum Lp-115 can inhibit H. pylori adhesion in an in vitro AGS cell line model and in vivo gastric mucosa of C57/BL6 mice, indicating preclinical evidence of these two strains for potential clinical use. However, the mechanisms by which NCFM and Lp-115 inhibit H. pylori colonization remains to be explored. Some studies have reported that probiotics’ effects are strain specific in inhibiting the colonization of H. pylori. For example, some Lactobacillus spp. such as L. acidophilus and L. bulgaricus have a high affinity for gastric epithelial cells, and they can protect the gastric mucosa by blocking or inhibiting the adhesion of H. pylori to gastric epithelial cells (de Klerk et al., 2016; Takeda et al., 2017; Zhao et al., 2018; Song et al., 2019). Some lactobacilli, such as L. plantarum and Ligilactobacillus salivarius, cannot compete with H. pylori for the binding site on the gastric mucosa but inhibit the activity of H. pylori through the antibacterial properties of metabolites, including Lactic acid and hydrogen peroxide (de Klerk et al., 2016; Takeda et al., 2017; Zhao et al., 2018; Song et al., 2019). Therefore, the immune system’s independent effects of probiotics against H. pylori infection may include affecting H. pylori gastric adhesive colonization or inhibiting H. pylori activity through the bacteriostatic properties of metabolites. These effects and other specific mechanisms need to be explored further.
H. pylori infection induces inflammation of gastric mucosa and expression of cytokines such as TNF-α or chemokines like CXCL8 (also known as IL-8) (Panpetch et al., 2016; Zhao et al., 2020; Tang et al., 2021). Previous studies have shown that L. rhamnosus GMNL-74 and L. acidophilus GMNL-185 reduce H. pylori induced gastric inflammation (Chen et al., 2019; Song et al., 2019). In the current study, we are showing for the first that L. acidophilus NCFM and/or L. plantarum Lp-115 inhibit H. pylori P12 induced IL-8 and TNF-α expression in vitro, which is consistent with previous studies analyzed other L. acidophilus strains (Ryan et al., 2009; Hwang et al., 2012). Previous studies have shown that H. pylori SS1 can induce T-helper cell type 1 (Th1) driven inflammation in C57BL/6 mice and that lactobacilli can suppress this response and may thus be involved in modulating Th1/Th2 balance (Boltin, 2016; Asgari et al., 2018; Asgari et al., 2020). In the H. pylori SS1 infected C57BL/6 mouse model, our study also confirmed that L. acidophilus NCFM and/or L. plantarum Lp-115 could inhibit Ifng but increase Il4 expression, consistent with the previous report that L. acidophilus can turn H. pylori induced Th1 type inflammation into Th2 type inflammation in C57BL/6mice(Boltin, 2016). Helper T cells (Th1, Th2) are essential factors in immunity and the main effector molecules of Th1 are IFN-γ and IL-12, but IL-8 (CXCL15 in mice) may also contribute to the inflammation. The primary effector molecule of Th2 mediated inflammation is IL-4, whereas IL-10 may contribute to inhibiting Th2 responses. The two kinds of helper T-cells regulate and inhibit each other by secreting different factors to maintain the balance of the Th1 and Th2 (Schmitt and Ueno, 2015; Jafarzadeh et al., 2018; Saravia et al., 2019). Previous studies demonstrated that lactobacilli strains could balance the Th1/Th2 immune response. Certain L. plantarum strains can maintain normal intestinal immune function by stimulating the secretion of cytokines and regulating the Th1/Th2 balance (Xie et al., 2015; Boltin, 2016; Meng et al., 2019). Conversely, one study showed that L. rhamnosus GG could increase the number of CD4+ T lymphocytes, assist in differentiating Th cells and enhance Th1 immune responses (Shi et al., 2020). In this study, the intervention of L. acidophilus NCFM and L. plantarum Lp-115 might alleviate H. pylori infection induced host inflammatory response by down regulating local Th1 immune response in the gastric mucosa (inhibiting proinflammatory factor IFN-γ) while promoting Th2 response to produce the anti-Th1 cytokine IL-4. Thus, L. acidophilus NCFM and L. plantarum Lp-115 may play an essential role in promoting the differentiation of T cells into Th2 cells to balance H. pylori induced Th1 inflammation.
Although the focus of the study was not safety related, it shows that while the tested strains have different efficacy, they are not negatively affecting the AGS cell line or the H. pylori infected mice, which is in line with earlier reports (Daniel et al., 2006; Morovic et al., 2017). Thus, their choice of probiotic strain and rational application must be seriously considered. Based on the classification of risk factors posed by individuals, the safest, most effective, and most affordable lactobacilli to manage H. pylori infection should be selected for further investigation.
In conclusion, probiotic health benefits are strain-specific; thus, data specific for strain and health benefits should be investigated. After screening several strains, we chose two safe lactobacilli candidate strains: L. acidophilus NCFM and L. plantarum Lp-115, which inhibit H. pylori adhesion and host inflammatory responses in cell line and mouse models. H. pylori has a high infection rate and high prevalence of drug resistance worldwide. The current study presented a unique value in managing H. pylori in vitro and in vivo. The clinical intervention study with the two probiotic strains or their combination is warranted.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by The Fifth Affiliated Hospital of Zhengzhou University.
SS: Data curation; Formal analysis; Investigation; Software; Validation; Visualization; Writing—original draft. FR: Formal analysis; Software; Investigation; Validation. HQ: Formal analysis; Software; Investigation; Validation. IB: Formal analysis; Writing—review & editing. JY: Funding acquisition. DG: Funding acquisition. ACO: Writing—review & editing. MJL: Writing—review & editing. PZ: Conceptualization; Project administration; Resources; Supervision. YM: Methodology; Project administration; Formal analysis; Writing—review & editing. All authors contributed to the article and approved the submitted version.
This study was fully funded by Danisco (China) Holding Co., Ltd. (a subsidiary of International Flavors & Fragrances, IFF), Shanghai, China, which was the study sponsor. No public funding was received to conduct this study.
JY and DG are former employees of Danisco China Holding Co. Ltd., part of International Flavors & Fragrances, IFF. ACO and MJL are employees of Danisco Sweeteners Oy, part of IFF. YM and PZ (principal investigators), SS, and FR are employed by The Fifth Affiliated Hospital of Zhengzhou University.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The sponsor participated in the design of the study, the interpretation of data, In review writing of the manuscript, and in the decision to publish the results.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1196084/full#supplementary-material
Asgari, B., Kermanian, F., Derakhshan, N., Asna-Ashari, M., Sadat, Z. R. N., Yaslianifard, S. (2018). Honey-derived lactobacillus rhamnosus alleviates helicobacter pylori-induced gastro-intestinal infection and gastric inflammation in C57bl/6 mice: an immuno-histologic study. Arq. Gastroenterol. 55 (3), 279–282. doi: 10.1590/S0004-2803.201800000-70
Asgari, B., Kermanian, F., Hedayat Yaghoobi, M., Vaezi, A., Soleimanifar, F., Yaslianifard, S. (2020). The Anti-Helicobacter pylori Effects of Lactobacillus acidophilus, L. plantarum and L. rhamnosus in Stomach Tissue of C57BL/6 Mice. Visc Med. 36 (2), 137–143. doi: 10.1159/000500616
Boltin, D. (2016). Probiotics in Helicobacter pylori-induced peptic ulcer disease. Best Pract. Res. Clin. Gastroenterol. 30 (1), 99–109. doi: 10.1016/j.bpg.2015.12.003
Chen, Y. H., Tsai, W. H., Wu, H. Y., Chen, C. Y., Yeh, W. L., Chen, Y. H., et al. (2019). Probiotic Lactobacillus spp. act Against Helicobacter pylori-induced Inflammation. J. Clin. Med. 8 (1), 90. doi: 10.3390/jcm8010090
Chey, W. D., Leontiadis, G. I., Howden, C. W., Moss, S. F. (2017). ACG clinical guideline: treatment of helicobacter pylori infection. Am. J. Gastroenterol. 112 (2), 212–239. doi: 10.1038/ajg.2016.563
Daniel, C., Poiret, S., Goudercourt, D., Dennin, V., Leyer, G., Pot, B. (2006). Selecting lactic acid bacteria for their safety and functionality by use of a mouse colitis model. Appl. Environ. Microbiol. 72 (9), 5799–5805. doi: 10.1128/AEM.00109-06
Dargenio, C., Dargenio, V. N., Bizzoco, F., Indrio, F., Francavilla, R., Cristofori, F. (2021). Limosilactobacillus reuteri Strains as Adjuvants in the Management of Helicobacter pylori Infection. Medicina (Kaunas) 57 (7), 733. doi: 10.3390/medicina57070733
de Klerk, N., Maudsdotter, L., Gebreegziabher, H., Saroj, S. D., Eriksson, B., Eriksson, O. S., et al. (2016). Lactobacilli reduce helicobacter pylori attachment to host gastric epithelial cells by inhibiting adhesion gene expression. Infect. Immun. 84 (5), 1526–1535. doi: 10.1128/IAI.00163-16
Fallone, C. A., Chiba, N., van Zanten, S. V., Fischbach, L., Gisbert, J. P., Hunt, R. H., et al. (2016). The toronto consensus for the treatment of helicobacter pylori infection in adults. Gastroenterology 151 (1), 51–69 e14. doi: 10.1053/j.gastro.2016.04.006
Fallone, C. A., Moss, S. F., Malfertheiner, P. (2019). Reconciliation of recent helicobacter pylori treatment guidelines in a time of increasing resistance to antibiotics. Gastroenterology 157 (1), 44–53. doi: 10.1053/j.gastro.2019.04.011
Fang, J. Y., Du, Y. Q., Liu, W. Z., Ren, J. L., Li, Y. Q., Chen, X. Y., et al. (2018). Chinese consensus on chronic gastritis, (2017, shanghai). J. Dig Dis. 19 (4), 182–203. doi: 10.1111/1751-2980.12593
Goderska, K., Agudo Pena, S., Alarcon, T. (2018). Helicobacter pylori treatment: antibiotics or probiotics. Appl. Microbiol. Biotechnol. 102 (1), 1–7. doi: 10.1007/s00253-017-8535-7
Hooi, J. K. Y., Lai, W. Y., Ng, W. K., Suen, M. M. Y., Underwood, F. E., Tanyingoh, D., et al. (2017). Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153 (2), 420–429. doi: 10.1053/j.gastro.2017.04.022
Hu, Y., Zhu, Y., Lu, N. H. (2017). Novel and effective therapeutic regimens for helicobacter pylori in an era of increasing antibiotic resistance. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00168
Hwang, S. W., Kim, N., Kim, J. M., Huh, C. S., Ahn, Y. T., Park, S. H., et al. (2012). Probiotic suppression of the H. pylori-induced responses by conjugated linoleic acids in a gastric epithelial cell line. Prostaglandins Leukot. Essent. Fatty Acids 86 (6), 225–231. doi: 10.1016/j.plefa.2012.04.002
Jafarzadeh, A., Larussa, T., Nemati, M., Jalapour, S. (2018). T cell subsets play an important role in the determination of the clinical outcome of Helicobacter pylori infection. Microb. Pathog. 116, 227–236. doi: 10.1016/j.micpath.2018.01.040
Jones, N. L., Koletzko, S., Goodman, K., Bontems, P., Cadranel, S., Casswall, T., et al. (2017). Joint ESPGHAN/NASPGHAN guidelines for the management of helicobacter pylori in children and adolescents (Update 2016). J. Pediatr. Gastroenterol. Nutr. 64 (6), 991–1003. doi: 10.1097/MPG.0000000000001594
Keikha, M., Karbalaei, M. (2021). Probiotics as the live microscopic fighters against Helicobacter pylori gastric infections. BMC Gastroenterol. 21 (1), 388. doi: 10.1186/s12876-021-01977-1
Kim, D. H., Son, B. K., Min, K. W., Han, S. K., Na, J. U., Choi, P. C., et al. (2020). Chronic gastritis is associated with a decreased high-density lipid level: histological features of gastritis based on the updated sydney system. J. Clin. Med. 9 (6), 1856. doi: 10.3390/jcm9061856
Lee, A., O'Rourke, J., De Ungria, M. C., Robertson, B., Daskalopoulos, G., Dixon, M. F. (1997). A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112 (4), 1386–1397. doi: 10.1016/s0016-5085(97)70155-0
Lin, C. C., Huang, W. C., Su, C. H., Lin, W. D., Wu, W. T., Yu, B., et al. (2020). Effects of multi-strain probiotics on immune responses and metabolic balance in helicobacter pylori-infected mice. Nutrients 12 (8), 2476. doi: 10.3390/nu12082476
Liong, M. T. (2008). Safety of probiotics: translocation and infection. Nutr. Rev. 66 (4), 192–202. doi: 10.1111/j.1753-4887.2008.00024.x
Liou, J. M., Malfertheiner, P., Lee, Y. C., Sheu, B. S., Sugano, K., Cheng, H. C., et al. (2020). Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut 69 (12), 2093–2112. doi: 10.1136/gutjnl-2020-322368
Liu, W. Z., Xie, Y., Lu, H., Cheng, H., Zeng, Z. R., Zhou, L. Y., et al. (2018). Fifth Chinese National Consensus Report on the management of Helicobacter pylori infection. Helicobacter 23 (2), e12475. doi: 10.1111/hel.12475
Lu, M., Yu, S., Deng, J., Yan, Q., Yang, C., Xia, G., et al. (2016). Efficacy of probiotic supplementation therapy for helicobacter pylori eradication: A meta-analysis of randomized controlled trials. PloS One 11 (10), e0163743. doi: 10.1371/journal.pone.0163743
Malfertheiner, P., Megraud, F., Rokkas, T., Gisbert, J. P., Liou, J. M., Schulz, C., et al. (2022). Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut 71, 17247–1762. doi: 10.1136/gutjnl-2022-327745
Maluf, S., Salgado, J. V., Cysne, D. N., Camelo, D. M. F., Nascimento, J. R., Maluf, B. V. T., et al. (2020). Increased glycated hemoglobin levels in patients with helicobacter pylori infection are associated with the grading of chronic gastritis. Front. Immunol. 11. doi: 10.3389/fimmu.2020.02121
McFarland, L. V., Huang, Y., Wang, L., Malfertheiner, P. (2016). Systematic review and meta-analysis: Multi-strain probiotics as adjunct therapy for Helicobacter pylori eradication and prevention of adverse events. United Eur. Gastroenterol. J. 4 (4), 546–561. doi: 10.1177/2050640615617358
Meng, Y., Wang, J., Wang, Z., Zhang, G., Liu, L., Huo, G., et al. (2019). Lactobacillus plantarum KLDS1.0318 ameliorates impaired intestinal immunity and metabolic disorders in cyclophosphamide-treated mice. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00731
Morovic, W., Roper, J. M., Smith, A. B., Mukerji, P., Stahl, B., Rae, J. C., et al. (2017). Safety evaluation of HOWARU((R)) Restore (Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium animalis subsp. lactis Bl-04 and B. lactis Bi-07) for antibiotic resistance, genomic risk factors, and acute toxicity. Food Chem. Toxicol. 110, 316–324. doi: 10.1016/j.fct.2017.10.037
Ouwehand, A. C., Invernici, M. M., Furlaneto, F. A. C., Messora, M. R. (2018). Effectiveness of multistrain versus single-strain probiotics: current status and recommendations for the future. J. Clin. Gastroenterol. 52 Suppl 1, S35–S40. doi: 10.1097/MCG.0000000000001052
Panpetch, W., Spinler, J. K., Versalovic, J., Tumwasorn, S. (2016). Characterization of Lactobacillus salivarius strains B37 and B60 capable of inhibiting IL-8 production in Helicobacter pylori-stimulated gastric epithelial cells. BMC Microbiol. 16 (1), 242. doi: 10.1186/s12866-016-0861-x
Robinson, K., Atherton, J. C. (2021). The spectrum of helicobacter-mediated diseases. Annu. Rev. Pathol. 16, 123–144. doi: 10.1146/annurev-pathol-032520-024949
Rokka, S., Myllykangas, S., Joutsjoki, V. (2008). Effect of specific colostral antibodies and selected lactobacilli on the adhesion of Helicobacter pylori on AGS cells and the Helicobacter-induced IL-8 production. Scand. J. Immunol. 68 (3), 280–286. doi: 10.1111/j.1365-3083.2008.02138.x
Ryan, K. A., O'Hara, A. M., van Pijkeren, J. P., Douillard, F. P., O'Toole, P. W. (2009). Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J. Med. Microbiol. 58 (Pt 8), 996–1005. doi: 10.1099/jmm.0.009407-0
Saravia, J., Chapman, N. M., Chi, H. (2019). Helper T cell differentiation. Cell Mol. Immunol. 16 (7), 634–643. doi: 10.1038/s41423-019-0220-6
Savoldi, A., Carrara, E., Graham, D. Y., Conti, M., Tacconelli, E. (2018). Prevalence of antibiotic resistance in helicobacter pylori: A systematic review and meta-analysis in world health organization regions. Gastroenterology 155 (5), 1372–1382 e17. doi: 10.1053/j.gastro.2018.07.007
Schmitt, N., Ueno, H. (2015). Regulation of human helper T cell subset differentiation by cytokines. Curr. Opin. Immunol. 34, 130–136. doi: 10.1016/j.coi.2015.03.007
Shah, S. C., Tepler, A., Chung, C. P., Suarez, G., Peek, R. M., Jr., Hung, A., et al. (2021). Host genetic determinants associated with helicobacter pylori eradication treatment failure: A systematic review and meta-analysis. Gastroenterology 161 (5), 1443–1459. doi: 10.1053/j.gastro.2021.07.043
Shi, C. W., Cheng, M. Y., Yang, X., Lu, Y. Y., Yin, H. D., Zeng, Y., et al. (2020). Probiotic lactobacillus rhamnosus GG promotes mouse gut microbiota diversity and T cell differentiation. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.607735
Shi, X., Zhang, J., Mo, L., Shi, J., Qin, M., Huang, X. (2019). Efficacy and safety of probiotics in eradicating Helicobacter pylori: A network meta-analysis. Med. (Baltimore) 98 (15), e15180. doi: 10.1097/MD.0000000000015180
Shmuely, H., Burger, O., Neeman, I., Yahav, J., Samra, Z., Niv, Y., et al. (2004). Susceptibility of Helicobacter pylori isolates to the antiadhesion activity of a high-molecular-weight constituent of cranberry. Diagn. Microbiol. Infect. Dis. 50 (4), 231–235. doi: 10.1016/j.diagmicrobio.2004.08.011
Simon, E., Calinoiu, L. F., Mitrea, L., Vodnar, D. C. (2021). Probiotics, prebiotics, and synbiotics: implications and beneficial effects against irritable bowel syndrome. Nutrients 13 (6), 2112. doi: 10.3390/nu13062112
Song, H., Zhou, L., Liu, D., Ge, L., Li, Y. (2019). Probiotic effect on Helicobacter pylori attachment and inhibition of inflammation in human gastric epithelial cells. Exp. Ther. Med. 18 (3), 1551–1562. doi: 10.3892/etm.2019.7742
Sousa, C., Ferreira, R., Azevedo, N. F., Oleastro, M., Azeredo, J., Figueiredo, C., et al. (2022). Helicobacter pylori infection: from standard to alternative treatment strategies. Crit. Rev. Microbiol. 48 (3), 376–396. doi: 10.1080/1040841X.2021.1975643
Suez, J., Zmora, N., Segal, E., Elinav, E. (2019). The pros, cons, and many unknowns of probiotics. Nat. Med. 25 (5), 716–729. doi: 10.1038/s41591-019-0439-x
Sugano, K., Tack, J., Kuipers, E. J., Graham, D. Y., El-Omar, E. M., Miura, S., et al. (2015). Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64 (9), 1353–1367. doi: 10.1136/gutjnl-2015-309252
Takeda, S., Igoshi, K., Tsend-Ayush, C., Oyunsuren, T., Sakata, R., Koga, Y., et al. (2017). Lactobacillus paracasei strain 06TCa19 suppresses inflammatory chemokine induced by Helicobacter pylori in human gastric epithelial cells. Hum. Cell 30 (4), 258–266. doi: 10.1007/s13577-017-0172-z
Tang, L., Tang, B., Lei, Y., Yang, M., Wang, S., Hu, S., et al. (2021). Helicobacter pylori-Induced Heparanase Promotes H. pylori Colonization and Gastritis. Front. Immunol. 12. doi: 10.3389/fimmu.2021.675747
Tharmalingam, N., Kim, S. H., Park, M., Woo, H. J., Kim, H. W., Yang, J. Y., et al. (2014). Inhibitory effect of piperine on Helicobacter pylori growth and adhesion to gastric adenocarcinoma cells. Infect. Agent Cancer 9 (1), 43. doi: 10.1186/1750-9378-9-43
Vieira, A. T., Teixeira, M. M., Martins, F. S. (2013). The role of probiotics and prebiotics in inducing gut immunity. Front. Immunol. 4. doi: 10.3389/fimmu.2013.00445
Wang, F., Feng, J., Chen, P., Liu, X., Ma, M., Zhou, R., et al. (2017). Probiotics in Helicobacter pylori eradication therapy: Systematic review and network meta-analysis. Clin. Res. Hepatol. Gastroenterol. 41 (4), 466–475. doi: 10.1016/j.clinre.2017.04.004
Xie, J., Yu, Q., Nie, S., Fan, S., Xiong, T., Xie, M. (2015). Effects of lactobacillus plantarum NCU116 on intestine mucosal immunity in immunosuppressed mice. J. Agric. Food Chem. 63 (51), 10914–10920. doi: 10.1021/acs.jafc.5b04757
Yang, J. Y., Kim, P., Jeong, S. H., Lee, S. W., Myung, Y. S., Baeg, M. K., et al. (2020). The effects of sulglycotide on the adhesion and the inflammation of helicobacter pylori. Int. J. Environ. Res. Public Health 17 (8), 2918. doi: 10.3390/ijerph17082918
Yoon, J. Y., Cha, J. M., Hong, S. S., Kim, H. K., Kwak, M. S., Jeon, J. W., et al. (2019). Fermented milk containing Lactobacillus paracasei and Glycyrrhiza glabra has a beneficial effect in patients with Helicobacter pylori infection: A randomized, double-blind, placebo-controlled study. Med. (Baltimore) 98 (35), e16601. doi: 10.1097/MD.0000000000016601
Zhao, K., Xie, Q., Xu, D., Guo, Y., Tao, X., Wei, H., et al. (2018). Antagonistics of Lactobacillus plantarum ZDY2013 against Helicobacter pylori SS1 and its infection in vitro in human gastric epithelial AGS cells. J. Biosci. Bioeng 126 (4), 458–463. doi: 10.1016/j.jbiosc.2018.04.003
Zhao, Q., Yin, W., Zhao, R., Wang, Y., Song, C., Wang, H., et al. (2020). Outer inflammatory protein of Helicobacter pylori impacts IL-8 expression, adherence, cell apoptosis and cell cycle of gastric cells independent of its copy number. Med. Microbiol. Immunol. 209 (5), 621–630. doi: 10.1007/s00430-020-00688-w
Keywords: Helicobacter pylori, Lactobacillus, adhesion, inflammation, probiotic
Citation: Shen S, Ren F, Qin H, Bukhari I, Yang J, Gao D, Ouwehand AC, Lehtinen MJ, Zheng P and Mi Y (2023) Lactobacillus acidophilus NCFM and Lactiplantibacillus plantarum Lp-115 inhibit Helicobacter pylori colonization and gastric inflammation in a murine model. Front. Cell. Infect. Microbiol. 13:1196084. doi: 10.3389/fcimb.2023.1196084
Received: 29 March 2023; Accepted: 07 July 2023;
Published: 09 August 2023.
Edited by:
Parth Sarthi Sen Gupta, D Y Patil International University, IndiaReviewed by:
Giuseppe Losurdo, University of Bari Medical School, ItalyCopyright © 2023 Shen, Ren, Qin, Bukhari, Yang, Gao, Ouwehand, Lehtinen, Zheng and Mi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengyuan Zheng, cHl6aGVuZ0B6enUuZWR1LmNu; Yang Mi, eWFuZ21pMTk4QHp6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.