- Department of Immunology and Molecular Microbiology, School of Dentistry and Dental Research Institute, Seoul National University, Seoul, Republic of Korea
To better understand the impact of gut dysbiosis on four autoimmune diseases [Sjögren’s syndrome (SS), systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and multiple sclerosis (MS)], this review investigated the altered gut bacteria in each disease and the shared ones among the four diseases. The enriched gut bacteria shared by three of the four autoimmune diseases were Streptococcus, Prevotella, and Eggerthella, which are associated with autoantibody production or activation of Th17 cells in immune-related diseases. On the other hand, Faecalibacterium comprises depleted gut bacteria shared by patients with SLE, MS, and SS, which is associated with various anti-inflammatory activities. The indexes of gut dysbiosis, defined as the number of altered gut bacterial taxa divided by the number of studies in SLE, MS, RA, and SS, were 1.7, 1.8, 0.7, and 1.3, respectively. Interestingly, these values presented a positive correlation trend with the standardized mortality rates —2.66, 2.89, 1.54, and 1.41, respectively. In addition, shared altered gut bacteria among the autoimmune diseases may correlate with the prevalence of polyautoimmunity in patients with SLE, SS, RA, and MS, that is, 41 percent, 32.6 percent, 14 percent, and 1–16.6 percent, respectively. Overall, this review suggests that gut dysbiosis in autoimmune diseases may be closely related to the failure of the gut immune system to maintain homeostasis.
1 Introduction
The etiology of autoimmune diseases is complex involving both genetic and environmental factors. Genetic risk factors for autoimmune diseases are composed of HLA and non-HLA genes expressed at different levels depending on the disease (Greiling et al., 2018; van der Meulen et al., 2019; Frazzei et al., 2022). On the other hand, environmental factors include smoking, lifestyle disorders, reduced sun exposure, and chronic stress (Frazzei et al., 2022). However, the scope of these factors to explain the cause of the rapid increase in autoimmune diseases over the decades is insufficient (Chen et al., 2017; Dinse et al., 2022; Frazzei et al., 2022). Recently, gut dysbiosis has attracted great attention as a risk factor for autoimmune diseases. However, it is unclear whether gut dysbiosis is a result or a cause of an autoimmune disease (Jubair et al., 2018). Autoimmune diseases such as primary Sjögren’s syndrome (SS), systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and multiple sclerosis (MS) have been linked to gut dysbiosis (Zhang et al., 2015; Chen et al., 2016a; Nikitakis et al., 2017; Luo et al., 2018; Bellando-Randone et al., 2021). The causes of gut dysbiosis include depletion of the mucus layer, rapid dietary changes, use of antibiotics, infection and inflammation, and gastrointestinal surgery (Van de Wiele et al., 2016). Chen et al. proposed the following five mechanisms by which gut dysbiosis contributes to autoimmune diseases: 1) dysregulation of TLR in antigen presenting cells (APCs) and imbalance of Treg/Th17 ratio; 2) generation of new autoantigens due to the modification of host proteins induced by microbial enzymes; 3) microbial components similar to self-peptides, activating autoreactive B and T cells; 4) induction of immunopathology through the transport of microbial components or metabolites throughout the host; and 5) autoantibody generation against curli-DNA composites (Chen et al., 2017). In this review, the altered gut bacteria in SLE, MS, RA, and SS were investigated to better understand the impact of gut dysbiosis on autoimmune diseases. First, we investigated whether there are common taxa in different studies of gut dysbiosis for each disease. Second, we investigated whether the four autoimmune diseases share altered gut bacteria and whether there are altered gut bacteria unique to each autoimmune disease. Third, the altered gut bacteria’s functions or mechanisms of action in immune-related diseases were investigated. Fourth, we explored whether the shared, altered gut bacteria are related to polyautoimmunity. Finally, we examined whether the degree of gut dysbiosis is related to the disease’s standardized mortality rate (SMR).
2 Gut dysbiosis in autoimmune diseases
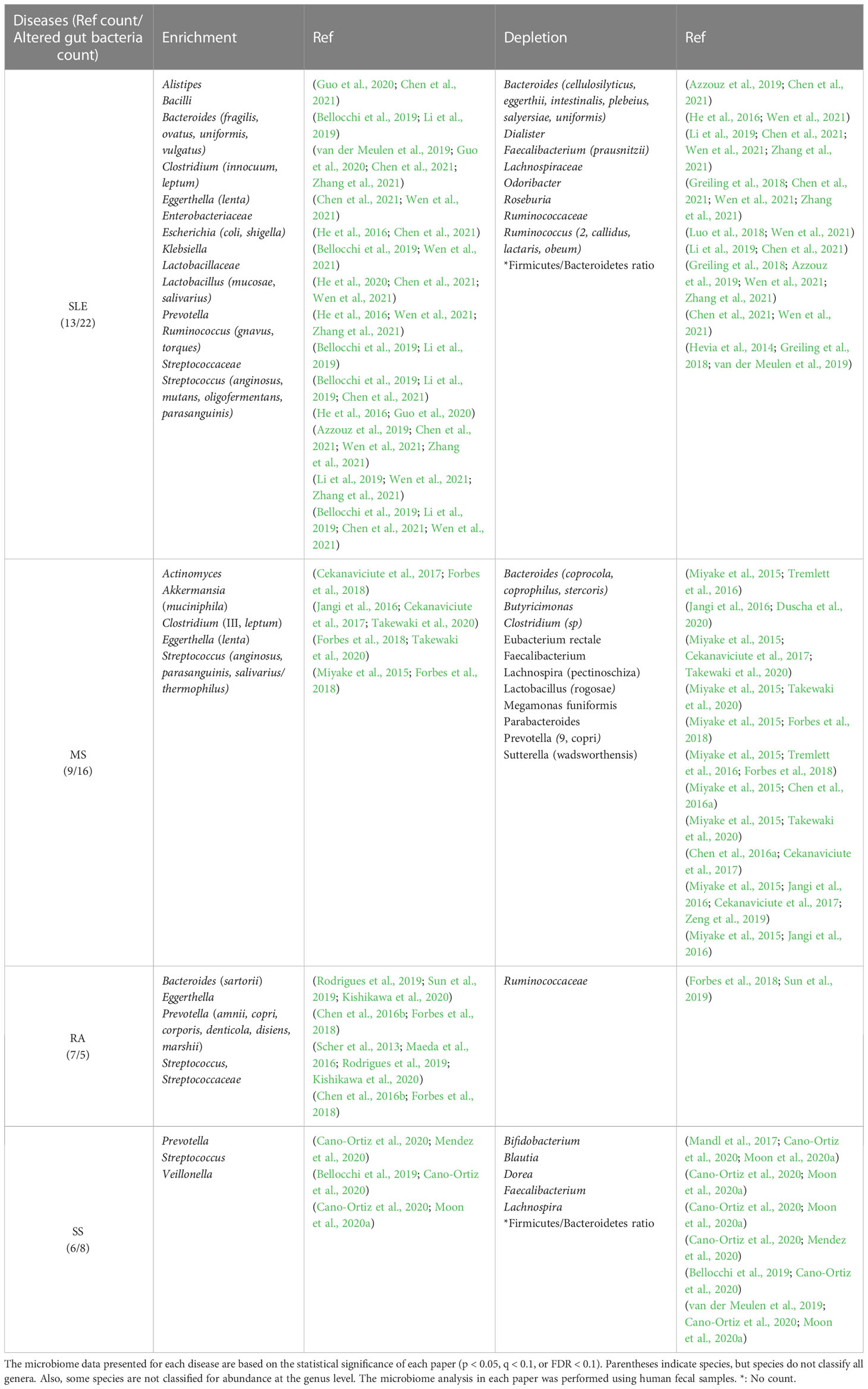
Altered gut bacteria refer to the taxa whose enrichment or depletion in the gut bacteriota has been cross-validated by at least two studies with statistical significance (p < 0.05, q < 0.1, or false discovery rate < 0.1). The taxonomic range of altered gut bacteria investigated in the four autoimmune diseases was at the family, genus, or species levels. Most altered bacteria were cross-validated at the genus or species levels because some papers only presented them at the genus or species levels.
2.1 Altered gut bacteria in SLE
SLE is a prototypical autoimmune disease associated with loss of self-tolerance of the immune system, abnormal antibody response to cytoplasmic antigens, persistent autoantibody production, and subsequent systemic inflammation (Chen et al., 2021). Its clinical signs include multiple symptoms, such as skin rash, glomerulonephritis, neurological disorders, and severe vasculitis, suggesting that the pathogenesis of SLE may be complex (Guo et al., 2020). A total of 22 altered gut bacterial genera/families were identified from the review of 13 papers published since 2014 (Table 1). While Alistipes, Bacilli, Bacteroides, Clostridium, Eggerthella, Escherichia, Klebsiella, Lactobacillus, Prevotella, Ruminococcus, and Streptococcus are enriched, Bacteroides, Dialister, Faecalibacterium, Odoribacter, Roseburia, and Ruminococcus are depleted in the gut of SLE patients. Interestingly, Bacteroides and Ruminococcus were reported to be enriched or depleted depending on studies, but at the species level, different species were enriched or depleted with the exception of B. uniformis, which was reported to be enriched or depleted in different studies (Table 1). Even in the same species, Bacteroides fragilis is classified into polysaccharide A-producing beneficial bacterium or enterotoxigenic bacterium, depending on strains (Nagao-Kitamoto and Kamada, 2017). Thus, B. fragilis enrichment in patients with SLE might be associated with enterotoxigenic strains. Among the 22 altered gut bacterial genera, Bacteroides, Escherichia, Ruminococcus, and Streptococcus have known functions associated with the induction of inflammatory response or autoimmunity in immune-related diseases (Vatanen et al., 2016; Cunningham, 2019; Henke et al., 2019; Qiu et al., 2019), whereas Faecalibacterium and Ruminococcus_2 have anti-inflammatory mechanisms of action (Houtman et al., 2022; Matsuoka et al., 2022). More details about this phenomenon have been described in Section 2.6.
2.2 Altered gut bacteria in MS
MS is an autoimmune disease in which the immune system destroys the myelin sheaths surrounding nerve axons in the central nervous system (CNS). MS is on the rise in developed countries and occurs three times higher in young women, for which an environmental factor, such as gut dysbiosis, than genetic factors seems to account (Miyake et al., 2015; Cekanaviciute et al., 2017; Duscha et al., 2020). This assumption is supported by the fact that the transfer of feces from MS patients exacerbates the disease in the animal models of MS (Berer et al., 2017). A total of 16 altered gut bacterial genera were found from the review of nine papers published since 2015 (Table 1). While Actinomyces, Akkermansia, Clostridium, Eggerthella, and Streptococcus are enriched, Bacteroides, Butyricimonas, Clostridium, Eubacterium, Faecalibacterium, Lachnospira, Lactobacillus, Megamonas, Parabacteroides, Prevotella, and Sutterella are depleted in the gut of MS patients. Eggerthella and Streptococcus are associated with the induction of autoimmunity (Valour et al., 2014; Ravindra et al., 2018; Cunningham, 2019; Alexander et al., 2022), whereas Butyricimonas and Faecalibacterium have reported anti-inflammatory functions (Jing et al., 2021; Bai et al., 2022; Houtman et al., 2022), as detailed in Section 2.6. Interestingly, the relative abundance of Prevotella_9 and Sutterella increased in experimental autoimmune encephalomyelitis mice after receiving fecal microbiota transplantation from healthy mice, which, in turn, improved clinical scores (Wang et al., 2021). These results suggest that beneficial bacteria in the host may maintain the homeostasis of the immune system in MS.
2.3 Altered gut bacteria in RA
RA is a chronic autoimmune disease that causes joint destruction and functional impairment. Recently, the etiology of RA has been hypothesized to be a combination of genetic factors and gut dysbiosis (Maeda et al., 2016; Jubair et al., 2018). Particularly, the concordance rate for RA in monozygotic twins studied in Europe is 15 percent, which is insufficient to solely explain its etiology by genetic influences (Aho et al., 1986; Silman et al., 1993). Autoantibody production against citrullinated peptides produced by Porphyromonas gingivalis is a mechanism to induce RA (Kishikawa et al., 2020). Anti-citrullinated protein antibodies (ACPAs) have been detected in all high-risk RA patients and 93 percent of patients with RA (Tong et al., 2019). A total of five altered gut bacterial genera/families were identified from the review of seven papers published since 2013 (Table 1). All four enriched genera, Bacteroides, Eggerthella, Prevotella, and Streptococcus, have known functions associated with the induction of inflammatory response or autoimmunity in immune-related diseases detailed in Section 2.6.
2.4 Altered gut bacteria in SS
SS is an autoimmune disease characterized by dry mouth and dry eyes (keratoconjunctivitis sicca). Eight genera were identified from the review of six papers published since 2017 (Table 1). While Prevotella, Streptococcus, and Veillonella are enriched, Bifidobacterium, Blautia, Dorea, Faecalibacterium, and Lachnospira are depleted in the gut bacteriota of SS patients. Among these, Prevotella and Streptococcus have known functions associated with the induction of inflammatory response or autoimmunity in immune-related diseases, whereas Bifidobacterium and Faecalibacterium have anti-inflammatory mechanisms of action or efficacy, as detailed in Section 2.6 (Maeda et al., 2016; O’Callaghan and van Sinderen, 2016; Cunningham, 2019; Yao et al., 2021; Houtman et al., 2022).
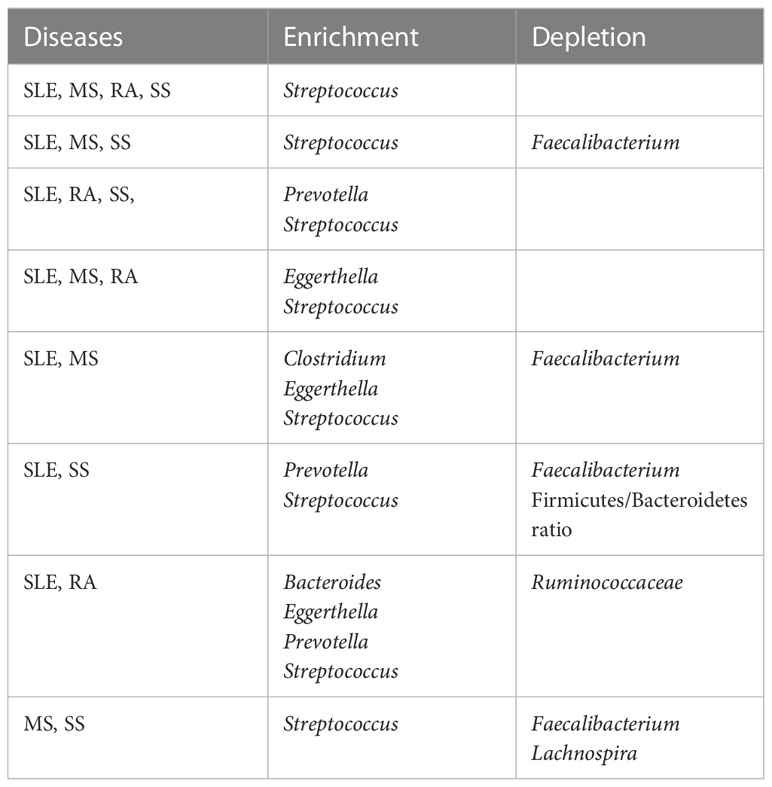
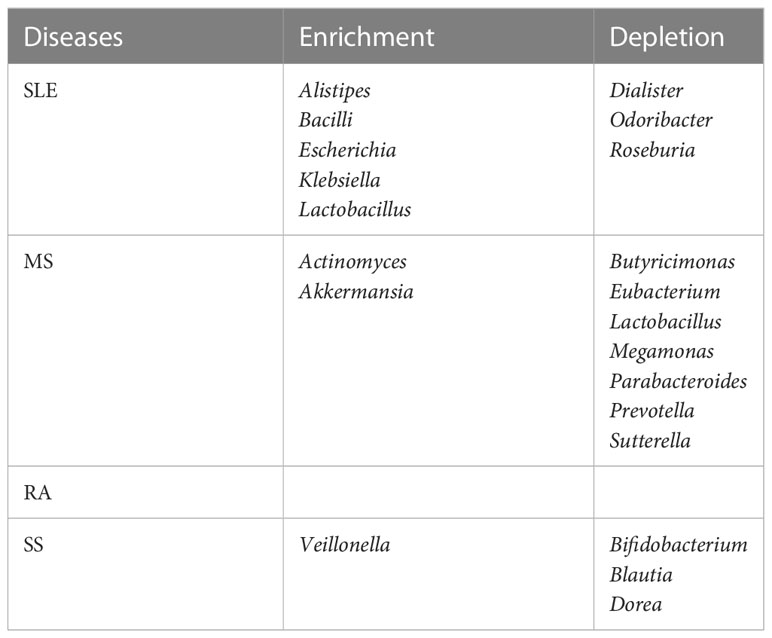
2.5 Altered gut bacteria shared among the four autoimmune diseases versus those unique to each disease
We posed the pertinent question of whether altered gut bacteria are shared among the four autoimmune diseases. The altered taxa listed in Table 1 were classified into those shared among the four autoimmune diseases (Table 2) and those unique to each autoimmune disease (Table 3).
Thereafter, we could identify taxa shared among four, three, and two diseases in various combinations (Table 2). Interestingly, Streptococcus was enriched in all four diseases. In addition to Streptococcus, Prevotella was commonly enriched in SLE, RA, and SS, and Eggerthella in SLE, MS, and RA. Meanwhile, SLE, MS, and SS shared the depletion of Faecalibacterium and the enrichment of Streptococcus. Comparing two autoimmune diseases, the SLE–MS, SLE–SS, and SLE–RA combinations shared at least four altered gut bacterial taxa, and the MS–SS combination shared three altered gut bacterial genera. Collectively, SLE shared altered gut bacteria (SAGB) through virtually all comparison groups.
In the case of uniquely altered gut bacteria in each autoimmunity, SLE, MS, and SS had eight, nine, and four genera, respectively, whereas RA had none (Table 3). The abundance of Lactobacillus was changed in SLE and MS, but in the opposite direction—enriched in SLE but depleted in MS. In addition, the depletion of Prevotella was unique to MS.
2.6 Potential contribution of altered gut bacteria to disease pathogenesis
To further understand the role of altered gut bacteria in the etiology of autoimmune diseases, we investigated whether the altered bacteria listed in Table 1 have known functions in immune-related diseases. Studies on gut bacteria’s role and mechanism of action of gut bacteria in human autoimmunity are still limited. Thus, studies on immune-related diseases were also included. We hypothesized that the altered gut bacteria shared among different diseases might be associated with common immunologic pathways of the diseases and that the bacteria unique to each disease may be associated with the specific characteristics of the diseases.
Streptococcus, enriched in all four autoimmune diseases, produces antigens that are cross-reactive with host-derived antigens (Cunningham, 2019). These cross-reactive antigens can activate T cells and contribute to autoantibody production through molecular mimicries—hallmarks of autoimmune diseases. This is the third model of immunopathology proposed by Chen et al. for autoimmune mechanisms (Chen et al., 2017). Streptococcus mutans/sanguinis bind to salivary proteins and glycoproteins to form biofilms (Matsumoto et al., 2004; Xie et al., 2020). In addition, bacterial biofilms are rich in bacterial extracellular DNA complexed with amyloid, which stimulates autoimmunity (Gallo et al., 2015; Andreasen et al., 2019; Qiu et al., 2019). Thus, DNA abundant in Streptococcus-induced biofilms might contribute to autoantibody production by forming a complex with E. coli-derived curli amyloid in the gut environment (Chen et al., 2019; Qiu et al., 2019; Barrasso et al., 2022). These results suggest that Streptococcus may be closely related to the development of autoimmune diseases through autoantibody production. However, further understanding of Streptococcus species and their strains involved in disease etiology is needed.
Eggerthella lenta is commonly enriched in SLE, MS, and RA (Tables 1, 2). In an inflammatory bowel disease (IBD) model, E. lenta activates Th17 cells through the cardiac glycoside reductase 2 (Cgr2) enzyme, which metabolizes endogenous Rorγt inhibitors (Alexander et al., 2022). However, the activation of Th17 cells by E. lenta is affected by two variables. First, a high concentration of dietary arginine (3 percent/kg) can inhibit the function of the Cgr2 enzyme (Alexander et al., 2022). Second, E. lenta does not express Cgr2 depending on the strain, and Cgr2- strains do not activate Th17 cells. This result indicates that the contribution of E. lenta to the development of autoimmune diseases may depend on host dietary factors and bacterial strains. This finding relates to the first immunopathology model proposed by Chen et al. for autoimmune mechanisms (Chen et al., 2017). In addition, E. lenta was enriched in the gut of mice exposed to cigarette smoke for seven months (Bai et al., 2022). Furthermore, the transplantation of feces from smoking-exposed mice into germ-free mice induced enrichment of E. lenta, an impairment of the gut barrier in the colonic epithelium, and an increase in proinflammatory cytokines IL-17 and TNF (Bai et al., 2022). Notably, smoking is a common risk factor for SLE, MS, and RA (Majka and Holers, 2006; Healy et al., 2009; Amador-Patarroyo et al., 2012; Mowry et al., 2012; Baka et al., 2009), and Th17 cells are involved in the pathogenesis of these three diseases (Yang et al., 2019; Moser et al., 2020; Robert and Miossec, 2020).
Prevotella is enriched in SLE, RA, and SS but depleted in MS. Specifically, P. copri is enriched in RA but depleted in MS (Table 1). Interestingly, the colonization of germ-free mice with fecal samples from RA patients dominated by P. copri induced a Th17 cell-dependent autoimmune arthritis, suggesting that gut dysbiosis with enriched P. copri contributes to the development of RA (Maeda et al., 2016). Kishikawa et al. also suggested that enriched multiple Prevotella spp. are associated with the etiology of RA in Japanese patients (Kishikawa et al., 2020). However, clinical trials of IL-17 blockers presented limited clinical efficacy in RA compared with their efficacies in psoriasis or psoriatic arthritis (Schett et al., 2013; Fauny et al., 2020). This suggests that the roles of Th17 cells and IL-17 in the etiology of RA may be multi-faceted, as the presence of Foxp3+IL-17+ T cells is observed in the subjects’ synovium (Komatsu et al., 2014; Dominguez-Villar and Hafler, 2018). Multiple Prevotella spp. have been suggested to be associated with the etiology of RA (Table 1); however, their mechanisms of action are more complex than previously recognized. Therefore, the roles of Th17 subtypes and multiple Prevotella spp. in the etiology of RA need to be clarified (Omenetti et al., 2019). Considering the role of Th17 cells in the pathogenesis of MS, further investigation is needed to determine the role of P. copri depletion in MS etiology.
Faecalibacterium is commonly depleted in SLE, MS, and SS (Tables 1, 2). Faecalibacterium maintains homeostasis of the gut immune system by secreting anti-inflammatory compounds such as (Houtman et al., 2022), salicylic acid (Miquel et al., 2015), and microbial anti-inflammatory molecules (Quevrain et al., 2016). In addition, F. prausnitzii and its supernatant effectively increase the function of Short-chain fatty acid (SCFA)-producing bacteria (Zhou et al., 2021). SCFAs are produced through the breakdown of various indigestible dietary fibers and complex carbohydrates catalyzed by the gut microbiota (Park and Kim, 2021). Beneficial bacteria in the oral cavity and gut of healthy individuals can modulate the inflammatory response through the secretion of immunomodulators such as SCFAs (acetate, butyrate, and propionate) (Feng et al., 2018; Dalile et al., 2019; Mendez et al., 2020; Vijay and Valdes, 2021; Houtman et al., 2022). In addition, Faecalibacterium, which secretes SCFAs such as butyrate, is well known for its anti-inflammatory properties (Van de Wiele et al., 2016).
The anti-inflammatory effect of SCFAs is mediated through the induction of Treg cells and the alleviation of disease symptoms (Machiels et al., 2014; Kim et al., 2017; Moon et al., 2020b; Vijay and Valdes, 2021). Specifically, among the three types of SCFAs, butyrate and propionate were effective in inducing Foxp3, but acetate was not [untreated 30.4 percent, acetate 31.4 percent, propionate 41.9 percent (p < 0.01), and butyrate 54.2 percent (p < 0.01)] (Furusawa et al., 2013). In patients with relapsing-remitting MS (RRMS), SCFA concentrations in the fecal samples were significantly reduced compared to controls (Takewaki et al., 2020). However, the hypersecretion of SCFAs may also lead to side effects, such as bacterial invasion associated with the reduced mucus layer and inflammation (Gaudier et al., 2009; Park et al., 2016; Clarke, 2020; Okumura et al., 2021). Butyrate enemas reduced the thickness of the adherent mucus layer by approximately two-fold when administered to mice (Gaudier et al., 2009). The fact that RA developed only in mice with increased gut permeability suggests that bacterial invasion may be associated with a decrease in the mucus layer (Clarke, 2020). These results suggest that the decrease and hypersecretion of SCFAs may be related to the etiology of autoimmune diseases, which are long-term chronic diseases. Thus, more detailed studies on the role of SCFAs in autoimmune diseases may be needed.
Bacteroides are enriched in SLE and RA (Table 1). The structure and function of Bacteroides-derived LPS have been shown in relation to the development of type 1 diabetes (T1D). The immunostimulatory efficacy of Bacteroides-derived LPS was four times lower than that of Escherichia coli-derived LPS. While the E. coli-derived LPS delayed the onset of T1D in non-obese diabetic mice by inducing endotoxin resistance, Bacteroides-derived LPS neither induced endotoxin resistance nor delayed the development of T1D (Vatanen et al., 2016). As a result, Bacteroides-derived LPS caused more inflammatory responses than E. coli. A similar mechanism may play a role in the pathogenesis of SLE and RA. However, SLE patients also have depleted species that belong to the Bacteroides genus. In this context, species-specific modulation of immune function by Bacteroides must be studied.
E. coli, enriched in SLE, can be divided into pathogenic and nonpathogenic strains (Palmela et al., 2018; Lorenz et al., 2020). Infection with E. coli expressing curli amyloid can induce the production of autoantibodies by forming a complex with DNA derived from bacteria or viruses. The amyloid/DNA complexes produce anti-nuclear autoantibodies and anti-dsDNA autoantibodies involved in SLE pathogenesis (Dema and Charles, 2016; Qiu et al., 2019). This was verified because curli amyloid-deficient mutant E. coli does not produce autoantibodies (Gallo et al., 2015). This finding may be related to the fifth model of immunopathology proposed by Chen et al. for autoimmune mechanisms (Chen et al., 2017).
Although Ruminococcus gnavus is a gram-positive bacterium, the complex glucorhamnan polysaccharide secreted from this bacterium induces TNFα through TLR4 in dendritic cells (Henke et al., 2019). In contrast, Ruminococcus_2 is associated with the improvement of metabolic dysfunction. For example, the consumption of barley for eight months in subjects with metabolic dysfunction improved blood sugar levels and cholesterol levels, which accompanied the enrichment of Ruminococcus_2 and Dialister in the subjects’ gut (Matsuoka et al., 2022). This suggests that the depletion of these commensal bacteria may be associated with the development of metabolic dysfunction. Abnormal metabolic reactions, such as elevations in glycolysis and mitochondrial oxidative metabolism, have also been reported in patients with SLE (Yin et al., 2015; He et al., 2020). Gut dysbiosis in patients with SLE includes enrichment of R. gnavus and depletion of Ruminococcus_2 and Dialister (Table 1). These results suggest that abnormal metabolism in SLE may be closely associated with gut dysbiosis.
A mouse model of spinal cord injury shows the neuroprotective effects of Butyricimonas, a genus depleted in patients with MS. Butyricimonas is depleted in mice with spinal cord injury but recovers by fecal microbiome transfer from healthy mice, which induces downregulated IL-1β and NF-κB signaling in the spinal cord (Jing et al., 2021). Therefore, these results suggest that the depletion of Butyricimonas in patients with MS may be closely related to its etiology (Table 1).
The Bifidobacterium genus was reported to be depleted in patients with SS in three papers (Table 1) (Mandl et al., 2017; Cano-Ortiz et al., 2020; Moon et al., 2020a). However, this commensal bacterium needed to be further classified for comparative analysis with other diseases because its relative abundance in gut dysbiosis differed depending on the species. For example, B. longum is effective in preventing IBD and treating diarrhea (O’Callaghan and van Sinderen, 2016; Yao et al., 2021). On the other hand, B. bifidum can induce the differentiation of Th17 cells (Rinaldi et al., 2019). Based on these results, the Bifidobacterium genus in SS, an autoimmune disease, is likely to be B. longum, but it remains a task to be identified at the species level in the future.
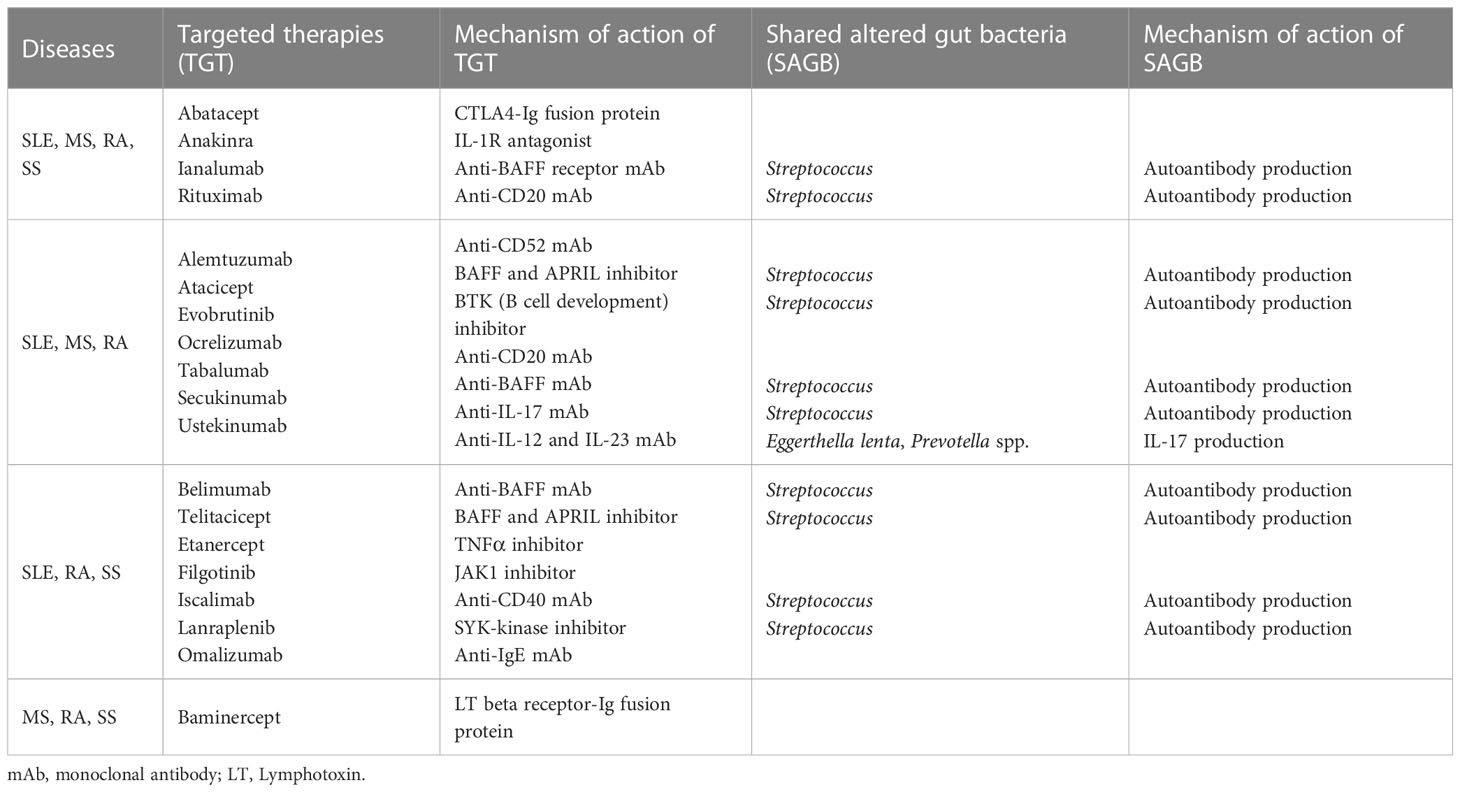
We also investigated how many targeted therapies are shared among the four autoimmune diseases. This is because the altered gut bacteria that may be associated with the etiology of the disease are shared in autoimmune diseases. Petitdemange et al. reported targeted therapies shared in autoimmune or inflammatory diseases (Petitdemange et al., 2020). Four targeted therapies (abatacept, anakinra, ianalumab, and rituximab) are shared among the four autoimmune diseases (Table 4). Among the targeted therapies shared by three diseases, seven (alemtuzumab, atacicept, evobrutinib, ocrelizumab, secukinumab, tabalumab, and ustekinumab) are shared among SLE, MS, and RA. Furthermore, seven (belimumab, etanercept, filgotinib, iscalimab, lanraplenib, omalizumab, and telitacicept) are shared among SLE, RA, and SS, and one (baminercept) is shared among MS, RA, and SS. These results suggest that targeted therapies in autoimmune diseases are related to overlapping immunological pathways due to common causes. It is tempting to say that the altered gut bacteria shared among diseases might be partially involved (Petitdemange et al., 2020). In particular, 52.6 percent (10 out of 19) of the treatments for these four diseases consisted of molecules that target B cells or antibody production, indicating that the altered gut bacteria may be closely related to autoantibody production.
2.7 Association between shared gut bacteria and polyautoimmunity
Polyautoimmunity can be defined as the coexistence of one or more autoimmune diseases in one patient (Rojas-Villarraga et al., 2012). Polyautoimmunity in patients with SLE, SS, and RA has a prevalence of 41 percent, 32.6 percent, and 14 percent, respectively (Ordonez-Canizares et al., 2022). Although data on overall polyautoimmunity in patients with MS are unavailable, the prevalence of coexisting SS has been suggested to be between 1 and 16.6 percent (Amador-Patarroyo et al., 2012). These results may be related to the fact that autoimmune diseases share altered gut bacteria associated with the failure to maintain immune homeostasis (Table 2). The enriched relative abundance of Bacteroides, Eggerthella, Prevotella, and Streptococcus, shared in autoimmune diseases, has been reported to be related to the promotion of immune responses in immune-related diseases. This is due to Bacteroides-derived LPS, metabolizing Rorγt inhibitors, Th17 cell induction, and antibodies to cross-reactive antigens, respectively (Maeda et al., 2016; Vatanen et al., 2016; Cunningham, 2019; Alexander et al., 2022). In particular, as aforementioned, Streptococcus, which is shared by all four autoimmune diseases, has been suggested to be involved in autoantibody formation (Cunningham, 2019). This result might also be partially related to the fact that many therapies for these four diseases involve the inhibition of autoantibody production (Table 4) (Petitdemange et al., 2020).
Meanwhile, SLE, MS, and SS patients showed a decreased abundance of Faecalibacterium abundance. The decrease of Faecalibacterium has the potential to significantly impact the etiology of autoimmune diseases because they secrete various immune modulators, such as butyrate, salicylic acid, and microbial anti-inflammatory molecules, as aforementioned (Miquel et al., 2015; Quevrain et al., 2016; Houtman et al., 2022). These results suggest that the altered gut bacteria shared between autoimmune diseases might contribute to the development of polyautoimmunity (De Luca and Shoenfeld, 2019; Xu et al., 2021). However, a direct causal relationship between the shared, altered gut bacteria and polyautoimmunity remains to be further elucidated.
2.8 Association between altered gut bacteria and mortality
The total number of altered gut bacteria in each autoimmune disease differed depending on the disease—22 in SLE, 16 in MS, 5 in RA, and 8 in SS (Table 1). As the number of altered taxa cross-validated across different studies can increase with the increased number of studies, we defined a gut dysbiosis index as the number of altered gut bacterial taxa divided by the number of studies. The gut dysbiosis indexes of SLE, MS, RA, and SA were 1.7, 1.8, 0.7, and 1.3, respectively. Based on these results, we investigated whether a higher degree of gut dysbiosis in SLE and MS was associated with mortality. The most recent papers from developed countries were used for similar comparative conditions for mortality due to each disease. The SMR of patients with MS was 2.89 [95 percent CI, 2.71 to 3.07; UK (period: 1980–2007)], indicating a 189 percent higher risk of death than the general population (Kingwell et al., 2012). The SMR of patients with SLE was 2.66 [95 percent CI, 2.09 to 3.39; Korea (period: 1990–2015)], indicating a 166 percent higher risk of death than the general population (Lee et al., 2016). However, the SMRs of patients with RA and SS were 1.54 [95 percent CI, 1.41 to 1.67; Netherlands (period: 1997–2012)] (van den Hoek et al., 2017) and 1.15 [95 percent CI, 0.86 to 1.50; USA (period: 2006–2015)] (Maciel et al., 2017), respectively. These values were slightly higher than or no different from the general population. Interestingly, the SMRs presented a positive correlation trend with the gut dysbiosis indexes (Figure 1), suggesting that a high degree of gut dysbiosis may adversely affect immune homeostasis and increase mortality rates.
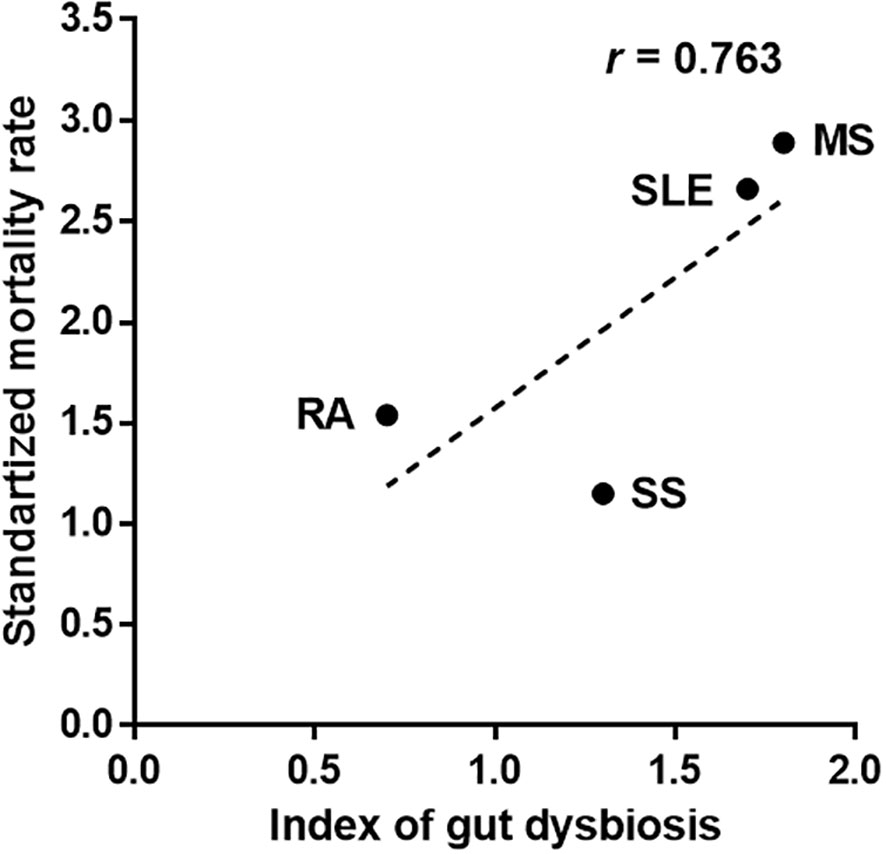
Figure 1 Association between the number of altered gut bacteria and mortality in patients with autoimmune diseases. The index of gut dysbiosis is defined as the number of altered gut bacterial taxa divided by the number of studies. R is Pearson’s correlation coefficient.
3 Conclusion
The importance of gut dysbiosis in the etiology of autoimmune diseases is increasing. Thus, to better understand the impact of gut dysbiosis, we first investigated the cross-validated altered gut bacteria in each disease and further analyzed the altered gut bacteria shared between autoimmune diseases. Interestingly, the shared, altered gut bacteria enriched in autoimmune diseases are partially related to autoantibody production or the activation of Th17 cells in reports of immune-related diseases. In particular, the decrease of Faecalibacterium shared in SLE, MS, and SS, which secretes various immunomodulatory substances, can greatly affect the failure to maintain immune homeostasis. The SMR in patients with SLE and MS was higher than that of RA and SS, which was shown to be positively correlated with the total number of altered gut bacteria in four autoimmune diseases. In taxonomic abundance analysis, Bifidobacterium, Bacteroides, Lactobacillus, Prevotella, and Ruminococcus should be classified at the species level, not the genus level, as their relative abundance may vary depending on the species. However, the abundance of Bacteroides fragilis and Prevotella copri varies according to diseases or strains, even if the species are the same, so further research is needed. In addition, the non-cross-validated microbiome, excluded from this study, is left for future tasks by accumulating more data. This review suggests that the altered gut bacteria in patients with autoimmune diseases may be closely related to abnormal immune activity and weakened anti-inflammatory activity. In addition, the increased number and sharing of altered gut bacteria are likely to be associated with disease exacerbation and polyautoimmunity, respectively. However, the direct causal relationship between altered gut bacteria and each autoimmune disease remains to be clarified.
Author contributions
S-HC and YC contributed the concept and design of the paper. S-HC and YC participated in the review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Research Foundation of Korea (Daejun, Korea) through grants 2018R1A5A2024418 and 2020R1A2C2007038 awarded to YC and grant 2020R1A2C1100163 awarded to S-HC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aho, K., Koskenvuo, M., Tuominen, J., Kaprio, J. (1986). Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 13 (5), 899–902.
Alexander, M., Ang, Q. Y., Nayak, R. R., Bustion, A. E., Sandy, M., Zhang, B., et al. (2022). Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host Microbe 30 (1), 17–30 e19. doi: 10.1016/j.chom.2021.11.001
Amador-Patarroyo, M. J., Arbelaez, J. G., Mantilla, R. D., Rodriguez-Rodriguez, A., Cardenas-Roldan, J., Pineda-Tamayo, R., et al. (2012). Sjogren’s syndrome at the crossroad of polyautoimmunity. J. Autoimmun 39 (3), 199–205. doi: 10.1016/j.jaut.2012.05.008
Andreasen, M., Meisl, G., Taylor, J. D., Michaels, T. C. T., Levin, A., Otzen, D. E., et al. (2019). Physical determinants of amyloid assembly in biofilm formation. mBio 10 (1), e02279-18. doi: 10.1128/mBio.02279-18
Azzouz, D., Omarbekova, A., Heguy, A., Schwudke, D., Gisch, N., Rovin, B. H., et al. (2019). Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum Dis. 78 (7), 947–956. doi: 10.1136/annrheumdis-2018-214856
Bai, X., Wei, H., Liu, W., Coker, O. O., Gou, H., Liu, C., et al. (2022). Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut 71 (12), 2436–2450. doi: 10.1136/gutjnl-2021-325021
Baka, Z., Buzas, E., Nagy, G.. (2009). Rheumatoid arthritis and smoking: putting the pieces together. Arthritis Res. Ther. 11 (4), 238. doi: 10.1186/ar2751
Barrasso, K., Chac, D., Debela, M. D., Geigel, C., Steenhaut, A., Rivera Seda, A., et al. (2022). Impact of a human gut microbe on vibrio cholerae host colonization through biofilm enhancement. Elife 11, e73010. doi: 10.7554/eLife.73010
Bellando-Randone, S., Russo, E., Venerito, V., Matucci-Cerinic, M., Iannone, F., Tangaro, S., et al. (2021). Exploring the oral microbiome in rheumatic diseases, state of art and future prospective in personalized medicine with an AI approach. J. Pers. Med. 11 (7), 625. doi: 10.3390/jpm11070625
Bellocchi, C., Fernandez-Ochoa, A., Montanelli, G., Vigone, B., Santaniello, A., Quirantes-Pine, R., et al. (2019). Identification of a shared microbiomic and metabolomic profile in systemic autoimmune diseases. J. Clin. Med. 8 (9), 1291. doi: 10.3390/jcm8091291
Berer, K., Gerdes, L. A., Cekanaviciute, E., Jia, X., Xiao, L., Xia, Z., et al. (2017). Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U.S.A. 114 (40), 10719–10724. doi: 10.1073/pnas.1711233114
Cano-Ortiz, A., Laborda-Illanes, A., Plaza-Andrades, I., Membrillo Del Pozo, A., Villarrubia Cuadrado, A., Rodriguez Calvo de Mora, M., et al. (2020). Connection between the gut microbiome, systemic inflammation, gut permeability and FOXP3 expression in patients with primary sjogren’s syndrome. Int. J. Mol. Sci. 21 (22), 8733. doi: 10.3390/ijms21228733
Cekanaviciute, E., Yoo, B. B., Runia, T. F., Debelius, J. W., Singh, S., Nelson, C. A., et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114 (40), 10713–10718. doi: 10.1073/pnas.1711235114
Chen, D., Cao, Y., Yu, L., Tao, Y., Zhou, Y., Zhi, Q., et al. (2019). Characteristics and influencing factors of amyloid fibers in s. mutans biofilm. AMB Express 9 (1), 31. doi: 10.1186/s13568-019-0753-1
Chen, J., Chia, N., Kalari, K. R., Yao, J. Z., Novotna, M., Paz Soldan, M. M., et al. (2016a). Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 6, 28484. doi: 10.1038/srep28484
Chen, B. D., Jia, X. M., Xu, J. Y., Zhao, L. D., Ji, J. Y., Wu, B. X., et al. (2021). An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol. 73 (2), 232–243. doi: 10.1002/art.41511
Chen, B., Sun, L., Zhang, X. (2017). Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J. Autoimmun 83, 31–42. doi: 10.1016/j.jaut.2017.03.009
Chen, J., Wright, K., Davis, J. M., Jeraldo, P., Marietta, E. V., Murray, J., et al. (2016b). An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 8 (1), 43. doi: 10.1186/s13073-016-0299-7
Clarke, J. (2020). Disease onset goes with its gut in RA. Nat. Rev. Rheumatol. 16 (7), 350. doi: 10.1038/s41584-020-0436-y
Cunningham, M. W. (2019). Molecular mimicry, autoimmunity, and infection: The cross-reactive antigens of group a streptococci and their sequelae. Microbiol. Spectr. 7 (4), 1–51. doi: 10.1128/microbiolspec.GPP3-0045-2018
Dalile, B., Van Oudenhove, L., Vervliet, B., Verbeke, K. (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16 (8), 461–478. doi: 10.1038/s41575-019-0157-3
De Luca, F., Shoenfeld, Y. (2019). The microbiome in autoimmune diseases. Clin. Exp. Immunol. 195 (1), 74–85. doi: 10.1111/cei.13158
Dema, B., Charles, N. (2016). Autoantibodies in SLE: Specificities, isotypes and receptors. Antibodies (Basel) 5 (1), 2. doi: 10.3390/antib5010002
Dinse, G. E., Parks, C. G., Weinberg, C. R., Co, C. A., Wilkerson, J., Zeldin, D. C., et al. (2022). Increasing prevalence of antinuclear antibodies in the united states. Arthritis Rheumatol. 74 (12), 2032–2041. doi: 10.1002/art.42330
Dominguez-Villar, M., Hafler, D. A. (2018). Regulatory T cells in autoimmune disease. Nat. Immunol. 19 (7), 665–673. doi: 10.1038/s41590-018-0120-4
Duscha, A., Gisevius, B., Hirschberg, S., Yissachar, N., Stangl, G. I., Eilers, E., et al. (2020). Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 180 (6), 1067–1080 e1016. doi: 10.1016/j.cell.2020.02.035
Fauny, M., Moulin, D., D’Amico, F., Netter, P., Petitpain, N., Arnone, D., et al. (2020). Paradoxical gastrointestinal effects of interleukin-17 blockers. Ann. Rheum Dis. 79 (9), 1132–1138. doi: 10.1136/annrheumdis-2020-217927
Feng, W., Ao, H., Peng, C. (2018). Gut microbiota, short-chain fatty acids, and herbal medicines. Front. Pharmacol. 9. doi: 10.3389/fphar.2018.01354
Forbes, J. D., Chen, C. Y., Knox, N. C., Marrie, R. A., El-Gabalawy, H., de Kievit, T., et al. (2018). A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 6 (1), 221. doi: 10.1186/s40168-018-0603-4
Frazzei, G., van Vollenhoven, R. F., de Jong, B. A., Siegelaar, S. E., van Schaardenburg, D. (2022). Preclinical autoimmune disease: A comparison of rheumatoid arthritis, systemic lupus erythematosus, multiple sclerosis and type 1 diabetes. Front. Immunol. 13. doi: 10.3389/fimmu.2022.899372
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504 (7480), 446–450. doi: 10.1038/nature12721
Gallo, P. M., Rapsinski, G. J., Wilson, R. P., Oppong, G. O., Sriram, U., Goulian, M., et al. (2015). Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 42 (6), 1171–1184. doi: 10.1016/j.immuni.2015.06.002
Gaudier, E., Rival, M., Buisine, M. P., Robineau, I., Hoebler, C. (2009). Butyrate enemas upregulate muc genes expression but decrease adherent mucus thickness in mice colon. Physiol. Res. 58 (1), 111–119. doi: 10.33549/physiolres.931271
Greiling, T. M., Dehner, C., Chen, X., Hughes, K., Iniguez, A. J., Boccitto, M., et al. (2018). Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 10 (434), eaan2306. doi: 10.1126/scitranslmed.aan2306
Guo, M., Wang, H., Xu, S., Zhuang, Y., An, J., Su, C., et al. (2020). Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes 11 (6), 1758–1773. doi: 10.1080/19490976.2020.1768644
He, J., Chan, T., Hong, X., Zheng, F., Zhu, C., Yin, L., et al. (2020). Microbiome and metabolome analyses reveal the disruption of lipid metabolism in systemic lupus erythematosus. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01703
He, Z., Shao, T., Li, H., Xie, Z., Wen, C. (2016). Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 8, 64. doi: 10.1186/s13099-016-0146-9
Healy, B. C., Ali, E. N., Guttmann, C. R., Chitnis, T., Glanz, B. I., Buckle, G., et al. (2009). Smoking and disease progression in multiple sclerosis. Arch. Neurol. 66 (7), 858–864. doi: 10.1001/archneurol.2009.122
Henke, M. T., Kenny, D. J., Cassilly, C. D., Vlamakis, H., Xavier, R. J., Clardy, J. (2019). Ruminococcus gnavus, a member of the human gut microbiome associated with crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 116 (26), 12672–12677. doi: 10.1073/pnas.1904099116
Hevia, A., Milani, C., Lopez, P., Cuervo, A., Arboleya, S., Duranti, S., et al. (2014). Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 5 (5), e01548–e01514. doi: 10.1128/mBio.01548-14
Houtman, T. A., Eckermann, H. A., Smidt, H., de Weerth, C. (2022). Gut microbiota and BMI throughout childhood: The role of firmicutes, bacteroidetes, and short-chain fatty acid producers. Sci. Rep. 12 (1), 3140. doi: 10.1038/s41598-022-07176-6
Jangi, S., Gandhi, R., Cox, L. M., Li, N., von Glehn, F., Yan, R., et al. (2016). Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015. doi: 10.1038/ncomms12015
Jing, Y., Yu, Y., Bai, F., Wang, L., Yang, D., Zhang, C., et al. (2021). Effect of fecal microbiota transplantation on neurological restoration in a spinal cord injury mouse model: Involvement of brain-gut axis. Microbiome 9 (1), 59. doi: 10.1186/s40168-021-01007-y
Jubair, W. K., Hendrickson, J. D., Severs, E. L., Schulz, H. M., Adhikari, S., Ir, D., et al. (2018). Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol. 70 (8), 1220–1233. doi: 10.1002/art.40490
Kim, D., Zeng, M. Y., Nunez, G. (2017). The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp. Mol. Med. 49 (5), e339. doi: 10.1038/emm.2017.24
Kingwell, E., van der Kop, M., Zhao, Y., Shirani, A., Zhu, F., Oger, J., et al. (2012). Relative mortality and survival in multiple sclerosis: Findings from British Columbia, Canada. J. Neurol. Neurosurg. Psychiatry 83 (1), 61–66. doi: 10.1136/jnnp-2011-300616
Kishikawa, T., Maeda, Y., Nii, T., Motooka, D., Matsumoto, Y., Matsushita, M., et al. (2020). Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum Dis. 79 (1), 103–111. doi: 10.1136/annrheumdis-2019-215743
Komatsu, N., Okamoto, K., Sawa, S., Nakashima, T., Oh-hora, M., Kodama, T., et al. (2014). Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 20 (1), 62–68. doi: 10.1038/nm.3432
Lee, Y. H., Choi, S. J., Ji, J. D., Song, G. G. (2016). Overall and cause-specific mortality in systemic lupus erythematosus: an updated meta-analysis. Lupus 25 (7), 727–734. doi: 10.1177/0961203315627202
Li, Y., Wang, H. F., Li, X., Li, H. X., Zhang, Q., Zhou, H. W., et al. (2019). Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin. Sci. (Lond) 133 (7), 821–838. doi: 10.1042/CS20180841
Lorenz, B., Ali, N., Bocklitz, T., Rosch, P., Popp, J. (2020). Discrimination between pathogenic and non-pathogenic e. coli strains by means of raman microspectroscopy. Anal. Bioanal Chem. 412 (30), 8241–8247. doi: 10.1007/s00216-020-02957-2
Luo, X. M., Edwards, M. R., Mu, Q., Yu, Y., Vieson, M. D., Reilly, C. M., et al. (2018). Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol. 84 (4), e02288-17. doi: 10.1128/AEM.02288-17
Machiels, K., Joossens, M., Sabino, J., De Preter, V., Arijs, I., Eeckhaut, V., et al. (2014). A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63 (8), 1275–1283. doi: 10.1136/gutjnl-2013-304833
Maciel, G., Crowson, C. S., Matteson, E. L., Cornec, D. (2017). Incidence and mortality of physician-diagnosed primary sjogren syndrome: Time trends over a 40-year period in a population-based US cohort. Mayo Clin. Proc. 92 (5), 734–743. doi: 10.1016/j.mayocp.2017.01.020
Maeda, Y., Kurakawa, T., Umemoto, E., Motooka, D., Ito, Y., Gotoh, K., et al. (2016). Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68 (11), 2646–2661. doi: 10.1002/art.39783
Majka, D. S., Holers, V. M. (2006) Cigarette smoking and the risk of systemic lupus erythematosus and rheumatoid arthritis. Ann. Rheum Dis. 65 (5), 561–563. doi: 10.1136/ard.2005.046052
Mandl, T., Marsal, J., Olsson, P., Ohlsson, B., Andreasson, K. (2017). Severe intestinal dysbiosis is prevalent in primary sjogren’s syndrome and is associated with systemic disease activity. Arthritis Res. Ther. 19 (1), 237. doi: 10.1186/s13075-017-1446-2
Matsumoto, N., Salam, M. A., Watanabe, H., Amagasa, T., Senpuku, H. (2004). Role of gene E2f1 in susceptibility to bacterial adherence of oral streptococci to tooth surfaces in mice. Oral. Microbiol. Immunol. 19 (4), 270–276. doi: 10.1111/j.1399-302X.2004.00151.x
Matsuoka, T., Hosomi, K., Park, J., Goto, Y., Nishimura, M., Maruyama, S., et al. (2022). Relationships between barley consumption and gut microbiome characteristics in a healthy Japanese population: a cross-sectional study. BMC Nutr. 8 (1), 23. doi: 10.1186/s40795-022-00500-3
Mendez, R., Watane, A., Farhangi, M., Cavuoto, K. M., Leith, T., Budree, S., et al. (2020). Gut microbial dysbiosis in individuals with sjogren’s syndrome. Microb. Cell Fact 19 (1), 90. doi: 10.1186/s12934-020-01348-7
Miquel, S., Leclerc, M., Martin, R., Chain, F., Lenoir, M., Raguideau, S., et al. (2015). Identification of metabolic signatures linked to anti-inflammatory effects of faecalibacterium prausnitzii. mBio 6 (2), e00300-15. doi: 10.1128/mBio.00300-15
Miyake, S., Kim, S., Suda, W., Oshima, K., Nakamura, M., Matsuoka, T., et al. (2015). Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PloS One 10 (9), e0137429. doi: 10.1371/journal.pone.0137429
Moon, J., Choi, S. H., Yoon, C. H., Kim, M. K. (2020a). Gut dysbiosis is prevailing in sjogren’s syndrome and is related to dry eye severity. PloS One 15 (2), e0229029. doi: 10.1371/journal.pone.0229029
Moon, J., Yoon, C. H., Choi, S. H., Kim, M. K. (2020b). Can gut microbiota affect dry eye syndrome? Int. J. Mol. Sci. 21 (22), 8443. doi: 10.3390/ijms21228443
Moser, T., Akgün, K., Proschmann, U., Sellner, J., Ziemssen, T. (2020). The role of TH17 cells in multiple sclerosis: Therapeutic implications. Autoimmun Rev. 19 (10), 102647. doi: 10.1016/j.autrev.2020.102647
Mowry, E. M., Waubant, E., McCulloch, C. E., Okuda, D. T., Evangelista, A. A., Lincoln, R. R., et al. (2012). Vitamin d status predicts new brain magnetic resonance imaging activity in multiple sclerosis. Ann. Neurol. 72 (2), 234–240. doi: 10.1002/ana.23591
Nagao-Kitamoto, H., Kamada, N. (2017). Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 17 (1), 1–12. doi: 10.4110/in.2017.17.1.1
Nikitakis, N. G., Papaioannou, W., Sakkas, L. I., Kousvelari, E. (2017). The autoimmunity-oral microbiome connection. Oral. Dis. 23 (7), 828–839. doi: 10.1111/odi.12589
O’Callaghan, A., van Sinderen, D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00925
Okumura, S., Konishi, Y., Narukawa, M., Sugiura, Y., Yoshimoto, S., Arai, Y., et al. (2021). Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 12 (1), 5674. doi: 10.1038/s41467-021-25965-x
Omenetti, S., Bussi, C., Metidji, A., Iseppon, A., Lee, S., Tolaini, M., et al. (2019). The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity 51 (1), 77–89 e76. doi: 10.1016/j.immuni.2019.05.004
Ordonez-Canizares, M. C., Mena-Vazquez, N., Redondo-Rodriguez, R., Manrique-Arija, S., Jimenez-Nunez, F. G., Urena-Garnica, I., et al. (2022). Frequency of polyautoimmunity in patients with rheumatoid arthritis and systemic lupus erythematosus. J. Clin. Rheumatol. 28 (1), e38–e43. doi: 10.1097/RHU.0000000000001574
Palmela, C., Chevarin, C., Xu, Z., Torres, J., Sevrin, G., Hirten, R., et al. (2018). Adherent-invasive escherichia coli in inflammatory bowel disease. Gut 67 (3), 574–587. doi: 10.1136/gutjnl-2017-314903
Park, J., Goergen, C. J., HogenEsch, H., Kim, C. H. (2016). Chronically elevated levels of short-chain fatty acids induce T cell-mediated ureteritis and hydronephrosis. J. Immunol. 196 (5), 2388–2400. doi: 10.4049/jimmunol.1502046
Park, J., Kim, C. H. (2021). Regulation of common neurological disorders by gut microbial metabolites. Exp. Mol. Med. 53 (12), 1821–1833. doi: 10.1038/s12276-021-00703-x
Petitdemange, A., Blaess, J., Sibilia, J., Felten, R., Arnaud, L. (2020). Shared development of targeted therapies among autoimmune and inflammatory diseases: A systematic repurposing analysis. Ther. Adv. Musculoskelet Dis. 12, 1759720X20969261. doi: 10.1177/1759720X20969261
Qiu, C. C., Caricchio, R., Gallucci, S. (2019). Triggers of autoimmunity: The role of bacterial infections in the extracellular exposure of lupus nuclear autoantigens. Front. Immunol. 10. doi: 10.3389/fimmu.2019.02608
Quevrain, E., Maubert, M. A., Michon, C., Chain, F., Marquant, R., Tailhades, J., et al. (2016). Identification of an anti-inflammatory protein from faecalibacterium prausnitzii, a commensal bacterium deficient in crohn’s disease. Gut 65 (3), 415–425. doi: 10.1136/gutjnl-2014-307649
Ravindra, N., Sadashiva, N., Mahadevan, A., Bhat, D. I., Saini, J. (2018). Central nervous system actinomycosis-a clinicoradiologic and histopathologic analysis. World Neurosurg. 116, e362–e370. doi: 10.1016/j.wneu.2018.04.205
Rinaldi, E., Consonni, A., Cordiglieri, C., Sacco, G., Crasa, C., Fontana, A., et al. (2019). Therapeutic effect of bifidobacterium administration on experimental autoimmune myasthenia gravis in Lewis rats. Front. Immunol. 10. doi: 10.3389/fimmu.2019.02949
Robert, M., Miossec, P. (2020). Interleukin-17 and lupus: enough to be a target? For which patients? Lupus 29 (1), 6–14. doi: 10.1177/0961203319891243
Rodrigues, G. S. P., Cayres, L. C. F., Goncalves, F. P., Takaoka, N. N. C., Lengert, A. H., Tansini, A., et al. (2019). Detection of increased relative expression units of bacteroides and prevotella, and decreased clostridium leptum in stool samples from Brazilian rheumatoid arthritis patients: A pilot study. Microorganisms 7 (10), 413. doi: 10.3390/microorganisms7100413
Rojas-Villarraga, A., Amaya-Amaya, J., Rodriguez-Rodriguez, A., Mantilla, R. D., Anaya, J. M. (2012). Introducing polyautoimmunity: Secondary autoimmune diseases no longer exist. Autoimmune Dis. 2012, 254319. doi: 10.1155/2012/254319
Scher, J. U., Sczesnak, A., Longman, R. S., Segata, N., Ubeda, C., Bielski, C., et al. (2013). Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2, e01202. doi: 10.7554/eLife.01202
Schett, G., Elewaut, D., McInnes, I. B., Dayer, J. M., Neurath, M. F. (2013). How cytokine networks fuel inflammation: Toward a cytokine-based disease taxonomy. Nat. Med. 19 (7), 822–824. doi: 10.1038/nm.3260
Silman, A. J., MacGregor, A. J., Thomson, W., Holligan, S., Carthy, D., Farhan, A., et al. (1993). Twin concordance rates for rheumatoid arthritis: Results from a nationwide study. Br. J. Rheumatol. 32 (10), 903–907. doi: 10.1093/rheumatology/32.10.903
Sun, Y., Chen, Q., Lin, P., Xu, R., He, D., Ji, W., et al. (2019). Characteristics of gut microbiota in patients with rheumatoid arthritis in shanghai, China. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00369
Takewaki, D., Suda, W., Sato, W., Takayasu, L., Kumar, N., Kimura, K., et al. (2020). Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 117 (36), 22402–22412. doi: 10.1073/pnas.2011703117
Tong, Y., Zheng, L., Qing, P., Zhao, H., Li, Y., Su, L., et al. (2019). Oral microbiota perturbations are linked to high risk for rheumatoid arthritis. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00475
Tremlett, H., Fadrosh, D. W., Faruqi, A. A., Zhu, F., Hart, J., Roalstad, S., et al. (2016). Gut microbiota in early pediatric multiple sclerosis: A case-control study. Eur. J. Neurol. 23 (8), 1308–1321. doi: 10.1111/ene.13026
Valour, F., Senechal, A., Dupieux, C., Karsenty, J., Lustig, S., Breton, P., et al. (2014). Actinomycosis: Etiology, clinical features, diagnosis, treatment, and management. Infect. Drug Resist. 7, 183–197. doi: 10.2147/IDR.S39601
van den Hoek, J., Boshuizen, H. C., Roorda, L. D., Tijhuis, G. J., Nurmohamed, M. T., van den Bos, G. A., et al. (2017). Mortality in patients with rheumatoid arthritis: A 15-year prospective cohort study. Rheumatol. Int. 37 (4), 487–493. doi: 10.1007/s00296-016-3638-5
van der Meulen, T. A., Harmsen, H. J. M., Vila, A. V., Kurilshikov, A., Liefers, S. C., Zhernakova, A., et al. (2019). Shared gut, but distinct oral microbiota composition in primary sjogren’s syndrome and systemic lupus erythematosus. J. Autoimmun 97, 77–87. doi: 10.1016/j.jaut.2018.10.009
Van de Wiele, T., Van Praet, J. T., Marzorati, M., Drennan, M. B., Elewaut, D. (2016). How the microbiota shapes rheumatic diseases. Nat. Rev. Rheumatol. 12 (7), 398–411. doi: 10.1038/nrrheum.2016.85
Vatanen, T., Kostic, A. D., d’Hennezel, E., Siljander, H., Franzosa, E. A., Yassour, M., et al. (2016). Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165 (4), 842–853. doi: 10.1016/j.cell.2016.04.007
Vijay, A., Valdes, A. M. (2021). Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr 76, 489–501. doi: 10.1038/s41430-021-00991-6
Wang, S., Chen, H., Wen, X., Mu, J., Sun, M., Song, X., et al. (2021). The efficacy of fecal microbiota transplantation in experimental autoimmune encephalomyelitis: Transcriptome and gut microbiota profiling. J. Immunol. Res. 2021, 4400428. doi: 10.1155/2021/4400428
Wen, M., Liu, T., Zhao, M., Dang, X., Feng, S., Ding, X., et al. (2021). Correlation analysis between gut microbiota and metabolites in children with systemic lupus erythematosus. J. Immunol. Res. 2021, 5579608. doi: 10.1155/2021/5579608
Xie, Y., Zhang, M., Zhang, W., Liu, X., Zheng, W., Jiang, X. (2020). Gold nanoclusters-coated orthodontic devices can inhibit the formation of streptococcus mutans biofilm. ACS Biomater. Sci. Eng. 6 (2), 1239–1246. doi: 10.1021/acsbiomaterials.9b01647
Xu, Q., Ni, J. J., Han, B. X., Yan, S. S., Wei, X. T., Feng, G. J., et al. (2021). Causal relationship between gut microbiota and autoimmune diseases: A two-sample mendelian randomization study. Front. Immunol. 12. doi: 10.3389/fimmu.2021.746998
Yang, P., Qian, F. Y., Zhang, M. F., Xu, A. L., Wang, X., Jiang, B. P., et al. (2019). Th17 cell pathogenicity and plasticity in rheumatoid arthritis. J. Leukoc. Biol. 106 (6), 1233–1240. doi: 10.1002/JLB.4RU0619-197R
Yao, S., Zhao, Z., Wang, W., Liu, X. (2021). Bifidobacterium longum: Protection against inflammatory bowel disease. J. Immunol. Res. 2021, 8030297. doi: 10.1155/2021/8030297
Yin, Y., Choi, S. C., Xu, Z., Perry, D. J., Seay, H., Croker, B. P., et al. (2015). Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 7 (274), 274ra218. doi: 10.1126/scitranslmed.aaa0835
Zeng, Q., Junli, G., Liu, X., Chen, C., Sun, X., Li, H., et al. (2019). Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem. Int. 129, 104468. doi: 10.1016/j.neuint.2019.104468
Zhang, S. X., Wang, J., Chen, J. W., Zhang, M. X., Zhang, Y. F., Hu, F. Y., et al. (2021). The level of peripheral regulatory T cells is linked to changes in gut commensal microflora in patients with systemic lupus erythematosus. Ann. Rheum Dis. 80 (11), e177. doi: 10.1136/annrheumdis-2019-216504
Zhang, X., Zhang, D., Jia, H., Feng, Q., Wang, D., Liang, D., et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21 (8), 895–905. doi: 10.1038/nm.3914
Keywords: autoimmunity, gut dysbiosis, Sjögren’s syndrome, rheumatoid arthritis, systemic lupus erythematosus, multiple sclerosis
Citation: Chang S-H and Choi Y (2023) Gut dysbiosis in autoimmune diseases: Association with mortality. Front. Cell. Infect. Microbiol. 13:1157918. doi: 10.3389/fcimb.2023.1157918
Received: 03 February 2023; Accepted: 15 March 2023;
Published: 31 March 2023.
Edited by:
Piyush Baindara, University of Missouri, United StatesReviewed by:
Shikha Negi, Cincinnati Children’s Hospital Medical Center, United StatesVijeta Sharma, Hackensack Meridian Health, United States
Nisha Singh, University of Maryland, United States
Copyright © 2023 Chang and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youngnim Choi, youngnim@snu.ac.kr
 Sung-Ho Chang
Sung-Ho Chang